Abstract
Cryo-electron microscopy (cryo-EM) has recently provided invaluable experimental data about the full-length cystic fibrosis transmembrane conductance regulator (CFTR) 3D structure. However, this experimental information deals with inactive states of the channel, either in an apo, quiescent conformation, in which nucleotide-binding domains (NBDs) are widely separated or in an ATP-bound, yet closed conformation. Here, we show that 3D structure models of the open and closed forms of the channel, now further supported by metadynamics simulations and by comparison with the cryo-EM data, could be used to gain some insights into critical features of the conformational transition toward active CFTR forms. These critical elements lie within membrane-spanning domains but also within NBD1 and the N-terminal extension, in which conformational plasticity is predicted to occur to help the interaction with filamin, one of the CFTR cellular partners.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2835-7) contains supplementary material, which is available to authorized users.
Keywords: CFTR, ABC exporter, Filamin, Comparative modeling, Metadynamics, Cryo-electron microscopy
Introduction
Cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), altering chloride efflux in epithelial cells [1]. CFTR belongs to the large ABC transporter superfamily, but is the only member known to function as an ion channel. Therefore, it has been considered as a “broken” ABC transporter, having an atrophied or uncoupled cytoplasmic-side gate [2]. However, the structural features associated with this specific evolution remained for a long time poorly understood in the absence of experimental 3D structure of the full-length protein. These experimental 3D structures were particularly hard to obtain due to the low solubility of the protein and its limited thermal stability [3]. Contributing to this are the dynamics fluctuations of the protein, exacerbated by intrinsic disorder associated with specific regions, especially the large regulatory (R) region, linking the two halves of the protein and regulating the protein activity through phosphorylation [4]. Understanding the CFTR 3D structure and its conformational landscape, within its interaction network, could, however, provide insights into the way the CFTR folding and functional defects could be rescued [5].
In the last year, a major breakthrough has been achieved in CF research with the publication of the first experimental 3D structures, at medium to high resolution, of the full-length zebrafish and human CFTR proteins [6, 7]. These 3D structures were solved by cryo-electron microscopy (cryo-EM) in an inward-facing conformation, representing a non-phosphorylated, ATP-free, quiescent (apo) state of the CFTR channel (Supplementary Data 1A). Although electron density was too weak to assign secondary structures, the regulatory (R) domain, which is unique to CFTR among the proteins belonging to the ABC superfamily, was observed in these quiescent structures between the two nucleotide-binding domains (NBDs), preventing their dimerization and channel opening. Even more recently, the cryo-EM 3D structure of a phosphorylated, ATP-bound conformation of the full-length zebrafish CFTR protein was also reported, in which the R region is disengaged from its inhibitory position and the NBDs form a head-to-tail dimer [8] (Supplementary Data 1B). However, despite the ATP-bound state of the protein, the channel remained blocked in a closed state at its extracellular surface, with a conformation of the membrane-spanning domains (MSDs) similar to that observed in the quiescent, apo conformation. In all these three cryo-EM structures [6–8], the N-terminal region preceding MSD1 adopts a “lasso” conformation, inserting partly into the membrane and packing against MSD transmembrane (TM) helices. Also, a discontinuity is observed at the level of helix TM8, accompanied by a displacement of helix TM7 from its usual position in ABC exporters. Some efforts have already been undertaken to further exploit these 3D structures through molecular dynamics simulations, highlighting the current need of additional studies, including theoretical approaches, to understand gating cycle [9, 10].
Before these cryo-EM data on CFTR, several 3D models of the CFTR MSD/NBD assembly have been built by comparative modeling, based on crystal 3D structures of ABC exporters ([11] for a review). Molecular dynamics (MD) simulations of models of the open form of the channel, starting from the Sav1866 ABC exporter template in an outward-facing conformation, globally agreed with the general channel properties, including a narrow bottleneck separating a wide inner vestibule and a distinct outer vestibule [12–14]. Models of the open and closed forms were also proposed based on the use of the McjD and TM287/288 ABC templates, respectively [15]. More recently, outward- and inward-facing models were also proposed using an integrated experimental-MD simulation over a significant time (200 ns) [16]. Comparison of these models with cryo-EM structures can be performed to reveal differences, which may reflect distinct, functionally relevant conformational states, setting aside potential problems regarding the resolution of experimental 3D structures or imprecision in the models. As regards to this last point, it is generally difficult to appreciate the accurateness of models, as they were built (at least for the MSDs) at very low level of sequence identity. This does not mean that models are not able to reflect the structure of the CFTR channel [17], but highlights the need of a prior, critical assessment through structural alignments, now possible since cryo-EM data are available.
In 2015, we reported a MD-refined model of the CFTR open form, based on a sequence alignment with the Sav1866 template optimized through the consideration of ABC exporter structural invariants. This model was characterized by a critical salt bridge, typical of the open form, between the residues R352 and D993 [18, 19] and revealed particular features of TM8, including less amino acids than other ABC exporters. It also suggested that opening of the CFTR channel was linked to the appearance of four lateral, horizontal portals at the level of the intracellular loops (ICLs) at the end of the TM helices, allowing ions to flow from the cytosol toward the central vertical pore [14] (Supplementary Data 1C). In this model, the movement of the MSD TM helices associated with channel opening was proposed to be linked to a conformational shift occurring at the level of the NBD1 alpha-subdomain, where F508 [the deletion of which (F508del) is the most frequent mutation in CF] is located. Moreover, subsequent further short MD simulations suggested that closure of the pore may occur via limited movement of the MSDs, with F337 playing a central role at the level of the extracellular gate [14] (Supplementary Data 1D). We herein designate this closed conformer the transient (t)-closed form of the CFTR channel, as it is quickly observed starting from the full-open conformation. The above-mentioned salt bridge between R352 and D993, typical of the open form [18, 19], was also observed here, although weaker. However, this conformation lacks the salt bridge between R347 and D924 [18, 20] observed in the experimental cryo-EM 3D structures, the apo quiescent ones, as well as the ATP-bound, yet closed conformation [6–8]. This last structure is called here the s-closed (stable-closed) form, as opposed to the t-closed (transient-closed) form observed in our model, a name which is supported by its particular stability during μs-long MD simulations and by its inability to spontaneously shift toward an open form of the channel [10]. A 100 ns MD simulation of the human CFTR model that we built based on this zebrafish s-closed conformation (see “Materials and methods”) also led to observe a stable conformation [root-mean-square deviation (RMSD) of only 2.68 Å for 1108 superimposed Cα atoms between the two extreme frames]. Of note, the ICL lateral portals and the extracellular gate of our models were further supported by patch clamp recording and substituted cysteine accessibility mutagenesis (SCAM), highlighting, for instance, some critical amino acids, such as K190, R248, R303, K370 (TM4–TM6 portal), K1041, and R1048 (TM10–TM12 portal), involved in electrostatic interactions with chloride ions [21–24]. These were also now supported by the very recent cryo-EM 3D structure of the phosphorylated, ATP-bound CFTR [8].
In the present work, we first provide further strong support to our 3D models of the open and closed forms of the CFTR channel (1) by performing metadynamics simulations, which allowed to estimate the stability of the models in the open and closed forms and gave new insights into the channel dynamics and thermodynamics, and (2) by providing a structural alignment of these refined models with the experimental cryo-EM 3D structures of CFTR, which showed that the positions of equivalent amino acids within the different TM helices are identical (or in the correct register) and thus allowed to unambiguously check the quality of the sequence alignment.
Next, the comparison of the cryo-EM apo, quiescent [7] and ATP-bound, yet closed (s-closed form) [8] conformers of CFTR with our model of the t-closed form highlighted some local conformational differences at the level of the TM7-TM8-TM9 three-helix block, suggesting that two distinct conformations of the closed channel might exist. A similar closed extracellular gate is present in these two closed forms but with a different network of salt bridges, the t-closed form, however, being able to fast evolve toward the open form and thus potentially allowing rapid flickering. We also indicate here that transition from inactive toward active forms (in other words, from the s-closed toward the t-closed and open forms) might be associated with significant conformational changes, not only at the level of MSDs but also at the level of the NBDs and the N-terminal region. Conformational transitions of the N-terminal region are here considered in the context of the interaction of CFTR with one of its cellular partners (filamin), which may help to stabilize the open form of the channel.
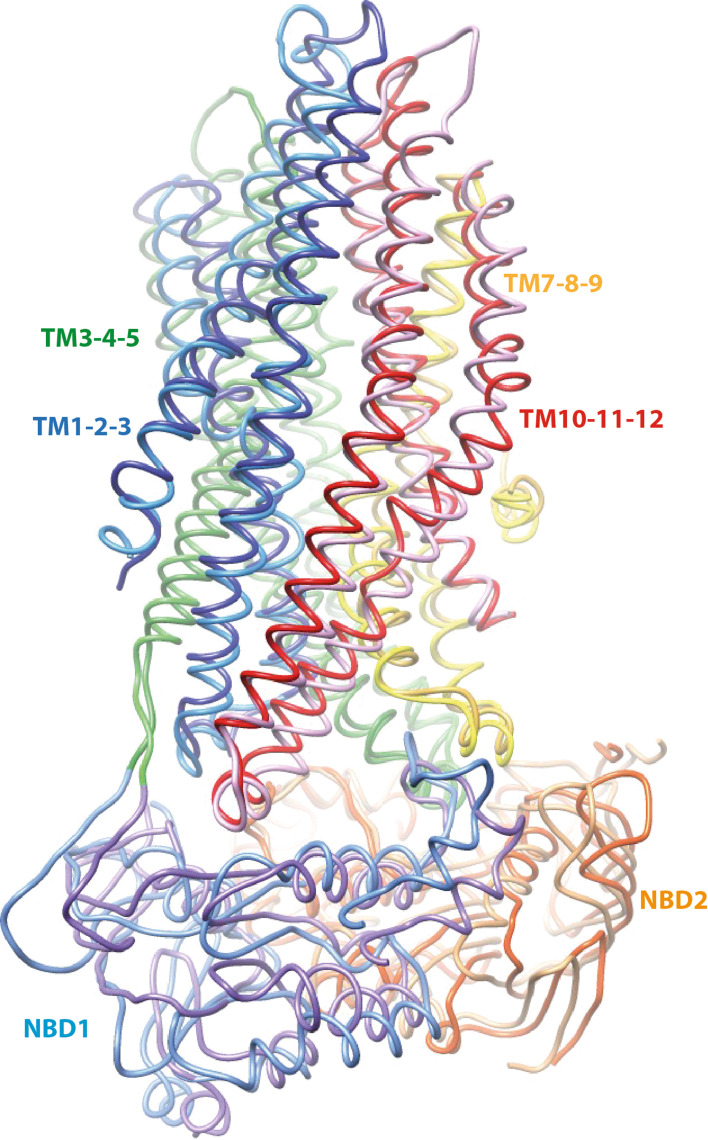
Finally, relying on the consistency of our models with experimental data, we show that the global transition between the quiescent and open forms of the CFTR channel (thus two extreme states) may be described through simple movements of four blocks of three consecutive TM helices. The different conformations can even be more generally described through the consideration of two stable blocks (as also recently proposed when comparing cryo-EM 3D structures [8]), each including NBD1 or NBD2, together with their interacting intracellular loops and associated TM helices.
Combining experimental and theoretical data, thus, provides a general framework to better understand and analyze the CFTR function, moreover giving atomic details which may be used for refining experimental 3D data acquired at medium resolution.
Materials and methods
Molecular modeling
To allow an easier comparison of the structures at the amino acid level, a model of the s-closed form of human CFTR was built based on the experimental 3D structure of the s-closed form of zebrafish CFTR [8] (PDBID:5W81), using Modeller 9v15 [25] and the alignment provided in Figure S1 of the Zhang’s article [8]. The two sequences share 55% identity, but the comparison of experimental 3D structures of CFTR in the dephosphorylated state has already shown that they are essentially identical [root–mean-square deviation (RMSD) of 1.9 Å for 1062 Cα positions] [7]. Accordingly, the RMSD observed here between the 3D structures of the model (human) and experimental (zebrafish) CFTR s-closed 3D structures is of 1.83 Å for 1060 Cα positions. Note that there is only one insertion in the zebrafish sequence (G64) within the available coordinates, located between Lh2 and elbow1, the other ones being located in the regulatory insertion (RI), in the regulatory region (R domain), within ECL4 (between TM7 and TM8), as well as in the linker insertion LI (no available density). To keep the conformation similar in the lasso region despite this insertion, we used the segment encompassing aa 61–77 of human CFTR in an apo conformation (perfectly superimposable to the corresponding segment of zebrafish CFTR both in an apo and s-closed conformations) to locally model the 3D structure of the human CFTR in the s-closed state (PDBID:5UAK), while the rest of the protein was modeled based on the experimental 3D structures of zebrafish CFTR in an s-closed state (PDBID:5W81).
A model of the Saccharomyces cerevisiae YOR1 was also built, using as template Sav1866 (PDBID:2HYD) and the previously published alignment [14], and submitted to molecular dynamics simulations (see below).
Molecular dynamics (MD) simulation
Details about the construction and molecular dynamics (MD) simulations of models of the human CFTR MSD:NBD assembly (which have led to models of the open and t-closed forms) are reported elsewhere [14]. These simulations have been performed without external constraints.
The same protocols were followed for MD simulations of CFTR models and CFTR-FlnA IG complexes for which the convergence has been checked and duplicates calculated (Supplementary Table 1).
Each model was embedded in a lipid bilayer consisting of POPC molecules using VMD or the charmm-gui platform [26–28]. The complexes were then immersed in an orthorhombic box filled with TIP3P water molecules [29]. Electric neutrality was achieved by the addition of Na+ [30] and Cl− [31] ions reaching a final concentration of 15 mM. The location of ions was optimized using a Monte Carlo approach.
The whole systems contain from 232,717 to 251,666 for CFTR models and from 258,460 to 544,929 atoms for CFTR-FlnA IG complexes. An additional MD was performed using CFTR intracellular N-terminal segment from 1 to 22 in complex with FlnA IG21 immersed in a cubic solvent box (48,773 atoms, Supplementary Table 1).
Once a system is prepared, equilibration steps were performed with the CHARMM36 force field [32] with the CMAP correction [33] in NAMD 2.9 [34]. The water molecule and ion positions were first minimized by 10,000 steps using the conjugate gradient method [35] and keeping the solute fixed. Harmonic restraints on the heavy atoms of the protein backbone and the ATP molecule (initially 500 kcal/mol/Å2) and the lipid heads (initially 800 kcal/mol/Å2) were then relaxed stepwise during a total of 3 ns equilibration using the NPT ensemble. The system was then released of any constraints, and a production phase was then conducted (the simulation time of each system is reported in Supplementary Table 1) at a temperature of 310 K and a pressure of 1 bar. Temperature was kept constant using Langevin dynamics [34], and a Nose–Hoover Langevin piston [36] was used for pressure control. Periodic boundary conditions were used to avoid artifacts at the box boundaries. Infinite-range Coulomb interactions were treated with the particle mesh Ewald approach [37]. For the van der Waals interactions, a switching function was applied at 10 Å and the cutoff was set to 12 Å. SHAKE was applied to constrain the bond lengths involving hydrogen atoms. The integration time step was 2 fs and coordinates were saved every 1000 steps (2 ps).
The RMSDs between the initial models and the last MD snapshots are given in Supplementary Table 1 and the figures legends. 10–25 ns appeared generally sufficient to reach stable models.
Metadynamics simulation setup
With the aim of obtaining a quantitative free-energy landscape connecting the open and t-closed CFTR conformations, a metadynamics simulation [38] was carried out using the same setup as above, employing NAMD 2.9 with Plumed version 2.3 [39]. A critical ingredient of free-energy calculations is the choice of a few collective variables: these are functions of Cartesian coordinates, meant to capture the slow degrees of freedom characterizing the transformation [40]. We adopted path collective variables (PCVs, [41]), S and Z, describing, respectively, the progress along, and the distance from, the CFTR closure/opening pathway:
where N is the number of reference configurations used to represent a putative pathway, D is a distance metric for structural similarity, and lambda is a parameter chosen as ~ 2.3/D(Rk,Rk+1) to obtain a smooth free-energy landscape. We defined D as the mean-square deviation among interatomic distances measured on a limited set of representative atoms involved in the conformational transitions of CFTR, namely 65 alpha carbons from the upper part of the CFTR central TM helices (Supplementary Table 2). These atoms display a displacement larger than 8 Å. As a reference pathway, we extracted 12 configurations along the CFTR closure pathway obtained from the unbiased MD simulation in [14].
Along the metadynamics trajectory, an external history-dependent bias potential is added to the Hamiltonian of the system. This potential is constructed as a sum of Gaussians periodically deposited in the PCVs space. At time t, the bias potential is written as:
where the total simulation time is 300 ns, W = 0.3 kJ/mol, σs = 0.08, and σz = 0.015 nm2, with a deposition stride τ = 2 ps (1000 steps). The deposited hills tend to fill the minima in the free-energy landscape: after an initial transient, observation of stationary conditions, with V(S,Z,t) growing approximately parallel to itself, is taken as evidence that the simulation reached convergence. At this point, a time average of the bias potential provides an unbiased estimate of the underlying free-energy surface F(S,Z) [42]. The landscape presented in Fig. 1d is obtained as the average V(S,Z,t) between 120 and 300 ns, with a statistical uncertainty estimated as 5 kJ/mol based on the deviation between F(S) profiles (obtained integrating the Z coordinate) averaged in the 120–210 and 210–300 ns intervals.
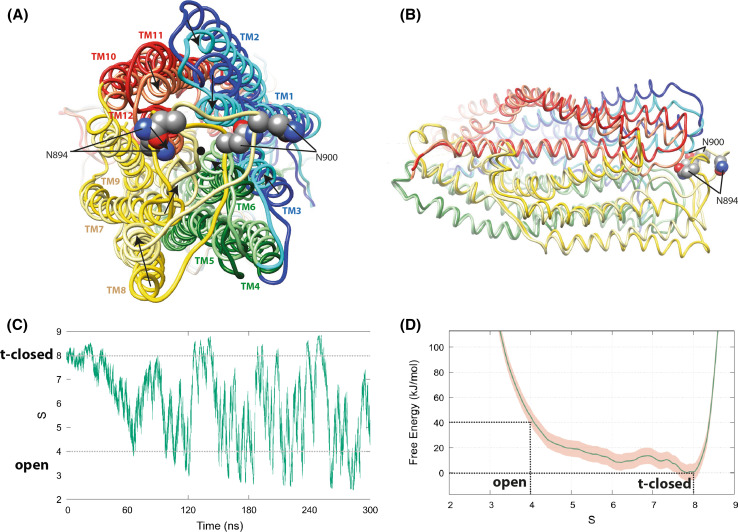
Fig. 1.
Models of the CFTR open and t-closed forms after metadynamics simulations. The initial metadynamics simulations were conducted with a CFTR sequence with a slight variation (D to N) in a non-conserved, likely non-critical position (249), but similar MD trajectories and reversible transition pathway were afterwards observed using the CFTR wild-type sequence. Coordinates of open and t-closed conformational states are provided for both simulations in our web site. a Superimposition (view from the extracellular side) of representative conformations of the open (dark colors) and t-closed (light colors) states after metadynamics simulations. Movements leading to the closure of the channel appear to be almost equally distributed on the upper parts of the TM helices TM1, TM2, TM3, TM6, TM7, TM8, TM11, and TM12, converging toward the pore center represented by a black dot. The asparagine N894 and N900 which are linked to glycans are highlighted. These positions suggest that glycans may be sometimes oriented toward the extracellular entrance of the pore. b Orthogonal view. For sake of clarity, the NBDs are not shown. The central and lower parts (ICLs, at left) of the TM helices are almost identical for the open and t-closed states (RMSD 1.83 Å for 405 superimposed Cα). c The progress along CFTR closure/opening pathway. The collective variable S is extracted from the metadynamics simulation and plotted in function of time. The variable tracks the similarity between each simulation snapshot and the reference configurations, using the mean-square deviation among interatomic distances on 65 alpha carbons from the upper part of the CFTR central TM helices (see “Materials and methods” section and Supplementary Table 2). d Free-energy surface F(S) from the metadynamics simulation. The profiles are obtained integrating the Z coordinate and averaging the bias potential in the 120–300 ns interval. The pink shaded region represents the deviation between F(S) profiles averaged in the 120–210 and 210–300 ns intervals
Molecular graphics and analyses
Molecular graphics and analyses were performed using DeepView/SwissPDBViewer [43] and the UCSF Chimera package [44]. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). In particular, the solvent accessible surfaces have been calculated by the DeepView/SwissPDBViewer tool, as well as side-chain rotamers. As energy minimization and MD simulations tend to fill cavities, it was sometimes necessary to choose different side-chain rotamers, while keeping the main chain unchanged, to obtain continuity in local areas (such as lateral portals).
Coordinates of models
The coordinates of representative open and t-closed conformations of CFTR obtained from metadynamics simulations are available at http://www.impmc.upmc.fr/~callebau/CFTR_2018.html.
Results
Further assessment of the models of the open and t-closed forms using metadynamics
First, we wanted to further assess the reliability of models of the open and t-closed forms, as the duration of the MD simulations performed to reach these models was relatively short [14]. We first performed a standard MD simulation starting from the same initial, open form of the model, after an independent equilibration step, to allow for the sampling of different initial conditions. This MD simulation led to a t-closed conformation similar to that observed before (Supplementary Data 2-A). Then, metadynamics was used for computing free energies associated with the models and for accelerating the occurrence of rare events (conformational changes). We considered here a set of collective variables (based on the positions of 65 Cα atoms from the upper part of the TM helices), capturing important features of the closure process (see “Materials and methods”). The underlying hypotheses are that CFTR parts not explicitly included in the variables would be able to follow the movement by virtue of mechanical connections, and that the sizable length of the simulation (300 ns) would allow degrees of freedom orthogonal to the biased variables to relax. In applying enhanced sampling techniques such as metadynamics to a system of the size and complexity of CFTR, it is mandatory to focus the biasing forces on a subset of atoms, to avoid ineffective dispersion of bias energy in the form of heat. The viability of our definition of collective variables appears supported by the fact that we observed reversible transition pathways (several channel opening/closing events), that are furthermore structurally homogeneous (both among them and with the spontaneous closing pathway [14]), resulting in a converged bias profile (Fig. 1). The statistical convergence of the latter has been assessed by estimating the degree of parallel growth of profiles averaged within different time intervals—a signature of stationary conditions resulting from the cancelation of the underlying free-energy profile [40, 42]. The reversible transition pathway was observed whatever the initial post-equilibration conformer of the open form is used as starting point (Supplementary Data 2-B).
Movements leading to the closure of the channel appear to be almost equally distributed on the upper parts of the TM helices TM1, TM2, TM3, TM6, TM7, TM8, TM11, and TM12, converging toward the pore center. The open state appears less stable from an energetic point of view, and, indeed, it rapidly evolves to the more stable t-closed conformation. The free-energy difference for S (the path collective variable describing the progress along the closure/opening pathway) values varying between S = 4 (open state) and S = 8 (t-closed state) is ~ 40 kJ/mol (Fig. 1d) and, hence, is comparable to the free energy released by the ATP hydrolysis (between ~ 20 and 40 kJ/mol) in solution [45–47]. This suggests that the transition observed here may not correspond to intra-burst flickering, but rather to inter-burst conformational transitions, as flicker closures occur with timescales that are not consistent with ATP hydrolysis [48]. Accordingly, flicker-closed conformations could correspond to intermediate states found in the proposed metadynamics cycle.
Reliability of the 3D models of the CFTR open and t-closed forms relative to cryoEM data
We then verified the reliability of the sequence alignment that we have made at the MSDs level with the Sav1866 template and its agreement with the observed, experimental 3D structures. We, thus, considered extreme open and t-closed conformers observed after metadynamics and compared them to the apo and s-closed conformations observed by cryo-EM. We considered more particularly the t-closed (model) and s-closed (cryo-EM) conformations, which represent the more similar conformational states, with respect to the apo and open states. To that aim, to allow an easier comparison at the amino acid level, we built a model of the s-closed form of human CFTR based on the experimental 3D structure of the s-closed form of zebrafish CFTR. Both 3D structures are essentially similar as assessed by a RMSD value of 1.83 Å for 1060 Cα positions (see “Materials and methods”).
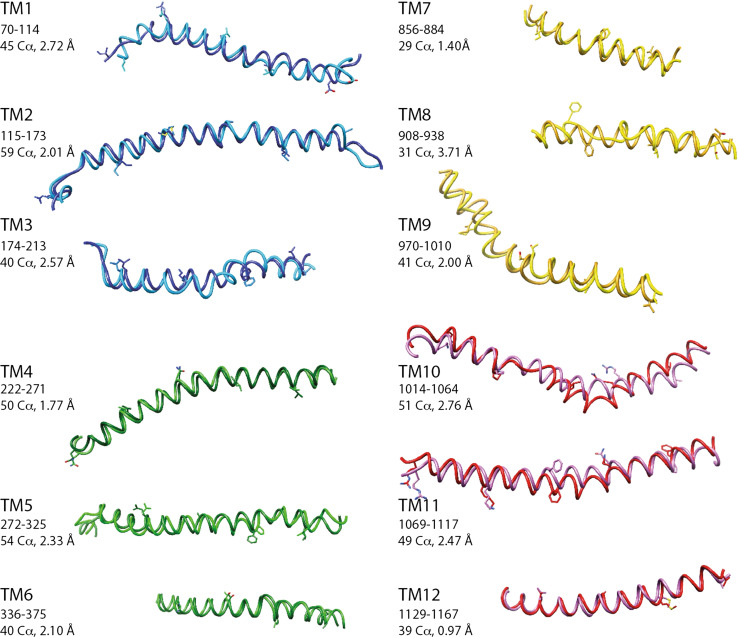
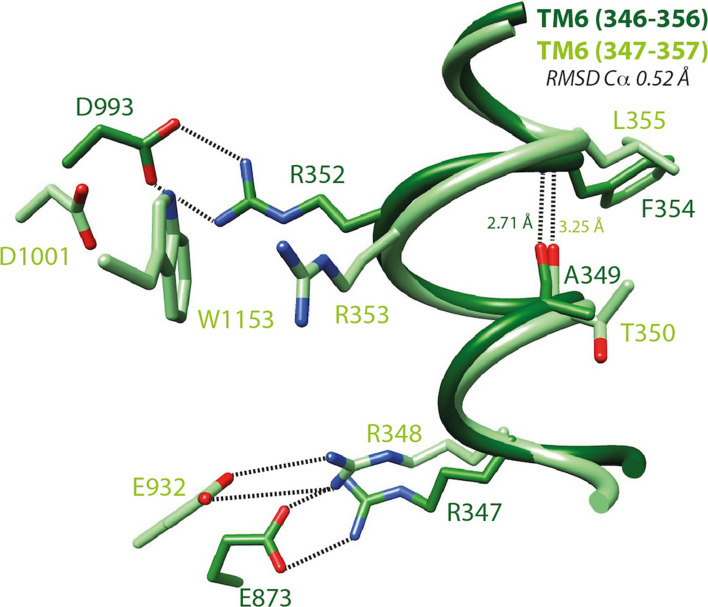
The overall match between the s-closed and t-closed structures is good (4.60 Å for 972 Cα positions, Fig. 2). At the level of the individual helices, the superimposition revealed a striking exact correspondence of amino acid positions, thus strongly supporting the reliability of our models (Fig. 3). This correspondence is observed within the frame of very similar conformations (RMS between 0.97 and 2.76 Å), except for TM8 (RMS 3.71 Å), in which unwinding is observed in both the experimental 3D structures and models, however, at different locations. A limited unwinding is also observed in TM10 for the t-closed conformation. It is important to note that this local difference in conformation is observed between positions that are perfectly in correspondence, and thus does not likely correspond to an error in sequence alignment, but rather may depict an alternative conformation (see below). Of note is that some geometrical peculiarities of TM helices, linked to critical functional features, were particularly well predicted in our models. This is, for instance, the case of a π-bulge within TM6 (with a i-i + 5 H-bond), located in the vicinity of R347 and R352 (zebrafish R348 and R353, respectively), both amino acids being involved in salt bridges critical for CFTR gating (Fig. 4, see below) [18].
Fig. 2.
S-closed/t-closed forms—comparison of the whole structures. Superimposition of the main 3D structure elements of the model of the human CFTR s-closed form (light colors, directly built from the zebrafish cryo-EM 3D structure [8]) and human CFTR t-closed form (dark colors). 4.60 Å RMSD for 972 Cα atoms (aa 70–402, 436–636, 846–856, 945–1167, and 1193–1426; the missing Cα atoms are those before aa 70 and after 1426, as well as those of RI, LI and a large part of TM7–TM8)
Fig. 3.
S-closed/t-closed forms—comparison of individual TM helices. Superimposition of the individual transmembrane (TM) helices of the human CFTR s-closed form (directly built from the zebrafish cryo-EM 3D structure [8], light colors) and t-closed form (dark colors). The considered segments and corresponding RMSD are indicated. Amino acid side chains are shown every ten residues, except for glycine [G930 (aa 931 is shown) and G1130 (aa 1129 is shown)]
Fig. 4.
S-closed/open forms—focus on TM6. Local superimposition of the TM6 segment including a π-bulge within the s-closed form (cryo-EM, zebrafish CFTR, PDBID:5W81 [8], light green) and the open form (model, human CFTR, this study, dark green) of CFTR. The depicted amino acids are equivalent between human and zebrafish CFTR, expect from E873 (human) and E932 (zebrafish), corresponding to E871 (zebrafish) and D924 (human), respectively. An H-bond between i (human A349/zebrafish T350) and i + 5 (human F354/zebrafish L355) stabilizes this local conformation, which is located in close vicinity of the two basic residues involved in salt bridges typical of the open (human R352/zebrafish R353; salt bridge with human D993/zebrafish D1001) and closed (human R347/zebrafish R348; salt bridge with human D924/zebrafish E932) forms, respectively. Of note are (1) the impossibility of forming directly a salt bridge typical of the open form in the reported s-closed conformation due to the presence of an intercalating tryptophane (zebrafish W1153, human W1145) and (2) the presence in the open form (model) of an alternate bond involving R347 and E873. Within this segment, the putative accessibility of V350 (V351 in zebrafish CFTR, not shown), discussed in a previous article [14], can be well understood regarding these data: V350 is totally buried in the open state of the channel and partially exposed to the solvent in the apo and s-closed states. V350 is also one of the four pivots described in Supplementary Data 16
This overall correspondence of conformation is also observed when considering four distinct blocks, each containing three consecutive TM helices (TM1–TM3, TM4–TM6, TM7–TM9, TM10–TM12) (Supplementary Data 3). This division of the MSDs into four 3-helix blocks is based on an internal symmetry that we reported previously, which can be considered for understanding the conformational transitions within ABC exporters [14]. The comparison of the t-closed, s-closed, and open forms also led to an overall good superimposition at the level of the three-helix blocks, as illustrated by low RMSD values. This additional comparison particularly well illustrates (1) the good match also existing at the level of the intra- and extra-cellular loops (ICLs and ECLs, respectively) and (2) that only small differences exist between these different conformational states. Again of note are the local conformational differences existing at the level of TM8, accompanied by a displacement of the TM7 position (see below).
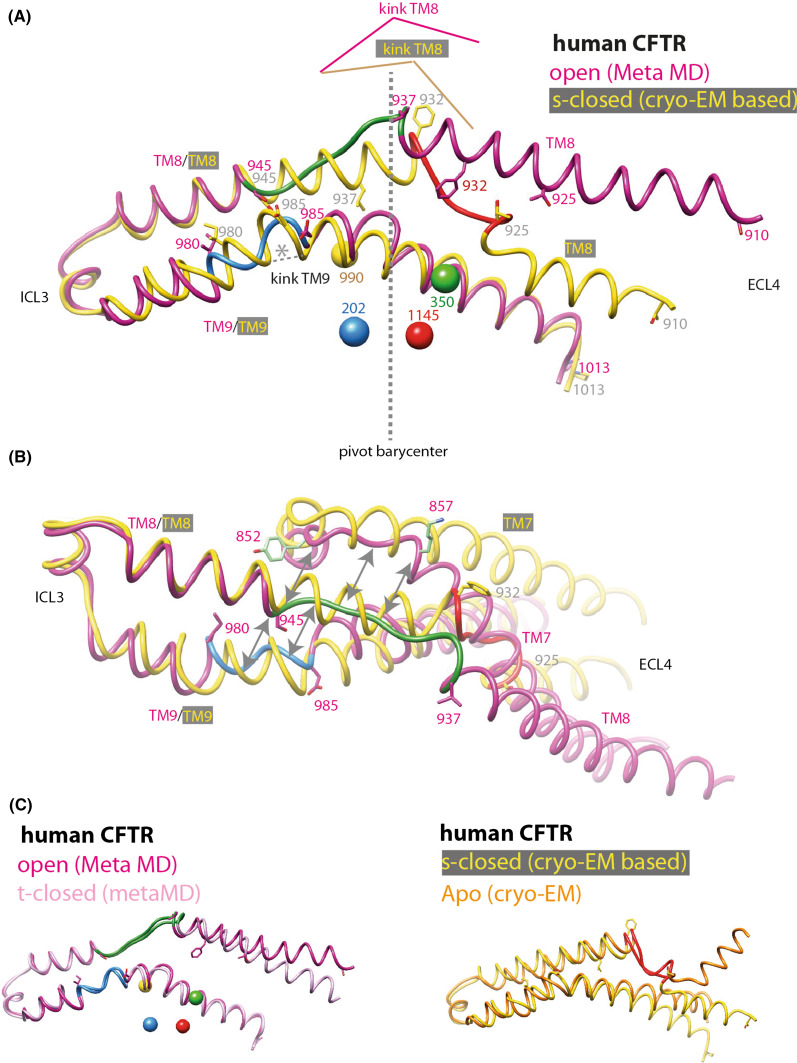
Focus on TM8
Having demonstrated the correspondence of the amino acid positions between the model and cryoEM 3D structure (see before), we noticed that we succeeded in identifying a striking feature of helix TM8, which is slightly shortened relative to the Sav1866 3D structure in terms of number of amino acids. This shortening in the three cryo-EM 3D structures is associated with an unwound sequence segment encompassing amino acids (aa) 925–932 (human CFTR numbering), while this sequence maintains a canonical helical character in our 3D models of the t-closed and open forms (Fig. 5). In contrast, in our models, it is the 937–945 sequence segment that is unwound, while having a canonical helical conformation in the cryo-EM 3D structures (Fig. 5). This unwinding is observed at the position where a two-amino acid deletion has been made relative to the Sav1866 template (aa 942–943), considering the best match with the ABC amino acid profile (see the corresponding alignment in the supplementary Data 1 of [14]). This unwinding did not propagate during MD simulations and did not lead to a change of orientation of TM8, in contrast to that observed in the cryo-EM structures. In contrast, the local perturbation of TM8 due to an additional one amino acid deletion introduced upstream (at the level of D924) was abolished upon MD simulation (data not shown), thereby weakening the possibility of bias associated with these local singularities. More interestingly, no orientation change was also observed after 20 ns MD simulations when the amino acid deletion was shifted upward (aa 931–932), at a location similar to that for which unwinding is observed in cryoEM data (data not shown). Worth noting is the recently reported cryo-EM 3D structure of chicken CFTR, however, solved at low resolution, but indicating no kinked conformation for TM8 [49]. This is in agreement with our model and strongly supports the overall plasticity of TM8, with an unwound segment, occurring in experimental and modeled TM8 3D structures at both sides of a kink located at similar positions (Fig. 5). Taking into account the usual cautions linked to modeling, this suggests that an orientation of TM8 different from that observed in cryo-EM (allowed by the flexibility and possible sliding associated with local unwinding around this kink) may be the key factor for evolving toward an active channel.
Fig. 5.
S-closed/open forms—focus on TM7–TM8–TM9. Local superimposition of the TM7–TM8–TM9 block (a, b two different orientations) within the model of the human CFTR s-closed form (directly derived from the cryo-EM 3D structure of zebrafish CFTR, PDBID:5W81 [8], yellow) and open form (pink). 68 Cα atoms (aa 945–1013) were superimposed, with a resulting RMSD of 2.02 Å. The unwound segments are reported in green (aa 937–945), blue (aa 980–985), and pink (aa 852–857) for the open form, in red (aa 925–932) for the s-closed form. Of note is the opposite situation for wound/unwound segments at the level of the kink of TM8 for the open and s-closed forms. The four pivots around which the conformational transitions can be described (aa 202, 350, 990, 1145) are also reported, with their barycenter located at the junction of the unwound segments between the open and s-closed forms. c Stability of the compared segments in the active (left) and inactive (right) conformations.
We also note that the kink is located at the level of the barycenter of the four pivots (aa 202, 350, 910, and 1145, colored balls in Fig. 5a), around which conformational transitions between the open and closed forms of the models can be described (see “Discussion”). Interestingly, the unwound, extended conformation of TM8 in the structure models (aa 937–945) appears to be correlated to extended conformations in both TM7 (852–857) and TM9 (980–985), while the secondary structure regularity in other regions remains unchanged (Fig. 5b). This feature likely excludes the possibility of a local artifact on the TM8 model. It should be noted that local winding/unwinding of ABC TM helices occurring during conformational transitions was very recently highlighted in the P-glycoprotein 3D structures, solved in inward- and outward-facing conformations [50, 51]. Indeed, as suggested here for the CFTR TM8 925–937 segment, a striking difference exists at the level of two symmetrical TM helices (TM4 and TM10) of the P-glycoprotein, being unwound in the inward-facing conformation, while wound as a regular helix in the outward-facing conformation (Supplementary Data 4). However, for CFTR, the situation appears more complex as a shift toward an unwound downstream segment is observed in the other conformation (Fig. 5a).
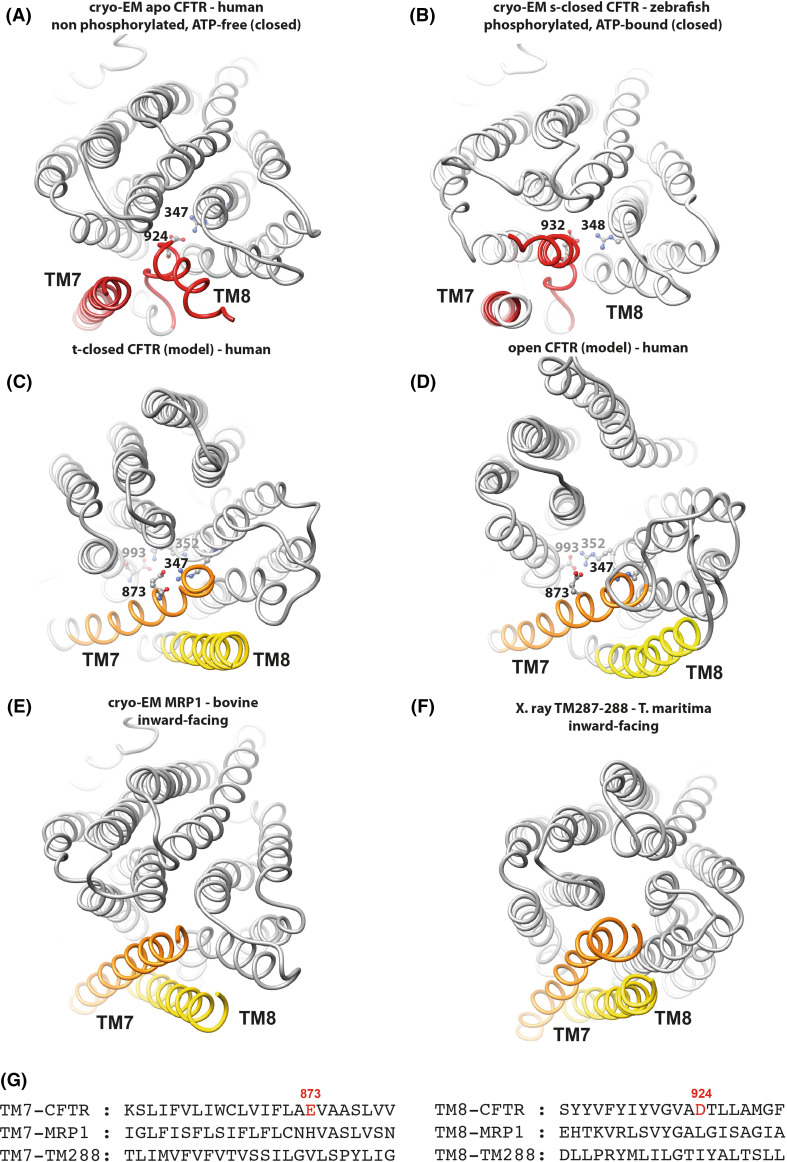
The alternate, canonical orientation of TM8 observed in our models (Fig. 6c,d) correlates with the presence of several markers of the active channel. In the cryo-EM 3D structures, TM8 unwinding allows D924 (zebrafish E932) in TM8 (black in Supplementary Data 5) to make a salt bridge with R347 (zebrafish R348) in TM6 (green), a bond that is more typical of the closed form of the channel [18], while the second part of TM7 (purple) moves outward. This salt bridge is shown in details in the right middle and bottom panels of Supplementary Data 5. The presence of an acidic amino acid at position 924 (establishing the salt bridge with R347) is specific of the CFTR sequence and may explain the TM8 unwinding and the unusual position of the TM7–TM8 couple (Fig. 6a,b), which does not exist in other ABC exporters whose 3D structures were solved in inward-facing conformations. This is exemplified in Fig. 6e, f with the recently published cryo-EM 3D structure of bovine MRP1 (PDBID:5UJ9 [52]) or the X-ray 3D structure of Thermotoga maritima TM287/288 (PDBID:4Q4A [53]).
Fig. 6.
The TM7-TM8 pair. Comparison of 3D structures, viewed from the extracellular side, of the membrane-spanning domains (MSDs) of CFTR from cryoEM 3D structures in closed conformations [a apo, quiescent form (human): PDBID:5UAK [7]. b s-closed form (zebrafish): PDBID:5W81 [8]. Amino acid (aa) 932 is equivalent to human CFTR aa 924, aa 348 to human CFTR347)] and from human CFTR 3D structure models in a t-closed form (c) and in an open form (d) [14]. These 3D structures were compared to two experimental 3D structures in inward-facing conformations (e bovine MRP1, PDBID:5UJ9 [52]; f T. maritima TM287/288, PDBID:4Q4A) [53], highlighting the atypical position of the TM7–TM8 helices in the two cryo-EM CFTR 3D structures. Amino acids participating in salt bridges in the different conformations are shown in a ball-and-stick representation (also see Supplementary Data 5). g Structural alignment of the sequences of the CFTR, MRP1, and TM288 TM helices 8 and 7, highlighting specific features in the CFTR sequence
The alternate conformation that we observe in our models, which lacks the salt bridge [human D924-R347] of the s-closed form, is stabilized by another salt bridge between R347 (in TM6) and E873 (in TM7) [Fig. 4 and Supplementary Data 5 (top right)] and, although the pore is still closed, it already displays the D993–R352 salt bridge typical of the open form [18]. These bonds are also observed in our model of the open form (Fig. 6c, d), however, being stronger in this last case. Figure 6a–d indicates that in the open and t-closed forms (models), E873 substitutes D924 [apo and t-closed forms (cryo-EM)] for interaction with R347, TM7 [open and t-closed forms (models) locally taking the place occupied by TM8 (apo and t-closed forms (cryo-EM)]. In the reported cryo-EM structure of the zebrafish CFTR s-closed form, the open state-specific salt bridge (D1001–R353, equivalent to human D993–R352) is unable to form, due to the presence of an intercalating tryptophan (W1153, equivalent to human W1145), which establishes a cation/π interaction with R353 (Fig. 4 and Supplementary Data 6). However, MD simulations performed on this s-closed form [either the zebrafish experimental 3D structure (μs-long MD simulation) [10] or a model of human CFTR, based on this last experimental 3D structure (100 ns MD simulation), see “Introduction”] highlighted a rapid formation of an optimal D993–R352 bridge (distances of 2.86 Å and 2.72 Å), due to a small movement of W1145, while the R347–D924 remains stable (distances of 2.77 and 2.66 Å). Finally, it is also noteworthy that channel closure involves a similar environment in the apo, s-closed, and t-closed forms of CFTR, around the highly critical F337 residue (Supplementary Data 7).
Thus, comparison of experimental and theoretical data strongly suggests that the t-closed conformer, observed in our MD simulations and whose reliability is further supported by metadynamics simulations, could be a required passage, allowing a fast transition toward the open state of the channel.
Comparison of the cryo-EM 3D structures of the apo, quiescent form and of the s-closed form led Zhang and colleagues [8] to propose that local movements of upper parts of TM8 and TM12 control channel opening. In the present work, the comparison of cryo-EM 3D structures and our models show that local differences do indeed exist at this level, with a progressive evolution of the beginning of TM8 (human 910–923, zebrafish 918–931) from an s-closed to an open conformation, through an intermediate t-closed state (Supplementary Data 3—block 3). However, in contrast to what is suggested by Zhang and colleagues [8], we propose that this should not be the unique factor implied. Indeed, other, possible complex conformational changes, involving repositioning of the TM7-TM8, are likely necessary to allow transition toward the open state, as observed in our models of the t-closed and open forms. The presence of a π-bulge in TM6, located in the vicinity of the critical residue R352 and shared by the s-closed form and our model of the open form (Fig. 4), is likely to play a critical role in this conformational plasticity, as reported in other systems [54, 55].
Finally, we note that the peculiar behavior of TM8 and its mobility may be associated with the fact that its extracellular end holds the unique long extracellular loop ECL4 of CFTR and its two glycosylation sites (Fig. 1a). Striking difference in orientation is observed for this upper part of TM8 not only between the cryo-EM apo and the s-closed conformers (Supplementary Data 7), but also between the open and t-closed forms in our MD simulations (Supplementary Data 2), thereby further highlighting its critical role.
Critical features of the open state of the CFTR channel, as deduced from modeling
Some features, specific of the open-channel model and related to the MSDs and NBDs, were reported previously [14], but are here further described as they shed new light on unexplored features of cryo-EM data. In addition, we present here novel predictive insights into the interactions that can be mediated by the CFTR N-terminal region, within the context of its potential plasticity. Of course, some of such predictions made outside the frame of known templates may be considered as speculative as no experimental 3D structure is yet available to support their reliability in a straightforward way. This explorative research was, however, supported by sound experimental data and may, thus, open new avenues for further experimental characterization.
Lateral openings
An important feature revealed by our model of the open form of the channel [14] was the presence of cytoplasmic portals, displayed at the level of the ICLs, which together form a tight two-layered bundle of helices. The diameters of these portals [typically twice the size of a chloride ion (~ 7 Å)] are sufficient to allow anions, such as Cl− or HCO3−, to access the central channel pore from the cytosol. The four equivalent portals, almost perpendicular to the central vertical channel pore itself, are arranged in a cross-shaped configuration, the entrance of each being located between the cytoplasmic extensions of two TM helices: TM5–TM8, TM2–TM11, TM10–TM12, and TM4–TM6 (Supplementary Data 8-A), and their shapes appear similar in this open form. Basic side chains are generally located at or near each portal entrance, probably attracting anions toward the central pore.
This prediction has been supported by experimental data based on patch clamp recording and substituted cysteine accessibility mutagenesis (SCAM) [21, 22]. Indeed, consistent with our model, the side chains of basic residues such as K190, R248, R303, K370, R1041, and R1048, located between the cytoplasmic extensions of TM4–TM6 and, to a lesser extent, TM10–TM12, were shown to play a role in the attraction of chloride ions [21]. Involvement of the TM5–TM8 and TM2–TM11 portals was, however, not studied in these experiments. A very recent study, based on additional experiments including channel blockers, pleads in favor of the critical role of the TM4–TM6 portal [56]. We show here that this regularity is broken in the t-closed form, in which only the TM4–TM6 portal appears to be present (Supplementary Data 8-B). Interestingly, we also show that similar portals can also be observed in the cryo-EM quiescent apo 3D structure at the same locations, provided that alternative rotamers of some side chains are considered (Supplementary Data 8-C). However, these “apo” portals do not exhibit the regularity observed in the model of the open state. In particular, the TM4–TM6 portal is transformed into a large entrance; it may, however, remain similar to what we observed with our open-state 3D structure, when adding a possible model of the R region (in agreement with the cryo-EM weak electron density). The channel linked to the TM5–TM8 portal is longer, a feature associated with the opening of the MSD toward the cytosolic side. In contrast, the two other portals are shorter and closed by elements typical of the apo structure, i.e., the R region for the TM10–TM12 portal and the N-ter lasso for the TM2–TM11 portal.
The opening between TM4 and TM6 is also observed in the cryo-EM 3D structure of phosphorylated, ATP-bound s-closed CFTR [8], but no other lateral portal has been clearly reported in this 3D structure, although the TM10–TM12 one is still present (Supplementary Data 8-D).
Thus, the cryo-EM data provide here the evidence that, although still inefficient, some sketches of portals already exist in these inactive forms, supporting the relevance of the four, functional ones observed in our model of the CFTR open form. It has to be noted here that the local unwinding of TM8 (see before) is located at the level of the entrance of one of the four portals in the CFTR open form, while the three other lateral openings are formed between unwound TM helices (Supplementary Data 9). Of note is that winding/unwinding of TM helices is also observed in the P-glycoprotein 3D structure (see before), for which a recent 20 μs coarse-grained simulation has shown that two lateral portals allow phospholipids to enter the central pocket [57]. The position of these portals is close to the TM4–TM6 and TM10–TM12 portals of CFTR, conserved between the CFTR open and s-closed conformers.
In conclusion, the presence of four similar, narrow portals only in the model of the open form of CFTR (Supplementary Data 8-A) suggests that this situation may increase ion flux, while inhibiting the entrance of larger substances.
Upward movement of the NBD1 F508 region in the CFTR open form
When compared to the experimental 3D structures of the inward-facing apo conformation [6, 7], our model of the open, outward-facing conformation of the CFTR channel is characterized by a significant upward movement of the NBD1 alpha-subdomain, carrying F508, along ICL4, from a position in close contact with R1070 toward another one, in contact with L1077, thus approximately two helical turns upward (Supplementary Data 10). This position of the NBD1 alpha-subdomain, also present after our metadynamics simulation, clearly differs from the two experimental 3D structures of other ABC exporters in outward-facing conformations (Sav1866 [58] and MsbA [59]). We, moreover, show here that this movement is also not observed when MD simulations similar to those performed on our Sav1866-based model of CFTR were performed on a model of the 3D structure of yeast YOR1, which was built using the same Sav1866 3D structure as template (and the alignment provided in the Supplementary Data of [14]) (Supplementary Data 11-A). YOR1, which works as a pump for the mitochondrial toxin oligomycin, is asymmetric, having like CFTR a long regulatory insertion and a degenerate ATP-binding site, and has an equivalent mutation to the human CFTR F508del, known as F670del, which like CFTR also leads to misfolding and degradation [60]. We have also analyzed the recent very long MD simulations which have been performed on the asymmetric TM287/288 experimental 3D structure [61]. In their work, Göddeke and colleagues have indeed made long all-atom MD simulations (500 ns each), starting from an inward-facing conformation, leading to clearly recognize three major, distinct conformers of ABC exporters (inward facing, occluded, and outward facing). They observed spontaneous conformational transitions toward an outward-facing state, similar to the Sav1866 3D structure (RMSD between 2 and 3 Å). We also observe that our model of the open form is very close to their TM287/288 outward-facing state (RMS 2.98 Å for 449 Cα), while the TM287/288 intermediate occluded state is similar to the conformation of the t-closed model (RMSD 3.15 Å for 450 Cα) and to that of the s-closed 3D structure (RMS 3.60 Å for 412 Cα). No upward movement of the NBD alpha-subdomains can, however, be observed in the outward-facing conformation after these MD simulations (Supplementary Data 11-B).
Upward movement is also not observed in the s-closed, ATP-bound form of zebrafish CFTR, which as already mentioned before, is locked in a closed conformation. It is, thus, possible that the movement of the NBD1 alpha-subdomain, together with the formation of the open form-specific, optimal R352–D993 salt bridge and unwinding at the level of the block TM7-8-9, may be linked to the opening of the channel. Of note is that the different positions of the NBD1 alpha-subdomain between the model of the open form and the cryo-EM 3D structures of the s-closed/apo forms are observed while the ICL4s are perfectly superimposable, suggesting that the alternate position may be relevant at the functional level.
We show here that this alternate conformation of the NBD1 alpha-subdomain is further stabilized by a specific network of local interactions (Supplementary Data 10-D). Indeed, one can observe: (1) a strong salt bridge (2.9 Å) between R1070 and E504, (2) a π–π interaction between the two antiparallel tyrosine Y512 and Y563, (3) a cation–π interaction between F1068 (within ICL4) and R560 (in NBD1) in a full parallel contact and another one between the same F1068 and K564 (in NBD1), and (4) an hydrophobic cluster around F508, including I507, Y1073, T1076, and L1077. Importantly, this new situation of F508 also leads to an anti-clockwise movement of the associated loop by about 90° (Supplementary Data 10), resulting at least in a close contact with its neighbor I507, as well as the formation of a salt bridge between E504 and R1070.
Some parallels can be drawn with ABC type 2 importers, which are different from type 1 exporters, the family to which CFTR belongs [62], but which share a similar ICL coupling helix within each MSD (corresponding in ABC type 1 exporters to ICL2 and ICL4 or ICL2′, depending on the fact that the exporter is encoded on a single or two separate chains). This coupling helix docks into a cleft between the NBD RecA-like and helical subdomains (Supplementary Data 12). The importance of the NBD α-helical subdomain for signal transmission toward the MSDs has been well described for MJ1267 [63], in which an outward rotation of this subdomain is coupled to the loss of its molecular contacts with the ATP and the glutamine of the Q loop. As detailed in Supplementary Data 13, similar contacts are observed between ICLs and the NBD amino acids equivalent to F508 [64, 65]. Most importantly, some ABC type 2 importers do possess a NBD-specific insertion (called the LIVG insert), linked by allostery to the ATP-binding site [66] and positioned in a similar “upward” position than that observed for NBD1 F508. This observation, thus, gives further, although indirect, credit to the likelihood of CFTR NBD1 alpha-subdomain “upward” position that we predicted here.
The N-terminal region
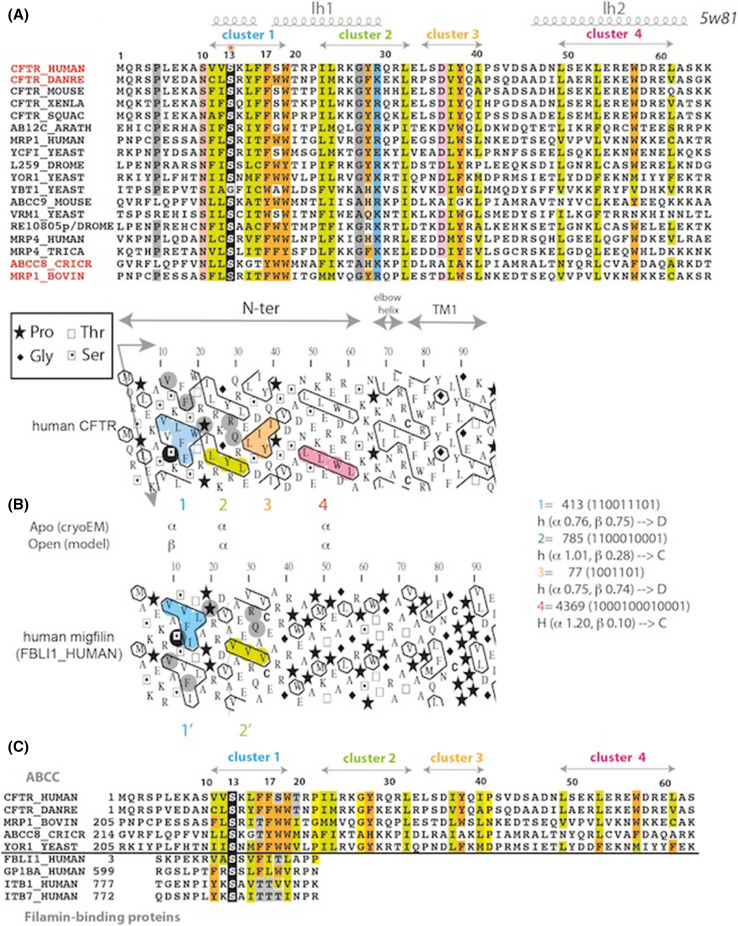
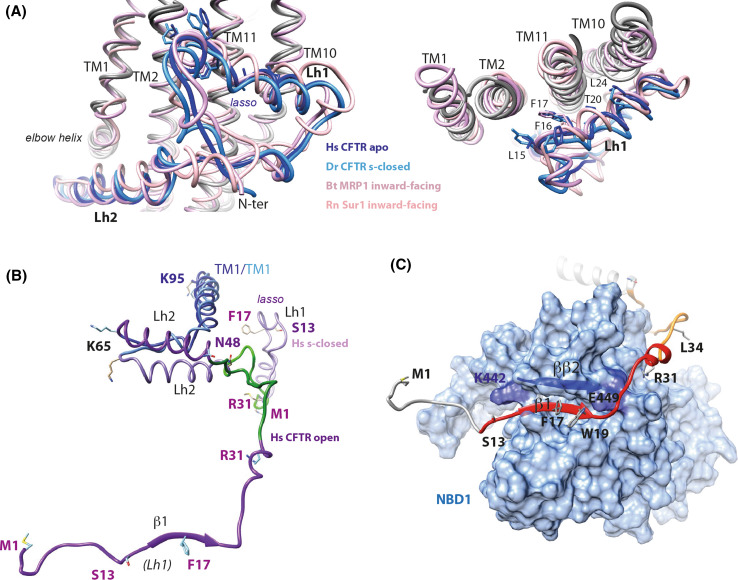
The N-terminal sequence of CFTR, upstream MSD1, is well conserved among ABC exporters of the ABCC subfamily (Fig. 7a), indicating that it may play an important structural and/or functional role. The three cryo-EM 3D structures of zebrafish and human CFTR [6–8] have revealed that these N-terminal extensions adopt almost identical “lasso” conformations, also present in the cryo-EM inward-facing 3D structures of bovine MRP1 [52] and of mouse/rat SUR1 (ABCC8) [67, 68] (Fig. 8a). The 40 first amino acids indeed constitute a circular “noose” structure, which includes lasso helix 1 (Lh1, aa 11–29, split in two orthogonal helices: aa 11–16 and aa 18–29) packed against TM helices (mainly TM1–TM2 and TM10–TM11) principally through hydrophobic interactions involving for CFTR L15, F16, F17, T20, L24, and Y28. The extended end of the lasso, long helix 2 (Lh2, aa 46–61), is tucked under the elbow helix (aa 66–72) (Fig. 8b).
Fig. 7.
Sequence of the N-terminal extension of human CFTR. a Alignment of the sequences of the CFTR N-terminal extension and from exporters of the ABCC subfamily. UniProt identifiers are indicated in front of the sequences, except for the sequence of D. melanogaster RE10805p (genbank identifier: ACD81872.1). Secondary structures, as observed from the cryo-EM data (PDBID:5W81), are reported up to the alignment, as well as the positions of hydrophobic clusters (b). b HCA plot of the N-terminal extension sequence of human CFTR. In brief, the sequence is shown on a duplicated alpha-helical net, on which the hydrophobic amino acids are contoured. These form clusters, the positions of which mainly correspond to regular secondary structure limits. This representation allows the comparison and alignment of highly divergent sequences, based on the fact that secondary structures are much more conserved than sequences. Horizontal clusters tend to form α-helices, vertical ones extended structures (β-strands). At right are indicated the propensities of the hydrophobic clusters (designated with their Peitsch and binary codes) for the α- and β-state [80]. Clusters 2 and 4 are mainly associated with α-helices (H and h correspond to strong and weak propensities, respectively). However, clusters 1 and 3, which have ambiguous shapes, harbor a discordant behavior [D as opposed to concordant (C) observed for clusters 2 and 4)], being associated even more frequently with the opposite secondary structure, depending on the amino acid composition and/or to the environment [81]. The HCA plot of the human CFTR N-terminal sequence is compared here to human migfilin (Uniprot Q8WUP2, FBLI1_HUMAN), a protein that serves as an anchoring site for filamin-containing actin filaments. In particular, it binds to the FlnA IG21 domain through its first hydrophobic cluster [69, 72, 73]. The serine residue highlighted in black is aligned with the CFTR serine residue S13, which is conserved among ABCC proteins. The similar shapes of clusters 1/1′ and 2/2′ are indicative of similar 2D structures. c Corresponding sequence alignment between the N-terminal extension of ABCC proteins and members of the migfilin family (designated using their UniProt identifiers)
Fig. 8.
Structures of the CFTR N-terminal extension: the lasso and a possible, alternate conformation. a Superimposition of the lasso conformations of the N-terminal extension of CFTR, in an apo, ATP-free state [human (Hs) CFTR apo [7], PDBID:5UAK, dark blue], in the s-closed, ATP-bound state [zebrafish (Dr) CFTR s-closed [8], PDBID:5W81, light blue], in the N-terminal extension of bovine (Bt) MRP1 [52], (PDBID:5UJA, dark pink) and in rat (Rn) SUR1 [68], (PDBID:5TWV, light pink). Two orthogonal views are displayed, highlighting the particular conformation of this sequence segment (left) and its contacts with TM1-TM2 and TM10-TM11 (right). b Structure of the CFTR N-terminal extensions (light purple) in the human s-closed conformation (“lasso”) and, as hypothesized, in the open conformation (dark purple, this study after 20 ns MD simulation). This view was done after superimposition of parts of the TM1 helices (blue, aa 65–95, RMSD 2.88 Å on 31 Cα positions). The lasso segment encompassing aa 32–48 was rotated around aa N48, in its native conformation, leading to directly allow the association of strands β1 and NBD1 ββ2, before MD simulation. c View of the contacts that the CFTR N-terminal region is proposed to make with NBD1 (solvent accessible surface in light blue; aa 382–645, with strand ββ2 highlighted in dark blue). The segments that were experimentally shown to make weak and strong contacts with a NBD1 construct (aa 373–589) are shown in orange and red, respectively [82]
The filamin context
A first issue relative to the CFTR N-terminal sequence concerns its ability to interact with partners of the membrane traffic machinery. Previous experimental work has shown that the CFTR 20 first amino acids interact with the cytoskeletal filamin (FlnA and FlnB) proteins, which are known to stabilize CFTR at the cell surface by anchoring it to the actin cytoskeleton [69]. Filamins are large dimers of ~ 280 kDa subunits, which possess amino-terminal actin-binding, calponin homology domains followed by 24 immunoglobulin-like (IG) repeats organized as long, linear rod-like strands, the most C-terminal repeat mediating dimerization [70, 71]. Two flexible hinges separate IG15 from IG16 and IG23 from IG24, respectively. Filamin proteins crosslink F-actin to form orthogonal networks that are responsible for cellular integrity and attachment to membrane receptors, including ion channels. Two studies [72, 73] have reported that IG19, IG21, and IG23 are major components of the interaction of FlnA with the N-terminal sequence of CFTR. It has been shown that IG23 is a major component, followed by IG19, IG21, and farther, IG17 [72]. Moreover, as highlighted in the experimental 3D structure of the first 20 amino acids of CFTR in complex with FlnA IG21 [73], the CFTR N-terminal sequence adopts a conformation made of a β-strand interacting in an antiparallel fashion with the FlnA IG21 strand βC. This conformation was observed between two FlnA IG21 domains, but the IG21(B)-CFTR interaction was thought to be an artifact, as a 1:1 stoichiometry is observed in IG titration experiments [73]. We here supported this 1:1 stoichiometry as we observed a stable CFTR extended conformation (aa 1–22) in complex with a single IG21(A) molecule after 50 ns of molecular dynamics simulation (RMSD between the initial and final extended conformations: 2.61 Å). This conformation is, thus, completely different from the helical “lasso” Lh1 observed in the cryo-EM structures. Of note is that FlnA IG19 and IG23 can compete with FlnA IG21 for CFTR N-terminus binding [73] and that the CFTR serine S13 (mutated in CF patients) plays a pivotal role in such an interaction.
CFTR s-closed conformer and filamin
The most probable partner of the CFTR N-terminal sequence within the context of the s-closed conformation would be the FlnA IG23 domain, as the last filamin IG24 domain is in contact with the inner leaflet of the cytoplasmic membrane [74] and homo-dimerize through its CD face. This hypothesis is supported by the recent cryo-EM structure of the Sur1/Kir6.2 complex [67]. Indeed, the 307–326 segment of Kir2.1 (equivalent to the 319–338 segment of Kir6.2) was previously shown to bind to part of the 2481–2647 fragment of FlnA (end of IG23 and IG24) [75]. On the 3D structure of the Sur1/Kir6.2 complex [67], the Kir6.2 317–334 loop (interacting with FlnA) is located at the immediate vicinity of the Sur1 lasso Lh1 helix. This observation, thus, provides strong support for the interaction of the Lh1 helix with the FlnA IG23 domain. The observation that IG23 is a major component of CFTR/FlnA interaction [72] also further supports this hypothesis.
Regarding this possible interaction with IG23, two hypotheses can furthermore be considered: either (1) the interaction occurs with the very N-terminal sequence (aa 1–21) in the conformation observed in the cryo-EM 3D structures or (2) it may involve partial flexibility of this sequence, while the other amino acids (aa 22–65) remained in the observed lasso conformation. Indeed, the first 21 amino acids are largely found in the cryo-EM 3D structures in coil conformations, with only aa 11–16 corresponding to the first part of the Lh1 helix, being partially anchored to the membrane. A particular flexibility of this last sequence can be predicted, as it includes a hydrophobic cluster which may fold as either an α-helix or a β-strand (see below) and as it has been observed in an extended conformation in IG21-CFTR complexes [73].
For further exploring our two hypotheses, we have manually docked the βCD face of the IG23 experimental 3D structure (PDBID:2K3T) onto CFTR aa 1–21, whether these amino acids are partly in helical conformation (as observed in the cryo-EM data) or in a β-strand conformation, as seen in the FLNa IG domain/CFTR complexes [73]. The βCD face of FlnA odd IG domains is indeed known to be sometimes masked by intramolecular interactions with strands βA strand of even-numbered domains [76] but is relieved through interactions with partners (such as the CFTR N-terminus in a β-strand conformation).
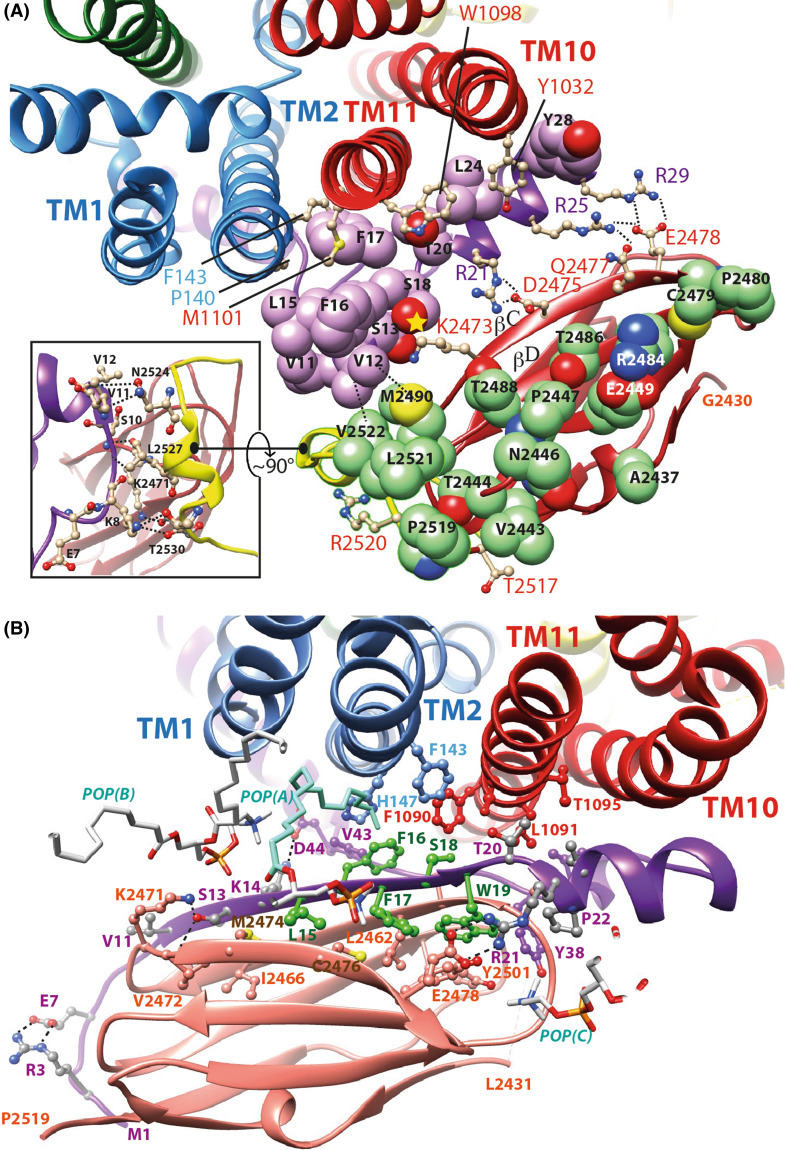
Unmodified lasso Regarding our first hypothesis (interaction of IG23 with intact lasso), we thus manually docked the IG23 domain on the CFTR lasso Lh1, as it exists in the cryo-EM 3D structures (Fig. 9a). The stability of the complex was further supported after MD simulations of 50 ns. These simulations did not lead to any steric clash and instead, revealed many contacts among which with the critical serine CFTR S13 in the vicinity of FlnA K2473 (Fig. 9a). Of note is that this last amino acid is the immediate neighbor of V2472, which has been reported to be critical for the interaction of CFTR in a β conformation with FlnA [73]. K2473 also interacts with CFTR S18, which has otherwise been reported to interact with the FlnA IG21 domain within the context of a CFTR N-terminal β-strand conformation [73].
Fig. 9.
Two possible models of the hypothetical association of the human filamin IG23 domain with CFTR N-terminus lasso. a Model of the FlnA IG23/CFTR Lh1 complex, which repeatedly showed a good stability after 50 ns of MD simulations. The carbon atoms of the CFTR Lh1 lasso are colored in purple, those of FlnA IG23 in green. Two main interaction regions are present in this model: at right polar contacts with Lh1 S13, S18, R21, R25, and R29 and at left hydrophobic contacts involving Lh1 V11, V12, V15, and F16 with IG23 V2522, M2490, and L2521. Amino acids of TM2, TM10, and TM11 interacting with the lasso Lh1 are depicted in ball and sticks. F143, P140, M1101, W1098, and Y1032 are at the same level with respect to the membrane than the IG23 amino acids depicted in green. In the inset are indicated the contacts between the lasso segment including aa K8–V11 and the beginning of the IG23–IG24 linker (in yellow), which may strengthen the stability of the complex. This region lies nearly perpendicular to the Lh1/IG23 main contact shown in this panel. b Model of the FlnA IG23/CFTR modified Lh1 (aa 1–21). Amino acids 1–21 are here considered in a different, extended conformation (β-strand between V11 and T20), all the other parts of the lasso remaining unmodified. The conformation presented here is stable after 50 ns MD simulation and displays favorable interactions with the TM1, TM2, and TM11 helices as well as with the membrane lipids. Three of them are depicted [POP(A), POP(B) and POP(C)]. The POP(A) chain colored in light blue (between TM2 and CFTR F16) stayed very stable along the MD simulation. The sequence L15–W19 is colored in green. Of note is that the zebrafish CFTR sequence (Y15–F16–F17–W18–W19), like several ABC exporters, possesses an aromatic amino acid at position 18. The L15Y and S18W substitutions are fully compatible with the present model, with the following immediate contacts [CFTR Y15 = FlnA M2474 and C2476, CFTR F17, POP(A), CFTR F16 = POP(A), CFTR V43, H147 and F1090, CFTR F17 = FlnA C2476 and Y2483, CFTR Y15 and W19, CFTR W18 = FlnA 2461, CFTR W19, F143, F1090, L1091, CFTR W19 = FlnA T2454, L2462 and Y2483, CFTR F17, and W18]. The orientation of FlnA IG23 domains is globally similar for the two models, with a relatively small translation toward TM1 and TM2 (~ 8 to 9 Å). IG23 E2478 establishes a salt bridge with R25/R29 in the first model (a) and with R21 in the second one (b)
The FlnA IG23 domain can not be stable in this hydrophobic environment while it is not essentially hydrophobic, as illustrated in Supplementary Data 14. FlnA IG23 indeed shares characteristics similar to those of the CFTR lasso Lh1. The contacts between the two proteins are tight and consistent: hydrophobic contacts between V11, V12, L15, F16 (CFTR) and L2521, V2522, M2490 (FlnA), interaction between CFTR S13/S18 and FlnA K2473, salt bridge between CFTR R21 and FlnA D2475, CFTR R25 and FlnA Q2477/E2578, and CFTR R29 and FlnA E2478 (Fig. 9a). One can also note the E2449–R2484 interaction, which allows the presence of these amino acids in a stable way in this hydrophobic environment. Importantly, the beginning of the IG23–IG24 linker appears to be tightly associated with the K8–V11 segment of the CFTR lasso (Fig. 9a), strengthening the stability of the complex. To the best of our knowledge, these particular sequence features of IG23, rendering it particularly suited to interact with a membrane environment, have not been reported so far. Supporting the model of the IG23/lasso-CFTR complex is the FlnA mutation M2474E, which abolishes interaction with CFTR [72]. M2474 is located in an hydrophobic environment and is the immediate neighbor of K2473 that, following our hypothesis, directly interacts with the CFTR lichpin S13.
N-ter-modified lasso The second hypothesis (interaction within the context of the lasso conformation, but with a N-terminal segment (aa 1–21) partially refolded in an extended structure, as present in the known CFTR-FlnA IG complexes [73]) involves a particular conformational flexibility of this region, which can be foreseen at the sequence level using hydrophobic cluster analysis (HCA) [77–79]. This approach allows the prediction of the probable, most stable secondary structure elements that are formed within proteins. Hydrophobic clusters defined using HCA generally well match the positions of regular secondary structures, and a lot of them have clear secondary preferences, correlating with their binary patterns and associated with 2D shapes [80, 81]. As shown in Fig. 7b, the CFTR N-terminus contains four hydrophobic clusters (labeled 1–4), showing the highest conservation of amino acids. The shapes of clusters 2 (Peitsch code 785, binary code 1100010001) and 4 (Peitsch code 4369, 1000100010001) are indicative of α-helices, while those of clusters 1 (Peitsch code 413, 110011101) and 3 (Peitsch code 77, 1001101) are far more ambiguous [80, 81]. In these cases of unclear binary pattern preference, the observed secondary structure is generally more dependent on the amino acid sequence and on the 3D environment. In particular, cluster 1, including the critical S13, might be associated either with an α-helix [as observed in cryo-EM data (Lh1, covering V11 to L15)] or with a long β-strand (as observed in the CFTR-FlnA complex [73]). Local sequence and hydrophobic cluster similarities are observed in this region between CFTR and filamin-binding motifs, as found in migfilin and β-integrins tails [69, 72, 73] (Fig. 7b, c). The strong hydrophobic character of cluster 1 must, however, be shielded in any cases by another hydrophobic area, as observed in the lasso conformation.
The “extended” hypothesis is further supported by the stability that we observed for this conformation in complex with a single IG domain after MD simulation (see before). As this strand should be in the CFTR-FlnA complex in direct contact with TM helices, the IG partner must fit this hydrophobic environment, a condition that the FlnA IG23 domain may particularly fulfills (see above). We, thus, modeled the N-terminal extended conformation, in which IG21 of the previously MD-refined IG21–CFTR complex was substituted by IG23. This hypothetical complex well fits the 3D structure of the s-closed CFTR 3D structure in this region and is stable along 50 ns MD simulation (Fig. 9b). Furthermore, the unusual amino acid composition between aa 15 and 19 (YFFWW in zebrafish CFTR) can also be understood in light of this complex, as the five aromatic amino acids are in contact either with IG23 or with the TM helices and/or membrane lipids (Fig. 9b).
The two hypothetical positions of FlnA IG23 reported here (within the context of the lasso in intact and modified conformations) are close to each other, E2428 establishing a salt bridge with CFTR R25/R29 in the first case and with CFTR R21 in the second one, both positions being distant from 8 to 9 Å by a similar orientation.
Hypothesis of an alternate path for the CFTR N-terminal extension
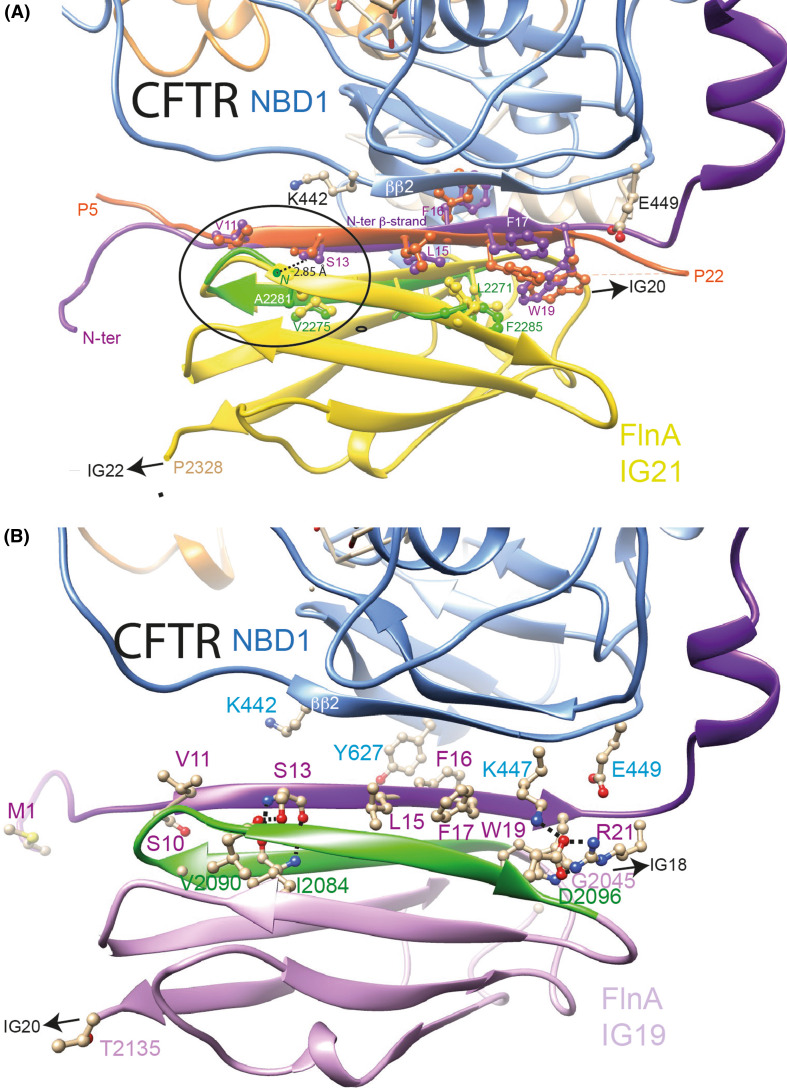
These two possible models for the interaction of FlnA IG23 and CFTR N-terminus within the lasso structure, however, likely do not account for the interactions that the CFTR N-terminus also makes with IG19, IG21, and farther, IG17 [72], especially as these IG domains possess hydrophilic surfaces. They also do not account for the interaction it makes with the NBDs, which were revealed by an overlay assay using an overlapping peptide library [82]. These multiple interactions are likely possible in the context of the full-length protein only if spatial flexibility (and not only local plasticity) occurs for the N-terminal segment. We, thus, here wanted to propose an alternate model of the N-terminus, which can be obtained through the displacement of Lh1 by rotating the segment included between Lh1 and Lh2 (aa 32–48) and keeping the position of Lh2 unchanged, as this helix is tightly associated with its neighbors. This model, developed here on the model of the CFTR open form, but which is also applicable on the cryo-EM 3D structure, takes into account structural and steric constraints, imposing to find an existing edge β-strand at the surface of the CFTR NBDs, able to interact with (and thus stabilize) the CFTR β-strand included in the [V11–R21] segment, while avoiding any steric clash between the so-formed CFTR N-ter/CFTR-NBD β-sheet and an FlnA IG domain. There are only two possibilities, the edge β-strands ββ2 in NBD1 (D443-I448) and the βc6 in NBD2 (K1420-I1427), this last one located at the opposite side of CFTR being, however, unreachable starting from the beginning of the N-terminal extension. Rotation of the lasso aa 32–48 segment around amino acid 48, maintaining its exact conformation observed in the lasso, led to directly propose a favorable interaction of cluster 1 (aa 11–19, modeled as a β-strand) with the edge ββ2 strand of NBD1 (D443-I448) (secondary structures were labeled as in [14]) through a groove that well adapts the polypeptidic chain of the displaced segment (Fig. 8b, c). Many favorable contacts are observed after 50 ns simulation, including a small β-sheet (aa I23–L24, in interaction with I448 and R450) preceding the K26-Q30 helical segment (RMSD of 3.11 Å between the initial and last frames for 74 superimposed Cα positions). The stability of this NBD1 ββ2 strand in complex with the N-terminus of CFTR was further assessed by repeated MD simulations (50 ns). This configuration also allows an adequate association with the FlnA IG21 domain (as present in the corresponding structure of the complex [73]). The stability of this ternary complex was also assessed by repeated MD simulations (over 10–20 ns), which, moreover, led to optimize or reveal some specific contacts. For instance, at the level of cluster 1, a longer β-strand (purple in Fig. 10a) is observed, which is stabilized by strong interaction between CFTR residue S13 and the FlnA IG21 A2281N atom, as also present in the experimental 3D structure of the complex [73]. A similar interaction was observed when MD simulation was performed on the same complex in which the FlnA IG21 was replaced by a homology model of IG19 (interaction between S13 and the oxygen atom of V2090, Fig. 10b). In this last case, a salt bridge is also observed between FlnA D2096 and CFTR R21 (N-terminus) and K447 (NBD1). Finally, a similar complex can eventually be also considered between the CFTR N-terminus in a β-strand conformation and IG23, which has also been demonstrated to interact with CFTR (Supplementary Data 15). Of note is that all the hydrophobic amino acids of CFTR N-terminus cluster 1 (aa 11–19) are interacting with hydrophobic amino acids belonging to either the FlnA IG domains or CFTR NBD1 (Fig. 10, Supplementary Data 15). The following contacts, relative to FlnA IG19 and to the zebrafish CFTR, are observed for CFTR Y15 (FlnA L2082. I2092, CFTR F17, N445), F16 (FlnA G2079, CFTR W18, Y627), F17 (FlnA L2080, T2094, CFTR Y15, W19, K447), W18 (CFTR F16, T20, K447, K615), and W19 (FlnA CFTR F17, R21, FlnA Y2077, L2080, C2102).
Fig. 10.
Models of human filamin in complex with CFTR N-terminus (β-strand conformation). a Complex of the CFTR open form (model) with the IG21 domain of FlnA. The analysis of the crystal structure of the FlnA IG21 (yellow) in complex with the CFTR N-terminus (peptide 5–22, orange) (PDBID:3ISW [73]) suggested that this last region of CFTR, observed in a β-strand conformation (and thus designated here as the N-ter β-strand), may form a β-sheet with NBD1 partners (strand ββ2, light blue). The CFTR 3D structure, in which the N-terminal region was modeled based on this observation, in complex with the FlnA IG21 domain, was then submitted to molecular dynamics (20 ns), leading to observe a longer N-ter β-strand (purple), which was stabilized by a strong interaction between S13 OG and A2281N atom. The green ribbon represents the FlnA IG21 β-hairpin 2271–2285 after MD simulations (RMSD between the two N-ter β-strands (orange (IG21/CFTR peptide complex, PDBID:3ISW [73]) and purple (full-length CFTR/IG21 complex after MD) and the two β-hairpin 2271-2285 (yellow (IG21/CFTR peptide complex) and green [full-length CFTR/IG21 complex after MD)] = 1.19 Å (24 Cα)). The circle indicates the region where the best superimposition is observed. b Complex of the CFTR open form (model) with the IG19 domain of FlnA. MD simulation (50 ns) was performed on the complex between IG19 (docked as IG21 on the CFTR N-ter β-strand) and CFTR. IG19 remained stable along the simulation (RMSD of 1.95 Å for 193 Cα atoms (IG19 + aa 1–22 of CFTR) between 0 and 10 ns). Among others, an H-bond is present between S13 hydroxyl and the V1290 carbonyl oxygen, as well as a salt bridge between FlnA D2096 and CFTR R21 and K447
Discussion
The nowadays available experimental 3D structures of the full-length CFTR protein at a relatively high resolution (mean 3.8–3.4 Å, [6–8]) constitute an important milestone, which opens new avenues for CF research [5]. However, in the phosphorylated, ATP-bound conformation (called here the s-closed conformation), the pore remains closed near the extracellular entrance [8]. Zhang and colleagues [8] have proposed that some local conformational changes in the extracellular ends of TM8 and TM12 would be sufficient to shift from a non-conducting state toward an open channel. There are some inconsistency with this hypothesis, one being the transient occurrence of a salt bridge typical of the open conformation (R352–D993), which on the reported s-closed structure does not directly form due to a too large distance and the presence of an intercalating tryptophane (Fig. 4). However, a 100 ns MD simulation that we performed on a human CFTR model, based on the zebrafish s-closed 3D structure [8], led to the stable formation of this salt bridge, while the channel remains closed (data not shown), suggesting that this feature is not limited to the open form and is in itself not sufficient to allow, at least at this time scale, the transition toward the open form. This salt bridge is also observed after μs-long MD simulations performed on the zebrafish s-closed 3D structure, while the channel still remained closed [10]. Other theoretical investigations using molecular dynamics have also pinpointed the fact that the experimental structures do not reconcile a chloride channel pathway for the active site [9] and importantly, μs-long simulations have shown that the s-closed conformation is particularly stable, including the unwound segment of TM8 [10], suggesting that the transition toward an open conformation may involve significant conformational change of the channel. Information about other, especially active forms of the channel, are thus still needed, and molecular models can offer insightful information in this context about the conformational landscape, provided that their reliability can be assessed. This was done for existing models first by comparison with experimental data or by adding constraints in the modeling process [11], and now, cryo-EM data permitted to directly check the quality of alignment used to model the CFTR structure on the ABC exporter template(s). This is critical as comparative modeling was challenging with the transmembrane helices of CFTR, forming the MSDs, which share only very low levels of sequence identity with the possible templates. It is important to recall here that the Sav1866 and MsbA 3D structures, used to model the open form of CFTR channel, are still now the only experimental templates available from type 1 exporters at high resolution in a true outward-facing conformation. Indeed, the recent cryo-EM 3D structure of Pgp in an outward-facing conformation [51] also clearly possesses features of occluded conformations, with intrinsic flexibility of the extracellular segments and a relative closure of the outward-facing cavity, similar to what is observed in the CFTR t-closed and s-closed conformations (RMSD 3.60 for 412 Cα positions).
We thus first checked here, considering the cryo-EM data [6–8], the quality of the HCA-based sequence alignment used for modeling the CFTR structure, resulting from a long, meticulous analysis of the whole ABC exporter family, and leading us to propose after non-constrained molecular dynamics simulations models of the open and closed (called here t-closed) conformations [14]. We observed a striking correspondence between the amino acids of the models and of the cryo-EM 3D structures after structural superimposition, providing thereby an unambiguous support of our theoretical studies. Based on this agreement, the knowledge of both the apo (cryo-EM) and open (model) forms of CFTR allows to describe an astonishing simple global conformational path linking the two extreme states. Indeed, the CFTR protein can be divided into two large blocks, named block-NBD1 and block-NBD2, whatever its conformation (open or closed) is. Such blocks can each be well superimposed between the experimental and theoretical structures and can be connected around pivot regions (centered on aa 202, 350, 990, and 1145) (Supplementary Data 16). Each block is constituted by the corresponding NBD, the two associated ICLs (ICL4 and ICL1 for NBD1, and ICL2 and ICL3 for NBD2) and the associated TM helices. Only a local difference is observed at the level of the alpha-subdomain of NBD1, which moves upward in the open form of the channel along ICL4. This different conformation, apparently specific to the open form, appears to be stabilized by strong interactions occurring within NBD1 and between NBD1 and ICL4. We then observed that each of the two extreme conformations of CFTR can be reconstituted by considering the blocks defined on the other conformation, with some local flexibility around the pivot regions, i.e., the open form being based on the blocks observed in the apo cryo-EM 3D structure and the apo form being based on the blocks predicted in the model of the open form (Supplementary Data 17). Thus, such a relatively simple mechanism may account for the conformational transitions between the different forms. Interestingly, Zhang and colleagues [8] also proposed to describe the conformational transition between their cryo-EM apo and s-closed conformations as rigid-body displacements of two halves, and exploited this framework to highlight the possible local conformational changes (at the level of the ends of TM8 and TM12) that may account for the ion path closure of the channel in the s-closed form.
The local conformational differences that we observed at the level of individual TM helices between cryo-EM and our models of the CFTR open and t-closed forms indicate key elements which may play a critical role in the transition of the channel into active forms, features which were not yet resolved when considering the very recent cryo-EM structure of a phosphorylated, ATP-bound form of CFTR. Without rank in order of importance, the presumed and perhaps linked key events within the MSD:NBD assembly appear to include: (1) a more canonical position of the TM8 helix, as this one, stabilized by tight interactions with TM6 and TM5, interferes with the anion flow in the current cryoEM 3D structures [10], (2) the displacement of F508 and the residues in its immediate neighborhood in the upward direction (in the direction of the membrane surface), the contact with ICL4 being maintained, and (3) the formation of four equivalent lateral portals at the level of the ICLs. These specific features, involving local unwinding of TM helices and NBD1 α subdomain movement, are observed without any other striking irregularities or instability, suggesting that they may be associated with specific functional features of the channel (as this was also suggested relative to the unwinding of TM8 observed in the cryo-EM 3D structure [8]). Of note are other recent theoretical studies on a top-closed/bottom-closed homology model, which have also highlighted one portal corresponding to the TM4–TM6 one described in our model [9, 15]. The presence of portals has not been discussed in another recent refined model of the outward-facing model based on cross-linking experiments, which was built together with an inward-facing one to understand the CFTR pore structure [16]. This pore structure shares many similarities around the selectivity filter (involving TM1, TM6, and TM12) with that observed in our t-closed model, as well as in other homology models [15].
The recent cryo-EM 3D structures (apo and s-closed forms [6–8]) have highlighted the particular “lasso” conformation adopted by the CFTR N-terminal sequence, specific for the ABCC subfamily members. The importance of this region is underscored by several CF mutations [83]. This region is also known to interact with multiple cellular partners (e.g., [84–88]) and is critical for maintaining a right balance between cytoskeletal tethering and endocytic trafficking. Of these partners, filamins (FlnA and FlnB) tether the CFTR protein to the membrane-proximal actin cytoskeleton and regulate its surface localization and dynamics [69]. In this context, we propose here that the CFTR N-terminus aa 11–19 may be able to interact with FlnA IG domains using different local conformations (α-helix or β-strand) and spatial positions (at the level at the MSDs or in contact with NBD1 strand ββ2), depending on the FlnA IG domain which is involved in the interaction. Such plasticity of the N-terminal extension was already suggested in an independent comment [89], proposing that the lasso conformation might rotate outward like an extending arm, and adopt different conformation(s), to be able to interact with partners of the membrane traffic machinery as well as with the R domain (reviewed in [4]). In particular, the intermolecular interaction of the N-terminus with the R domain suggested by others [90] is not observed in the present cryo-EM 3D structures, where there is no direct contact with the electron density corresponding to this last region [6]. Thus, these multiple and exclusive interactions, possibly involving distinct filamin IG domains (IG23, IG21, IG19) and different conformations of the CFTR N-terminal extension, may ensure a good stability of the CFTR protein at the membrane, regardless of the conformational state of the channel. Concerning this particular issue, it appears that the modeled open conformation of CFTR may not be reached in the presence of such a “lasso” conformation, as present in the cryo-EM 3D structures of CFTR. Indeed, the open conformation is characterized, relative to the cryo-EM 3D structures, by large movements of the TM7–TM8 pair of helices (linked by the large extracellular loop ECL4, containing the two N-glycosylation sites N894 and N900) as well as of the pseudo-symmetric TM1–TM2 pair (Supplementary Data 18). These movements may, thus, compromise the position of the N-terminal sequence in a lasso conformation at the interface with the membrane-spanning domains, and it is, thus, possible that the transition toward an open form is associated either with a local rearrangement or by a more drastic reorganization, as proposed here with the NBD1 strand ββ2 interaction.
We observe that several sensitive regions (R347, R352, W1145, TM8 unwound segments, Lh1 lasso) lie within the membrane at the level of the four pivots (aa 202, 350, 990, and 1145), therefore supporting their mutual importance (Supplementary Data 19).
Finally, one can also hypothesize that the IG domain string should remain in quasi-constant contact with the protein, independently of the association with the CFTR N-terminus. Of note is that the hydrophobic character of a part of the FlnA IG23 domain (predicted here to interact with the lasso conformation) might be associated with a stable anchoring at the membrane, even in the case of a possible conformational change leading to an alternate, NBD1 ββ2 associated position of the N-terminal extension. In this context, FlnA IG21 may maintain a possible interaction with the lasso Lh2. In contrast, when the CFTR N-terminal extension leaves this NBD1 ββ2 associated position, the FlnA IG domain (here thought to correspond to IG19) is left without partner. However, a direct association of an FlnA IG β-strand with CFTR strand ββ2 remains possible (the two strands are parallel and only 3 Å distant from each other), as further supported by the stability of the so-constructed complexes observed after a 10 ns MD simulation. This additional interaction might, thus, contribute to stabilize CFTR in the absence of the N-terminal extension.
Consistent with the two main possible positions of the CFTR N-terminal extensions (lasso and NBD1 ββ2) and associated FlnA domains, it is possible to speculate about hypothetical linear paths of FlnA IG repeats arranged in a head-to-tail manner, at the surface of NBD1 and linking IG19 (in contact with the CFTR N-terminal extension associated with NBD1 strand ββ2) to IG23 (in contact with the lasso Lh1, close to the MSDs) (Supplementary Data 20). IG21, located between IG19 and IG23, may be able to interact with the lasso Lh2 helix in both the main conformations of the N-terminal extension, an interaction involving CFTR aa 72–80, Supplementary Data 21–22). The 35 amino acid-long hinge between IG23 and IG24 is clearly made of two different parts. The first one possesses a relatively long, extended, well-conserved sequence (VFVDSLT, aa 2533–2539), while the second one is predicted as a highly flexible loop. With respect to the position of this hinge between IG23 and IG24, the extended, β-strand conformation is here predicted to be associated with the CD face of the IG22 domain, positioning the IG24 domains (from both subunits of the dimer) in contact with the polar leaflet of the membrane. Thus, in this model, the FlnA IG21, IG22, and IG23 domains may be favorably associated with the lasso conformation (Supplementary Data 22). It should be noted that only NBD1, but not NBD2, is involved in these multiple contacts, meaning that the separation of the two NBDs in the apo conformation should not interfere with filamin binding.
Finally, it is worth noting that the predicted, alternate position of the N-terminus sequence wrapping around NBD1 might offer some avenues to be explored for stabilizing this domain, which is known to be particularly thermally unstable when carrying the F508del mutation [91–93]. Qualitatively, the present hypothesis is consistent with the observed strength of CFTR-FlnA interaction (IG23, IG19, IG21, and IG17, in a decreasing order [72]). Indeed, IG23 displays many contacts, followed by IG19 and then IG21. The possible association of IG17 remains presently unknown at the structural level.
These considerations about the N-terminus conformational versatility might also apply to the whole ABCC family, in which this sequence is uniformly conserved. Along these lines, it is also noteworthy that the access to the edge strand ββ2 from NBD1 β-subdomain (or a newly formed one upon binding of CFTR N-terminal extension) is also possible for the N-terminal extension of SUR1, even in the presence of the preceding TMD0 domain (specific for this protein), due to the flexibility of the L0 segment separating TMD0 from TMD1 [aa 190–220 (data not shown)]. A similar situation might occur for MRP1, even though this region, which was described as critical for the protein function [94], is presently described with fewer atomic details. Indeed, starting from P205 at the beginning of the lasso in the 3D structure of MRP1, it is also possible to build a β-strand (aa I219–W223) which may contact the linear segment (H622–L625) within the linker between TMD1 and NBD1 (data not shown), leaving unchanged the TMD0 domain, which was in contrast demonstrated to be required neither for the MRP1 transport function nor for its targeting to the plasma membrane [94]. Also, within the complex structure of yeast Yor1p, a linker between amino acids 159 and 195 may likely play a similar role (data not shown).
Metadynamics simulations have already been applied to the study of complex mechanism of ion permeation and gating in other ion channels [95]. Here, these allowed us to further explore the path linking the open and t-closed conformers of CFTR, which involves only limited movements of transmembrane helices. Interestingly, our metadynamics simulations led us to observe that the t-closed form was energetically favored over the open form, the free-energy difference between the two conformations being similar to that provided by the hydrolysis of ATP into ADP. This suggests that the CFTR structure might spontaneously move relatively quick from an open to a closed state, leading to small movements of the NBD1 helical sub-domain favoring hydrolysis at the canonical ATP-binding site (closer to the F508 region than the non-canonical site). After ATP hydrolysis, a new ATP molecule may immediately enter the canonical ATP-binding site and restore the CFTR open state. Our metadynamics simulations, thus, open new perspectives to understand structural changes at the ATP-binding site associated with the gating cycle. Metadynamics simulations will also be performed in the future to understand the accessibility of the open/s-closed conformation from the s-closed cryo-EM 3D structures and get more insights into the particular behavior of TM8.
Metadynamics simulations also allowed to sample additional relevant conformations, as this is exemplified here with the amino acid R117 (within the TM1–TM2 loop). The R117H mutation, associated with a mild form of the disease, is characterized by a moderate reduction of conductance but strong gating defects, indicating gating-state-dependent interactions [96, 97]. This amino acid forms, in both the open and t-closed forms analyzed here and extracted from the metadynamics simulation, a stable and strong salt bridge with E1124 (within the TM11–TM12 loop, main distance below 3 Å) and, therefore, maintains contact between the two MSD blocks. In our initial model [14], these distances were longer (between 5.5 and 5.8 Å). In the apo and s-closed forms, this amino acid is not implicated in such contacts but alternatively establish multiple local bonds with the carbonyl main-chain atoms of the TM1-TM2 loops (Y109, D110, N113, E115 and H114, E115, E117 for human apo and zebrafish s-closed conformations, respectively). Of note, TM1, TM2, and TM11 are the transmembrane helices in contact with the lasso. This clearly different behavior is in good agreement with the recent comments of the Csanady’s team [98].
In conclusion, although there are still many unanswered questions, the now available structural data may allow us to speculate about a possible scenario for the CFTR gating cycle. Experimental work has proposed a sequential mechanism in which NBD1/NBD2 dimerization in the presence of ATP is followed by pore opening [98, 99]. Accordingly, CFTR phosphorylation and ATP binding may first conduct to an intermediate, occluded state identical or similar to the s-closed state. Then, large conformational changes at the level of the TM7–TM8 and TM1–TM2 helices, perhaps associated with flexibility of the N-terminal extension, as well as with an upward movement of the F508-containing NBD1 α-subdomain could permit to reach “active conformations”, quickly shifting between open and t-closed forms via, respectively, ATP hydrolysis and ATP binding at the canonical ATP-binding site. The return to an s-closed/apo state would intervene only after ATP hydrolysis at the non-canonical-binding site. Obviously, sophisticated experimental work is still needed to support these predictions and gain further insights in the complex, dynamic behavior of the CFTR channel.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the French Association Vaincre La Mucoviscidose (Paris). It was granted access to the HPC resources of IDRIS/CINES under the allocations 2014-077206, 2015-077206, 2016-077206, and 0020707206 made by GENCI.
Footnotes
Jean-Paul Mornon and Isabelle Callebaut are co-senior authors.
Brice Hoffmann and Ahmad Elbahnsi contribute equally to the work.
References
- 1.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 2.Gadsby DC. Ion channels versus ion pumps: the principal difference, in principle. Nat Rev Mol Cell Biol. 2009;10:344–352. doi: 10.1038/nrm2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aleksandrov LA, Jensen TJ, Cui L, Kousouros JN, He L, Aleksandrov AA, Riordan JR. Thermal stability of purified and reconstituted CFTR in a locked open channel conformation. Protein Expr Purif. 2015;116:159–166. doi: 10.1016/j.pep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozoky Z, Krzeminski M, Chong P, Forman-Kay JD. Structural changes of CFTR R region upon phosphorylation: a plastic platform for intramolecular and intermolecular interactions. FEBS J. 2013;280:4407–4416. doi: 10.1111/febs.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callebaut I, Hoffmann B, Mornon J-P. The implications of CFTR structural studies for cystic fibrosis drug development. Curr Opin Pharmacol. 2017;34:112–118. doi: 10.1016/j.coph.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Chen J. Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell. 2016;167:1586–1597. doi: 10.1016/j.cell.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Zhang Z, Csanády L, Gadsby DC, Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Liu F, Chen J. Conformational changes of CFTR upon phosphorylation and ATP binding. Cell. 2017;170:483–491. doi: 10.1016/j.cell.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 9.Tordai H, Leveles I, Hegedus T. Molecular dynamics of the cryo-EM CFTR structure. Biochem Biophys Res Commun. 2017;491:986–993. doi: 10.1016/j.bbrc.2017.07.165. [DOI] [PubMed] [Google Scholar]
- 10.Corradi V, Gu R-X, Vergani P, Tieleman D. Structure of transmembrane helix 8 and possible membrane defects in CFTR. Biophys J. 2018;114:1751–1754. doi: 10.1016/j.bpj.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callebaut I, Hoffmann B, Lehn P, Mornon J-P. Molecular modelling and molecular dynamics of CFTR. Cell Mol Life Sci. 2017;74:3–22. doi: 10.1007/s00018-016-2385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton J, Kalid O, Schushan M, Ben-Tal N, Villà-Freixa J. New model of cystic fibrosis transmembrane conductance regulator proposes active channel-like conformation. J Chem Inf Model. 2012;52:1842–1853. doi: 10.1021/ci2005884. [DOI] [PubMed] [Google Scholar]
- 13.Norimatsu Y, Ivetac A, Alexander C, O’Donnell N, Frye L, Sansom MS, Dawson DC. Locating a plausible binding site for an open-channel blocker, GlyH-101, in the pore of the cystic fibrosis transmembrane conductance regulator. Mol Pharmacol. 2012;82:1042–1055. doi: 10.1124/mol.112.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mornon J-P, Hoffmann B, Jonic S, Lehn P, Callebaut I. Full-open and closed CFTR channels, with lateral tunnels from the cytoplasm and an alternative position of the F508 region, as revealed by molecular dynamics. Cell Mol Life Sci. 2015;72:1377–1403. doi: 10.1007/s00018-014-1749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corradi V, Vergani P, Tieleman DP. Cystic fibrosis transmembrane conductance regulator (CFTR): closed and open state channel models. J Biol Chem. 2015;290:22891–22906. doi: 10.1074/jbc.M115.665125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das J, Aleksandrov AA, Cui L, He L, Riordan JR, Dokholyan NV. Transmembrane helical interactions in the CFTR channel pore. PLoS Comput Biol. 2017;13:e1005594. doi: 10.1371/journal.pcbi.1005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linsdell P. Functional architecture of the CFTR chloride channel. Mol Membr Biol. 2014;31:1–16. doi: 10.3109/09687688.2013.868055. [DOI] [PubMed] [Google Scholar]
- 18.Cui G, Freeman CS, Knotts T, Prince CZ, Kuang C, McCarty NA. Two salts bridges differentially contribute to the maintenance of cystic fibrosis transmembrane conductance regulator (CFTR) channel function. J Biol Chem. 2013;288:20758–20767. doi: 10.1074/jbc.M113.476226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui G, Zhang ZR, O’Brien AR, Song B, McCarty NA. Mutation at arginine 352 alters the pore architecture of CFTR. J Membr Biol. 2008;222:91–106. doi: 10.1007/s00232-008-9105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotten JF, Welsh MJ. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR. Evidence for disruption of a salt bridge. J Biol Chem. 1999;274:5429–5435. doi: 10.1074/jbc.274.9.5429. [DOI] [PubMed] [Google Scholar]
- 21.El Hiani Y, Linsdell P. Functional architecture of the cytoplasmic entrance to the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem. 2015;290:15855–15865. doi: 10.1074/jbc.M115.656181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Hiani Y, Negoda A, Linsdell P. Cytoplasmic pathway followed by chloride ions to enter the CFTR channel pore. Cell Mol Life Sci. 2016;73:1917–1925. doi: 10.1007/s00018-015-2113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, Hwang TC. Localizing a gate in CFTR. Proc Natl Acad Sci USA. 2015;112:2461–2466. doi: 10.1073/pnas.1420676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negoda A, El Hiani Y, Cowley EA, Linsdell P. Contribution of a leucine residue in the first transmembrane segment to the selectivity filter region in the CFTR chloride channel. Biochem Biophys Acta. 2017;1859:1049–1058. doi: 10.1016/j.bbamem.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 26.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Yl Qi. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 30.Lamoureux G, Roux B. Absolute hydration free energy scale for alkali and halide ions established from simulations with a polarizable force field. J Phys Chem B. 2006;110:3308–3322. doi: 10.1021/jp056043p. [DOI] [PubMed] [Google Scholar]
- 31.Beglov D, Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J Chem Phys. 1994;100:9050–9063. doi: 10.1063/1.466711. [DOI] [Google Scholar]
- 32.Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, Mackerell AD. Optimization of the CHARMM additive force field for DNA: improved treatment of the BI/BII conformational equilibrium. J Chem Theory Comput. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackerell AD. Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fletcher R (ed) (2000) Practical methods of optimization: fletcher/practical methods of optimization. Wiley, Chichester
- 36.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. doi: 10.1063/1.470648. [DOI] [Google Scholar]
- 37.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 38.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci. 2002;99(99):12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G. PLUMED 2: new feathers for an old bird. Comput Phys Commun. 2014;185:604–613. doi: 10.1016/j.cpc.2013.09.018. [DOI] [Google Scholar]
- 40.Pietrucci F. Strategies for the exploration of free energy landscapes: unity in diversity and challenges ahead. Rev Phys. 2017;2:32–45. doi: 10.1016/j.revip.2017.05.001. [DOI] [Google Scholar]
- 41.Branduardi D, Gervasio FL, Parrinello M. From A to B in free energy space. J Chem Phys. 2007;126:054103. doi: 10.1063/1.2432340. [DOI] [PubMed] [Google Scholar]
- 42.Crespo Y, Marinelli F, Pietrucci F, Laio A. Metadynamics convergence law in a multidimensional system. Phys Rev E Stat Nonlinear Soft Matter Phys. 2010;81:055701. doi: 10.1103/PhysRevE.81.055701. [DOI] [PubMed] [Google Scholar]
- 43.Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinform. 2012;13:173. doi: 10.1186/1471-2105-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Harrisson CB, Schulten K. Quantum and classical dynamics simulations of ATP hydrolysis in solution. J Chem Theory Comput. 2012;8:2328–2335. doi: 10.1021/ct200886j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Meis L. Role of water in the energy of hydrolysis of phosphate compounds—energy transduction in biological membranes. Biochem Biophys Acta. 1989;973:333–349. doi: 10.1016/s0005-2728(89)80440-2. [DOI] [PubMed] [Google Scholar]
- 47.George P, Witonsky RJ, Trachtman M, Wu C, Dorwart W, Richman L, Richman W, Shurayh F, Lentz B. “Squiggle-H2O”. An enquiry into the importance of solvation effects in phosphate ester and anhydride reactions. Biochem Biophys Acta. 1970;223:1–15. doi: 10.1016/0005-2728(70)90126-x. [DOI] [PubMed] [Google Scholar]
- 48.Vergani P, Nair AC, Gadsby D. On the machanism of MgATP-dependent gating of CFTR Cl− channels. J Gen Physiol. 2003;121:17–36. doi: 10.1085/jgp.20028673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fay JF, Aleksandrov LA, Jensen TJ, Cui LL, Kousouros JN, He L, Aleksandrov AA, Gingerich DS, Riordan J, Chen JZ (2018) Cryo-EM visualization of an active high open probability CFTR ion channel. bioRxiv 274316 [DOI] [PubMed]
- 50.Alam A, Küng R, Kowal J, McLeod R, Tremp N, Broude E, Roninson I, Stahlberg H, Locher K. Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc Natl Acad Sci USA. 2018;115:E1973–E1982. doi: 10.1073/pnas.1717044115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim Y, Chen J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science. 2018;359:915–919. doi: 10.1126/science.aar7389. [DOI] [PubMed] [Google Scholar]
- 52.Johnson ZL, Chen J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell. 2017;68:1075–1085. doi: 10.1016/j.cell.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 53.Hohl M, Hürlimann LM, Böhm S, Schöppe J, Grütter MG, Bordignon E, Seeger MA. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci USA. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooley RB, Arp DJ, Karplus PA. Evolutionary origin of secondary structures: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J Mol Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riek RP, Graham RM. The elusive π-helix. J Struct Biol. 2011;173:153–160. doi: 10.1016/j.jsb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Li M, Cowley E, El Hiani Y, Linsdell P. Functional organization of cytoplasmic portals controlling access to the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel pore. J Biol Chem. 2018;293:5649–5658. doi: 10.1074/jbc.RA117.001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barreto-Ojeda E, Corradi V, Gu RX, Tieleman DP. Coarse-grained molecular dynamics simulations reveal lipid access pathways in P-glycoprotein. J Gen Physiol. 2018 doi: 10.1085/jgp.201711907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 59.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katzmann D, Epping E, Moye-Rowley W. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol Cell Biol. 1999;19:2998–3009. doi: 10.1128/MCB.19.4.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Göddeke H, Timachi M, Hutter C, Galazzo L, Seeger M, Karttunen M, Bordignon E, Schäfer L. Atomistic mechanism of large-scale conformational transition in a heterodimeric ABC exporter. J Am Chem Soc. 2018;140:4543–4551. doi: 10.1021/jacs.7b12944. [DOI] [PubMed] [Google Scholar]
- 62.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 63.Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure. 2001;9:571–586. doi: 10.1016/S0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 64.Naoe Y, Nakamura N, Doi A, Sawabe M, Nakamura H, Shiro Y, Sugimoto H. Crystal structure of bacterial haem importer complex in the inward-facing conformation. Nat Commun. 2016;7:13411. doi: 10.1038/ncomms13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woo J, Zeltina A, Goetz B, Locher K. X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat Struct Mol Biol. 2012;19:1310–1315. doi: 10.1038/nsmb.2417. [DOI] [PubMed] [Google Scholar]
- 66.Wang C, Karpowich N, Hunt J, Rance M, Palmer A. Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. J Mol Biol. 2004;342:525–537. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Li N, Wu JX, Ding D, Cheng J, Gao N, Chen L. Structure of a pancreatic ATP-sensitive potassium channel. Cell. 2017;168:101–110. doi: 10.1016/j.cell.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 68.Martin GM, Yoshioka C, Rex EA, Fay JF, Xie Q, Whorton MR, Chen JZ, Shyng SL. Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. Elife. 2017;6:e24149. doi: 10.7554/eLife.24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thelin WR, Chen Y, Gentzsch M, Kreda SM, Sallee JL, Scarlett CO, Borchers CH, Jacobson K, Stutts MJ, Milgram SL. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J Clin Investig. 2007;117:364–374. doi: 10.1172/JCI30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura F, Stossel TP, Hartwig JH. The filamins. Cell Adhes Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Playford MP, Nurminen E, Pentikäinen OT, Milgram SL, Hartwig JH, Stossel TP, Nakamura F. Cystic fibrosis transmembrane conductance regulator interacts with multiple immunoglobulin domains of filamin A. J Biol Chem. 2010;285:17156–17165. doi: 10.1074/jbc.M109.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith L, Page RC, Xu Z, Kohli E, Litman P, Nix J, Ithychanda SS, Liu J, Qin J, Misra S, et al. Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin. J Biol Chem. 2010;285:17166–17176. doi: 10.1074/jbc.M109.080911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Light S, Sagit R, Ithychanda SS, Qin J, Elofsson A. The evolution of filamin-a protein domain repeat perspective. J Struct Biol. 2015;179:289–298. doi: 10.1016/j.jsb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sampson LJ, Leyland ML, Dart C. Direct interaction between the actin-binding protein filamin-A and the inwardly rectifying potassium channel, Kir2.1. J Biol Chem. 2003;278:41988–41997. doi: 10.1074/jbc.M307479200. [DOI] [PubMed] [Google Scholar]
- 76.Sethi R, Seppälä J, Tossavainen H, Ylilauri M, Ruskamo S, Pentikäinen OT, Pentikäinen U, Permi P, Ylänne J. A novel structural unit in the N-terminal region of filamins. J Biol Chem. 2014;289:8588–8598. doi: 10.1074/jbc.M113.537456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 78.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J-P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bitard-Feildel T, Callebaut I. Exploring the dark foldable proteome by considering hydrophobic amino acids topology. Sci Rep. 2017;7:41425. doi: 10.1038/srep41425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eudes R, Le Tuan K, Delettré J, Mornon J-P, Callebaut I. A generalized analysis of hydrophobic and loop clusters within globular protein sequences. BMC Struct Biol. 2007;7:2. doi: 10.1186/1472-6807-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rebehmed J, Quintus F, Mornon J-P, Callebaut I. The respective roles of polar/nonpolar binary patterns and amino acid composition in protein regular secondary structures explored exhaustively using hydrophobic cluster analysis. Proteins. 2016;84:624–638. doi: 10.1002/prot.25012. [DOI] [PubMed] [Google Scholar]
- 82.Wang W, He Z, O’Shaughnessy TJ, Rux J, Reenstra WW. Domain-domain associations in cystic fibrosis transmembrane conductance regulator. Am J Physiol Cell Physiol. 2002;282:C1170–C1180. doi: 10.1152/ajpcell.00337.2001. [DOI] [PubMed] [Google Scholar]
- 83.Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, Hong JS, Pollard HB, Guggino WB, Balch WE, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell. 2016;27:424–433. doi: 10.1091/mbc.e14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang SY, Di A, Naren AP, Palfrey HC, Kirk KL, Nelson DJ. Mechanisms of CFTR regulation by syntaxin 1A and PKA. J Cell Sci. 2002;115:783–791. doi: 10.1242/jcs.115.4.783. [DOI] [PubMed] [Google Scholar]
- 85.Cormet-Boyaka E, Di A, Chang SY, Naren AP, Tousson A, Nelson DJ, Kirk KL. CFTR chloride channels are regulated by a SNAP-23/syntaxin 1A complex. Proc Natl Acad Sci USA. 2002;99:12477–12482. doi: 10.1073/pnas.192203899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li C, Roy K, Dandridge K, Naren AP. Molecular assembly of cystic fibrosis transmembrane conductance regulator in plasma membrane. J Biol Chem. 2004;279:24673–24684. doi: 10.1074/jbc.M400688200. [DOI] [PubMed] [Google Scholar]
- 87.Ameen N, Silvis M, Bradbury NA. Endocytic trafficking of CFTR in health and disease. J Cyst Fibros. 2007;6:1–14. doi: 10.1016/j.jcf.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peters KW, Qi J, Johnson JP, Watkins SC, Frizzell RA. Role of snare proteins in CFTR and ENaC trafficking. Pflugers Arch. 2001;443:S65–S69. doi: 10.1007/s004240100647. [DOI] [PubMed] [Google Scholar]
- 89.Ford RC. ABC7/CFTR. In: Goerge A, editor. ABC transporters—40 years on. New York: Springer International Publishing; 2016. pp. 319–340. [Google Scholar]
- 90.Naren AP, Cormet-Boyaka E, Fu J, Villain M, Blalock JE, Quick MW, Kirk KL. CFTR chloride channel regulation by an interdomain interaction. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- 91.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt J, Brouillette CG. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010;19:1917–1931. doi: 10.1002/pro.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Spencer Emtage J, Wetmore DR, Brouillette CG, et al. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010;19:1932–1947. doi: 10.1002/pro.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He L, Aleksandrov AA, An J, Cui L, Yang Z, Brouillette CG, Riordan JR. Restauration of NBD1 thermal stability is necessary and sufficient to correct DF508 CFTR folding and assembly. J Mol Biol. 2015;427:106–120. doi: 10.1016/j.jmb.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bakos E, Evers R, Szakács G, Tusnády GE, Welker E, Szabó K, de Haas M, van Deemter L, Borst P, Váradi A, et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- 95.Furini S, Domene C. Computational studies of transport in ion channels using metadynamics. Biochim Biophys Acta. 2016;1858:1733–1740. doi: 10.1016/j.bbamem.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 96.Sheppard DN, Rich DP, Ostedgaard LS, Gregory RJ, Smith AE, Welsh MJ. Mutations in CFTR associated with mild-disease-form Cl- channels with altered pore properties. Nature. 1993;362:160–164. doi: 10.1038/362160a0. [DOI] [PubMed] [Google Scholar]
- 97.Yu YC, Sohma Y, Hwang TC. On the mechanism of gating defects caused by the R117H mutation in cystic fibrosis transmembrane conductance regulator. J Physiol. 2016;594:3227–3244. doi: 10.1113/JP271723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sorum B, Töröcsik B, Csanády L. Asymmetry of movements in CFTR’s two ATP sites during pore opening serves their distinct functions. Elife. 2017;6:e29013. doi: 10.7554/eLife.29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sorum B, Czégé D, Csanády L. Timing of CFTR pore opening and structure of its transition state. Cell. 2015;163:724–733. doi: 10.1016/j.cell.2015.09.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.