Abstract
Doxorubicin is one of the most effective drugs for the first-line treatment of high-grade osteosarcoma. Several studies have demonstrated that the major cause for doxorubicin resistance in osteosarcoma is the increased expression of the drug efflux transporter ABCB1/P-glycoprotein (Pgp). We recently identified a library of H2S-releasing doxorubicins (Sdox) that were more effective than doxorubicin against resistant osteosarcoma cells. Here we investigated the molecular mechanisms of the higher efficacy of Sdox in human osteosarcoma cells with increasing resistance to doxorubicin. Differently from doxorubicin, Sdox preferentially accumulated within the endoplasmic reticulum (ER), and its accumulation was only modestly reduced in Pgp-expressing osteosarcoma cells. The increase in doxorubicin resistance was paralleled by the progressive down-regulation of genes of ER-associated protein degradation/ER-quality control (ERAD/ERQC), two processes that remove misfolded proteins and protect cell from ER stress-triggered apoptosis. Sdox, that sulfhydrated ER-associated proteins and promoted their subsequent ubiquitination, up-regulated ERAD/ERQC genes. This up-regulation, however, was insufficient to protect cells, since Sdox activated ER stress-dependent apoptotic pathways, e.g., the C/EBP-β LIP/CHOP/PUMA/caspases 12-7-3 axis. Sdox also promoted the sulfhydration of Pgp that was subsequently ubiquitinated: this process further enhanced Sdox retention and toxicity in resistant cells. Our work suggests that Sdox overcomes doxorubicin resistance in osteosarcoma cells by at least two mechanisms: it induces the degradation of Pgp following its sulfhydration and produces a huge misfolding of ER-associated proteins, triggering ER-dependent apoptosis. Sdox may represent the prototype of innovative anthracyclines, effective against doxorubicin-resistant/Pgp-expressing osteosarcoma cells by perturbing the ER functions.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2967-9) contains supplementary material, which is available to authorized users.
Keywords: Osteosarcoma, P-glycoprotein, H2S-releasing doxorubicin, Endoplasmic reticulum-associated protein degradation, Endoplasmic reticulum stress
Introduction
Osteosarcoma is the most frequent bone tumor. High-grade osteosarcomas, localized in the extremities in patients younger than 40 years without evidence of metastases at diagnosis, are usually treated with pre- and post-operative polychemotherapy, in association with surgical removal of the tumor. Doxorubicin (dox) is one of the leader drugs, together with cisplatin and methotrexate. Unfortunately, this multimodal treatment achieves control disease in no more than 60% of osteosarcoma patients. Despite alternative therapeutic strategies, the prognosis of osteosarcoma has not significantly improved in the last decades [1, 2].
One of the main factors hampering dox success is the presence of ATP-binding cassette B1/P-glycoprotein (ABCB1/Pgp), which effluxes dox, reducing its intracellular accumulation and cytotoxicity [3, 4]. Pgp is indeed predictive of poor response to dox treatment in osteosarcoma patients [5–7]. Another factor reducing dox’s efficacy is the onset of a dose-dependent cardiotoxicity [8], which limits the cumulative dose of the drug that can be administered.
To circumvent Pgp-mediated resistance, several natural and synthetic inhibitors have been tested: although successful in vitro, they often failed in vivo for the low specificity, the side-effects and the unexpected toxicity due to inhibition of the physiological ABC transporters-mediated drug detoxification in normal tissues [9]. Recently, libraries of small molecules with selective cytotoxicity against Pgp-overexpressing cells—i.e., molecules inducing the so-called “collateral sensitivity”—have been identified, but the molecular bases of their selective cytotoxicity are poorly understood [10]. Pgp-silencing vectors, used alone or in combination with chemotherapeutic drugs [11], inhibitors of transcription factors that up-regulate Pgp [12, 13], inhibitors of surface proteins necessary for Pgp activity [14] have been successfully experimented in vitro and in preclinical models, but the application of these strategies to osteosarcoma patients remains uncertain.
Different approaches that reduce the oxidative damage produced by anthracyclines have been considered to overcome dox-related cardiotoxicity. Recently, it has been shown that hydrogen sulfide (H2S) protects cardiomyocytes from dox-induced toxicity [15] by scavenging reactive oxygen species (ROS) [16]. Starting from these observations, we recently synthesized a library of doxs conjugated with H2S-releasing groups: most compounds resulted less cardiotoxic than dox since the presence of H2S prevented the increase of ROS [17].
The role of H2S in tumor biology is controversial, depending on the cancer cell line, the amount and the kinetics of H2S release: some experimental evidences supported a pro-tumor effect of H2S [18, 19], while an increasing number of works demonstrated the opposite effect [20–22], fostering the synthesis of H2S-releasing molecules with potential oncological applications [20, 21, 23]. Interestingly, all the H2S-releasing doxs of our library were more cytotoxic than dox against human dox-resistant osteosarcoma cells. Curiously, H2S-releasing doxs inhibited topoisomerase II and increased ROS levels less than dox [17], suggesting that their higher cytotoxicity did not rely on these two classical effects of dox [24].
In the present work, we investigated the molecular bases of the high efficacy of H2S-releasing doxs against Pgp-expressing human osteosarcoma cells, to identify the properties that make these compounds noteworthy of future evaluation in preclinical models of osteosarcoma. To this aim, we focused on the most effective H2S-releasing dox, i.e., the compound 10 in [17], here termed Sdox.
Materials and methods
Chemicals
Fetal bovine serum (FBS) and culture medium were from Invitrogen Life Technologies (Carlsbad, CA, USA). Plasticware for cell cultures was from Falcon (Becton–Dickinson, Franklin Lakes, NJ, USA). The protein content in cell extracts was assessed with the BCA kit from Sigma Chemical Co. (St. Louis, MO, USA). Electrophoresis reagents were obtained from Bio-Rad Laboratories (Hercules, CA, USA). Dox was purchased by Sigma Chemical Co. Sdox was synthesized as described in [17]. Unless otherwise specified, all the other reagents were purchased from Sigma Chemical Co.
Cell lines
Human dox-sensitive osteosarcoma U-2OS and Saos-2 cell lines were purchased from ATCC (Manassas, VA, USA). The corresponding variants with increasing resistance to dox (U-2OS/DX30, U-2OS/DX100, U-2OS/DX580, Saos-2/DX30, Saos-2/DX100, Saos-2/DX580), selected by culturing parental cells in medium containing progressively increased dox dosages, 30, 100, 580 ng/ml dox, respectively, were generated as reported in [25], and continuously cultured in the presence of dox. All cell lines were authenticated by microsatellite analysis, using the PowerPlex kit (Promega Corporation, Madison, WI, USA; last authentication: January 2017). Primary non-transformed osteoblasts were obtained as reported in [26]. Primary human fibroblasts were a kind gift of Prof. Francesco Novelli, Department of Molecular Biotechnology and Health Sciences, University of Torino, Italy. Rat H92c cells were from ATCC. Cells were maintained in medium supplemented with 10% v/v FBS, 1% v/v penicillin–streptomycin, 1% v/v l-glutamine.
Doxorubicin accumulation
Dox content was measured fluorimetrically as detailed previously [27], using a Synergy HT Multi-Detection Microplate Reader (Bio-Tek Instruments, Winoosky, MT, USA). The results were expressed as nmoles dox/mg cell proteins, according to a titration curve previously set. In competition assays with verapamil, 250 × 105 cells were incubated with dox or Sdox, alone or in combination with verapamil. The intracellular fluorescence was detected using a Guava® easyCyte flow cytometer (Millipore, Bedford, MA, USA) equipped with the InCyte software (Millipore). The results were expressed as mean fluorescence intensity (MFI).
Cell cytotoxicity
The extracellular release of lactate dehydrogenase (LDH), considered an index of cell damage and necrosis, was measured as reported in [27]. The results were expressed as percentage of extracellular LDH versus total (intracellular plus extracellular) LDH.
Cell apoptosis
1 × 106 cells were stained with the Annexin V-FITC Apoptosis Detection kit (Sigma Chemicals Co.), using either Annexin-V-fluorescein isothiocyanate (FITC) and propidium iodide (PI), as per manufacturer’s instructions. Samples were analyzed using Guava® easyCyte flow cytometer (Millipore) and InCyte software (Millipore).
Cell viability
Cell viability was measured by the ATPlite Luminescence Assay System (PerkinElmer, Waltham, MA, USA), as per manufacturer’s instructions, using a Synergy HT Multi-Detection Microplate Reader to measure the relative luminescence units (RLU). The RLUs of untreated cells were considered as 100% viability; the results were expressed as a percentage of viable cells versus untreated cells. To determine IC50, 2 × 103 cells were incubated for 72 h with increasing concentrations of dox or Sdox (from 10 nM to 0.5 mM). IC50 was considered the concentration of the drug that reduced cell viability to 50% using the CalcuSyn software (http://www.biosoft.com/w/calcusyn.htm; BioSoft, Cambridge, UK). The fold change of dox or Sdox sensitivity was determined by calculating the ratios of the IC50 of DX-resistant variants to that of their corresponding parental cell lines.
Clonogenic assay
To have a comparable cloning efficiency, clonogenic assay was performed by seeding in triplicate monocellular suspensions of 3200 cells (parental cell lines) or 12,500 cells (DX-resistant variants) in 60-mm dishes. After 24 h, the medium was changed with fresh medium containing the following increasing concentrations of dox or Sdox: 0, 0.01, 0.05, 0.10, 0.50, 1, 2.5, 5, 10, 15, 20, 30, 40, 50, 60, 70, 80, 90, 100 µM. After additional 120 h of culture, cells were fixed with methanol:acetic acid (3:1) and stained with Giemsa. Colonies containing at least 30 cells were counted. The number of colonies in drug-treated dishes was related to those of the appropriate control to calculate the percentages of clonogenic inhibition. Percentages of clonogenic inhibition were then used to estimate the drug concentration resulting in 50% inhibition of colonies formation (IC50) using the CalcuSyn software. The fold change of dox or Sdox sensitivity was determined by calculating the ratios of the IC50 of DX-resistant variants to that of their corresponding parental cell lines.
DNA damage
5 × 105 cells were seeded onto glass coverslips. Cells were fixed using 4% w/v paraformaldehyde for 15 min, washed with PBS, permeabilized with 1% Triton X-100 for 5 min, washed with PBS and incubated for 1 h with an antibody detecting p(Ser139)-γH2AX (1:1000, Abcam, Cambridge, UK), considered a marker of DNA damage [28]. Samples were washed five times with PBS and incubated for 1 h with a Alexa488-conjugated secondary antibody (1:100, Abcam). After washing steps, cells were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), diluted 1:1000 in PBS for 5 min, washed four times with PBS and once with deionized water. The cover slips were mounted with Gel Mount Aqueous Mounting and examined with a Leica DC100 fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany). For each experimental point, a minimum of five microscopic fields were examined.
Intracellular drug localization
5 × 105 cells were grown on sterile glass coverslips and transfected with the green fluorescence protein (GFP)-KDEL fused-calreticulin expression vector (Cell Light BacMan 2.0, Invitrogen Life Technologies) to label endoplasmic reticulum (ER). After 24 h cells were incubated with 5 µM dox or Sdox for 6 h. Samples were rinsed with PBS, fixed with 4% w/v paraformaldehyde for 15 min, washed three times with PBS and once with water, mounted with 4 μl of Gel Mount Aqueous Mounting. Slides were analyzed Leica DC100 fluorescence microscope. For each experimental condition, a minimum of five microscopic fields were examined.
PCR arrays
Total RNA was extracted and reverse-transcribed using iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories). The PCR arrays were performed on 1 μg cDNA, using the Unfolded Protein Response Plus PCR Array (Bio-Rad Laboratories), as per manufacturer’s instructions. Data analysis was performed with PrimePCR™ Analysis Software (Bio-Rad Laboratories).
Immunoblotting
Cells were rinsed with ice-cold lysis buffer (50 mM Tris, 10 mM EDTA, 1% v/v Triton-X100; pH 7.5), supplemented with the protease inhibitor cocktail set III (80 μM aprotinin, 5 mM bestatin, 1.5 mM leupeptin, 1 mM pepstatin; Calbiochem, San Diego, CA, USA), 2 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4. Cells were then sonicated (10 bursts of 10 s, 4 °C, 100 W, using a Labsonic sonicator, Hielscher, Teltow, Germany) and centrifuged at 13,000×g for 10 min at 4 °C. 20 μg of protein extracts were subjected to 4–20% gradient SDS-PAGE and probed with the following antibodies: anti-ABCB1/Pgp (1:500, Calbiochem); anti-ER degradation enhancing α-mannosidase-like protein 1 (EDEM1; 1:500, Abcam); anti-UDP-glucose glycoprotein glucosyltransferase 1 (UGGT1; 1:250, Abcam); anti-SEC62 homolog/preprotein translocation factor (SEC62; 1:250, Abcam); anti-valosin containing protein (VCP; 1:1000, Abcam); anti-glucose-regulated protein 78/binding immunoglobulin protein (GRP78/BiP; 1:500, Abcam), anti-inositol requiring kinase-1α (IRE1α; 1:500, Thermo Scientific Inc., Rockford, IL), anti-X-box binding protein 1 (XBP1; 1:1000, Abcam), anti-protein kinase-like endoplasmic reticulum kinase (PERK; 1:500, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), anti-phospho(Ser51)eukariotic initiation factor-2α (peIF2α; 1:500, Abcam), anti-eIF2α (1:1000, Abcam), anti-activating transcription factor 4 (ATF4; 1:1000, Abcam), anti-activating transcription factor 6 (ATF6; 1:500, Abcam), anti-CCAAT-enhancer-binding protein-β (C/EBP-β, recognizing either LAP or LIP isoform; 1:500, Santa Cruz Biotechnology Inc.), anti-C/EBP homologous protein/growth arrest and DNA damage 153 (CHOP/GADD153; 1:500, Santa Cruz Biotechnology Inc.), anti-Tribbles homolog 3 (TRB3; 1:500, Proteintech, Chicago, IL, USA), anti-p53 up-regulated modulator of apoptosis (PUMA; 1:1000, Cell Signaling Technology, Danvers, MA); anti-caspase 12 (1:500, Abcam), anti-caspase 7 (1:1000, Abcam), anti-caspase 3 (1:500, GeneTex, Hsinhu City, Taiwan), anti-β-tubulin (1:1000, Santa Cruz Biotechnology Inc.). The membranes were then incubated with peroxidase-conjugated secondary antibodies (1:3000, Bio-Rad Laboratories) and washed with Tris-buffered saline-Tween 0.1% v/v solutions. Protein bands were detected by enhanced chemiluminescence (Bio-Rad Laboratories). Nuclear extracts were prepared using the Nuclear Extract kit (Active Motif, La Hulpe, Belgium). Nuclear proteins were separated by SDS-PAGE and probed with anti-CHOP or anti-TATA box binding protein antibodies (TBP, 1:500, Santa Cruz Biotechnology Inc.). Microsomal fractions were prepared using the Endoplasmic Reticulum Isolation Kit (Sigma Chemicals. Co), as per manufacturer’s instructions. 100 μg of microsomal proteins were resolved by SDS-PAGE and probed with an anti-mono/poly-ubiquitin antibody (1:1000, Axxora, Lausanne, Switzerland) or with an anti-calreticulin antibody (1:1000, Affinity Bioreagents, Rockford, IL, USA). To measure ubiquitinated Pgp, 100 μg of microsomal proteins were immunoprecipitaed with the anti-Pgp antibody, using 25 μl of PureProteome Magnetic Beads (Millipore), then probed with the anti-mono/poly-ubiquitin antibody.
Microsomal protein sulfhydration
The sulfhydration of microsomal proteins (100 μg) or immunopurified Pgp (50 μg) was measured according to [29], with minor modifications. Samples were re-suspended in 600 μl solubilization buffer (150 mM NaCl, 0.5% v/v Tween 20, 50 mM Tris 7.5, 1 mM EDTA) and incubated for 2 h at 4 °C with 2 μM Alexa Fluor 680 conjugated C2 maleimide (Thermo Fisher Scientific, Waltham, MA), to label either sulfhydrated and unsulfhydrated cysteines. Samples were vortexed every 20 min. 300 μl were transferred in a new series of tubes and incubated for 1 h at 4 °C with 1 mM dithiothreitol (DTT) that removes maleimide adducts from sulfhydrated cysteines only, increasing the labelling specificity. The samples’ fluorescence (λ excitation = 684 nm; λ emission = 714 nm) was read with a Synergy HT Multi-Detection Microplate Reader. The fluorescence of samples treated with maleimide and DTT treated samples was divided per the fluorescence of the same samples without DTT [29]. The results were expressed as relative fluorescence units (RFU)/mg of microsomal proteins or Pgp protein.
Microsomal protein ubiquitination
The ubiquitination of microsomal proteins (100 μg) or immunopurified Pgp (50 μg) was measured with the E3Lite Customizable Ubiquitin Ligase kit (Life-Sensors Inc., Malvern, PA). Samples were diluted in 100 μl of ubiquitination assay buffer (1 M Tris/HCl, 500 mM MgCl2, 10 mM DTT; pH 8), and incubated for 30 min at 37 °C, in the presence of 5 nM E1 activating enzyme provided by the kit, 100 nM E2 conjugating enzyme Ube2g2 (LifeSensors Inc.), 200 μM ATP, 6 mM human recombinant ubiquitin. Samples were washed twice with PBS-Tween 0.1% v/v containing 5% w/v bovine serum albumin and incubated with the biotinylated anti-ubiquitin antibody of the kit, followed by streptavidin/horseradish peroxidase-conjugated polymer and enhanced chemiluminescence detection. The chemiluminescent signal was read using a Synergy HT Multi-Detection Microplate Reader. The results were expressed as RLU/mg of microsomal proteins or Pgp protein.
Flow cytometry analysis
ABCB1/Pgp amount on cell surface was measured as reported in [26]. Samples were analyzed using Guava® easyCyte flow cytometer (Millipore) and InCyte software (Millipore). Control experiments included incubation of cells with not-immune isotypic antibody, followed by secondary antibody.
Statistical analysis
All data in the text and figures are provided as mean ± SD. The results were analyzed by a one-way analysis of variance (ANOVA) and Tukey’s test. p < 0.05 was considered significant.
Results
H2S-releasing doxorubicin exerts cytotoxic effects against drug-resistant osteosarcoma cells
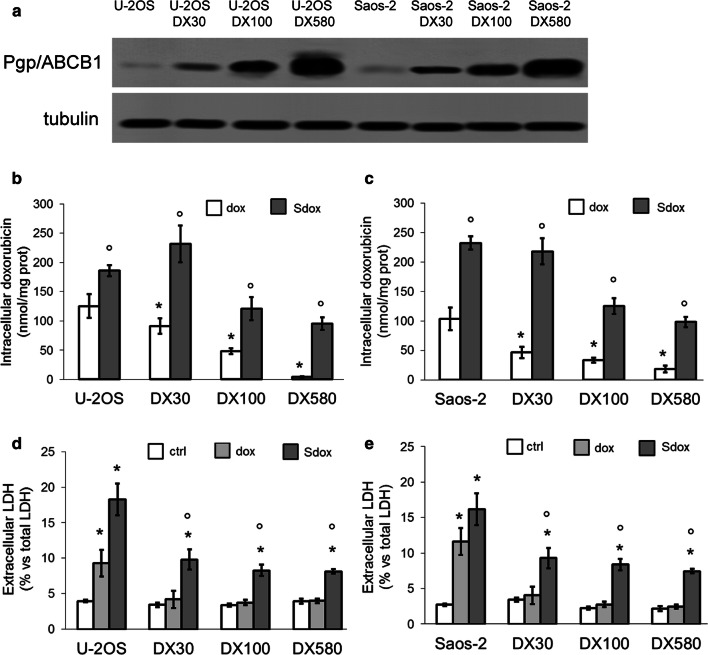
We compared the cytotoxic and anti-proliferative effects of dox and Sdox in U-2OS and Saos-2 cells, and in the respective variants with increasing expression of ABCB1/Pgp (Fig. 1a) and resistance to dox. As expected, dox was well retained in U-2OS and Saos-2 cells, but it was progressively less accumulated in resistant variants (Fig. 1b, c) where it did not induce cell damage, measured as release of LDH (Fig. 1d, e). Although the accumulation and the release of LDH were also progressively decreased in the resistant cells treated with Sdox, such decrease was less pronounced: overall, Sdox was significantly more accumulated and induced significantly more damage than dox in all resistant variants (Fig. 1b–e).
Fig. 1.
H2S-releasing doxorubicin is effective against resistant osteosarcoma cells. a Expression of ABCB1/Pgp in human doxorubicin-sensitive U-2OS cells and Saos-2 cells, and their doxorubicin-resistant variants (DX30, DX100, DX580) by immunoblotting. The β-tubulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. b, c Intracellular doxorubicin accumulation, measured after a 6 h incubation with 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox), by a fluorimetric assay in duplicates. Data are mean ± SD (n = 6 independent experiments). *p < 0.05 for DX cells vs. parental U-2OS or Saos-2 cells; °p < 0.001 for Sdox vs. dox. d, e Extracellular release of LDH measured spectrophotometrically in triplicates, in cells grown 24 h in drug-free medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). Data are mean ± SD (n = 6 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for Sdox vs. dox
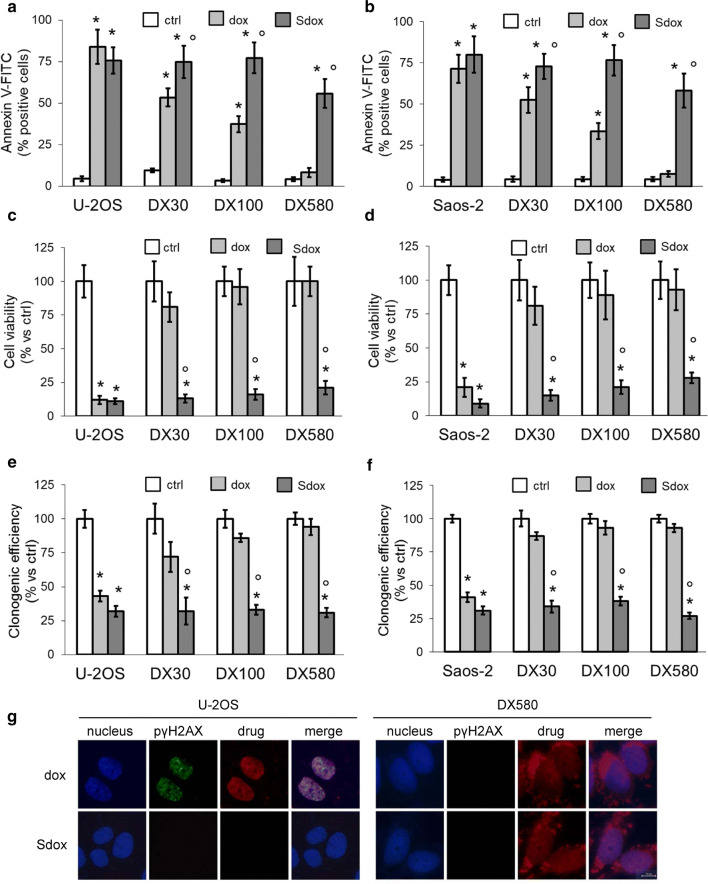
To verify whether the cell damage was followed by cell death, we first measured the percentage of apoptotic cells after a 24 h treatment with dox or Sdox. Both drugs induced a significant increase of Annexin V-FITC-positive U-2OS and Saos-2 cells (Fig. 2a, b), indicating the induction of early apoptosis, and of Annexin V-FITC/PI-positive cells (Supplementary Fig. 1, Online Resource 1), suggesting the progression towards late apoptosis and/or necrosis. Dox progressively reduced its pro-apoptotic effect in the DX30 and DX100 sublines of both U-2OS and Saos-2 cells, and was completely ineffective in the DX580 subpopulation (Fig. 2a, b; Supplementary Fig. 1, Online Resource 1). In line with this result, dox decreased cell viability (Fig. 2c, d) and clonogenic potential (Fig. 2e, f) in U-2OS and Saos-2 cells only, and lost its anti-proliferative efficacy in resistant sublines. By contrast, Sdox-reduced cell viability and clonogenic potential to the same extent in sensitive and resistant cells (Fig. 2c–f).
Fig. 2.
H2S-releasing doxorubicin induces apoptosis and clonogenic arrest without eliciting DNA damage. Human doxorubicin-sensitive U-2OS cells and Saos-2 cells and their resistant sublines DX30, DX100 and DX580 were incubated 24 h (a, b), 72 h (c, d) or 120 h (e, f) in fresh medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). a, b Percentage of annexin V-FITC-positive cells, measured by flow cytometry in duplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.02 for Sdox vs. dox. c, d Percentage of viable cells, measured by a chemiluminescence-based assay in quadruplicates. Data are mean ± SD (n = 6 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for Sdox vs. dox. e, f Clonogenic assay, measured in triplicates after staining colonies with Giemsa and counting by optical microscopy. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for Sdox vs. dox. g Representative immunofluorescence analysis of p(Ser139)-γH2AX in U-2OS cells and its DX580 variant after 24 h of drug treatment. Magnification: ×63 objective lens (1.10 numerical aperture); ×10 ocular lens. Bar: 10 µM. The micrographs are representative of one out of three experiments
The IC50 of dox calculated with the clonogenic assay (Table 1) or viability assay (Supplementary Table 1, Online Resource 2; Supplementary Fig. 2, Online Resource 3) indicated that all cell lines were sensitive to Sdox compared to dox. Moreover, DX-resistant variants showed a decrease of sensitivity to dox treatment, accordingly to their dox-resistance level, which was not present for Sdox, confirming the absence of cross-resistance to this drug.
Table 1.
IC50 (μM) to doxorubicin and H2S-releasing doxorubicin according to the clonogenic assay
| Cell line | IC50 dox | IC50 dox: fold change compared to parental cells | IC50 Sdox | IC50 Sdox: fold change compared to parental cells |
|---|---|---|---|---|
| U-2OS | 1.04 | 1.02 | ||
| U-2OS/DX30 | 7.89* | 7.59 | 1.09° | 1.07 |
| U-2OS/DX100 | 10.54* | 10.13 | 0.86° | 0.84 |
| U-2OS/DX580 | 21.60* | 20.77 | 0.91° | 0.89 |
| Saos-2 | 0.89 | 0.79 | ||
| Saos-2/DX30 | 2.29* | 2.57 | 0.66° | 0.84 |
| Saos-2/DX100 | 6.56* | 7.37 | 0.59° | 0.75 |
| Saos-2/DX580 | 9.53* | 10.71 | 0.89° | 1.13 |
Cells were incubated for 5 days with fresh medium or increasing concentrations (10 nM–100 µM) of doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). The number of colonies and the corresponding IC50 was calculated as reported in the “Materials and methods” section. Data are mean ± SD (n = 3 independent experiments)
*p < 0.005 for dox-resistant variants vs. their parental cells; °p < 0.001 for Sdox versus dox
Differently from dox, Sdox was not cytotoxic, in terms of increase in LDH release and reduction in cell viability, against not-transformed cells, such as human osteoblasts and fibroblasts, and rat cardiomyocytes (Supplementary Fig. 3, Online Resource 4).
Interestingly, whereas dox accumulation was significantly increased in U-2OS/DX580 cells treated with the Pgp inhibitor verapamil, Sdox accumulation was slightly but not significantly increased. As expected because of the very low amount of Pgp, verapamil had no effect on drug accumulation (Supplementary Fig. 4a, b, Online Resource 5), LDH release (Supplementary Fig. 4c, d, Online Resource 5) and cell viability (Supplementary Fig. 4e, f, Online Resource 5) in U-2OS cells treated with dox and Sdox, but it increased dox accumulation and cytotoxicity in the DX580 variant. Again, verapamil did not significantly increase the cytotoxicity elicited by Sdox in resistant cells (Supplementary Fig. 4d, f, Online Resource 5).
To investigate the mechanisms at the basis of Sdox cytotoxicity and proliferation arrest, we analyzed if the compound damaged DNA, eliciting a peculiar effect of dox [24]. Indeed, dox induced the appearance of p(Ser139)-γH2AX foci in the nucleus of U-2OS cells, whereas it was ineffective in DX580 variant (Fig. 2g). By contrast, Sdox did not induce the appearance of p(Ser139)-γH2AX foci in sensitive or resistant cell lines (Fig. 2g), leading to hypothesize a mechanism of action completely different from dox. Interestingly, this experimental set also revealed a different intracellular localization of dox and Sdox.
H2S-releasing doxorubicin localizes within ER and up-regulates ERAD/ERQC-related genes
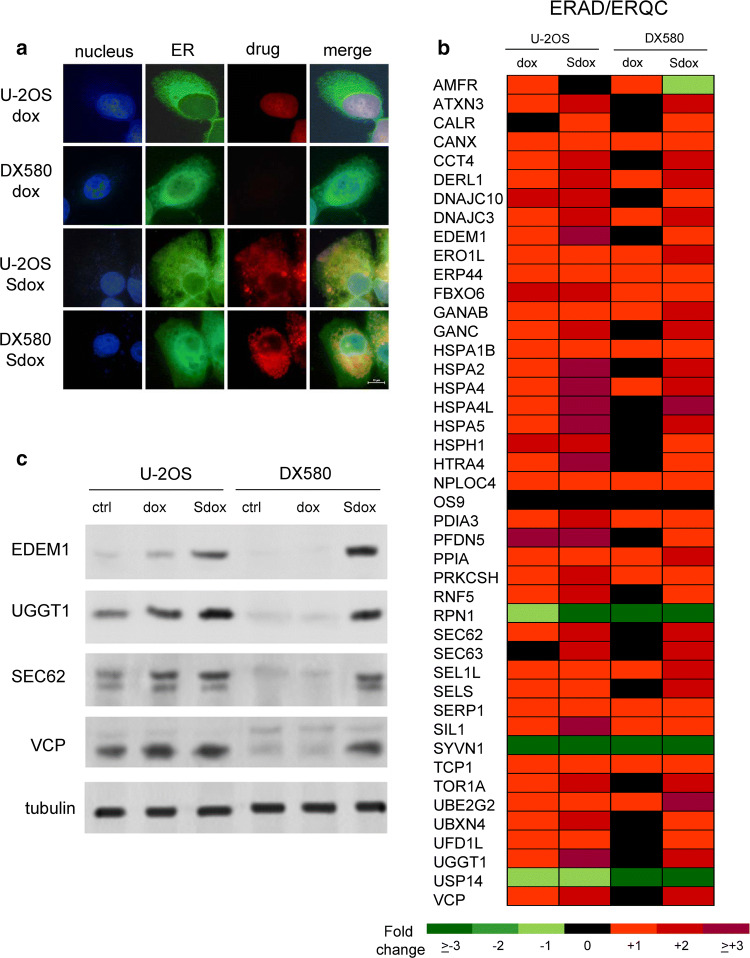
The analysis of such differential localization indicated that dox had a prevalent nuclear localization in U-2OS cells after 24 h, Sdox was mainly localized within ER (Fig. 3a). As expected on the basis of the low intracellular retention (Fig. 1b), dox was undetectable within U-2OS/DX580 cells. By contrast, Sdox resulted accumulated in the ER also in resistant cells (Fig. 3a). Of note, 20 min after incubation, dox was detectable in the ER, as well as in the nucleus, of U-2OS cells (Supplementary Fig. 5, Online Resource 6); after this time point the drug was completely localized within the nucleus, as observed at 24 h (Fig. 3a)
Fig. 3.
H2S-releasing doxorubicin localizes within endoplasmic reticulum and up-regulates ERAD/ERQC system. a U-2OS and U-2OS/DX580 cells were incubated for 24 h with the GFP-KDEL-calreticulin expression vector to label endoplasmic reticulum (ER), then treated with 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox) for the last 6 h. The intracellular localization of the drugs was analyzed by fluorescence microscopy. Magnification: ×63 objective lens (1.42 numerical aperture); ×10 ocular lens. Bar: 10 μm. The micrographs are representative of four experiments with similar results. b Hitmap of ER-associated degradation/endoplasmic reticulum quality control (ERAD/ERQC)-related genes in U-2OS and U-2OS/DX580 cells, after 24 h treatment with drug-free medium, 5 µM dox or Sdox. The figure reports genes up-or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown in the figure). The whole list of genes analyzed is reported in the Supplementary Tables 3, 4 (Online Resources 9, 10). c Expression of the indicated ERAD/ERQC-related proteins measured by immunoblotting in U-2OS and U-2OS/DX580 cells, treated as in b. The β-tubulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. ctrl drug-free medium
We thus investigated whether the cytotoxic effects of Sdox were mediated by alterations of ER functions.
A baseline gene profiling of U2-OS and resistant clones revealed a variable up- or down-regulation of genes controlling the unfolded protein response (UPR; Supplementary Table 2, Online Resource 7; Supplementary Fig. 6a, Online Resource 8), a series of events triggered by misfolded/unfolded proteins accumulated within ER lumen, leading to protein degradation and cell survival, or ER stress and cell death [30, 31]. Genes mediating cell death or survival consequent to UPR were either up- or down-regulated in untreated sensitive and resistant cells (Supplementary Table 2, Online Resource 7; Supplementary Fig. 6b, Online Resource 8). By contrast, a remarkable number of genes related to ER-associated protein degradation (ERAD) and ER-quality control (ERQC), two processes checking protein folding and removing irreversibly damaged proteins [30, 31], were progressively down-regulated with the increase of dox-resistance (Supplementary Table 2, Online Resource 7; Supplementary Fig. 6c, Online Resource 8). Both dox and Sdox up-regulated most of ERAD/ERQC-related genes in U-2OS cells (Fig. 3b; Supplementary Table 3, Online Resource 9). As expected as a consequence of the low intracellular accumulation, dox produced minor changes in the expression of this gene set in U-2OS/DX580 cells. By contrast, Sdox still up-regulated most ERAD/ERQC-related genes in drug-resistant cells (Fig. 3b; Supplementary Table 4, Online Resource 10).
The key proteins of ERAD pathway EDEM1, UGGT1, SEC62 and VCP were lower in U-2OS/DX580 cells compared to U-2OS cells (Fig. 3c). In keeping with the gene profiling, dox increased these proteins in drug-sensitive but not in drug-resistant cells, Sdox increased them in both cell populations (Fig. 3c).
These data suggest that Sdox forces resistant cells to up-regulate the ERAD/ERQC apparatus, likely as a consequence of an increased burden of unfolded/misfolded proteins in the ER.
H2S-releasing doxorubicin increases sulfhydrated proteins of ER and promotes their ubiquitination
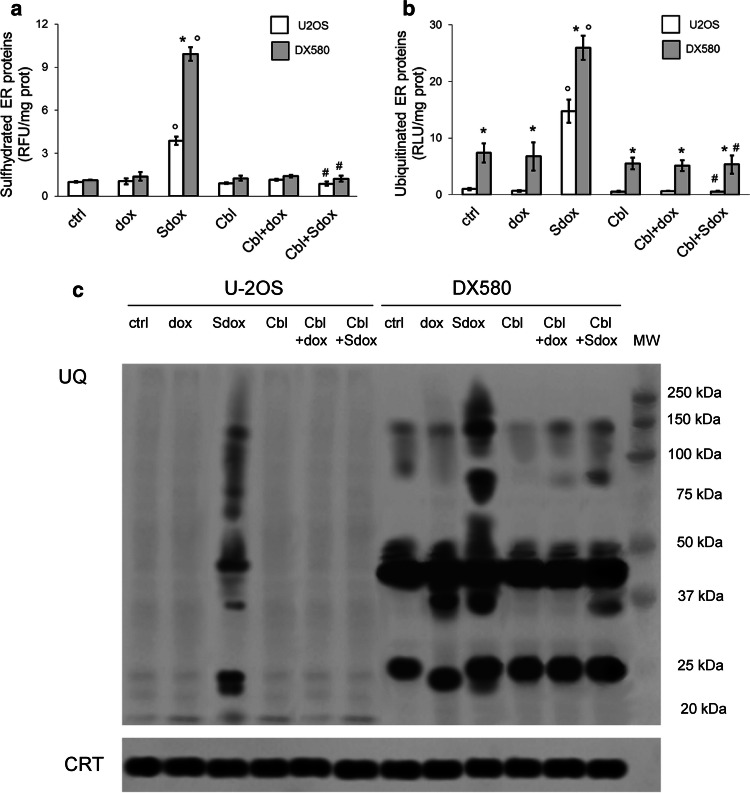
Since Sdox releases H2S [17] that sulfhydrates cysteines and impairs the formation of disulfide bonds [32], we hypothesized that this property induced a significant misfolding of proteins synthesized within the ER. We did not detect any appreciable difference of protein sulfhydration between U-2OS and U-2OS/DX580 cells, either under baseline conditions or after dox treatment (Fig. 4a). By contrast, Sdox significantly increased the amount of sulfhydrated proteins in both sensitive and resistant cells: this effect was due to the release of H2S, since it was abrogated by the co-incubation with the H2S-scavenger hydroxy-cobalamin (Fig. 4a).
Fig. 4.
H2S-releasing doxorubicin increases sulfhydration and ubiquitination of ER-associated proteins. U-2OS and U-2OS/DX580 cells were grown in drug-free medium (ctrl) or in the presence of 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox) for 24 h. When indicated, the H2S-scavenger hydroxy-cobalamin (100 µM, Cbl) was co-incubated. Cells were lysed and microsomal fractions were isolated. a Fluorimetric analysis of sulfhydrated microsomal proteins, performed in triplicates. Data are mean ± SD (n = 5 independent experiments). *p < 0.001 for U-2OS/DX580 cells vs. U-2OS cells; °p < 0.001 for Sdox-treated cells vs untreated (ctrl) or dox-treated cells; #p < 0.001 for Cbl-treated cells vs. corresponding cells without Cbl. b Ubiquitination of microsomal proteins, measured by a chemiluminescence-based assay in triplicates. Data are mean ± SD (n = 5 independent experiments). *p < 0.001 for U-2OS/DX580 cells vs. U-2OS cells; °p < 0.001 for Sdox-treated cells vs untreated (ctrl) or dox-treated cells; #p < 0.001 for Cbl-treated cells vs. corresponding cells without Cbl. d. Microsomal proteins were resolved by SDS-PAGE and probed with an anti-mono/poly-ubiquitin (UQ) antibody. The caltreticulin (CRT) expression was used as control of equal protein loading. MW molecular weight. The figure is representative of one out of four experiments
U-2OS cells had low basal ubiquitination of ER-associated proteins; this ubiquitination rate was not affected by dox (Fig. 4b, c). By contrast, the microsomal ubiquitinated proteins of U-2OS/DX580 cells were significantly higher, either in untreated or in Sdox-treated cells (Fig. 4b, c), likely as a consequence of the low activity of ERAD/ERQC apparatus in the resistant population. Sdox increased ubiquitination of ER-associated proteins and this effect was abolished by H2S-scavenging (Fig. 4b, c), suggesting that sulfhydration was necessary to promote protein ubiquitination. The extent of ubiquitination exerted by Sdox was greater in U-2OS/DX580 cells than in U-2OS cells (Fig. 4b, c), providing a further confirmation of the lower efficiency of ERAD/ERQC system in U-2OS/DX580 cells.
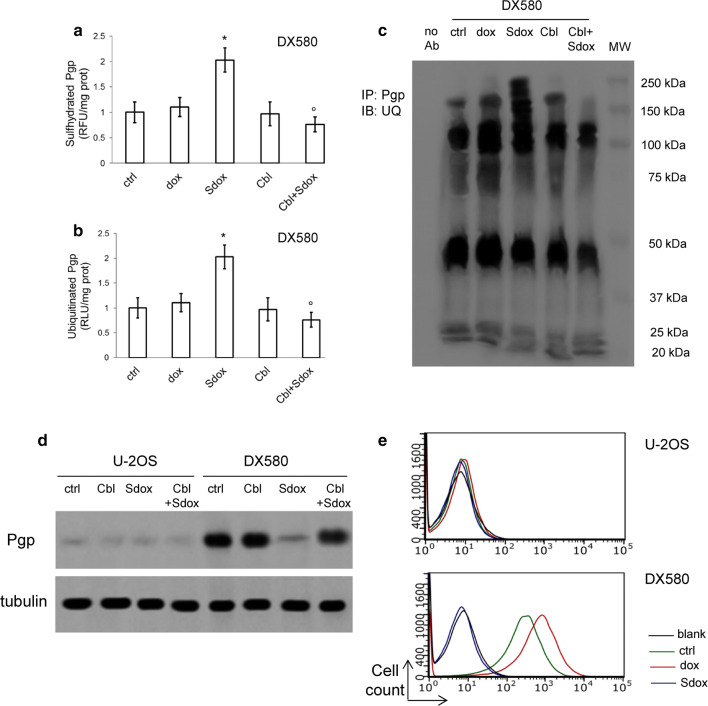
H2S-releasing doxorubicin decreases Pgp amount by promoting its sulfhydration and ubiquitination in resistant osteosarcoma cells
Pgp is synthesized in the ER where it undergoes to proper folding and formation of disulfide bonds. This process is relevant for Pgp functions, since several cysteine residues are critical to preserve the structure and catalytic activity of the protein [33]. Sdox strongly increased the sulfhydration of Pgp extracted from the ER of U-2OS/DX580 cells (Fig. 5a). This process was followed by Pgp ubiquitination (Fig. 5b, c) and resulted in a decrease of Pgp amount in whole cell lysates (Fig. 5d). The abrogation of all these effects by hydroxy-cobalamin (Fig. 5a–d) indicated that H2S is responsible for the sulfhydration and subsequent ubiquitination of Pgp. As a result, the amount of Pgp on cell surface, that was higher in U-2OS/DX580 cells compared to U-2OS cells, returned to the same levels of sensitive cells in the resistant cells treated with Sdox (Fig. 5e).
Fig. 5.
H2S-releasing doxorubicin decreases Pgp by inducing its sulfhydration and ubiquitination. U-2OS/DX580 cells were grown in drug-free medium (ctrl) or in the presence of 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox) for 24 h. When indicated, the H2S-scavenger hydroxy-cobalamin (100 µM, Cbl) was co-incubated. a Pgp was isolated by immunoprecipitation; the amount of sulfhydrated Pgp was measured fluorimetrically in triplicates. Data are mean ± SD (n = 5 independent experiments). *p < 0.001 for Sdox-treated cells vs untreated (ctrl) or dox-treated cells; °p < 0.001 for Cbl-treated cells vs. corresponding cells without Cbl. b Ubiquitinated Pgp, measured by a chemiluminescence-based assay in triplicates. Data are mean ± SD (n = 5 independent experiments). *p < 0.001 for Sdox-treated cells vs untreated (ctrl) or dox-treated cells; °p < 0.001 for Cbl-treated cells vs. corresponding cells without Cbl. c Microsomal proteins were immunoprecipitated (IP) with an anti-Pgp antibody, then immunoblotted (IB) with an anti-mono/poly-ubiquitin (UQ) antibody. The caltreticulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. No Ab: untreated U-2OS/DX580 cell lysate immunoprecipitated in the absence of antibody, to check the specificity of the procedure. MW: molecular weight. The figure is representative of one out of four experiments. d Expression of Pgp in whole lysate derived from cells treated as reported in a. U-2OS cells were used as control of lowly expressing Pgp. The β-tubulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. e Surface Pgp in U-2OS and U-2OS/DX580 cells, treated as reported in a, measured as per flow cytometry in duplicates. Blank: cells incubated with not-immune isotypic antibody. Histograms are representative of one out of five experiments
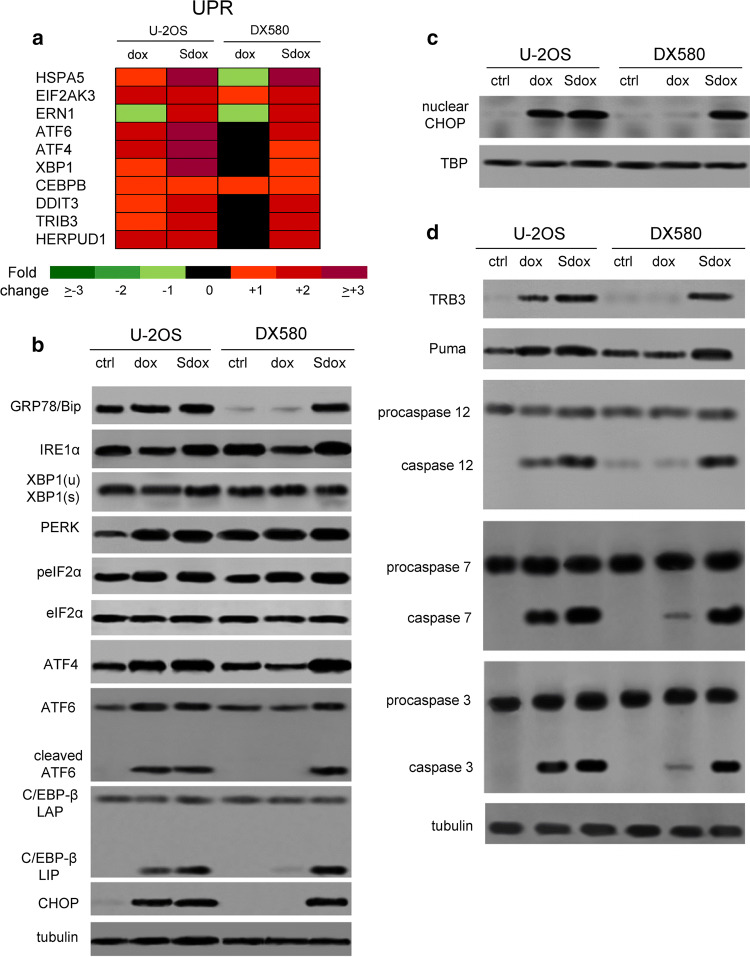
H2S-releasing doxorubicin triggers ER-dependent apoptosis in drug-resistant osteosarcoma cells
Since UPR may trigger either cell death or survival [30, 31], we finally investigated which type of response was induced by Sdox. As shown in Supplementary Tables 3, 4 (Online Resources 9, 10) and Fig. 6a, Sdox up-regulated the expression of UPR sensors (e.g., HSPA5/GRP78, EIF2AK3/PERK, ERN1/IRE1α, ATF6) and effectors (e.g., ATF4, XBP1, CEBPB, DDIT3/CHOP, TRIB3, HERPUD1) in both U-2OS and U2-OS/DX580 cells. A similar trend was elicited by dox in sensitive cells, but not in resistant ones (Supplementary Tables 3, 4, Online Resources 9, 10; Fig. 6a). Sdox-treated cells increased the expression of the GRP78/Bip, an upstream sensor of UPR [31] (Fig. 6b), suggesting that cells exposed to drug exhibited ER stress. The increase of GRP78/Bip was paralleled by the increase of the main ER stress effectors, as demonstrated by the activation of IRE1α/XBP1 axis, PERK/phospho(Ser51) eiF2α/ATF4 axis and ATF6 (Fig. 6b). ATF6 was present in the cleaved form (Fig. 6b) that up-regulates several ERAD-related and UPR-related genes [30].
Fig. 6.
H2S-releasing doxorubicin triggers ER-dependent pro-apoptotic pathways in resistant osteosarcoma cells. a Hitmap of unfolded protein response (UPR)-related genes in U-2OS and U-2OS/DX580 cells, after 24 h treatment with drug-free medium, 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). The figure reports genes up- or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown). The whole list of genes analyzed is reported in Supplementary Tables 3, 4 (Online Resources 9, 10). b Cells treated as in a were lysed and probed with the indicated antibodies. The β-tubulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. XBP1(u): un-spliced; XBP1(s): spliced. c CHOP amount measured in nuclear extracts of cells treated as reported in a. The TBP expression was used as control of equal protein loading. The figure is representative of one out of four experiments. d Whole cell lysates from cells treated as indicated in a were probed with the indicated antibodies. The β-tubulin expression was used as control of equal protein loading. The figure is representative of one out of four experiments. ctrl drug-free medium
A prolonged activation of UPR effectors increases the expression of the pro-apoptotic isoform of C/EBP-β (i.e., C/EBP-β LIP) and of its downstream effector CHOP [34, 35]. CHOP is known to activate TRB3 [36] and PUMA [37] that trigger caspase-mediated apoptosis [37, 38]. In U-2OS/DX580 cells, Sdox increased the amount of C/EBP-β LIP and CHOP levels (Fig. 6b), the nuclear translocation of CHOP (Fig. 6c), the expression of TRB3 and PUMA, and the cleavage of caspase 12, 7 and 3 (Fig. 6d). Interestingly, all Sdox effects were exerted with the same extent in dox-sensitive and dox-resistant cells. By contrast, dox activated these pathways only in the sensitive population (Fig. 6).
The ER stress-triggered apoptosis induced by S-dox was confirmed by the up-regulation of several cell death-inducing genes (e.g., BAX, BEX2, CREB3, ERN2, HTRA2, UHRF1), coupled with the down-regulation of pro-survival genes (e.g., DNAJB9, MAPK8, PCNA, PPP1R15A, RRM2) (Supplementary Tables 3, 4, Online Resources 9, 10; Supplementary Fig. 7, Online Resource 11) in both sensitive and resistant osteosarcoma cells. Once again, dox induced a gene signature similar to Sdox in sensitive cells, while its effects in resistant cells were not univocal, since it increased either cell death-related or cell survival-related genes (Supplementary Tables 3, 4, Online Resources 9, 10; Supplementary Fig. 7, Online Resource 11).
Discussion
In this work, we showed that impairing ER functions with a ER-targeting dox, namely a H2S-releasing dox, was effective against Pgp-expressing osteosarcoma cells.
The results with the Pgp-inhibitor verapamil—that slightly but non significantly increased the intracellular accumulation and cytotoxicity of Sdox—suggested that this compound is less effluxed by Pgp than dox. Our hypothesis is supported by at least two experimental findings. First, Sdox intracellular accumulation had a little decrease in DX30, DX100 and DX580 sublines, where the amount of Pgp hugely increased compare to parental U-2OS and Saos-2 cells. Second, the efflux kinetics in U-2OS/DX580 revealed higher Km and lower Vmax of Sdox compared to dox [17]. This kinetics suggests that: (i) Sdox has a lower affinity for Pgp than dox; (ii) the amount of active Pgp is decreased in Sdox-treated cells. Although resistant cells dramatically decreased the amount of Pgp after Sdox treatment, as discussed below, the residual amount of the protein can be still inhibited by verapamil, justifying the small increase in Sdox intracellular retention and cytotoxicity induced by verapamil. Of note, Pgp has been detected in the cytoplasm and perinuclear region of U-2OS/DX580 cells [39]: this localization is compatible with the presence of Pgp in the ER surrounding the nucleus. Since Sdox has a peculiar accumulation within ER, we might speculate that the progressive decrease in Sdox accumulation in DX variants might be due to the residual Pgp not completely sulfhydrated by Sdox and still active, either on ER (where it pumps Sdox from the ER lumen to the cytosol) or on cell surface (where it pumps the drug outside the cell). The residual activity of Pgp and the efflux of Sdox through Pgp, however, were minimal, since Sdox was significantly more accumulated and significantly more cytotoxic than dox in all the resistant variants.
Indeed dox produced less cell damages, apoptosis and/or necrosis with the progressive increase of resistance: the damages elicited by dox did not produce any significant reduction of cell viability and clonogenic potential of resistant cells. By contrast, Sdox retained all these properties in both sensitive and resistant cell lines, where the cell damage, apoptosis and necrosis were followed by a strong reduction of cell viability and colonies-forming ability.
Notably, Sdox was not toxic in non-transformed cells, confirming previous data showing that Sdox did not damage cardiomyocytes, one of the cell types most sensitive to dox [24]. We thus hypothesized that Sdox efficacy relies on the ability of targeting pathways which are specifically active and biologically relevant in neoplastic cells.
The apoptosis consequent to DNA damage is a peculiar mechanism of dox cytotoxicity [24, 28]. However, Sdox did not induce DNA damage in osteosarcoma cells. This feature can be explained by the lack of nuclear localization of Sdox and/or by the lack of topoisomerase II inhibition [17]. These results suggest that the cytotoxicity and inhibition of clonogenic potential elicited by Sdox against resistant cells was independent from nuclear localization of the drug.
By reducing oxidative damage [16, 29] that is elicited by many chemotherapeutic drugs, H2S can potentially induce chemoresistance. In contrast with these observations, we found that Sdox was cytotoxic notwithstanding the reduction of intracellular ROS and that the drug’s cytotoxicity was reduced by H2S scavenging [17]. Differently from most works using H2S donors, we used a hybrid compound containing a H2S-releasing group linked with dox. Dox moiety can trigger redox cycles and increase the synthesis of reactive nitrogen species such as nitric oxide [40]. Under these conditions, H2S shifts from an anti-oxidant to a pro-oxidant molecule, promotes the synthesis of radical species and induces cell damage [41]. In line with our data, H2S-releasing valproic acid exerted cytotoxic effects in lung cancer cells, by impairing the mitochondrial redox balance [42].
Altering the homeostasis of intracellular compartments of drug-resistant cells is an effective strategy to overcome resistance. Indeed, we recently demonstrated that a mitochondrial-targeted dox [43] overcame Pgp-mediated dox-resistance in osteosarcoma cells by decreasing mitobiogenesis, reducing the ATP supply for Pgp, depolarizing mitochondria and triggering a mitochondria-dependent apoptosis [26].
Sdox had a peculiar localization within ER, raising the hypothesis that it can be cytotoxic by activating ER-dependent cell death pathways. We thus investigated if sensitive and resistant osteosarcoma cells differ for specific ER-related functions and how these functions can be affected by Sdox. Indeed, multiple linkages exist between ER functions and chemoresistance. On the one hand, cells with constitutive or acquired chemoresistance due to Pgp expression are refractory to ER stress-triggered cell death, because they do not activate ER-dependent apoptotic pathways [35]. On the other hand, cancer cells adapted to survive during chronic ER stressing conditions acquire a chemoresistant phenotype [44]. Cells surviving in stressing conditions moderately activate the ERQC program and/or promote ERAD pathways to remove misfolded proteins. If these integrated programs fail, ER stress induces apoptotic pathway such as the C/EBP-β/CHOP/TRB3/caspase 3 axis [35, 45, 46].
We found that the progressive increase of dox-resistance in osteosarcoma cells was associated with the progressive down-regulation of ERAD/ERQC-related genes. This signature is similar to that found in dox-resistant colon and lung cancer cells [35], suggesting that different tumor types resistant to dox share defects in protein folding and/or misfolded protein degradation. We hypothesized that these defects make resistant cells more susceptible to the apoptosis triggered by elevated levels of unfolded proteins within ER, as it occurs in VCP-defective cells upon stress induction [47].
Dox is known to induce ER stress [48], but this response is limited to sensitive cells [35]. In U-2OS cells, indeed, dox up-regulated ERAD/ERQC system genes and proteins, suggesting that it likely damages ER proteins that must be extracted from ER and destroyed. Such compensatory response, however, was not sufficient to protect sensitive cells that—upon dox treatment—activated ER-dependent pro-apoptotic pathways. While dox was localized in the nucleus after 24 h, in the first 20 min after cell exposure, it was found within the ER of U-2OS cells. After this time point, the nuclear localization became predominant and dox resulted undetectable in other compartments. This result suggests that during the uptake process the drug had a transient delivery to ER, providing the rationale basis to explain the ER stress triggered by dox and the consequent ER-dependent apoptosis in sensitive cells. Overall, we suggest that at least two mechanisms—i.e., DNA damage-dependent and ER stress-dependent apoptosis—mediate the cytotoxicity of dox in osteosarcoma sensitive cells. By contrast, the progressive decrease of intracellular dox retention in resistant variants prevented the induction of apoptosis.
Differently from dox, Sdox produced ER-dependent apoptosis in both sensitive and resistant cells. Multiple reasons may explain Sdox efficacy in both cell populations.
First, the predominant accumulation within the ER in sensitive and resistant cells increased the amount of H2S and dox sequestered within this compartment.
Second, Sdox induces a huge sulfhydration of ER-associated proteins. By changing the pattern of disulfide bonds and the consequent tertiary structures of proteins, sulfhydration can either activate or inhibit the target proteins [29, 32]. In our model, sulfhydration was paralleled by increased ubiquitination of ER-associated proteins, suggesting that Sdox increased misfolded/unfolded proteins that are primed for the degradation. Pgp has several cysteines whose mutations can alter catalytic function, ATP binding and protein stability [33, 49]. In U-2OS/DX580 cells, Sdox-induced sulfhydration and ubiquitination of Pgp reduced the protein’s amount to the same level of U-2OS cells: this event provides an additional mechanism explaining why Sdox was well accumulated in Pgp-expressing U-2OS/DX580 and Saos-2/DX580 variants. The result also explains the lower Vmax of Sdox compared to dox [17], indicative of a reduced amount of Pgp on cell surface.
Interestingly, Sdox induced a higher amount of sulfhydrated and ubiquitinated proteins in resistant cells than in sensitive cells. In the case of Pgp, this difference can be justified by the higher amount of the protein in U-2OS/DX580 cells. In the case of other proteins, the defective ERAD/ERQC pathways, which represents an “Achille’s heel” of resistant osteosarcoma cells, may explain the different phenotype. As dox did in sensitive cells, Sdox increased the expression of ERAD/ERQC-related genes in both sensitive and resistant cells, likely as a physiological consequence of the increased misfolded/unfolded proteins. Since resistant cells have a defective ERQC/ERAD system, the attempt to compensate the misfolding produced by Sdox was much more difficult than in sensitive cells. Therein, although Sdox was less accumulated in resistant cells, it was sufficient to trigger a ER-dependent apoptosis consequent to the burden of misfolded/ubiquitinated proteins. We believe that this is the main mechanism responsible of Sdox cytotoxicity.
ER stress is usually associated to conditions promoting oxidative stress, such as radiotherapy, chemotherapy, ionizing radiations, viral infections [50]. Our work suggests that also a strongly reductive environment within ER lumen, like the environment produced by H2S, induces ER stress.
It has been reported that H2S activates the UPR sensor PERK by sulfhydrating the PERK activator protein tyrosine phosphatase 1B [51]. Moreover, H2S modulates the expression of genes involved in UPR, metabolism, cell death and survival, by activating ATF4-dependent pathways [52]. Consistently, Sdox up-regulated UPR-related genes and increased the ratio cell death-related genes/cell survival-related genes. The ER-dependent pro-apoptotic pathways induced by Sdox in sensitive and resistant cells were the same pathways induced by dox in sensitive cells, suggesting that both drugs activated the same ER stress-dependent cell death effectors, although they have different upstream mechanisms, i.e., the induction of oxidative stress for dox, the induction of reductive stress for Sdox [35, 46].
In summary, we characterized the pharmacodynamic properties of a H2S-releasing dox, whose efficacy against dox-resistant osteosarcoma cells relies on multiple factors. First, Sdox has low affinity for Pgp and is localized within ER: these two factors may limit the drug efflux through the Pgp present on cell surface. Second, Sdox exploits a peculiar phenotype of dox-resistant osteosarcoma cells, i.e., the defective ERAD/ERQC system: by increasing the amount of misfolded proteins, it overflows the ERAD/ERQC buffering capacity of resistant cells and triggers ER stress-dependent apoptosis, notwithstanding its lower accumulation in these cells. Third, since Pgp is misfolded and ubiquitinated upon Sdox treatment, we may hypothesize that Sdox triggers a “virtuous circle”: by promoting Pgp degradation, Sdox enhances its own accumulation; this step further amplifies the anti-tumor efficacy of Sdox against resistant cells.
The Sdox characterized in this work can be considered the prototype of a new family of H2S-releasing anthracyclines that kill both dox-sensitive and resistant human osteosarcoma cells. After being validated in pre-clinical models of dox-resistant osteosarcoma, a step that is currently ongoing, this lead compound may represent a significant advancement in the treatment of dox-resistant/Pgp-overexpressing osteosarcomas, where therapeutic improvements are urgently needed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIFF 7569 kb) Supplementary Fig. 1 (Online Resource 1) Representative dot plots of apoptotic/necrotic cells. Doxorubicin-sensitive U-2OS and Saos-2 human osteosarcoma cell lines and their resistant variants (DX30, DX100 and DX580) were incubated 24 h with drug-free medium, 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox), then the percentage of cells positively stained for Annexin V-FITC or propidium iodide (PI) was measured by flow cytometry in duplicates. Dot plots are representative of 1 out of 3 experiments
Supplementary material 3 (TIFF 1895 kb) Supplementary Fig. 2 (Online Resource 3) Dose–response effect of doxorubicin and H2S-releasing doxorubicin on cell viability. Human doxorubicin-sensitive U-2OS cells and Saos-2 cells and their resistant sublines DX580 were incubated for 72 h with increasing concentrations (from 10 nM to 0.5 mM) of doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). Cell viability measured by a chemiluminescence-based assay in quadruplicates. Data are mean ± SD (n = 6 independent experiments)
Supplementary material 4 (TIFF 1229 kb) Supplementary Fig. 3 (Online Resource 4). Effects of doxorubicin and H2S-releasing doxorubicin on non-transformed cells. Human osteoblasts, human fibroblasts and rat H9c2 cardiomyocytes were grown in drug-free medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox) for 24 h (panels a, c, e) or 72 h (panels b, d, f). a, c, e Extracellular release of LDH measured spectrophotometrically in triplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for Sdox vs. dox. b, d, f Cell viability measured with a chemiluminescence-based method in quadruplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.002 for treated vs. untreated cells; °p < 0.05 for Sdox vs. dox
Supplementary material 5 (TIFF 1530 kb) Supplementary Fig. 4 (Online Resource 5) Effects of verapamil on H2S-releasing doxorubicin accumulation and cytotoxicity. Doxorubicin-sensitive U-2OS human osteosarcoma cell line and its U-2OS/DX580 resistant variant were cultured for 6 h (panels a-b), 24 h (panels c-d) or 72 h (panels e–f) in drug-free medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox), in the absence (-) or presence (+) of 50 µM verapamil (ver). a-b. Intracellular drug accumulation, measured by flow cytometry in duplicates, and expressed as mean fluorescence intensity (MFI). Data are mean ± SD (n = 6 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for verapamil-treated vs. verapamil-untreated cells. c-d Extracellular release of LDH measured spectrophotometrically in triplicates. Data are mean ± SD (n = 4 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.01 for verapamil-treated vs. verapamil-untreated cells. e–f Cell viability measured with a chemiluminescence-based method in quadruplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for verapamil-treated vs. verapamil-untreated cells
Supplementary material 6 (TIFF 4630 kb) Supplementary Fig. 5 (Online Resource 6) Early intracellular localization of doxorubicin within sensitive osteosarcoma cells. U-2OS cells were incubated for 24 h with the GFP-KDEL-calreticulin expression vector to label endoplasmic reticulum (ER), then treated with 5 µM doxorubicin (dox) for 10 min, 20 min, 30 min, 1 h, 3 h, 6 h, 24 h. The intracellular localization of the drug was analyzed by fluorescence microscopy. Magnification: 63 × objective lens (1.42 numerical aperture); 10 × ocular lens Bar: 7.5 μm. The micrographs are representative of the dox localization after 20 min, corresponding to the time point with the highest accumulation within the ER, and are representative of 3 experiments with similar results
Supplementary material 8 (TIFF 1184 kb) Supplementary Fig. 6 (Online Resource 8) Expression of ERAD/ERQC and UPR-related genes in doxorubicin-sensitive and doxorubicin-resistant osteosarcoma cells. a-c Hitmap of unfolded protein response (UPR)-related genes, cell death/survival related genes, ER-associated degradation/endoplasmic reticulum quality control (ERAD/ERQC)-related genes in U-2OS/DX30, U-2OS/DX100 and U-2OS/DX580 cells. The figure reports genes up-or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown). The whole list of genes analyzed is reported in Supplementary Table 2 (Online Resource 7)
Supplementary material 11 (TIFF 338 kb) Supplementary Fig. 7 (Online Resource 11) Expression of genes related to cell death and survival in doxorubicin-sensitive and doxorubicin-resistant osteosarcoma cells. Hitmap of genes related to cell death/survival in U-2OS and U-2OS/DX580 cells, after 24 h treatment with drug-free medium, 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). The figure reports genes up-or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown). The whole list of genes analyzed is reported in Supplementary Tables 3-4 (Online Resources 9-10)
Acknowledgements
The work was supported by Italian Association for Cancer Research (IG15232 to CR); Italian Ministry of University and Research (RBFR12SOQ1 to C.R.); Istituto Ortopedico Rizzoli I.R.C.C.S. (5 x mille contributions to the Rizzoli Institute). We are grateful to Dr. Maria Alessandra Contino, Department of Pharmacy, University of Bari “Aldo Moro”, Bari, Italy, for the fruitful discussion, to Dr. Maria Pia Patrizio, Istituto Ortopedico Rizzoli I.R.C.C.S., for the help for the IC50 calculations and to Mr. Costanzo Costamagna, Department of Oncology, University of Torino, for the technical assistance.
Abbreviations
- dox
Doxorubicin
- ABCB1/Pgp
ATP binding cassette B1/P-glycoprotein
- H2S
Hydrogen sulfide
- ROS
Reactive oxygen species
- Sdox
H2S-releasing doxorubicin
- FBS
Fetal bovine serum
- MFI
Mean fluorescence intensity
- LDH
Lactate dehydrogenase
- FITC
Fluorescein isothiocyanate
- PI
Propidium iodide
- RLU
Relative luminescence units
- DAPI
4′,6-Diamidino-2-phenylindole dihydrochloride
- GFP
Green fluorescence protein
- ER
Endoplasmic reticulum
- EDEM1
ER degradation enhancing α-mannosidase like protein 1
- UGGT1
UDP-glucose glycoprotein glucosyltransferase 1
- SEC62
SEC62 homolog/preprotein translocation factor
- VCP
Valosin-containing protein
- GRP78/BiP
Glucose-regulated protein 78/binding immunoglobulin protein
- IRE1α
Inositol requiring kinase-1α
- XBP1
X-box binding protein 1
- PERK
Protein kinase-like endoplasmic reticulum kinase
- eIF2α
Eukariotic initiation factor-2α
- ATF4
Activating transcription factor 4
- ATF6
Activating transcription factor 6
- C/EBP-β
CCAAT-enhancer-binding protein-β
- CHOP/GADD153
C/EBP homologous protein/growth arrest and DNA damage 153
- TRB3
Tribbles homolog 3
- PUMA
p53 up-regulated modulator of apoptosis
- TBP
TATA box binding protein antibodies
- DTT
Dithiothreitol
- RFU
Relative fluorescence units
- UPR
Unfolded protein response
- ERAD
Endoplasmic reticulum-associated protein degradation
- ERQC
Endoplasmic reticulum quality control
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Hattinger CM, Fanelli M, Tavanti E, Vella S, Ferrari S, Picci P, Serra M. Advances in emerging drugs for osteosarcoma. Expert Opin Emerg Drugs. 2015;20:495–514. doi: 10.1517/14728214.2015.1051965. [DOI] [PubMed] [Google Scholar]
- 2.Hattinger CM, Fanelli M, Tavanti E, Vella S, Riganti C, Picci P, Serra M. Doxorubicin-resistant osteosarcoma: novel therapeutic approaches in sight? Future Oncol. 2017;13:673–677. doi: 10.2217/fon-2016-0519. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 4.Fanelli M, Hattinger CM, Vella S, Tavanti E, Michelacci F, Gudeman B, Barnett D, Picci P, Serra M. Targeting ABCB1 and ABCC1 with their specific inhibitor CBT-1® can overcome drug resistance in osteosarcoma. Curr Cancer Drug Targets. 2016;16:261–274. doi: 10.2174/1568009616666151106120434. [DOI] [PubMed] [Google Scholar]
- 5.Baldini N, Scotlandi K, Barbanti-Bròdano G, et al. Expression of P-glycoprotein in high-grade osteosarcomas in relation to clinical outcome. N Engl J Med. 1995;333:1380–1385. doi: 10.1056/NEJM199511233332103. [DOI] [PubMed] [Google Scholar]
- 6.Serra M, Scotlandi K, Reverter-Branchat G, Ferrari S, Manara MC, Benini S, Incaprera M, Bertoni F, Mercuri M, Briccoli A, Bacci G, Picci P. Value of P-glycoprotein and clinicopathologic factors as the basis for new treatment strategies in high-grade osteosarcoma of the extremities. J Clin Oncol. 2003;21:536–542. doi: 10.1200/JCO.2003.03.144. [DOI] [PubMed] [Google Scholar]
- 7.Serra M, Pasello M, Manara MC, Scotlandi K, Ferrari S, Bertoni F, Mercuri M, Alvegard TA, Picci P, Bacci G, Smeland S. May P-glycoprotein status be used to stratify high-grade osteosarcoma patients? Results from the Italian/Scandinavian Sarcoma Group 1 treatment protocol. Int J Oncol. 2006;29:1459–1468. doi: 10.3892/ijo.29.6.1459. [DOI] [PubMed] [Google Scholar]
- 8.Lipshultz SE, Karnik R, Sambatakos P, Franco VI, Ross SW, Miller TL. Anthracycline-related cardiotoxicity in childhood cancer survivors. Curr Opin Cardiol. 2014;29:103–112. doi: 10.1097/HCO.0000000000000034. [DOI] [PubMed] [Google Scholar]
- 9.Callaghan R, Luk F, Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab Dispos. 2014;42:623–631. doi: 10.1124/dmd.113.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szakács G, Hall MD, Gottesman MM, Boumendjel A, Kachadourian R, Day BJ, Baubichon-Cortay H, Di Pietro A. Targeting the Achilles heel of multidrug-resistant cancer by exploiting the fitness cost of resistance. Chem Rev. 2014;114:5753–5774. doi: 10.1021/cr4006236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsouris V, Joo MK, Kim SH, Kwon IC, Won YY. Nano carriers that enable co-delivery of chemotherapy and RNAi agents for treatment of drug-resistant cancers. Biotechnol Adv. 2014;32:1037–1050. doi: 10.1016/j.biotechadv.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Doublier S, Belisario DC, Polimeni M, Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A, Sapino A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: a potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer. 2012;12:e4. doi: 10.1186/1471-2407-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roncuzzi L, Pancotti F, Baldini N. Involvement of HIF-1α activation in the doxorubicin resistance of human osteosarcoma cells. Oncol Rep. 2014;32:389–394. doi: 10.3892/or.2014.3181. [DOI] [PubMed] [Google Scholar]
- 14.Kopecka J, Rankin GM, Salaroglio IC, Poulsen SA, Riganti C. P-glycoprotein-mediated chemoresistance is reversed by carbonic anhydrase XII inhibitors. Oncotarget. 2016;7:85861–85875. doi: 10.18632/oncotarget.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo R, Lin J, Xu W, Shen N, Mo L, Zhang C, Feng J. Hydrogen sulfide attenuates doxorubicin-induced cardiotoxicity by inhibition of the p38 MAPK pathway in H9c2 cells. Int J Mol Med. 2013;31:644–650. doi: 10.3892/ijmm.2013.1246. [DOI] [PubMed] [Google Scholar]
- 16.Sen S, Kawahara B, Gupta D, Tsai R, Khachatryan M, Roy-Chowdhuri S, Bose S, Yoon A, Faull K, Farias-Eisner R, Chaudhuri G. Role of cystathionine β-synthase in human breast cancer. Free Radic Biol Med. 2015;86:228–238. doi: 10.1016/j.freeradbiomed.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Chegaev K, Rolando B, Cortese D, Gazzano E, Buondonno I, Lazzarato L, Fanelli M, Hattinger CM, Serra M, Riganti C, Fruttero R, Ghigo D, Gasco A. H2S-donating doxorubicins may overcome cardiotoxicity and multidrug resistance. J Med Chem. 2016;59(10):4881–4889. doi: 10.1021/acs.jmedchem.6b00184. [DOI] [PubMed] [Google Scholar]
- 18.Szabo C, Coletta C, Chao C, Módis K, Szczesny B, Papapetropoulos A, Hellmich MR. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci USA. 2013;110:12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellmich MR, Coletta C, Chao C, Szabo C. The therapeutic potential of cystathionine β-synthetase/hydrogen sulfide inhibition in cancer. Antioxid Redox Signal. 2015;22:424–448. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma K, Liu Y, Zhu Q, Liu CH, Duan JL, Tan BK, Zhu YZ. H2S donor, S-propargylcysteine, increases CSE in SGC-7901 and cancer-induced mice: evidence for a novel anti-cancer effect of endogenous H2S? PLoS One. 2011;6:e20525. doi: 10.1371/journal.pone.0020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu S, Gao Y, Huang X, Wang X. GYY4137, a hydrogen sulfide (H2S) donor, shows potent anti-hepatocellular carcinoma activity through blocking the STAT3 pathway. Int J Oncol. 2014;44:1259–1267. doi: 10.3892/ijo.2014.2305. [DOI] [PubMed] [Google Scholar]
- 22.Lv M, Li Y, Ji MH, Zhuang M, Tang JH. Inhibition of invasion and epithelial-mesenchymal transition of human breast cancer cells by hydrogen sulfide through decreased phospho-p38 expression. Mol Med Rep. 2014;10:341–346. doi: 10.3892/mmr.2014.2161. [DOI] [PubMed] [Google Scholar]
- 23.Kashfi K, Olson KR. Biology and therapeutic potential of hydrogen sulfide and hydrogen sulfide-releasing chimeras. Biochem Pharmacol. 2013;85:689–703. doi: 10.1016/j.bcp.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granados-Principal S, Quiles JL, Ramirez-Tortosa CL, Sanchez-Rovira P, Ramirez-Tortosa MC. New advances in molecular mechanisms and the prevention of adriamycin toxicity by antioxidant nutrients. Food Chem Toxicol. 2010;48:1425–1438. doi: 10.1016/j.fct.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Serra M, Scotlandi K, Manara MC, Maurici D, Lollini PL, De Giovanni C, Toffoli G, Baldini N. Establishment and characterization of multidrug-resistant human osteosarcoma cell lines. Anticancer Res. 1993;13(2):323–329. [PubMed] [Google Scholar]
- 26.Buondonno I, Gazzano E, Jean SR, Audrito V, Kopecka J, Fanelli M, Salaroglio IC, Costamagna C, Roato I, Mungo E, Hattinger CM, Deaglio S, Kelley SO, Serra M, Riganti C. Mitochondria-targeted doxorubicin: a new therapeutic strategy against doxorubicin-resistant osteosarcoma. Mol Cancer Ther. 2016;15:2640–2652. doi: 10.1158/1535-7163.MCT-16-0048. [DOI] [PubMed] [Google Scholar]
- 27.Riganti C, Miraglia E, Viarisio D, Costamagna C, Pescarmona G, Ghigo D, Bosia A. Nitric oxide reverts the resistance to doxorubicin in human colon cancer cells by inhibiting the drug efflux. Cancer Res. 2005;65:516–525. [PubMed] [Google Scholar]
- 28.Ikeda M, Kurose A, Takatori E, Sugiyama T, Traganos F, Darzynkiewicz Z, Sawai T. DNA damage detected with gammaH2AX in endometrioid adenocarcinoma cell lines. Int J Oncol. 2010;36:1081–1088. doi: 10.3892/ijo_00000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim I, Xu W, Reed J. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 31.Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Rose P, Moore PK. Hydrogen sulfide and cell signaling. Annu Rev Pharmacol Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- 33.Pan L, Aller SG. Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics. Sci Rep. 2015;5:e7880. doi: 10.1038/srep07880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiribau C, Gaccioli F, Huang C, Yuan C, Hatzoglou M. Molecular symbiosis of chop and c/ebp beta isoform lip contributes to endoplasmic reticulum stress-induced apoptosis. Mol Cell Biol. 2010;30:3722–3731. doi: 10.1128/MCB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riganti C, Kopecka J, Panada E, Barak S, Rubinstein M. The role of C/EBP-β LIP in multidrug resistance. J Natl Cancer Inst. 2015;107(5):djv046. doi: 10.1093/jnci/djv046. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Su L, Zhong N, Hao X, Zhong D, Singhal S, Liu X. Salinomycin induces cell death with autophagy through activation of endoplasmic reticulum stress in human cancer cells. Autophagy. 2013;9:1057–1068. doi: 10.4161/auto.24632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cazanave SC, Elmi NA, Akazawa Y, Bronk SF, Mott JL, Gores GJ. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G236–G243. doi: 10.1152/ajpgi.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 39.Maraldi NM, Zini N, Santi S, Scotlandi K, Serra M, Baldini N. P-glycoprotein subcellular localization and cell morphotype in MDR1 gene-transfected human osteosarcoma cells. Biol Cell. 1999;91:17–28. doi: 10.1111/j.1768-322X.1999.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 40.Simunek T, Sterba M, Popelova O, Adamcova M, Hrdina R, Gersl V. Anthracycline-induced cardiotoxicity: overview of studies examining the roles of oxidative stress and free cellular iron. Pharmacol Rep. 2009;61:154–171. doi: 10.1016/S1734-1140(09)70018-0. [DOI] [PubMed] [Google Scholar]
- 41.Bruce King S. Potential biological chemistry of hydrogen sulfide (H2S) with the nitrogen oxides. Free Radic Biol Med. 2013;55:1–7. doi: 10.1016/j.freeradbiomed.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tesei A, Brigliadori G, Carloni S. Organosulfur derivatives of the HDAC inhibitor valproic acid sensitize human lung cancer cell lines to apoptosis and to cisplatin cytotoxicity. J Cell Physiol. 2012;227:3389–3396. doi: 10.1002/jcp.24039. [DOI] [PubMed] [Google Scholar]
- 43.Chamberlain GR, Tulumello DV, Kelley SO. Targeted delivery of doxorubicin to mitochondria. ACS Chem Biol. 2013;8:1389–1395. doi: 10.1021/cb400095v. [DOI] [PubMed] [Google Scholar]
- 44.Salaroglio IC, Panada E, Moiso E, Buondonno I, Provero P, Rubinstein M, Kopecka J, Riganti C. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol Cancer. 2017;16:e91. doi: 10.1186/s12943-017-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meir O, Dvash E, Werman A, Rubinstein M. C/ebp-beta regulates endoplasmic reticulum stress-triggered cell death in mouse and human models. PLoS One. 2010;5:e9516. doi: 10.1371/journal.pone.0009516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 47.Bastola P, Neums L, Schoenen FJ, Chien J. VCP inhibitors induce endoplasmic reticulum stress, cause cell cycle arrest, trigger caspase-mediated cell death and synergistically kill ovarian cancer cells in combination with Salubrinal. Mol Oncol. 2016;10:1559–1574. doi: 10.1016/j.molonc.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panaretakis T, Kepp O, Brockmeier U, Tesniere A, Bjorklund AC, Chapman DC, Durchschlag M, Joza N, Pierron G, van Endert P, Yuan J, Zitvogel L, Madeo F, Williams DB, Kroemer G. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. Embo J. 2009;28:578–590. doi: 10.1038/emboj.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swartz DJ, Mok L, Botta SK, Singh A, Altenberg GA, Urbatsch IL. Directed evolution of P-glycoprotein cysteines reveals site-specific, non-conservative substitutions that preserve multidrug resistance. Biosci Rep. 2014;34:e00116. doi: 10.1042/BSR20140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 51.Krishnan N, Fu C, Pappin DJ, Tonks NK. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, Wang B, Bebek G, Evans CR, Fox PL, Gerson SL, Hoppel CL, Liu M, Arvan P, Hatzoglou M. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (TIFF 7569 kb) Supplementary Fig. 1 (Online Resource 1) Representative dot plots of apoptotic/necrotic cells. Doxorubicin-sensitive U-2OS and Saos-2 human osteosarcoma cell lines and their resistant variants (DX30, DX100 and DX580) were incubated 24 h with drug-free medium, 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox), then the percentage of cells positively stained for Annexin V-FITC or propidium iodide (PI) was measured by flow cytometry in duplicates. Dot plots are representative of 1 out of 3 experiments
Supplementary material 3 (TIFF 1895 kb) Supplementary Fig. 2 (Online Resource 3) Dose–response effect of doxorubicin and H2S-releasing doxorubicin on cell viability. Human doxorubicin-sensitive U-2OS cells and Saos-2 cells and their resistant sublines DX580 were incubated for 72 h with increasing concentrations (from 10 nM to 0.5 mM) of doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). Cell viability measured by a chemiluminescence-based assay in quadruplicates. Data are mean ± SD (n = 6 independent experiments)
Supplementary material 4 (TIFF 1229 kb) Supplementary Fig. 3 (Online Resource 4). Effects of doxorubicin and H2S-releasing doxorubicin on non-transformed cells. Human osteoblasts, human fibroblasts and rat H9c2 cardiomyocytes were grown in drug-free medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox) for 24 h (panels a, c, e) or 72 h (panels b, d, f). a, c, e Extracellular release of LDH measured spectrophotometrically in triplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for Sdox vs. dox. b, d, f Cell viability measured with a chemiluminescence-based method in quadruplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.002 for treated vs. untreated cells; °p < 0.05 for Sdox vs. dox
Supplementary material 5 (TIFF 1530 kb) Supplementary Fig. 4 (Online Resource 5) Effects of verapamil on H2S-releasing doxorubicin accumulation and cytotoxicity. Doxorubicin-sensitive U-2OS human osteosarcoma cell line and its U-2OS/DX580 resistant variant were cultured for 6 h (panels a-b), 24 h (panels c-d) or 72 h (panels e–f) in drug-free medium (ctrl) or in medium containing 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox), in the absence (-) or presence (+) of 50 µM verapamil (ver). a-b. Intracellular drug accumulation, measured by flow cytometry in duplicates, and expressed as mean fluorescence intensity (MFI). Data are mean ± SD (n = 6 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for verapamil-treated vs. verapamil-untreated cells. c-d Extracellular release of LDH measured spectrophotometrically in triplicates. Data are mean ± SD (n = 4 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.01 for verapamil-treated vs. verapamil-untreated cells. e–f Cell viability measured with a chemiluminescence-based method in quadruplicates. Data are mean ± SD (n = 3 independent experiments). *p < 0.001 for treated vs. untreated cells; °p < 0.001 for verapamil-treated vs. verapamil-untreated cells
Supplementary material 6 (TIFF 4630 kb) Supplementary Fig. 5 (Online Resource 6) Early intracellular localization of doxorubicin within sensitive osteosarcoma cells. U-2OS cells were incubated for 24 h with the GFP-KDEL-calreticulin expression vector to label endoplasmic reticulum (ER), then treated with 5 µM doxorubicin (dox) for 10 min, 20 min, 30 min, 1 h, 3 h, 6 h, 24 h. The intracellular localization of the drug was analyzed by fluorescence microscopy. Magnification: 63 × objective lens (1.42 numerical aperture); 10 × ocular lens Bar: 7.5 μm. The micrographs are representative of the dox localization after 20 min, corresponding to the time point with the highest accumulation within the ER, and are representative of 3 experiments with similar results
Supplementary material 8 (TIFF 1184 kb) Supplementary Fig. 6 (Online Resource 8) Expression of ERAD/ERQC and UPR-related genes in doxorubicin-sensitive and doxorubicin-resistant osteosarcoma cells. a-c Hitmap of unfolded protein response (UPR)-related genes, cell death/survival related genes, ER-associated degradation/endoplasmic reticulum quality control (ERAD/ERQC)-related genes in U-2OS/DX30, U-2OS/DX100 and U-2OS/DX580 cells. The figure reports genes up-or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown). The whole list of genes analyzed is reported in Supplementary Table 2 (Online Resource 7)
Supplementary material 11 (TIFF 338 kb) Supplementary Fig. 7 (Online Resource 11) Expression of genes related to cell death and survival in doxorubicin-sensitive and doxorubicin-resistant osteosarcoma cells. Hitmap of genes related to cell death/survival in U-2OS and U-2OS/DX580 cells, after 24 h treatment with drug-free medium, 5 µM doxorubicin (dox) or H2S-releasing doxorubicin (Sdox). The figure reports genes up-or down-regulated at least twofold, in at least one cell line, compared to untreated U-2OS cells (n = 6 independent experiments). The expression of each gene in U-2OS cells was considered 1 (not shown). The whole list of genes analyzed is reported in Supplementary Tables 3-4 (Online Resources 9-10)