Abstract
Sea anemone venoms have long been recognized as a rich source of peptides with interesting pharmacological and structural properties, but they still contain many uncharacterized bioactive compounds. Here we report the discovery, three-dimensional structure, activity, tissue localization, and putative function of a novel sea anemone peptide toxin that constitutes a new, sixth type of voltage-gated potassium channel (KV) toxin from sea anemones. Comprised of just 17 residues, κ-actitoxin-Ate1a (Ate1a) is the shortest sea anemone toxin reported to date, and it adopts a novel three-dimensional structure that we have named the Proline-Hinged Asymmetric β-hairpin (PHAB) fold. Mass spectrometry imaging and bioassays suggest that Ate1a serves a primarily predatory function by immobilising prey, and we show this is achieved through inhibition of Shaker-type KV channels. Ate1a is encoded as a multi-domain precursor protein that yields multiple identical mature peptides, which likely evolved by multiple domain duplication events in an actinioidean ancestor. Despite this ancient evolutionary history, the PHAB-encoding gene family exhibits remarkable sequence conservation in the mature peptide domains. We demonstrate that this conservation is likely due to intra-gene concerted evolution, which has to our knowledge not previously been reported for toxin genes. We propose that the concerted evolution of toxin domains provides a hitherto unrecognised way to circumvent the effects of the costly evolutionary arms race considered to drive toxin gene evolution by ensuring efficient secretion of ecologically important predatory toxins.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2897-6) contains supplementary material, which is available to authorized users.
Keywords: Neurotoxin, Ion channel, Mass spectrometry imaging, 3D structure, Concerted evolution, Extreme resolution mass spectrometry imaging, On-tissue reduction alkylation
Introduction
Venoms are complex cocktails of bioactive molecules that disrupt the physiology of envenomated prey [1, 2]. Although these toxins include a wide range of molecules such as proteins, peptides, polyamines, and salts, the impressive molecular diversity of most invertebrate venoms is due to disulfide-rich peptides [3]. In animals that rely on venom for prey capture, diet and foraging ecology are thought to be major drivers of toxin evolution, with the acquisition of resistance in prey countered by diversifying selection acting on toxin genes in the predator [4]. As a result, the venoms of predatory animals tend to be highly diverse, often containing hundreds to thousands of unique bioactive toxins [3]. One such group is sea anemones, which are benthic, sessile cnidarians that use venom for a variety of ecological functions, including prey capture, defence, digestion, and inter- and intraspecific competition.
Given the ecological importance of venom in sea anemones, and the fact that the cnidarian venom system has been evolving for > 700 million years [5], it is not surprising that sea anemones have evolved a rich variety of venom toxins including enzymes, cytolysins, and neurotoxins [6, 7]. Of these, disulfide-rich peptide neurotoxins constitute the largest molecular diversity. According to the classification system proposed by Mikov and Kozlov [8], at least 17 different peptide folds have been identified in sea anemone venoms [9], although recent proteomics studies suggest that they likely contain 30 or more [10].
In addition to being the most diverse components of sea anemone venoms, neuroactive peptides are also the most well studied. They have been used as tools for probing ion channel structure and function, and for developing novel therapies [7]. For example, ShK, a venom peptide from the sea anemone Stichodactyla helianthus, recently completed phase 1 clinical trials for treatment of autoimmune disease [11]. Neurotoxins from sea anemone venoms act on a diverse range of ion channels, including acid-sensing ion channels (ASIC), transient receptor potential ion (TRP) channels, and voltage-gated sodium (NaV) and potassium (KV) channels. Of these, KV toxins are the most diverse group, comprising 136 of the 320 annotated sea anemone toxins in UniProtKB. These KV toxins are currently divided into five distinct types based on their sequence, disulfide-bridge pattern, and activity [12].
Here we describe the structure, activity, function and evolution of a new, sixth type of sea anemone KV toxin. κ-Actitoxin-Ate1a (henceforth Ate1a), from venom of the Waratah sea anemone Actinia tenebrosa, is the shortest sea anemone toxin reported to date, and it adopts a novel β-hairpin-like 3D fold. In contrast with many β-hairpin peptides, Ate1a lacks antimicrobial activity and instead serves a predatory function via potent inhibition of prey KV channels. While most families of predatory toxins evolve via bursts of extensive duplication and diversification, this is not the case for the Ate1a toxin family, whose members remain remarkably well conserved despite their ancient evolution. Our data suggest that this extreme conservation is due to intra-gene concerted evolution, a process that has to our knowledge not been previously reported for any toxin family and which we propose is a hitherto unrecognised mechanism of maintaining efficient secretion of ecologically important toxins.
Materials and methods
Venom collection and fractionation
Sea anemones were housed in aquaria at The University of Queensland. Animals were kept for no longer than 2 months prior to any experiments, and the average conditions for the system where the animals were kept were: 10 h light, 14 h dark; salinity 35.9 ppt; pH 8.22, temperature 29.19 °C. Venom was obtained by electrical stimulation [13] and fractionated using reverse-phase HPLC as described previously [10].
Mass spectrometry
Peptide masses in lyophilised HPLC fractions were analysed using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (AB SCIEX 5800 MALDI-TOF/TOF mass spectrometer). HPLC fractions were mixed 1:1 (v/v) with α-cyano-4-hydroxy-cinnamic acid (7.5 mg/mL in 50/50 acetonitrile (ACN)/H2O, 0.1% trifluoroacetic acid. Mass spectra were collected in reflector positive mode. 1,5-Diaminonaphthalene was used as a reductive matrix [14] to sequence intact Ate1a using in-source dissociation (ISD) MS. The sample was mixed 1:1 (v/v) with 1,5-diaminonaphthalene (15 mg/mL in 50/50 ACN/H2O, 1% formic acid, and spectra were interpreted manually.
MALDI MS imaging (MSI) was performed according to published protocols [15, 16] using an UltraFlex III mass spectrometer (Bruker-Daltonik, Bremen, Germany). On-tissue reduction and alkylation of cystines were carried out on de-paraffinised tissue sections using a volatile reaction protocol as described [17], but with a 3.5 mL reaction volume in a 50 mL Corning Falcon tube (Thermo Fisher Scientific).
For ultra-high mass resolution MSI we used a SolariX XR 7T FT-ICR mass spectrometer (Bruker-Daltonik, Bremen, Germany) operated in positive ion mode. Data size was set to 1 M across the mass range 400–6000 m/z. MALDI source was set to a laser power 50%, a total of 500 shots per scan at a frequency of 2 kHz, and smart walk was enabled with a width of 90 µm. The collision cell radiofrequency (RF) was set to 1.4 MHz, collision RF amplitude 1100 Vpp, transfer optics time of flight 1.5 ms at a frequency of 2 MHz with RF amplitude 400 Vpp. The sweep excitation was set to 20%. For isotopic fine structure analysis, data size was set to 4 M across the mass range 200–3000 m/z. Data were collected and averaged across eight scans. MALDI source was set to a laser power 50%, with a total of 5000 shots per scan at frequency of 2 kHz, and the laser was manually moved across sample area. For isolation the quadrupole was set to 1890.00 with an isolation window of 5 m/z. The collision cell RF was set to 2 MHz and collision RF amplitude 1200 VPP. The transfer optics were set to a time of flight = 1.5 ms, with RF set to 4 MHz and RF amplitude 400 Vpp. Sweep excitation was set to 19%. For data analysis, we used Bruker (Bruker-Daltonik, Bremen, Germany) DataAnalysis 5.0 and for image analysis Bruker FlexImaging 5.0 and Bruker SCiLS Lab 2017a were used.
Transcriptomics
Total RNA extraction by TRIzol, cDNA library preparation, transcriptome sequencing, read trimming, and assembly was performed as described previously [10]. Raw sequence reads (SRA) and Trinity-assembled contigs were deposited with links to BioProject accession number PRJNA414357 in the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/). Coding sequences (CDSs) were identified using the Galaxy tool ‘Get open reading frames or coding sequences’ [18]. The Ate1a sequence determined using ISD-MALDI-MS was used to search the translated transcriptome using NCBI BLAST + blastp [19].
Evolution of Ate1a
The Ate1a prepropeptide sequence was used to search for homologues in the UniProtKB, NCBI nr and EST databases, as well as a tentacle transcriptome of S. haddoni [10], using NCBI BLAST + blastp. Nucleotide sequences were retrieved and aligned using mafft v7.304b [20], domains were extracted using CLC Main Workbench v7.6.1, and maximum likelihood phylogenies reconstructed with IQ-Tree v1.5.5 [21] for each domain type. The evolutionary model (FLU + G4) was determined using ModelFinder [22], and support values estimated by ultrafast bootstrap using 10,000 iterations [23].
Ate1a synthesis
Ate1a (H-RCKTCSKGRCRPKPNCG-NH2) was assembled using Fmoc chemistry (0.1 mmol scale) on a Symphony automated peptide synthesiser (Protein Technologies) on Fmoc-Rink-amide polystyrene resin. Amino acid couplings, Fmoc deprotections, and removal of side-chain protecting groups were achieved using published protocols [24]. Side-chain protecting groups were: Arg(Pbf), Asn(Trt), Cys(Trt), Lys(Boc), Ser(tBu) and Thr(tBu). The crude product (electrospray ionisation (ESI)–MS m/z: calc. (avg) 632.1 [M + 3H]3+, found 632.0) was oxidatively folded by stirring in 0.1 M NH4HCO3 (pH 8.1) at room temperature for 3 days to give a single major isomer that was isolated by preparative HPLC (ESI–MS m/z: calc. (avg) 630.8 [M + 3H]3+, found 631.0).
NMR structure determination of Ate1a
The solution structure of Ate1a was determined using 2D NMR spectroscopy. Spectra of synthetic Ate1a (1 mM in 20 mM sodium phosphate, pH 6, 5% D2O) were acquired at 10 °C on a cryoprobe-equipped Avance 600 MHz spectrometer (Bruker BioSpin). Resonance assignments were made using 2D 1H-1H TOCSY, 2D 1H-1H NOESY spectra, and natural abundance 2D 1H-15N HSQC and 1H-13C HSQC spectra. Spectra were analysed using CcpNmr Analysis v2.4.1 [25]. Resonance assignments (97.8% complete) have been deposited in BioMagResBank (accession number 30342). Dihedral-angle restraints were derived using TALOS-N [26] and the restraint range was set to twice the estimated standard deviation. The NOESY spectrum was manually peak-picked, then 96.3% of the peak list was automatically assigned, and structures calculated using CYANA v3.97 [27] The final structure was calculated using 66 interproton distance restraints, 6 disulfide-bond restraints and 23 dihedral-angle restraints. 200 structures were calculated and then the 20 with highest stereochemical quality as judged by MolProbity were used to represent the solution structure of Ate1a. Atomic coordinates are available from the Protein Data Bank (accession code 6AZA).
Electrophysiological characterisation of Ate1a
The pharmacological effect of Ate1a was analysed by heterologous expression of rKV1.1, rKV1.2, hKV1.3, rKV1.4, rKV1.5, rKV1.6, Shaker IR, rKV2.1, hKV3.1, rKV4.2, hKV7.2, hKV11.1, rNaV1.2, rNaV1.3, rNaV1.4, hNaV1.5, mNaV1.6, hNaV1.7, rNaV1.8, rASIC1a, rASIC1b, rASIC2a, and rASIC3 in Xenopus laevis oocytes, with lowercase r, m and h indicating the channel is of rat, mouse or human origin, respectively. The linearized plasmids were transcribed using the T7 or SP6 mMESSAGE-mMACHINE transcription kit (Ambion, Waltham, MA, USA). The KV1.1 triple mutant channel was constructed as previously described [28]. Oocytes were injected with 50 nL of cRNA at a concentration of 0.05–1 ng/nL using a micro-injector (Drummond Scientific, Broomall, PA, USA). Injected oocytes were stored at 19 °C in an ND96 solution (in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 2 MgCl2 and 5 HEPES; pH 7.4), supplemented with 50 μg/mL gentamicin sulphate.
Two-electrode voltage-clamp recordings were performed at room temperature (18–22 °C) using a Geneclamp 500 or Axoclamp 900A amplifier (Molecular Devices, Sunnyvale, CA, USA) controlled by a pClamp data acquisition system (Axon Instruments, Union City, CA, USA). Whole-cell currents from oocytes were recorded 1–5 days after injection, when a whole-cell current could be observed. The bath solution composition was ND96 (in mM: 2 NaCl, 96 KCl, 1.8 CaCl2, 2 MgCl2 and 5 HEPES; pH 7.4). Voltage and current electrodes were filled with KCl (3 M). The resistance of both electrodes was kept between 0.8 and 1.0 MΩ. KV currents were filtered at 0.5 or 2 kHz using a four-pole low-pass Bessel filter and leak subtraction were performed using a − P/4 protocol. KV1.1 − KV1.6 and Shaker IR currents were evoked by 500 ms depolarization to 0 mV followed by a 500 ms pulse to − 50 mV, from a holding potential of − 90 mV. KV2.1, KV3.1, KV4.2 and KV4.3 currents were elicited by 500 ms pulses to + 20 mV from a holding potential of − 90 mV. Current traces of hERG channels were elicited by applying a + 40 mV pulse for 2.5 s followed by a step to − 120 mV for 2.5 s. NaV currents were evoked by 100 ms depolarization pulse from a holding potential of −90 to −20 mV, with the exception of NaV1.8 which were pulsed to 0 mV. NaV data were digitized at 20 kHz; leak and background conductance were identified by blocking channels with tetrodotoxin and subtracted from currents. ASIC currents were acquired (digitized 2 kHz and filtered at 0.01 Hz) and elicited by a drop in pH from 7.45 to 6.5, 5.5, 4.5, and 6.3 (for ASIC1a, ASIC1b, ASIC2a, and ASIC3, respectively).
Due to the lack of a hNaV1.1 clone for oocyte screening, activity on this channel was assessed via manual patch-clamp electrophysiology. HEK293 cells heterologously expressing hNaV1.1 (SB Drug Discovery, Glasgow, UK) were maintained at 37 °C in a humidified 5% CO2 incubator in minimal essential medium supplemented with 10% FBS v/v, 2 mM l-glutamine and selection antibiotics as recommended by the manufacturer. Cells were grown to 70–80% confluence and passaged every 2–4 days using Detachin (Genlantis, San Diego, CA, USA). Whole-cell patch-clamp recordings were obtained using a MultiClamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA). Patch pipettes were pulled from standard wall borosilicate glass capillaries (1.5 mm × 0.86 mm, OD/ID; SDR Scientific, Sydney, AUS) using a microelectrode puller (P-97; Sutter Instrument Co., Novato, CA, USA) and had a resistance of 1.2–1.5 MW when filled with pipette solution. The pipette solution was composed of (in mM) 150 CsCl, 1 EGTA, 10 HEPES and was adjusted to pH 7.2 with CsOH. The external bath solution consisted of (in mM) 140 NaCl, 4 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES and adjusted to pH 7.4 with NaOH. Currents were monitored for at least 5 min after establishing whole-cell configuration to allow currents to stabilise. The pulse protocol consisted of cells being held at − 90 mV for 10 s, followed by a hyperpolarizing step to − 120 mV for 200 ms, then a depolarizing step to − 15 mV for 50 ms. Series resistance and prediction compensation between 50 and 75% was applied to reduce voltage errors. Recorded currents were acquired with a Digidata 1550B (Molecular Devices) converter at 50 kHz after passing through a low-pass Bessel filter of 10 kHz. A P/6 subtraction protocol provided by the Clampex (Molecular Devices) acquisition software was used to remove linear leak and residual capacitance artefacts.
Ate1a concentration–response relationships data were fitted with the Hill equation y = 100/[1 + (IC50/[toxin])h], where y is the amplitude of the toxin-induced effect, IC50 is the toxin concentration at half-maximal efficacy, [toxin] is the toxin concentration and h is the Hill coefficient. To investigate the current–voltage (I–V) relationship, current traces were evoked by 10 mV depolarization steps from a holding potential of − 90 mV. The values of IK were plotted as function of voltage and fitted using the Boltzmann equation IK/Imax = [1 + exp(Vg − V)/k]−1, where Imax represents maximal IK, Vg is the voltage corresponding to half-maximal current and k is the slope factor. To assess the concentration dependence of the Ate1a induced inhibitory effects, a concentration–response curve was constructed in which the percentage of current inhibition was plotted as a function of toxin concentration. Data were fitted with the Hill equation. All data represent at least three independent experiments (n ≥ 3) and are presented as mean ± standard error. Comparison of two sample means was made using a paired Student’s t test (P < 0.05). All data were analysed using Clampfit 10.3 (Molecular Devices) and origin 7.5 software (Origin Lab., Northampton, MA, USA).
Antimicrobial activity of Ate1a
Antimicrobial screening was performed by the Community for Antimicrobial Drug Discovery (CO-ADD) (http://www.co-add.org). Inhibition of growth was measured against five bacteria: Escherichia coli (ATCC 25922), Klebsiella pneumoniae (ATCC 700603), Acinetobacter baumannii (ATCC 19606), Pseudomonas aeruginosa (ATCC 27853) and Staphylococcus aureus (MRSA; ATCC 43300), and two fungi: Candida albicans (ATCC 90028) and Cryptococcus neoformans (ATCC 208821), following protocols previously described [Page 5, Supplementary Information in 29].
Susceptibility of red blood cells and cultured cells to Ate1a
Hemolytic assays employed erythrocytes isolated from fresh human blood collected from healthy donors using protocols approved by the Human Research Ethics Committee at The University of Queensland. Hemolysis was quantified as described [30] by measuring haemoglobin release at 405 nm, with melittin and cyclic gomesin as controls. The cytotoxicity of Ate1a was determined against HeLa, MCF-7, and HFF-1 cell cultures using a resazurin colorimetric assay [31], with melittin and cyclic gomesin as positive controls.
Interactions of Ate1a with lipid bilayers
Surface plasmon resonance (SPR) was used to monitor the affinity of Ate1a for lipid membranes using a Biacore 3000 instrument (GE Healthcare) and an L1 chip at 25ºC. Synthetic POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine) (Avanti polar lipids) were used to prepare small unilamellar vesicles (SUVs, 50 nm diameter) composed of POPC or POPC/POPS (4:1 molar ratio) dispersed in HEPES buffer (10 mM HEPES containing 150 mM NaCl, pH 7.4) and homogenized by extrusion. The L1 chip possesses a dextran coat modified with alkyl chains to allow attachment of liposomes and formation of lipid bilayers. SUVs were injected onto an L1 chip for 40 min at a flow rate of 2 μL/min; the signal reached a steady state below 10,000 response units in the four flow channels confirming coverage of the chip surface and formation of stable bilayers. Serial two-fold dilutions of Ate1a, starting from 64 μM, were injected over deposited lipid bilayers for 180 s at a flow rate of 5 μL/min (association phase); dissociation was followed for 600 s [32, 33]. An N-to-C cyclized version of gomesin (UniProtKB P82358) was included for comparison. The chip was regenerated as before [34]. All solutions were freshly prepared and filtered using a 0.22 μm filter; HEPES buffer was used as running buffer. Response units were normalized to peptide-to-lipid ratio (P/L) as previously described [32].
Toxicity of Ate1a in vivo
Toxicity of Ate1a to brine shrimps (A. salina) and amphipods (family Talitridae) was examined as previously described [35, 36]. For assays where the toxin was dissolved in medium, synthetic Ate1a was dissolved to a concentration of 0.5 mg/mL in filtered artificial seawater. Assays were performed in 24-well plates for shrimps and 6-well plates for amphipods (Thermo Fisher Scientific). Paralysis and lethality were assessed by microscopic observation and responsiveness to contact with a 10 µL plastic tip. Bovine serum albumin (5 mg/mL) was dissolved in the medium to be used as control (no toxicity). For injection assays, Ate1a was diluted with a physiological solution for crustaceans (in mM: NaCl 470.4, KCl 8.0, CaCl2 18.0, MgCl2 1.5, NaHCO3 6.0 and glucose 5.6). The injection volume was 9.4 nL. Groups of ten amphipods (4.35–11.53 mg) were challenged with 5.3 mM of toxin and observed for mortality or paralysis up to 4 h. Controls were amphipods that were not injected or were injected with crustacean physiological solution.
Results
Discovery of Ate1a
Specimens of A. tenebrosa were collected off the coast of North Stradbroke Island, Queensland, Australia (27°15S, 153°15E), and venom was obtained by electrical stimulation [10]. Fractionation of venom using reversed-phase chromatography revealed a conspicuous early eluting peak containing an unusually low-mass component (Fig. 1a), which we confirmed to be a disulfide-rich peptide by de novo sequencing using ISD-MALDI MS (Fig. 1b). The toxin, which we named Ate1a, is a 17-residue peptide with two disulfide bonds and an amidated C-terminus (RCKTCSKGRCRPKPNCG-NH2), yielding a monoisotopic mass of 1887.93 Da. Ate1a is a novel peptide with no BLAST hits in the UniProtKB or NCBI databases.
Fig. 1.
Isolation and sequencing of Ate1a. a C18 RP-HPLC chromatogram showing fractionation of crude A. tenebrosa venom. The early eluting peak containing Ate1a is highlighted in red. Inset shows average mass and isotope family for Ate1a. b De novo sequencing of Ate1a using ISD-MALDI MS
To confirm the amino acid sequence of Ate1a and identify any venom homologues, we generated a transcriptome from tentacles actively regenerating venom, as described previously [10]. De novo assembly with Trinity yielded 87,485 contigs, which translated to 457,470 potential CDSs. A BLAST search was used to identify the transcript encoding Ate1a, and this returned a single contig containing a partial CDS with multiple copies of a peptide domain encoding a sequence identical to that determined by ISD-MALDI MS. Analysis of remapped reads revealed that this contig represents two unique transcripts whose CDS differ by two synonymous and two non-synonymous mutations in the propeptide regions of the Ate1a preproprotein (Figure S1, NCBI SRA accession SRR6282389).
Ate1a belongs to a novel peptide family that evolves by intra-gene concerted evolution
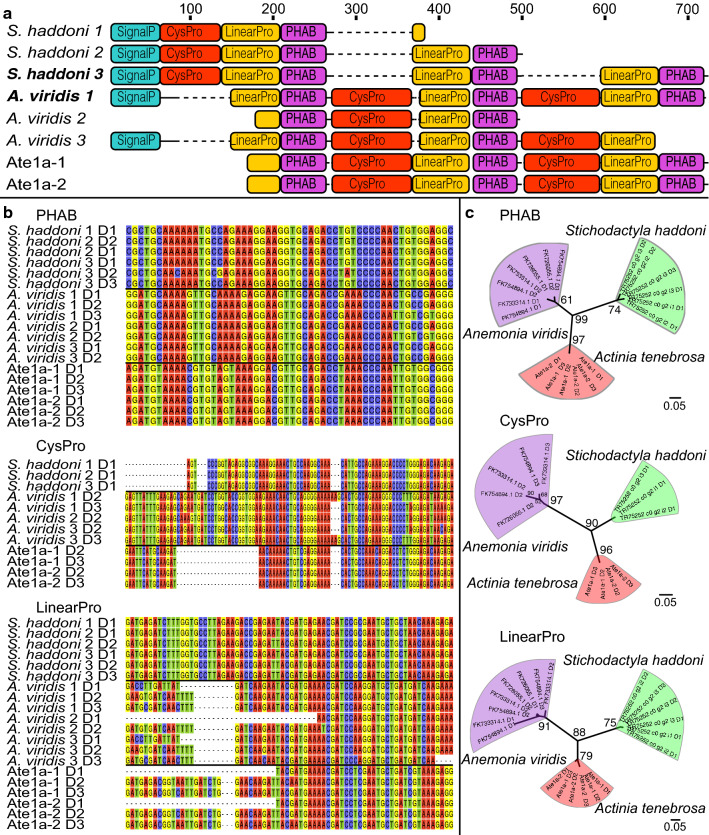
We used the complete sequence of the Ate1a precursor to search for related sequences. A BLAST search against the UniProtKB and GenBank nr databases returned no significant hits, but BLAST searches against the NCBI expressed sequence tag (EST) database returned one full-length and two partial Ate1a-like prepropeptide sequences in Anemonia viridis. Similarly, a BLAST search against our published tentacle transcriptome from Stichodactyla haddoni [10] yielded three unique contigs, including one full-length prepropeptide. These species include two separate families (Actiniidae and Stichodactylidae), suggesting that the Ate1a toxin gene family arose in a common ancestor of the superfamily Actinioidea [37].
All identified Ate1a-like prepropeptides are comprised of the same set of domains separated by dibasic cleavage sites: a signal peptide, one or two cysteine-containing propeptide domains, and three cysteine-free propeptide domains that each precedes an Ate1a-like domain (Fig. 2a). The domain architecture is also the same for all prepropeptides except for the position of the cysteine-containing propeptide, which in Stichodactylidae (S. haddoni) is found as a single copy immediately following the signal peptide, but in Actiniidae (A. tenebrosa and A. viridis) exists as two copies that each follow the first two Ate1a-like domains. This suggests that the Ate1a precursor gene underwent early domain duplication events followed by either multiple domain deletions or convergent deletions and duplications. Strikingly, however, the domains share almost 100% nucleotide identity with corresponding domains within the same prepropeptide (Fig. 2b). Phylogenetic analysis revealed that domains from each species form well-supported clades with respect to those from other species (Fig. 2c). This suggests that the extreme sequence conservation observed in domains of the Ate1a gene family is due to concerted evolution, an evolutionary phenomenon that, to our knowledge, has previously only been demonstrated for a single animal toxin gene family [38], but never for within-gene toxin domains.
Fig. 2.
Domain architecture and evolution of Ate1a precursors. a Domain architecture of Ate1a and Ate1a-like contigs. Prepropeptides are composed of a signal peptide (SignalP), one or two cysteine-containing propeptide domains (CysProP), and three cysteine-free propeptide domains (LinearProP) that each precedes an Ate1a-like PHAB domain. b Nucleotide sequence alignments for each domain. c Maximum likelihood phylogenetic reconstructions for each domain. Bootstrap support values are shown at the nodes, while horizontal bars indicate genetic distance. Sequence accessions are for S. haddoni (1–3) TR75252_c0_g2_i1–3 and A. viridis (1) FK754894, (2) FK726055, and (3) FK733314. See also Figure S1
Ate1a defines a new peptide fold
The unique primary structure of Ate1a prompted us to characterise its three-dimensional (3D) structure using NMR spectroscopy. We synthesised Ate1a using solid-phase peptide synthesis and confirmed correct folding of the synthetic product by HPLC co-elution with native peptide (Fig. S2).
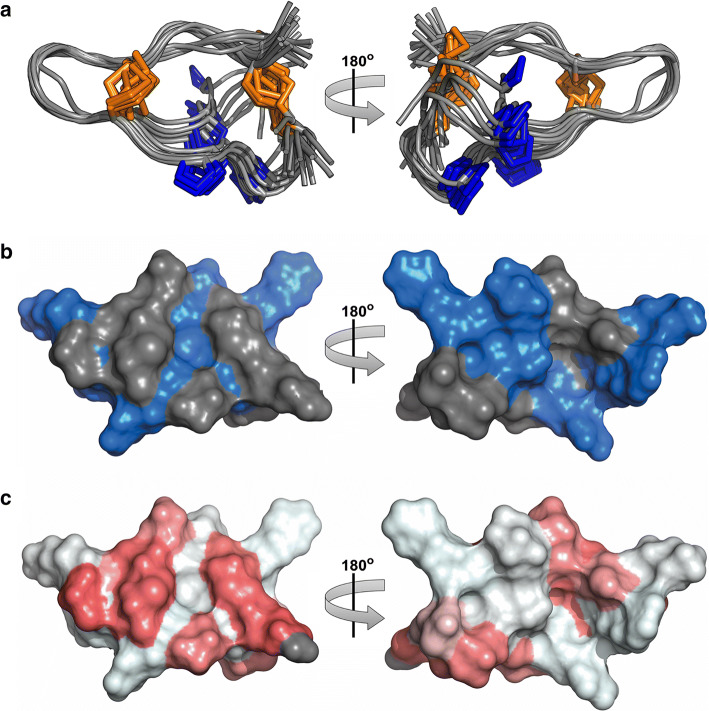
The 3D structure of Ate1a (Fig. 3) was determined using homonuclear NMR methods, and statistics for the ensemble of structures are shown in Supplementary Table 1. MolProbity analysis [39] revealed that the structure has excellent stereochemical quality, with no steric clashes and ~ 90% of residues in the most favoured Ramachandran regions. The precision of the structure, however, is not very high (backbone RMSD 1.11 ± 0.28 Å), suggesting that it is highly dynamic, particularly within the longest intercystine loop (loop 3). This may be due the presence of two proline residues, which leads Ate1a to adopt two distinct conformations (Fig. 3a). Both conformations adopt a fold similar to that of β-hairpin-like peptides [40], where the C- and N- termini are connected via two semi-parallel disulfide bonds (C1–C4 and C2–C3). One face of the toxin has a high proportion of positively charged residues (Fig. 3b) whereas the opposite face is rich in hydrophobic residues (Fig. 3c).
Fig. 3.
3D structure of Ate1a. a Solution structure of Ate1a (ensemble of 20 structures; PDB code 6AZA). Disulfide bonds are highlighted in orange and proline side chains are shown in blue. b Surface representation of Ate1a with cationic and uncharged residues shown in blue and grey, respectively. c Surface representation of Ate1a showing relative hydrophobicity, which increases from white to red. See also Figure S2 and Table S1
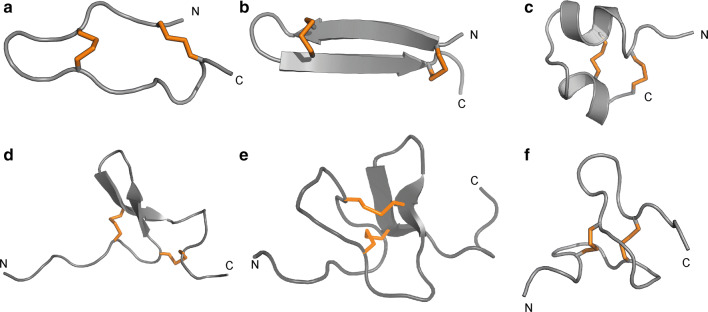
Although Ate1a displays a hairpin-like structure (Fig. 4a), it is neither a true hairpin scaffold nor similar to any other previously described hairpin-like peptide fold. The two-disulfide-enclosed loops of Ate1a differ substantially in length, with loop 1 containing just two residues compared to five in loop 3. In combination with the two prolines in loop 3, this asymmetry prevents the formation of secondary structures characteristic of other disulfide-enclosed hairpin-like structures such as β-hairpin antimicrobial peptides (AMPs) [40] (Fig. 4b) or the cystine-stabilised α/α (CSαα) fold [41] (Fig. 4c). Ate1a also differs from other hairpin-like folds found in animal toxins, such as the boundless β-hairpin (BBH) [42] (Fig. 4d) and disulfide-directed hairpin (DDH) fold [43] (Fig. 4e), or the two-disulfide fold of RhTx from venom of the centipede Scolopendra subspinipes [44] (Fig. 4f). Thus, Ate1a is the prototypic member of a previously undescribed peptide fold that we coined the proline-hinged asymmetric β-hairpin-like (PHAB) fold.
Fig. 4.
Ate1a is the first member of the new PHAB fold. Comparison of the PHAB fold with other peptide folds containing two disulfide bonds and a similar number of residues (16–29 residues). Disulfide bonds are shown as orange tubes and N- and C-termini are labelled. a Ate1a; b β-hairpin fold represented by the spider peptide gomesin (PDB 1KFP); c CS α/α motif represented by the scorpion toxin κ-hefutoxin1 (PDB 1HP9); d boundless β-hairpin motif represented by sea anemone toxin π-AnmTX Ugr 9a-1 (PDB: 2LZO); e disulfide-directed hairpin represented by scorpion toxin U1-Liotoxin-Lw1a (PDB 2KYJ); f unstructured two-disulfide peptide fold represented by centipede toxin RhTx (PDB 2MVA)
Ate1a represents a new type of sea anemone KV toxin
Many Arg/Lys-rich, disulfide-stabilised β-hairpin peptides (e.g., gomesin and tachyplesin-1) function as AMPs in the innate immune system. They often have high affinity for lipid membranes and possess both anticancer and antimicrobial activity [29]. Although Ate1a does not adopt a typical β-hairpin fold, it is highly positively charged. However, Ate1a had no antimicrobial activity at concentrations up to 256 μg/mL. Similarly, Ate1a was not cytotoxic or cytolytic against cultured human cancer cell lines or erythrocytes (Fig. S3). Consistent with these results, Ate1a displayed only weak affinity for, and rapid dissociation from, lipid membranes compared to gomesin [45] (Fig. S4). Taken together, Ate1a’s lack of antimicrobial and cytolytic activity, as well as its low affinity for lipid membranes suggests that it does not play a role in defence against pathogens.
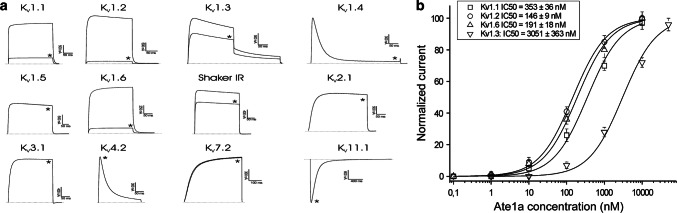
Ion channels are the most common molecular target of disulfide-rich venom peptides, and we therefore used electrophysiology to screen Ate1a against 8 NaV channels, 12 KV channels, and 4 ASIC subtypes. Ate1a was found to selectively target several members of the Shaker subfamily of KV channels; at 3 μM it inhibited currents mediated by KV1.1 (84 ± 4%), KV1.2 (94 ± 3%), KV1.3 (38 ± 4%), KV1.6 (92 ± 2%), and Shaker IR (23 ± 2%) channels (Fig. 5a). No activity was observed on other channels at the same concentration (Fig. S5). Fitting of the Hill equation to concentration–response curves for KV1.1, KV1.2, KV1.3 and KV1.6 yielded IC50 values of 353 nM, 146 nM, 3051 nM and 191 nM, respectively (Fig. 5b). Thus, given its unique sequence and structure, Ate1a represents a new, sixth type of sea anemone KV toxin.
Fig. 5.
Electrophysiological characterization of KV isoforms inhibited by Ate1a. a Representative whole-cell current traces obtained from KV channels expressed in Xenopus oocytes in the absence (control) and presence (*) of 3 μM Ate1a. b Concentration–response curves obtained by plotting current inhibition as a function of increasing Ate1a concentration. See also Figures S3–S5 and Table S2
Ate1a is a toxin with a predatory function
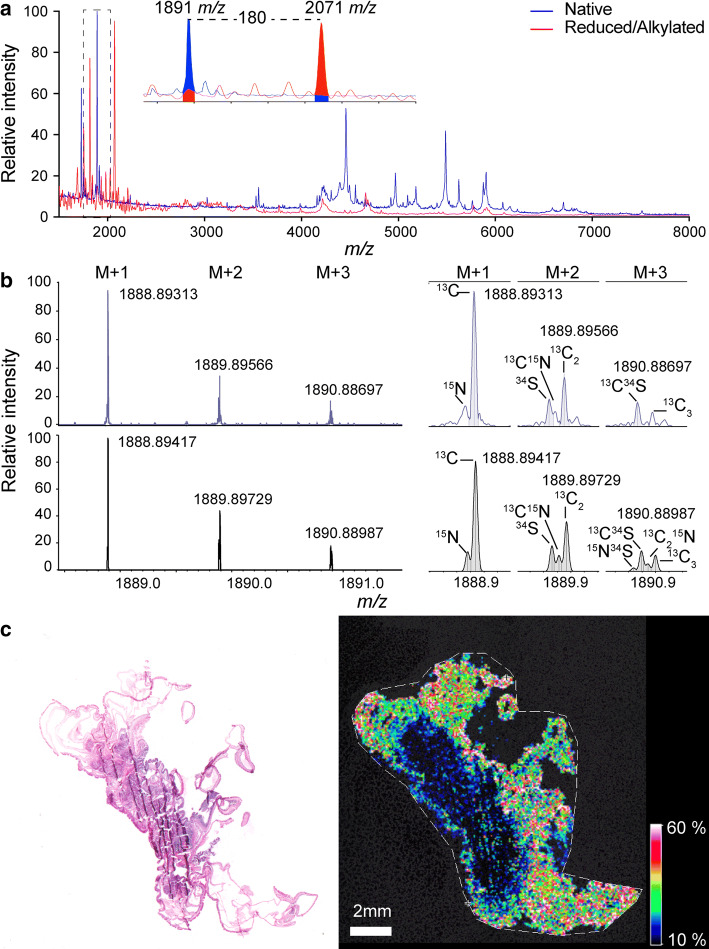
Although the pharmacological activity of a toxin can provide clues to its ecological function, it is not by itself definitive. However, the near-universal distribution of nematocytes in the epithelium of sea anemones means that toxin function can be inferred from tissue distribution [46–48]. We therefore investigated the tissue distribution of Ate1a using MSI, which allows visualization of the spatial distribution of unlabelled low-mass biomolecules (1–20 kDa) [15, 16, 49]. A peak corresponding to the average mass of Ate1a was clearly observed in MALDI-TOF-MSI spectra acquired from cross-sectioned A. tenebrosa. The identity of this peak was further supported by on-tissue gas-phase reduction and alkylation, which resulted in a peak shift matching the alkylation of four cystines (Fig. 6a). Finally, ultra-high mass resolution analysis using MALDI-FT-ICR-MSI allowed us to fit the predicted isotope structure of Ate1a to the observed spectra and confirm its identity (Fig. 6b). MSI revealed that Ate1a is non-uniformly distributed within the body of A. tenebrosa, with almost exclusive localization in tentacles (Fig. 6c), suggesting that it is involved in prey capture. Ate1a mass signals were weak or absent in actinopharynx, mesenterial filaments, and gastrovascular cavity, indicating that Ate1a does not play a role in prey digestion. Moreover, A. tenebrosa normally retracts its tentacles in response to disturbances, and thus the weak Ate1a signal in the trunk region indicates it is not primarily involved in defence.
Fig. 6.
Tissue distribution of Ate1a determined using MSI. a MSI linear positive mode spectra acquired from a cross-sectioned animal, with peaks corresponding to Ate1a filled in. The spectrum of native tissue is blue while the spectrum obtained after on-tissue gas-phase reduction and alkylation is shown in red. Inset shows a mass difference of 180 Da, corresponding to ethanolylation of four cysteine residues. b Ultra-high mass resolution analysis of the peak corresponding to Ate1a acquired using MALDI-FT-ICR-MSI at a resolution of 16,000,000 and resolving power at 1890 m/z of > 500,000, showing observed (top) and calculated (bottom) spectra. c Left: histological image of the sea anemone section used for MSI experiments, stained with hematoxylin and eosin. Right: distribution of the peak corresponding to the average mass of Ate1a as observed by MALDI-TOF MSI
In addition to nematocytes, sea anemones also produce toxins in ectodermal gland cells [35]. Unlike nematocytes, which are stinging cells that inject venom, gland-cell toxins are released into the water and absorbed by prey. To determine which cell type produces Ate1a, we conducted toxicity bioassays using brine shrimp and amphipods, the latter being a major prey item of Actinia spp. [50, 51]. Injection of Ate1a into amphipods resulted in impaired swimming followed by contractile paralysis (Supplementary material S1). In contrast, Ate1a did not affect either species when dissolved into the medium (artificial sea water) (Supplemental videos S1 and S2). Taken together, our data suggest that Ate1a is a neurotoxin produced in nematocytes and used primarily for prey capture.
Discussion
Although sea anemone venoms are a rich source of bioactive peptides, recent omics studies have highlighted how little we still know about their composition, function, and evolution [10, 48]. Here we described the discovery and functional characterization of a new peptide class from venom of the sea anemone A. tenebrosa, one of the most commonly encountered sea anemones in intertidal zones around Australia and New Zealand [52]. Ate1a has a primary structure unlike any previously described peptide, and assumes a unique 3D fold that is reminiscent of β-hairpin AMPs [40].
In contrast to β-hairpin AMPs, the 3D structure of Ate1a is devoid of regular secondary structure. Instead, the asymmetry of the two sides of the β-hairpin-like structure of Ate1a prevents β-sheet formation, and distinguishes the 3D structure from previously described two-disulfide peptide folds (Fig. 4). The longer of the two ‘loops’ is also highly dynamic (Fig. 3a), a property facilitated by the presence of two prolines that are conserved in all identified Ate1a homologues. Proline-containing peptides have the ability to populate two discrete conformations, and this cis–trans conformational switch works like a hinge that can potentially serve as a precise regulator of biological function [53, 54]. While proline hinges play a diversity of roles in protein biology, one of these roles is reorienting surface loops to modulate protein binding surfaces and in turn ligand recognition [55]. Thus, we predict that the proline-hinged loop of Ate1a represents a region that is important for the function of this toxin family. Given the structural and likely functional importance of this structural feature, we named this new structural scaffold the “proline-hinged asymmetric β-hairpin-like” (PHAB) fold.
Reflecting the structural distinctiveness of the PHAB fold from β-hairpin-like peptides, Ate1a neither has antimicrobial, antifungal, or cytolytic activity, nor strong affinity for lipid membranes (Fig. S2). Instead, it is a potent inhibitor of Shaker-type KV channels, with nanomolar potency on KV1.1, KV1.2 and KV1.6. KV channels play crucial roles in neuronal signalling, muscle contraction, and secretion [56], and hence they are a common target of animal toxins. Many venomous taxa have convergently evolved toxins that target KV channels to induce paralysis, general hyperexcitability, cardiac disorders, convulsions and death [1]. This is also the case in sea anemones, where KV toxins are represented by five unique peptide folds: ShK (type 1), Kunitz-domain (type 2), β-defensin-like (type 3), boundless β-hairpin (type 4), and an unknown fold predicted to form an inhibitor cystine knot (type 5) [12]. The PHAB fold is unlike any of these structural scaffolds, and therefore, it represents a new, sixth type of sea anemone KV toxin.
Although Ate1a is a novel KV toxin, correlating toxin activity with ecological function is often not straightforward [57]. However, like other cnidarians, sea anemones lack a centralised venom delivery system, and instead rely on localised production of toxins to complement their functional anatomy [46–48]. In A. tenebrosa, toxins are produced in five tissues and regions that have distinct ecological functions: acrorhagi (aggressive intraspecific encounters), tentacles (prey capture and immobilisation), mesenteric filaments (used principally in digestion), column (external defence after retracting tentacles), and actinopharynx (prey immobilisation and digestion). In addition, sea anemones produce toxins in two distinct cell types that deliver venom by either injection (nematocytes) or absorption following secretion into the water column (gland cells) [35]. Ate1a is found predominantly in the tentacles of A. tenebrosa (Fig. 6), which is suggestive of a predatory function. Moreover, Ate1a impaired swimming and led to paralysis and death when injected in amphipods, a major prey of Actinia species, but had no effect when dissolved into the medium. We conclude that Ate1a is a predatory toxin that cannot reach its KV targets without being inoculated into prey by nematocysts.
Venom proteins are thought to evolve via toxin recruitment events, whereby a gene encoding a normal body protein is duplicated and expressed in the venom-producing tissue [4]. Functionally important toxin types are reinforced through duplication and diversification, and this is considered a hallmark of toxin evolution in predatory venoms, where toxins evolve continuously to counter acquisition of prey resistance [4]. Although recent research suggests that venoms evolve via a two-step process in which initial rapid toxin diversification is followed by periods of purifying selection due to the metabolic costs of diversifying selection [58, 59], predatory toxins nevertheless tend to be part of large, highly diverse gene families. Strikingly, however, this diversity is entirely absent in the PHAB gene family, despite their likely role in predation. Instead, its members are highly conserved and consist of just 2–3 almost identical copies in each species (Fig. 2b).
The sequence conservation at the nucleotide level is not limited to between-gene copies of each species, but extends to the domains encoded by each transcript. Despite the emergence and domain duplication of the PHAB fold in an actinioidean ancestor, all four domain type (signal peptide, two propeptide domains, and PHAB domains seen in Fig. 2a) are remarkably well conserved. Furthermore, the nucleotide sequences encoding each domain type are more similar to the respective domains contained on the same transcript than to corresponding domain copies in other species (Fig. 2c). This form of domain conservation is likely to have occurred by concerted evolution, an evolutionary process driven by continuous recombination that results in homogenisation of genetic variance across gene copies and so-called ‘horizontal evolution’ [60]. Although concerted evolution has been described for a number of gene families, including NaV type I toxins from Nematostella vectensis and Actinia equina [38], it is considered rare for intra-gene protein domain repeats [61], and has never previously been reported within toxin gene domains.
In contrast with the general view of gene duplication as a facilitator of toxin gene diversification, recent studies have suggested that gene duplication may be of immediate importance for increased expression levels rather than generation of sequence diversity [62]. Similarly, the concerted evolution of NaV Type I toxins from N. vectensis and A. equina has been suggested to confer a selective advantage through a ‘dosage’ effect of gene expression [38]. In the PHAB gene family, this lack of high gene-copy numbers is compensated for by encoding multiple, identical toxin precursors, thereby effectively multiplying toxin expression levels. Concerted evolution may also facilitate ‘transmission’ of advantageous mutations from a single toxin gene locus to other loci, or preventing the loss of highly effective toxins. Reflecting this, concerted evolution of protein domain repeats has been proposed to be triggered by arms race-type co-evolution [61].
Interestingly, similarly conserved domain repeats were also identified by Honma et al. [63] for the BBH-like AmeI (GenBank accession AB180685) from the venom of the sea anemone Antheopsis maculata, which is encoded on the same transcript as six repeats that share near-identical nucleotide sequences. Although further work is required to determine whether these domains evolve by concerted evolution, their identical nature suggests that intra-gene concerted evolution may in anemones not be restricted to PHAB toxins. Thus, concerted evolution of toxin-domain repeats may provide a hitherto unrecognised mechanism of circumventing the effects of the metabolically expensive evolutionary arms race typically considered to drive toxin gene evolution. In the case of Ate1a and other members of the PHAB family, this has led to efficient secretion through high domain copy numbers of structurally unusual, but ecologically important, predatory KV toxins.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Brazilian Government (Science Without Borders PhD scholarship to BM), Australian Research Council (DECRA Fellowship DE160101142 to EABU, Future Fellowship FT150100398 to STH, ARC Linkage Grant LP140100832 to BRH and GFK), and National Health & Medical Research Council (Principal Research Fellowship APP1044414 to GFK). We thank Jason Cockington and Gillian Lawrence for maintenance of sea anemones, and Dr. Alun Jones for help with mass spectrometry experiments. Antimicrobial screens were performed by CO-ADD, funded by the Wellcome Trust (UK) and The University of Queensland (Australia). We thank Dr. Lachlan Rash for assistance with ASIC clone acquisition; Prof. John Wood for ASIC1a, ASIC2a, and ASIC3 clones; Prof. Stefan Gründer for the ASIC1b clone; Prof. Alan Goldin for the NaV1.6 clone; and Prof. Frank Bosmans for the NaV1.7 clone.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Contributor Information
Glenn F. King, Email: glenn.king@imb.uq.edu.au
Eivind A. B. Undheim, Email: e.undheim@uq.edu.au
References
- 1.Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JD, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, Renjifo C, de la Vega RC. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genom Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 2.Jenner R, Undheim E. Venom: the secrets of nature’s deadliest weapon. London: Natural History Museum; 2017. [Google Scholar]
- 3.King GF. Venoms to drugs: venom as a source for the development of human therapeutics. London: Royal Society of Chemistry; 2015. [Google Scholar]
- 4.Casewell NR, Wuster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 5.Van Iten H, Marques AC, Leme JDM, Pacheco MLAF, Simões MG. Origin and early diversification of the phylum Cnidaria Verrill: major developments in the analysis of the taxon’s Proterozoic-Cambrian history. Palaeontology. 2014;57:677–690. doi: 10.1111/pala.12116. [DOI] [Google Scholar]
- 6.Frazao B, Vasconcelos V, Antunes A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview. Mar Drugs. 2012;10:1812–1851. doi: 10.3390/md10081812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jouiaei M, Yanagihara AA, Madio B, Nevalainen TJ, Alewood PF, Fry BG. Ancient venom systems: a review on cnidaria toxins. Toxins. 2015;7:2251–2271. doi: 10.3390/toxins7062251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikov AN, Kozlov SA. Structural features of cysteine-rich polypeptides from sea anemone venoms. Russian J Bioorg Chem. 2015;41:455–466. doi: 10.1134/S1068162015050088. [DOI] [PubMed] [Google Scholar]
- 9.Logashina YA, Solstad RG, Mineev KS, Korolkova YV, Mosharova IV, Dyachenko IA, Palikov VA, Palikova YA, Murashev AN, Arseniev AS, Kozlov SA, Stensvag K, Haug T, Andreev YA. New disulfide-stabilized fold provides sea anemone peptide to exhibit both antimicrobial and TRPA1 potentiating properties. Toxins. 2017;9:154. doi: 10.3390/toxins9050154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madio B, Undheim EAB, King GF. Revisiting venom of the sea anemone Stichodactyla haddoni: omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J Proteomics. 2017;166:83–92. doi: 10.1016/j.jprot.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Tarcha EJ, Olsen CM, Probst P, Peckham D, Munoz-Elias EJ, Kruger JG, Iadonato SP. Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: a randomized phase 1b trial. PLoS One. 2017;12:e0180762. doi: 10.1371/journal.pone.0180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orts DJB, Moran Y, Cologna CT, Peigneur S, Madio B, Praher D, Quinton L, De Pauw E, Bicudo JEPW, Tytgat J, de Freitas JC. BcsTx3 is a founder of a novel sea anemone toxin family of potassium channel blocker. FEBS J. 2013;280:4839–4852. doi: 10.1111/febs.12456. [DOI] [PubMed] [Google Scholar]
- 13.Malpezzi EL, de Freitas JC, Muramoto K, Kamiya H. Characterization of peptides in sea anemone venom collected by a novel procedure. Toxicon. 1993;31:853–864. doi: 10.1016/0041-0101(93)90220-D. [DOI] [PubMed] [Google Scholar]
- 14.Fukuyama Y, Iwamoto S, Tanaka K. Rapid sequencing and disulfide mapping of peptides containing disulfide bonds by using 1,5-diaminonaphthalene as a reductive matrix. J Mass Spectrom. 2006;41:191–201. doi: 10.1002/jms.977. [DOI] [PubMed] [Google Scholar]
- 15.Undheim EAB, Sunagar K, Hamilton BR, Jones A, Venter DJ, Fry BG, King GF. Multifunctional warheads: diversification of the toxin arsenal of centipedes via novel multidomain transcripts. J Proteomics. 2014;102:1–10. doi: 10.1016/j.jprot.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell ML, Hamilton BR, Madio B, Morales RAV, Tonkin-Hill GQ, Papenfuss AT, Purcell AW, King GF, Undheim EAB, Norton RS. The use of imaging mass spectrometry to study peptide toxin distribution in Australian sea anemones. Austr J Chem. 2017;70:1235–1237. doi: 10.1071/CH17228. [DOI] [Google Scholar]
- 17.Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem. 2004;333:174–181. doi: 10.1016/j.ab.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Cock PJ, Gruning BA, Paszkiewicz K, Pritchard L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ. 2013;1:e167. doi: 10.7717/peerj.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST + : architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardoso FC, Dekan Z, Smith JJ, Deuis JR, Vetter I, Herzig V, Alewood PF, King GF, Lewis RJ. Modulatory features of the novel spider toxin mu-TRTX-Df1a isolated from the venom of the spider Davus fasciatus. Br J Pharmacol. 2017;174:2528–2544. doi: 10.1111/bph.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 28.Tytgat J, Debont T, Carmeliet E, Daenens P. The alpha-dendrotoxin footprint on a mammalian potassium channel. J Biol Chem. 1995;270:24776–24781. doi: 10.1074/jbc.270.42.24776. [DOI] [PubMed] [Google Scholar]
- 29.Edwards IA, Elliott AG, Kavanagh AM, Zuegg J, Blaskovich MA, Cooper MA. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect Dis. 2016;2:442–450. doi: 10.1021/acsinfecdis.6b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YH, Colgrave ML, Clark RJ, Kotze AC, Craik DJ. Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J Biol Chem. 2010;285:10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torcato IM, Huang YH, Franquelim HG, Gaspar D, Craik DJ, Castanho MA, Troeira Henriques S. Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim Biophys Acta. 2013;1828:944–955. doi: 10.1016/j.bbamem.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Henriques ST, Huang YH, Castanho MA, Bagatolli LA, Sonza S, Tachedjian G, Daly NL, Craik DJ. Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J Biol Chem. 2012;287:33629–33643. doi: 10.1074/jbc.M112.372011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henriques ST, Huang YH, Rosengren KJ, Franquelim HG, Carvalho FA, Johnson A, Sonza S, Tachedjian G, Castanho MA, Daly NL, Craik DJ. Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J Biol Chem. 2011;286:24231–24241. doi: 10.1074/jbc.M111.253393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henriques ST, Pattenden LK, Aguilar MI, Castanho MA. PrP(106–126) does not interact with membranes under physiological conditions. Biophys J. 2008;95:1877–1889. doi: 10.1529/biophysj.108.131458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran Y, Genikhovich G, Gordon D, Wienkoop S, Zenkert C, Ozbek S, Technau U, Gurevitz M. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc Biol Sci. 2012;279:1351–1358. doi: 10.1098/rspb.2011.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira JS, Zaharenko AJ, Ferreira WA, Jr, Konno K, Shida CS, Richardson M, Lucio AD, Beirao PS, de Freitas JC. BcIV, a new paralyzing peptide obtained from the venom of the sea anemone Bunodosoma caissarum. A comparison with the Na + channel toxin BcIII. Biochim Biophys Acta. 2006;1764:1592–1600. doi: 10.1016/j.bbapap.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez E, Barbeitos MS, Brugler MR, Crowley LM, Grajales A, Gusmao L, Haussermann V, Reft A, Daly M. Hidden among sea anemones: the first comprehensive phylogenetic reconstruction of the order Actiniaria (Cnidaria, Anthozoa, Hexacorallia) reveals a novel group of hexacorals. PLoS One. 2014;9:e96998. doi: 10.1371/journal.pone.0096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran Y, Weinberger H, Sullivan JC, Reitzel AM, Finnerty JR, Gurevitz M. Concerted evolution of sea anemone neurotoxin genes is revealed through analysis of the Nematostella vectensis genome. Mol Biol Evol. 2008;25:737–747. doi: 10.1093/molbev/msn021. [DOI] [PubMed] [Google Scholar]
- 39.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panteleev PV, Balandin SV, Ivanov VT, Ovchinnikova TV. A therapeutic potential of animal beta-hairpin antimicrobial peptides. Curr Med Chem. 2017;24:1724–1746. doi: 10.2174/0929867324666170424124416. [DOI] [PubMed] [Google Scholar]
- 41.Chagot B, Pimentel C, Dai L, Pil J, Tytgat J, Nakajima T, Corzo G, Darbon H, Ferrat G. An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis. Biochem J. 2005;388:263–271. doi: 10.1042/BJ20041705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osmakov DI, Kozlov SA, Andreev YA, Koshelev SG, Sanamyan NP, Sanamyan KE, Dyachenko IA, Bondarenko DA, Murashev AN, Mineev KS, Arseniev AS, Grishin EV. Sea anemone peptide with uncommon β-hairpin structure inhibits acid-sensing ion channel 3 (ASIC3) and reveals analgesic activity. J Biol Chem. 2013;288:23116–23127. doi: 10.1074/jbc.M113.485516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JJ, Hill JM, Little MJ, Nicholson GM, King GF, Alewood PF. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc Natl Acad Sci USA. 2011;108:10478–10483. doi: 10.1073/pnas.1103501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang S, Yang F, Wei N, Hong J, Li B, Luo L, Rong M, Yarov-Yarovoy V, Zheng J, Wang K, Lai R. A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat Commun. 2015;6:8297. doi: 10.1038/ncomms9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henriques ST, Lawrence N, Chaousis S, Ravipati AS, Cheneval O, Benfield AH, Elliott AG, Kavanagh AM, Cooper MA, Chan LY, Huang YH, Craik DJ. Redesigned spider peptide with improved antimicrobial and anticancer properties. ACS Chem Biol. 2017;12:2324–2334. doi: 10.1021/acschembio.7b00459. [DOI] [PubMed] [Google Scholar]
- 46.Basulto A, Perez VM, Noa Y, Varela C, Otero AJ, Pico MC. Immunohistochemical targeting of sea anemone cytolysins on tentacles, mesenteric filaments and isolated nematocysts of Stichodactyla helianthus. J Exp Zool Part A Comp Exp Biol. 2006;305:253–258. doi: 10.1002/jez.a.256. [DOI] [PubMed] [Google Scholar]
- 47.Beckmann A, Ozbek S. The nematocyst: a molecular map of the cnidarian stinging organelle. Int J Dev Biol. 2012;56:577–582. doi: 10.1387/ijdb.113472ab. [DOI] [PubMed] [Google Scholar]
- 48.Macrander J, Brugler MR, Daly M. A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genom. 2015;16:221. doi: 10.1186/s12864-015-1417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Undheim EAB, Hamilton BR, Kurniawan ND, Bowlay G, Cribb BW, Merritt DJ, Fry BG, King GF, Venter DJ. Production and packaging of a biological arsenal: evolution of centipede venoms under morphological constraint. Proc Natl Acad Sci USA. 2015;112:4026–4031. doi: 10.1073/pnas.1424068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chintiroglou C, Koukouras A. The feeding habits of three Mediterranean sea anemone species, Anemonia viridis (Forskål), Actinia equina (Linnaeus) and Cereus pedunculatus (Pennant) Helgol Mar Res. 1992;46:53–68. [Google Scholar]
- 51.Kruger LM, Griffiths CL. Sources of nutrition in intertidal sea anemones from the south-western Cape, South Africa. S Afr J Zool. 1996;31:110–119. doi: 10.1080/02541858.1996.11448402. [DOI] [Google Scholar]
- 52.Ottaway JR. Population ecology of the intertidal anemone Actinia tenebrosa: pedal locomotion and intraspecific aggression. Austr J Mar Freshw Res. 1978;29:787–802. doi: 10.1071/MF9780787. [DOI] [Google Scholar]
- 53.Robson SA, King GF. Domain architecture and structure of the bacterial cell division protein DivIB. Proc Natl Acad Sci USA. 2006;103:6700–6705. doi: 10.1073/pnas.0601397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 55.Andreotti AH. Opening the pore hinges on proline. Nat Chem Biol. 2006;2:13–14. doi: 10.1038/nchembio0106-13. [DOI] [PubMed] [Google Scholar]
- 56.Hille B. Ion channels of excitable membranes. 3. Sunderland: Sinauer; 2001. [Google Scholar]
- 57.Bende NS, Dziemborowicz S, Mobli M, Herzig V, Gilchrist J, Wagner J, Nicholson GM, King GF, Bosmans F. A distinct sodium channel voltage-sensor locus determines insect selectivity of the spider toxin Dc1a. Nat Commun. 2014;5:4350. doi: 10.1038/ncomms5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sunagar K, Moran Y. The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015;11:e1005596. doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Undheim EAB, Mobli M, King GF. Toxin structures as evolutionary tools: using conserved 3D folds to study the evolution of rapidly evolving peptides. BioEssays. 2016;38:539–548. doi: 10.1002/bies.201500165. [DOI] [PubMed] [Google Scholar]
- 60.Brown DD, Wensink PC, Jordan E. A comparison of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972;63:57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- 61.Schüler A, Bornberg-Bauer E. Evolution of protein domain repeats in metazoa. Mol Biol Evol. 2016;33:3170–3182. doi: 10.1093/molbev/msw194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Margres MJ, Bigelow AT, Lemmon EM, Lemmon AR, Rokyta DR. Selection to increase expression, not sequence diversity, precedes gene family origin and expansion in rattlesnake venom. Genetics. 2017;206:1569–1580. doi: 10.1534/genetics.117.202655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honma T, Hasegawa Y, Ishida M, Nagai H, Nagashima Y, Shiomi K. Isolation and molecular cloning of novel peptide toxins from the sea anemone Antheopsis maculata. Toxicon. 2005;45:33–41. doi: 10.1016/j.toxicon.2004.09.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.