Abstract
True Bugs (Insecta: Heteroptera) produce venom or saliva with diverse bioactivities depending on their feeding strategies. However, little is known about the molecular evolution of the venom toxins underlying these biological activities. We examined venom of the giant fish-killing water bug Lethocerus distinctifemur (Insecta: Belostomatidae) using infrared spectroscopy, transcriptomics, and proteomics. We report 132 venom proteins including putative enzymes, cytolytic toxins, and antimicrobial peptides. Over 73% (96 proteins) showed homology to venom proteins from assassin bugs (Reduviidae), including 21% (28 proteins from seven families) not known from other sources. These data suggest that numerous protein families were recruited into venom and diversified rapidly following the switch from phytophagy to predation by ancestral heteropterans, and then were retained over > 200 my of evolution. In contrast, trophic switches to blood-feeding (e.g. in Triatominae and Cimicidae) or reversions to plant-feeding (e.g., in Pentatomomorpha) were accompanied by rapid changes in the composition of venom/saliva, including the loss of many protein families.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2768-1) contains supplementary material, which is available to authorized users.
Keywords: Venom, Saliva, Heteroptera, Belostomatidae, Nepomorpha, Venom evolution, Trophic shift
Introduction
The composition of an animal’s venom evolves dynamically within its larger ecological context. This process is well documented at the sequence level of toxin-encoding genes, which frequently exhibit accelerated rates of duplication and hyperdivergence, due to both increased positive selection and reduced negative selection [1–4]. Diversifying selection may be particularly prevalent following niche shifts such as changes in prey specialisation or hunting behaviour [5–7]. Complete loss or gain of venom use (i.e., changes in whether a secretion is used for envenomation or not) unsurprisingly entails marked consequences for the evolution of its composition over time. For example, diversifying selection is most rapid in taxa that only recently evolved venom use [8], whereas loss of venom use results in accumulation of frame-shift and other mutations in toxin-encoding genes, as observed in the egg-eating marbled sea snake Aipysurus eydouxii [9, 10]. Beyond the level of sequence mutation, dramatic changes in venom activity and composition may reflect rapid changes in expression levels of toxin-encoding genes, i.e., in the recruitment of new proteins into venom or their loss or downregulation. For example, differences in prey type drive the presence or absence of particular venom proteins in pit viper venom [11]; tetragnathid spiders that use the ‘wandering’ hunting strategy rather than building webs show reduced levels of peptide toxins [12]; and in parasitoid wasps, the rapid changes in venom composition that accompany changes in host specificity reflect rapid mutation of the cis-regulatory elements that control toxin expression levels [13].
Heteropteran insects are an ideal group in which to explore how large changes in trophic strategy—for example between phytophagy, entomophagy, and haematophagy, and including complete loss or gain of venom use—co-evolve with the composition of a venom/saliva secretion. Heteroptera diverged from phytophagous hemipteran taxa such as cicadas (Auchenorrhyncha), aphids (Sternorrhyncha), and moss bugs (Coleorrhyncha) around 293–262 mya (95% confidence interval) [14]. Since the vast majority of ‘lower Heteroptera’ [15] (Nepomorpha, Enicocephalomorpha, Leptopodomorpha, Gerromorpha, and Dipsocoromorpha) are predatory and venomous, a trophic switch to predation likely occurred closely following or accompanying the divergence of Heteroptera from other hemipterans [15–19]. Thus, the salivary systems of plant-feeding bugs were adapted to form venom systems, and their oral secretions acquired new bioactivities such as the ability to paralyse prey [20–23]. The predatory heteropterans diversified and some groups subsequently transitioned to different trophic strategies. The Pentatomomorpha (stink bugs and allies) and some Cimicomorpha such as Miridae (plant bugs) returned to phytophagy; whereas the kissing bugs (Triatominae) and bed and bat bugs (Cimicidae and Polyctenidae) independently became ectoparasitic blood-feeders [15]. These trophic changes were accompanied by alterations in the bioactivity of the venom/saliva secretion [17]: instead of immobilising and liquefying prey, the venom of blood-feeders adapted to counteract blood clotting and vasoconstriction [24], while the saliva of plant-feeding species is thought to subvert host-plant defences [25]. Although blood- and plant-feeders constitute a minority of heteropteran families, their venom/saliva secretion is the best characterised due to their economic and medical importance [24, 26]. We recently reported the venom proteome of the predaceous assassin bug, Pristhesancus plagipennis (Cimicomorpha: Reduviidae). The venom of this species contains a wide range of peptides and proteins, packaged into two separate complex venoms in different gland lumens [23, 27]. Venom produced by the posterior main gland (PMG) quickly paralyses prey, and is rich in proteases, putative cytolytic toxins, and peptides. In contrast, the smaller anterior main gland (AMG) secretes a venom rich in ‘haemolysin-like’ proteins and uncharacterised proteins; this venom has little effect on insect prey and appears to be used defensively [23].
Characterisation of venom from giant water bugs (Nepomorpha: Belostomatidae) has the potential to inform multiple aspects of venom evolution in Heteroptera. Belostomatids diverged early in the radiation of Heteroptera from the major terrestrial infraorders (Pentatomomorpha + Cimicomorpha = Terheteroptera) [15, 28], probably between 254 and 226 mya [14], and recognisable Belostomatidae were abundant fauna of Triassic lakes > 200 mya [29]. Belostomatids have several distinguishing features compared to most other venomous Heteroptera. They are adapted to an aquatic lifestyle, and their raptorial forearms used in prey capture are particularly well developed [30]. Some belostomatids, such as Lethocerus sp., grow exceedingly large (up to 12 cm long), allowing them to prey on vertebrates including fish, amphibians, turtles, and small birds [31, 32]. Therefore, examination of nepomorphan venoms has the potential to provide unique insights about venom evolution in Heteroptera and might additionally uncover novel toxins adapted to modulate vertebrate targets.
The previous investigations into the composition of belostomatid venoms focussed on lysophospholipids, reported to be the major component of Belostoma anurum venom [22]. Lysophospholipids, which can be formed through enzymatic digestion of phospholipids by phospholipase A2 (PLA2), may exert toxic effects via modification of nerve terminals that disrupt neurotransmission [33]. Both purified lysophosphatidylcholine and B. anurum venom block twitch contractions in a mammalian nerve-muscle preparation and cause paralysis when injected into zebrafish [22]. However, while PLA2 enzymes are commonly found in predatory venoms from other animal taxa [34, 35], the inclusion of lysophospholipids themselves as the principle toxins in venom has not been reported for any other predatory venomous animal. In any case, studies combining transcriptomics and proteomics to study nepomorphan venoms are lacking. This fact, combined with the wide variety of enzymatic activities reported in belostomatid venom (e.g., PLA2, hyaluronidase, protease, amylase, esterase, glucosidase, glucosaminidase, invertase, lipase, nuclease, and phosphatase) [22, 36–38], suggests that peptides and proteins may be important but undescribed components of belostomatid venom.
In this study, we use vibrational spectroscopy, transcriptomics, and mass spectrometry to characterise venom of the Australian giant fish-killing water bug Lethocerus distinctifemur. We found a little evidence for lysophospholipids in the venom of this species but instead report evidence of rich protein content. Many detected giant water bug venom proteins show sequence homology to the venom proteins of distantly related reduviid heteropterans, which are separated by > 200 million years of evolution but share a predaceous trophic strategy descended from a common ancestor. By comparing data from diverse hemipteran insects with a range of trophic strategies, we find evidence for rapid alterations in the composition of the heteropteran venom/saliva secretion over evolutionary time. Mapping these data against established phylogeny suggests rapid evolution: (a) accompanying the transition to predatory envenomation by ancestral heteropterans and (b) accompanying the subsequent independent transitions to haematophagy or phytophagy by multiple heteropteran subgroups.
Results
Rich protein content of belostomatid venom
We examined venoms from representative species of each of the two major subfamilies of Belostomatidae, the giant fish-killing water bug L. distinctifemur (Lethocerinae; Fig. 1) and the smaller water bug Diplonychus eques (Belostomatinae). Venom was extracted from individuals of each species by electrostimulation (see “Materials and methods”). In the previous studies, belostomatid venom collected by electrostimulation was described as ‘white and viscous’ or ‘milky’, and this was linked to the high lipid content of the venom [22]. We found that both L. distinctifemur and D. eques produced venom that was sometimes clear and sometimes milky; individual animals often produced both clear venom and milky venom during the course of a single venom extraction. As far as possible, clear venom samples were kept separate from samples containing milky material, resulting in two samples from each species for analysis.
Fig. 1.

Giant fish-killing water bug (Lethocerus sp.) envenomating a goldfish (Carassius auratus) held in its raptorial forearms
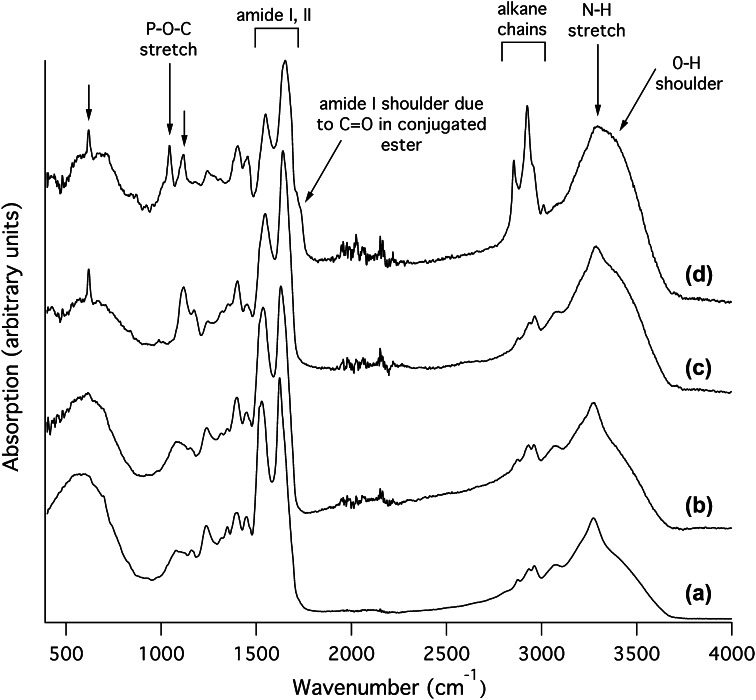
To investigate their protein and lipid contents, we examined each belostomatid venom sample using Fourier transform infrared spectroscopy (FTIR; Fig. 2). For both the ‘clear’ and ‘milky’ venom samples from L. distinctifemur, and the ‘clear’ sample from D. eques, the FTIR spectra obtained were similar to those of pure protein [39, 40], being dominated by the amide I–III regions (maxima 1622–1655, 1526–1552, and 1235–1245 cm−1 respectively) and N–H stretch region (maxima 3272–3286 cm−1). In contrast, the spectrum of the ‘milky’ D. eques sample showed features indicating a mixture of protein plus a lipid component similar to a lysophospholipid. Strong CH2 peaks at 2854 and 2923 cm−1 (symmetric and asymmetric stretches, respectively) with weaker CH3 absorptions near 2958 and 2872 cm−1 (symmetric and asymmetric stretches, respectively) indicate the presence of long alkane chains [41]. In addition, the D. eques milky venom spectrum (but not other spectra) displayed a broad shoulder from 3300 to 3400 cm−1 attributed to O–H stretch; an amide I shoulder from 1710–1740 cm−1 attributed to a conjugated C=O ester; and a sharp peak at 1043 cm−1 attributed to a P–O–C stretch [41]. These results are consistent with a strong content of lysophospholipids in the milky venom sample obtained from D. eques, but not other samples. Both samples from D. eques (but not those from L. distinctifemur) additionally contain peaks at 1118 and 618 cm−1 (indicated by arrows in Fig. 2) that may originate from a sulfur- or phosphorus-containing group [41]. These results support the notion that belostomatine water bugs such as D. eques—members of the same subfamily as Belostoma anurum [22] —can produce lysophospholipid-rich venom. However, D. eques can also produce primarily protein-based venom under some circumstances. For L. distinctifemur, our results suggest that the venom consists predominantly of proteins.
Fig. 2.
Infrared absorption spectra of belostomatid venoms. Both the milky (a) and clear (b) samples of Lethocerus distinctifemur venom as well as the clear sample of Diplonychus eques venom (c) are dominated by absorption bands typical of protein, including the amide I and II regions and N–H stretch. The milky sample of D. eques venom (d) contained additional bands consistent with the presence of a lysophospholipid or similar compound with long alkane chains. Each spectrum is normalised and offset
Venom proteome of the giant fish-killing water bug
We combined RNA-Seq analysis of venom glands with venom proteomics to gain a holistic overview of the venom system of L. distinctifemur. For RNA-Seq experiments, we dissected out venom glands from one adult of each gender. These glands are anatomically complex [15] and they were separated into four different compartments: the posterior main gland (PMG) lobe, anterior main gland (AMG) lobe, ‘storage sacks’ (SS) [42, 43], and accessory glands (AG). Isolation of mRNA yielded 43.2, 58.9, 6.0, and 0.08 ng mRNA per mg tissue, respectively. These results suggest that the PMG and AMG are active secretory tissues, whereas the storage sacks and accessory glands are not. Yields of mRNA from the PMG and AMG were sufficient to prepare a TruSeq cDNA library. Sequencing of this library using next-generation sequencing on an Illumina HiSeq platform yielded 17,083,410 and 22,958,470 paired-end reads for the PMG and AMG, respectively. All reads were assembled together (see “Materials and methods”) to yield 78,238 contigs, from which all open-reading frames (ORFs) > 90 bp were extracted and translated to produce a library of possible protein-coding sequences.
Mass spectral signatures of venom proteins were obtained by LC–MS/MS analysis of trypsin-digested crude venom and subsets of venom components separated by 1D SDS-PAGE (Fig. 3, left two lanes) or acid precipitation (see “Materials and methods”). A search of all venom-derived mass spectra against translated ORFs from RNA-Seq experiments using the Paragon and ProtGroup algorithms in ProteinPilot resulted in identification of 102 protein and peptide sequences (Supplementary Dataset S1; Fig. 4). To these 102 sequences identified confidently from MS analysis of venom, we added 30 putative venom proteins and peptides that matched the following criteria: proteins confidently detected by MS of venom-gland extracts that additionally possess predicted signal peptides and stop codons (19 sequences); proteins detected by MS of venom samples, with confidence at the peptide level of > 95%, but only 10–95% at the protein level (9 sequences); and peptides encoded by transcripts that possess signal peptides and are among the 20 most highly expressed transcripts in venom glands (either PMG or AMG; see “Venom proteins are expressed and accumulated in a compartment-specific manner”) but which were not detected by MS (2 sequences). This resulted in a venom proteome of 132 proteins and peptides.
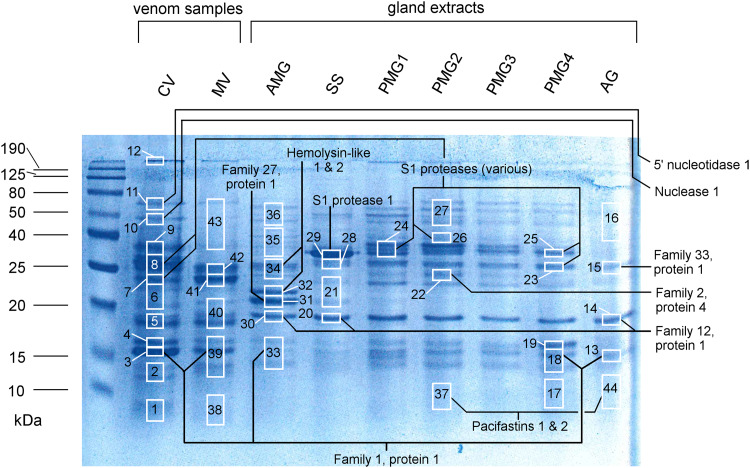
Fig. 3.
SDS-PAGE and LC–MS/MS sampling of venom and venom gland extracts. CV clear venom, MV milky venom, AMG anterior main gland, SS storage sacks, PMG1–4 extracts from the anterior (PMG1) to posterior quarter (PMG4) of the posterior main gland, AG accessory gland. Left, molecular-weight markers. White rectangles show portions of the gel used for LC–MS/MS protein identification experiments (number indicates sample number). Annotations show protein identifications with precursor counts > 30% of total non-contaminant precursor counts
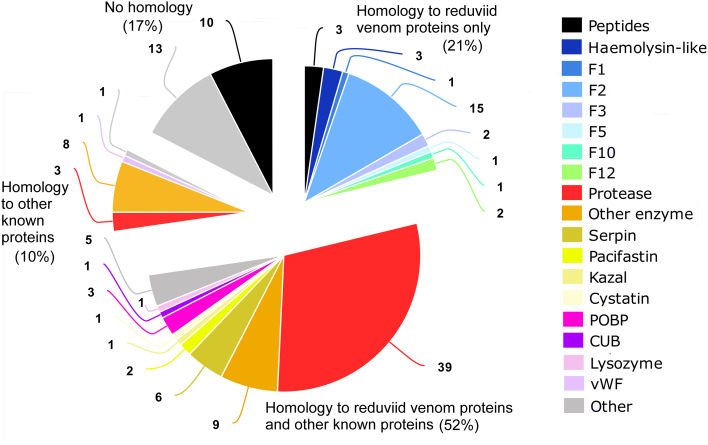
Fig. 4.
Protein families detected in the venom of Lethocerus distinctifemur. F1–F12 heteropteran venom families 1–12, vWF von Willebrand factor
We annotated detected sequences using the HMMER algorithm against the Pfam protein family database and BLASTp searches against (a) UniProt’s UniRef90 database, (b) venom proteins detected in venoms from L. distinctifemur in this study, and (c) venom proteins detected in assassin bug venoms in the previous studies [23, 27]. The resulting annotations (Supplementary Dataset S1) show that 96 (73%) of detected L. distinctifemur venom proteins have homology to previously described assassin bug venom proteins (89 or 67% of which have a BLASTp hit, E < 0.01, to assassin bug venom proteins). Furthermore, 28 of these (21%) showed no detectable homology except to proteins reported in assassin bug venom. A further 23 sequences are novel (i.e., no detectable homology with any known sequences). Major protein classes evident in L. distinctifemur venom are proteases (42 sequences), other enzymes (17), members of the cystatin, pacifastin, serpin, and Kazal protease inhibitor families (10), odorant-binding proteins (OBPs; 3), and haemolysin-like proteins (3). In addition, we identified 15 venom peptides (those with a predicted mature sequence < 100 residues). Of these venom peptides, 12 are devoid of cysteine residues and three are annotated by HMMER as possessing an antimicrobial domain. One peptide (Ld14a, predicted mass 9.6 kDa) is disulfide-rich, with 12 cysteine residues that likely form six disulfide bonds.
Venom proteins are expressed and accumulate in a compartment-specific manner
Several studies have reported that the individual gland compartments of heteropteran venom glands produce different secretions [23, 38, 44]. We investigated the secretory activity of the L. distinctifemur PMG and AMG by examining the most highly expressed contigs in each compartment (i.e., 59 contigs with fragments per kilobase million (FPKM) of 350–22,844 in PMG and 31 contigs with FPKM of 350–35,742 in AMG). These data were compared to the protein content of the glands (PMG, AMG, AG, and SS) as determined by LC–MS/MS analysis of gland extracts either in their liquid form or after separation by 1D SDS-PAGE (Fig. 3, lanes 3–9).
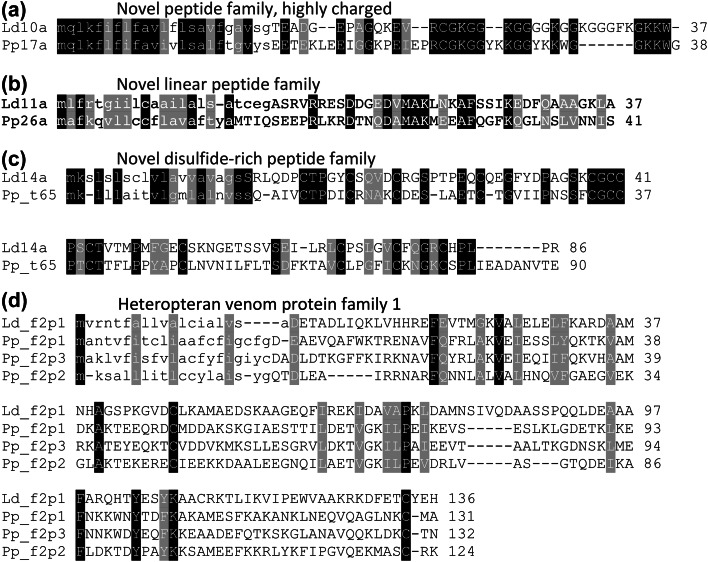
The two most highly expressed contigs in the PMG encode proteins homologous to venom protein family 1 from the assassin bug P. plagipennis [27]. Most of the highly expressed contigs in PMG encode either proteases (20) or proteins homologous to venom protein family 2 of P. plagipennis (11). In contrast, the most abundant transcript in the AMG encodes the peptide Ld10a (Fig. 5a), a putative orthologue of P. plagipennis venom peptide Pp17a. Other abundant transcripts in the AMG encode pacifastins and haemolysin-like proteins (Supplementary Dataset S2).
Fig. 5.
Alignments of homologous proteins occur in venoms of distantly related heteropterans. Ld10a, Lethocerus distinctifemur venom peptide 10a; Pp17a, Pristhesancus plagipennis (assassin bug) venom peptide 17a. Ld11a, L. distinctifemur venom peptide 11a; Pp26a, P. plagipennis venom peptide 26a; Ld14a, L. distinctifemur venom peptide 14a; Pp_t65, P. plagipennis peptide encoded by venom gland transcript 65; Ld_f2p1, L. distinctifemur venom protein family 1, protein 1; Pp_f2p, P. plagipennis venom protein family 2
These differing patterns of protein expression were confirmed by LC–MS/MS analysis of extracts of each gland compartment separated by SDS-PAGE (Fig. 3; Supplementary Table S1). The AMG extract yielded intense gel bands of approximately 20, 22, and 25 kDa that were not observed in other compartments. LC–MS/MS analysis revealed that the 22 and 25 kDa bands correspond to members of the haemolysin-like protein family, while the 20 kDa band represents a novel protein designated venom protein family 27. In contrast, PMG extracts yielded intense bands at 13–17 and 24–35 kDa. The most posterior segment analysed, PMG4, showed increased levels of OBP1 and reduced levels of proteases compared to other PMG extracts. However, all PMG samples had a similar protein profile, with abundant proteins belonging to heteropteran venom protein families 1 and 2 and S1 proteases, consistent with our transcriptomic analysis. Venom collected by electrostimulation more closely resembles the secretion of the PMG (Fig. 3). However, numerous proteins that appear to be expressed preferentially in the AMG were identified (Supplementary Dataset S1, final two columns).
Overall, the gland-specific expression patterns observed for the giant water bug L. distinctifemur are similar to those we reported previously for assassin bugs [23]: the PMG preferentially expresses venom family 1 and 2 proteins and S1 proteases, whereas the AMG preferentially expresses haemolysin-like proteins, pacifastins, and peptides such as Ld10a.
Ancient recruitment and radiation of heteropteran venom proteins
Protein families confidently identified in both reduviid and belostomatid venom, but unknown from other sources, include the haemolysin-like proteins [23, 45], heteropteran venom protein families 1, 2, 5, 10, and 12, and CUB domain family proteins (defined as proteins with CUB domains < 300 amino acids in length but without protease or other known domains). In addition, both venoms contain S1 proteases, serpins, inositol phosphate phosphatases, histidine phosphatases, OBPs, cystatins, transferrins, nucleases, lipases, and lysozymes. To quantitatively investigate how the composition of the venom/saliva secretion varies across Heteroptera, we compared the venom proteome of L. distinctifemur with sialomes from diverse species, focussing only on proteins detected by mass spectrometry, in order to avoid weakly expressed proteins. Comparison data sets included assassin bug venom [23, 27], kissing bug venoms [45–49], bed bug venom [50], stink bug saliva [26], plant bug saliva [51], and aphid saliva [52]. Water bug venom is most similar to assassin bug venom, with 89 (67%) of the 132 water bug venom proteins documented in this study having homologues in venom of the assassin bug P. plagipennis (BLASTp E < 0.01; Table 1). Venom and saliva from blood- or plant-feeding groups were less similar, containing homologues for < 35% of water bug venom proteins (E < 0.01). If S1 proteases are disregarded, 35% of water bug venom proteins have homologues in assassin bug venom, 13% in bed bug venom, and < 7% for all other groups (Table 1).
Table 1.
Detected homologues of L. distinctifemur venom proteins
| Species | Pristhesancus plagipennis | Cimex lectularius | Rhodnius prolixus | Panstrongylus herreri | Triatoma lectularia | Halyomorpha halys | Triatoma pallidipennis | Triatoma infestans | Lygus hesperus | Triatoma infestans | Rhodnius robustus and Rhodnius breviceps | Sitobion avenae and Metopolophium dirhodum |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylogeny | Heteroptera: Cimicomorpha: Reduviidae: Harpactorinae | Heteroptera: Cimicomorpha: Cimicidae: Cimicinae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Pentatomomorpha: Pentatomidae: Pentatominae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Cimicomorpha: Miridae: Mirinae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Heteroptera: Cimicomorpha: Reduviidae: Triatominae | Sternorrhyncha: Aphidoidea: Aphididae: Aphidinae |
| Trophic strategy | Predation (invertebrates) | Haematophagy | Haematophagy | Haematophagy | Haematophagy | Phytophagy | Haematophagy | Haematophagy | Phytophagy (facultative predation) | Haematophagy | Haematophagy | Phytophagy |
| Study | Walker et al. (2017, 2018) | Francischetti et al. (2010) | Montandon et al. (2016) | Montandon et al. (2016) | Montandon et al. (2016) | Peiffer et al. (2014) | Hernandez-Vargas et al. (2017) | Assumpcao et al. (2008) | Cooper et al. (2013) | Charneau et al. (2007) | Costa et al. (2011) | Rao et al. (2013) |
| Methods | Venom gland transcriptomics and MS/MS of venom | Venom gland transcriptomics and MS/MS of venom glands | MS/MS of venom (against previous transcriptomics) | MS/MS of venom (against previous transcriptomics) | MS/MS of venom (against previous transcriptomics) | MS/MS of saliva (against previous transcriptomics) | Venom gland transcriptomics and MS/MS of venom | Venom gland transcriptomics and MS/MS of venom | MS/MS of saliva (against previous transcriptomics) | MS/MS of venom (against previous transcriptomics) | Venom gland transcriptomics and MS/MS of venom | MS/MS of saliva (against previous transcriptomics of different species) |
| No. BLASTp hitsa | 82 | 46 | 39 | 36 | 5 | 37 | 7 | 6 | 6 | 3 | 1 | 1 |
| No. non-S1 protease BLASTp hitsa | 34 | 13 | 3 | 2 | 4 | 2 | 6 | 6 | 6 | 3 | 1 | 1 |
| All BLASTp hitsa (%) | 62.1 | 34.8 | 29.5 | 27.3 | 3.8 | 28 | 5.3 | 4.5 | 4.5 | 2.3 | 0.8 | 0.8 |
| Non-S1 protease BLASTp hitsa (%) | 34.7 | 13.3 | 3.1 | 2 | 4.1 | 2 | 6.1 | 6.1 | 6.1 | 3.1 | 1 | 1 |
Proteomes of venom/saliva secretions of other heteropteran insects determined by transcriptomics and proteomics were BLAST searched using L. distinctifemur venom proteins as queries
aBLAST hits with E < 0.01
Belostomatid and reduviid venom proteins belonging to the same family might have been convergently recruited in each lineage due to similar selection pressures imposed by the trophic strategy of predatory envenomation. Alternatively, the presence of some protein families in venom would represent synapomorphies if belostomatid and reduviid venom proteins are orthologues, descended from a venom protein present in a common ancestor. We considered that independent recruitment from multigenic families might result in belostomatid and reduviid venom proteins having closer homologues among non-heteropteran hemipterans than to each other. To test this, we used the tBLASTn algorithm to search the genomes of the sternorrhynchans Pachypsylla venusta (Aphalaridae), Diuraphis noxia (Aphididae), Diaphorina citri (Liviidae), Pseudococcus longispinus (Pseudococcidae), and the auchenorrhynchan Philaenus spumarius (Aphrophoridae), using as queries venom protein families identified with high confidence by MS in both L. distinctifemur and P. plagipennis venom (Table 2). Nucleotide sequences expressed in L. distinctifemur and P. plagipennis venom glands encoding the queries were appended to the largest database (P. spumarius). We observed the following pattern: for proteins not reported from any other source except heteropteran venoms, no close homologues (E < 0.01) were found among non-heteropteran hemipterans, whereas belostomatid and reduviid sequences were highly similar (E < 10−5 except for families 1 and 5, where homology signals are likely limited by the short sequence length). For proteins previously reported from other sources, highly similar sequences were found in both heteropteran and non-heteropteran species, but the belostomatid and reduviid sequences were always most closely related to each other. Thus, we did not observe any evidence of independent recruitment of these shared venom protein families using this method.
Table 2.
BLAST searches of hemipteran genomes for genes encoding venom protein homologues
| Protein family | With giant water bug sequences as query | With assassin bug sequences as query | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pristhesancus plagipennis | Pachypsylla venusta | Diuraphis noxia | Philaenus spumarius | Diaphorina citri | Pseudococcus longispinus | Lethocerus distinctifemur | Pachypsylla venusta | Diuraphis noxia | Philaenus spumarius | Diaphorina citri | Pseudococcus longispinus | |
| High confidence ID in L. distinctifemur and P. plagipennis venom but not reported elsewhere | ||||||||||||
| CUB domain | 2E−12 | 2.2 | > 10 | > 10 | 1.3 | > 10 | 1E−06 | 0.032 | 0.41 | 1.2 | 0.52 | 0.71 |
| Haemolysin-like | 2E−21 | 0.21 | > 10 | 0.99 | 1.1 | 0.011 | 5E−20 | 0.022 | 3.1 | 1.4 | 0.48 | > 10 |
| Venom family 1 | 0.001 | 9.5 | > 10 | > 10 | > 10 | > 10 | 0.002 | 1.3 | 0.84 | 5.1 | 4.4 | 0.35 |
| Venom family 2 | 1E−10 | 0.16 | 0.21 | 0.1 | 7E−03 | 0.01 | 3E−06 | 0.54 | 0.61 | 1.3 | 0.75 | 1.5 |
| Venom family 5 | 0.039 | 1.3 | > 10 | > 10 | 1.6 | 3.3 | 0.061 | 1.6 | 9.7 | 0.15 | 2.4 | 1.1 |
| Venom family 10 | 1E−06 | 0.5 | 6.9 | > 10 | 0.77 | > 10 | 1E−06 | > 10 | 10 | 3.2 | 6.3 | > 10 |
| Venom family 12 | 3E−13 | > 10 | > 10 | > 10 | > 10 | > 10 | 1E−113 | 4.7 | > 10 | 0.87 | 1.5 | 2.7 |
| High confidence ID in L. distinctifemur and P. plagipennis venom and previously known | ||||||||||||
| CRiSP | 5E−37 | 1E−04 | 0.11 | 6E−06 | 6E−04 | 4E−06 | 1E−28 | 2E−06 | 0.002 | 7E−06 | 6E−04 | 2E−06 |
| Cystatin | 5E−09 | 2E−05 | > 10 | 0.05 | 5E−05 | 0.98 | 1E−09 | 2.2 | 4.1 | 0.82 | 0.29 | 0.5 |
| IPPase | 1E−77 | 8E−29 | 1E−32 | 2E−07 | 5E−09 | 4E−31 | 1E−78 | 1E−28 | 1E−35 | 1E−09 | 1E−08 | 7E−32 |
| Lipase | 3E−64 | 2E−18 | 4E−26 | 3E−13 | 2E−15 | 3E−29 | 5E−63 | 7E−17 | 2E−33 | 3E−21 | 6E−17 | 2E−55 |
| Lysozyme | 5E−24 | 6E−04 | > 10 | 3E−04 | 0.003 | 3.5 | 3E−25 | 2E−09 | > 10 | 3E−12 | 1E−11 | 2.4 |
| Nuclease | 3E−139 | 2E−23 | 2E−11 | 2E−25 | 2E−22 | 1E−52 | 3E−133 | 8E−20 | 4E−12 | 9E−19 | 8E−15 | 3E−49 |
| Pacifastin | 4E−25 | 0.056 | > 10 | 0.17 | 0.12 | > 10 | 2E−23 | 0.009 | > 10 | 0.36 | 0.075 | > 10 |
| POBP | 3E−37 | 8E−15 | 5E−10 | 1E−13 | 1E−14 | 1E−11 | 4E−37 | 3E−13 | 6E−12 | 3E−13 | 2E−13 | 7E−13 |
| S1 protease | 6E−124 | 7E−17 | 2E−22 | 4E−14 | 1E−11 | 4E−19 | 4E−132 | 9E−79 | 3E−60 | 1E−86 | 2E−76 | 1E−91 |
| Serpin | 6E−127 | 2E−24 | 8E−34 | 6E−35 | 1E−20 | 1E−59 | 9E−116 | 2E−05 | 1E−26 | 8E−13 | 2E−07 | 2E−35 |
| Transferrin | 0 | 9E−172 | 1E−22 | 3E−19 | 1E−139 | 3E−07 | 0 | 3E−164 | 2E−23 | 5E−18 | 5E−129 | 9E−07 |
In each case examined, L. distinctifemur and P. plagipennis venom proteins are more similar to each other than any non-heteropteran sequences
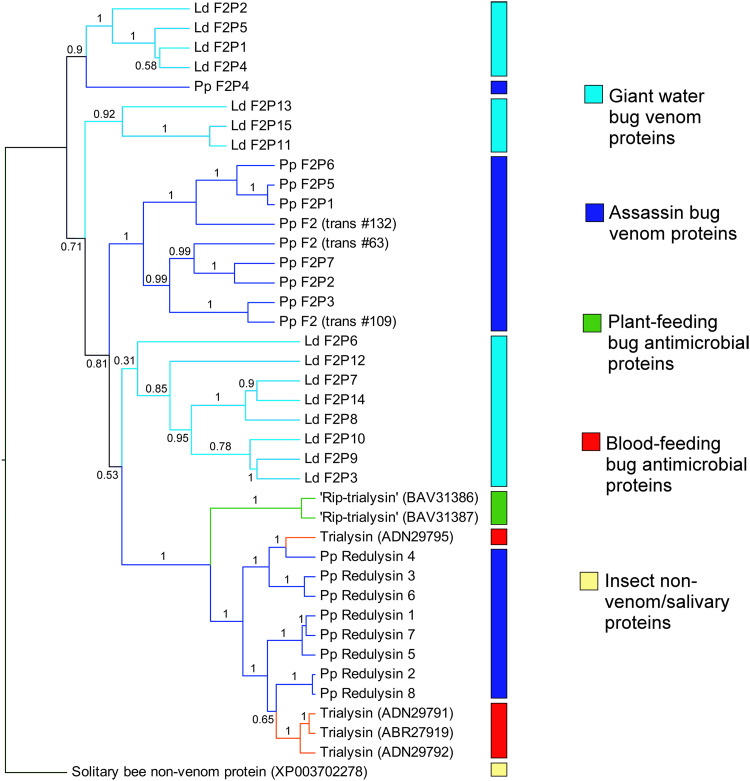
Heteropteran venom family 2 is a particularly large group of venom proteins, with 15 and 7 representatives detected by MS in the venoms of L. distinctifemur and P. plagipennis, respectively. Despite close homologues not being detected among non-heteropteran hemipterans (Table 2), highly similar sequences exist in the GenBank nr database originating from diverse bilaterian and cnidarian animals (BLASTp E < 10−9), often containing Pfam DUF4773. This suggests that family 2 venom proteins were recruited into heteropteran venoms from an undescribed but phylogenetically widespread secreted non-venom protein. Redulysin/trialysin proteins probably also belong to this family, to which they display weak evidence of homology by BLAST search (lowest E-value = 0.004) and a shared pattern of 8–10 cysteine residues in the C-terminal domain (Supplementary Fig. S1). We considered that if family 2 proteins were independently recruited into belostomatid and reduviid lineages before undergoing gene duplication, L. distinctifemur and P. plagipennis proteins should form separate phylogenetic clades. To test this hypothesis, we reconstructed the phylogeny of this family using Bayesian inference, taking a homologous predicted protein sequence from the solitary bee Megachile rotundata as outgroup. However, belostomatid and reduviid venom do not form separate phylogenetic clusters; instead, proteins from both L. distinctifemur and P. plagipennis appear in several separate, well-supported clades (Fig. 6). This result suggests that family 2 proteins were recruited into venom prior to the divergence of the nepomorphan and belostomatid lineages. Since we attempted to but did not find any evidence for independent recruitment of shared protein families, and because postulating single rather than multiple recruitment events are preferable by parsimony, we propose that the presence of some protein families in venoms of giant water bugs and assassin bugs are synapomorphous.
Fig. 6.
Phylogeny of redulysin/trialysin/family 2 venom proteins estimated by Bayesian inference. Branches are coloured to indicate the trophic strategy of the producing species with blue = predatory, green = plant-feeding, red = hematophagous. An uncharacterised putative secreted product from the solitary bee Megachile rotundata is the designated outgroup. Node labels indicate posterior probabilities. Note that clades of water bug (light blue) and assassin bug (dark blue) sequences are interspersed, suggesting duplication of family 2 proteins prior to the divergence of Cimicomorpha and Nepomorpha. Sequence reference numbers in brackets refer to either transcriptome-derived sequences (‘trans#’ refers to transcriptome indices in Supplementary Figure S1, 21) or GenBank Accession numbers
Discussion
In contrast to the previous studies that emphasised the presence of bioactive lysophospholipids in belostomatid venom [22], we found that water bug venoms are rich in proteins. Only one sample of cloudy venom, from the belostomatine D. eques, yielded an infrared absorption spectrum consistent with a significant lysophospholipid content, and no evidence of lysophospholipids was found in venom of the lethocerine bug L. distinctifemur. This suggests that the presence of a significant lysophospholipid content in belostomatid venom may be a derived characteristic in Belostoma anurum [22] or has been lost in L. distinctifemur.
The venom systems of giant water bugs and assassin bugs display numerous features in common, both at the level of protein primary structure and in gland-specific expression patterns. We propose that some or all of these similarities represent synapomorphies, i.e., that these traits were present in the common ancestor of Belostomatidae and Reduviidae and were retained in both groups. Furthermore, we propose that the similarity of belostomatid and reduviid venoms, compared with the saliva/venom of previously investigated blood- and plant-feeding heteropteran species, is best explained as adaptations to their shared trophic strategy of predatory envenomation. Below, we argue that such a hypothesis is consistent with the current theories of the phylogeny and evolutionary history of feeding strategies in Heteroptera, as well as previous studies focussing on the molecular evolution of venom and saliva secretions.
Heteroptera as a monophyletic group is strongly supported [28, 53]. However, other aspects of heteropteran evolution remain to be clarified. For example, the sister group to Heteroptera might either be Coleorrhyncha (moss bugs) [28] or Coleorrhyncha + Sternorrhyncha + Auchenorrhyncha [53]. The best estimates of the date of divergence of Heteroptera from the other hemipteran suborders, based on molecular data calibrated with fossil evidence [14], are between 293 and 262 mya (95% confidence interval; see also [53, 54]), though the true date is likely to be older if Coleorrhyncha is not sister to Heteroptera. Despite the precise relationships between Nepomorpha and the other Lower Heteroptera infraorders being uncertain [15], all major hypotheses of heteropteran phylogeny agree that Nepomorpha diverged from the terheteropteran lineage (including assassin bugs) early, around 254–226 mya [14]. In any case, fossil evidence indicates that Belostomatidae had emerged as a family > 200 mya [29]. Predation and envenomation (i.e., the injection of salivary secretions into prey to subdue and liquefy them) are practised by almost all Lower Heteroptera infraorders, strongly suggesting that the last common ancestor of extant Heteroptera was also predatory and venomous [15, 16, 18, 19]. The salient point of this phylogenetic discussion for the current study is that the adoption of a predatory and venomous lifestyle preceded divergence of the belostomatid and reduviid lineages by a length of time reasonably estimated as between 5 and 100 million years, a plausible window of time to accumulate numerous adaptations at the molecular level [3, 8]. The ancestral shift from phytophagy to predation is likely to have strongly altered selection pressures on the secretory output of the oral glands, resulting in increased expression of polypeptides that facilitate prey paralysis and/or tissue liquefaction [5, 8, 13]. The presence of these proteins in venom may have been retained in the belostomatid and reduviid lineages, because they facilitate predation, which is likely to have continued as the trophic strategy in both lineages—unbroken between the last common ancestor of Heteroptera and today [15, 18, 19].
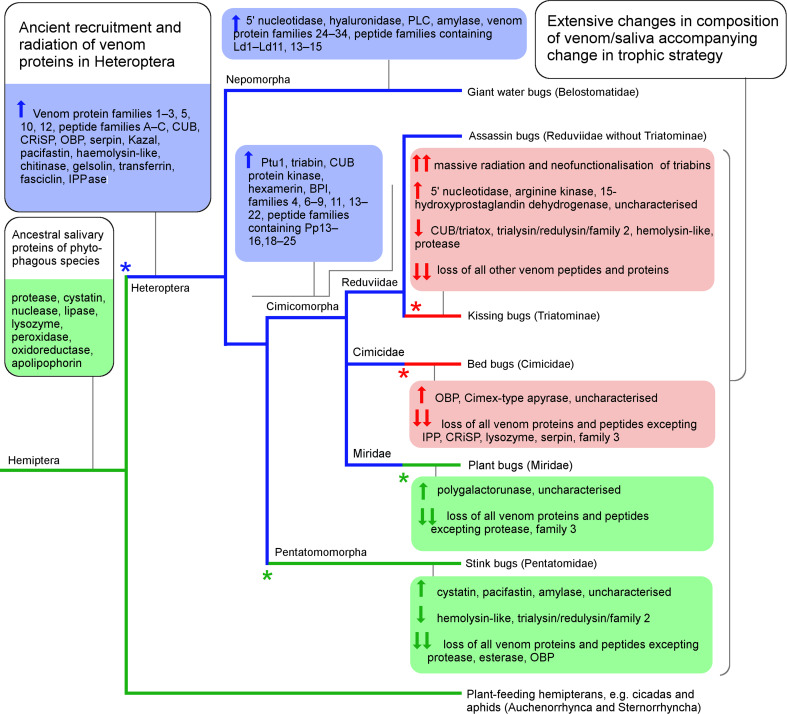
In contrast, multiple other heteropteran subgroups subsequently transitioned to either parasitic blood-feeding or reverted to plant-feeding; these trophic switches are strongly supported by phylogenetic evidence [15, 18, 19]. Some venom proteins of predaceous heteropterans are likely to have been unnecessary and possibly disadvantageous when applied to other trophic strategies. For example, venoms of belostomatids and predatory reduviids induce strong pain sensations when injected into vertebrates [55, 56], whereas the venom of blood-feeding reduviids has an analgesic effect [57]. Blood-feeding reduviids have likely undergone active selection to lose pain-causing molecules from their venom to evade host detection, accompanying their transition to haematophagy 24–38 mya [58]. Consistent with this notion, numerous venom protein families shared by Belostomatidae and Reduviidae appear to have been lost or downregulated in Triatominae, and replaced by other protein families that have expanded via gene duplication and neofunctionalisation. For example, the triabin family of proteins is a minor component of venoms from predaceous reduviids [27] but dominate the venoms of blood-feeding reduviids, in which they function to combat haemostasis by multiple mechanisms [24]. In consequence, the venoms of predaceous and blood-feeding reduviids that diverged recently (24–38 mya [58]) are less similar in terms of protein families represented than the venoms of predaceous reduviids and belostomatids, which diverged > 200 mya but share a conserved trophic strategy. Our results suggest that the oral secretions of other groups that have transitioned away from predation such as the bed and bat bugs (Cimicidae + Polyctenidae), the plant-feeding stink bugs (Pentatomidae) and plant bugs (Miridae) have undergone similar rapid transitions in protein composition. One limitation of this result is that the proteomes we employed for comparison were conducted independently and the completeness of the reported proteomes varies, which might result in false-negatives for the presence of protein families in venom/saliva in some taxa. Nevertheless, the available data, including high-coverage proteomes from blood-feeding triatomine reduviids [48], support massive loss of venom protein families accompanying trophic shifts away from predation. An alternative or complementary hypothesis to the ancient recruitment of heteropteran venom proteins and their loss in blood- and plant-feeding species is that some or all protein families represented in venoms of both belostomatid and predaceous reduviids were recruited into venom convergently, due to similar selection pressures. Further information on the composition of the venom/saliva secretion of additional heteropteran and hemipteran species is required to resolve these possibilities in detail. One scenario that is consistent with currently available data is presented in Fig. 7.
Fig. 7.
Ancient recruitment and radiation of heteropteran venom proteins. Highly simplified heteropteran phylogeny showing probable locations of trophic switches and changes in venom composition. Branches are coloured green for predominantly phytophagous lineages, blue for predatory lineages, and red for haematophagous lineages (after [15]). Major changes in trophic strategy are marked with an asterisk. Possible recruitment and loss events for different protein families in the venom/saliva secretion are based on comparison with previously reported proteomes. Upward arrow: protein recruited or abundance in venom increased. Downward arrow: protein lost or abundance in venom diminished
Trophic strategy in general is unlikely to explain all aspects of venom composition. For example, some snake venoms contain 5′ nucleotidase enzymes with apyrase activity, degrading ATP/ADP and thereby inhibiting ATP/ADP-induced platelet aggregation [59, 60]. This enzyme is also found in venom from the blood-feeding reduviid Triatoma infestans, where it combats platelet aggregation during blood-feeding [61]. A 5′ nucleotidase was the most confidently identified protein in venom from the giant water bug L. distinctifemur, but has not been reported from predatory reduviids [23, 27]. This finding is consistent with the notion that belostomatid venom 5′ nucleotidase inhibits ATP/ADP-induced thrombocyte aggregation, which occurs in fish as well as other vertebrates [62] but has not been reported in insects [63]. Therefore, prey range may also affect venom composition in Heteroptera.
We found that L. distinctifemur produces different sets of venom proteins in separate gland lumens, and that gland-specific expression patterns present another similarity to reduviids [23]. In particular, we observed high expression of the haemolysin-like protein family in the AMG and high expression of proteases and family 1 and 2 venom proteins in the PMG, which is also the case for the reduviid P. plagipennis. This result suggests that giant water bugs share the ability of P. plagipennis to modulate their venom composition in a context-dependent manner [23], and that this ability represents another synapomorphy of the heteropteran venom system. For P. plagipennis, we have suggested that venom from the PMG performs prey capture and digestion, whereas venom from the AMG is used defensively [23]. However, the function of the two compartments of the main gland in belostomatids requires further investigation. Our results also contrast with a report that describes protease activity existing in the accessory gland of belostomatids but not the main gland [38], as the reverse arrangement was observed in our study. This difference might be partially explained by the different methods employed, since we looked at protein secretion and storage, whereas the study of Swart and colleagues measured substrate cleavage [38].
The present study is consistent with reports that rapid changes in venom composition in response to changes in venom use are achieved primarily through evolution in the expression levels of genes encoding venom proteins [5, 12, 13]. Moreover, our results may assist the functional characterisation of heteropteran venom constituents, as proteins that are strongly conserved in predaceous groups, but readily lost in species that feed on blood or plants, may be specifically related to prey capture or the breakdown of animal tissues. This study provides insights into the evolutionary history of venom production in Heteroptera.
Materials and methods
Insects and venom collection
Giant fish-killer water bugs (L. distinctifemur) were purchased from Australian Insect Farm (Innisfail, Australia). For L. distinctifemur, all venom extractions and dissections occurred less than 2 weeks after insects were collected from the wild by the suppliers using light traps. Smaller belostomatine water bugs (D. eques) were collected from ponds in Regency Downs, Queensland, Australia, and fed water snails collected from the same location. Venom was collected non-lethally from adult bugs by applying electrostimulation (20 ms pulses of 40 V, 5 Hz) to the ventral or dorsal surface between the head and thorax, whilst the proboscis was inserted into a P200 pipette tip. Venom was immediately stored on ice and clarified by centrifugation (17,000 rcf, 10 min, 4 °C), and the supernatant stored at − 20 °C until analysis.
Fourier transform infrared spectroscopy
FTIR spectra acquired over the frequency range 400–4000 cm−1 were obtained on a Nicolet 5700 FTIR spectrometer (Thermo Fisher) with a diamond attenuated total reflectance (ATR) attachment controlled by OMNIC software. Resolution was 2 cm−1 and each spectrum was averaged over 64 scans. For each sample, 4 µl diluted venom (concentration ~ 1–5 mg/ml) was pipetted directly onto the ATR attachment and allowed to dry under a N2 gas stream. Spectra for each sample shown are the result of averaging three different dried spots.
Transcriptomics
For RNA-Seq experiments, insects were anaesthetised by CO2 for 10 min, and the main gland complex and accessory glands were dissected out and cleaned. Each gland compartment (PMG, AMG, SS, and AG) was then carefully separated. Tissue was pooled between individuals and each side of the body (33, 9, 4, and 3 mg of tissue, respectively). Tissue was stored at − 20 °C until further analysis in > 10-fold; the tissue volume of RNAlater. Total RNA was extracted using a DNeasy kit (Qiagen) and mRNA purified using a Dynabeads mRNA Direct kit (1426, 530, 24, and 0.24 ng of mRNA, respectively) according to the manufacturer’s instructions. Nucleic acid concentrations were quantified by A260 on a Nanodrop spectrophotometer (Thermo Scientific).
Construction and sequencing of RNA-Seq libraries were performed by the Institute for Molecular Bioscience Sequencing Facility at The University of Queensland, Australia. For each of the posterior and anterior lobes of the main gland, a TruSeq library was constructed from ~ 300 ng mRNA. The multiplexed libraries were then sequenced as a part of a high-output run on an Illumina HiSeq instrument, yielding 17,083,410 and (posterior lobe) and 22,958,470 (anterior lobe) 150 bp paired-end reads. A combined transcriptome of the anterior and posterior lobe was then assembled as described previously [27] using Trinity v2.2.0, CLC Genomics Workbench (CLC) and CD-HIT-EST [64]. Contigs were refined by re-mapping all reads with the ‘update contigs’ option selected in CLC Genomics Workbench. A library of possible protein sequences was generated using GetORF [65] with length > 90 bp. Gene expression data were obtained using the RNA-Seq Gene Expression module in CLC Genomics Workbench.
Proteomics
To obtain gland protein extracts, a single adult male was anaesthetised by CO2 for 10 min and dissected under phosphate-buffered saline (PBS). After obtaining the venom gland complex, glands were washed and then carefully cut into their separate compartments (PMG, AMG, SS, and AG). The gland region from each side of the body was then pooled into 30 µl PBS; the sample was then vortexed for 10 s and centrifuged (17,000 rcf, 30 s) to empty the gland lumens. Glandular tissue was removed with tweezers, the extract was clarified by centrifugation (17,000 rcf, 1 min), and the supernatant analysed. For electrophoresis, 10 µg (A280 equivalent) of venom or protein extract was denatured in SDS-PAGE sample buffer containing 100 mM dithiothreitol for 2 min at 95 °C, run on a 12.5% PAGE gel under denaturing conditions, and stained with Coomassie R250.
Tandem mass spectrometry (LC–MS/MS) of whole venom and SDS-PAGE bands was performed as previously described [27]. Briefly, tryptic digests of protein samples were separated on a Nexera Nano system coupled to an SCIEX 5600 mass spectrometer equipped with a Turbo V ion source. The resulting mass spectra in WIFF format were then compared to a library of possible protein sequences generated from RNA-Seq experiments (see “Transcriptomics”) together with a list of common MS contaminants using a Paragon search within ProteinPilot (SCIEX) software. Minimum criteria for protein identification were three distinct peptides observed with > 95% confidence, or one peptide with > 95% confidence plus a signal peptide with a D-score > 0.7 in SignalP 4.1.
Bioinformatics
Protein sequences were annotated by BLAST search against the UniProt UniRef90 database, as well as in-house databases of assassin bug venom proteins. Further annotation of protein domains was performed using SignalP 4.1 [66] plus a HMMER search against the Pfam database [67].
For phylogenetic analysis, sequences were aligned using MAFFT [68] using the G-INS-I algorithm recommended for less than 200 sequences with a global homology and BLOSSUM62 as the substitution matrix. ProtTest3 software [69] determined the best model as LG + I (Invariant sites) + G (Gamma), with discrete gamma distribution in four categories. Bayesian inference analysis was implemented in BEAST version 1.8.3 [70]. The analysis ran for 5 million cycles, with sampling every 1000 generations. The burn-in option on TreeAnnotator was used to discard the initial 10% of trees.
For comparing protein sequences with venom/saliva secretions of other heteropterans, we located studies from diverse heteropteran groups that used tandem mass spectrometry to identify proteins with high confidence against a transcriptome database prepared from the same species [23, 26, 27, 45, 49–52]. For each species, a FASTA file containing the identified sequences was prepared based on the published data and used as a database for a local BLAST search.
Hemipteran genomes searched (Table 2) comprised one representative from each family of non-heteropteran hemipterans available, AZLD00000000.1 (Pachypsylla venusta), JOTR00000000.1 (Diuraphis noxia), MPAX00000000.1 (Philaenus spumarius), AWGM00000000.1 (Diaphorina citri), and FIZU00000000.1 (Pseudococcus longispinus). L. distinctifemur and P. plagipennis sequences were appended to the largest genome, that of P. spumarius, so that the cited E-values for the venomous species are the most conservative possible.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Dataset S1: Identification and annotation of Lethocerus distinctifemur venom proteins (XLSX 207 kb)
Supplementary Dataset S2: Most abundant transcripts in each compartment of Lethocerus distinctifemur venom glands (XLSX 90 kb)
Supplementary Fig. S1: Alignment of amino acid sequences of family 2 venom proteins, redulysins and trialysins (PDF 77 kb)
Supplementary Table S1: Abundant venom proteins detected by LC–MS/MS (PDF 36 kb)
Acknowledgements
We thank the Australian Insect Farm for acquiring insects, Alun Jones for assistance with proteomics experiments, Idriss Blakey for assistance with FTIR spectroscopy, Christiane Weirauch for discussions of phylogeny, and Eivind Undheim for assistance with animals and sequencing costs. This work was supported by a University of Queensland Postdoctoral Fellowship to A.A.W. and a Principal Research Fellowship to G.F.K. from the Australian National Health and Medical Research Council. Sequences discovered in this project were deposited to GenBank with identifiers of MF683255–MF683386.
Contributor Information
Andrew A. Walker, Email: a.walker@uq.edu.au
Glenn F. King, Email: glenn.king@imb.uq.edu.au
References
- 1.Aird SD, Arora J, Barua A, Qiu L, Terada K, Mikheyev AS. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol Evol. 2017;9:2640–2649. doi: 10.1093/gbe/evx199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kordiš D, Gubenšek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261:43–52. doi: 10.1016/S0378-1119(00)00490-X. [DOI] [PubMed] [Google Scholar]
- 3.Olivera BM, et al. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Ann N Y Acad Sci. 1999;870:223–237. doi: 10.1111/j.1749-6632.1999.tb08883.x. [DOI] [PubMed] [Google Scholar]
- 4.Pineda SS, et al. Diversification of a single ancestral gene into a successful toxin superfamily in highly venomous Australian funnel-web spiders. BMC Genom. 2014;15:177. doi: 10.1186/1471-2164-15-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duda TF, Palumbi SR. Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc R Soc Lond B Biol Sci. 2004;271:1165–1174. doi: 10.1098/rspb.2004.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson T, et al. Rapid radiations and the race to redundancy: an investigation of the evolution of Australian elapid snake venoms. Toxins. 2016;8:309. doi: 10.3390/toxins8110309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch VJ. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunagar K, Moran Y. The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015;11:e1005596. doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Fry BG, Kini RM. Putting the brakes on snake venom evolution: the unique molecular evolutionary patterns of Aipysurus eydouxii (marbled sea snake) phospholipase A2 toxins. Mol Biol Evol. 2005;22:934–941. doi: 10.1093/molbev/msi077. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Fry BG, Kini RM. Eggs-only diet: its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii) J Mol Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. [DOI] [PubMed] [Google Scholar]
- 11.Daltry JC, Wuster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 12.Binford GJ. Differences in venom composition between orb-weaving and wandering Hawaiian Tetragnatha (Araneae) Biol J Linn Soc. 2001;74:581–595. doi: 10.1111/j.1095-8312.2001.tb01415.x. [DOI] [Google Scholar]
- 13.Martinson EO, Mrinalini, Kelkar YD, Chang C-H, Werren JH. The evolution of venom by co-option of single-copy genes. Curr Biol. 2017;27:2007–2013. doi: 10.1016/j.cub.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y-H, et al. Phylogenetic divergences of the true bugs (Insecta: Hemiptera: Heteroptera), with emphasis on the aquatic lineages: the last piece of the aquatic insect jigsaw originated in the Late Permian/Early Triassic. Cladistics. 2016;32:390–405. doi: 10.1111/cla.12137. [DOI] [PubMed] [Google Scholar]
- 15.Walker AA, Weirauch C, Fry BG, King GF. Venoms of heteropteran insects: a treasure trove of diverse pharmacological toolkits. Toxins. 2016;8:43. doi: 10.3390/toxins8020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobben RH. On the original feeding habits of the Hemiptera (Insecta): a reply to Merrill Sweet. Ann Entomol Soc Am. 1979;72:711–715. doi: 10.1093/aesa/72.6.711. [DOI] [Google Scholar]
- 17.Cohen AC. Plant feeding by predatory Heteroptera: Evolutionary and adaptational aspects of trophic switching. In: Alomar O, editor. Zoophytophagous Heteroptera: implication for life history and integrated pest management. Lanham: Thomas Say Publications in Entomology; 1996. pp. 1–7. [Google Scholar]
- 18.Weirauch C, Schuh RT, Cassis G, Wheeler WC. Revisiting habitat and lifestyle transitions in Heteroptera (Insecta: Hemiptera): insights from a combined morphological and molecular phylogeny. Cladistics. 2018 doi: 10.1111/cla.12233. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Leavengood JM, Chapman EG, Burkhardt D, Song F, Jiang P, Liu J, Zhou X, Cai W (2017) Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc R Soc Lond B: Biol Sci 284(1862). 10.1098/rspb.2017.1223 [DOI] [PMC free article] [PubMed]
- 20.Edwards JS. The action and composition of the saliva of an assassin bug Platymeris rhadamanthus Gaerst. (Hemiptera, Reduviidae) J Exp Biol. 1961;38:61–77. [Google Scholar]
- 21.Maran SPM, Ambrose DP. Paralytic potential of Catamiarus brevipennis (Serville), a potential biological control agent (Insecta: Heteroptera: Reduviidae) In: Ignacimuth A, Sen A, Janarthanan S, editors. Biotechnological applications for integrated pest management. New Delhi: Oxford Publishing; 2000. pp. 125–131. [Google Scholar]
- 22.Silva-Cardoso L, et al. Paralytic activity of lysophosphatidylcholine from saliva of the waterbug Belostoma anurum. J Exp Biol. 2010;213:3305–3310. doi: 10.1242/jeb.041954. [DOI] [PubMed] [Google Scholar]
- 23.Walker AA et al (2018) The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat Commun. 10.1038/s41467-018-03091-5 [DOI] [PMC free article] [PubMed]
- 24.Ribeiro JMC, Assumpção TC, Francischetti IMB. An insight into the sialomes of bloodsucking Heteroptera. Psyche (Stuttg.) 2012;2012:1–16. [Google Scholar]
- 25.Miles PW. The saliva of Hemiptera. In: Treherne JE, Berridge MJ, Wigglesworth VB, editors. Advances in insect physiology. New York: Academic Press; 1972. pp. 183–255. [Google Scholar]
- 26.Peiffer M, Felton GW. Insights into the saliva of the brown marmorated stink bug Halyomorpha halys (Hemiptera: Pentatomidae) PLoS One. 2014;9:e88483. doi: 10.1371/journal.pone.0088483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker AA, Madio B, Jin J, Undheim EA, Fry BG, King GF. Melt with this kiss: paralyzing and liquefying venom of the assassin bug Pristhesancus plagipennis (Hemiptera: Reduviidae) Mol Cell Proteom. 2017;16:552–566. doi: 10.1074/mcp.M116.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weirauch C, Schuh RT. Systematics and evolution of Heteroptera: 25 years of progress. Annu Rev Entomol. 2011;56:487–510. doi: 10.1146/annurev-ento-120709-144833. [DOI] [PubMed] [Google Scholar]
- 29.Fraser NC, Grimaldi DA, Olsen PE, Axsmith B. A Triassic Lagerstätte from eastern North America. Nature. 1996;380:615. doi: 10.1038/380615a0. [DOI] [Google Scholar]
- 30.Schuh RT, Slater JA. True bugs of the world (Hemiptera: Heteroptera): classification and natural history. New York: Cornell University Press; 1995. [Google Scholar]
- 31.Ohba S-Y. Field observation of predation on a turtle by a giant water bug. Entomol Sci. 2011;14:364–365. doi: 10.1111/j.1479-8298.2011.00450.x. [DOI] [Google Scholar]
- 32.Ohba S-Y, Nakasuji F. Dietary items of predacious aquatic bugs (Nepoidea: Heteroptera) in Japanese wetlands. Limnology. 2006;7:41–43. doi: 10.1007/s10201-006-0161-5. [DOI] [Google Scholar]
- 33.Caccin P, Rigoni M, Bisceglie A, Rossetto O, Montecucco C. Reversible skeletal neuromuscular paralysis induced by different lysophospholipids. FEBS Lett. 2006;580:6317–6321. doi: 10.1016/j.febslet.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 34.Kordiš D. Evolution of phospholipase A2 toxins in venomous animals. Acta Chim Slov. 2011;58:638–646. [PubMed] [Google Scholar]
- 35.Šribar J, Križaj I. Secreted phospholipases A2—not just enzymes. Acta Chim Slov. 2011;58:678–688. [PubMed] [Google Scholar]
- 36.Rastogi SC. On the salivary enzymes of some phytophagous and predaceous heteropterans. Sci Cult. 1962;28:479–480. [Google Scholar]
- 37.Rees AR, Offord RE. Studies on the protease and other enzymes from the venom of Lethocerus cordofanus. Nature. 1969;221:675–677. doi: 10.1038/221675a0. [DOI] [PubMed] [Google Scholar]
- 38.Swart CC, Deaton LE, Felgenhauer BE. The salivary gland and salivary enzymes of the giant waterbugs (Heteroptera; Belostomatidae) Comp Biochem Physiol A Mol Integr Physiol. 2006;145:114–122. doi: 10.1016/j.cbpa.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 40.Fen N, DeOliveira DB, Trumble WR, Sarkar HK, Singh BR. Secondary structure estimation of proteins using the amide III region of Fourier transform infrared spectroscopy: application to analyze calcium-binding-induced structural changes in calsequestrin. Appl Spectrosc. 1994;48:1432–1441. doi: 10.1366/0003702944028065. [DOI] [Google Scholar]
- 41.Coates J. Interpretation of infrared spectra, a practical approach. In: Meyers RA, editor. Encyclopedia of analytical chemistry. Chichester: Wiley; 2000. pp. 10815–10837. [Google Scholar]
- 42.Baptist BA. The morphology and physiology of the salivary glands of Hemiptera-Heteroptera. Q J Microsc Sci. 1941;s2-83:91–139. [Google Scholar]
- 43.Swart CC, Felgenhauer BE. Structure and function of the mouthparts and salivary gland complex of the giant waterbug, Belostoma lutarium (Stål) (Hemiptera: Belostomatidae) Ann Entomol Soc Am. 2003;95:870–882. doi: 10.1603/0013-8746(2003)096[0870:SAFOTM]2.0.CO;2. [DOI] [Google Scholar]
- 44.Haridass ET, Ananthakrishnan TN. Functional morphology of the salivary system in some reduviids (Insecta-Heteroptera-Reduviidae) Proc Indian Acad Sci. 1981;90:145–160. doi: 10.1007/BF03186026. [DOI] [Google Scholar]
- 45.Assumpção TCF, Francischetti IM, Andersen JF, Schwarz A, Santana JM, Ribeiro JM. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas’ disease. Insect Biochem Mol Biol. 2008;38:213–232. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charneau S, et al. The saliva proteome of the blood-feeding insect Triatoma infestans is rich in platelet-aggregation inhibitors. Int J Mass Spectrom. 2007;268:265–276. doi: 10.1016/j.ijms.2007.05.004. [DOI] [Google Scholar]
- 47.Costa CM, Sousa MV, Ricart CA, Santana JM, Teixeira AR, Roepstorff P, Charneau S. 2-DE-based proteomic investigation of the saliva of the Amazonian triatomine vectors of Chagas disease: Rhodnius brethesi and Rhodnius robustus. J. Proteom. 2011;74:1652–1663. doi: 10.1016/j.jprot.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 48.Hernández-Vargas MJ, Gil J, Lozano L, Pedraza-Escalona M, Ortiz E, Encarnación-Guevara S, Alagón A, Corzo G. Proteomic and transcriptomic analysis of saliva components from the hematophagous reduviid Triatoma pallidipennis. J Proteom. 2017;162:30–39. doi: 10.1016/j.jprot.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 49.Montandon CE, Barros E, Vidigal PM, Mendes MT, Anhê ACBM, de Oliveira Ramos HJ, de Oliveira CJF, Mafra C. Comparative proteomic analysis of the saliva of the Rhodnius prolixus, Triatoma lecticularia and Panstrongylus herreri triatomines reveals a high interspecific functional biodiversity. Insect Biochem Mol Biol. 2016;71:83–90. doi: 10.1016/j.ibmb.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Francischetti IM, et al. Insight into the sialome of the bed bug, Cimex lectularius. J Proteome Res. 2010;9:3820–3831. doi: 10.1021/pr1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooper WR, Nicholson SJ, Puterka GJ. Salivary proteins of Lygus hesperus (Hemiptera: Miridae) Ann Entomol Soc Am. 2013;106:86–92. doi: 10.1603/AN12096. [DOI] [Google Scholar]
- 52.Rao SAK, Carolan JC, Wilkinson TL. Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS One. 2013;8:e57413. doi: 10.1371/journal.pone.0057413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Tian Y, Zhao Y, Bu W. Higher level phylogeny and the first divergence time estimation of Heteroptera (Insecta: Hemiptera) based on multiple genes. PLoS One. 2012;7:e32152. doi: 10.1371/journal.pone.0032152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haddad V, Schwartz EF, Schwartz CA, Carvalho LN. Bites caused by giant water bugs belonging to Belostomatidae family (Hemiptera, Heteroptera) in humans: a report of seven cases. Wilderness Environ Med. 2010;21:130–133. doi: 10.1016/j.wem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Zerachia T, Bergmann F, Shulov A. Pharmacological activities of the venom of the predaceous bug Holotrichius innesi H. (Heteroptera, Reduviidae) In: Kaiser E, editor. Animal and plant toxins. Munich: Goldman; 1973. pp. 143–146. [Google Scholar]
- 57.Lavoipierre MM, Dickerson G, Gordon RM. Studies on the methods of feeding of blood-sucking arthropods. I. The manner in which triatomine bugs obtain their blood-meal, as observed in the tissues of the living rodent, with some remarks on the effects of the bite on human volunteers. Ann Trop Med Parasitol. 1959;53:235–250. doi: 10.1080/00034983.1959.11685921. [DOI] [PubMed] [Google Scholar]
- 58.Hwang WS, Weirauch C. Evolutionary history of assassin bugs (Insecta: Hemiptera: Reduviidae): insights from divergence dating and ancestral state reconstruction. PLoS One. 2012;7:e45523. doi: 10.1371/journal.pone.0045523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhananjaya BL, Nataraju A, Rajesh R, Raghavendra Gowda CD, Sharath BK, Vishwanath BS, D’Souza CJM. Anticoagulant effect of Naja naja venom 5′ nucleotidase: demonstration through the use of novel specific inhibitor, vanillic acid. Toxicon. 2006;48:411–421. doi: 10.1016/j.toxicon.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 60.Hart ML, Köhler D, Eckle T, Kloor D, Stahl GL, Eltzschig HK. Direct treatment of mouse or human blood with soluble 5′-nucleotidase inhibits platelet aggregation. Arterioscler Thromb Vasc Biol. 2008;28:1477–1483. doi: 10.1161/ATVBAHA.108.169219. [DOI] [PubMed] [Google Scholar]
- 61.Faudry E, et al. Triatoma infestans apyrases belong to the 5′-nucleotidase family. J Biol Chem. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- 62.Tavares-Dias M, Ragonha Oliveira S. A review of the blood coagulation system of fish. Br J Biosci. 2009;7:205–224. [Google Scholar]
- 63.Dushay MS. Insect hemolymph clotting. Cell Mol Life Sci. 2009;66:2643–2650. doi: 10.1007/s00018-009-0036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 66.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 67.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset S1: Identification and annotation of Lethocerus distinctifemur venom proteins (XLSX 207 kb)
Supplementary Dataset S2: Most abundant transcripts in each compartment of Lethocerus distinctifemur venom glands (XLSX 90 kb)
Supplementary Fig. S1: Alignment of amino acid sequences of family 2 venom proteins, redulysins and trialysins (PDF 77 kb)
Supplementary Table S1: Abundant venom proteins detected by LC–MS/MS (PDF 36 kb)