Abstract
Women with triple-negative breast cancer (TNBC) are generally treated by chemotherapy but their responsiveness may be blunted by DNA double-strand break (DSB) repair. We previously reported that IGFBP-3 forms nuclear complexes with EGFR and DNA-dependent protein kinase (DNA-PKcs) to modulate DSB repair by non-homologous end-joining (NHEJ) in TNBC cells. To discover IGFBP-3 binding partners involved in chemoresistance through stimulation of DSB repair, we analyzed the IGFBP-3 interactome by LC–MS/MS and confirmed interactions by coimmunoprecipitation and proximity ligation assay. Functional effects were demonstrated by DNA end-joining in vitro and measurement of γH2AX foci. In response to 20 µM etoposide, the DNA/RNA-binding protein, non-POU domain-containing octamer-binding protein (NONO) and its dimerization partner splicing factor, proline/glutamine-rich (SFPQ) formed complexes with IGFBP-3, demonstrated in basal-like TNBC cell lines HCC1806 and MDA-MB-468. NONO binding to IGFBP-3 was also shown in a cell-free biochemical assay. IGFBP-3 complexes with NONO and SFPQ were blocked by inhibiting EGFR with gefitinib or DNA-PKcs with NU7026, and by the PARP inhibitors veliparib and olaparib, which also reduced DNA end-joining activity and delayed the resolution of the γH2AX signal (i.e. inhibited DNA DSB repair). Downregulation of the long noncoding RNA in NHEJ pathway 1 (LINP1) by siRNA also blocked IGFBP-3 interaction with NONO–SFPQ. These findings suggest a PARP-dependent role for NONO and SFPQ in IGFBP-3-dependent DSB repair and the involvement of LINP1 in the complex formation. We propose that targeting of the DNA repair function of IGFBP-3 may enhance chemosensitivity in basal-like TNBC, thus improving patient outcomes.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03033-4) contains supplementary material, which is available to authorized users.
Keywords: IGF binding protein, TNBC, P54NRB, PSF, lncRNA, PARP inhibitors
Background
Radiotherapy and some chemotherapies act by causing DNA double-strand breaks (DSBs), potentially the most lethal form of DNA damage. Cancer cells typically respond by initiating DNA repair, cell death, or senescence [1]. Since repairing damaged DNA reverses the effect of DNA-damaging treatments, it is an important mechanism by which cancer cells oppose the effects of these therapies. Overcoming treatment resistance is a major goal to improve cancer therapy. The two predominant DSB repair mechanisms that maintain genomic integrity are homology-directed recombination (HR), which is accurate but only occurs from late S to G2 phase of the cell cyle, and non-homologous end-joining (NHEJ), more error-prone but active at all cell cycle stages. NHEJ involves the assembly and sequential reorganization of protein complexes at the site of DNA strand breaks, including the DNA-dependent protein kinase (DNA-PK) complex comprising DNA-PK catalytic subunit and the Ku70–Ku80 heterodimer [2]. Inhibitors of both HR and NHEJ pathways have been developed in attempts to overcome DNA-repair-mediated resistance [1].
In triple-negative breast cancers (TNBC), which are unresponsive to estrogen receptor- or HER2-directed treatments, cytotoxic chemotherapy is a front-line treatment, and no targeted treatment is yet widely accepted [3]. Inhibitors of poly(ADP-ribose) polymerase-1 (PARP1) have been extensively trialed, mainly in cases with BRCA1 mutation or dysregulation [3]. PARP1, which adds multiple ADP-ribose units to proteins (known as PARylation), is involved in coordinating single-strand DNA repair [4]; its inhibition results in increased DNA single-strand breaks which, upon replication, are typically repaired by homologous recombination pathways involving BRCA1 and BRCA2.
There is substantial evidence that PARP inhibitors disrupt DNA repair by NHEJ [5]. For example, the EGF receptor (EGFR) is known to translocate to the cell nucleus as part of the NHEJ process [6], and the PARP inhibitor veliparib (ABT-888) has been shown to inhibit nuclear EGFR translocation and DNA repair in response to radiotherapy [7]. PARP1 has also been shown to PARylate the DNA- and RNA-binding protein NONO (non-POU domain-containing octamer-binding protein), which is then recruited to DNA damage sites and stimulates NHEJ while inhibiting HR [8]. This effect was also blocked by veliparib. The involvement of NONO and its binding partner SFPQ (splicing factor, proline and glutamine rich) in NHEJ is increasingly recognized, with a role in DNA pairing during the ligation phase, and potentially additional functions [9].
We previously reported that insulin-like growth factor binding protein-3 (IGFBP-3) is involved in DSB repair by NHEJ, forming nuclear complexes with both EGFR and DNA-PKcs in response to DNA-damaging chemotherapy [10]. This effect was unexpected because in some cancers IGFBP-3 acts as a tumor suppressor [11]. We now report that in TNBC cell lines, both NONO and SFPQ form PARP-dependent complexes with IGFBP-3 after exposure to DNA-damaging chemotherapy, and that preventing these interactions inhibits the DNA damage response. These findings present novel opportunities for drug intervention in DNA damage repair by NHEJ, with the potential to modulate treatment resistance in TNBC.
Materials and methods
Etoposide was obtained from Sigma-Aldrich (St. Louis, MO, USA). Gefitinib (Iressa) and NU7026 (LY293646) were from MedChem Express (Monmouth Junction, NJ, USA). NU7441 was from Tocris Bioscience, Bristol, UK. Veliparib (ABT-888) was from Selleckchem, Houston, TX, USA and olaparib from AdooQ Bioscience, Irvine, CA. Rabbit antiserum R-100 against full-length human IGFBP-3, and recombinant human IGFBP-3 expressed in human cells, were prepared in-house. Recombinant human NONO, Myc-DDK-tagged (TP326567) was obtained from Origene, Rockville, MD, USA. FLAG antibody plates (L00455C) were from GenScript, Piscataway, NJ, USA. LINP1 TaqMan probes were designed using the Custom TaqMan Assay Design Tool (ThermoFisher) and purchased from Life Technologies, Mulgrave, VIC, Australia. Goat anti-rabbit IgG-HRP (ab97080) was from Abcam, Melbourne, VIC, Australia, and 1-Step Turbo TMB-ELISA substrate solution was from ThermoFisher, Scoresby, VIC, Australia.
Cell culture
The human basal-like TNBC cell lines MDA-MB-468 and HCC1806 were obtained from ATCC, Manassas, VA and maintained in RPMI 1640 medium containing 5% FBS and 10 µg/mL bovine insulin under standard conditions. Cryopreserved stocks were established within 1 month of receipt (in 2010), and fresh cultures for use in experiments were established from these stocks every 2–3 months. All cell lines tested negative for mycoplasma. Inhibitor treatments were carried out for 24 h with veliparib (20 µM), olaparib (10 µM), gefitinib (10 µM), or NU7026 (20 µM), followed by etoposide (20 µM).
siRNA mediated transient knockdown
IGFBP-3 was downregulated using siRNAs from Qiagen (Hilden, Germany) (Table 1). DNA-PKcs was downregulated using siRNA (Hs_PRKDC_6 FlexiTube) from Qiagen (Melbourne, VIC, Australia). LINP1 was downregulated using custom siRNAs from Dharmacon (Lafayette, CO, USA) (Table 1). Transfection was performed by electroporation (Amaxa Nucleofector, Lonza, Cologne, Germany). In brief, the cells were harvested by trypsinization and resuspended at 1 × 106 cells in 100 µL Transfection Reagent solution V (Lonza), and mixed with 100 nM targeting siRNA or AllStars negative control siRNA (Qiagen). Immediately after electroporation, cells were transferred to complete medium and plated for analysis. Knockdown was confirmed by qRT-PCR as previously described [12] using Taqman probe Hs00181211_m1 for IGFBP-3, a custom synthesized probe for LINP1, and hydroxymethylbilane synthase (HMBS; Hs00609297_m1) as an internal control (Applied Biosystems, Foster City, CA, USA).
Table 1.
IGFBP-3 and LINP1 siRNAs
| Name | Sense strand |
|---|---|
| IGFBP-3 | |
| siRNA #1a | 5′ GUUGACUACGAGUCUCAGAUU 3′ |
| siRNA #2b | 5′ AGGUUAAUGUGGAGCUCAAUU 3′ |
| LINP1 | |
| siRNA #1c | 5′ UGAUUCAGCUGCAUAAAUAUU 3′ |
| siRNA #2c | 5′ GAUAGGAACCCCAGGGAAUUU 3′ |
| siRNA #3d | 5′ CACUCUCCAGCCUGCAAGAUU 3′ |
aDesigned by Qiagen (Hilden, Germany)
bCatalog No. SI02780589, Qiagen
cDesigned by Dharmacon (Lafayette, CO)
dReported by Zhang et al. [18]
Coimmunoprecipitation and western blotting
Immunoprecipitation of IGFBP-3 complexes using anti-IGFBP-3 IgG (Fab fraction) coupled to agarose beads was performed as previously described [10]. For immunoprecipitations using NONO and DNA-PK antibodies, cells (~ 1 × 106) were lysed in 1 mL ice-cold RIPA lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease (cOmplete™ Mini) and phosphatase (PhosSTOP™) inhibitors (Roche; Sigma-Aldrich, Sydney, Australia) at 4 °C for 1 h and spun at 10,000 × g for 10 min to pellet cell debris. Lysates were precleared by mixing with 20 µL of Protein A agarose beads (Roche; Sigma-Aldrich) for 1 h at 4 °C. Pre-cleared lysates were mixed overnight with specific antibodies and Protein A agarose beads (blocked by mixing with 1% BSA in RIPA buffer for 1 h at 4 °C). Antibodies used for IP were: NONO [N-terminal] (Sigma-Aldrich #N8789), 2.5 µg per sample; total DNA-PK (Santa Cruz Biotechnology #sc-9051), 2 µg per sample; phospho-DNA-PK[S2056] (Abcam #ab-18192), 2 µg per sample.
To prepare nuclear extracts for coIP, cellular fractionation was performed according to the manufacturer’s protocol for the NE-PER Nuclear and Cytoplasmic Extraction Kit (ThermoFisher).
Immunoprecipitated samples were resuspended in Laemmli sample buffer containing 50 mM dithiothreitol, heated at 95–100 °C for 6 min, and fractionated on 12% SDS-PAGE gels. Proteins were transferred to Protran® supported nitrocellulose membranes (Amersham, UK) at 160 mA for 2 h. Membranes were blocked in 50 g/L skim milk powder and probed with primary antibodies (SFPQ [EPR11847], 1:10,000, Abcam #ab177149; NONO (as above), 1:2000; IGFBP-3 [C19], 1:750, Santa Cruz Biotechnology #sc-6003; pEGFR [Y1068], 1:2000, Cell Signaling #2234; pDNA-PK (as above), 1:2000; total DNA-PK (as above), 1:1000; GAPDH [14C10], 1:2000, Cell Signaling #2118; Lamin B1, 1:2000, Abcam #ab16048) at 4 °C for 16 h. Immunoreactive bands were visualized as previously described [10].
PLA
PLA was performed using the Duolink Detection Kit (Olink Bioscience Uppsala, Sweden) as previously described [10]. Briefly, cells were grown on 8-mm glass coverslips to 50% confluency, treated, and prepared for microscopy by fixing, permeabilizing and blocking. Coverslips were incubated with primary antibody pairs (raised in different species) targeting the proteins under investigation overnight at 4 °C: SFPQ (as above), 1:500; NONO (as above), 1:500; IGFBP-3 (as above), 1:100. This was followed by incubation with PLA probes MINUS and PLUS for 1 h at 37 °C, probe ligation for 30 min at 37 °C and amplification over 100 min at 37 °C. Interactions were detected as amplified far-red signals using a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany) and quantitated using Image J software.
γH2AX immunofluorescence
Cells grown on 8-mm glass coverslips were washed three times with PBS, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 5 min and blocked with 2% BSA for 1 h. Cells were then incubated with rabbit anti-phospho-histone H2A.X (Ser139) (1:200; Cell Signaling Technology, #9718) overnight at 4 °C, washed, and further incubated with anti-rabbit secondary antibody, tagged with Alexa Fluor 594 (Life Technologies, Carlsbad, CA, USA). For controls, cells were treated with isotype-matched IgG from the same species. Slides were mounted using ProLong Gold Antifade Reagent (Life Technologies). Fluorescence images were captured by confocal laser scanning microscope. γH2AX fluorescence was quantitated in 5–6 fields for each condition using ImageJ (NIH, Bethesda, MD), and corrected for the number of nuclei per field (average = 14), visualized by DAPI staining. Data were calculated from three replicate experiments.
Discovery of IGFBP-3-interacting proteins
MDA-MB-468 cells were grown to 90% confluence in T75 flasks in RPMI 1640 medium containing 5% fetal calf serum and 10 µg/mL bovine insulin, then exposed to 20 µM etoposide, or medium alone for control cells, for 2 h. Medium was removed, and cells were washed twice in PBS, then lysed with 1 mL ice-cold RIPA buffer supplemented with protease and phosphatase inhibitors (as above) at 4 °C for 30 min. After centrifugation to remove insoluble material, the supernatant was incubated overnight with anti-IGFBP-3 IgG (Fab fraction) conjugated to agarose beads as previously described (10). Control precipitations used agarose beads without antibody. Beads were pelleted by centrifugation, washed 4 times in ice-cold PBS, resuspended in 50 µL 0.1% solution of RapiGest SF surfactant (Waters, Rydalmere, NSW, Australia) in 20 mM Tris–HCl buffer, pH 7.4. After boiling for 5 min to dissociate immunoprecipitated proteins, supernatants were collected by centrifugation and stored at − 80 °C before analysis. For proteomic analysis, tris(2-carboxyethyl)phosphine was added to 5 mM final concentration, samples were heated at 60 °C for 30 min, then cooled to room temperature. Iodoacetamide was added to 15 mM and reacted for 30 min in the dark. Trypsin Gold (MS grade; Promega, Alexandria, NSW, Australia) was added at 1:50 by protein weight, the solutions were incubated overnight at 37 °C, and TFA was added to 0.5% final. After 45 min at 37 °C, samples were immersed in liquid nitrogen to precipitate the RapiGest, then centrifuged for 10 min, and the supernatants collected. Samples were fractionated on an UltiMate 3000 nanoLC (Thermo Scientific) and spotted onto a Bruker MTP 384 AnchorChip target plate (Bruker, Preston, VIC, Australia) using a Proteineer fc II fraction collector (Bruker) as described previously [13]. MS/MS data were acquired on an UltrafleXtreme MALDI TOF/TOF mass spectrometer (Bruker) with a smart beam laser run at 2 kHz, with data processing and peptide identification performed as previously described [13].
NONO-IGFBP-3 binding assay
NONO was diluted in 50 mM sodium phosphate, 0.05% BSA, pH 7.4, and incubated 16 h at indicated concentrations in wells of FLAG (i.e. DDK) antibody plates. All incubations were at 22 °C in 100 µL of 0.1 M Tris–HCl, 0.05% BSA, pH 7.4 (incubation buffer) unless noted otherwise. After 4 washes with 250 µL cold incubation buffer, wells were incubated for 2 h at 22 °C with recombinant human IGFBP-3 at indicated concentrations in incubation buffer containing 1% BSA. After 4 washes as above, wells were incubated 2 h with anti-human IGFBP-3 antiserum R-100 at 1:25,000, washed 4 times, incubated 1 h with goat anti-rabbit IgG-HRP at 1:20,000, washed 4 times, and incubated 30 min with 100 µL TMB solution. Reactions were stopped by adding 100 µL 1 M H2SO4 and absorbance read at 450 nm.
DNA end-joining assay
Nuclear extraction and end-joining assay was performed as described by Andrin et al. [14, 15] with slight modifications. Briefly, HCC1806 cells were grown in flasks and treated with inhibitors for 24 h followed by etoposide treatment for 2 h as described in Materials and Methods. After isolation of nuclei by centrifugation through a buffer containing 300 mM sucrose, the washed nuclear pellet was extracted into high-salt buffer (20 mM Hepes, pH 7.5, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 1.5 mM MgCl2) for 30 min on ice, and insoluble material was removed by centrifugation. The soluble nuclear extract was used in the end-joining assay. Restriction enzymes NheI and EcoRI (New England Biolabs, Ipswich, MA, USA) were used to digest a EGFP-C1 plasmid (Clontech, Mountain View, CA, USA) to generate a DNA fragment of 4 kb with non-homologous ends. The linearized plasmid was separated by 0.8% agarose gel electrophoresis, purified using a DNA gel extraction kit (Qiagen), and used as the substrate for end-joining assays. Nuclear extract (2 µg) was mixed with end-joining assay buffer (7.5 mM Tris pH 8.0, 0.2 mM CaCl2, 10 mM MgCl2, 50 mM KCl, 1.2 mM ATP and 0.5 mM DTT) and allowed to stand for 30 min at 22 °C. Repair was initiated by adding 100 ng of prepared linearized DNA and incubated at 25 °C for 30 min, stopped by the addition of 0.5 M EDTA, 0.5% SDS and 10 mg/mL Proteinase K. DNA bands were separated on a 0.7% agarose gel, stained with SYBR Gold (Life Technologies), and visualized on a BioRad ChemiDoc imaging system.
Statistics
ANOVA with post hoc Fisher’s LSD test (SPSS v.22 for Mac; IBM Corp, Armonk, NY, USA) was used for multiple group comparisons.
Results
We previously reported that the formation of nuclear complexes involving IGFBP-3, DNA-PKcs and EGFR peaked 2–4 h after etoposide treatment in MDA-MB-468 basal-like breast cancer cells [10]. To discover other proteins involved in these interactions, we undertook an unbiased proteomic screen for proteins that interact with IGFBP-3 2 h after etoposide treatment. Examination by LC–MALDI–TOF/TOF mass spectrometry of proteins co-precipitating with IGFBP-3 from whole cell lysates consistently revealed both NONO and SFPQ as putative IGFBP-3 binding partners (Online Resource 1). Unique peptides for each protein, identified by mass spectrometry from IGFBP-3-coimmunoprecipitation (coIP) experiments, are shown in Online Resource 2. These interactions, and their stimulation by chemotherapy treatment, were confirmed by coIP and western blotting, and by proximity ligation assay (PLA). Figure 1a shows western blots of whole cell lysates from MDA-MB-468 cells treated with etoposide for 0–4 h, after immunoprecipitation using agarose-immobilized anti-human IGFBP-3 IgG (Fab fragment) or control, non-immune agarose beads. CoIP of both NONO and SFPQ typically peaked after 2 h exposure to etoposide, although the time-course was variable among experiments, with earlier (1 h) or later (4 h) peaks seen in some experiments. This variability may be related to the passage number of the cells, with the peak time tending to increase with extended passages after thawing. Weak bands for both antigens were also seen in some control IPs.
Fig. 1.
IGFBP-3 forms a complex with NONO and SFPQ in response to etoposide treatment. a MDA-MB-468 basal-like TNBC cells were exposed to 20 µM etoposide (Etop) for the indicated times, and IGFBP-3-interacting proteins precipitated from cell lysates by anti-IGFBP-3 antiserum (Fab fraction) coupled to agarose beads. Uncoupled agarose beads were used for IP controls. Samples were blotted for NONO and SFPQ after fractionation by SDS-PAGE. Panels on right show blots of whole-cell lysates without IP. Molecular weight markers are shown on the left. b IGFBP-3 was downregulated in MDA-MB-468 cells by siRNA, and cell lysates IP’d with IGFBP-3-Fab beads 2 h after etoposide stimulation. Precipitates were blotted for NONO, SFPQ, and IGFBP-3. c MDA-MB-468 cells were treated with etoposide, and nuclear extracts were prepared and immunoprecipitated with IGFBP-3-Fab beads. Panels on left show SFPQ, NONO, lamin B1 (nuclear marker), and GAPDH (cytoplasmic marker) in whole nuclear extracts (5% of immunoprecipitated sample). GAPDH in the whole cytoplasmic fraction, run on the same gel, is also shown for comparison. Panels on right show the same proteins after IP. Only 0 and 4 h time points are shown from a 4-h timecourse. For each analyte, all samples (input and IP) were run on the same gel. For SFPQ and NONO, but not Lamin B1 or GAPDH, the input blots shown were from shorter exposures, to avoid saturating the images. d Similar experiment to that shown in Fig. 1a, but in HCC1806 basal-like TNBC cells. e Quantitation of bands immunoblotted for NONO and SFPQ in HCC1806 cells. Data are mean band density ± SEM from 5 experiments. *P < 0.05 vs. time 0 by post hoc Fisher’s LSD test after ANOVA. f Binding of recombinant IGFBP-3 to immobilized recombinant NONO, measured in an ELISA format in which bound IGFBP-3 is immunodetected and quantitated colorimetrically at 450 nm. See Methods for details. Panels show dose–response curves for NONO (left) and IGFBP-3 (right)
When IGFBP-3 was downregulated transiently in MDA-MB-468 cells by siRNA, the amount of NONO and SFPQ detectable after IP with anti-human IGFBP-3, 2 h after etoposide treatment, was greatly reduced compared to that from cells treated with control non-silencing siRNA (Fig. 1b), supporting the interpretation that these proteins were precipitating in a complex containing IGFBP-3. Immunoprecipitated IGFBP-3 was detected as a diffuse band around 40 kDa (known to be a mixture of glycosylation isoforms) plus a weak band, probably proteolyzed IGFBP-3, below 30 kDa. An increase in IGFBP-3-associated NONO and SFPQ after etoposide treatment was similarly observed in isolated nuclear extracts rather than whole cell lysates (Fig. 1c). Similar to MDA-MB-468 cells, IGFBP-3-associated NONO and SFPQ also increased in HCC1806 basal-like breast cancer cells in response to etoposide, typically peaking 1–2 h after etoposide treatment (Fig. 1d). A representative image of immunoprecipitated IGFBP-3, measured in most coIP experiments, is also shown in Fig. 1d. In Fig. 1e, the association of NONO and SFPQ with IGFBP-3 in HCC1806 cells is quantitated for 5 experiments, the broad peaks representing the somewhat variable time-courses.
IGFBP-3 interaction with NONO was also examined in a cell-free system using a direct binding assay in which IGFBP-3 bound to immobilized NONO was detected in an ELISA format. Figure 1f shows dose–response curves for a fixed IGFBP-3 concentration (10 ng/100 µL; approx. 2.5 nM) bound to increasing concentrations of immobilized NONO, and for increasing IGFBP-3 concentrations bound to a fixed amount of NONO (25 ng/100 µL; approx. 4.6 nM). The NONO-IGFBP-3 interaction appears dose-dependent and saturable, consistent with NONO forming a specific protein–protein interaction with IGFBP-3.
Figure 2 confirms the association of NONO and SFPQ with IGFBP-3 in breast cancer cells by proximity ligation assay (PLA). Biomolecular interactions between IGFBP-3 and either binding partner are minimal before etoposide treatment, typically peaking 2 h after exposure to 20 µM etoposide and decreasing again at 4 h. In control PLA experiments, in which either detection antibody was omitted, no signal was observed (not shown). These independent approaches confirm that both NONO and SFPQ form transient nuclear complexes with IGFBP-3 in basal-like TNBC cells treated with etoposide.
Fig. 2.
IGFBP-3 complexes with NONO and SFPQ visualized by proximity ligation assay. a MDA-MB-468 and b HCC1806 TNBC cells were exposed to 20 µM etoposide (Etop) for the indicated times, and bimolecular interactions, shown as red dots, between IGFBP-3 and NONO or SFPQ, as indicated, were measured by PLA as described in the Methods. All SFPQ images in MDA-MB-468 cells have been equally enhanced using ImageJ software to make the red dots more visible. Bar 25 µm
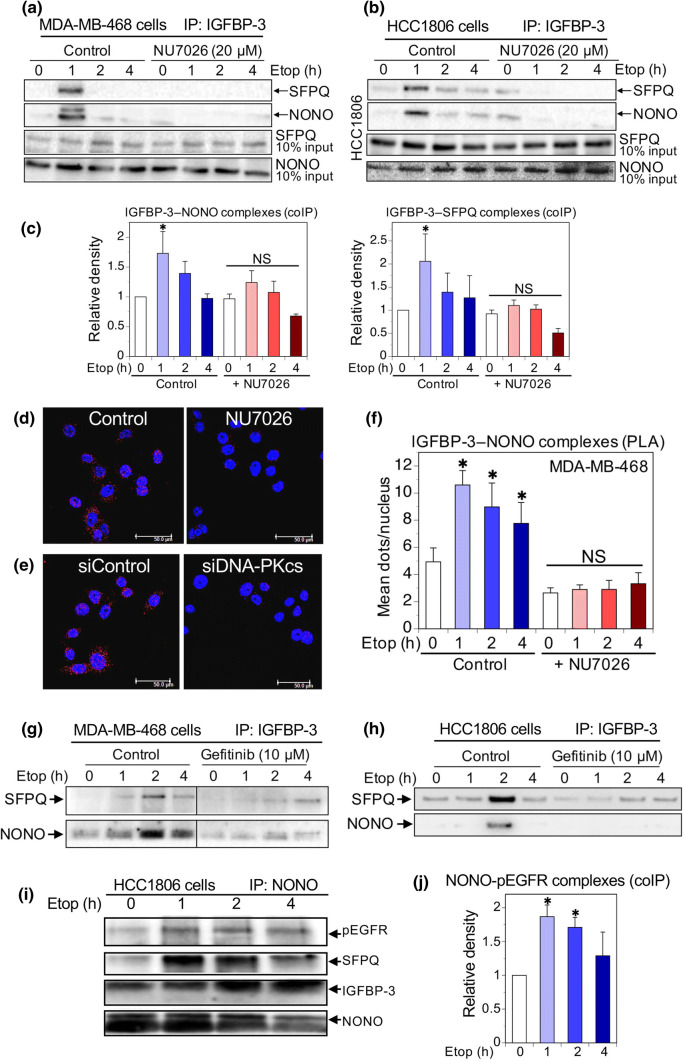
We previously reported that complex formation involving IGFBP-3, DNA-PKcs and EGFR in response to DNA-damaging chemotherapy requires autophosphorylation of DNA-PKcs as well as EGFR kinase activity [10]. To determine whether the IGFBP-3 complexes with NONO and SFPQ require DNA-PKcs activation, we used the inhibitor NU7026, which blocks DNA-PKcs activity with an IC50 of 0.23 µM [16]. As shown in Fig. 3a, preincubation of MDA-MB-468 cells overnight with 20 µM NU7026 blocked the ability of etoposide to increase complex formation between IGFBP-3 and NONO/SFPQ, as determined by IGFBP-3 coIP experiments. A similar inhibitory effect was seen using another DNA-PKcs inhibitor, NU7441 (not shown). Inhibition by NU7026 was also observed in HCC1806 cells (Fig. 3b). Summary data for four experiments in MDA-MB-468 cells are shown in Fig. 3c, showing a significant stimulatory effect of etoposide on complex formation (with a mean peak time of 1 h in these experiments), abolished in cells preincubated with NU7026. This inhibitory effect was also visualized by PLA (Fig. 3d), in which preincubation with 10 µM NU7026 essentially abolishes NONO-IGFBP-3 interaction in MDA-MB-468 cells. Similarly, in Fig. 3e, downregulation of DNA-PKcs with siRNA also abolished the NONO-IGFBP-3 interaction. Figure 3f summarizes the inhibition of IGFBP-3–NONO complexes, detected by PLA, by NU7026 for three experiments.
Fig. 3.
Inhibition of the IGFBP-3 interaction with NONO and SFPQ. a Western blots of NONO and SFPQ in MDA-MB-468 cell lysates immunoprecipitated with IGFBP-3-Fab beads, showing that incubation overnight with 20 µM NU7026 blocked the formation of IGFBP-3 complexes with NONO and SFPQ formed in response to 20 µM etopside. b Similar experiment in HCC1806 cells. c Quantitation of IGFBP-3–NONO and IGFBP-3–SFPQ complexes in MDA-MB-468 cells: mean relative band density (normalized to time 0 controls) ± SEM from 4 experiments. *P < 0.05 vs. the corresponding time 0 by post hoc Fisher’s LSD test after ANOVA. NS, not significant. d, e Proximity ligation assays in MDA-MB-468 cells, after 2 h etoposide treatment showing that NONO-IGFBP-3 complexes (red dots) are inhibited in cells preincubated overnight with DNA-PKcs inhibitor NU7026, 10 µM (d), or 48 h after DNA-PKcs knockdown with siRNA (e). Blue = nuclei (DAPI). Bar 50 µm. f Quantitation of inhibition by NU7026 of IGFBP-3 interaction with NONO measured by PLA; 5 fields (~ 20 nuclei/field) counted for each condition and each time-point in each experiment. Means ± SEM for 3 replicate experiments. *P < 0.05 vs. the corresponding time 0 by post hoc Fisher’s LSD test after ANOVA. NS, not significant. g, h Western blots of NONO and SFPQ in MDA-MB-468 and HCC1806 cell lysates, respectively, IP’d with IGFBP-3-Fab beads, showing that incubation overnight with 10 µM gefitinib blocked the formation of IGFBP-3 complexes with NONO and SFPQ in response to 20 µM etopside. i Western blots showing NONO, SFPQ, IGFBP-3, and pEGFR (Y1068) in HCC1806 cell lysates immunoprecipitated with NONO Ab. NONO bands are partly obscured by strong IgG heavy-chain bands. j Quantitation of pEGFR immunoprecipitated in NONO complexes in HCC1806 cells: mean relative band density (normalized to time 0 controls) ± SEM from 4 experiments. *P < 0.05 vs. time 0 values by post hoc Fisher’s LSD test after ANOVA
To support the concept that phospho-DNA-PKcs (pDNA-PKcs) and NONO–SFPQ form part of the same complex, we undertook further immunoprecipitations using antibodies against total DNA-PKcs (t-DNA-PKcs) and pDNA-PKcs. As seen in Online Resource 3, Suppl Fig. 1, IP with t-DNA-PKcs Ab from HCC1806 cell lysates showed an apparent increase in complexed SFPQ over 4 h following etoposide treatment, but this was not statistically significant by ANOVA. In contrast, IP with pDNA-PKcs Ab showed a stronger peak of complexed SFPQ, reproducibly occurring 4 h after etoposide exposure (P < 0.005 vs. all other time points), even though the peak of DNA-PK phosphorylation appeared earlier, at 2 h. Complementing the inhibitor experiments in Fig. 3, this suggests that DNA-PKcs must preferentially be phosphorylated to participate in this complex. Why the pDNA-PKcs-SFPQ interaction appears to peak later than the peak of DNA-PKcs phosphorylation remains unclear.
EGFR phosphorylation was also necessary for these complexes to form. As shown in Fig. 3g, h, preincubation with the EGFR kinase inhibitor gefitinib before exposing cells to etoposide also prevented IGFBP-3–NONO/SFPQ complexes, measured by coIP in both MDA-MB-468 and HCC1806 cells. To further demonstrate that these proteins appear part of a multi-protein complex, we undertook immunoprecipitations from etoposide-treated HCC1806 cells using NONO antibody. Figure 3i shows NONO, SFPQ, IGFBP-3, and pEGFR (Tyr1068) in these immunoprecipitates, with NONO-pEGFR complexes quantitated from 4 experiments in Fig. 3j. In conjunction with our earlier findings [10], these experiments are consistent with the involvement of IGFBP-3 in a multi-protein DNA repair complex that involves activated DNA-PKcs and EGFR as well as the DNA- and RNA-binding proteins, NONO and SFPQ.
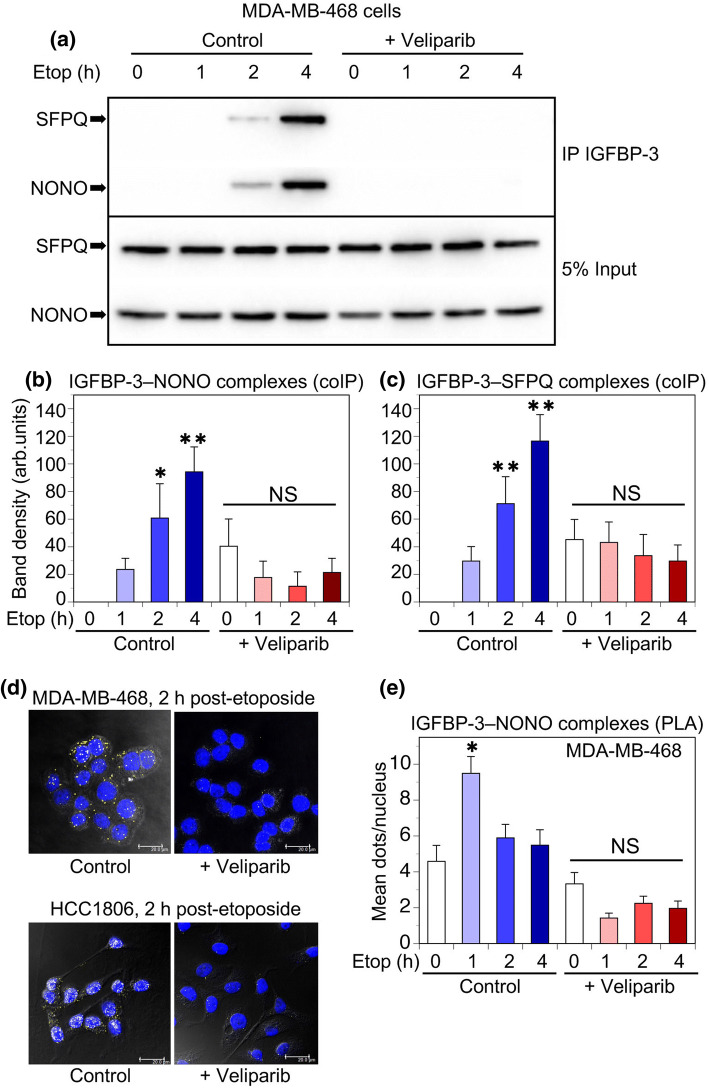
Since NONO recruitment to DNA damage sites is reported to be PARP-dependent [8], we examined the effect of PARP inhibition on IGFBP-3 interactions with both NONO and SFPQ. Figure 4a shows in MDA-MB-468 cells that IGFBP-3 complexes with NONO and SFPQ, determined by immunoblotting after coIP, were abolished if cells were preincubated with the PARP1 and PARP2 inhibitor veliparib (20 µM) for 24 h prior to exposure to etoposide. Data for 3 experiments in MDA-MB-468 cells are summarized in Fig. 4b, c for IGFBP-3–NONO and IGFBP-3–SFPQ interactions, respectively. A similar inhibitory effect was seen after preincubation with a second PARP inhibitor, olaparib, at 10 µM (Online Resource 3, Suppl Fig. 2). The inhibitory effect of veliparib on complex formation was confirmed by PLA in both MDA-MB-468 and HCC1806 cells (Fig. 4d), showing the increase in IGFBP-3–NONO complexes 2 h after etoposide treatment was abolished by preincubation with 20 µM veliparib. Figure 4e shows the quantitation of 3 replicate experiments in MDA-MB-468 cells, with the effect of veliparib highly significant by ANOVA (P < 0.001). Therefore, the formation of EGFR-dependent complexes between IGFBP-3 and NONO/SFPQ in basal-like TNBC cell lines exposed to DNA-damaging chemotherapy requires PARP activity.
Fig. 4.
PARP inhibition blocks IGFBP-3 complex formation with NONO and SFPQ. a MDA-MB-468 cells were incubated 24 h with 20 µM veliparib (ABT-888) before exposure to 20 µM etoposide. NONO and SFPQ complexes were IP’d with IGFBP-3-Fab beads and detected by immunoblotting. b, c Quantitation of bands immunoblotted for NONO and SFPQ in MDA-MB-468 cells. Data are mean band density ± SEM from 4 experiments. *P < 0.05, **P < 0.005 vs. the corresponding time 0 by post hoc Fisher’s LSD test after ANOVA. NS, not significant. d PLA showing interactions (yellow dots) between IGFBP-3 and NONO in MDA-MB-468 cells (above) and HCC1806 cells (below) after 2 h treatment with 20 µM etoposide, following 24 h preincubation ± 20 µM veliparib. Blue = nuclei (DAPI). Bar 20 µm. Confocal images superimposed over phase contrast images. e Quantitation of inhibition by 20 µM veliparib of IGFBP-3 interaction with NONO over 4 h of etoposide treatment in MDA-MB-468 cells, measured by PLA; 5 fields (~ 20 nuclei/field) counted for each condition and each time-point in each experiment. Data are mean values ± SEM from 3 experiments. *P < 0.001 vs. the corresponding time 0 by post hoc Fisher’s LSD test after ANOVA. NS, not significant
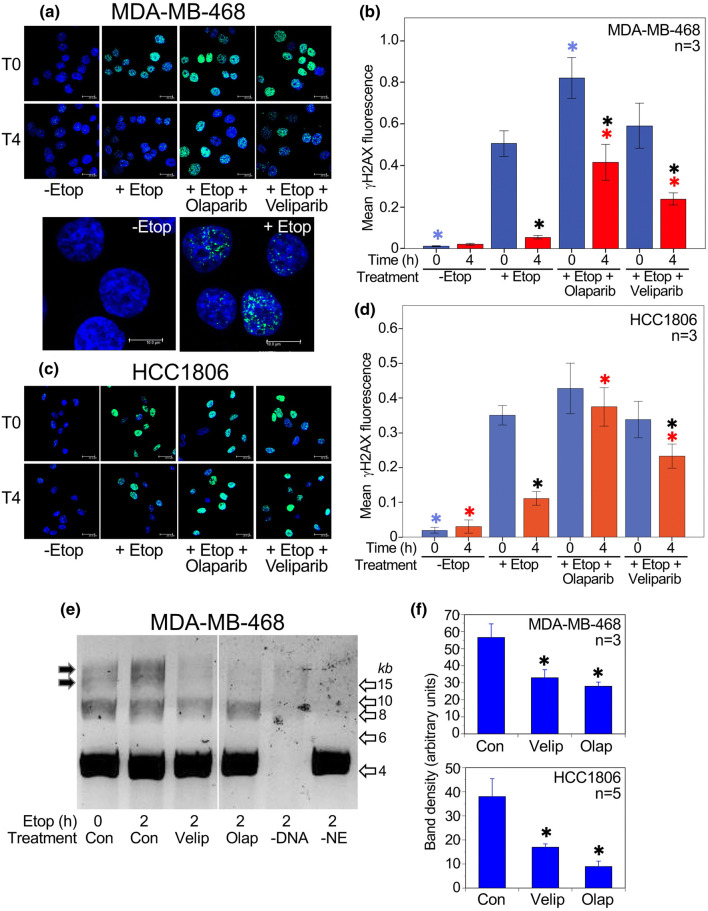
Consistent with this, DNA repair activity in TNBC cell lines was inhibited by PARP inhibitors. As shown in Fig. 5a, c, treatment of either MDA-MB-468 or HCC1806 cells with etoposide for 1 h (T0) caused a significant increase in foci of histone H2AX phosphorylated on serine 139 (γH2AX), which accumulates at sites of DNA double-strand breaks [17]. This signal had substantially declined after 4 h (T4), consistent with DNA repair over this period. The addition of either PARP inhibitor, olaparib or veliparib, significantly prevented the loss of γH2AX signal, indicating that both drugs were inhibitory to DSB repair in these cell lines, neither of which has a mutation in BRCA1 or BRCA2. Data for both cell lines are quantitated in Fig. 5b, d. Etoposide treatment also increased activity in a direct DNA end-joining assay using nuclear extracts from treated cells (Fig. 5e). In extracts from cells treated with either PARP inhibitor, end-joining activity was inhibited by approximately 50% in both MDA-MB-468 and HCC1806 cells, as shown quantitatively in Fig. 5f.
Fig. 5.
PARP inhibition decreases DNA end-joining in TNBC cells. a Upper panels: γH2AX immunofluorescence in MDA-MB-468 cells at time 0 (i.e. 1 h after exposure to etoposide) and after 4 h of recovery (T4). Cells were pre-treated with olaparib (10 µM) or veliparib (20 µM) as indicated. Bar 20 µm. Lower panels: representative images at higher power of T0 cells ± etoposide, to illustrate the punctate γH2AX fluorescence. Bar 10 µm. b Mean fluorescence values (arbitrary units) ± SEM are shown from 3 experiments. c, d γH2AX immunofluorescence in HCC1806 cells, with quantitation from 3 experiments, details as for panels a and b. Bar 20 µm. ANOVA with Fisher’s LSD post hoc LSD test: *(blue) P < 0.05 vs. T0 + etoposide; *(red) P < 0.05 vs. T4 + etoposide; *P < 0.05 vs. the corresponding T0 value. e DNA end-joining assay: cells were treated with inhibitors (20 µM veliparib or 10 µM olaparib) or no inhibitor (Con) for 24 h, then exposed to 20 µM etoposide for 2 h. In control lanes (right), DNA or nuclear extract (NE) has been omitted. After adding nuclear extract for 30 min, substrate DNA was added and end-joining proceeded for 30 min at 25 °C. A representative gel is shown for MDA-MB-468 cells. Black arrows show the bands quantitated. Open arrow show size markers in kb. All lanes are from a single gel. f Upper panel: Quantitation of end-joining activity 2 h after etoposide in MDA-MB-468 cells, mean ± SEM, n = 3. Lower panel: Quantitation of end-joining activity 2 h after etoposide in HCC1806 cells, mean ± SEM, n = 5. *P < 0.05 vs. control by post hoc Fisher’s LSD test after ANOVA
NONO and SFPQ are RNA-binding proteins known to interact with several lncRNAs [18]. Since there is evidence for the role of lncRNAs in DNA DSB repair [19, 20], we questioned whether lncRNAs were involved in EGFR-dependent IGFBP-3 complexes with NONO/SFPQ in TNBC. A recently identified lncRNA, LINP1, was reported to be highly expressed in TNBC, regulated by EGFR signaling, and involved in DNA-PK binding during NHEJ [21]; therefore, we investigated whether LINP1 might be involved in IGFBP-3-NONO–SFPQ complexes associated with NHEJ in TNBC cell lines. Data in Online Resource 4, Suppl Fig. 3a, shows the relative expression of LINP1, measured by qPCR, in four TNBC cell lines and the triple-negative, phenotypically normal mammary epithelial cell line, MCF-10A. Relative to MDA-MB-468 cells, MDA-MB-231, Hs578T and HCC1806 cells each have 15- to 40-fold higher LINP1 expression, whereas MCF-10A has similar expression to MDA-MB-468.
Since LINP1 was reported as being upregulated by EGFR in MDA-MB-468 and MCF10A cells [21], we examined the effect of exposure to EGF and the EGFR inhibitor, gefitinib, which prevents the interaction between IGFBP-3 and NONO/SFPQ (Fig. 3g). Online Resource 4 (Suppl Fig. 3c) shows that in MDA-MB-468 cells, which express low LINP1, exposure to 50 ng/mL EGF for 4 h caused a mean 4.3-fold increase in LINP1 expression. Similarly, in the low-expressing MCF-10A cells, EGF stimulated LINP1 expression 2.7-fold (Online Resource 4, Suppl Fig. 3d). In contrast, in Hs578T, MDA-MB-231, and HCC1806 cells, with high basal LINP1 expression, EGF did not further increase LINP1 (Online Resource 4, Suppl Fig. 3e–g). In the highest-expressing HCC1806 cell line EGF actually caused a significant 23% decline in LINP1 expression over 4 h (Online Resource 4, Suppl Fig. 3g). The inverse regulation by EGFR signaling in HCC1806 cells was also seen when cells were exposed to 10 µM gefitinib, resulting in a 40% increase in LINP1 expression (Online Resource 4, Suppl Fig. 3h). Together these findings suggest that the ability of EGF to stimulate LINP1 expression is inversely related to basal LINP1 expression, as represented by the significant curve fit shown in Online Resource 4, Suppl Fig. 3b (R2 = 0.956).
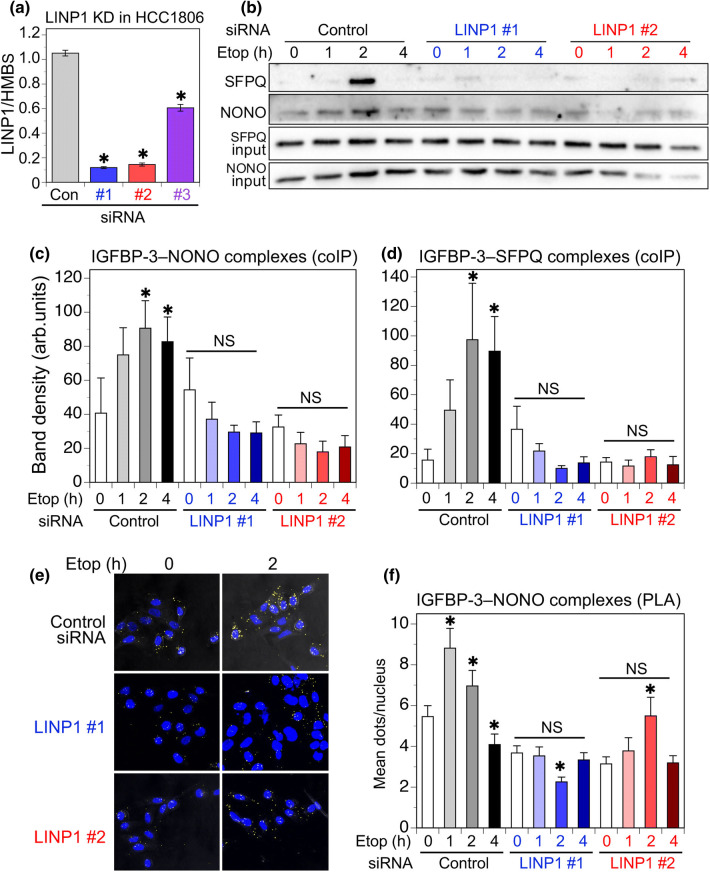
To investigate whether LINP1 has a role in DNA repair complexes involving IGFBP-3, siRNAs were designed to transiently downregulate LINP1 in the high-expressing cell line, HCC1806. A published LINP1 siRNA [21] was also evaluated. The sequences are shown in Table 1. As shown in Fig. 6a, both custom-designed siRNAs (#1 and #2) downregulated LINP1 by 85–90%, 48 h after transfection, whereas the published siRNA (#3) was less effective under our conditions. Figure 6b shows IGFBP-3 complexes with NONO and SFPQ in HCC1806 cells treated with 20 µM etoposide, 24 h after LINP1 downregulation by siRNA #1 or #2. Both IGFBP-3–NONO and IGFBP-3–SFPQ complexes appear to require LINP1 as their formation is significantly inhibited when LINP1 is downregulated. Quantitation from 5 experiments is shown in Fig. 6c, d. The effect of LINP1 downregulation was also observed by PLA for IGFBP-3–NONO interactions (Fig. 6e). Quantitation from 4 experiments (Fig. 6f) again shows a significant inhibition of IGFBP-3–NONO interaction in response to etoposide, after LINP1 downregulation. The studies point to a facilitating role of LINP1 in the DNA repair complexes involving IGFBP-3 that form in response to DNA-damaging chemotherapy.
Fig. 6.
Effect of LINP1 downregulation in HCC1806 cells. aLINP1 expression, measured by qPCR relative to HMBS, after transient transfection of two custom siRNAs (#1, #2), and a previously reported [21] siRNA (#3). Mean values ± SEM, 3 experiments in duplicate. *P < 0.001 vs. non-silencing control. b IGFBP-3 complexes with NONO and SFPQ following etoposide (Etop) stimulation, measured by coIP and immunoblotting, were decreased in cells with LINP1 downregulation. c, d Quantitation of IGFBP-3–NONO and IGFBP-3–SFPQ coIP complexes: mean band density ± SEM from 5 experiments. *P < 0.05 vs. corresponding time 0, by post hoc Fisher’s LSD test after ANOVA. NS: not significantly different from time 0. e IGFBP-3 complexes with NONO measured by PLA were decreased in cells with LINP1 downregulation. Representative images are from 2 h etoposide-treated cells previously transfected with non-silencing control siRNA or LINP1 siRNA #1 or #2. f Quantitation of PLA dots, 5 fields (average 13.7 nuclei/field) counted for each condition and time-point in each experiment. Means ± SEM for 4 replicate experiments. *P < 0.001 vs. the corresponding time 0, by post hoc Fisher’s LSD test after ANOVA. NS: not significantly different from time 0
Discussion
Controlling the cancer cell response to DNA damage induced by chemotherapy or radiotherapy is an important tool in overcoming treatment resistance. By implicating the DNA- and RNA-binding heterodimerization partners, NONO and SFPQ, in a DNA repair complex that also includes IGFBP-3, EGFR, and DNA-PKcs [10], this study provides further support for the role of IGFBP-3 in the PARP-dependent DNA damage response in TNBC cells. Since the repair of DSBs opposes the effects of DNA-damaging therapies [22], its inhibition has been extensively studied as an approach to enhancing cell sensitivity to DNA lesions caused by these treatments [1]. NHEJ, which can occur at any stage of the cell cycle, is regarded as the most commonly used mechanism of DSB repair in mammalian cells, including during late S to G2 phase when homologous recombination repair may also occur [23, 24]. In this study, we induced DNA damage with the topoisomerase II poison, etoposide, which generates DSBs that are predominantly repaired by NHEJ [25]. Among the possible mechanisms that may influence the balance between HR and NHEJ repair pathways [24], binding of the NONO–SFPQ complex was found to stimulate end-joining activity [26] and, explicitly, the recruitment of NONO–DNA damage sites, following its PARylation by PARP1, has been shown to enhance NHEJ and suppress HR activity [8].
Our proteomic search for proteins that associate with IGFBP-3 in response to etoposide treatment of TNBC cells discovered that NONO and SFPQ appear to form part of a nuclear complex that, together with IGFBP-3, also involves EGFR and DNA-PKcs. In the two basal-like TNBC cell lines examined, formation of this complex peaked approximately 2 h after exposure to etoposide, with a range of 1–4 h. NONO and SFPQ are multifunctional proteins with a predominantly nuclear localization [27], although extranuclear functions have also been described [27, 28]. We observed nuclear complexes between NONO/SFPQ and IGFBP-3 by both PLA and coIP from nuclear extracts, with some PLA complexes also apparent extranuclearly. SFPQ extranuclear localization has been associated with doxorubicin sensitivity [28], whereas its nuclear involvement in DNA damage repair [9] would promote chemoresistance.
The lncRNA LINP1 appears to facilitate the interaction between NONO/SFPQ and IGFBP-3, at least in HCC1806 cells, since its downregulation by two siRNAs blocked complex formation as measured by coIP and PLA. LINP1 was investigated because it was reported as a regulator of DSB repair by NHEJ in TNBC cell lines [21]. Subsequently, LINP1 has been shown to affect both chemosensitivity [29] and radiosensitivity [30] through its role in DSB repair. However, the regulation of LINP1 by EGFR signaling in breast cancer cell lines appeared more complex than originally described [21], since cell lines with low LINP1 expression (MDA-MB-468, MCF-10A) showed upregulation by EGF as reported, whereas cell lines with intermediate LINP1 expression (Hs578T, MDA-MB-231) were unaffected by EGF, and in the highest-expressing cell line (HCC1806), LINP1 was significantly downregulated by EGF. Because LINP1 expression is extremely low in MDA-MB-468 cells, we only undertook downregulation experiments in HCC1806, showing inhibition of the interaction between NONO/SFPQ and IGFBP-3 when LINP1 was downregulated. The modest upregulation of LINP1 in HCC1806 cells by the EGFR inhibitor gefitinib might, therefore, be expected to have a positive effect on NONO/SFPQ interaction with IGFBP-3, whereas we found that gefitinib in fact prevents these interactions. This is consistent with our previous observation that EGFR kinase inhibition blocks the interaction of IGFBP-3 with DNA-PKcs [10], and supports the importance of autophosphorylated EGFR as a component of the DNA-PK-dependent DNA repair complex [31].
NONO and SFPQ are RNA-binding proteins, and their interaction with several lncRNAs has been reported, including NEAT1 [18, 32], MALAT1 [33], and GAPLINC [34]. The functional role we have demonstrated for LINP1 in IGFBP-3 complexes with NONO/SFPQ suggests that LINP1 may also interact directly with these proteins, although a direct interaction was not investigated. LINP1 was reported to act as a scaffold between DNA-PKcs and Ku80 [21], and the recent demonstration that the NONO/SFPQ-binding lncRNA, NEAT1, is involved in a complex involving DNA-PKcs and Ku70/Ku80, as well as NONO and SFPQ [32], provides a parallel to the DNA repair complex we propose involving IGFBP-3, EGFR, and DNA-PKcs in addition to NONO/SFPQ and LINP1.
Our discovery that IGFBP-3 is involved in DNA repair by NHEJ [10] was unexpected because IGFBP-3 is pro-apoptotic in many contexts. This is, in part, mediated by its ability to inhibit pro-survival IGF1R signaling, but IGF1R-independent effects have also been shown in many laboratories [35–39]. In contrast, IGFBP-3 can promote cell survival by enhancing GRP78-dependent autophagy [40], stimulating sphingosine kinase 1 activity [41], and activating MAPK signaling through integrin β1 binding [42]. IGFBP-3 is known to translocate to the cell nucleus under some conditions, and its nuclear functions have been reviewed previously [43]. Although this report is the first to describe NONO and SFPQ as IGFBP-3 binding partners, they are known to interact with the DNA binding domain of the retinoid X receptor (RXR) and the thyroid receptor [44], nuclear receptors that also bind IGFBP-3 [43]. Since IGFBP-3 regulates the transcriptional activity of these and other nuclear receptors [43, 45], it is possible that NONO and SFPQ are involved in other nuclear pathways involving IGFBP-3 in addition to DNA repair by NHEJ.
Inhibition of PARP1, which acts in the repair of DNA single-strand breaks (SSBs) by the base excision repair pathway, causes an accumulation of SSBs. During DNA replication these lead to stalling at replication forks, generating DSBs that would be repaired by homology-directed repair; accordingly, PARP1 inhibitors are lethal in cells deficient in HR [5, 46]. Although PARP1 inhibitors are generally used clinically for this purpose (e.g. for BRCA-mutated tumors), they are now recognized as also affecting repair by DNA end-joining [5]. This may in part involve alternative end-joining pathways [47], but there is also evidence of a role for PARP1 in classical NHEJ [5]. As noted earlier, the PARP1 inhibitor veliparib (ABT-888) was found to inhibit NHEJ by blocking the recruitment of PARylated NONO to DNA-PKcs [8], and DNA-PKcs itself has also been described as a PARP substrate, its activity stimulated by PARP-mediated ADP-ribosylation [48]. The repair complex we describe in this study, involving IGFBP-3 and the NONO/SFPQ heterodimer, requires active (autophosphorylated) DNA-PKcs for its formation, as it was inhibited by both the ATP-competitive DNA-PK inhibitor NU7026 and by DNA-PKcs downregulation by siRNA.
We have previously shown that IGFBP-3 binds to DNA-PKcs in response to etoposide treatment in TNBC cell lines [10], and that IGFBP-3 is a substrate for DNA-PKcs kinase activity [49]; this phosphorylation appears necessary for IGFBP-3 to exert a pro-survival effect in retinal endothelial cells [50]. In TNBC cells, IGFBP-3 downregulation inhibits DNA-PKcs autophosphorylation at Ser2056, DNA-PKcs interaction with EGFR, and DNA end-joining activity [10]. In the present study, etoposide-stimulated complex formation was also blocked by the PARP inhibitors olaparib and veliparib in our cell lines, as was the loss of γH2AX foci and DNA end-joining activity. Veliparib has previously been shown to decrease nuclear EGFR and attenuate NHEJ in response to radiation [7], and combining EGFR inhibition with veliparib prolongs γH2AX foci in TNBC cell lines, and inhibits TNBC tumor growth in a mouse xenograft model [51]. Given that etoposide-induced DNA damage is most likely to be repaired by NHEJ [25], our evidence is consistent with the involvement of IGFBP-3 in the classical NHEJ pathway with blockade by PARP inhibition, although the involvement of DNA-PKcs in the repair of stalled DNA replication forks [52] illustrates the complexity of cross-talk among various DNA repair pathways.
Conclusions
This study advances understanding of the complexity of DNA double-strand break repair in response to chemotherapy in TNBC cell lines, by demonstrating the involvement of the DNA- and RNA-binding heterodimerization partners, NONO and SFPQ, and the facilitating role of the lncRNA, LINP1, in the nuclear protein complex previously shown to include IGFBP-3, EGFR, and DNA-PKcs [10]. Both IGFBP-3 and EGFR are abundant in basal-like TNBC, although generally expressed poorly in other breast cancer subtypes. Women with TNBC typically have lower 5-year survival rates than those with other breast cancer types, and relapse early after treatment is a common feature [53]. We propose that disruption of the proposed nuclear repair complex, by identifying and blocking one or more key protein–protein interactions, may complement the inhibition of key post-translational modifications (e.g. DNA-PKcs or EGFR autophosphorylation, or protein PARylation) to provide a novel approach to enhancing sensitivity to chemo- and radiotherapy in TNBC, and so offer an opportunity to improve patient survival rates.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Grants DP140100137 from the Australian Research Council and IN-17-040 from the National Breast Cancer Foundation, Australia, to RCB.
Author contributions
The study was designed by RCB. Data were collected and analysed by HCdS, MZL, LP, JLM and RCB. The manuscript was drafted by RCB. All authors revised the article critically for intellectual content and gave their approval of this version to be published.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khanna A. DNA damage in cancer therapeutics: a boon or a curse? Cancer Res. 2015;75:2133–2138. doi: 10.1158/0008-5472.CAN-14-3247. [DOI] [PubMed] [Google Scholar]
- 2.Rulten SL, Grundy GJ. Non-homologous end joining: common interaction sites and exchange of multiple factors in the DNA repair process. BioEssays. 2017;39:1600209. doi: 10.1002/bies.201600209. [DOI] [PubMed] [Google Scholar]
- 3.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer—the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 4.Abbotts R, Wilson DM., 3rd Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 7.Nowsheen S, Bonner JA, Yang ES. The poly(ADP-Ribose) polymerase inhibitor ABT-888 reduces radiation-induced nuclear EGFR and augments head and neck tumor response to radiotherapy. Radiother Oncol. 2011;99:331–338. doi: 10.1016/j.radonc.2011.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krietsch J, Caron MC, Gagne JP, Ethier C, Vignard J, Vincent M, Rouleau M, Hendzel MJ, Poirier GG, Masson JY. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucl Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaafar L, Li Z, Li S, Dynan WS. SFPQ*NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining. Nucl Acids Res. 2017;45:1848–1859. doi: 10.1093/nar/gkw1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin MZ, Marzec KA, Martin JL, Baxter RC. The role of insulin-like growth factor binding protein-3 in the breast cancer cell response to DNA-damaging agents. Oncogene. 2014;33:85–96. doi: 10.1038/onc.2012.538. [DOI] [PubMed] [Google Scholar]
- 11.Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer. 2014;14:329–341. doi: 10.1038/nrc3720. [DOI] [PubMed] [Google Scholar]
- 12.Martin JL, de Silva HC, Lin MZ, Scott CD, Baxter RC. Inhibition of insulin-like growth factor-binding protein-3 signaling through sphingosine kinase-1 sensitizes triple-negative breast cancer cells to EGF receptor blockade. Mol Cancer Therap. 2014;13:316–328. doi: 10.1158/1535-7163.MCT-13-0367. [DOI] [PubMed] [Google Scholar]
- 13.Hunt NJ, Phillips L, Waters KA, Machaalani R. Proteomic MALDI-TOF/TOF-IMS examination of peptide expression in the formalin fixed brainstem and changes in sudden infant death syndrome infants. J Proteom. 2016;138:48–60. doi: 10.1016/j.jprot.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Andrin C, Hendzel MJ. F-actin-dependent insolubility of chromatin-modifying components. J Biol Chem. 2004;279:25017–25023. doi: 10.1074/jbc.M401805200. [DOI] [PubMed] [Google Scholar]
- 15.Andrin C, McDonald D, Attwood KM, Rodrigue A, Ghosh S, Mirzayans R, Masson JY, Dellaire G, Hendzel MJ. A requirement for polymerized actin in DNA double-strand break repair. Nucleus. 2012;3:384–395. doi: 10.4161/nucl.21055. [DOI] [PubMed] [Google Scholar]
- 16.Hollick JJ, Golding BT, Hardcastle IR, Martin N, Richardson C, Rigoreau LJ, Smith GC, Griffin RJ. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-dependent protein kinase (DNA-PK) Bioorg Med Chem Lett. 2003;13:3083–3086. doi: 10.1016/S0960-894X(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 17.Scully R, Xie A. Double strand break repair functions of histone H2AX. Mutat Res. 2013;750:5–14. doi: 10.1016/j.mrfmmm.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dianatpour A, Ghafouri-Fard S. The role of long non coding RNAs in the repair of DNA double strand breaks. Int J Mol Cell Med. 2017;6:1–12. [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Wang Y. Studies of lncRNAs in DNA double strand break repair: what is new? Oncotarget. 2017;8:102690–102704. doi: 10.18632/oncotarget.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, Yuan J, Shan W, Li C, Hu X, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai A, Yan Y, Gerson SL. Advances in therapeutic targeting of the DNA damage response in cancer. DNA Repair (Amst) 2018;66–67:24–29. doi: 10.1016/j.dnarep.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end joining for repair of DNA double-strand breaks. J Biol Chem. 2018;293:10512–10523. doi: 10.1074/jbc.TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. Excli J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udayakumar D, Dynan WS. Characterization of DNA binding and pairing activities associated with the native SFPQ. NONO DNA repair protein complex. Biochem Biophys Res Commun. 2015;463:473–478. doi: 10.1016/j.bbrc.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knott GJ, Bond CS, Fox AH. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucl Acids Res. 2016;44:3989–4004. doi: 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren S, She M, Li M, Zhou Q, Liu R, Lu H, Yang C, Xiong D. The RNA/DNA-binding protein PSF relocates to cell membrane and contributes cells’ sensitivity to antitumor drug, doxorubicin. Cytometr A. 2014;85:231–241. doi: 10.1002/cyto.a.22423. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Li Y, Song X, Zhang N, Sang Y, Zhang H, Liu Y, Chen B, Zhao W, Wang L, et al. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol Ther. 2018;19:120–131. doi: 10.1080/15384047.2017.1394543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Liu H, Shi L, Yu X, Gu Y, Sun X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle. 2018;17:439–447. doi: 10.1080/15384101.2018.1442625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodemann HP, Dittmann K, Toulany M. Radiation-induced EGFR-signaling and control of DNA-damage repair. Int J Radiat Biol. 2007;83:781–791. doi: 10.1080/09553000701769970. [DOI] [PubMed] [Google Scholar]
- 32.Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, Dubois E, Schwartz O, Bennasser Y, Benkirane M. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67:387–399. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 33.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 36.Lee KW, Cohen P. Nuclear effects: unexpected intracellular actions of insulin-like growth factor binding protein-3. J Endocrinol. 2002;175:33–40. doi: 10.1677/joe.0.1750033. [DOI] [PubMed] [Google Scholar]
- 37.Ingermann AR, Yang YF, Han J, Mikami A, Garza AE, Mohanraj L, Fan L, Idowu M, Ware JL, Kim HS, et al. Identification of a novel cell death receptor mediating IGFBP-3-induced anti-tumor effects in breast and prostate cancer. J Biol Chem. 2010;285:30233–30246. doi: 10.1074/jbc.M110.122226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agostini-Dreyer A, Jetzt AE, Stires H, Cohick WS. Endogenous IGFBP-3 mediates intrinsic apoptosis through modulation of Nur77 phosphorylation and nuclear export. Endocrinology. 2015;156:4141–4151. doi: 10.1210/en.2015-1215. [DOI] [PubMed] [Google Scholar]
- 39.Hollowood AD, Lai T, Perks CM, Newcomb PV, Alderson D, Holly JM. IGFBP-3 prolongs the p53 response and enhances apoptosis following UV irradiation. Int J Cancer. 2000;88:336–341. doi: 10.1002/1097-0215(20001101)88:3<336::AID-IJC3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 40.Grkovic S, O’Reilly VC, Han S, Hong M, Baxter RC, Firth SM. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene. 2013;32:2412–2420. doi: 10.1038/onc.2012.264. [DOI] [PubMed] [Google Scholar]
- 41.Granata R, Trovato L, Garbarino G, Taliano M, Ponti R, Sala G, Ghidoni R, Ghigo E. Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: involvement of the sphingolipid signaling pathways. FASEB J. 2004;18:1456–1458. doi: 10.1096/fj.04-1618fje. [DOI] [PubMed] [Google Scholar]
- 42.Burrows C, Holly JM, Laurence NJ, Vernon EG, Carter JV, Clark MA, McIntosh J, McCaig C, Winters ZE, Perks CM. Insulin-like growth factor binding protein 3 has opposing actions on malignant and nonmalignant breast epithelial cells that are each reversible and dependent upon cholesterol-stabilized integrin receptor complexes. Endocrinology. 2006;147:3484–3500. doi: 10.1210/en.2006-0005. [DOI] [PubMed] [Google Scholar]
- 43.Baxter RC. Nuclear actions of insulin-like growth factor binding protein-3. Gene. 2015;569:7–13. doi: 10.1016/j.gene.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B, Lee HY, Weinzimer SA, Powell DR, Clifford JL, Kurie JM, Cohen P. Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J Biol Chem. 2000;275:33607–33613. doi: 10.1074/jbc.M002547200. [DOI] [PubMed] [Google Scholar]
- 46.Sonnenblick A, de Azambuja E, Azim HA, Jr, Piccart M. An update on PARP inhibitors–moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 47.Sallmyr A, Tomkinson AE. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem. 2018;293:10536–10546. doi: 10.1074/jbc.TM117.000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 49.Schedlich LJ, Nilsen T, John AP, Jans DA, Baxter RC. Phosphorylation of insulin-like growth factor binding protein-3 by deoxyribonucleic acid-dependent protein kinase reduces ligand binding and enhances nuclear accumulation. Endocrinology. 2003;144:1984–1993. doi: 10.1210/en.2002-220798. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Steinle JJ. DNA-PK phosphorylation of IGFBP-3 is required to prevent apoptosis in retinal endothelial cells cultured in high glucose. Invest Ophthalmol Vis Sci. 2013;54:3052–3057. doi: 10.1167/iovs.12-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PLoS One. 2012;7:e46614. doi: 10.1371/journal.pone.0046614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying S, Chen Z, Medhurst AL, Neal JA, Bao Z, Mortusewicz O, McGouran J, Song X, Shen H, Hamdy FC, et al. DNA-PKcs and PARP1 bind to unresected stalled DNA replication forks where they recruit XRCC1 to mediate repair. Cancer Res. 2016;76:1078–1088. doi: 10.1158/0008-5472.CAN-15-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.