Abstract
Despite continuing advances in the development of biomacromolecules for therapeutic purposes, successful application of these often large and hydrophilic molecules has been hindered by their inability to efficiently traverse the cellular plasma membrane. In recent years, cell-penetrating peptides (CPPs) have received considerable attention as a promising class of delivery vectors due to their ability to mediate the efficient import of a large number of cargoes in vitro and in vivo. However, the lack of target specificity of CPPs remains a major obstacle to their clinical development. To address this issue, researchers have developed strategies in which chemotherapeutic drugs are conjugated to cancer targeting peptides (CTPs) that exploit the unique characteristics of the tumor microenvironment or cancer cells, thereby improving cancer cell specificity. This review highlights several of these strategies that are currently in use, and discusses how multi-component nanoparticles conjugated to CTPs can be designed to provide a more efficient cancer therapeutic delivery strategy.
Keywords: Hypoxia, Low pH, Metabolism, Nanoplatforms, Proteases, Targeted therapy, Tumors
Introduction
In recent years, the use of biomacromolecules, such as proteins and nucleic acids, as therapeutic agents has become widely recognized as one of the most promising areas of drug development research [1]. The size and strong hydrophilic nature of protein- and nucleic acid-based therapeutics, however, makes it very challenging to effectively transport them across the semi-permeable plasma membrane. To overcome this obstacle, various approaches have been developed for cellular delivery of proteins and nucleic acids, such as liposomes, electroporation, and microinjection [2]. Although these methods offer several advantages for cargo transport, they all possess major drawbacks, such as low efficiency for delivery in vivo, lack of tissue and cell specificity, poor bioavailability and extensive toxicity, all of which limit their successful application in a clinical setting [2, 3]. In view of these issues, researchers have continued to search for more effective transport systems, which have led to the discovery and development of a variety of short peptides that constitute a new class of delivery vectors. These peptides, most commonly known as cell-penetrating peptides (CPPs) or protein transduction domains (PTDs), have the ability to efficiently enter a wide range of cell types without damaging the cell membrane, and have been found to be very effective in overcoming various bio-barriers at different tissue levels [4–6]. As a result, CPPs constitute a promising approach for the intracellular delivery of therapeutics, including anticancer drugs [7].
The history of this class of peptides dates back to the late 1980s when the trans-activator of transcription protein (TAT) of the human immunodeficiency virus and the Drosophila Antennapedia homeodomain were first recognized for their cell-penetrating propensities [7, 8]. The spontaneous cellular internalization of both proteins led to extensive structural and functional studies to define the minimal amino acid sequence needed for membrane translocation, and thus develop effective transport vectors [9]. This resulted in the discovery of the first CPPs, the TAT peptide and Antp (or penetratin) (see Table 1 for peptide sequences), which paved the way for generating a large number of protein-derived and chimeric CPPs, as well as synthetically preparing a wide variety of CPP analogues [6, 9, 10].
Table 1.
Primary sequences of the cell-penetrating peptides (CPPs) and cancer-targeting CPPs (CTPs) discussed in this review
One of the earliest studies to demonstrate the delivery efficacy of a CPP in vivo was carried out by Schwarze et al. where, following an intraperitoneal injection in mice of β-galactosidase fused to the 11-amino acid TAT peptide, the fusion protein was detected in all tissues, including the brain, revealing that the chimeric protein can cross the blood–brain barrier [11]. The potential of CPPs in increasing the delivery efficiency of nanoparticles was also first confirmed by Josephson et al. [12]. Their findings revealed that dextran-coated superparamagnetic iron oxide nanoparticles functionalized with the TAT peptide were internalized into lymphocytes over 100 times more efficiently than non-modified nanoparticles. Since then, many studies have reported the use of CPPs both in vitro and in vivo to efficiently deliver colloidal carriers, such as liposomes and polymeric nanoparticles, as well as different types of cargo, ranging from nucleic acids, polymers and imaging agents to low molecular weight drugs [13–17].
CPPs: an overview
CPPs are typically composed of less than 30 amino acids and often carry a net positive charge at physiological pH due to the presence of cationic arginine and lysine residues [17, 18]. This overall positive charge promotes electrostatic interactions with negatively charged cell-surface constituents, such as heparan sulfate proteoglycans, which are believed to play an important role in initiating the uptake of CPPs [19, 20]. Several studies have shown that CPPs containing a large number of arginine residues have superior internalization efficiency compared to lysine-rich peptides; this difference is attributed to the guanidinium headgroup of arginine residues, which forms stable bidentate hydrogen bonds with cell membrane components, as opposed to the ammonium cations of lysines that can only form one hydrogen bond [21, 22]. Interestingly, negatively charged CPPs have also been reported. An example is a variant of sweet arrow peptide (SAP, derived from the N-terminus of the maze storage protein γ-zein VHL(PPP)8), in which the arginine residues were replaced by glutamate residues [24]. This variant, denoted SAP(E), adopts a polyproline II helical secondary structure and carries a net negative charge [23]. Surprisingly, the cellular internalization efficiency of SAP(E) was comparable to that of the cationic parent SAP [24]. The uptake mechanism of the modified SAP(E) is thought to be mediated by conformational transitions due to either refolding or locally aggregating on the cell membrane [24].
Studies have demonstrated that the presence and specific positioning of hydrophobic residues also influence the ability of the CPP to interact with and traverse the plasma membrane [17, 25]. For instance, tryptophan was shown to play a key role in the cellular uptake of CPPs due to the aromatic side chain exhibiting a favorable free energy of insertion into the plasma membrane [23]. Rydberg et al. designed six CPPs, all containing arginine residues but differing in number and positioning of tryptophan residues [26]. The uptake efficiency of these CPPs was directly proportional to the number tryptophan residues present, and positioning of the tryptophans in the middle of, or evenly distributed along, the peptide’s amino acid sequence increased its cellular internalization.
Other factors that influence CPP internalization include chirality and amphipathicity [27]. Since natural l-amino acids are susceptible to proteases, thereby limiting their successful application in vivo, various strategies have been utilized to enhance the stability of CPPs. One such approach has been to replace the l-amino acids with their d-analogues [23]. Although incorporation of d-amino acids resulted in a marked resistance of CPPs to degradation, the use of non-natural amino acids was reported to lower the peptides’ internalization efficiency in a cell-dependent manner [23]. Furthermore, the ability of amphipathic CPPs to adopt α-helices and β-sheets was shown to enhance their cellular uptake efficiency [23]. Thus, the primary peptide structure, both in terms of amino acid content as well as the specific location of the residues within the sequence, is likely to play a role in mediating the cellular uptake of CPPs [25].
Despite numerous studies, the exact mechanisms by which CPPs enter the cells remain poorly understood. Two major cellular uptake mechanisms, nevertheless, have been put forward to explain how these peptides are able to efficiently deliver various kinds of cargoes into cells [4, 25]. The first mechanism entails direct translocation through the lipid bilayer via an energy-independent manner, while the second involves an energy-dependent endocytic process [4]. Various forms of endocytosis, from macropinocytosis (a nonspecific form of fluid phase endocytosis), to clathrin- and caveolae-mediated endocytosis, have been proposed as the cellular entry route of CPPs [17].
Currently, most CPPs are believed to be taken up by cells, at least in part, through endocytosis. However, evidence for an endocytosis-independent, direct translocation pathway has also been presented for several peptides, such as CADY, MPG and Pep-1, which are able to translocate across the plasma membrane at temperatures as low as 4 °C that typically inhibit energy-dependent endocytosis [28–31]. Although the exact nature of the direct translocation pathway remains unclear, various models have been proposed to describe this mechanism, including transient pore formation, inverted micelle formation, a detergent-like ‘carpet’ mechanism or membrane thinning [32]. Energy-independent direct translocation is generally favourable at high peptide concentrations (10- to 20-fold molar excess of CPPs) and for hydrophobic or amphipathic CPPs [33, 34]. The initial step in all proposed direct translocation models is the interaction between the positively charged amino acids of CPPs, such as arginine and lysine, and the negatively charged membrane’s components, such as heparan sulfate and the phospholipid headgroups, which results in membrane structure destabilization [17]. Furthermore, in certain cases contradictory observations of both endocytic and energy-independent uptake are reported for the same CPP. It has been suggested that these observations may be due to different factors that affect the cellular uptake mechanisms, including the properties of the CPP (sequence, charge, size and concentration), as well as the characteristics of the cargo and nature of the coupling to the CPP [17, 29].
Despite these controversies, studies have shown that CPPs are highly efficient in facilitating the in vitro cell uptake of different cargo molecules. The in vivo use of CPPs, however, has been proven to be more complex, and ultimately less efficient, mainly due to lack of cell specificity; most CPPs will non-specifically interact with the membranes of a range of cell types due to the presence of heparan sulfate proteoglycans and other cell-surface components [19]. This, in turn, poses a serious challenge for using CPPs to facilitate the delivery of therapeutics, such as anticancer drugs [11, 35]. To address this obstacle, researchers have developed strategies to deliver CPPs to a particular site (tissue or organ), one of which is localized injection of CPPs attached to cargo molecules. Although this approach has demonstrated effective delivery of the CPP-cargo to tumor cells, the main drawback is that it is only applicable to solid, localized tumors or easily accessible organs [36]. In light of this, alternative strategies have been developed in which chemotherapeutic drugs are conjugated to cancer targeting peptides (CTPs), e.g., activatable CPPs and pH triggered membrane peptides, that exploit the unique characteristics of the tumor microenvironment or cancer cells, thereby improving cancer cell targeting [37, 38].
Cancer targeting strategies
Extracellular acidity
pH (low) insertion peptides (pHLIPs)
Cancer cells often exhibit elevated levels of aerobic glycolysis (Warburg effect), which serves as their primary energy-generating pathway [39]. This increased rate of glycolysis, in turn, leads to a higher production of lactate and protons, resulting in an acidic extracellular environment that has been shown to play a role in enhancing metastasis and local invasion of tumors [40]. Several studies have used this particular tumor characteristic to deliver chemotherapeutic drugs to cancer cells [41]. In particular, there is a growing body of research focused on the pH (low) insertion peptides (pHLIPs) family (see Table 1 for sequence) and their utility in tumor imaging [42–45] and cargo/drug delivery [45–48] since their discovery by Hunt et al. [49]. pHLIPs are water soluble, moderately hydrophobic bacteriorhodopsin C helix-derived peptides [50, 51], which have received considerable attention due to their pH-dependent targeting of cancer cells [52–55]. Consequently, pHLIPs provide a strategy for delivering therapeutic cargoes and combating diseases that are characterized by extracellular acidity, including cancer [46, 56, 57].
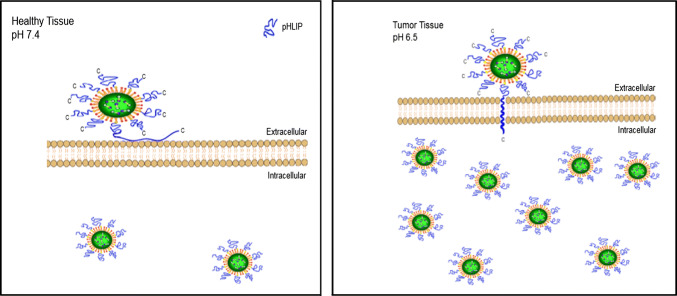
Since the tumor-targeting ability of pHLIPs was thought to be based on its pH-dependent membrane insertion property [52–54], several thermodynamics and kinetics studies were done to evaluate the membrane interactions of the peptides using a variety of biophysical techniques, including circular dichroism (CD) spectroscopy, steady-state fluorescence spectroscopy [58–65], solid-state NMR spectroscopy [66–68], as well as small angle X-ray scattering (SAXS) [69]. Employing the aforementioned methods have revealed that the wild-type (WT) pHLIP family transitions have three prominent forms or states, which range from random coil to helix. State I refers to pHLIP WT existing as a stable, unstructured monomer in solution at physiological pH (pH 7.4) [69], whereas State II takes place when pHLIP WT is bound to the surface of lipid bilayers, but remains in a coiled-like conformation. Finally, upon acidification, state III is achieved in which the negatively charged residues, such as aspartates, are protonated, thereby increasing the hydrophobicity of pHLIP WT and allowing it to insert into the bilayer as a monomeric transmembrane α-helix. Adopting this helical conformation has been shown to be accompanied by a release of free energy (about 2 kcal/mol), which is utilized to translocate the peptide across the targeted membrane and deliver conjugated cargo molecules [53]. The drug delivery capacity of these peptides was successfully demonstrated by Moshnikova et al., where pHLIP with a single cysteine at the C- or N-terminus conjugated to the toxin α-amanitin translocated into target cancer cells in a pH-dependent manner [47]. Moreover, coating a drug delivery system with pHLIP was shown to enhance its tumor targeting capability [70]. A schematic representation of pHLIP-mediated delivery of nanoparticles into healthy tissue and tumor tissue is shown in Fig. 1.
Fig. 1.
Schematic representation of pHLIP-mediated delivery of nanoparticles into healthy tissue (left) and tumor tissue (right). The acidity of the tumor microenvironment results in greater entry of the pHLIP-coated nanoparticles into cancer cells compared to cells in healthy tissue where the extracellular space is at a physiological pH [70]
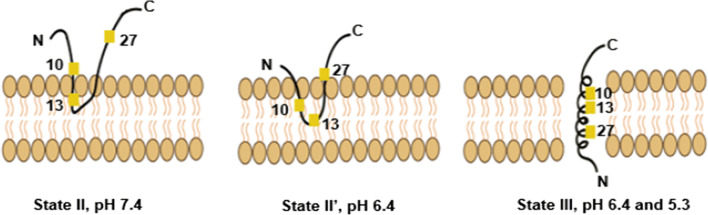
The exact insertion mechanism of pHLIP into membranes remains unclear. Protonation of the carboxylic side chains of the central aspartic residues (D14 and D25) was always thought to be crucial for the peptide’s folding and insertion in response to acidity [52]. However, solid-state nuclear magnetic resonance (ssNMR) analysis conducted by Shu et al. and Hanz et al. on an isotope-labelled pHLIP variant has suggested that the C-terminal aspartic residues, D31 and D33, may also play a role in pHLIP’s pH-dependent targeting. At pH 6.4, and prior to complete insertion, pHLIP peptides were found to co-exist in both unstructured-peripheral (state II; approximately 70%) and α-helical-inserted (state III; approximately 30%) structures, suggesting a new membrane-embedded state of transition: state II′ that lies between states II and III and differs in terms of structure and membrane location [65, 67, 68]. Briefly, Shu et al. and Hanz et al. proposed that during state II (at pH 7.4), the N-terminal half alanine residues, A10 and A13, bind to the headgroup phosphates of the outer-leaflet phospholipid bilayer. During state II′ (at pH 6.4), residues A10 and A13 bury deeper (but still peripherally) into the membrane bilayer, pulling A27 and the polar C-terminal half (including D25, D31, D33) downward and closer to the 31P nuclei of the phospholipid. During the state II–state II′ transition, the C-terminus aspartates (D31, D33) are protonated first, leading to pHLIP hydrophobicity enhancement and deep adsorption into the hydrophobic membrane interior, which in turn triggers protonation of D25 and D14 and complete membrane insertion [67, 68, 71]. The mechanism proposed by Shu et al. and Hanz et al. is summarized in Fig. 2.
Fig. 2.
Schematic representation of the structure and membrane locations of pHLIP at neutral and acidic pH. In state II at neutral pH, the N-terminal alanine residues, A10 and A13, interact with the headgroup phosphates, while the C-terminal A27 is located extracellularly. During state II′ at pH 6.4, residues A10 and A13 are buried deeper into the membrane, pulling A27 closer to the bilayer surface. Finally, during State III, at pH 6.4 and 5.3, A10 and A13 are buried deeper into the membrane than in State II′, with A27 located below the headgroups
(adapted from Ref. [67])
Gupta and Mertz recently examined the protonation of pHLIP WT’s residues (D14, D25, D31, and D33) during the transition from state I to state II [72]. They revealed that titration of these residues under acidic conditions led to both α-helical and β-strand conformations of the peptide. Although it was previously reported that protonation of both D14 and D25 increased the interactions between N- and C-terminal regions, a reduction in the helicity of the peptide was observed upon protonation of both these aspartates. Conversely, protonation of either single residue (D14 or D25), or all aspartic acid residues, resulted in a more helical conformation. However, protonation of all aspartic acid residues (D14, D25, D31, and D33) abolished interaction between the pHLIP’s N and C-terminus [72], indicating that an increase in interactions between the N- and C-terminus of pHLIP due to protonation has no direct correlation with the peptide’s helical formation [72]. In agreement with these findings, Karabadzhak et al. tested different pHLIP variants by changing the sequence of the peptide [66, 73]. Their findings also revealed that truncation of pHLIP’s protonatable residues in the C-terminus half has no effect on the helical conformation, but is correlated with the folding-insertion and unfolding-exiting time [66]. Increasing the pH stimulates the exit process through deprotonation of the transmembrane pHLIP residues and, hence, unfolding of the peptide [66].
The main limitation, however, of the aforementioned studies is that the model membranes used were composed solely of phosphatidylcholine (POPC); the plasma membrane consists of a more complex lipid composition [74] that will potentially have a different effect on pHLIP’s membrane interaction. Vasquez-Montes’s group confirmed this by examining the impact of a biological membrane’s lipid composition on the insertion process of pHLIP WT and the pHLIP-P20G variant [75]. Inserting pHLIP into large unilamellar vesicles (LUVs) composed of anionic lipids, cholesterol, or phosphoethanolamine (POPE) led to elimination of state II, which they referred to as “shallow” penetration (state IIS), at neutral pH [75]. State IIS was not characterized by a spectral shift of pHLIP’s tryptophan fluorescence nor a conformational change in both pHLIP WT and pHLIP-P20G peptides [76]; however, a reduction in acrylamide quenching was observed indicating partitioning into the bilayer. Formation of state IIS was suggested to be a result of the repulsive electrostatic interactions between the charged residues of pHLIP and the anionic phospholipid headgroups [75, 76]. In the case of neutral lipids, such as POPE, pHLIP’s interfacial partitioning was suggested to be influenced by the tighter phospholipid packing due to ethanolamine forming hydrogen bonds in the interfacial region of the bilayer [75]. These findings raise questions concerning the mechanism of pHLIP-membrane interaction and whether the results in model membrane systems are representative of the complex biological membrane.
Acidity-triggered rational membrane (ATRAM)
Although pHLIP has been shown to insert into membranes of solid tumors upon acidification and has been successfully used for imaging of cancer cells, studies have shown that it has several limitations [40, 77]. These include modest targeting of mildly acidic tumors and a strong propensity to aggregate. In view of these issues, Nguyen et al. designed the acidity-triggered rational membrane (ATRAM) peptide by modifying pHLIP’s sequence (23.5% sequence similarity between both peptides) (see Table 1 for sequence) [40]. Negatively charged glutamate residues rather than aspartates were incorporated into the sequence of the ATRAM peptide as they have higher pKa values (6.5), increasing the peptide’s efficiency in sensing and targeting acidic diseased tissues. A central proline residue was also included to increase solubility, as well as glycine resides since they tend to favour the formation of α-helices in membranes and minimize aggregation in solution. Additionally, a single tryptophan was introduced to study the properties of the peptide at different pH conditions using intrinsic fluorescence experiments [40].
These modifications resulted in ATRAM being very soluble at high concentrations (> 200 µM), as opposed to pHLIP which aggregates at low concentrations (< 7 μM), and enhancing ATRAM’s membrane targeting under acidic conditions [40]. At neural pH, ATRAM was shown to lie parallel to the membrane plane in an unstructured conformation, whereas under acidic conditions similar to the peritumoral pH, it would undergo a conformational change forming a transmembrane α-helix that drives membrane insertion [40]. Moreover, cell viability studies in two different human cell lines showed that ATRAM is non-toxic under physiological conditions, indicating that it does not damage cell membranes [40]. Malignant melanoma (A375) and bronchi alveolar carcinoma (H358) cell lines were then used to compare the pH-dependent targeting of cells by ATRAM and pHLIP. Both peptides bound to cancer cells in a pH-dependent manner, but ATRAM had a stronger interaction with cancer cells under acidic conditions, thereby underlining its superior cell targeting propensities [40]. In a recently published study, a comparison of the performance of pHLIP variants and ATRAM was carried out using a wide range of assays; the results confirmed that ATRAM exhibits a significantly higher membrane affinity and stronger membrane insertion compared to pHLIP WT and most of its variants [77].
Tumor microenvironment hypoxia
Another characteristic of tumor microenvironments is their hypoxic nature [78], which drives malignant progression in cancer cells [79]. The inherent hypoxia of tumors has been exploited to improve the cancer-targeting specificity of CPPs. A central component of the hypoxic response of tumor cells is the transcription factor hypoxia-inducible factor-1α (HIF-1α), which plays a role in mediating the expression of a variety of genes involved in angiogenesis, oxygen homeostasis, metabolism and cell viability [80]. The oxygen-dependent degradation (ODD) domain of HIF-1α is composed of ~ 200 amino acids and regulates the stability of the transcription factor. Under normoxic conditions, the ODD domain was shown to interact with the von Hippel–Lindau tumor suppressor, which results in degradation of HIF-1α by the ubiquitin–proteasome pathway [81].
Harada et al. constructed TAT fusion proteins (TAT-β-gal and the TAT-caspase-3) fused to a segment of the ODD of HIF-1α to target tumors in vivo [5, 81]. The reasoning was that a TAT-ODD fusion protein would be stable in the hypoxic regions of tumors, but would be degraded and non-functional in the normoxic environment of normal tissues. After intraperitoneal injection of TAT-ODD-β-gal into tumor-bearing mice, the fusion protein accumulated only in the hypoxic regions of solid tumors and was absent from healthy tissue. Conversely, the same treatment with TAT-β-gal resulted in the parent protein accumulating in all tissues. In addition, intraperitoneal injection of the TAT-ODD-caspase-3 protein into tumor-bearing mice lead to a significant reduction of tumor growth, and consequently the size of the tumors, without any of the obvious toxic side effects that would be normally be a consequence of delivering active caspase-3 to a mouse [81]. As a result, cytotoxic TAT-ODD fusion proteins can potentially be used as a novel therapy for targeting solid tumors.
Cancer-associated proteases
Cancer-associated proteases (CAPs) include cathepsins, urokinases, caspases and matrix metalloproteases (MMPs), such as gelatinases MMP-2 and MMP-9 [82]. These proteases are overexpressed in most types of cancer tissues, but are either absent or found in lower concentrations in healthy tissues [83, 84]. MMPs, a family of secreted and transmembrane proteins, are known to play a critical role in promoting cancer invasion and metastasis by degrading the extracellular matrix and activating both growth factors and angiogenesis [37, 85]. Several studies have also shown a correlation between MMP overexpression and poor clinical outcome in several types of cancers, including breast, colon, gastric, oesophageal, and small and non-small cell lung cancers [86, 87]. As a result, the expression of specific MMPs serves as both a prognostic indicator of clinical outcome and a marker of tumor progression in a wide range of tumors.
Charge masking strategy
Roger Tsien’s group exploited the overexpression of MMPs in tumors to generate cancer targeting CPPs [37]. These so-called activatable cell-penetrating peptides (ACPPs) are composed of a polycationic CPP covalently attached to a neutralizing polyanionic domain via a cleavable peptide linker, thereby adopting a hairpin conformation [25]. This particular conformation and the ionic interactions between the CPP and inhibitory anionic counterpart masks the cell-penetrating functionality of the CPP by reducing the overall charge to nearly zero, which in turn minimizes adsorption and uptake into cells unless the linker is cleaved [88, 89]. Furthermore, the peptide linker was created to be specific to MMP-2 and MMP-9 to target a wide range of tumors [37]. Upon exposure to cancer cells, their membrane-bound and secreted MMPs can cleave the linker, dissociating the inhibitory peptide and activating the CPP; the released CPP along with its cargo would then bind to, and enter the tumor cells [88]. Moreover, due to the wide variety of CAPs overexpressed in tumor microenvironments, ACPPs with cleavable linkers specific to other proteases can be readily designed [90].
Shi et al. highlighted the therapeutic potential of this targeting method when they designed an ACPP (see Table 1 for peptide sequence) that was covalently attached to the anticancer drug, doxorubicin (ACPP-DOX); their findings revealed that incubating fibrosarcoma cells with ACPPs resulted in reduced cytotoxicity compared to CPPs, whereas ACPP-DOX effectively inhibited fibrosarcoma cell proliferation [91]. The uptake of ACPP-DOX was also observed to be higher in fibrosarcoma cells, which overexpress MMPs, compared to breast cancer cells, which express much lower levels of MMPs. Likewise, Aguilera et al. demonstrated that, in addition to improved specificity, ACPPs are less toxic than CPPs: CPPs caused severe toxicity in mice at a dose 4 times lower than the MMP-cleavable ACPPs [89].
Despite their promise, several issues have restricted the use of ACPPs in cancer therapy, including endosomal entrapment and significant off-target uptake by other tissues in vivo [92]. Additionally, it has been reported that MMP targeting is not very specific as it is likely caused by cleavage in the vascular compartment as opposed to tumor-specific cleavage [88]. As a result, ACPPs have been primarily utilized as molecular imaging probes for visualizing enzymatic reactions, or to differentiate between tumor and healthy tissues [88, 93]. The first in vivo study to demonstrate the use of radiolabeled ACPPs for the detection of MMP activity in tumors was carried out by van Duijnhoven et al. [88]. Furthermore, using mouse models, Nguyen et al. showed that both fluorescently labelled free ACPPs and ACPPs conjugated to dendrimers (ACPPDs) could improve the precision of tumor resection [93]; removal of tumors using ACPPs or ACPPDs led to fewer residual cancer cells left behind, and increased tumor-free survival, compared to cases where tumors were removed with traditional bright-field illumination. Although antibodies are suitable for fluorescence-guided surgeries, their use is fairly limited as they can only be applied to certain types of tumors that express specific markers. Each epitope can also only bind one antibody at a time resulting in a weak signal amplification [93]. ACPPs, on the other hand, appear to be better agents for in vivo imaging of tumors as they are smaller in size, which increases their solid tumor penetration efficiency beyond blood vessels. Each active MMP can also cleave many substrate ACPPs, offering a stronger amplification signal [85].
Steric hindrance strategy
In addition to a polynanionic masking domain, many studies have utilized long chain polymers, such as poly(ethylene glycol) (PEG), as steric hindrance to shield the strong positive charge of CPPs [94]. Although PEG has been shown to effectively reduce cellular uptake by healthy cells, the main drawback is that it can also block or reduce uptake in target cells. Thus, it is crucial that PEG is efficiently removed at the target site to activate the CPP and allow it to traverse the plasma membrane. With this in mind, the Leroux group reported an ACPP strategy in which CPP charge shielding was achieved by using a self-immolative azobenzene linkage for the delivery of nucleic acid-based cargoes [95]. In this approach, lysine residues of a CPP-peptide nucleic acid (PNA) construct were conjugated to PEGylated azobenzene groups. Upon exposure to bacterial azoreductase secreted by microbiota present in colon mucosa, the azobenzene bonds could be cleaved resulting in the detachment of PEG and the uptake of the PNA cargo into the colonic epithelial cells to inhibit expression of the protein of interest (Fig. 3). In vitro studies in the absence of azoreductase revealed that PEGylation resulted in a complete loss of activity of the construct, and PEGylated CPP-PNA did not adversely affect cell viability up to 12 μM, whereas the CPP-PNA conjugate alone was toxic at this concentration, thereby demonstrating the effectiveness of this shielding strategy to ‘inactivate’ the CPP moiety [95]. Thus, this activatable PEGylated CPP-nucleic acid conjugate could potentially be used as a delivery vector for the treatment of inflammatory bowel diseases and colon cancer.
Fig. 3.
Schematic illustration of ACPP-mediated delivery of peptide nucleic acid (PNA) based on CPP inactivation using a self-immolative azobenzene moiety. Upon reductive cleavage of the azobenzene bonds, the PEG chains are detached and the CPP promotes the uptake of the PNA cargo into the target cells
(adapted from Ref. [95])
Minimal side chain modification strategy
Another method to temporarily inactivate a CPP is by carrying out a minimal side chain modification rather than inactivating it via the peptide main chain. Through an alanine scan of the TAT peptide (see Table 1 for peptide sequence), Wender et al. showed that modifying the side chain of either of the lysine residues present in the sequence can inhibit the peptide’s cellular uptake [96]. This finding was then used by Jin et al. to develop pH-activatable TAT peptides [97]. In this approach, the TAT lysine residues were amidized into succinyl amides, which significantly inhibited the cellular uptake of the resulting peptide presumably due to its reduced number of positive charges. However, upon exposure to acidic pH, the modified peptide was able to enter cells due to hydrolysis of the succinyl amides. Functionalizing poly(ethylene glycol)-block-poly(ε-caprolactone) (PEG-PCL) micelles with these modified TAT peptides resulted in accumulation of the micelles in tumor tissue and successful delivery of the doxorubicin cargo to this tissue [97]. A similar approach was carried out by Bode et al. where they attached enzyme-cleavable domains to one or both lysine side chains of the TAT peptide [98]. These domains were specific to aminopeptidase and dipeptidyl peptidase as both enzymes are overexpressed in many types of cancer cells. As a result, attaching either of these domains temporarily blocked the activity of the TAT peptide, whereas in the presence of the respective enzyme, the inactive alanine modification was cleaved, thereby restoring the activity of the CPP. Bode et al.’s findings revealed that modifying only one lysine residue resulted in near complete inhibition of the peptide’s cellular uptake, indicating that this strategy can be useful in controlling the activity of CPPs [98].
Cancer cell’s metabolic activity
An alternative approach for improving CPP specificity entails taking advantage of the internal biological state of targeted cells. As an example of such a strategy, Vocero-Akbani et al. reported killing HIV-infected cells by exploiting their HIV protease activity [99]. In this study, a chimeric CPP was constructed by fusing the TAT CPP to a caspase-3 precursor protein, a crucial mediator of apoptosis, and replacing the endogenous cleavage sites of the construct with HIV proteolytic cleavage sites. Despite this construct entering both HIV-infected and uninfected cells, caspase-3 induced apoptosis only occurred in HIV-infected cells. This was due to the construct being processed into its active form only when cleaved by the HIV protease that is exclusively present in HIV-infected cells. Uninfected cells, on the other hand, did not undergo apoptosis as the chimeric CPP construct remained in an inactive form.
Many diseases, such as cancer and autoimmune disorders, are also characterized by abnormal enzymatic activity, which can be exploited to design an ‘active targeting strategy’ that is able to differentiate between healthy and diseased cells [90]. In addition to an overexpression of MMPs, most highly aggressive cancer cells also exhibit elevated levels of glycolysis, which serves as their primary energy-generating pathway, irrespective of oxygen availability, a phenomenon termed “the Warburg effect” [100]. The high glycolytic rates are due, in part, to the overexpression and increased activity of mitochondria-bound type II hexokinase (HKII) [101]. HKII binds to the outer mitochondrial membrane (OMM) through the pore-like voltage-dependent anion channel (VDAC), a major channel for transport of ions and metabolites [102]. Interaction with VDAC occurs via the N-terminal 15 amino acid of HKII, which allows the enzyme to access mitochondria-generated ATP and phosphorylate glucose to glucose-6 phosphate, a vital metabolite that is necessary not only for glycolysis, but also for generating nucleic acids, lipids, and proteins that are required for growth and cell proliferation [103]. Additionally, binding of HKII to VDAC suppresses apoptosis in cancer cells by maintaining the integrity of the outer mitochondrial membrane (OMM) and inhibiting the release of key apoptogenic molecules, such as cytochrome c, from the intermembrane space [104]. Thus, overexpression of HKII is required for tumor initiation and maintenance, as well as promotion of metastasis [105]. Consequently, several groups are currently developing novel strategies that target this enzyme to induce apoptosis in cancer cells; these may involve directly inhibiting the synthesis of HKII or disrupting the HKII-VDAC interaction [101, 103].
Woldetsadik et al. designed a cancer-specific CPP composed of pHK, a peptide that corresponds to the VDAC binding N-terminal 15 amino acid of HKII (see Table 1 for peptide sequence), covalently attached to a short penetrating–accelerating sequence (PAS; GKPILFF) [103]. Since pHK is poorly cell permeable, the short PAS sequence was incorporated into the CPP design to enhance its cellular uptake and endosomal escape [106, 107]. Attachment of PAS to pHK resulted in up to sevenfold greater cellular uptake compared to pHK alone, and the efficient cellular internalization of pHK-PAS was attributed to the CPP utilizing by both macropinocytosis and energy-independent mechanisms. Once in the cytosol, pHK-PAS localized to mitochondria, where it bound to VDAC, competitively dissociating the endogenous full-length HKII in the process. The disruption of the HKII–VDAC interaction in cancer cells resulted in mitochondrial membrane potential depolarization, inhibition of mitochondrial respiration and glycolysis, depletion of intracellular ATP levels, release of cytochrome c and finally, apoptosis [103]. Significantly, pHK-PAS was considerably less toxic to non-cancerous cells, in which displacement of HKII from mitochondria did not lead to apoptosis, indicating that the peptide exhibited selective HKII-mediated cytotoxicity against cancer cells.
Tumor lineage CPPs
Phage display and mRNA display technologies are two useful tools for screening and identifying CPPs from peptide libraries. However, owing to the smaller molecular size of mRNA display-based complex and the larger number of peptides displayed, mRNA display technology is more suitable and efficient for isolating CPPs than phage display. Kondo et al. used mRNA display to construct a random peptide library, which was screened to obtain ten novel CPPs. The tumor lineage specificity of each newly synthesized CPP was then determined through functional screening [108]. Kondo et al. reported that the amino acid sequences of these CPPs contained ~ 30% positively charged amino acids and lacked any identity to previously reported CPPs and other recorded mammalian proteins, thereby indicating that these CPPs may encode artificial sequences. Significantly, these tumors lineage-homing CPPs could noninvasively target their corresponding tumors while exhibiting minimal localization to normal tissues. For instance, CPP2 was shown to target and easily penetrate primary colon adenocarcinoma cells while showing minimal targeting of healthy tissues, and CPP44 was capable of invading hepatic tumor cells and myelogenous leukemia with a low level of cellular uptake in the liver and other organs. As a result, tumor lineage-homing CPPs generated by mRNA display technology are potentially useful tools for developing peptide-based molecular delivery systems [108].
A multi-component nanoplatform
Cancer nanomedicine has the potential to overcome the intrinsic limitations of conventional cancer therapies, including poor solubility, toxicity to normal cells, and short circulation half-life [109]. However, the use of nanoparticles in a clinical setting has been hindered by certain drawbacks, including low efficiency of intracellular delivery in vivo and an inability to cross the blood–brain barrier [3]. Moreover, despite greater accumulation of nanoparticles in tumor tissue relative to healthy tissues [109] due to the enhanced permeability and retention (EPR) effect, recent reports indicate that only 0.7% of administered nanoparticle doses actually end up in tumors [109] Several studies have reported increased cellular uptake of nanoparticles functionalized with CPPs, such as TAT, polyarginine, pAntp (or penetratin) and low molecular weight protamine (LMWP) [110–112]. Furthermore, coating with CTPs has been shown to improve the tumor targeting of nanoparticles [113]. Therefore, combining drug-loaded nanoparticles with CTPs can potentially lead to more selective and efficient delivery systems for cancer therapy, resulting in improved clinical outcomes.
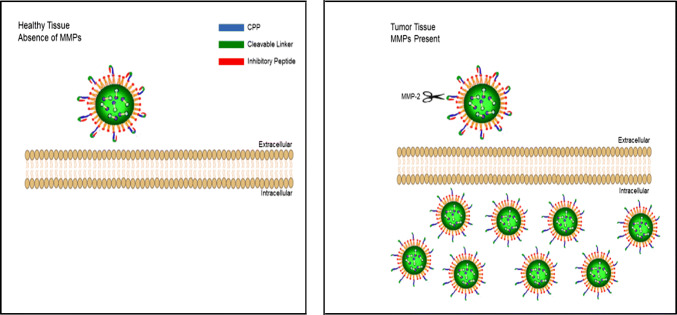
A ‘modularized concept’ for designing a peptide-based nanosystem has been proposed by Qin et al. [113]. The different components of nanoformulations were classified into different categories, known as modules, based on their functions. Thus, the desired modules and encapsulated therapeutic molecules can be selected and integrated when designing a well-controlled nanosystem, thereby targeting the tumor microenvironment with increased specificity, on-demand response, enhanced cellular uptake, and improved tumor therapy. This ‘multi-responsive’ system for cancer therapy was applied by Huang et al. in devising a nanoparticle system for gene delivery that exploited both the peritumoral pH and increased MMP activity [38]. These nanoparticles were modified with a ‘dual-triggered’ ACPP composed of a pH-responsive polyanionic peptide, an MMP-cleavable linker, and a polycationic CPP (Fig. 4). The pH-responsive polyanionic peptide was used to inhibit the CPP until the nanoparticles entered the acidic peritumoral environment. Within this low pH environment, the pH-sensitive peptide became uncharged or positively charged allowing the overexpressed MMPs to cleave the linker, and the released CPP then facilitated the uptake of the nanoparticle into cancer cells.
Fig. 4.
Schematic representation of ACPP-mediated delivery of nanoparticles into healthy tissue (left) and tumor tissue (right). Upon exposure to cancer cells, their membrane-bound and secreted MMPs can cleave the linker and activate the CPPs, thereby facilitating cellular uptake of the nanoparticles
Yoo et al. reported an alternative anticancer drug delivery system, which exploited the elevated levels of extracellular MMPs and intracellular reactive oxygen species (ROS) within tumor tissue [114]. A protease-activatable CPP was designed containing an ROS-responsive methionine, a cell permeable lysine chain and an MMP-cleavable linker connected to PEG chains. This CPP, denoted MLMP, was used to form a micellar nanoparticle that encapsulated within its core the hydrophobic anticancer drug doxorubicin (DOX). The reasoning was that upon exposure to MMPs present in the extracellular matrix of cancer cells, the MMP-linker would be cleaved, thereby releasing the PEG chains and exposing the poly-l-lysine chains. Due to the cell-penetrating propensity of the poly-l-lysine chains, the micellar structure would then be able to efficiently enter the cancer cells. The high levels of ROS in cancer cells would convert the hydrophobic thioether groups within the methionine chains to hydrophilic sulfoxide groups, which would destroy the micellar MLMP structure and release the encapsulated DOX intracellularly. Compared to free DOX, MLMP (DOX) exhibited higher DOX delivery efficacy and greater tumor inhibition efficiency, coupled with lower nonspecific toxicity, underlining the potential of dual stimuli-MLMP as a drug delivery system for cancer therapy [114].
Conclusion
The tumor microenvironment and a cancer cell’s biological state provide can be exploited to facilitate the selective delivery of chemotherapeutic drugs. In recent years, several in vitro and in vivo studies have reported the efficient delivery of cancer therapeutics using CTPs designed to exploit the peritumoral environment or cancer cell surface markers. Rapid progress in the field of nanotechnology has also provided the necessary tools for the design of well-defined, multi-component drug delivery nanoplatforms. By carefully selecting the building blocks and modulating the physicochemical properties of different nanoformulations, enhanced delivery efficacy and improved therapeutic outcomes can potentially be achieved through synergistically targeting multiple key features of cancer cells or the tumor microenvironment. CTPs are currently used as building blocks for multi-component nanoplatforms due to their biocompatibility, chemical versatility and ability in recognizing key components of tumor tissues, which can potentially lead to more selective and efficient delivery systems for cancer therapy.
Acknowledgements
The authors thank Professor David Male (The Open University) for critical reading of the manuscript. This work was supported by funding from NYU Abu Dhabi (Research Grant) and an ADEK Award for Research Excellence Grant (AARE17-089) to M.M.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Juliano R. Challenges to macromolecular drug delivery. London: Portland Press Limited; 2007. [DOI] [PubMed] [Google Scholar]
- 2.Bolhassani A. Potential efficacy of cell-penetrating peptides for nucleic acid and drug delivery in cancer. Biochim Biophys Acta (BBA) Rev Cancer. 2011;1816(2):232–246. doi: 10.1016/j.bbcan.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 3.McErlean EM, McCrudden CM, McCarthy HO. Multifunctional delivery systems for cancer gene therapy. In: Hashad D, editor. Gene therapy—principles and challenges. London: InTech; 2015. [Google Scholar]
- 4.Regberg J, Srimanee A, Langel Ü. Applications of cell-penetrating peptides for tumor targeting and future cancer therapies. Pharmaceuticals. 2012;5(9):991–1007. doi: 10.3390/ph5090991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magzoub M, Gräslund A. Cell-penetrating peptides: small from inception to application. Q Rev Biophys. 2004;37(2):147–195. doi: 10.1017/S0033583505004014. [DOI] [PubMed] [Google Scholar]
- 6.Reissmann S. Cell penetration: scope and limitations by the application of cell-penetrating peptides. J Pept Sci. 2014;20(10):760–784. doi: 10.1002/psc.2672. [DOI] [PubMed] [Google Scholar]
- 7.Derossi D, et al. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269(14):10444–10450. [PubMed] [Google Scholar]
- 8.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272(25):16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 9.Zorko M, Langel Ü. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57(4):529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Snyder EL, Dowdy SF. Cell penetrating peptides in drug delivery. Pharm Res. 2004;21(3):389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 11.Schwarze SR, et al. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 12.Josephson L, et al. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconjug Chem. 1999;10(2):186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Q-Y, et al. Gene delivery to tumor cells by cationic polymeric nanovectors coupled to folic acid and the cell-penetrating peptide octaarginine. Biomaterials. 2011;32(29):7253–7262. doi: 10.1016/j.biomaterials.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Asai T, et al. Cell-penetrating peptide-conjugated lipid nanoparticles for siRNA delivery. Biochem Biophys Res Commun. 2014;444(4):599–604. doi: 10.1016/j.bbrc.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 15.Lehto T, Kurrikoff K, Langel Ü. Cell-penetrating peptides for the delivery of nucleic acids. Expert Opin Drug Deliv. 2012;9(7):823–836. doi: 10.1517/17425247.2012.689285. [DOI] [PubMed] [Google Scholar]
- 16.Juliano RL, et al. Cell-targeting and cell-penetrating peptides for delivery of therapeutic and imaging agents. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1(3):324–335. doi: 10.1002/wnan.4. [DOI] [PubMed] [Google Scholar]
- 17.Bechara C, Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013;587(12):1693–1702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Raucher D, Ryu JS. Cell-penetrating peptides: strategies for anticancer treatment. Trends Mol Med. 2015;21(9):560–570. doi: 10.1016/j.molmed.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Foged C, Nielsen HM. Cell-penetrating peptides for drug delivery across membrane barriers. Expert Opin Drug Deliv. 2008;5(1):105–117. doi: 10.1517/17425247.5.1.105. [DOI] [PubMed] [Google Scholar]
- 20.Subrizi A, et al. Tat (48-60) peptide amino acid sequence is not unique in its cell penetrating properties and cell-surface glycosaminoglycans inhibit its cellular uptake. J Control Release. 2012;158(2):277–285. doi: 10.1016/j.jconrel.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Åmand HL, et al. Stimulated endocytosis in penetratin uptake: effect of arginine and lysine. Biochem Biophys Res Commun. 2008;371(4):621–625. doi: 10.1016/j.bbrc.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell DJ, et al. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res. 2000;56(5):318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 23.Kalafatovic D, Giralt E. Cell-penetrating peptides: design strategies beyond primary structure and amphipathicity. Molecules. 2017;22(11):1929. doi: 10.3390/molecules22111929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franz J, et al. SAP (E)—a cell-penetrating polyproline helix at lipid interfaces. Biochim Biophys Acta (BBA) Biomembr. 2016;1858(9):2028–2034. doi: 10.1016/j.bbamem.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Kristensen M, Birch D, Mørck Nielsen H. Applications and challenges for use of cell-penetrating peptides as delivery vectors for peptide and protein cargos. Int J Mol Sci. 2016;17(2):185. doi: 10.3390/ijms17020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rydberg HA, et al. Effects of tryptophan content and backbone spacing on the uptake efficiency of cell-penetrating peptides. Biochemistry. 2012;51(27):5531–5539. doi: 10.1021/bi300454k. [DOI] [PubMed] [Google Scholar]
- 27.Borrelli A, et al. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules. 2018;23(2):295. doi: 10.3390/molecules23020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rydström A, et al. Direct translocation as major cellular uptake for CADY self-assembling peptide-based nanoparticles. PLoS One. 2011;6(10):e25924. doi: 10.1371/journal.pone.0025924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Z, et al. Cell-penetrating peptides: possible transduction mechanisms and therapeutic applications. Biomed Rep. 2016;4(5):528–534. doi: 10.3892/br.2016.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simeoni F, et al. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31(11):2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Copolovici DM, et al. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8(3):1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 32.Avci F, Sariyar Akbulut B, Ozkirimli E. Membrane active peptides and their biophysical characterization. Biomolecules. 2018;8(3):77. doi: 10.3390/biom8030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mussbach F, et al. Transduction of peptides and proteins into live cells by cell penetrating peptides. J Cell Biochem. 2011;112(12):3824–3833. doi: 10.1002/jcb.23313. [DOI] [PubMed] [Google Scholar]
- 34.Brock R. The uptake of arginine-rich cell-penetrating peptides: putting the puzzle together. Bioconjug Chem. 2014;25(5):863–868. doi: 10.1021/bc500017t. [DOI] [PubMed] [Google Scholar]
- 35.Vivès E, Schmidt J, Pèlegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta (BBA) Rev Cancer. 2008;1786(2):126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 36.LeCher JC, Nowak SJ, McMurry JL. Breaking in and busting out: cell-penetrating peptides and the endosomal escape problem. Biomol Concepts. 2017;8(3–4):131–141. doi: 10.1515/bmc-2017-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang T, et al. Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc Natl Acad Sci USA. 2004;101(51):17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, et al. Tumor targeting and microenvironment-responsive nanoparticles for gene delivery. Biomaterials. 2013;34(21):5294–5302. doi: 10.1016/j.biomaterials.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 39.Jiang B. Aerobic glycolysis and high level of lactate in cancer metabolism and microenvironment. Genes Dis. 2017;4(1):25–27. doi: 10.1016/j.gendis.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen VP, et al. A novel soluble peptide with pH-responsive membrane insertion. Biochemistry. 2015;54(43):6567–6575. doi: 10.1021/acs.biochem.5b00856. [DOI] [PubMed] [Google Scholar]
- 41.Gao W, Chan JM, Farokhzad OC. pH-responsive nanoparticles for drug delivery. Mol Pharm. 2010;7(6):1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daumar P, et al. Efficient 18F-labeling of large 37-amino-acid pHLIP peptide analogues and their biological evaluation. Bioconjug Chem. 2012;23(8):1557–1566. doi: 10.1021/bc3000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reshetnyak YK, et al. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol. 2011;13(6):1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vāvere AL, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Can Res. 2009;69(10):4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karabadzhak AG, et al. pHLIP-FIRE, a cell insertion-triggered fluorescent probe for imaging tumors demonstrates targeted cargo delivery in vivo. ACS Chem Biol. 2014;9(11):2545–2553. doi: 10.1021/cb500388m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An M, et al. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci. 2010;107(47):20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moshnikova A, et al. Antiproliferative effect of pHLIP-amanitin. Biochemistry. 2013;52(7):1171–1178. doi: 10.1021/bi301647y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reshetnyak YK, et al. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci. 2006;103(17):6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunt JF, et al. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 1997;36(49):15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 50.Andreev OA, Engelman DM, Reshetnyak YK. Targeting diseased tissues by pHLIP insertion at low cell surface pH. Front Physiol. 2014;5:1–7. doi: 10.3389/fphys.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao L, et al. pHLIP peptide targets nanogold particles to tumors. Proc Natl Acad Sci. 2013;110(2):465–470. doi: 10.1073/pnas.1219665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreev OA, Engelman DM, Reshetnyak YK. Targeting acidic diseased tissue: new technology based on use of the pH (Low) Insertion Peptide (pHLIP) Chim Oggi. 2009;27(2):34. [PMC free article] [PubMed] [Google Scholar]
- 53.Andreev OA, Engelman DM, Reshetnyak YK. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol. 2010;27(7):341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deacon JC, Engelman DM, Barrera FN. Targeting acidity in diseased tissues: mechanism and applications of the membrane-inserting peptide, pHLIP. Arch Biochem Biophys. 2015;565:40–48. doi: 10.1016/j.abb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zoonens M, Reshetnyak YK, Engelman DM. Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors. Biophys J. 2008;95(1):225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Y, et al. pH-responsive pHLIP (pH low insertion peptide) nanoclusters of superparamagnetic iron oxide nanoparticles as a tumor-selective MRI contrast agent. Acta Biomater. 2017;55:194–203. doi: 10.1016/j.actbio.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 57.Wyatt LC, Lewis JS, Andreev OA, Reshetnyak YK, Engelman DM. Applications of pHLIP technology for cancer imaging and therapy. Trends Biotechnol. 2017;35(7):653–664. doi: 10.1016/j.tibtech.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weerakkody D, et al. Family of pH (low) insertion peptides for tumor targeting. Proc Natl Acad Sci. 2013;110(15):5834–5839. doi: 10.1073/pnas.1303708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reshetnyak YK, et al. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc Natl Acad Sci. 2008;105(40):15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wijesinghe D, et al. pH dependent transfer of nano-pores into membrane of cancer cells to induce apoptosis. Sci Rep. 2013;3:3560. doi: 10.1038/srep03560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Q, et al. A smart tumor targeting peptide–drug conjugate, pHLIP-SS-DOX: synthesis and cellular uptake on MCF-7 and MCF-7/Adr cells. Drug Deliv. 2016;23(5):1734–1746. doi: 10.3109/10717544.2015.1028601. [DOI] [PubMed] [Google Scholar]
- 62.Kyrychenko A. NANOGOLD decorated by pHLIP peptide: comparative force field study. Phys Chem Chem Phys. 2015;17(19):12648–12660. doi: 10.1039/C5CP01136A. [DOI] [PubMed] [Google Scholar]
- 63.Anderson MD (2015) Development of new tools for study of tumor microenvironment. Open access Dissertations. Paper 359
- 64.Demoin DW, et al. PET imaging of extracellular pH in tumors with 64Cu-and 18F-labeled pHLIP peptides: a structure–activity optimization study. Bioconjug Chem. 2016;27(9):2014–2023. doi: 10.1021/acs.bioconjchem.6b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andreev OA, et al. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc Natl Acad Sci. 2010;107(9):4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karabadzhak AG, et al. Modulation of the pHLIP transmembrane helix insertion pathway. Biophys J. 2012;102(8):1846–1855. doi: 10.1016/j.bpj.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shu NS, et al. Residue-specific structures and membrane locations of pH-low insertion peptide by solid-state nuclear magnetic resonance. Nat Commun. 2015;6:1–10. doi: 10.1038/ncomms8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanz SZ, et al. Protonation-driven membrane insertion of a pH-low insertion peptide. Angew Chem Int Ed. 2016;55(40):12376–12381. doi: 10.1002/anie.201605203. [DOI] [PubMed] [Google Scholar]
- 69.Narayanan T, et al. pHLIP peptide interaction with a membrane monitored by SAXS. J Phys Chem B. 2016;120(44):11484–11491. doi: 10.1021/acs.jpcb.6b06643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniels JL, et al. Synthesis and characterization of pHLIP® coated gold nanoparticles. Biochem Biophys Rep. 2017;10:62–69. doi: 10.1016/j.bbrep.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott HL, et al. The negative charge of the membrane has opposite effects on the membrane entry and exit of pH-low insertion peptide. Biochemistry. 2015;54(9):1709–1712. doi: 10.1021/acs.biochem.5b00069. [DOI] [PubMed] [Google Scholar]
- 72.Gupta C, Mertz B. Protonation Enhances the Inherent Helix-Forming Propensity of pHLIP. ACS Omega. 2017;2(11):8536–8542. doi: 10.1021/acsomega.7b01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sosunov EA, et al. pH (low) insertion peptide (pHLIP) targets ischemic myocardium. Proc Natl Acad Sci. 2013;110(1):82–86. doi: 10.1073/pnas.1220038110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vasquez-Montes V, et al. Comparison of lipid-dependent bilayer insertion of pHLIP and its P20G variant. Biochim Biophys Acta (BBA) Biomembr. 2018;1860(2):534–543. doi: 10.1016/j.bbamem.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kyrychenko A, et al. Lipid headgroups modulate membrane insertion of pHLIP peptide. Biophys J. 2015;108(4):791–794. doi: 10.1016/j.bpj.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wyatt LC, et al. Peptides of pHLIP family for targeted intracellular and extracellular delivery of cargo molecules to tumors. Proc Natl Acad Sci. 2018;115(12):E2811–E2818. doi: 10.1073/pnas.1715350115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrova V, et al. The hypoxic tumour microenvironment. Oncogenesis. 2018;7(1):10. doi: 10.1038/s41389-017-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim Y, et al. Hypoxic tumor microenvironment and cancer cell differentiation. Curr Mol Med. 2009;9(4):425–434. doi: 10.2174/156652409788167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harada H, Hiraoka M, Kizaka-Kondoh S. Antitumor effect of TAT-oxygen-dependent degradation-caspase-3 fusion protein specifically stabilized and activated in hypoxic tumor cells. Can Res. 2002;62(7):2013–2018. [PubMed] [Google Scholar]
- 82.Nalla AK, et al. Targeting MMP-9, uPAR, and cathepsin B inhibits invasion, migration and activates apoptosis in prostate cancer cells. Cancer Gene Ther. 2010;17(9):599. doi: 10.1038/cgt.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polette M, et al. Tumour invasion and matrix metalloproteinases. Crit Rev Oncol Hematol. 2004;49(3):179–186. doi: 10.1016/j.critrevonc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 84.Weidle UH, Tiefenthaler G, Georges G. Proteases as activators for cytotoxic prodrugs in antitumor therapy. Cancer Genom Proteom. 2014;11(2):67–79. [PubMed] [Google Scholar]
- 85.Olson ES, et al. In vivo characterization of activatable cell penetrating peptides for targeting protease activity in cancer. Integr Biol. 2009;1(5–6):382–393. doi: 10.1039/b904890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hofmann H-S, et al. Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res. 2005;11(3):1086–1092. [PubMed] [Google Scholar]
- 87.Gong Y, Chippada-Venkata UD, Oh WK. Roles of matrix metalloproteinases and their natural inhibitors in prostate cancer progression. Cancers. 2014;6(3):1298–1327. doi: 10.3390/cancers6031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Duijnhoven SM, et al. Tumor targeting of MMP-2/9 activatable cell-penetrating imaging probes is caused by tumor-independent activation. J Nucl Med. 2011;52(2):279–286. doi: 10.2967/jnumed.110.082503. [DOI] [PubMed] [Google Scholar]
- 89.Aguilera TA, et al. Systemic in vivo distribution of activatable cell penetrating peptides is superior to that of cell penetrating peptides. Integr Biol. 2009;1(5–6):371–381. doi: 10.1039/b904878b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson CF, Cui H. Protease-sensitive nanomaterials for cancer therapeutics and imaging. Ind Eng Chem Res. 2017;56(20):5761–5777. doi: 10.1021/acs.iecr.7b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi N-Q, et al. Enhancing cellular uptake of activable cell-penetrating peptide–doxorubicin conjugate by enzymatic cleavage. Int J Nanomed. 2012;7:1613. doi: 10.2147/IJN.S30104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang T, et al. Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc Natl Acad Sci. 2004;101(51):17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen QT, et al. Surgery with molecular fluorescence imaging using activatable cell-penetrating peptides decreases residual cancer and improves survival. Proc Natl Acad Sci. 2010;107(9):4317–4322. doi: 10.1073/pnas.0910261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malhotra M, et al. Development and characterization of chitosan-PEG-TAT nanoparticles for the intracellular delivery of siRNA. Int J Nanomed. 2013;8:2041. doi: 10.2217/nnm.13.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee SH, et al. Activatable cell penetrating peptide–peptide nucleic acid conjugate via reduction of azobenzene PEG chains. J Am Chem Soc. 2014;136(37):12868–12871. doi: 10.1021/ja507547w. [DOI] [PubMed] [Google Scholar]
- 96.Wender PA, et al. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc Natl Acad Sci. 2000;97(24):13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin E, et al. Acid-active cell-penetrating peptides for in vivo tumor-targeted drug delivery. J Am Chem Soc. 2013;135(2):933–940. doi: 10.1021/ja311180x. [DOI] [PubMed] [Google Scholar]
- 98.Bode SA, et al. Enzyme-activatable cell-penetrating peptides through a minimal side chain modification. Bioconjug Chem. 2015;26(5):850–856. doi: 10.1021/acs.bioconjchem.5b00066. [DOI] [PubMed] [Google Scholar]
- 99.Vocero-Akbani AM, et al. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat Med. 1999;5(1):29. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 100.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mathupala S, Ko YA, Pedersen P. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25(34):4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arzoine L, et al. Voltage-dependent anion channel 1-based peptides interact with hexokinase to prevent its anti-apoptotic activity. J Biol Chem. 2009;284(6):3946–3955. doi: 10.1074/jbc.M803614200. [DOI] [PubMed] [Google Scholar]
- 103.Woldetsadik AD, et al. Hexokinase II–derived cell-penetrating peptide targets mitochondria and triggers apoptosis in cancer cells. FASEB J. 2017;31(5):2168–2184. doi: 10.1096/fj.201601173R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277(9):7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 105.Patra KC, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24(2):213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rezgui R, et al. Precise quantification of cellular uptake of cell-penetrating peptides using fluorescence-activated cell sorting and fluorescence correlation spectroscopy. Biochim Biophys Acta (BBA) Biomembr. 2016;1858(7):1499–1506. doi: 10.1016/j.bbamem.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 107.Takayama K, et al. Effect of the attachment of a penetration accelerating sequence and the influence of hydrophobicity on octaarginine-mediated intracellular delivery. Mol Pharm. 2012;9(5):1222–1230. doi: 10.1021/mp200518n. [DOI] [PubMed] [Google Scholar]
- 108.Kondo E, et al. Tumour lineage-homing cell-penetrating peptides as anticancer molecular delivery systems. Nat Commun. 2012;3:951. doi: 10.1038/ncomms1952. [DOI] [PubMed] [Google Scholar]
- 109.Shi J, et al. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zuo HD, et al. The effect of superparamagnetic iron oxide with iRGD peptide on the labeling of pancreatic cancer cells in vitro: a preliminary study. BioMed Res Int. 2014;2014:1–8. doi: 10.1155/2014/852352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shirazi AN, et al. Cyclic peptide-capped gold nanoparticles for enhanced siRNA delivery. Molecules. 2014;19(9):13319–13331. doi: 10.3390/molecules190913319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li H, Tsui T, Ma W. Intracellular delivery of molecular cargo using cell-penetrating peptides and the combination strategies. Int J Mol Sci. 2015;16(8):19518–19536. doi: 10.3390/ijms160819518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qin H, et al. Tumor microenvironment targeting and responsive peptide-based nanoformulations for improved tumor therapy. Mol Pharmacol. 2017;1:1. doi: 10.1124/mol.116.108084. [DOI] [PubMed] [Google Scholar]
- 114.Yoo J, et al. Protease-activatable cell-penetrating peptide possessing ROS-triggered phase transition for enhanced cancer therapy. J Control Release. 2017;264:89–101. doi: 10.1016/j.jconrel.2017.08.026. [DOI] [PubMed] [Google Scholar]