Abstract
Alternative splicing is a regulatory process of gene expression based on the flexibility in the selection of splice sites. In this manuscript we present the characterisation of an alternative splicing of the NF1 pre-mRNA induced by cold-shock conditions. We demonstrate that the accuracy of the splicing mechanism was perturbed after keeping samples for a short period of time at room temperature, resulting in the insertion of a 31-bp cryptic exon between exons 4a and 4b of the NF1 mRNA. This alternative splicing is not cell type specific and is not induced by other stress conditions such as heat shock or hyper-osmolarity. The alternative spliced mRNA is efficiently transported to the cytoplasm and it is proven to belong to the poly A+ mRNA fraction. Previous misleading interpretations about this transcript, together with our finding relating its presence to cold shock and not to the NF1 disease, strongly indicate that this phenomenon should be taken into account in genetic testing when RNA methodology is used for mutation detection. This is the first description of an alternative splicing induced by cold shock in a human pre-mRNA and should provide further insights into the factors that control alternative splicing.
INTRODUCTION
RNA splicing is the cellular mechanism responsible for the removal of intronic sequences from the primary transcript and the fusion of exonic sequences. Alternative splicing of pre-mRNA is a common process of differential gene expression whereby different exon combinations are included in transcripts from the same gene during RNA processing (1–3). It is regulated by factors that modify the relative rates of splice site recognition. Pre-mRNA structure including splice-site strength, exon size and RNA secondary structure strongly influence splice-site selection (4,5). Several cis-acting regulatory elements, including intron and exon sequences, and trans-acting factors are necessary to control alternative splicing in vertebrates (6–8). Trans-acting factors bind to cis-acting regulatory elements in the pre-mRNA to regulate interactions between pre-mRNA and spliceosome machinery and influence splice site choice (1,3,9–11).
The use of alternative splice sites has been shown to be dependent on stress conditions in vitro. In plants, temperature has been found to regulate alternative splicing. Pre-mRNA from an invertase gene in potato was subject to exon skipping induced by cold stress (12) and in granule-bound starch synthase (GBSS) pre-mRNA the selection of leader intron 5′ splice sites is also affected by temperature (13). In mouse, the heat shock protein 47 (HSP47) has been found to present an alternative splicing induced by heat shock (14). In rat, the pattern of splice site selection of the tropomyosin I pre-mRNA in vitro varies depending on the ionic conditions (15), as previously found studying E1A transcripts (16). In humans, only one case of temperature-dependent alternative splicing has been described in fibroblasts of a patient affected by Ehlers–Danlos syndrome (EDS) type VII (17). This patient bore a mutation in the last nucleotide of exon 6 of the COL1A1 gene, which produced an aberrant spliced transcript not detectable at 31°C which gradually increased up to 39°C.
Neurofibromatosis type 1 (NF1) is a common genetic autosomal dominant neurocutaneous disorder, with an incidence of around 1 in 3500 newborns. At the clinical level it is mainly characterised by the presence of café-au-lait spots, neurofibromas and Lisch nodules (18). The NF1 gene is located in the human chromosome 17 (17q11.2), contains 60 exons, some of them alternatively spliced, and produces a 11–13 kb ubiquitously expressed transcript (19–23). The NF1 product, neurofibromin, is a member of the GTPase-activating proteins acting as a downregulator of Ras (24–27). To date, eight NF1 alternative transcripts expressed in humans, in addition to the original first described, have been reported. Four of them contain supplementary coding regions through inclusion of additional exons in frame (22,28–30). The remaining four introduce a frame shift giving truncated transcripts, three of them by skipping constitutive exons (31,32). In this paper, we describe that cold shock induces the insertion of a cryptic exon in the NF1 mRNA, located between exons 4a and 4b. To our knowledge, this is the first report of alternative splicing regulated by cold stress in humans.
MATERIALS AND METHODS
Samples and nucleic acid extraction
Peripheral lymphocytes were isolated from 5 ml of whole blood by centrifugation on Ficoll-Paque (Pharmacia, Upsala, Sweden). Fibroblasts and the osteoblastoma cell line U2OS (American Type Culture Collection, MD, USA) were grown in DMEM supplemented with 10% foetal calf serum, 50 U/ml penicillin, 50 µg/ml streptomycin and 2 mM l-glutamine at 37°C in an atmosphere of 5% CO2. RNA isolation was performed on peripheral blood lymphocytes, on EBV-transformed lymphoblastoid cell lines, on primary cultured fibroblasts from normal skin, on the U2OS cell line and on freshly frozen human tissues using the Tripure isolation reagent (Boehringer Mannheim, Mannheim, Germany), following the conditions recommended by the manufacturer. Cytoplasmic RNA was prepared using RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Poly A+ RNA was purified from cytoplasmic RNA using the Oligotex Direct mRNA kit (Qiagen) as recommended. DNA from peripheral blood lymphocytes of each patient was extracted by the ‘salting out’ method (33).
cDNA synthesis, PCR amplification, single strand conformation polymorphism/heteroduplex (SSCP/HD) analysis and sequencing
Total RNA (2–5 µg) was reverse transcribed into cDNA using random hexamers and Superscript II reverse transcriptase (Gibco BRL, Life Technologies, Paisley, UK) according to the manufacturer’s instructions. Amplification of the 5′ end of the NF1 cDNA was performed (1 min at 94°C, 1 min at 58°C and 1 min at 74°C for 40 cycles) using primers I1D and I1R designed by Hoffmeyer et al. (34). SSCP/HD of this PCR product was performed as described elsewhere (35). Sequencing was performed using an automatic DNA analyser (ABI PRISMTM 377 DNA Sequencer).
Genomic structure and characterisation of exon 4a-2
The characterisation of the genomic region surrounding the cryptic exon was performed by sequencing two PCR products derived from long PCRs using the Expand long template system (Boehringer Mannheim). These fragments were obtained by utilising primers containing sequence of the alternative exon 4a-2 in both senses against primers located in exons 4a and 4b. The amplification conditions were 35 cycles at 94°C for 10 s, 56°C for 30 s and 68°C for 5 min. The primer sequences were as follows: forward E4aD2: 5′-GCTGGTCAAACAGTTGCTGC-3′ (corresponding to exon 4a), reverse insR: 5′-GAGGGAAGTATCATCAAGG-3′ (corresponding to exon 4a-2 in antisense orientation), forward insD: 5′-CCTTGATGATACTTCCCTC-3′ (corresponding to exon 4a-2 in sense orientation) and the reverse primer E4bR (corresponding to exon 4b) designed by Li et al. (23). To amplify exon 4a-2 from genomic DNA we designed intronic primers surrounding this cryptic exon, forward primer IVS4aD: 5′-CTTTATCACAAGATTTTAGGTTAC-3′ and reverse primer IVS4aR: 5′-AGTGAAACTAATATAAATTCTGAAG-3′.
Alternative splicing
To assay for alternative splicing events involving exon 4a-2 we amplified a smaller region by RT–PCR. The primers used were E4aD2 and I1R (94°C for 30 s, 58°C for 30 s and 74°C for 1 min for 25 cycles) yielding a product of 308 bp in the absence of exon 4a-2 and of 339 bp in the presence of this alternative exon. This PCR was performed using total RNA, total cytoplasmic RNA and cytoplasmic poly A+ RNA as a template, the latter two to check if the alternative transcript was efficiently transported to the cytoplasm. Histone H1 (forward primer H1D: 5′-ACTGCTCCACTTGCTCCTAC-3′ and reverse primer H1R: 5′-GCTTCCTAGGCTTGGCTGC-3′) and GAPDH (glyceraldehyde phosphate dehydrogenase) (36) cDNA fragments were amplified as controls of poly A– and poly A+ mRNA, respectively.
Stress conditions, specificity and reversibility
Whole blood, EBV-transformed lymphoblastoid cells, primary fibroblasts in culture and U2OS cell line were exposed to several different stress conditions. Cold shock (15, 17, 20, 25 and 32°C) and heat shock (42°C) were assayed for different periods of time (0, 3, 6, 12, 18, 24, 36, 48 and 72 h). Hyperosmolarity (200 and 500 mM sorbitol) for 30 and 60 min and deprivation of serum for 48 h were also tested. To study the reversibility of the cold shock induction, fibroblasts were held for 3, 6 and 24 h at room temperature, and then recovered at 37°C for 3 days.
RESULTS
Identification and characterisation of exon 4a-2
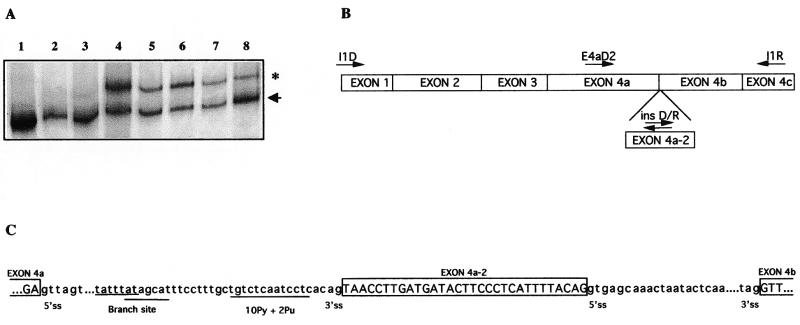
In the process of the identification of germline mutations in NF1 patients by cDNA-SSCP/HD analysis (35) we detected an abnormal band at the most 5′ fragment, corresponding to nucleotides 1–632 of the NF1 cDNA (nucleotide 1 denotes start site of translation). This abnormal band, with an intensity similar to that corresponding to the normal one, was first observed in the analysis of blood samples shipped to our laboratory from an NF1 family of the Balearic Islands and was present in all members of the kindred, not co-segregating with the disease (Fig. 1A). Cloning and sequencing of this fragment revealed an insertion of 31 bp between nucleotides 479 and 480 of the NF1 cDNA. Since this insertion occurs at the junction between exons 4a and 4b, we named this cryptic exon 4a-2 (Fig. 1B). This 31-bp insertion was out of frame and would lead to a truncated NF1 protein containing only 183 residues, with the last 23 being different to the wild-type NF1 protein. The existence of the 31-bp sequence at the genomic level was confirmed by hybridising against PAC clones containing the 5′ NF1 region (data not shown), indicating that this cDNA insertion could arise from intron 4a. Since the genomic sequence for this intron was not available in public databases at time of analysis, we performed several long PCRs and were able to estimate the size of this intron to be ~6–7 kb and to situate the 31-bp insertion 672 bp 3′ from exon 4a. Interestingly, the nucleotide sequences located at the boundaries of exon 4a-2 conform to the consensus GT-AG splicing rule (Fig. 1C). By using the Splice Site by Prediction Neural Network (http://www.fruitfly.org/seq_tools/splice.html ), the scores for the donor and acceptor sites of exon 4a-2 are of 0.99 and 0.51, respectively. Whilst preparing this manuscript, a sequence of 119 kb belonging to the 5′ end of the NF1 gene has been introduced in GeneBank (AC0004222). This sequence confirmed our data about the genomic organisation of this region of the NF1 gene and evidenced that this exon is embedded in a mammalian interspersed repetitive (MIR) element (http://repeatmasker.genome.washington.edu/cgi-bin ) located in intron 4a.
Figure 1.
(A) SSCP/HD analysis of the NF1 cDNA fragment containing exons 1 to 4c from lymphocyte RNA of different individuals. The arrowhead indicates the normal NF1 fragment in the heteroduplex and the asterisk highlights the fragment corresponding to the alternative splicing. Lanes 1–3 are from subjects in which RNA was extracted immediately after venipuncture and lanes 4–8 show samples shipped to our laboratory in which this transcript was first visualised. Lanes 2, 3, 6 and 8 correspond to NF1 patients. (B) Structure of the 5′ end of the NF1 cDNA. Arrows indicate the orientation of the primers used for the different PCR amplifications. The cryptic exon 4a-2 is shown. (C) Partial genomic sequence of the region surrounding the alternative exon 4a-2 in which is indicated the conserved consensus sequence for splicing. In uppercase letters is shown the coding sequence and in lowercase the intronic sequences.
To test whether the insertion of exon 4a-2 was associated with any change in genomic DNA, we amplified and sequenced the genomic region containing exon 4a-2 and intron boundaries in all members of the family in which the exon was first detected and in a set of 25 controls and 25 other NF1 patients. Sequence analysis did not reveal any change in any of the samples, allowing us to conclude that exon 4a-2 was an alternative exon.
Pattern of expression of exon 4a-2 in different tissues and cell lines
To discern whether the expression of the alternative exon 4a-2 could have any association with NF1, we analysed the presence of this exon by RT–PCR using primers E4aD2/IR in RNA from lymphocytes from 60 subjects (with and without the NF1 phenotype), 20 cutaneous neurofibromas, one plexiform neurofibroma, one café-au-lait spot, one astrocytoma cell line, and a set of different normal tissues: skin, testis, foetal and adult brain, foetal and adult kidney, foetal liver and lung. In addition, EBV-transformed lymphoblastoid cell lines were established from four of the individuals, two NF1 and two non-NF1, in which the alternative exon was initially identified. We observed that exon 4a-2 only appeared in some blood samples. The presence of this cryptic exon did not correlate with NF1 but with the origin of the sample, all specimens with the insertion were shipped to our laboratory and RNA was prepared one or several days after blood extraction.
Alternative splicing of exon 4a-2 is induced by cold stress and is not specific of blood lymphocytes
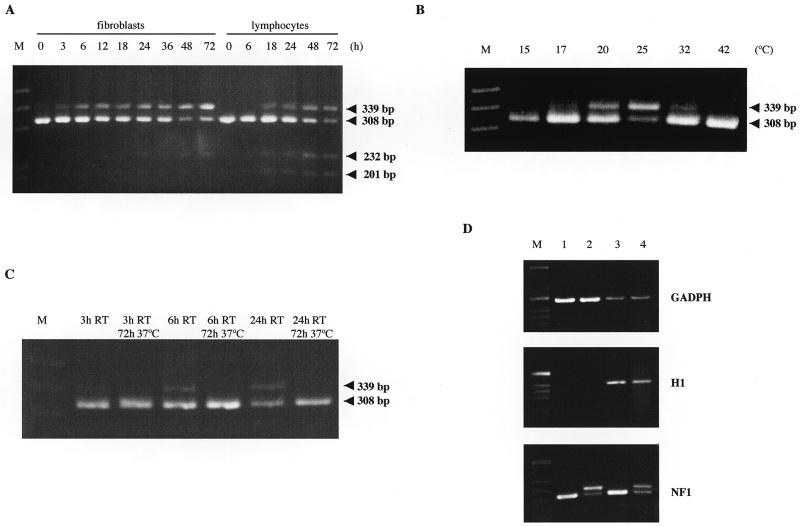
Temperature is a likely variable between RNA from freshly drawn blood versus RNA from blood that is shipped. To test whether the alternative splicing of this exon could be induced by cold stress, we held the blood after venipuncture at room temperature (~25°C) for different periods of time (Fig. 2A). After 18 h at room temperature two major products were detected, one of 308 bp and a larger of 339 bp, the latter being the product of alternative splicing of exon 4a-2. The amount of this second product increased with time of exposure to cold stress.
Figure 2.
RT–PCR analysis of alternative splicing of the NF1 mRNA using primers E4aD2 and I1R (A) from total RNA of fibroblasts and lymphocytes maintained at room temperature for the indicated time (h); (B) from total RNA of fibroblasts maintained 48 h at the indicated temperature (°C); (C) from total RNA of fibroblasts treated as indicated and (D) from poly A+ cytoplasmic RNA (lanes 1 and 2) and total cytoplasmic RNA (lanes 3 and 4) of control fibroblasts (lanes 1 and 3) and of cold shocked (lanes 2 and 4). Amplifications of GADPH and H1 were used as controls of poly A+ and poly A–, respectively. M, the molecular weight marker; RT, room temperature. The PCR product of 308 and 339 bp correspond to the normally spliced and the alternative transcript with exon 4a-2, respectively. The 201-bp product shows skipping of exon 4b and the upper band of 232 bp presented the same skipping plus the incorporation of the cryptic exon described above (4a-2).
To analyse the specificity of the alternative splicing phenomenon we have studied its occurrence in other cell types and under different stress conditions. RNA was extracted from primary fibroblasts in culture that had been held at room temperature for different periods of time (Fig. 2A). RT–PCR amplified the product containing the cryptic exon in all samples except the control. The same assay was performed using an osteoblastoma cell line and the same results were obtained (data not shown). These findings indicated that the alternative splicing was not specific to blood lymphocytes but can occur in other cell types.
In order to analyse the range of temperatures in which the alternative exon appeared, we exposed primary cultured fibroblasts to 15, 17, 20, 25 and 32°C for 48 h (Fig. 2B). We found that the expression of the alternative transcript was not detectable in fibroblasts held at 15°C, increased gradually up to 25°C and decreased at 32°C.
Reversibility of the alternative splicing and other stress conditions
To test if the splicing of this alternative exon is a reversible phenomenon we kept primary fibroblasts in culture at room temperature for 3, 6 and 24 h. The cells were then recovered at 37°C for 3 days. No evidence of the larger product was detected after the recovery period demonstrating that the splicing of the cryptic exon is a reversible process (Fig. 2C).
To analyse different stress conditions primary fibroblasts were (i) exposed to a 42°C heat shock for 48 h (Fig. 2B), (ii) treated with 200 and 500 mM sorbitol for 1 h and (iii) deprived of serum for 48 h. In none of these three conditions was exon 4a-2 detected by RT–PCR (data not shown).
The alternative transcript belongs to the cytoplasmic poly A+ mRNA fraction
In order to study if the RNA containing exon 4a-2 is a bona fide mRNA, cytoplasmic RNA was extracted from fibroblasts held at room temperature for 0 (control) and 48 h and poly A+ RNA was purified. We identified the presence of the alternative exon in both fractions, total and poly A+, confirming that this transcript is polyadenylated and efficiently transported to the cytoplasm (Fig. 2D).
Characterisation of additional bands present in the RT–PCR products
The agarose gel electrophoresis of the RT–PCR products revealed the presence of one faint extra band in cell controls (0 h) and two in cells submitted to cold shock (Fig. 2A). In order to investigate their nature, these bands were excised from the gel and sequenced. The 201-bp product showed skipping of exon 4b and was not associated to cold shock. The upper band of 232 bp presented the skipping of exon 4b plus the incorporation of the cryptic exon described above (4a-2), and was induced by cold shock.
DISCUSSION
We describe here a cold shock-induced alternative splicing consisting in the insertion of a cryptic exon (4a-2) that disrupts the open reading frame of the NF1 mRNA. This alternative spliced mRNA was efficiently transported to the cytoplasm and has been proven to belong to the poly A+ mRNA fraction, indicating that it could be translated into a truncated NF1 protein of 183 amino acids. This alternative splicing is not specific of blood lymphocytes and is not induced by other tested stress conditions such as heat shock or hyperosmolarity. Moreover, a low proportion of another alternative splicing consisting of exon 4b skipping was also identified. Its presence was not associated with temperature conditions but a transcript lacking exon 4b and bearing the insertion of the cryptic 4a-2 exon was also identified under cold stress.
In a gene as large as NF1, RNA splicing is a highly complex stage of gene expression. We recently described the importance of splicing defects in NF1, since in at least 32% of a set of 80 NF1 patients the disease was caused by splicing alterations (35). Several NF1 alternative transcripts expressed in humans have been identified (22,28–32). Illegitimate splicing of some constitutive NF1 exons has also been reported (37). These illegitimate transcripts were detected in all cell types examined but in all cases at very low levels, similar to the exon 4b skipping detected here. Furthermore, an alternative splicing consisting in the insertion of a cryptic exon (10a-2) between exons 10a and 10b was detected in different cell types (38), although to our knowledge it has not been investigated whether its expression is related to temperature or other stress conditions.
The cryptic exon 4a-2 has also been found independently by other groups (39–42). Park et al. (39) described an NF1 patient with five different aberrantly spliced transcripts, some of them including the insertion of the cryptic 31-bp exon and the skipping of exon 4b. These authors reported this complex aberrant splicing as the cause of the disease in this patient although they did not detect any genomic defect. Likewise, Osborn et al. (41) described skipping of exon 4b plus the 31-bp insertion as a cDNA change causing NF1 in a patient without characterising the mutation at the genomic level. Moreover, Wallace et al. (40) found the transcript containing exon 4a-2 in 15 cutaneous and plexiform neurofibromas, two neurofibrosarcomas and trace levels in other normal tissues, and suggested that this out of frame insertion could contribute to the inactivation of the NF1 locus in these tumours. Our results in neurofibromas do not support this finding and suggest that studies performed immediately after neurofibroma excision should be necessary in order to clarify the putative involvement of this transcript in these tumours. Finally, Messiaen et al. (42) detected this cryptic exon at low levels in different cell types in NF1 patients as well as in normal persons and suggested that it was not associated with the disease. These data and our results that link the presence of this cryptic exon to cold shock and not to the NF1 disease, indicate that it would be important to take into account this phenomenon in molecular NF1 diagnosis when RNA is used for mutation detection. Therefore, it is important to confirm RNA alterations in genomic DNA (35) to elucidate whether RNA changes represent an alternative transcript or reflect the result of a genomic mutation.
The length of the internal exons plays an important role in splice site selection (4), with an average size for vertebrates of 137 nt (43). Dominiski et al. (44) performed experiments modifying the size of internal constitutive exons and found that exons shortened to <51 nt were generally ignored by the splicing machinery, being spliced in the resulting transcript. Alternative exons identified in NF1 transcripts have lengths of around this critical size: 30 bp exon 9br, 63 bp exon 23a and 54 bp exon 48a. The 31-bp exon that we describe here could be skipped in non-cold stress conditions since the proximity of its 3′ and 5′ splice site may impair the assembly of the splicing machinery. It has largely been reported that temperature influences RNA secondary structure (45). In order to explain the insertion of exon 4a-2 under cold stress, we hypothesise that the temperature decrease could modify pre-mRNA secondary structure favouring the interaction between the splicing machinery and the consensus splicing sequences. This hypothesis could be extended to other cases, supporting the idea that temperature changes could alter pre-mRNA secondary structure either favouring this interaction, or making it difficult, depending on the local sequence of the pre-mRNA. An example for the latter could be the potato invertase pre-mRNA described previously (12). Another possibility could be that temperature regulates the activity of a trans-acting factor facilitating the recognition of this cryptic exon in cold shock conditions.
To date, the only well-defined function of neurofibromin is the down-regulation of the Ras proteins (24–27). It is well established that Ras participates in the best characterised mitogen-activated protein kinase (MAPK) cascade, the Ras-Raf-ERK (extracellular signal-regulated protein kinase) pathway (46). It is also recognised that MAPK cascades can mediate responses to stress related stimuli, via c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) and p38 pathways (47). With the data we have up to now we are not able to discern whether the phenomenon described here also occurs in any physiological condition. If so, it could be speculated that this alternative splicing would be a way of NF1 inactivation which would lead to the activation of the stress related pathways.
Although to our knowledge this is the first report of an alternative splicing induced by cold stress in a human gene, it is possible that this phenomenon also affects other genes. Further studies are needed to elucidate whether this is a common mechanism in the human genome and to investigate the possibility of a physiological function for this putative truncated NF1 protein.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to the patients who participated in the study. We thank Helena Kruyer for help in the preparation of the manuscript; Rafael de Cid for making cell lines; Carles Pucharcós and Félix Rueda for helpful comments and suggestions. This work was supported by grants of the Fondo de Investigaciones Sanitarias de la Seguridad Social (98-0992), the Institut Català de la Salut, the Ministerio de Educación y Ciencia (CICYT/SAF96-1787-E) and the Generalitat de Catalunya (CIRIT/1997SGR-00085). E.A. is a fellow of the Comissió Interdepartamental de Recerca i Innovació Tecnològica of the Generalitat de Catalunya. S.L. is supported by the Spanish Ministry of Education and Science.
REFERENCES
- 1.Smith C.W.J., Patton,J.G. and Nadal-Ginard,B. (1989) Annu. Rev. Genet., 23, 527–577. [DOI] [PubMed] [Google Scholar]
- 2.Cooper T.A. and Mattox,W. (1997) Am. J. Hum. Genet., 61, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez A.J. (1998) Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 4.Sterner D.A. and Berget,S.M. (1993) Mol. Cell. Biol., 13, 2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman T.P. and Roesser,J.R. (1998) Biochemistry, 37, 15941–15950. [DOI] [PubMed] [Google Scholar]
- 6.Reed R. and Maniatis,T. (1986) Cell, 46, 681–690. [DOI] [PubMed] [Google Scholar]
- 7.Del Gatto F. and Breathnach,R. (1995) Mol. Cell. Biol., 15, 4825–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlo T., Sterner,D.A. and Berget,S.M. (1996) RNA, 2, 342–353. [PMC free article] [PubMed] [Google Scholar]
- 9.Reed R. (1996) Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- 10.Adams M.D., Rudner,D.Z. and Rio,D.C. (1996) Curr. Opin. Cell. Biol., 8, 331–339. [DOI] [PubMed] [Google Scholar]
- 11.Krecic A.M. and Swanson,M.S. (1999) Curr. Opin. Cell. Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 12.Bournay A.S., Hedley,P.E., Maddison,A., Waugh,R. and Machray,G.C. (1996) Nucleic Acids Res., 24, 2347–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin P.D. and Park,W.D. (1999) Plant Mol. Biol., 40, 719–727. [DOI] [PubMed] [Google Scholar]
- 14.Takechi H., Hosokawa,N., Hirayoshi,K. and Nagata,K. (1994) Mol. Cell. Biol., 14, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helfman D.M., Ricci,W.M. and Finn,L.A. (1988) Genes Dev., 2, 1627–1638. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt P., Gattoni,R., Keohavong,P. and Stevenin,J. (1987) Cell, 50, 31–39. [DOI] [PubMed] [Google Scholar]
- 17.Weil D., D’Alessio,M., Ramirez,F., Steinmann,B., Wirtz,M.K., Glanville,R.W. and Hollister,D.W. (1989) J. Biol. Chem., 264, 16804–16809. [PubMed] [Google Scholar]
- 18.Riccardi V.M. (1992) Neurofibromatosis: Phenotype, Natural History and Pathogenesis, 2nd Edn. Johns Hopkins University Press, Baltimore.
- 19.Cawthon R.M., Weiss,R., Xu,G., Viskochil,D., Culver,M., Stevens,J., Robertson,M., Dunn,D., Gesteland,R., O’Connell,P. and White,R. (1990) Cell, 62, 193–201. [DOI] [PubMed] [Google Scholar]
- 20.Viskochil D., Buchberg,A.M., Xu,G., Cawthon,R.M., Stevens,J., Wolff,R.K., Culver,M., Carey,J.C., Copeland,N.G., Jenkins,N.A., White,R. and O’Connell,P. (1990) Cell, 62, 187–192. [DOI] [PubMed] [Google Scholar]
- 21.Wallace M.R., Marchuk,D.A., Andersen,L.B., Letcher,R., Odeh,H.M., Saulino,A.M., Fountain,J.W., Brereton,A., Nicholson,J., Mitchell,A.L., Brownstein,B.H. and Collins,F.S. (1990) Science, 249, 181–186. [DOI] [PubMed] [Google Scholar]
- 22.Danglot G., Regnier,V., Fauvet,D., Vassal,G., Kujas,M. and Bernheim,A. (1995) Hum. Mol. Genet., 4, 915–920. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., O’Connell,P., Breidenbach,H.H., Cawthon,R., Stevens,J., Xu,G., Neil,S., Robertson,M., White,R. and Viskochil,D. (1995) Genomics, 25, 9–18. [DOI] [PubMed] [Google Scholar]
- 24.Martin G.A., Viskochil,D. and Bollag,G. (1990) Cell, 63, 843–849. [DOI] [PubMed] [Google Scholar]
- 25.Xu G., O’Connell,P., Viskochil,D., Cawthon,R., Robertson,M., Culver,M., Dunn,D., Stevens,J., Gesteland,R., White,R. and Weiss,R. (1990) Cell, 62, 599–608. [DOI] [PubMed] [Google Scholar]
- 26.Xu G., Lin,B., Tanaka,K., Dunn,D., Wood,D., Gesteland,R., White,R., Weiss,R. and Tamanoi,F. (1990) Cell, 63, 835–841. [DOI] [PubMed] [Google Scholar]
- 27.Ballester R., Marchuk,D., Boguski,M., Saulino,A., Letcher,R., Wigler,M. and Collins,F.S. (1990) Cell, 63, 851–859. [DOI] [PubMed] [Google Scholar]
- 28.Nishi T., Lee,P.S., Oka,K., Levin,V.A., Tanase,S., Morino,Y. and Saya,H. (1991) Oncogene, 6, 1555–1559. [PubMed] [Google Scholar]
- 29.Gutmann D.H., Andersen,L.B., Cole,J.L., Swaroop,M. and Collins,F.S. (1993) Hum. Mol. Genet., 2, 989–992. [DOI] [PubMed] [Google Scholar]
- 30.Skuse G.R. and Cappione,A.J. (1997) Hum. Mol. Genet., 6, 1707–1712. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H., Takahashi,K., Kubota,Y. and Shibahara,S. (1992) Biochem. Biophys. Res. Commun., 187, 984–990. [DOI] [PubMed] [Google Scholar]
- 32.Park V.M., Kenwright,K.A., Sturtevant,D.B. and Pivnick,E.K. (1998) Hum. Genet., 103, 382–385. [DOI] [PubMed] [Google Scholar]
- 33.Miller S.A., Dykes,D.D. and Polesky,H.F. (1988) Nucleic Acids Res., 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmeyer S., Assum,G., Kaufmann,D., Schwenk,K. and Krone,W. (1994) Hum. Genet., 94, 97–100. [DOI] [PubMed] [Google Scholar]
- 35.Ars E., Serra,E., García,J., Kruyer,H., Gaona,A., Lázaro,C. and Estivill,E. (2000) Hum. Mol. Genet., 9, 237–247. [DOI] [PubMed] [Google Scholar]
- 36.Strehlau J., Pavlakis,M., Lipman,M., Shapiro,M., Vasconcellos,L., Harmon,W. and Strom,T.B. (1997) Proc. Natl Acad. Sci. USA, 94, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlelt B., Hoffmeyer,S., Kruse,P., Leistner,W., Krone,W. and Kaufmann,D. (1999) Medizinische Genetik, 3, 487. [Google Scholar]
- 38.Kaufmann D., Kruse,P. and Barlelt,B. (1999) Medizinische Genetik, 3, 487. [Google Scholar]
- 39.Park V.M. and Pivnick,E.K. (1998) J. Med. Genet., 35, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace M., Trimpert,R.E., Thomson,S.A.M., Fishbein,L. and Abernathy,C.R. (1998) Am. J. Hum. Genet., S63, A90. [Google Scholar]
- 41.Osborn M.J. and Upadhyaya,M. (1999) Hum. Genet., 105, 327–332. [DOI] [PubMed] [Google Scholar]
- 42.Messiaen L., Callens,T., Roux,K., Mortier,G., De Paepe,A., Abramowicz,M., Pericak-Vance,M., Vance,J. and Wallace,M. (1999) Medizinische Genetik, 3, 480. [DOI] [PubMed] [Google Scholar]
- 43.Hawkins J.D. (1988) Nucleic Acids Res., 16, 9893–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominski Z. and Kole,R. (1991) Mol. Cell. Biol., 11, 6075–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M., Jaeger,J.A. and Turner,D.H. (1991) Nucleic Acids Res., 19, 2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall C.J. (1996) Curr. Opin. Cell. Biol., 8, 197–204. [DOI] [PubMed] [Google Scholar]
- 47.Robinson M.J. and Cobb,M.H. (1997) Curr. Opin. Cell. Biol., 9, 180–186. [DOI] [PubMed] [Google Scholar]