Abstract
Safe and efficient genome editing has been an unmitigated goal for biomedical researchers since its inception. The most prevalent strategy for gene editing is the use of engineered nucleases that induce DNA damage and take advantage of cellular DNA repair machinery. This includes meganucleases, zinc-finger nucleases, transcription activator-like effector nucleases, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9) systems. However, the clinical viability of these nucleases is marred by their off-target cleavage activity (≥ 50% in RNA-guided endonucleases). In addition, in vivo applications of CRISPR require systemic administration of Cas9 protein, mRNA, or DNA, which presents a significant delivery challenge. The development of nucleic acid probes that can recognize specific double-stranded DNA (dsDNA) regions and activate endogenous DNA repair machinery holds great promise for gene editing applications. Triplex-forming oligonucleotides (TFOs), which were introduced more than 25 years ago, are among the most extensively studied oligomeric dsDNA-targeting agents. TFOs bind duplex DNA to create a distorted helical structure, which can stimulate DNA repair and the exchange of a nearby mutated region—otherwise leading to an undesired phenotype—for a short single-stranded donor DNA that contains the corrective nucleotide sequence. Recombination can be induced within several hundred base-pairs of the TFO binding site and has been shown to depend on triplex-induced initiation of the nucleotide excision repair pathway and engagement of the homology-dependent repair pathway. Since TFOs do not possess any direct nuclease activity, their off-target effects are minimal when compared to engineered nucleases. This review comprehensively covers the advances made in peptide nucleic acid-based TFOs for site-specific gene editing and their therapeutic applications.
Keywords: PNA, PLGA nanoparticles, Gamma PNA, Gene editing, Anemia
Introduction
Genome engineering offers the promise of remediating disease phenotypes by manipulating their underlying genotypes [1]. This prospect is most alluring in hematological disorders wherein, inherited genetic variations result in abnormal expression of genes crucial for the viability of hematopoietic lineages or exogenous pathogenic agents threaten the survival of hematopoietic subpopulations vital to immune function. Indeed, these disorders possess many of the features required to reap the potential benefits of this burgeoning technology, particularly: monogenicity, implying that palliative effects could be imparted by appropriate modifications to a single gene; compartmentalization, as genetic manipulation of cells in a single tissue is usually sufficient to remediate disease; and growth selectivity of even a small fraction of correctly modified cells in the population of interest—a useful feature for a field populated by reagents with substantial diversity in efficacy and safety profiles.
The predominant reagents currently available to genetic engineers are the exogenous nucleases that are engineered to bind target genomic loci and introduce double-strand breaks (DSB) [2]. DSBs are structural aberrations that trigger endogenous repair mechanisms which strive to restore the structural integrity of the DNA duplex. In the context of hematology, activation of DNA repair can facilitate various outcomes. It can correct the pathological mutation in the disease-associated gene, direct the targeted insertion of coding sequences for the deficient factors, disrupt genes encoding repressors of surrogates for the defective gene product, or perturb coding sequences for surface epitopes that render some hematopoietic lineages labile to destruction by invading pathogens [3]. While nuclease-based strategies have been effective in principle, important challenges remain in the prevailing methods for targeted reagent delivery and, perhaps more concerning, the avidity and activity of these reagents for/at off-target loci [4–6].
In most applications, nuclease reagents (or their precursory plasmids) are effectively delivered in vitro by electroporation [7]. Though successful for the introduction of diverse cargo to a variety of clinically relevant primary cell types, it remains unfeasible in vivo, and adversely affects survival of treated cells when used ex vivo. The later limitation is especially important for some hematological disorders where as a consequence of depletion in relevant hematopoietic lineages, there exist low basal levels of relevant primary cells available for harvesting and modification ex vivo.
Even when efficient intracellular transfer of the requisite nucleases is achieved, the destruction of the DNA duplex stimulates repair pathways that produce a wide spectrum of molecular outcomes. A majority off-repair events are mediated by non-homologous end joining (NHEJ), a pathway that prioritizes DNA structure over the DNA sequence; while a minority of the modifications are mediated by homology-directed repair (HDR) that preserves the sequence of the DNA template. Although both NHEJ and HDR are required for genome engineering in hematological disorders, the stochastic distribution of molecular outcomes resulting from combination of both these homologous regions of duplex DNA targets, with resultant local helical distortion—as reagents for provoking gene modification [8, 9]. However, the utility of these oligomers is hindered by their lability to cellular nuclease.
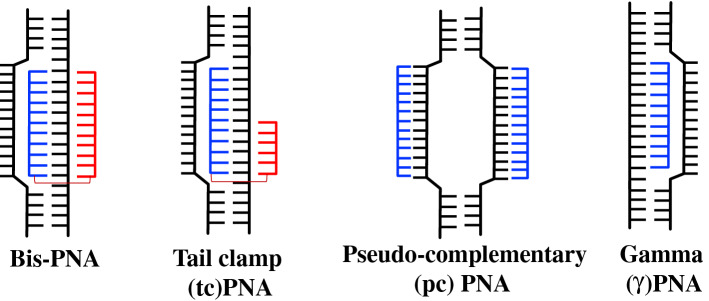
Peptide nucleic acids (PNAs) are synthetic nucleic acid analogs with pseudo-peptide backbone which imparts resistance to nuclease enzymes. PNAs consist of N-(2-aminoethyl)-glycine units-based backbone. Further nucleobases [Adenine (A), Guanine (G), Cytosine (C) and Thymine (T)] are attached to the backbone by a methylene carbonyl linkage (Fig. 1). PNAs charge-neutral property enables strong binding with DNA and RNA targets via Watson–Crick (WC) base pairing. This binding could be further increased by introducing cationic functionalities to PNA oligomer [10, 11]. It has been demonstrated that different designs of PNA can invade double-stranded DNA (dsDNA) and activate DNA repair and recombination events in the mammalian cells to induce site specific gene editing. In addition, PNAs circumvent many of the aforementioned limitations associated with nuclease and other class of oligonucleotide-based gene editing reagents. A variety of PNA designs and combinations have been developed for gene editing applications, including bis-PNAs [12, 13], tail clamp PNAs (tcPNAs) [14], pseudo-complementary PNAs (pcPNAs) [15] and new generation gamma PNAs (γPNAs) [16] (Fig. 2).
Fig. 1.
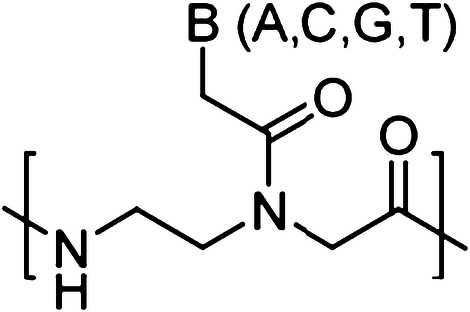
Chemical structure of regular PNA. B denotes nucleobases (A, C, G and T)
Fig. 2.
DNA binding modes of different PNAs
Poor cellular uptake properties of PNAs limit their broader clinical application. In past, several attempts have been made for increasing cellular uptake of PNAs. These strategies include conjugation with cell-penetrating peptides (CPP) [17] like penetratin, nuclear localization signal (NLS) and conjugation with pH low insertion peptide (pHLIP) targeting tumor microenvironment [18]. Few promising strategies include inclusion of cell transduction domain (guanidinium) onto PNAs to increase its uptake [19]. Recently, nanoconstruct-based approach has garnered great attention to deliver therapeutically active PNAs both ex vivo as well as in vivo. Several polymeric nanoparticles (NPs) have been used for PNAs delivery as enlisted in Table 1.
Table 1.
Nanoparticle-based strategies for enhanced delivery of PNAs
| Polymeric nanoparticles | Application | References |
|---|---|---|
| N,N,N-trimethyl-O-alkyl chitosans (TMACs) NPs | Drug delivery | [38] |
| PEGylated nanosized graphene oxide(PEG-nGO) constructs | Antisense (Cancer) | [39] |
| Mesoporous Silica NPs | Antisense (Cancer) | [40, 41] |
| Membrane penetrating oxidized carbon (MPOCs) NPs | Gene therapy | [42] |
| Porous-silicon (PSi) films | Drug delivery and biosensing | [43, 44] |
| Zeolite-l-nanocrystals | Drug delivery | [45] |
| Poly-lactic-co-glycolic acid (PLGA) NPs | Antisense, Antigene | [8, 46–48] |
| Poly-beta-amino-esters (PBAE) NPs | Antisense, Antigene | [33] |
| Surface modified PLGA/PBAE NPs | Antisense, Antigene | [49] |
| Peptide coated PLGA NPs | Antigene | [50] |
| Cationic shell-cross-linked knedel-like (cSCK) NPs | Drug Delivery | [51] |
| Avidin-labeled protein nanoparticles | Antisense (HIV infections) | [52] |
Bis-PNA
Bis-PNAs consist of two PNA strands linked via a flexible poly diethylene glycol linker and targets only homopurine region of genomic DNA. One PNA strand binds to the target DNA via WC base pairing in antiparallel orientation, whereas another PNA strand binds to the homopurine region of DNA via Hoogsteen base pairing forming a PNA/DNA/PNA triplex clamp (Fig. 3). In bis-PNA, pseudoisocytosine (also called J nucleobase) is used instead of cytosine for pH-independent base pairing to G in Hoogsteen binding domain to form a stable triplex structure at physiological pH [20]. Triplex clamp formed by bis-PNAs are highly stable and exhibit thermal denaturation temperature (Tm) > 70 °C. Triplex clamp results in displacement of homologous DNA strand forming D-loop (Fig. 2). Glazer and co-worker have demonstrated that triplex helix created by bis-PNA activates the nucleotide excision repair (NER) mechanism in cells and induce homologous recombination of donor DNA strand (containing the correct sequence/base) at the mutated site [12, 13]. The potential of bis-PNAs to induce recombination was studied in plasmid vector pSupFG1/G144C in vitro. Recombination frequencies were compared between bis-PNA–donor DNA conjugate, a mixture of bis-PNA with donor DNA, and bifunctional TFO–donor DNA (A-AG30). Bis-PNA–donor DNA conjugate induced higher recombination frequency (62 × 10−5) in comparison to TFO–donor DNA conjugate (38 × 10−5). However, maximum recombination frequency (81 × 10−5) was observed in mixture of bis-PNA and donor DNA that was fivefold higher than the donor DNA alone. These results demonstrated that a triplex clamp created by bis-PNA stimulates recombination of donor DNA at the target site.
Fig. 3.
Schematic of the PNA–DNA system. The 50–60-mer donor DNA is homologous to the gene target of choice except for a several base-pair mutation. The bis-PNA binds near the target and catalyzes homologous recombination of the donor strand into the target. Allele-specific PCR (AS-PCR) can distinguish between modified (mutant) and unmodified (wild-type) genomic DNA
Further the role of nucleotide exclusion factor, Xeroderma Pigmentosum (XPA) in NER pathway was assessed by comparing recombination frequency in XPA-depleted (−XPA) cell extracts and extracts treated with recombinant XPA protein supplement (+XPA) [21]. The extracts were depleted of XPA by rabbit polyclonal antibody against recombinant human XPA protein followed by immunoprecipitation. UV-exposed samples acted as a control because DNA repair due to UV damage is mediated by XPA. XPA depletion resulted in reduced DNA repair both in bis-PNA and UV-treated extracts and addition of XPA protein restored the DNA repair indicating that XPA plays an important role in DNA repair activity induced by bis-PNAs. Further, XPA depletion resulted in 14% decrease in recombination activity for mixture of bis-PNA and donor DNA, while 39% decrease in activity was reported for bis-PNA–donor DNA conjugate establishing that DNA repair is mediated by NER pathway [13].
Prior work has demonstrated that triplex-forming bis-PNAs effectively bind the beta (β)-globin gene and stimulate modification at a β-thalassemia-associated site in human CD34+ hematopoietic stem cells (HSCs) without loss of pluripotency [13]. Glazer and co-worker designed several triplex-forming bis-PNAs that mediate recombination at the first position of intron 2 (IVS2-1) of the β-globin gene and achieved recombination frequencies of 0.1–0.5% in a CHO cell GFP/β-globin gene fusion model with gene editing verified at the protein, mRNA (by qRT-PCR), and genomic DNA (by direct sequencing) levels. Primary CD34+ HSCs transfected with bis-PNAs and donor DNAs showed the gene editing at the β-thalassemia locus, with the presence of the mutation detected in HSC-derived cells grown in erythroid and neutrophil differentiating conditions. Transfection was accomplished via the Amaxa nucleofector [22], which although useful for proof-of-principle studies, is toxic to hematopoietic cells and is applicable only for ex vivo research applications.
In another study, it was demonstrated that Poly (lactic co-glycolic acids) (PLGA) NP-based delivery of bis-PNAs/donor DNA combination can lead to site-specific gene editing of CD34+ HSCs [14]. PLGA is a commonly used biodegradable polymer for drug delivery systems and medical devices. It has been approved by both the US Food and Drug Administration (USFDA) and European Medical Agency in a variety of clinical applications. An appealing feature of PLGA is that it degrades by hydrolysis into endogenous non-toxic metabolites (lactic acid and glycolic acid), which enhances its biocompatibility for in vivo delivery. Dye-loaded PLGA NPs showed surprisingly efficient uptake in CD34+ HSCs. Bis-PNAs/donor DNA were formulated into 150 nm spherical PLGA NPs, with ample loading of nucleic acids (250–450 pmol/mg NPs). Further, the PLGA NPs loaded with bis-PNAs/donor DNA combinations stimulated genomic recombination to modify the IVS2-1 splice site within the β-globin gene. Allele-specific PCR confirmed that NPs-delivered bis-PNA/donor DNA mediate site-specific modification in CD34+ HSCs. Importantly, the PLGA NPs with bis-PNA/donor DNA are not toxic to the progenitor cells. Progenitor cells that were genetically modified with NPs were differentiated into both erythroid and neutrophil populations, without loss of the gene modification.
Pseudo-complementary PNAs (pcPNAs)
Unlike bis-PNAs, pcPNAs consists of two PNA strands where each PNA binds to the complementary DNA strand in a sequence unrestricted manner via double duplex invasion-based mechanism (Fig. 2). Since, PNA–PNA duplex binding is stronger than PNA–DNA duplex binding, to prevent the self-quenching between two strands of pcPNAs, 2,6 diaminopurine (D) and 2-thiouracil (U) modified nucleobase are used instead of regular A and T nucleobases [20]. Due to the presence of modified nucleobases, the two PNA strands do not form a stable PNA–PNA duplex [23]. It was indicated that pcPNAs can bind to the dsDNA containing mixed purine and pyrimidine sequences (~ 40% AT rich sequences). In gene editing-based experiments, pcPNAs induced higher gene editing frequency (0.65%) in supF reporter gene in comparison to TFO (0.14%) and bis-PNA (0.21%). However, in CHO-GFP/IVS2-1 reporter cell, pcPNAs/donor DNA combinations could stimulate the recombination at β-thalassemic mutation site (IVS2-1) only at frequency of 0.012%. Further pre-treatment with histone deacetylase (HDAC) inhibitor (SAHA) improved the gene editing frequency to 0.17% that was threefold higher than only donor DNA [15].
Tail Clamp PNAs (tcPNAs)
Another promising PNA design has been used for gene editing-based application called tail clamp PNAs (tcPNAs). In tcPNAs, the WC binding domain is extended so that it can bind beyond the homopurine region and bind to a longer target site and thereby enhance the binding specificity. This creates an even larger helical distortion by increasing the length of the strand invasion and P-loop complex (Fig. 2). In binding studies, tcPNAs show greater affinity and specificity compared to bis-PNAs. Importantly, high binding affinity to DNA by tcPNAs does not require a long homopurine run; in fact, homopurine stretches as short as 5 bp are sufficient.
In a collaborative effort, it has been demonstrated that tcPNAs/donor DNA combination, delivered via modified PLGA NPs, demonstrated significant gene editing in F508del mutation in cystic fibrosis transmembrane conductance regulator (CFTR) gene in cystic fibrosis (CF) [9]. CF is a multi-system genetic disease affecting the respiratory, gastrointestinal and reproductive tracts [24]. Although the average life expectancy is 37 years, 50% of individuals with CF die in childhood from respiratory failure making it a very serious pediatric health problem. CF is most commonly caused by a three base-pair deletion (F508del) mutation in CFTR, an ion channel that mediates chloride transport [25]. Although CF is one of the most rigorously characterized genetic diseases, current treatment of patients with CF focuses on symptomatic management rather than primary correction of the genetic defect. CFTR is considered not readily amenable to gene therapy because of challenges including in vivo gene delivery, inflammatory reactions, and transient gene expression. NPs formulated from a blend of PLGA and PBAE and further surface modified with the nuclear localization sequence-containing cell-penetrating peptide MPG (modified PLGA/PBAE/MPG NPs) demonstrated superior gene editing efficiency as compared to PLGA NPs alone. Intranasal administration of modified PLGA NPs encapsulating tcPNA/donor DNA in CF mice showed nasal potential difference (NPD) similar to wild-type mice. Gene editing frequencies were reported to be > 5% in nasal epithelium and > 1% in lungs. Further 10% editing was noticed in human CFBE cells treated in vitro with modified PLGA NPs.
Gamma PNAs (γPNAs)
In addition, Ly and co-workers have developed another novel class of PNAs called gamma PNAs (γPNAs) [26, 27]. Compared to classical PNAs, γPNAs are highly water soluble; they neither aggregate nor adhere to surfaces or other macromolecules in a nonspecific manner (Fig. 4) [28, 29]. As individual strands, γPNAs adopt a right-handed helical motif—as confirmed by circular dichroism (CD), NMR, and X-ray crystallography—and hybridize to DNA or RNA strands with unusually higher affinity and sequence specificity [30]. In addition, prior studies also revealed that on average each γ-backbone modification stabilizes a PNA–DNA duplex by 4 °C [31]. They are the only class of oligonucleotide molecules developed to date that has been shown to be capable of invading any sequence of double helical genomic DNA at physiological conditions, with recognition occurring through WC base-pairing. γPNAs have been exploited in a number of biological and biomedical applications, from electronic barcoding of single gene [32] to gene correction (achieving clinically acceptable correction frequencies with extremely low off-target effects, as compared to that of zinc-finger nucleases or CRISPR/Cas system) [16, 33, 34]. In this section, we discussed the recent gene editing results of γPNAs-based probes.
Fig. 4.
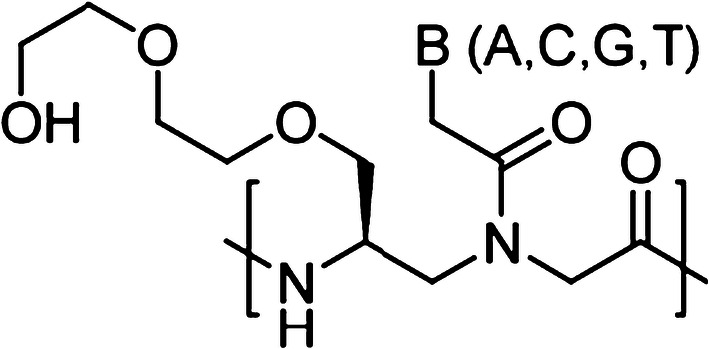
Chemical structure of gamma PNA (γPNA) containing an ethylene glycol (MP) at the γ-position. B denotes nucleobases (A, C, G and T)
PLGA NPs containing ss γPNAs for site-specific gene editing
Effectiveness of ss γPNAs/donor DNA combination encapsulated in PLGA NPs for successful gene editing have been assessed in green fluorescent protein (GFP) transgenic mouse model. This transgenic mouse model has a β-globin/GFP fusion transgene consisting of human β-globin intron 2 carrying a thalassemia-associated IVS2-654 (C → T) mutation embedded within the GFP coding sequence. The presence of IVS2-654 (C → T) mutation results in improper splicing of β-globin/GFP mRNA and lack of GFP expression [35]. This model allows for robust quantification of gene editing frequencies [33]. To test whether ss γPNAs can induce gene editing, series of γPNA oligomers were designed and synthesized. Further, γPNA/donor DNA was encapsulated in PLGA NPs using double emulsion solvent evaporation technique. Due to the presence of ethylene glycol units in γPNAs, γPNA/donor DNA complex formed a clear solution at room temperature and higher total nucleic acid loading was observed in γPNA/donor DNA in comparison to regular PNA/donor DNA samples. NPs containing γPNA/donor DNA resulted in gene editing frequency of 0.1% in ex vivo studies using bone marrow cells of GFP transgenic mice treated with 2 mg/mL of NPs, which was higher than the levels observed with regular PNA/donor DNA NPs (0.02%). PBAE/PLGA NPs with 15% PBAE (poly (beta-amino) ester) and 85% PLGA led to improved loading of γPNA/donor DNA combination and sustained release of nucleic acids. Significantly higher gene editing frequency (0.8%) was observed in PBAE/PLGA NPs containing γPNA/donor DNA combination. Further, these PNAs were able to induce gene editing up to a distance of ~ 100 bp (0.43%) and ~ 250 bp (0.23%) away from the donor DNA binding site. SS γPNA/donor DNA combination was found to be non-toxic without any impact on differentiation potential of hematopoietic progenitor cells. Further in vivo studies were conducted by administering four retro-orbital injections of 2 mg PBAE/PLGA NPs containing regular PNA or γPNA/donor DNA combinations. Deep sequencing analysis resulted in gene editing frequency of 0.077% in bone marrow cells of mice treated with PBAE/PLGA NPs containing γPNA/donor DNA with minimum off-target effects (≤ 0.01%). To study the inflammatory response, RT-PCR analysis was performed on bone marrow cells to determine levels of inflammatory markers like interleukin 6 (IL-6) and tumor necrosis factor alpha (TNFα) and no significant difference was observed in their levels between PBAE/PLGA NPs-treated cells and untreated cells. Overall, it was successfully demonstrated that ss γPNA encapsulated in PBAE/PLGA NPs exhibit higher loading, binding affinity and gene editing abilities without any sequence restriction as established via both ex vivo and in vivo studies in transgenic mice model.
Enhanced gene editing by gamma-modified tcPNAs (γtcPNAs)
In our published work, we showed that tcPNAs with poly diethylene glycol substitution at γ position results in higher gene editing frequencies in transgenic GFP mouse model. PNA-mediated triplex formation induces DNA repair and recombination of the genomic site with a 60-nucleotide ss donor DNA that is homologous to a portion of the β-globin intron 2 sequences except for providing a wild-type nucleotide at the IVS2-654 position in transgenic GFP mouse model. Via recombination, the splice-site mutation is corrected resulting in expression of functional GFP. Hence, GFP expression provides a direct phenotypic assessment of genome editing frequencies that can be quantified by flow cytometry.
In a recent study, we designed a series of tcPNAs to bind to the β-globin intron 2 near the IVS2-654 (C → T) mutation [16]. These PNAs were combined with the donor DNA and formulated into PLGA NPs. These NPs were added to the culture medium of bone marrow (BM) cells from GFP mice, and 2 days later, the cells were scored for gene editing by flow cytometry to quantify GFP expression. We identified the most active tcPNA oligomer. Further, we performed chemical modifications onto regular tcPNA with every other residue in the WC domain substituted with γPNA. A scrambled sequence control was also made with the same base composition as that of test γtcPNA.
We treated mouse BM cells with PLGA NPs containing regular tcPNA and γtcPNA, as well as its scrambled control. We also sorted the BM cells based on cell surface markers, so that we could interrogate the extent of gene editing in individual stem and progenitor cell populations. We made two key findings: (1) The gene editing occurred primarily in CD117+ cells. CD117 is the product of the c-Kit gene and is a receptor tyrosine kinase that marks stem cell populations. This was encouraging because results demonstrated that HSCs may be particularly susceptible to PNA-mediated gene editing, and HSCs are the most desirable cell population to edit. (2) The NPs containing γtcPNA gave substantially higher levels of gene editing (up to 9% in a single treatment) compared to regular tcPNA (about 2.5%) indicating the superior activity of the γPNAs. There was no effect with scrambled control. We attribute this increased efficacy of γtcPNA to the enhanced DNA binding properties of γPNAs, which take on a pre-organized helical conformation enforced by the γ substitution.
Gene editing in mice with thalassemia and amelioration of the disease phenotype
Prompted by these results in reporter mice, we tested gene editing in a β-thalassemia mouse model [36]. These mice carry the human β-globin gene replacing the mouse β-globin gene locus and containing the same β-thalassemia splicing mutation at IVS2-654 as in the GFP reporter mice. We treated these mice via simple IV injection with γtcPNA/donor DNA NPs given four times at two-day intervals. Because we had determined that activation of the c-Kit pathway with stem cell factor (SCF) boosts gene editing (an effect associated with increased DNA repair in the c-Kit + cells), we also treated the mice with SCF. This regimen yielded gene editing at a frequency of 7% in Lin-Sca1 + cKit + CD150 + CD135 cells, a population that is highly enriched for long-term HSCs [16]. We also observed gene editing in other progenitor population cells. This treatment produced amelioration of the disease phenotype in the thalassemic mice with sustained reversal of the anemia, normalization of hemoglobin concentrations, and decrease in reticulocytosis. In parallel, there was also reduced extramedullary hematopoiesis and marked reduction in splenomegaly with improvement in splenic architecture on histologic examination.
Gene editing ability was further studied in human CD34+ cells, after introducing the mutation at IVS2-654 of normal human CD34+ cells. These cells were then treated with blank PLGA NPs, SCF plus the γtcPNA/donor DNA containing PLGA NPs. Deep sequencing analysis showed gene editing frequency of 5% at IVS2-654 position in case of SCF plus the γtcPNA/donor DNA and low off-target effects; 400,000-fold lower when compared to the 5% frequency at the targeted site. Bone marrow transplantation studies in NOD-scid IL2rgnull mice with γtcPNA/donor DNA nanoparticles and SCF treated cells showed 3.4% gene editing frequency.
Conclusion
Overall, this review present various novel approaches for site-specific gene editing based on nanoparticle-delivered PNA-based strategies. It has been well documented that PNAs can invade double duplex DNA creating a loop in double- stranded DNA which instigates the NER pathway allowing the homologous donor DNA to provide the correct genetic sequence at target site. Each PNA invades the dsDNA via a specific mechanism (Fig. 2 and Table 2) resulting in gene editing of mutated region. Further chemical modifications of PNAs at γ position led to more efficient γPNAs which can bind specifically to the target site without any sequence restriction.
Table 2.
Advantages and gene editing efficiencies of PNAs
| PNA | Advantages | Gene editing efficiency |
|---|---|---|
| Bis-PNA |
PNA/DNA/PNA clamps are stable and can target only homopurine regions Exhibit high binding affinity and specificity |
60-fold higher gene correction than the background in SupFG1 reporter gene in vitro [12] 0.2% of gene editing reported in CHO/IVS2-1 reporter cell assay using bisPNA targeting IVS2-194 in combination with donor DNA in S-Phase of cell cycle [13] 0.2% of gene editing observed in CD34+ hematopoietic progenitor cells treated with PLGA NPs encapsulating bis-PNA (targeting IVS2-194) and donor DNA [14] |
| pcPNA |
Targets mixed sequence containing ≥ 40% A:T bp High binding affinity and specificity Modified 2,6-diaminopurine and 6-thiouracil bases prevent formation of PNA:PNA duplex Binds to target DNA strand only via WC base pairing |
0.65% of gene editing in an episomal target site using supF reporter gene [15] 0.012% gene editing in CHO/IVS2-1 reporter cell assay after single transfection [15] Increased gene editing frequency observed in S-Phase (0.19%) and combination of S-Phase with chloroquine (0.25%) and histone deacetylase inhibitor, SAHA (0.78%) [15] |
| tcPNA |
Targets homopurine regions of dsDNA Extended WC domain increases the specificity Binds to target site via both WC and Hoogsteen base pairing |
2.8% CCR5 gene editing in primary human CD34+ cells with transfection of tcPNA and donor DNA together in order to impart HIV-1 resistance [9] 5.7% CFTR gene editing in nasal epithelium using PBAE/PLGA/MPG NPs in CF mouse model [9] |
| γPNA |
Targets mixed DNA sequence without any sequence restriction Capable of targeting 0–100% GC rich DNA sequence Higher aqueous solubility Improved loading in NPs |
0.8% of gene editing frequency in bone marrow cells of GFP mouse model treated with PBAE/PLGA NPs [33] Up to 4.0% gene editing in β-thalassemic mice with marked improvement in the phenotypic features [16] |
The critical difference between PNAs and nuclease-based gene editing is important since off-target strand breaks caused by nucleases could lead to leukemias and other malignancies. This technology is known as “minimally invasive” gene repair, as gene editing occurs in situ via recruitment of the cells’ own DNA repair machinery and without the need for viral vectors. Because this approach seeks to edit the mutated gene at the specific mutation site, it avoids the risk of deleterious ectopic integration in the genome that has been seen with virus-mediated gene therapy [37].
Acknowledgements
This work was supported by University of Connecticut startup funds. In addition, this material is based upon work supported by the State of Connecticut under the Regenerative Medicine Research Fund and Cooley’s Anemia Foundation Research Fellowship Grant application. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut or Connecticut Innovations, Incorporated.
Footnotes
Shipra Malik and Stanley Oyaghire contributed equally to this work.
References
- 1.Bak RO, Gomez-Ospina N, Porteus MH. Gene editing on center stage. Trends Genet. 2018;34(8):600–611. doi: 10.1016/j.tig.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Song B, Fan Y, He W, Zhu D, Niu X, Wang D, Ou Z, Luo M, Sun X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24(9):1053–1065. doi: 10.1089/scd.2014.0347. [DOI] [PubMed] [Google Scholar]
- 4.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quijano E, Bahal R, Ricciardi A, Saltzman WM, Glazer PM. Therapeutic peptide nucleic acids: principles, limitations, and opportunities. Yale J Biol Med. 2017;90(4):583–598. [PMC free article] [PubMed] [Google Scholar]
- 7.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNeer NA, Chin JY, Schleifman EB, Fields RJ, Glazer PM, Saltzman WM. Nanoparticles deliver triplex-forming pnas for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol Ther. 2011;19(1):172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeer NA, Anandalingam K, Fields RJ, Caputo C, Kopic S, Gupta A, Quijano E, Polikoff L, Kong Y, Bahal R, Geibel JP, Glazer PM, Saltzman WM, Egan ME. Correction of F508del CFTR in airway epithelium using nanoparticles delivering triplex-forming PNAs. Nat Commun. 2015;6:6952. doi: 10.1038/ncomms7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature (London) 1993;365(6446):566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science (Washington, D. C., 1883) 1991;254(5037):1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 12.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci USA. 2002;99(26):16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR, Nielsen PE, Kole R, Glazer PM. Correction of a splice-site mutation in the beta-globin gene stimulated by triplex-forming peptide nucleic acids. Proc Natl Acad Sci USA. 2008;105(36):13514–13519. doi: 10.1073/pnas.0711793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeer NA, Schleifman EB, Cuthbert A, Brehm M, Jackson A, Cheng C, Anandalingam K, Kumar P, Shultz LD, Greiner DL, Mark Saltzman W, Glazer PM. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013;20(6):658–669. doi: 10.1038/gt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonkar P, Kim K-H, Kuan JY, Chin JY, Rogers FA, Knauert MP, Kole R, Nielsen PE, Glazer PM. Targeted correction of a thalassemia-associated β-globin mutation induced by pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2009;37(11):3635–3644. doi: 10.1093/nar/gkp217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahal R, Ali McNeer N, Quijano E, Liu Y, Sulkowski P, Turchick A, Lu YC, Bhunia DC, Manna A, Greiner DL, Brehm MA, Cheng CJ, Lopez-Giraldez F, Ricciardi A, Beloor J, Krause DS, Kumar P, Gallagher PG, Braddock DT, Mark Saltzman W, Ly DH, Glazer PM. In vivo correction of anaemia in beta-thalassemic mice by gammaPNA-mediated gene editing with nanoparticle delivery. Nat Commun. 2016;7:13304. doi: 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiraishi T, Nielsen PE. Cellular delivery of peptide nucleic acids (PNAs) Methods Mol Biol. 2014;1050:193–205. doi: 10.1007/978-1-62703-553-8_16. [DOI] [PubMed] [Google Scholar]
- 18.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas SM, Sahu B, Rapireddy S, Bahal R, Wheeler SE, Procopio EM, Kim J, Joyce SC, Contrucci S, Wang Y, Chiosea SI, Lathrop KL, Watkins S, Grandis JR, Armitage BA, Ly DH. Antitumor effects of EGFR antisense guanidine-based peptide nucleic acids in cancer models. ACS Chem Biol. 2013;8(2):345–352. doi: 10.1021/cb3003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egholm M, Christensen L, Dueholm KL, Buchardt O, Coull J, Nielsen PE. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995;23(2):217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc Natl Acad Sci USA. 2002;99(9):5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurisse R, De Semir D, Emamekhoo H, Bedayat B, Abdolmohammadi A, Parsi H, Gruenert DC. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 2010;10:9. doi: 10.1186/1472-6750-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc Natl Acad Sci USA. 1999;96(21):11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis PB. Cystic fibrosis. Pediatr Rev. 2001;22(8):257–264. doi: 10.1542/pir.22-8-257. [DOI] [PubMed] [Google Scholar]
- 25.Davis P. Cystic fibrosis since 1938. Am J Respir Crit Care Med. 2006;173(5):475–482. doi: 10.1164/rccm.200505-840OE. [DOI] [PubMed] [Google Scholar]
- 26.Rapireddy S, Bahal R, Ly DH. Strand invasion of mixed-sequence, double-helical B-DNA by γ-peptide nucleic acids containing G-Clamp nucleobases under physiological conditions. Biochemistry. 2011;50(19):3913–3918. doi: 10.1021/bi2002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahal R, Sahu B, Rapireddy S, Lee C-M, Ly DH. Sequence-unrestricted, Watson-Crick recognition of double helical B-DNA by (R)-MiniPEG-γPNAs. ChemBioChem. 2012;13(1):56–60. doi: 10.1002/cbic.201100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand invasion of extended, mixed-sequence B-DNA by γPNAs. J Am Chem Soc. 2009;131(34):12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahu B, Sacui I, Rapireddy S, Zanotti KJ, Bahal R, Armitage BA, Ly DH. Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing γ-peptide nucleic acids with superior hybridization properties and water solubility. J Org Chem. 2011;76(14):5614–5627. doi: 10.1021/jo200482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A simple γ-backbone modification preorganizes peptide nucleic acid into a helical structure. J Am Chem Soc. 2006;128(31):10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 31.Sahu B, Chenna V, Lathrop KL, Thomas SM, Zon G, Livak KJ, Ly DH. Synthesis of conformationally preorganized and cell-permeable guanidine-based gamma-peptide nucleic acids (gammaGPNAs) J Org Chem. 2009;74(4):1509–1516. doi: 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer A, Rapireddy S, Ly DH, Meller A. Electronic barcoding of a viral gene at the single-molecule level. Nano Lett. 2012;12(3):1722–1728. doi: 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahal R, Quijano E, McNeer NA, Liu Y, Bhunia DC, Lopez-Giraldez F, Fields RJ, Saltzman WM, Ly DH, Glazer PM. Single-stranded gammaPNAs for in vivo site-specific genome editing via Watson-Crick recognition. Curr Gene Ther. 2014;14(5):331–342. doi: 10.2174/1566523214666140825154158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricciardi AS, Bahal R, Farrelly JS, Quijano E, Bianchi AH, Luks VL, Putman R, Lopez-Giraldez F, Coskun S, Song E, Liu Y, Hsieh WC, Ly DH, Stitelman DH, Glazer PM, Saltzman WM. In utero nanoparticle delivery for site-specific genome editing. Nat Commun. 2018;9(1):2481. doi: 10.1038/s41467-018-04894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sazani P, Gemignani F, Kang S-H, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat Biotechnol. 2002;20(12):1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 36.Svasti S, Suwanmanee T, Fucharoen S, Moulton HM, Nelson MH, Maeda N, Smithies O, Kole R. RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice. Proc Natl Acad Sci USA. 2009;106(4):1205–1210. doi: 10.1073/pnas.0812436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Goncalves MA. Engineered viruses as genome editing devices. Mol Ther. 2016;24(3):447–457. doi: 10.1038/mt.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Wang J, Huang S, Yu L, Wang Y, Chen H, Wang D. Self-assembled nanoparticles for cellular delivery of peptide nucleic acid using amphiphilic N,N,N-trimethyl-O-alkyl chitosan derivatives. J Mater Sci Mater Med. 2018;29(8):114. doi: 10.1007/s10856-018-6120-y. [DOI] [PubMed] [Google Scholar]
- 39.Baek A, Baek YM, Kim HM, Jun BH, Kim DE. Polyethylene glycol-engrafted graphene oxide as biocompatible materials for peptide nucleic acid delivery into cells. Bioconjug Chem. 2018;29(2):528–537. doi: 10.1021/acs.bioconjchem.8b00025. [DOI] [PubMed] [Google Scholar]
- 40.Bertucci A, Prasetyanto EA, Septiadi D, Manicardi A, Brognara E, Gambari R, Corradini R, De Cola L. Combined delivery of temozolomide and anti-miR221 PNA using mesoporous silica nanoparticles induces apoptosis in resistant glioma cells. Small. 2015;11(42):5687–5695. doi: 10.1002/smll.201500540. [DOI] [PubMed] [Google Scholar]
- 41.Ma X, Devi G, Qu Q, Toh DF, Chen G, Zhao Y. Intracellular delivery of antisense peptide nucleic acid by fluorescent mesoporous silica nanoparticles. Bioconjug Chem. 2014;25(8):1412–1420. doi: 10.1021/bc5002714. [DOI] [PubMed] [Google Scholar]
- 42.Arayachukiat S, Seemork J, Pan-In P, Amornwachirabodee K, Sangphech N, Sansureerungsikul T, Sathornsantikun K, Vilaivan C, Shigyou K, Pienpinijtham P, Vilaivan T, Palaga T, Banlunara W, Hamada T, Wanichwecharungruang S. Bringing macromolecules into cells and evading endosomes by oxidized carbon nanoparticles. Nano Lett. 2015;15(5):3370–3376. doi: 10.1021/acs.nanolett.5b00696. [DOI] [PubMed] [Google Scholar]
- 43.Beavers KR, Mares JW, Swartz CM, Zhao Y, Weiss SM, Duvall CL. In situ synthesis of peptide nucleic acids in porous silicon for drug delivery and biosensing. Bioconjug Chem. 2014;25(7):1192–1197. doi: 10.1021/bc5001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beavers KR, Werfel TA, Shen T, Kavanaugh TE, Kilchrist KV, Mares JW, Fain JS, Wiese CB, Vickers KC, Weiss SM, Duvall CL. Porous silicon and polymer nanocomposites for delivery of peptide nucleic acids as anti-MicroRNA therapies. Adv Mater. 2016;28(36):7984–7992. doi: 10.1002/adma.201601646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertucci A, Lulf H, Septiadi D, Manicardi A, Corradini R, De Cola L. Intracellular delivery of peptide nucleic acid and organic molecules using zeolite-L nanocrystals. Adv Healthc Mater. 2014;3(11):1812–1817. doi: 10.1002/adhm.201400116. [DOI] [PubMed] [Google Scholar]
- 46.Cheng CJ, Saltzman WM. Polymer nanoparticle-mediated delivery of microRNA inhibition and alternative splicing. Mol Pharm. 2012;9(5):1481–1488. doi: 10.1021/mp300081s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bahal R, McNeer NA, Ly DH, Saltzman WM, Glazer PM. Nanoparticle for delivery of antisense gammaPNA oligomers targeting CCR5. Artif DNA PNA XNA. 2013;4(2):49–57. doi: 10.4161/adna.25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A, Quijano E, Liu Y, Bahal R, Scanlon SE, Song E, Hsieh WC, Braddock DE, Ly DH, Saltzman WM, Glazer PM. Anti-tumor activity of miniPEG-gamma-modified PNAs to inhibit MicroRNA-210 for cancer therapy. Mol Ther Nucleic Acids. 2017;9:111–119. doi: 10.1016/j.omtn.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fields RJ, Quijano E, McNeer NA, Caputo C, Bahal R, Anandalingam K, Egan ME, Glazer PM, Saltzman WM. Modified poly(lactic-co-glycolic acid) nanoparticles for enhanced cellular uptake and gene editing in the lung. Adv Healthc Mater. 2015;4(3):361–366. doi: 10.1002/adhm.201400355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109(26):E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang H, Zhang K, Shen G, Wooley KL, Taylor JS. Cationic shell-cross-linked knedel-like (cSCK) nanoparticles for highly efficient PNA delivery. Mol Pharm. 2009;6(2):615–626. doi: 10.1021/mp800199w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langer K, Coester C, Weber C, von Briesen H, Kreuter J. Preparation of avidin-labeled protein nanoparticles as carriers for biotinylated peptide nucleic acid. Eur J Pharm Biopharm. 2000;49(3):303–307. doi: 10.1016/S0939-6411(00)00068-0. [DOI] [PubMed] [Google Scholar]