Abstract
Various stress factors leading to protein damage induce the activation of an evolutionarily conserved cell protective mechanism, the heat shock response (HSR), to maintain protein homeostasis in virtually all eukaryotic cells. Heat shock factor 1 (HSF1) plays a central role in the HSR. HSF1 was initially known as a transcription factor that upregulates genes encoding heat shock proteins (HSPs), also called molecular chaperones, which assist in refolding or degrading injured intracellular proteins. However, recent accumulating evidence indicates multiple additional functions for HSF1 beyond the activation of HSPs. Here, we present a nearly comprehensive list of non-HSP-related target genes of HSF1 identified so far. Through controlling these targets, HSF1 acts in diverse stress-induced cellular processes and molecular mechanisms, including the endoplasmic reticulum unfolded protein response and ubiquitin–proteasome system, multidrug resistance, autophagy, apoptosis, immune response, cell growth arrest, differentiation underlying developmental diapause, chromatin remodelling, cancer development, and ageing. Hence, HSF1 emerges as a major orchestrator of cellular stress response pathways.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2836-6) contains supplementary material, which is available to authorized users.
Keywords: Ageing, Apoptosis, Autophagy, Cancer, Cell cycle, Circadian rhythm, Development, Differentiation, Heat shock factor 1, Heat shock proteins, Heat shock response, Immune response, Multidrug resistance, Oxidative stress, Proteasome, Unfolded protein response
The heat shock response
Intracellular proteins have been evolutionarily optimized to function in a relatively tight temperature range. Even a small scale but long-acting change in the ambient temperature can severely perturb protein homeostasis (proteostasis), thereby compromising cellular processes and leading to accelerated ageing and the incidence of various proteotoxicity-triggered disorders. In humans, such pathologies include diverse neurodegenerative diseases, heart failure, cancer, diabetes, tissue atrophy and fibrosis, and immune deficiency [1–7]. Heat shock and other cellular stress factors, such as oxidizing agents, toxins, heavy metals, and infective microbes, can induce a conserved cell defence mechanism, the heat shock response (HSR), to maintain proteostasis in eukaryotic cells [8]. The HSR primarily involves the expression of heat shock proteins (HSPs), also termed molecular chaperones, which facilitate the synthesis and ensure the structural stability of other intracellular proteins. HSPs can also mediate the refolding or degradation of damaged intracellular proteins. This cell protective mechanism enables the cell to survive under harsh environmental conditions, predominantly at elevated temperatures. The activation of HSPs is mainly achieved by the heat shock transcription factor HSF1. Hence, HSF1 functions as a central regulator of the HSR.
HSF1 controls the HSR
HSF1 is an evolutionarily conserved transcription factor that is known to be primarily activated in response to heat stress. When triggered, HSF1 becomes trimerised and phosphorylated, and then translocated into the nucleus where it binds to conserved heat shock-responsive DNA elements (HSEs) to upregulate genes coding for HSPs. HSEs are generally located in the upstream untranslated region of HSF1 target genes. As each DNA-binding domain of the HSF1 homotrimer recognises an nGAAn pentameric sequence motif (where “n” indicates any nucleotide), a stable association of HSF1 to its binding element requires three pentameric sequences with alternating orientation, TTCnnGAAnnTTC [9–12]. Both in vitro and in vivo studies have shown that HSF1 prefers HSEs with tandem, properly orientated repeats [10]. At the same time, a ChIP-seq (chromatin-immunoprecipitation associated with deep sequencing) analysis on Drosophila has revealed that HSF1 can bind only a subset of its potential HSEs in vivo, and in the surroundings of these occupied binding elements active chromatin marks are located [13]. Furthermore, a recent PRO-seq (precision nuclear run-on sequencing) analysis performed on human K562 erythroleukemic cell line recognised numerous heat stress-responsive genes, and, according to the results obtained, HSF1 binding was detectable in the vicinity of coding regions in only 29 and 2% of the heat shock-activated and heat shock-repressed genes identified, respectively [14]. This indicates that beyond the HSE, the local chromatin structure and interaction of different transcription factors may also play an important role in transcriptional response to heat stress.
HSF1 activation is accomplished at the level of protein–protein interaction and posttranslational modification [8]. Certain HSF1-induced HSPs, such as HSPA1A/B/L (HSP70), HSPA1A (HSP72), and HSPC1 (HSP90), directly inhibit HSF1 via binding to its trimerisation domain. In addition, a cytoplasmic histone deacetylase (HDAC6) and a valosin-containing protein (VCP, a highly conserved member of the AAA—ATPases associated with a variety of cellular activities—family proteins) participate in repressing the HSF1–HSP complex [15]. This negative autoregulatory feedback loop ensures that HSF1-mediated stress response occurs precisely according to the actual level of protein damage [16]. HSF1 activity is also influenced by the phosphorylation, sumoylation or acetylation status of certain amino acids of the protein [8, 17, 18].
In both nematodes (Caenorhabditis elegans) and mammals, HSF1 activity can be modified by certain stress-induced kinases that represent components of conserved signal transduction systems including the insulin/IGF1 (insulin-like growth factor receptor 1) and cyclic guanosine monophosphate-mediated (cGMP) pathways [19, 20]. Some of these kinases like p38-Map (p38 mitogen-activated protein kinase), c-Jun (Jun proto-oncogene), Gsk3 (glycogen synthase kinase 3), and Erk1 (extracellular signal-regulated kinase 1) inhibit, while others including PI3K (phosphatidylinositol 3-kinase), Akt (Akt serine/threonine kinase), and cAMP-dependent PKA (protein kinase A) promote HSF1 activity [1, 8, 21, 22].
90-kDa molecular chaperone family has multiple roles in eukaryotic cells. For example, HSPC1 (HSP90) binds damaged proteins and transmits them to HSPD1 (HSP60) and HSPA1A/B/L (HSP70) for further processing [23, 24]. It also plays a diverse role in the functioning of signal transduction systems [25–27].
HSF1 is an essential gene in yeast, flies and nematodes [28–30]. The lethal phenotype of HSF1(–) mutant yeast cells can be suppressed by constitutively co-expressing HSPA1A/B/L (Hsp70) and HSPC1 (Hsp90) [31]. In mammals, however, HSF1 is not essential for viability and dispensable for the basal expression of molecular chaperones [32]. This may be due to the fact that in these organisms the basal expression of chaperones is also influenced by other transcription factors including STAT1 (signal transducer and activator of transcription), STAT3, and NF-IL6 (nuclear factor for interleukin-6 expression) [33]. In addition, while most invertebrate genomes contain only a single HSF1 gene, at least 9 HSF paralogues have been identified in vertebrates, HSF1-5, Hsfx1/2 and Hsfy1/2 [8]. It is possible that the expression of mammalian Hsp genes is controlled redundantly by HSF paralogues. In humans, for example, HSF1 coordinates the HSR, but HSF2 and HSF4 also participate in Hsp activation. Due to the functional diversity of HSF proteins, distinguishing between the HSP-dependent and -independent functions of HSF1 is quite difficult in mammals. Therefore, a systematic identification of HSF1 target genes in divergent genetic model systems appears to be particularly important in understanding how HSF1 integrates various stress response pathways.
Roles of HSF1 beyond the HSR
Until the beginning of this century, HSF1 was generally considered a heat shock-induced transcription factor that regulates the expression of HSPs (Table S1). Since then, however, HSF1 has also been revealed to influence numerous developmental events and cellular processes, and implicated in various molecular stress-induced pathologies [1–8]. In the last 15 years, several single gene-based and genome-scale studies were conducted to identify novel targets of HSF1 in genetic model organisms and cell cultures [34–40]. Based on these works, it has become clear that following heat stress several genes become up- or downregulated independently of HSF1, and that HSF1 controls the transcription of numerous genes encoding proteins others than HSPs, largely in a temperature-independent manner. These so-called non-HSP target genes of HSF1 are implicated in various physiological and stress-induced cellular processes and molecular mechanisms including protein modification and degradation, cell cycle, programmed cell death, ageing, the endoplasmic reticulum (ER) unfolded protein response (UPR), multidrug resistance, and immune function. It is intriguing that the regulatory sequence of some HSF1 target genes does not contain a conserved HSE element [35, 36], and that the presence of a conserved HSE does not necessarily accompany an HSF1-dependent transcriptional regulation [33]. Moreover, the transcriptional activity of several HSF1 targets is repressed by the transcription factor, and this negative regulatory interaction occurs independently of temperature. For example, in C. elegans, HSF-1 influences the transcription of genes encoding collagens involved in cuticle formation [35]. These data imply that the functional spectrum of HSF1 is much broader than previously assumed.
HSF1 in stress-induced cellular processes and molecular mechanisms
HSF1 and the endoplasmic reticulum unfolded protein response
The accumulation of un- and misfolded proteins in the lumen of ER, where newly translated proteins are stored and become modified, causes ER stress, which leads to the activation of genes encoding ER-resident proteins required for protein folding [41]. This molecular information is transmitted from the ER lumen to the nucleus by a conserved intracellular signalling pathway, the UPR. The UPR actually consists of three parallel intracellular signal transduction systems; the first is mediated by IRE1α/XBP1 (inositol-requiring protein 1α/spliced X box-binding protein 1) proteins, the second involves PERK/ATF4 (protein kinase RNA-like endoplasmic reticulum kinase/activating transcription factor 4) proteins, while the third relies on ATF6/ATF6f (activating transcription factor 6/cytosolic domain fragment of ATF6) proteins. The UPR functions to reduce the damaging effect of harmful proteins through restoring protein folding capacity in the affected cell. Together with the HSR, the UPR protects eukaryotic cells from the damaging effect of proteotoxicity. These two highly conserved stress-response systems, the HSR and UPR, are activated in the cytosol and ER, respectively.
In yeast, the expression of an HSE-containing reporter system is significantly increased in response to ER stress, and this transcriptional upregulation depends on Ire1/IRE1 and Hac1/XBP1 activity [42]. Thus, it is possible that ER stress triggers HSF1 to participate in the UPR. Consistent with these results, a constitutively active HSF1 increases tolerance to ER stress in both wild-type and ire1(–) mutant yeast cells [43, 44]. Indeed, a genome-wide gene expression analysis revealed that HSF1 activates both ER-resident and cytosolic chaperones.
In human cells, HSF1-regulated chaperones are implicated in the control of UPR [45]. For example, HSPC1 (HSP90) was shown to interact with and stabilize the cytosolic domain of IRE1α, thereby influencing the UPR. HSPA1A (Hsp72) is expressed at basal levels under physiological conditions. In cells constitutively overexpressing HSPA1A (Hsp72), this chaperone activates the IRE1α/XBP1 branch of the UPR through forming a stable complex with the cytoplasmic domain of IRE1α [46]. Heat shock also leads to an UPR-like response, in which ER-associated chaperones and their regulatory transcription factors are triggered simultaneously [47]. Using a dominant negative HSF1 mutation, transcript levels of certain ER-resident chaperones, such as HSPA5 (BiP) and DNAJB9, were shown to become elevated following heat shock. Thus, genes encoding these chaperones do not serve as targets for HSF1. Vertebrate genomes, however, code for multiple HSF1 paralogues, which certainly function redundantly with HSF1. This suggests that the role of specific HSF paralogues cannot be excluded from the heat shock-induced UPR.
The yeast ERO1 gene encodes an ER oxidoreductin, which is required for the formation of protein disulphide bonds [48]. ERO1 was shown to be upregulated by HSF1 in response to various stress factors (Table 1; Table S1) [49]. This regulatory interaction is particularly interesting, because ERO1 transcription is also activated by the bZIP (basic-leucine zipper) transcription factor Hac1 involved in the UPR. Hac1, the yeast orthologue of Xbp1, binds an XBP1 responsive cis-regulatory element in the promoter of UPR-regulated genes [50]. Therefore, ERO1 serves as a genetic factor where HSF1 and the UPR directly interact with each other. In other words, HSF1 may directly activate the UPR by controlling ERO1 under conditions of proteotoxicity.
Table 1.
Selected list of HSF1 target genes identified so far
| Target gene | Organism/cell culture | Function | Regulation | References |
|---|---|---|---|---|
| ERO1 | Yeast | Unfolded protein response | Up | [47] |
| SIS1 | Yeast | Oxidative stress response | Up | [51] |
| CUP1 | Yeast | Oxidative stress response | Up | [51] |
| BTN2 | Yeast | Oxidative stress response | Up | [51] |
| SGT2 | Yeast | Oxidative stress response | Up | [51] |
| SSA3 | Yeast | Oxidative stress response | Up | [51] |
| BAG3 | HeLa | Chaperones | Up | [116] |
| HeLa | Chaperones | Up | [117] | |
| Fas | A549 cells | Apoptosis | Up | [123] |
| Tdag51 | Mice | Apoptosis | Up | [121] |
| p21 | U2OS cells | Apoptosis | Up | [127] |
| gadd45 | U2OS cells | Apoptosis | Up | [127] |
| miR-34a | Mice, Sca-1+ cells | Apoptosis | Down | [118] |
| p62 | RKO, A549 and MCF-7 cells | Autophagy | Up | [55] |
| C57BL/6J mice, HeLa and HEK293 cells | Autophagy | Up | [56] | |
| ATG10 | Tomato | Autophagy | Up | [67] |
| ATG18f | Tomato | Autophagy | Up | [67] |
| Atg7 | MDA-MB-231 and -436 cells | Autophagy | Up | [65] |
| lgg-1 | C. elegans | Autophagy | Down | [38] |
| lgg-2 | C. elegans | Autophagy | Down | [38] |
| epg-9 | C. elegans | Autophagy | Down | [38] |
| UBB | HepG2 | Ubiquitin–proteasome system | Up | [77] |
| UBC | HepG2 | Ubiquitin–proteasome system | Up | [77] |
| let-70 | C. elegans | Ubiquitin–proteasome system | Up | [79] |
| ubi-1 | C. elegans | Ubiquitin–proteasome system | Up | [79] |
| rpn-3 | C. elegans | Ubiquitin–proteasome system | Up | [79] |
| cyp-35B1 | C. elegans | Ageing | Up | [166] |
| sip-1 | C. elegans | Ageing | Up | [166] |
| daf-7 | C. elegans | Ageing, development | Down | [17] |
| daf-9 | C. elegans | Ageing, development | Up | [17] |
| maoc-1 | C. elegans | Dauer development | Up | [150] |
| daf-22 | C. elegans | Dauer development | Up | [150] |
| ABCB1/MDR | Mouse melanoma B16F10 cells | Multidrug resistance | Up | [83] |
| HeLa cells | Multidrug resistance | Up | [81] | |
| U2-OS cells, HepG2 | Multidrug resistance | Up | [82] | |
| Sat III | HeLa cells |
Pericentric long non-coding RNA Nuclear stress bodies |
Up | [100–102] |
| TERRA | HeLa cells |
Subtelomeric long non-coding RNA Long non-coding RNA |
Up | [116] |
| FUT4 | Human breast cancer cells (MCF-7, MDA-MB-231) | Cell growth, cell cycle | Up | [140] |
| IER5 | HeLa cells | Cell growth, cell cycle | Up | [141] |
| JUN | HeLa cells | Cell growth, cell cycle | Up | [136] |
| FoxM1 | Mouse embryonic fibroblast cells | Cell growth, cell cycle | Up | [139] |
| HuR | Mouse cell culture, MCF7, MCF10A, SKBR-3, MDA-MB-231, SK-OV-3 | Hypoxia | Up | [142] |
| FOS1 | Human and rodent cells | Cell growth, cell cycle | Up | [133] |
| Human and rodent cells | Cell growth, cell cycle | Up | [134] | |
| G-CSF | Mice | Immunity | Down | [175] |
| IL-6 | Mouse embryo fibroblast (MEF) cells | Immunity | Chromatin opening | [85] |
| Mouse embryo fibroblast (MEF) cells | Immunity | Chromatin opening | [59] | |
| Mice spleen cells | Immunity | Up | [87] | |
| TNFα | Mice | Immunity | Down | [174] |
| Human peripheral blood monocytes | Immunity | Down | [177] | |
| Mice, murine macrophages | Immunity | Down | [173] | |
| CXC chemokine genes (CXCL-1, -2, -5, -8) | Human pulmonary epithelial-like A549 cells | Immunity | Up, down | [178] |
| c-fms | Chinese hamster ovaricytes | Immunity | Down | [176] |
| IL-1beta | Chinese hamster ovaricytes | Immunity | Down | [176] |
| Human monocyte cell line THP-1 | Immunity | Up | [191] | |
| LIF1 | Mice | Immunity | Down | [191] |
| HLA-G | Human melanoma cell line M8 (HLA-A1, -A2, -B12, and B40/male) | Immunity | Up | [172] |
| ATF3 | MEFs | Immunity | Up | [59] |
| SPI1/PU.1 | Mice and monocyte | Immunity | Up | [182] |
| COX-2 | Primary human umbilical vein endothelial cells | Immunity | Up | [179] |
| tkt-1 | C. elegans | Metabolism | Up | [38] |
| aldo-2 | C. elegans | Metabolism | Up | [38] |
| R05F9.6 | C. elegans | Metabolism | Up | [38] |
| CKS2 | human breast cancer cell line | Cancer, cell cycle | Up | [150] |
| LY6K | Human breast cancer cell line | Cancer, signalling | Up | [150] |
| RBM23 | Human breast cancer cell line | Cancer, gene regulation | Up | [150] |
| CCT6A | Human breast cancer cell line | Cancer, protein folding | Down | [150] |
| CKS1B | Human breast cancer cell line | Cancer, cell cycle | Up | [150] |
| ST13 | Human breast cancer cell line | Cancer, protein folding | Down | [150] |
| EIF4A2 | Human breast cancer cell line | Cancer, translation | Up | [150] |
Non-HSP-related targets involved in cellular stress response are shown only. For a comprehensive list of HSF1 target genes, see Table S1
HSF1 and oxidative stress
Cellular response to oxidative stress is considered a kind of HSR. Indeed, the potent oxidizing agent hydrogen peroxide (H2O2) also evokes HSF1 homotrimerisation [40]. Two Cys residues, Cys35 and Cys105, were identified in the DNA-binding domain of HSF1, the transformation of which to Ser inhibits the ability of the protein to become homotrimerised and to bind DNA. Moreover, unlike in wild-type cells, C35S and C105S mutant HSF1 proteins remain in the cytoplasm following oxidative stress. Based on these results, it can be concluded that HSF1, through these Cys residues, is able to directly sense the cellular redox state, which greatly influences the structure and function of intracellular proteins. Indeed, misfolded proteins accumulate following oxidative stress. In model organisms, HSP genes become upregulated in response to oxidative stress [51, 52]. In yeast exposed to H2O2, Hsf1 binds to the promoter of CUP1, BTN2, SIS1, HSP1, SGT2, and SSA3 genes, and upregulates their transcriptional activity (Table 1; Table S1) [53]. A proteomic analysis has also indicated that menadione provoking oxidative stress induces the activation of various proteins, including metabolic enzymes, antioxidant enzymes, as well as chaperones and their co-factors in a Hsf1-dependent manner [54]. Thus, HSF1 is capable of inducing several cell protective mechanisms in response to oxidative stress.
Nrf2 (Nf E2-related factor 2) acts as a key regulator of cellular response to oxidative stress, and controls the transcription of genes that function to attenuate the effect of cellular stress caused by reactive oxygen species (ROS) [55]. HSF1 and Nrf2 share several target genes, including HMOX1 (hemoxygenase) and SQSTM1 (sequestosome)/p62, [56–58], Atf3 (activating transcription factor) [59, 60], HSPA1A/B/L (Hsp70), and HSPB1 (Hsp25/27) [61]. This implies that the two stress-induced transcription factors collaborate with each other under certain circumstances. Furthermore, certain small HSPs (sHSPs), such as HSPB1 (HSP25) and HSPB5 (αB-crystallin), are known to participate in maintaining cellular redox state, thereby reducing the level of oxidative damage. Due to the overexpression of these sHSPs, the concentration of the antioxidant glutathione (GSH) increases, together with glucose-6-phosphate-dehydrogenase (G6PD) activity, which contributes to the reduced state of GSH [61].
At physiological temperatures, the expression of sHSPs also relies on HSF1 [62]. In HSF1−/− mutant mice, HSPB1 (HSP25) and HSPB5 (αB-crystallin) levels are significantly lowered, as compared with wild-type (HSF1+/+) mice. In this mutant genetic background, the GSH/GSSG (glutathione disulphide) ratio is considerably decreased, and heart muscle cells contain higher levels of ROS as a consequence of reduced G6PD activity.
HSF1 can trigger autophagy
Autophagy (cellular self-eating) is a highly regulated self-degradation process of eukaryotic cells, during which parts of the cytoplasm are delivered into the lysosomal compartment for enzymatic breakdown. The autophagic process plays a fundamental role in maintaining cellular proteostasis and organellar integrity through ensuring normal macromolecule and organelle turnover and eliminating (degrading) damaged, largely toxic proteins [63, 64]. It has been demonstrated in several mammalian cell lines and in vivo models that autophagy is induced following heat shock [65, 66]. Surprisingly, a constitutively active HSF1 mutant protein was shown to inhibit both heat shock-induced and basal autophagy, and in the absence of HSF1 activity, starvation- and rapamycin-induced autophagy become elevated [65, 67]. Another study, however, reported that treatment of tumorous cells with the chemotherapeutic agent carboplatin leads to an increased amount of autophagic structures, and this change fails to occur when HSF1 is silenced [68]. These data suggest that HSF1 and the HSR may also play a role in the regulation of autophagy under certain conditions. Indeed, HSF1 is triggered by carboplatin and able to bind to a HSE found in the regulatory region of the autophagy gene Atg7 (Table 1; Table S1). This way, HSF1 may directly induce autophagy at the transcriptional level by upregulating certain key Atg genes.
In plants, which contain a complex family of dozens of HSF members [69], HsfA1a plays an orthologous function to HSF1 [70]. In tomato, HsfA1a was shown to confer tolerance to drought by inducing autophagy through directly promoting the expression of at least two Atg genes, Atg10 and Atg18 [71]. Their expression and the formation of autophagic structures were elevated in plants overexpressing HsfA1a, but were compromised in plants defective for HsfA1a.
SQSMT1/p62, which encodes a specific adaptor for selective autophagy (the protein actually delivers substrates for autophagy and also participates in autophagosome nucleation), was also identified as an HSF1 target gene in MCF-7 cells [57]. In this experimental paradigm, HSF1 conferred resistance to inhibitors of HSPC1 (HSP90), which plays an important role in various types of cancer, by promoting SQSMT1/p62 expression and the autophagic flux. Another study also showed that HSF1 can induce the autophagic clearance of protein aggregates through regulating SQSMT1/p62 activity [58]. According to this analysis, HSF1 inhibition blocked autophagosome formation and, hence, the elimination of damaged proteins.
Unlike severe stress, which is almost always harmful to the organism, mild stress can be favourable for health and survival. This phenomenon is called hormesis. In C. elegans, Atg genes were recently demonstrated to be required for longevity and increased thermotolerance caused by hormetic heat stress or HSF-1 overexpression [72]. Although direct regulatory interaction was not examined, mild heat stress could induce the expression of multiple Atg genes including those involved in the formation of autophagic structures and lysosomal breakdown [73–75]. Many of these Atg genes contain at least one putative HSE in their upstream regulatory sequence. In good accordance with these observations, HSF-1 overexpression was also capable of inducing the amount of autophagic structures and the expression of these Atg genes [72]. According to a recent ChIP-seq analysis performed on C. elegans, HSF-1 directly represses certain autophagy-related genes including lgg-1/Atg8, lgg-2/Atg8, and epg-9 during larval development under physiological conditions (Table 1; Table S1) [36]. Hence, autophagy is induced by HSF1 under certain cellular conditions.
Interaction between HSF1 and the ubiquitin–proteasome system
Beside autophagy, the ubiquitin–proteasome system (UPS) can also participate in the degradation of intracellular proteins. The UPS-targeted proteins are first labelled by ubiquitination, that is, conjugated with the small, highly conserved protein ubiquitin, and then transferred into the 26S proteasome complex, in which they are digested by enzymatic proteolysis to oligopeptides. The primary role of UPS is to control protein activity, but compromised and toxic proteins can also be eliminated by this molecular machinery. Defects in the UPS can lead to HSF1 and HSF2 overactivation, which in turn elevates HSP expression [76–79]. These data support an intimate relationship between HSF1 and the UPS in eliminating misfolded cytoplasmic proteins. It may seem reasonable that certain HSF proteins directly upregulate specific UPS genes. Indeed, a strong correlation was shown between HSF2 deficiency and the transcriptional activity of two ubiquitin genes, Ubb and Ubc, in mouse embryonic fibroblasts cells [77]. In addition, the expression of several components of the 20S and 19S proteasome subunits was also lowered in a genetic background defective for HSF2.
A ChIP-seq analysis on human cell lines has also revealed that both HSF1 and HSF2 are capable of binding the upstream regulatory region of Ubb and Ubc, and this interaction causes a significant transcriptional upregulation of these targets (Table 1; Table S1) [80]. By another study, Ubb gene was also identified as a direct HSF1 target in HepG2 cells [81]. The authors have found that the anti-inflammatory pyrrolidine dithiocarbamate promotes HSF1 binding to the Ubb promoter. Based on these results, it can be assumed that in response to proteotoxicity HSF proteins ensure the increased levels of ubiquitin and proteasomal proteins required for the effective operation of the UPS.
Co-activation of HSF1 and the UPS can also be observed during development. For instance, in C. elegans males, an apoptosis-independent programmed cell death process eliminates the so-called linker cells, and during this process, HSF1 increases the transcription of three UPS-related genes, ubi-1 (ubiquitin), rpn-3 (a proteasome subunit), and let-70 (E2 ligase) [82]. In this paradigm, HSF1 contributes to cell death rather than protecting cells from undergoing demise.
HSF1 in multidrug resistance
In cancer therapy, a significant problem of treatment effectiveness often results from the development of drug resistance that tumorous cells acquire against the chemical agent applied. This phenomenon is mainly caused by the activity of ABC (ATP-binding cassette) transmembrane proteins that pump the drug out from the cell at the expense of ATP hydrolysis. Behind drug resistance, there are some underlying mechanisms of chemoresistance, including those influencing the passage of drugs into the target cell, the efflux of drugs from the target cell, drug metabolism, and the loss of the affected cell by apoptosis. HSF1 can influence chemoresistance in multiple ways. One of them results from its anti-apoptotic (cell protective) effect, which is mainly achieved through HSP upregulation [83]. In addition, the role of HSF1 was also shown in the regulation of a specific ABC transporter [84–87]. Accordingly, HSF1 overexpression leads to an increased activity of ABCB1 (also called MDR1 or P-pg), which causes resistance against the chemotherapeutic agent doxorubicin [84–86]. However, another study reported ABCB1 hyperactivity under condition of HSF1 deficiency [87]. Thus, the regulatory interplay between HSF1 and ABCB1 is far from being clear. HSF1 may directly activate ABCB6 expression [85], or the regulation of ABCB1 by HSF1 may occur at the posttranscriptional level [84]. The latter alternative was supported by the finding that ABCB1 activation can happen even in an HSF1 mutant background defective for the transactivation domain of the protein [86]. It is possible that the effect of HSF1 on ABCB1 activity depends on cell type or transcription factors by which HSF1 interacts, or that HSF1 simply maintains an open chromatin state at the ABCB1 promoter to enable the recruitment of other transcription factors to this particular regulatory sequence. A similar role for HSF1 was previously reported. Accordingly, HSF1 binds the IL-6 (interleukin 6) promoter, allowing the recruitment of specific enhancers and inhibitors to this locus [88].
HSF1 in non-coding RNA transcription and chromatin remodelling
In primates, following proteotoxic stress electrodense structures, also called nuclear stress bodies (nSBs), become apparent in the nucleus [89, 90]. In addition to ribonucleoproteins and RNAs, HSF1 represents a major component of nSBs. In humans, nSBs are localised to specific loci on chromosomes 9, 12, and 15, where HSF1 and HSF2 attach to pericentric satellite III (Sat III) repeats [91–93]. At these specific heterochromatic regions, the two transcription factors promote the expression of long non-coding RNAs (Table 1; Table S1) [94, 95]. The exact role of nSBs in the HSR, however, remains unclear. Since nSBs contain several splicing factors and RNA-processing enzymes, it is possible that these proteins are stored in an inactive form in these compartments during stress [96, 97]. It was shown recently that Sat III transcripts recruit essential factors required for transcription in nSBs. This suggests that nSBs contribute to transcriptional inhibition under proteotoxic stress [98].
Accumulating evidence indicates that the expression of non-coding RNAs of telomeric origin also increases under heat stress [99–101]. Telomeric repeat-containing RNAs (TERRA) are chromosome-specific, long non-coding RNAs transcribed in the subtelomeric regions of chromosomes [100, 102]. Telomeres also become shortened (damaged) upon heat stress [100, 103]. Following TRF2 (telomeric repeat-binding factor 2) depletion, elevated TERRA transcription may play a key role in DNA damage response protecting naked telomeric sequences [104]. Furthermore, several HSEs have been identified in TERRA promoters. In accordance with these results, HSF1 was shown to directly induce the transcription of certain TERRAs (3p and 10p–18p), thereby lowering the level of telomere damage under condition of heat stress (Table 1; Table S1) [105]. Together, HSF1 participates in protecting telomere structure.
Numerous publications have reported that HSF1 influences stress-induced HSP transcription in collaboration with chromatin remodelling factors [106–113]. Such factors may promote the transition of HSEs into accessible (open) chromatin regions [13]. In C. elegans, for example, E2F/DP transcription factors support HSF1 binding to the promoter of genes that participate in controlling larval development [36]. In mouse fibroblast cells, HSF1 maintains the open chromatin state in the proximity of IL-6 gene even in the absence of cellular stress, thereby endorsing the accessibility of other transcription factors to this regulatory region [88]. These results imply that HSF1 is also involved in the regulation of chromatin state.
HSF1 and apoptosis
Genomics studies have revealed that HSF1 is able to upregulate several genes with an inhibitory effect on apoptosis [34–39]. The anti-apoptotic function of HSF1 contributes to the survival of hyperproliferating cells. In tumorous cells, this role of HSF1 is mainly achieved by upregulating HSPB1 (Hsp27) and HSPA1A/B/L (HSP70). These HSPs negatively interact with members, such as death receptors, caspases, and mitochondrial factors, of the apoptotic cell death pathway [10, 114–118]. Furthermore, HSF1 was shown to induce directly the transcriptional activity of BAG3, which encodes a co-chaperone for HSPA1A/B/L (HSP70) [119, 120]. In Sca-1+ (stem cell antigen) stem cells, it also binds the promoter region and represses the expression of MiR-34a gene (HSF1 actually increases H3K27me3 levels at this locus), which, in turn, blocks HSPA1A/B/L (HSP70) [121]. These data imply that HSF1 can prevent normal and tumorous cells from undergoing apoptosis.
In contrast, both heat stress and HSF1 overexpression can trigger the apoptotic cell death program through modulating the Fas pathway [122, 123]. Fas is a death receptor that becomes activated upon ligand binding to trigger the extrinsic part of the apoptotic pathway. In germline cells of male mice, HSF1 directly associates with the regulatory region of Tdag51 (T-cell death-associated gene) to activate its transcription in response to heat stress [124]. Tdag51 codes for a Pleckstrin homology domain protein that induces apoptosis in T-lymphocytes and neurons, and triggers the activity of the pro-apoptotic Fas gene in T-cell hybridoma cells [125]. At high temperatures, HSF1, hence, can induce apoptosis through activating the Fas-mediated pathway in a Tdag-51-dependent fashion.
Later, human FAS1 was identified as a direct transcriptional target of HSF1 (Table 1; Table S1) [126]. The death-associated protein kinase DAPK exerts a pro-apoptotic effect, and its transcription is elevated by certain cytokines such as Fas and TNFα (tumour necrosis factor alpha) [127, 128]. During mild TNFα-mediated inflammatory stress, HSF1 directly upregulates DAPK which leads to the apoptotic loss of colorectal tumour cells [129]. Together, these data show that, under certain circumstances, HSF1 is capable of inducing the extrinsic part of the apoptotic cell death pathway.
HSF1 also play a regulatory role in p53-induced gene expression. Indeed, HSF1 is required for p53-mediated cell cycle arrest and apoptosis [130], and its activity is essential for the expression of several p53 target genes under both normal and genotoxic stress-induced conditions. HSF1 may operate as a p53 co-factor. First, according to ChIP experiments, the parallel binding of HSF1 and p53 is detectable on the promoter of certain p53 target genes such as p21 and gadd45 (Table 1; Table S1). Second, the silencing of HSF1 significantly reduces p53 binding to its targets, as compared with the control genetic background. Other studies have raised the possibility that HSF1 also participates in controlling nuclear translocation of p53 [8, 131]. Based on these data, one may conclude that HSF1 can trigger apoptotic cell death through promoting p53-induced gene expression.
HSF-1 in cell growth and proliferation
Above physiological temperatures, the cell cycle process can become blocked. Several regulatory proteins involved in cell cycle control are stabilized by molecular chaperones. As an example, Cdc2 (cyclin-dependent kinase) forms a complex with HSPC1 (HSP90) to mediate signal transduction [132–134]. Thus, HSF1 may control cell cycle and proliferation via inducing HSP chaperones. Indeed, double knockout of HSF1 and HSF3 leads to a decreased expression of HSPC1 (Hsp90a) in chicken DT40 cells even under physiological conditions. This causes Cdc2 instability and the affected cell sticks in the G2 phase upon heat stress. Hyper-activation of HSPC1 (Hsp90a) can rescue normal cell cycle progression even in the presence of HSF1/3 deficiency. In this context, HSF1 deficiency suppresses RAS oncogene- and p53 tumour suppressor mutation-induced tumour development in mice [135].
AP-1 (activator protein) complex consists of a Jun and a Fos family member. The complex controls the expression of numerous genes implicated in cell cycle regulation, as well as cell migration and death. Upon proteotoxicity, HFS1 binds to an HSE located in the Fos1 promoter, and Fos1 expression is increased in a HFS1-dependent manner, (Table 1; Table S1) [136, 137]. HSF2 also activates Fos1 transcription [138]. In HeLa cells exposed to stress, HSF1 interacts with regulatory sequences of the proto-oncogene Jun and promotes its transcription (Table 1; Table S1) [139]. FoxM1 (Forkhead box M1) transcription factor acts as a master regulator of cell cycle, and its dysregulation is strongly linked to tumour development and progression. FoxM1 is required for the survival of cells exposed to heat stress and for G2/M transition. Its direct transcriptional targets include the cell cycle genes cyclin B1, Cdc20, and Cdc2 [140, 141]. In response to proteolytic stress, HSF1 binds the promoter of FoxM1 to stimulate its expression [142]. Therefore, HSF1 controls cell cycle in both direct and indirect ways.
HSF1 also directly controls the transcriptional activity of FUT4 (Fucosyltransferase IV), which plays an important regulatory role in proliferation (Table 1; Table S1) [143]. The serum- and growth factor-induced IER5 (immediate early response gene) is also under the control of HSF1 (Table 1; Table S1) [144]. Based on these data, one can conclude that HSF1 plays a key role in the control of cell growth and division, and, accordingly, it serves as an important drug target in cancer therapy [7].
HIF1 (hypoxia-inducible factor) mediates the effect of low oxygen concentration (hypoxia) on cell growth and proliferation. Its translation is mediated by the mRNA-binding protein HuR (human antigen R), the transcription of which, in turn, directly relies on HSF1 [145]. Therefore, HSF1 promotes tumour growth in an oxygen-depleted environment by triggering angiogenesis. HuR also interacts with the long non-coding RNA lincRNA-p21, a major mediator of p53-dependent apoptosis, which inhibits the translation of CTNNB1 mRNA encoding beta catenin, a key regulator of cell division [146].
In addition to regulating gene expression, HSF1 can control cell growth and proliferation via other mechanisms. For example, it influences translation and cell growth by modulating the JNK-mTOR (c-Jun N-terminal kinase-mammalian target of rapamycin) pathway [147]. HSF1 is likely to promote mTOR-mediated protein synthesis by inhibiting JNK at the posttranscriptional level. It also directly binds the upstream regulatory region of Hgf gene encoding hepatocyte growth factor (Table 1; Table S1). Furthermore, in case of elevated mTOR activity, HSF1 translation and HSP transcription become markedly increased. Thus, HSF1 is capable of controlling proteotoxic stress response at the level of both transcription and translation.
HSF1 in cancer
Since HSF1 is implicated in controlling cell growth and proliferation, as well as apoptosis (see chapters above), its potential effect on cancer development should be also discussed briefly. Through regulating its target genes, HSF1 modulates several cellular processes and molecular mechanisms (e.g., autophagy, the UPR, oxidative stress response, and multidrug resistance) that are involved in the survival of terminally differentiated cells. HFS1 primarily supports the survival of cells by increasing their stress tolerance. It also elevates the ability of cancer cells to resist against various stress factors. Accordingly, increased HSF1 activity has been detected in numerous types of cancer [6, 7, 148, 149]. In malignant breast tumour cells, HSF1 was shown to control the expression of cancer-specific target genes, such as CKS2 (cyclin-dependent kinases regulatory subunit 2), LY6K (lymphocyte antigen 6 family member K), RBM23 (RNA-binding motif protein), CCT6A (chaperonin containing TCP1 subunit 6A), CKS1B (CDC28 protein kinase regulatory subunit 1B), ST13 (suppression of tumorigenicity) and EIF4A2 (eukaryotic translation initiation factor 4A2) (Table 1; Table S1), that are distinct from those involved in heat shock response [150].
Several oncoproteins, such as ERBB2/HER2, c-MET, CYCLIND1, CDK4, BRAF, AKT, and TP53, require HSF1 for maintaining their structural stability. On the other hand, signalling system involved in cell growth and proliferation is also affected by HSF1. The best examples include the RAS/RAF/MEK (Ras/rapidly accelerated fibrosarcoma/MAPK-ERK kinase), TGFβ (transforming growth factor-beta), and PI3K/AKT/mTOR (phosphatidylinositol 3-kinase) signalling pathways [19, 149]. Cancer cells are also frequently exposed to hypoxia. HIF1 is an important factor in tumour growth and metastasis [151]. HSF1 promotes HIF1 translation and, hence, tumour angiogenesis by inducing the transcription of the mRNA-binding protein HuR (human antigen R) [152]. Thus, HSF1 influences both tumorigenesis and tumour growth.
HSF-1 controls stress-induced developmental diapause
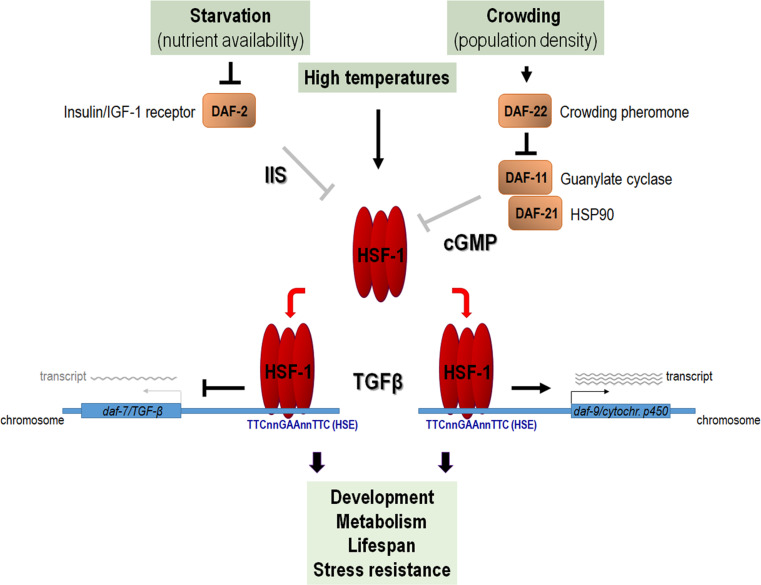
Grown under normal conditions, the hatched C. elegans embryo develops into a reproductive adult through four larval stages, L1–L4, which are separated by moults. However, under harsh environmental conditions such as starvation, high temperatures and elevated population density (crowding), the L1 larva develops into a highly stress-resistant dauer larval stage, which is a non-ageing developmental diapause [153]. On exposure to normal conditions, the animal exits from the dauer stage and resumes development. Mutations causing constitutive dauer larval development even under normal conditions confer increased stress resistance to the adult animal and significantly extend its lifespan. Concentration of the C. elegans dauer pheromone is proportional with population density as each animal secretes a nearly equal amount of substance to the environment. The crowding pheromone encoded by daf-22 (dauer formation defective), together with its ligand DAF-6, inhibits the guanylate cyclase DAF-11 and HSPC1/HSP90-like DAF-21 proteins, which, in turn, activate DAF-7, a TGFβ-like ligand. High levels of DAF-22 thereby hamper TGFβ signalling required for normal reproductive growth and promote dauer development. HSF-1 has been shown to mediate the stimulatory effect of DAF-11 and DAF-21 on daf-7 transcription [19]. DAF-11 and DAF-21 inhibit HSF-1, while HSF-1 directly represses daf-7 (Table 1; Table S1). This molecular interaction shows how HSF-1 becomes activated by a hormonal cue, the dauer pheromone, and how it promotes a stress-induced developmental event, the decision to form dauer larvae instead of reproductive adults. Moreover, HSF-1 upregulates directly another key component of the TGFβ signalling pathway, daf-9, which encodes a cytochrome P450, thereby fine-tuning the decision between dauer development vs. reproductive growth [19]. These results demonstrate that HSF-1 can sense and respond to at least three different stress factors, food deprivation, elevated temperatures, and crowding (Fig. 1).
Fig. 1.
Signal integration by HSF-1 in response to various stress factors in C. elegans. HSF-1 is activated by diverse environmental stress factors, including starvation, high temperatures, and crowding. This activation is mediated by the insulin/IGF-1 (IIS) and cyclic guanosine monophosphate (cGMP) signalling pathways. HSF-1 activity then influences the expression of key components (daf-7 and daf-9) of transforming growth factor-beta (TGFβ) signalling, which regulates development, metabolism, ageing, and stress resistance in this organism. This way, HSF-1 intertwines the insulin/IGF-1, cGMP, and TGFβ signalling systems in orchestrating cellular stress response. Arrows indicate activation; bars represent inhibitory interactions
The insulin/IGF-1 signalling pathway, which similarly affects metabolism, development, stress resistance, and lifespan, also lowers HSF-1 activity [20]. In nematodes defective for the insulin/IGF-1 receptor DAF-2, daf-7 transcript levels are decreased in an HSF-1-dependent manner, as compared with the wild type [19]. DAF-2 deficiency attenuates daf-7 expression, while, in daf-2(-); hsf-1(-) double mutant animals, daf-7 is expressed at nearly normal levels. This implies that HSF-1 influences development, metabolism, lifespan, and stress resistance through intertwining three signal transduction systems, cGMP/GC (cyclic guanosine monophosphate/guanylate cyclase), insulin/IGF-1, and TGFβ signalling (Fig. 1).
The influence of HSF-1 on stress-induced dauer development is further supported by the fact that its several target genes are involved in the synthesis of ascaroside pheromones, also called daumones. acox-1 (acyl-coenzyme A oxidase), dhs-28 (dehydrogenases), maoc-1 (maoc-like dehydratase) and daf-22 expression are each controlled by HSF-1 (Table 1; Table S1) [153]. Accordingly, under condition of heat stress, dauer pheromone extracts isolated from wild-type animals trigger dauer development more effectively than those isolated from hsf-1(–) mutants. Hence, dauer pheromone synthesis at higher temperatures is regulated by HSF-1.
HSF1 and ageing
During the lifespan of an organism, damaged macromolecules, in particular misfolded, aggregated and oxidized proteins, progressively accumulate in the cytoplasm, which leads to massive levels of cell death and eventually a tissue dysfunction at advanced ages [63, 154–158]. In ageing cells, proteostasis becomes gradually compromised, implying that molecular pathways ensuring protein homeostasis also participate in the ageing process. Indeed, the capacity of cellular stress-response (maintenance and repair) mechanisms including the HSR and UPR, as well as autophagy, markedly declines as the organism ages [159–163]. By inducing these mechanisms and pathways, HSF1 preserves proteostasis at advanced ages and hence promotes longevity [164–167].
In C. elegans, elevated dosages of HSF-1 increase both stress tolerance and lifespan [168, 169]. Furthermore, worms depleted for HSF-1 live significantly shorter than untreated control [170]. These studies have shown that HSF1 activity is required for longevity in daf-2(–) mutant nematodes defective for insulin/IGF-1 signalling. The FOXO (Forkhead box O) transcription factor DAF-16, the terminal effector of the pathway, regulates several genes in collaboration with HSF-1. These targets include sHSP genes [168], sip-1 (stress-induced protein), and cyp-35B1/dod-13 (cytochrome P450 family) [171] encoding a cytochrome P450 that is also involved in controlling lipid storage (Table 1; Table S1). HSF-1 also affects certain TGFβ pathway components including daf-7/TGFβ and daf-9/p450, which inhibit dauer larval development [19]. These results demonstrate that HSF-1 controls the ageing process at least in part through functioning downstream of insulin/IGF-1 signalling.
In Drosophila, overexpressing mitochondrial Hsp22 promotes longevity [172]. Mice mutant for a co-chaperone (CHIP) exhibit a faster rate of the ageing process, while, in long-lived mutant mouse strains certain Hsp genes become upregulated [173]. Together, the effect of HSF1 on ageing is established in a complex manner: first, through the upregulation of HSP genes, second, via modulating other stress/repair pathways that determine lifespan (e.g., autophagy), and, third, by influencing signalling systems (e.g., IIS) that affect the rate at which the cells age.
HSF1 in immune response
Studies on invertebrate and vertebrate genetic systems have demonstrated that HSF1 enables the normal function of the immune system [174–177]. In C. elegans, for example, HSF-1 is required for tolerance against several pathogenic agents, while in mice defective for HSF1 IgG generation is also compromised.
Certain cytokines promoting fever and inflammation are inhibited by HSF1. The interaction between HSF1 and TNFα (tumour necrosis factor alpha) appears to be direct as HSF1 binds to its regulatory region [178]. The expression of other cytokine- or cytokine receptor-encoding genes, such as IL-6, c-fms (colony-stimulating factor-1 receptor) and m-scf (macrophage colony-stimulating factor), G-CSF (granulocyte-colony-stimulating factor) and IL-1b (interleukin 1 beta), is also repressed by HSF1 (Table 1; Table S1) [60, 175, 178–181]. In case of IL-6, it has been noted that HSF1 binds its promoter region and maintains an open chromatin state in order to recruit transcription factors with inhibitory (e.g., ATF3) or stimulatory (e.g., NF-κB; nuclear factor kappa B) effects. These cytokines play a role in provoking inflammation. In addition, the regulatory role of HSF1 was shown in the activity of several other cytokines such as CXCL1, 2, 5, and 8 (chemokine ligand), as well as IL-10 [182, 183]. HSF1 also stimulates the transcription of COX-2 (cyclooxygenase), a key regulator of inflammatory processes [184].
HSF1 regulates the hypothalamic-specific expression of the heat-inducible ion channel gene TRPV1 (transient receptor potential vanilloid) [185]. Since this protein plays an important role in the control of body temperature, HSF1 has a protective effect during fever. It inhibits certain cytokines to reduce fever and to protect against chronic inflammation. Consistent with the potential anti-inflammatory effect of HSF1, treatment with heat shock can lower the level of death caused by sepsis [186].
The function of HSF1 was also revealed in haematopoiesis [187]. It directly controls the expression of SPI1/PU.1 (PU-box) transcription factor, which influences macrophage differentiation. As a competitive inhibitor for NF-IL-6, HSF1 blocks G-CSF (granulocyte-colony-stimulating factor) transcription, thereby promoting myeloid differentiation [178].
During acute viral infection, HSPs are generally induced. Following infection by HIV, HSPB1 (Hsp27), DNAJB1 (Hsp40) and HSPA1A/B/L (HSP70) expression becomes rapidly elevated [188, 189]. Consistent with this change, the activity of HSF1 also increases, and the protein directly binds HIV-1LTR promoter to trigger expression [190].
HFS1 in development and physiology
Among the HSF1 targets identified so far, several genes are involved in the control of normal developmental events (Table 1; Table S1). This suggests that HSF1 functions, contrary to its traditional name “heat shock”, not only under stress-induced circumstances, but also in normal biological processes.
Larval development
In C. elegans, a strong hsf-1(–) mutation, the deletional allele ok600 affecting both regulatory and transactivation domains of the encoded protein, arrests development at the L2/L3 larval stages [29, 36]. Another hsf-1 allele, the nonsense mutation sy441 altering the transactivation domain at the C-terminus, enables the organism to develop into adulthood under stress-free conditions [191]. In hsf-1(sy441) mutants, HSP induction is also compromised. Thus, larval development and reproductive growth in nematodes rely on HSF-1 activity. Indeed, a recent ChIP-seq analysis has identified around 70 target genes for HSF-1 which mediate larval development in this organism (Table 1; Table S1) [36]. The promoter of the development-related HSF1 targets often contains a partial HSE and a GC-rich motif (Table 1; Table S1). This raises the possibility that HSF1 binds partial HSEs with the help of a co-factor. For example, numerous developmental genes are coordinately regulated by HSF1 and the E2F/DP coactivator complex [36].
In Drosophila, larval development also requires HSF1 function; flies defective for HSF1 arrest development at the late L2-to-early L3 larval stages [28]. Using a conditional (thermosensitive) hsf1 mutant starin, it has been revealed that HSF1 deficiency from the onset of the L2 larval stage allows the animal to develop as an adult. Moreover, HSF1 plays a role in oogenesis as its defects cause maternal effect sterility. HSF1 probably affects larval development independently of HSPs (under stress-free conditions, the expression levels of HSPs do not significantly differ between HSF1 mutants and wild-type animals) [28].
Neuronal development
In mice, HSF1 participates in the development of the central nervous system. The absence of HSF1 activity can lead to impaired hippocampal spino- and neurogenesis, and severe perturbations in behaviour, such as depression and aggressivity [192]. It has been revealed that HSF1 directly controls the expression of two polysialyltransferase genes, St8siaII and St8siaIV, in the hippocampus, thereby influencing levels of PSA-NCAM (polysialylated-neural cell adhesion molecule), which is essential for synapse development [192]. Moreover, HSF1 displays a cell protective role during neuronal development via upregulating Syt1 and Vamp2 genes in response to cellular insults (Table S1) [193, 194]. Dp71 gene encoding dystrophin was also identified as a direct HSF1 target [195]. As dystrophin has a key role in the development of the nervous system, particularly that of hippocampus, it is possible that reduced expression of Dp71 contributes to behavioural abnormalities in mice deficient in HSF1 function.
In the mouse neuroepithelium, proliferation and differentiation of sensory neurons are regulated by several cytokines. One of them is called LIF1 (leukaemia inhibitory factor), the expression of which is promoted by HSF1, but inhibited by HSF4 [196]. LIF1 overexpression triggered by HSF4 deficiency may contribute to neuroepithelial atrophy and smell disorders.
HSF1 and HSF4 also operate antagonistically in regulating the differentiation of eye lens epithelial cells [197]. As demonstrated, HSF4 hampers fibroblast growth factor (FGF) genes, such as FGF1, 4, and 7, which drive this differentiation event. FGF7 was identified as a direct HSF4 target. Unlike HSF4, HSF1 activates these targets (Table S1). In good accordance with these results, the HSF1:HSF4 double mutant genotype allows a nearly normal development of the lens epithelium of mice.
Gamete differentiation
HSF1 has a dual role in spermatogenesis. First, it prevents immature germ cells from undergoing death [198]. Second, it participates in the elimination of defective spermatocytes during meiotic prophase (pachiten checkpoint) [199]. Although the above roles of HSF1 occur through HSP regulation, in mice HSF1 and HSF2 directly control several sex chromosome-specific multicopy genes in meiotic spermatocytes and postmitotic haploid round spermatids [132]. These genes located on chromosome X or Y are required for the proper packaging of DNA into the sperm.
HSF1 is also essential for oocyte meiosis in Drosophila [28] and in other animal species. In HSF1 knockout mice, oocytes do not develop further; these cells arrest the early development at phase I or II of meiosis [200]. Presumably, lowered HSPC1 (HSP90) levels are responsible for this phenomenon, because, in the absence of HSF1, HSPC1 (Hsp90) becomes downregulated, and HSPC1- (Hsp90) specific inhibitors cause a very similar phenotype. Using a comparative transcriptome analysis of wild-type and HSF1 defective mouse oocytes, it has been demonstrated that HSF1 influences the transcription of several genes during oocyte meiosis [201]. In HSF1−/− mutant oocytes, the synaptonemal complex, recombination nodules and DNA repair are all compromised. Thus, HSF1 may function as an important meiotic transcription factor as well.
Circadian rhythm
Circadian rhythm is the endogenous oscillation of biological processes of about 24 h. Cells have an inner circadian clock driving this 24-h rhythm, which rapidly becomes desynchronized when cells are maintained in cultures [202]. Certain stimuli, such as temperature, however, are capable of resetting and synchronizing this clock; rhythmic alternations of temperature lead to rhythmic changes of clock genes [130, 203]. The core clock gene Per2 can be induced by heat shock, and its promoter contains functional HSEs [204, 205]. Moreover, both nuclear localization and phosphorylation levels of HSF1 display a circadian rhythm, which are likely to be driven by changes in temperature [206]. In chicken epiphysis, for example, HSF1 activity is triggered by light [207].
Conclusions
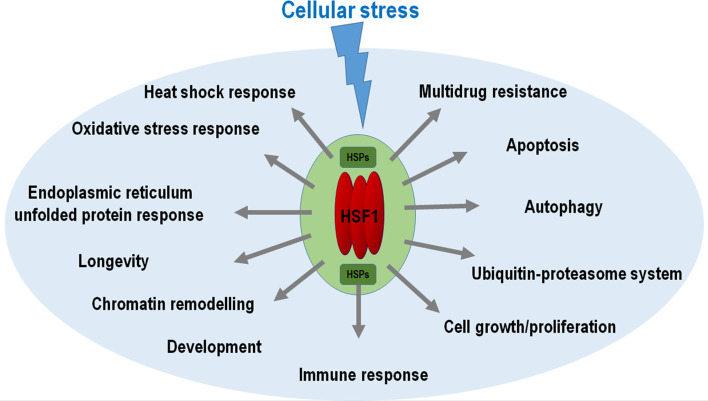
Results from the last 2 decades have clearly indicated that the roles of HSF1 are not limited to the control of the HSR. Through influencing the transcriptional activity of target genes encoding diverse proteins other than HSPs, this transcription factor influences basic cellular maintenance processes and molecular mechanisms like autophagy, the UPR and UPS, multidrug resistance, programmed cell death, cell cycle arrest, chromatin structure, and immune response (Fig. 2). These functions of HSF1 are often achieved in a complex manner. For instance, HSF1 affects cell growth and proliferation by directly regulating specific target genes involved in these processes (Table 1; Table S1), via influencing (an)other intermediate process(es) (e.g., autophagy [135, 208, 209]) or mechanism(s), or through upregulating HSP genes that participate in that particular processes (Fig. 2). Another impressive example is the ageing process, the rate of which is also modulated by HSF1 (Fig. 2). In C. elegans, HSF-1 controls several DAF-16 target genes (DAF-16 acts an effector of insulin/IGF-1 signalling that plays a key regulatory role in lifespan determination); among them, certain Hsp genes [168] and Atg genes [71] are required for normal lifespan [208]. HSF1 similarly affects various developmental events including decision between normal reproductive growth and dauer larval formation (in nematodes), gamete differentiation (in vertebrates), generation of various immune factors (in organisms ranging from nematodes to mammals), and synapse formation (in mammals) (Fig. 2). Besides HSPs, direct transcriptional targets of HSF1 include genes that encode for example signalling components, ABC transporters, immune proteins, Atg proteins, proteasome-specific proteins, chromatin remodellers, and factors involved in cell growth and proliferation (Table 1; Table S1). These data imply that HSF1 is able to sense and respond to diverse cellular stress factors leading to proteotoxicity, such as high temperatures, starvation, crowding, adverse chemical agents (toxins and heavy metals), and potentially harmful metabolic factors (ROS). These data imply that HSF1 functions as a major orchestrator of distinct cellular stress response mechanisms (Fig. 2).
Fig. 2.
Co-regulation of cellular stress response mechanisms and processes by HSF1. HSF1 is activated by various cellular stress factors (represented by the blue swallow). HSF1 activity then upregulates key components of diverse stress response pathways and processes. Thus, HSF1 acts as a major orchestrator of cellular stress response pathways and processes
Accumulating evidence indicates that, during development, HSF1 also controls a transcriptional program distinct from the HSR. For example, in C. elegans, it interacts with E2F and DP transcription factors to regulate larval development [36]. It is intriguing that, even in the absence of transactivation domain, HSF1 can mediate development and thermotolerance by upregulating various non-HSP target genes [210]. In murine neuroblastoma HT22 cells, overexpressing HSF1 lacking trimerisation and transactivation domains exerts a neuroprotective effect, as the mutant protein is not able to bind to classical HSEs [211]. Thus, this effect of HSF1 does not rely on HSP upregulation. An important issue in this field will certainly be uncovering the mechanisms, whereby HSF1 influences these processes. It is possible, for instance, that HSF1 can interact with other transcription factors through its trimerisation domain. Alternatively, it can open local chromatin structures to recruit other transcription factors to target genes or function as a co-factor for other transcription factors. Addressing these questions will open new avenues in this field.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the grants OTKA (Hungarian Scientific Research Fund) NK78012 and K115378, MEDinPROT Protein Science Research Synergy Program (provided by the Hungarian Academy of Sciences; HAS), and VEKOP (VEKOP-2.3.2-16-2017-00014). B.J and T.V. are also supported by the MTA-ELTE Genetics Research Group (01062).
Author contributions
Each author (JB, PC, and TV) has participated in collecting non-HSP targets of HSF1, characterising the role of HSF1 in functions other than the HSR, and writing the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interest.
References
- 1.Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19(1):4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellaye PS, Burgy O, Causse S, Garrido C, Bonniaud P. Heat shock proteins in fibrosis and wound healing: good or evil? Pharmacol Ther. 2014;143(2):119–132. doi: 10.1016/j.pharmthera.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Hooper PL, Balogh G, Rivas E, Kavanagh K, Vigh L. The importance of the cellular stress response in the pathogenesis and treatment of type 2 diabetes. Cell Stress Chaperon. 2014;19(4):447–464. doi: 10.1007/s12192-014-0493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willis MS, Patterson C. Hold me tight: role of the heat shock protein family of chaperones in cardiac disease. Circulation. 2010;122(17):1740–1751. doi: 10.1161/CIRCULATIONAHA.110.942250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q. Role of heat shock proteins in atherosclerosis. Arterioscler Thromb Vasc Biol. 2002;22(10):1547–1559. doi: 10.1161/01.ATV.0000029720.59649.50. [DOI] [PubMed] [Google Scholar]
- 6.Dai C, Sampson SB. HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 2016;6(1):17–28. doi: 10.1016/j.tcb.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 10.Acunzo J, Katsogiannou M, Rocchi P. Small heat shock proteins HSP27 (HspB1), alphaB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int J Biochem Cell Biol. 2012;44(10):1622–1631. doi: 10.1016/j.biocel.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8(9):3761–3769. doi: 10.1128/MCB.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaeger AM, Makley LN, Gestwicki JE, Thiele DJ. Genomic heat shock element sequences drive cooperative human heat shock factor 1 DNA binding and selectivity. J Biol Chem. 2014;289(44):30459–30469. doi: 10.1074/jbc.M114.591578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6(9):e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun. 2017;8(1):255. doi: 10.1038/s41467-017-00151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pernet L, Faure V, Gilquin B, Dufour-Guerin S, Khochbin S, Vourc’h C. HDAC6-ubiquitin interaction controls the duration of HSF1 activation after heat shock. Mol Biol Cell. 2014;25(25):4187–4194. doi: 10.1091/mbc.E14-06-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperon. 2004;9(2):122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323(5917):1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;103(1):45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barna J, Princz A, Kosztelnik M, Hargitai B, Takacs-Vellai K, Vellai T. Heat shock factor-1 intertwines insulin/IGF-1, TGF-beta and cGMP signaling to control development and aging. BMC Dev Biol. 2012;12:32. doi: 10.1186/1471-213X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148(1–2):322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderwood SK, Xie Y, Wang X, Khaleque MA, Chou SD, Murshid A, Prince T, Zhang Y. Signal transduction pathways leading to heat shock transcription. Sign Transduct Insights. 2010;2:13–24. doi: 10.4137/STI.S3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi HS, Li B, Lin Z, Huang E, Liu AY. cAMP and cAMP-dependent protein kinase regulate the human heat shock protein 70 gene promoter activity. J Biol Chem. 1991;266(18):11858–11865. [PubMed] [Google Scholar]
- 23.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 24.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40(2):253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Buchner J. Hsp90 and Co.—a holding for folding. Trends Biochem Sci. 1999;24(4):136–141. doi: 10.1016/S0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 26.Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79(2):129–168. doi: 10.1016/S0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 27.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18(3):306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 28.Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16(9):2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton EA, Lamitina T. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell. 2013;12(1):112–120. doi: 10.1111/acel.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54(6):855–864. doi: 10.1016/S0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 31.Solis EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell. 2016;63(1):60–71. doi: 10.1016/j.molcel.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18(21):5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephanou A, Latchman DS. Transcriptional modulation of heat-shock protein gene expression. Biochem Res Int. 2011;2011:238601. doi: 10.1155/2011/238601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6(7):R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genom. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Chauve L, Phelps G, Brielmann RM, Morimoto RI. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 2016;30(18):2062–2075. doi: 10.1101/gad.283317.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell. 2016;62(1):63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24(12):5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15(3):1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn SG, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17(4):516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 42.Weindling E, Bar-Nun S. Sir2 links the unfolded protein response and the heat shock response in a stress response network. Biochem Biophys Res Commun. 2015;457(3):473–478. doi: 10.1016/j.bbrc.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27(7):1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou J, Tang H, Liu Z, Osterlund T, Nielsen J, Petranovic D. Management of the endoplasmic reticulum stress by activation of the heat shock response in yeast. FEMS Yeast Res. 2014;14(3):481–494. doi: 10.1111/1567-1364.12125. [DOI] [PubMed] [Google Scholar]
- 45.Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1alpha. Mol Cell Biol. 2002;22(24):8506–8513. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010;8(7):e1000410. doi: 10.1371/journal.pbio.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heldens L, Hensen SM, Onnekink C, van Genesen ST, Dirks RP, Lubsen NH. An atypical unfolded protein response in heat shocked cells. PLoS One. 2011;6(8):e23512. doi: 10.1371/journal.pone.0023512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zito E. ERO1: a protein disulfide oxidase and H2O2 producer. Free Radic Biol Med. 2015;83:299–304. doi: 10.1016/j.freeradbiomed.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Takemori Y, Sakaguchi A, Matsuda S, Mizukami Y, Sakurai H. Stress-induced transcription of the endoplasmic reticulum oxidoreductin gene ERO1 in the yeast Saccharomyces cerevisiae. Mol Genet Genom. 2006;275(1):89–96. doi: 10.1007/s00438-005-0065-9. [DOI] [PubMed] [Google Scholar]
- 50.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 51.Chuang YY, Chen Y, Gadisetti Chandramouli VR, Cook JA, Coffin D, Tsai MH, DeGraff W, Yan H, Zhao S, Russo A, Liu ET, Mitchell JB. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62(21):6246–6254. [PubMed] [Google Scholar]
- 52.Lee J, Bruce-Keller AJ, Kruman Y, Chan SL, Mattson MP. 2-Deoxy-d-glucose protects hippocampal neurons against excitotoxic and oxidative injury: evidence for the involvement of stress proteins. J Neurosci Res. 1999;57(1):48–61. doi: 10.1002/(SICI)1097-4547(19990701)57:1<48::AID-JNR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto A, Ueda J, Yamamoto N, Hashikawa N, Sakurai H. Role of heat shock transcription factor in Saccharomyces cerevisiae oxidative stress response. Eukaryot Cell. 2007;6(8):1373–1379. doi: 10.1128/EC.00098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim IS, Kim H, Kim YS, Jin I, Yoon HS. HSF1-mediated oxidative stress response to menadione in Saccharomyces cerevisiae KNU5377Y3 by using proteomic approach. Adv Biosci Biotechnol. 2013;4:44–54. doi: 10.4236/abb.2013.41007. [DOI] [Google Scholar]
- 55.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samarasinghe B, Wales CT, Taylor FR, Jacobs AT. Heat shock factor 1 confers resistance to Hsp90 inhibitors through p62/SQSTM1 expression and promotion of autophagic flux. Biochem Pharmacol. 2014;87(3):445–455. doi: 10.1016/j.bcp.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe Y, Tsujimura A, Taguchi K, Tanaka M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy. 2017;13(1):133–148. doi: 10.1080/15548627.2016.1248018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim KH, Jeong JY, Surh YJ, Kim KW. Expression of stress-response ATF3 is mediated by Nrf2 in astrocytes. Nucleic Acids Res. 2010;38(1):48–59. doi: 10.1093/nar/gkp865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takii R, Inouye S, Fujimoto M, Nakamura T, Shinkawa T, Prakasam R, Tan K, Hayashida N, Ichikawa H, Hai T, Nakai A. Heat shock transcription factor 1 inhibits expression of IL-6 through activating transcription factor 3. J Immunol. 2010;184(2):1041–1048. doi: 10.4049/jimmunol.0902579. [DOI] [PubMed] [Google Scholar]
- 61.Mehlen P, Kretz-Remy C, Preville X, Arrigo AP. Human hsp27, Drosophila hsp27 and human alphaB-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNFalpha-induced cell death. EMBO J. 1996;15(11):2695–2706. doi: 10.1002/j.1460-2075.1996.tb00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 2002;21(19):5164–5172. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16(1):94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 64.Vellai T, Takacs-Vellai K, Sass M, Klionsky DJ. The regulation of aging: does autophagy underlie longevity? Trends Cell Biol. 2009;19(10):487–494. doi: 10.1016/j.tcb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dokladny K, Zuhl MN, Mandell M, Bhattacharya D, Schneider S, Deretic V, Moseley PL. Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J Biol Chem. 2013;288(21):14959–14972. doi: 10.1074/jbc.M113.462408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nivon M, Richet E, Codogno P, Arrigo AP, Kretz-Remy C. Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy. 2009;5(6):766–783. doi: 10.4161/auto.8788. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y, Gong S, Shunmei E, Zou J. Induction of macroautophagy by heat. Mol Biol Rep. 2009;36(8):2323–2327. doi: 10.1007/s11033-009-9451-4. [DOI] [PubMed] [Google Scholar]
- 68.Desai S, Liu Z, Yao J, Patel N, Chen J, Wu Y, Ahn EE, Fodstad O, Tan M. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7) J Biol Chem. 2013;288(13):9165–9176. doi: 10.1074/jbc.M112.422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002;16(12):1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017;22(1):53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Cai S, Yin L, Shi K, Xia X, Zhou Y, Yu J, Zhou J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015;11(11):2033–2047. doi: 10.1080/15548627.2015.1098798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumsta C, Chang JT, Schmalz J, Hansen M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat Commun. 2017;8:14337. doi: 10.1038/ncomms14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sigmond T, Barna J, Toth ML, Takacs-Vellai K, Pasti G, Kovacs AL, Vellai T. Autophagy in Caenorhabditis elegans. Methods Enzymol. 2008;451:521–540. doi: 10.1016/S0076-6879(08)03230-8. [DOI] [PubMed] [Google Scholar]
- 74.Fodor E, Sigmond T, Ari E, Lengyel K, Takacs-Vellai K, Varga M, Vellai T. Methods to study autophagy in Zebrafish. Methods Enzymol. 2017;588:467–496. doi: 10.1016/bs.mie.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 75.Varga M, Fodor E, Vellai T. Autophagy in zebrafish. Methods. 2015;75:172–180. doi: 10.1016/j.ymeth.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 76.Kawazoe Y, Nakai A, Tanabe M, Nagata K. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur J Biochem. 1998;255(2):356–362. doi: 10.1046/j.1432-1327.1998.2550356.x. [DOI] [PubMed] [Google Scholar]
- 77.Lecomte S, Desmots F, Le Masson F, Le Goff P, Michel D, Christians ES, Le Drean Y. Roles of heat shock factor 1 and 2 in response to proteasome inhibition: consequence on p53 stability. Oncogene. 2010;29(29):4216–4224. doi: 10.1038/onc.2010.171. [DOI] [PubMed] [Google Scholar]
- 78.Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18(9):5091–5098. doi: 10.1128/MCB.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pirkkala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20(8):2670–2675. doi: 10.1128/MCB.20.8.2670-2675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vihervaara A, Sergelius C, Vasara J, Blom MA, Elsing AN, Roos-Mattjus P, Sistonen L. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci USA. 2013;110(36):E3388–E3397. doi: 10.1073/pnas.1305275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song S, Kole S, Precht P, Pazin MJ, Bernier M. Activation of heat shock factor 1 plays a role in pyrrolidine dithiocarbamate-mediated expression of the co-chaperone BAG3. Int J Biochem Cell Biol. 2010;42(11):1856–1863. doi: 10.1016/j.biocel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kinet MJ, Malin JA, Abraham MC, Blum ES, Silverman MR, Lu Y, Shaham S. HSF-1 activates the ubiquitin proteasome system to promote non-apoptotic developmental cell death in C. elegans. eLife. 2016;5:73. doi: 10.7554/elife.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beere HM. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J Clin Investig. 2005;115(10):2633–2639. doi: 10.1172/JCI26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tchenio T, Havard M, Martinez LA, Dautry F. Heat shock-independent induction of multidrug resistance by heat shock factor 1. Mol Cell Biol. 2006;26(2):580–591. doi: 10.1128/MCB.26.2.580-591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vilaboa NE, Galan A, Troyano A, de Blas E, Aller P. Regulation of multidrug resistance 1 (MDR1)/P-glycoprotein gene expression and activity by heat-shock transcription factor 1 (HSF1) J Biol Chem. 2000;275(32):24970–24976. doi: 10.1074/jbc.M909136199. [DOI] [PubMed] [Google Scholar]
- 86.Vydra N, Toma A, Glowala-Kosinska M, Gogler-Piglowska A, Widlak W. Overexpression of Heat Shock Transcription Factor 1 enhances the resistance of melanoma cells to doxorubicin and paclitaxel. BMC Cancer. 2013;13:504. doi: 10.1186/1471-2407-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krishnamurthy K, Vedam K, Kanagasabai R, Druhan LJ, Ilangovan G. Heat shock factor-1 knockout induces multidrug resistance gene, MDR1b, and enhances P-glycoprotein (ABCB1)-based drug extrusion in the heart. Proc Natl Acad Sci USA. 2012;109(23):9023–9028. doi: 10.1073/pnas.1200731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem. 2007;282(45):33210–33217. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- 89.Cotto J, Fox S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110(Pt 23):2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- 90.Biamonti G, Vourc’h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2(6):a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alastalo TP, Hellesuo M, Sandqvist A, Hietakangas V, Kallio M, Sistonen L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J Cell Sci. 2003;116(Pt 17):3557–3570. doi: 10.1242/jcs.00671. [DOI] [PubMed] [Google Scholar]
- 92.Denegri M, Moralli D, Rocchi M, Biggiogera M, Raimondi E, Cobianchi F, De Carli L, Riva S, Biamonti G. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol Biol Cell. 2002;13(6):2069–2079. doi: 10.1091/mbc.01-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, Morimoto RI, Vourc’h C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J Cell Biol. 2002;156(5):775–781. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164(1):25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15(2):543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12(11):3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Metz A, Soret J, Vourc’h C, Tazi J, Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci. 2004;117(Pt 19):4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- 98.Goenka A, Sengupta S, Pandey R, Parihar R, Mohanta GC, Mukerji M, Ganesh S. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J Cell Sci. 2016;129(19):3541–3552. doi: 10.1242/jcs.189803. [DOI] [PubMed] [Google Scholar]
- 99.Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe’er D, Ruppin E, Sharan R, Kupiec M. Environmental stresses disrupt telomere length homeostasis. PLoS Genet. 2013;9(9):e1003721. doi: 10.1371/journal.pgen.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10(2):228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 101.Martinez-Guitarte JL, Diez JL, Morcillo G. Transcription and activation under environmental stress of the complex telomeric repeats of Chironomus thummi. Chromosome Res. 2008;16(8):1085–1096. doi: 10.1007/s10577-008-1260-4. [DOI] [PubMed] [Google Scholar]
- 102.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 103.Maeda T, Guan JZ, Koyanagi M, Makino N. Alterations in the telomere length distribution and the subtelomeric methylation status in human vascular endothelial cells under elevated temperature in culture condition. Aging Clin Exp Res. 2013;25(3):231–238. doi: 10.1007/s40520-013-0045-6. [DOI] [PubMed] [Google Scholar]
- 104.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koskas S, Decottignies A, Dufour S, Pezet M, Verdel A, Vourc’h C, Faure V. Heat shock factor 1 promotes TERRA transcription and telomere protection upon heat stress. Nucleic Acids Res. 2017;45(11):6321–6333. doi: 10.1093/nar/gkx208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17(11):1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomson S, Hollis A, Hazzalin CA, Mahadevan LC. Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol Cell. 2004;15(4):585–594. doi: 10.1016/j.molcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 108.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol. 2001;21(17):5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shivaswamy S, Iyer VR. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol. 2008;28(7):2221–2234. doi: 10.1128/MCB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367(6463):525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 111.Morgan WD. Transcription factor Sp1 binds to and activates a human hsp70 gene promoter. Mol Cell Biol. 1989;9(9):4099–4104. doi: 10.1128/MCB.9.9.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83(1):29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 113.Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 2016;30(15):1731–1746. doi: 10.1101/gad.284430.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stankiewicz AR, Lachapelle G, Foo CP, Radicioni SM, Mosser DD. Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J Biol Chem. 2005;280(46):38729–38739. doi: 10.1074/jbc.M509497200. [DOI] [PubMed] [Google Scholar]
- 115.Chauhan D, Li G, Hideshima T, Podar K, Mitsiades C, Mitsiades N, Catley L, Tai YT, Hayashi T, Shringarpure R, Burger R, Munshi N, Ohtake Y, Saxena S, Anderson KC. Hsp27 inhibits release of mitochondrial protein Smac in multiple myeloma cells and confers dexamethasone resistance. Blood. 2003;102(9):3379–3386. doi: 10.1182/blood-2003-05-1417. [DOI] [PubMed] [Google Scholar]
- 116.Paul C, Manero F, Gonin S, Kretz-Remy C, Virot S, Arrigo AP. Hsp27 as a negative regulator of cytochrome C release. Mol Cell Biol. 2002;22(3):816–834. doi: 10.1128/MCB.22.3.816-834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rocchi P, Jugpal P, So A, Sinneman S, Ettinger S, Fazli L, Nelson C, Gleave M. Small interference RNA targeting heat-shock protein 27 inhibits the growth of prostatic cell lines and induces apoptosis via caspase-3 activation in vitro. BJU Int. 2006;98(5):1082–1089. doi: 10.1111/j.1464-410X.2006.06425.x. [DOI] [PubMed] [Google Scholar]
- 118.Voss OH, Batra S, Kolattukudy SJ, Gonzalez-Mejia ME, Smith JB, Doseff AI. Binding of caspase-3 prodomain to heat shock protein 27 regulates monocyte apoptosis by inhibiting caspase-3 proteolytic activation. J Biol Chem. 2007;282(34):25088–25099. doi: 10.1074/jbc.M701740200. [DOI] [PubMed] [Google Scholar]
- 119.Du ZX, Zhang HY, Meng X, Gao YY, Zou RL, Liu BQ, Guan Y, Wang HQ. Proteasome inhibitor MG132 induces BAG3 expression through activation of heat shock factor 1. J Cell Physiol. 2009;218(3):631–637. doi: 10.1002/jcp.21634. [DOI] [PubMed] [Google Scholar]
- 120.Franceschelli S, Rosati A, Lerose R, De Nicola S, Turco MC, Pascale M. Bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol. 2008;215(3):575–577. doi: 10.1002/jcp.21397. [DOI] [PubMed] [Google Scholar]
- 121.Feng Y, Huang W, Meng W, Jegga AG, Wang Y, Cai W, Kim HW, Pasha Z, Wen Z, Rao F, Modi RM, Yu X, Ashraf M. Heat shock improves Sca-1 + stem cell survival and directs ischemic cardiomyocytes toward a prosurvival phenotype via exosomal transfer: a critical role for HSF1/miR-34a/HSP70 pathway. Stem Cells. 2014;32(2):462–472. doi: 10.1002/stem.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tran SE, Meinander A, Holmstrom TH, Rivero-Muller A, Heiskanen KM, Linnau EK, Courtney MJ, Mosser DD, Sistonen L, Eriksson JE. Heat stress downregulates FLIP and sensitizes cells to Fas receptor-mediated apoptosis. Cell Death Differ. 2003;10(10):1137–1147. doi: 10.1038/sj.cdd.4401278. [DOI] [PubMed] [Google Scholar]
- 123.Xia W, Voellmy R, Spector NL. Sensitization of tumor cells to fas killing through overexpression of heat-shock transcription factor 1. J Cell Physiol. 2000;183(3):425–431. doi: 10.1002/(SICI)1097-4652(200006)183:3<425::AID-JCP16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 124.Hayashida N, Inouye S, Fujimoto M, Tanaka Y, Izu H, Takaki E, Ichikawa H, Rho J, Nakai A. A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J. 2006;25(20):4773–4783. doi: 10.1038/sj.emboj.7601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neef R, Kuske MA, Prols E, Johnson JP. Identification of the human PHLDA1/TDAG51 gene: down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 2002;62(20):5920–5929. [PubMed] [Google Scholar]
- 126.Shunmei E, Zhao Y, Huang Y, Lai K, Chen C, Zeng J, Zou J. Heat shock factor 1 is a transcription factor of Fas gene. Mol Cell. 2010;29(5):527–531. doi: 10.1007/s10059-010-0065-4. [DOI] [PubMed] [Google Scholar]
- 127.Cohen O, Inbal B, Kissil JL, Raveh T, Berissi H, Spivak-Kroizaman T, Feinstein E, Kimchi A. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol. 1999;146(1):141–148. doi: 10.1083/jcb.146.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rennier K, Ji JY. Shear stress regulates expression of death-associated protein kinase in suppressing TNFalpha-induced endothelial apoptosis. J Cell Physiol. 2012;227(6):2398–2411. doi: 10.1002/jcp.22975. [DOI] [PubMed] [Google Scholar]
- 129.Benderska N, Ivanovska J, Rau TT, Schulze-Luehrmann J, Mohan S, Chakilam S, Gandesiri M, Ziesche E, Fischer T, Soder S, Agaimy A, Distel L, Sticht H, Mahadevan V, Schneider-Stock R. DAPK-HSF1 interaction as a positive-feedback mechanism stimulating TNF-induced apoptosis in colorectal cancer cells. J Cell Sci. 2014;127(Pt 24):5273–5287. doi: 10.1242/jcs.157024. [DOI] [PubMed] [Google Scholar]
- 130.Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 2000;14(6):645–649. [PMC free article] [PubMed] [Google Scholar]
- 131.Akerfelt M, Vihervaara A, Laiho A, Conter A, Christians ES, Sistonen L, Henriksson E. Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J Biol Chem. 2010;285(45):34469–34476. doi: 10.1074/jbc.M110.157552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aligue R, Akhavan-Niak H, Russell P. A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J. 1994;13(24):6099–6106. doi: 10.1002/j.1460-2075.1994.tb06956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Munoz MJ, Jimenez J. Genetic interactions between Hsp90 and the Cdc2 mitotic machinery in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1999;261(2):242–250. doi: 10.1007/s004380050963. [DOI] [PubMed] [Google Scholar]
- 134.Nakai A, Ishikawa T. Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J. 2001;20(11):2885–2895. doi: 10.1093/emboj/20.11.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, Regos A, Orosz L, Kovacs AL, Vellai T. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177(1):655–660. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ishikawa T, Igarashi T, Hata K, Fujita T. c-fos induction by heat, arsenite, and cadmium is mediated by a heat shock element in its promoter. Biochem Biophys Res Commun. 1999;254(3):566–571. doi: 10.1006/bbrc.1998.9979. [DOI] [PubMed] [Google Scholar]
- 137.Ishikawa T, Sekine N, Hata K, Igarashi T, Fujita T. Prostaglandin A1 enhances c-fos expression and activating protein-1 activity. Mol Cell Endocrinol. 2000;164(1–2):77–85. doi: 10.1016/S0303-7207(00)00241-0. [DOI] [PubMed] [Google Scholar]
- 138.Wilkerson DC, Skaggs HS, Sarge KD. HSF2 binds to the Hsp90, Hsp27, and c-Fos promoters constitutively and modulates their expression. Cell Stress Chaperon. 2007;12(3):283–290. doi: 10.1379/CSC-250.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sawai M, Ishikawa Y, Ota A, Sakurai H. The proto-oncogene JUN is a target of the heat shock transcription factor HSF1. FEBS J. 2013;280(24):6672–6680. doi: 10.1111/febs.12570. [DOI] [PubMed] [Google Scholar]
- 140.Zhao YY, Gao XP, Zhao YD, Mirza MK, Frey RS, Kalinichenko VV, Wang IC, Costa RH, Malik AB. Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J Clin Investig. 2006;116(9):2333–2343. doi: 10.1172/JCI27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leung TW, Lin SS, Tsang AC, Tong CS, Ching JC, Leung WY, Gimlich R, Wong GG, Yao KM. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507(1):59–66. doi: 10.1016/S0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- 142.Dai B, Gong A, Jing Z, Aldape KD, Kang SH, Sawaya R, Huang S. Forkhead box M1 is regulated by heat shock factor 1 and promotes glioma cells survival under heat shock stress. J Biol Chem. 2013;288(3):1634–1642. doi: 10.1074/jbc.M112.379362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang X, Wang J, Liu S, Yan Q. HSF1 and Sp1 regulate FUT4 gene expression and cell proliferation in breast cancer cells. J Cell Biochem. 2014;115(1):168–178. doi: 10.1002/jcb.24645. [DOI] [PubMed] [Google Scholar]
- 144.Ishikawa Y, Sakurai H. Heat-induced expression of the immediate-early gene IER5 and its involvement in the proliferation of heat-shocked cells. FEBS J. 2015;282(2):332–340. doi: 10.1111/febs.13134. [DOI] [PubMed] [Google Scholar]
- 145.Gabai VL, Meng L, Kim G, Mills TA, Benjamin IJ, Sherman MY. Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR. Mol Cell Biol. 2012;32(5):929–940. doi: 10.1128/MCB.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chou SD, Murshid A, Eguchi T, Gong J, Calderwood SK. HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene. 2015;34(17):2178–2188. doi: 10.1038/onc.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Su KH, Cao J, Tang Z, Dai S, He Y, Sampson SB, Benjamin IJ, Dai C. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol. 2016;18(5):527–539. doi: 10.1038/ncb3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92(19):1564–1572. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 149.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schulz R, Streller F, Scheel AH, Ruschoff J, Reinert MC, Dobbelstein M, Marchenko ND, Moll UM. HER2/ErbB2 activates HSF1 and thereby controls HSP90 clients including MIF in HER2-overexpressing breast cancer. Cell Death Dis. 2014;5:e980. doi: 10.1038/cddis.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 152.Almeida DV, Nornberg BF, Geracitano LA, Barros DM, Monserrat JM, Marins LF. Induction of phase II enzymes and hsp70 genes by copper sulfate through the electrophile-responsive element (EpRE): insights obtained from a transgenic zebrafish model carrying an orthologous EpRE sequence of mammalian origin. Fish Physiol Biochem. 2010;36(3):347–353. doi: 10.1007/s10695-008-9299-x. [DOI] [PubMed] [Google Scholar]
- 153.Joo HJ, Park S, Kim KY, Kim MY, Kim H, Park D, Paik YK. HSF-1 is involved in regulation of ascaroside pheromone biosynthesis by heat stress in Caenorhabditis elegans. Biochem J. 2016;473(6):789–796. doi: 10.1042/BJ20150938. [DOI] [PubMed] [Google Scholar]
- 154.Takács-Vellai K, Bayci A, Vellai T. Autophagy in neuronal cell loss: a road to death. BioEssays. 2006;28(11):1126–1131. doi: 10.1002/bies.20489. [DOI] [PubMed] [Google Scholar]
- 155.Vellai T, Tóth ML, Kovács AL. Janus-faced autophagy: a dual role of cellular self-eating in neurodegeneration? Autophagy. 2007;3(5):461–463. doi: 10.4161/auto.4282. [DOI] [PubMed] [Google Scholar]
- 156.Vellai T, Takács-Vellai K. Regulation of protein turnover by longevity pathways. Adv Exp Med Biol. 2010;694:69–80. doi: 10.1007/978-1-4419-7002-2_7. [DOI] [PubMed] [Google Scholar]
- 157.Sturm A, Ivics Z, Vellai T. The mechanism of ageing: primary role of transposable elements in genome disintegration. Cell Mol Life Sci. 2015;72(10):1839–1847. doi: 10.1007/s00018-015-1896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sturm A, Perczel A, Ivics Z, Vellai T. The Piwi-piRNA pathway: road to immortality. Aging Cell. 2017;16(5):906–911. doi: 10.1111/acel.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180(6):1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 163.Dues DJ, Andrews EK, Schaar CE, Bergsma AL, Senchuk MM, Van Raamsdonk JM. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging. 2016;8(4):777–795. doi: 10.18632/aging.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Papp D, Kovacs T, Billes V, Varga M, Tarnoci A, Hackler L, Jr, Puskas LG, Liliom H, Tarnok K, Schlett K, Borsy A, Padar Z, Kovacs AL, Hegedus K, Juhasz G, Komlos M, Erdos A, Gulyas B, Vellai T. AUTEN-67, an autophagy-enhancing drug candidate with potent antiaging and neuroprotective effects. Autophagy. 2016;12(2):273–286. doi: 10.1080/15548627.2015.1082023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rattan SI. Hormetic modulation of aging and longevity by mild heat stress. Dose Response. 2006;3(4):533–546. doi: 10.2203/dose-response.003.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kovacs T, Billes V, Komlos M, Hotzi B, Manzeger A, Tarnoci A, Papp D, Szikszai F, Szinyakovics J, Racz A, Noszal B, Veszelka S, Walter FR, Deli MA, Hackler L, Jr, Alfoldi R, Huzian O, Puskas LG, Liliom H, Tarnok K, Schlett K, Borsy A, Welker E, Kovacs AL, Padar Z, Erdos A, Legradi A, Bjelik A, Gulya K, Gulyas B, Vellai T. The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci Rep. 2017;7:42014. doi: 10.1038/srep42014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Billes V, Kovacs T, Hotzi B, Manzeger A, Tagscherer K, Komlos M, Tarnoci A, Padar Z, Erdos A, Bjelik A, Legradi A, Gulya K, Gulyas B, Vellai T. AUTEN-67 (Autophagy Enhancer-67) hampers the progression of neurodegenerative symptoms in a Drosophila model of Huntington’s disease. J Huntingtons Dis. 2016;5(2):133–147. doi: 10.3233/JHD-150180. [DOI] [PubMed] [Google Scholar]
- 168.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 169.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15(2):657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Walker GA, Thompson FJ, Brawley A, Scanlon T, Devaney E. Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J. 2003;17(13):1960–1962. doi: 10.1096/fj.03-0164fje. [DOI] [PubMed] [Google Scholar]
- 171.Iser WB, Wilson MA, Wood WH, 3rd, Becker K, Wolkow CA. Co-regulation of the DAF-16 target gene, cyp-35B1/dod-13, by HSF-1 in C. elegans dauer larvae and daf-2 insulin pathway mutants. PLoS One. 2011;6(3):e17369. doi: 10.1371/journal.pone.0017369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18(3):598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 173.Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28(12):4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nagarsekar A, Hasday JD, Singh IS. CXC chemokines: a new family of heat-shock proteins? Immunol Investig. 2005;34(3):381–398. doi: 10.1081/IMM-200067648. [DOI] [PubMed] [Google Scholar]
- 175.Inouye S, Izu H, Takaki E, Suzuki H, Shirai M, Yokota Y, Ichikawa H, Fujimoto M, Nakai A. Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem. 2004;279(37):38701–38709. doi: 10.1074/jbc.M405986200. [DOI] [PubMed] [Google Scholar]
- 176.Singh V, Aballay A. Heat shock and genetic activation of HSF-1 enhance immunity to bacteria. Cell Cycle. 2006;5(21):2443–2446. doi: 10.4161/cc.5.21.3434. [DOI] [PubMed] [Google Scholar]
- 177.Ibrahim EC, Morange M, Dausset J, Carosella ED, Paul P. Heat shock and arsenite induce expression of the nonclassical class I histocompatibility HLA-G gene in tumor cell lines. Cell Stress Chaperon. 2000;5(3):207–218. doi: 10.1379/1466-1268(2000)005<0207:HSAAIE>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Singh IS, He JR, Calderwood S, Hasday JD. A high affinity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-alpha gene is a transcriptional repressor. J Biol Chem. 2002;277(7):4981–4988. doi: 10.1074/jbc.M108154200. [DOI] [PubMed] [Google Scholar]
- 179.Ambade A, Catalano D, Lim A, Mandrekar P. Inhibition of heat shock protein (molecular weight 90 kDa) attenuates proinflammatory cytokines and prevents lipopolysaccharide-induced liver injury in mice. Hepatology. 2012;55(5):1585–1595. doi: 10.1002/hep.24802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang L, Yang M, Wang Q, Liu M, Liang Q, Zhang H, Xiao X. HSF1 regulates expression of G-CSF through the binding element for NF-IL6/CCAAT enhancer binding protein beta. Mol Cell Biochem. 2011;352(1–2):11–17. doi: 10.1007/s11010-010-0624-1. [DOI] [PubMed] [Google Scholar]
- 181.Xie Y, Zhong R, Chen C, Calderwood SK. Heat shock factor 1 contains two functional domains that mediate transcriptional repression of the c-fos and c-fms genes. J Biol Chem. 2003;278(7):4687–4698. doi: 10.1074/jbc.M210189200. [DOI] [PubMed] [Google Scholar]
- 182.Ferat-Osorio E, Sanchez-Anaya A, Gutierrez-Mendoza M, Bosco-Garate I, Wong-Baeza I, Pastelin-Palacios R, Pedraza-Alva G, Bonifaz LC, Cortes-Reynosa P, Perez-Salazar E, Arriaga-Pizano L, Lopez-Macias C, Rosenstein Y, Isibasi A. Heat shock protein 70 down-regulates the production of toll-like receptor-induced pro-inflammatory cytokines by a heat shock factor-1/constitutive heat shock element-binding factor-dependent mechanism. J Inflamm (Lond) 2014;11:19. doi: 10.1186/1476-9255-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Maity TK, Henry MM, Tulapurkar ME, Shah NG, Hasday JD, Singh IS. Distinct, gene-specific effect of heat shock on heat shock factor-1 recruitment and gene expression of CXC chemokine genes. Cytokine. 2011;54(1):61–67. doi: 10.1016/j.cyto.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rossi A, Coccia M, Trotta E, Angelini M, Santoro MG. Regulation of cyclooxygenase-2 expression by heat: a novel aspect of heat shock factor 1 function in human cells. PLoS One. 2012;7(2):e31304. doi: 10.1371/journal.pone.0031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fan-xin M, Li-mei S, Bei S, Xin Q, Yu Y, Yu C. Heat shock factor 1 regulates the expression of the TRPV1 gene in the rat preoptic-anterior hypothalamus area during lipopolysaccharide-induced fever. Exp Physiol. 2012;97(6):730–740. doi: 10.1113/expphysiol.2011.064204. [DOI] [PubMed] [Google Scholar]
- 186.Hotchkiss R, Nunnally I, Lindquist S, Taulien J, Perdrizet G, Karl I. Hyperthermia protects mice against the lethal effects of endotoxin. Am J Physiol. 1993;265(6 Pt 2):R1447–R1457. doi: 10.1152/ajpregu.1993.265.6.R1447. [DOI] [PubMed] [Google Scholar]
- 187.Jego G, Lanneau D, De Thonel A, Berthenet K, Hazoume A, Droin N, Hamman A, Girodon F, Bellaye PS, Wettstein G, Jacquel A, Duplomb L, Le Mouel A, Papanayotou C, Christians E, Bonniaud P, Lallemand-Mezger V, Solary E, Garrido C. Dual regulation of SPI1/PU.1 transcription factor by heat shock factor 1 (HSF1) during macrophage differentiation of monocytes. Leukemia. 2014;28(8):1676–1686. doi: 10.1038/leu.2014.63. [DOI] [PubMed] [Google Scholar]
- 188.Kumar M, Mitra D. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J Biol Chem. 2005;280(48):40041–40050. doi: 10.1074/jbc.M508904200. [DOI] [PubMed] [Google Scholar]
- 189.Santoro MG. Heat shock proteins and virus replication: hsp70 s as mediators of the antiviral effects of prostaglandins. Experientia. 1994;50(11–12):1039–1047. doi: 10.1007/BF01923459. [DOI] [PubMed] [Google Scholar]
- 190.Rawat P, Mitra D. Cellular heat shock factor 1 positively regulates human immunodeficiency virus-1 gene expression and replication by two distinct pathways. Nucleic Acids Res. 2011;39(14):5879–5892. doi: 10.1093/nar/gkr198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168(4):1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Uchida S, Hara K, Kobayashi A, Fujimoto M, Otsuki K, Yamagata H, Hobara T, Abe N, Higuchi F, Shibata T, Hasegawa S, Kida S, Nakai A, Watanabe Y. Impaired hippocampal spinogenesis and neurogenesis and altered affective behavior in mice lacking heat shock factor 1. Proc Natl Acad Sci USA. 2011;108(4):1681–1686. doi: 10.1073/pnas.1016424108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Varodayan FP, Harrison NL. HSF1 transcriptional activity mediates alcohol induction of Vamp2 expression and GABA release. Front Integr Neurosci. 2013;7:89. doi: 10.3389/fnint.2013.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Varodayan FP, Pignataro L, Harrison NL. Alcohol induces synaptotagmin 1 expression in neurons via activation of heat shock factor 1. Neurosci. 2011;193:63–71. doi: 10.1016/j.neuroscience.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tan J, Tan S, Zheng H, Liu M, Chen G, Zhang H, Wang K, Zhou J, Xiao XZ. HSF1 functions as a transcription regulator for Dp71 expression. Cell Stress Chaperon. 2015;20(2):371–379. doi: 10.1007/s12192-014-0558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takaki E, Fujimoto M, Sugahara K, Nakahari T, Yonemura S, Tanaka Y, Hayashida N, Inouye S, Takemoto T, Yamashita H, Nakai A. Maintenance of olfactory neurogenesis requires HSF1, a major heat shock transcription factor in mice. J Biol Chem. 2006;281(8):4931–4937. doi: 10.1074/jbc.M506911200. [DOI] [PubMed] [Google Scholar]
- 197.Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23(21):4297–4306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Izu H, Inouye S, Fujimoto M, Shiraishi K, Naito K, Nakai A. Heat shock transcription factor 1 is involved in quality-control mechanisms in male germ cells. Biol Rep. 2004;70(1):18–24. doi: 10.1095/biolreprod.103.020065. [DOI] [PubMed] [Google Scholar]
- 199.Widlak W, Benedyk K, Vydra N, Glowala M, Scieglinska D, Malusecka E, Nakai A, Krawczyk Z. Expression of a constitutively active mutant of heat shock factor 1 under the control of testis-specific hst70 gene promoter in transgenic mice induces degeneration of seminiferous epithelium. Acta Biochim Polon. 2003;50(2):535–541. [PubMed] [Google Scholar]
- 200.Metchat A, Akerfelt M, Bierkamp C, Delsinne V, Sistonen L, Alexandre H, Christians ES. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90alpha expression. J Biol Chem. 2009;284(14):9521–9528. doi: 10.1074/jbc.M808819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Le Masson F, Razak Z, Kaigo M, Audouard C, Charry C, Cooke H, Westwood JT, Christians ES. Identification of heat shock factor 1 molecular and cellular targets during embryonic and adult female meiosis. Mol Cell Biol. 2011;31(16):3410–3423. doi: 10.1128/MCB.05237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Leise TL, Wang CW, Gitis PJ, Welsh DK. Persistent cell-autonomous circadian oscillations in fibroblasts revealed by six-week single-cell imaging of PER2::LUC bioluminescence. PLoS One. 2012;7(3):e33334. doi: 10.1371/journal.pone.0033334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26(6):567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tamaru T, Hattori M, Honda K, Benjamin I, Ozawa T, Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One. 2011;6(9):e24521. doi: 10.1371/journal.pone.0024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22(3):331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hatori M, Hirota T, Iitsuka M, Kurabayashi N, Haraguchi S, Kokame K, Sato R, Nakai A, Miyata T, Tsutsui K, Fukada Y. Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Proc Natl Acad Sci USA. 2011;108(12):4864–4869. doi: 10.1073/pnas.1015959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tóth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 209.Vellai T, Bicsák B, Tóth ML, Takács-Vellai K, Kovács AL. Regulation of cell growth by autophagy. Autophagy. 2008;4(4):507–509. doi: 10.4161/auto.5670. [DOI] [PubMed] [Google Scholar]
- 210.Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR, 3rd, Manning G, Dillin A. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346(6207):360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Verma P, Pfister JA, Mallick S, D’Mello SR. HSF1 protects neurons through a novel trimerization- and HSP-independent mechanism. J Neurosci. 2014;34(5):1599–1612. doi: 10.1523/JNEUROSCI.3039-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.