Abstract
Fibrillin microfibrils are ubiquitous elements of extracellular matrix assemblies that play crucial roles in regulating the bioavailability of growth factors of the transforming growth factor beta superfamily. Recently, several “a disintegrin and metalloproteinase with thrombospondin motifs” (ADAMTS) proteins were shown to regulate fibrillin microfibril function. Among them, ADAMTS17 is the causative gene of Weill-Marchesani syndrome (WMS) and Weill-Marchesani-like syndrome, of which common symptoms are ectopia lentis and short stature. ADAMTS17 has also been linked to height variation in humans; however, the molecular mechanisms whereby ADAMTS17 regulates skeletal growth remain unknown. Here, we generated Adamts17-/- mice to examine the role of Adamts17 in skeletogenesis. Adamts17-/- mice recapitulated WMS, showing shorter long bones, brachydactyly, and thick skin. The hypertrophic zone of the growth plate in Adamts17-/- mice was shortened, with enhanced fibrillin-2 deposition, suggesting increased incorporation of fibrillin-2 into microfibrils. Comprehensive gene expression analysis of growth plates using laser microdissection and RNA sequencing indicated alteration of the bone morphogenetic protein (BMP) signaling pathway after Adamts17 knockout. Consistent with this, phospho-Smad1 levels were downregulated in the hypertrophic zone of the growth plate and in Adamts17-/- primary chondrocytes. Delayed terminal differentiation of Adamts17-/- chondrocytes, observed both in primary chondrocyte and primordial metatarsal cultures, and was prevented by BMP treatment. Our data indicated that Adamts17 is involved in skeletal formation by modulating BMP-Smad1/5/8 pathway, possibly through inhibiting the incorporation of fibrillin-2 into microfibrils. Our findings will contribute to further understanding of disease mechanisms and will facilitate the development of therapeutic interventions for WMS.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03188-0) contains supplementary material, which is available to authorized users.
Keywords: Adamts17, Fibrillin, Microfibril, Skeletal formation
Introduction
Endochondral ossification is an essential process for skeletal formation [1]. In the first step, mesenchymal progenitor cells are recruited into condensations, where they differentiate into chondrocytes that produce cartilage-specific matrix proteins, such as type II collagen (Col2a1) and aggrecan. The cartilage then enlarges through chondrocyte proliferation and matrix production. Later, chondrocytes cease proliferation and undergo hypertrophic differentiation, characterized by the secretion of type X collagen (Col10a1). Finally, hypertrophic chondrocytes undergo apoptotic cell death and the cartilage matrix is degraded by proteases, including matrix metalloproteinase 13 (Mmp13), for the proceeding calcification and bone formation [1].
During the endochondral ossification, the extracellular matrix (ECM) microenvironment surrounding the chondrocytes plays an important role not only in providing structural and mechanical support, but also in regulating chondrocyte behavior [2]. Among the macromolecules that constitute the ECM, “a disintegrin and metalloproteinase with thrombospondin motifs” (ADAMTS) proteins are members of a large superfamily of 19 secreted ADAMTS proteases and 7 ADAMTS-like (ADAMTSL) proteins [3]. ADAMTS proteases are characterized by highly homologous N-terminal catalytic and disintegrin-like domains and a C-terminal region that contains thrombospondin repeats, which are believed to bind to the extracellular matrix (ECM) [3]. ADAMTSL proteins lack the N-terminal protease domain, but are homologous to the ancillary domain of ADAMTS proteases [3]. Weill-Marchesani syndrome (WMS), a rare connective tissue disorder characterized by short stature, brachydactyly, joint stiffness, thick skin, and lens abnormalities, is caused by autosomal dominant fibrillin-1 (FBN1) mutations (OMIM 608328) [4, 5] or autosomal recessive ADAMTS10 mutations (OMIM 277600) [6]. Recently, Mularczyk et al. have reported that mice with a truncation mutation of ADAMTS10 found in WMS patients recapitulate the short stature phenotype of WMS and show developmental changes in the growth plate [7]. Abnormalities in the ciliary apparatus were also observed, coupled with altered distribution of Fbn1 and fibrillin-2 (Fbn2) [7]. The skeletal muscle mass increases in the mutant mice and BMP signaling is reduced in the skeletal muscle and embryonic fibroblasts [7].
Autosomal recessive mutations of ADAMTS17, which has a similar structure to ADAMTS10, cause Weill-Marchesani-like syndrome (WMLS; OMIM 607511) [8]. WMLS patients lack some features of WMS patients, such as brachydactyly and thick skin. Meanwhile, ADAMTS17 mutations in an Indian family were recently shown to cause the WMS phenotype, including short stature, ectopia lentis, and brachydactyly, suggesting an overlap of WMS and WMLS [9]. Moreover, ADAMTS17 has also been reported to be associated with height variation in humans [10, 11], indicating that Adamts17 is involved in skeletal growth. However, there have been no reports of Adamts17 knockout mice and the underlying molecular mechanisms remain unknown. Here, we have generated Adamts17 null mice, and analyzed their general phenotype and skeletal development. We further performed gene expression screening analysis in Adamts17-null growth plates by laser microdissection (LMD) and RNA sequencing (RNA-seq), and examined alterations in signaling pathways caused by Adamts17 deletion.
Materials and methods
Animals
All mouse experiments were performed according to protocols approved by the Animal Care and Use Committee of the University of Tokyo (approval number: M-P12-131). In each experiment, we used the genotypes of littermates maintained in a C57BL/6 J background as controls. CAG-Cre [12] mice were provided by RIKEN BRC (Saitama, Japan). Col2a1-Cre [13] was purchased from the Jackson Laboratory (Bar Harbor, ME, USA.) To generate a conditional null allele of Adamts17, a segment of exon 4 was flanked with loxP sequences to generate a frame-shift mutation by Cre recombination, leading to disruption of the zinc-dependent metalloprotease domain of Adamts17 and its downstream sequences (Figs. 1a, S1a). To generate Adamts17-flox mice, targeting vector, including the neo-expressing cassette, was used to induce recombination between homologous regions of the targeting vector and the Adamts17 locus (targeted allele) (Fig. 1a). Adamts17-flox mice generation was followed by flippase recombinase target-mediated recombination of the Adamts17-targeted allele (floxed allele) (Fig. 1a).
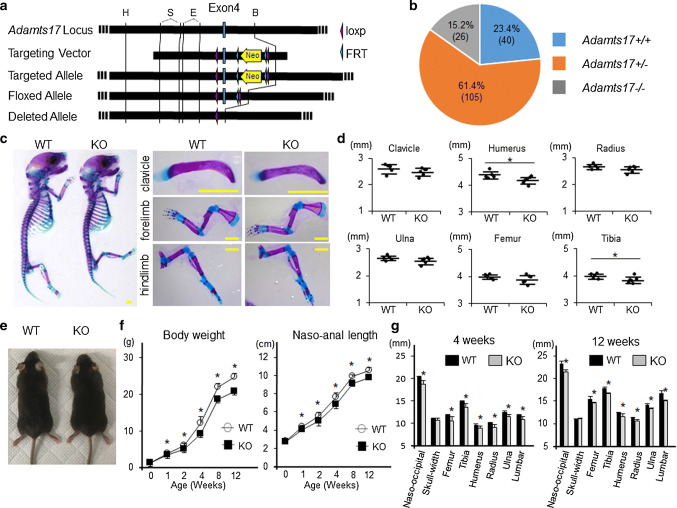
Fig. 1.
Adamts17-/- mice display postnatal impairment of skeletal growth. a Generation of Adamts17-flox mice. The diagrams show the Adamts17 genomic locus with exon 4 (Adamts17 locus); Adamts17 targeting vector including the neo-cassette (Targeting Vector); recombination between homologous regions of the targeting vector and the Adamts17 locus, leading to the generation of the Adamts17-targeted allele (Targeted Allele); floxed Adamts17 allele following flippase recombinase target (FRT)-mediated recombination of the Adamts17-targeted allele (Floxed allele); and Adamts17-deleted allele following Cre-mediated recombination of the floxed Adamt17 allele (Deleted allele). H, HindIII; S, SmaI; E, EcoRV; B, BamHI. b Pie chart of the genotypes detected at 7 days of age from intercrosses of Adamts17 ± mice on the C57BL/6 strain. Note the reduced viability of Adamts17-/- mice. c Alizarin Red and Alcian Blue double-staining of the whole skeleton (left), clavicle, upper extremities, and lower extremities (right) of WT and Adamts17-/- (KO) littermates at P0. Scale bars, 2 mm. d Length of clavicles and long bones of WT (n = 5) and KO (n = 6) littermates at P0. e Gross appearance of 12-week-old WT and KO mice. f Time-course of naso-anal length and body weight of male WT and KO mice (n = 6 per genotype). g Bone length of 4-week-old and 12-week-old male WT and KO mice (n = 5 per genotype). Statistical significance was calculated using a two-tailed unpaired Student’s t test. *P < 0.05 between genotypes
To generate Adamts17-/- mice, we first generated CAG-Cre; Adamts17fl/fl by mating Adamts17fl/fl mice with CAG-Cre mice. Adamts17 ± mice were obtained by crossing CAG-Cre; Adamts17fl/fl mice and wild-type (WT) mice. Thereafter, Adamts17-/- mice were generated by crossing Adamts17 ± mice. Sequences of the primers used for genotyping are shown in Table S1.
Evaluation of skeletal formation
Radiographs were obtained using a soft X-ray apparatus (CMB-2; Softex Co., Tokyo, Japan). After fixation in 99.5% ethanol and acetone, double-staining of newborn mouse skeletons was performed with a solution containing Alizarin Red S (Sigma-Aldrich, St Louis, MO, USA) and Alcian Blue 8GX (Sigma-Aldrich).
Histological analyses and immunohistochemistry
Mouse limbs and skin samples were fixed in 4% paraformaldehyde, buffered with phosphate-buffered saline (PBS, pH 7.4), at 4 °C for 1 day. Specimens were embedded in paraffin and 5-µm thick sagittal sections were cut. Hematoxylin and eosin (H & E), Safranin-O, and Masson’s trichrome staining were performed according to standard protocols. Postnatal day 7 (P7) knee joints were decalcified in 10% ethylenediaminetetracetic acid (EDTA), pH 7.4, for 1 week. We examined the sagittal sections of the knee joint at the depth where posterior cruciate ligament attach to the tibia as reported previously [14]. The lengths of the proliferative and hypertrophic zones were measured along the midline of the tibial growth plate at this depth using NIH Image J software. The proliferative zone was defined as the region with flattened, stack-forming chondrocytes and the hypertrophic zone was defined as the region with rounded and enlarged chondrocytes. For immunohistochemistry, sections were incubated with antibodies against Adamts17 (1:100; H00170691-M01; Novus Biologicals, CO, USA), Fibrillin-1 (1:100; MAB1919; Chemicon, Burlington, MA, USA), Fibrillin-2 (1:100; LS-C410948; LSbio, Seattle, WA, USA), Col2 (200:1; MAB8887; Millipore Sigma, St. Louis, USA), Col10 (1:100;14-9771-82; eBioscience, San Diego, CA, USA), Mmp13 (1:100; P18165-1-AP; Proteintech, Rosemont, IL, USA), Ihh (1:100; 13388-1-AP; Proteintech, Rosemont, IL, USA), PTHrP (1:200; 10817-1-AP; Proteintech, Rosemont, IL, USA), and phosphorylated Smad1 (pSmad1, 1:100, ab73211, Abcam). As for the antibody against Adamts17, Adamts17-/- samples were used as negative control. In case of the antibodies against Adamts17, Fibrillin-1, and Fibrillin-2, anti-mouse Alexa Fluor Plus 488 (1:1,000; A32723; Invitrogen, Carlsbad, CA, USA), and anti-rabbit Alexa Fluor Plus 488 (1:1,000; A32731; Invitrogen) secondary antibodies were used for visualization. For the antibodies against Col2, Col10, Ihh, and PTHrP, the sections were subsequently treated with anti-mouse (Col2 and Col10) or anti-rabbit (Ihh and PTHrP) EnVision-Plus System-HRP (DAKO, Glostrup, Denmark) for 30 min. Peroxidase labeling was visualized using peroxidase substrate (3,3′-diaminobenzide) and was counterstained with Carrazzi’s hematoxylin. For pSmad1 detection, a CSA Biotin-Free Tyramide Signal Amplification System (Dako, Carpinteria, CA, USA) was used according to the manufacturer’s instructions. For nucleotide 5-ethynyl-2′-deoxyuridine (EdU) labeling, mice at age of 7 days were intraperitoneally injected with EdU at a dose of 50 μg/g body weight 2 h before killing, as previously described [15]. EdU detection was performed using a Click-iT EdU imaging kit (Invitrogen), according to the manufacturer’s instructions. EdU-positive cells were visualized using an Alexa Fluor 594 azide (Invitrogen). For TUNEL staining, an in situ Apoptosis Detection Kit (Takara Bio, Otsu, Japan) was used.
Laser microdissection and RNA sequencing
We performed LMD using frozen sections prepared according to Kawamoto’s film method [16], as previously described [17]. Briefly, the adhesive surface of Cryofilm Type IIC (Section-lab, Hiroshima, Japan) was attached to the specimen and 10-µm thick sections were cut. Microdissections of prehypertrophic and hypertrophic zone samples of P7 tibia were performed using a Leica LMD 6500 laser microdissection system (Leica Microsystems, Wetzlar, Germany). RNA extraction was performed using the Picopure RNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) and was amplified by two rounds of in vitro transcription using the Arcturus RiboAmp HS PLUS Kit (Thermo Fisher Scientific), according to the manufacturer’s protocols. A cDNA library was constructed according to the TruSeq RNA Sample Preparation V2 Guide Rev. C (Illumina, San Diego, CA, USA) and was sequencing on a HiSeq 2000 instrument (Illumina). We then normalized sequence data by the trimmed mean of M (TMM) values method [18]. Three pairs of Adamts17-/- mice and their WT littermates were used. Differentially expressed genes were determined as having expression levels that were significantly increased or decreased (p < 0.05) in Adamts17 KO growth plates by more than twice or less than half that of WT growth plates. These differentially expressed genes were entered into gene ontology (GO) analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources (david.ncifcrf.gov). Raw and processed data are available in the Gene Expression Omnibus (GEO) database under accession number GSE123076.
Primary chondrocyte culture
Primary chondrocytes were prepared from costal cartilage of P2 mice, as previously described [19]. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/HG (Sigma-Aldrich), containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Sigma-Aldrich). When cells reached confluence (day 0), differentiation was initiated using culture medium containing 0.05 mg/mL ascorbic acid (Wako, Osaka, Japan) and 10 mM β-glycerophosphate (Sigma-Aldrich). The medium was changed every 2 day. At day 20, mineral deposits were visualized by Alizarin Red staining and were quantified by NIH Image J software (National Institutes of Health, Bethesda, MD). Immunofluorescence analysis of pSmad1 was performed at day 4 in primary chondrocytes. Chondrocytes were cultured in 2% FBS and subsequently, fixed and incubated with antibodies against pSmad1 at 1:100 dilution. An Alexa Fluor Plus 488 anti-rabbit IgG secondary antibody was used for detection. In some experiments, cell culture media was supplemented with 100 ng/mL of recombinant human BMP-2 (rhBMP-2; Peprotech, Rocky Hill, NJ, USA). As for pellet culture, 5 × 105 primary chondrocytes were transferred into a 15 ml tube (Falcon) and centrifuged at 500 g for 10 min as reported previously [20]. After 3 days incubation in (DMEM)/HG, containing 10% FBS and 1% penicillin/streptomycin, the pellets were transferred into petri dishes and incubated in the same medium for a further 3 weeks.
Quantitative RT-PCR (qRT-PCR)
Total RNA was purified using an RNeasy Mini Kit (Qiagen). One microgram of total RNA was reverse transcribed using a ReverTraAce qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). Each PCR reaction contained 1 × THUNDERBIRD SYBR qPCR Mix (Toyobo), 0.3 mM specific primers, and 20 ng of cDNA. mRNA levels of target genes were normalized to those of β-actin. Expression levels for each target gene were calculated using the 2−ΔCt method [21]. All reactions were run in triplicate on a Thermal Cycler Dice instrument (Takara Bio, Otsu, Japan). Primer sequences are shown in Table S2.
Western blotting
Cells were lysed in Mammalian Protein Extraction Reagent (M-PER, Thermo Scientific) containing Complete, Mini Protease Inhibitor Cocktail Tablets (Roche, Basel, Switzerland; 11836153001) and PhosSTOP Phosphatase Inhibitor Cocktail Tablets (Roche, 04906845002). Cell lysates were subjected to SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). After blocking with 6% skim milk, membranes were incubated with primary antibodies against Smad1 (1:1000; 9743S; Cell Signaling Technology, Danvers, MA, USA), pSmad1/5 (1:500, Cell Signaling Technology, 9516S), and actin (1:2000; Sigma-Aldrich; A4700). Membranes were incubated with a horseradish peroxidase-conjugated antibody (Promega, Madison, WI, USA), and immunoreactive proteins were visualized with ECL Prime Western Blotting Detection Reagent (GE Healthcare, Chicago, IL, USA) and an AE-6981 Light Capture II instrument (ATTO, Tokyo, Japan). Signal intensity was quantified with NIH Image J software and presented as the ratio of pSmad1/Smad1 signal intensity.
Metatarsal organ culture
Metatarsal organ culture was performed as previously described [22]. Briefly, metatarsal rudiments were dissected from Adamts17 ± and Adamts17-/- littermate embryos at E15.5. Metatarsal rudiments were cultured in α-modified essential medium without nucleosides (Wako), supplemented with 0.05 mg/mL ascorbic acid, 1 mM β-glycerophosphate, and 10% FBS in a humidified atmosphere of 5% CO2 in air at 37℃ for 24 h. Rudiments were then incubated in 300 µL of the same medium, with or without 500 ng/mL of rhBMP2 for an additional 4 day. For immunohistological analyses, rudiments were sectioned at day 5. Areas of mineralized, Col10-positive, and Mmp13-positive cartilage were measured using NIH Image J software. For gene expression analyses, total RNA was extracted from two pooled metatarsal rudiments from each embryo.
Statistical analyses
With the exception of RNA-seq analyses, statistical significance between two groups were analyzed using an unpaired Student’s t test. For RNA-seq analyses, a paired t test was applied due to the large variations in gene expression among each littermate [17]. Data were expressed as the mean ± SD. Data analyses were performed using Microsoft Excel (2013) and a p value < 0.05 was considered significant. All tests were two-tailed.
Results
Adamts17-/- mice recapitulated Weill-Marchesani-syndrome and showed postnatal dwarfism
We first generated Adamts17-flox mice in which exon 4 was flanked with loxP sites (Fig. 1a). The floxed allele was designed to generate a frame-shift mutation by Cre recombination, leading to disruption of the zinc-dependent metalloprotease domain of Adamts17 and its downstream sequences (Fig. 1a). Adamts17-flox mice, which were confirmed by genotyping tail DNA (Fig. S1a), were mated with CAG-Cre mice, which ubiquitously express Cre recombinase [12]. Since CAG-Cre; Adamts17fl/fl mice were viable and fertile, we further generated Adamts17-/- mice by crossing CAG-Cre; Adamts17fl/fl with WT mice. In addition to genotyping tail DNA, we used RT-PCR to confirm the deletion of exon 4 in cDNA derived from various tissues of Adamts17-/- mice (Fig. S1b, c). Intercrosses of Adamts17 ± mice provided a reduced birth rate of Adamts17-/- mice compared with the expected Mendelian ratio (Fig. 1b), as has also been reported for Adamts10-/- mice [23]. Although Adamts17-/- neonates showed reduced length only in the humerus and tibia (Fig. 1c, d), they displayed apparent dwarfism after 1 week (Fig. 1e, f). Radiographic analyses at 4 and 12 weeks of age revealed that the longitudinal lengths of the skull, long bones, and vertebrae, all of which are formed through endochondral ossification, were 7–9% and 6–10% decreased by Adamts17 deficiency, respectively (Fig. 1g).
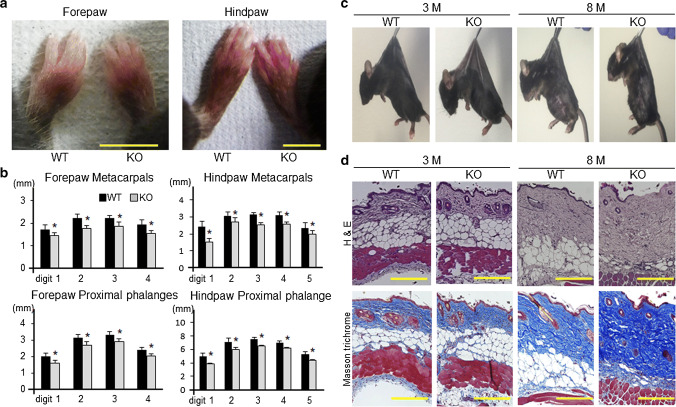
Because ADAMTS17 is the responsible gene for both WMS and WMLS [8, 9], we further investigated the precise phenotypes of the digits and skin. At 8 months of age, brachydactyly was observed in Adamts17-/- fore- and hindlimbs (Fig. 2a). Metacarpal and proximal phalange length were significantly reduced in the fore- and hindpaws of Adamts17-/- mice, compared with age- and sex-matched WT mice (Fig. 2b). Gross examination suggested decreased skin elasticity in Adamts17-/- mice at 8 months, although no apparent differences at 3 months (Fig. 2c). H&E and Masson’s trichrome staining showed increased collagen deposition and diminished epidermal fat in the skin of Adamts17-/- mice at 8 months (Fig. 2d). All these data indicate that Adamts17-/- mice recapitulated WMS.
Fig. 2.
Adamts17-/- mice recapitulate Weill-Marcehsani syndrome. a Distal forepaws and hindpaws of WT and Adamts17-/- (KO) mice at 8 months of age. Scale bars, 5 mm. b Bone length of forepaws and hindpaws of 8-month-old male WT (n = 7) and KO (n = 5) mice. *P < 0.05 versus WT. c Gross inspection of skin stiffness of 3- and 8-month-old WT and KO mice. Mice were anesthetized, shaved, and suspended by forceps. d Hematoxylin and eosin (H&E) and Masson’s trichrome staining of skin from 3- and 8-month-old WT and KO mice. Scale bars, 300 µm. Statistical significance was calculated using a two-tailed unpaired Student’s t test
Adamts17 deletion leads to reduced length of the hypertrophic chondrocyte zone, accompanied by microfibril dysregulation
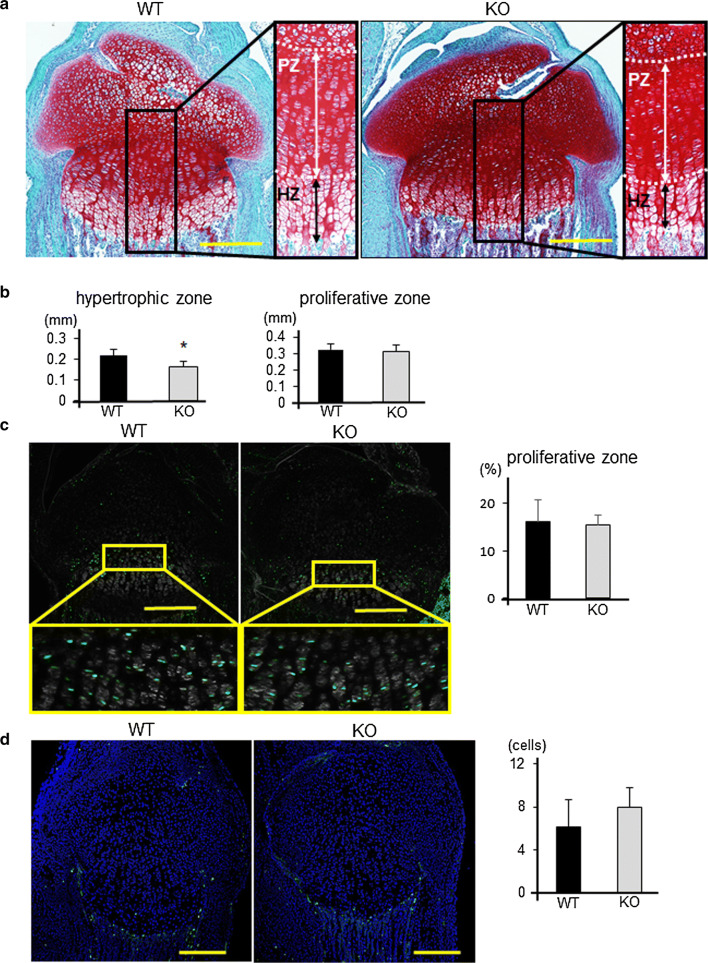
We next evaluated endochondral ossification in Adamts17-/- mice. Since skeletal dysplasia of Adamts17-/- mice started at 1 week of age, we examined the histology of P7 proximal tibias. Safranin-O staining showed that the length of the hypertrophic zone was significantly reduced in tibias from Adamts17-/- mice (Fig. 3a, b). Chondrocyte proliferation and apoptosis, as determined by EdU labeling and TUNEL staining, respectively, were not significantly different between genotypes (Fig. 3c, d). There were no apparent differences in distribution of marker proteins, such as Col2, Col10, Ihh, and PTHrP between WT and Adamts17-/- growth plates (Fig. S2).
Fig. 3.
Histological analyses of growth plates from WT and Adamts17-/- mice. a Safranin-O staining of proximal tibiae from WT and Adamts17-/- (KO) mice at P7; right panels show higher magnifications of the boxed areas in the left panels. The white broken lines indicate the boundary between resting zone and proliferative zone (PZ), or the boundary between PZ and hypertrophic zone (HZ). Scale bars, 300 µm. b Length of hypertrophic and proliferative zones in WT and KO mice at P7 (n = 5 per genotype). *P < 0.05 versus WT. c EdU labeling (green) and phase contrast images of proximal tibiae from WT and KO mice at P7 and rates of EdU-positive cells in PZ (n = 5 per genotype). Inset boxes in the upper panels indicate the regions of enlarged images in the lower panels. Scale bars, 300 μm. d TUNEL staining of proximal tibiae from WT and KO mice at P7 and number of TUNEL-positive cells in the chondro-osseous junction (n = 4 per genotype). Scale bars, 300 µm. Statistical significance was calculated using a two-tailed unpaired Student’s t test
To investigate whether the skeletal phenotype of Adamts17-/- mice was caused by loss-of-function of Adamts17 in chondrocytes, we further examined the skeletal phenotype of Col2a1-Cre; Adamts17fl/fl mice, in which Adamts17 is ablated in chondrocytes. In addition to genotyping cartilage DNA, RT-PCR confirmed the deletion of exon 4 in cDNA derived from Col2a1-Cre; Adamts17fl/fl cartilage (Fig. S1a, c). The lengths of the skull, long bones, and vertebrae were decreased 4–9% by Adamts17 deficiency in chondrocytes at 12 weeks of age (Fig. S3a, b).
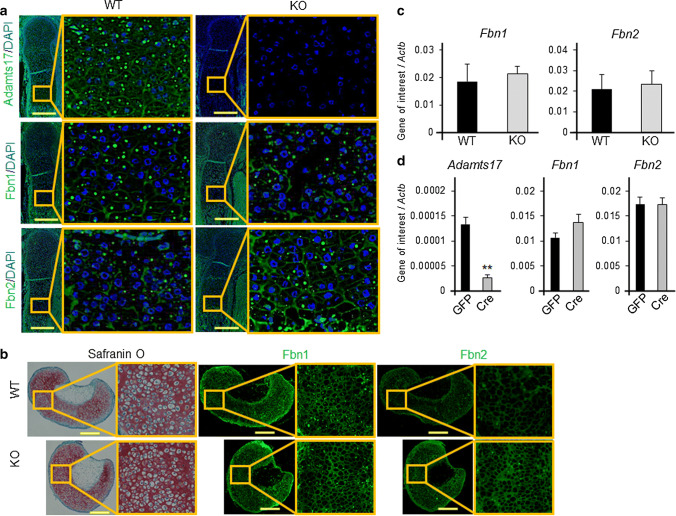
We then examined the expression and localization of Adamts17 and fibrillin microfibrils in the epiphyseal cartilage of mouse embryos. We investigated growth plates at embryonic stage, because a previous study has reported that the regions of fibrillin-2 are masked by microfibril structure in postnatal tissues [24]. Adamts17 protein was localized to the extracellular matrix of the growth plate (Fig. 4a). Microfibrils, as assessed by Fbn1 immunofluorescence, localized particularly in the hypertrophic zone (Fig. 4a). Because Adamts10 and Adamtsl2 are involved in the regulation of microfibril composition [7, 23, 25], we examined the expression of Fbn1 and Fbn2 in growth plates from Adamts17-/- and WT mice. Notably, Fbn2 signal intensity was enhanced by Adamts17 deletion, although Fbn1 was not obviously affected (Fig. 4a). To further confirm this result, we investigated the microfibril deposition of the pellets obtained from WT or Adamts17-/- chondrocytes [20]. As expected, enhanced Fbn2 deposition was also observed in this system (Fig. 4b). Fbn1 and Fbn2 mRNA expression in femoral epiphyseal cartilage of P0 mice was not significantly different between genotypes (Fig. 4c). In primary chondrocytes isolated from Adamts17fl/fl neonates, mRNA levels were also not altered by adenoviral transfection of Cre (Fig. 4d).
Fig. 4.
Perichondrial microfibril dysregulation in the growth plate of Adamts17-/- mice.a Immunofluorescent staining of Adamts17, Fibrillin-1 (Fbn1), and Fibrillin-2 (Fbn2) in humeral distal growth plates of WT mice at E16.5. The inset box in the left panels indicates the location of the enlarged image shown on the right panels. Scale bars, 300 µm. b Semiserial histological sections of cultured pellets of primary chondrocytes were stained with safranin-O and immunostained for Fbn1 and Fbn2. The inset box in the left panels indicates the location of the enlarged image shown on the right panels. Scale bars, 300 µm. c Fbn1 and Fbn2 mRNA levels in femoral epiphyseal cartilage of WT (n = 6) and KO (n = 5) mice at P0. d Adamts17, Fbn1, and Fbn2 mRNA levels in Adamts17fl/fl primary chondrocytes that were adenovirally transfected with GFP or Cre and cultured for an additional 3 day (n = 3 per well). mRNA levels were normalized to β-actin mRNA levels (Actb). **P < 0.01 versus GFP. Statistical significance was calculated using a two-tailed unpaired Student’s t test
BMP signaling is reduced in growth plates of Adamts17-/- mice
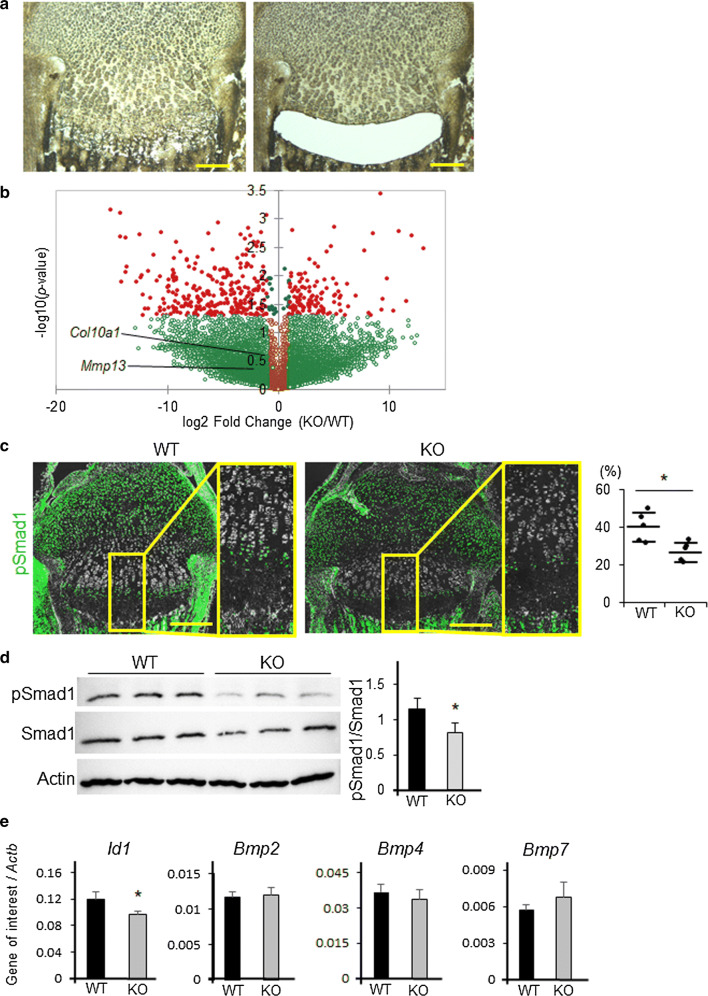
To further examine the underlying mechanisms, we performed gene expression screening in Adamts17-/- P7 proximal tibial epiphyseal cartilages using LMD and RNA-seq (Fig. 5a). We microdissected prehypertrophic and hypertrophic zones because there was a significant difference in the length of these zones between Adamts17-/- and WT growth plates. A Volcano plot clearly revealed that there were more downregulated genes (276 genes) than upregulated genes (122 genes) by Adamts17 KO among the genes with significant (p < 0.05) differential expression (Fig. 5b). Hypertrophic markers, such as Col10a1 and Mmp13, tended to decrease in Adamts17 KO growth plate though it did not reach statistical significance (Fig. 5b). Among downregulated genes, enriched analyses of the GO biological process showed that the BMP signaling pathway is one of the processes significantly affected by Adamts17 knockout (Table 1), suggesting that BMP signaling pathway is inactivated by Adamts17 KO. To confirm this result, we performed immunohistochemistry of pSmad1, a downstream effector of canonical BMP signaling pathway. In the WT mice, pSmad1 positive cells were observed mainly in the prehypertrohic zone (Fig. 5c). In contrast, the number of pSmad1-positive cells was significantly decreased in the prehypertrophic zone of Adamts17-/- mice (Fig. 5c), as compared with that in WT mice.
Fig. 5.
BMP signaling is reduced in the growth plates of Adamts17-/- mice. a Sections of proximal tibial epiphyseal cartilages before and after prehypertrophic and hypertrophic zones were microdissected by LMD. Extracted mRNA was amplified and analyzed by RNA sequencing. Scale bars, 200 µm. b Volcano plot for RNA-seq data obtained from laser microdissected samples. Red dots indicate genes that were significantly (p < 0.05) and differentially expressed in Adamts17 KO growth plates by more than twice or less than half those of WT growth plates. c Immunofluorescent staining of phosphorylated Smad1 (pSmad1) (green) and phase contrast images in proximal tibiae of WT and Adamts17-/- (KO) mice at P7 and the percentage of pSmad1-positive cells in the prehypertrophic and hypertrophic zones of WT and KO (n = 5 per genotype) mice. The inset box in the left panels indicates the location of the enlarged image shown on the right panels. Scale bars, 300 µm. d Immunoblotting of pSmad1, Smad1, and actin in protein extracts from primary chondrocytes from WT and KO mice and the ratio of pSmad1/Smad1 signal intensity (n = 3 wells per genotype). e Id1,Bmp2, Bmp4, and BMP7 mRNA levels in primary chondrocytes from WT and KO mice (n = 3 wells per genotype). mRNA levels were normalized to β-actin mRNA levels (Actb). Statistical significance was calculated using a two-tailed unpaired Student’s t test. *P < 0.05 versus WT
Table 1.
The top five enriched gene ontology biological processes in downregulated 276 genes
| Go term BP | Count | P value |
|---|---|---|
| Motor neuron axon guidance | 4 | 0.0049 |
| Regulating of bone remodeling | 3 | 0.0064 |
| Branching morphogenesis of an epithelial tube | 4 | 0.0099 |
| Positive regulation of BMP signaling pathway | 4 | 0.0099 |
| Cellular response to cholesterol | 3 | 0.011 |
We then investigated the association between BMP-Smad1/5/8 signaling activity and Adamts17 using primary chondrocytes. Immunofluorescence analysis showed that the nuclear accumulation of pSmad1 was suppressed in Adamts17-/- chondrocytes (Fig. S4). Western blotting also showed a significant decrease in pSmad1 protein in Adamts17-/- chondrocytes (Fig. 5d). In support of decreased amount of pSmad1 protein, the expression of Id1, a BMP-responsive gene, was also decreased in Adamts17-/- chondrocytes (Fig. 5e). Meanwhile, mRNA levels of representative BMP ligands were not decreased in Adamts17-/- chondrocytes (Fig. 5e).
Delayed terminal differentiation of Adamts17-/- chondrocytes is prevented by BMP treatment
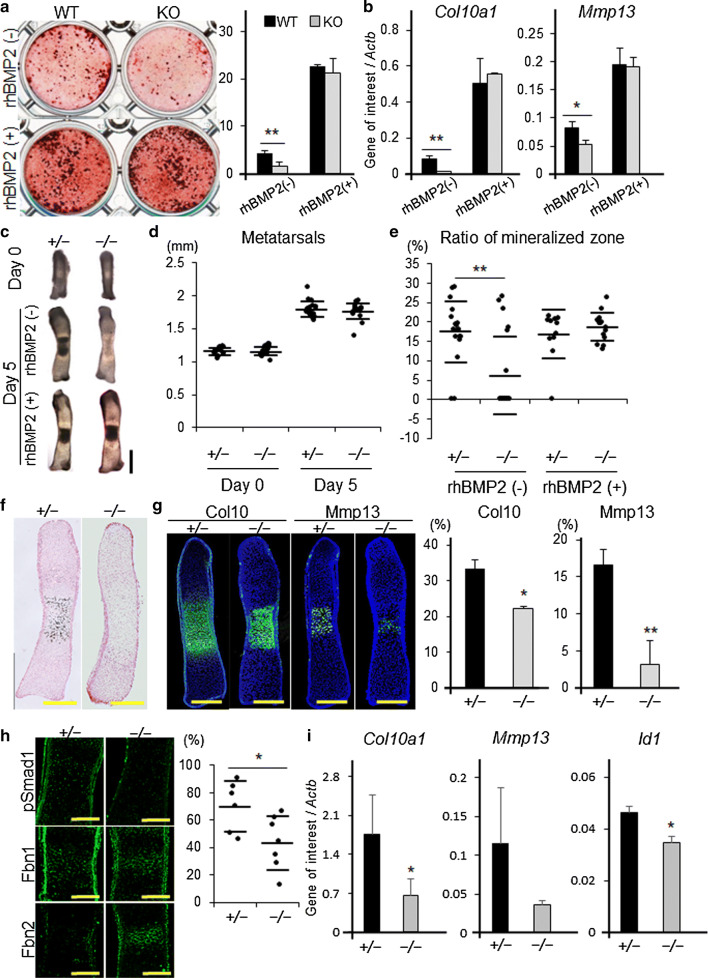
To investigate the effect of Adamts17 deletion on the terminal differentiation of chondrocytes, primary chondrocytes derived from Adamts17-/- and WT embryos were cultured with maturation medium [19]. Adamts17-/- chondrocytes yielded fewer mineralized nodules than WT controls (Fig. 6a). Col10a1 and Mmp13, representative terminal differentiation markers, were markedly decreased in Adamts17-/- chondrocytes (Fig. 6b). Notably, the inhibition of differentiation of Adamts17-/- chondrocytes was prevented by administration of rhBMP2 (Fig. 6a, b).
Fig. 6.
Delayed terminal differentiation of Adamts17-/- chondrocytes is prevented by BMP treatment. a Alizarin Red staining of primary chondrocytes, from WT and Adamts17-/- (KO) mice, cultured with or without 100 ng/mL of rhBMP2 for 20 days and the ratio of Alizarin Red-positive area (n = 3 wells per genotype). b Col10a1 and Mmp13 mRNA levels in chondrocytes from WT and KO mice, cultured with or without 100 ng/mL of rhBMP2. mRNA levels were normalized to β-actin mRNA levels (Actb). c Gross appearance of metatarsal rudiments of Adamts17 ± and Adamts17-/- mice at day 0 and after 5 days of culture, with or without 500 ng/mL of rhBMP2. Scale bars, 500 µm. d The entire length of metatarsal rudiments of Adamts17 ± (n = 18) and Adamts17-/- (n = 20) mice at day 0 and day 5. e The ratio of the mineralized zone of metatarsal rudiments from Adamts17 ± (rhBMP2 [+], n = 11; rhBMP2 [−], n = 18) and Adamts17-/- (rhBMP2 [+], n = 15; rhBMP2 [−], n = 20) mice at day 5. f von Kossa staining of metatarsal rudiments of Adamts17 ± and Adamts17-/- mice at day 5. Scale bars, 300 µm. g Immunofluorescent staining of Col10 and Mmp13 in metatarsal rudiments of Adamts17 ± and Adamts17-/- mice at day 5 and the ratio of Col10a1- and Mmp13-positive areas (n = 3 per genotype). Scale bars, 300 µm. *P < 0.05, **P < 0.01 versus Adamts17 ± . h Semiserial histological sections of cultured metatarsals cultured for 5 days were immunostained for phosphorylated Smad1 (pSmad1), Fbn1 and Fbn2, and the percentage of pSmad1-positive cells in the hypertrophic zones of Adamts17 ± (n = 6) and Adamts17-/- mice (n = 7) was shown. *P < 0.05 versus Adamts17 ± . i Col10a1, Mmp13, and Id1 mRNA levels in cultured metatarsals from Adamts17 ± and Adamts17-/- mice (n = 4 per genotype). mRNA levels were normalized to β-actin mRNA levels (Actb). Statistical significance was calculated using a two-tailed unpaired Student’s t test. *P < 0.05 versus Adamts17 ±
Finally, we used organ culture to examine the association of Adamts17 with BMP-Smad1/5/8 pathway in the terminal differentiation of chondrocytes. We prepared primordial metatarsal cartilage from E15.5 embryos of Adamts17-/- mice and also from Adamts17 ± mice, which showed normal skeletal growth. Although the length of the metatarsal rudiments was not different between genotypes at day 0 or day 5, mineralization was delayed more frequently and the ratio of the mineralized zone was significantly reduced in the metatarsal rudiments of Adamts17-/- mice (Fig. 6c–e). Therefore, we performed immunohistological analyses at day 5 to investigate whether terminal differentiation was actually inhibited. von Kossa staining showed apparently delayed mineralization in metatarsal rudiments from Adamts17 -/- mice (Fig. 6f). This was coupled with reduced expression of Col10a1 and Mmp13 (Fig. 6g). These findings were associated with reduced BMP signaling, as evidenced by a reduced ratio of pSmad1-positive cells, and Adamts17-/- metatarsals were accompanied with enhanced Fbn2 deposition (Fig. 6h). The expressions of Col10a1 and Id1 were significantly decreased whereas the expression of Mmp13 tended to be decreased, but did not reach statistical significance (p = 0.07) (Fig. 6i). Furthermore, the reduction of the mineralized zone in metatarsal rudiments from Adamts17-/- mice was completely prevented by the administration of rhBMP2 (Fig. 6c, e). Considering these in vitro and ex vivo results, we conclude that the loss of Adamts17 impairs chondrocyte terminal differentiation due to reduced BMP-Smad1/5/8 signaling.
Discussion
In the present study, we showed that Adamts17 knockout leads to postnatal impairment of skeletogenesis, which is a phenocopy of WMS. In adult Adamts17-/- mice, the axial length of bones was shortened and WMS-specific features, including brachydactyly and thick skin were also observed, although the skin phenotype was not investigated in depth in the present study. As for skeletal phenotype, the length of the hypertrophic zone was reduced and Fbn2 deposition was enhanced in the growth plate of Adamts17-/- mice. Comprehensive expression analysis using LMD and RNA-seq indicated alterations of the BMP signaling pathway by Adamts17 knockout. In Adamts17-/- chondrocytes, BMP signaling was reduced without affecting mRNA levels of BMP ligands. The delayed terminal differentiation of Adamts17-/- chondrocytes was prevented by BMP treatment in experiments using primary chondrocytes and primordial metatarsal cartilage. All these findings indicate that Adamts17 is involved in skeletal formation through the modulation of BMP signaling activity.
Reports that WMS is caused by mutations in FBN1, ADAMTS10, and ADAMTS17 suggest a close association of these genes, and experimental data has accumulated regarding the molecular interaction between microfibrils and ADAMTS/L proteins. Some ADAMTS/L proteins, such as ADAMTS10 and ADAMTSL2, modify the composition of microfibrils [7, 23, 25]. Furthermore, ADAMTS10, ADAMTSL4, and ADAMTSL6 have been shown to accelerate microfibril formation in vitro and in vivo [26-28]. Notably, chondrocyte-specific Adamtsl6 transgenic mice show enhanced Fbn1 deposition in rib cartilage, indicating that a strong interaction between microfibrils and ADAMTS/L proteins also exists in cartilage [29].
WMS and WMLS share common phenotypes and both disorders are caused by mutations in FBN1, ADAMTS10, or ADAMTS17, which are associated with microfibril biogenesis. In the current study, Adamts17-/- mice showed brachydactyly and thick skin, in addition to skeletal dysplasia. Humans with WMS-like syndrome, which is caused by ADAMTS17 mutations, typically only show short stature and eye abnormalities [8], although some ADAMTS17 mutations do cause brachydactyly [9]. In addition, ADAMTS10 deficiency in mice does not lead to extra-ocular manifestations of WMS [23]. The presence of two fibrillins (Fbn1, 2) in mice, instead of the three (FBN1-3) present in humans, provides a possible explanation for different phenotypes between humans with WMLS and Adamts17-/- mice.
Fbn2 deposition was increased both in the growth plates of Adamts17-/- mice and the pellets from Adamts17 -/- chondrocytes without affecting the level of Fbn2 mRNA. Fbn2 accumulation has also been reported in the skeletal muscle and ciliary zonules of Adamts10-/- mice [7, 23] and in bronchial smooth muscle cells of Adamtsl2-/- mice [25]. Although the actual mechanism by which Adamts17 selectively regulates Fbn2 incorporation into microfibrils is unclear, two possible hypotheses have been postulated. The first is that Fbn2 is a substrate of Adamts17 and Fbn2 accumulation is caused by a decrease in its degradation in Adamts17-/- mice. The second hypothesis is that Adamts17 negatively regulates Fbn2 through properties other than its protease activity. The first hypothesis is less likely because recent biochemical analyses have shown that Adamts17 does not cleave Fbn2 in vitro [30], although Adamts10 can cleave Fbn2 [23]. Furthermore, Adamtsl2-/- mice also display Fbn2 accumulation. Nevertheless, Adamtsl2 lacks the protease domain that is present in both Adamts10 and Adamts17 [3]. Adamts10, 17, and Adamtsl2 have a highly homologous C-terminal ancillary domain, which is thought to play an important role in the assembly or integrity of the extracellular matrix [3]. In addition, an ADAMTS17 mutation that retained an intact catalytic domain also caused WMLS [8], indicating that the catalytic domain of Adamts17 is not involved in the pathophysiology of WMLS. Taken together, it seems reasonable that Adamts17 negatively regulates Fbn2 by functioning as an Adamtsl protein rather than a protease.
In the present study, BMP-Smad1/5/8 signaling was reduced by Adamts17 knockout both in vivo and in vitro. Similarly, BMP signaling has been shown to be suppressed in mouse embryonic fibroblast cultures from Adamts10 WMS mice, which harbor a truncation mutation seen in WMS patients [7]. On the other hand, BMP signaling is activated in the skeletal muscle of Fbn2-knockout mice [31]. Considering these results, Fbn2 may negatively regulate BMP signaling and WMS may represent an Fbn2 gain-of-function phenotype [32]. A recent study reported that Fbn2 can sequester BMP complexes in a latent state [31]. This finding is consistent with our data where BMP-Smad1/5/8 signaling activity was reduced in Adamts17-/- chondrocytes without significantly changing the mRNA expression of BMP ligands. Though there was no significant difference in the expression of BMP ligands in chondrocyte cultures, it is possible that the change in the expression of BMP ligands in tissues other than chondrocytes, such as periosteum or perichondrium, where BMP ligands are reported to be highly expressed [33], may be attributed to the phenotype observed in Adamts17-/- mice.
WMS is ascribed to the acromelic dysplasia group of disorders, which are characterized by short stature, short hands and feet, and muscular build. The acromelic dysplasia group comprises geleophysic dysplasia [34], acromicric dysplasia [4], and Myhre syndrome [35], in addition to WMS. WMS is caused by mutations in microfibril-related genes and microfibrils play important roles in tissue homeostasis through their interaction with TGF-β and BMP [36-38]. Meanwhile, Myhre syndrome patients have a mutation in SMAD4, which results in a stabilized, but nonfunctional, mutant [35]. SMAD4 is known to be a mediator of both TGF-β and BMP signaling [39] and the expression of TGF-β and BMP target genes is actually decreased in fibroblasts from Myhre syndrome patients [34]. These data also support the idea that the impairment of TGF-β superfamily signaling causes the skeletal dysplasia observed in the acromelic dysplasia group.
In conclusion, Adamts17-/- mice recapitulated the WMS phenotypes of short stature, brachydactyly, and thick skin. We showed, for the first time, that the skeletal impairment observed in Adamts17-/- mice was associated with reduced BMP-Smad1/5/8 signaling and enhanced Fbn2 deposition in the growth plate, which may be attributed to this altered BMP signaling. Further studies on the role of ADAMTS/L proteins in skeletogenesis may provide insights into the pathomechanism of the acromelic dysplasia group of disorders.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank J. Sugita for technical assistance. This study was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (16K10810, 17K10924, 17H04311).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All experiments performed in this study comply with the current laws of Japan.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/16/2019
In the published article, the Fig. 4a was published incorrectly.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Melrose J, Shu C, Whitelock JM, Lord MS. The cartilage extracellular matrix as a transient developmental scaffold for growth plate maturation. Matrix Biol. 2016;52–54:363–383. doi: 10.1016/j.matbio.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Hubmacher D, Apte SS. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 2015;47:34–43. doi: 10.1016/j.matbio.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Goff C, Mahaut C, Wang LW, Allali S, Abhyankar A, Jensen S, Zylberberg L, Collod-Beroud G, Bonnet D, Alanay Y, Brady AF, Cordier MP, Devriendt K, Genevieve D, Kiper PO, Kitoh H, Krakow D, Lynch SA, Le Merrer M, Megarbane A, Mortier G, Odent S, Polak M, Rohrbach M, Sillence D, Stolte-Dijkstra I, Superti-Furga A, Rimoin DL, Topouchian V, Unger S, Zabel B, Bole-Feysot C, Nitschke P, Handford P, Casanova JL, Boileau C, Apte SS, Munnich A, Cormier-Daire V. Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet. 2011;89(1):7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, Le Merrer M, Collod-Beroud G, Boileau C, Munnich A, Cormier-Daire V. In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J Med Genet. 2003;40(1):34–36. doi: 10.1136/jmg.40.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, Dollfus H, Alembik Y, Munnich A, Legeai-Mallet L, Cormier-Daire V. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am J Hum Genet. 2004;75(5):801–806. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mularczyk EJ, Singh M, Godwin ARF, Galli F, Humphreys N, Adamson AD, Mironov A, Cain SA, Sengle G, Boot-Handford RP, Cossu G, Kielty CM, Baldock C. ADAMTS10-mediated tissue disruption in Weill-Marchesani Syndrome. Hum Mol Genet. 2018;27:3657–3687. doi: 10.1093/hmg/ddy276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, Al-Rajhi A, Alkuraya FS, Meyer BF, Al Tassan N. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85(5):558–568. doi: 10.1016/j.ajhg.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah MH, Bhat V, Shetty JS, Kumar A. Whole exome sequencing identifies a novel splice-site mutation in ADAMTS17 in an Indian family with Weill-Marchesani syndrome. Mol Vis. 2014;20:790–796. [PMC free article] [PubMed] [Google Scholar]
- 10.van Duyvenvoorde HA, Lui JC, Kant SG, Oostdijk W, Gijsbers AC, Hoffer MJ, Karperien M, Walenkamp MJ, Noordam C, Voorhoeve PG, Mericq V, Pereira AM, Claahsen-van de Grinten HL, van Gool SA, Breuning MH, Losekoot M, Baron J, Ruivenkamp CA, Wit JM. Copy number variants in patients with short stature. Eur J Hum Genet. 2014;22(5):602–609. doi: 10.1038/ejhg.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura H, Hasuwa H, Inoue N, Ikawa M, Okabe M. Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem Biophys Res Commun. 2004;321(2):275–279. doi: 10.1016/j.bbrc.2004.06.139. [DOI] [PubMed] [Google Scholar]
- 13.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26(2):145–146. doi: 10.1002/(SICI)1526-968X(200002)26:2<145::AID-GENE14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Mack SA, Maltby KM, Hilton MJ. Demineralized murine skeletal histology. Methods Mol Biol. 2014;1130:165–183. doi: 10.1007/978-1-62703-989-5_12. [DOI] [PubMed] [Google Scholar]
- 15.Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods. 2009;177(1):122–130. doi: 10.1016/j.jneumeth.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Kawamoto T, Kawamoto K. Preparation of thin frozen sections from nonfixed and undecalcified hard tissues using Kawamot's film method (2012) Methods Mol Biol. 2014;1130:149–164. doi: 10.1007/978-1-62703-989-5_11. [DOI] [PubMed] [Google Scholar]
- 17.Mori Y, Chung UI, Tanaka S, Saito T. Determination of differential gene expression profiles in superficial and deeper zones of mature rat articular cartilage using RNA sequencing of laser microdissected tissue specimens. Biomed Res. 2014;35(4):263–270. doi: 10.2220/biomedres.35.263. [DOI] [PubMed] [Google Scholar]
- 18.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirando AJ, Dong Y, Kim J, Hilton MJ. Isolation and culture of murine primary chondrocytes. Methods Mol Biol. 2014;1130:267–277. doi: 10.1007/978-1-62703-989-5_20. [DOI] [PubMed] [Google Scholar]
- 20.Yahara Y, Takemori H, Okada M, Kosai A, Yamashita A, Kobayashi T, Fujita K, Itoh Y, Nakamura M, Fuchino H, Kawahara N, Fukui N, Watanabe A, Kimura T, Tsumaki N. Pterosin B prevents chondrocyte hypertrophy and osteoarthritis in mice by inhibiting Sik3. Nat Commun. 2016;7:10959. doi: 10.1038/ncomms10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin I, Jakob M, Schafer D, Dick W, Spagnoli G, Heberer M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthr Cartil. 2001;9(2):112–118. doi: 10.1053/joca.2000.0366. [DOI] [PubMed] [Google Scholar]
- 22.Houston DA, Staines KA, MacRae VE, Farquharson C. Culture of murine embryonic metatarsals: a physiological model of endochondral ossification. J Vis Exp. 2016;118:54978. doi: 10.3791/54978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LW, Kutz WE, Mead TJ, Beene LC, Singh S, Jenkins MW, Reinhardt DP, Apte SS. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 2018;77:117–128. doi: 10.1016/j.matbio.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charbonneau NL, Jordan CD, Keene DR, Lee-Arteaga S, Dietz HC, Rifkin DB, Ramirez F, Sakai LY. Microfibril structure masks fibrillin-2 in postnatal tissues. J Biol Chem. 2010;285(26):20242–20251. doi: 10.1074/jbc.M109.087031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubmacher D, Wang LW, Mecham RP, Reinhardt DP, Apte SS. Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia—a novel mouse model providing insights into geleophysic dysplasia. Dis Model Mech. 2015;8(5):487–499. doi: 10.1242/dmm.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, Sakai LY, Keene DR, Apte SS. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J Biol Chem. 2011;286(19):17156–17167. doi: 10.1074/jbc.M111.231571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriel LA, Wang LW, Bader H, Ho JC, Majors AK, Hollyfield JG, Traboulsi EI, Apte SS. ADAMTSL4, a secreted glycoprotein widely distributed in the eye, binds fibrillin-1 microfibrils and accelerates microfibril biogenesis. Invest Ophthalmol Vis Sci. 2012;53(1):461–469. doi: 10.1167/iovs.10-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito M, Kurokawa M, Oda M, Oshima M, Tsutsui K, Kosaka K, Nakao K, Ogawa M, Manabe R, Suda N, Ganjargal G, Hada Y, Noguchi T, Teranaka T, Sekiguchi K, Yoneda T, Tsuji T. ADAMTSL6beta protein rescues fibrillin-1 microfibril disorder in a Marfan syndrome mouse model through the promotion of fibrillin-1 assembly. J Biol Chem. 2011;286(44):38602–38613. doi: 10.1074/jbc.M111.243451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsui K, Manabe R, Yamada T, Nakano I, Oguri Y, Keene DR, Sengle G, Sakai LY, Sekiguchi K. ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J Biol Chem. 2010;285(7):4870–4882. doi: 10.1074/jbc.M109.076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubmacher D, Schneider M, Berardinelli SJ, Takeuchi H, Willard B, Reinhardt DP, Haltiwanger RS, Apte SS. Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci Rep. 2017;7:41871. doi: 10.1038/srep41871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengle G, Carlberg V, Tufa SF, Charbonneau NL, Smaldone S, Carlson EJ, Ramirez F, Keene DR, Sakai LY. Abnormal activation of BMP signaling causes myopathy in FBN2 null mice. PLoS Genet. 2015;11(6):e1005340. doi: 10.1371/journal.pgen.1005340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai Lynn Y., Keene Douglas R. Fibrillin protein pleiotropy: Acromelic dysplasias. Matrix Biology. 2019;80:6–13. doi: 10.1016/j.matbio.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Salazar VS, Capelo LP, Cantu C, Zimmerli D, Gosalia N, Pregizer S, Cox K, Ohte S, Feigenson M, Gamer L, Nyman JS, Carey DJ, Economides A, Basler K, Rosen V (2019) Reactivation of a developmental Bmp2 signaling center is required for therapeutic control of the murine periosteal niche. eLife 8:e42386. doi:10.7554/eLife.42386. [DOI] [PMC free article] [PubMed]
- 34.Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, Bauer F, Flori E, Prost-Squarcioni C, Krakow D, Ge G, Greenspan DS, Bonnet D, Le Merrer M, Munnich A, Apte SS, Cormier-Daire V. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40(9):1119–1123. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Goff C, Mahaut C, Abhyankar A, Le Goff W, Serre V, Afenjar A, Destree A, di Rocco M, Heron D, Jacquemont S, Marlin S, Simon M, Tolmie J, Verloes A, Casanova JL, Munnich A, Cormier-Daire V. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet. 2011;44(1):85–88. doi: 10.1038/ng.1016. [DOI] [PubMed] [Google Scholar]
- 36.Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bachinger HP, Sakai LY. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283(20):13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wohl AP, Troilo H, Collins RF, Baldock C, Sengle G. Extracellular regulation of bone morphogenetic protein activity by the microfibril component fibrillin-1. J Biol Chem. 2016;291(24):12732–12746. doi: 10.1074/jbc.M115.704734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, Rifkin DB. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J Cell Physiol. 2012;227(12):3828–3836. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Chen G, Li YP. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.