Abstract
In the post-embryonic stage of Arabidopsis thaliana, roots can be initiated from the vascular region of the existing roots or non-root organs; they are designated as lateral roots (LRs) and adventitious roots (ARs), respectively. Some root-like organs can also be initiated from the vasculature. In tissue culture, auxin-induced callus, which is a group of pluripotent root-primordium-like cells, is formed via the rooting pathway. The formation of feeding structures from the vasculature induced by root-knot nematodes also borrows the rooting pathway. In this review, we summarize and discuss recent progress on the role of LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16; also known as ASYMMETRIC LEAVES2-LIKE18, ASL18), a member of the LBD/ASL gene family encoding plant-specific transcription factors, in roots and root-like organ initiation. Different root and root-like organ initiation processes have distinct priming mechanisms to specify founder cells. All these priming mechanisms converge to activate LBD16 expression in the primed founder cells. The activation of LBD16 expression leads to organ initiation via promotion of cell division and establishment of root-primordium identity. Therefore, LBD16 might play a common and pivotal role in root and root-like organ initiation.
Keywords: Root founder cell, Lateral root, Adventitious root, LBD16, Callus, Root-knot nematodes
Introduction
In the post-embryonic stage of Arabidopsis thaliana, many types of roots can originate from the vasculature. Adventitious roots (ARs) can form from non-root organs, such as detached leaf or stem explants and the hypocotyl [1–8]. In a developing root, acropetal lateral roots (simply referred to LR in this review) form from the xylem-pole pericycle cells under the guidance of an oscillating auxin flux derived from the root cap [9–14]. An existing root can also produce adventitious lateral roots, mainly via the AR formation pathway [15–23].
Some root-like organs can also be initiated from the vasculature in A. thaliana. In tissue culture, callus can be induced on auxin-rich callus-inducing medium (CIM) [24–32]. Callus forms via the rooting pathway and the cellular nature of the newly formed callus is a group of root-primordium-like cells [33–38]. Callus cells are pluripotent, because they are competent for shoot regeneration on cytokinin-rich shoot-inducing medium (SIM) or root regeneration on root-inducing medium (RIM). In addition, root-knot nematodes can induce the formation of feeding structures from the vasculature of A. thaliana in either roots or detached leaf explants [39, 40]. The formation of feeding structures has been proposed to borrow the rooting pathway [40–48]. Therefore, it is possible that feeding structures induced by root-knot nematodes are also root-like organs.
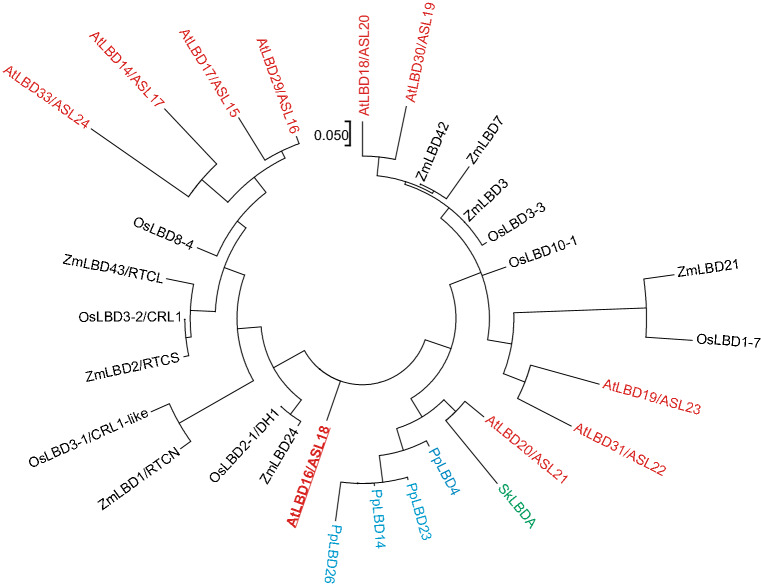
LATERAL ORGAN BOUNDARIES DOMAIN (LBD; also known as ASYMMETRIC LEAVES2-LIKE, ASL) transcription factor genes belong to a plant-specific family that is present in diverse taxa ranging from algae to seed plants. There are at least six classes of LBD genes in A. thaliana [49–54]. The class-IB, also known as the root-associated LBD/ASL clade, has ten members in A. thaliana [53, 54] (Fig. 1), and they are involved in many aspects of plant development. Here, we review recent progress in research on the role of A. thaliana LBD16 (Fig. 1), which belongs to class-IB LBD genes, in root and root-like organ initiation.
Fig. 1.
Phylogeny of class-IB LBD genes in land plants. Phylogenetic analysis of protein sequences of class-IB LBDs from a bryophyte Physcomitrella patens (Pp, in blue), a lycophytes Selaginella kraussiana (Sk, in green), a dicot Arabidopsis thaliana (At, in red), and monocot Oryza sativa (Os, in black) and Zea mays (Zm, in black). Sequence of SkLBDA was obtained by analyzing RNA-seq and DNA-seq data published previously [91] (GenBank, MH107252). Other sequences were obtained from the published data [53]. Phylogenetic tree of full length of the LBD proteins was constructed using the maximum-likelihood method (the Poisson model) with MEGA7.0 and default parameters [92]
LBD16 in LR initiation
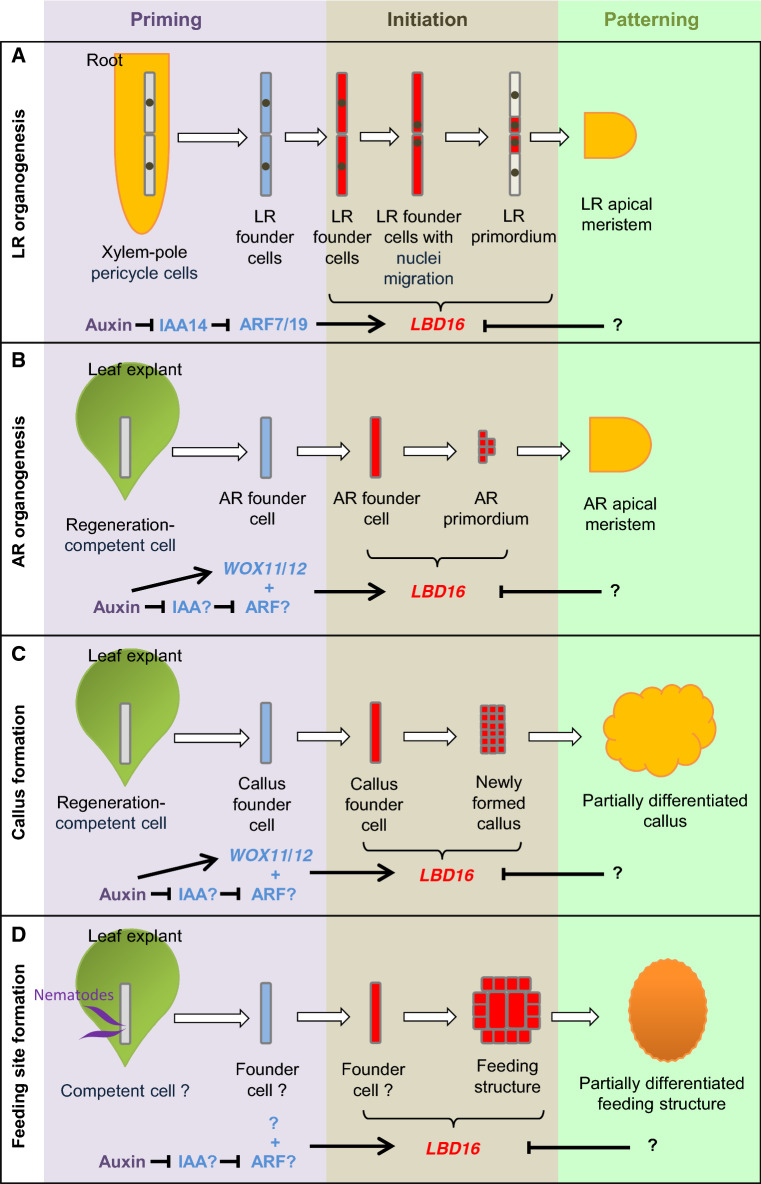
When A. thaliana is grown vertically on the medium, the primary root usually produces LRs by an acropetal sequence. The formation of LR in A. thaliana involves successive steps of cell fate transitions [13, 14, 55–57] (Fig. 2a). Auxin is transported into and accumulates in the xylem-pole pericycle cells in the priming step for specification of a group of LR founder cells. After priming, the nuclei of the LR founder cells migrate to start the initiation step, and pairs of LR founder cells undergo asymmetric cell division to form the LR primordium. The LR primordium undergoes continuous cell division, and then, the LR primordium forms the LR apical meristem with functional domains in the patterning step [55].
Fig. 2.
Model of LBD16 in root and root-like organ formation. Model summarizing the common role of A. thaliana LBD16 in LR formation from primary root (a), AR formation from detached leaf explants (b), callus formation from detached leaf explant (c), and feeding structure formation from detached leaf explant (d). Cells and genes that are involved in the priming step are in blue, and those in the initiation step are in red
The expression of LBD16 is specifically induced in the primed LR founder cells before the nuclei migrate and asymmetric division occurs [58] (Fig. 2a). During the initiation step, LBD16 expression continues in the LR primordium, but its expression gradually decreases during the patterning step to form the LR apical meristem ([9, 20, 58] and our unpublished data) (Fig. 2a). The lbd16 mutant and LBD16-SRDX transgenic lines (in which the LBD16 pathway was blocked) formed fewer LRs from the primary root than did wild type [9, 20, 58–60]. Specifically, blocking of the LBD16 pathway resulted in defective polar nuclei migration in LR founder cells and defective asymmetrical cell division, but it did not affect the specification of LR founder cells with auxin accumulation in the priming step [58]. Therefore, LBD16 seems to be specifically involved in the initiation of the LR primordium.
In LR formation, LBD16 is activated by AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 (ARF7/19), two functionally redundant transcription factors [9, 58, 59, 61] (Fig. 2a). At a low auxin level, ARF7/19 form a protein complex with the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) protein IAA14 (also known as SOLITARY-ROOT, SLR) [62]. The interaction with IAA14 represses the transcriptional activation activity of ARF7/19. In the priming step of lateral rooting, auxin is transported into and highly accumulates in LR founder cells. This results in the degradation of the IAA14 protein and, therefore, the release of ARF7/19. The release of ARF7/19 derepresses their transcriptional activator role. In the primed founder cells, ARF7/19 can directly bind to the AUXIN RESPONSE ELEMENTs (AuxREs) on the promoter of LBD16 to activate transcription [9, 58, 59, 63]. The arf7 arf19 double mutant showed defective LR initiation and a loss of LBD16 expression when the plants were vertically cultured on growth medium. Overexpression of LBD16 in the arf7 arf19 double mutant background partially rescued the LR formation defect, suggesting that LBD16 acts downstream of ARF7/19 [9].
Overall, during LR organogenesis, ARF7/19 activate LBD16 expression in the primed LR founder cells, and then, LBD16 promotes cell division and fate transition during the initiation of LR primordium.
LBD16 in AR initiation
In A. thaliana, ARs can form from many tissues after wounding or in response to environmental signals [1–8]. By studying de novo root regeneration from detached A. thaliana leaf explants [64], the role of LBD16 in AR initiation was analyzed [8, 20, 36, 65] (Fig. 2b). After detachment of leaf explants, endogenous auxin is produced and polar-transported into regeneration-competent cells (i.e., procambium cells and some vascular parenchyma cells) near the wounded site. Accumulation of auxin in a regeneration-competent cell activates the expression of two partially redundant transcription factor genes, WUSCHEL-RELATED HOMEOBOX11 (WOX11) and WOX12 (WOX11/12), resulting in the priming step, i.e., cell fate transition from a regeneration-competent cell to an AR founder cell [36]. In the initiation step, the AR founder cell divides to form the AR primordium. To date, there is no solid evidence that nucleus migration and asymmetrical cell division of pairs of founder cells occur during the initiation of the AR primordium. Further analysis of the first round of cell division in AR formation from A. thaliana leaf explants will clarify how the AR primordium is initiated in the future.
LBD16 is specifically induced in the primed AR founder cell during the initiation step ([65] and our unpublished data) (Fig. 2b). LBD16 expression continues in the AR primordium and gradually decreases when the AR primordium enters the patterning step to differentiate into the AR apical meristem [20, 36, 65] (Fig. 2b). The lbd16 mutant showed defects in AR organogenesis from leaf explants [20], suggesting that LBD16 is required for AR initiation.
During AR formation from leaf explants, LBD16 is not activated by ARF7/19 [20, 36]. The arf7 arf19 double mutant was able to form ARs from detached leaf explants, suggesting that AR initiation from leaf explants is not strictly dependent on ARF7/19, in contrast to LR initiation [20]. Therefore, other upstream activators may induce LBD16 expression during AR formation. WOX11/12 were proposed to directly activate LBD16 [20]. In the WOX11–SRDX transgenic lines, LBD16 expression was not upregulated in the primed AR founder cells or the AR primordium. There are two WOX-binding elements on the LBD16 promoter, and WOX11 can directly bind to these elements. Mutations in the WOX-binding elements resulted in the loss of the LBD16 expression during AR initiation, suggesting that WOX11/12 are key upregulators of LBD16 in AR formation. Activation of LBD16 by WOX11/12 requires the presence of auxin, suggesting that the upregulation of LBD16 in the primed AR founder cells may require ARF proteins other than ARF7/19. Those ARFs may cooperate with WOX11/12 to upregulate LBD16 during AR primordium initiation from leaf explants [8] (Fig. 2b).
Although ARF7/19 do not regulate LBD16 expression during AR formation from leaf explants, the two ARFs have been shown to control AR formation from hypocotyls [66], indicating that different priming mechanisms might be involved in AR formations from different organs. It will be interesting to determine whether the auxin signaling pathways involved in AR formation differ between leaf explants and hypocotyls in the future.
WOX11/12 did not regulate LBD16 during LR formation when A. thaliana was vertically grown on the medium [20]. WOX11/12 are not expressed in LR founder cells; therefore, they cannot be responsible for LBD16 upregulation during LR initiation. Interestingly, ectopic WOX11-SRDX expression under the control of the 35S promoter in LR founder cells did not block LBD16 activation by ARF7/19, suggesting that there is some unknown mechanism to abolish the function of the WOX11 protein in the LR founder cells [20].
The A. thaliana primary root can produce not only LRs but also adventitious lateral roots [20–23]. The priming of founder cells during adventitious lateral rooting is more likely to follow the AR pathway than the LR pathway, i.e., involving WOX11/12 but not ARF7/19 [20]. It is possible that LBD16 could also be involved in the initiation step of adventitious lateral rooting, as it is in AR formation. Further research on the role of LBD16 in adventitious lateral root formation is required to test this hypothesis.
Overall, during AR organogenesis from leaf explants, WOX11/12 together with ARFs activate LBD16 expression in the primed AR founder cells. LBD16 then promotes the initiation of AR primordium.
LBD16 in callus initiation and pluripotency acquisition
De novo shoot regeneration (also known as de novo shoot organogenesis) in tissue culture usually experiences two phases: first, callus forms from detached explants on auxin-rich CIM; then, callus is moved to cytokinin-rich SIM for adventitious shoot induction [24–32]. The results of recent studies have suggested that the newly formed callus on CIM is a group of root-primordium-like cells and the process of callus initiation borrows the rooting pathway [33–38].
Using callus formation from A. thaliana leaf explants as an example, the priming and initiation steps of callus formation are similar to those that occur during AR formation [34, 36, 38, 67–70] (Fig. 2c). In the priming step, exogenous auxin in the medium activates WOX11/12 expression to trigger fate transition from a regeneration-competent cell to a callus founder cell [36, 38, 69]. Then, the callus founder cell undergoes division to become the newly formed callus cells, which have root-primordium-like features at the molecular level [36–38, 69]. When callus is cultured on CIM for a long time, partial differentiation into root apical meristem-like tissue can occur (i.e., the formation of partially differentiated callus) [69].
LBD16 expression is specifically induced in the primed callus founder cell in the initiation step on CIM ([34, 38] and our unpublished data) (Fig. 2c). LBD16 expression continues in the root-primordium-like newly formed callus [38]. The lbd16 mutant exhibited slowed cell division during callus initiation, indicating that LBD16 is required for proper cell division [34, 38]. When the callus is continuously cultured on the CIM for a long time, LBD16 expression level decreases in root apical meristem-like partially differentiated callus ([34, 69] and our unpublished data) (Fig. 2c).
When newly formed callus is moved to SIM, LBD16 expression sharply decreases and then disappears, suggesting that cytokinin might have a role in repressing LBD16 expression on SIM [38]. Interestingly, the callus of the lbd16 mutant showed defective shoot formation on SIM, suggesting that LBD16 is also required for the acquisition of pluripotency in the newly formed callus during its initiation [38]. The cellular basis of pluripotency in the newly formed callus was proposed to be the root-primordium identity [37, 38]. Therefore, LBD16 promotes establishment of the root-primordium cell fate identity in newly formed callus.
During callus formation from A. thaliana detached leaf explants on CIM, LBD16 expression is upregulated by auxin together with WOX11/12 [34, 38] (Fig. 2c), similar to the case during AR formation. The previous studies have indicated that ARF7/19 might be involved in activating LBD16 during callus initiation [34, 71]. However, LBD16 expression was not completely abolished in the arf7 arf19 double mutant background when callus was initiated from leaf explants (our unpublished data), suggesting that at least some other ARF proteins are also involved in the activation of LBD16 during this process (Fig. 2c). In addition, the chromatin factor ARABIDOPSIS TRITHORAX-RELATED 2 (ATXR2), a putative histone methyltransferase, acts as a co-activator with ARFs to upregulate LBD16 [71]. ATXR2 might regulate LBD16 expression via deposition of histone H3 lysine 36 trimethylation (H3K36me3) epigenetic markers on the LBD16 locus, thereby facilitating its transcription. Consistent with the molecular role of ATXR2, the atxr2 mutant showed partially defective callus formation from leaf explants, and the LBD16 expression level was lower in atxr2 leaf explants than in the wild-type leaf explants during callus initiation. Therefore, WOX11/12 and the auxin-mediated ARF pathway together with chromatin factors could act together to upregulate LBD16 during callus initiation on CIM. A recent study using enhanced yeast one-hybrid (eY1H) screening showed that LBD16 is involved in a complex gene regulatory network involving multiple key transcription factors related to regeneration [72].
The AtbZIP59 transcription factor interacts with LBD16 during callus formation [70]. Like LBD16 overexpression [34], AtbZIP59 overexpression induced autonomous callus formation in the absence of exogenous auxin [70]. The AtbZIP59–LBD16 complex was shown to directly regulate the expression of FAD-BINDING BERBERINE (FAD-BD), which encodes a BBE-like enzyme involved in cell wall metabolism during LR emergence [70, 73].
Therefore, callus initiation from leaf explants may borrow the AR formation pathway. Similarly, it is possible that callus formation from root explants may borrow the acropetal lateral rooting pathway or adventitious lateral rooting pathway. Regardless of the founder cells from different rooting pathways, LBD16 is the pivotal gene to be induced during callus initiation, and is required for the establishment of pluripotency (i.e., root-primordium identity) in the newly formed callus.
LBD16 in root-knot nematode feeding site formation
Root-knot nematodes, a type of plant endoparasitic nematode, can induce the formation of feeding structures from the vasculature in roots or in other organs (e.g., detached leaf explants) [39, 40]. Giant cells, the most commonly formed feeding cells in the feeding structures, are initiated from the vasculature and undergo repeated mitosis with aborted cytokinesis induced by nematode effectors [48].
Using A. thaliana roots and detached leaf explants as the model systems, a series of studies revealed the role of LBD16 in the formation of the feeding structures [40, 47, 48] (Fig. 2d). At the transcriptome level, the early stages of the formation of the feeding structure gall in A. thaliana roots resemble those of the formation of the root primordium [47]. In addition, in A. thaliana leaf explants, root-knot nematodes can feed on giant cells within a callus-like structure induced from the vasculature, similar to the gall induced in roots [40]. The formation of giant cells within these feeding structures (galls in roots or callus-like structures in leaf explants) is likely to borrow the root organogenesis pathways. The cells that are initiated to form the feeding structures have high auxin levels and express LBD16 [40, 47, 48] (Fig. 2d). Mutation of LBD16 led to partially defective feeding structure formation in roots and leaf explants [40, 47, 48]. The LBD16 expression level decreases when the feeding structures are partially differentiated [40, 47] (Fig. 2d). These observations suggested that the auxin signaling pathway and the LBD16-mediated molecular network are adopted for the formation of feeding structures induced by root-knot nematodes.
Interestingly, ARF7/19 are not strictly involved in the formation of giant cells and feeding structures. Primary roots of the arf7 arf19 double mutant were able to form feeding structures in response to root-knot nematodes [40]. Thus, upregulation of LBD16 and the formation of feeding structures either from the roots or leaf explants may occur mainly through the AR pathway instead of the LR pathway. At present, it is unclear which ARFs are involved in this process (Fig. 2d).
It is plausible to speculate that the initiation step of feeding structures is based on the AR formation mechanism involving auxin-induced LBD16 expression. In future research, it will be interesting to test whether WOX11/12 are also involved in this process.
Conclusion and perspectives
The common role of LBD16 in root and root-like organ formation is at the initiation step (see models in Fig. 2). Expression of LBD16 is induced in the specified founder cells after the priming step, and it continues to be expressed in the root primordium, the newly formed callus, or feeding structures. Its expression gradually decreases when the patterning step begins. The roles of LBD16 in root and root-like organ initiation may be to control cell division and to determine root-primordium identity.
Although LBD16 is upregulated in the initiation step during the formation of roots and root-like organs, the upstream mechanisms that activate LBD16 differ depending on the process. First, different ARFs are required to fulfill the auxin-mediated signaling pathway: ARF7/19 for LRs and other ARFs for ARs or AR-like organs (e.g., adventitious lateral roots from the primary root, callus from leaf explants, or feeding structures). Second, WOX11/12 are required for LBD16 upregulation in ARs or AR-like organs, but not LRs. The different molecular events’ upstream of LBD16 in the priming step might be due to the different status of founder cells in LR or AR organogenesis. While LR organogenesis requires several pairs of founder cells with nuclei migration and asymmetric founder cell division, there is no conclusive evidence that these factors are required for AR organogenesis or AR-like organ formation. Therefore, different molecular mechanisms operate in different LR and AR founder cells. One hypothesis was that WOX11/12-mediated AR organogenesis could be inherited from the ancient ability of intermediate-clade WOX (IC-WOX) genes in root founder cells [74]; because similar IC-WOX-mediated root organogenesis was also observed in a fern [75].
In summary, in the formation of root and root-like organs, diverse upstream priming mechanisms for the specification of founder cells may converge to activate LBD16 expression to achieve cell division and fate transition in organ initiation (see models in Fig. 2).
Many questions about LBD16 remain unanswered. First, it is largely unclear how other class-IB LBD genes interact with LBD16 during root and root-like organ formation. It will be interesting to test whether class-IB LBD genes have partially redundant or unique functions. For example, LBD29 is expressed in the root primordium during LR and AR formation [9, 36, 60, 76, 77], and its homologs CROWN ROTLESS1 (CRL1) in rice and ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS (RTCS) in maize are involved in adventitious root (crown root) formation [78–82]. In addition, many class-IB LBD genes are upregulated during LR formation from the primary root or during callus formation on CIM [34, 36, 59, 73, 77, 83–87]. Furthermore, class-IB LBD proteins may form complexes to regulate downstream targets [88]. Further research is required to explore the shared and common roles of class-IB LBD genes. Second, it will be interesting to determine which ARF protein upregulates LBD16 expression in AR organogenesis and AR-like organ formation. Why different ARF proteins function in different types of founder cells is an interesting question. Third, the genome-wide target analysis of LBD16 will provide new insights into how it regulates organ initiation. Fourth, the mechanism that downregulates LBD16 expression in the patterning step is unclear. Fifth, further analysis of the protein complex involving LBD16 will provide new information about how LBD16 regulates gene expression in the specific context of organ initiation. Sixth, it will be important to determine how the LBD16 pathway acts synergistically with other pathways, for example the PLETHORAs (PLTs) and WOX5/7 pathways, in root and root-like organ formation. PLT3/5/7 and WOX5/7 are all expressed in the root primordium or newly formed callus [33, 36, 37]. Mutations in either PLT3/5/7 or WOX5/7 resulted in abnormal root-primordium development or the loss of pluripotency in callus cells ([37, 89, 90] and our unpublished data). It is unclear how LBD16, PLT3/5/7, and WOX5/7 form a regulatory network in this process [38]. Seventh, it will be interesting to explore how class-IB LBD genes have evolved and how and when were they recruited into root organogenesis, together with other rooting-related genes (e.g., ARFs and WOXs) [51–54, 74]. Answers to these questions will improve our understanding of root and root-like organ initiation and evolution.
Acknowledgements
We apologize for references not cited due to space limitations. This work was supported by grants from the National Natural Science Foundation of China (31630007), National Basic Research Program of China (973 Program, 2014CB943500), the Key Research Program of CAS (QYZDB-SSW-SMC010), the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” of CAS (XDPB0403), and National Key Laboratory of Plant Molecular Genetics.
Abbreviations
- AR
Adventitious root
- ARF
AUXIN RESPONSE FACTOR
- ASL
ASYMMETRIC LEAVES2-LIKE
- ATXR2
ARABIDOPSIS TRITHORAX-RELATED 2
- Aux/IAA
AUXIN/INDOLE-3-ACETIC ACID
- AuxRE
AUXIN RESPONSE ELEMENT
- CIM
Callus-inducing medium
- CRL1
CROWN ROTLESS1
- eY1H
Enhanced yeast one-hybrid
- FAD-BD
FAD-BINDING BERBERINE
- LBD
LATERAL ORGAN BOUNDARIES DOMAIN
- LR
Lateral root
- PLT
PLETHORA
- RIM
Root-inducing medium
- RTCS
ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS
- SIM
Shoot-inducing medium
- SLR
SOLITARY-ROOT
- WOX
WUSCHEL-RELATED HOMEOBOX
Compliance with ethical standards
Conflict of interest
No conflicts of interest declared.
Footnotes
Wu Liu and Jie Yu have contributed equally to this work.
References
- 1.Falasca G, Altamura MM. Histological analysis of adventitious rooting in Arabidopsis thaliana (L.) Heynh seedlings. Plant Biosyst. 2003;137(3):265–274. doi: 10.1080/11263500312331351511. [DOI] [Google Scholar]
- 2.da Costa CT, de Almeida MR, Ruedell CM, Schwambach J, Maraschin FS, Fett-Neto AG. When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front Plant Sci. 2013;4:133. doi: 10.3389/fpls.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson JA, Rasmussen A, Traini R, Voss U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ. Branching out in roots: uncovering form, function, and regulation. Plant Physiol. 2014;166(2):538–550. doi: 10.1104/pp.114.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellini C, Pacurar DI, Perrone I. Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol. 2014;65:639–666. doi: 10.1146/annurev-arplant-050213-035645. [DOI] [PubMed] [Google Scholar]
- 5.Verstraeten I, Schotte S, Geelen D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front Plant Sci. 2014;5:495. doi: 10.3389/fpls.2014.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnbaum KD. How many ways are there to make a root? Curr Opin Plant Biol. 2016;34:61–67. doi: 10.1016/j.pbi.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Steffens B, Rasmussen A. The physiology of adventitious roots. Plant Physiol. 2016;170(2):603–617. doi: 10.1104/pp.15.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L. De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr Opin Plant Biol. 2018;41:39–45. doi: 10.1016/j.pbi.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19(1):118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329(5997):1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xuan W, Audenaert D, Parizot B, Moller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mahonen AP, Vanneste S, Beeckman T. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol. 2015;25(10):1381–1388. doi: 10.1016/j.cub.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 12.Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Moller BK, Skorzinski N, Njo MF, De Rybel B, Audenaert D, Nowack MK, Vanneste S, Beeckman T. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science. 2016;351(6271):384–387. doi: 10.1126/science.aad2776. [DOI] [PubMed] [Google Scholar]
- 13.Du Y, Scheres B. Lateral root formation and the multiple roles of auxin. J Exp Bot. 2018;69(2):155–167. doi: 10.1093/jxb/erx223. [DOI] [PubMed] [Google Scholar]
- 14.Stoeckle D, Thellmann M, Vermeer JE. Breakout-lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol. 2018;41:67–72. doi: 10.1016/j.pbi.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Esau K. Plant anatomy. 2. New York: Wiley; 1965. [Google Scholar]
- 16.Barlow PW. Adventitious roots of whole plants: their forms, functions, and evolution. In: Jackson MB, editor. New root formation in plants and cuttings. Hingham: Martinus Nijhoff; 1986. pp. 67–110. [Google Scholar]
- 17.Charlton WA. Lateral root initiation. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots: the hidden half. 2. New York: Marcel Dekker Inc.; 1996. pp. 149–173. [Google Scholar]
- 18.Paolillo DJ, Jr, Zobel RW. The formation of adventitious roots on root axes is a widespread occurrence in field-grown dicotyledonous plants. Am J Bot. 2002;89(9):1361–1372. doi: 10.3732/ajb.89.9.1361. [DOI] [PubMed] [Google Scholar]
- 19.Hou G, Hill JP, Blancaflor EB. Developmental anatomy and auxin response of lateral root formation in Ceratopteris richardii. J Exp Bot. 2004;55(397):685–693. doi: 10.1093/jxb/erh068. [DOI] [PubMed] [Google Scholar]
- 20.Sheng L, Hu X, Du Y, Zhang G, Huang H, Scheres B, Xu L. Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development. 2017;144(17):3126–3133. doi: 10.1242/dev.152132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu D, Miao J, Yumoto E, Yokota T, Asahina M, Watahiki M. YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant Cell Physiol. 2017;58(10):1710–1723. doi: 10.1093/pcp/pcx107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baesso B, Chiatante D, Terzaghi M, Zenga D, Nieminen K, Mahonen AP, Siligato R, Heliariutta Y, Scippa GS, Montagnoli A. PRE3 and WOX11 transcription factors are involved in the formation of new lateral roots from secondary growth taproot in A. thaliana. Plant Biol (Stuttg) 2018 doi: 10.1111/plb.12711. [DOI] [PubMed] [Google Scholar]
- 23.Ge Y, Fang X, Liu W, Sheng L, Xu L. Adventitious lateral rooting: the plasticity of root system architecture. Physiol Plant. 2018 doi: 10.1111/ppl.12741. [DOI] [PubMed] [Google Scholar]
- 24.Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS. De novo shoot organogenesis: from art to science. Trends Plant Sci. 2011;16(11):597–606. doi: 10.1016/j.tplants.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011;21(4):212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Ikeuchi M, Sugimoto K, Iwase A. Plant callus: mechanisms of induction and repression. Plant Cell. 2013;25(9):3159–3173. doi: 10.1105/tpc.113.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su YH, Zhang XS. The hormonal control of regeneration in plants. Curr Top Dev Biol. 2014;108:35–69. doi: 10.1016/B978-0-12-391498-9.00010-3. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Huang H. Genetic and epigenetic controls of plant regeneration. Curr Top Dev Biol. 2014;108:1–33. doi: 10.1016/B978-0-12-391498-9.00009-7. [DOI] [PubMed] [Google Scholar]
- 29.Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143(9):1442–1451. doi: 10.1242/dev.134668. [DOI] [PubMed] [Google Scholar]
- 30.Kareem A, Radhakrishnan D, Sondhi Y, Aiyaz M, Roy MV, Sugimoto K, Prasad K. De novo assembly of plant body plan: a step ahead of Deadpool. Regeneration (Oxf) 2016;3(4):182–197. doi: 10.1002/reg2.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K, Seo PJ. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018;23(3):235–247. doi: 10.1016/j.tplants.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Sang YL, Cheng ZJ, Zhang XS. iPSCs: a comparison between animals and plants. Trends Plant Sci. 2018 doi: 10.1016/j.tplants.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010;18(3):463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012;22(7):1169–1180. doi: 10.1038/cr.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He C, Chen X, Huang H, Xu L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012;8(8):e1002911. doi: 10.1371/journal.pgen.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 2014;26(3):1081–1093. doi: 10.1105/tpc.114.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, Prasad K. PLETHORA genes control regeneration by a two-step mechanism. Curr Biol. 2015;25(8):1017–1030. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Hu X, Qin P, Prasad K, Hu Y, Xu L. The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018;59(4):734–743. doi: 10.1093/pcp/pcy010. [DOI] [PubMed] [Google Scholar]
- 39.Kyndt T, Vieira P, Gheysen G, de Almeida-Engler J. Nematode feeding sites: unique organs in plant roots. Planta. 2013;238(5):807–818. doi: 10.1007/s00425-013-1923-z. [DOI] [PubMed] [Google Scholar]
- 40.Olmo R, Cabrera J, Moreno-Risueno MA, Fukaki H, Fenoll C, Escobar C. Molecular transducers from roots are triggered in Arabidopsis leaves by root-knot nematodes for successful feeding site formation: a conserved post-embryogenic de novo organogenesis program? Front Plant Sci. 2017;8:875. doi: 10.3389/fpls.2017.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barthels N, van der Lee FM, Klap J, Goddijn OJ, Karimi M, Puzio P, Grundler FM, Ohl SA, Lindsey K, Robertson L, Robertson WM, Van Montagu M, Gheysen G, Sijmons PC. Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell. 1997;9(12):2119–2134. doi: 10.1105/tpc.9.12.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koltai H, Dhandaydham M, Opperman C, Thomas J, Bird D. Overlapping plant signal transduction pathways induced by a parasitic nematode and a rhizobial endosymbiont. Mol Plant Microbe Interact. 2001;14(10):1168–1177. doi: 10.1094/MPMI.2001.14.10.1168. [DOI] [PubMed] [Google Scholar]
- 43.Favery B, Complainville A, Vinardell JM, Lecomte P, Vaubert D, Mergaert P, Kondorosi A, Kondorosi E, Crespi M, Abad P. The endosymbiosis-induced genes ENOD40 and CCS52a are involved in endoparasitic–nematode interactions in Medicago truncatula. Mol Plant Microbe Interact. 2002;15(10):1008–1013. doi: 10.1094/MPMI.2002.15.10.1008. [DOI] [PubMed] [Google Scholar]
- 44.Mathesius U. Conservation and divergence of signaling pathways between roots and soil microbes-the Rhizobium-legume symbiosis compared to the development of lateral roots, mycorrhizal interactions and matode-induced galls. Plant Soil. 2003;255:105–119. doi: 10.1023/A:1026139026780. [DOI] [Google Scholar]
- 45.Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inze D, Beeckman T, Gheysen G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008;148(1):358–368. doi: 10.1104/pp.108.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quentin M, Abad P, Favery B. Plant-parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front Plant Sci. 2013;4:53. doi: 10.3389/fpls.2013.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cabrera J, Diaz-Manzano FE, Sanchez M, Rosso MN, Melillo T, Goh T, Fukaki H, Cabello S, Hofmann J, Fenoll C, Escobar C. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014;203(2):632–645. doi: 10.1111/nph.12826. [DOI] [PubMed] [Google Scholar]
- 48.Cabrera J, Fenoll C, Escobar C. Genes co-regulated with LBD16 in nematode feeding sites inferred from in silico analysis show similarities to regulatory circuits mediated by the auxin/cytokinin balance in Arabidopsis. Plant Signal Behav. 2015;10(3):e990825. doi: 10.4161/15592324.2014.990825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43(5):467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 50.Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129(2):747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006;39(1):248–262. doi: 10.1016/j.ympev.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16(1):47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Coudert Y, Dievart A, Droc G, Gantet P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol Biol Evol. 2013;30(3):569–572. doi: 10.1093/molbev/mss250. [DOI] [PubMed] [Google Scholar]
- 54.Kong Y, Xu P, Jing X, Chen L, Li L, Li X. Decipher the ancestry of the plant-specific LBD gene family. BMC Genom. 2017;18(Suppl 1):951. doi: 10.1186/s12864-016-3264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14(7):399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18(8):450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 57.Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ, Guyomarc’h S. Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development. 2016;143(18):3363–3371. doi: 10.1242/dev.135319. [DOI] [PubMed] [Google Scholar]
- 58.Goh T, Joi S, Mimura T, Fukaki H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012;139(5):883–893. doi: 10.1242/dev.071928. [DOI] [PubMed] [Google Scholar]
- 59.Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009;151(3):1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Z, Zhu J, Du X, Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta. 2012;236(4):1227–1237. doi: 10.1007/s00425-012-1673-3. [DOI] [PubMed] [Google Scholar]
- 61.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 63.Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. Auxin-dependent compositional change in mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad Sci USA. 2016;113(23):6562–6567. doi: 10.1073/pnas.1600739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Qu Y, Sheng L, Liu J, Huang H, Xu L. A simple method suitable to study de novo root organogenesis. Front Plant Sci. 2014;5:208. doi: 10.3389/fpls.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu X, Xu L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016;172(4):2363–2373. doi: 10.1104/pp.16.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43(1):118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 67.Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226(5):1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- 68.Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57(4):626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 69.Yu J, Liu W, Liu J, Qin P, Xu L. Auxin control of root organogenesis from callus in tissue culture. Front Plant Sci. 2017;8:1385. doi: 10.3389/fpls.2017.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu C, Cao H, Zhang Q, Wang H, Xin W, Xu E, Zhang S, Yu R, Yu D, Hu Y. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat Plants. 2018;4(2):108–115. doi: 10.1038/s41477-017-0095-4. [DOI] [PubMed] [Google Scholar]
- 71.Lee K, Park OS, Seo PJ. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci Signal. 2017;10(507):eaan0316. doi: 10.1126/scisignal.aan0316. [DOI] [PubMed] [Google Scholar]
- 72.Ikeuchi M, Shibata M, Rymen B, Iwase A, Bagman AM, Watt L, Coleman D, Favero DS, Takahashi T, Ahnert SE, Brady SM, Sugimoto K. A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol. 2018 doi: 10.1093/pcp/pcy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013;73:212–224. doi: 10.1111/tpj.12013. [DOI] [PubMed] [Google Scholar]
- 74.Liu W, Xu L. Recruitment of IC-WOX genes in root evolution. Trends Plant Sci. 2018;23(6):490–496. doi: 10.1016/j.tplants.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 75.Nardmann J, Werr W. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol Biol. 2012;78(1–2):123–134. doi: 10.1007/s11103-011-9851-4. [DOI] [PubMed] [Google Scholar]
- 76.Feng Z, Sun X, Wang G, Liu H, Zhu J. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann Bot. 2012;110(1):1–10. doi: 10.1093/aob/mcs019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, Casimiro I, Hill K, Benkova E, Fukaki H, Brady SM, Scheres B, Peret B, Bennett MJ. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development. 2016;143(18):3340–3349. doi: 10.1242/dev.136283. [DOI] [PubMed] [Google Scholar]
- 78.Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17(5):1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taramino G, Sauer M, Stauffer JL, Jr, Multani D, Niu X, Sakai H, Hochholdinger F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 2007;50(4):649–659. doi: 10.1111/j.1365-313X.2007.03075.x. [DOI] [PubMed] [Google Scholar]
- 80.Majer C, Xu C, Berendzen KW, Hochholdinger F. Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.) Philos Trans R Soc Lond B Biol Sci. 2012;367(1595):1542–1551. doi: 10.1098/rstb.2011.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coudert Y, Le VA, Adam H, Bes M, Vignols F, Jouannic S, Guiderdoni E, Gantet P. Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of post-embryonic root formation. New Phytol. 2015;206(1):243–254. doi: 10.1111/nph.13196. [DOI] [PubMed] [Google Scholar]
- 82.Xu C, Tai H, Saleem M, Ludwig Y, Majer C, Berendzen KW, Nagel KA, Wojciechowski T, Meeley RB, Taramino G, Hochholdinger F. Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol. 2015;207(4):1123–1133. doi: 10.1111/nph.13420. [DOI] [PubMed] [Google Scholar]
- 83.Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, Magyar Z, Kamei CL, Koncz C, Bogre L, Persiau G, De Jaeger G, Friml J, Simon R, Beeckman T, De Veylder L. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell. 2011;23(10):3671–3683. doi: 10.1105/tpc.111.088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang NY, Lee HW, Kim J. The AP2/EREBP gene PUCHI Co-Acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in Arabidopsis. Plant Cell Physiol. 2013;54(8):1326–1334. doi: 10.1093/pcp/pct081. [DOI] [PubMed] [Google Scholar]
- 85.Lee HW, Kim J. EXPANSINA17 upregulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013;54(10):1600–1611. doi: 10.1093/pcp/pct105. [DOI] [PubMed] [Google Scholar]
- 86.Lee HW, Cho C, Kim J. Lateral Organ Boundaries Domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015;168(4):1792–1806. doi: 10.1104/pp.15.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jeon E, Young Kang N, Cho C, Joon Seo P, Chung Suh M, Kim J. LBD14/ASL17 positively regulates lateral root formation and is involved in ABA response for root architecture in Arabidopsis. Plant Cell Physiol. 2017;58(12):2190–2201. doi: 10.1093/pcp/pcx153. [DOI] [PubMed] [Google Scholar]
- 88.Lee HW, Kang NY, Pandey SK, Cho C, Lee SH, Kim J. Dimerization in LBD16 and LBD18 transcription factors is critical for lateral root formation. Plant Physiol. 2017;174(1):301–311. doi: 10.1104/pp.17.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol. 2013;23(11):956–962. doi: 10.1016/j.cub.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 90.Du Y, Scheres B. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc Natl Acad Sci USA. 2017;114(44):11709–11714. doi: 10.1073/pnas.1714410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ge Y, Liu J, Zeng M, He J, Qin P, Huang H, Xu L. Identification of wox family genes in Selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front Plant Sci. 2016;7:93. doi: 10.3389/fpls.2016.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]