Abstract
GAGA factor of Drosophila melanogaster (DmGAF) is a multifaceted transcription factor with diverse roles in chromatin regulation. Recently, ThPOK/c-Krox was identified as its vertebrate homologue (vGAF), which has a basic domain structure similar to DmGAF and is decorated with a number of post-translationally modified residues. In vertebrate genomes, vGAF associates with purine-rich GAGA sequences and performs diverse chromatin-mediated functions, viz., gene activation, repression and enhancer blocking. Expansion of regulatory chromatin proteins with the acquisition of PTMs appears to be the general trend that facilitated the evolution of complexity in vertebrates. Here, we compare the structural and functional features of vGAF with those of DmGAF and also assess the possible functional redundancy among paralogues of vGAF. We also discuss the underlying mechanisms which aid in the diverse and context-dependent functions of this protein.
Keywords: GAGA factor, ThPOK, c-Krox, ZBTB, Chromatin regulation
Introduction
Transcriptional regulation is fundamental to all life processes. In eukaryotes, the expression of transcriptional units is regulated by an array of chromatin-associated proteins, CAPs [1]. CAPs regulate transcription at various hierarchal levels starting from modulating the local chromatin environment for promoter accessibility to the initiation of the transcription and, subsequently, transcriptional elongation and termination. Many of the CAPs that are involved in these hierarchal processes either activate or repress transcription based on specific structural domains associated with them [2–4]. Contrary to this, there are several CAPs, which perform context-dependent functions using their isoform, specific protein interacting partners or post-translational modifications [5, 6]. One such multifunctional protein is GAGA factor (GAF) which binds to GA-rich DNA sequences. Drosophila melanogaster GAF, DmGAF, is encoded by the Trithorax-like (Trl) gene [7]. DmGAF was first identified as a sequence-specific activator of the Ubx promoter [8]. Subsequent studies showed that DmGAF not only associates with developmentally important genes, but it is also present on the promoters of many housekeeping and inducible genes [9–11]. Further studies showed that DmGAF is implicated in various nuclear processes like gene activation, polycomb-mediated silencing, enhancer blocking, position effect variegation and chromosomal segregation [7, 8, 12–15]. Despite playing a prominent role in transcriptional regulation and genome organization, the vertebrate homologue of GAF remained elusive for a long time until the identification of ThPOK/c-Krox, which is encoded by the zbtb7b gene in mice and humans [16].
Here, we present an overview of various nuclear processes where vGAF is implicated along with a parallel comparison of DmGAF. Further, we collate the existing studies on vGAF to discuss possible mechanisms behind the associated nuclear processes and show its potential role in gene regulation and cell lineage maintenance.
Structure of GAF proteins
Domain structure
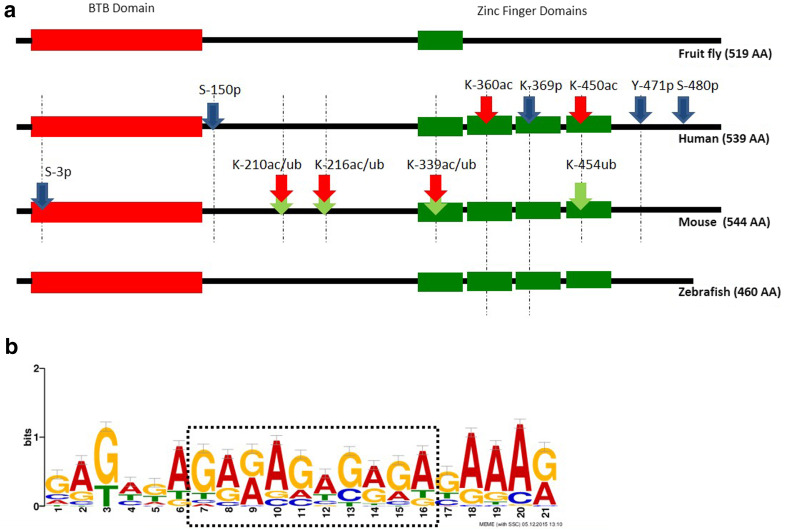
The zbtb7b gene encodes the GAF protein in vertebrates. While the length of the vGAF in humans (539 amino acid), mice (544 amino acid), zebrafish (460 amino acid), fugu (541 amino acid) and other vertebrate species is different [16, 17], the basic domain structure is well conserved. All the vGAF proteins have an N-terminal BTB/POZ domain and four zinc finger domains at their C-termini (Fig. 1a). Out of four zinc finger domains, the first three are C2H2 type, while in the fourth the last histidine is not conserved. Multiple sequence alignment of GAF from various vertebrate species shows that the first two zinc fingers have a very high level of sequence conservation in comparison to the last two zinc fingers. Similarly, the N-terminal BTB/POZ domain is conserved in these proteins except that one of the paralogues in mammals (ZBTB7B) has a 10–14 amino acid insertion in the BTB/POZ domain, which is otherwise absent in lower vertebrates [16]. Like vGAF, DmGAF has the same domain structure except that it has only one zinc finger domain and an additional polyglutamine tail at its C-terminus [18]. The nuclear magnetic resonance (NMR) structure of DNA-binding domain of DmGAF with 16 bp oligonucleotide shows that a single zinc finger domain and two basic regions BR1 and BR2, present on the N-terminal side of the zinc finger, provide specific contacts to all the bases in the GAGAG pentamer [19]. Molecular modelling of the DNA-binding domain of zebrafish GAF (DrGAF) shows that four zinc fingers adapt a structure which allows the recognition of polypurines in a continuous stretch of 12 bp DNA [16]. Sequence analysis of 33 known binding sites of vGAF in mouse genome also shows a long consensus sequence of 21 bp with a central (GA) × 5 motif embedded in the purine-rich DNA strand (Fig. 1b).
Fig. 1.
a GAF domain structure and post-translational modifications: all GAF proteins contain a BTB domain (red); DmGAF contains one zinc finger domain, while vGAF contains four zinc finger domains (green). PTM site phosphorylation (blue), acetylation (red), and ubiquitination (green) are shown by arrows marked with amino acid residue position. Dotted lines show the extent of conservation of these amino acid residue in human, mouse and zebrafish proteins. b The DNA-binding motif of vGAF proteins. Thirty-three known binding sites of vGAF (ThPOK) in mouse genome were taken from literature [21, 39, 51]. MEME analysis done on these sequences shows a 21 bp long binding motif with a central polypurine-rich sequence
Multimerization of GAF
The BTB/POZ domain is a protein–protein interaction motif found in a large number of proteins across eukaryotes. The presence of this domain at the N-terminus of the GAF proteins raises the possibility of higher-order protein structures through di/oligomerization. Indeed, DmGAF has been shown to oligomerize and form up to octamers on a DNA substrate. Contrary to this, ThPOK (vGAF) has only been shown to form dimers, both in vivo and in vitro [20, 21]. Though the oligomerization of DmGAF helps in the co-operative binding of protein on the promoter, such effects of vGAF dimerization are yet to be understood [22, 23].
Post-translational modifications
Post-translational modifications (PTMs) on transcription factors influence their sub-cellular localization, stability, sequence-specific DNA binding, and their interaction with other proteins [6]. GAF is one such transcription factor where at least the DNA-binding functions and stability of the protein are regulated by an array of PTMs. In D. melanogaster, GAF is one of the targets of casein kinase 2 (CK2) and undergoes phosphorylation at S378 and S388. Similarly, P300/CBP-associated factor (PCAF) also targets the DmGAF and acetylates the protein at K325 and K373. These residues are in the DNA-binding domain of the protein, and therefore modification of these residues decreases the affinity of the protein for DNA [24, 25]. On the other hand, proteome-wide studies in human cells show that vGAF is a phospho-protein with at least five phosphorylated Ser/Thr residues which are conserved in the mouse (Fig. 1a) [26–28]. Other than phosphorylation, PTMs like acetylation and ubiquitination can also decorate vGAF. TIP60 (Tat-interactive protein, 60 kDa) is shown to associate with human vGAF and acetylate the protein at the K360 residue resulting in the higher stability of the protein [29]. Similarly, the histone acetyltransferase, p300, differentially acetylates mouse vGAF (MmGAF) protein at K210, K216, and K339, in a cell-type-specific manner. Acetylation of MmGAF at these sites competes with ubiquitination and results in differential stability of the protein in cell types where p300 expression is low [30].
Functions of GAF proteins
As discussed above, GAF proteins have two structural domains, viz., zinc fingers and BTB domain. The BTB domain is a protein–protein interaction domain, while zinc fingers bind to DNA in a sequence-specific manner. These domains endow GAF with an ability to bind at multiple loci in the genome and recruit proteins of diverse functions, thus enabling GAF to perform context-dependent regulatory functions [31, 32]. These diverse regulatory functions finally contribute towards important biological processes, viz., cellular differentiation and maintenance of cell-type identity. Here, we describe various regulatory functions of vertebrate GAF protein and discuss in brief the biological processes associated with them.
GAF in gene activation
GAF has multifaceted roles in gene regulation which are due to the context-dependent binding in the genome, different interacting partners and post-translational modifications. Although the role of DmGAF in gene activation is well studied for several genes such as Ubx, hsp26, and alpha-tubulin [8, 10, 13], the functional roles of vGAF in such nuclear processes are still emerging. The first report on the regulatory potential of vGAF came from a study where ThPOK/c-Krox was first identified as a tissue-specific activator of type I collagen gene in the mouse dermis [33, 34]. This regulatory function is well conserved in humans and depends on the sequence-specific binding at GAGA sites [35]. Recent studies have shown that vGAF-binding sites are also present in promoters of the SOCS (suppressor of cytokine signalling) genes Socs1 and Cish. Transgenic mouse lines overexpressing vGAF show an increase in transcription of these genes, suggesting a direct role of vGAF in their activation [36]. Similarly, vGAF has been shown to mediate the activation of TNF-alpha alleles in LPS-stimulated macrophages [37].
GAF in gene repression
Several studies suggest a role of vGAF in gene activation, but it is also shown that depending on the context vGAF can create and maintain repressed chromatin states and thus negatively influence transcription. Recent studies on the regulation of the collagen gene in normal and scleroderma human fibroblast cells show that vGAF not only activates the type I collagen gene, but is also essential for recruiting the p65 subunit of NF-kappa-B which decreases transcription from type I collagen promoters, probably to fine-tune protein levels of collagen [38]. Similarly, it is also known that vGAF can act as negative regulator of genes such as UDP glucose dehydrogenase, eomesodermin, cd8, Igh, and cyp3a [21, 29, 39, 40]. Interestingly, vGAF-mediated silencing tends to associate the affected loci with transcriptionally inactive nuclear compartments, such as heterochromatin and LAD (lamina-associated domains), further suggesting its importance in chromatin compaction and repression [21, 39]. In D. melanogaster, GAF is known to associate with polycomb response elements (PRE) [14, 41]. DmGAF co-immunoprecipitates with PRC1 and also facilitates the recruitment of PHO on the chromatin [42, 43]. These studies demonstrate its involvement in polycomb-mediated repression. Also, in vertebrates, sequence analysis of mammalian PREs shows specific enrichment of GAF motifs as compared to random DNA sequences [44]. Indeed, a recent study demonstrates that vGAF binds at the evx2-hoxd13 PRE as well [45]. Another study on the transcriptional regulation of 4q35 genes through D4Z4 repeat elements reported that these repeats have sequence motifs similar to Drosophila PREs and recruit members of PRC1 (RING1B, BMI1), PRC2 (EZH2, SUZ12), and H2Aub1. Intriguingly, vGAF binds at this locus with another mammalian PRC2 recruiter Jarid2 [46]. Similarly, in CHO cell lines, vGAF binds at an evolutionarily conserved site in hoxd11 promoter along with PRC1/PRC2 members, H2Aub1 and Jarid2 [46]. Although these reports indicate the importance of vGAF in the PcG system, further studies are required to understand its precise role in PcG recruitment.
GAF in enhancer blocking activity
Polycomb-mediated repressive memory keeps genes of non-specific lineages in a silent state throughout subsequent mitotic generations after a particular cell type differentiates from pluripotent cells. Another layer of regulation is laid by cell-type-specific enhancers which allow the activation of lineage-specific genes in specific chromatin domains. These chromatin domains are marked by insulator elements which prevent the inter-domain cross talk among regulatory elements. DmGAF associates with insulator elements and is essential for their function in D. melanogaster [47]. Interestingly, DmGAF-associated insulator elements play an important role in the regulation of the Hox cluster of D. melanogaster. The SF1 element in the antennapedia complex insulates regulatory sequences of the scr and ftz genes, while the Fab7 element in the bithorax complex ensures autonomous function of iab6 and iab7 regulatory domains. Both these elements have GAGA sequences and show the involvement of DmGAF in their enhancer blocking activity [15, 48]. The role of GAGA factor-associated insulators in the regulation of Hox genes is conserved during evolution of vertebrates as well. An evolutionarily conserved GAGA-rich DNA fragment between the mouse evx2 and hoxd13 genes shows insulator activity in transgenic assays [49, 50]. Interestingly, the same element binds to DmGAF when tested in transgenic Drosophila, while in mammalian cell culture it associates with vGAF. Moreover, several intergenic elements in Hox clusters of mice that show enhancer blocking activity in mammalian cell culture associate with vGAF and not with CTCF, the only known boundary factor in vertebrates. This further suggests the evolutionarily conserved role of GAF in the regulation of Hox clusters [51].
GAF in the maintenance of cell lineage and mitotic memory
A closer look at the genes which are known to be regulated by vGAF suggests that most of them are cell lineage specific and developmentally important. Down-regulation of vGAF in differentiated CD4+ T cells results in spurious activation of genes belonging to the CD8+ cell lineage [52]. Similarly, in murine fibroblasts, knockdown of vGAF results in the release of developmentally important loci from the repressive environment of the nuclear lamina [39]. These results suggest that vGAF is important for the maintenance of cell lineage after differentiation. Furthermore, vGAF remains attached to its interphase-binding sites on the chromosome during mitosis [39]. The concept wherein a sequence-specific DNA-binding factor remains attached to condensed mitotic chromosome during cell division is termed “mitotic bookmarking”. It has been suggested that these transcription factors remain bound to DNA during cell division and, upon mitotic exit, stably silence gene expression [53, 54]. We speculate that binding of vGAF at certain developmentally important loci throughout the cell cycle ensures the stable silencing of these genes and thereby contributes towards the maintenance of cell lineage. Interestingly, a recent study shows that vGAF can act as an oncogene, as constitutive expression of this protein in mouse T-lymphocytes leads to aggressive metastatic lymphomas which are clonally derived from DN4 T-lymphocytes [55]. This suggests that the forced expression of vGAF in these cells alters the cell lineage identity and imparts the self-renewing capacity to DN4 T-lymphocytes which otherwise do not express vGAF. This idea is consistent with the observation that vGAF expressing DN4 lymphoma cells show increased expression of hematopoietic stem cell markers such as sca1 and c-kit [55]. Taken together, these observations indicate that vGAF is essential for maintaining cellular lineages and its misexpression can lead to disease conditions.
Paralogues of vertebrate GAF protein and functional redundancy
The loss of function homozygous mutant of vGAF shows 75% embryonic lethality in mice. Surviving adults show CD4+ T cell deficiency, low fecundity in females and corneal defects in old mice. However, heterozygous mutants do not show any detectable phenotype [56, 57]. Later studies focusing on lineage commitment in T cells report vGAF/ThPOK knockout mice exhibiting a similar CD4+ T-cell deficiency. Contrary to the expectations from a gene involved in diverse nuclear processes, vGAF knockout mice do not show severe phenotypes in multiple tissues. However, it is observed that in mammals, knockouts of seemingly very important genes sometimes do not show severe phenotypes. This is due to a resilient transcriptional network, constructed through paralogous genes which is the result of genome duplication and selective retention of developmentally important genes during vertebrate evolution. For example, invertebrates generally possess one gene for each member of the PRC1 and PRC2 complex, while in vertebrates the expansion of these complexes leads to the presence of multiple paralogues for each PcG member. Bioinformatic and nuclear localization studies suggest that each of these paralogues has acquired novel non-redundant functions and contributes to the complexity of the epigenetic machinery in vertebrates [58]. Multiple paralogues within PRC1 and PRC2, however, also show a degree of functional redundancy during embryonic development and lymphoid differentiation, respectively [59, 60]. On the same lines, DmGAF is encoded by a single gene in D. melanogaster, while in vertebrates vGAF (ZBTB7B/ThPOK) has two known paralogues, namely, ZBTB7A (LRF/Pokemon) and ZBTB7C (Apm1). These paralogues of vGAF have acquired context-dependent novel functions in gene regulatory processes [61–63]. ZBTB7A is a key transcription factor that regulates the B versus T lymphoid cell fate decision, while ZBTB7C has recently been identified as the molecular switch for transcription of the matrix metalloproteinase genes [64–66]. Multiple sequence alignment of all three paralogues from different vertebrate species shows a high sequence similarity in both BTB and zinc finger domains. However, a small insertion in the BTB domain of mammalian ZBTB7B suggests that this mammalian GAF may have acquired unique features of specific protein–protein interaction compared to the other two paralogues (Fig. 2a). Interestingly, key residues in the DNA-binding zinc finger domains of all three paralogues of vGAF show very high sequence conservation, indicating the commonality in DNA-binding preferences of these proteins (Fig. 2b). Taken together, these studies imply that the high degree of sequence conservation in structural domains of these proteins indicate a degree of functional redundancy among these proteins, in addition to the unique functions that they acquired during evolutionary diversification. Initial studies show that all the three paralogues can modulate the promoter activity of multiple extracellular matrix genes in NIH3T3 cells, suggesting a redundant function of these proteins in regulating the genes of the extracellular matrix [67]. Furthermore, ZBTB7A co-immunoprecipitates with vGAF [30] and these two proteins perform redundant functions in the maintenance of CD4+ T cells and their differentiation into helper effector T cells [52, 68]. These observations suggest that closely related paralogues of vGAF may have overlapping functions and thus can compensate for vGAF deficiency at least in certain tissue types.
Fig. 2.
Multiple sequence alignment of a BTB and b zinc finger domains of all paralogues of vGAF from different species of vertebrate using CLUSTAL omega. All paralogues show a high degree of alignment except mammalian ZBTB7B proteins, which have an insertion within the BTB domain. Red arrows represent the positions of amino acid residues conserved in all paralogues which make contact with DNA base pairs, while green arrows represent the zinc ion-binding amino acid residues
GAF protein: a protein for multiple functions
Zinc fingers and the BTB domain are two structural modules in GAF proteins that confer sequence-specific DNA-binding and protein–protein interaction ability to them. Sequence-specific DNA binding leads to tethering of GAF molecules at genomic regions enriched in GAGA sequences, while the BTB domain recruits other functionally diverse proteins at these regions of the genome (Fig. 3a). Interaction of GAF with diverse proteins might further be fine-tuned by an array of PTMs present on the GAF protein, while its targeting to different loci may also depend on the genomic context of its binding, viz., nature of the surrounding chromatin environment and the local cis-regulatory elements [24, 25, 30–32]. For example, DmGAF recruits the protein lola-like at the iab7 PRE which further helps in assembling polycomb-repressive complexes at this locus, while it recruits TAF3 at the Ubx promoter leading to activation of gene expression [31, 32]. These findings suggest that the regulatory outcome at a locus is defined by the type of transcription factors recruited by DmGAF. It is highly probable that specific PTMs of GAF add more selectivity to its DNA-binding ability as well as protein interactors. Studies on vGAF also suggest that this protein associates with a variety of nuclear factors which are involved both in activation and repression. Many of the histone deacetylases (HDACs) such as HDAC3, HDAC4, HDAC5, and HDAC10 co-immunoprecipitate with vGAF [21, 39]. Functionally also, vGAF-dependent silencing of cd8, IgH, and cyp3a loci is sensitive to TSA (an HDAC class II inhibitor), suggesting HDAC-mediated gene silencing by vGAF at these loci [21, 39]. Not only HDACs, but also vGAF can interact and recruit many other proteins which act as co-repressors, such as BCL6, ZBTB7A, ZBTB5, and p65 [38, 67, 69]. Interestingly, vGAF also interacts with another set of nuclear factors which are involved in gene activation including sp1, sp3, CEBPZ, and MORF4L2 [30, 35, 69]. There are a number of other nuclear proteins as well, which are not directly linked to gene activation or repression but still associate with vGAF [70]. These divergent protein interactions of vGAF suggest that it could be a part of the protein complexes performing diverse functions. Furthermore, cell-type-specific availability of its interacting partners may add another layer of regulation on the type of protein complexes recruited by vGAF at its binding sites in the genome [29, 30].
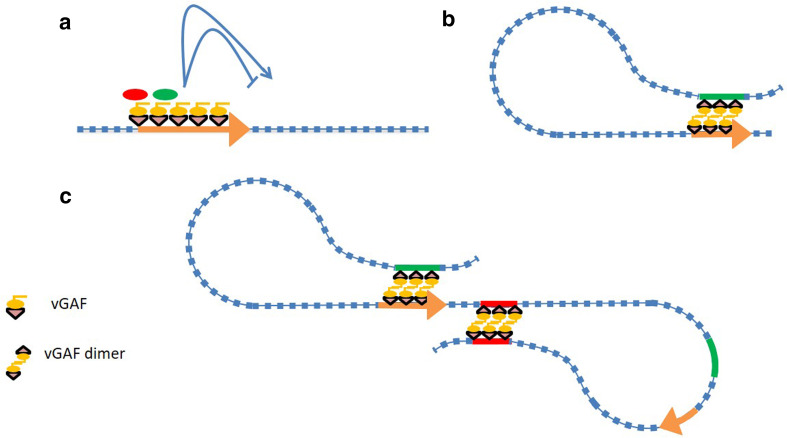
Fig. 3.
Mechanistic model of vGAF-mediated gene regulation. a vGAF molecules tethered to promoters (orange arrows) can recruit diverse proteins (red and green ovals) through heterologous protein interaction that can result in either gene activation or repression. b vGAF containing protein complexes assembled at DRE (green bars)/promoter (orange arrows) can interact through vGAF dimerization or through heterologous protein interaction between vGAF and other proteins. This may result in activation or repression at the promoter, based on the nature of the DRE involved. c vGAF molecules tethered to the insulator element (red bars) can come close and form dimers, thereby bringing these insulator elements together. This interaction between insulators results in looping out of the intervening DNA sequence, which can form a chromatin domain wherein possible interactions within the loop are allowed, while the interaction from outside of the loop is not allowed
GAF proteins can regulate gene expression through long-range DNA interactions as well. DmGAF has been shown to mediate the linking of an enhancer with its cognate promoter through self-oligomerization [71]. Interestingly, vGAF can also form dimers both in vivo and in vitro and has been shown to be involved in long-range DNA interaction of TNF-alpha alleles [20, 21, 37]. We speculate that certain aspects of vGAF-mediated gene activation, repression and enhancer blocking may involve heterologous/homologous protein–protein interactions and long-range DNA interactions. Protein complexes recruited by vGAF at distant regulatory elements (DRE) can physically interact with promoters via dimerization of vGAF molecules or through vGAF interacting proteins tethered to the promoter. Such interactions can bring both the promoter and the DRE in close proximity, which in turn facilitates the recruitment of activator or repressive complexes at promoters depending on the type of DREs involved (Fig. 3b). However, certain vGAF-associated DREs can interact mutually leading to the formation of distinct chromatin loops, thereby insulating the enhancer–promoter interaction within the loop from the regulatory elements present outside the loop (Fig. 3c).
DmGAF has been studied in different contexts and has been shown to play a role in diverse chromatin-associated processes. Multiple studies showed that DmGAF is involved in nucleosome rearrangement and creation of nucleosome-free regions through its interaction with NURF, FACT, or PBAP complexes [72–77]. Nucleosome-free regions created by DmGAF at promoters of multiple genes are essential for promoter-proximal pausing of PolII [78–80]. Studies on vGAF also suggest occupancy of this protein at nucleosome-free regions [51]; however, the chromatin remodelling proteins used in this process and the regulatory significance of these nucleosome-free regions are yet to be elucidated.
GAF protein on GAGA-repetitive DNA
GAF proteins preferentially bind to closely spaced (GA)n repeats. Interestingly, in the human genome more that 18% of genes have at least one simple sequence repeat (SSR) present in the promoter region [81, 82]. GA repeats in human gene promoters are shown to adopt secondary structures, and an increase in repeat number is associated with hyperacetylation of nucleosomes at promoters [83]. A bioinformatic analysis of the distribution of GA repeats 120 bp upstream of the TSS found that more than 2% of protein-coding genes in humans have at least one (GA)3 element in their promoters, excluding those promoters where GA repeat is present along with other classes of SSR [84]. Some of these genes are developmentally important and a few also show conservation of repeat association in their promoters across species. In addition, the number of repeat units is closely linked to the transcriptional output of the promoter [85, 86], pointing to the role of these repeat units in regulating the local chromatin structure. The in vivo relevance of this observation is linked to the unravelling of a GAGA repeat binding factor which could translate the number of GA repeat units into the expression output. Although such a protein factor remains to be identified, vGAF appears to be the most probable candidate as it binds to GAGA nucleotides and has the ability to multimerize. It will be interesting to see if vGAF actually modulates the activity of these promoters through their GA repeat elements.
Conclusions
The vertebrate GAGA factor has a domain structure similar to that of DmGAF and has the ability to form dimers both in vivo and in vitro. GAF is decorated with cell-type-specific post-translational modifications and interacts with a diverse set of nuclear factors. Being an important CAP, it plays a role in gene activation, repression, and enhancer blocking. vGAF has also been shown to be involved in the maintenance of cell lineage. Like many other important CAPs, vGAF has also expanded during vertebrate evolution and has two other paralogues, viz. ZBTB7A and ZBTB7C. These proteins have acquired novel functions and play important roles during development and differentiation. We propose that vGAF performs multiple context-dependent functions by virtue of its differential PTMs and through the recruitment of other regulatory protein at different loci in the genome. Furthermore, its ability to form dimers can play an important role in its involvement in long-range interactions. While ThPOK is very well studied in T-cell development and functions related to the immune system, its functional roles as vGAF in the development of other organs and processes are largely unexplored. The protein holds a potential to emerge as a regulator of chromatin organization, with regulatory consequences in multiple developmental and differentiation-related processes. To understand the cell-type-specific functions of vGAF, it is important to study the dynamics of genome-wide binding profiles of vGAF across different cell types and the genomic features associated with these sites. Also, the effect of overexpression and knockdown of vGAF on the transcriptome may further help in understanding the significance of genomic binding of vGAF. Examining the vGAF knockout mice for phenotypes other than the reported CD4+ T-cell deficiency may also reveal its functional features. In addition, it will be informative to understand the functional redundancy among vGAF paralogues and their contribution towards gene regulatory networks.
Acknowledgements
AS acknowledges the SRF fellowship from CSIR. RKM is thankful to CSIR (BSC0118) for the financial assistance. The authors thank Raghunand R. Tirumalai for critical reading of the manuscript and valuable suggestions.
Abbreviations
- BTB
Broad-complex, tramtrack and bric a brac
- BMI1
B lymphoma Mo-MLV insertion region 1 homologue
- CAP
Chromatin-associated proteins
- c-Krox
C-Kruppel-related zinc finger protein
- DmGAF
Drosophila melanogaster GAF
- DrGAF
Danio rerio GAF
- EZH2
Enhancer of zeste 2
- FACT
Facilitates chromatin transcription
- GAF
GAGA factor
- LPS
Lipopolysaccharides
- MmGAF
Mus musculus GAF
- NURF
Nucleosome remodelling factor
- PTM
Post-translational modification
- POZ
POxvirus and zinc finger domain
- PcG
Polycomb group
- PRC1
Polycomb repressive complex 1
- PRC2
Polycomb repressive complex 2
- PRE
Polycomb repressive element
- PBAP
Polybromo-associated Brm
- SOCS
Suppressor of cytokine signalling
- SUZ12
Suppressor of zeste 12
- ThPOK
Th-inducing POK
- TSA
Trichostatin A
- vGAF
Vertebrate GAF
References
- 1.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 2.Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 4.Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21(3):928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez AJ. Developmental role of transcription factor isoforms generated by alternative splicing. Dev Biol. 1995;172(2):396–411. doi: 10.1006/dbio.1995.8050. [DOI] [PubMed] [Google Scholar]
- 6.Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci. 2014;35(2):76–85. doi: 10.1016/j.tips.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371(6500):806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 8.Biggin MD, Tjian R. Transcription factors that activate the ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 9.Benyajati C, Ewel A, McKeon J, Chovav M, Juan E. Characterization and purification of Adh distal promoter factor 2, Adf-2, a cell-specific and promoter-specific repressor in Drosophila. Nucleic Acids Res. 1992;20(17):4481–4489. doi: 10.1093/nar/20.17.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odonnell KH, Wensink PC. Gaga factor and Tbf1 bind DNA elements that direct ubiquitous transcription of the Drosophila alpha-1-tubulin gene. Nucleic Acids Res. 1994;22(22):4712–4718. doi: 10.1093/nar/22.22.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Steensel B, Delrow J, Bussemaker HJ. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci USA. 2003;100(5):2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122(4):1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 13.Lu Q, Wallrath LL, Granok H, Elgin SCR. (Ct)N. (Ga)N repeats and heat-shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila-Hsp26 gene. Mol Cell Biol. 1993;13(5):2802–2814. doi: 10.1128/MCB.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21(4):1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168(3):1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J Mol Biol. 2010;400(3):434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Nagamine R, Korenaga H, Sakai M, Secombes CJ, Kono T. Characterization and expression analysis of Th-POK from the Japanese pufferfish, Takifugu rubripes . Comp Biochem Physiol B Biochem Mol Biol. 2013;164(2):124–132. doi: 10.1016/j.cbpb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Chromatin. Ga-ga over GAGA factor. Curr Biol. 1995;5(3):238–241. doi: 10.1016/S0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- 19.Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. The solution structure of a specific GAGA factor–DNA complex reveals a modular binding mode. Nat Struct Biol. 1997;4(2):122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 20.Bonchuk A, Denisov S, Georgiev P, Maksimenko O. Drosophila BTB/POZ domains of “ttk Group” can form multimers and selectively interact with each other. J Mol Biol. 2011;412(3):423–436. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 21.Rui J, Liu H, Zhu X, Cui Y, Liu X. Epigenetic silencing of CD8 genes by ThPOK-mediated deacetylation during CD4 T cell differentiation. J Immunol. 2012;189(3):1380–1390. doi: 10.4049/jimmunol.1201077. [DOI] [PubMed] [Google Scholar]
- 22.Espinas ML, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem. 1999;274(23):16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 23.Katsani KR, Hajibagheri MAN, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18(3):698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonet C, Fernandez I, Aran X, Bernues J, Giralt E, Azorin F. The GAGA protein of Drosophila is phosphorylated by CK2. J Mol Biol. 2005;351(3):562–572. doi: 10.1016/j.jmb.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 25.Aran-Guiu X, Ortiz-Lombardia M, Oliveira E, Costa CB, Odena MA, Bellido D, Bernues J. Acetylation of GAGA factor modulates its interaction with DNA. Biochemistry. 2010;49(43):9140–9151. doi: 10.1021/bi1004427. [DOI] [PubMed] [Google Scholar]
- 26.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4(179):rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(D1):D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YY, Tsun A, Gao ZM, Han ZJ, Gao YY, Li ZY, Lin F, Wang Y, Wei G, Yao ZJ, Li B. 60-kDa Tat-interactive protein (TIP60) positively regulates Th-inducing POK (ThPOK)-mediated repression of eomesodermin in human CD4(+) T cells. J Biol Chem. 2013;288(22):15537–15546. doi: 10.1074/jbc.M112.430207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Zhang JL, Rui JX, Liu XL. p300-mediated acetylation stabilizes the Th-inducing POK factor. J Immunol. 2010;185(7):3960–3969. doi: 10.4049/jimmunol.1001462. [DOI] [PubMed] [Google Scholar]
- 31.Chopra VS, Srinivasan A, Kumar RP, Mishra K, Basquin D, Docquier M, Seum C, Pauli D, Mishra RK. Transcriptional activation by GAGA factor is through its direct interaction with dmTAF3. Dev Biol. 2008;317(2):660–670. doi: 10.1016/j.ydbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Mishra K, Chopra VS, Srinivasan A, Mishra RK. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech Dev. 2003;120(6):681–689. doi: 10.1016/S0925-4773(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 33.Galera P, Musso M, Ducy P, Karsenty G. C-Krox, a transcriptional regulator of type-I collagen gene-expression, is preferentially expressed in skin. Proc Natl Acad Sci USA. 1994;91(20):9372–9376. doi: 10.1073/pnas.91.20.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galera P, Park RW, Ducy P, Mattei MG, Karsenty G. c-Krox binds to several sites in the promoter of both mouse type I collagen genes. Structure/function study and developmental expression analysis. J Biol Chem. 1996;271(35):21331–21339. doi: 10.1074/jbc.271.35.21331. [DOI] [PubMed] [Google Scholar]
- 35.Kypriotou M, Beauchef G, Chadjichristos C, Widom R, Renard E, Jimenez SA, Korn J, Maquart FX, Oddos T, Von Stetten O, Pujol JP, Galera P. Human collagen Krox up-regulates type I collagen expression in normal and scleroderma fibroblasts through interaction with Sp1 and Sp3 transcription factors. J Biol Chem. 2007;282(44):32000–32014. doi: 10.1074/jbc.M705197200. [DOI] [PubMed] [Google Scholar]
- 36.Luckey MA, Kimura MY, Waickman AT, Feigenbaum L, Singer A, Park JH. The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat Immunol. 2014;15(7):638–645. doi: 10.1038/ni.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stratigi K, Kapsetaki M, Aivaliotis M, Town T, Flavell RA, Spilianakis CG. Spatial proximity of homologous alleles and long noncoding RNAs regulate a switch in allelic gene expression. Proc Natl Acad Sci USA. 2015;112(13):E1577–E1586. doi: 10.1073/pnas.1502182112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beauchef G, Bigot N, Kypriotou M, Renard E, Poree B, Widom R, Dompmartin-Blanchere A, Oddos T, Maquart FX, Demoor M, Boumediene K, Galera P. The p65 subunit of NF-kappaB inhibits COL1A1 gene transcription in human dermal and scleroderma fibroblasts through its recruitment on promoter by protein interaction with transcriptional activators (c-Krox, Sp1, and Sp3) J Biol Chem. 2012;287(5):3462–3478. doi: 10.1074/jbc.M111.286443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149(7):1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 40.Beauchef G, Kypriotou M, Chadjichristos C, Widom RL, Poree B, Renard E, Moslemi S, Wegrowski Y, Maquart FX, Pujol JP, Galera P. c-Krox down-regulates the expression of UDP-glucose dehydrogenase in chondrocytes. Biochem Biophys Res Commun. 2005;333(4):1123–1131. doi: 10.1016/j.bbrc.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Hodgson JW, Argiropoulos B, Brock HW. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol Cell Biol. 2001;21(14):4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmoudi T, Zuijderduijn LM, Mohd-Sarip A, Verrijzer CP. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Res. 2003;31(14):4147–4156. doi: 10.1093/nar/gkg479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20(9):3187–3197. doi: 10.1128/MCB.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauer M, Trupke J, Ringrose L. The quest for mammalian Polycomb response elements: are we there yet? Chromosoma. 2015 doi: 10.1007/s00412-015-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasanthi D, Nagabhushan A, Matharu NK, Mishra RK. A functionally conserved Polycomb response element from mouse HoxD complex responds to heterochromatin factors. Sci Rep. 2013;3:3011. doi: 10.1038/srep03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12(21):3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22(12):3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagishi T, Ozawa M, Ohtsuka C, Ohyama-Goto R, Kondo T. Evx2-Hoxd13 intergenic region restricts enhancer association to Hoxd13 promoter. PLoS One. 2007;2(1):e175. doi: 10.1371/journal.pone.0000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasanthi D, Anant M, Srivastava S, Mishra RK. A functionally conserved boundary element from the mouse HoxD locus requires GAGA factor in Drosophila. Development. 2010;137(24):4239–4247. doi: 10.1242/dev.058701. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava S, Puri D, Garapati HS, Dhawan J, Mishra RK. Vertebrate GAGA factor associated insulator elements demarcate homeotic genes in the HOX clusters. Epigenetics Chromatin. 2013;6(1):8. doi: 10.1186/1756-8935-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, Bosselut R. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nat Immunol. 2014;15(10):947–956. doi: 10.1038/ni.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150(4):725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104(9):3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HO, He X, Mookerjee-Basu J, Zhongping D, Hua X, Nicolas E, Sulis ML, Ferrando AA, Testa JR, Kappes DJ. Disregulated expression of the transcription factor ThPOK during T-cell development leads to high incidence of T-cell lymphomas. Proc Natl Acad Sci USA. 2015;112(25):7773–7778. doi: 10.1073/pnas.1424104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kappes DJ, He X, He X. Role of the transcription factor Th-POK in CD4:CD8 lineage commitment. Immunol Rev. 2006;209:237–252. doi: 10.1111/j.0105-2896.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 57.He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- 58.Sowpati DT, Ramamoorthy S, Mishra RK. Expansion of the polycomb system and evolution of complexity. Mech Dev. 2015;138(Pt 2):97–112. doi: 10.1016/j.mod.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Lanzuolo C, Orlando V. Memories from the polycomb group proteins. Annu Rev Genet. 2012;46:561–589. doi: 10.1146/annurev-genet-110711-155603. [DOI] [PubMed] [Google Scholar]
- 60.Avgustinova A, Benitah SA. Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol. 2016;17(10):643–658. doi: 10.1038/nrm.2016.76. [DOI] [PubMed] [Google Scholar]
- 61.Lee DK, Kang JE, Park HJ, Kim MH, Yim TH, Kim JM, Heo MK, Kim KY, Kwon HJ, Hur MW. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J Biol Chem. 2005;280(30):27783–27791. doi: 10.1074/jbc.M504909200. [DOI] [PubMed] [Google Scholar]
- 62.Jeon BN, Kim MK, Choi WI, Koh DI, Hong SY, Kim KS, Kim M, Yun CO, Yoon J, Choi KY, Lee KR, Nephew KP, Hur MW. KR-POK interacts with p53 and represses its ability to activate transcription of p21WAF1/CDKN1A. Cancer Res. 2012;72(5):1137–1148. doi: 10.1158/0008-5472.CAN-11-2433. [DOI] [PubMed] [Google Scholar]
- 63.Jeon BN, Kim YS, Choi WI, Koh DI, Kim MK, Yoon JH, Kim MY, Hur B, Paik PD, Hur MW. Kr-pok increases FASN expression by modulating the DNA binding of SREBP-1c and Sp1 at the proximal promoter. J Lipid Res. 2012;53(4):755–766. doi: 10.1194/jlr.M022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeon BN, Yoon JH, Kim MK, Choi WI, Koh DI, Hur B, Kim K, Kim KS. Hur MW (2016) Zbtb7c is a molecular ‘off’ and ‘on’ switch of Mmp gene transcription. Biochim Biophys Acta. 1859;11:1429–1439. doi: 10.1016/j.bbagrm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316(5826):860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakurai N, Maeda M, Lee SU, Ishikawa Y, Li M, Williams JC, Wang L, Su L, Suzuki M, Saito TI, Chiba S, Casola S, Yagita H, Teruya-Feldstein J, Tsuzuki S, Bhatia R, Maeda T. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Investig. 2011;121(7):2583–2598. doi: 10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widom RL, Lee JY, Joseph C, Gordon-Froome I, Korn JH. The hcKrox gene family regulates multiple extracellular matrix genes. Matrix Biol. 2001;20(7):451–462. doi: 10.1016/S0945-053X(01)00167-6. [DOI] [PubMed] [Google Scholar]
- 68.Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, Naik S, dos Santos L, Wei L, Jenkins MK, O’Shea JJ, Belkaid Y, Bosselut R. The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity. 2012;37(4):622–633. doi: 10.1016/j.immuni.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabasi AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21(7):1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367(6463):525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 73.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83(6):1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 74.Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17(13):1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okada M, Hirose S. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol Cell Biol. 1998;18(5):2455–2461. doi: 10.1128/MCB.18.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lomaev D, Mikhailova A, Erokhin M, Shaposhnikov AV, Moresco JJ, Blokhina T, Wolle D, Aoki T, Ryabykh V, Yates JR, 3rd, Shidlovskii YV, Georgiev P, Schedl P, Chetverina D. The GAGA factor regulatory network: identification of GAGA factor associated proteins. PLoS One. 2017;12(3):e0173602. doi: 10.1371/journal.pone.0173602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakayama T, Shimojima T, Hirose S. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development. 2012;139(24):4582–4590. doi: 10.1242/dev.083246. [DOI] [PubMed] [Google Scholar]
- 78.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28(10):3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6(2):284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 80.Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 2015;11(3):e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolton KA, Ross JP, Grice DM, Bowden NA, Holliday EG, Avery-Kiejda KA, Scott RJ. STaRRRT: a table of short tandem repeats in regulatory regions of the human genome. BMC Genom. 2013;14:795. doi: 10.1186/1471-2164-14-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramamoorthy S, Garapati HS, Mishra RK. Length and sequence dependent accumulation of simple sequence repeats in vertebrates: potential role in genome organization and regulation. Gene. 2014;551(2):167–175. doi: 10.1016/j.gene.2014.08.052. [DOI] [PubMed] [Google Scholar]
- 83.Han YJ, de Lanerolle P. Naturally extended CT. AG repeats increase H-DNA structures and promoter activity in the smooth muscle myosin light chain kinase gene. Mol Cell Biol. 2008;28(2):863–872. doi: 10.1128/MCB.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heidari A, Nariman Saleh Fam Z, Esmaeilzadeh-Gharehdaghi E, Banan M, Hosseinkhani S, Mohammadparast S, Oladnabi M, Ebrahimpour MR, Soosanabadi M, Farokhashtiani T, Darvish H, Firouzabadi SG, Farashi S, Najmabadi H, Ohadi M. Core promoter STRs: novel mechanism for inter-individual variation in gene expression in humans. Gene. 2012;492(1):195–198. doi: 10.1016/j.gene.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 85.Valipour E, Kowsari A, Bayat H, Banan M, Kazeminasab S, Mohammadparast S, Ohadi M. Polymorphic core promoter GA-repeats alter gene expression of the early embryonic developmental genes. Gene. 2013;531(2):175–179. doi: 10.1016/j.gene.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 86.Morris EE, Amria MY, Kistner-Griffin E, Svenson JL, Kamen DL, Gilkeson GS, Nowling TK. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res Ther. 2010;12(6):R212. doi: 10.1186/ar3189. [DOI] [PMC free article] [PubMed] [Google Scholar]