Abstract
The organization of the nuclear periphery is crucial for many nuclear functions. Nuclear lamins form dense network at the nuclear periphery and play a substantial role in chromatin organization, transcription regulation and in organization of nuclear pore complexes (NPCs). Here, we show that TPR, the protein located preferentially within the nuclear baskets of NPCs, associates with lamin B1. The depletion of TPR affects the organization of lamin B1 but not lamin A/C within the nuclear lamina as shown by stimulated emission depletion microscopy. Finally, reduction of TPR affects the distribution of NPCs within the nuclear envelope and the effect can be reversed by simultaneous knock-down of lamin A/C or the overexpression of lamin B1. Our work suggests a novel role for the TPR at the nuclear periphery: the TPR contributes to the organization of the nuclear lamina and in cooperation with lamins guards the interphase assembly of nuclear pore complexes.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03037-0) contains supplementary material, which is available to authorized users.
Keywords: Translocated promoter region, TPR, Lamina, Lamins, Nuclear pore complex, Super-resolution imaging, Image analysis, Nucleus
Introduction
In metazoan cells, the nuclear periphery represents a complex environment with many functions including chromatin organization, transcription regulation, transport or communication with the cytoplasm. It is composed of a nuclear envelope (inner- and outer nuclear membrane enriched with a variety of associated proteins) underlain with a nuclear lamina and perforated with nuclear pore complexes (NPCs). The nuclear lamina forms a dense filamentous mesh at the inner face of the nuclear envelope. The lamina is composed of intermediate filament proteins, A- and B-type lamins. Whereas B-type lamins (lamin B1 and lamin B2) are encoded by two genes LMNB1 and LMNB2, A-type lamins, lamin A and lamin C, by a single gene, LMNA [1]. Both embryonic and adult mammalian cells express at least one B-type lamin, whereas the expression of A-type lamins is developmentally regulated [2]. Super-resolution microscopy analysis revealed that in mammalian nuclei, each of the lamin species (lamin A, C, B1 and B2) form distinct but overlapping networks. The feature of interconnections between individual networks is unclear and appears indirect [3–6].
The lamina defines the mechanical characteristics of the nucleus [2, 7] with a dominant role of A-type lamins in mechanical stiffness and strain resistance and B-type lamins contributing to the nuclear integrity and nuclear shape [8, 9]. The nuclear lamina further regulates the organization of chromatin, which is crucial for many biological functions such as transcription regulation, DNA replication and repair [1].
Both A- and B-type lamin networks are connected with NPCs [5, 6], which represent over 100 MDa large assemblies regulating transport of macromolecules via the nuclear envelope (NE). The functional characteristics of the inter-connections of NPCs and lamina are not fully understood. In mammalian cells, synchronous movement of NPCs with B-type lamins suggested a tight connection between B-type lamins and NPCs [10]. In addition, lamin depletion or mutation caused clustering and abnormal NPC distribution in C. elegans and in D. melanogaster [11, 12]. These data suggest the importance of B-type lamins for NPC anchoring and distribution. Experiments in mouse fibroblasts suggested that both A-type lamins as well as lamin B1, when expressed at sufficient levels, function redundantly to maintain an even distribution of NPCs over the NE [13]. A slight degree of NPC clustering has been shown as a feature of A-type lamin-deficient mouse embryonic fibroblasts (MEFs) [14]. Adopting combinatorial approach of super-resolution microscopy and BioID (proximity dependent biotinylation), lamin C but not lamin A was shown to be preferentially involved in NPCs attachment and distribution in MEF cells possibly via the interaction with the TPR [6]. Overall, the data show a potential role of both A- and B-type lamins in NPC anchoring and distribution.
In metazoan cells, NPCs are assembled by two different mechanisms in two cell-cycle stages, during nuclear assembly post anaphase and during nuclear growth in interphase. NPC-free islands typical for G1 cells that dispersed gradually during the progression of the cell to the S-phase were observed by Maeshima and colleagues [15]. These NPC-free zones result from the ‘core’ regions, where the nuclear membrane sealing is locally delayed in mitosis due to removal of spindle microtubules from DNA surface [16]. Thus, instead of rapid post-mitotic NPC assembly, these areas are populated by NPCs during G1 by de novo interphase assembly process. These NPC-free islands are void of lamin B but enriched in lamin A/C. Knock-down of lamin A/C resulted in a disappearance of NPC-free islands, whereas lamin A/C overexpression stabilized this feature implicating that lamin A/C played the essential role in NPC distribution. [17].
How are the NPCs interconnected with the nuclear lamina is also intensively studied. The NPCs are composed of approximately 30 proteins called nucleoporins (Nups) in multiple copies that fulfill various functions within the NPC [18]. The best described is the interaction of lamina and vertebrate Nup153 that along with translocated promoter region protein (TPR) creates a basket of the NPCs [19]. Nup153 has been shown to interact with C. elegans lamin [20] and with both, A- and B-type lamins in vertebrates [21]. Importantly, a depletion of Nup153 from Xenopus nuclei led to clustering of NPCs, loss of TPR from nuclear baskets of NPCs and to the mobility of the NPCs within the NE suggesting that NPCs lacking Nup153 loose the connection that anchors the NPCs within the NE [19].
TPR is a large protein of ~ 265 kDa. It contains several coiled-coil domains of which the initial N-terminal segment was shown to be able to form dimers [22]. Since its discovery, the TPR has been shown to function in many nuclear processes, namely in regulation of mRNA export [23–25], as a scaffolding element for signaling proteins [26], in mitotic processes [25, 27], in regulation of normal spindle assembly check point response [28, 29], and in regulation of G0–G1 arrest [30]. The TPR has been detected in oncogenic fusions with kinase domains of the trk, met and raf proto-oncogenes [31]. It is required for establishing heterochromatin exclusion zones in mammalian cells [32]. By chromatin immunoprecipitation combined with microarray hybridization, the TPR and Nup153 were shown to associate with active transcription regions and were employed in regulation of transcription and dosage compensation [33]. The TPR was shown to interact with lamin C (and to a lesser extent with lamin A); however, the functional implications of such partnership remain unclear [6]. Finally, the TPR was implicated in regulation of NPC density at the nuclear surface of mammalian cells. Whereas Funasaka and colleagues [34] showed reduction on NPC density, McCloskey and team [35] found dramatic increase in NPC density after TPR depletion in mammalian cells. Overall, with its strategic position at nuclear baskets, the TPR emerges as a multifunctional protein of a great importance.
Interactions of TPR and lamina and their functional significance remain unclear. In our work, we show that TPR co-immunoprecipitates with both lamin A/C and lamin B1 in mammalian HeLa cells. We used stimulated emission depletion (STED) to discern the lamina network organization and its changes after TPR depletion. The knock-down of TPR affected mRNA levels of lamin B1 within 48 h. Three and more weeks of TPR knock-down were followed by drop of lamin B1 and lamin A/C protein levels. In addition, lower levels of TPR resulted in altered NPCs distribution that could be rescued by simultaneous reduction of lamin A or by overexpression of lamin B1. The data suggest that TPR along with lamins functions as an important regulatory component of lamina which along with A- and B-type lamins controls NPC distribution.
Results
Nucleoporin TPR co-immunoprecipitates with lamin B1 and affects lamin B1 network organization
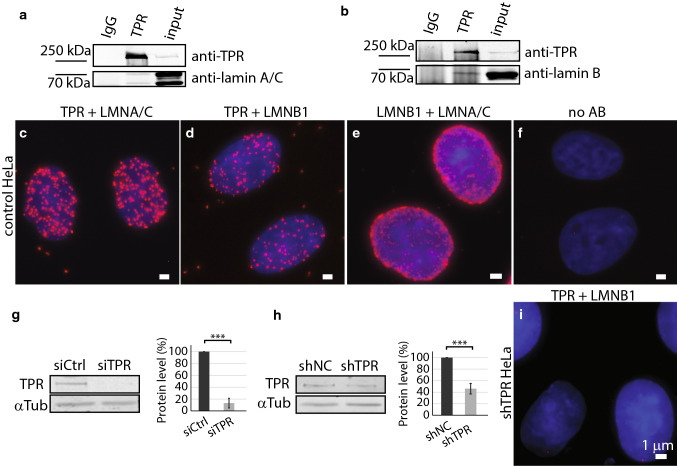
Nuclear pore basket protein TPR was shown to interact with both lamin C as well as lamin A [6]. By means of immunoprecipitation (IP), we tested the interaction of TPR and lamin B1. Consistently with previously published data, the TPR co-immunoprecipitated with lamin A/C (Fig. 1a), but in addition, we also detected the co-immunoprecipitation of TPR and lamin B1 (Fig. 1b). To confirm TPR and lamin B1 complex in vivo, we used the proximity ligation assay (PLA). Similarly to IPs, results proved very close proximity of TPR and lamin A/C (Fig. 1c) as well as TPR and lamin B1 (Fig. 1d), though the frequency of the other was lower. As a positive control, we combined antibodies against lamin A/C and lamin B1 (Fig. 1e); as a negative control, the primary antibodies were omitted (Fig. 1f). We also tested if the association between TPR and lamin B1 decreases in cells with reduced TPR amount. Thus, we depleted TPR protein levels by small interfering RNA (siTPR) as well as by short hairpin RNA complementary to mRNA sequence of TPR (shTPR) stably expressed in HeLa cells. The levels of TPR protein were specifically reduced after 48 h of siRNA treatment (Fig. 1g) as well as in cells with stably integrated shRNA (Fig. 1h). Importantly, the abundance of the TPR–lamin B1-positive spots decreased dramatically in cells with depleted TPR (Fig. 1i shows the result of PLA assay in HeLa cells with stably depleted TPR—shTPR).
Fig. 1.
TPR co-immunoprecipitates with lamin B1. a Immunoprecipitation of lamin A/C by anti-TPR antibody. b Immunoprecipitation of lamin B1 by anti-TPR antibody. Proximity ligation assay shows the close distance between TPR and lamin A/C (c), TPR and lamin B1 (d), lamin B1 and lamin A/C (e) and negative control with no primary antibody used (f). Western blotting and quantification show the reduction of TPR amount in HeLa cells with siTPR for 48 h: Student t test, T = 12.35, df = 5, P < 0.001 (g) or in stable HeLa cell line expressing shTPR: Student t test, T = 11.01, df = 7, P < 0.001. Graphs represent data from three independent biological replicates. 3.5 × 106 cells were used as a starting material. 10 μg of total protein per each lane was loaded for the analysis (h). i Proximity ligation assay detected reduced abundance of the TPR–lamin B1-positive signal in Hela cells with reduced TPR protein levels by shRNA. Scale bar 1 µm. ***P < 0.001
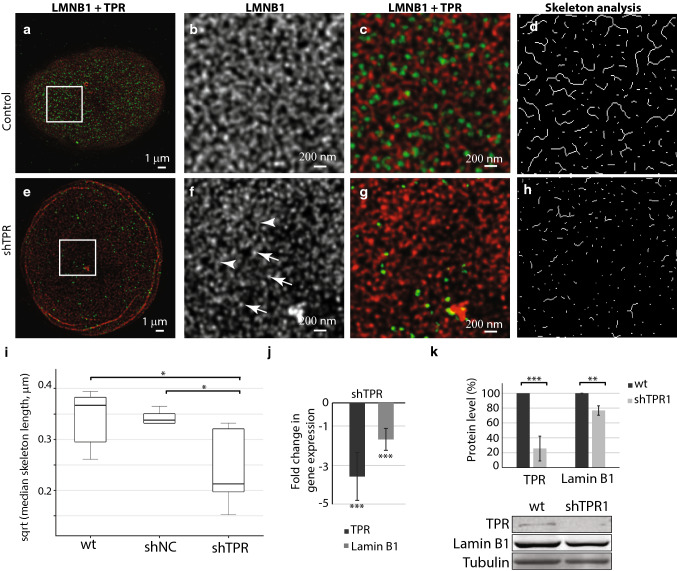
We were interested in how the TPR is involved in the organization of nuclear lamina network. We investigated the structure of the nuclear lamina formed by lamin B1 and A-type lamins in cells with reduced TPR levels and in control cells by means of super-resolution microscopy (STED). In agreement with previous studies [6, 36], lamin B1 formed filaments of similar intensity that appeared randomly organized into a dense meshwork in control HeLa cells (Fig. 2a–c and Fig. S1a–S1c). In contrast, silencing of TPR expression in both siTPR/shTPR HeLa cells caused defects in the meshwork of lamin B1 (Fig. 2e–g and Fig. S1e–g). In TPR-depleted cells, lamin B1 filaments appeared fragmented as reflected by brighter spots of lamin B1 (Fig. 2f and Fig. S1f, arrows) and much less intense filaments of lamin B1 (Fig. 2f and Fig. S1f, arrowheads). To analyze the lamina network in more detail, we used Skeleton plugin in Fiji (see “Materials and methods”). Briefly, we applied a thinning algorithm on segmented and binarized lamina filaments (Fig. 2d, h and Fig. S1d and S1h; for more details, see “Materials and methods”) and assessed the median length of skeletonized branches of lamin B1 in control and TPR-depleted HeLa cells (Fig. 2i and Fig. S1i). In shTPR/siTPR cell lines, the branches of lamin B1 as detected by Skeleton plugin were significantly shorter in comparison to control cells (shTPR: wt × shTPR T = 2.44, df = 6.98, P = 0.04; shNC × shTPR T = 2.79, df = 4.26, P = 0.047; siTPR: wt_si0 × siTPR T = − 0.22, df = 14.84, P = 0.82, siCtrl × siTPR T = 3.84, df = 15.98, P = 0.001). Such results indicate that TPR is important for the organization of lamin B1 network within nuclear lamina.
Fig. 2.
Lamin B1 network is affected by long-term (3 weeks) reduction of TPR amount. The lamin B1 network in control cell nucleus (a–d) and in a nucleus with reduced TPR (e–h) as visualized by STED. Cells were immunolabeled with rabbit anti-lamin B1 and mouse anti-TPR antibodies. Lamin B1 is shown in red, TPR is depicted in green. a, e Peripheral z-sections show the overview of lamin B1 organization. Insets in a and e correspond to b–d and f–h, respectively. Lamin B1 forms dense network of randomly organized filaments of similar intensity (b) that appear fragmented and spotted after TPR knock-down (f). Arrows point to brighter lamin B1 spots, arrowheads demarcate lamin B1 filaments of reduced intensity. c, g Merged image of TPR (in green) and lamin B1 (in red). d, h The graphical visualisation of skeleton analysis of b and f, respectively. i The box plots show square root transformed median lengths of skeletonized lamin B1 filaments in wild type (wt) HeLa cells, in cells expressing shNonCoding RNA (shNC) as well as in shTPR cell line. Wt × shTPR: T = 2.44, df = 6.98, P = 0.04; shNC × shTPR: T = 2.79, df = 4.26, P = 0.047; j fold change in mRNA levels of lamin B1 in shTPR cell line. TPR fold change: T = 8.0, df = 5; P < 0.001; LMNB1 fold change: T = 8.0, df = 5, P < 0.001. k Quantification of lamin B1 protein amount after TPR knock-down. TPR protein levels: Student t test: T = 21, df = 8, P < 0.001; LMNB1 protein levels: T = 4.7, df = 8, P < 0.01. Graph represents data from three independent biological replicates analyzed in triplicates. Lower panel shows representative western blots stained with anti-TPR, anti-lamin B1 and anti-tubulin antibodies in control and in shTPR cells. *P < 0.05, **P < 0.01, ***P < 0.001. 15 μg of total protein per each lane was loaded
Because the silencing of TPR expression caused partial fragmentation of lamin B1 network, in the next step, we tested if this lamin B1 network alteration resulted from reduced lamin B1 protein levels. We inspected both, mRNA and protein levels of lamin B1 after short- (48 h) or long (at least 3 weeks)-term TPR depletion (Fig. S1j and Fig. 2j). mRNA levels of lamin B1 decreased almost twice after 48 h of siRNA transfection (Fig. S1j) and remained reduced also after 3 weeks of TPR attenuation (Fig. 2j). Interestingly, we did not detect any changes in protein levels of lamin B1 within 48 h of TPR knock-down using siRNA (Fig. S1k). Because the lamin B1 protein has been reported to have life time of about 5–7 days [37], we hypothesized that the reduction of protein levels of lamin B1 might become prominent in 2–3 weeks after TPR knock-down. Indeed, when the TPR was depleted for a longer time period (at least 14 days), we detected a decrease in protein levels of lamin B1 (Fig. 2k). Reducing the TPR levels by shRNA resulted in a drop of lamin B1 amount down to approximately 77% of the lamin B1 in the control (Fig. 2k), which is in accordance with the decreased intensity of lamin B1 filaments analyzed by ScanR in shTPR cells but not in siTPR cells (Fig. S2). Taken together, these data indicate that TPR affects the lamin B1 network and may be involved in the regulation of lamin B1 gene expression.
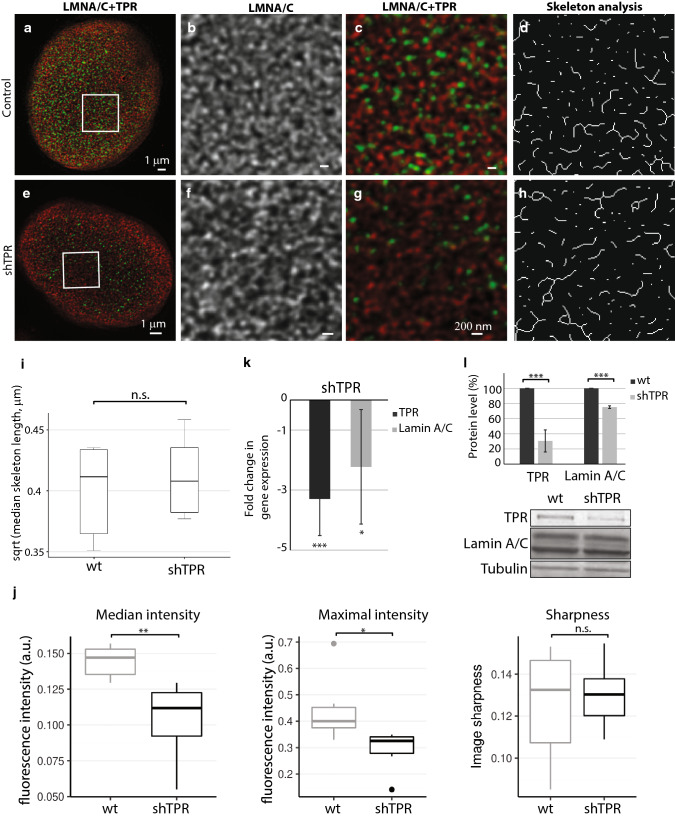
Because TPR interacts with A-type lamins, we also inspected the arrangement of lamin A/C network in cells with a reduced amount of TPR (Fig. 3 and Fig. S3). In the control cells, the lamin A/C network was composed of randomly organized branched filaments of high density that were clearly distinct at the darker background (Fig. 3a–c and Fig. S3a–c). In HeLa cells with reduced TPR levels, the visual appearance of lamin A/C network did not seem to differ from the control using nor short term (siTPR, Fig. S3e–h) nor long term silencing of TPR expression (shTPR, Fig. 3e–h). Indeed, the analysis of segmented and skeletonized (Skeleton plugin in Fiji) lamin A/C filaments in siTPR/shTPR cells did not show any difference in comparison to the control HeLa cells (Fig. 3i: T = − 0.51, df = 9.91, P = 0.58; Fig S3i: wt × siTPR: T = − 2.31, df = 3.55, P = 0.09; siCtrl × siTPR: T = − 0.79, df = 4.74, P = 0.46). To capture most of the qualitative changes in lamin A/C network, we inspected the median and the maximal intensity of acquired images as well as sharpness of images after TPR depletion using shRNA. In four out of five experiments, three and more weeks of TPR depletion by shRNA resulted in significant decrease of the maximal and the median fluorescent intensity of lamin A/C; the sharpness of the image, however, did not change (Fig. 3j; median intensity W = 35.5, P = 0.004, maximal intensity W = 33, P = 0.015, sharpness W = 17, P = 0.93). Accordingly, both protein and mRNA levels of lamin A/C dropped after long-term TPR depletion (Fig. 3k, i) but not in the cells treated with siTPR for 48 h where we only observed the reduction of mRNA of lamin A (Fig. S3j and S3k). Thus, the data suggest that TPR does not significantly affect the organization of lamin A/C network but it might participate in the regulation of the expression of the lamin A/C.
Fig. 3.
Lamin A/C network is not affected by long-term (3 weeks) reduced TPR amount by stable expression of shTPR. The lamin A/C network in control HeLa cell nucleus (a–d) and in a cell nucleus with reduced TPR amount (e–h) as visualized by STED. Cells were immunolabeled with mouse anti-lamin A/C (in red) and rabbit anti-TPR (in green). a, e Peripheral z-sections show the overview of lamin A/C organization. Insets in a and e correspond to b–d and f–h, respectively. Lamin A/C forms dense network of randomly organized filaments (b) that does not change in response to reduced TPR amount (f). c, g Merged image of lamin A/C (in red) and TPR (in green). d, h The graphical visualisation of skeleton analysis of the insets b and f, respectively. i Box plot shows square root transformed median lengths of skeletonized lamin A/C filaments in wild-type (wt) HeLa cells and in shTPR. Wt × shTPR: Student t test: T = − 0.61, df = 9.91, P = 0.6. j Comparison of descriptive statistics of median and maximal intensity and sharpness of the images. Intensity in sharpness was scaled to fit the range of 0–1. TPR depletion decreased both median (Wilcoxon exact test, W = 35.5, P = 0.004) and maximal (W = 33, P = 0.015) fluorescent intensity. k Fold change in mRNA levels of lamin A/C in shTPR cell line. TPR fold change: T = 7.2, df = 6, P < 0.001; LMNA/C fold change: T = 3.1, df = 6, P < 0.05. l Quantification of lamin A/C protein amount after TPR knock-down. TPR protein levels: T = 20.6, df = 8, P < 0.001; LMNA/C protein levels: T = 8.3, df = 8, P < 0.001. Graph represents data from three independent biological replicates analyzed in triplicates. Lower panel shows representative western blots stained with anti-TPR, anti-lamin A/C and anti-tubulin antibodies in control and in shTPR cells. ***P < 0.001, **P < 0.01, *P < 0.05, n.s. non-significant. 15 μg of total protein per each lane was loaded
As the nuclear pore basket is formed along with TPR also by Nup153, and Nup153 has been shown to interact with both types of lamins [21], next we tested if TPR attenuation affects Nup153. We compared the protein level of Nup153 in control and TPR-depleted cells and also checked the recruitment of Nup153 to nuclear pore basket. Neither in siTPR nor in shTPR cells, did we observe any significant changes in Nup153 protein level (Fig. S3l and Fig. S3m). We also did not observe any changes in the localization of Nup153 to nuclear pores as a consequence of TPR depletion (Fig. S3n). These data confirm that the observed defects in lamina network organization are only caused by loss of TPR.
The distribution of nuclear pore complexes is affected in TPR-depleted cells and can be rescued by the overexpression of lamin B1 or the reduction of lamin A
Funasaka and colleagues [34] showed by electron microscopy that reduction of TPR by siRNA causes severe reduction in NPC number after only 72 h of transfection. It remained possible that the reduction in NPC numbers seen by this study was due to more frequent observations of G1 cells, for instance. Also it remained unanswered whether the reduction of NPCs occurred over the whole nuclear surface or within the smaller areas of the nuclear surface. Therefore, we used light microscopy (wide-field as well as STED/SIM) to re-examine the matter. We acquired a number of peripheral z-sections of control and shTPR cells stained with Nup62 to visualize NPCs distribution and PCNA to detect S-phase cells and cells synchronized in G2 phase. In accordance with Maeshima and colleagues [17], we observed different populations of cells with NPCs distributed either homogenously (mostly cells in S and G2 stage based on both the PCNA pattern and nuclear size, Fig. 4a and Fig. S4a, black arrows) or with NPC-free zones (cells in G1 stage but also a portion of S-phase cells, Fig. 4b, c, gray and white arrows in Fig. S4a). We analyzed the number of nuclei with NPCs dispersed (1) homogenously (Fig. 4a, black arrows in Fig. S4a), (2) with small NPC-free zones that covered less than approximately 1/10 of the nuclear surface (Fig. 4b, gray arrows in Fig. S4a), and (3) with large NPC-free zones covering at least 1/10 of the nuclear surface (Fig. 4c, white arrows in Fig. S4a) in control and shTPR cells. Figure 4d shows that in TPR-depleted cells, the number of nuclei with NPC-free zones increased significantly (χ2 = 11.41, df = 2, P < 0.001, see also Fig. S4 and “Materials and methods”) in comparison to control wild-type HeLa cells as well as HeLa cells stably expressing short hairpin targeting non-coding genomic region (shNC). To exclude the possibility of measuring cells in G1 phase of the cell cycle where all pores are not fully assembled, we measured the NPC-free zones also in cells synchronized in G2 phase. In agreement with the previous result, we observed a significantly higher number of NPC-free zones in TPR-depleted cells than in controls (Fig. 4e).
Fig. 4.
A distribution of NPCs is affected in TPR-depleted HeLa cells (shTPR HeLa cell line). a–c The distribution of NPCs in control HeLa cells as viewed by immunostaining with anti-Nup62 antibody and wide-field Leica DM6000 microscope. a Homogenous distribution. b Small NPC-free zones. c Large NPC-free zones comprehend more than 1/10 of nuclear surface. d The ratio of homogenous distribution, small and large NPC-free zones in control wild-type S-G2 cells (wt), in cells stably expressing shRNA targeting non-coding regions (shNC) and TPR (shTPR cells). In TPR-depleted cells, the number of nuclei with NPC-free zones increased significantly (χ2 = 11.41, df = 2, P < 0.001, Chi squared test), **P < 0.01, ***P < 0.001, n.s. not significant. e The ratio of homogenous, small and large NPC-free zones in control wild type cells (wt) and in cells stably expressing shRNA targeting non-coding regions (shNC) and TPR (shTPR) synchronized in G1/S and in G2 phase of the cell cycle. The number of NPC-free zones significantly increased in TPR-depleted cells (***P < 0.001, n.s. not significant). f–i Recruitment of nucleoporins Nup107, Nup133 and central FG-Nups into NPC-free zones is impaired in TPR-depleted cells. f The ratio of distribution of NPC-free zones in shNC and shTPR cells stained with Nup153 antibody (χ2 = 7.08, df = 2, P = 0.029). g The ratio of homogenous distribution, small and large NPC-free zones in shNC and shTPR cells transfected with GFP–Nup107 construct (χ2 = 9.713, df = 2, P = 0.008). h The ratio of homogenous distribution, small and large NPC-free zones in shNC and shTPR cells overexpressing GFP–Nup133 (χ2 = 23.855, df = 2, P < 0.001). i The ratio of distribution of NPC-free zones in shNC and shTPR cells stained with antibody recognizing central FG-Nups (χ2 = 138.5, df = 2, P < 0.0001)
Recently, it was shown that the assembly of NPCs during interphase depends on Nup153, which further recruits nucleoporins of the central part of NPC [38]. Thus, we examined how TPR knock-down affects recruitment of Nup153 to NPCs-free regions. We immunodetected Nup153 and Nup62 in control and TPR-depleted cells and found that Nup153 staining was distributed almost homogenously throughout the nuclear envelope and large NPCs-free regions (deficient for Nup62 staining) were filled with Nup153 (Fig. 4f and Fig. S4b). Thus, Nup153 localization to NPC-free regions is independent of TPR. Further, we tested the recruitment of nucleoporins of the outer ring and the central part of NPCs. We overexpressed constructs coding GFP-tagged Nup107 or Nup133 in control and shTPR cells and examined their recruitment to NPC-free regions. The data revealed that both Nup107 (Fig. 4g and Fig. S4c) and Nup133 (Fig. 4h and Fig. S4d) displayed significantly impaired recruitment to NPC-free zones in shTPR cells in comparison to the control cells. Similarly, the central FG-Nups (detected by antibody Mab414) were not properly recruited to NPC-free zones devoid of Nup62 (Fig. 4i and Fig. S4e). Thus, we conclude that new NPCs start to assemble in NPCs-free zones but these pores represent only assembly intermediates and TPR depletion leads to defects in complete NPCs assembly in NPC-free regions during interphase.
Further, we analyzed the density of NPCs over the whole nuclear surface in control HeLa as well as in cells with depleted TPR levels. In the control cells, the average NPC density was 7.3 (SD 0.5) NPC/µm2. In shTPR cells, the average density decreased to 6.4 (SD 0.7), if the whole nuclear surface of TPR-depleted cells was analyzed. However, if we analyzed the area of a rather homogenous NPC distribution separately from the NPC-free zones, we found out that the density of NPCs in TPR-depleted cells did not differ from the density of NPCs in the control cells and reached 8.2 (SD 0.8) NPC/µm2 or 8.1 (SD 0.3) NPC/µm2 in control and TPR-depleted cells, respectively. Thus, our data support the previously published observation that TPR knock-down reduces the NPC numbers. However, our data favor the hypothesis that the reduction in NPC number does not occur over the whole nuclear surface. Instead, large NPCs-free zones (typical for G1 control cells) remain in S-G2 nuclei of TPR-depleted cells.
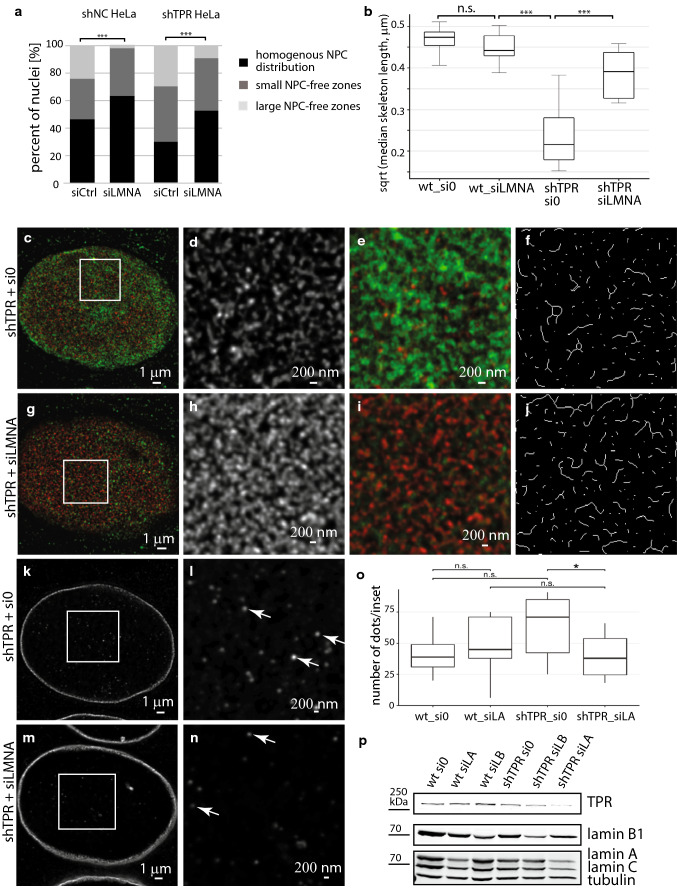
To reveal the mechanism of how TPR may regulate the NPCs distribution in the cell nucleus, we also focused on the TPR–lamin association. It has been shown that both lamin B1 and lamins A/C participate in either NPC anchoring or distribution [6, 12, 14]. As TPR was not proven to be necessary for the NPC assembly [39], we speculated that the reduction of lamin B1 amount and the aberrant lamina structure, as a consequence of TPR depletion, might be the reason for the aberrant NPC distribution of shTPR cells. To confirm this, we tested two potential mechanisms of how the NPC distribution could be affected. First, we examined if the percentage of cells with NPC-free zones in shTPR cells could be increased by simultaneous knock-down of lamin B1 (to further decrease lamin B1 levels) and, secondly, if the regular NPC distribution in shTPR cells could be rescued by lamin B1 overexpression. We used siRNA against lamin B1 to knock down the respective protein in control HeLa as well as in shTPR cells. Interestingly, the reduction of lamin B1 levels constrained the homogenous NPC distribution in control cells and resulted in a significant increase of nuclei with large NPC-free zones (χ2 = 9.65, df = 2, P < 0.01, Fig. 5a and Fig. S5a and S5b). The simultaneous knock-down of lamin B1 along with TPR resulted in a slightly higher number of nuclei with large NPC-free zones, in average 36% of nuclei had large NPC-free zones in cells with knock-down of both TPR and lamin B1 in comparison to 29% of nuclei in shTPR cells (χ2 = 4.79, df = 2, P < 0.1, Fig. 5a, Table S1). The overexpression of lamin B1 did not affect the NPC distribution in control HeLa cells (Fig. 5b) which suggests that the endogenous lamin B1 levels are sufficient for the normal NPC distribution. On the other hand, the elevated levels of lamin B1 partially rescued the homogenous distribution of NPCs within the shTPR HeLa cell population (30% of cells had homogenously distributed NPCs in shTPR cells compared to 42% in shTPR transfected with GFP–lamin B1; χ2 = 13.36, df = 2, P < 0.01; Fig. 5b, Fig. S5c and Table S1). This suggested that an exactly regulated amount of lamin B1 is crucial for the NPC distribution.
Fig. 5.
Lamin B1 levels influence NPC distribution in HeLa cell population with reduced TPR amount. a Knock-down of lamin B1 prevents homogenous distribution of NPCs in HeLa cells expressing RNA against non-coding genomic region (shNC) (test of interaction in Poisson mixed-effects model, χ2 = 9.65, df = 2, P < 0.05). Reduction of lamin B1 in shTPR cells slightly increases the numbers of nuclei with NPC-free zones (χ2 = 4.79, df = 2, P < 0.1). b GFP–lamin B1 overexpression did not affect the NPC distribution of the entire population of control HeLa cells. Overexpression of GFP–lamin B1 partially rescued the normal proportions of homogenous NPC distributions in shTPR cells (χ2 = 13.39, df = 2, P < 0.01). ***P < 0.001, *P < 0.05, •P < 0.1, n.s. not significant
Maeshima and colleagues [17] showed that high levels of lamin A/C restricted homogenous NPC distribution over the nuclear surface, whereas the reduction of lamin A/C caused a disappearance of NPC-free islands. Thus, we investigated if the reduction of lamin A could rescue homogenous NPC distribution in shTPR HeLa cells and, thus, compensate for TPR absence. The proportion of control cells with homogenous NPC distribution did not increase after lamin A knock-down (χ2 = 0.39, df = 2, P = 0.82, Fig. 6a and Fig. S5d). Importantly, reduction of lamin A levels in shTPR HeLa cells rescued the number of nuclei with homogenous NPC distribution to control levels (χ2 = 8.57, df = 2, P < 0.05, Fig. 6a and Fig. S5e, Table S1), where up to 52% of nuclei comprehend homogenously distributed NPCs and only 10% of nuclei had large NPC-free zones. Thus, low levels of lamin A/C result in the homogenous NPC distribution and can compensate for the TPR absence.
Fig. 6.
Reduction of lamin A levels can rescue the NPC distribution as well as an organization of lamin B1 peripheral network in shTPR HeLa cells. a Knock-down of lamin A favors homogenous NPC distribution in shTPR cells (test of interaction in Poisson mixed-effects model; shNC: χ2 = 0.39, df = 2, P = 0.82; shTPR: χ2 = 8.57, df = 2, P < 0.05). b–j Reduction of lamin A levels in shTPR HeLa cells rescues the lamin B1 network organization. b Box plot shows square root transformed median lengths of the skeletonized lamin B1 filaments in control cells as well as in cells with reduced TPR and lamin A levels. The lamin A knock-down results in elongation of lamin B1 filaments in shTPR cells in comparison to shTPR only. Wt_si0 × wt_siLMNA: Student t test: T = 1.1, df = 14, P = 0.29; wt_siLMNA × shTPR_si0: T = 8.6, df = 14, P < 0.001; shTPR_si0 × shTPR_siLMNA: T = − 5.1, df = 16, P < 0.001. The lamin B1 network in cell with reduced TPR levels (c–f) and in cell with reduced both, TPR and lamin A amount (g–j) as visualized by STED. The cells were immunostained with mouse anti-lamin A/C (shown in green) and rabbit anti-lamin B1 (shown in red). c, g Peripheral z-sections show the overview of lamin A/C//B1 organization. Insets in c and g correspond to d–f and h–j, respectively. Lamin B1 appears fragmented in shTPR cells (d) whereas it seems more polymerized in shTPR cells with reduced lamin A levels (h). e, i Merged images of lamin A/C and lamin B1. f, j The graphical visualisation of skeleton analysis of the insets d and h, respectively. k–n STED images of immunofluorescently detected lamin B1 in central z-sections of shTPR cells (k, l) and shTPR cells with reduced lamin A levels (m, n). Insets in k and m correspond to l and n, respectively. Arrows in l and n point to the intranuclear lamin B1 dots. o Analysis of number of lamin B1 dots in central z-sections of STED images of HeLa nuclei immunolabeled with lamin A/C and lamin B1. The number of lamin B1 intranuclear dots decreased significantly under co-depletion of TPR and lamin A (Wilcoxon exact test, W = 48.5, P = 0.045). p Western blotting of lamin levels in control cells and in cells with reduced TPR (shTPR), lamin A (siLA) and lamin B1 (siLB) levels shows that lamin B1 amount did not increase in response to lamin A reduction in shTPR cells. *P < 0.05, ***P < 0.001, n.s. non-significant. 3.5 × 106 cells were used as a starting material. 20 μg of total protein per each lane was loaded
We showed that TPR depletion caused fragmentation of lamin B1 network (Fig. 2 and Fig. S1). Because lamin A depletion re-established NPC distribution, we pondered whether lamin A depletion could also re-establish lamin B1 network in shTPR cells. Thus, we immunodetected lamin B1 in shTPR HeLa cells treated with siRNA against lamin A and analyzed the lengths of thresholded skeletonized lamin B1 filaments at the nuclear periphery in control and in siLMNA-treated shTPR cells as previously (see “Materials and methods”). Results of four biological replicates showed that lamin B1 filaments were longer in shTPR cells with depleted lamin A and resembled lamin B1 network in cells transfected with control siRNA and siLMNA (Fig. 6b; wt_si0 × wt_siLMNA: T = 1.1, df = 14, P = 0.29, wt_siLMNA × shTPR_si0: T = 8.6, df = 14, P < 0.001, shTPR_si0 × shTPR_siLMNA: T = − 5.1, df = 16, P < 0.001). Whereas lamin B1 network was fragmented in shTPR cells transfected with control siRNA (Fig. 6c–f), it restored to an almost normal pattern if lamin A was depleted along with TPR (Fig. 6g–j). We hypothesized that lamin B1 network polymerized either from nucleoplasmic pool of lamin B1 or, alternatively, levels of lamin B1 elevated in response to lamin A drop. To find the answer, we compared lamin B1 distribution at central z-sections of shTPR HeLa cells transfected with control siRNA with shTPR cells transfected with siLMNA. The nucleoplasm of shTPR HeLa cells transfected with control siRNA contained many lamin B1 dots of variable fluorescence intensity (Fig. 6k, l, arrows). In contrast, the nucleoplasm of shTPR cells treated with siLMNA contained less lamin B1 dots (Fig. 6n, arrows). We analyzed the number of lamin B1 intranuclear dots in control and shTPR cells treated with control siRNA and with siLMNA. Figure 6o shows that the number of lamin B1 intranuclear dots decreased significantly when TPR was depleted along with lamin A (W = 48.5, P = 0.045). We also checked the protein levels of lamin B1 in control HeLa as well as in shTPR cells treated with control siRNA, siLMNA and siLMNB. Western blot analysis confirmed that the levels of lamin B1 protein did not change when TPR was depleted simultaneously with lamin A (Fig. 6p). Thus, our data may implicate that the decrease of lamin A causes redistribution of lamin B1 from the nucleoplasm to the nuclear periphery, where it polymerizes to form lamin B1 network. Further, our data suggest that polymerized lamin B1 network is necessary for homogenous NPC distribution over the nuclear periphery and that both proteins TPR and lamin A can influence such process via affecting lamin B1 network.
Materials and methods
Cell line, plasmid transfection and siRNA transfection
Adherent human cervical carcinoma (HeLa) cells were grown on glass coverslips (rectangle, 18 mm in diameter, 170 ± 5 µm thickness; Marienfeld, Lauda-Königshofen, Germany) in D-MEM (Sigma-Aldrich, St. Louis, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, USA) at 37 °C in 5% CO2-humidified atmosphere.
Cells were transfected with plasmid DNA using Lipofectamine® 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s recommendations. GFP–TPR plasmid was obtained from Addgene. GFP–lamin B1 was a gift from Prof. C. J. Hutchoson (Durham University, Durham, UK), GFP–lamin A/C was kindly provided by Prof. J. E. Eriksson (Åbo Akademi University, Turku, Finland). GFP–Nup107 (#391) and GFP–Nup133 (#392) constructs were kindly provided by Dr. Valerie Doye, Institute Jaques Monod (CNRS, Université Paris Diderot [40]. GFP–TPR-M4 and GFP–TPR-17 constructs were kindly provided by Dr. Tomas Vomastek (Institute of Microbiology, ASCR, Prague, Czech Republic). Transfections with siRNA were performed using Lipofectamine® RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocol. MISSION esiRNAs targeting lamin A (EHU063791), lamin B1 (EHU057911) and TPR (EHU129911) were from Sigma-Aldrich (St. Louis, MO, USA).
Stable HeLa cell line with depleted TPR was prepared by lentiviral knock-down using pLKO.1 vectors (Sigma-Aldrich) expressing shRNAs targeting TPR transcripts in different regions (targeted sequence 1—shTPR: AGAAGAAAGTACTGATGGAGA) as well as non-targeting shRNA (SHC016-1EA, Sigma-Aldrich). Lentiviral particles were produced in HEK 293T cells. HEK 293T cells of 30% density were plated on 15-cm petri dish and 24 h later co-transfected with 22.5 µg of shRNA vector, 10 µg of pMD2.G and 17.5 µg of dR8.91 packaging plasmids using polyethylenenimine (Polysciences, 23966). The viral particles were collected 48 and 72 h after transfection, precipitated by 10% PEG 6000 and used to transduce HeLa cells. HeLa cells of 20% density were plated in 24-well plates and 24 h later infected by adding all isolated lentiviral particles. The transduction medium was replaced with fresh culturing medium 6 h after infection. Transduced cells were selected 24 h after transduction by adding puromycin at a final concentration of 4 µg/ml for 10 days. Western blotting and quantitative RT-PCR were performed to screen transduced cells for effective depletion of the TPR protein and mRNA, respectively, and two lines with the strongest TPR depletion were used for further experiments.
Antibodies
Primary antibodies were used as follows:
anti-TPR mouse monoclonal (nTPR, ab58344, Abcam, Cambridge, UK, 1:300), anti-TPR rabbit polyclonal (cTPR, ab84516, Abcam, Cambridge, UK, 1:300), anti-lamin B1 rabbit polyclonal IgG (ab16048, Abcam, Cambridge, UK, 1:100), anti-lamin A/C mouse monoclonal (SAB4200236, Sigma, Sigma-Aldrich, St. Louis, USA, 1:100), anti-Nup 62 rat monoclonal IgG (ab188413, Abcam, Cambridge, UK, 1:500); anti-Nup153 rabbit polyclonal antibody (NB100-93329, Novus Biologicals, 1:200), Nuclear pore complex proteins (ab24609, Abcam Cambridge, UK, 1:200), anti-GFP mouse monoclonal (ab1218, Abcam, Cambridge, UK, 1:200, anti-GFP rabbit polyclonal (1828014, Molecular probes at Thermofisher Scientific, Waltham, MA USA, 1:200), anti-PCNA human polyclonal, anti-tubulin α (N-terminal structural domain, TU-01, aa 65–79, 1:100) mouse monoclonal was kindly provided by Dr. Pavel Draber (Institute of Molecular Genetics of the ASCR, v.v.i., Prague, Czech Republic).
Secondary antibodies were as follows:
Goat anti-rabbit IgG (H + L) antibody conjugated with Alexa Fluor 488 (A11034, Invitrogen, Carlsbad, USA), goat anti-rat IgG (H + L) antibody conjugated with Alexa Fluor 488 (A21434, Invitrogen, Carlsbad, USA), goat anti-mouse IgG (H + L) antibody conjugated with Alexa Fluor 488 (A21236, Invitrogen, Carlsbad, USA), Goat anti-rabbit IgG (H + L) antibody conjugated with Alexa Fluor 555 (A21429, Invitrogen, Carlsbad, USA), Goat anti-rabbit IgG (H + L) antibody conjugated with Alexa Fluor 647 (A21245, Invitrogen, Carlsbad, USA), goat anti-rat IgG (H + L) antibody conjugated with Alexa Fluor 488 (A11006, Invitrogen, Carlsbad, USA).
Secondary antibodies for western blotting were goat anti-mouse IRDye® 680RD Goat anti-Mouse IgG (Licor, Lincoln, NE, USA) and IRDye® 680RD Goat anti-Rabbit IgG (Licor, Lincoln, NE, USA).
Proximity ligation assay (PLA)
For the proximity ligation assay, the Duolink in situ reagents Oragene kit (Merck KGaA, Darmstadt, Germany) was used according to the manufacturer’s instructions. Primary antibodies were used in following combinations: rabbit polyclonal anti-Lamin B1 (ab16048, Abcam, Cambridge, UK, 1:100) + mouse monoclonal anti-TPR (nTPR, ab58344, Abcam, Cambridge, UK, 1:300); mouse monoclonal anti-lamin A/C (SAB4200236, Sigma, Sigma-Aldrich, St. Louis, USA, 1:100) + rabbit polyclonal anti-TPR (cTPR, ab84516, Abcam, Cambridge, UK, 1:300). As a positive control, a combination of rabbit polyclonal anti-Lamin B1 (specified above) and mouse monoclonal anti-lamin A/C (specified above) was used. As a negative control, no primary antibody was used.
Western blotting
HeLa cells grown on petri dish were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris–HCl, pH 8.0, 5 mM EDTA, pH 8.0, 1% NP-40, 0.1% SDS) and clarified by centrifugation (15,000g, 15 min, 4 °C). Cleared lysates were boiled in 1 × Laemli Sample buffer for 8 min, separated with gradient SDS-PAGE (4–20%) and transferred to nitrocellulose membrane (Life Sciences). Membranes were blocked with 5% bovine serum albumin in PBS for 1 h in room temperature and incubated with primary antibodies in 1% bovine serum albumin in PBS—0.05% Tween (1 h in room temperature or in 4 °C overnight). Membranes were probed with IRDye secondary antibodies (LI-COR) in PBS—0.05% Tween and proteins were detected using multicolor detection system Odyssey (LI-COR). The protein level in samples was quantified using Image studio Lite version 5.2. Level of lamin B1, Lamin A/C and TPR was normalized to tubulin level as loading control. In control cells, level of lamins and TPR was set as 100% and assessed as percentage change in TPR-depleted cells. The statistical data evaluation was performed by paired t test using GraphPad Prism software. The effectivity of TPR knock-down and quantification of TPR, lamin B1, lamin A/C and Nup153 level was analyzed from three independent biological experiments. Individual samples—total protein lysates—were always prepared from 3.5 × 106 cells. 10 μg, 15 μg or 20 μg of total protein per each sample was loaded on the gel; the exact amount of protein used for western blot is indicated within the respective figure legend.
Immunoprecipitation
HeLa cells grown on petri dish were washed twice with ice-cold PBS, scraped from the dish, and centrifuged at 1000g for 10 min. Harvested cells were resuspended and lysed in ice-cold lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100) supplemented with protease inhibitor cocktail (EMD Millipore) and than pulse-sonicated on ice (30 pulses; 0.5 s for each pulse at 50% of maximum energy). The cell lysate was centrifuged at 16,000g, and the supernatant was immunoprecipitated for 2 h at 4 °C with anti-TPR (ab58344, Abcam, Cambridge, UK, 4 µg) and control anti-mouse IgM antibody (ab81032, Abcam, Cambridge, UK, 4 µg) prebound at protein G magnetic beads (50 µl of slurry, Pierce, Thermo Scientific). After five washings with lysis buffer, the immunoprecipitated proteins were resuspended in 40 μl of 2 × sample buffer, separated by gradient (4–20%) SDS-PAGE and transferred to a nitrocellulose membrane (Life Sciences). Membranes were blocked, probed with primary and secondary antibodies and detected as described above (“Western blotting” section). The co-immunoprecipitations were performed in four biological replicates. 20 × 106 cells were used as a starting material for co-immunoprecipitations. 2–5 mg of total protein per sample was used for the incubation with bead-bound antibody. 35 μg of total protein was loaded as input.
RNA isolation and reverse transcription
Total RNA was isolated using TRI Reagent (Sigma) according to manufacturer’s protocol. Briefly, cells were lysed in TRI Reagent and incubated 5 min in room temperature. Chloroform was added and the mixture was incubated for 15 min in room temperature and then centrifuged 15 min at 12,000g. To the aqueous phase 100% isopropanol was added, the mixture was incubated for 10 min and centrifuged 10 min at 12,000g. Pellet was washed with 75% ethanol, dried and resuspended in RNAse-free water. The cDNAs were synthesized using oligo(dT)20 primers from SuperScript III First-Strand Synthesis SuperMix (Invitrogen) as recommended in manufacturer’s protocol.
qPCR
Amplifications were carried out using Power SYBR Green PCR master mix and 7300 Real Time PCR System (Applied Biosystems) with the following primers: TPR primer 5′-TCATGCCCCACCTCAGGAGT, 3′ primer 5′-GGTGGCCCAAACCGGAATCT, Lamin B1 5′ primer, 5′-CCGTTCCTCTAAACGCCAGC-3′, 3′ primer, 5′-CTTAGCATAGTTGAGGAGCAGC-3′, Lamin A 5′ primer, 5′-GTCGGTGACTCAGTGTTCGC-3′ and 3′ primer 5′-CACGCAGCTCCTCACTGTAG-3′. Reactions were performed at 25 μl solution containing 1 μl cDNA, 12.5 μl SYBR Green and 1 μM primers according to manufacturer’s protocol under these conditions: 10 min at 95 °C followed by 45 cycles of 95 °C for 15 s and 60 °C for 30 s. The reactions were performed in triplicates from at least three independent experiments. Levels of mRNA were evaluated using method and normalized to actin as a reference gene. In control cells, mRNA level of lamins and TPR was set as 0 and assessed as a fold change in TPR-depleted cells. The statistical evaluation was performed by paired t test using GraphPad Prism software.
Immunofluorescence and microscopy
Immunofluorescence staining was performed according to the protocol described in Ref. [41]. For routine observations and for the analysis of NPC distribution over the nuclear surface, fully automated upright microscope Leica DM6000 (Leica Microsystems) with HCX PL Apo 100 ×/1.40 oil objective was used. The microscope was equipped with fluorescent cubes for DAPI (Ex 360/40, Em 470/40), FITC (Ex 480/40, Em 527/30), TRITC (Ex 546/12, Em 600/40) and Cy5 (Ex 620/60, Em 700/75), Leica DFC 350FX Camera and LAS X software.
Structured-illumination microscopy was performed on the Delta Vision OMX Blaze (Applied Precision) imaging system equipped with 3D structured illumination (3D-SIM) and a 60 × oil immersion objective (NA 1.42, PlanApo N) and with 4 × pco.edge sCMOS camera. Images were illuminated by lasers of wavelength of 405 nm, 488 nm and 568 nm and collected using filter sets for blue (395.5/435.5), green (477.0/528.0) and red (571.0/609.0) fluorescence. For z-stacks, 0.125 μm optical sections were acquired. For the image reconstruction and registration, the SoftWorx (Applied Precision) software implemented with the 3D SI Reconstruction function was used. Wiener filter values were set to 0.01 for DAPI and 0.001 for green and red channel. Samples were mounted to 2,2′-thiodiethanol–n-propyl gallate antifade mounting (Life Technologies, Carlsbad, CA, USA). Prior to image analysis, the quality of reconstructed images was checked using SIMCheck toolbox [42]. The DeltaVision OMX system’s 3D-SIM provides lateral resolution of approximately 110 nm ± 5 nm in 405 channel, 120 nm ± 5 nm in 488 channel and 135 ± 5 nm in 568 channel. Prior to imaging, the lateral resolution as well as quality of registration has been checked using TetraSpeck Microspheres (Cat. No. T2729, Thermo Fisher Scientific, Waltham, MA, USA) of defined width of 100 nm.
Stimulated emission depletion (STED) was performed on the TCS SP8 STED 3X (Leica Microsystems) equipped with 660 nm STED laser, STED White Objective CS 100 ×/1.40 OIL. Image acquisition was performed using depletion laser of 660 nm and 3X 3D STED, gating 0.4–6 ns for Alexa-555 detection and 0.2–8 for Alexa-488 detection. The Leica Microsystems STED provides lateral resolution of about 80 nm ± 5 nm in 488 channel and 50 nm ± 5 nm in 555 channel. Images were acquired using LAS X software and deconvolved by Huygens Professional software (algorithm CMLE, theoretical PSF, signal to noise ratio 10).
Image analysis
To analyze the microscopy data of the same quality (and to avoid, for instance, comparing blurred data resulting from slightly curved areas at the nuclear surface), we cut a square of each acquired image of the same dimensions (232 × 214 pixels) and analyzed peripheral z-sections (the z-section showing the nuclear periphery) of the individual insets.
Skeleton analysis was performed in FIJI (ImageJ). The images were binarized using a thresholding method described by [43]. The centerlines of the objects were computed by applying a thinning algorithm on the segmented binary images with the use of built-in plugins Skeletonize (2D/3D) and Analyze Skeleton (2D/3D). The plugins are implementations of a 3D thinning algorithm from [44]. Results were obtained in micrometers, median values were calculated, square root transformed, summarized and visualized graphically in Matlab software (Release 2015a, The MathWorks, Inc., Natick, MA, USA).
Image sharpness was estimated using eigenvalues of the image covariance matrix [45] and normalized by dividing by the sample maximum for visualization purposes.
NPC density was measured by macro in FIJI (ImageJ) in peripheral z-sections of images acquired by Delta Vision OMX (Applied Precision) imaging system equipped with 3D structured illumination (3D-SIM) and reconstructed with the implemented SoftWorx (Applied Precision) software. Briefly, NPCs were thresholded, segmented and the information about all segmented object (NPCs) was collected by the Particle Analyzer in Fiji/ImageJ. The ratio of object number and area was calculated to determine the NPC number/µm2.
NPC density per unit was measured as described previously [35]. Briefly, NPC numbers were measured using SR-SIM acquired images in the rectangular volume 150 × 150 pixels. NPCs were segmented and data were collected by Fiji/ImageJ.
Statistics
For statistical analysis of the lengths of skeletonized lamins the data were square root transformed and analyzed by Student’s t test. To compare the mean intensity, the maximal intensity and the sharpness of the images between treatments, Wilcoxon exact test was used. We analyzed four independent experiments, ten image sections per a variant were analyzed in each experiment.
Lamin B1 dots inside the nucleoplasm were segmented using a threshold separating low-intensity background from high-intensity lamin dots. We selected a threshold that was as low as possible (to detect most of the lamin dots), but not too low to detect large artificial low-intensity regions, defined to be larger than 200 px. The choice of 200 px was empirically found to balance the sensitivity and specificity of the segmentation and yield sensible results. The number of dots and the median dot pixel intensity were compared using Wilcoxon exact test across control and/or treatment groups. We analyzed ten images per sample in three independent experiments.
To compare the observed counts of NPC distribution in the three categories (homogenous/small NPC-free areas/large NPC-free areas) between control and treated cells, the Poisson mixed-effects models with Chi squared model–submodel test of the treatment–category interaction were used. Statistical analyses were carried out in R (R Core Team 2017). We analyzed four independent experiments, the number of nuclei analyzed in each category of differentially treated Hela cells is summarized in Supplemental Table 1.
Discussion
Here, we show that TPR, a protein of nuclear pore basket, co-immunoprecipitates with lamin B1 and participates in lamin B1 network organization. Further, it is involved in regulation of amounts of lamin A/C and lamin B1. Finally, TPR along with A- and B-lamins engages in NPC distribution over the nuclear surface. Our work, thus, uncovers another piece of complex interaction network at the nuclear periphery.
Importantly, we provide data showing structural features of the lamin B1 as well as lamin A/C network organization in HeLa cells by means of STED. Our data clearly show the filamentous arrangement of A- and B-type lamins. Previous works detecting nuclear lamins by means of super-resolution microscopy revealed randomly organized filaments with a punctate pattern [5, 6, 13]. Our data support the finding that the lamina network is composed of randomly arranged filaments; moreover, our images show a very dense network of clearly defined A- and B-type filaments and as such provide another insight into the mammalian nuclear lamina organization.
The short-term (48 h) loss of TPR results in disintegration of lamin B1 network (but not lamin A/C network) suggesting that the disintegration of the TPR–lamin B1 interaction is sufficient for alteration of lamin B1 network and the TPR–lamin B1 interaction itself is important for lamin B1 network organization. Whereas roles of lamins in NPC organization have been shown previously [10–12, 14, 17], the opposite phenomena, where nuclear lamina would be influenced structurally by a protein of nuclear pores, have not been described yet. TPR is a large protein with several coiled-coil domains at its N-terminal part which were shown to be able to form dimers or tetramers [22]. Being part of the nuclear pore basket, TPR locates to a close proximity to the nuclear lamina. We can speculate that the coiled-coil domains of TPR might interact with coiled-coil domains of lamins.
We also considered the potential role of Nup153 in nuclear lamina organization. Along with TPR, Nup153 forms the nuclear basket [19]. Similarly to TPR, Nup153 interacts with both lamin types [21] and, thus, changes in Nup153 amount or localization as a potential effect of TPR silencing may also contribute to the phenotype we described here. Nevertheless, we demonstrate that silencing of TPR alter significantly neither Nup153 protein level nor Nup153 localization to nuclear pores. In addition, these data are in agreement with previously published data [39]. Thus, we assume that the observed alterations in lamina organization are particularly due to the intense TPR depletion.
Further, our data suggest that TPR may be involved in regulation of mRNA levels of both lamin B1 and lamin A/C as these levels drop within 48 h after the TPR depletion with siRNA and stay at lower levels also during the long-term TPR depletion in stable shTPR HeLa cells. The protein levels of lamin B1 drop eventually in longer time periods (in at least a 2-week time) presumably because lamins are proteins with longer turnover. A relative stability of lamin B1 has been reported to be about 5–7 days in human primary skin fibroblasts [37], but up to 9 months in mouse embryos [46]. The TPR has been previously implicated in regulation of amounts of several nucleoporins, namely Nup62 and Nup98, the mechanism has not been revealed, though [34]. The Nup153 also participates in the regulation of gene expression as shown in several cell types [47–49], and both nucleoporins, TPR and Nup153, have been shown to associate with active transcription regions and to regulate the transcription in Drosophila [33]. Thus, it is possible that nuclear pore basket proteins also regulate the expression of lamins. Importantly, a depletion of TPR resulted in elevated levels of p53 and a cellular senescence [34]. In turn, the activation of p53 was shown to induce lamin B1 loss via decrease in mRNA stability and this, again, led to a cellular senescence [50]. Altogether, these data suggest that TPR might regulate lamin B1 levels via activation of p53, which needs experimental confirmation. Other regulation mechanisms are also possible, for instance, via kinases that interact with TPR [26], and thus could phosphorylate substrates in close proximity. Nevertheless, data supporting these hypotheses are missing.
Our data further show that the TPR depletion affects NPCs assembly and distribution in interphase nuclei. Large NPC-free zones were detected previously by Maeshima et al. [17] and correspond to areas devoid of post-mitotic NPC assembly due to the presence of spindle microtubules that need to be removed from DNA surface before nuclear envelope and NPC assembly [16]. These NPC-poor zones are populated by newly assembled NPCs during G1/S phases of the interphase [51]. Reduced density of NPCs after only 72 h of transfection with siRNA against TPR was described by means of electron microscopy [34]. The work, however, did not consider the differences in the NPC organization during the cell cycle (specifically, the presence of large zones of NPC exclusion in G1 [34], neither has it shown if the whole nuclear surface was affected. Finally, electron microscopy is not the favorable technique for statistical analysis of large portion of the data.
Here, we show that in cells depleted of TPR, large zones of NPC exclusion remain at the nuclear surface also during S and G2 cell stages. This suggests that the process of de novo NPC assembly within the NPC-free zones became disturbed. For interphase NPC assembly, several Nups were shown essential for either curvature of membrane (Nup153, Nup133, reviewed in Ref. [52]) or for targeting of other proteins to assembling NPC. Nup153 is essential for recruitment of Nup107–160 complex to the inner nuclear membrane [38]. In our experimental settings, levels and localization of Nup153 remained almost unchanged and thus, targeting of Nup107–160 complex via Nup153 should not be affected. However, in shTPR cells, the recruitment of Nup107 and Nup133 to NPC-free zones was impaired and we also observed defects in NPCs assembly using Nup62 staining visualizing the central part of NPCs. These data point to important role of TPR in the early steps of interphase NPC assembly. Indeed, its role has recently been suggested [35]. In this work, TPR and ERK signaling were described as negative regulators of NPCs assembly and TPR depletion caused higher density of NPCs. However, we did not observe any significant changes in NPCs density neither in shTPR (Fig. S6a) nor in cells transfected with siRNA targeting TPR (Fig. S6b). We also did not detect any changes in cells overexpressing phosphorylation-deficient mutant of TPR or TPR mutant deficient for ERK binding (Fig. S6c; constructs described previously in Ref. [26]).
Non-homogenous NPC distribution in HeLa cells with reduced TPR levels might result from inappropriate NPC assembly. The TPR itself is not required for the mitotic NPC assembly [39]; however, the TPR protein was shown necessary for interphase NPC assembly [35]. TPR is also substrate of other kinases [25] which might regulate the recruitment of Nup107–160 complex and central FG-Nups to NPC-free zones. Nevertheless, the mechanism remains uncovered.
The role of both lamins, lamin A and lamin B1, in distribution of NPCs was also well studied. Thus, changes in lamin networks caused by TPR depletion are more likely to be responsible for aberrant NPC distribution. Previous works offered multiple evidence for the importance of both A- and B-type lamins in NPC anchoring and stabilization [6, 10–14, 20, 21]; however, the interaction with lamina involved Nup153. The role of TPR via lamins in NPC distribution has been suggested [6]; no confirmatory data have been shown though.
We speculated that the disruption of the (direct or indirect) interaction of TPR with either A- or B-type lamins might have been a cause for an aberrant NPC distribution. Our data, however, indirectly unfavor such hypothesis. The normal distribution of NPCs in the shTPR HeLa cell population could be rescued by simultaneous overexpression of lamin B1 or depletion of lamin A in shTPR cells. Thus, in our experimental settings, the TPR–lamin B1 interaction would not be rescued because of constantly reduced TPR levels in shTPR HeLa cells.
Our data suggest that the reduced levels of lamin B1 and fragmented lamin B1 network caused NPC distribution phenotype by so far uncharacterized mechanism. The NPC distribution phenotype could be caused by lamin B1 knock-down using siRNA, or by reduction of TPR levels (Fig. 7a, b). And vice versa, recovery of lamin B1 network in shTPR cells, both directly by overexpression of lamin B1–GFP and indirectly by reduction of lamin A levels, rescued NPC distribution (Fig. 7b, c). For the NPC assembly and distribution, the right levels of lamins were shown necessary [13, 17]. Lower amounts of lamin A/C resulted in uniform NPC distribution [17]; on the other hand, lower levels of lamin B1 caused uneven NPC distribution [13]. Our data also confirmed these conclusions. Among others, we detected reduction of both A- and B-lamin types in response to TPR depletion. Knock-down of lamin A/C and lamin B1 has the opposite effect on NPC distribution; however, we detected the phenotype corresponding to reduction of lamin B1. We think that this is due to the fact the reduction of lamin A/C in shTPR cells is far less striking in comparison to lamin A reduction with siLMNA.
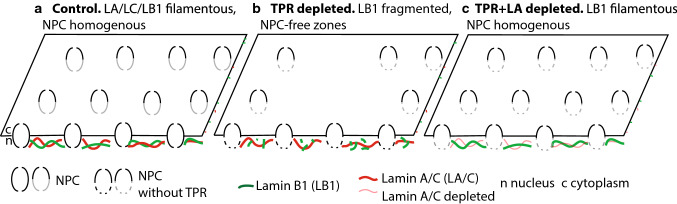
Fig. 7.
A scheme showing interdependence of lamin A/C, lamin B1 and TPR organization and NPC distribution in HeLa cells. a In control cells, lamin A/C and lamin B1 are fully polymerized, TPR forms nuclear baskets of NPCs that are homogenously distributed at the nuclear envelope. b Long-term TPR depletion results in lamin B1 (but not lamin A/C) network fragmentation, reduction of lamin B1 levels and the presence of NPC-free zones. c Co-depletion of TPR and lamin A restores both lamin B1 network organization and NPC homogenous distribution
Our results further showed that lamin A was able to potentiate or hinder lamin B1 network formation (Figs. 6, 7). This might occur via control of soluble pool of lamin B1 in the nucleoplasm. In our experiments, such soluble lamin B1 pool could be released to recover peripheral lamin B1 network in the lamin A absence. Thus, polymerized lamin B1 is necessary to secure appropriate NPC distribution and could be influenced by TPR as well as lamin A.
To conclude, we show that the TPR is along with Nup153 in complex inter-connecting both A- and B-type lamins. Further, our data disclose the role of lamins in NPC distribution in later G1 and point to a close interplay between A- and B-type lamins and TPR to secure the interphase NPCs assembly. Thus, the TPR is in the center of complex mechanism that controls proper function of the nuclear periphery. The details, however, are missing. It would be interesting to define lamin-binding region(s) within the TPR protein as well as to investigate the mechanism of regulation of lamin expression by the TPR to offer further insight into the complex functioning of the nuclear periphery.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank to Iva Jelinkova for the help with immunofluorescence and HeLa cell culture. We thank the Microscopy Centre, Light Microscopy Core Facility, IMG ASCR, Prague, Czech Republic for the help with imaging, especially to Ivan Novotny for the help with microscope settings and acquisitions by SIM and STED.
Author contributions
JF was responsible for conception, JF and MM for design of the experiments, JF, MM, KF and PH for the biological data analysis; JF, MM, JU and LS for the experimental execution; ME for the Matlab programming, TS for statistical analysis; JF and MC for the image analysis, JF for drafting the article and JF, MM, TS, ME, MC, JU, LS, KF and PH for editing the article prior to submission.
Funding
This work was supported by the Grant Agency of the Czech Republic: [15-08835Y], [16-03346S], [16-03403S], [17-09103S], [18-19714S], by the institutional support of long-term conceptual support of development of the scientific organization (RVO: 68378050) and by the project “BIOCEV—Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University” (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund. M.M. was supported by grant of ASCR (L200521801). J.U. was supported by GAUK (930218). We acknowledge the Light and Electron Microscopy Core Facility, IMG CAS supported by the MEYS CR (LM2015062), OPPK (CZ.2.16/3.1.00/21547), NPU I (LO1419) and ERDF (Project no. CZ.02.1.01/0.0/0.0/16_013/0001775).
Compliance with ethical standards
Conflict of interest
The authors declare no competing or financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jindřiška Fišerová and Miloslava Maninová contributed equally to the work.
References
- 1.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 2.Naetar N, Ferraioli S, Foisner R. Lamins in the nuclear interior—life outside the lamina. J Cell Sci. 2017;130(13):2087–2096. doi: 10.1242/jcs.203430. [DOI] [PubMed] [Google Scholar]
- 3.de Leeuw R, Gruenbaum Y, Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320(5881):1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell. 2015;26(22):4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, Stewart C, Burke B. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol. 2016;26(19):2651–2658. doi: 10.1016/j.cub.2016.07.049. [DOI] [PubMed] [Google Scholar]
- 7.Cho S, Irianto J, Discher DE. Mechanosensing by the nucleus: from pathways to scaling relationships. J Cell Biol. 2017;216(2):305–315. doi: 10.1083/jcb.201610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281(35):25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 9.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341(6149):1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154(1):71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137(5):1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11(11):3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell. 2014;25(8):1287–1297. doi: 10.1091/mbc.e13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, Nakatomi R, Yahata K, Imamoto F, Hashikawa T, Yokota H, Imamoto N. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol. 2010;17(9):1065–1071. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- 16.Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522(7555):231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 17.Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, Imamoto N. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci. 2006;119(Pt 21):4442–4451. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 18.Hurt E, Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr Opin Cell Biol. 2015;34:31–38. doi: 10.1016/j.ceb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20(20):5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 2000;19(15):3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Haboubi T, Shumaker DK, Koser J, Wehnert M, Fahrenkrog B. Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus. 2011;2(5):500–509. doi: 10.4161/nucl.2.5.17913. [DOI] [PubMed] [Google Scholar]
- 22.Pal K, Bandyopadhyay A, Zhou XE, Xu Q, Marciano DP, Brunzelle JS, Yerrum S, Griffin PR, Vande Woude G, Melcher K, Xu HE. Structural basis of TPR-mediated oligomerization and activation of oncogenic fusion kinases. Structure. 2017;25(6):867–877 e863. doi: 10.1016/j.str.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coyle JH, Bor YC, Rekosh D, Hammarskjold ML. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA. 2011;17(7):1344–1356. doi: 10.1261/rna.2616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontoura BM, Dales S, Blobel G, Zhong H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci USA. 2001;98(6):3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajanala K, Sarkar A, Jhingan GD, Priyadarshini R, Jalan M, Sengupta S, Nandicoori VK. Phosphorylation of nucleoporin Tpr governs its differential localization and is required for its mitotic function. J Cell Sci. 2014;127(Pt 16):3505–3520. doi: 10.1242/jcs.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vomastek T, Iwanicki MP, Burack WR, Tiwari D, Kumar D, Parsons JT, Weber MJ, Nandicoori VK. Extracellular signal-regulated kinase 2 (ERK2) phosphorylation sites and docking domain on the nuclear pore complex protein Tpr cooperatively regulate ERK2–Tpr interaction. Mol Cell Biol. 2008;28(22):6954–6966. doi: 10.1128/MCB.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano H, Funasaka T, Hashizume C, Wong RW. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem. 2010;285(14):10841–10849. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1–Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22(21):2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweizer N, Ferras C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J Cell Biol. 2013;203(6):883–893. doi: 10.1083/jcb.201309076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David-Watine B. Silencing nuclear pore protein Tpr elicits a senescent-like phenotype in cancer cells. PLoS One. 2011;6(7):e22423. doi: 10.1371/journal.pone.0022423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snow CJ, Paschal BM. Roles of the nucleoporin Tpr in cancer and aging. Adv Exp Med Biol. 2014;773:309–322. doi: 10.1007/978-1-4899-8032-8_14. [DOI] [PubMed] [Google Scholar]
- 32.Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29(10):1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6(2):e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funasaka T, Tsuka E, Wong RW. Regulation of autophagy by nucleoporin Tpr. Sci Rep. 2012;2:878. doi: 10.1038/srep00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCloskey A, Ibarra A, Hetzer MW. Tpr regulates the total number of nuclear pore complexes per cell nucleus. Genes Dev. 2018;32(19–20):1321–1331. doi: 10.1101/gad.315523.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22(24):3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell. 2005;16(6):2872–2881. doi: 10.1091/mbc.e04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmer B, Lorenz M, Moreno-Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S, Antonin W. Nup153 recruits the Nup107–160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev Cell. 2015;33(6):717–728. doi: 10.1016/j.devcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 39.Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14(5):1923–1940. doi: 10.1091/mbc.e02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154(6):1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiserova J, Efenberkova M, Sieger T, Maninova M, Uhlirova J, Hozak P. Chromatin organization at the nuclear periphery as revealed by image analysis of structured illumination microscopy data. J Cell Sci. 2017;130(12):2066–2077. doi: 10.1242/jcs.198424. [DOI] [PubMed] [Google Scholar]
- 42.Ball G, Demmerle J, Kaufmann R, Davis I, Dobbie IM, Schermelleh L. SIMcheck: a toolbox for successful super-resolution structured illumination microscopy. Sci Rep. 2015;5:15915. doi: 10.1038/srep15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai W. Moment-preserving tresholding: a new approach. Comput Vis Graph Image Process. 1985;29:377–393. doi: 10.1016/0734-189X(85)90133-1. [DOI] [Google Scholar]
- 44.Arganda-Carreras I, Fernandez-Gonzalez R, Munoz-Barrutia A, Ortiz-De-Solorzano C. 3D reconstruction of histological sections: application to mammary gland tissue. Microsc Res Tech. 2010;73(11):1019–1029. doi: 10.1002/jemt.20829. [DOI] [PubMed] [Google Scholar]
- 45.Wee CY, Parmesran R. Measure of image sharpness using eigenvalues. Inf Sci. 2007;177(12):2533–2552. doi: 10.1016/j.ins.2006.12.023. [DOI] [Google Scholar]
- 46.Razafsky D, Ward C, Potter C, Zhu W, Xue Y, Kefalov VJ, Fong LG, Young SG, Hodzic D. Lamin B1 and lamin B2 are long-lived proteins with distinct functions in retinal development. Mol Biol Cell. 2016;27(12):1928–1937. doi: 10.1091/mbc.e16-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29(12):1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nanni S, Re A, Ripoli C, Gowran A, Nigro P, D’Amario D, Amodeo A, Crea F, Grassi C, Pontecorvi A, Farsetti A, Colussi C. The nuclear pore protein Nup153 associates with chromatin and regulates cardiac gene expression in dystrophic mdx hearts. Cardiovasc Res. 2016;112(2):555–567. doi: 10.1093/cvr/cvw204. [DOI] [PubMed] [Google Scholar]
- 49.Toda T, Hsu JY, Linker SB, Hu L, Schafer ST, Mertens J, Jacinto FV, Hetzer MW, Gage FH. Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell. 2017;21(5):618–634 e617. doi: 10.1016/j.stem.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell. 2012;23(11):2066–2075. doi: 10.1091/mbc.e11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife. 2016;5:e19071. doi: 10.7554/eLife.19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otsuka S, Steyer AM, Schorb M, Heriche JK, Hossain MJ, Sethi S, Kueblbeck M, Schwab Y, Beck M, Ellenberg J. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat Struct Mol Biol. 2018;25(1):21–28. doi: 10.1038/s41594-017-0001-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.