Abstract
Gut microbiota has evolved along with their hosts and is an integral part of the human body. Microbiota acquired at birth develops in parallel as the host develops and maintains its temporal stability and diversity through adulthood until death. Recent developments in genome sequencing technologies, bioinformatics and culturomics have enabled researchers to explore the microbiota and in particular their functions at more detailed level than before. The accumulated evidences suggest that though a part of the microbiota is conserved, the dynamic members vary along the gastrointestinal tract, from infants to elderly, primitive tribes to modern societies and in different health conditions. Though the gut microbiota is dynamic, it performs some basic functions in the immunological, metabolic, structural and neurological landscapes of the human body. Gut microbiota also exerts significant influence on both physical and mental health of an individual. An in-depth understanding of the functioning of gut microbiota has led to some very exciting developments in therapeutics, such as prebiotics, probiotics, drugs and faecal transplantation leading to improved health.
Keywords: Gut microbiota, Functions, Health, Therapeutics
Introduction
The life forms on this earth can be clustered into three broad domains: namely Archaea, Bacteria and Eukaryota [1]. All life has evolved from a simple unicellular common ancestor over billion years of evolution giving rise to a complexity of cells within an organism. The human is a superorganism that functions in harmony with trillions of symbiotic bacteria and eukaryotic cells. The host and its symbionts together are called a “holobiont,” and their collective genome is known as “hologenome”. Variation in the hologenome either by changes in the host genome or the microbiome may occur with reasonable fidelity maintaining plasticity of the holobiont [2]. In 2001, the human genome project was completed after which it was correctly argued that the “crowning achievement” in biology would be incomplete until the synergistic activities between human and microbes are understood [3–5]. Subsequently, several scientific efforts were initiated to understand the relationships between human and human-associated microbial communities. Discoveries of the Human Microbiome Project (HMP) and the Metagenome of Human Intestinal Tract (MetaHIT) opened new horizons in microbiome research for an enhanced understanding of host–microbe interactions at four major colonisation sites of the human body; viz. oral, gut, vagina and skin. Of these four sites, the human gut microbiota has drawn the attention of microbiologists for its clinical significance. Several gut microbiome projects including the Australian Gut Project, the American Gut project, the British gut project, the Canadian Microbiome Initiative, the Human MetaGenome Consortium Japan, the My NewGut project of the European Union and the International Human Microbiome Consortia, etc. were undertaken for a better understanding of the complex gut ecosystem and its role in health and diseases. The human gut (200–300 m2 of mucosa) is the “secret garden” of ten trillion diverse symbionts (50 bacterial phyla and about 100–1000 bacterial species), collectively known as the ‘microbiota’. Microbiota are ten times more abundant than our somatic and germ line cells of the body. The collective genes of microbiota are known as the ‘microbiome’ which is 150 times larger than the human genome [6, 7]. In an individual, 150–170 bacterial species predominate and get benefits from the warm nutrient rich environment of the gut and perform protective, metabolic and structural functions. In this review, we have summarised (a) the general features of the gut microbiota, the laws of symbiosis and its development along with its distribution in the gastrointestinal (GI) tract; (b) the relationships between the gut microbiota in a metabolic, immunological and structural landscapes of the human body and (c) promising therapeutic approaches including prebiotics, probiotics, drugs and faecal microbiota transplantation that involve in modulation of the gut microbial ecology.
Methods to study gut microbiota
Culture-dependent
In 1881, Robert Koch introduced the plating technique both to culture and identify microorganisms by characterising their biochemical and physiological properties. A culture-based method detects only 30–50% of the bacteria inhabiting in the intestine. Artificial cultural conditions provide a less favourable environment for the growth of uncultivable bacteria. Conventionally, different selective and nutrient agar media are used to isolate and culture the bacteria. Improved methods, such as the enrichment culture technique and pre-incubation of faeces in blood culture bottles, rumen fluid and sterile stool extract were introduced in different culture and physicochemical conditions. These cultural modifications act as natural simulants and facilitate the isolation of previously uncultivable bacteria [8–11]. However, the growth of predominant bacteria in faeces masks the isolation of less dominant ones. Further modifications including the use of antibiotics, bacteriophages and filtration overcome these difficulties. For example, the dominant population of Escherichia coli under Proteobacteria phylum hindered the isolation and identification of new bacterial species. The addition of lytic bacteriophages in the culture plate lysed E. coli and facilitated growth of an unknown bacterial species (Enterobacter massiliensis) of the Enterobacteriaceae family that was not previously detected using classical culture methods. An extensive study on the culturomics approach highlighted the gap between culture-dependent and independent studies and estimated the number of the uncultured microbes from the different microbiome projects [9]. They introduced three modifications to isolate uncultured bacteria with multiple cultural conditions viz. (1) preincubation of a sample in a blood culture bottle, (2) the addition of rumen fluid and (3) the addition of sheep blood that enhanced the isolation of bacterial species by 56, 40 and 25%, respectively. This culture-based approach also used the matrix-assisted laser desorption/ionization based time-of-flight mass spectrometry to discriminate a large number colonies and 16S rRNA sequencing for bacterial identification. This culturing approach had identified 1057 prokaryotic species that include (1) 531 species to the human gut repertoire of which 146 bacteria were previously not reported in the gut, (2) 187 bacteria and 1 archaea (Haloferax alexandrinus) not previously isolated from humans and (3) 197 potentially new species. Therefore, the application of culturomics is significant in exploring gut microbial diversity as well as in understanding their causative or curative roles in health and disease.
Culture-independent
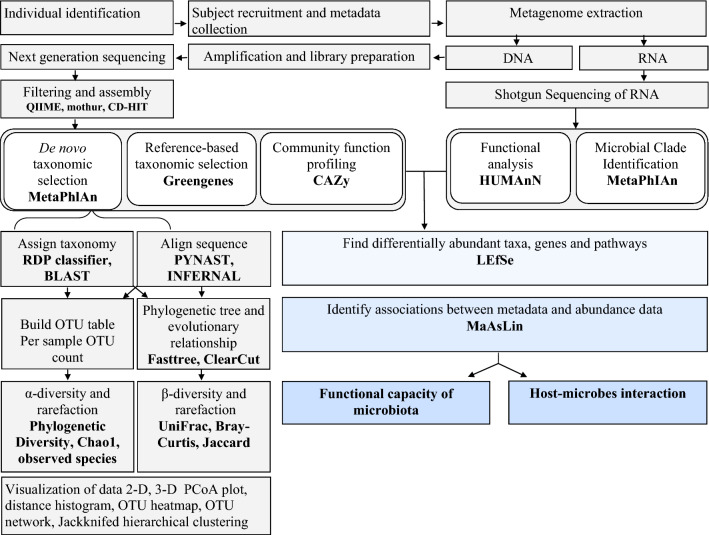
With the advent of Sanger sequencing in 1970, Carl Woese and his colleagues introduced 16S rRNA to investigate the bacterial taxa and their phylogeny based on conserved sequences of hypervariable (V1–V9) region [12]. In the following 30 years, polymerase chain reaction (PCR)-based methods, such as denaturing/temperature gradient gel electrophoresis, single-strand-conformation polymorphism, restriction fragment length polymorphism, terminal restriction fragment length polymorphism and quantitative PCR have been developed to study the diversity of microorganisms. Likewise, non-PCR-based molecular techniques, such as microarray and fluorescence in situ hybridization have also been adopted to aid the exploration of microbial diversity [13]. In the 1990s, preparation of a clonal library and sequencing of the metagenome by Sanger’s method paved the way to identify taxonomic composition and phylogeny of a microbial community in an ecological niche. In the following years, a number of improvements were made to Sanger’s sequencing method which mostly involved the replacement of phosphor tritium-radiolabelling with fluorescently tagged bases and its fluorometric detection through capillary-based electrophoresis. About a decade later in 2005, high-throughput sequencing technique (next generation of sequencing called NGS) was developed to overcome the difficulties identified in Sanger’s method in term of ease, cost and time [13]. In this technique, adapter-bracketed DNA molecules are passed over a lawn of complementary oligonucleotides bound to a flow-cell. Thereafter, the bound DNA fragment undergoes a process known as bridge amplification and produces a cluster of clonal populations. During amplification, each fluorescent labelled deoxyribonucleotide triphosphate is detected during sequential cycles of DNA synthesis and identified by fluorophore excitation in a massive and parallel fashion. A typical workflow involved in a culture-independent study of the microbiota is presented in Fig. 1. In 2010, catalogues of 3.3 million non-redundant faecal microbial genes were reported using NGS. This number of genes was 200 times more than any previous studies [6]. Thereafter, a catalogue of 9.8 million non-redundant microbial genes was published from the metagenomic data with 1267 samples; including 760 European from MetaHIT, 139 American from the HMP and 368 from China (large diabetic study). Each sample consisted of 750,000 genes of which 300,000 genes were similar in more than 50% of the individuals [14]. A Danish study of 540 adults reported the concept of high and low gene counts of the gut microbiome related to obesity [15]. In the recent years, sequencing of the whole genome by NGS involves a fragmentation step before sequencing and then assembling into one long contiguous sequence in silico. Moreover, DNA extraction, library preparation, choice of primer and short read lengths of DNA could be prone to errors [16]. Now, the third generation of DNA sequencing techniques such as PacBio RS II and the MinION nanopore device have shown substantial advancement over second-generation DNA sequencing. In 2011, the PacBio developed a technology based on single molecule real-time sequencing platform using zero-mode waveguides properties. MinION was developed by Oxford Nanopore’s technology in which DNA is passed through a nanoscale pore structure to measure the changes of electrical field surrounding the pore which is base dependent. Both technologies can produce longer reads over the NGS and can cover the whole genome. Hence, these techniques can bypass the computational challenges of genome assembly and transcript reconstruction. In addition, epigenetic modifications such as DNA methylation at CpG site and histone modification are detected directly without tagging, breaking and filtering of the DNA. Moreover, the third generation of sequencers are potable and require minimal pre-processing. Therefore, data are collected and analysed in real time for efficient diagnosis and rapid remedial actions. For example, Ebola virus (EBV) was detected within 44 s using Oxford Nanopore’s MinION.
Fig. 1.
Bioinformatics work flow of culture independent approach. QIIME quantitative insights into microbial ecology, MG-RAST metagenomics rapid annotation using subsystem technology, CAZy carbohydrate active-enzymes, MetaPhlAn metagenomic phylogenetic analysis, KEGG Kyoto encyclopaedia for genes and genomics, COG clusters of orthologous group, PICRUst phylogenetic investigation of communities by reconstruction of unobserved states, HUMAnN The Human Microbiome Project Unified Metabolic Analysis Network, LEfSe Linear Discriminate Analysis with Effect Size, MaAsLin Multivariate Association with Linear Models, MetaPhlAn Metagenomic Phylogenetic Analysis, PICRUSt Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, rRNA ribosomal RNA
Features of GI microbiota
Symbiosis and stability
The gut microbial community is dynamic and has adapted to colonize in the GI. Some specific characteristics are pivotal for colonization, such as an arsenal of enzymes to utilize available nutrients, the right cell-surface molecular pattern to attach at the “right” habitat, ability to evade bacteriophages and fitness to the reaction-ready immune system of host. A microbe should withstand the physical and chemical stresses in the gut to proliferate rapidly without washing out [17]. Finally, it should survive in the dry and toxic stresses of the “ex-host” environment when making a jump to other hosts [18]. The relationship between host and microbes is commensal (one partner benefits while the other seems unaffected) rather than mutualistic (both partners benefited). The homeostasis of gut microbiota is maintained through feedback mechanisms. Positive feedback disrupts the cooperation of the microbial community in which two microbial species increase in their abundance and can generate a runaway effect [19, 20]. For example, cooperation between two species to increase their abundances in which upsurges of one species increases the abundance of the second and so on. The upsurges of both microbes disrupt the overall diversity and abundance of a community in an ecological niche. Three negative feedback mechanisms have been proposed by which host can create a selective pressure between the cooperating microbes to maintain their homeostasis in the gut. First, the host immune response shapes the microbial community by provoking a specific immune response. Second, spatial segregation of microbial species as these can grow in separate locations as their interaction will be weakened. Finally, supplementation of microbes with alternative carbon sources in such a way that these species no longer rely on their cooperative partners [21]. Mostly, two levels of natural selection of an individual maintain the law of symbiosis and its stability. At the host level, “top-down” selection on the microbial community favours stable societies with a high degree of functional redundancy. An opposing “bottom-up” selection pressure originates from the microbes to allow them to become functionally specialized [17]. Existing relationships between the host and adaptation of the microbial community maintains the gut ecological niche by nurturing its richness and diversity.
Diversity and integrity
The human gut microbiota consists of several types of microbes including bacteria, archaea, eukarya, viruses and parasites [19]. The gut microenvironment mainly favours the growth of bacteria from seven predominant divisions (Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia and Cyanobacteria) [22]. Among these seven divisions, the Bacteroidetes and Firmicutes constitute more than 90% of the total population. Most of the species under the phylum Bacteroidetes belong to the genera of Bacteroides and Prevotella. Bacterial species under the phylum Firmicutes such as Clostridium clusters IV and XIVa which include the genera Clostridium, Eubacterium and Ruminococcus are predominant in gut. Methanobrevibacter smithii a hydrogen-consuming methanogen and halophilic Haloferax alexandrinus and Haloferax massiliensis spp. nov. from Archaea had been reported from human gut [9]. Apart from taxonomic classification, the human microbiome is classified into three discrete enterotypes: Bacteroides or Prevotella or Ruminococcus. Enterotype 1 is characterised by dominance of Bacteroides with saccharolytic and proteolytic activities [23]. Enterotype 2 is Prevotella dominant and acts as a mucin glycoprotein degrader. Enterotypes 1 involved in synthesis of biotin, riboflavin, pantothenate and ascorbate, while enterotype 2 involved in thiamine and folate synthesis. Enterotype 3 is characterised by dominance of Ruminococcus with mucin degrading activities and membrane transportation of sugars. However, the concept enterotypes is debated due to the observation of a high degree of variation between individuals and some data showing more of a continuum rather than three discrete clusters, i.e. the debate is not related to diet differences, though clearly diet drives different composition types [24]. Intake of animal protein and fat for a long period enriched for Bacteroides enterotype while carbohydrate-enriched diet encouraged Prevotella. Sometimes, the enterotypes Ruminococcus and Bacteroides are overlapped and found to be indistinguishable [25]. A group of researchers from Taiwan classified enterotypes into Bacteroides, Prevotella and Enterobacteriaceae and also correlated with their dietary habits. They also claimed that Enterobacteriaceae could be a new sub-enterotypes in the Asian population [26]. Whatever may be the enterotypes, the abundance of bacterial phyla may vary significantly from individual to individual in where some microbial members function as “core microbiota”, while others act more like a “flexible pool”. The “core microbiota” adapts for reproducibility the “flexible pool” helps in adaptation of a host. The flexible pool is generally acquired from ingested food, water and various components from the environment [27]. The microbial species, Faecalibacterium prausnitzii, Roseburia intestinalis and Bacteroides uniformis are considered as core bacterial species (< 0.5% relative abundance). Sometimes, exchange of genetic material between core and flexible pool confers the fitness to host for adaptation either to an environment or to a dietary habit. For example, the genes of porphyranases, agarases and its associated proteins were found in the marine bacterium, Zobellia galactanivorans living outside the gut. These genes has been transferred to the gut bacterium Bacteroides plebeius in Japanese individuals due to consumption of seaweeds that are absent in the North American [28]. In spite of the exchange of genes, different individuals share similar metabolic and functional pathways, including fructose/mannose metabolism, amino-sugar metabolism and N-glycan degradation [29].
Gut microbiota composition
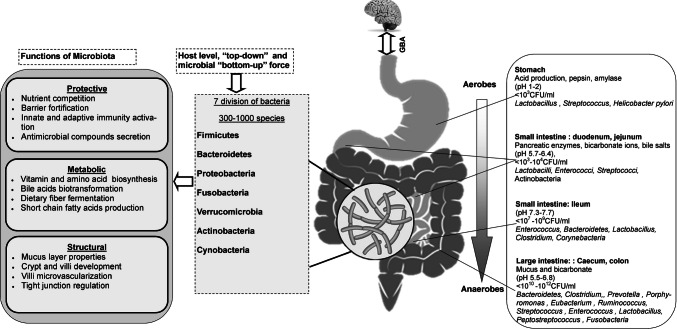
The GI tract is divided functionally and anatomically into the stomach, small and large intestine (LI). The distinct microenvironment and physiochemical barrier of each compartment selects the growth of specific microbiota (Fig. 2).
Fig. 2.
Distribution of normal gut flora in different parts of intestine and its functional activities and GBA. GBA gut brain axis
Stomach
Previously the stomach was thought to be sterile and unreceptive to bacterial growth due to a bactericidal barrier, reflux of bile acids, thickness of the mucus layer and gastric peristalsis. In 1981, the Lancet reported a large number of acid-resistant bacterial strains, such as Streptococcus, Neisseria and Lactobacillus in the stomach. While in 1982, Campylobacter pyloridis was discovered by Robin Warren and Barry Marshall and later in 1984, it was named as Helicobacter pylori. In stomach, more than 65% of phylotypes originated from the mouth. These mouth originated bacteria such as Veillonella, Lactobacillus and Clostridium were found to be acid-resistant and transient [30]. Healthy human stomach is normally inhabited by five major phyla viz. Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria and Proteobacteria and the bacterial genera Prevotella, Streptococcus, Veillonella, Rothia and Haemophilus [31].
Small intestine (SI)
The SI majorly helps in digestion and absorption of foods and nutrients. The SI is divided into three parts: duodenum, jejunum and ileum. Duodenum microenvironment is characterised by bile acids, pancreatic secretions, and antimicrobials agents where faster transit of food and plenty of oxygen limits the bacterial density (103–4 CFU/ml) and diversity. Firmicutes and Actinobacteria are the predominant phyla in the duodenum [32]. The jejunum supports the bacterial colonisation in terms of diversity and density (103–7 CFU/ml) and mostly supports the growth of Gram-positive aerobes and facultative anaerobes (103–7 CFU/ml) including Lactobacilli, Enterococci and Streptococci [32]. In the transition to the ileum, the bacterial density reaches up to 109 CFU/ml with predominance of aerobic species. In contrast, the distal part of ileum close to the ileocecal valve is populated with anaerobes and Gram-negative organisms similar to the colon.
Large intestine (LI)
The LI majorly consists of ascending, transverse and descending colon and cecum. It is a predominant site of water absorption and fermentation of undigested food due to the slower transit of food and its anaerobic condition. In the LI, the number of anaerobes outnumbers the aerobes by a factor of 100–1000. The bacterial density reaches to 1012 CFU/ml and is mainly dominated by Firmicutes and Bacteroidetes [33]. The ratio of these two bacterial phyla may alter in different stages of life and even in various pathophysiological conditions. The ratio of Firmicutes and Bacteroidetes is considered as a predictive marker of health and diseases [34]. In the lumen of LI, bacterial genera such as Bacteroides, Bifidobacterium, Streptococcus, Enterobacteriaceae, Enterococcus, Clostridium, Lactobacillus and Ruminococcus are predominant, whereas Clostridium, Lactobacillus, Enterococcus and Akkermansia are associated with the mucosa. In addition, a few pathogens including Campylobacter jejuni, Salmonella enterica, Vibrio cholera, E coli and Bacteroides fragilis may present in the LI with lower abundance (0.1%) [35].
Change of microbiota with age
The human microbiota has evolved along with its host and is an integral part of human body. Microbiota is acquired at birth and develops in parallel with the host and plays an important role in the body through adulthood until death. A century of research had claimed that the womb, placenta, amniotic fluid and meconium are sterile and microbiota starts to be acquired after birth. However, recent PCR and DNA sequencing-based methods suggested the presence of bacterial communities in the placenta, amniotic fluid and meconium [36]. Although these advanced techniques overcome the limitations of the cultivable approach, however, they have some inherent limitations. First, the detected bacterial DNA may come from either live or dead bacteria. For example, Bifidobacterium and Lactobacillus were detected in placenta by the molecular approach; however, the same bacteria were unculturable even though these are readily cultivable organisms [37]. Second, DNA-based assessments of a sample with low microbial biomass are extremely prone to error and contamination in a clinical setting. There is controversy over two opposing hypotheses “sterile womb” and “in utero colonisation” [36]. Apart from this controversy, various maternal factors including prenatal stress, antibiotic therapies and prolong gestation have roles in colonisation of the gut microbiota of a new-born baby. A few studies showed the changes of hormonal profiles during pregnancy that encouraged the growth of Lactobacillus (iners, crispatus, jensenii and johnsonii) and Clostridiales, Bacteroidales and Actinomycetales in the vagina [38, 39]. During vaginal birth, a baby acquires these microbes in their gut. In contrast, a baby born by caesarean section (C-section) acquires microbes from the mother’s skin including Staphylococcus, Corynebacterium and Propionibacterium spp. Lower microbial diversity and delayed colonisation of Bacteroidetes in the C-section baby makes the baby vulnerable to certain pathogens and atopic diseases [40]. After birth, breast-fed babies consume more lysozyme, immunoglobins, lactoferrin, glycans, sialylated and other complex oligosaccharides through mother’s milk than formula-fed babies. As a result, gut microflora of breast fed babies are dominated by Bifidobacterium and Lactobacillus sp. whereas formula fed baby are dominated with Clostridium, Granulicatella, Citrobacter, Enterobacter and Bilophila [7, 41]. These evidences indicate that vaginal birth and breast-fed babies have healthier gut microbes compared to the formula-fed babies. Thereafter by the age of 3–5 years, the unstable structure and composition of microbiota starts to differentiate and acquires similarity (40–60%) to that of adult [7]. During this period, the gut microbiome also changes in parallel from the earliest lactate utilization to plant polysaccharide digestion, vitamin biosynthesis and xenobiotic degradation [42]. The composition and functions of the established microbiota remain the same if there is no change in long-term dietary habits, antibiotics treatment, stress and pathophysiology in adulthood. In China, research on longevity revealed that the genera Roseburia and E. coli were significantly higher, whereas Lactobacillus, Faecalibacterium, Parabacteroides, Butyricimonas, Coprococcus, Megamonas, Mitsuokella, Sutterella, and Akkermansia were significantly less in centenarians [43]. In Italy, research on centenarians showed a decline in the abundance of core microbiota (Ruminococcaceae, Lachnospiraceae, and Bacteroidaceae) with the prevalence of subdominant genera and families (Eggerthella, Akkermansia, Anaerotruncus, Synergistaceae, Bilophila, and Christensenella) along with age. Abundance of Akkermansia, Bifidobacterium and Christensenella has been identified as a putative signature in the gut ecosystem for healthy ageing and longevity [43–45]. In the centenarians, lifespan decreased due to changes within the phylum Firmicutes and enrichment of ‘pathobionts’ and proinflammatory response mediated by the cytokines TNF-α, IL-6, MCP-1 and IL-8 [46]. In contrast, microbial secreted colanic acid, a polysaccharide that has been found to regulate fission and fusion dynamics of mitochondria for tuning longevity of Caenorhabditis elegans [47].
Genetic variation
Host genetics can influence gut microbiota and its metabolic phenotype. A study in the UK analysed > 1000 faecal samples from 416 twin pairs and showed that host genetics shapes the gut microbiome and obesity phenotype. They found that Christensenellaceae family is the heritable taxon and formed a co-occurrence network with other heritable bacteria and methanogenic Archaea. Transplantation of Christensenella minuta in germ-free mice altered the gut microbiota and reduced adiposity and weight in the recipient mice [48]. In a transcriptional analysis, it was found that the expression of 6000 genes of the colonic epithelia was linked to gut microbiota. They identified 12 allele-specific single-nucleotide polymorphism (SNPs) linked to gut microbiota and 8 were found to be associated with diseases including colorectal cancer, Type 2 diabetes (T2D) and obesity [49]. A large-scale cohort study on 1561 healthy individuals indicated that one-third of the faecal bacterial taxa were heritable and 58 SNPs in 1098 subjects were associated with the relative abundance of 33 taxa. Among these, four loci were replicated in the second cohort of 463 subjects related to Rikenellaceae, Faecalibacterium, Lachnospira, and Eubacterium [50]. A genome-wide study on Hutterites, a religiously isolated group of people in North America showed that at least eight bacterial taxa were associated with SNPs of the host. The bacterial genus Akkermansia was found to be related to obesity and body mass index. Besides, a gender wise variation of gut microbiota was also observed due to difference of their social practices and daily activities [51]. Gut microbial composition also depends on secretor status of ABH antigen in mucosa. The enzyme fucosyltransferase 2 encoded by FUT2 gene that converts type 1 N-acetyllactosamine glycan into H antigen which functions as a precursor for the A, B and Lewis b antigens. Non-secretor individual is unable to secrete ABH antigen in their mucus due to a non-sense mutation in the FUT2 gene. Bifidobacterial diversity and richness, particularly B. bifidum, B. adolescentis and B. catenulatum/pseudocatenulatum was found to be reduced in the non-secretor individual [52]. Research from Israel showed that environmental factors dominate over genetics in shaping the gut microbiota. A cohort of 1046 healthy individuals showed that genetically unrelated individuals had similar gut microbiota composition rather than their relatives who did not have a history of household sharing. They also analysed the previous data of 2252 twins from the UK and found that only 1.9–8.1% of microbiome taxa was heritable. In contrast, 20% of the inter-person microbiome variability is associated with factors such as diet, drugs, anthropometric parameters and lifestyle [53].
Geographical location
The ‘Baas-Becking’ hypothesis proposed that all microbes distributed everywhere and the local environment is the determinant of the microbial biodiversity [54]. For example, the diet and foraging lifestyle of the hunter-gatherer group (Hadza of Tanzania) determined microbial richness and diversity in the gut in comparison with urban Italian and the African rural farming groups [57]. The dominance of Prevotella, Treponema and unclassified Bacteroidetes in the Hadza aided in the digestion of plant originated fibrous foods [55]. The functionalities study of microbiome on the same group showed that metabolic pathways involved in digestion of a broad-spectrum of carbohydrates, branched-chain amino acid degradation and aromatic amino acid biosynthesis. In contrast, microbiomes of the urban Italians had resistome functionality that was similar to antibiotic resistance genes found in their environment [56]. In the Western lifestyle, the intake of more amino acids, lipids, cholesterol and dairy supports the growth of Faecalibacterium, Ruminococcus, Bifidobacterium, Bacteroides, Blautia, Bilophila and Alistipes while consumption of sugar and complex carbohydrates by the non-industrialized ethnic societies supports the growth of Prevotella [57]. Microbial profiling of rural and urban cohorts from seven ethnic groups of 9 provinces in China reported nine core bacteria (Balutia, Clostridium, Ruminococcus, Faecalibacterium, Subdoligranulum, Roseburia, Coproccus, Bacteroides and Phascolarctobacterium) linked to their ethnicities/geographies and lifestyles [58]. In India, we have reported six core bacteria (Faecalibacterium, Eubacterium, Clostridium, Blautia, Ruminococcus, and Roseburia) from 15 ethnicities of four geographical locations. The gut bacterial profiles of Indian ethnicities were similar to the Mongolian population [59]. However, faecal metabolites were dissimilar due to a degree of association of gut microbiota across the ethnicities [60]. Exposure to extreme environments such as high altitude (above 1493 m) is also a factor responsible for altering the gut microbiota due to lower pressure of atmospheric oxygen. As a result, high-altitude visitors like pilgrims, trekkers, climbers and military personnel suffer from non-specific GI complications [61–63]. The cold environmental stress is also a factor for changing of the gut microbiota and energy homeostasis. The transplantation of gut microbiota from cold stress mice to the germ-free mice promoted the conversion of the white fat into more brown fat. In addition, gut microbiota enhanced the absorption of nutrients during stress by increasing the length of villi and microvilli of the intestine to satisfy the demand of the energy in cold condition [64].
Functions of microbiota
Gut microbiota perform their function on four different landscapes in the human body: metabolic, protective structural and neurological.
Metabolic
Dietary fibres
The GI of humans digests about 85% of carbohydrates, 66–95% of proteins and all fats. In the colon, gut microbiota scavenge ~ 10–30% of energy and the rest is excreted as faeces [65]. Dietary fibres including lignin, non-starch polysaccharides, resistant starch (RS) and oligosaccharides [e.g. fructo-oligosaccharides (FOS) and galacto-oligosaccharides (GOS)] are resistant to digestion by host digestive enzymes [66]. Gut microbes have an array of enzymes for utilisation of these diverse carbohydrates. carbohydrate active enzyme (CAZy) a database (http://www.cazy.org/) documented all of the enzymes and categorised these into four families of glycosidases such as glycoside hydrolases (153 subfamilies), glycosyltransferases (106 subfamilies), polysaccharide lyases (28 subfamilies) and carbohydrate esterases (16 subfamilies) [67]. Members of the phyla Bacteroidetes and Firmicutes have the largest set of GH encoding genes in their genomes for utilization of different polysaccharides as carbon sources. Gut microbiota of the LI ferments all of the dietary fibres, resulting in the releases of gases (methane, hydrogen and carbon dioxide), SCFAs (formate, acetate, propionate, butyrate, valerate, isovalerate and hexanoate), smaller amounts of organic acids (lactate and succinate) and alcohols (methanol and ethanol). Gut microbial species such as Roseburia spp., Eubacterium rectale and Faecalibacterium prausnitzii and Clostridium groups IV and XIVa are the main producers SCFAs [68]. Among the SCFAs acetate, propionate and butyrate are prevalent and present in cecum in the molar ratio of 40:40:20 to 75:15:10 depending on diet. SCFAs (90–95%) are absorbed in the colon through passive diffusion or by ion exchange process and meets about 10% of caloric demand of the human [69]. Acetate is found in the blood (0.10–0.15 mM) and serves as an energy source to peripheral tissues and lipogenesis and cholesterol biosynthesis in liver. Butyrate serves as an energy source of colonocytes and produces ketone bodies and carbon dioxide. Apart from being energy source, butyrate also regulates energy homeostasis by stimulating gut enteroendocrine cells for the production of leptin from adipocytes including the production Glucagon-like peptide-1 in L cells [70, 71]. It also reduces the effect of harmful metabolites of bile acids and phenols. Propionate generally traverses the colonocytes and transports to the liver where it functions as acetate [72].
Protein and amino acids
In the LI, undigested protein is broken into peptides, amino acids and other metabolites via extracellular bacterial proteases and peptidases. These metabolites can be broadly classified into neuroactive compounds (nitric oxide, the tryptamine and phenethylamine), sulphide-containing metabolites (H2S), aromatic compounds (phenol, p-cresol and indole), polyamines (spermine, spermidine and cadaverine), SCFAs (isobutyrate, 2-methylbutyrate and isovalerate from branch chain amino acids) and ammonia. Some of the metabolites such as phenol, p-cresol and indole have been shown to induce IBD, colorectal cancer and kidney dysfunction [70]. The MEROPS database documented the peptidase and protease enzymes of the gut bacteria [73]. According to this database Clostridium spp. Bacteroides spp., Lactobacillus spp., etc. have a large diversity of proteases. [74]. The sources of dietary protein have a distinct effect on gut microbial composition. For example, a beef meat rich diet increased the population of Bacteroides and Clostridia but had a lower abundance of Bifidobacterium adolescentis [75]. In contrast, pea and whey proteins encouraged the growth of both Bifidobacterium and Lactobacillus, while whey additionally suppressed the pathogenic Bacteroides fragilis and Clostridium perfringens [76, 77]. Interestingly, protein and amino acids in association with the commensals also influence food choice, behaviour and reproduction. In Drosophila melanogaster, commensal bacteria elicited specific appetites for amino acid rich food. It was observed that two gut bacteria, Acetobacter pomorum and Lactobacilli, influenced the preference of flies for sucrose in absence of isoleucine in a food [78]. In addition, availability of dietary protein to carbohydrate ratio determines the catabolic pathways of the gut microbiota. Catabolism resulted in the production of GABA (Lactobacillus spp., Bifidobacterium spp. and Lactococcus lactis), norepinephrine (Escherichia spp. and Bacillus spp.), dopamine (Bacillus spp.), histamine and serotonin (Streptococcus spp., Escherichia spp. and Enterococcus spp.) [74]. These catabolic products may have an influential role in modulating the gut–brain axis or in maintaining the nitrogen balance of a host.
Lipid
A high-fat diet (HFD) induces microbial dysbiosis, enhances adiposity and low-grade inflammation in the adipose tissue. In rodents, feeding of HFD promoted an increase in Bilophila wadsworthia, a sulphite-reducing pathobiont. Further, increase in the population of B. wadsworthia exerted a pro-inflammatory response mediated by TH1 cell of genetically susceptible Il10(−/−), but not in wild-type mice [79]. Experimental evidences suggested that consumption of HFD caused a decrease in Bacteroidetes and an increase in both Firmicutes and Proteobacteria [80]. Gut microbiota transplantation from obese mice to germ-free mice induced some obesity-associated phenotypes, i.e. increased intestinal permeability and inflammation enhanced lipogenesis and adipogenesis. Diets rich in saturated fat increased the population of Bacteroides, Bilophila and F. prausnitzii, while unsaturated fatty acid increased the Lactic acid bacteria, Bifidobacteria and A. muciniphila [81, 82]. African Americans consume higher animal protein and fat and lower fibre diet than rural Africans. This dietary practice elevated the level of colonic secondary bile acids, lowered colonic short-chain fatty acid and increased the risk of colon cancer in the African Americans [83].
Bile salt
Bile acid is a steroid acid that is synthesized by at least 17 different enzymes in the liver for the excretion of excess cholesterol. Hydroxylation of cholesterol initiates synthesis of primary bile via the action of cholesterol 7α hydroxylase and produces chenodeoxycholic acid (45%) and cholic acid (31%). Before secretion of primary bile acids into the canalicular lumen, they are conjugated via an amide bond at the terminal carboxyl group of glycine or taurine. The glycoconjugates and tauroconjugates are more amphipathic in nature to facilitate the metabolism of dietary fat, fat-soluble vitamins and cholesterol [84]. The released conjugated bile salt in the duodenum is reabsorbed (95%) from the distal ileum via sodium-dependent bile transporter present in the brush border membrane of the enterocytes. A small portion of the unabsorbed bile salt undergoes deconjugation by the action of bile salt hydrolase of gut microbiota and produces deoxycholate, ursodeoxycholate and lithocholate [70, 85]. These secondary bile acids are passively absorbed in the colon and are transported back to the liver through enterohepatic circulation and the rest is excreted through faeces. The transformation of bile acids is mainly performed by anaerobic bacteria of the genera Bacteroides, Eubacterium and Clostridium (Clusters XIVa and XI) and a minor fraction of aerobic bacteria such as Actinobacteria and Proteobacteria. Bile salt hydrolase is also found in the human gut including members of Lactobacilli and Bifidobacteria. Deconjugation by 7αβ-dehydroxylation of bile salts increases their hydrophobicity and lowers its absorption and showed pathological effects including obesity and cancer [86, 87].
Choline
Choline is an essential water-soluble nutrient and a constituent of lecithin, which is present in many plants and animal organs. Choline and its metabolites help to maintain the structural integrity and signalling roles of cell membranes, cholinergic neurotransmission (acetylcholine synthesis), and a source for methyl groups via its metabolite, trimethylglycine (betaine), which participates in the biosynthesis of S-adenosylmethionine. The gut microbiota metabolizes choline into trimethylamine (TMA) which is absorbed in the gut. Moreover, TMA is converted to trimethylamine-N-oxide (TMAO) by flavin monooxygenases 1 and 3 (FMO1 and FMO3) in the liver. The formation of TMAO exacerbates hepatic insulin signalling and glucose tolerance and also promotes adipose tissue inflammation, atherosclerosis and cardiovascular diseases [88]. Now, the microbial conversion of dietary choline is an emerging metabolic hallmark of cardiovascular diseases. Eight gut bacteria including C. sporogenes, Anaerococcus hydrogenalis, Providencia rettgeri, C. asparagiforme, C. hathewayi, E. fergusonii, Proteus penneri and Edwardsiella tarda have been identified to produce TMA due to the consumption of a choline-rich diet [89]. Two enzymes (CutC and CutD) were reported in Desulfovibrio desulfuricans that can convert the choline into TMA and acetaldehyde [90].
Polyphenol
Polyphenols are secondary metabolites formed in a variety of plants through shikimate/phenylpropanoid and/or polyketide pathways which contain more than one phenolic unit and are broadly classified into two main groups, flavonoids and non-flavonoids. The majority of the polyphenols are present as glycosides with conjugation of different sugars such as glucose, galactose, rhamnose, ribulose, arabinopyrinose and arabinofuranose units. Inactive polyphenols including flavones, flavonols, flavanones, flavanols, catechins, anthocyanins, isoflavones, hydroxycinnamic acids, lignans, etc. are transformed into active compounds after removal of the sugar by the gut microbiota in the SI [91]. The polyphenols such as hydroxycinnamic acids are commonly esterified to sugars, organic acids and lipids which are transformed into aglycone by microbial esterases. The aglycones are found to be absorbed in the SI or colon and metabolized into hydroxyphenylacetic acids. Another dietary polyphenol such as ellagic acid and ellagitannins are transformed into urolithin A (an anti-inflammatory metabolite) by Gordonibacter spp. [92]. Polyphenols can also detoxify gut metabolites and repress the growth of pathogenic bacteria, such as Clostridium perfringens, Clostridium difficile and Bacteroides spp. [70]. Experimental evidence suggested that supplementation of epigallocatechin-3-gallate (GCG) and resveratrol (RES) altered the human gut microbial composition. Supplementation with GCG and RES increased oxidation of fat with reduced abundance of Bacteroidetes and F. prausnitzii in men but not in women [93].
Protective
The GI tract is the interactional scaffold of gut microbiota and gut immune system [94]. The first layer of the gut immune system including gut-associated lymphoid and Peyer’s patches developed due to fundamental intertwining with gut commensal. The developed immune barrier and a set of immune mechanisms limit the direct contact of commensal to the epithelial cell surface. A second layer of immunity rapidly detects and kills the bacteria without penetration to intestinal tissue. A third set of immune responses localized within the mucosa without activating the systemic immune system. The innate immune barrier consists of mucus, antimicrobial peptides and secretory IgA.
Mucus layer
The mucus layer consists of mucin glycoprotein secreted from goblet cell and assembled into a viscous gel-like layer on the intestinal epithelia that protects from the attachment of microbes directly to epithelia. It also acts as a lubricant and transports the luminal contents without hampering the epithelial lining. Chemically, mucin backbone consists of a peptide with alternating O-linked glycosylated (70–80%) and non-glycosylated domains. N-Acetylglucosamine, N-acetylgalactosamine, fucose and galactose are the four primary mucin oligosaccharides with either terminal sialic acid or sulfate groups. The sulfated (sulfomucins) or nonsulfated (sialomucins) mucins are acidic in nature, expressed throughout the LI epithelia. The mucus layer is about 150 µm thick epithelia and forms two distinct strata of alternating sialo- and sulphomucins. The acidic sulfated mucin is proximal to epithelia and is less degradable by bacterial glycosidases and host proteases. The acidic mucin prevents direct adherence of commensal to colonic epithelial cells [95]. The mucin not only acts as a barrier but also provides a direct source of carbohydrates and peptides for commensals. For example, Bacteroides thetaiotaomicron produces multiple fucosidases that cleave fucose from glycans and activate the goblet cells in secretion of mucin. Besides, Bacteroides fragilis synthesizes fucosylated capsular polysaccharides, a component of the bacterial outer membrane which confers advantages for competitive and preferential colonisation [96]. Bifidobacterium bifidum can produce endo-α-N-acetylgalactosaminidase and 1, 2-α-l-fucosidase to use mucins as an energy source in vitro [97]. In contrast, mucolytic bacteria such as Ruminococcus torques, Ruminococcus gnavus may also increase in intestinal bowel disease (IBD) and provide the substrate to other non-mucolytic bacteria [98]. Some of the other pathogenic bacteria are genetically equipped and try to cross the mucus barrier to induce the infection. For example, H. pylori produce urease to increase the pH and to lower viscosity of mucus for the penetration of epithelial cell surface [99]. Pathogens such as Campylobacter jejuni and Salmonella spp. use their flagella to cross the mucus barrier. E. coli and Shigella flexneri do so by secreting a mucin-binding serine protease which rapidly digests mucus and also induce its hyper secretion to compete against indigenous bacteria [100].
Antimicrobial peptides (AMPs)
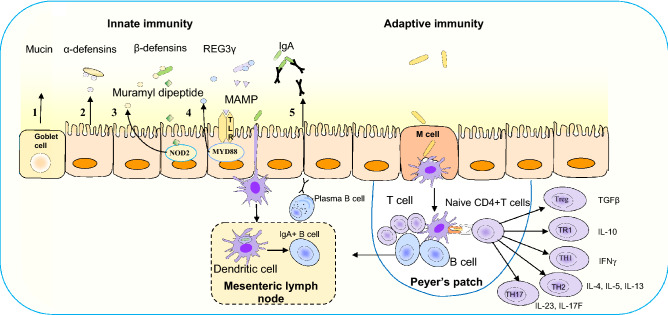
Antimicrobial peptides (AMP) are secreted by intestinal epithelial cells including enterocytes, Goblet and Paneth cells which restrict the access of commensals as well as pathogens to epithelia. The AMPs including defensins, cathelicidins and C-type lectins are a group of conserved small (20–40 amino acids) cationic proteins virtually retained in the mucus layer. These can bind either with negatively charged microbial cell membranes or enzymatically attack the cell wall to disrupt the outer or inner membrane. Defensins are major antimicrobial peptide classified into α, β and θ defensins. α-Defensin expressed constitutively as pro-cryptdin irrespective of a microbial signal (Fig. 3). Later it is converted into mature-cryptdin by matrix metalloproteinase-7 (MMP-7) to defend microbial adherence to mucosa [101]. Mice lacking of the MMP-7 gene are unable to produce mature cryptdin and are highly susceptible to Salmonella typhimurium infection [102]. β-Defensin is expressed both in SI and LI epithelia through the signalling of nucleotide-binding oligomerization domain-containing protein 2 (NOD2) and depends on a microbial signal derived from the lumen. NOD2 deficiency in mice resulted in an increased susceptibility to Listeria monocytogenes infection due to lower expression of defensins [103]. θ-Defensin has only been reported in the leukocytes of rhesus macaques. Cathelicidin-related antimicrobial peptide (mCRAMP) is encoded by the gene Cnlp and its expression is restricted to surface colonic epithelia. Colonic epithelial cell extracts from Cnlp +/+ and mutant Cnlp−/− mice showed that lack of mCRAMP expression increased the adherence of enteric pathogen Citrobacter rodentium to intestinal epithelial cells [104]. The C-type lectins are a superfamily consisting of more than 1000 proteins having one or more characteristic C-type lectin-like domains. C-type lectins play an important role in innate and adaptive immunity to control the gut bacterial infections. Microorganism-associated molecular patterns activate toll-like receptors (TLRs) recruiting cytosolic adaptor myeloid differentiation primary-response protein 88 (MYD88) to restrict the penetration of bacteria through epithelial layer (Fig. 3). For example, RegIIIγ, a secreted antibacterial lectin effective against Gram-positive bacteria and maintaining a ~ 50 μm zone that physically separate the microbiota from the epithelial surface of SI [105].
Fig. 3.
Immune mechanisms for limiting the commensals within the epithelial layer. Innate immunity: (1) mucin glycoproteins of the goblet cells form a layer over the epithelia to restrict bacterial adhesion; (2) constitutive expression of α-defensins from epithelial cells without microbial signal; (3_ β-defensin expression by muramyl dipeptide of Gram positive bacteria mediated by cytosolic nucleotide-binding oligomerization domain-containing protein 2 (NOD2); (4) C-type lectin regenerating islet-derived protein 3γ (REG3γ) via microorganism-associated molecular patterns (MAMP) activates toll-like receptors (TLRs) recruiting cytosolic adaptor myeloid differentiation primary-response protein 88 (MYD88) to restrict bacterial penetration through epithelial layer; (5) dendritic cells (DC) located beneath the epithelial dome of Peyer’s patches take up bacteria and migrate to mesenteric lymph node and induce B cells to differentiate into IgA+ plasma cells for secretion of IgA. Transcytosis of IgA across the epithelial cell layer limits bacterial association with the epithelium. Adaptive immunity: transcytosis of bacteria through M cells and luminal antigen activates DC in the Peyer′s patch and helps to differentiate the naive CD4+ T cells into Treg and TR1 characterized by expression of anti-inflammatory cytokines transforming growth factor (TGFβ) and IL10. The TH1, TH2, and TH17 cells are characterized by proinflammatory cytokines including (1) IFNγ, (2) IL-4, 5 and 13 and (3) IL-23 and IL-17F, respectively
IgA
Secretory IgA (SIgA) serves as the third line of defence and protects the intestinal epithelia from enteric toxins and pathogenic microorganisms. Differential immune recognition of pathogens and commensals maintains immune tolerance and activation of the cells present in Pyers’ patches, lymphoid follicle or lamina propia. The macrophage-like non-migratory CX3CR1+ dendritic cells (DCs) of Peyer’s patches randomly capture luminal antigens through transepithelial transport. The captured antigen is transferred to migratory CD103+ DCs which move to the interfollicular area of Payer’s patches [106, 107]. The CD103+ DCs interacts with Treg cells to promote B cells for IgA-induction with the assistance of retinoic acid, transforming growth factor-β (TGF-β), IL-10 and cytokine thymic stromal lymphopoietin. Activation of Treg cells by commensals induces B cell to differentiate into plasma B cells for producing of IgA. Thereafter, the secreted IgA translocate from lymphoid sites to the intestinal lamina propria and transcytosis across the epithelial cell layer. Transcytosis of IgA in the lumen entraps bacteria in mucus to neutralize toxins and pathogens as blocking their access to epithelial receptors. It also facilitates their removal by peristaltic and mucociliary activities [108]. The population density of commensals vs. pathobionts or its associated endogenous molecules determine the IgA secretion in the gut. For example, endogenous polysaccharide A from a commensal, Bacteroides fragilis mediated an anti-inflammatory response characterized by induction of Treg cells and IL-10 and TGF-β. In addition, metabolites such as butyrate made by commensals can also activate the same anti-inflammatory response mediated by Treg cells. In dysbiotic condition, a certain subsets of commensal are increased in their abundance and act as “pathobionts” [94]. The bloom of pathobionts and breakdown of tolerance to immune system activate the pro-inflammatory response. For example, in experimental colitis H. hepaticus activated the TH17 cell which is mediated by inflammatory cytokines such as IL-17, IL-23 and TNF-α. The malnourishment of children is also a factor to alter the gut microbial composition and IgA responses. Malnourishment phenotype of Malawian infants can be transferred after the transplantation of 11 IgA(+) bacterial consortium to gnotobiotic mice. The transplantation of the bacterial consortium caused a rapid disruption of the intestinal epithelial barrier, weight loss and sepsis. These sepsis conditions were prevented by the administration of two IgA-targeted bacterial species from a healthy microbiota [109].
Innate and adaptive immunity
The innate immune response can discriminate between commensals and pathogens via germline-encoded pattern-recognition receptors. Toll like receptors (TLRs) identify the conserved microbial-associated molecular patterns including bacterial-derived lipopolysaccharide (LPS), lipoprotein, flagellin and unmethylated CpG-containing DNA of pathogens. The subfamilies of TLR1, TLR2 and TLR6 recognise lipids, whereas TLR7, TLR8 and TLR9 recognise nucleic acids. However, TLR5 identify flagellin and TLR4 can recognise several structurally unrelated ligands such as lipopolysaccharide and plant diterpene paclitaxel [110]. The cytosolic antigens including γ-d-glutamyl-meso-diaminopimelic acid and muramyl dipeptide of bacterial peptidoglycan layer are recognised by the NOD like receptors [111, 112]. Antigen-presenting cells like DCs and macrophages also express TLRs which ingest and degrade pathogens and express co-stimulatory molecules and cytokines. These cytokines help to differentiate the naive CD4+ T into T helper 1 (TH1), TH2, TH17 and Treg and TR1 cells (Fig. 3). A balance of Treg cells and the CD4+ effector T cells in the intestinal mucosa discriminate commensal and pathogenic microbial constituents. Microbial dysbiosis may induce inflammatory response mediated by TH1, TH2 and TH17 cells. The subset of TH1 and TH2 cell activation is characterized by the expression of pro-inflammatory cytokines including IL-4, IL-5, IFN-γ and IL-13. TH17 cell is characterized by the synthesis of IL-17 which stimulates stromal cells to express the pro-inflammatory cytokines including IL-6 and IL-8 and IL-22 [113, 114].
Microbiota produced metabolites in immunity
Short chain fatty acids (SCFAs)
The carbohydrates that escape from the digestion and absorption in SI are utilized by a number of microbes present in LI. The saccharolytic activities of the microbes present in LI ferment the undigested carbohydrates resulting in SCFAs production, viz. acetate, propionate and butyrate. SCFAs are either transported or diffused into host cells and sensed by binding with G protein-coupled receptors such as GPR41, GPR43 and GPR109A on epithelial and immune cells [71]. Butyrate binds with GPR43 and activates anti-inflammatory cytokine production such as TGFβ and IL-10 as well as upregulates the FOXP3 of Treg cells. Butyrate also inhibits histone deacetylase activity and down regulates the nuclear factor-κβ mediated inflammatory response. Experimental evidence indicated that acetate had greater binding affinity to GPR43 than butyrate. Feeding of acetate stimulated in secretion of IgA in the intestine of wild type mice but not in GPR43−/− knockout mice. Acetate induced expression of Aldh1a2 in DC which converts vitamin A into retinoic acid mediating the signalling of B and goblet cells in secretion of mucins and IgA to fortify the barrier function [115]. The knockout of the receptor GPR43 in mice showed an increased susceptibility to dextran sodium sulfate-induced colitis by increasing chemotaxis of neutrophils and inflammatory gene expression. Propionate and butyrate in combination also effective to inhibit LPS induced inflammation by activating Treg cell and reduced production of inflammatory cytokines such as IL-6 and IL-12 [116].
Aryl hydrocarbon receptor (AHR)
AHR is a cytoplasmic ligand-induced receptor expressed by epithelial and some tumour cells. Kynurenine, a metabolite of tryptophan is a ligand of AHR. Three groups of microbial metabolites, viz. (1) tryptophan derived (indole, indole-3-acetate and tryptamine), (2) bacterial virulence factors (phenazine, naphthoquinone and phthiocol) and (3) short chain fatty acids also binds to AHR. Some dietary components including flavonoids, stilbenes, carotenoids and indoles from plants are also ligands of AHR [71, 117]. AHR ligands prevent the infection of Citrobacter rodentium and Candida albicans by snatching of metal ions. Supplementation of dietary synthetic AHR ligands and transfusion of intraepithelial lymphocytes to Ahr−/− mice help to restore the epithelial barrier function and normalized dysbiotic bacterial population [118].
Polyamines
The polyamines are polycationic in nature and found in all the life forms. There are three major sources of polyamines, viz. food, cellular biosynthesis and microbial biosynthesis in the gut. The major polyamines are putrescine, spermidine and spermine synthesized from arginine by the enzyme arginase 1 (which converts arginine to ornithine). The ornithine decarboxylase (which synthesizes putrescine from ornithine) and other enzymes sequentially interconverts putrescine to spermidine and spermine [71]. Arginase 1 and nitric oxide synthase compete for arginine to produce either polyamines or nitric oxide to balance immune responses. Administration of arginine in combination with B. animalis subsp. lactis LKM512 was shown to increase colonic polyamines which lowered the colonic and circulatory TNF-α and IL-6 [119]. Apart from arginine supplementation, spermine can also inhibit the activation of M1 macrophage by suppressing the expression of ornithine decarboxylase in the synthesis of pro-inflammatory cytokines without modifying the expression anti-inflammatory mediators TGFβ and IL-10 [120]. Polyamine enrichment to breast feed rat pups accelerated maturation of intraepithelial CD8+ and CD4+ T cells with early appearance of B cells in spleen [121]. Higher production of mucus and secretory IgA was also observed in the SI that closely resembled to that of breast-fed mice. These findings indicated that polyamines in the diet provide immunological benefits to the host.
Structural
Hippocrates (400 bc) stated that “bad digestion is at the root of all evil” and “death sits in the bowels” suggesting that the gut may act as a “motor” of critical illness. This hypothesis states that overgrowth and imbalance of commensal flora increases gut permeability which may cause systemic sepsis that may lead to death [122]. The intestinal epithelium is a single layer of columnar cells which are tightly bound together by intercellular junctional complexes that regulate the paracellular permeability. The junctional complexes consist of tight junction (ZO; zonula occludens), adherens junction (zonula adherens) and desmosome. The adherens junctions are located beneath the tight junction and both together make the apical junctional complexes that are associated with actin cytoskeleton. A bridging between apical junctional complexes and actin filaments involved in cell–cell adhesion and intracellular signalling whereas adherens junctions and desmosomes provide the adhesive forces necessary for the maintenance of intercellular interactions [123]. The tight junction barrier exhibits both size and charge selectivity with two distinct routes across an intact epithelial monolayer, termed as the ‘pore’ and ‘leak’ pathways. The pore pathway refers to a high-capacity, size-selective and charge-selective route, whereas the leak pathway is a low-capacity pathway that has more limited selectivity [124]. The enterotoxins of some bacterial pathogens such as enteropathogenic E. coli, C. difficile and C. perfringens are capable of weakening the barrier functions of the tight junctions [123, 125, 126]. Cytokines are produced in the intestine including IL-4, IL-6, IL-13, IL-1β, TNF-α and IFN-γ promote to lose the tight junctions and increase the permeability. Cell culture and animal model based studies revealed that cytokines including IL-10, IL-17 and TGF-β renovate the intestinal barrier to decrease permeability [124, 127]. Nutrients and food factors such as glutamine deprivation, fatty acid molecules (eicosapentaenoic, docosahexaenoic, c-linoleic acids, capric acid and lauric acid), ethanol and acetaldehyde increase intestinal permeability. The amino acids glutamine and tryptophan and other nutritional factors including casein peptide, SCFAs vitamin A and D and polyphenolic molecules (quercetin and curcumin) restrict the infiltration of luminal content [128]. Furthermore, treatments of probiotics such as L. plantarum DSM 2648 and E. coli Nissle 1917 on Caco-2 cells have been shown to upregulate the expression of tight junction proteins [129, 130].
Neurological
The bidirectional communication between the gut and the brain is known as gut-brain axis (GBA). The gut is connected to the brain through the enteric nervous system (ENS) involving parallel outflow of the parasympathetic (vagus nerve) and sympathetic (prevertebral ganglia) nervous systems and hypothalamic- pituitary- adrenal axis (HPA) (Fig. 2) [131, 132]. The ENS is regarded as the “second brain” of the gut and consists of millions of neurons and is divided into two types of ganglia (myenteric and submucosal plexuses). Activities of gut microbiota can control the ENS as well as CNS via (1) production, expression and turnover of neurotransmitters and neurotrophic factors, (2) maintaining the intestinal barrier and tight junction integrity, (3) modulating the enteric sensory afferents, (4) production of bacterial metabolites and (5) mucosal immune regulation. The ENS can sense more than 30 neurotransmitters most of which are found in the CNS, such as acetylcholine, dopamine and serotonin. More than 90% of serotonin and 50% of dopamine originate in the gut which are mainly produced by the gut microbiota. These two neurotransmitters play an important role in transmission of “fight to flight” message to the brain in controlling of mood, happiness and pleasure of an individual [133]. Bacterial metabolites, particularly SCFAs were shown to induce tryptophan hydroxylase 1 in enterochromaffin cells to release serotonin in the gut. Serotonin can stimulate the sympathetic nervous system to influence memory and learning process. Emerging data supports the fact that dysbiosis in gut microbiota during functional GI disorders disrupts the GBA and leads to mood disorders. Experimental evidences also suggested that probiotic can induce brain derived neurotrophic factors in the hippocampus and cerebral cortex and regulate cognitive functions as well as muscle repair, regeneration, and differentiation [131]. In parallel, probiotics can modulate the HPA to reduce the release of cortisol from the adrenal glands. The attenuated level of cortisol and a complex interaction with the amygdala, hippocampus and hypothalamus to lower anxiety-like behaviour and stress reactivity [131]. A study on IBS grouped the IBS population into two subgroups, IBS1 (microbial profile distinct from healthy control) and healthy control like-IBS (microbial profile indistinguishable from healthy control). The microbial genera Blautia, Streptococcus and Bacteroides differentiated gut microbiota of the IBS1 patients and healthy controls. Such dysbiosis in gut microbiota was associated with the volume of the brain region including sensory- and salience-related regions and with a history of early life trauma [134]. A study from China showed a relationship between the gut microbiota with major depressive-like disorder (MDD). They found that abundance of Bacteroidetes, Proteobacteria and Actinobacteria and inadequacy of Firmicutes was linked to “active major depressive disorder” (high score of clinically significant depression) and “responded MDD” (reduction of MDD after 4 weeks of treatment) [135]. Treatment with efficacious probiotics like Bifidobacterium longum NCC3001 reduced depression in IBS patients and improved their quality of life. Although the probiotic bacterium had no effect on IBS symptoms, they however reduce the response of negative emotional stimuli in multiple regions of brain, including amygdala and fronto-limbic region [136]. The neurodegenerative disease Alzheimer’s is characterized by extracellular amyloid beta (Aβ) deposits and plaque formation in the brain. Gut microbial profiling of Aβ-precursor protein (APP) of transgenic mice exhibited a remarkable shift as compared to non-transgenic wild-type mice. There was a reduced abundance of Allobaculum, Akkermansia and unclassified genera in Rikenellaceae and S24-7 was found as compared to wild-type mice. Subsequently, they found that transplantation of gut microbiota from conventionally raised APP transgenic mice to germ-free APP transgenic mice increased the cerebral Aβ pathology unlike the transplantation of microbiota from wild-type mice [137].
Therapeutic interventions
Probiotics
Élie Metchnikoff (1905) believed that some Bulgarian peasants who consumed a fermented milk product called ‘kefir’ had better health and longer lives. Subsequently, the bacterium Lactobacillus delbrueckii ssp. subsp. bulgaricus was isolated from ‘kefir’ and has since been used in commercial curd preparations. During the First World War, the physician Alfred Nissle isolated a strain of E. coli (E. coli Nissle 1917) from a healthy soldier from an army suffering from infectious diarrhoea and has since been used as a commercial probiotic. Such empirical observations introduced the concept of probiotics, presently defined as “live microorganisms which, when administered in adequate amounts confer health benefit to the host” [138, 139]. Commercially available probiotics are commonly of Bifidobacterium, Lactobacillus and Streptococcus genera as well as yeasts of the Saccharomyces genus either singly or in combinations. To date, the mechanisms of action of probiotics are ascribed to their adhesion to the intestinal–lumen interface, competition with pathogens for nutrients and receptor binding, fortification of mucosal barrier, promotion of innate and adaptive immune responses of host, production of bacteriocins, production of signalling molecules via CNS and modulation of cell kinetics [140]. For adhesion to the mucosal surface of the intestine, probiotics such as Lactococcus lactis ssp. lactis BGKP1 exhibit auto-aggregation and its mucin binding surface protein attaches to the gastric mucin protein MUC5AC [141]. The protein “Transaldolase” of B. bifidum has also been reported to act as an important colonisation factor. Recently, a group of proteins called moonlighting proteins including elongation factor Tu (EF-Tu), α-enolase, glyceraldehyde-3-phosphate dehydrogenase, etc. have been shown to play an important role in colonisation of human gut by degradation of extracellular matrix or by facilitating a close contact with the epithelium [142, 143]. Directly, probiotics exhibit antimicrobial effect against pathogens by secreting an array of antibacterial substances which includes organic acids, hydrogen peroxide and bacteriocins. These compounds mainly help to reduce the viable count of pathogens as well as their metabolisms and toxin production through the reduction of luminal pH and production of SCFAs. Over 60% of the human population suffers from lactose intolerance due to lower level and activity of lactase enzyme. The lactose intolerance depends on several factors, such as age, race and integrity of the SI membrane and transit time. The probiotics can produce lactase which promote the digestion of unabsorbed lactose and prevent acid and gas formation [144]. Probiotics also prevent the cardiovascular disease by regulating the metabolism of lipid, viz. (1) decreasing the absorption of cholesterol through co-precipitation with the deconjugated bile salts, (2) incorporating and assimilating the cholesterol in the cell membrane, (3) converting the cholesterol into coprostanol and (4) inhibiting the expression of cholesterol transporter in the enterocytes [145]. Recently, researchers are exploring the role of probiotics in modulating of inflammatory disease, obesity, type 2 diabetes, cardiovascular disease, autoimmuno psoriasis, arthritis and cancer. A randomized double-blind placebo-controlled study of a formulation of probiotics with fermented milk containing (S. salivarius subsp. thermophilus, E. faecium, L. rhamnosus GG, L. acidophilus, L. plantarum, L. paracasei, L. delbrueckii subsp. Bulgaricus, B. breve and B. animalis subsp. lactis) and fructooligosaccharides (FOS) was found to be effective in management of constipation associated with Parkinson’s disease [146]. Another formulation of probiotics in which consumption of milk (200 ml/day) containing L. acidophilus, L. casei, B. bifidum and L. fermentum (2 × 109 CFU/g for each) for 12 weeks was shown to improve cognitive function in elderly Alzheimer’s patients without effecting the biomarkers of oxidative stress and inflammation, fasting plasma glucose and lipid profiles [147]. Based on more than 38 studies, the probiotic bacteria Bifidobacteria and Lactobacilli have been reported to improve psychiatric disorder including anxiety, depression, autism spectrum disorder, obsessive–compulsive disorder and memory abilities (spatial and non-spatial) [148]. A large-scale trial in India has shown that administration of L. plantarum and FOS to new-borns within 7 days of birth reduced neonatal sepsis and morbidity rate to 5.4% as compared to 9% in their first two months of life [149]. In Finland, results of 4 trials conducted between 1997 and 2012 (with 10–15 years of follow-up) showed that perinatal administration of probiotic L. rhamnosus GG singly or in combinations with B. lactis Bb-12, L. paracasei ST11 and B. longum BL999 reduced allergic diseases [150]. Treatment with Saccharomyces boulardii for 12 weeks in HIV patients decreased population of Clostridiaceae family and reduced chronic inflammation [151].
Prebiotics
Gibson and Roberfroid (1995) defined prebiotics as a class of compounds that stimulate the growth and/or activity of a limited number of beneficial bacteria (Lactobacilli and/or Bifidobacteria) in the colon to improve host health [152]. However, this definition has now been broadened and included carbohydrate and non-carbohydrate substances that confer microbiota-mediated health benefits [153]. The European Union has approved prebiotics (inulin, FOS and GOS) as safe food ingredients whereas the FDA has not yet recognised these. In the GI tract, prebiotics can resist gastric acidity and can reach in LI where their selective utilization by microbiota produces SCFAs. Prebiotics such as FOS and GOS including the patented GOS produced by Bimuno (B-GOS) modulate neural growth factors such as brain-derived neurotrophic factor, neurotransmitters and synaptic proteins including synaptophysin and NMDA receptor subunits [154, 155]. Hence, prebiotic utilization by gut microbiota may confer health benefits elsewhere in the body. For example, GOS stimulated growth of Bifidobacteria in the mouse gut that led to modulation of cortical IL-1β and 5-HT2A receptor expression that reduced anxiety levels and increased brain barrier function in obese mice [156, 157]. Experimental evidence also suggested that prebiotic supplementation was effective against stunted memory, Alzheimer’s disease and dementia. In rats, improved memory was observed due to administration of polydextrose/GOS mixture and oligofructose-enriched inulin [158, 159]. Previously, it was reported that the metabolism of sialic acid of sialyllactose enhanced memory [160]. Recently, deficiency of sialylated human milk oligosaccharides of Malawian mothers have shown to cause stunted infants. Transplantation of faecal microbiota of a 6-month-old stunted baby to germ-free animal model on Malawian diet prototype with sialylated bovine milk oligosaccharides promoted quality composition of gut microbiota for anabolic process, lean body mass gain and brain metabolism [161]. A similar study on infant formula showed that bovine milk-derived oligosaccharides and B. animalis ssp. lactis (CNCM I-3446) improved the GI health markers (microbiota pattern, faecal IgA and stool pH) as compared to breastfed infants. No difference has been found in diarrhoea and febrile infection across the studied population [162].
Faecal microbiota transplantation (FMT)
Faecal microbiota transplantation (FMT) refers to a “stool transplant” into the GI tract from a healthy individual to a patient for the treatment of specific diseases. During Dong-jin dynasty in the fourth century, the benefits of FMT were recognised in Chinese medicine. The doctor prescribed the use of faecal suspension orally for patients who suffered from food poisoning or severe diarrhoea. In the sixteenth century, FMT was known as “yellow soup” and a medical miracle that brought patients back from the brink of death. In the seventeenth century, an Italian anatomist, Fabricius Aquapendente, introduced faecal therapy in veterinary medicine for compelling rumination. In United States, the FDA has considered human faeces as an experimental drug since 2013. Now, FMT has been used experimentally to treat GI diseases including colitis, constipation, IBD, and chronic fatigue syndrome, Parkinson’s disease, multiple sclerosis [163, 164]. In modern medicine, FMT has been shown to be more effective against pseudomembranous colitis and C. difficile infection (CDI) than neurological and other complications. The success rate of FMT on CDI patients was found to be over 90% who had failed to respond to antibiotic treatment [164, 165]. In recent techniques of FMT, enemas, nasogastric intubation, enteric intubation, colonoscopes and gastroscopes are used for faecal transplantation. The treatment of CDI with antibiotics including metronidazole, vancomycin, or fidaxomicin may cause recurrent infection, antibiotics resistance and dysbiosis of normal gut microbiota [166]. A study on CDI patients showed profound dysbiosis commonly characterized by complete disappearance of Bacteroidetes with a marked reduction in Firmicutes and massive increases in the relative abundances of Proteobacteria [167]. FMT increased the abundance of the families Bacteroidaceae, Rikenellaceae and Porphyromonadaceae under Bacteroidetes and Ruminococcaceae, Lachnospiraceae, Verrucomicrobiaceae and unclassified Clostridiales under Firmicutes and reduced the population of Proteobacteria in CDI patients. In addition to dysbiosis of normal microbiota in CDI, presence of primary bile salts such as taurocholate, cholate and chenodeoxycholic acid stimulate the germination of C. difficile spore. In contrast, post-FMT increased secondary biles (lithocholic acid and deoxycholate) of donor samples that inhibit spore germination as well as vegetative growth of C. difficile [168]. A systematic review on FMT based on 18 studies with 611 patients highlighted that primary cure, overall recurrence, early and late recurrence rates of CDI were 91.2, 5.5, 2.7 and 1.7%, respectively. The adverse events such as IBD flare, infectious and autoimmune diseases did not have any significant relation with FMT [169]. Moreover, in-depth study on understanding of the mechanisms of FMT, optimisation of methodologies for FMT including donor screening, stool preparation and routes of administration may improve the efficacy and safety for a better clinical application.
Drugs
The gut microbial enzymes process pharmaceutical compounds and alter pharmacokinetics, absorption, distribution, metabolism and elimination [170, 171]. Drug efficacy may vary from individual to individual due to variation in the microbial composition of their gut. In 1971, a preclinical study in rats showed that oral administration of prontosil or Neoprontosil transformed to inactive sulphanilamide by gut microbiota before absorption [172]. Similarly, in the 1980s, researchers discovered that the bacterium Eggerthella lenta (Actinobacteria phyla) inactivated digoxin (a cardiac glycoside) and its bioavailability, therapeutic index and induced toxicity [173]. Cardiac glycoside reductase is the homolog of FAD-dependent fumarate reductases responsible for the reduction and inactivation of digoxin [174]. Paracetamol, a common analgesic and antipyretic drug is detoxified in the liver by glucuronidation and sulfation. The microbial metabolite p-cresol produced by C. difficile acts as competitive inhibitor of hepatic sulfotransferases for sulfonation that elevate the risk of hepatotoxicity [175]. Irinotecan (CPT-11) is a chemotherapeutic drug that is transformed into the active form (SN-38) by carboxylesterases in the host tissues and acts as an inhibitor of topoisomerase I in colon tumor cells. Hepatic UDP-glucuronosyltransferases conjugate SN-38 into nontoxic SN-38-G before secreting into the SI. However, conversion of the nontoxic SN-38-G to SN-38 by β-glucuronidase activity of E. coli, Bacteroides vulgatus and C. ramosum leads to diarrhoea, inflammation and anorexia [171, 176]. Levodopa, a dopamine precursor undergoes decarboxylation within the central nervous system and exerts dopaminergic effect against Parkinson’s disease. Gut microbiota can transform levodopa into m-tyramine and m-hydroxyphenylacetic by dehydroxylation that leads to decreased bioavailability and induces H. pylori infection [176]. Metformin is a popular drug and used in controlling T2D. The treatment of metformin increased the abundance of butyrate producers such as Roseburia spp., Subdoligranulum spp. and a cluster of butyrate-producing Clostridiales spp. as compared to a metformin-untreated group. Microbial shifts under metformin treatment enhanced its efficacy to control glucose level in plasma [177]. Similarly, Gegen Qinlian Decoction is a traditional Chinese herbal formula used for treatment of T2D patients that increased F. prausnitzii and short-chain fatty acid production [178]. Berberine is a Chinese herb (Coptis chinensis) that also induces the growth of Allobaculum and Blautia, as well as SCFA levels in HFD-fed rats [179]. These findings suggest a role for the gut microbiota in drug metabolism that needs to be considered as a new parameter for assessment of a drug’s therapeutic index to treat diseases.
Future perspective
Modern lifestyle and the growing environmental stresses are bringing new challenges to the human race to sustain on this planet. The human race cannot evolve quickly to adapt to such changes. However, the hidden design in the “hologenome” of human body may help to adapt to the changing environment. Nowadays, there is evidence that diets with smart microbial communities may assist in human-microbe symbiosis for better health. An in-depth understanding of the functioning of microbiota in different health and disease conditions may further help to formulate a strategy for targeted intervention as and when required.
Acknowledgements
The authors are thankful to Department of Science and Technology, India and author A.A. is thankful to Department of Biotechnology, India for supporting with fellowship under Unit of Excellence Project (BT/550/NE/U-Excel/2014). The authors are also thankful to Dr. Josephine Brennan (Scientist, Department of Agriculture, Food and Marine, Ireland) for help in improving the English of the manuscript.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. MBio. 2016;7(2):e01395-01315. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J. In a map for human life, count the microbes, too. Science. 2001;291(5512):2316. doi: 10.1126/science.291.5512.2316b. [DOI] [PubMed] [Google Scholar]
- 4.NHW Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001;9(5):206–208. doi: 10.1016/S0966-842X(01)02041-8. [DOI] [PubMed] [Google Scholar]
- 6.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 10.Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28(1):237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, et al. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 2016;8(1):72. doi: 10.1186/s13073-016-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15(12):1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 15.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Li Y, Li S, Hu N, He Y, Pong R, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Lederberg J. Infectious history. Science. 2000;288(5464):287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 19.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allesina S, Tang S. Stability criteria for complex ecosystems. Nature. 2012;483(7388):205–208. doi: 10.1038/nature10832. [DOI] [PubMed] [Google Scholar]
- 21.McNally L, Brown SP. Microbiome***: Ecology of stable gut communities. Nat Microbiol. 2016;1:15016. doi: 10.1038/nmicrobiol.2015.16. [DOI] [PubMed] [Google Scholar]
- 22.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 23.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, et al. Rethinking “enterotypes”. Cell Host Microbe. 2014;16(4):433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang C, Tseng HC, Chen HM, Wang WC, Chiu CM, Chang JY, et al. Diversity and enterotype in gut bacterial community of adults in Taiwan. BMC Genom. 2017;18(Suppl 1):932. doi: 10.1186/s12864-016-3261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapira M. Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol Evol. 2016;31(7):539–549. doi: 10.1016/j.tree.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41(2):558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardone G, Compare D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United Eur Gastroenterol J. 2015;3(3):255–260. doi: 10.1177/2050640614566846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Aidy S, van den Bogert B, Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr Opin Biotechnol. 2015;32:14–20. doi: 10.1016/j.copbio.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9(1):123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146(6):1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Muñoz ME, Arrieta M-C, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(1):48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol. 2009;48(1):8–12. doi: 10.1111/j.1472-765X.2008.02475.x. [DOI] [PubMed] [Google Scholar]
- 38.Farage MA, Miller KW, Sobel JD. Dynamics of the vaginal ecosystem–hormonal influences. Infect Dis Res Treat. 2010;3:1. [Google Scholar]
- 39.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. 2015;3(1):419–445. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F, Yu T, Huang G, Cai D, Liang X, Su H, et al. Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J Microbiol Biotechnol. 2015;25(8):1195–1204. doi: 10.4014/jmb.1410.10014. [DOI] [PubMed] [Google Scholar]
- 44.Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S. Inflammaging and ‘Garb-aging’. Trends Endocrinol Metab. 2017;28:199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, et al. Microbial genetic composition tunes host longevity. Cell. 2017;169(7):1249–1262 e1213. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richards AL, Burns MB, Alazizi A, Barreiro LB, Pique-Regi R, Blekhman R, et al. Genetic and transcriptional analysis of human host response to healthy gut microbiota. mSystems. 2016;1(4):e00067-16. doi: 10.1128/mSystems.00067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48(11):1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 51.Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. PLoS One. 2015;10(11):e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One. 2011;6(5):e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 54.Baas-Becking LGM (1934) Geobiologie; of inleiding tot de milieukunde. WP Van Stockum and Zoon NV
- 55.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25(13):1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 57.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015;9(9):1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dehingia M, Devi KT, Talukdar NC, Talukdar R, Reddy N, Mande SS, et al. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci Rep. 2015;5:18563. doi: 10.1038/srep18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dehingia M, Sen S, Bhaskar B, Joishy TK, Deka M, Talukdar NC, et al. Ethnicity influences gut metabolites and microbiota of the tribes of Assam, India. Metabolomics. 2017;13(6):69. doi: 10.1007/s11306-017-1206-y. [DOI] [Google Scholar]
- 61.Adak A, Maity C, Ghosh K, Pati BR, Mondal KC. Dynamics of predominant microbiota in the human gastrointestinal tract and change in luminal enzymes and immunoglobulin profile during high-altitude adaptation. Folia Microbiol. 2013;58(6):523–528. doi: 10.1007/s12223-013-0241-y. [DOI] [PubMed] [Google Scholar]
- 62.Adak A, Maity C, Ghosh K, Mondal KC. Alteration of predominant gastrointestinal flora and oxidative damage of large intestine under simulated hypobaric hypoxia. Z Gastroenterol. 2014;52(2):180–186. doi: 10.1055/s-0033-1336007. [DOI] [PubMed] [Google Scholar]
- 63.Adak A, Ghosh K, Mondal KC. Modulation of small intestinal homeostasis along with its microflora during acclimatization at simulated hypobaric hypoxia. Indian J Exp Biol. 2014;52(11):1098–1105. [PubMed] [Google Scholar]
- 64.Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell. 2015;163(6):1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Bergman EN. Energy contributions of volatile fatty-acids from the gastrointestinal-tract in various species. Physiol Rev. 1990;70(2):567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 66.Anderson JW, Baird P, Davis RH, Jr, Ferreri S, Knudtson M, Koraym A, et al. Health benefits of dietary fiber. Nutr Rev. 2009;67(4):188–205. doi: 10.1111/j.1753-4887.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 67.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 69.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73(4):1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 71.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2017;46(D1):D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Portune KJ, Beaumont M, Davila AM, Tome D, Blachier F, Sanz Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: the two sides of the coin. Trends Food Sci Technol. 2016;57:213–232. doi: 10.1016/j.tifs.2016.08.011. [DOI] [Google Scholar]
- 75.Hentges DJ, Maier BR, Burton GC, Flynn MA, Tsutakawa RK. Effect of a high-beef diet on the fecal bacterial flora of humans. Cancer Res. 1977;37(2):568–571. [PubMed] [Google Scholar]
- 76.Swiatecka D, Narbad A, Ridgway KP, Kostyra H. The study on the impact of glycated pea proteins on human intestinal bacteria. Int J Food Microbiol. 2011;145(1):267–272. doi: 10.1016/j.ijfoodmicro.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 77.Meddah AT, Yazourh A, Desmet I, Risbourg B, Verstraete W, Romond MB. The regulatory effects of whey retentate from bifidobacteria fermented milk on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) J Appl Microbiol. 2001;91(6):1110–1117. doi: 10.1046/j.1365-2672.2001.01482.x. [DOI] [PubMed] [Google Scholar]
- 78.Leitao-Goncalves R, Carvalho-Santos Z, Francisco AP, Fioreze GT, Anjos M, Baltazar C, et al. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 2017;15(4):e2000862. doi: 10.1371/journal.pbio.2000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary fat-induced taurocholic acid production promotes pathobiont and colitis in IL-10−/− mice. Gastroenterology. 2012;142(5):S-12. doi: 10.1016/S0016-5085(12)60043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk. Curr Opin Clin Nutr Metab Care. 2015;18(5):515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benítez-Páez A, Del Pulgar EMG, Kjølbæk L, Brahe LK, Astrup A, Larsen L, et al. Impact of dietary fiber and fat on gut microbiota re-modeling and metabolic health. Trends Food Sci Technol. 2016;57:201–212. doi: 10.1016/j.tifs.2016.11.001. [DOI] [Google Scholar]
- 82.Vaughn AC, Cooper EM, DiLorenzo PM, O’Loughlin LJ, Konkel ME, Peters JH, et al. Energy-dense diet triggers changes in gut microbiota, reorganization of gut-brain vagal communication and increases body fat accumulation. Acta Neurobiol Exp. 2017;77(1):18–30. doi: 10.21307/ane-2017-033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7(8):678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 85.Selwyn FP, Csanaky IL, Zhang Y, Klaassen CD. Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab Dispos. 2015;43(10):1544–1556. doi: 10.1124/dmd.115.065276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 87.Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med. 2017;56:54–65. doi: 10.1016/j.mam.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 88.Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, Ferguson D, et al. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Rep. 2017;19(12):2451–2461. doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6(2):e02481-02414. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thibodeaux CJ, van der Donk WA. Converging on a mechanism for choline degradation. Proc Natl Acad Sci USA. 2012;109(52):21184–21185. doi: 10.1073/pnas.1219534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marin L, Miguelez EM, Villar CJ, Lombo F. Bioavailability of dietary polyphenols and gut microbiota metabolism: antimicrobial properties. Biomed Res Int. 2015;2015:905215. doi: 10.1155/2015/905215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Romo-Vaquero M, Garcia-Villalba R, Gonzalez-Sarrias A, Beltran D, Tomas-Barberan FA, Espin JC, et al. Interindividual variability in the human metabolism of ellagic acid: contribution of Gordonibacter to urolithin production. J Funct Foods. 2015;17:785–791. doi: 10.1016/j.jff.2015.06.040. [DOI] [Google Scholar]
- 93.Most J, Penders J, Lucchesi M, Goossens GH, Blaak EE. Gut microbiota composition in relation to the metabolic response to 12-week combined polyphenol supplementation in overweight men and women. Eur J Clin Nutr. 2017;71(9):1040–1045. doi: 10.1038/ejcn.2017.89. [DOI] [PubMed] [Google Scholar]
- 94.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 97.Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74(6):1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105(11):2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 99.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106(34):14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Navarro-Garcia F, Gutierrez-Jimenez J, Garcia-Tovar C, Castro LA, Salazar-Gonzalez H, Cordova V. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect Immun. 2010;78(10):4101–4109. doi: 10.1128/Iai.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275(51):40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 102.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, et al. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286(5437):113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 103.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104(3):997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iimura M, Gallo RL, Hase K, Miyamoto Y, Eckmann L, Kagnoff MF. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J Immunol. 2005;174(8):4901–4907. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 105.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303(5664):1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 107.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2(4):361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101(7):1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7(276):276ra224–276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 111.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 112.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307(5710):731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 113.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22(1):283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 114.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10(4):946. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147(3):629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 119.Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep. 2014;4:4548. doi: 10.1038/srep04548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang M, Wang H, Tracey KJ. Regulation of macrophage activation and inflammation by spermine: a new chapter in an old story. Crit Care Med. 2000;28(4 Suppl):N60–66. doi: 10.1097/00003246-200004001-00007. [DOI] [PubMed] [Google Scholar]
- 121.Perez-Cano FJ, Gonzalez-Castro A, Castellote C, Franch A, Castell M. Influence of breast milk polyamines on suckling rat immune system maturation. Dev Comp Immunol. 2010;34(2):210–218. doi: 10.1016/j.dci.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 122.Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock. 2007;28(4):384–393. doi: 10.1097/shk.013e31805569df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, et al. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 2015;347(6223):775–778. doi: 10.1126/science.1261833. [DOI] [PubMed] [Google Scholar]
- 124.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14(1):9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yuhan R, Koutsouris A, Savkovic SD, Hecht G. Enteropathogenic Escherichia coli-induced myosin light chain phosphorylation alters intestinal epithelial permeability. Gastroenterology. 1997;113(6):1873–1882. doi: 10.1016/S0016-5085(97)70006-4. [DOI] [PubMed] [Google Scholar]
- 126.Hecht G, Koutsouris A, Pothoulakis C, LaMont JT, Madara JL. Clostridium difficile toxin B disrupts the barrier function of T 84 monolayers. Gastroenterology. 1992;102(2):416–423. doi: 10.1016/0016-5085(92)90085-D. [DOI] [PubMed] [Google Scholar]
- 127.Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res. 2015;13(1):11–18. doi: 10.5217/ir.2015.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141(5):769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 129.Anderson RC, Cookson AL, McNabb WC, Kelly WJ, Roy NC. Lactobacillus plantarum DSM 2648 is a potential probiotic that enhances intestinal barrier function. FEMS Microbiol Lett. 2010;309(2):184–192. doi: 10.1111/j.1574-6968.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- 130.Putaala H, Salusjarvi T, Nordstrom M, Saarinen M, Ouwehand AC, Bech Hansen E, et al. Effect of four probiotic strains and Escherichia coli O157:H7 on tight junction integrity and cyclo-oxygenase expression. Res Microbiol. 2008;159(9–10):692–698. doi: 10.1016/j.resmic.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 131.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28(2):203. [PMC free article] [PubMed] [Google Scholar]
- 132.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain–gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6(5):306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sarkar A, Lehto SM, Harty S, Dinan TG, Cryan JF, Burnet PW. Psychobiotics and the manipulation of bacteria–gut–brain signals. Trends Neurosci. 2016;39(11):763–781. doi: 10.1016/j.tins.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Labus JS, Hollister EB, Jacobs J, Kirbach K, Oezguen N, Gupta A, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017;5(1):49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21(6):786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 136.Pinto-Sanchez MI, Hall GB, Ghajar K, Nardelli A, Bolino C, Lau JT, et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: a pilot study in patients with irritable bowel syndrome. Gastroenterology. 2017;153:448–459. doi: 10.1053/j.gastro.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 137.Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy K, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep. 2017;7:41802. doi: 10.1038/srep41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nissle A. Old and new experiences on therapeutic successes by restoration of the colonic flora with mutaflor in gastrointestinal diseases. Die Med Welt. 1961;29:1519. [PubMed] [Google Scholar]
- 139.Podolsky SH. Cultural divergence: Elie Metchnikoff’s Bacillus bulgaricus therapy and his underlying concept of health. Bull Hist Med. 1998;72(1):1–27. doi: 10.1353/bhm.1998.0056. [DOI] [PubMed] [Google Scholar]
- 140.Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194(9):4081–4087. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lukic J, Strahinic I, Jovcic B, Filipic B, Topisirovic L, Kojic M, et al. Different roles for lactococcal aggregation factor and mucin binding protein in adhesion to gastrointestinal mucosa. Appl Environ Microbiol. 2012;78(22):7993–8000. doi: 10.1128/AEM.02141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gonzalez-Rodriguez I, Sanchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, et al. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol. 2012;78(11):3992–3998. doi: 10.1128/AEM.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19(8):415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- 144.Oak SJ, Jha R. The effects of probiotics in lactose intolerance: a systematic review. Crit Rev Food Sci Nutr. 2018 doi: 10.1080/10408398.2018.1425977. [DOI] [PubMed] [Google Scholar]
- 145.Cho YA, Kim J. Effect of probiotics on blood lipid concentrations: a meta-analysis of randomized controlled trials. Medicine. 2015;94(43):e1714. doi: 10.1097/MD.0000000000001714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Barichella M, Pacchetti C, Bolliri C, Cassani E, Iorio L, Pusani C, et al. Probiotics and prebiotic fiber for constipation associated with Parkinson disease: an RCT. Neurology. 2016;87(12):1274–1280. doi: 10.1212/WNL.0000000000003127. [DOI] [PubMed] [Google Scholar]
- 147.Akbari E, Asemi Z, Daneshvar Kakhaki R, Bahmani F, Kouchaki E, Tamtaji OR, et al. Effect of probiotic supplementation on cognitive function and metabolic status in Alzheimer’s disease: a randomized, double-blind and controlled trial. Front Aging Neurosci. 2016;8:256. doi: 10.3389/fnagi.2016.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang H, Lee IS, Braun C, Enck P. Effect of probiotics on central nervous system functions in animals and humans: a systematic review. J Neurogastroenterol Motil. 2016;22(4):589–605. doi: 10.5056/jnm16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548(7668):407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 150.Lundelin K, Poussa T, Salminen S, Isolauri E. Long-term safety and efficacy of perinatal probiotic intervention: evidence from a follow-up study of four randomized, double-blind, placebo-controlled trials. Pediatr Allergy Immunol. 2017;28(2):170–175. doi: 10.1111/pai.12675. [DOI] [PubMed] [Google Scholar]
- 151.Villar-García J, Güerri-Fernández R, Moya A, González A, Hernández JJ, Lerma E, et al. Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: a double-blind, randomised, placebo-controlled trial. PLoS One. 2017;12(4):e0173802. doi: 10.1371/journal.pone.0173802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125(6):1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 153.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 154.Savignac HM, Corona G, Mills H, Chen L, Spencer JP, Tzortzis G, et al. Prebiotic feeding elevates central brain derived neurotrophic factor, N-methyl-d-aspartate receptor subunits and d-serine. Neurochem Int. 2013;63(8):756–764. doi: 10.1016/j.neuint.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Williams S, Chen L, Savignac HM, Tzortzis G, Anthony DC, Burnet PW. Neonatal prebiotic (BGOS) supplementation increases the levels of synaptophysin, GluN2A-subunits and BDNF proteins in the adult rat hippocampus. Synapse. 2016;70(3):121–124. doi: 10.1002/syn.21880. [DOI] [PubMed] [Google Scholar]
- 156.Savignac HM, Couch Y, Stratford M, Bannerman DM, Tzortzis G, Anthony DC, et al. Prebiotic administration normalizes lipopolysaccharide (LPS)-induced anxiety and cortical 5-HT2A receptor and IL1-beta levels in male mice. Brain Behav Immun. 2016;52:120–131. doi: 10.1016/j.bbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Cossio LF, Fourrier C, Sauvant J, Everard A, Capuron L, Cani PD, et al. Impact of prebiotics on metabolic and behavioral alterations in a mouse model of metabolic syndrome. Brain Behav Immun. 2017;64:33–49. doi: 10.1016/j.bbi.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 158.Messaoudi M, Rozan P, Nejdi A, Hidalgo S, Desor D. Behavioural and cognitive effects of oligofructose-enriched inulin in rats. Brit J Nutr. 2005;93(Suppl 1):S27–30. doi: 10.1079/BJN20041348. [DOI] [PubMed] [Google Scholar]
- 159.Waworuntu R, Hain H, Chang Q, Thiede L, Hanania T, Berg B. Dietary prebiotics improve memory and social interactions while reducing anxiety when provided early in life to normally developing rodents (6375) FASEB J. 2014;28(1 Suppl):637.635. [Google Scholar]
- 160.Wang B, Yu B, Karim M, Hu H, Sun Y, McGreevy P, et al. Dietary sialic acid supplementation improves learning and memory in piglets. Am J Clin Nutr. 2007;85(2):561–569. doi: 10.1093/ajcn/85.2.561. [DOI] [PubMed] [Google Scholar]
- 161.Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164(5):859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Radke M, Picaud JC, Loui A, Cambonie G, Faas D, Lafeber HN, et al. Starter formula enriched in prebiotics and probiotics ensures normal growth of infants and promotes gut health: a randomized clinical trial. Pediatr Res. 2017;81(4):622–631. doi: 10.1038/pr.2016.270. [DOI] [PubMed] [Google Scholar]
- 163.Rossen NG, MacDonald JK, de Vries EM, D’Haens GR, de Vos WM, Zoetendal EG, et al. Fecal microbiota transplantation as novel therapy in gastroenterology: a systematic review. World J Gastroenterol. 2015;21(17):5359–5371. doi: 10.3748/wjg.v21.i17.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol. 2012;107(11):1755. doi: 10.1038/ajg.2012.251. [DOI] [PubMed] [Google Scholar]
- 165.Eiseman Á, Silen W, Bascom G, Kauvar A. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44(5):854–859. [PubMed] [Google Scholar]
- 166.Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA. 2015;313(4):398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, et al. Ursodeoxycholic acid inhibits Clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol. 2016;50(8):624–630. doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G310–319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li YT, Cai HF, Wang ZH, Xu J, Fang JY. Systematic review with meta-analysis: long-term outcomes of faecal microbiota transplantation for Clostridium difficile infection. Aliment Pharmacol Ther. 2016;43(4):445–457. doi: 10.1111/apt.13492. [DOI] [PubMed] [Google Scholar]
- 170.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 171.Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7(2):123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 172.Gingell R, Bridges JW, Williams RT. The role of the gut flora in the metabolism of prontosil and neoprontosil in the rat. Xenobiotica. 1971;1(2):143–156. doi: 10.3109/00498257109044386. [DOI] [PubMed] [Google Scholar]
- 173.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305(14):789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 174.Koppel N, Bisanz J, Turnbaugh P, Balskus EP. Characterization of a cardiac drug-inactivating enzyme from the prominent human gut microbe. Eggerthella lenta. FASEB J. 2017;31(1 Suppl):608.603. [Google Scholar]
- 175.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–222. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9(3):552–562. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7(8):e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]