Abstract
“Cellular reprogramming” facilitates the generation of desired cellular phenotype through the cell fate transition by affecting the mitochondrial dynamics and metabolic reshuffle in the embryonic and somatic stem cells. Interestingly, both the processes of differentiation and dedifferentiation witness a drastic and dynamic alteration in the morphology, number, distribution, and respiratory capacity of mitochondria, which are tightly regulated by the fission/fusion cycle, and mitochondrial clearance through autophagy following mitochondrial fission. Intriguingly, mitophagy is said to be essential in the differentiation of stem cells into various lineages such as erythrocytes, eye lenses, neurites, myotubes, and M1 macrophages. Mitophagy is also believed to play a central role in the dedifferentiation of a terminally differentiated cell into an induced pluripotent cell and in the acquisition of ‘stemness’ in cancer cells. Mitophagy-induced alteration in the mitochondrial dynamics facilitates metabolic shift, either into a glycolytic phenotype or into an OXPHOS phenotype, depending on the cellular demand. Mitophagy-induced rejuvenation of mitochondria regulates the transition of bioenergetics and metabolome, remodeling which facilitates an alteration in their cellular developmental capability. This review describes the detailed mechanism of the process of mitophagy and its association with cellular programming through alteration in the mitochondrial energetics. The metabolic shift post mitophagy is suggested to be a key factor in the cell fate transition during differentiation and dedifferentiation.
Keywords: Cellular reprogramming, Mitophagy, Metabolic shift, Stemness, Differentiation
Introduction
The term “cellular reprogramming” refers to the conversion of a specific cell type into another. This concept came into existence after John Gurdon’s landmark experiments on Xenopus laevis, which described the significance of nuclear differentiation during embryonic development, following the discovery of the method for testing the potentialities of nuclei from embryonic cells by King and Briggs [1, 2]. Later, in the late twentieth century, Wilmut et al. demonstrated that modifying somatic cell fate is possible even in mammals, after the successful birth of Dolly, a cloned sheep [3]. Specifically, it has also been demonstrated that cellular pluripotency can be induced in a differentiated somatic cell by direct reprogramming in which a somatic cell type such as a fibroblast is converted to a pluripotent cell type—the induced pluripotent stem cell (iPSC). Encouraged from these studies, Takahashi and Yamanaka in [4] revolutionized stem cell biology when they partially reprogrammed the mouse embryonic fibroblasts into iPSCs by the overexpression of four mouse transcription factors—Oct4, Sox2, Klf4, and c-Myc (mOSKM)—using retroviral vectors, without requiring oocyte cytoplasm [4]. Moreover, extensive research has led to the generation of completely reprogrammed mouse iPSC lines that have functional equivalence with mouse embryonic stem cells (ESCs) [5, 6]. Follow-up work further re-established the reprogramming process in human cells, when iPSCs were derived by the ectopic expression of hOSKM transcription factors or by replacement with Klf4 and c-Myc with NANOG and Lin28 [7, 8].
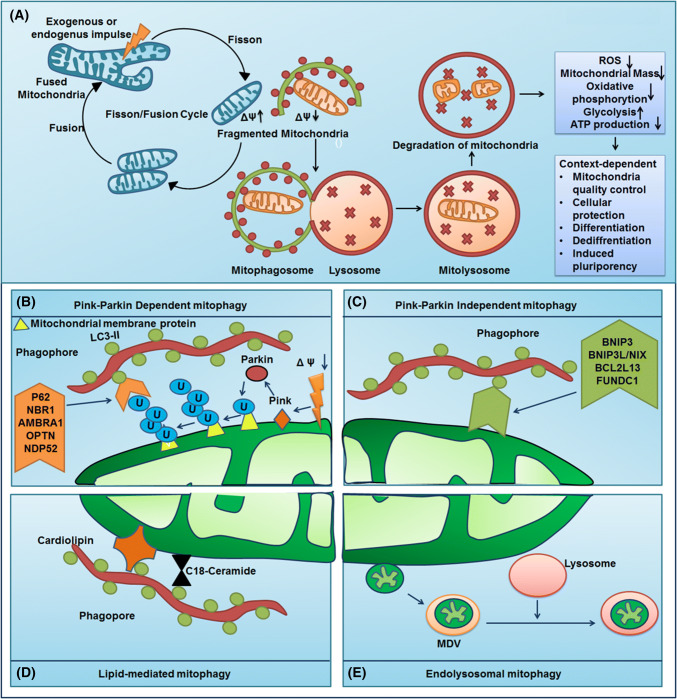
In contrast to non-selective autophagy or bulk autophagy, selective autophagy or cargo-specific autophagy might occur under nutrient-rich conditions to mediate the removal of long-lived protein aggregates or superfluous or damaged organelles. The term mitophagy was coined by Lemasters and colleagues to describe the sequestration of mitochondria into autophagosomal vesicles coated with a yeast Atg8 homolog protein, viz. microtubule-associated protein 1 light chain 3 (MAP1LC3), which occurred within 5 min of starvation and photodamage [9]. However, the first study on mitophagy-like events using electron microscopy of mammalian cells acknowledged an increased mitochondrial sequestration in lysosomes during glucagon-mediated catabolic stimulation of hepatocytes [10]. The selective autophagolysosomal degradation maintains the quality of mitochondria and homeostasis. In the context of mitophagy activation, the machinery involved in mitochondrial morphology and dynamics regulated by fission/fusion has a great impact (Fig. 1a). Mitochondrial fusion is induced by the protein optic atrophy 1 (OPA1), which mediates fusion of the inner membrane, whereas the proteins Mitofusins 1 and 2 (MFN1 and MFN2) mediate fusion of the outer membrane. Similarly, mitochondrial fission is regulated by the GTPase dynamin-related protein 1 (DRP1) [11]. The division or fission of mitochondria generates two structurally and functionally distinct daughter mitochondria with polarized and depolarized phenotypes. The polarized daughter mitochondria undergo fusion, forming a mitochondrial network, while the depolarized mitochondria are passed on for autophagolysosomal culling (Fig. 1a) [12, 13]. Mitochondrial dynamics are facilitated by selective mitochondrial degradation, which is required for cellular function and development. Broadly, mitophagy can be categorized into quality control-related mitophagy and developmentally induced mitophagy. While quality control-related mitophagy is involved in the housekeeping functions of respiring eukaryotic cells that carry out lysosomal/vacuolar degradation of superfluous mitochondria or damaged mitochondria, developmentally induced mitophagy is involved in developmental processes such as maturation of reticulocytes. However, little information is available regarding key mechanisms regulating mitochondrial dynamics and biogenesis within pluripotent stem cells (PSCs). Furthermore, the association between reprogramming-associated reduction of mitochondria and activation of mitophagy needs to be described in more detail. This document elucidates the role of mitophagy in the regulation of cell fate transition.
Fig. 1.
Mitophagy mechanism and its role in cellular homeostasis. a AMPK might act as a mitophagy sensor and N-myristoylation of AMPKβ by the type-I N-myristoyltransferase 1 (NMT1) facilitates AMPK recruitment to the damaged mitochondria and further AMPK-mediated recruitment of ATG16-ATG5-12 and VPS34 to form autophagosome around mitochondria. Mitochondria structural and functional integrity is routinely managed by the fission/fusion cycle. The post-mitochondrial fission, the mitochondrial with lost membrane polarity undergo autophagic degradation via the formation of mitophagosome and mitolysosome which serves great implication in cellular physiological events such as cell survival, cell death, and cellular fate determination. The canonical mitophagy pathway requires the LC3 decorated autophagosome for mitochondrial selection and degradation which can be: b PINK–Parkin-dependent mitophagy involves the stabilization of PINK1 in the depolarized mitochondrial membrane leading to the recruitment of Parkin. Parkin ubiquitinates the mitochondrial membrane protein which in turn interact with the autophagy adaptor molecules such as p62, NBR1, AMBRA1, OPTN, and NDP52 to form the mitophagosome; c PINK–Parkin independent mitophagy do not require the autophagy adaptor molecules rather the outer mitochondrial receptors for mitophagy-like BNIP3, BNIP3L/Nix, BCL2L13, and FUNDC1 directly interacts with LC3 to form mitophagosome; d lipid-mediated mitophagy involves lipid molecules such as C18 ceramide and cardiolipin as mitophagy receptor. e The non-canonical mitophagy do not involve the LC3-studded autophagosome rather it performs the endo-lysosomal degradation of mitochondria-derived vesicle
Stem cells and mitochondria
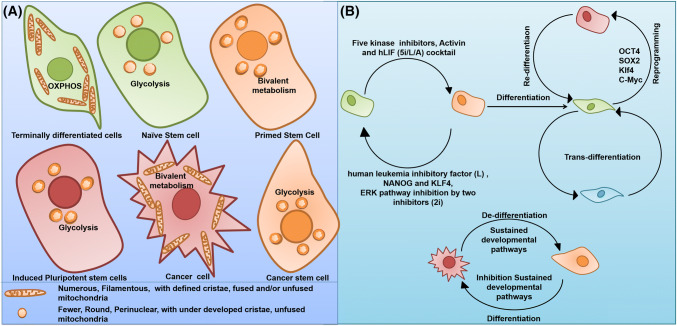
Mitochondria are the essential regulators of all nucleated cells and govern the generation of cellular ATP by oxidative phosphorylation (OXPHOS), heme biosynthesis, calcium homeostasis, cell signaling, and apoptosis. Interestingly, many studies suggest the pivotal role of mitochondrial homeostasis in the regulation of stem cell maintenance and differentiation [14–16]. During the process of stem cell differentiation and reprogramming, mitochondria undergo opposite and reversible alterations in the structural and functional integrity. Reverse remodeling of the mitochondrial network—immature mitochondrial network—was observed in the process of reprogramming of human and mouse somatic cells into iPSCs, whereas the elongation of the mitochondrial network was observed during the differentiation of human embryonic stem cells (hESCs) [16–18]. The structural analysis indicated that undifferentiated human and mouse ESCs have spherical mitochondria with poorly developed cristae, fewer copies of mitochondrial DNA (mtDNA), and perinuclear distribution compared to their long, tubular-shaped, branched, and cristae-rich somatic cell counterparts (Fig. 2a) [19]. However, besides the morphological and ultrastructural changes during differentiation, there occurs an increase in the mitochondrial mass, mitochondrial DNA copy number, oxygen consumption, respiratory reserve capacity, and reactive oxygen species (ROS), while the rates of glycolysis and lactate production are reduced in mouse embryonic stem cells (mESCs), hESCs, and iPSCs (Fig. 2a) [19–23]. An in vivo study on spermatogonia, inner cell mass and early embryos suggests that relatively immature-appearing mitochondrial network and comparatively lesser mitochondrial activity appear to be prospective features of ‘stemness’ [24]. As mitochondria undergo opposite and reversible alterations in morphology and metabolism, the plasticity of mitochondrial networks is regularly maintained by a constant balance between the mitochondrial fusion/fission, the modulation of fusion/fission balance could perform an important role in the sustenance and maintenance of ESCs [25]. Moreover, the subcellular content, configuration, and distribution of mitochondria are highly variable during the cell fate transition. Furthermore, the spectacular reduction of mitochondrial number and mass in iPSCs compared to their parental somatic cells tends to indicate an involvement of mitophagy-like events during the process of cell fate conversion [26, 27].
Fig. 2.
Mitochondrial dynamics in different cell types and cellular interconversion. a Mitochondria shows a tremendous alteration in morphology and bioenergetics in different in cell types including stem cells, terminally differentiated cells, cancer cell, and cancer stem cells which is necessary to maintain the required energy demand of specific cell type. b The cell types can undergo cell fate transition in specific set of conditions: (i) naïve stem cells and primed stem cells could ectopically inter-convert. (ii) “Differentiation” is the process where the pluripotent stem cells are transitioned into mature cells that are terminally differentiated and “Dedifferentiation” is conversion of a terminally differentiated cell into undifferentiated condition. (iii) Pluripotency can be induced in the adult somatic cells through somatic “reprogramming” to generate induced pluripotent stem cells (iPSCs) via the use of transcription factor cocktail such as OCT4, SOX2, Klf4, and c-Myc and again, iPSCs can “redifferentiate” into a terminally differentiated cell type. (iv) The terminally differentiated cells can convert into another terminally differentiated cell types without any undifferentiated intermediate through a process called “transdifferentiation”. (v) The cancer cells also can dedifferentiate into cancer stem cells and vice versa
Mitophagy and mechanism
The timing to target the mitochondria for selective autophagy is the first major concern faced by a cell in the context of mitophagy. An evolutionarily conserved protein kinase adenosine monophosphate-activated protein kinase (AMPK), a potent sensor of intracellular energy, is activated in response to nutrient depletion and other endogenous and exogenous stress [28]. Egan et al. reported that AMPK-mediated phosphorylation of UNC-51-like kinase (ULK1) in a yeast ATG1 homolog connects energy sensing to mitophagy. Their study reported that the loss of AMPK or ULK1 may lead to anomalous accumulation of the autophagic adaptor p62 and impaired mitophagy. Moreover, it was found that in contrast to wild-type controls, expression of the autophagic adaptor p62 and the mitochondrial marker protein cytochrome c oxidase subunit IV (COX IV) were significantly elevated in AMPK- or ULK1-deficient hepatocytes, suggesting that AMPK-phosphorylated ULK1 is highly essential for mitochondrial homeostasis and connects the sensing of cellular energy to mitophagy [29]. A spatiotemporal model for selective mitochondrial recruitment in mitophagy was proposed to explain this. N-Myristoylation of AMPKβ by the type-I N-myristoyltransferase 1 (NMT1) is required for recruitment of AMPK to the mitochondria. Moreover, mitochondrial damage leads to the physical association of AMPK with ATG16-ATG5-12 and AMPK-dependent recruitment of the vacuolar protein sorting 34 (VPS34) and ATG16 complexes within the mitochondria (Fig. 1a) [30]. The next major concern is to determine how a mitochondrion can be exclusively targeted to the autophagosome. It has been observed that the ubiquitin-like protein LC3 plays an active role in the biogenesis of autophagosome and participates in cargo-specific recruitment [24]. The protein LC3 contains an evolutionarily conserved W and L hydrophobic pocket that helps in the docking of proteins containing LC3 interacting region (LIR) motif, Atg8-family-interacting motif (AIM), or LC3 recognition sequence (LRS) [31–36]. This motif comprises a core consensus sequence of [W/F/Y]xx[L/I/V] having an aromatic residue followed by a hydrophobic residue and preceded by negatively charged residues that help the positively charged residues on the proteins to interact with LC3 [31, 37–39]. Moreover, the phosphorylation of autophagic receptor of serine/threonine residues in the LIR preceding region is crucial for the regulation of autophagy [40]. In this context, two modes of mitophagy, namely canonical and non-canonical mitophagy, can be described. Canonical mitophagy relies on autophagy-dependent degradation, whereas non-canonical autophagy involves autophagy-independent endo-lysosomal degradation of mitochondria.
Canonical mitophagy
The LIR motif-containing receptors, localized on the surface of the outer mitochondrial membrane (OMM), guide the cargo to interact with LC3 containing autophagosome and form the basis of mitophagy [31]. Based on the manner of targeting mitochondria, two groups of receptor systems exist. The first group of receptors, including sequestosome-1 (SQSTM1/p62), Neighbor of Brca1 (NBR1), Activating molecule in BECN1 regulated autophagy protein 1 (AMBRA1), and optineurin (OPTN), contains a ubiquitin-binding domain which localizes them to Parkin-ubiquitinated mitochondria (Fig. 1b) [32, 37–41]. On the other hand, the second group of receptors including BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), BCL2/adenovirus E1B 19 kDa interacting protein 3-like/NIP3-like protein X (BNIP3L/NIX), Bcl-2-like protein 13 (Bcl-rambo) (BCl2L13), and FUN14 domain-containing protein 1 (FUNDC1) contains a characteristic mitochondria-targeting C-terminal trans-membrane (TM) domain and a Bcl-2 homology 3 (BH3) domain (Fig. 1c) [42–49]. Rather than direct interaction, the first group of receptors interacts with LC3 via the ubiquitin-binding adaptor protein containing LIR motifs. However, the second group of receptors can directly interact with LC3 via their LIR motif. Interestingly, there also exists a third group of receptor system, which involves lipids as mitophagy receptors (Fig. 1d) [50, 51].
PINK/Parkin-dependent/ubiquitin-binding domain-containing receptor-mediated mitophagy
PINK/Parkin-dependent mitophagy requires the core autophagic machinery, as it has been shown that blocking the activities of ATG3, ATG5, ATG7, and class III PI3K impedes selective elimination of mitochondria [32]. The serine/threonine kinase-PTEN-induced putative kinase 1 (PINK1)-E3 ligase Parkin1-mediated mitophagy is the best-understood mode of mitophagy, wherein PINK1 acts as a molecular sensor for polarization of mitochondria. Upon polarization, the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) complex in healthy mitochondria constitutively import PINK1 to the inner mitochondrial membrane, wherein the presence of mitochondrial targeting sequence allows it to be localized on the mitochondrial membrane. After successful import to the inner membrane, PINK1 is readily cleaved by mitochondrial-processing protease (MPP) and the inner membrane presenilin-associated rhomboid-like (PARL) protease [52–54]. However, under depolarized conditions, the loss of mitochondrial membrane potential prevents the import of PINK1 to the inner membrane and promotes its stabilization on the outer mitochondrial membrane. Accumulation of PINK1 promotes the translocation of Parkin from the cytosol to damaged mitochondria (Fig. 1b) [54, 55]. Recruitment of Parkin may occur in two ways. One method involves the phosphorylation of MFN2 at Ser (S) 442 and Thr (T) 111, rendering the phosphorylated MFN2 as a receptor to attract Parkin [56], and the second method involves the phosphorylation of both ubiquitin and ubiquitin-like domain of Parkin at Ser (S) 65 by PINK1, which drives the recruitment of Parkin to the OMM [57–59]. Following this, the Parkin appears to stimulate the formation of two types of polyubiquitin chains: Lys (K) 48 linkage associated with proteasomal degradation of the substrate, and Lys (K) 63 and 27 linkage associated with autophagic degradation (Fig. 1b) [60, 61]. The generation of Lys (K) 63- or 27-linked ubiquitin chains on outer mitochondrial membrane activates mitophagy rather than induction of proteasomal degradation [62]. Outer mitochondrial proteins such as MFN2, voltage-dependent anion channel 1 (VDAC1), and mitochondrial Rho GTPase 1 (MIRO1) are identified as the Parkin substrates [32, 37]. However, it has been observed that the LIR domain of the inner mitochondrial membrane protein Prohibitin2 binds to LC3 and leads to Parkin-dependent mitophagy in response to mitochondrial depolarization [63].
The ubiquitin-binding adaptor proteins are recruited to the ubiquitinated receptors on depolarized mitochondria, facilitating the delivery of damaged mitochondria to autophagosomes by binding to LC3 [30, 56]. Moreover, the ubiquitination of OMM proteins also leads to the recruitment of different LIR-containing autophagic receptors, which bind to ubiquitin-tagged OMM proteins such as p62, optineurin, and NBR1 (Fig. 1b) [37, 40, 64]. However, the mitochondrial clustering proteins p62 and VDAC1 have been reported to be dispensable in the downstream mitochondrial degradation [65]. The Beclin 1-PI3K complex can directly interact with PINK1 and Parkin [66]. Moreover, AMBRA1, an activator of the Beclin 1 complex, may be recruited by Parkin to depolarized mitochondria, leading to the activation of the Beclin 1 complex and nucleation of the pre-autophagosomal membrane around damaged mitochondria [67]. AMBRA1, with its LIR motif, locally activates and directly captures the autophagosomes at depolarized mitochondria [68, 69]. Furthermore, the cytoplasmic E3 ubiquitin ligase SMAD-ubiquitin regulatory factor1 (SMURF1) can trigger Parkin-dependent mitophagy by assisting the delivery of mitochondria to the autophagosome [70]. In Parkin-overexpressing cells, PINK1-recruited autophagic receptors such as nuclear dot protein 52 kDa (NDP52) and OPTN further recruit initiating factors for autophagy, including ULK1, double FYVE-containing protein 1 (DFCP1), and WIPI-1, independently of LC3, onto depolarized mitochondrial membrane, suggesting that autophagosomes are produced in damaged mitochondria and processing of LC3 can be localized for the downstream engulfment of mitochondria into autophagosome [38, 70]. Again, the cellular energy sensor AMPK is spatially localized to mitochondria, providing a potential sensing mechanism to signal the local production of autophagosomes (Fig. 1a) [69].
PINK1/Parkin-independent/mitochondrial targeting TM domain and BH domain-containing receptor-mediated mitophagy
The overexpression of Parkin in PINK1-null mutant Drosophila was shown to rectify defects in mitochondrial integrity, suggesting that PINK1 is not essential for Parkin function [71] and mitophagy can occur independently of PINK–Parkin (Fig. 1c). The PINK1/Parkin-independent mitophagy receptor system involves a group of LIR-containing receptors that contain mitochondria targeting the TM domain, which helps them to constitutively localize at the OMM. Report suggests that BH domains are required for fragmentation of mitochondria [48]. It is worth mentioning that TM domains containing OMM-localized mitophagy receptors contain conserved serine/threonine preceding the LIR and the LIR phosphorylation status characterizes a vital mitophagy receptor system allowing them to co-localize with the autophagosome mediated by LC3. Several types of mitophagy receptors or receptor-related factors have been identified in mammalian cells, including NIX/BNIP3L, FUNDC1, and BCL2L13 (Fig. 1b) [72, 73].
BNIP3 and its homolog BNIP3L/NIX are a BCL-2 subfamily of BH3-only proteins localized to OMM via their C-terminal TM domains and play the role of mitophagy receptor via the cytoplasm-oriented typical N-terminal LIR motif which helps in its association with the autophagosome and regards it as a mitophagy receptor [44, 45, 74]. As mentioned earlier, the activity of BNIP3 depends on the phosphorylation of Ser (S) 17 and 24 residues flanking the LIR region of the polypeptide, and stimulates its binding to LC3B and Golgi-associated ATPase enhancer of 16 kDa (GATE-16) [43]. Like BNIP3, Ser (S) 34/35 phosphorylation of SWxxL LIR motif of NIX promotes mitophagy [75]. Moreover, the interaction of Rheb with NIX was reported to induce mitophagy to balance the demand for mitochondrial quality control [76]. Furthermore, FUNDC1, with three TM domains and a typical N-terminal LIR Y (18) xxL23 motif exposed to the cytosol, is a promising mitophagy receptor located on the OMM. The interaction between FUNDC1 and LC3 is highly dependent on the conserved Tyr (Y) 18 and Leu (L) 21 residues. Mutations in this motif were described to inhibit the interaction between FUNDC1, LC3, and mitophagy [46]. Dephosphorylation of both Ser (S) 13 and Tyr (Y) 18 of FUNDC1 in response to hypoxic stress in the LIR motif and Ser13 induces mitophagy [77]. Moreover, the mitochondria-localized phosphatase phosphoglycerate mutase family 5 (PGAM5) mediates the dephosphorylation of FUNDC1 at Ser (S) 13, which facilitates better interaction of LC3 and FUNDC1, resulting in selective incorporation in the autophagosome and subsequent mitophagy [75, 78]. ULK1 is also involved in the phosphorylation of Ser (S) 17 of the FUNDC1 LIR motif, facilitating the binding of LC3 and mitophagy [47]. The presence of a WxxL LIR motif in BCL2L13 and its similarity to the yeast mitophagy receptor Atg32 help in identifying it as a mitophagy receptor. It gets anchored to OMM via its C-terminal TM domain [48]. Its BH domain carries out mitochondrial fragmentation independent of DRP1 and directs fragmented mitochondria to autophagosomes and endo-lysosomes [75]. BCL2L13 has two WxxL/I LIR motifs at positions 147–150 and 273–276, of which the motif at residues 273–276 is a functional LIR motif that interacts with LC3 [48]. Increased LIR activation via the phosphorylation of Ser (S) 272 residue of BCL2L13 promotes the mitochondria to co-localize with autophagosomes [79]. Recently, our group had reported that p53 upregulated the modulator of apoptosis (PUMA), a BH3-only protein which contains LIR at the C-terminal end and interacts with LC3 to induce mitophagy [80].
Lipid-mediated mitophagy
Like the protein mitophagy receptors, certain lipid molecules such as ceramide and cardiolipin act as mitophagy receptors when localized to the OMM. These lipids can directly interact with LC3 to induce mitophagy (Fig. 1d). Cardiolipin, a negatively charged phospholipid that generally resides within the inner mitochondrial membrane, gets externalized to OMM by phospholipid scramblase-3 (PLS3) during mitochondrial stress [51]. Cardiolipin undergoes electrostatic interactions with positively charged R11 and R10 residues in the cardiolipin-binding region of LC3A and LC3B [45]. Moreover, although the LC3-deletion mutant of the cardiolipin-binding site could not induce mitophagy, it could induce non-selective autophagy [51]. Similarly, C18-ceramide at the OMM can directly bind with I35 and F52 of LC3B to induce DRP1-dependent mitophagy [36, 50].
Non-canonical mitophagy
In contrast to canonical mitophagy, the non-canonical modes of mitophagy do not require LC3-decorated autophagosomes; instead, they exhibit direct inter-organellar interactions between the mitochondria and endo-lysosomes (Fig. 1d) [81–83]. In this case, despite the presence of whole mitochondria, the PINK1- and Parkin-induced mitochondria-derived vesicles (MDVs) are targeted to lysosomes independent of autophagy and function as a homeostatic quality control mechanism for small-scale levels of damage that do not require the sacrifice of an entire mitochondrion [81]. The X-linked inhibitor of apoptosis (XIMP), E3 ligase-mediated activation of ubiquitylation at the OMM, and inner mitochondrial membrane (IMM) promote autophagy-independent movement of endo-lysosomal machinery into mitochondria [83, 84]. Moreover, the p53-induced mitochondria-eating protein (Mieap) promotes the endo-lysosomal degradation of mitochondria in an autophagy- and Parkin-independent manner [82, 85]. Interestingly, BNIP3 or NIX LIR mutants bereft of mitophagic function also activate the non-canonical mitophagy pathway [83]. The most important advantage of non-canonical mitophagy is the involvement of direct interactions between endo-lysosomes and mitochondria which saves time.
Mitophagy regulates the stem cell fate
Mitophagy promotes stem cell survival
Mitochondrial activity and metabolism influence the lifespan of stem cells. However, the mechanism of decline in the functionality of stem cells with respect to age remains unclear. It has been reported that the pro-apoptotic protein p53 could deplete the stem cells under telomere attrition or oncogenic stimuli [86]. It has also been demonstrated that mitophagy-dependent removal of p53 allows the expression of NANOG and promotes cell survival and hepatic carcinogenesis [87]. Again, the reduction of mitophagy-related genes PINK or Parkin induces intestinal stem cell senescence [88]. A recent report has described an interesting phenomenon, wherein foreign somatic cell-derived mitochondria were removed in a mitophagy-dependent manner from mesenchymal stem cells (MSCs), which induced the expression of cytoprotective enzymes heme oxygenase-1 (HO-1) and facilitated mitochondrial biogenesis. MSCs donate their mitochondria to the neighboring injured cells and facilitate their survival from oxidative stress-induced death [89]. MSCs are also reported to outsource mitophagy via arrestin domain-containing protein 1-mediated MVs (ARMMs) to unload mitochondria to be engulfed by macrophages, re-utilized to increase bioenergetics, and shuttle microRNAs through extracellular vesicles that promote cell survival [90]. Further studies also report that the deletion of the autophagy protein ATG7 leads to the accumulation of mitochondria due to the inefficient removal by mitophagy, resulting in the loss of normal HSC functions and death of mice within weeks [91].
Mitophagy regulates cellular potency
Mitophagy and stemness
Self-renewal and pluripotency are the key characteristics of stem cells. Stem cells are broadly classified into embryonic stem cells (inner mass of blastocyst) and non-embryonic “somatic” or “adult” stem cells. ESCs are pluripotent, which means that they may give rise to all cell types of the body, whereas adult stem cells, somatic stem cells, or tissue-specific stem cells (multipotent or unipotent) are more specialized than ESCs and can generate various cell types within specific tissues or organs. In particular, stem cells have two phases of pluripotency: Naïve and Primed [92]. Embryonic cells remain pluripotent within the inner cell mass of pre-implantation embryos, but during the course of development, they lose potency as they start to commit for differentiation into specific somatic lineages during post-implantation development. ESCs in the pre-implantation embryos are referred to as “Naïve”, while they become ‘Primed’ during post-implantation development in the embryo. The naïve ESCs are the developmental ground state, whereas the primed ESCs represent a relatively more mature stage. The conversion from Naïve to Primed can be achieved using a mixture of five kinase inhibitors (5i), human leukemia inhibitory factor (hLIF), and growth factor activin [93]. The reverse conversion from Primed to Naïve can be achieved through ectopic expression of NANOG and KLF4, and inhibition of the ERK pathway by two inhibitors (2i) (Fig. 2b) [94]. Moreover, somatic cell reprogramming into iPSCs can be done with OSKM (OCT4, SOX2, KLF4, c-Myc) [95] (Fig. 2b). Recent studies have suggested that mitophagy plays a pivotal role in the maintenance and differentiation of stem cells. Cell reprogramming faces the challenge of balancing stability and plasticity and must overcome critical barriers such as cell-cycle checkpoints, epithelial–mesenchymal transition (EMT), and metabolic reprogramming, to progress cell fate conversion from a stochastic early phase to pluripotency. Although several reports explain the role of mitophagy in somatic cell physiology, its proposed critical role in maintaining the archetypal properties of stem cells is now rudimentary.
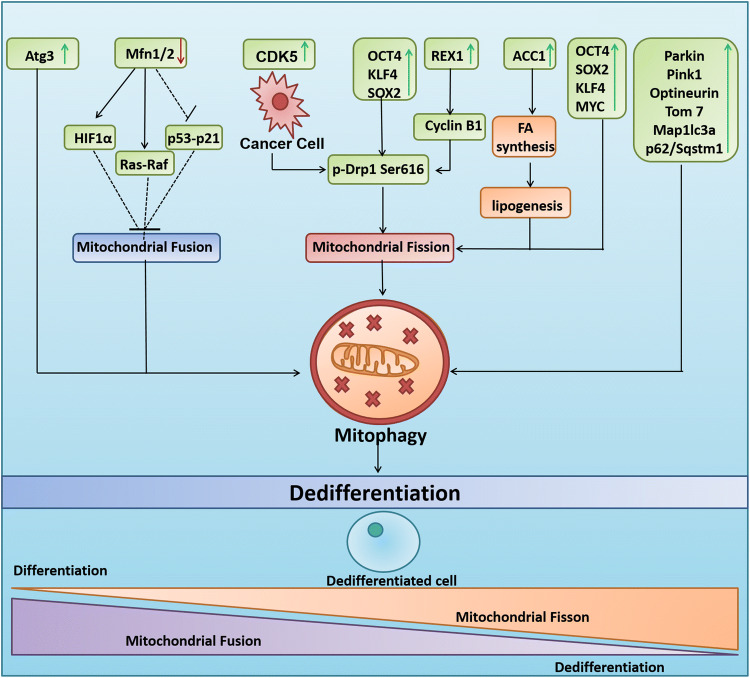
Developmentally associated mitophagy has been reported to maintain the archetypal properties of stem cells. Reprogramming of somatic cells to iPSCs has been proposed to bio-energetically take advantage of mitophagy. In this regard, the loss of PINK1-dependent mitophagy was observed to reduce iPSC reprogramming, indicating that mitophagy regulates cell fate plasticity and maintains pluripotency [96]. Another study has described the mechanism of regulation of induced pluripotency by mitophagy using a pharmacological mitochondrial division inhibitor 1 (mdivi-1) that prevented the self-assembly of DRP1. Moreover, mdivi-1 is also reported to inhibit mitochondrial complex I-dependent O2 consumption reversibly and inverse electron transfer-mediated ROS generation at 50 μM concentration that aim mitochondrial fission [97]. It was also observed that the inhibition of mitophagy by inhibiting DRP1 negatively affected the reprogramming efficiency of mouse embryonic fibroblasts when transduced with the Yamanaka three-factor cocktail (OCT4, KLF4, and SOX2) (Fig. 3). The study also reported that if mdivi-1 was treated at the early stages of reprogramming, complete inhibition of somatic cell reprogramming into pluripotent cells was achieved, pointing to the potential involvement of mitophagy in the regulation of cell fate [98]. Furthermore, the somatic cell reprogramming by SKP/SKO (Sox2, Klf4, and Pou5f1/Oct4) and SKPM/SKOM (SKP/SKO with Myc/c-Myc) was also reported to involve BNIP3L-dependent mitophagy [99]. Similarly, the deletion of Atg3 was found to be associated with a reduction in the removal of mitochondria through autophagy, reprogramming efficiency, induction of pluripotency, and accumulation of abnormal mitochondria in established iPSCs (Fig. 3) [100]. Moreover, hematopoietic stem cells (HSCs) positive for the angiopoietin receptor Tie2 were found to significantly enrich mitophagy-related genes including Parkin, PINK1, OPTN, TOM7, MAPLC3a, and p62/SQSTM1, which confirms the relationship between mitophagy and pluripotency [101]. The higher occurrence of mitophagy reduces mitochondrial mass and differentiation of stem cells and facilitates the retention of “Stemness Status”, resulting in various disease conditions. In this context, it was reported that increased mitophagy occurs in mesenchymal stem cells associated with progressive supranuclear palsy, inhibits their differentiation into adipocytes, and retains their stem-like features [102]. Again, selective mitophagy in adipose stem cells has been reported to reduce the chondrogenic differentiation potential in equine metabolic syndrome, allowing them to maintain their “stemness” status [103].
Fig. 3.
Mitophagy regulates cellular pluripotency. Induction of mitophagy via the activation of mitochondrial fission through the OCT4, KLF4 and SOX2 cocktail, cyclin b1 dependent DRP1 Ser (S) 616 phosphorylation, ACC1 enzyme mediated lipogenesis leads to the dedifferentiation of somatic cells into stem cells. Moreover, the inhibition of mitochondrial fusion by reduced expression of MFN1 leads to activation of HIF1α and Ras–Raf and inhibition of p53–p21 which further leads to mitophagy-mediated acquisition of pluripotency. REX1, ACC1, Parkin, PINK1, Optineurin, TOM7, MAP1LC3a, p62/Sqstm1 and ATG3 are also implicated in inducing pluripotency through the activation of mitophagy. The activation of CDK5 in cancer cells leads to the phosphorylation of DRP1 at Ser (S) 616 leading to mitochondrial fission, mitophagy and cancer stemness
Mitochondrial dynamics and stemness
Although the mitochondrial fission/fusion cycle dictates mitochondrial dynamics, it is also involved in the process of mitophagy. The process of fission and fusion reciprocally regulates the process of selective mitochondrial elimination [104]. Mitochondrial fission divides the fused and elongated mitochondria into pieces of a manageable size to be engulfed by the autophagosome for quality control and selective removal by mitophagy. Mitochondrial fission is considered to be one of the prerequisites for the initiation of mitophagy [105–107]. The activation of mitochondrial fission protein DRP1 to p-DRP1-(S)-616 due to phosphorylation by cyclin B1 in REX1-overexpressed condition is associated with the mitochondrial fission-associated pluripotency (Fig. 3) [108]. Moreover, the erv1-like growth factor (Gfer) modulates the levels of DRP1 for maintenance of mESC mitochondrial morphology, function, and preservation of the expression of pluripotency marker in these primitive cells during homeostasis [109]. Furthermore, it has been described that the Yamanaka four-factor cocktail OSKM-mediated cell reprogramming prompts mitochondrial fission and any impairment in this process leads to a reduction in cellular reprogramming by interfering with cell-cycle progression in a DNA damage-independent manner (Fig. 3) [110]. In addition, it has now been confirmed that the critical involvement of fatty acid synthesis pathway promotes ESC pluripotency and iPSC formation through regulation of mitochondrial fission [111]. Unlike mitochondrial fission, mitochondrial fusion is supposed to inhibit mitophagy [104]. In this context, the depletion of mitochondrial fusion proteins MFN1/2 is reported to reciprocally inhibit the p53–p21 pathway that promotes both the conversion of somatic cells to a pluripotent state and the maintenance of pluripotency (Fig. 3) [112]. Moreover, the induction of mitochondrial fusion has been correlated with the inhibition of mitophagy and reduction of somatic cell reprogramming to pluripotency [98].
Mitophagy and cancer stemness
The role of mitophagy in developing stemness in cancer cells has recently become a subject of intense discussion. Cancer stem cells (CSCs), cancer initiating cells (CICs), or tumor initiating cells (TICs) are a small subpopulation of intra-tumor cells that possess the unique ability of exclusive self-renewal, tumorigenesis, and metastatic potential. Like normal stem cells, CSCs have an inherent ability of enhanced resistance to DNA damage and apoptosis. However, unlike the bulk tumor cells, CSCs are capable of unlimited self-renewing hierarchical differentiation and tumorigenicity. Moreover, enhanced invasive capacity, metastatic proficiency, and EMT in CSCs confer resistance to therapy, resulting in the relapse of cancer [113, 114]. Our group is actively working on the acquisition of stemness and chemoresistance in cancer stem cells in oral cancer. Recently, we have reported that the mitophagic flux in cisplatin-resistant oral cancer cells is higher, and that these resistant cells possess higher stemness than their parental counterparts [115]. Several lines of evidence reveal that the fission factor DRP1 is closely associated with CSCs and its fate determination. Brain tumor initiating cells (BTICs) contain fragmented mitochondria, suggesting increased mitochondrial fission. The phosphorylation of p-DRP1Ser616, catalyzed by cyclin-dependent kinase 5 (CDK5), regulates mitochondrial fission which plays an active role in the maintenance of stemness by downregulating the activation of AMPKα (Fig. 3). A study reported that cyclooxygenase-2 (COX-2) maintained the stemness of nasopharyngeal carcinoma (NPC) by enhancing the activity of DRP1 both in vitro and in vivo (Fig. 3) [116]. Perinuclear distribution of mitochondria and mitochondrial fission in NPC causes a switch from oxidative phosphorylation to glycolysis, which is considered to be a key event in the acquisition of stem-like characteristics in cancer [117]. Moreover, Parkin-dependent mitophagy generates EMT-mediated CD44 high transformed esophageal keratinocytes, which is associated with the pathogenesis of esophageal squamous cell carcinoma (ESCC), characterized by invasion, metastasis, and resistance to treatment [118]. Recently, it has been reported that mitophagy in hepatic cancer cells inhibits the binding of p53 to the promotor of NANOG—a transcription factor associated with stemness and self-renewal of CSCs, thereby maintaining hepatic CSC population [87]. Similarly, mitophagy flux and BNIP3L expression have been shown to be significantly higher in CD133+/CD44+ HCT8 human colorectal cancer-derived CSCs [119]. The relevance of mitophagy in CSC phenotypes is under-explored. A detailed experimental investigation of the implications of mitophagy in the acquisition and maintenance of stemness is required to control the cancer stemness, tumorigenicity, and recurrence.
Mitophagy and differentiation
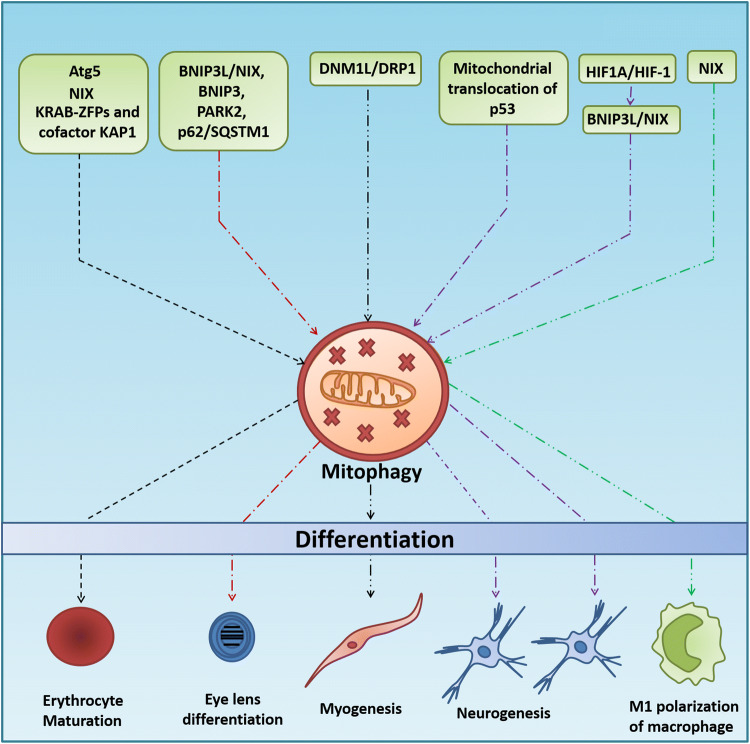
Apart from quality control and maintenance of pluripotency, mitophagy also participates in the cellular differentiation during specialized developmental stages in mammalian cells. Developmentally controlled mitophagy promotes cellular differentiation in several distinct developmental contexts. Autophagy-mediated selective removal of mitochondria is implicated in differentiating reticulocytes. Erythrocyte maturation involves canonical autophagic degradation of mitochondria. Knocking out Atg5 was observed to inhibit mitochondrial degradation and reduce reticulocyte differentiation (Fig. 4). NIX knockout animals showed 30–50% persistence of mitochondria in differentiated erythrocytes, suggesting the involvement of mitophagy in the maturation of reticulocytes (Fig. 4) [45, 72, 73]. Another finding by Barde et al. indicated that KRAB-containing zinc finger proteins (KRAB-ZFPs), together with their cofactor KAP1, controlled the timely elimination of mitochondria from maturing erythroblasts through mitophagy, further establishing the association between mitophagy and differentiation (Fig. 4) [120]. Similarly, enhanced mitophagy in CD34+ hematopoietic progenitor cells (HPCs) was observed during erythrocyte differentiation in β-thalassemia [121].
Fig. 4.
Role of mitophagy in cell differentiation. ATG5, NIX, and KRAB-ZFPs and cofactor KAP1-induced elimination of mitochondria govern the differentiation of reticulocytes. The BNIP3L/NIX, BNIP3, PARK2 and p62/SQSTM1 mediated mitochondrial clearance regulates eye lens differentiation, whereas DNM1L/DRP1-induced mitophagy could activate the myogenic differentiation. Hypoxia-mediated activation of BNIP3L-NIX and mitochondrial translocation of p53 leads to mitophagy-dependent neurogenic differentiation. NIX-dependent mitophagy induces the M1 polarization of macrophages
Mitophagy is also involved in the myogenesis or differentiation of primitive myoblasts into mature myotubes. This is only possible due to the DNM1L/DRP1-mediated fragmentation, followed by the removal of mitochondria via p62/SQSTM1-mediated mitophagy (Fig. 4) and repopulation of mitochondria via peroxisome proliferator-activated receptor gamma and coactivator 1 alpha (PPARGC1A/PGC-1α)-mediated biogenesis with established mitochondrial networks that are better primed for OXPHOS [122, 123]. Similarly, the stable overexpression of a dominant negative mutant DRP1 (K38A) and mdivi-1 treatment was shown to dramatically reduce the myogenic differentiation into myotube formation in both C2C12 cells and myogenic differentiation of primary myoblasts (Fig. 4). Furthermore, mitophagy is also induced during the differentiation of mouse myoblasts in developmental transitions in the muscle tissue, and essential metabolic transitions that occur during the differentiation of cardiomyocytes is proven to be mitophagy-dependent [122, 124].
Similar to erythrocyte maturation and myogenesis, the process of neurogenesis is strictly dependent on the selective removal of mitochondria. Importantly, mitochondrial translocation of p53 during the early stages of neural differentiation was shown to promote mitophagy which decreases the oxidative stress and promotes neurogenic potential and outgrowth of neurites (Fig. 4) [125]. Furthermore, it was reported that differentiation of embryogenesis-associated mouse retinal ganglion cells (RGCs) depends on BNIP3L/NIX-mediated mitophagy (Fig. 4). NIX deficiency in retina leads to increased mitochondrial mass and decreased neuronal differentiation (Fig. 4) [126]. Moreover, hypoxia triggers the stabilization of HIF1A/HIF-1, resulting in increased BNIP3L-dependent mitophagy and RGC neurogenesis (Fig. 4) [127].
Moreover, the expression levels of genes for autophagic proteins ATG3, ATG4B, BECN1 (beclin-1), FYVE and coiled-coil domain-containing protein 1 (FYCO1), WD repeat domain phosphoinositide-interacting protein 1 (WIPI1) and mitophagic proteins BNIP3L/NIX, BNIP3, Parkinson Juvenile Disease Protein 2 (PARK2), and p62/SQSTM1 (Fig. 4) indicate a comprehensive array of mitochondrial regulatory and degradation pathways to maintain mitochondrial populations in the lens epithelium in maturing lens fiber cells [128]. Dual-label confocal imaging and electron microscopic analysis by Costello et al. confirmed the presence of mitophagosomes (autophagic vesicles containing mitochondria) in lens epithelial cells, i.e., immature lens fiber cells during early stages of differentiation of lens fiber cells, suggesting the pivotal role of mitophagy in the differentiating lens fiber cells for lens development [129]. Moreover, the inhibition of mitophagy was demonstrated to abrogate fibroblast differentiation [130]. Similarly, the inactivation of adipocyte mitophagy both in vitro and in vivo resulted in adipocytes with atypical morphology and with several small lipid droplets and large numbers of mitochondria in post-differentiated cells, implicating the importance of mitophagy in the pre-adipocyte differentiation into white adipocytes [131].
In some cases, it was reported that the conversion of one terminal phenotype into another is dependent on mitophagy. In this context, NIX-dependent mitophagy was shown to contribute to macrophage polarization toward the pro-inflammatory and more glycolytic M1 phenotype, but not the OXPHOS phenotypic M2 polarization (Fig. 4) [126]. Moreover, the transition of adipocytes from beige to white is regulated by autophagy-dependent clearance of mitochondria [132].
Metabolic remodeling: a key factor in mitophagy-mediated cellular reprogramming
Metabotypes of stem cells
Due to the essential role of mitochondria in the bioenergetics and depending upon the energy demand of the concerned cell, they undergo structural and functional remodeling. Such remodeling facilitates alterations in the respiratory function and determines the fate of the concerned cell. It was reported that different respiratory phenotypes determine the differentiation status of the cell and that mitophagy acts as a key mechanism for manipulation of the cell fate through the remodeling of metabolic pathways. The human pluripotent stem cells (hPSCs) display lower OXPHOS with perinuclear mitochondria that are less fused into a filamentous network structure with swollen, less mature appearing cristae folds of the inner membrane (Fig. 2a). The hPSCs are metabolically similar to the developmentally more mature, glycolytic mouse epiblast stem cells (mEpiSCs), rather than mESCs, which show a bivalent metabolism that can dynamically switch between glycolysis and OXPHOS on demand [133] (Fig. 2a). The human ESCs (hESCs) or EpiSCs, due to their low COX IV expression, have lower mitochondrial respiratory capacity despite having a more developed and expanding mitochondrial content (Fig. 2a). Moreover, EpiSCs/hESCs are functionally comparable to the glycolytic phenotype in cancer (Warburg effect) (Fig. 2a) [95, 133]. Naïve hPSCs or ground state hPSCs show higher ATP production through OXPHOS as compared to the more mature “primed” hPSCs, which are more glycolytic (Fig. 2a) [95]. Furthermore, fibroblasts are more oxidative than primed hPSCs and factors inducing glycolysis inhibit OXPHOS to promote iPSC reprogramming. Mitochondrial integrity and functional energetics cause relative shifts in metabolism, from Naïve through “Primed” pluripotent states and finally to lineage-directed differentiation [95].
Metabolic transition and cell fate
Extensive metabolic reconfiguration is a crucial phenomenon reported to occur during transition of cell fate. Somatic cell reprogramming to iPSCs primarily involves the transition from OXPHOS to glycolysis. This process is highly coordinated through the epi-transcriptional networks that promote the upregulation of glycolytic genes and downregulation of OXPHOS genes. The structural and morphological conversion into immature mitochondria makes them ineligible for the OXPHOS. Moreover, induced pluripotency could be achieved by the activation of pyruvate dehydrogenase kinase 1 (PDK1) in combination with only one transcription factor OCT4 that facilitates the metabolic transition from OXPHOS to glycolysis [18]. The glycolytic genes precede the expression of pluripotent genes metabolic remodeling could be regarded as essential pre-requisites during the dedifferentiation process (Fig. 5b) [18]. Intriguingly, the role of mitophagy in metabolic transition and its subsequent effect on cell fate is poorly understood, making it a prospective aspect worth exploring. However, the few available studies indicate that mitophagy significantly influences the mitochondrial dynamics and a cell’s decision to remain pluripotent or undergo differentiation. Mitophagy may proceed to glycolysis or OXPHOS depending on the demand, and the cell fate is regulated accordingly.
Fig. 5.
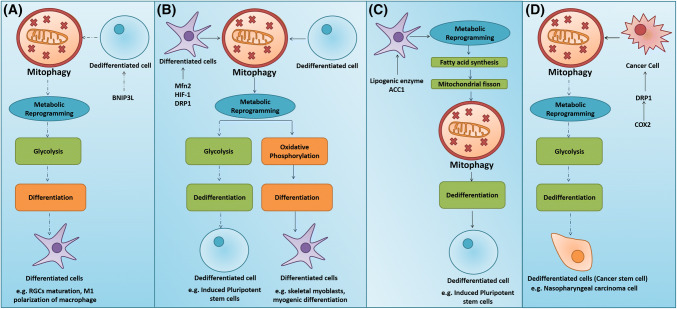
Mitophagy-mediated metabolic reshuffling regulates cell fate transition. a Mitophagy-mediated metabolic reprogramming towards glycolysis in dedifferentiated cells induces differentiation. b Mitophagy-mediated metabolic reprogramming towards glycolysis induces differentiation in terminally differentiated cells stimulates dedifferentiation, whereas mitophagy-mediated metabolic reprogramming towards OXPHOS induces differentiation of dedifferentiated cells. c Metabolic reprogramming into increased lipogenesis in terms of fatty acid synthesis induces mitophagy-dependent dedifferentiation. d Mitophagy-induced metabolic reprogramming towards glycolysis promotes dedifferentiation of cancer cells and offers a stemness status
The bioenergetic changes from OXPHOS to glycolysis critically regulate nuclear reprogramming, as it has been reported that inadequate bioenergetic metabotype cannot deliver the attributes of self-renewal and pluripotency to a cell. Moreover, quite interestingly, the cellular content and activity of the mitochondrial H+-ATPase synthase, a core component of OXPHOS in iPSCs, is dramatically limited. It can be observed that H+-ATPase synthase activity is directly correlated with OXPHOS and inversely associated with the rate of glucose utilization by aerobic glycolysis [134–136]. Furthermore, a significant downregulation of the catalytic β1-F1-ATPase subunit and an increase in the expression of ATPase inhibitor factor 1 (IF1) in adult stem cell (ASC) markers were reported to be associated with the maintenance of aerobic glycolysis [137]. In this scenario, mitophagy-driven metabolic transition from OXPHOS to glycolysis induces pluripotency. A report indicates that depletion of MFN1/2 activates the Ras–Raf and hypoxia-inducible factor 1α (HIF1α) signaling to facilitate the glycolytic metabolic transition at an early stage of reprogramming. The reduction of MFN1/2 facilitates pluripotency by restructuring the mitochondrial dynamics and bioenergetics [112]. The rejuvenation of mitochondria, which contributes to the ability of iPSCs to suppress differentiation, is due to the mitophagy-driven bioenergetic transition and metabolome remodeling. To confirm this, the mitophagy-deficient iPSC colonies were observed to exhibit impaired glycolysis [96]. Moreover, in nasopharyngeal cancer models, the DRP1-mediated mitochondrial fission was reported to induce a metabolic switch from OXPHOS to glycolysis and promote cancer stemness (Fig. 5d) [117].
Unlike the dedifferentiation process, the differentiation of stem cells into a terminally differentiated mature phenotype requires the metabolic reprogramming into an oxidative phenotype, concomitantly with a shift from cytosolic anaerobic glycolysis to mitochondrial respiration to meet the increased energy demand [103]. To fulfill this requirement, the mitochondria undergo substantial modifications in terms of number, structure, morphology, and distribution, which enable them to undergo OXPHOS. For instance, during differentiation, the skeletal myoblasts specifically shift from a highly glycolytic state to a phenotype relying predominantly on OXPHOS (Fig. 5b). In this case, it appears that mitophagy-driven metabolic transition into OXPHOS induces the differentiation of stem cells (Fig. 5b). The metabolic remodeling into OXPHOS that promotes the terminal differentiation from the progenitor cells to a particular cell type is believed to be critically dependent on mitophagy. During early myogenic differentiation, mitochondrial fusion protein OPA1 is rapidly upregulated after dynamin-1-like protein (DNM1L)/DRP1-mediated mitophagy which results in the repopulation of mitochondria via peroxisome proliferator-activated receptor-gamma coactivator (PGC)-1alpha (PPARGC1A)/PGC-1α-mediated biogenesis, resulting in the reformation of mitochondrial networks that are better primed for OXPHOS than myoblasts (Fig. 5b) [138]. In this case, it is apparent that the differentiating cell, upon receiving the signal for differentiation, initially gets rid of the immature mitochondria through mitophagy to form new mitochondria that can meet the energy demand.
Extensive metabolic resetting, wherein the mitochondrial activity is higher in most cell types, precedes cellular differentiation. In contrast, some studies reported that during neurogenesis, the achievement of hypoxia-induced BNIP3L-dependent mitophagy-mediated metabolic shift toward glycolysis (not OXPHOS) regulates the numbers of RGCs and selects the first neurons to differentiate in the retina as well as decide which axons form the optic nerve (Fig. 5a). Moreover, mitophagy-mediated metabolic reprogramming to the glycolytic phenotype causes differentiation of macrophages toward M1 polarization to develop a rapid immune response during inflammation (Fig. 5a) [127]. The transition to glycolysis facilitates the faster supply of energy for a rapid immune response. Hence, in this scenario, mitophagy-driven metabolic transition into glycolysis induces stem cell differentiation. Similarly, a metabolic switch from mitochondrial fatty acid (FA) oxidation and pyruvate oxidation to glycolysis and glutaminolysis in the quiescent skeletal muscle stem cells or satellite cells (SCs) occur during the transition to activation and proliferation state [139, 140]. Fine-tuning the flux of mitochondrial FA oxidation and histone acetylation by NAD+-sirtuin-1 (SIRT1)-PGC1α axis might orchestrate a delicate balance of quiescence, proliferation, and senescence of skeletal muscle stem cells [138, 139].
The acquisition of stemness can be attributed to the ability of the cell to channelize the intermediates of the mitochondrial tricarboxylic acid (TCA) cycle to lipid biosynthesis (lipogenesis), which is an important switch required for the self-renewal and differentiation of ESCs and iPSCs. Many predominant lipid metabolite molecules such as diacylglycerol, arachidonic acid, and prostaglandins are also found in iPSCs [141]. The inhibition of lipogenic enzymes acetyl-CoA carboxylase (ACACA) and fatty acid synthase (FASN) has been reported to decrease the reprogramming efficiency. Moreover, ACACA and FASN are highly expressed in iPSCs [142]. Pharmacological inhibition of ACACA and FASN has been reported to inhibit the formation of mammospheres in a fatty acid-dependent manner, which strongly advocates the role of de novo lipogenesis, lipid metabolites, and lipid catabolism in the self-renewal and survival of CSCs, and gives rise to a lipogenic state of stem cells [134, 141]. The FASN-driven lipogenic switch is the key metabolic event influencing the dedifferentiation process during the somatic cell reprogramming to iPSCs, by coupling the Warburg effect to anabolic metabolism [142]. Although, earlier reports emphasize that de novo lipogenesis is a crucial phenomenon in dedifferentiation, its correlation with mitophagic events is poorly understood. A recent study has reported that mitochondrial fission-dependent lipogenesis in terms of FA synthesis is crucial for the maintenance of stem cell pluripotency as well as during cellular reprogramming (Fig. 5c). Experimentally, the lipogenic enzyme ACC1 was observed to regulate de novo FA synthesis, resulting in increased mitochondrial fission. The mitochondrial fission occurs through the consumption of acetyl coenzyme A (AcCoA) leading to the degradation of acetylation-mediated mitochondrial fission 1 (FIS1) protein ubiquitin–proteasome and generation of lipid products favoring the mitochondrial dynamic equilibrium toward fission, which in turn is critical for the maintenance of stem cell pluripotency [111].
Conclusions and future perspectives
Apart from the quality control for cellular homeostasis, mitophagy also plays a crucial role in maintaining the balance between stemness and differentiation during the processes of cellular development and maturation. Mitochondrial division is tightly associated with cellular reprogramming to control the pluripotency of different stem cells. In addition, autophagy-dependent mitochondrial clearance influences the structural and functional aspects of mitochondria to determine cell fate and maintain homeostasis. Moreover, mitochondrial energy metabolism has been linked to mitophagy which influences the differentiation and commitment of cell types. Mitophagy-regulated metabolic shift plays an essential role in facilitating the restoration of differentiated cells to a pluripotent state. Although the details are yet to be explained, the role of mitophagy in maintaining cancer stem cell pool and promoting cancer progression is well known. As the levels of metabolites change remarkably during the cell fate transition, metabolic remodeling could be regarded as a prerequisite for cellular reprogramming. However, there are still few issues which need to be addressed: (1) how does mitophagy occur independently of bulk autophagy; (2) how are mitochondria selectively targeted to autophagosome; and (3) when and how does mitophagy come into play in stem cell biology? Furthermore, it is still unclear how a cell identifies the healthy and damaged mitochondria to execute mitophagy. Moreover, the key machinery that acts as a mitophagy sensor still remains elusive. Future studies should focus on elucidating the ability of mitophagy to direct metabolic shift on the acquisition and maintenance of stem cell pluripotency and differentiation to modulate different diseases, including aging and cancer.
Acknowledgements
Research support was partly provided by Department of Biotechnology [Grant number: BT/PR7791/BRB/10/1187/2013; Science and Technology Department, Government of Odisha; the Board of Research in Nuclear Sciences (BRNS) [number: 37(1)/14/38/2016-BRNS/37276], Department of Atomic Energy (DAE); Science and Engineering Research Board (SERB) [number: EMR/2016/001246], Department of Science and Technology.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 2.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilmut I, Schnieke AE, Mcwhir J, et al. Viable offspring derived from fetal and adult mammalian cells. Cloning Stem Cell. 2007;9:3–7. doi: 10.1089/clo.2006.0002. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Wernig M, Meissner A, Cassady JP, et al. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Brambrink T, Foreman R, Welstead GG, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Duve C, Wattiaux R. Functions of lysosomes. Ann Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 11.Nasrallah CM, Horvath TL. Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol. 2014;10:650. doi: 10.1038/nrendo.2014.160. [DOI] [PubMed] [Google Scholar]
- 12.Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–59. doi: 10.1016/S0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 13.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Duan S, Yi F, et al. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Prigione A, Lichtner B, Kuhl H, et al. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell–like metabolic reprogramming. Stem Cell. 2011;9:1338–1348. doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- 16.Fang D, Yan S, Yu Q, Chen D, et al. Mfn2 is required for mitochondrial development and synapse formation in human induced pluripotent stem cells/hiPSC derived cortical neurons. Sci Rep. 2016;6:31462. doi: 10.1038/srep31462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanet A, Arnould T, Najimi M, et al. Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Facucho-Oliveira JM, Alderson J, Spikings EC, et al. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 20.Prigione A, Ruiz-Pérez MV, Bukowiecki R, et al. Metabolic restructuring and cell fate conversion. Cell Mol Life Sci. 2015;72:1759–1777. doi: 10.1007/s00018-015-1834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukowiecki R, Adjaye J, Prigione A. Mitochondrial function in pluripotent stem cells and cellular reprogramming. Gerontology. 2014;60:174–182. doi: 10.1159/000355050. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Khvorostov I, Hong JS, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varum S, Rodrigues AS, Moura MB, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell. 2010;21:1225–1236. doi: 10.1091/mbc.e09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youle RJ, Van Der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prigione A, Fauler B, Lurz R, et al. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cell. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong L, Tilgner K, Saretzki G, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cell. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 28.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 29.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang J, Xu ZX, Ding Z, et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat Commun. 2015;6:7926. doi: 10.1038/ncomms8926. [DOI] [PubMed] [Google Scholar]
- 31.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif–crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 32.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cell. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 35.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 36.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 37.Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamark T, Kirkin V, Dikic I, et al. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 40.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in Parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxid Redox Signal. 2012;17:794–802. doi: 10.1089/ars.2011.4407. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Massen S, Terenzio M, et al. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanna RA, Quinsay MN, Orogo AM, et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 47.Wu W, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakawa T, Yamaguchi O, Hashimoto A, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otsu K, Murakawa T, Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. 2015;11:1932–1933. doi: 10.1080/15548627.2015.1084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sentelle RD, Senkal CE, Jiang W, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin SM, Lazarou M, Wang C, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meissner C, Lorenz H, Weihofen A, et al. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 54.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 59.Ordureau A, Sarraf SA, Duda DM, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan NC, Salazar AM, Pham AH, et al. Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 62.Geisler S, Holmström KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y, Chiang WC, Sumpter R, et al. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168(224–38):e10. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hollville E, Carroll RG, Cullen SP, et al. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell. 2014;55:451–466. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Narendra D, Kane LA, Hauser DN, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michiorri S, Gelmetti V, Giarda E, et al. The Parkinson-associated protein PINK1 interacts with beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 67.Van Humbeeck C, Cornelissen T, Hofkens H, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strappazzon F, Vietri-Rudan M, Campello S, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strappazzon F, Nazio F, Corrado M, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orvedahl A, Sumpter R, Jr, Xiao G, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J, Lee SB, Lee S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 72.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamaguchi O, Murakawa T, Nishida K, et al. Receptor-mediated mitophagy. J Mol Cell Cardiol. 2016;95:50–56. doi: 10.1016/j.yjmcc.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 74.Hamacher-Brady A, Brady N, Logue S, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 75.Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73:775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Melser S, Chatelain EH, Lavie J, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013;17:719–730. doi: 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 77.Chen G, Han Z, Feng D, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 78.Wu H, Xue D, Chen G, et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014;10:1712–1725. doi: 10.4161/auto.29568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bian Y, Song C, Cheng K, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteom. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 80.Panda PK, Naik PP, Meher BR, et al. PUMA dependent mitophagy by Abrus agglutinin contributes to apoptosis through ceramide generation. Biochim Biophys Acta Mol Cell Res. 2018;1865:480–495. doi: 10.1016/j.bbamcr.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 81.McLelland GL, Soubannier V, Chen CX, et al. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miyamoto Y, Kitamura N, Nakamura Y, et al. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS One. 2011;6:e16054. doi: 10.1371/journal.pone.0016054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hamacher-Brady A, Choe S, Krijnse-Locker J, et al. Intramitochondrial recruitment of endolysosomes mediates Smac degradation and constitutes a novel intrinsic apoptosis antagonizing function of XIAP E3 ligase. Cell Death Differ. 2014;21:1862. doi: 10.1038/cdd.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamacher-Brady A, Brady NR. Bax/Bak-dependent, Drp1-independent targeting of XIAP into inner-mitochondrial compartments counteracts Smac-dependent effector caspase activation. J Biol Chem. 2015;M115:643064. doi: 10.1074/jbc.M115.643064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kitamura N, Nakamura Y, Miyamoto Y, et al. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One. 2011;6:e16060. doi: 10.1371/journal.pone.0016060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Begus-Nahrmann Y, Lechel A, Obenauf AC, et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- 87.Liu K, Lee J, Kim JY, et al. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 2017;68(281–92):e5. doi: 10.1016/j.molcel.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koehler CL, Perkins GA, Ellisman MH, et al. Pink1 and Parkin regulate Drosophila intestinal stem cell proliferation during stress and aging. J Cell Biol. 2017;216:2315–2327. doi: 10.1083/jcb.201610036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mahrouf-Yorgov M, Augeul L, Da Silva CC, et al. Mesenchymal stem cells sense mitochondria released from damaged cells as danger signals to activate their rescue properties. Cell Death Differ. 2017;24:1224. doi: 10.1038/cdd.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Phinney DG, Di Giuseppe M, Njah J, et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. doi: 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mortensen M, Soilleux EJ, Djordjevic G, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 93.Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Teslaa T, Teitell M. Pluripotent stem cell energy metabolism: an update. EMBO J. 2015;34:138–153. doi: 10.15252/embj.201490446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vazquez-Martin A, Van den Haute C, Cufí S, et al. Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging. 2016;8:1330. doi: 10.18632/aging.100976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bordt EA, Clerc P, Roelofs BA, et al. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev Cell. 2017;40:583–594. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vazquez-Martin A, Cufí S, Corominas-Faja B, et al. Mitochondrial fusion by pharmacological manipulation impedes somatic cell reprogramming to pluripotency: new insight into the role of mitophagy in cell stemness. Aging. 2012;4:393. doi: 10.18632/aging.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xiang G, Yang L, Long Q, et al. BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy. 2017;13:1543–1555. doi: 10.1080/15548627.2017.1338545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu K, Zhao Q, Liu P, et al. ATG3-dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy. 2016;12:2000–2008. doi: 10.1080/15548627.2016.1212786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ito K, Turcotte R, Cui J, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354:1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Angelova PR, Barilani M, Lovejoy C, et al. Mitochondrial dysfunction in parkinsonian mesenchymal stem cells impairs differentiation. Redox Biol. 2017;14:474–484. doi: 10.1016/j.redox.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marycz K, Kornicka K, Grzesiak J, et al. Macroautophagy and selective mitophagy ameliorate chondrogenic differentiation potential in adipose stem cells of equine metabolic syndrome: new findings in the field of progenitor cells differentiation. Oxid Med Cell Longev. 2016;2017:3861790. doi: 10.1155/2017/3861790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song M, Mihara K, Chen Y, et al. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–285. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mao K, Klionsky DJ. Mitochondrial fission facilitates mitophagy in Saccharomyces cerevisiae. Autophagy. 2013;9:1900–1901. doi: 10.4161/auto.25804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frank M, Duvezin-Caubet S, Koob S, et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim et Biophys Acta (BBA) Mol Cell Res. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 107.Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Son MY, Choi H, Han YM, et al. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem Cells. 2013;31:2374–2387. doi: 10.1002/stem.1509. [DOI] [PubMed] [Google Scholar]
- 109.Todd LR, Gomathinayagam R, Sankar U. A novel Gfer-Drp1 link in preserving mitochondrial dynamics and function in pluripotent stem cells. Autophagy. 2010;6:821–822. doi: 10.4161/auto.6.6.12625. [DOI] [PubMed] [Google Scholar]
- 110.Prieto J, León M, Ponsoda X, et al. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle. 2016;15:3240–3250. doi: 10.1080/15384101.2016.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L, Zhang T, Wang L, et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330–1347. doi: 10.15252/embj.201695417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Son M, Kwon Y, Son M, et al. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ. 2015;22:1957–1969. doi: 10.1038/cdd.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 114.Naik PP, Das DN, Panda PK, et al. Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncol. 2016;62:122–135. doi: 10.1016/j.oraloncology.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 115.Naik PP, Mukhopadhyay S, Panda PK, et al. Autophagy regulates cisplatin-induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 2018;51:e12411. doi: 10.1111/cpr.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou TJ, Zhang SL, He CY, et al. Downregulation of mitochondrial cyclooxygenase-2 inhibits the stemness of nasopharyngeal carcinoma by decreasing the activity of dynamin-related protein 1. Theranostics. 2017;7:1389. doi: 10.7150/thno.17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen YA, Wang CY, Hsieh YT, et al. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Whelan KA, Chandramouleeswaran PM, Tanaka K, et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36:4843–4858. doi: 10.1038/onc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan C, Luo L, Guo CY, et al. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 120.Barde I, Rauwel B, Marin-Florez RM, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu L, Xu W, Xu L, et al. Mitophagy is increased during erythroid differentiation in β-thalassemia. Int J Hematol. 2017;105:162–173. doi: 10.1007/s12185-016-2114-z. [DOI] [PubMed] [Google Scholar]
- 122.Sin J, Andres AM, Taylor DJ, et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2016;12:369–380. doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim B, Kim JS, Yoon Y, et al. Inhibition of Drp1-dependent mitochondrial division impairs myogenic differentiation. Am J Physiol Regul Integr Comp Physiol. 2013;305:R927–R938. doi: 10.1152/ajpregu.00502.2012. [DOI] [PubMed] [Google Scholar]
- 124.Gong G, Song M, Csordas G, Kelly DP, et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xavier JM, Morgado AL, Sola S, et al. Mitochondrial translocation of p53 modulates neuronal fate by preventing differentiation-induced mitochondrial stress. Antioxid Redox Signal. 2014;21:1009–1024. doi: 10.1089/ars.2013.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Esteban-Martínez L, Sierra-Filardi E, McGreal RS, et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017;36:1688–1706. doi: 10.15252/embj.201695916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Esteban-Martinez L, Boya P. BNIP3L/NIX-dependent mitophagy regulates cell differentiation via metabolic reprogramming. Autophagy. 2017;14:915–917. doi: 10.1080/15548627.2017.1332567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chauss D, Basu S, Rajakaruna S, et al. Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens. G3 Genes Genomes Genet. 2014;4:1515–1527. doi: 10.1534/g3.114.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Costello MJ, Brennan LA, Basu S, et al. Autophagy and mitophagy participate in ocular lens organelle degradation. Exp Eye Res. 2013;116:141–150. doi: 10.1016/j.exer.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Larson-Casey JL, Deshane JS, Ryan AJ, et al. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity. 2016;44:582–596. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goldman SJ, Zhang Y, Jin S. Autophagic degradation of mitochondria in white adipose tissue differentiation. Antioxid Redox Signal. 2011;14:1971–1978. doi: 10.1089/ars.2010.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Altshuler-Keylin S, Shinoda K, Hasegawa Y, et al. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016;24:402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou W, Choi M, Margineantu D, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Corominas-Faja B, Cuyàs E, Gumuzio J, et al. Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget. 2014;5:8306. doi: 10.18632/oncotarget.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sánchez-Cenizo L, Formentini L, Aldea M, et al. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Willers IM, Cuezva JM. Post-transcriptional regulation of the mitochondrial H+-ATP synthase: a key regulator of the metabolic phenotype in cancer. Biochim Biophys Acta (BBA) Bioenerget. 2011;1807:543–551. doi: 10.1016/j.bbabio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 137.Sánchez-Aragó M, García-Bermúdez J, Martínez-Reyes I, et al. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013;14:638–644. doi: 10.1038/embor.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wenz T, Rossi SG, Rotundo RL, et al. Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 139.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ryall JG, Dell’Orso S, Derfoul A, et al. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Panopoulos AD, Yanes O, Ruiz S, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vazquez-Martin A, Corominas-Faja B, Cufi S, et al. The mitochondrial H+-ATP synthase and the lipogenic switch: new core components of metabolic reprogramming in induced pluripotent stem (iPS) cells. Cell Cycle. 2013;12:207–218. doi: 10.4161/cc.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]