Abstract
Accurate determination of microRNA expression levels is a prerequisite in using these small non-coding RNA molecules as novel biomarkers in disease diagnosis and prognosis. Quantitative PCR is the method of choice for measuring the expression levels of microRNAs. However, a major obstacle that affects the reliability of results is the lack of validated reference controls for data normalization. Various non-coding RNAs have previously been used as reference controls, but their use may lead to variations and lack of comparability of microRNA data among the studies. Despite the growing number of studies investigating microRNA profiles to discriminate between healthy and disease stages, robust reference controls for data normalization have so far not been established. In the present article, we provide an overview of different reference controls used in various diseases, and highlight the urgent need for the identification of suitable reference controls to produce reliable data. Our analysis shows, among others, that RNU6 is not an ideal normalizer in studies using patient material from different diseases. Finally, our article tries to disclose the challenges to find a reference control which is uniformly and stably expressed across all body tissues, fluids, and diseases.
Keywords: MicroRNA normalization, Reference controls, Tissue, Plasma, Serum, Benign and malignant diseases
Introduction
MicroRNAs (miRNAs) are single-stranded, non-coding RNA molecules that play a pivotal role in post-transcriptional regulation of gene expression. They act as translational repressors by binding to complementary sequences in the 3′-untranslated region (UTR) of their target mRNAs [1]. To date, numerous reports have documented that miRNAs are involved in multiple pathways that play a role in cell differentiation, proliferation, and apoptosis. Moreover, aberrant expression of miRNAs has been detected in a wide range of pathological conditions including cancers [2–5], neurodegenerative disorders [6, 7], and cardiovascular diseases [8]. This detection points to disease-specific miRNA expression profiles that correlate with patient diagnosis, prognosis, and responses to treatment [9]. Thus, measuring the expression levels of miRNAs is of scientific and clinical significance.
Several methods, such as real-time PCR, northern blotting, next-generation sequencing, and microarray assays, have been described for miRNA quantification [10]. Real-time PCR is the gold standard method for measuring miRNA expression due to its high sensitivity, specificity, reproducibility, and low template requirements [11]. The use of reference controls for data normalization, technical and handling variations introduced by differences in the amount and quality of starting material, as well as efficiency and performance of the technical platforms are expected to be corrected for an accurate miRNA quantification [12]. Thus, an optimal selection of reference controls is critical for miRNA expression studies. Several RNA species, including small nuclear RNAs (snRNAs), nucleolar RNAs (snoRNAs), ribosomal RNAs (rRNAs), miRNAs, and synthetic RNAs have been used as endogenous and exogenous reference controls so far.
Despite an increasing number of miRNA studies, there is no current consensus on reference controls for miRNA analyses in various diseases. The use of “pseudo” reference controls seems to be one of the major reasons for the discrepancies in miRNA expression levels among the previously published studies [13]. For example, Hong et al. reported that miR-21 is up-regulated by 550-fold in pancreatic cancer tissues if normalized to RNU6 [14], while using RNU6 and 5S rRNA as reference controls, du Rieu et al. detected a 20-fold up-regulation in miR-21 levels in the same malignant tissues [15]. This is a tremendous discrepancy and does not make the data convincing. In the following, we introduce different reference controls used for the miRNA quantification in different sources and diseases, and discuss their impact on data normalization.
For our literature research, we applied PubMed, and used the following keywords: reference control, normalizer, internal control plus serum, and plasma, tissue (Table 1).
Table 1.
Summary of non-coding RNAs used in different studies and their eligibility as data normalizers
| Disease | RNA type | Source | Sample size (patient/control) | Suitability | References |
|---|---|---|---|---|---|
| Hepatitis B | RNU6 | Serum | 52/57 | No | [28] |
| RNU6 | Serum-derived exosomes | 50/50 | No | [29] | |
| RNU6 | Plasma | 20/– | No | [30] | |
| The combination of miR-22*, miR-26a, and miR-221 | Serum | 52/57 | Yes | [28] | |
| miR-221/miR-22* | Serum-derived exosomes | 50/50 | Yes/no | [29] | |
| miR-221/miR-106a/miR-21 | Plasma | 20/– | No/yes/yes | [30] | |
| Chronic kidney disease | miR-16/miR-92a | Urine | 33/5 | Yes/no | [33] |
| Coronary artery disease | miR-6090/miR-4516/RNU6 | Plasma | 111/111 | Yes/yes/no | [36] |
| Hypertension | miR-92a-3p/miR-16-5p/miR-21-5p | Plasma | 18/10 | Yes/yes/yes | [38] |
| Tuberculosis | miR-93 | Plasma | 36/24 | Yes | [39] |
| Major depressive disorder | The combination of miR-101-3p and miR-93-5p/RNU6 | Plasma | 16/14 | Yes/no | [40] |
| Parkinson’s disease | The combination of RNU24 and Z30 snoRNA/RNU6B/miR-103a-3p | Blood | 38/38 | Yes/no/no | [41] |
| Colorectal cancer | The combination of miR-193a-5p, miR-27a and let-7g/RNU6B | Tissue | 53/53 | Yes/no | [44] |
| RNU6/miR-191-5p | Serum | 173/100 | Yes/yes | [45] | |
| miR-106b-5p/miR-25-3p/miR-93-5p | Serum | 30/30 | Yes/yes/yes | [47] | |
| the combination of miR-345 and miR-16 | Tissue | 35/39 | Yes | [48] | |
| miR-520d/miR-1228/miR-345 | Exosome, plasma, tissue | 20/20 | Yes/yes/yes | [51] | |
| Breast cancer | miR-425/miR-16/RNU6B | Blood | 40/20 | Yes/yes/no | [52] |
| The combination of miR-16 and let-7a | Tissue | 26/– | Yes | [53] | |
| miR-16/5S rRNA/RNU6 | Serum | 15/15 | No/no/no | [54] | |
| RNU6 | Tissue | 20/20 | No | [58] | |
| Bladder cancer | The combination of miR-151-5p, miR-125a-5p, miR-148b and miR-101/the combination of miR-148b, miR-874, and miR-181b/RNU6B | Tissue | 58/58 | Yes/yes/no | [60] |
| miR-193a-5p/miR-16-5p | Serum | 60/35 | Yes/yes | [62] | |
| Renal cell carcinoma | The combination of miR-28, miR-103 and miR-106a/RNU6B/RNU44 | Tissue | 57/57 | Yes/no/no | [63] |
| RNU6B/miR-16 | Serum | 34/23 | No/yes | [64] | |
| miR-145 | Tissue | 34/34 | Yes | [64] | |
| Cervical cancer | miR-23a/miR-191/RNU6 | Tissue | 23/23 | Yes/yes/no | [66] |
| RNU6/RNU6B/miR-423 | Tissue | 20/10 | No/no/yes | [68] | |
| Hepatocellular carcinoma | the combination of miR-221, let-7a, and miR-26a/miR-16/RNU6/5S rRNA | Serum | 33/33 | Yes/no/no/no | [70] |
| The combination of miR-221, let-7a, and miR-26a/RNU6 | Serum-derived exosomes | 50/50 | Yes/no | [29] | |
| The combination of miR-21 and miR-106a/RNU6 | Plasma | 20/– | Yes/no | [30] | |
| Prostate cancer | miR-130b/miR-16/RNU6B | Tissue | 76/76 | Yes/no/yes | [72] |
| RNU6B/RNU24/RNU43 | Tissue | 19/19 | No/yes/no | [73] | |
| miR-191 | Urine | 35/26 | Yes | [74] | |
| Pancreatic cancer | U91 snoRNA/RNU6/miR-16 | Tissue | 24/- | Yes/no/no | [76] |
| RNU6B | Serum-derived exosomes | 41/8 | Yes | [77] | |
| Gastric cancer | miR-16/miR-93 | Serum | 40/20 | Yes | [80] |
| Endometrioid endometrial carcinoma | The combination of RNU48, RNU44 and SNORD75/miR-92a/miR-26b | Tissue |
30/15 (fresh samples) 44/14 (FFPEa samples) |
Yes/no/no | [81] |
| Ovarian cancer | RNU48/miR-16-5p/miR-92a-3p | Tissue | 75/30 | Yes/no/no | [84] |
aFormalin-fixed paraffin-embedded
Technical challenges
Accurate and comparable miRNA quantification depends on pre-analytical and analytical factors. Several aspects should be considered, starting from the selection and quality of the matrix, the technical platform and method applied for miRNA extraction, amplification and detection, and, finally, the strategy of data quantification and normalization [13, 16, 17]. MiRNAs can be extracted from cultured cells, conditioned cell culture media, as well as fresh, frozen and fixed tissues, and fresh and stored body fluids, e.g., whole blood, plasma, serum, and urine. To avoid molecular changes in miRNA levels leading to global miRNA instability and causing enrichment or depletion of particular miRNAs in the human samples, their collection requires the preservation by different methods, such as blood coagulation prevention, tissue freezing, fixing, and paraffin-embedding [18]. However, the administration of archival tissue blocks with formalin and paraffin may lead to low yields of amplifiable RNA, false-positive data, and poor reproducibility.
Although miRNAs circulate stable in liquid biopsies, their levels are known to be affected by several pre-analytical factors. Initially, blood collection by anticoagulants, such as heparin, acid citrate dextrose or EDTA, requires a rapid processing of blood. There are also tubes (PAX) which stabilize circulating miRNAs and prevent cell lysis. Thus, they allow a prolonged storage and facilitate transport at ambient temperature between the clinic and a laboratory. Notably, plasma should be shipped. Blood cell count also significantly affects miRNA levels in serum/plasma; therefore, samples from patients with inflammatory status, who usually have high white blood cell counts, should be excluded from the analysis. In addition, hemolytic samples should also be excluded. Hemolysis can be measured by hemoglobin quantification by spectral analysis. For the preparation of serum/plasma, a low- and high-speed step centrifugation should be carried out to avoid a contamination with cells. For long-term storage, miRNA samples should be rather stored at temperatures of − 80 °C than − 20 °C [16].
MiRNAs are challenging molecules to quantify, mainly because of their very short length, their GC content, similarities in sequences among miRNAs of the same family, and the low abundance in the body fluids [16]. Moreover, miRNAs only represent a small part of total RNA, and exist in three forms: the short, linear mature miRNA, the hairpin pre-miRNA, and the long pri-miRNA [19]. MiRNA recovery also depends on their GC content and the free energy (ΔG) of their most stable secondary structure. Structured miRNAs that fold into a stable secondary structure display a low ΔG. For example, phenol-based isolation techniques result in a poor recovery rate of structured miRNAs with a low GC content in samples with low RNA concentrations [20].
The gold standard for detection of particular miRNAs is real-time PCR which is specific and sensitive, allowing the detection of small quantities of miRNAs. The most common RT-PCR uses stem-loop-shaped RT-primer TaqMan assays (Applied Biosystems), assays using locked nucleic acid primers (Exiqon), and assays with poly-A tailing primers (Qiagen). However, using these techniques, only a limited set of miRNAs can be tested in a single reaction [21]. In contrast, the TaqMan real-time PCR-based arrays using format microfluidic cards on which preloaded PCR primers can simultaneously profile hundreds of known miRNAs [22] provide a better comparison of the levels of several miRNAs. Finally, massively parallel sequencing (next-generation sequencing and RNA-seq) technology has enabled profiling of know and new miRNAs.
Unfortunately, most studies have only investigated miRNAs in populations with a low patient number. These small-sized cohorts may introduce biases resulting in data misinterpretation, and influence a confident detection and validation of disease biomarkers. A previously published meta-analysis demonstrated that the majority of the studies have only dealt with populations below 100 individuals with a median size of 69 subjects for the detection of miRNAs in non-neoplastic diseases [23]. To conclude, these common factors in performing miRNA analyses may contribute to lacking comparability of miRNA data among the studies and exacerbate reproducibility of the studies.
Eligibility of a reference miRNA for data normalization
To accurately determine the levels of analyzed miRNAs, their expression data should be normalized relatively to endogenous and exogenous reference genes. As listed in the following sections, different studies use different normalization strategies. This leads to ambiguous data interpretation and impairing comparisons between studies. So far, no optimal normalization strategy seems to have reached consensus status for the scientific community. To find the best endogenous reference miRNAs, different algorithms including geNorm and Normfinder were created. For each miRNA, geNorm calculates a stability score as the average of a miRNA with all other miRNAs. The less stable miRNA is removed, and calculation is repeated until the most stable pair has been obtained. Normfinder estimates the inter-group and intra-group variances of the log-transformed expression ratios, and then combines them into a stability value [24]. However, the reference miRNA selected by these algorithms needs a further validation in human tissues to examine their steady expression across the samples derived from patients as well as healthy individuals. A stable expressed miRNA folds into a stable secondary structure displays a short hairpin structure and exhibits a low free energy. Accordingly, a stable reference miRNA for an accurate data normalization should harbor the feature of a housekeeping gene which is uniformly, consistently, and highly expressed among the samples, and less degraded by RNA nucleases.
Small nuclear, nucleolar, ribosomal RNAs, and miRNAs as reference controls
The non-coding snRNAs, snoRNAs, and rRNAs do not belong to the family, but they are frequently used for miRNA data normalization. For example, snRNAs are divided into Sm- and Lsm-classes and involved in pre-mRNA intron splicing. SnoRNAs which are also divided into two classes have diverse functions, of which 2′-O-methylation and pseudouridylation of rRNAs, tRNAs, and snRNAs are the main functions. rRNAs are the major components of the ribosome complex, and are essential for translation in all living organisms [25, 26].
Liver disorders
The small non-coding RNU6 is the most frequently used reference gene for data normalization. However, RNU6 is not a miRNA, and therefore, miRNA data normalization with a different RNA type should always be scrutinized [27]. In this regard, Zhu et al. evaluated the reliability of RNU6 as a reference control for miRNA data normalization in Hepatitis B (HBV). Analyzing RNU6 levels in the serum of 52 HBV patients and 57 healthy controls, they found large variations in RNU6 expression (10 cycles) and a different expression between the two groups. The stability of RNU6 was further determined using geNorm and NormFinder algorithms, and both programs identified RNU6 as the least steadily expressed gene [28]. Likewise, Li et al. found that expression of RNU6 was highly unsteady in serum-derived exosomes of 50 HBV patients and 50 healthy controls. They also detected that data normalization with RNU6 introduced bias into the analysis, resulting in the misinterpretation of miR-21 expression levels in HBV patients compared with hepatocellular carcinoma patients [29]. Moreover, Tang et al. showed that RNU6 expression lacked a constant level in the plasma samples of 20 HBV patients [30]. In a benign liver disorder, Benz et al. indicated that RNU6 levels were highly variable and down-regulated in the serum of 64 liver fibrosis patients [31]. Finally, we also demonstrated that the reliability of RNU6 as a reference gene for serum miRNA data normalization needs further verification [32].
In addition, numerous studies have normalized their miRNA data to miRNAs. The use of miRNAs as normalizer seems to be more appropriate data normalization, since this normalization strategy with the same family of RNA molecules comprises the same methods, such as extraction, reverse transcription, and PCR. In this regard, Zhu et al. selected ten miRNAs from the TaqMan low-density array that had expression levels equal to the mean levels as potential reference controls, from which five miRNAs (miR-22*, miR-26a, miR-221, miR-16, and miR-30e) were used for further evaluation as they were detected with the same high levels in all serum samples. Then, GeNorm algorithm specified that the optimal number of genes for an accurate normalization was 3, and recommended to use the combination of miR-22*, miR-26a, and miR-221 for miRNA normalization. The high stability of this miRNA panel was also proved by NormFinder algorithm. Using a cohort of 52 HBV patients and 57 healthy controls, these scientists found that none of the three miRNAs was differentially expressed between the two groups due to their steady expression among all subjects independent from the cohort [28]. The high stability of miR-221 in patients with HBV was also proved in the study conducted by Li et al. However, in this study, miR-22* was identified as a highly unstable reference control in exosomes from the serum of HBV patients [29]. In contrast, Tang et al. identified miR-221 as one of the least stable reference genes in the plasma samples of HBV patients, and showed that miR-106a and miR-21 were highly stable in their samples [30].
Kidney disorders
Lange and coworkers analyzed miR-16, miR-21, miR-92a, and miR-124a as endogenous controls in expression studies on urinary exosomal miRNAs of patients with chronic kidney disease (CKD). They introduced miR-16 as the best candidate, since four algorithms demonstrated its high stability, as well as the mean value of miR-16 in urine samples did not significantly differ between 33 CKD patients and 5 healthy subjects. They also indicated that though miR-92a was the most stable candidate, it is not suitable for miRNA normalization because of its differential expression between the two groups [33].
Cardiovascular diseases
Mase et al. evaluated the stability of five widely used reference genes in the atrial tissue specimens from 18 patients using geNorm, NormFinder, BestKeeper, and ΔCt assays. RNU48 and RNU6 were the most and least stable reference genes, respectively. The impact of adopting different normalization strategies was demonstrated in the case of the quantification of miR-499a-5p expression in the study population. Using RNU48 as a normalizer, miR-499a-5p was significantly over-expressed in the patients with atrial fibrillation compared to the sinus rhythm patients being in line with a previous study [34], whereas normalization with RNU6 led to the loss of any subgroup expression difference [35].
Zhang et al. investigated the suitability of ten potential reference controls for plasma miRNA quantification in stable coronary artery disease (CAD). Using NormFinder and BestKeeper algorithms, they identified miR-6090 and miR-4516 as the most stable candidates in their study. They confirmed the reliability of miR-6090 and miR-4516 for miRNA normalization by indicating that their levels were not significantly different between CAD patients and healthy individuals in two independent cohorts comprising 21 and 90 pairs of plasma samples. They also proved the lack of RNU6 as a normalizer for the quantification of miRNAs in the plasma of CAD patients as RNU6 was the least stable reference gene according to the NormFinder and BestKeeper algorithms. Its use as a normalizer led to the misinterpretation of miR-21 expression. When the expression of miR-21 was normalized to the validated endogenous controls of miR-6090 and miR-4516, significant differences were detected between CAD patients and controls. However, normalization with RNU6 did not reveal these differences [36].
Wang et al. assessed the stability of 7 common normalizers in the serum of 62 cases, of which 25 were patients with heart failure and 10 were patients with hypertension. The results of BestKeeper and ΔCt algorithms showed that miR-16 and let-7i were the most stable reference genes with a low standard deviation (SD) [37].
Solayman et al. showed that miR-92a-3p, miR-16-5p, and miR-21-5p were suitable normalizers for plasma miRNA expression in hypertension studies. Using plasma samples from 18 hypertensive patients and 10 healthy controls, they found that the expression levels of none of these miRNAs were statistically different between the two groups, and these three endogenous controls were highly stable according to the geNorm and NormFinder programs [38].
Tuberculosis
Barry et al. reported that miR-93 was a suitable reference control for normalizing miRNA levels in tuberculosis (TB) patients. They showed that its levels were steadily expressed in plasma of 12 Chinese TB patients and 12 healthy controls as well as in 24 Australian TB patients and 12 healthy controls. This high stability of miR-93 in the different populations was also demonstrated by the geNorm and Normfinder algorithms [39]. The study also indicated that ethnic differences should be kept in mind when investigating reliable reference controls, because the expression of miRNAs may differ significantly between different nations.
CNS disorders
Major depressive disorder
Liu et al. analyzed 1425 miRNAs on a microarray, and selected miR-320d, miR-101-3p, miR-106a-5p, miR-423-5p, and miR-93-5p for searching a suitable plasma-based reference genes for miRNA normalization in major depressive disorder (MDD). They subsequently measured plasma expression levels of these candidates in a cohort that consisted of 16 patients and 14 controls, and found that all 5 miRNAs displayed no significant differences between the two groups. They assessed the stability of the candidates using four algorithms, and the results of the merged data revealed that miR-101-3p and miR-93-5p were the most stable miRNAs. As the optimal number of reference genes for a proper normalization was two, the combination of miR-101-3p and miR-93-5p could be used for plasma miRNA normalization in MDD. Liu et al. also evaluated the expression of RNU6 in the plasma samples of both cohorts. They found that although RNU6 was steadily expressed between the two groups, it was also one of the least stable reference controls in comparison to the validated endogenous controls of miR-101-3p and miR-93-5p. Thus, RNU6 may not be suitable for miRNA normalization. Finally, they normalized the expression levels of miR-147b and miR-30c-1-3p to the validated reference controls and demonstrated that these two miRNAs were down-regulated in the patients with MDD which was accordant with the microarray data [40].
Parkinson’s disease
Due to their literature research, Serafin et al. [41] investigated RNU24, RNU6B, and Z30 snoRNA as normalizers in 38 pairs of blood samples from patients with Parkinson’s disease (PD) and healthy controls. They identified the combination of RNU24 and Z30 snoRNA as a reliable reference panel for data normalization in their experimental cohort, as its high stability was proved by three different algorithms. Furthermore, they considered RNU6B as an inappropriate reference gene in their study because of its low PCR amplification efficiency and low stability. They also found that miR-103a-3p was the least stable reference gene in the blood samples of PD patients according to the geNorm and the comparative ΔCt algorithms. They selected miR-29a-3p and miR-30b-5p, two dysregulated miRNAs in PD [42, 43], to test the effect of using different combinations of reference genes on the relative expression values. The results showed that these two target miRNAs were up-regulated in PD patients when their levels were normalized to the stable pair of endogenous controls of RNU24 and Z30 snoRNA, while the target miRNAs were down-regulated using miR-103a-3p as a normalizer.
Cancers
In particular, there is a wealth of literature on miRNA quantification in different cancer types with different normalization strategies, resulting in frequently non-comparable miRNA data in the same tumor type and stage.
Colorectal cancer
To identify the most stably expressed miRNAs in their study, Eriksen et al. profiled a panel of more than 750 miRNAs on 10 pairs of rectal cancer and adjacent tissues, and applied a global mean expression normalization strategy. MiR-645, miR-193a-5p, miR-27a, and let-7g were selected as candidate reference controls. The high stability of these miRNAs was further proved using the NormFinder algorithm. Subsequent real-time PCR-based validation of these candidates was performed in two serial steps with 25 and 28 pairs of samples, and showed that the levels of miR-193a-5p, miR-27a, and let-7g were not significantly different between the rectal cancer and adjacent tissues in both experimental steps, allowing the combination of them to be as a reliable normalizer for miRNA expression analyses in rectal cancer. They also assessed the suitability of RNU6B as a normalizer, since it has been frequently used in the miRNA expression studies on rectal cancer. Using NormFinder algorithm, they demonstrated that RNU6B showed a ten times lower expression steadiness in comparison to the validated combination of miR-193a-5p, miR-27a, and let-7g. Using a two-phase real-time PCR-based validation with at first 50 and then 56 samples, they found that RNU6B was differentially expressed between rectal cancer and adjacent tissue, indicating that RNU6B is not appropriate for miRNA normalization in rectal cancer [44]. In contrast, Zheng et al. suggested that RNU6 may be suitable for serum miRNA normalization in colorectal cancer as it was ranked as the second and third best normalizers in the geNorm and NormFinder algorithms, respectively. Using two separate cohorts of 45 patients plus 40 controls and of 128 patients plus 60 controls, they demonstrated that RNU6 was steadily expressed between patient and healthy groups. Furthermore, they observed no differential expression of RNU6 among the four tumor stages of colorectal cancer [45].
To identify a set of reliable miRNA references in colorectal cancer, Niu et al. established an miRNA profiling assay, and assessed 485 endogenous miRNAs. They found that miR-106b-5p, miR-25-3p, and miR-93-5p met the criteria of suitable normalizers. Measuring their expression in the serum of 30 colorectal cancer patients and 30 healthy individuals, they confirmed that the three miRNAs were steadily expressed in the diseased and healthy groups. NormFinder and geNorm algorithms proved that these miRNAs are highly stable in the cohorts analyzed. Accordingly, the use of miR-106b-5p, miR-25-3p, and miR-93-5p for data normalization led to similar results. For instance, miR-144-3p whose prognostic value was described in colorectal cancer [46] was significantly up-regulated in the patient group, when data were normalized to the three normalizers [47].
Conducting profiling of 380 miRNAs in ten pairs of colorectal tumor and normal tissues, Chang et al. [48] identified eight candidate reference genes including miR-345 as a reference gene with an expression profile closest to the mean expression value. Using real-time PCR and a larger cohort comprising tissues from 35 patients and 39 controls, they did not found any evidence for a differential expression of all candidate reference genes between tumor and normal tissues. In this regard, Ct values of miR-16 showed the least variability. Stability evaluation was further assessed using the two algorithms. NormFinder and geNorm identified miR-345 and miR-16 as the most stable reference genes. In addition, both programs determined that the combination of miR-345 and miR-16 is the most stable reference panel. Finally, Chang et al. found a significant up-regulation of miR-21 and miR-31, two well-established oncogenic miRNAs in colorectal cancer [49] when they used this panel for data normalization. However, the utility of miR-16 as a normalizer in colorectal cancer needs further evaluation, since it is well known for its deregulation in this cancer type [50].
MiR-191-5p was also identified as a reliable endogenous control for serum miRNA normalization in colorectal cancer, as it was the most stable reference gene as disclosed by the geNorm and NormFinder software. By applying two independent cohorts of 45 patients plus 40 controls and 128 patients plus 60 controls, Zheng et al. showed that the Ct values of miR-191-5p were constant between both groups and cancer stage-independent [45]. Conversely, Danese et al. reported that the expression levels of miR-191 were significantly higher in exosomes and plasma of colorectal cancer patients than in those of healthy controls [51]. In this respect, this laboratory carried out a literature research, and evaluated 12 miRNAs as internal controls, of which five miRNAs were selected for further analysis, as they showed no features of a known oncogene or tumor suppressor gene. They determined that miR-520d, miR-1228 and miR-345 were suitable endogenous controls for miRNA normalization in different matrices, as their Ct values were steady between exosome, plasma and tissue samples of 20 colorectal cancer patients and 20 healthy controls. Accordingly, these three miRNAs were also the most stable candidates in all matrices when NormFinder and BestKeeper algorithms were performed [51].
Breast cancer
McDermott et al. performed miRNA profiling on the blood samples of ten breast cancer patients and ten healthy controls. Ten miRNAs with expression profiles closest to their mean value were identified as potential endogenous controls. Among them, miR-425 was selected for a further analysis as the other miRNAs were reported by the previous articles to be deregulated in breast cancer. They also evaluated miR-16, miR-142-3p and miR-484, because they were frequently used as normalizers in the literature. GeNorm algorithm determined that miR-425 and miR-16 were the most stable reference genes among the candidates. The expression of miR-425 and miR-16 was further verified in a larger cohort of 40 breast cancer patients and 20 healthy women. The results confirmed the steady expression of miR-16 and miR-425 in both groups [52]. Likely, using geNorm and NormFinder programs, Davoren et al. also identified miR-16 as one of the most stable reference controls in their experimental cohort of 26 breast cancer patients with different tumor grades. They also showed that miR-16 was equivalently expressed between the malignant and benign tumor groups. They validated miR-16 in combination with let-7a as the best normalization strategy for miRNA analysis [53]. Conversely, Appaiah et al. found that miR-16 levels were significantly higher in the serum of 15 disease-free breast cancer patients’ than 15 healthy controls with or without performing data normalization. They also observed that serum levels of miR-16 were elevated in breast cancer patients with active metastasis [54]. To date, numerous studies have shown that miR-16 is a biomarker of breast cancer and, thus, lacks the features of a normalizer in this disease [55, 56].
In their study, McDermott et al. also quantified RNU6B, RNU44, and RNU48 in the same cohorts, and found that these snRNAs were the least stable reference genes as derived from their microarray data set consisting of 380 miRNAs. They showed that the aberrantly expressed RNU6B as a normalizer introduced significant analysis biases. When they normalized their data of miR-93 which is usually not dysregulated in breast cancer [57] with RNU6B, they detected that miR-93 was overexpressed in the cancer group, but was constantly expressed when they carried out the normalization with their validated endogenous controls of miR-425 and miR-16 [52]. Similarly, Lou et al. showed that RNU6 levels were significantly higher in breast carcinoma tissue than in adjacent normal tissue of 20 breast cancer patients. They also found that tissue levels of RNU6 exhibited a high inter-individual variability [58].
In their study, Appaiah et al. also quantified RNU44 in the serum of 55 healthy controls and 69 breast cancer patients, and found that the levels of RNU44 were similar between both groups. Thus, RNU44 could be used as a normalizer in normalizing not only miRNA but also 5S rRNA and RNU6 data. In addition, calculation of the average Ct values of 5S rRNA and RNU6 showed that their levels were significantly higher in the serum of 39 cancer patients than of 40 healthy subjects. A further analysis in this cohort showed that 5S rRNA levels were increased in the serum of estrogen receptor-/progesterone receptor-negative (ER−/PR−) patients, while RNU6 levels were elevated in both ER−/PR− and ER+/PR+ patients, when RNU44 was used as an internal control. The use of further cohorts of 15 healthy controls, 15 symptom-free breast cancer patients and 15 patients with overt metastasis showed that RNU6 was up-regulated in the serum of symptom-free and metastatic patients, when its levels were normalized to RNU44. These results revealed that RNU6 was significantly over-expressed in women who had breast cancer, irrespective of disease activity [54].
Finally, Gee et al. showed that snoRNAs had highly variable expression levels in breast cancer patients and were associated with their clinicopathological factors. For example, the amounts of RNU48 were negatively correlated with tumor grade, those of RNU48 and RNU43 were inversely correlated with proliferation score, and lower levels of RNU44 were an adverse prognostic factor for overall survival [59], suggesting that these snoRNAs have tumor suppressive features.
Bladder cancer
Ratert et al. identified 16 putative miRNAs as reference controls using miRNA microarray data derived from 24 tissue specimens of bladder cancer patients and healthy controls. The stability value of these putative reference genes was determined using three algorithms. GeNorm recommended a panel of four miRNAs including miR-151-5p, miR-125a-5p, miR-148b, and miR-101 for an accurate normalization, while miR-148b, miR-874, and miR-181b were the most stable reference genes in the NormFinder program. In addition, miR-874 and miR-151-5p were ranked at second and third positions by BestKeeper, respectively. The expression of these stable reference genes which was further verified in 58 normal and malignant samples sustained the findings [60]. However, since miR-148b is an established plasma biomarker of bladder cancer [61], its use as a normalizer in this disease needs further evaluation.
Ratert et al. also demonstrated that RNU6B is one of the least stable reference genes in their investigations, as it was ranked at the 10th position by geNorm, 12th position by NormFinder, and 9th position by BestKeeper. They used miR-20a, an miRNA up-regulated in malignant bladder tissues as detected in the microarray data, as a target miRNA of interest, to examine the effect of their reference control selection on the levels of this miRNA. As expected, they found that miR-20a was up-regulated in the malignant tissues using their validated endogenous controls (the combination of miR-151-5p, miR-125a-5p, miR-148b, and miR-101 or the combination of miR-148b, miR-874, and miR-181b) as normalizers, but its expression was not differentially expressed between the malignant and control groups, when RNU6B was used as a normalizer. In other words, RNU6B was unable to detect the expression changes in miR-20a levels in bladder cancer [60].
Wang et al. identified miR-193a-5p and miR-16-5p as reliable reference controls for the quantification of serum miRNAs in bladder cancer because of their steady expression in the serum of 60 patients and 35 controls. Both miRNAs were also ranked as the most stable reference genes according to the geNorm and NormFinder algorithms. Using a further cohort, these scientists supported their findings that these two internal controls were, in fact, stably expressed between both groups. Finally, they normalized the expression levels of miR-148b, an established biomarker of bladder cancer [61], to their validated panel of reference controls, and found that miR-148b is a biomarker for bladder cancer [62].
Renal cell carcinoma
To identify reliable reference controls for renal cell carcinoma (RCC), Wotschofsky et al. performed a microarray including 117 miRNAs, from which 6 miRNAs were considered to be stably expressed. The stability of these six miRNAs was evaluated using geNorm and NormFinder algorithms. MiR-28, miR-103, miR-106a, and miR-151 were selected for further analysis in 57 samples of non-malignant and malignant tissues from RCC patients. With the exception of miR-151, these miRNAs were steadily expressed between both cohorts, suggesting that their combination can be used as a normalizer [63]. They also evaluated RNU6B, RNU44, and RNU48 as internal controls for miRNA normalization in RCC because of their frequent use in the previous studies. Using the same specimen pairs, they observed significant differences in RNU6B and RNU44 levels between the non-malignant and malignant tissues. GeNorm and NormFinder algorithms showed that RNU6B and RNU44 were the least stable reference genes in the cohorts. Finally, these authors demonstrated that using RNU6B as a reference gene for the relative quantification led to erroneous results, since normalization with RNU6B resulted in miR-19b over-expression in the RCC tissues, whereas miR-19b was under-expressed in the malignant tissues when its levels were normalized to the validated endogenous controls of the combination of miR-28, miR-103, and miR-106a [63]. Likely, Iwamoto et al. confirmed these data, and found that Ct values of RNU6B were significantly lower in the serum of RCC patients than in healthy controls. However, they found constant Ct values of miR-145 in tissue and miR-16 in serum between RCC patients and healthy controls by examining their expression profiles in 34 pairs of tumor and normal tissues and 34 serum samples of RCC patients plus 23 of healthy subjects [64]. Nevertheless, Sanders et al. showed that other members rather than RNU6B and RNU44; for example, RNU43 and RNU1-4 were suitable reference genes for serum miRNA normalization in RCC [65]. Therefore, evaluation and comparison of the suitability of all members of this family would be recommendable for data normalization in a large study.
Cervical cancer
Shen et al. investigated nine candidate reference controls for miRNA normalization in cervical cancer tissues as detected by the literature research and their microarray data. Both geNorm and NormFinder algorithms determined miR-23a and miR-191 as the most stable candidates. Furthermore, miR-23a and miR-191 were equivalently expressed between 23 cervical cancer and 23 normal tissues. The Ct values of both miRNAs remained constantly in a large cohort of 108 samples that represented the full pathological spectrum of cervical cancer samples [66]. The high stability of miR-191 and miR-23a in cervical cancer tissues was also proved in a study by Leitao et al. [67]. The suitability of RNU6 was also investigated in the study conducted by Shen et al. Although RNU6 was equivalently expressed in a cohort of 23 pairs of cervical cancer and healthy tissues, it was identified as a reference gene with low stability (seventh position out of nine candidates) by the geNorm algorithm. Using miR-424 as the target miRNA of interest, the authors showed that RNU6 is not appropriate for tissue miRNA normalization in cervical cancer, since the use of RNU6 as a normalizer provided contradictory values of miR-424 levels in human papillomavirus (HPV)-negative and -positive tissues, whereas normalization to the validated endogenous controls of miR-23a and miR-191 and their microarray data indicated that miR-424 levels were significantly different between both cohorts [66]. Likely, Babion et al. demonstrated that RNU6 and RNU6B were the least stable reference genes in the geNorm, NormFinder, and BestKeeper algorithms, and their use as normalizers lacked to detect the statistically significant changes in miR-100 and miR-15b levels between the normal and malignant tissue of cervical cancer patients. In contrast, they observed that miR-423 is a reliable endogenous control for miRNA expression studies in cervical cancer as it was the most stable reference gene in the geNorm, NormFinder, and BestKeeper algorithms, and displayed steady expression levels in 20 cancerous and 10 normal tissues [68]. Finally, Hansen et al. showed that RNU6 was up-regulated 15-fold in the cervical cancer tissues compared with normal tissues, and its levels were associated with the stage of cervical cancer [69].
Hepatocellular carcinoma
Li et al. selected ten candidate reference genes from the previous research articles, and assessed their suitability for serum miRNA normalization in hepatocellular carcinoma (HCC). RefFinder identified miR-221, let-7a, and miR-26a as the most stable endogenous controls. The Ct values showed their steady expression in 33 pairs of pre- and post-operative serum samples, suggesting that their combination is appropriate for data normalization [70]. This panel of miRNAs was also appropriate for data normalization in the serum-derived exosomes of HCC patients [29]. The researchers also showed that miR-16 is not reliable for miRNA analysis in HCC as normalization with miR-16 caused the down-regulation of miR-122 in the serum of post-operative patients, whereas normalization with the combination of miR-221, let-7a, and miR-26a showed similar serum levels of miR-122 in 40 pre- and post-operative patients [70]. In addition, Li et al. found that Ct values of RNU6 and 5S rRNA significantly differed between pre- and post-operative patients, and that RNU6 and 5S rRNA were the least stable reference genes in their experimental cohort [70]. The lack of RNU6 as a normalizer in HCC was also reported in other studies. According to the geNorm and NormFinder algorithms, RNU6 was the least stable reference gene in exosomes from the serum of 50 HCC patients and 50 healthy subjects [29]. Ding et al. showed that its serum levels decreased by fivefold in HCC patients compared to healthy controls [71]. Consistent with these results, Tang et al. identified RNU6 as the least stable reference gene in the plasma samples of 30 HCC patients and 20 healthy individuals using a comprehensive gene stability assay. They recommended the combined use of miR-21 and miR-106a as an endogenous control for miRNA expression studies in HCC as their Ct values did not significantly differ between the plasma samples [30]. However, the use of miR-21, one of the most analyzed miRNAs in cancer, as a normalizer is very questionable, because miR-21 has been shown to be deregulated in most cancer types.
Prostate cancer
Schaefer et al. identified miR-130b as a reliable endogenous control for miRNA normalization in prostate cancer because of its steady expression in cancerous and normal tissues. They calculated equivalent values in 76 pairs of malignant and non-malignant samples. GeNorm and Normfinder software proved miR-130b as the most stable gene in their analyses. In addition, this laboratory showed that miR-16 is a bad normalizer in prostate cancer, as it was significantly under-expressed in the malignant tissues, and the fold changes of miR-96, miR-125b, miR-205, and miR-375 normalized to miR-16 were significantly different from those related to miR-130b. They also documented the steady expression of RNU6B between 76 pairs of malignant and normal tissues and its high stability. Thus, they approved the reliability of RNU6B for tissue miRNA normalization in prostate cancer [72]. In contrast, Carlsson et al. identified RNU6B as the least stable reference gene in their experimental cohort using NormFinder and BestKeeper software. However, they proposed RNU24 as a suitable reference gene for tissue miRNA normalization in prostate cancer as its levels were not significantly different between 19 pairs of malignant and normal tissues, and were identified as the most stable reference gene using NormFinder and BestKeeper algorithms. Conversely, RNU43 was not suitable for miRNA normalization in this type of cancer due to its low stability and differential expression between the cancerous and normal tissues [73]. Egidi et al. recommended the use of miR-191 for urine miRNA normalization in patients with prostate cancer as its Ct values were constant in the urine sediments of 35 prostate cancer patients and 26 controls with benign prostatic hyperplasia. MiR-191 was also selected as the most stable reference gene by the BestKeeper algorithm [74].
Esophageal squamous cell carcinoma
Chen et al. identified RNU48 as an ideal reference gene for real-time PCR in esophageal squamous cell carcinoma due to its stable expression in 50 pairs of cancerous and normal tissues. They also found that 5S rRNA, RNU6, and RNU6B were no appropriate reference genes in this setting as their Ct values varied between esophageal normal and squamous cell carcinoma samples. Moreover, they were the least stable genes in the geNorm and NormFinder algorithms. However, Chen et al. determined miR-28-5p, miR-34a-5p, and miR-186-5p as suitable internal controls for data normalization in esophageal squamous cell carcinoma, as their Ct values were steady in both tissues and these miRNAs were highly stable according to the geNorm and NormFinder software [75].
Pancreatic cancer
Popov et al. identified U91 snoRNA as a new internal control for accurate miRNA normalization in pancreatic cancer. They showed that this snoRNA was highly stable and had the lowest standard deviation in the tissues analyzed. Furthermore, they quantified the expression of six target miRNAs including miR-21, miR-96, miR-148a, miR-155, miR-196a, and miR-217 using alien spike in and U91 snoRNA as normalizers, and detected that the difference between the spike in and U91 snoRNA was statistically insignificant for all miRNAs analyzed except miR-217, suggesting a similar behavior of U91 snoRNA to the alien spike in. They also showed an overexpression of RNU6 in the pancreatic cancer tissues and significant differences of miRNA expression, when normalized to RNU6 in comparison to the artificial spike in [76], whereas RNU6B was steadily expressed in the serum exosomes of 41 pancreatic cancer patients as reported by Que et al. [77]. In addition, Popov et al. found that miR-16 was not trustworthy for miRNA normalization in pancreatic cancer, as its tissue levels varied from − 2.94 up to 7.38-fold between malignant and normal pancreatic tissues. It was also the least stable reference gene using NormFinder algorithm. This laboratory also demonstrated that miR-16 introduced bias to the miRNA quantification, since the average fold change in miR-21, miR-96, miR-148a, miR-155, and miR-196a differed significantly between the normalization with miR-16 and the artificial spike in [76]. These findings were also supported by our laboratory [78].
Neuroendocrine tumors
Sperveslage et al. analyzed nine small RNAs with regard to their applicability as reference controls for tissue miRNA normalization in ileal neuroendocrine tumors (NETs), and revealed that RNU61 and RNU95 were the most suitable candidates as they were the most stable RNAs according to the comprehensive gene ranking and not differentially expressed between ten pairs of primary tumors and metastases. Furthermore, they indicated that RNU95 was also a reliable reference control for miRNA quantification in pancreatic NETs [79].
Gastric cancer
Song et al. selected six miRNAs as potential endogenous controls by literature research. The stability value of the six miRNAs was determined using geNorm and NormFinder programs. Both programs identified miR-16 and miR-93 as the most stable reference genes for data normalization. The expression of both miRNAs was measured in a cohort of serum samples from 40 gastric cancer patients and 20 healthy controls. Neither miR-16 nor miR-93 was differentially expressed between the both cohorts, suggesting them as appropriate normalizers in gastric cancer [80].
Endometrioid endometrial carcinoma
Torres et al. showed that the combination of RNU48, RNU44, and SNORD75 may be used for miRNA normalization in endometrioid endometrial carcinoma (EEC) using real-time PCR, as they were the most stable reference panel in NormFinder and geNorm algorithms, and no significant difference was observed in their mean Ct values in 45 fresh tissue samples, of which 30 samples were obtained from EEC patients and 15 samples were obtained from healthy controls. However, the reliability of RNU48 and SNORD75 as normalizers needs to be further verified as the Ct values of these two snoRNAs were statistically different between the formalin-fixed paraffin-embedded (FFPE) tissue specimens of 44 EEC patients and 14 healthy controls. They also showed that miR-92a was not reliable for tissue miRNA normalization in EEC as it was differentially expressed with a high inter-group variation between healthy controls and EEC patients. Normalization with miR-92a led to the down-regulation of miR-9, miR-141, miR-183, miR-200a, and miR-200c in the ECC tissue samples, while these miRNAs were up-regulated in ECC when the data were normalized to the validated reference controls of RNU48, RNU44, and SNORD75. Furthermore, the study showed that miR-26b was also not suitable as a normalizer in real-time PCR due to its low stability in geNorm and NormFinder algorithms and because of its high inter-group variation between the malignant and normal tissues [81].
Ovarian cancer
Bignotti et al. reported that let-7a-5p and miR-191-5p were among the most stable internal controls in a cohort of 105 tissue samples, of which 75 samples were obtained from high-grade serous ovarian carcinoma patients and 30 samples were obtained from healthy individuals. They also revealed that the expression of let-7a-5p and miR-191-5p were not significantly different between the two groups. This interesting study was also accounted for the presence of potential outliers, by fitting weighted least squares with weights computed by M-estimation [82]. To test non-difference of small non-coding RNA expression among groups, Bignotti et al. additionally used the two one-sided test (TOST) approach, a type of intersection union test [83]. By adopting the strict equivalence range of ± 0.36 and using TOST, miR-191-5p emerged as the most equivalent candidate in the tumor sample cohort. Furthermore, they demonstrated that miR-16-5p and miR-92a-3p were unsuitable normalizers in ovarian cancer as they were the least stable reference genes in geNorm and NormFinder algorithms, and their expression statistically differed between the cancerous and normal groups. Similar negative results were obtained for SNORD72. On the other hand, RNU48 was identified as the best reference gene for data normalization in this study, since its levels were not statistically different between 75 malignant and 30 normal tissues as detected by its steady expression in both ovarian tissue types. RNU48 was also the most stable reference control in the NormFinder software. It was also shown that RNU48 was the only candidate that falls within the fixed equivalence range between malignant and non-malignant samples [84].
Assortment of different cancer types
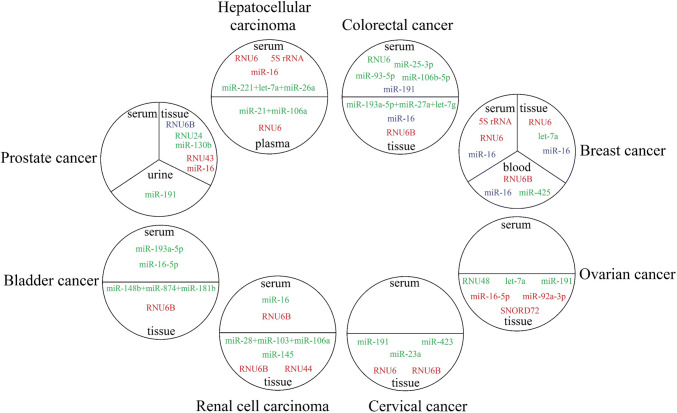
To find a stable endogenous control for quantifying circulating miRNAs in different cancer types, Hu et al. preformed microarray analysis and assessed 723 miRNAs in the plasma samples of a cohort of 80 controls (34 healthy subjects, 21 HBV patients, and 25 cirrhosis patients) and 171 cancer patients (57 HCC, 41 CRC, and 73 lung cancer). They selected seven miRNAs based on the NormFinder findings, of which miR-1228 was the most stable candidate. Using an independent cohort of 184 plasma samples from the same population subgroups, they verified the steady expression of miR-1228 that displayed the lowest coefficient of variability (CV). Moreover, no statistical difference between early and late tumor stages in HCC, CRC, and lung cancer was found Quantification of miR-1228 levels in additional 109 patients with esophageal, gastric, renal, prostate, and breast cancers sustained the low CV value of miR-1228. These findings suggest that miR-1228 is one of the rare normalizers for miRNA quantification in several various cancer types [85]. Further studies that introduced stable reference controls in an assortment of cancer types are specified in Table 2 and Fig. 1.
Table 2.
Stable reference controls in different cancer types
| First author | Sample type | Screening phase | Validation phase | |||||
|---|---|---|---|---|---|---|---|---|
| Number of assessed miRNAs in the microarray | Cases and control samples | Candidate reference genes | Cases and control samples | Assessment of the differential expression using RT-qPCR | Method used to assess miRNAs stability | Most stably expressed miRNAs | ||
| Peltier [86] | Tissue | 287 |
13 Flash-frozen normal tissue 5 Pairs of flash-frozen cancerous and adjacent normal tissues (lymphoid, colon, prostate, lung, and esophagus) |
miR-191, miR-93, miR-106a, miR-17-5p, miR-25, miR-16, let-7a, miR-24, miR-103, miR-99a, 5S, RNU6 |
12 Pairs of flash-frozen lung cancer and adjacent normal tissues 16 Pairs of FFPE lung cancer and adjacent normal tissues |
– | geNorm, NormFinder |
Normal tissues: miR-191 Cancerous tissues: miR-103 |
| Rice [87] | Plasma | 380 |
20 CRC patients 10 Patients with breast cancer, lung cancer, pancreatic cancer 11 Patients with colorectal adenoma, 12 healthy controls |
let-7a, let-7d, let-7g, miR-16, RNU6, RNU48, miR-191, miR-223, miR-484, miR-520d-5p | – | – | The mean Ct value, SD | miR-520d-5p, RNU6 |
| Chen [88] | Serum | Not applicable |
30 NSCLC patients 20 Patients with gastric cancer, esophageal cancer, breast cancer 10 Patients with pancreatic cancer, CRC, HCC, cervical cancer 100 Healthy controls |
25 miRNAs (SBS technology) + GAPDH, β-actin, RNU6, RNU44, RNU48, miR-16, miR-191, miR-103, miR-23a |
278 Cancer patients, 200 patients with inflammatory diseases, 320 patients with type 2 diabetes 1313 Healthy controls |
✓ | geNorm, NormFinder | let-7d/g/i |
| Hu [89] | Serum | Not applicable |
60 Patients with lung cancer, 48 patients with breast cancer, 40 patients with gastric cancer, 30 patients with HCC, and cervical cancer 96 healthy controls |
miR-484, miR-320, miR-191, miR-16 | 45 Cancer patients, 5 healthy controls | ✓ | NormFinder | miR-484 + miR-191 |
| Xiang [90] | Serum | Not applicable |
20 Nasopharyngeal cancer patients 20 Healthy controls |
miR-16, miR-24, miR-142-3p, miR-19b, miR-192, RNU6 |
20 Patients with gastric cancer, nasopharyngeal cancer, CRC, breast cancer 30 Healthy controls |
✓ | The difference of ΔCt values | miR-16 |
| Inada [91] | FFPE lymph-node tissue | 71 |
41 Patients with various metastatic cancers 16 Heathy controls |
miR-103a, miR-24, let-7a, miR-152, miR-191, miR-34a, miR-92a, miR-148b, miR-16, miR-21 | – | – | geNorm, NormFinder, BestKeeper, comparative ΔCt method | miR-103a + miR-24 + let-7a |
Fig. 1.
Preliminary test of reference controls in different sources and diseases as performed by different studies. Reference genes are illustrated in green if the respective study found them to be suitable for data normalization, but they were not validated. Reference genes are illustrated in red if the study found them unsuitable. Reference genes with controversial results are displayed in blue
Healthy individuals
Lamba et al. performed miRNA profiling and evaluated the expression of 22 miRNAs in the liver specimens from 50 healthy individuals, of which 14 miRNAs were selected for stability analysis using geNorm and NormFinder programs. MiR-23b was determined as the most stable reference gene in geNorm analysis, while it ranked at the fourth position in NormFinder. In addition, miR-152 was the most stable candidate in NormFinder ranking and ranked at the second position in geNorm. Using geNorm, it was shown that two endogenous controls were sufficient for miRNA normalization, suggesting that the combination of miR-23b and miR-152 were reliable for miRNA quantification in hepatic tissue samples. Lamba et al. also investigated the reliability of RNU6 and RNU6B as reference genes for hepatic miRNA quantification. Both geNorm and NormFinder identified RNU6 as the least stable reference control in the tissue samples. Besides, RNU6B was also not among the top genes in stability. Additional analyses showed that RNU6 levels were higher in males than in females, indicating its high inter-group variability. Finally, the impact of different normalization approaches was documented by miR-150, miR-138, and miR-34a that displayed gender differences. The differences in expression levels between males and females were significant when data were normalized to the validated endogenous controls of the combination of miR-23b and miR-152, but not when RNU6 or RNU6B were used for normalization [92]. Measuring RNU6B levels in the serum of 44 healthy controls, Benz et al. also demonstrated that serum levels of RNU6B harbored a high inter-individual variability of up to eight cycles in real-time PCR [31].
Exogenous controls
To ensure that miRNA quantification is not affected by the technical variability that may be introduced at different analysis steps, synthetic, non-human spike-in miRNAs should be used. The Caenorhabditis elegans miRNA cel-miR-39 as an exogenous control is frequently used for data normalization [13, 93, 94], but cel-miR-54 [95], synthetic miRNAs Quanto EC1 and Quanto EC2 [96], as well as the simian virus gene SV40 [97] have also been used. The addition of these exogenous miRNAs to the samples before reverse transcription of RNA documents the uniform handling of the samples and RT-PCR efficiencies. This spike-in method can additionally eliminate deviations of the experimental process and, thus, provides more reliable results. However, the use of such a spike-in control does not correct for deviations in sampling or quality of the samples. Therefore, data normalization should always be carried out by a combination of an endogenous and an exogenous control miRNA, to warrant that such differences in miRNA detection may be compensated [98].
Regrettably, the purpose of an exogenous reference gene is frequently misunderstood. In this regard, Niu et al. wrongly presumed that cel-miR-54-5p may be a suitable alternative in the absence of an endogenous control, because the normalization of the relative expression of miR-27a, miR-144, and miR-223 to the exogenous cel-miR-54-5p and to the endogenous controls of miR-93-5p, miR-25-3p, and miR-106b-5p showed similar values in the serum of CRC patients and healthy controls [47]. However, exogenous controls can never replace endogenous controls. To date, numerous studies have utilized cel-miR-54 as a normalizer, e.g., Luque et al. added cel-miR-54 to the serum samples of patients with early preeclampsia to check their handling in sample-to-sample variations [99]. Likely, the expression levels of serum miRNAs of CRC patients and plasma miRNAs of pancreatic cancer were normalized to the spiked-in cel-miR-54 in the study conducted by Zhang et al. [100] and Xu et al. [101], respectively. Sohn et al. spiked synthetic cel-miR-39 in the serum of hepatocellular carcinoma samples for the normalization of their exosomal miRNA panel [102], while Appourchaux et al. added cel-miR-39 to the serum samples of patients with chronic hepatitis B and C [103]. We usually applied cel-miR-39 in our assays and found appropriate Ct values of 21 [104, 105]. Mmu-miR-295 and cel-miR-238 that are derived from Mus musculus and C. elegans, respectively, are further examples of exogenous controls [106, 107].
Conclusion
MiRNAs have become a challenging area of intense research in vitro and in vivo systems to investigate their utility as novel biomarkers of timely disease diagnosis, prognosis, and response to therapy. However, before introducing in the clinic, some obstacles have to overcome. Accuracy and efficiency of miRNA quantification depend on several technical steps and platforms, such as RNA isolation, reverse transcription, real-time-PCR using probes and/or SYBR Green, miRNA arrays, and next-generation sequencing. In addition, the input material, e.g., tissue, plasma, serum, and urine, has also to be considered, along with the age and storing of the samples. Finally, the obtained data have to be corrected by both, endogenous and exogenous reference controls. To date, several types of RNA molecules from different RNA families have been used as reference controls. However, this diversity is one of the main factors that contribute to the differences in the relative values of miRNAs among the studies. Whereas some studies showed that certain miRNAs are significantly up-regulated, other studies reported the same miRNAs to be only weakly up-regulated or even down-regulated in the same disease, population, disease stage, and source.
These controversial results in miRNA expression, summarized in Table 1, have drawn particular attention to the effect of reference controls in data normalization and highlight the urgent need for a proper reference control [13, 108, 109]. Approaching several previously published articles, we suggest that RNU6 which is frequently used as a normalizer is not eligible for data normalization in many diseases, e.g., HBV, breast cancer, cervical cancer, HCC, and different cardiovascular diseases. In addition, RNU6 does not belong to the miRNA family. Therefore, data normalized to such a RNA molecule should be critically interpreted. Instead of normalizing with a single reference gene, the combination of several endogenous reference controls as well as with an exogenous reference control should be preferred [110].
To summarize, our findings document the controversial data on a normalizer in the same biological material, but also show that not always a suitable normalizer for tissue is also suitable for plasma or serum of the same disease. Unfortunately, this suggests that each experimental study should verify the uniform expression of a reference control in relation to the specimen type and disease under investigation. Reference controls cannot be simply transferred from one study to another without their previous validation. The most accurate approach entails a selection of stably expressed reference controls by means of high-throughput technologies, such as microarray and sequencing, and subsequent validation of their expression and stability in single analyses. However, this approach is appropriate for extensive studies and is often difficult to carry out due to the substantial cost and amount of work. Therefore, multicenter projects that carry out such experiments and compare the multitude of potential reference controls in different sources of large populations are urgently needed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: the importance of miR-21 and miR-331. Br J Cancer. 2017;117(2):266. doi: 10.1038/bjc.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin H-M, Mahon KL, Spielman C, Gurney H, Mallesara G, Stockler MR, et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br J Cancer. 2017;116(8):1002. doi: 10.1038/bjc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahraman M, Röske A, Laufer T, Fehlmann T, Backes C, Kern F, et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci Rep. 2018;8(1):11584. doi: 10.1038/s41598-018-29917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L, Doecke JD, Sharples R, Villemagne VL, Fowler CJ, Rembach A, et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2015;20(10):1188. doi: 10.1038/mp.2014.127. [DOI] [PubMed] [Google Scholar]
- 7.Roser AE, Gomes LC, Schünemann J, Maass F, Lingor P. Circulating miRNAs as diagnostic biomarkers for Parkinson’s disease. Front Neurosci. 2018;12:625. doi: 10.3389/fnins.2018.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou S-S, Jin J-P, Wang J-Q, Zhang Z-G, Freedman JH, Zheng Y, et al. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin. 2018;39(7):1073. doi: 10.1038/aps.2018.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Liu J, Chen J, Wang H, Yang L, Chen F, et al. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat Commun. 2018;9(1):1614. doi: 10.1038/s41467-018-03537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55(4):623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 11.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nat Protoc. 2008;3(6):1101. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 12.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong TH, Park IY. MicroRNA expression profiling of diagnostic needle aspirates from surgical pancreatic cancer specimens. Ann Surg Treat Res. 2014;87(6):290–297. doi: 10.4174/astr.2014.87.6.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 16.Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V. Challenges in using circulating miRNAs as cancer biomarkers. BioMed Res Int. 2015;2015:731479. doi: 10.1155/2015/731479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzenbach H. Methods for quantification and characterization of microRNAs in cell-free plasma/serum, normal exosomes and tumor-derived exosomes. Transl Cancer Res. 2017;7(2):S253–S263. [Google Scholar]
- 18.Ibberson D, Benes V, Muckenthaler MU, Castoldi M. RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol. 2009;9(1):102. doi: 10.1186/1472-6750-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, et al. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11(9):1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D-J, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JY, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn. 2012;14(1):71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50(4):244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One. 2014;9(2):e89565. doi: 10.1371/journal.pone.0089565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marabita F, de Candia P, Torri A, Tegner J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2015;17(2):204–212. doi: 10.1093/bib/bbv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/S0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 26.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8(3):209. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 27.Qi R, Weiland M, Gao XH, Zhou L, Mi QS. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55(5):1640–1642. doi: 10.1002/hep.25558. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H-T, Dong Q-Z, Wang G, Zhou H-J, Ren N, Jia H-L, et al. Identification of suitable reference genes for qRT-PCR analysis of circulating microRNAs in hepatitis B virus-infected patients. Mol Biotechnol. 2012;50(1):49–56. doi: 10.1007/s12033-011-9414-6. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Zhang L, Liu F, Xiang G, Jiang D, Pu X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis Markers. 2015 doi: 10.1155/2015/893594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang G, Shen X, Lv K, Wu Y, Bi J, Shen Q. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med Sci Monit. 2015;21:617. doi: 10.12659/MSM.891028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benz F, Roderburg C, Cardenas DV, Vucur M, Gautheron J, Koch A, et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med. 2013;45(9):e42. doi: 10.1038/emm.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madadi S, Soleimani M. The crucial need of internal control validation in the normalization of circulating microRNAs. Dig Liver Dis. 2019;51:610–611. doi: 10.1016/j.dld.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Lange T, Stracke S, Rettig R, Lendeckel U, Kuhn J, Schlüter R, et al. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS One. 2017;12(8):e0183435. doi: 10.1371/journal.pone.0183435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling T-Y, Wang X-L, Chai Q, Lau T-W, Koestler CM, Park SJ, et al. Regulation of the SK3 channel by microRNA-499—potential role in atrial fibrillation. Heart Rhythm. 2013;10(7):1001–1009. doi: 10.1016/j.hrthm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masè M, Grasso M, Avogaro L, D’Amato E, Tessarolo F, Graffigna A, et al. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep. 2017;7:41127. doi: 10.1038/srep41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Tang W, Peng L, Tang J, Yuan Z. Identification and validation of microRNAs as endogenous controls for quantitative polymerase chain reaction in plasma for stable coronary artery disease. Cardiol J. 2016;23(6):694–703. doi: 10.5603/CJ.2016.0109. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Zhang X, Yuan J, Wu J, Deng X, Peng J, et al. Evaluation of the performance of serum miRNAs as normalizers in microRNA studies focused on cardiovascular disease. J Thorac Dis. 2018;10(5):2599–2607. doi: 10.21037/jtd.2018.04.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solayman MHM, Langaee T, Patel A, El-Wakeel L, El-Hamamsy M, Badary O, et al. Identification of suitable endogenous normalizers for qRT-PCR analysis of plasma microRNA expression in essential hypertension. Mol Biotechnol. 2016;58(3):179–187. doi: 10.1007/s12033-015-9912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barry SE, Chan B, Ellis M, Yang Y, Plit ML, Guan G, et al. Identification of miR-93 as a suitable miR for normalizing miRNA in plasma of tuberculosis patients. J Cell Mol Med. 2015;19(7):1606–1613. doi: 10.1111/jcmm.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Zhang L, Cheng K, Wang X, Ren G, Xie P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J Affect Disord. 2014;163:133–139. doi: 10.1016/j.jad.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Serafin A, Foco L, Blankenburg H, Picard A, Zanigni S, Zanon A, et al. Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Res Notes. 2014;7(1):715. doi: 10.1186/1756-0500-7-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsońs disease. J Biotechnol. 2011;152(3):96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 43.Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, et al. Convergence of miRNA expression profiling, α-synuclein interacton, and GWAS in Parkinson’s disease. PLoS One. 2011;6(10):e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eriksen AHM, Andersen RF, Pallisgaard N, Sørensen FB, Jakobsen A, Hansen TF. MicroRNA expression profiling to identify and validate reference genes for the relative quantification of microRNA in rectal cancer. PLoS One. 2016;11(3):e0150593. doi: 10.1371/journal.pone.0150593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;8(12):e83025. doi: 10.1371/journal.pone.0083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 47.Niu Y, Wu Y, Huang J, Li Q, Kang K, Qu J, et al. Identification of reference genes for circulating microRNA analysis in colorectal cancer. Sci Rep. 2016;6:35611. doi: 10.1038/srep35611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10(1):173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bandrés E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5(1):29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madadi S, Soleimani M. Comparison of miR-16 and cel-miR-39 as reference controls for serum miRNA normalization in colorectal cancer. J Cell Biochem. 2019;120:4802–4803. doi: 10.1002/jcb.28174. [DOI] [PubMed] [Google Scholar]
- 51.Danese E, Minicozzi A, Benati M, Paviati E, Lima-Oliveira G, Gusella M, et al. Reference miRNAs for colorectal cancer: analysis and verification of current data. Sci Rep. 2017;7(1):8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDermott AM, Kerin MJ, Miller N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One. 2013;8(12):e83718. doi: 10.1371/journal.pone.0083718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol. 2008;9(1):76. doi: 10.1186/1471-2199-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Appaiah HN, Goswami CP, Mina LA, Badve S, Sledge GW, Liu Y, et al. Persistent upregulation of U6: SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13(5):R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stückrath I, Rack B, Janni W, Jäger B, Pantel K, Schwarzenbach H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6(15):13387. doi: 10.18632/oncotarget.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madadi S, Soleimani M. Evaluation of miR-16 as an internal control in the patients with breast cancer. Hum Pathol. 2019;85:329. doi: 10.1016/j.humpath.2018.10.034. [DOI] [PubMed] [Google Scholar]
- 57.McDermott AM, Miller N, Wall D, Martyn LM, Ball G, Sweeney KJ, et al. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS One. 2014;9(1):e87032. doi: 10.1371/journal.pone.0087032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lou G, Ma N, Xu Y, Jiang L, Yang J, Wang C, et al. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int J Mol Med. 2015;36(5):1400–1408. doi: 10.3892/ijmm.2015.2338. [DOI] [PubMed] [Google Scholar]
- 59.Gee H, Buffa F, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104(7):1168. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ratert N, Meyer H-A, Jung M, Mollenkopf H-J, Wagner I, Miller K, et al. Reference miRNAs for miRNAome analysis of urothelial carcinomas. PLoS One. 2012;7(6):e39309. doi: 10.1371/journal.pone.0039309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adam L, Wszolek MF, Liu C-G, Jing W, Diao L, Zien A, et al., editors. Plasma microRNA profiles for bladder cancer detection. Urologic Oncology: Seminars and Original Investigations. Amsterdam: Elsevier; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Liu Y, Du L, Li J, Jiang X, Zheng G, et al. Identification and validation of reference genes for the detection of serum microRNAs by reverse transcription-quantitative polymerase chain reaction in patients with bladder cancer. Mol Med Rep. 2015;12(1):615–622. doi: 10.3892/mmr.2015.3428. [DOI] [PubMed] [Google Scholar]
- 63.Wotschofsky Z, Meyer H-A, Jung M, Fendler A, Wagner I, Stephan C, et al. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal Biochem. 2011;417(2):233–241. doi: 10.1016/j.ab.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Iwamoto H, Kanda Y, Sejima T, Osaki M, Okada F, Takenaka A. Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int J Oncol. 2014;44(1):53–58. doi: 10.3892/ijo.2013.2169. [DOI] [PubMed] [Google Scholar]
- 65.Sanders I, Holdenrieder S, Walgenbach-Brünagel G, von Ruecker A, Kristiansen G, Müller SC, et al. Evaluation of reference genes for the analysis of serum miRNA in patients with prostate cancer, bladder cancer and renal cell carcinoma. Int J Urol. 2012;19(11):1017–1025. doi: 10.1111/j.1442-2042.2012.03082.x. [DOI] [PubMed] [Google Scholar]
- 66.Shen Y, Li Y, Ye F, Wang F, Wan X, Lu W, et al. Identification of miR-23a as a novel microRNA normalizer for relative quantification in human uterine cervical tissues. Exp Mol Med. 2011;43(6):358. doi: 10.3858/emm.2011.43.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leitao MCG, Coimbra EC, de Lima RCP, de Lima Guimaraes M, de Andrade Heraclio S, Neto JCS, et al. Quantifying mRNA and microRNA with qPCR in cervical carcinogenesis: a validation of reference genes to ensure accurate data. PLoS One. 2014;9(11):e111021. doi: 10.1371/journal.pone.0111021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babion I, Snoek BC, van de Wiel MA, Wilting SM, Steenbergen RD. A strategy to find suitable reference genes for miRNA quantitative PCR analysis and its application to cervical specimens. J Mol Diagn. 2017;19(5):625–637. doi: 10.1016/j.jmoldx.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Hansen CN, Ketabi Z, Rosenstierne MW, Palle C, Boesen HC, Norrild B. Expression of CPEB, GAPDH and U6snRNA in cervical and ovarian tissue during cancer development. APMIS. 2009;117(1):53–59. doi: 10.1111/j.1600-0463.2008.00015.x. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Xiang GM, Liu LL, Liu C, Liu F, Jiang DN, et al. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol Med Rep. 2015;12(3):4683–4691. doi: 10.3892/mmr.2015.3919. [DOI] [PubMed] [Google Scholar]
- 71.Ding X, Ding J, Ning J, Yi F, Chen J, Zhao D, et al. Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Rep. 2012;5(6):1428–1432. doi: 10.3892/mmr.2012.838. [DOI] [PubMed] [Google Scholar]
- 72.Schaefer A, Jung M, Miller K, Lein M, Kristiansen G, Erbersdobler A, et al. Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med. 2010;42(11):749. doi: 10.3858/emm.2010.42.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carlsson J, Helenius G, Karlsson M, Lubovac Z, Andrén O, Olsson B, et al. Validation of suitable endogenous control genes for expression studies of miRNA in prostate cancer tissues. Cancer Genet Cytogenet. 2010;202(2):71–75. doi: 10.1016/j.cancergencyto.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 74.Egidi MG, Cochetti G, Guelfi G, Zampini D, Diverio S, Poli G, et al. Stability assessment of candidate reference genes in urine sediment of prostate cancer patients for miRNA applications. Dis Markers. 2015 doi: 10.1155/2015/973597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L, Jin Y, Wang L, Sun F, Yang X, Shi M, et al. Identification of reference genes and miRNAs for qRT-PCR in human esophageal squamous cell carcinoma. Med Oncol. 2017;34(1):2. doi: 10.1007/s12032-016-0860-7. [DOI] [PubMed] [Google Scholar]
- 76.Popov A, Szabo A, Mandys V. Small nucleolar RNA U91 is a new internal control for accurate microRNAs quantification in pancreatic cancer. BMC Cancer. 2015;15(1):774. doi: 10.1186/s12885-015-1785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11(1):219. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Madadi S, Soleimani M. Plasma microRNA investigation: the impact of selecting a suitable internal control on data normalization in pancreatic cancer. J Hepato-Biliary-Pancreatic Sci. 2019;26(2):E1. doi: 10.1002/jhbp.599. [DOI] [PubMed] [Google Scholar]
- 79.Sperveslage J, Hoffmeister M, Henopp T, Klöppel G, Sipos B. Establishment of robust controls for the normalization of miRNA expression in neuroendocrine tumors of the ileum and pancreas. Endocrine. 2014;46(2):226–230. doi: 10.1007/s12020-014-0202-5. [DOI] [PubMed] [Google Scholar]
- 80.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57(4):897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 81.Torres A, Torres K, Wdowiak P, Paszkowski T, Maciejewski R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol Oncol. 2013;130(3):588–594. doi: 10.1016/j.ygyno.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 82.Huber PJ. Robust statistics. Berlin: Springer; 2011. [Google Scholar]
- 83.Schuirmann D, editor. On hypothesis-testing to determine if the mean of a normal-distribution is contained in a known interval. Biometrics. Washington DC: International Biometric Society; 1981. [Google Scholar]
- 84.Bignotti E, Calza S, Tassi RA, Zanotti L, Bandiera E, Sartori E, et al. Identification of stably expressed reference small non-coding RNA s for micro RNA quantification in high-grade serous ovarian carcinoma tissues. J Cell Mol Med. 2016;20(12):2341–2348. doi: 10.1111/jcmm.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135(5):1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- 86.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14(5):844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rice J, Roberts H, Rai SN, Galandiuk S. Housekeeping genes for studies of plasma microRNA: a need for more precise standardization. Surgery. 2015;158(5):1345–1351. doi: 10.1016/j.surg.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 88.Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, et al. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8(11):e79652. doi: 10.1371/journal.pone.0079652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu Z, Dong J, Wang L-E, Ma H, Liu J, Zhao Y, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33(4):828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 90.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210–214. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 91.Inada K, Okoshi Y, Cho-Isoda Y, Ishiguro S, Suzuki H, Oki A, et al. Endogenous reference RNAs for microRNA quantitation in formalin-fixed, paraffin-embedded lymph node tissue. Sci Rep. 2018;8(1):5918. doi: 10.1038/s41598-018-24338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lamba V, Ghodke-Puranik Y, Guan W, Lamba JK. Identification of suitable reference genes for hepatic microRNA quantitation. BMC Res Notes. 2014;7(1):129. doi: 10.1186/1756-0500-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8(2):61–81. doi: 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 94.Stevic I, Müller V, Weber K, Fasching PA, Karn T, Marmé F, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018;16(1):179. doi: 10.1186/s12916-018-1163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuhlmann JD, Baraniskin A, Hahn SA, Mosel F, Bredemeier M, Wimberger P, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin Chem. 2014;60(1):206–213. doi: 10.1373/clinchem.2013.213066. [DOI] [PubMed] [Google Scholar]
- 96.Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H, et al. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One. 2014;9(10):e109347. doi: 10.1371/journal.pone.0109347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anadol E, Schierwagen R, Elfimova N, Tack K, Schwarze-Zander C, Eischeid H, et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology. 2015;61(1):46–55. doi: 10.1002/hep.27369. [DOI] [PubMed] [Google Scholar]
- 98.Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 99.Luque A, Farwati A, Crovetto F, Crispi F, Figueras F, Gratacós E, et al. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci Rep. 2014;4:4882. doi: 10.1038/srep04882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Raju GS, Chang DW, Lin SH, Chen Z, Wu X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer. 2018;124(4):785–796. doi: 10.1002/cncr.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu Y-F, Hannafon BN, Zhao YD, Postier RG, Ding W-Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8(44):77028. doi: 10.18632/oncotarget.20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sohn W, Kim J, Kang SH, Yang SR, Cho J-Y, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47(9):e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Appourchaux K, Dokmak S, Resche-Rigon M, Treton X, Lapalus M, Gattolliat C-H, et al. MicroRNA-based diagnostic tools for advanced fibrosis and cirrhosis in patients with chronic hepatitis B and C. Sci Rep. 2016;6:34935. doi: 10.1038/srep34935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meng X, Joosse SA, Müller V, Trillsch F, Milde-Langosch K, Mahner S, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358. doi: 10.1038/bjc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 107.Sode J, Krintel SB, Carlsen AL, Hetland ML, Johansen JS, Hørslev-Petersen K, et al. Plasma microRNA profiles in patients with early rheumatoid arthritis responding to adalimumab plus methotrexate vs methotrexate alone: a placebo-controlled clinical trial. J Rheumatol. 2018;45(1):53–61. doi: 10.3899/jrheum.170266. [DOI] [PubMed] [Google Scholar]
- 108.Madadi S, Soleimani M. Study of serum microRNA expression in an amyotrophic lateral sclerosis patient: challenge of selecting suitable internal control for normalization. Muscle Nerve. 2019;59:E2–E3. doi: 10.1002/mus.26370. [DOI] [PubMed] [Google Scholar]
- 109.Madadi S, Soleimani M. U6 as a microRNA normalizer in serum of patients with hepatocellular carcinoma. Ann Clin Biochem. 2018;2018:4563218808820. doi: 10.1177/0004563218808820. [DOI] [PubMed] [Google Scholar]
- 110.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]