Abstract
Apical–basal polarity is an important characteristic of epithelia and Drosophila neural stem cells. The conserved Par complex, which consists of the atypical protein kinase C and the scaffold proteins Baz and Par6, is a key player in the establishment of apical–basal cell polarity. Membrane recruitment of Baz has been reported to be accomplished by several mechanisms, which might function in redundancy, to ensure the correct localization of the complex. However, none of the described interactions was sufficient to displace the protein from the apical junctions. Here, we dissected the role of the oligomerization domain and the lipid-binding motif of Baz in vivo in the Drosophila embryo. We found that these domains function in redundancy to ensure the apical junctional localization of Baz: inactivation of only one domain is not sufficient to disrupt the function of Baz during apical–basal polarization of epithelial cells and neural stem cells. In contrast, mutation of both domains results in a strongly impaired protein stability and a phenotype characterized by embryonic lethality and an impaired apical–basal polarity in the embryonic epithelium and neural stem cells, resembling a baz-loss of function allele. Strikingly, the binding of Baz to the transmembrane proteins E-Cadherin, Echinoid, and Starry Night was not affected in this mutant protein. Our findings reveal a redundant function of the oligomerization and the lipid-binding domain, which is required for protein stability, correct subcellular localization, and apical–basal cell polarization.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2792-1) contains supplementary material, which is available to authorized users.
Keywords: Cell polarity, Par3, Adherens junctions, Drosophila, Lipid binding
Introduction
Apical–basal polarity is a hallmark of epithelial tissues and is essential during development and tissue homeostasis. In monolayered epithelial cells, the apical plasma membrane domain faces towards the outer environment or a lumen and the basal domain contacts the basement membrane. This polarity is achieved by the distinct position of conserved protein complexes along the apical–basal axis: The Crumbs complex (consisting of the transmembrane protein Crumbs and the adaptor proteins PATJ and Stardust) and the Par complex determine the apical domain identity, whereas the Scribble/Lethal giant larvae/Discs large complexes counterbalance their activity at the baso-lateral domain [1, 2]. Both apical junctional complexes, the Crumbs and the Par complex, overlap in the so-called subapical region and at the adherens junctions (AJs) [3–6].
Bazooka (Baz), the Drosophila homolog of C. elegans and vertebrate Par3, is a scaffold protein and forms together with Par6 and aPKC, the Par complex [7–9]. Moreover, binding of the small GTPase CdcC42 to Par6 is essential for the apical accumulation of Baz and the formation of the AJs in the Drosophila epidermis [10]. In C. elegans, Par6 and aPKC shuttle between Par3 and Cdc42 to form distinct complexes, where aPKC is inactive in the Par3 complex, but becomes active in complex with Cdc42 to polarize the embryo [11, 12]. In line with these observations, Baz localizes subjacent to Par6 and aPKC at the AJs in Drosophila and binding of Cdc42 to Par6 promotes the segregation of Par6 and aPKC towards the sub apical membrane in photoreceptor cells [3, 13–17]. Par3/Baz acts as an apical cue to establish the AJ by positioning Drosophila E-Cadherin (DE-Cad) and mediates the formation of the tight junctions in cultured mammalian cells [18–21]. Furthermore, Par3 acts as an exocyst receptor to regulate the delivery of membrane proteins [22, 23].
In addition, Baz is required to establish apical–basal polarity and correct spindle orientation of Drosophila neural stem cells (neuroblasts, NBs), which is essential for their asymmetric cell division [24–26]. NBs originate from the embryonic neuroectoderm and initially inherit their apical–basal polarization. During their asymmetric cell divisions, the Par complex localizes to the apical cortex of the NB, which maintains its stem cell identity after asymmetric division, whereas proteins of the basal domain are segregated into the second daughter cell, the ganglion mother cell, which further differentiates into two neurons or glia cells [27–29].
In epithelial cells, the kinase Par1 inhibits the formation of the Par complex in the baso-lateral region by phosphorylating Baz at two conserved residues (Ser151 and Ser1085). Binding of 14-3-3 proteins to these phosphorylated residues prevents the oligomerization and association with aPKC [30, 31]. In NBs, the protein phosphatase PP2A counteracts the Par1 phosphorylation and thereby promotes the maturation of the Par complex [32]. Vice versa, at least in mammalian cells, Par1 is excluded at the apical domain by aPKC-mediated phosphorylation [33]. Furthermore, aPKC phosphorylates and inhibits Lgl at the apical cortex of NBs, which leads to a release of Par6 and aPKC from Lgl to promote the formation of the mature Par complex and the asymmetric localization of Numb and Miranda to the basal region [34, 35]. The mutual antagonism of basal and apical protein complexes maintains a border between both regions in epithelia and neural stem cells.
Within the Par complex, Par6 activates aPKC by replacing its pseudosubstrate domain [36], whereas the aPKC-binding region of Baz inhibits aPKC kinase activity [37]. The phosphorylation of Ser980 of Baz by aPKC leads to the dissociation of Baz/PAR-3 and aPKC, whereupon Crumbs outcompetes phosphorylated Baz for binding to aPKC [3, 16, 17, 38]. Furthermore, Baz recruits the Crumbs adaptor Stardust during the early embryogenesis, which is released upon aPKC-mediated phosphorylation of Baz [4, 5]. Thus, Baz/Par3 functions as an important polarity cue, recruiting the Par complex to the apical junctions.
In the Drosophila epithelium, the small GTPase Rap1 and Canoe are essential to regulate the apical positioning of Baz during cellularization [39]. Vice versa, Baz and aPKC also contribute to the localization of Canoe [39]. Notably, how exactly Baz/Par3 itself localizes to the plasma membrane is still not fully clarified: Baz/PAR-3 contains an oligomerization domain (OD) at its N-terminus [40–44] and three PDZ (PSD-95, Disc Large, ZO-1) domains, which interact with the cell adhesion molecule Echinoid (Ed) and Armadillo (Arm), the Drosophila homologue of β-catenin, which in turn stabilizes DE-Cad and thereby localizes Baz to the AJs [45]. The PDZ domains of Baz/Par3 have been suggested to directly bind to phospholipids of the plasma membrane [46, 47] and we have demonstrated previously, that a C-terminal phosphoinositide lipid-binding (LB) domain of Baz directly binds to PtdIns(4,5)P2 (PIP2) and PtdIns(3,4,5)P3 (PIP3) to tether Baz to the cell cortex [48], which was confirmed independently in Drosophila and for mammalian Par3, too [49, 50]. Furthermore, the OD and the PDZ domains have been described to redundantly contribute to the apical localization of Baz in an overexpression system [51]. However, deletion of none of these domains in Baz/Par3 on its own or in combination disrupts the localization of the protein [4, 47, 48, 50, 51].
In this study, we report that impaired oligomerization of Baz enhances degradation of the protein, but affects only mildly the rescue capacity of the mutant protein. We confirm that the OD and LB motifs mediate the correct localization of the protein in redundancy. Consequently, loss of both domains results in Baz degradation, which leads to the disruption of apical–basal cell polarity in epithelial cells of the embryonic epidermis and embryonic NBs and consequently embryonic lethality.
Materials and methods
DNA and constructs
Cloning of Baz pENTR was described before [32]. For expression plasmids, we recloned Baz pENTR variants into UGW, UWS, and PWG vectors (modified from UGW, UWG, and PWG, which were obtained from the Drosophila Genomic Resource Center as described before [52]) using the gateway technology (Life Technologies). The following primers were used to introduce the mutants in Baz pENTR:
Baz1-968: 5′-ACAAACTCGGGCTGAGGATCCGGAGGTCACGCCTCCAAGGTG-3′
BazV14D-F: 5′-GGCGACGTTCGCATTCTG GAT CCCTGTGGTTCCGGC-3′
BazD68K-F: 5′-GTCCGCGACGTGGCC AAA GATCGGGAGCAGATATTG-3′
BazK1173K1174A-F: 5′-AAGTCGTCGCGGGCCGCGGCGCCAAGCATACTGCGC-3′
Fly stocks and genetics
In all experiments, we used the baz815-8 allele, which is a null allele. baz germline clones were generated with baz815-8 FRT19A using the dominant female sterile technique [53]. Homozygous mutant embryos were identified by loss of mCherry signal (from FM7-sqh::mCherry) in Western Blots. For immunofluorescence of germline clones, male embryos were selected by the absence of Sxl staining.
Ubi::GFP-Baz, Ubi::Baz-One-Strep, and UASp::Baz-GFP transgenes were generated using phiC31-mediated germline transformation; and attP40 was used as landing site [54]. For overexpression of Baz during the early embryogenesis, we used mat-tub::GAL4 (#6356) (obtained from the Bloomington Drosophila Stock Center).
Lethality test
To test the lethality of embryos, 100 embryos of each genotype were tested in three biological replicates. Embryos derived from germline clones were selected against mCherry. The embryos were kept at 25 °C on apple juice agarose plates; and the amount of dead embryos, larval stage 1/2 (L1/2), larval stage 3 (L3), pupae, and survivors was counted.
Cuticle preparation
Cuticle preparations were done as described previously [55]. The cuticle phenotypes were classified into the four categories: wild type, shrunken with holes, holes, and cuticle rest.
Real-time PCR analysis
Embryos from overnight apple juice agar plates were used to isolate total RNA with TRIzol (Life Technologies) according to the manufacturer’s instructions. To convert the RNA into cDNA, 1 µg total RNA was used for reverse transcription with the qScript cDNA Synthesis Kit (Quantabio). Real-time PCR was performed using the SensiFAST™ SYBR No-ROX Kit (Bioline) and the LightCycler 480 II (Roche). Relative expression levels of genes of interest were calculated as ΔCt values normalized to the rp49 control. The following primers were used: Baz qPCR F 5′-GTCCGTTTGTGACGCAGGTG-3′, Baz qPCR R 5′-GGTCGGCGCGCCCACCCTTC-3′, rp49 F 5′-GCGGGTGCGCTTGTTCGATCC-3′, rp49 R 5′-CCAAGGACTTCATCCGCCACC-3′.
Cell culture
Drosophila S2R cells were kept at 25 °C in Drosophila Schneider medium supplemented with 10% FCS and 1% penicillin and streptomycin and passaged every 3–4 days.
Cells were transfected with FuGene HD (Promega) according to the manufacturer’s instructions and allowed to grow for additional 3–4 days after transfection.
Antibody production
To produce sera against the N-terminus of Baz, a rabbit and a guinea pig were immunized with the recombinant GST-Baz1-318 (Eurogentec Inc.).
Immunofluorescence
Embryos were fixed in 4% formaldehyde, phosphate buffer pH 7.4 as previously described [32]. The following primary antibodies were used for immunofluorescence: chicken anti GFP (1:2.000, Aves Labs Inc., #GFP-1020), mouse anti Dlg (1:25, DSHB, #4F3), rat anti DE-Cad (1:5, DSHB, #DCAD2), rabbit anti Baz (1:1.000, this study), Gp anti Baz (1:500, this study), rat anti Mir (1:1.000, gift from A. Wodarz), rabbit anti aPKC (1:500, Santa Cruz, #sc-216), and mouse anti Sxl (1:25, DSHB, #M114). Secondary antibodies conjugated with Alexa 488, Alexa 568, and Alexa 647 (Life Technologies) were used at 1:400. Images were taken with a Zeiss LSM710 and processed and analyzed with FIJI [56]. To quantify the co-localization of Baz with aPKC or DE-Cad, the Coloc 2 plugin was used. Epithelia of three representative embryos per genotype were analyzed and the Pearson correlation coefficient is shown.
Embryonic lysates, immunoprecipitation, and western blotting
For embryonic lysates, embryos from overnight apple juice agar plates were collected and dechorionated in 50% bleach. The embryos were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 50 mM Tris–HCl, pH 7.5) supplemented with protease inhibitors. After incubation for 20 min at 4 °C, the lysates were centrifuged and SDS sample buffer was added before boiling at 95 °C for 5 min.
For immunoprecipitation, transfected S2R+ cells were lysed in lysis buffer supplemented with protease inhibitors. After centrifugation, cell lysates were added to StrepTactin beads for precipitation of One-Strep tagged Baz proteins for 45 min at 4 °C. The beads were washed three times in lysis buffer and 2× SDS sample buffer was added before boiling at 95 °C for 5 min followed by western blotting. Western blotting was performed according to the standard protocols. The following primary antibodies were used in this study: mouse anti GFP (1:500, Santa Cruz #sc-9996), rabbit anti GST (1:5000, Sigma #G7781), mouse anti c-Myc (1:100, DSHB, #9E10), mouse anti actin (1:1000, Santa Cruz #sc-47778), and rabbit anti Baz (1:1000, a gift from A. Wodarz). Quantification of Western blots was done with FIJI [56] of three biological replicates. For Baz blots, the whole lane was quantified and normalized towards actin.
In vitro crosslinking
The first 81 amino acids of wt or BazV14D D68K were fused to GST and purified from E. coli (strain BL21*) using glutathione beads (Macherey-Nagel). For in vitro crosslinking experiments, 5 µM recombinant protein was incubated in PBS for 1 h on ice. Subsequently, formaldehyde was added to a final concentration of 2% and incubated for 10 min at room temperature. The reaction was quenched by adding Tris to a final concentration of 250 mM. Then, 5× SDS sample buffer was added before boiling at 65 °C for 5 min. The crosslinked proteins were analyzed by western blotting.
Statistical analysis
Data were analyzed using one-way ANOVA followed by Turkey’s post hoc test with Graphpad Prism 6. All plots are expressed as the mean ± standard deviation (SD).
Results
Oligomerization and lipid binding promote Baz localization redundantly
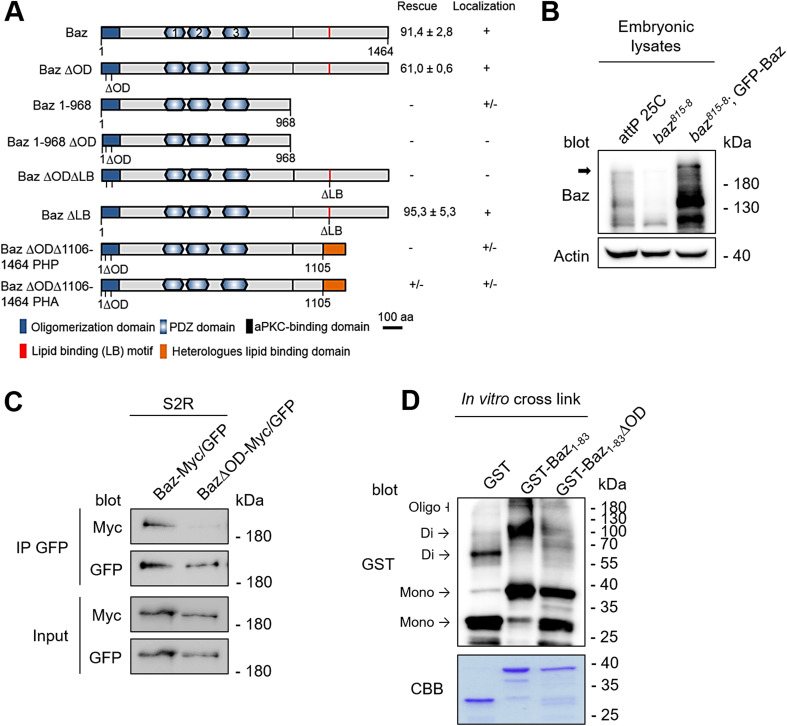
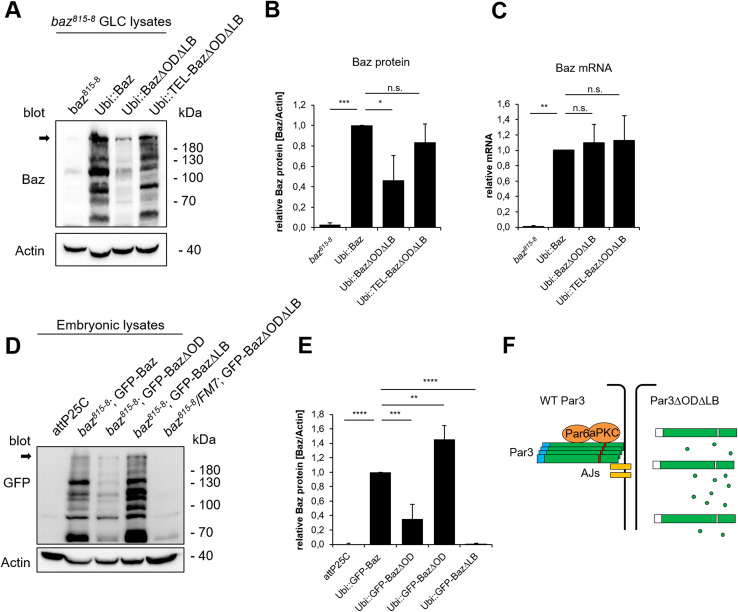
Baz functions on the top of a hierarchy regulating apical–basal polarization in the epidermis of the developing Drosophila embryo. However, it is still not fully understood, how Baz itself is recruited to the membrane. To analyze the contribution of structural domains of Baz to its localization in the Drosophila embryonic epithelium, we generated transgenic flies that express GFP-tagged baz transgenes (Fig. 1a). The baz constructs were expressed with the ubiquitin promoter, which resulted in a weak overexpression of the full-length protein (Fig. 1b). GFP-Baz was expected to run at ~ 190 kDa, but we have observed that the endogenous as well as the ectopically expressed protein display a higher molecular mass in western blot, which might result from posttranslational modifications such as phosphorylation by aPKC, Par1, or Rho-kinase [16, 30, 50]. Moreover, we always detected several smaller specific Baz fragments (Fig. 1a, c, d) [4, 32], which presumably result from (proteolytic) processing of Baz.
Fig. 1.
Oligomerization of Baz is dispensable for viability. a Schematic representation of different Baz deletion constructs. All constructs were expressed from the same genomic locus (attP40) with an N-terminal GFP-tag under the control of the ubiquitin promotor. The ∆OD mutation (V14DD68K) prevents oligomerization and the ∆LB mutation (K1173-74A) abolishes membrane binding. The ability to rescue the embryonic lethality of baz815-8 germ lines clones was quantified and the localization determined, where “+” indicates the wild-type situation, “+/−” indicates a lateral cortical localization and “−” indicates a cytoplasmic localization. b Expression of GFP-Baz in the baz815-8 mutant background results in a weak overexpression compared towards endogenous Baz. Full-length Baz is indicated with an arrow. c Co-immunoprecipitation of GFP- and Myc-tagged Baz or Baz∆OD in S2R cells. Mutations of the OD domain strongly attenuate the capacity of Baz to self-associate. d In vitro crosslinking of the Baz OD domain (aa 1–83). Mutations of the OD domain (V14DD68K) strongly impair the formation of oligomers in vitro
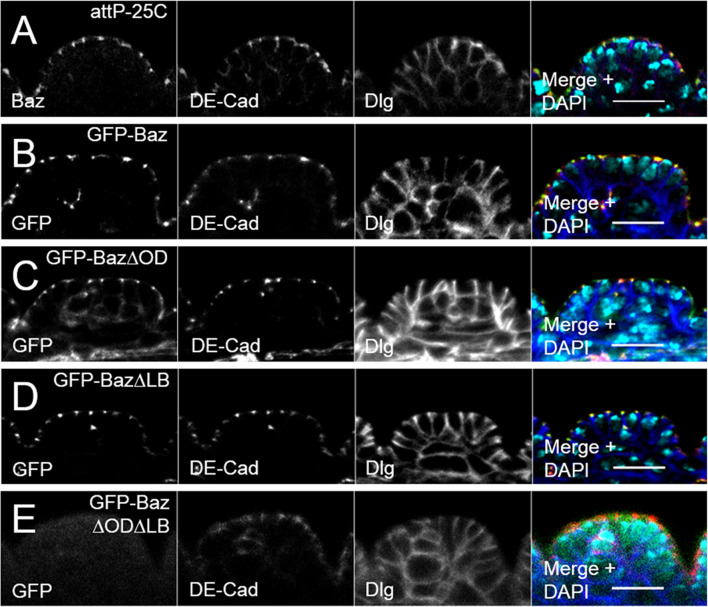
GFP-Baz co-localizes with the AJ marker DE-Cad at the AJs (Fig. 2b), similar to endogenous Baz (Fig. 2a) and can fully rescue the embryonic lethality of the baz815-8 null allele (91.4 ± 2.8% hatched L1 larvae) (Fig. 1a).
Fig. 2.
Oligomerization and lipid-binding promote Baz localization redundantly. a In immunostainings of the Drosophila embryonic epithelium, endogenous Baz (green) co-localizes with DE Cadherin (DE-Cad, red) at the apical junctions. Disc large (Dlg, blue) was stained as a lateral marker. b Localization of GFP-Baz is indistinguishable from the endogenous protein. c Oligomerization-deficient GFP-Baz∆OD displays an accumulation at the AJ, overlapping with DE-Cad. d The localization of a lipid-binding deficient GFP-Baz∆LB protein is similar to wild-type Baz. e GFP-Baz∆OD∆LB double mutant is absent from the cell cortex and displays a diffuse cytoplasmic localization. All transgenes were expressed in a wild-type background. Scale bars are 10 µm
To analyze the function of the N-terminal oligomerization domain, we generated a oligomerization-deficient version of Baz, Baz V14D D68K (hereafter Baz∆OD), which has been reported to abolish the self-association of two N-terminal monomers in rat Par3 [42]. Indeed, Baz∆OD had a strongly attenuated capacity to self-associate in vivo and in vitro (Fig. 1c, d). Surprisingly, in lethality tests with baz815-8 germ line clones (GLCs), which are deprived of maternal Baz mRNA and protein, GFP-Baz∆OD rescued the embryonic lethality of the baz815-8 null allele almost as efficient as wild-type Baz (61.0 ± 0.6%) (Fig. 1a). Furthermore, GFP-Baz∆OD localizes to the AJs in epithelial cells of the embryonic epithelium indistinguishable from its wild-type counterpart (Fig. 2c compared to Fig. 2a, b).
Baz is capable of binding to the phospholipids PIP2 and PIP3 [48]. However, mutation of the lipid-binding domain of Baz (GFP-BazK1173-1174A = Baz∆LB, Fig. 6b) did not attenuate the apical junctional localization of GFP-Baz∆LB or its rescue capacity (95.3 ± 5.3%) (Figs. 1a, 2d). In contrast, a variant of Baz which cannot oligomerize or bind to phospholipids (GFP-Baz∆OD∆LB) displayed a cytoplasmic localization and failed to rescue the baz815-8 mutant (Figs. 1a, 2e). Thus, Baz oligomerization and binding to phospholipids function in redundancy to target the protein to the apical junctions and ensure its function. This is confirmed by the finding that a Baz variant encoding the first 968 amino acids (GFP-Baz1-968), which includes the oligomerization domain, is localized to the apical junctions with a slight baso-lateral mislocalization (Suppl. Figure 1). Mutation of the oligomerization domain in GFP-Baz1-968 (GFP-Baz1-968∆OD) abolishes its cortical localization (Suppl. Figure 1). Notably, GFP-Baz1-968 did not rescue the embryonic lethality of baz815-8, most likely due to a lack of the C-terminal part of the protein, which includes the aPKC-binding region.
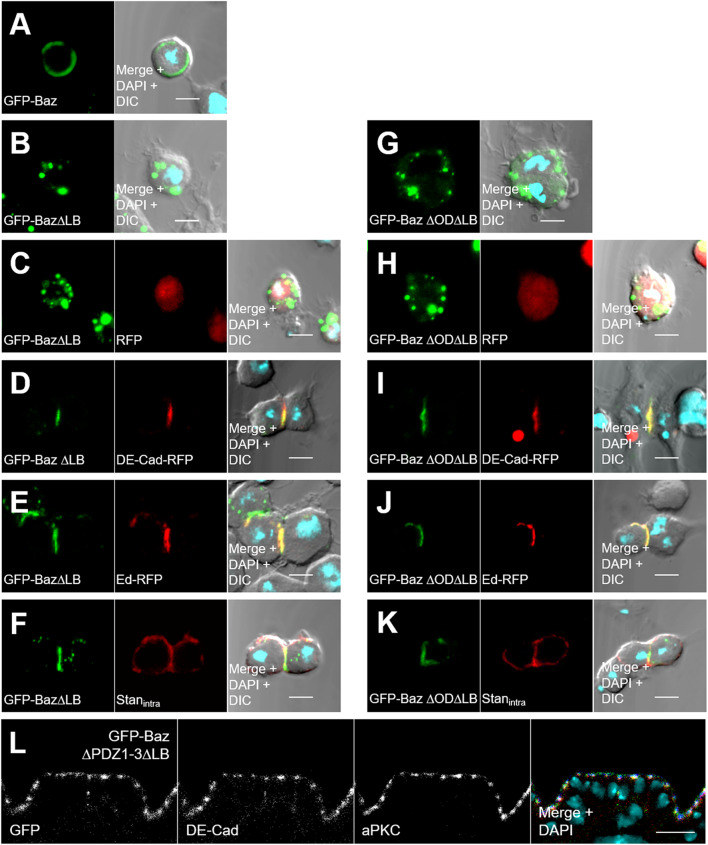
Fig. 6.
Recruitment of Baz by DE-Cad, Ed, and Stan does not depend on self-association or lipid-binding of Baz. a Wild-type Baz-GFP was expressed with the ubiquitin promoter in S2R cells and localizes at the plasma membrane. b Baz∆LB-GFP accumulates in cytoplasmic aggregates. c RFP displays a diffuse cytoplasmic localization and cannot recruit Baz∆LB-GFP to the cortex. d, e DE-Cad-RFP and Ed-RFP recruit Baz∆LB-GFP to artificial cell–cell contacts. f Similar, the intracellular domain of Stan fused to the extracellular and transmembrane domain of DE-Cad (DE-Cad∆intra-Stan∆extra) targets Baz∆LB-GFP to cell–cell contacts. DE-Cad∆intra-Stan∆extra was detected with an anti DE-Cad antibody, which recognizes the extracellular domain of DE-Cad. g–k In the same experimental setup with Baz∆OD∆LB-GFP, the double mutant was efficiently recruited by DE-Cad, Ed, and Stan without apparent differences. l Deletion of all three PDZ domains in Baz∆LB does not affect the localization of the mutant protein at the apical junctions (green = GFP-Baz∆PDZ1-3 ∆LB, red = DE-Cad, blue = aPKC). DIC differential interference contrast. Scale bars are 5 μm in a–k and 10 μm in l
Taken together, the N-terminal oligomerization domain and the C-terminal LB motif of Baz contribute redundantly to the localization of the protein. Based on the rescue capacity of the Baz variants, we found that neither the oligomerization nor the binding to phospholipids is essential for viability.
Binding to phospholipids is not sufficient for the function of Baz
To further investigate the role of the LB motif regarding the localization and function of Baz, we substituted the C-terminus including the intrinsic LB motif of GFP-Baz∆OD by the Pleckstrin homology (PH) domains of either PLCδ (Baz∆OD∆1106-1464-PHP) or Akt1 (Baz∆OD∆1106-1464-PHA) (Fig. 1a). The PH domain of PLCδ specifically binds to PtdIns(4,5)P2 (PIP2) [57], whereas the PH domain of Akt1 binds to PtdIns(3,4,5)P3 (PIP3) [58]. Baz itself binds both, PIP2 and PIP3 in vitro [48].
In the epithelium of transgenic embryos, both GFP-Baz∆OD∆1106-1464-PHP and GFP-Baz∆OD∆1106-1464-PHA had a cortical localization and punctual enrichments at the AJ where they co-localized with DE-Cad (Suppl. Figure 1). However, only GFP-Baz∆OD∆1106-1464-PHA rescued occasionally the zygotic baz815-8 allele (< 1%), but not embryos, which have been depleted for the maternal and zygotic protein expression (GLCs). Moreover, expression of both variants together, GFP-Baz∆OD∆1106-1464-PHP and GFP-Baz∆OD∆1106-1464-PHA, in a baz-mutant background did not produce surviving animals, indicating that either simultaneous binding of one Baz molecule to PIP2 and PIP3 is essential or that the C-terminus (aa 1106–1464) is essential for Baz’ function in our experimental setup, which differs from the previous studies, which used proteins overexpressed by the UAS/GAL4 system [48, 51].
Baz∆OD∆LB fails to polarize the epithelium of the embryonic epidermis
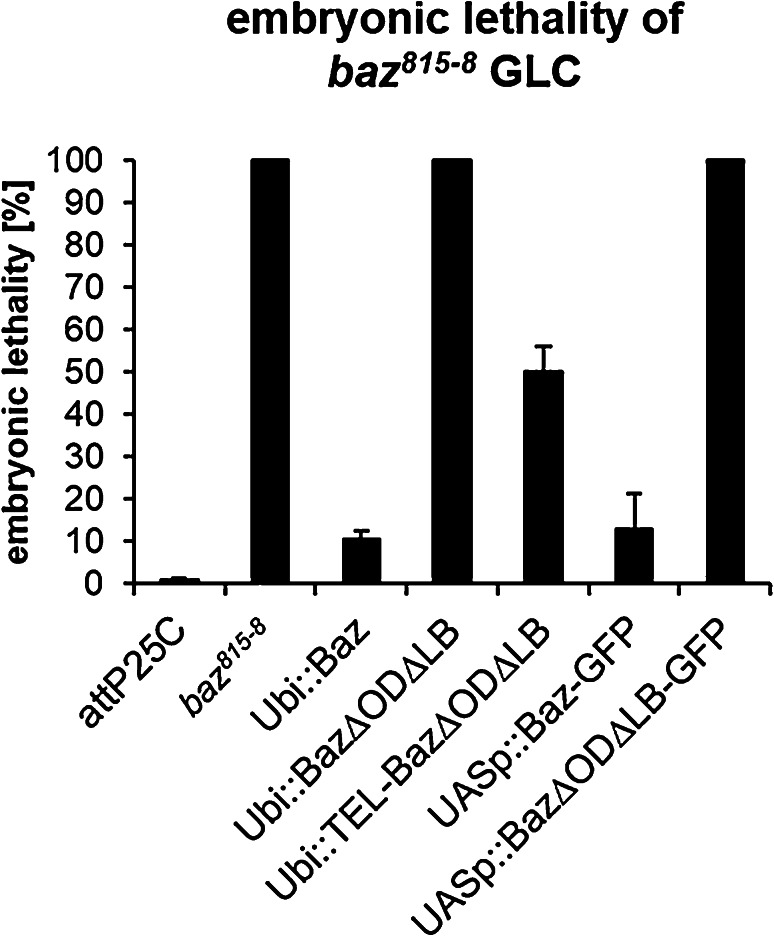
To better understand why Baz∆OD∆LB failed to rescue baz815-8 mutant embryos, we analyzed the epithelium of GLCs. To exclude the possibility that the N-terminal GFP-tag interferes with the function of Baz, we created transgenic flies, which express Baz transgenes fused with a small One-Strep-tag (OneS) at the C-terminus under the control of the ubiquitin promotor. As the GFP variants, Baz-OneS displayed a strong rescue capacity of the embryonic lethality in baz815-8 GLCs, whereas Baz∆OD∆LB-OneS displayed a complete embryonic lethality (Fig. 3). To test if the overexpression of Baz∆OD∆LB might help to overcome the embryonic lethality of baz815-8 GLCs, we expressed UASp::Baz-GFP and UASp::Baz∆OD∆LB-GFP with mat-Tub::Gal4 in baz815-8 GLCs (Fig. 5a). Baz-GFP rescues the embryonic lethality of GLCs to the same extend as Baz-OneS (87.3 ± 8.5 and 89.5 ± 1.8%, respectively), whereas Baz∆OD∆LB-GFP expressing GLCs failed to escape embryonic lethality (Fig. 3).
Fig. 3.
Loss of oligomerization and lipid-binding causes embryonic lethality. Baz variants that carry a C-terminal One-Strep (OneS) tag were expressed under the ubiquitin promoter and tested for their capacity to rescue baz818-8 germ line clones. Baz-OneS efficiently rescues the lethality of the baz818−8 allele (10.5 ± 1.8% embryonic lethality). Baz∆OD∆LB-OneS failed to rescue the embryonic lethality. Fusion of the oligomerization domain of the human TEL protein (aa 45–115) to the N-terminus of Baz restores its function and rescues embryonic lethality of baz815-8 to a large extent (50.0 ± 6.1% embryonic lethality). Similarly, wild-type Baz-GFP overexpressed with mat-Tub::Gal4 using the Gal4/UAS-system had comparable efficiencies as the constitutively expressed variant (12.7 ± 8.5%), whereas overexpression of Baz∆OD∆LB-GFP failed to rescue (100% embryonic lethality). Bars represent the mean ± SD, n = 300 each
Fig. 5.
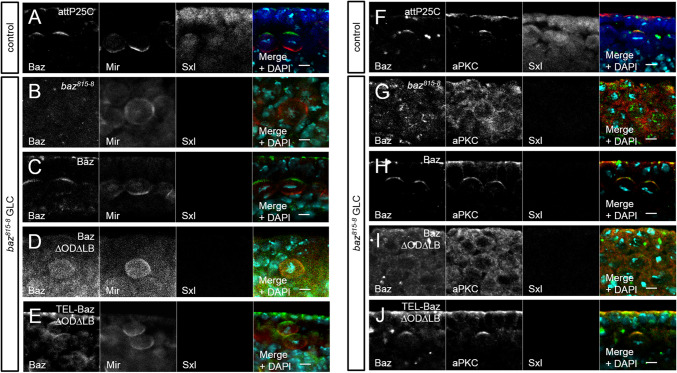
Neuroblast polarity requires Baz’s capacity to either self-associate or to bind lipids. a Immunostaining of endogenous Baz (green), Mir (red), and Sxl (blue) in embryonic metaphase NBs. Baz localizes at the apical cortex, whereas Mir accumulates basally. b–e Immunostaining of Baz variants (green) and endogenous Mir (red) in embryonic NBs of baz818-8 germ line clones during metaphase. Hemizygous mutant embryos were identified by the absence of Sxl staining. b Loss of Baz in baz818-8 germ line clones disrupts NB polarity and Mir localization. c Baz-OneS rescues NB polarity, whereas Baz∆OD∆LB-OneS does not localize at the cortex and fails to polarize NBs (d). e TEL-Baz∆OD∆LB-OneS restores apical–basal polarity in metaphase NBs, such as wild-type Baz. f Immunostaining of endogenous Baz (green), aPKC (red), and Sxl (blue) in embryonic metaphase NBs. Baz recruits aPKC to the apical pole of metaphase NBs. g aPKC accumulates in the cytoplasm in metaphase NBs of baz818-8 germ line clones. h, j Baz-OneS and TEL-Baz∆OD∆LB-OneS both show a comparable localization and recruit aPKC to the apical pole. i In contrast, Baz∆OD∆LB-OneS localizes in the cytoplasm, such as endogenous aPKC. Scale bars are 5 µm
Next, we evaluated the phenotypes of embryonic cuticles of baz GLCs expressing the different rescue constructs. The Cuticle is secreted from the epidermis and allows drawing a conclusion of its integrity, in particular the formation of a function apical domain. We divided the observed phenotypes in five groups [hatched (= normal cuticle), wild type (= dead but cuticle without obvious defects), cuticle rest, holes and shrunken with holes]. As expected, the baz815-8 mutant displayed large cuticle hole or some cuticle rest (Suppl. Figure 2). In contrast to the null allele, most animals of Baz-OneS hatch and the dead embryos had either a wild-type or shrunken cuticle phenotype (4.8 or 5.7%, respectively, Suppl. Figure 2). Baz∆OD∆LB-OneS partially rescued the baz815-8 phenotype, because some embryos developed further and had either a wt or shrunken with holes phenotype (2.3 or 23.3%, respectively, Suppl. Figure 2).
Interestingly, fusion of the heterologous oligomerization domain of the human TEL protein (residues 45–115 [59]) to the N-terminus of Baz∆OD∆LB-OneS (TEL-Baz∆OD∆LB-OneS) partly restores the rescue capacity of the mutant Baz protein. Furthermore, TEL-Baz∆OD∆LB-OneS embryos had a milder phenotype, as most dead embryos had a wild-type or shrunken cuticle phenotype (21.9 and 17.9%, respectively, Suppl. Figure 2).
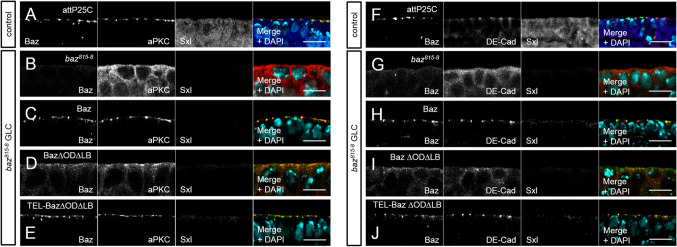
Next, we scored for the localization of polarity markers in the embryonic epithelium by immunostainings. Hemizygous mutant embryos derived from GLC were identified by the lack of Sex lethal (Sxl) staining. As expected, in the epithelium of baz815-8 GLCs, we did not detect a signal for Baz, whereas aPKC exhibited a cytoplasmic localization (Fig. 4b). Baz-OneS and TEL-Baz∆OD∆LB-OneS both had a robust apical localization and recruited aPKC to the apical junctions, similar to endogenous Baz (Fig. 4a, c, e; Suppl. Figure 3B, D, F). In contrast, Baz∆OD∆LB-OneS showed a cytoplasmic mislocalization (Fig. 4d). Nevertheless, Baz∆OD∆LB-OneS managed to recruit some aPKC to the apical junctions, but the majority of the aPKC protein still accumulates in the cytoplasm (Fig. 4d). The amount of aPKC which co-localized with Baz∆OD∆LB-OneS was significantly reduced, but was to a large extent rescued by TEL-Baz∆OD∆LB-OneS (Suppl. Figure 3L). In general, the overall structure of the epithelium was disrupted in Baz∆OD∆LB-OneS embryos.
Fig. 4.
Epithelial polarization requires the functional redundancy of the OD and LB domains. a Immunostaining of endogenous Baz (green), aPKC (red), and Sxl (blue) in the embryonic epidermis. b–e Immunostaining of Baz variants and endogenous aPKC in the embryonic epidermis of baz818-8 germ line clones. Hemizygous mutant embryos were identified by the absence of Sxl staining. b Loss of Baz in the embryonic epidermis of disrupts epithelial polarization and aPKC accumulates in the cytoplasm. c Baz-OneS efficiently recruits aPKC to the apical junctions, such as endogenous Baz, whereas Baz∆OD∆LB-OneS displays a cytoplasmic mislocalization. d Baz∆OD∆LB-OneS rescues targets some aPKC to the apical junctions, but the majority of the protein still accumulates in the cytoplasm. e TEL-Baz∆OD∆LB-OneS localizes at the apical junctions and is capable of recruiting aPKC to rescue the defects of Baz∆OD∆LB-OneS. f–j Immunostaining of Baz (green), DE-Cad (red), and Sxl (blue) in the embryonic epidermis demonstrates a loss of AJ in baz815-8 germ line clones, which can be rescued by wild-type Baz (h) and TEL-Baz∆OD∆LB-OneS (j) but not by Baz∆OD∆LB-OneS (i). Scale bars are 10 µm
Then, we examined the assembly of intact AJ by scoring for the localization of DE-Cad in baz815-8 GLCs (Fig. 4f–j). Baz-OneS and TEL-Baz∆OD∆LB-OneS both showed an apical junctional targeting of DE-Cad comparable to the wild-type control (Fig. 4f, h, j; Suppl. Figure 3G, I, K). Unlike as for aPKC, we did not observe a localization of DE-Cad in the apical region or at the plasma membrane in Baz∆OD∆LB-OneS expressing baz815-8 GLCs (Fig. 4i; Suppl. Figure 3L).
In summary, Baz∆OD∆LB-OneS has only a weak capability to polarize the embryonic epithelium and displayed strong cuticle defects, as well as an impaired function, since aPKC was inefficiently and DE-Cad not at all recruited to the apical junctions. Restoring the oligomerization capacity of Baz by fusing the oligomerization domain of TEL to Baz∆OD∆LB-OneS restores its functionality to a large extent.
Apical–basal polarity of embryonic NBs is disrupted in Baz∆OD∆LB embryos
Similar to polarization of the epithelium of the embryonic epidermis, Baz is also required for the establishment of apical–basal polarity of dividing NBs. Hence, we investigated if Baz∆OD∆LB-OneS had similar phenotypes in embryonic NBs as in the epithelium. Baz accumulates at the apical cortex, recruiting aPKC and Par6, whereas the scaffold protein Miranda (Mir) is restricted to the basal region of metaphase NBs (Fig. 5a). Basal segregation of Mir depends on the apical formation of the Par complex [60, 61]. Therefore, we analyzed the localization of Baz variants and Mir in NBs of baz815-8 GLCs. We found that Baz∆OD∆LB-OneS phenocopied the baz null allele, as Mir is not restricted to the basal region of NBs in both genotypes but can be found more or less all around the cortex (Fig. 5b, d). Baz∆OD∆LB-OneS also failed to localize to the apical membrane of NBs, but rather displayed a weak cytoplasmic localization (Fig. 5d). Like in the epithelium, Baz-OneS and TEL-Baz∆OD∆LB-OneS rescued the asymmetric distribution of Mir and localized to the apical membrane in metaphase NBs, such as the control (Fig. 5a, c, e).
In contrast to the epithelium where aPKC displayed at least a minimal polarization in Baz∆OD∆LB-OneS GLC, its localization in NBs is cytoplasmic, similar to the baz null allele (Fig. 5g, i). As expected, Baz-OneS and TEL-Baz∆OD∆LB-OneS recruit aPKC to the apical cortex comparable to endogenous protein (Fig. 5f, h, j).
Thus, the functional redundancy of the OD and the LB motif are essential to polarize embryonic NBs, since neither Mir nor aPKC exhibited a correct localization in Baz∆OD∆LB-OneS GLC.
Baz∆OD∆LB is still recruited to the cortex by transmembrane proteins
To elucidate the mechanism why Baz∆OD∆LB-OneS fails to accumulate at the apical junctions and why it is non-functional, we tested whether the membrane recruitment by its described interacting transmembrane proteins is disturbed. Therefore, we used Drosophila S2R cells as a model system, because they do not exhibit a polarity or detectable amounts of polarity proteins [48]. In S2R cells, Baz-GFP clearly localizes to the cell cortex (Fig. 6a), whereas Baz∆LB-GFP appears in cytoplasmic aggregates (Fig. 6b), indicating that in S2R cells, cortical localization of Baz depends exclusively on the binding to phospholipids. Co-transfection of Baz∆LB-GFP with DE-Cad-RFP or Ed-RFP led to the formation of cell–cell contacts of transfected cells. Both proteins recruited Baz∆LB-GFP to the artificial cell–cell contacts, but not RFP alone (Fig. 6c–e). In addition, the intracellular domain of the atypical cadherin Starry night (Stan) fused to the extracellular domain of DE-Cad also recruited Baz∆LB-GFP to ectopic cell–cell contacts (Fig. 6f). By contrast, the Stan isoform Flamingo, which lacks the C-terminal PDZ-binding motif, did not recruit lipid-binding deficient Baz (data not shown). Expression of Baz∆OD∆LB-GFP alone or together with DE-Cad, Ed, or Stan showed that Baz∆OD∆LB-GFP was recruited to the cell–cell contacts, such as its oligomerizing counterpart (Fig. 6g–k). Surprisingly, deleting the PDZ domains of GFP-Baz∆LB, which facilitate binding to Stan, Ed, and Arm/DE-Cad, does not disturb its localization at the apical junctions in the embryonic epidermis (Fig. 6l).
Cortical targeting protects Baz from degradation
Despite its capacity to interact with transmembrane proteins, Baz∆OD∆LB-OneS is non-functional and does not localize to the apical junctions. Therefore, we tested, whether simultaneous loss of oligomerization and lipid-binding affects the protein stability, we blotted baz815-8 GLC rescued with GFP-Baz and Baz-OneS variants to detect the exogenous Baz protein. Indeed, we observed that loss of the oligomerization domain caused a reduced amount of Baz protein in the embryo, which is further enhanced upon the loss of the LB motif (Fig. 7a, b, d, e). In contrast, mutation of the LB motif alone did not reduce the amount of protein, but rather seemed to elevate it (Fig. 7d, e), whereas the amount of Baz∆OD∆LB-OneS protein is drastically reduced in lysates of GLC (Fig. 7a, b). Notably, introduction of an ectopic oligomerization capacity in (TEL-Baz∆OD∆LB-OneS) restored protein stability (Fig. 7a, b). The reduced amount of Baz∆OD∆LB-OneS was not due to impaired gene expression, because all transgenes were expressed from the same promoter and genomic location and exhibited comparable mRNA levels with no significant differences (Fig. 7c).
Fig. 7.
Oligomerization and lipid binding are crucial for Baz’ stabilization. a Lysates of baz818-8 germ line clones that express different Baz variants were blotted against Baz. Actin was used as loading control. Full-length Baz is indicated with an arrow. Loss of Baz oligomerization and lipid binding (Baz∆OD∆LB) strongly decreases the amount of Baz protein. TEL-Baz∆OD∆LB-OneS rescues the degradation of Baz∆OD∆LB-OneS. b Quantification of Baz-OneS variants in baz818-8 germ line clones. The whole Baz lanes were quantified and normalized towards actin from three biological replicates. c qPCR of total RNA from the baz818-8 germ line clones shows that all transgenes were expressed without significant differences. d Embryonic lysates of different GFP-Baz variants in a baz818-8 genetic background were blotted against GFP. Full-length Baz is indicated with an arrow. Actin was used as loading control. Loss of oligomerization reduces the stability of GFP-Baz∆OD, which is drastically enhanced upon the mutation of the lipid-binding motif in GFP-Baz∆OD∆LB. However, mutation of the lipid-binding motif alone (GFP-Baz∆LB) does not affect the protein stability. e Quantification of GFP-Baz variants. The whole GFP lanes were quantified and normalized towards actin from three biological replicates. f Scheme of the functional redundancy between the OD and LB domains. Bars represent the mean ± SD. Statistics were one-way ANOVA followed by Tukey’s post hoc test, n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Finally, we tested whether the phenotypes observed in Baz∆OD∆LB rescued embryos are only due to protein degradation of the mutant protein. Strikingly, overexpression of wild-type Baz-GFP but not Baz∆OD∆LB-GFP rescued the embryonic lethality of baz815-8 (Fig. 3), although both proteins are expressed at comparable levels (Suppl. Figure 4A). Moreover, Baz∆OD∆LB-GFP is still cytoplasmic, whereas its wild-type counterpart localizes to the apical junctions (Suppl. Figure 4C in comparison to B). These data suggest that Baz∆OD∆LB fails to localize to the apical junctions, accumulates in the cytoplasm, and is degraded.
Discussion
Taken together, the oligomerization domain of Baz is not essential for viability of the Drosophila embryo, but contributes to the stability of the protein; and the functional redundancy of the oligomerization domain and the LB motif are indispensable for the function of Baz during Drosophila embryogenesis (Fig. 7f).
The previous studies reported either an important role of the OD for Baz/Par3 localization in Drosophila and mammalian cells [40, 41] or found only a minor influence of the OD on Baz localization [47, 51], which we could confirm. This discrepancy might be explained by the different setups: In contrast to the previous studies [40, 48, 51] using overexpressed proteins with the UAS/GAL4 system in rescue experiments, we used a constitutive expression, which resulted in a rather mild overexpression. Nonetheless, the slightly reduced rescue capacity (61% of Baz∆OD in contrast to 91% for wild-type Baz) and the redundant function of the OD underline the importance of Baz self-association. The fact that the heterologous oligomerization domain of TEL can rescue the defects of Baz∆OD∆LB suggests that the OD promotes indeed self-association instead of interaction with other binding partners.
In line with the previous results [48, 51], we found that the lipid-binding domain of Baz/Par3 is dispensable for the localization and function of the protein—however, it can, to a far extent, compensate the loss of the OD. Notably, in contrast to McKinley et al. [51], we found that deletion of the PDZ domains does not affect the localization of GFP-Baz (or other apical determinants), which might be explained by the fact that, in that study, the authors used the UAS/GAL4-System to (over)express a C-terminal-tagged Baz (in which the GFP might be cleaved due to processing of the C-terminus, which can be frequently observed) in both, live-imaging and fixed samples, whereas we utilized a constitutively expressed transgene and an N-terminal tag only in fixed tissues. Although the PDZ domains as well as the aPKC-binding region certainly contribute to the fine-tuning of Baz localization, our data demonstrate that the OD and the LB domain are the crucial two domains which regulate the membrane-tethering of the protein. We further demonstrate here that protein stability of Baz depends on membrane localization, as Baz∆OD∆LB is degraded, whereas fusion of heterologous lipid-binding domains or an oligomerization domain rescues the protein stability (Suppl. Figure 1E; Fig. 7a, b). A reduced amount of Baz has been reported to localize at the apical domains upon deletion of the OD [51], which might be explained by our observation of the decreased protein stability of Baz∆OD.
Finally, one important question remains: How does the OD contribute to the localization of Baz? One likely possibility is that Baz forms oligomers, which are then recruited to the plasma membrane by a transmembrane- or membrane-associated protein. The previous studies have identified three transmembrane proteins (DE-Cad (via Arm), Ed, Stan, and Canoe) as interaction partners of Baz, which might be capable of recruiting the protein to the membrane [39, 45, 62]. The interaction of Baz with Arm, Ed, and Stan has been described to be mediated by the PDZ domains of Baz [45, 51, 62]. However, deletion of all three PDZ domains together with the lipid-binding motif does not substantially affect the apical–junctional localization of the mutant protein (Fig. 6l). Given that Baz∆OD∆LB does not display an apical accumulation, the functional redundancy of the OD and LB motif might be essential for the initial localization to the plasma membrane. Here, it seems to be not important, whether Baz binds to PIP2 or PIP3, as both chimeric rescue constructs restore protein stability and (at least to some extent) the localization of Baz∆OD∆LB (Supplementary Fig. 1C–E). Work from Harris and Peifer nicely demonstrated that Baz functions upstream of at least DE cadherin in the polarization of the embryonic epidermis [19]. However, it is still unclear, if other Baz-binding proteins, such as Ed, Stan, or Canoe, could contribute to the initial recruitment of Baz to the plasma membrane of epithelial cells during the early embryonic development [39, 45, 62]. Therefore, we tried to abolish the expression of DE Cadherin, Ed, and Stan in the early embryos using triple GLCs, which unfortunately did not produce any eggs (data not shown), indicating that these three genes are involved in oogenesis, too. Nonetheless, we observed that Baz∆OD∆LB is still able to interact with DE-Cad, Ed and Stan (Fig. 6). This is surprising as in vivo, none of these interaction partners seem to be capable of targeting the mutant protein to the apical junctions, although they are all expressed in the embryonic epidermis. Thus, we can exclude DE-Cad, Ed, and Stan to be involved in the initial recruitment of Baz. Although Canoe is important for the apical positioning of Baz, loss of Canoe does not prevent the membrane association of Baz [39]. Moreover, the fact that deletion of all three PDZ domains in BazΔLB does not disturb the correct apical junctional localization of the mutant protein (Fig. 6l) suggests that another domain is essential for the recruitment of Baz oligomers. A possible model for the correct recruitment of Baz could be that in the absence of lipid binding, oligomerized Baz is targeted to the apical junctions by several weak mechanisms and interactions with proteins, independently (or redundantly) of the PDZ domains. The multiplicity of backup mechanisms for Baz localization further underlines the critical role of the localization of this polarity regulator in establishing apical–basal polarity in the embryonic epidermis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1: Localization of Baz variants in the embryonic epidermis. (A-D) Immunostainings of different GFP-Baz variants in the embryonic epidermis. All transgenes were expressed with the ubiquitin promotor in a wild-type background. GFP (green), DE-Cad (red), and Dlg (blue) were stained. (A) GFP-Baz1-968 localizes mainly at the apical junctions with some baso-lateral mislocalization. (B) Mutation of the OD domain in this truncated protein causes a cytoplasmic accumulation of GFP-Baz1-968∆OD. (C, D) Chimeric proteins that carry the pleckstrin homology (PH) domains of either human PLCδ or Akt1 fused to the C-terminus GFP-Baz∆OD1-1105 promote a cortical localization in the embryonic epidermis. (E) Western blot of embryonic lysates of the Baz variants (A-D). The Baz variants were detected with a GFP antibody and Actin was used as a loading control. Scale bars are 10 µm (TIFF 857 kb)
Supplemental Fig. 2: Cuticle phenotypes of Baz variants. (A) Cuticle phenotypes were classified into the four categories wt, shrunken with holes, holes, and cuticle rests. (B) The cuticle phenotypes of baz818−8 germ line clones that express different Baz variants with the ubiquitin promoter were quantified (n = 300 per genotype). Embryos that normally developed and hatched are also included as “hatched”. baz818−8 germ line clones display holes or cuticle rests. Embryos that express Baz-OneS hatch to a large extend (89,5%) or display either wt or shrunken with hole phenotypes. No embryos hatch upon the expression of Baz∆OD∆LB-OneS. Most embryos have either cuticle rests or holes phenotypes (58,1 and 16,3%, respectively). Nevertheless, some embryos develop further and have a wt or shrunken with holes phenotype (2,3 and 23,3%, respectively). The chimeric TEL-Baz∆OD∆LB-OneS protein rescues the Baz∆OD∆LB phenotypes to a large extent as half of the embryos hatch (50,2%) or have either wt or shrunken with holes phenotypes (21,9 and 17,9%, respectively). Scale bars are 200 µm (TIFF 417 kb)
Supplemental Fig. 3: Quantification of Baz-OneS constructs in the embryonic epithelium. (A) Scheme of the area (33 × 4 µm) of the apical region of all immunostainings in Fig. 4, which has been quantified with FIJI Plot Profile. The gray values of either Baz/aPKC (B-F) or Baz/DE-Cad (G-K) in the apical region were plotted against the distance. (L) The Pearson correlation coefficient of the co-localization of either Baz/aPKC or Baz/DE-Cad has been determined with the Coloc2 Plugin in FIJI for all genotypes in Fig. 4. For the quantification, three different embryos of each genotype have been analyzed. Bars represent the mean ± S.D. Statistics were one-way ANOVA followed by Turkey’s post hoc test, n.s. p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 (TIFF 1777 kb)
Supplemental Fig. 4: Expression of Baz variants with the UAS/Gal4-system. (A) Western blot of embryonic lysates from embryos that express Baz-GFP and Baz∆OD∆LB-GFP with the UASp promotor, driven by mat-Tub::Gal4. The Baz variants were detected with a GFP antibody and actin was used as a loading control. Full-length Baz is indicated with an arrow. Note that the pattern of specific bands is different in Baz∆OD∆LB (e.g., the band around 130 kDa is much weaker), which might be due to differences in the posttranslational processing of full-length Baz (presumably by cleavage). (B, C) Immunostaining of Baz variants (green), DE-Cad (red), and aPKC (blue) in the embryonic epidermis. (B) Baz-GFP localizes at the apical junction, (C) whereas Baz∆OD∆LB-GFP displays a diffuse cytoplasmic localization. Scale bars are 10 µm (TIFF 902 kb)
Acknowledgements
We thank the Bloomington Drosophila stock center at the University of Indiana (USA) and the Developmental Studies Hybridoma Bank at the University of Iowa (USA) and Frank Sprenger for providing reagents. This work was supported by Grants of the German Research Foundation (DFG) to M. P. K. (DFG3901/1-2, SFB1348-A05).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
References
- 1.Goldstein B, Macara IG. The PAR proteins. Fundamental players in animal cell polarization. Dev Cell. 2007;13(5):609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pocha SM, Knust E. Complexities of Crumbs function and regulation in tissue morphogenesis. Curr Biol. 2013;23(7):R289–R293. doi: 10.1016/j.cub.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Harris TJC, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170(5):813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krahn MP, Bückers J, Kastrup L, Wodarz A. Formation of a Bazooka–Stardust complex is essential for plasma membrane polarity in epithelia. J Cell Biol. 2010;190(5):751–760. doi: 10.1083/jcb.201006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen A, Sun R, Krahn MP. Localization and function of Pals1-associated tight junction protein in Drosophila is regulated by two distinct apical complexes. J Biol Chem. 2015;290(21):13224–13233. doi: 10.1074/jbc.M114.629014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch L, Feicht S, Sun R, Sen A, Krahn MP. Domain-specific functions of Stardust in Drosophila embryonic development. R Soc Open Sci. 2016;3(11):160776. doi: 10.1098/rsos.160776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki A, Ohno S. The PAR-aPKC system. Lessons in polarity. J Cell Sci. 2006;119(Pt 6):979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 8.Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3(1):43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 9.Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150(6):1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutterer A, Betschinger J, Petronczki M, Knoblich JA. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev Cell. 2004;6(6):845–854. doi: 10.1016/j.devcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez J, Peglion F, Martin J, Hubatsch L, Reich J, Hirani N, Gubieda AG, Roffey J, Fernandes AR, St Johnston D, Ahringer J, Goehring NW. aPKC cycles between functionally distinct PAR protein assemblies to drive cell polarity. Dev Cell. 2017;42(4):400–415.e9. doi: 10.1016/j.devcel.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S-C, Low TYF, Nishimura Y, Gole L, Yu W, Motegi F. Cortical forces and CDC-42 control clustering of PAR proteins for Caenorhabditis elegans embryonic polarization. Nat Cell Biol. 2017;19(8):988–995. doi: 10.1038/ncb3577. [DOI] [PubMed] [Google Scholar]
- 13.Nam S-C, Choi K-W. Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development. 2003;130(18):4363–4372. doi: 10.1242/dev.00648. [DOI] [PubMed] [Google Scholar]
- 14.Vogelmann R, Nelson WJ. Fractionation of the epithelial apical junctional complex. Reassessment of protein distributions in different substructures. Mol Biol Cell. 2005;16(2):701–716. doi: 10.1091/mbc.e04-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128(2):383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morais-de-Sá E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141(3):509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr Biol. 2010;20(12):1065–1074. doi: 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 18.Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T, Suzuki Y, Yamanaka T, Suzuki A, Mizuno K, Ohno S. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J Cell Sci. 2002;115(Pt 12):2485–2495. doi: 10.1242/jcs.115.12.2485. [DOI] [PubMed] [Google Scholar]
- 19.Harris TJC, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167(1):135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7(3):262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, Yonemura S, Ohno S. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci. 2009;122(Pt 10):1595–1606. doi: 10.1242/jcs.043174. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed SM, Macara IG. The Par3 polarity protein is an exocyst receptor essential for mammary cell survival. Nat Commun. 2017;8:14867. doi: 10.1038/ncomms14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalli G. RalA and the exocyst complex influence neuronal polarity through PAR-3 and aPKC. J Cell Sci. 2009;122(Pt 10):1499–1506. doi: 10.1242/jcs.044339. [DOI] [PubMed] [Google Scholar]
- 24.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402(6761):544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 25.Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402(6761):548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 26.Kuchinke U, Grawe F, Knust E. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr Biol. 1998;8(25):1357–1365. doi: 10.1016/S0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- 27.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132(4):583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Wodarz A. Molecular control of cell polarity and asymmetric cell division in Drosophila neuroblasts. Curr Opin Cell Biol. 2005;17(5):475–481. doi: 10.1016/j.ceb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18(1):4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115(6):691–704. doi: 10.1016/S0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 31.Hurd TW, Fan S, Liu C-J, Kweon HK, Hakansson K, Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13(23):2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 32.Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical–basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16(6):901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol. 2004;14(8):736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422(6929):326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 35.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division. Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135(1):161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graybill C, Wee B, Atwood SX, Prehoda KE. Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J Biol Chem. 2012;287(25):21003–21011. doi: 10.1074/jbc.M112.360495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soriano EV, Ivanova ME, Fletcher G, Riou P, Knowles PP, Barnouin K, Purkiss A, Kostelecky B, Saiu P, Linch M, Elbediwy A, Kjær S, O’Reilly N, Snijders AP, Parker PJ, Thompson BJ, McDonald NQ. aPKC inhibition by Par3 CR3 flanking regions controls substrate access and underpins apical-junctional polarization. Dev Cell. 2016;38(4):384–398. doi: 10.1016/j.devcel.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7(11):1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 39.Choi W, Harris NJ, Sumigray KD, Peifer M. Rap1 and Canoe/afadin are essential for establishment of apical–basal polarity in the Drosophila embryo. Mol Biol Cell. 2013;24(7):945–963. doi: 10.1091/mbc.e12-10-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003;13(15):1330–1334. doi: 10.1016/S0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- 41.Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278(33):31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- 42.Feng W, Wu H, Chan L-N, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. EMBO J. 2007;26(11):2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Kim H, Beers M, Kemphues K. Different domains of C. elegans PAR-3 are required at different times in development. Dev Biol. 2010;344(2):745–757. doi: 10.1016/j.ydbio.2010.05.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Wang W, Chen J, Zhang K, Gao F, Gao B, Zhang S, Dong M, Besenbacher F, Gong W, Zhang M, Sun F, Feng W. Structural insights into the intrinsic self-assembly of Par-3 N-terminal domain. Structure. 2013;21(6):997–1006. doi: 10.1016/j.str.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Wei S-Y, Escudero LM, Yu F, Chang L-H, Chen L-Y, Ho Y-H, Lin C-M, Chou C-S, Chia W, Modolell J, Hsu J-C. Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev Cell. 2005;8(4):493–504. doi: 10.1016/j.devcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Feng W, Chen J, Chan L-N, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28(5):886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 47.Yu CG, Harris TJC. Interactions between the PDZ domains of Bazooka (Par-3) and phosphatidic acid. In vitro characterization and role in epithelial development. Mol Biol Cell. 2012;23(18):3743–3753. doi: 10.1091/mbc.e12-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krahn MP, Klopfenstein DR, Fischer N, Wodarz A. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr Biol. 2010;20(7):636–642. doi: 10.1016/j.cub.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 49.Horikoshi Y, Hamada S, Ohno S, Suetsugu S. Phosphoinositide binding by par-3 involved in par-3 localization. Cell Struct Funct. 2011;36(1):97–102. doi: 10.1247/csf.11005. [DOI] [PubMed] [Google Scholar]
- 50.Simões SdM, Blankenship JT, Weitz O, Farrell DL, Tamada M, Fernandez-Gonzalez R, Zallen JA. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell. 2010;19(3):377–388. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKinley RFA, Yu CG, Harris TJC. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J Cell Sci. 2012;125(Pt 5):1177–1190. doi: 10.1242/jcs.091884. [DOI] [PubMed] [Google Scholar]
- 52.Sen A, Nagy-Zsvér-Vadas Z, Krahn MP. Drosophila PATJ supports adherens junction stability by modulating Myosin light chain activity. J Cell Biol. 2012;199(4):685–698. doi: 10.1083/jcb.201206064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144(4):1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wieschaus E, Nusslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila: a practical approach. Oxford: IRL Press; 1986. pp. 199–227. [Google Scholar]
- 56.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji. An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains. Calcium- and agonist-induced dynamic changes and relationship to myo-3Hinositol-labeled phosphoinositide pools. J Cell Biol. 1998;143(2):501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.James SR, Downes CP, Gigg R, Grove SJ, Holmes AB, Alessi DR. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem J. 1996;315(Pt 3):709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20(15):4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rolls MM, Albertson R, Shih H-P, Lee C-Y, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163(5):1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19(9):723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wasserscheid I, Thomas U, Knust E. Isoform-specific interaction of Flamingo/Starry Night with excess Bazooka affects planar cell polarity in the Drosophila wing. Dev Dyn. 2007;236(4):1064–1071. doi: 10.1002/dvdy.21089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1: Localization of Baz variants in the embryonic epidermis. (A-D) Immunostainings of different GFP-Baz variants in the embryonic epidermis. All transgenes were expressed with the ubiquitin promotor in a wild-type background. GFP (green), DE-Cad (red), and Dlg (blue) were stained. (A) GFP-Baz1-968 localizes mainly at the apical junctions with some baso-lateral mislocalization. (B) Mutation of the OD domain in this truncated protein causes a cytoplasmic accumulation of GFP-Baz1-968∆OD. (C, D) Chimeric proteins that carry the pleckstrin homology (PH) domains of either human PLCδ or Akt1 fused to the C-terminus GFP-Baz∆OD1-1105 promote a cortical localization in the embryonic epidermis. (E) Western blot of embryonic lysates of the Baz variants (A-D). The Baz variants were detected with a GFP antibody and Actin was used as a loading control. Scale bars are 10 µm (TIFF 857 kb)
Supplemental Fig. 2: Cuticle phenotypes of Baz variants. (A) Cuticle phenotypes were classified into the four categories wt, shrunken with holes, holes, and cuticle rests. (B) The cuticle phenotypes of baz818−8 germ line clones that express different Baz variants with the ubiquitin promoter were quantified (n = 300 per genotype). Embryos that normally developed and hatched are also included as “hatched”. baz818−8 germ line clones display holes or cuticle rests. Embryos that express Baz-OneS hatch to a large extend (89,5%) or display either wt or shrunken with hole phenotypes. No embryos hatch upon the expression of Baz∆OD∆LB-OneS. Most embryos have either cuticle rests or holes phenotypes (58,1 and 16,3%, respectively). Nevertheless, some embryos develop further and have a wt or shrunken with holes phenotype (2,3 and 23,3%, respectively). The chimeric TEL-Baz∆OD∆LB-OneS protein rescues the Baz∆OD∆LB phenotypes to a large extent as half of the embryos hatch (50,2%) or have either wt or shrunken with holes phenotypes (21,9 and 17,9%, respectively). Scale bars are 200 µm (TIFF 417 kb)
Supplemental Fig. 3: Quantification of Baz-OneS constructs in the embryonic epithelium. (A) Scheme of the area (33 × 4 µm) of the apical region of all immunostainings in Fig. 4, which has been quantified with FIJI Plot Profile. The gray values of either Baz/aPKC (B-F) or Baz/DE-Cad (G-K) in the apical region were plotted against the distance. (L) The Pearson correlation coefficient of the co-localization of either Baz/aPKC or Baz/DE-Cad has been determined with the Coloc2 Plugin in FIJI for all genotypes in Fig. 4. For the quantification, three different embryos of each genotype have been analyzed. Bars represent the mean ± S.D. Statistics were one-way ANOVA followed by Turkey’s post hoc test, n.s. p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 (TIFF 1777 kb)
Supplemental Fig. 4: Expression of Baz variants with the UAS/Gal4-system. (A) Western blot of embryonic lysates from embryos that express Baz-GFP and Baz∆OD∆LB-GFP with the UASp promotor, driven by mat-Tub::Gal4. The Baz variants were detected with a GFP antibody and actin was used as a loading control. Full-length Baz is indicated with an arrow. Note that the pattern of specific bands is different in Baz∆OD∆LB (e.g., the band around 130 kDa is much weaker), which might be due to differences in the posttranslational processing of full-length Baz (presumably by cleavage). (B, C) Immunostaining of Baz variants (green), DE-Cad (red), and aPKC (blue) in the embryonic epidermis. (B) Baz-GFP localizes at the apical junction, (C) whereas Baz∆OD∆LB-GFP displays a diffuse cytoplasmic localization. Scale bars are 10 µm (TIFF 902 kb)