Abstract
The solution structure and hydration of the chimeric duplex [d(CGC)r(aaa)d(TTTGCG)]2, in which the central hybrid segment is flanked by DNA duplexes at both ends, was determined using two-dimensional NMR, simulated annealing and restrained molecular dynamics. The solution structure of this chimeric duplex differs from the previously determined X-ray structure of the analogous B-DNA duplex [d(CGCAAATTTGCG)]2 as well as NMR structure of the analogous A-RNA duplex [r(cgcaaauuugcg)]2. Long-lived water molecules with correlation time τc longer than 0.3 ns were found close to the RNA adenine H2 and H1′ protons in the hybrid segment. A possible long-lived water molecule was also detected close to the methyl group of 7T in the RNA–DNA junction but not with the other two thymines (8T and 9T). This result correlates with the structural studies that only DNA residue 7T in the RNA–DNA junction adopts an O4′-endo sugar conformation, while the other DNA residues including 3C in the DNA–RNA junction, adopt C1′-exo or C2′-endo conformations. The exchange rates for RNA C2′-OH were found to be ~5–20 s–1. This slow exchange rate may be due to the narrow minor groove width of [d(CGC)r(aaa)d(TTTGCG)]2, which may trap the water molecules and restrict the dynamic motion of hydroxyl protons. The minor groove width of [d(CGC)r(aaa)d(TTTGCG)]2 is wider than its B-DNA analog but narrower than that of the A-RNA analog. It was further confirmed by its titration with the minor groove binding drug distamycin. A possible 2:1 binding mode was found by the titration experiments, suggesting that this chimeric duplex contains a wider minor groove than its B-DNA analog but still narrow enough to hold two distamycin molecules. These distinct structural features and hydration patterns of this chimeric duplex provide a molecular basis for further understanding the structure and recognition of DNA·RNA hybrid and chimeric duplexes.
INTRODUCTION
DNA·RNA hybrid duplexes are formed during transcription of DNA sequences into RNA and also during reverse transcription of RNA sequences into DNA [reviewed by Ogawa and Okazaki (1)]. The structure and recognition of DNA·RNA hybrid duplexes and DNA·RNA chimeric duplexes have been the focus of numerous studies because of their crucial roles in transcription, reverse transcription and replication (2–38). The structural studies of DNA·RNA hybrid duplexes, mainly by nuclear magnetic resonance (NMR) spectroscopy, have revealed that DNA·RNA hybrid duplexes adopt neither an A-form nor a B-form structure in solution, but an intermediate heteromerous duplex structure (25). The sugars of the RNA strand have the normal N-type C3′-endo conformation, but those of the DNA strand have an intermediate O4′-endo conformation or have multiple conformers in dynamic exchange (26,33,34). The minor groove width of DNA·RNA hybrid duplexes was found to be between that of A- and B-form duplexes values (23). These structural features were used to explain the mechanism whereby RNase H discriminates between DNA·RNA hybrid duplexes and pure RNA or DNA duplexes (23).
X-ray crystallographic studies found that even the introduction of one ribonucleotide into the DNA strand transforms the whole duplex to the A-form geometry with all the sugars in the C3′-endo conformation (18–20). Similar results have been found for structures of several Okazaki fragments determined by X-ray crystallography (7,20,22). On the other hand, NMR studies have revealed that for Okazaki fragments, the DNA·RNA hybrid section assumes a conformation similar to that found for pure hybrid duplexes, while the DNA section assumes a conformation closer to B-form DNA (10–12,14,16). However, when one RNA was inserted into a DNA strand, it was found that the overall conformation of this duplex remains as B-form DNA, except that the sugar pucker of the RNA became C3′-endo (17). When a DNA duplex is flanked by DNA·RNA hybrids, the central DNA fragment was again found to adopt a B-form conformation and the transition from A-form RNA to B-form DNA involves only one nucleotide step (13).
Recently, Kochetkov and co-workers have found that RNA/DNA chimeras can be used as templates for HIV-1 reverse transcriptase (39). Hogrefe et al. have reported that DNA·RNA hybrid flanked by DNA duplexes can be recognized by Escherichia coli RNase H (40). Also, it has been proposed that RT-associated RNaseH domain, which degrades the RNA template of DNA·RNA hybrid chimeric duplexes, may distinguish double-strand RNA, RNA·DNA junctions and hybrid duplexes according to their distinct hydration patterns (41). The possible correlation between the structure and function of DNA·RNA hybrid and chimeric duplexes and their interactions with important enzymes, such as HIV reverse transcriptase and RNase H, prompted us to study the conformational properties of a chimeric duplex [d(CGC)r(aaa)d(TTTGCG)]2 in which the hybrid junctions are flanked by DNA duplexes at both ends. We have used two-dimensional NMR, simulated annealing and restrained molecular dynamics to determine the three-dimensional solution structure and hydration of this dodecamer for which the DNA and RNA analogs have been extensively studied by X-ray crystallography (42) and NMR (43).
MATERIALS AND METHODS
Sample preparations
The d(CGC)r(aaa)d(TTTGCG) dodecamer was prepared on an Applied Biosystems 380B DNA synthesizer in 6 µmol scale using solid-phase phosphoramidite chemistry (3,4). The purified sample (15 mg) was dissolved in 0.3 ml of 90% H2O:10% D2O containing 20 mM sodium phosphate, 200 mM sodium chloride and 0.05 mM EDTA, pH 7.0. For non-exchangeable proton studies, the sample was repeatedly dried from 99.96% D2O in a speed vacuum, and then dissolved in 0.3 ml of 99.996% D2O.
NMR spectroscopy
Phase sensitive NOESY and ROESY spectra in water (90% H2O:10% D2O) were recorded at 273, 278, 283 and 300 K under identical conditions except at different mixing times of 50 and 100 ms, respectively. The intense bulky water signal was suppressed using the Watergate method (44) with a short delay τ of 139 µs, resulting in optimum excitation near 1.7, 7.7 and 13.7 p.p.m. These spectra were collected into 2048 complex points in the t2 dimension and 800 complex points in the t1 dimension with relaxation delay of 1.4 s between each scan. The data was further apodized with a sin-square window function and zero-filled to 4096 complex points in the t2 dimension and 1024 complex points in the t1 dimension. For non-exchangable protons, NOESY spectra at four different mixing times of 60, 120, 240 and 480 ms were acquired during a 4-day period without removing the sample from the probe at 300 K. These phase-sensitive NOESY spectra were collected into 2048 complex points in the t2 dimension and 512 points in the t1 dimension with 32 scans per t1 experiment and a relaxation delay of 2 s between each scan. The acquired data were transferred to an IRIS Indigo 2 workstation and processed using the XWIN-NMR program (Bruker). The NOESY data sets were apodized with a shifted-sinebell window function for the first 512 points and zero filled to 2048 points in both the t2 and t1 dimensions. The DQF-COSY spectrum was recorded in the phase-sensitive mode with time-proportional phase incrementation.
Distance restraint determinations
Proton–proton distances were calculated using the NOE initial rate two-spin approximation at short mixing times normalized to the cross-relaxation rate of the cytosine H5-H6 proton pairs in NOESY spectra collected in D2O (dH5-H6 = 2.5 Å) (45). To guard against spin-diffusion errors, in the present study we initially used the qualitative NOE pattern to establish that this duplex is in the broad family of right-handed structures, and then used this information to ascertain which spin pairs are likely to have an intervening third spin that might produce errors in the calculated distance (46). For non-exchangeable protons, typical error limits for the upper and lower bound restraints were ±0.4 Å for distances <3.0 Å, ±0.8 Å for distances <4.0 Å and ±1.4 Å for distances >4.0 Å. For exchangeable protons, including imino, amino and RNA C2′-OH protons, distance restraints were set loosely to 2.0–4.0 Å for strong cross-peaks and 3.0–6.0 Å for weak cross-peaks in NOESY spectra collected in water. The restraints for Watson–Crick base pairing H-bond were defined as follows: G(N1)-C(N3) = 2.95 ± 0.2 Å, G(N2)-C(O2) = 2.86 ± 0.2 Å, G(O6)-C(N4) = 2.95 ± 0.2 Å, A(N1)-T(N3) = 2.82 ± 0.2 Å, A(N6)-T(O4) = 2.94 ± 0.2 Å.
Sugar and backbone conformations
To maintain a right-handed helix structure between A- and B-form conformation, six backbone dihedral angles restraints (α = –65° ± 60°, β = 170° ± 60°, γ = 55° ± 60°, δ = 115° ± 60° for DNA and 90° ± 60° for RNA residues, ɛ = 180° ± 60° and ζ = –75° ± 60°) were employed for each residue to exclude unreasonable geometry. Individual sugar and backbone conformation was further confirmed via a combination of NOESY distances and J-coupling data, as outlined by Reid and co-workers (10,47). This combined strategy to constrain the backbone has been applied to other DNA·RNA chimeras (11–14,16) and described in more detail elsewhere (46,47). In addition, two sugar pucker dihedral angle restraints (from a combination of NOESY and DQF-COSY data) and one χ angle restraint were also added to maintain the B-type and A-type sugar conformation for DNA and RNA residues, respectively.
Structure calculations and analysis
Three-dimensional structures of [d(CGC)r(aaa)d(TTTGCG)]2 were calculated using 530 NOE distance restraints, 30 hydrogen bond restraints, and 208 backbone, sugar and glycosidic dihedral angle restraints. Distance restraints were derived from 288 intranucleotide, 146 sequential, 6 cross-strand and 90 exchangeable proton NOEs. Initial A- and B-form DNA·RNA chimeric structures were generated using Insight II (Molecular Simulations Inc.) program. First, the simulated annealing protocol in the program X-PLOR 3.851 (A. T. Brünger, Yale University) was applied to the starting structures at 1000 K and slowly cooled to 300 K with a time step of 1.5 ps per 50 K. From the 50 simulated annealing structures, 10 structures with the lowest energy were chosen for further optimization. Restrained molecular dynamics were then carried out in vacuum with a distance-dependent dielectric constant. The dynamics were initiated at 300 K and the temperature was gradually increased to 1000 K with a time step of 0.5 ps per 50 K and then evolved for 20.0 ps at 1000 K. Subsequently, the system was cooled to 300 K in 14.0 ps and equilibrated for 9.0 ps. The coordinates saved after every 0.5 ps in the last 2.0 ps of the equilibrium were averaged, and the averaged structure was subjected to a further conjugate gradient minimization of 500 steps using Powell algorithm until a final gradient of 0.1 kcal mol–1 was reached. All the structures were finally refined by relaxation matrix based on NOE intensity (48). NOE volumes of 124 cross-peaks were integrated for each NOESY spectrum at three different mixing times (60, 120 and 240 ms) and used as restraints with uniform upper and lower bonds of ±10%. These restraints were doubled to 248 for two degenerate strands. With the incorporation of distance and dihedral restraints, the R1/6 factor (49,50) was minimized during the refinement. A cut-off of 5.5 Å was applied for relaxation calculation and an isotropic correlation time of 5.1 ns was used based on a grid search. During dynamics, the temperature of the system was slowly heated from 100 to 1000 K as the force constant of relaxation was scaled up to a final value of 400. Simultaneously, the force constants of distance restraints of non-exchangeable protons were scaled down to zero. The system was then cooled to 300 K and the final structure was subjected to conjugate gradient minimization until a final gradient of 0.1 kcal mol–1 was reached. The hydrogen bonding of base pairing and exchangeable proton distance restraints was maintained throughout the refinement.
Ten final refined structures were selected for further structural analysis. The helical parameters and torsion angles were analyzed by CURVE 5.2 (51). The family of 10 structures, together with the NMR restraints used in their determination, has been deposited with the Protein Data Bank. The chemical shifts have been deposited to the Biomagnetic Resonance Bank at the University of Wisconsin.
RESULTS
Resonance assignments and analysis of NMR data
The numbering system for [d(CGC)r(aaa)d(TTTGCG)]2 is
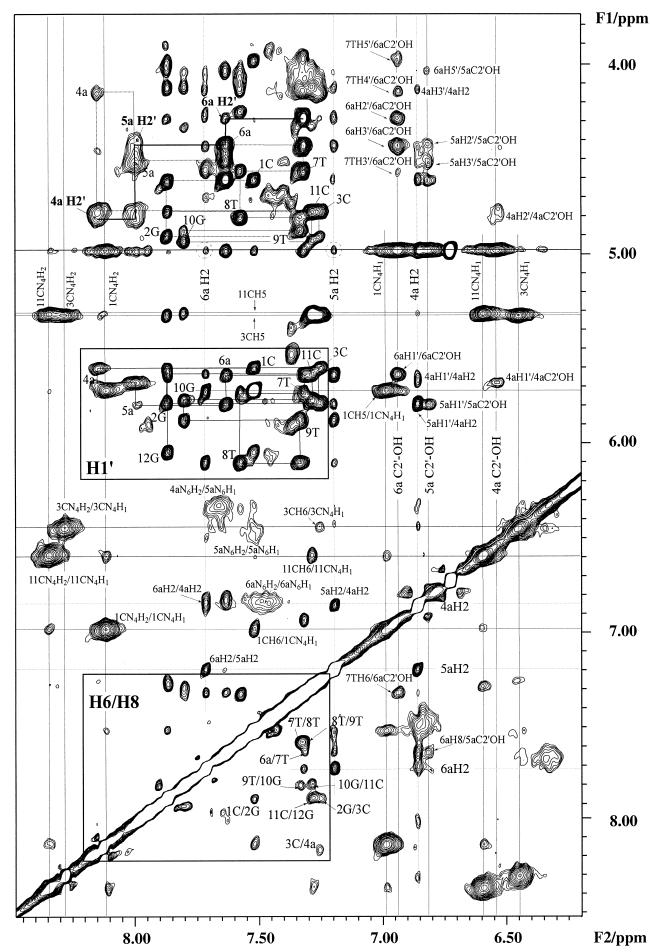
where the lower-case letters designate RNA residues. Figure 1 shows the expanded region of the 2D NOESY spectrum of [d(CGC)r(aaa)d(TTTGCG)]2 in H2O recorded at 278 K, 14.1 T. Its exchangeable and non-exchangeable proton cross-peaks were assigned using NOESY, ROESY and TOCSY spectra. The H1′ protons were assigned by standard sequential assignment method, followed by the assignment of H2′ and H2′′ protons of DNA, as well as H3′ and H4′ protons of all residues (52,53). The H2′ proton of the three RNA adenines were assigned using the H2′-H3′ cross-peaks in the TOCSY spectrum. All the three RNA adenine H2 protons showed both interresidue and intraresidue cross-peaks, particularly in the H6/H8-H1′ region (Fig. 1). The cross-peaks between RNA adenine H2 protons and water along the F2 dimension were clearly identified except the overlapped cross-peak of 4a H2 which was buried in the intense cross-peaks of hydroxyl protons. The chemical shifts of the exchangeable and non-exchangeable protons of [d(CGC)r(aaa)d(TITGCG)]2 are provided in the Supplementary Material (Table S1) available at NAR Online.
Figure 1.
Expansion of the 100 ms water NOESY spectrum of [d(CGC)r(aaa)d(TTTGCG)]2 at 278 K, 600 MHz. For the amino protons, cross-peaks are labeled according to their assignments. In the base to H1′ region, the intraresidue H6/H8-H1′ and H6/H8-H3′ cross-peaks are labeled with their residue numbers. Two dashed circles show the hydration cross-peaks of H2 protons of 5a and 6a while 4a H2 is buried under the C2′-OH cross-peaks. Three C2′-hydroxyl protons are labeled according to their chemical shifts.
In addition to the exchangeable imino and amino proton assignments, we have observed three sets of NOEs from the three labile protons resonating at 6.2–6.6 p.p.m. to the ribose protons (Fig. 1). Since the amino and imino protons of the bases have been assigned, we were left with the only remaining possibility being that the three slowly exchanging labile protons at 6.2–6.6 p.p.m. must belong to RNA 2′OH hydroxyl groups. These assignments were further supported from their cross-peaks with the RNA H1′ and H2′ protons and even the H3′, H4′, H5′/H5′′ protons (Fig. 1). These labile protons did not show any detectable NOEs to any imino protons, which was also consistent with their assignments as ribose 2′OH protons. Their chemical shifts (6.2–6.6 p.p.m.) are in agreement with previously published assignments of ribose hydroxyl protons (24,54,55).
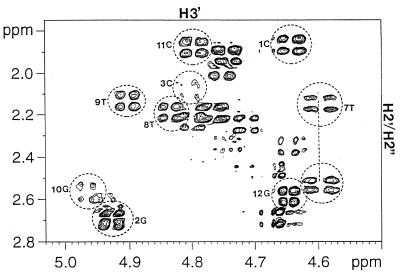
In the NOESY data collected in D2O for the H8/H6 to the DNA H2′/H2′′ region (data not shown), the (n)H6/H8 to (n)H2′ peaks were the most intense and were stronger than the (n)H6/H8 to (n-1)H2′′ peaks, which in turn were uniformly stronger than the (n)H6/H8 to (n)H2′′ and (n)H6/H8 to (n-l)H2′ peaks. These results indicate that none of the DNA residue in this chimeric duplex assumes a pure A-form conformation in solution. Furthermore, all the DNA sugar residues have quite respectable H1′-H2′ and H3′-H4′ cross peaks in the DQF-COSY spectrum indicating that sugar conformations for the DNA residues are not A-form (data not shown). The DNA residue 7T in the RNA–DNA junction, however, was found to have medium to strong H2′′-H3′ DQF-COSY cross peak (Fig. 2). The other DNA residues, including residue 3C in the DNA–RNA junction, showed no detectable H2′′-H3′ cross peaks in the DQF-COSY spectrum. The above results indicate that residue 7T in the RNA–DNA junction adopts an O4′-endo sugar conformation, while the other DNA residues adopt C1′-exo or C2′-endo conformations (10). The H1′-H2′ DQF-COSY cross peak of RNA residue 4a at the DNA–RNA junction was found to be detectable but less intense than that of the DNA residues. No detectable H1′-H2′ DQF-COSY cross peaks for RNA residues 5a and 6a were observed. Also, strong (n)H8 to (n–1)H2′ NOESY cross peaks were found for the RNA residues indicating that the RNA segment in the chimeric strand assumes an A-form conformation.
Figure 2.
Expansion of the H2′/H2′′-H3′ region of the DQF-COSY spectrum of [d(CGC)r(aaa)d(TTTGCG)]2 showing the H2′/H2′′-H3′ cross peak for DNA residue 7T. The other DNA residues, including residue 3C in the DNA–RNA junction, showed no detectable H2′′-H3′ DQF-COSY cross-peaks.
Hydration of [d(CGC)r(aaa)d(TTTGCG)]2
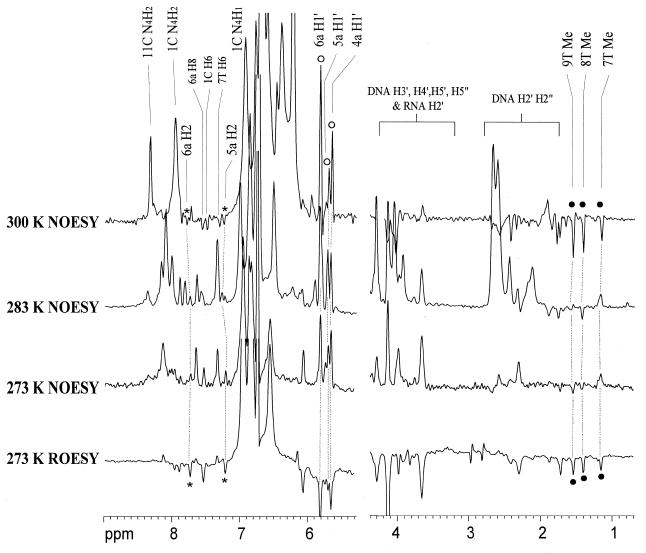
The qualitative and quantitative water hydration residence time was determined by the cross-relaxation rate constants using NOESY and ROESY spectra (56,57). Comparison of the 1D cross-sections of NOESY and ROESY spectra at the water chemical shift along the F2 dimension showed the evidences for hydration (Fig. 3). It is important to note that coincident chemical shifts, especially H3′ and H4′ protons of nucleic acids, can also give cross-peaks along the water chemical shift. This coincidence was ruled out by performing experiments under different temperatures, which gave different water chemical shifts to confirm the water signals. The phase sensitive NOESY and ROESY spectra recorded in water at 273, 283 and 300 K were used to identify the water chemical shift and hence the hydration peaks. Long-lived water molecules (correlation time τc >0.3 ns), with opposite signs of NOE and ROE, were found close to all H2 and H1′ protons of the RNA adenines, except for 4a H2, which was buried under the intense hydroxyl signals at 6.1–6.7 p.p.m. Similar water-H1′ cross-peaks in RNA were also observed previously (55). In contrast, cross-peaks to the H1′ protons are usually not detected in DNA except at the ends of the duplex or at much longer mixing times. The increase in water-H1′ cross peak intensities with increasing temperature could be explained as NOEs with the 2′OH which are relayed to the water resonance by chemical exchange (Fig. 3). The methyl group of 7T exhibited a positive NOE to water at 273 K (Fig. 3), i.e. with a water correlation time longer than 0.3 ns, then changed to a negative NOE when the temperature was raised to 300 K (Fig. 3). However, NOE peaks of the other two methyl groups (8T and 9T), more distant from the center of the duplex, were negative under all temperatures. Such positive NOE peak to water of the 7T methyl group was not observed in pure DNA duplexes (58–61).
Figure 3.
Cross-sections through the water chemical shift along the F1 dimension of [d(CGC)r(aaa)d(TTTGCG)]2 at 273, 283 and 300 K with mixing time of 100 and 50 ms respectively for NOESY and ROESY spectra. The hydration peaks of adenine H2 and H1′ protons are labeled with asterisks and open circles respectively. The hydration peaks of thymine methyl protons are labeled with filled circles.
Orientation and exchange rate of C2′-hydroxyl protons
Molecular modeling studies of the RNA duplexes and the DNA·RNA hybrids showed that there are three stable rotamers for the RNA C2′-OH hydroxyl groups (62). In these rotamers, the C2′-OH can have hydrogen bonds with O3′ of the 3′-phosphate [φ = 90°, when the C2′-OH-H1′ distance is large, ~3.5 Å, and 3JH2′-C2′-OH <4 Hz], with O4′ of either the neighboring 3′-ribose or in the same sugar [φ = 180°, when the C2′-OH-H1′ distance is short, <2.5 Å, and 3JH2′-C2′-OH = 10–12 Hz], or toward the attached base [φ = 0 to –30°, when the C2′-OH-H1′ distance <2.5 Å, and 3JH2′-C2′-OH = 4–7 Hz] (63). Among these orientations, the O3′ direction is the most favorable for hydrogen bonding framework (62). As we can see from Figure 1, the observed 4a 2′OH-H1′, 5a 2′OH-H1′ and 6a 2′OH-H1′ NOE distances were all close to or even larger than 3.5 Å using the cytosine H5-H6 distance (2.5 Å) as a standard. Thus, all of the three RNA C2′-OH groups tend to orient toward the O3′ direction and hence the small 3JH2′-C2′-OH values. This was further confirmed by the correlation spectroscopy. We did not observe any cross-peak of the three RNA 2′OH protons from TOCSY spectra (mixing times of 20, 46, 200 and 500 ms at pH 5.5–7.5, 273 and 278 K) due to the possible O3′ domain conformation (3JH2′-C2′-OH <4 Hz). Besides, many cross-peaks were detected along the C2′-OH chemical shift in the NOESY spectrum recorded at 278 K (Fig. 1). There were more and stronger cross-peaks along the chemical shift of 6a C2′-OH than of 5a and 4a, which in turn had few, weak cross-peaks. Presumably, the hydration pattern and geometry of the grooves, especially the minor groove in the hybrid segment may be the cause of this trend. Since we have also observed distinct hydration pattern in the center of the hybrid segment (i.e. a water correlation time was observed longer than 0.3 ns at the 7T methyl group but not 8T and 9T methyl groups which are more distant from the center of the duplex), there may be long-lived water molecules between 5a and 6a 2′OH protons to mediate stronger NOEs to the ribose protons. Although the possibility of multiple rotamers existence cannot be ruled out for 2′OH groups, the O3′ orientation is still the most preferred one compared with O4′ and base orientations, due to the large distances between 2′OH and H1′ protons.
The exchange rate of the C2′-hydroxyl protons with solvent water was obtained from measurement of the line width of the hydroxyl proton cross-peaks in NOESY spectra along the water chemical shift in the F2 dimension (63). As the C2′-OH peaks were partially overlapped, we first did decomposition for overlapping to obtain individual line-widths by assuming all peaks had Lorentzian line-shapes. However, due to overlapping and other unresolved scalar coupling, the decomposed line-widths gave us only the estimation of the upper limit of the exchange rate. By performing analysis at several temperatures ranging from 273 to 288 K, we obtained the C2′-OH exchange rate with an overall range of ~5–20 s–1. This exchange rate is lower than other DNA·RNA hybrid duplexes studied previously (k = 26.7 ± 13.8 s–1), which had r(gaga) sequence in the central region (63).
Structural features of [d(CGC)r(aaa)d(TTTGCG)]2
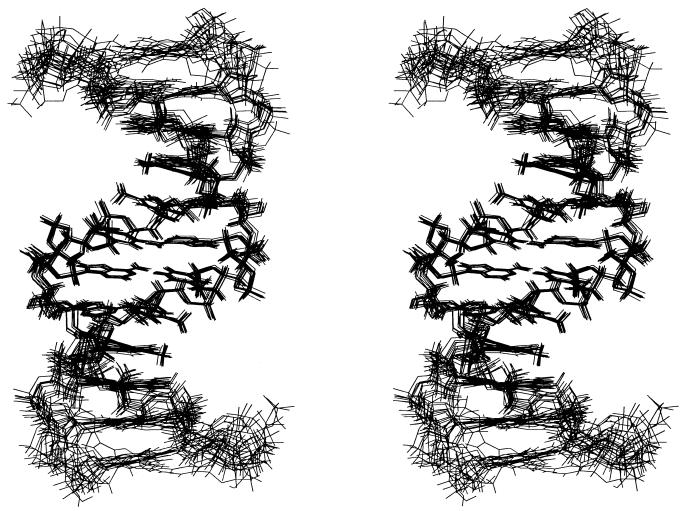
A total of 10 final simulated annealing, restrained molecular dynamics, and NOE relaxation matrix refined structures (five from initial A-form and five from initial B-form structures) were superimposed and are shown in Figure 4. The energetic and geometric statistics of the 10 best-refined structures are given in Table 1. All of the final structures were well defined. For the hybrid segment, the average pairwise r.m.s.d. (heavy atoms) was found to be 0.43 ± 0.08 Å and 1.25 ± 0.36 Å for the overall structures (Table 1). The average R1/6 factor was 0.0266 ± 0.0003, indicating the results correlate well with the experimental data.
Figure 4.
Stereoview of the superimposed 10 final refined structures of [d(CGC)r(aaa)d(TTTGCG)]2.
Table 1. Structure statistics for the 10 final refined structures of [d(CGC)r(aaa)d(TTTGCG)]2.
| NOE restraints | 530 | ||
| intranucleotidea | 288 | ||
| sequentiala | 146 | ||
| cross-strand | 6 | ||
| involving exchangeable protonsb | 90 | ||
| Torsion angle restraintsc | 208 | ||
| Hydrogen bond restraints | 30 | ||
| Relaxation matrix refinement | |||
| number of peak integrals used at each mixing timed | 248 | ||
| average R1/6 factor | 0.0266 ± 0.0003 | ||
| Refinement statistics (10 final lowest-energy structures) | |||
| NOE violation >0.5 Å | <2 | ||
| dihedral angle violation >5° | 0 | ||
| average pairwise r.m.s.d. | |||
| all heavy atom | 1.25 ± 0.36 | ||
| without 5′ and 3′ terminus | 0.81 ± 0.26 | ||
| hybrid segmente | 0.43 ± 0.08 | ||
| average r.m.s.d. from ideal covalent geometry | |||
| bond lengths (Å) | 0.0085 ± 0.0001 | ||
| bond angles (˚) | 3.31 ± 0.12 | ||
| impropers (˚) | 0.34 ± 0.01 |
aThe NOE restraints were for the two degenerate strands.
bTwenty-two imino proton to non-exchangeable proton NOEs, 20 amino proton to exchangeable proton NOEs, 18 imino and amino proton NOEs, and 30 C2′-hydroxyl proton to non-exchangeable proton NOEs.
cSee Materials and Methods.
dCross-peak volumes were measured in three NOESY spectra with mixing times of 60, 120 and 240 ms. Number of peaks were for the two degenerate strands.
eHybrid segment is defined as 4a to 6T, i.e. [r(aaa)d(TTT)]2.
Previously, the X-ray structure of the DNA duplex, [d(CGCAAATTTGCG)]2, was found to be of B-type (42) and the structure of the RNA duplex, [r(cgcaaauuugcg)]2, was determined to be an A-type by NMR spectroscopy (43). The r.m.s.d. values between the X-ray derived B-DNA duplex (A3T3), the NMR derived A-RNA duplex (A3U3), and the NMR derived chimeric duplex (a3T3, this study) are listed in Table 2. The global conformation of [d(CGC)r(aaa)-d(TTTGCG)]2 appears to be between its A-RNA and B-DNA analogs. However, some of the local structural parameters of [d(CGC)r(aaa)d(TTTGCG)]2 were more similar to its B-DNA analog than to its A-RNA analog, particularly with respect to the minor groove width (Fig. 5A). The overall final refined structures of [d(CGC)r(aaa)d(TTTGCG)]2 do not assume an A- or B-type conformation which is consistent with the earlier studies of DNA–RNA chimeric duplexes (10–17).
Table 2. r.m.s.d. values between different structures (Å).
| Structure | a3T3 | A3T3 | A3U3 | A-DNA | B-DNA | A-RNA |
|---|---|---|---|---|---|---|
| a3T3 | ||||||
| A3T3 | 4.21 ± 0.15 | |||||
| A3U3 | 4.39 ± 0.21 | 4.82 | ||||
| A-DNA | 6.18 ± 0.21 | 6.55 | 3.67 | |||
| B-DNA | 4.86 ± 0.10 | 2.83 | 5.39 | 6.50 | ||
| A-RNA | 5.43 ± 0.23 | 6.12 | 3.33 | 1.30 | 5.98 |
r.m.s.d. values were calculated pairwise using the best 10 structures. a3T3 represents [d(CGC)r(aaa)d(TTTGCG)]2, A3T3 represents [d(CGCAAATTTGCG)]2, A3U3 represents [r(cgcaaauuugcg)]2, and A-DNA, B-DNA, A-RNA represent the standard A- and B-form DNA and A-form RNA.
Figure 5.
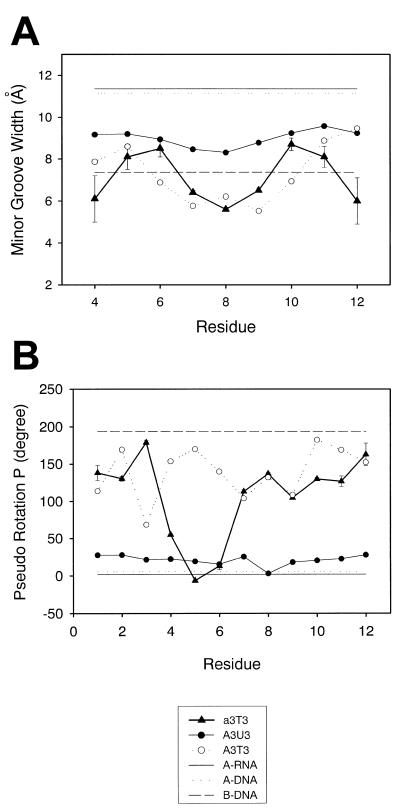
(A) Minor groove width as a function of position of the residue along the [d(CGC)r(aaa)d(TTTGCG)]2 duplex (triangles). The width is represented by the (n)P-(m+3)P phosphorus distance minus the phosphorus van der Waals radius of 5.8 Å. (B) Pseudo rotational phase angles (P) in the 10 final refined structures (triangles). The corresponding values for its X-ray derived B-DNA (open circles) and NMR derived A-RNA (closed circles) analogs and for B-DNA (dashed line), A-DNA (dotted line) and A-RNA (continuous line) are also plotted for comparison.
Figure 5B shows the sugar puckers for the final NMR refined solution structures compared to its DNA and RNA analogs as well as standard B- and A-form DNA and A-form RNA. The chimeric feature of the structure of the hybrid chimeric duplex in solution is apparent from Figure 5B. The sugar conformation of DNA residue 7T in the RNA–DNA junction was in the O4′-endo range which was consistent with the spectral analysis. The sugar P value of RNA residue 4a in the DNA–RNA junction was located around 60°, which was also consistent with the DQF-COSY data. The sugar conformation of the rest of the DNA residues in this chimeric duplex is in the S-type range (C1′-exo to C2′-endo) while the RNA residues are in the N-type range (C3′-endo). Other structural features (i.e. helical twists, X-displacement, rise, etc.) of [d(CGC)r(aaa)d(TTTGCG)]2 are shown in Table 3.
Table 3. Helical parameters for [d(CGC)r(aaa)d(TTTGCG)]2.
| Base | X disp (Å) | Buckle (°) | Pr. twist (°) | Incl. (°) | Twist (°) | Rise (Å) |
|---|---|---|---|---|---|---|
| 1C | –2.1 ± 0.4 | 5.4 ± 4.8 | –18.7 ± 3.6 | 0.6 ± 6.9 | 34.0 ± 6.2 | 3.5 ± 0.4 |
| 2G | –1.7 ± 0.5 | –1.2 ± 6.3 | –6.0 ± 4.1 | –2.6 ± 6.6 | 35.7 ± 1.1 | 3.3 ± 0.1 |
| 3C | –2.0 ± 0.4 | 0.5 ± 3.0 | –5.5 ± 2.1 | –4.3 ± 5.9 | 28.4 ± 0.9 | 3.0 ± 0.2 |
| 4a | –1.6 ± 0.3 | –4.2 ± 1.6 | –16.6 ± 2.2 | –2.0 ± 4.7 | 34.7 ± 1.9 | 3.0 ± 0.1 |
| 5a | –2.0 ± 0.3 | 4.5 ± 2.1 | –36.6 ± 1.4 | 0.4 ± 3.6 | 31.8 ± 1.0 | 3.1 ± 0.1 |
| 6a | –1.6 ± 0.3 | –4.5 ± 1.0 | –33.5 ± 0.8 | –1.7 ± 3.2 | 30.1 ± 0.3 | 2.6 ± 0.1 |
| 7T | –1.6 ± 0.3 | 3.9 ± 1.1 | –33.3 ± 1.0 | –1.7 ± 3.2 | 33.1 ± 0.9 | 3.6 ± 0.1 |
| 8T | –2.0 ± 0.3 | –6.0 ± 1.6 | –36.4 ± 1.7 | 0.7 ± 3.6 | 42.3 ± 0.5 | 2.7 ± 0.1 |
| 9T | –1.6 ± 0.3 | 3.8 ± 1.4 | –15.5 ± 2.6 | –1.5 ± 4.5 | 22.9 ± 1.1 | 3.3 ± 0.1 |
| 10G | –2.0 ± 0.4 | 0.6 ± 3.3 | –6.8 ± 3.0 | –3.6 ± 5.6 | 38.2 ± 1.4 | 2.9 ± 0.2 |
| 11C | –1.7 ± 0.6 | 2.5 ± 4.9 | –8.9 ± 2.7 | –1.7 ± 6.4 | 28.3 ± 4.6 | 3.5 ± 0.6 |
| 12G | –2.2 ± 0.4 | –10.6 ± 13.6 | –21.1 ± 8.5 | 1.7 ± 6.4 | – | – |
| Average | –1.8 ± 0.1 | –0.4 ± 3.6 | –19.9 ± 2.1 | –1.3 ± 1.4 | 32.7 ± 1.9 | 3.1 ± 0.2 |
| A3T3 | –0.3 ± 0.3 | 1.0 ± 7.4 | –16.0 ± 11.0 | 0.7 ± 3.2 | 36.0 ± 4.2 | 3.4 ± 0.2 |
| A3U3 | –4.8 ± 0.1 | 1.8 ± 5.2 | –24.7 ± 4.0 | 10.7 ± 4.9 | 32.3 ± 1.8 | 2.7 ± 0.4 |
| A-RNA | –5.3 | 0.0 | 14.4 | 15.9 | 31.5 | 3.5 |
| A-DNA | –5.4 | 0.0 | 13.7 | 19.1 | 30.9 | 3.4 |
| B-DNA | –0.7 | 0.0 | 3.7 | –5.9 | 36.0 | 3.4 |
Helical parameters were calculated using the program Curves v.5.2. A3T3 represents [d(CGCAAATTTGCG)]2, A3U3 represents [r(cgcaaauuugcg)]2, and A-DNA, B-DNA, A-RNA represent the standard A- and B-form DNA and A-form RNA.
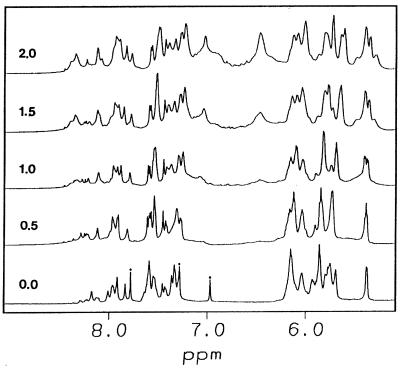
The width of the minor groove, as measured by the cross-strand phosphate–phosphate separation, was also significantly affected by the conformation of both strands. The minor groove width for [d(CGC)r(aaa)d(TTTGCG)]2 was wider than its B-DNA analog, but narrower than that of the RNA analog (Fig. 5A). Wemmer and co-workers observed that two distamycin molecules can bind in an antiparallel, side-by-side fashion to the minor groove of DNA duplexes (reviewed in 64–66). Their studies indicate that the drug affinity was affected by the width of the minor groove. Also, it was observed that there was no binding of distamycin with RNA duplexes due to their wide and shallow minor groove (67). Thus, the binding mode of distamycin to various nucleic acid duplexes can be used as a probe for the minor groove width. We have examined the binding mode of distamycin with [d(CGC)r(aaa)d(TTTGCG)]2, for which the minor groove is wider than its DNA analog but narrower than its RNA analog. It is evident from Figure 6 that the titration of this chimeric duplex [d(CGC)r(aaa)-d(TTTGCG)]2 with distamycin results in an increase in broadness of NMR signals at the binding sites of the ligand and the duplex (especially for the 4a, 5a and 6a H2 protons). This is probably due to slow hopping between the sites within the duplex or from slow dissociation of the complex (or both) (64). The possible 2:1 binding mode found in the titration experiments suggests that [d(CGC)r(aaa)d(TTTGCG)]2 has an intrinsically wider minor groove than its B-DNA analog but still narrow enough to hold two distamycin molecules.
Figure 6.
Titration of [d(CGC)r(aaa)d(TTTGCG)]2 with distamycin at 30°C. The ligand:duplex ratio is indicated for each spectrum. The asterisks indicate the adenosine H2 resonances.
DISCUSSION
The solution structure of [d(CGC)r(aaa)d(TITGCG)]2 is different from either the uniformly A-type RNA duplex [r(cgcaaauuugcg)]2 found in the solution state (43), or from the related B-type DNA duplex [d(CGCAAATTTGCG)]2 in crystalline state (42). The structure of [d(CGC)r(aaa)d(TTTGCG)]2 exhibits properties of a chimeric mixture of A-form and B-form, similar to other chimeric duplexes determined in solution (10–17).
With respect to the DNA duplex–hybrid duplex–DNA duplex junctions, only the 7T·18a (or the symmetrical 19T·6a) DNA base pair in the hybrid junction is involved in the structural transition. Sugar conformation of 7T (19T) was found to be in the O4′-endo conformation based on both NOESY and DQF-COSY spectra (Fig. 2). Also, quite high buckle and propeller twist values were observed between the junction base pairs and the hybrid segment (Table 3). A similar highly buckled structure was observed for the junction residues 4g·21C and 5T·20A in the solution structure of [r(cgcg)d(TATACGCG)]2 (13). Sugar conformation of DNA residue 3C (15C) in the DNA duplex–hybrid duplex junction, however, was found to be in the normal C1′-exo to C2′-endo conformation (Fig. 5A). Thus, structural parameters of DNA residue at 5′-end of a DNA–hybrid junction are not affected by the A-type RNA at 3′-end. Instead, changes in the structural parameters were observed for DNA residue at 3′-end of a hybrid junction by the A-type RNA at 5′-end. This influence was however involved in only one step.
The width of the minor groove is an important structural parameter for DNA·RNA hybrid and chimeric duplexes. Previous studies have shown that the minor groove width of DNA·RNA hybrid and chimeric duplexes was 8.5 Å compared to that of 11 Å for A-form and 7.5 Å for B-form duplexes (23) and was responsible for the recognition and cleavage activity by RNase H (23). In the present study we observed that although the global structure of [d(CGC)r(aaa)d(TTTGCG)]2, was in between its A-type RNA and B-type DNA analogs (Table 2), its minor groove width was found to be closer to the B-DNA analog than to the A-RNA duplex. This was further confirmed by the distamycin titration data that [d(CGC)r(aaa)d(TTTGCG)]2 may have an intrinsically wider minor groove than its B-DNA analog but still narrow enough to hold two distamycin molecules.
In addition to the structural features, we have also studied the hydration and dynamics of RNA C2′-OH in [d(CGC)r(aaa)d(TTTGCG)]2. Long-lived water molecules with correlation time τc longer than 0.3 ns were found close to the RNA adenine H2 and H1′ protons in the hybrid segment. A possible long-lived water molecule was also detected close to the methyl group of 7T in the RNA–DNA junction. It is reported in an X-ray structural study of the RNA duplex [r(CCCCGGGG)]2 that the ribose 2′OH group of the 5′-residue which lock the sugar-phosphate backbone in a C3′-endo conformation allows water molecules to bridge adjacent phosphates in the backbone and hence stabilize the major groove hydration network (68). In the present study, the hydration network in the major groove of DNA residue 7T at the RNA–DNA junction is stabilized by a C3′-endo conformation of the preceding 5′-RNA. This explains why a long-lived water molecule was detected close to the methyl group of 7T but not to the other two thymines (8T and 9T). This specific hydration pattern also correlates well with the structural studies that only DNA residue 7T at the RNA–DNA junction adopts an O4′-endo sugar conformation, while the other DNA residues including 3C in the DNA–RNA junction, adopt C1′-exo or C2′-endo conformations. Based on the NOE cross-peak patterns and the small coupling constants from TOCSY spectra (3JH2′-C2′-OH <4 Hz), we have found that the RNA C2′-OH groups tends to orient toward the O3′ direction. Molecular modeling studies of the RNA duplexes and the DNA·RNA hybrids have also shown that in the O3′ direction, the RNA C2′-OH may form a possible hydrogen bond with the 3′-phosphate group (62). The exchange rates for RNA C2′-OH were found to be around 5–20 s–1, compared to 26.7 ± 13.8 s–1 reported previously for the other DNA·RNA hybrid duplex (63). This slow exchange rate may be due to the narrow minor groove width of [d(CGC)r(aaa)-d(TTTGCG)]2, which may trap the water molecules and restrict the dynamic motion of hydroxyl protons.
In conclusion, we have determined the solution structure of the chimeric duplex [d(CGC)r(aaa)d(TTTGCG)]2, where the central hybrid region is flanked by DNA duplexes at both ends. The characteristic structural features and hydration patterns of this chimeric duplex may provide a molecular basis for further understanding the structure and recognition of DNA·RNA hybrid and chimeric duplexes.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Moti L. Jain for revising the English for us, Professor Shan-Ho Chou for synthesizing the samples, Professor Dinshaw J. Patel, Dr Chin H. Lin and Dr Andrew N. Lane for helpful discussions. This study was carried out on the 600 MHz NMR spectrometer at the Regional Instrument Center at Hsinchu, National Science Council, Taiwan, Republic of China. We thank the National Science Council of the Republic of China for research grants.
REFERENCES
- 1.Ogawa T. and Okazaki,T. (1980) Annu. Rev. Biochem., 49, 421–457. [DOI] [PubMed] [Google Scholar]
- 2.Arnott S., Chandrasekaran,R., Millane,R.P. and Park,H.-S. (1986) J. Mol. Biol., 188, 631–640. [DOI] [PubMed] [Google Scholar]
- 3.Chou S.-H., Flynn,P. and Reid,B.R. (1989) Biochemistry, 28, 2422–2435. [DOI] [PubMed] [Google Scholar]
- 4.Chou S.-H., Flynn,P., Wang,A. and Reid,B. (1991) Biochemistry, 30, 5248–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katahira M., Lee,S.J., Kobayashi,Y., Sugeta,H., Kyogoku,Y., Iwai,S., Ohtsuka,E., Benevides,J.M. and Thomas,G.J.J. (1990) J. Am. Chem. Soc., 112, 4508–4512. [Google Scholar]
- 6.Steely H.T.J., Gray,D.M. and Ratliff,R.L. (1986) Nucleic Acids Res., 14, 10071–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang A.H.-J., Fujii,S., van Boom,J.H., van der Marel,G.A., van Boeckel,S.A.A. and Rich,A. (1982) Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- 8.Hall K.B. and McLaughlin,L.W. (1991) Biochemistry, 30, 10606–10613. [DOI] [PubMed] [Google Scholar]
- 9.Haasnoot C.A.G., Westerink,H.P., van der Marel,G.A. and van Boom,J.H. (1983) J. Biomol. Struct. Dyn., 1, 131–149. [DOI] [PubMed] [Google Scholar]
- 10.Salazar M., Champoux,J.J. and Reid,B.R. (1993) Biochemistry, 32, 739–744. [DOI] [PubMed] [Google Scholar]
- 11.Fedoroff O.Y., Salazar,M. and Reid,B.R. (1996) Biochemistry, 35, 11070–11080. [DOI] [PubMed] [Google Scholar]
- 12.Salazar M., Fedoroff,O.Y. and Reid,B.R. (1996) Biochemistry, 35, 8126–8135. [DOI] [PubMed] [Google Scholar]
- 13.Zhu L., Salazar,M. and Reid,B.R. (1995) Biochemistry, 34, 2372–2380. [DOI] [PubMed] [Google Scholar]
- 14.Salazar M., Fedoroff,O.Y., Zhu,L. and Reid,B.R. (1994) J. Mol. Biol., 241, 440–455. [DOI] [PubMed] [Google Scholar]
- 15.Nishizaki T., Iwai,S., Ohtsuka,E. and Nakamura,H. (1997) Biochemistry, 36, 2577–2585. [DOI] [PubMed] [Google Scholar]
- 16.Nishizaki T., Iwai,S., Ohkubo,T., Kojima,C., Nakamura,H., Kyogoku,Y. and Ohtsuka,E. (1996) Biochemistry, 35, 4016–4025. [DOI] [PubMed] [Google Scholar]
- 17.Jaishree T.N., van der Marel,G.A., van Boom,J.H. and Wang,A.H.J. (1993) Biochemistry, 32, 4903–4911. [DOI] [PubMed] [Google Scholar]
- 18.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1994) J. Mol. Biol., 236, 275–285. [DOI] [PubMed] [Google Scholar]
- 19.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1994) Nucleic Acids Res., 22, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egli M., Usman,N. and Rich,A. (1993) Biochemistry, 32, 3221–3237. [PubMed] [Google Scholar]
- 21.Usman N., Egli,M. and Rich,A. (1992) Nucleic Acids Res., 20, 6695–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egli M., Usman,N., Zhang,S. and Rich,A. (1992) Proc. Natl Acad. Sci. USA, 89, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedoroff O.Y., Salazar,M. and Reid,B.R. (1993) J. Mol. Biol., 233, 509–523. [DOI] [PubMed] [Google Scholar]
- 24.Fedoroff O.Y., Ge,Y. and Reid,B.R. (1997) J. Mol. Biol., 269, 225–239. [DOI] [PubMed] [Google Scholar]
- 25.Salazar M., Fedoroff,O.Y., Miller,J.M., Ribeiro,N.S. and Reid,B.R. (1993) Biochemistry, 32, 4207–4215. [DOI] [PubMed] [Google Scholar]
- 26.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- 27.Hashem G.M., Pham,L., Vaughan,M.R. and Gray,D.M. (1998) Biochemistry, 37, 61–72. [DOI] [PubMed] [Google Scholar]
- 28.Ratmeyer L., Vinayak,R., Zhong,Y.Y., Zon,G. and Wilson,W.D. (1994) Biochemistry, 33, 5298–5304. [DOI] [PubMed] [Google Scholar]
- 29.Cross C.W., Rice,J.S. and Gao,X. (1997) Biochemistry, 36, 4096–4107. [DOI] [PubMed] [Google Scholar]
- 30.Rice J. and Gao,X. (1997) Biochemistry, 36, 399–411. [DOI] [PubMed] [Google Scholar]
- 31.Mujeeb A., Reynolds,M.A. and James,T.L. (1997) Biochemistry, 36, 2371–2379. [DOI] [PubMed] [Google Scholar]
- 32.Gyi J.I., Conn,G.L., Lane,A.N. and Brown,T. (1996) Biochemistry, 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez C., Stec,W., Reynolds,M.A. and James,T.L. (1995) Biochemistry, 34, 4969–4982. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez C., Stec,W., Kobylanska,A., Hogrefe,R.I., Reynolds,M. and James,T.L. (1994) Biochemistry, 33, 11062–11072. [DOI] [PubMed] [Google Scholar]
- 35.Gao X. and Jeffs,P.W. (1994) J. Biomol. NMR, 4, 367–384. [DOI] [PubMed] [Google Scholar]
- 36.Maltseva T.V., Agback,P., Repkova,M.N., Venyaminova,A.G., Ivanova,E.M., Sandstrom,A., Zarytova,V.F. and Chattopadhyaya,J. (1994) Nucleic Acids Res., 22, 5590–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebel S., Brown,T. and Lane,A.N. (1994) Eur. J. Biochem., 220, 703–715. [DOI] [PubMed] [Google Scholar]
- 38.Lane A.N., Ebel,S. and Brown,T. (1993) Eur. J. Biochem., 215, 297–306. [DOI] [PubMed] [Google Scholar]
- 39.Gudima S.O., Kazantseva,E.G., Kostyuk,D.A., Shchaveleva,I.L., Grishchenko,O.I., Memelova,L.V. and Kochetkov,S.N. (1997) Nucleic Acids Res., 25, 4614–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogrefe H.H., Hogrefe,R.I., Walder,R.Y. and Walder,J.A. (1990) J. Biol. Chem., 265, 5561–5566. [PubMed] [Google Scholar]
- 41.Szyperski T., Gotte,M., Billeter,M., Perola,E., Cellai,L., Heumann,H. and Wuthrich,K. (1999) J. Biomol. NMR, 13, 343–355. [DOI] [PubMed] [Google Scholar]
- 42.Edwards K.J., Brown,D.G., Spink,N., Skelly,J.V. and Neidle,S. (1992) J. Mol. Biol., 226, 1161–1173. [DOI] [PubMed] [Google Scholar]
- 43.Conte M.R., Conn,G.L., Brown,T. and Lane,A.N. (1997) Nucleic Acids Res., 25, 2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piotto M., Saudek,V. and Sklenar,V. (1992) J. Biomol. NMR, 2, 661–665. [DOI] [PubMed] [Google Scholar]
- 45.Noggle J.H. and Schirmer,R.E. (1971) The Nuclear Overhauser Effect. Academic Press, New York, NY.
- 46.Cheng J.-W., Chou,S.-H., Salazar,M. and Reid,B.R. (1992) J. Mol. Biol., 228, 118–137. [DOI] [PubMed] [Google Scholar]
- 47.Kim S.-G., Lin,L.-J. and Reid,B.R. (1991) Biochemistry, 31, 3564–3574. [DOI] [PubMed] [Google Scholar]
- 48.Yip P. and Case,D.A. (1989) J. Magn. Reson., 83, 643–648. [Google Scholar]
- 49.Nilges M., Habazettl,J., Brunger,A.T. and Holak,T.A. (1991) J. Mol. Biol., 219, 499–510. [DOI] [PubMed] [Google Scholar]
- 50.White S.A., Nilges,M., Huang,A. and Brunger,A.T. (1992) Biochemistry, 31, 1610–1621. [DOI] [PubMed] [Google Scholar]
- 51.Lavery R. and Sklenar,H. (1989) J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 52.Hare D.R., Wemmer,D.E., Chou,S.-H., Drobny,G. and Reid,B.R. (1983) J. Mol. Biol., 171, 319–336. [DOI] [PubMed] [Google Scholar]
- 53.Wuthrich K. (1986) NMR of Proteins and Nucleic Acids. John Wiley & Sons, New York, NY.
- 54.Leroy J.L., Broseta,D. and Gueron,M. (1985) J. Mol. Biol., 184, 165–178. [DOI] [PubMed] [Google Scholar]
- 55.Conte M.R., Conn,G.L., Brown,T. and Lane,A.N. (1996) Nucleic Acids Res., 24, 3693–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otting G., Liepinsh,E. and Wuthrich,K. (1991) Science, 254, 974–980. [DOI] [PubMed] [Google Scholar]
- 57.Lane A.N., Jenkins,T.C. and Frenkiel,T.A. (1997) Biochim. Biophys. Acta, 1350, 205–220. [DOI] [PubMed] [Google Scholar]
- 58.Jacobson A., Leupin,W., Liepinsh,E. and Otting,G. (1996) Nucleic Acids Res., 24, 2911–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liepinsh E., Otting,G. and Wuthrich,K. (1992) Nucleic Acids Res., 20, 6549–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liepinsh E., Leupin,W. and Otting,G. (1994) Nucleic Acids Res., 22, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sunnerhagen M., Denisov,V.P., Venu,K., Bonvin,A.M.J.J., Carey,J., Halle,B. and Otting,G. (1998) J. Mol. Biol., 282, 847–858. [DOI] [PubMed] [Google Scholar]
- 62.Auffinger P. and Westhof,E. (1997) J. Mol. Biol., 274, 54–63. [DOI] [PubMed] [Google Scholar]
- 63.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Nucleic Acids Res., 26, 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wemmer D.E. and Dervan,P.B. (1997) Curr. Opin. Struct. Biol., 7, 355–361. [DOI] [PubMed] [Google Scholar]
- 65.Fagan P. and Wemmer,D.E. (1992) J. Am. Chem. Soc., 114, 1080–1081. [Google Scholar]
- 66.Pelton J.G. and Wemmer,D.E. (1990) J. Am. Chem. Soc., 112, 1393–1399. [Google Scholar]
- 67.Wilson W.D., Ratmeyer,L., Zhao,M., Strekowski,L. and Boykin,D. (1993) Biochemistry, 32, 4098–4104. [DOI] [PubMed] [Google Scholar]
- 68.Egli M., Portmann,S. and Usman,N. (1996) Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.