Abstract
Cellular protein quality control (PQC) plays a significant role in the maintenance of cellular homeostasis. Failure of PQC mechanism may lead to various neurodegenerative diseases due to accumulation of aberrant proteins. To avoid such fatal neuronal conditions PQC employs autophagy and ubiquitin proteasome system (UPS) to degrade misfolded proteins. Few quality control (QC) E3 ubiquitin ligases interplay an important role to specifically recognize misfolded proteins for their intracellular degradation. Leucine-rich repeat and sterile alpha motif-containing 1 (LRSAM1) is a really interesting new gene (RING) class protein that possesses E3 ubiquitin ligase activity with promising applications in PQC. LRSAM1 is also known as RING finger leucine repeat rich (RIFLE) or TSG 101-associated ligase (TAL). LRSAM1 has various cellular functions as it modulates the protein aggregation, endosomal sorting machinery and virus egress from the cells. Thus, this makes LRSAM1 interesting to study not only in protein conformational disorders such as neurodegeneration but also in immunological and other cancerous disorders. Furthermore, LRSAM1 interacts with both cellular protein degradation machineries and hence it can participate in maintenance of overall cellular proteostasis. Still, more research work on the quality control molecular functions of LRSAM1 is needed to comprehend its roles in various protein aggregatory diseases. Earlier findings suggest that in a mouse model of Charcot–Marie–Tooth (CMT) disease, lack of LRSAM1 functions sensitizes peripheral axons to degeneration. It has been observed that in CMT the patients retain dominant and recessive mutations of LRSAM1 gene, which encodes most likely a defective protein. However, still the comprehensive molecular pathomechanism of LRSAM1 in neuronal functions and neurodegenerative diseases is not known. The current article systematically represents the molecular functions, nature and detailed characterization of LRSAM1 E3 ubiquitin ligase. Here, we review emerging molecular mechanisms of LRSAM1 linked with neurobiological functions, with a clear focus on the mechanism of neurodegeneration and also on other diseases. Better understanding of LRSAM1 neurobiological and intracellular functions may contribute to develop promising novel therapeutic approaches, which can also propose new lines of molecular beneficial targets for various neurodegenerative diseases.
Keywords: LRSAM1, Chaperone, Misfolded proteins, Neurodegeneration
Introduction
The proteins are the major stakeholders of the cell and are responsible for several critical cellular functions such as signaling, transport across the cell, cell division, nucleic acid organization and ATP generation [12, 19, 110]. To perform the above tasks, proteins are continuously synthesized by an arduous working machinery of ribosomes [76]. However, an over-production of proteins can burden the protein degradation machinery of cell and may induce protein aggregation [104]. Cells have developed a highly efficient system of cellular PQC to counter the abnormal inclusions of protein aggregates [20]. The PQC has three components, i.e., chaperones, UPS and autophagy [5]. The primary role of PQC machinery is to properly fold or separate the misfolded proteins from the healthy protein molecules, as a failure of folding may induce protein degradation [120]. From a very nascent stage of their synthesis, polypeptide chains are highly prone to misfold and can form aggregates, but they are rescued from the event of misfolding by a special class of protein known as chaperones [49].
Proteins which are not properly folded by the chaperones are deleterious to the cellular health and so must be removed by the machinery of autophagy and UPS [130]. Autophagy is bulk cellular waste degradation machinery that regulates many cellular mechanisms such as proliferation, survival, death, cell growth and immunity [31, 61]. Autophagy is primarily categorized into three different types, i.e., macroautophagy (or in few special cases also known as selective autophagy), microautophagy and chaperone-mediated autophagy [43]. During autophagy, a double-membranous structure surrounds the cellular waste and is decorated with various proteins such as microtubule-associated protein 1 light chain 3 (LC3), autophagy-related (ATG) proteins, uncoordinated 51-like kinase 1 (ULK1), Beclin1 proteins, which helps it in targeting the cellular waste to lysosome for their degradation by the lysosomal hydrolases, a process known as macroautophagy [64, 71, 75, 83, 102, 118]. However, in the past several years with the identification of molecules such as p62, ubiquitin in the process of autophagy, the selective nature of autophagy in protein degradation is also confirmed [69, 72]. Autophagy is also observed to be modulated by chaperones and with the help of specific receptors of lysosomal membrane it may target the substrate protein to the lysosome for degradation [65]. A defective autophagy can produce pathological changes in the cell that may finally lead to many types of disorders such as infections, cancer, neurodegeneration, aging and thus maintaining a healthy autophagy is crucial for cellular survival [73].
UPS is the other important cellular protein degradation machinery of the cell and was discovered while searching for answers on the theme of specific degradation of the protein [26]. The system is responsible for targeting a larger number of intracellular proteins for degradation [27], and it depends upon reaction between substrate protein, a approximately 8 kDa protein, i.e., ubiquitin and a series of enzymatic reactions carried out by E1-ubiquitin activating enzyme, E2-ubiquitin conjugating enzyme and E3-ubiquitin ligating enzyme [91]. The degradation mechanism of UPS involves first, the activation of ubiquitin molecules by ATP hydrolysis and its subsequent binding with E1 enzyme by thioester linkage. The activated ubiquitin then gets transferred to E2-ubiquitin conjugating enzyme by the thioester linkage and finally E2 transfers the activated ubiquitin directly or indirectly to the substrate protein, where the indirect transfer occurs with the help of E3 ubiquitin ligases [125]. The misfolded or aberrant protein is finally transferred to a complex protease of the cell, i.e., proteasome [91]. The complete process of ubiquitin conjugation and degradation by proteasome needs protein recognition functions of E3 ubiquitin ligases [129]. E3 ubiquitin ligases can contain either RING finger domains that recruit the substrate protein and help in passage of ubiquitin from the cysteine site of E2 to the misfolded protein or posses homologous to the E6-AP carboxyl terminus (HECT) domain which can form a complex with the activated ubiquitin and later transfer the same to the client protein [6]. Some reports also indicate that E2 ubiquitin conjugating enzyme can directly transfer the ubiquitin to the substrate protein without the assistance of E3 ubiquitin ligases but that requires further verifications [32, 112].
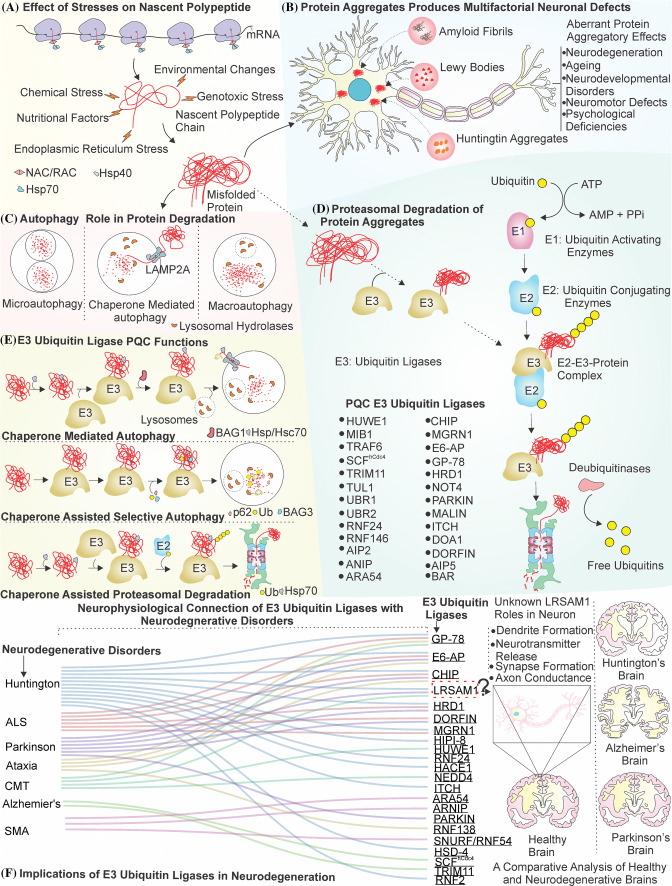
The E3 ubiquitin ligases modulate several cellular mechanisms such as cell division, differentiation, adhesion, migration, ATP production, apoptosis and are crucial for the overall cellular physiology and cell survival [6, 36, 56]. Dysfunctions of the E3 ubiquitin ligases are linked with different pathologies of cancer, neurodevelopment, neurodegeneration and metabolic disorders [119, 121]. The E3 ubiquitin ligases maintain the proteostasis state of the cell by selectively ubiquitinating and then further targeting misfolded proteins for degradation and thus they prevent the protein aggregation and the associated pathologies [53]. Some of them, which are identified and well characterized, are E6-associated protein (E6-AP), carboxy terminus of Hsc70 interacting protein (CHIP), Mahogunin ring finger-1 (MGRN1) and HMG-CoA reductase degradation 1 homolog (HRD1); and they assist in maintaining cellular proteostasis [23, 62, 87, 119, 131]. These E3 ubiquitin ligases are not only specifically restricted to use the machinery of proteasome for protein degradation but can also interact with chaperones and autophagy to remove the proteinaceous toxic inclusions of the cells [63]. Future research needs to focus on understanding the importance of several unexplored and novel E3 ubiquitin ligases which can maintain the proteostasis condition of the cell during various types of cellular mechanisms such as immunological response and cellular transport and that may elucidate their contributions to the pathologies of aging, neurodegeneration and cancer. A clear representation of the protein quality control machinery for maintenance of protein homeostasis in neurons is illustrated in Fig. 1. Along with it, the figure also shows how QC E3 ubiquitin ligases modulate cellular proteostasis and a lack of their functions results in different neurodegenerative and neurodevelopmental disorders.
Fig. 1.
Representation of E3 ubiquitin ligases in cellular protein quality control of neurons. a Nascent polypeptide chains are under constant threat of misfolding from various stresses and are prevented from aggregation by a concerted action of various chaperones. b The misfolded polypeptides are highly prone to form aggregates in the neurons. The aggregate inclusions observed in Alzheimer’s, Parkinson and Huntington’s disease disturb the functions of neuron and can generate several kinds of protein aggregatory effects. c, d Cells remove these toxic proteinaceous inclusions by two different components of protein quality control machinery, i.e., autophagy (c) and UPS (d). e E3 ubiquitin ligases were first observed to be central in the mechanism of UPS (d) for the removal of these misfolded proteins. However, later several studies indicated that E3 ubiquitin ligases with the assistance of chaperone such as Hsp70 and other co-chaperones can communicate with both autophagy and UPS to perform the task of removing misfolded protein aggregates from the cell. f Many different types of neurodegenerative disorders are linked with a functional loss of E3 ubiquitin ligases in the cell. LRSAM1 has shown potential to act as promising E3 ubiquitin ligase with the elucidation of its functions in several neurodegenerative disorders and in neuronal physiology
In our review, a less explored RING finger E3 ubiquitin ligase, i.e., leucine-rich repeat and sterile alpha motif-containing 1 (LRSAM1) protein has been investigated with respect to its multifactorial functions during cellular receptor endocytosis, in protein quality control mechanisms and in different neurological disorders. Detailed information of the LRSAM1 gene, its exons, introns and of its isoforms are given in the text. A comparative analysis of the leucine-rich domain of this E3 ubiquitin ligase in between few species has indicated the significance of several conserved amino acids in the functions of LRSAM1. This text analyzes the role of LRSAM1 in various cellular pathways. In the review, it is also discussed how a defect in the function of LRSAM1 can affect the normal cellular physiology. The knowledge acquired from these studies will help scientists to better explain how LRSAM1 functions are regulated during the pathology of CMT or why the expression level of LRSAM1 gets deregulated in a neurodegenerative condition [48, 113]. Information on the mechanism of action of LRSAM1, its substrate, associated pathways for protein degradation and its importance in the cell is presented in the text. The last section of the review discusses various unanswered questions on LRSAM1 such as finding its modulators, its functions in neurodevelopment and PQC, its importance in the pathomechanism of neurological and immunological disorders and that may have possible futuristic implications in finding therapeutic interventions against these pathologies.
LRSAM1 molecular nature: gene structure, isoforms and encoded protein
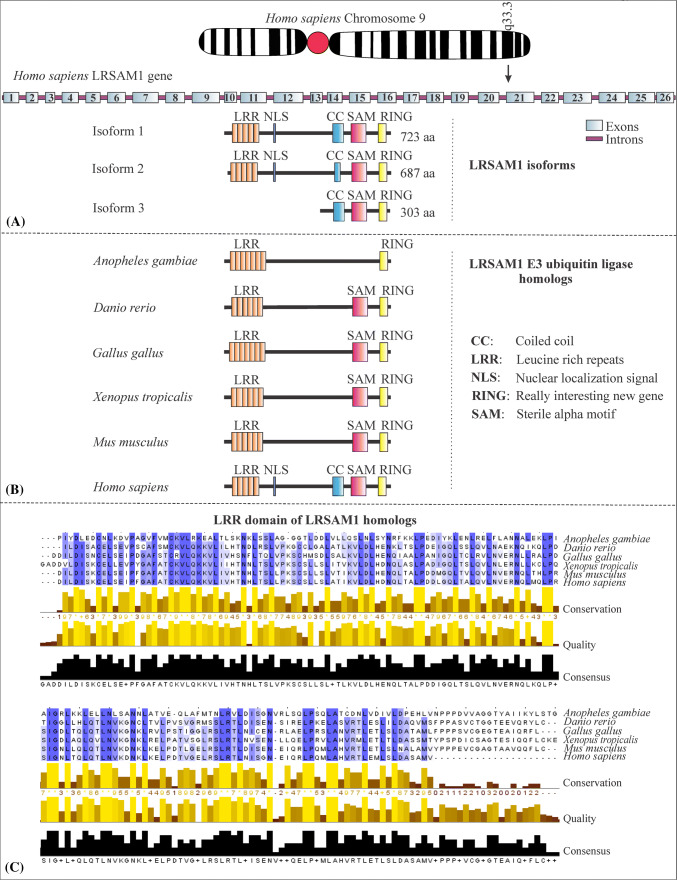
LRSAM1 was first identified as a leucine-rich repeat (LRR), nuclear localization signal and zinc finger-containing protein that regulates cell adhesion molecules [74]. The discovery of E3 ubiquitin ligase activity of LRSAM1 and elucidation of its different domains and functions are credited to [Amit et al. [4]]. The team isolated LRSAM1 as a interacting partner of tumor susceptibility gene 101 (TSG101) and named it as TSG101-associated ligase which is associated with ubiquitylating TSG101 and inactivates its ability to direct endocytic cargoes for endolysosomal degradation [4]. The same group further predicted the structure of protein to consist of leucine-rich repeats (LRR), coiled-coil (CC) region, ezrin–radixin–moesin (ERM), sterile alpha motif (SAM), two Pro-Thr-Ala-Pro (PTAP) sites and RING finger domain [4]. Along with TAL, LRSAM1 has another name, i.e., RIFLE [4, 74]. A full reconstruction of LRSAM1 gene with exons, the interspersed introns and different domains is given in Fig. 2. The LRSAM1 gene of Homo sapiens is located on chromosome 9 at position 9q33.3 and the protein is annotated to have three different isoforms differing at their N-terminal end, where its most highly expressed isoform I encodes a protein with molecular weight 84 kDa and has 26 exons (Fig. 2a). LRSAM1 of humans is expressed in a major amount in the motor neurons and spinal cord [37]. In mouse, LRSAM1 gene has 25 exons and is detected primarily in the motor neurons of the ventral horn, has a molecular weight of 84 kDa and localizes just behind the Golgi complex in fibroblast cell culture [11]. Furthermore, LRSAM1 is present in a large proportion in the mouse dorsal ganglion and spinal cord and hence LRSAM1 may have implications in various kinds of motor functions [11]. A complete description of different domains and motif of LRSAM1 such as LRR, NLS, CC, SAM and RING are provided in Fig. 2a. Figure 2a illustrates that LRSAM1 at its C terminus contains LRR repeats, whereas other domains such as CC, SAM (for protein–protein interaction) and RING (for ubiquitination) are present few bases downstream from the LRR repeat domains.
Fig. 2.
Schematic illustrations of Homo sapiens LRSAM1 gene, protein, domain organization, isoforms and the homologs in different vertebrates along with a sequence alignment performed for LRR domains. a LRSAM1 gene in Homo sapiens is located on the q arm of chromosome 9 and its locus position is at 9q33.3. The gene is made of 26 exons in humans and encodes LRSAM1 protein. The LRSAM1 protein exists in three different isoforms where the most commonly expressed form is 723 aa long. The different domains are arranged in the LRSAM1 protein and are explained in the text (LRSAM1 gene and proteins). b LRSAM1 protein is not observed in non-vertebrates species. In various vertebrates such as Danio rerio (NP_001093474.1), Anopheles gambiae (XP_001230957.2), Gallus gallus (XP_415540.3), Xenopus tropicalis (XP_012825245.1), Mus musuclus (NP_955006.1) and Homo sapiens (NP_001005373.1), LRSAM1 protein domains are compared with each other to indicate the variations of the LRSAM1 from lower to higher vertebrates. c The LRR domains of LRSAM1 proteins from different vertebrates as explained above were used for sequence alignment via using the tools of Clustal omega and the results of the same were then visualized by Jalview [29, 108]. A default coloring scheme was applied for Clustal omega results in Jalview software. The number 0–9 with plus sign shows the conservation of amino acids in LRR domain and an asterisk sign denotes complete conservation of amino acids
LRR repeat as observed in LRSAM1 are also present on innate immune activators, and provides immune system ability to recognize a specific pattern of proteins or sequences on the surface of pathogenic microorganisms [92]. Similarly, in LRSAM1, the LRR domain helps it to recognize Salmonella typhimurium and later build an immune response against the bacterial infection by activating autophagy [57, 92]. Autophagy of bacteria also requires its recognition by other immune proteins such as nuclear domain 10 protein 52 (NDP52) which along with LC3 helps it in delivery of ubiquitinated bacteria to the autophagosome for further destruction [126]. LRSAM1 interacts with NDP52 by its LRR domain and the binding is independent of ubiquitin binding of NDP52 [57]. The motif SAM1 is required for protein–protein interactions, self-associations and to bind different types of non-SAM-containing proteins; however, the complete function of SAM1 in LRSAM1 is not fully understood [68]. But the self-ubiquitination ability of LRSAM1 may be attributed to its SAM1 domain [10, 57].
Domains such as CC and PTAP motif of LRSAM1 helps LRSAM1 to bind with its substrate, i.e., TSG101, where CC domain is important in specifying ubiquitination target in TSG101 and PTAP–PSAP motif is required for mediating this interaction [4, 84]. The ubiquitination of TSG101 depends upon RING domain at the C’ terminus of LRSAM1 and mutations of the same not only affect the TSG101 ubiquitination, but it has shown to be the cause of pathological disorder such as CMT [4, 37, 48]. Other than ubiquitination, the RING domain is also critical for regulating functions such as transduction, signaling and transcription [13, 14, 78]. RING domain containing E3s are coded by more than 600 genes and they participate in a variety of cellular pathways [85]. The same domain is characterized by a specific sequence of amino acids such as C–X2–C–X(9–39)–C–X(1–3)–H–X(2–3)–C–X2–C–X(4–48)–C–X2–C, where C is cysteine, H is histidine and X any amino acid [40]. Cysteine and histidine are highly conserved amino acids of the RING domains of E3 ubiquitin ligases and they bind to two zinc atoms to maintain the structure of E3 ubiquitin ligases. The same cysteine residue also has significance in the biology of LRSAM1, as a mutational change in the cysteine residue near RING domain of LRSAM1 leads to the pathology of CMT2P [55].
Along with LRR, the RING domain of LRSAM1 also helps it to recognize bacteria and induce ubiquitination without the assistance of other accessory proteins [57]. The process of ubiquitination by LRSAM1 may involve its interaction with E1 enzyme, i.e., UBA1, E2 enzymes: UBE2D2, UBE2D3 and K6 or K27 types of polyubiquitination [57]. A copy of the LRSAM1 protein is present throughout the vertebrate genome but is not present in any of the non-chordate phylum and many of the domains of the LRSAM1 are highly conserved in vertebrates indicating a vertebrate lineage for LRSAM1 [4]. We performed a multiple sequence alignment for the leucine-rich repeat domains of various types of annotated LRSAM1 for Homo sapiens (NP_001005373.1), Mus musuclus (NP_955006.1), Gallus gallus (XP_415540.3), Xenopus tropicalis (XP_012825245.1), Danio rerio (NP_001093474.1) and Anopheles gambiae (XP_001230957.2). The results of the multiple sequence alignment are aligned in Fig. 2c with the help of Clustal omega and were visualized using Jalview software [29, 108]. The results of the sequence alignment shows conserved leucine amino acids of the LRR domains, which may play role in the generation of cellular immunological responses against pathological infections.
The discoveries of the several functions of LRSAM1 protein and its different domains that have been revealed in the last few years show the importance of LRSAM1 protein in maintaining cellular transport, cell development, immunological responses, ubiquitination, neurological and motor disorders. However, more research to search for novel roles of LRSAM1 domains in cellular physiology will serve to better explain the relevance of these structures in LRSAM1 and provide clues for their significance in overall cellular health and fitness.
Molecular functions of LRSAM1 in endosomal sorting of receptors
Endocytosis is a mechanism by which cell internalizes a majority of extracellular material, few ligands and the lipid portion of plasma membrane [35]. It also helps cells to degrade the cellular waste by lysosomal machinery and thus assists in ATP generation, signal transduction, cells division and cellular adhesion [45, 99]. Internalization of receptors by the endosomal mechanism is important in maintaining a homeostatic state of receptor populations in the cell. The recycling of receptors by the cell regulates the responsiveness of cell to the ligand binding, i.e., either activation or re-sensitization in case of an over-exposure of the ligands [58]. The sorting of the endosomes either for degradation or for recycling depends upon the ubiquitin signaling. The degradation by lysosomal machinery only occurs for the ubiquitinated endosome; the endosomes after ubiquitination is also known as multi-vesicular bodies that contain the vesicles made up of different proteins that need to be degraded by lysosomes [50]. The ubiquitinated cargo of endosomes is recognized by special machinery known as endosomal sorting complex required for transport-I (ESCRT-I).
In yeast, proteins of the ESCRT-I complex are known as Vps family of proteins, while the mammalian homologue of the same is called as TSG101 [7]. A TSG101 mutant lacks ability to sort lysosomal hydrolase, cathepsin D, mannose-6-phosphate and EGFR receptors which can have deleterious consequences in the cell like a prolonged signaling similar to the one present in tumor cells [7]. TSG101 is also critical for modulating the budding of HIV-1, as it binds with the PTAP motif of the L-domain present in the p6 protein of HIV-1 using its ubiquitin E2 variant (UEV) domain and a depletion of TSG101 by siRNA knockdown causes arrest of HIV-1 budding [41]. The above work shows how efficiently the virus machinery overtakes the host mechanism of receptor endocytosis and degradation to exit from the cell. However, a separate report suggests that an overexpression of a N-terminal domain, i.e., TSG-5’ of TSG101 abolishes virus assembly and interferes with the exit of virus from the cell and thus exerts an inhibitory effect on the budding of HIV-1 [33].
TSG101 can self-modulate its level in the cell and the same is observed in a study where an overexpression of TSG101 down-regulates the endogenous TSG101 level [38]. TSG101 in the cell is also regulated by LRSAM1, as it polyubiquitinates lysine residues of an uncomplexed TSG101 at the C terminus inducing its proteasomal degradation [84]. The interaction between TSG101 and LRSAM1 is of bimodal type and the first of these interactions is present in between the UEV domain of TSG101 and PTAP–PSAP1 motif of LRSAM1 and the other one occurs in between CC domain of LRSAM1 and the steadiness box of TSG101 [4]. The RING domain and the E3 ubiquitin ligase activity of LRSAM1 are important in modulating the level of TSG101 in the cell and point mutations in RING finger domain prevents the ubiquitination of TSG101 [4, 84]. The importance of UEV and RING finger domain for binding between TSG101 and LRSAM1 was further confirmed by TSG101 UEV and RING finger mutant, which lacks ability to bind LRSAM1 [84].
TSG101 is monoubiquitinated multiple times by MGRN1 E3 ubiquitin ligase and is targeted for degradation by the machinery of proteasome; however, LRSAM1 performs both monoubiquitination and polyubiquitination of TSG101 and affects its endolysosomal trafficking and proteasomal degradation, respectively [4, 67, 84]. LRSAM1 specifically ubiquitinates lysine residues present at the steadiness box C terminus of the TSG101 and the same is affected by mutations of the lysine residues to the arginine in the steadiness box. Interestingly, the binding is prevented by an ESCRTI complex protein, i.e., VPS28, as it masks the lysine residue of TSG101 from ubiquitination and further degradation [84]. This indicates how steadiness box at the C terminus of TSG101 affects the degradation of TSG101 [4, 84].
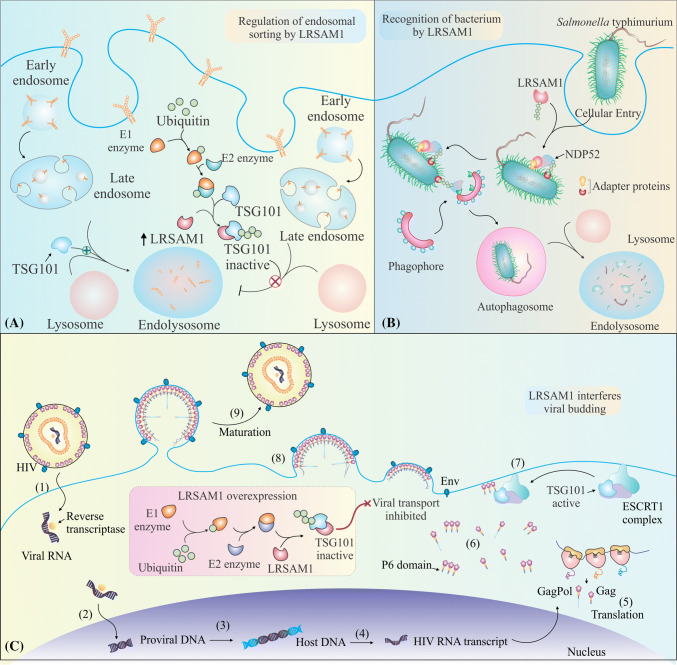
TSG101 controls the endosomal sorting of epidermal growth factor receptor (EGFR) and the same is affected by the multiple monoubiquitination of TSG101 by LRSAM1 and not by its polyubiquitination [4]. Multiple monoubiquitination of TSG101 by LRSAM1 inactivates the functions of late ESCRTI complex to sort proteins [4]. Deactivation of TSG101 by LRSAM1 prevents the endocytic degradation of EGFR by ESCRTI complex and leads to accumulation of EGFR. Catalytically RING inactive form of LRSAM1 induces the degradation of EGFR in the cell due to the stabilization of EGFR–TSG101 complex in the ESCRTI [4]. In Fig. 3a, we have explained how the LRSAM1-mediated ubiquitination of TSG101 interferes with the endosomal sorting functions of TSG101 and may inhibit the endolysosomal degradation of EGFR proteins in the cell.
Fig. 3.
LRSAM1 modulates endosomal protein sorting, xenophagy and virus budding. a LRSAM1 regulates the sorting of receptor proteins by selectively ubiquitinating an endosomal complex protein, i.e., TSG101. Ubiquitination of TSG101 reduces its ability to sort the endosomal complex of receptors proteins and that results in a failure of endolysosomal degradation of receptors. b LRSAM1 generates ubiquitin signals around the bacteria Salmonella typhimurium and targets the bacteria for autophagic degradation (or xenophagy). Different adaptor proteins such as NDP52 and also p62 are attracted towards the ubiquitin coat of the bacterial cell and assist in the process of selective autophagic degradation of bacteria. c The HIV virus binds to TSG101 protein of the host cell using the p6 domain of Gag proteins and thus utilizes the host cell endosomal machinery to egress from the cell. Overexpression of LRSAM1 leads to the inactivation of TSG101 and abolishes the formation of ESCRT complex I which effectively reduces HIV budding from the infected cell
Endosomal sorting in the cell is important for recycling and for the effective removal of different cellular wastes. A defective endosomal machinery generates various types of pathologies such as mucolipidosis II or N-acetylglucosamine l-cell disease, Hermansky–Pudlak syndrome, Chediak–Higashi syndrome, Charcot–Marie–Tooth type 2 and tuberous sclerosis [101]. LRSAM1-mediated ubiquitination and inactivation of TSG101 disrupt the endosomal trafficking of protein such as EGFR and may have implications in these above-mentioned pathologies and also in cancer, where EGFR level is deregulated [117]. The regulatory functions of LRSAM1 in endosomal trafficking may have further consequences in the mechanism of a disruptive endosomal trafficking disease such as CMT2P, where LRSAM1 ubiquitination functions are severely affected [37, 48]. Hence, LRSAM1 must be explored for its connection with these endosomal trafficking and also various cancer-related diseases.
TSG101 is also involved in the exit of retrovirus HIV1 virus-like particles from the cell [41]. Virus egress from the cell is a vital step in the life cycle of virus infection [97]. Several host proteins in the past are identified with their ability to assist virus in the process of virus budding from the cell. The PTAP motif in the p6 region of the gag protein of the virus binds with the UEV domain of the TSG101 and recruits TSG101 and other proteins of ESCRTI at the site of virus budding from the cell [82, 124]. LRSAM1 co-localizes with the gag protein of the retrovirus HIV1 in a complex with TSG101, the gag protein may likely recruit the complex of TSG101 and LRSAM1 to the site of virus budding and can cause insertion of LRSAM1 into the virus like particles, which is already known for TSG101 [4, 90]. The egress of the virus from the cell was higher for a LRSAM1 ubiquitin ligase mutant, suggesting a deubiquitinated TSG101 has an increased efficiency to get inserted into the virus-like particles, then an ubiquitinated TSG101. Thus, ubiquitination of TSG101 prevents the insertion of TSG101 into the complex of virus-like particles and blocks virus egress from the cell. In Fig. 3c, a cycle of virus entry and budding from the infected cell is shown. Further the same figure illustrates how LRSAM1-induced ubiquitination and inactivation of TSG101 causes a defective virus budding.
HIV1 virus has been observed to very cleverly use the host machinery for its budding from the cell as it promotes the activity of other E3 ubiquitin ligases such as neural precursor cell expressed developmentally down-regulated protein 4 (NEDD4) and membrane-associated ring-CH-type finger 8 (MARCH8) for endolysosomal degradation of anti-viral tetherin protein which helps in its release from the cell [105]. The knowledge on the blockage of HIV1 virus release from the infected cells via modulating LRSAM1 can be useful if it can be develop at the commercial level for treatment of AIDS patients, as the current anti-retroviral therapies available in market is based on usage of anti-retroviral drugs which after long-term usage in HIV-positive patients became ineffective since the patients develop drug resistance [89]. Identifying such new strategies, which can be used in treatment of HIV-infected patients, will be very effective in prolonging the life span of these immunocompromised patients.
Dynamic role of LRSAM1 in xenophagy
The bulk of cellular waste is removed from the cell by the machinery of autophagy, it is a degradation machinery which is conserved in many aspects of its functions from yeasts to higher mammals [88]. The substrates of autophagy may range from old and defective organelles such as endoplasmic reticulum, mitochondria, ribosomes, nucleus to misfolded or aberrant proteins and infectious agents [9]. These wastes and toxic materials are packaged into a double-membrane vesicular structure known as autophagosome, which is delivered to the lysosomes for final degradation [9, 134]. The process of autophagy seems to get activated under various situations of stresses such as proteotoxicity, heat, pH, nutrient, microbial infections and helps to protects the cells against these deleterious conditions [34].
As compared to popular notion, in past few decades, several reports have confirmed the selective nature of autophagy. Many proteins are characterized such as p62 and NDP52 that can regulate or execute the process of selective autophagy. Proteins selectively degraded by autophagy are recognized with the help of autophagy cargo-receptors such as p62, neighbor of BRCA1 gene 1 (NBR1), NDP52, and toll-interacting protein 1 (TOLLIP1), which recognize a specific signal like ubiquitin chain present on the substrate protein and sort them for the degradation by autophagy [66, 135]. The entry of the cargo receptor and the ubiquitinated proteins into the autophagic cascade is promoted by specific class of proteins known as ATG 8 family proteins [1]. There are two sub-families present in the ATG8 family proteins, i.e., LC3 and gamma-aminobutyric acid type A receptor-associated protein (GABARAP) group [1]. ATG8 family proteins binds with the autophagy cargo receptor proteins by their LC3-interacting region (LIR) domain and thus modulates the mechanism of selective autophagy [95].
Ubiquitination is a key to selective autophagic degradation of proteins, performed with the help of lysosome. Different E3 ubiquitin ligases such as CHIP, MGRN1, and E6-AP are implicated in the process of protein ubiquitination and later in their degradation by selective autophagy [22, 137]. These E3 ubiquitin ligases degrade many of the disease causing misfolded proteins which if starts to aggregate may produce deleterious consequences. LRSAM1 is involved in the process of autophagy and eliminates the bacterial cells by the process of selective autophagy known as xenophagy. Similarly, E3 ubiquitin ligases such as SMAD-specific E3 ubiquitin protein ligase 1 (smurf1), parkin, RING finger protein 5 (RNF5) and NEDD4 are also associated with the process of xenophagy and modulate host cell response against bacterial infection [39, 70, 77, 81]. Xenophagy is a poorly understood mechanism; the knowledge garnered from several years of research indicates that the mechanism is useful for the innate immunity system to remove bacterial pathogens, which can subvert the primary immunological response of the host.
Salmonella typhimurium is a intracellular pathogen, which causes gut infections, diarrhea, fever and inflammation in the intestine [42]. S. typhimurium can escape the response of extracellular immune system and replicates within the autophagic vacuole of the host cells [133]. The bacteria from the vacuole escapes into the cytoplasm of cell and can spread its pathology [42, 114]. However, autophagic system of the host cell in cytoplasm can capture S. typhimurium in a double-membranous structure known as autophagosome, which finally fuses with the lysosome for the degradation (Fig. 3b). The autophagic capturing of the bacteria depends upon the coat of a polyubiquitin chain on the S. typhimurium, recruitment of different autophagic adaptor proteins such as NDP52 and p62 and activation of autophagy (Fig. 3b) [107, 116]. LRSAM1 can effectively ubiquitinate the S. typhimurium cell wall coat and directs the bacteria for autophagic degradation [57]. This research also showed that LRSAM1 gets co-localized with the S. typhimurium post-infection and a knockdown of LRSAM1 increases the number of S. typhimurium in the host cell indicating a higher risk of bacterial infection.
LRSAM1 gets co-localized to the bacteria along with ubiquitin and generates a K6 or K27 type of ubiquitin signals associated with bacterial cell with the help of E1-UBA1 and E2-UBE2D2 or UBE2D3. The localization and ubiquitination of LRSAM1 with the bacterial cell requires the assistance of LRR and RING domains of LRSAM1, respectively [57]. The event of localization is followed by the recruitment of autophagic receptor protein NDP52, which interacts with LRSAM1 via its SKIP carboxyl homology (SKICH) domain; however, this binding has no effect on LRSAM1 localization to bacterial cell [57]. But the recruitment of autophagic receptors such as NDP52 and p62 at the bacterial infection site depends upon the ubiquitination of S. typhimurium by LRSAM1 and a knockdown of LRSAM1 leads to the failure of autophagic receptors to get localized at the site of bacterial infection [57]. Different in vitro experiments performed during the above-described study also shows a co-localization of LRSAM1 to the bacterial pellets of Shigella, Listeria and AEIC. Anti-bacterial property of LRSAM1 is further confirmed by finding that in CMT disease, LRSAM1-null patient-derived lymphoblasts are unable to mount an anti-bacterial response after exposure to a strain of S. typhimurium bacteria [57]. The above findings indicate that the CMT patients are more susceptible to S. typhimurium infection, as they lack a functional LRSAM1 protein. A complete illustration of LRSAM1 role in bacterial recognition and in targeting the S. typhimurium to autophagy is illustrated in Fig. 3b.
Autophagy is essential in maintaining cellular proteostasis and the same is involved in protecting the cells from pathogenesis of bacterial infection [8]. Several previous findings have shown how autophagy is being utilized by the host cells to mount appropriate responses against bacterial infections. E3 ubiquitin ligases such as parkin, NEDD4, smurf1 are found to participate in the mechanism of autophagy by performing the degradation of intracellular bacteria, which evades the primary immunogenic response of the cell [39, 77, 81]. The use of autophagy by LRSAM1 for targeting bacteria to autophagic degradation makes LRSAM1 an interesting option to treat bacterial infections of bacterial-infected and immune-compromised patients. However, elaboration of the LRSAM1 functions and its coordination with several autophagic proteins shall be first understood prior to its use as a possible therapeutic strategy in the treatment of bacterial diseases. More research on the questions of functions of LRSAM1 in xenophagy will add knowledge on how LRSAM1 is able to recognize several kinds of bacteria and thus consequently induce autophagic response. LRSAM1 possess LRR domain, which provides its ability to recognize specific patterns and signals, displayed by different pathogens and allergens and thus may subsequently regulate the innate immunity body response. Identifying more such functions of LRSAM1 will help us to better understand the LRSAM1 contributions in immunological responses.
LRSAM1-linked molecular pathways in neurodegenerative diseases
E3 ubiquitin ligases are characterized for their functions in development and maintenance of the cell. They participate in many cellular developmental mechanisms such as in neural stem cell differentiation and organogenesis [111, 132]. In the maintenance of cellular functions, E3 ubiquitin ligases modulate various mechanisms such as cell sorting, cell signaling, metabolism and transcriptions [2, 56, 127]. Further, the role of E3 ubiquitin ligases in the removal of toxic proteinaceous aggregates from the cell over past several decades has made them an excellent therapeutic and possible option to treat several types of protein aggregatory disorders such as Parkinson’s, Huntington’s and amyotrophic lateral sclerosis [21, 28].
Development, maintenance and modulation of components of brain is critical, a tedious process and is important for the normal health. E3 ubiquitin ligases are involved in these mentioned mechanisms and are often associated with neurodevelopmental and neurodegenerative disorders [121]. Finding out novel E3 ubiquitin ligases with functions in these above-explained mechanisms will help to develop an understanding on the significance of these E3 ubiquitin ligases for a healthy brain. LRSAM1 has shown the potential to be really important in many of the above facets of brain development and later in normal functioning of the brain.
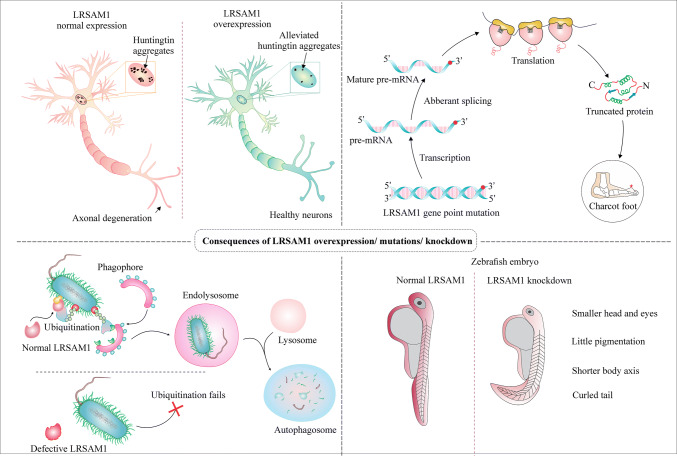
LRSAM1 is one of the fundamental genes, which was found to be mutated in the patients of CMT disease [46]. CMT is a hereditary polyneuropathy, which is often characterized by the sensory loss in the peripheral and distal muscles, muscle loss [54]. CMT is further classified as demyelinating, axonal and intermediate types (CMT1, CMT2, CMT4, respectively) [93]. CMT2 mutations are mostly inherited as autosomal dominant type and are of CMT 1 and CMT 2 types, whereas CMT 4 type mutations are transmitted as autosomal recessive types [115, 122]. Many reports in last few years from various parts of the world have identified mutations in LRSAM1 for CMT 2 phenotype and associated axonal degeneration (Fig. 4) [37]. Peripheral muscles are the worst affected organs in the CMT 2 patients because of the axonal degeneration. The disease is also known as CMT2P because of the wasting of peripheral muscles in CMT patients.
Fig. 4.
Effect of LRSAM1 overexpression and lack of functions on cellular physiology. a Neurons are highly susceptible to the toxic proteinaceous inclusions such as of huntingtin protein and that can lead to axonal degeneration. Ectopic expression of LRSAM1 has shown to alleviate neuronal cells from the toxic proteinaceous inclusions of expanded polyglutamine huntingtin protein. b Studies have suggested how a point mutation in LRSAM1 produces an abnormal protein and causes peripheral myopathies such as Charcot–Marie–Tooth type 2P (CMT2P) disease, a pathology of motor neurons that shows degeneration of axons. c LRSAM1-mediated production of ubiquitin signals around the bacterial cell is required for the removal of bacteria from the cells by the mechanism of autophagy. d LRSAM1 can have critical functions in the development of nervous system and other organs of the body. A morpholino oligonucleotide-induced knockdown of LRSAM1 of Zebrafish leads to abnormally small head, tails and produces little pigmentation, results in shorter body axis and a curved tail
LRSAM1 mutation such as a splice mutation produces a premature codon, which stops 20 bp prior to the final exon and produces a truncated protein that has a loss in function and leads to a phenotype of autosomal recessive axonal CMT or AR-CMT 2 [46]. A separate report showed frameshift mutation in the RING domain of LRSAM1 is inherited as an autosomal dominant type and cause CMT2P pathology. The results of the study also proved that LRSAM1 is needed for the neurodevelopment, tail formation and movement in embryo of Drosophila melanogaster (Fig. 4) [128]. Similarly in another work, an autosomal dominant pattern of inheritance was observed for LRSAM1 mutation which leads to frameshift and generates a stop codon in the new reading frame and produces a premature LRSAM1 protein in CMT phenotypes [93]. The results of LRSAM1 mutations and their consequences for CMT are validated in CMT mouse model also, where exposure to a neurotoxic agent such as acrylamide causes progressive axonal degeneration [11].
Additionally, a novel mutation of LRSAM1 was identified by Engelholm et al. to be the cause of CMT2P where extra base pairs were observed in the LRSAM1 transcript and a subsequent disruption occurred in the E3 ligase activity of LRSAM1 [37]. Recently, a novel missense mutation of Cys694Arg of LRSAM1 was found to be a causative factor of the CMT2P in a family [138]. The cysteine is a highly conserved amino acid in the RING domain of most of the E3 ubiquitin ligases and any alteration in this amino acid can have significant impact on the ubiquitination activity of LRSAM1 and may lead to pathology of CMT2P [138]. A missense mutation in the LRSAM1, which replaces the conserved cysteine residue is responsible for CMT 2G and leads to reclassification of CMT2G as CMT2P [98]. In another interesting recent study done in a Chinese family for CMT2P pathological analysis, LRSAM1 CMT2P mutation was observed to be present near to the RING finger domain of LRSAM1 and may affect LRSAM1 E3 ligase activity [136].
All the above reports indicate that different LRSAM1 mutations either frameshift or missense in the pathology of the CMT2P is predominantly located in the RING domain or in its vicinity. The importance of RING domain of LRSAM1 in the pathogenesis of CMT2P is described by a study explaining the loss of ubiquitination function after a RING domain mutation [48]. The RING domain of LRSAM1 is critical for the ubiquitination of TSG101 and has implications in the process of endosomal protein trafficking and this can affect endosomal protein trafficking and consequently lead to CMT2P disease. Thus, LRSAM1 may induce CMT2P pathology by dysregulating enodosomal protein trafficking [15]. Hence, before using LRSAM1 as a possible therapeutic target for the patients of CMT2P, the strategy needs its effective validation in cell culture, different animal models and then later in clinical studies.
LRSAM1 is also observed in several other neurobiological disorder mechanisms triggered by protein aggregation or protein misfolding. A study indicates that LRSAM1 overexpression can protect neuronal cells from the proteotoxicity of expanded polyglutamine proteins of Huntington’s disease by an unknown mechanism and can be a future research theme to be studied [113]. The same work also showed an elevated level of LRSAM1 in the transgenic mouse of Huntington’s and can also be crucial in the pathology of Huntington’s disease, as many E3 ubiquitin ligases in past such as E6-AP and MGRN1 have altered expression in Huntington’s transgenic mice [24, 80].
A very recent publication shows how the ubiquitin ligase function of LRSAM1 is required for the healthy functioning of transcriptional machinery of the cell and LRSAM1-mediated ubiquitination alters DNA-binding protein fused-in sarcoma (FUS) functions in the cell [55]. The results of the work describe how an interaction between LRSAM1 and FUS is needed for modulating the level of FUS in the cell and that regulates the cellular transcriptional machinery function. The RING domain in LRSAM1 is required for this interaction and a mutation in the RING domain disrupts this binding [55]. Since FUS is observed to be mutated in amyotrophic lateral sclerosis, binding of LRSAM1 with FUS may have significance in the pathology of amyotrophic lateral sclerosis and can be an area of future research work [51]. In a very current work, patients of CMT2P with LRSAM1 mutations have shown to develop symptoms of Parkinson’s disease at a later stage of their life [3]. Thus, It will be very interesting to check the association of LRSAM1 function with Parkinson’s pathology, as LRSAM1 is expressed primarily in the motor neurons and in spinal cord while the pathology of Parkinson’s has been basically associated with the defects observed in the substantia nigra region of the brain [3].
The various discussed research so far clearly suggests the involvement of LRSAM1 with the pathological mechanism of some of the detrimental neurodegenerative disorders such as Huntington’s, Parkinson’s, CMT and ALS. One of these studies explained that the functions of LRSAM1 are needed for neurodevelopment, head and tail movement in Drosophila melanogaster embryo [128]. This study clearly strengthens LRSAM1 role in neurodevelopment. As the mechanism of neurodevelopment is often associated with neuronal proliferation, LRSAM1-mediated modulation of neuronal proliferation further substantiates the importance of LRSAM1 in neurodevelopment [86]. Thus, these studies performed with LRSAM1 E3 ubiquitin ligase functions provide enough evidence to probe the importance of LRSAM1 in the regulation of different neurobiological mechanisms such as neurogenesis, neuronal growth, neurodevelopment, axonal transport, synapse formation and neurotransmitter release [86, 100, 106]. Since many of these mentioned mechanisms are severely affected in neurodegenerative disorders of Huntington’s, Parkinson’s and ALS; hence, the future research of LRSAM1 importance in these neurodegenerative disorders will make LRSAM1 a viable molecular therapeutic agent.
In our review, we have comprehended that LRSAM1 is essential for the maintenance of few crucial cellular functions such as endosomal protein sorting or in generating autophagic response against bacterial infection and for regulating several neurobiological disorders. All these identified roles of LRSAM1 indicate that it is very useful for cell especially neuronal cell in protecting it from different stresses produced internally or externally. LRSAM1 communicates with both UPS and autophagy and hence can modulate the PQC of the cell using either of these mechanisms. The contribution of LRSAM1 to the PQC mechanism should be examined in future studies. Many such facets of LRSAM1 should be critically analyzed.
Key questions, challenges and future prospective
LRSAM1 has shown to participate in several cellular physiological mechanisms, which are needed for transport, defense and proteostasis maintenance in the cell. Identifying such more novel functions of LRSAM1 in cells can be helpful in selectively targeting LRSAM1 for therapeutic interventions in the neurological and immunological disorders. The problems associated with the use of LRSAM1 as a therapeutic strategy should be characterized and discussed to develop a detailed molecular understanding of LRSAM1 functions in various cellular mechanisms. In this section of the review, few of these questions that are linked with the LRSAM1 study are systemically discussed.
The study of Amit et al. clearly suggests how LRSAM1 regulates endosomal trafficking of receptors and thus provides direction in identifying importance of LRSAM1 in mediating the endolysosomal degradation of other cellular receptors such as G protein-coupled receptors (GPCR), fibroblast growth factor receptors (FGFR), insulin and insulin-like growth factor receptors (IR and IGFR) of the cell [44, 59]. The proposed hypothesis can be fruitful in understanding how LRSAM1 may affect endosomal trafficking of receptors and consequently modulate their expression level in the cell. The findings from these studies will be useful in understanding the pathologies of several cancers where a continued receptor activation is observed [25, 103].
Different neurodegenerative diseases are shown to be caused by the result of protein aggregation. The cellular system has developed a productive clearance system to remove these toxic protein inclusions from the cells. Several quality control E3 ubiquitin ligases such as E6-AP, MGRN1 and CHIP are previously observed to be a central component of this clearance system [21, 119]. LRSAM1 may act as a QC E3 ubiquitin ligase, which can modulate the turnover of some of the important proteins implicated in the pathogenesis of neurodegenerative disease. Overexpression of LRSAM1 has neuroprotective functions to protect the neural cell population against the pathogenicity of the expanded huntingtin polyglutamine proteins in cell culture model [113]. The same neuroprotective effect of LRSAM1 in pathologies of Parkinson’s, ALS and Alzheimer’s must be checked for the QC efficacy of LRSAM1 in protein aggregatory diseases.
LRSAM1 degrades TSG101 by the machinery of proteasome and is also involved in autophagy for bacterial autophagy; this clearly suggests that LRSAM1 is capable of utilizing both the cellular protein degradation machinery for different functions and thus may modulate cellular proteostasis by both the cellular mechanisms [57, 84]. Identification and characterization of novel protein substrates of LRSAM1 and their degradation either by the usage of proteasome or autophagy in the cells will assist us to better comprehend LRSAM1 functions in several cellular mechanisms and gain further knowledge on the pathologies associated with LRSAM1 lack of functions.
Involvement of LRSAM1 in the mechanism of neurodevelopment indicates that LRSAM1 has the potential to modulate the proteome of the developing neurons. As in one study, LRSAM1 loss of function severely impairs the formation of tail and vertebrae in the Zebrafish model [128]. There are several possibilities where LRSAM1 may interact with different proteins of a developing neuron and hence regulate their cellular level. In previous research, various E3 ubiquitin ligases are observed to regulate the neuronal growth and hence development by regulating the level of few critical growth- and development-related proteins [121]. In one report, F-box/WD repeat-containing protein 7 (Fbw7), a type of Skp, Cullin, F-box-containing complex (SCF) E3 ubiquitin ligase, modulates neuronal stem cell differentiation by regulating the levels of transcription factors, i.e., neurogenic locus notch homolog protein 1 (Notch1) and c-Jun [52]. Similarly, in a separate work, E3 ubiquitin ligase msel-10 was observed to negatively regulate Notch signaling and may affect the developmental process of neural system, where Notch signaling is involved [79, 96]. Likewise, other studies have identified E3 ubiquitin ligases such as Mind bomb, Ligand of Numb Protein-X (LNX), Suppressor of deltex gene, that selectively target Notch pathway and hence affect the developmental mechanism of the nervous system [30, 60, 94]. All these findings indicate how E3 ubiquitin ligases play a central role in the neuronal development by controlling various protein levels in the cell; hence, similarly LRSAM1 may have a interesting contribution in vertebrate development by modulating developmental protein levels of neuron.
Several research studies in the past have indicated that E3 ubiquitin ligase can serve as a potential target for drugs in treating different pathologies. E3 ubiquitin ligase such as mouse double minute 2 (MDM2) is targeted by a small molecule antagonist such as Nutlin-3a which effectively binds MDM2 and causes a transcriptional upregulation of MDM2 substrate p53 and leads to cell cycle arrest, cell death and apoptosis in cancerous cells [123]. In few other reports, many members of cullin–RING E3 ubiquitin ligases which are involved in crucial cellular mechanism like cellular proliferation and are thus critical in pathologies of cancer are modulated using regulators such as Probe 8, VL111 and VH298 [16, 17, 109]. Similarly regulators, i.e., either inhibitor or activator for LRSAM1 can be characterized in future studies, which can be useful in modulating LRSAM1 activity and consequently in controlling dysregulated pathways associated with the biology of LRSAM1 such as endosomal cell sorting and protein quality control mechanism. But previous studies have identified that correctly characterizing modulators of these E3 ubiquitin ligases family such as MDM2 and CUL3 are mostly based upon the result of the ligand–enzyme structural studies and is obtained from the crystal structure of these E3 ubiquitin ligases [18, 123]. Thus, to characterize LRSAM1 modulators, it is also necessary to first elucidate the crystal structure of LRSAM1 E3 ubiquitin ligase. Efforts are already made in this direction where in one instance, the group of Guo Y et al. has successfully purified recombinant LRSAM1 for research purpose, which can be used in future studies to elucidate the crystal structure of LRSAM1 and thus in identifying its modulator [47].
The participation of LRSAM1 in the protein quality control has to be clearly defined and should be further investigated. LRSAM1 may possess the ability to interact with both degradation machineries of the cell, i.e., autophagy and proteasome and also has the potential to modulate cellular proteostasis making it interesting to investigate the functions of LRSAM1 in cellular protein quality control. Further other molecular functions of LRSAM1 in the protein quality control especially its capability to interact with different members of the protein quality control such as chaperone and thus to modulate protein turnover should be explored in future research. The research should also concentrate on to check for a possible role of LRSAM1 to facilitate a cross-talk between components of protein quality control for maintaining cellular proteostasis, which often gets disturbed in different protein conformational disorders.
LRSAM1 is an E3 ubiquitin ligase, which is involved in several cellular mechanisms such as endocytosis, immunological response and cellular adhesion and in maintaining proteostasis. We have examined the importance of LRSAM1 in various cellular pathways and observe its functions to be fundamental for cell survival and a disruption in the same may has deleterious consequences that can lead to pathological outcomes. More research on LRSAM1 should focus on utilizing LRSAM1 either for therapeutic intervention or for finding cure against many neurodegenerative, neurodevelopmental and immunological disorders.
Acknowledgements
Support for this work was obtained from Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India grant to (AM) EMR/2016/000716. AU received research fellowship in the duration of the work from University Grants Commission, Council for Scientific and Industrial Research, Government of India. The authors would like to thank Mr. Bharat Pareek for his technical assistance and entire lab management during the manuscript preparation.
Abbreviations
- ATG
Autophagy-related proteins
- CHIP
Carboxy terminus of Hsc70-interacting protein
- CMT
Charcot–Marie–Tooth
- CC
Coiled-coil
- E6-AP
E6-associated protein
- ESCRT-I
Endosomal sorting complex required for transport-I
- EGFR
Epidermal growth factor receptor
- ERM
Ezrin–radixin–moesin
- Fbw7
F-box/WD repeat-containing protein 7
- FUS
Fused-in sarcoma
- GABARAP
Gamma-aminobutyric acid type A receptor-associated protein
- HECT
Homologous to the E6-AP carboxyl terminus
- HRD1
HMG-CoA reductase degradation 1 homolog
- LNX
Ligand of numb protein-X
- LIR
LC3-interacting region
- LRR
Leucine-rich repeat
- LC3
Microtubule-associated protein 1 light chain 3
- LRSAM1
Leucine-rich repeat and sterile alpha motif-containing 1
- MARCH8
Membrane-associated ring-CH-type finger 8
- MDM2
Mouse double minute 2
- MGRN1
Mahogunin ring finger-1
- NBR1
Neighbor of BRCA1 gene 1
- NDP52
Nuclear domain 10 protein 52
- Notch1
Neurogenic locus notch homolog protein 1
- PI3K
Phosphatidylinositol 3-kinase
- PQC
Protein quality control
- PTAP
Pro-Thr-Ala-Pro
- RING
Really interesting new gene
- RNF5
RING finger protein 5
- SAM
Sterile alpha motif
- SCF
Skp, cullin, F-box-containing complex
- SKICH
SKIP carboxyl homology
- SMURF1
SMAD-specific E3 ubiquitin protein ligase 1
- TOLLIP1
Toll-interacting protein 1
- TSG101
Tumor susceptibility gene 101
- UEV
Ubiquitin E2 variant
- UBL
Ubiquitin-like protein
- UPS
Ubiquitin proteasome system
- ULK1
Uncoordinated 51-like kinase 1
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abdollahzadeh I, Schwarten M, Gensch T, Willbold D, Weiergräber OH. The Atg8 family of proteins—modulating shape and functionality of autophagic membranes. Front Genet. 2017;8:109. doi: 10.3389/fgene.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Aerts MB, Weterman MA, Quadri M, Schelhaas HJ, Bloem BR, Esselink RA, Baas F, Bonifati V, van de Warrenburg BP. A LRSAM1 mutation links Charcot-Marie-Tooth type 2 to Parkinson’s disease. Ann Clin Transl Neurol. 2016;3:146–149. doi: 10.1002/acn3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U, Yarden Y. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin–proteasome system. Biochim Biophys Acta (BBA) Mol Cell Res. 2014;1843:182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 6.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/bse0410015. [DOI] [PubMed] [Google Scholar]
- 7.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic (Copenhagen, Denmark) 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 8.Bah A, Vergne I. Macrophage autophagy and bacterial infections. Front Immunol. 2017;8:1483. doi: 10.3389/fimmu.2017.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. Mammalian autophagy: how does it work? Annu Rev Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 10.Bian W, Guo Y, Zhang Y, Li H. The self-association and activity regulation of LRSAM1 E3 ligase. Biochem Biophys Res Commun. 2017;485:95–101. doi: 10.1016/j.bbrc.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 11.Bogdanik LP, Sleigh JN, Tian C, Samuels ME, Bedard K, Seburn KL, Burgess RW. Loss of the E3 ubiquitin ligase LRSAM1 sensitizes peripheral axons to degeneration in a mouse model of Charcot–Marie–Tooth disease. Dis Models Mech. 2013;6:780–792. doi: 10.1242/dmm.010942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonora M, Patergnani S, Rimessi A, De Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, Wieckowski MR, Pinton P. ATP synthesis and storage. Purinergic Signal. 2012;8:343–357. doi: 10.1007/s11302-012-9305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 14.Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/S0959-440X(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 15.Bucci C, Bakke O, Progida C. Charcot–Marie–Tooth disease and intracellular traffic. Prog Neurobiol. 2012;99:191–225. doi: 10.1016/j.pneurobio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, Noblin DJ, Jorgensen WL, Ciulli A, Crews CM. Targeting the von Hippel–Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF–1alpha interaction. J Am Chem Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulatov E, Ciulli A. Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem J. 2015;467:365–386. doi: 10.1042/BJ20141450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulatov E, Zagidullin A, Valiullina A, Sayarova R, Rizvanov A. Small molecule modulators of RING-Type E3 ligases: MDM and cullin families as targets. Front Pharmacol. 2018;9:450. doi: 10.3389/fphar.2018.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casem ML (2016) Chapter 3—proteins. In: Casem ML (ed) Case studies in cell biology. Academic Press, Boston, pp 23–71
- 20.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harbor Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chhangani D, Joshi AP, Mishra A. E3 ubiquitin ligases in protein quality control mechanism. Mol Neurobiol. 2012;45:571–585. doi: 10.1007/s12035-012-8273-x. [DOI] [PubMed] [Google Scholar]
- 22.Chhangani D, Mishra A. Mahogunin ring finger-1 (MGRN1) suppresses chaperone-associated misfolded protein aggregation and toxicity. Sci Rep. 2013;3:1972. doi: 10.1038/srep01972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chhangani D, Nukina N, Kurosawa M, Amanullah A, Joshi V, Upadhyay A, Mishra A. Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. Biochem Biophys Acta. 2014;1842:1472–1484. doi: 10.1016/j.bbadis.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Chhangani D, Nukina N, Kurosawa M, Amanullah A, Joshi V, Upadhyay A, Mishra A. Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. BBA Mol Basis Dis. 2014;1842:1472–1484. doi: 10.1016/j.bbadis.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Ciardiello F. An update of new targets for cancer treatment: receptor-mediated signals. Ann Oncol. 2002;13:29–38. doi: 10.1093/annonc/mdf635. [DOI] [PubMed] [Google Scholar]
- 26.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 27.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Exp Biol Med (Maywood, N.J.) 2006;231:1197–1211. doi: 10.1177/153537020623100705. [DOI] [PubMed] [Google Scholar]
- 28.Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 30.Cornell M, Evans DA, Mann R, Fostier M, Flasza M, Monthatong M, Artavanis-Tsakonas S, Baron M. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics. 1999;152:567–576. doi: 10.1093/genetics/152.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das G, Shravage BV, Baehrecke EH. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harbor Perspect Biol. 2012;4:4. doi: 10.1101/cshperspect.a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 35.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 36.Du Z, He F, Yu Z, Bowerman B, Bao Z. E3 ubiquitin ligases promote progression of differentiation during C. elegans embryogenesis. Dev Biol. 2015;398:267–279. doi: 10.1016/j.ydbio.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engeholm M, Sekler J, Schöndorf DC, Arora V, Schittenhelm J, Biskup S, Schell C, Gasser T. A novel mutation in LRSAM1 causes axonal Charcot–Marie–Tooth disease with dominant inheritance. BMC Neurol. 2014;14:118. doi: 10.1186/1471-2377-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng GH, Lih C-J, Cohen SN. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Can Res. 2000;60:1736–1741. [PubMed] [Google Scholar]
- 39.Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, Levine B. The ubiquitin-ligase Smurf1 functions in selective autophagy of M. tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017;21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freemont PS. RING for destruction? CB. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 41.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 42.Gart EV, Suchodolski JS, Welsh TH, Alaniz RC, Randel RD, Lawhon SD. Salmonella typhimurium and multidirectional communication in the Gut. Front Microbiol. 2016;7:1827. doi: 10.3389/fmicb.2016.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5:a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guernsey DL, Jiang H, Bedard K, Evans SC, Ferguson M, Matsuoka M, Macgillivray C, Nightingale M, Perry S, Rideout AL, Orr A, Ludman M, Skidmore DL, Benstead T, Samuels ME (2010) Mutation in the gene encoding ubiquitin ligase LRSAM1 in patients with Charcot–Marie–Tooth disease. PLoS Genet 6 [DOI] [PMC free article] [PubMed]
- 47.Guo Y, Bian W, Zhang Y, Li H. Expression in Escherichia coli, purification and characterization of LRSAM1, a LRR and RING domain E3 ubiquitin ligase. Protein Expr Purif. 2017;129:158–161. doi: 10.1016/j.pep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Hakonen JE, Sorrentino V, Avagliano Trezza R, de Wissel MB, van den Berg M, Bleijlevens B, van Ruissen F, Distel B, Baas F, Zelcer N, Weterman MAJ. LRSAM1-mediated ubiquitylation is disrupted in axonal Charcot–Marie–Tooth disease 2P. Hum Mol Genet. 2017;26:2034–2041. doi: 10.1093/hmg/ddx089. [DOI] [PubMed] [Google Scholar]
- 49.Hendrick JP, Hartl FU. The role of molecular chaperones in protein folding. FASEB J. 1995;9:1559–1569. doi: 10.1096/fasebj.9.15.8529835. [DOI] [PubMed] [Google Scholar]
- 50.Henne William M, Buchkovich Nicholas J, Emr Scott D. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 51.Hill SJ, Mordes DA, Cameron LA, Neuberg DS, Landini S, Eggan K, Livingston DM. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci USA. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoeck JD, Jandke A, Blake SM, Nye E, Spencer-Dene B, Brandner S, Behrens A. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 53.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 54.Hoyle JC, Isfort MC, Roggenbuck J, Arnold WD. The genetics of Charcot–Marie–Tooth disease: current trends and future implications for diagnosis and management. Appli Clin Genet. 2015;8:235–243. doi: 10.2147/TACG.S69969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu B, Arpag S, Zuchner S, Li J. A novel missense mutation of CMT2P alters transcription machinery. Ann Neurol. 2016;80:834–845. doi: 10.1002/ana.24776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang C. Roles of E3 ubiquitin ligases in cell adhesion and migration. Cell Adhes Migr. 2010;4:10–18. doi: 10.4161/cam.4.1.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular salmonella typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irannejad R, Tsvetanova NG, Lobingier BT, von Zastrow M. Effects of endocytosis on receptor-mediated signaling. Curr Opin Cell Biol. 2015;35:137–143. doi: 10.1016/j.ceb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irannejad R, von Zastrow M. GPCR signaling along the endocytic pathway. Curr Opin Cell Biol. 2014;27:109–116. doi: 10.1016/j.ceb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/S1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 61.Jacquin E, Apetoh L (2018) Cell-intrinsic roles for autophagy in modulating CD4 T cell functions. Front Immunol 9 [DOI] [PMC free article] [PubMed]
- 62.Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 63.Joshi V, Amanullah A, Upadhyay A, Mishra R, Kumar A, Mishra A. A decade of boon or burden: what has the CHIP ever done for cellular protein quality control mechanism implicated in neurodegeneration and aging? Front Mol Neurosci. 2016;9:93. doi: 10.3389/fnmol.2016.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 67.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18:1129–1142. doi: 10.1091/mbc.e06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 69.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 70.Kuang E, Okumura C, Sheffy-Levin S, Varsano T, Chih-Wen Shu V, Qi J, Niesman I, Yang HJ, López-Otín C, Yuan Yang W, C Reed J, Broday L, Nizet V, Ronai ZE (2012) Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection [DOI] [PMC free article] [PubMed]
- 71.Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics. 1998;51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- 72.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 73.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Su Y, Ryder J, Yan L, Na S, Ni B. RIFLE: a novel ring zinc finger-leucine-rich repeat containing protein, regulates select cell adhesion molecules in PC12 cells. J Cell Biochem. 2003;90:1224–1241. doi: 10.1002/jcb.10674. [DOI] [PubMed] [Google Scholar]
- 75.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 76.Liljas A. Ribosomes. In: Brenner S, Miller JH, editors. Encyclopedia of genetics. New York: Academic Press; 2001. pp. 1723–1730. [Google Scholar]
- 77.Lin Q, Dai Q, Meng H, Sun A, Wei J, Peng K, Childress C, Chen M, Shao G, Yang W (2017) The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. 130:3839–3850 [DOI] [PubMed]
- 78.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 80.Maheshwari M, Samanta A, Godavarthi SK, Mukherjee R, Jana NR. Dysfunction of the ubiquitin ligase Ube3a may be associated with synaptic pathophysiology in a mouse model of huntington disease. J Biol Chem. 2012;287:29949–29957. doi: 10.1074/jbc.M112.371724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. PARKIN ubiquitin ligase mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 83.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/S0378-1119(97)00084-X. [DOI] [PubMed] [Google Scholar]
- 84.McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol Biol Cell. 2008;19:754–763. doi: 10.1091/mbc.e07-09-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Minaidou A, Nicolaou P, Christodoulou K. LRSAM1 depletion affects neuroblastoma SH-SY5Y cell growth and morphology: the LRSAM1 c.2047-1G > A Loss-of-function variant fails to rescue the phenotype. Cell J. 2018;20:340–347. doi: 10.22074/cellj.2018.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mishra A, Dikshit P, Purkayastha S, Sharma J, Nukina N, Jana NR. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem. 2008;283:7648–7656. doi: 10.1074/jbc.M706620200. [DOI] [PubMed] [Google Scholar]
- 88.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mtambo A, Chan K, Shen A, Lima V, Hogg R, Montaner J, Moore D. Treatment limitations imposed by antiretroviral drug resistance mutations: implication for choices of first line regimens in resource-limited settings. HIV medicine. 2012;13:141–147. doi: 10.1111/j.1468-1293.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 90.Myers EL, Allen JF. Tsg101, an inactive homologue of ubiquitin ligase E2, interacts specifically with human immunodeficiency virus type 2 Gag polyprotein and results in increased levels of ubiquitinated gag. J Virol. 2002;76:11226–11235. doi: 10.1128/JVI.76.22.11226-11235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21:245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ng A, Xavier RJ. Leucine-rich repeat (LRR) proteins: integrators of pattern recognition and signaling in immunity. Autophagy. 2011;7:1082–1084. doi: 10.4161/auto.7.9.16464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicolaou P, Cianchetti C, Minaidou A, Marrosu G, Zamba-Papanicolaou E, Middleton L, Christodoulou K. A novel LRSAM1 mutation is associated with autosomal dominant axonal Charcot–Marie–Tooth disease. Eur J Hum Genet. 2013;21:190–194. doi: 10.1038/ejhg.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21:93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noda NN, Inagaki F (2014). Chapter 2—selective autophagy: role of interaction between the Atg8 family interacting Motif and Atg8 family proteins. In: Hayat MA (ed) Autophagy: cancer, other pathologies, inflammation, immunity, infection, and aging. Academic Press, Amsterdam, pp 39–48
- 96.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 97.Oldham RA, Berinstein EM, Medin JA. Lentiviral vectors in cancer immunotherapy. Immunotherapy. 2015;7:271–284. doi: 10.2217/imt.14.108. [DOI] [PubMed] [Google Scholar]
- 98.Peeters K, Palaima P, Pelayo-Negro AL, Garcia A, Gallardo E, Garcia-Barredo R, Mateiu L, Baets J, Menten B, De Vriendt E, De Jonghe P, Timmerman V, Infante J, Berciano J, Jordanova A. Charcot–Marie–Tooth disease type 2G redefined by a novel mutation in LRSAM1. Ann Neurol. 2016;80:823–833. doi: 10.1002/ana.24775. [DOI] [PubMed] [Google Scholar]
- 99.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Przedborski S, Vila M, Jackson-Lewis V. Series introduction: neurodegeneration: what is it and where are we? J Clin Investig. 2003;111:3–10. doi: 10.1172/JCI200317522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Puertollano R (2006) Endocytic trafficking and human diseases. In: Endosomes. Springer, New York, pp 119–131
- 102.Pyo J-O, Nah J, Jung Y-K. Molecules and their functions in autophagy. Exp Mol Med. 2012;44:73–80. doi: 10.3858/emm.2012.44.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Regad T. Targeting RTK signaling pathways in cancer. Cancers. 2015;7:1758–1784. doi: 10.3390/cancers7030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10:S10. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 105.Roy N, Pacini G, Berlioz-Torrent C, Janvier K. Characterization of E3 ligases involved in lysosomal sorting of the HIV-1 restriction factor BST2. J Cell Sci. 2017;130:1596–1611. doi: 10.1242/jcs.195412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Santiago JA, Bottero V, Potashkin JA. Dissecting the molecular mechanisms of neurodegenerative diseases through network biology. Front Aging Neurosci. 2017;9:166. doi: 10.3389/fnagi.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soares P, Gadd MS, Frost J, Galdeano C, Ellis L, Epemolu O, Rocha S, Read KD, Ciulli A (2018) Group-based optimization of potent and cell-active inhibitors of the von Hippel-Lindau (VHL) E3 ubiquitin ligase: structure-activity relationships leading to the chemical probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298). 61:599–618 [DOI] [PMC free article] [PubMed]
- 110.Spies M, Smith BO. Protein–nucleic acids interactions: new ways of connecting structure, dynamics and function. Biophys Rev. 2017;9:289–291. doi: 10.1007/s12551-017-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stegmuller J, Bonni A (2010) Destroy to create: E3 ubiquitin ligases in neurogenesis. F1000 Biol Rep 2 [DOI] [PMC free article] [PubMed]
- 112.Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res. 2016;26:423–440. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tang B, Seredenina T, Coppola G, Kuhn A, Geschwind DH, Luthi-Carter R, Thomas EA. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lengths reveals genes associated with disease onset and progression in Huntington’s disease. Neurobiol Dis. 2011;42:459–467. doi: 10.1016/j.nbd.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tattoli I, Sorbara MT, Philpott DJ, Girardin SE. Bacterial autophagy: the trigger, the target and the timing. Autophagy. 2012;8:1848–1850. doi: 10.4161/auto.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tazir M, Bellatache M, Nouioua S, Vallat JM. Autosomal recessive Charcot–Marie–Tooth disease: from genes to phenotypes. JPNS. 2013;18:113–129. doi: 10.1111/jns5.12026. [DOI] [PubMed] [Google Scholar]
- 116.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 119.Upadhyay A, Amanullah A, Chhangani D, Mishra R, Mishra A. Selective multifaceted E3 ubiquitin ligases barricade extreme defense: potential therapeutic targets for neurodegeneration and ageing. Age Res Rev. 2015;24:138–159. doi: 10.1016/j.arr.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 120.Upadhyay A, Amanullah A, Chhangani D, Mishra R, Prasad A, Mishra A. Mahogunin ring Finger-1 (MGRN1), a multifaceted ubiquitin ligase: recent unraveling of neurobiological mechanisms. Mol Neurobiol. 2016;53:4484–4496. doi: 10.1007/s12035-015-9379-8. [DOI] [PubMed] [Google Scholar]
- 121.Upadhyay A, Joshi V, Amanullah A, Mishra R, Arora N, Prasad A, Mishra A. E3 ubiquitin ligases neurobiological mechanisms: development to degeneration. Front Mol Neurosci. 2017;10:151. doi: 10.3389/fnmol.2017.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vallat JM, Mathis S, Funalot B. The various Charcot–Marie–Tooth diseases. Curr Opin Neurol. 2013;26:473–480. doi: 10.1097/WCO.0b013e328364c04b. [DOI] [PubMed] [Google Scholar]
- 123.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, N.Y.) 303:844–848 [DOI] [PubMed]
- 124.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vilchez D, Saez I, Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 126.von Muhlinen N, Thurston T, Ryzhakov G, Bloor S, Randow F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy. 2010;6:288–289. doi: 10.4161/auto.6.2.11118. [DOI] [PubMed] [Google Scholar]
- 127.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 128.Weterman MA, Sorrentino V, Kasher PR, Jakobs ME, van Engelen BG, Fluiter K, de Wissel MB, Sizarov A, Nurnberg G, Nurnberg P, Zelcer N, Schelhaas HJ, Baas F. A frameshift mutation in LRSAM1 is responsible for a dominant hereditary polyneuropathy. Hum Mol Genet. 2012;21:358–370. doi: 10.1093/hmg/ddr471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Willis MS, Schisler JC, Patterson C. Appetite for destruction: E3 ubiquitin-ligase protection in cardiac disease. Future Cardiol. 2008;4:65–75. doi: 10.2217/14796678.4.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wong E, Cuervo AM. Integration of Clearance mechanisms: the proteasome and autophagy. Cold Spring Harbor Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang H, Zhong X, Ballar P, Luo S, Shen Y, Rubinsztein DC, Monteiro MJ, Fang S. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp Cell Res. 2007;313:538–550. doi: 10.1016/j.yexcr.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 132.Yokomizo T, Dzierzak E. Fine-tuning of hematopoietic stem cell homeostasis: novel role for ubiquitin ligase. Genes Dev. 2008;22:960–963. doi: 10.1101/gad.1669908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu HB, Croxen MA, Marchiando AM, Ferreira RBR, Cadwell K, Foster LJ, Finlay BB (2014) Autophagy facilitates salmonella replication in HeLa cells. mBio 5:e00865–00814 [DOI] [PMC free article] [PubMed]
- 134.Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao G, Song J, Yang M, Song X, Liu X (2018). A novel mutation of LRSAM1 in a Chinese family with Charcot–Marie–Tooth disease. 23:55–59 [DOI] [PubMed]
- 137.Zhou J, Zhang Y, Qi J, Chi Y, Fan B, Yu JQ, Chen Z. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi: 10.1371/journal.pgen.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhu M, Arpag S, Hamilton A, Yawn R, Zuchner S, Li J (2015) Charcot–Marie–Tooth disease type-2P is associated with a missense mutation in the RING domain of LRSAM1 (P2.033). Neurology 84