Abstract
Activation of Kupffer cells (KCs) induced that inflammatory cytokine production plays a central role in the pathogenesis of HBV infection. The previous studies from our and other laboratory demonstrated miRNAs can regulate TLR-inducing inflammatory responses to macrophage. However, the involvement of miRNAs in HBV-associated antigen-induced macrophage activation is still not thoroughly understood. Here, we evaluated the effects and mechanisms of miR-155 in HBV-associated antigen-induced macrophage activation. First, co-culture assay of HepG2 or HepG2.2.15 cells and RAW264.7 macrophages showed that HepG2.2.15 cells could significantly promote macrophages to produce inflammatory cytokines. Furthermore, we, respectively, stimulated RAW264.7 macrophages, mouse primary peritoneal macrophages, or healthy human peripheral blood monocytes with HBV-associated antigens, including HBcAg, HBeAg, and HBsAg, and found that only HBeAg could steadily enhance the production of inflammatory cytokines in these cells. Subsequently, miRNAs sequencing presented the up- or down-regulated expression of multiple miRNAs in HBeAg-stimulated RAW264.7 cells. In addition, we verified the expression of miR-155 and its precursors BIC gene with q-PCR in the system of co-culture or HBeAg-stimulated macrophages. Meanwhile, the increased miR-155 expression was positively correlation with serum ALT, AST, and HBeAg levels in AHB patients. Although MAPK, PI3K, and NF-κB signal pathways were all activated during HBeAg treatment, only PI3K and NF-κB pathways were involved in miR-155 expression induced by HBeAg stimulation. Consistently, miR-155 over-expression inhibited production of inflammatory cytokines, which could be reversed by knocking down miR-155. Moreover, we demonstrated that miR-155 regulated HBeAg-induced cytokine production by targeting BCL-6, SHIP-1, and SOCS-1. In conclusion, our data revealed that HBeAg augments the expression of miR-155 in macrophages via PI3K and NF-κB signal pathway and the increased miR-155 promotes HBeAg-induced inflammatory cytokine production by inhibiting the expression of BCL-6, SHIP-1, and SOCS-1.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2753-8) contains supplementary material, which is available to authorized users.
Keywords: HBeAg, miR-155, Macrophage, NF-κB, PI3K
Introduction
Hepatitis B is a viral infection that attacks the liver and leads to both acute and chronic inflammatory response in the liver. Nearly, 257 million people are livingly infected with hepatitis B virus, defined as HBsAg positive, in the world. According to the survey, complications of hepatitis B, including liver cirrhosis and hepatocellular carcinoma, cause more than 887,000 people deaths in 2015 [1–3]. The prevalence of HBV used to be ≥ 8% in China and once a nationwide HBV general survey showed that HBsAg carrier rate was 9.75% in population [4]. Hence, hepatitis B is one of the major health burden worldwide, especially in China. It is well known that HBV can replicate within hepatocytes without causing direct cell damage. Nevertheless, the host immune response is not only indispensable to control the spread of HBV infection, but it is also responsible for the inflammatory events causing liver injury [5]. We and others have reported that macrophages play an important role in the pathogenesis of HBV-induced immune-mediated liver injury [6–8]. However, the role of HBV-associated antigens, including HBcAg, HBeAg, and HBsAg, in activating macrophages has not been completely elucidated.
MicroRNAs (miRNA) are a class of endogenous, highly conserved, small non-coding RNA molecules and originate from genome of eukaryotic organisms and even kinds of virus. miRNAs have emerged as a major kind of gene expression regulators at post-transcriptional level and are involved in a wide variety of biological processes. Mature miRNA interacts with corresponding specific mRNA through imperfectly base pairing between the miRNA sequence and the 3′-untranslated region (3′-UTR) of specific mRNA, and leads to translational repression or degradation of target mRNA. Emerging evidences have shown that miRNAs play a critical role in TLR-induced macrophage activation and HBV replication in hepatocyte. For example, miR-146, 147, 155, and 210, which were induced by TLRs’ stimulation in macrophages, functioned as a regulator for TLR-induced inflammatory responses through targeting IRAK-1 and TRAF6, SHIP-1, or NF-κB1, respectively [9–12]. Ectopic expression of miR-155 enhanced the expression of several IFN-inducible anti-viral genes by suppressing SOCS-1 to promote JAK/STAT signaling pathway in human hepatoma cells [13]. Up-regulation of miR-199a-3p and miR-210 in HepG2.2.15 cells compared to HepG2 cells may play a role in regulating HBV replication and maintenance of a suitable level of virion production in persistent infection by targeting crucial HBV genes [14]. However, the effect and mechanism of HBV-infection-regulated macrophage miRNAs expression remain ill-defined.
In the present study, we demonstrated that co-culture of HepG2.2.15 cells and RAW264.7 macrophages could significantly promote inflammatory cytokine production. Furthermore, we verified that HBeAg, but not HBcAg and HBsAg, played a predominant role in HBV promoted inflammatory cytokine production from macrophages. Meanwhile, the miRNAs sequencing and q-PCR assay found that the expression of miR-155 was augmented in HBeAg-stimulated macrophages. In addition, over-expression of miR-155 dramatically increased cytokines production. In contrast, knockdown of miR-155 had a reverse effect. Besides, miR-155 expression was increased in peripheral blood monocytes from AHB patients, which was positively correlated with ALT, AST, and HBeAg levels in the serum. Mechanistically, we unveiled that NF-κB and PI3K signal pathway mediated miR-155 expression induced by HBeAg stimulation in macrophages and the inductive miR-155 raised the production of inflammatory cytokines by targeting BCL-6, SHIP-1 and SOCS-1 after HBeAg stimulation.
Materials and methods
Healthy controls and patients
A total of 31 subjects, including 8 healthy controls, 10 patients with AHB, and 13 patients with CHB, were enrolled from Shandong Provincial Hospital Affiliated to Shandong University and the Second Hospital of Shandong University, during April 2016 to March 2017 in this study. The criteria established the National Viral Hepatitis Conference of China (2015) were used to diagnosis AHB and CHB. Eight healthy individuals from the Healthy Physical Examination Center of Shandong Provincial Hospital Affiliated to Shandong University served as control group. Clinical characteristics of these enrolled subjects are summarized in Table 1. They were all negative for antibodies to HAV, HCV, HDV, HEV, and HIV. None of them were all autoimmune liver diseases and taking immunosuppressive drugs or anti-viral therapy more than 3 months. The study protocol and consent program were approved by the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University.
Table 1.
Clinical characteristics of enrolled subjects
| Group | Healthy controls | AHB | CHB |
|---|---|---|---|
| Case | 8 | 10 | 13 |
| Sex (male) | 8 | 7 | 11 |
| BMIa | 22.69 ± 1.65 | 24.08 ± 2.09 | 25.69 ± 1.81 |
| Age (years)a | 42 ± 14.59 | 45.5 ± 14.16 | 48 ± 13.60 |
| ALT (U/l)a | 17 ± 7.53 | 1568 ± 1168.66 | 343 ± 741.71 |
| AST (U/l)a | 21 ± 5.48 | 1457.5 ± 1176.76 | 243 ± 464.07 |
| HBV-DNA positive (> 103 copies/ml) | 0 | 10 (100%) | 11 (84.62%) |
| IIBsAg positive | 0 | 10 (100%) | 13 (100%) |
| HBeAg positive | 0 | 10 (100%) | 9 (69.23%) |
AHB patients with acute hepatitis B, CHB patients with chronic hepatitis B, BMI body mass index
aMedian ± SD
Mice, cells, and reagents
C57BL/6J mice were obtained from the Animal Research Committee of the Institute of Biology and Cell Biology (Shanghai, China) and housed in a specific pathogen-free environment. The animal room was kept at 20–22 °C under a 12 h light/dark cycle. All animal experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of the Shandong Provincial Hospital Affiliated to Shandong University, Jinan, Shandong Province, China. Mouse primary peritoneal macrophages were prepared as we described before [6]. Briefly, after 1 ml sterile 6% starch solution was injected into the peritoneal cavity for 72 h, serum-free DMEM medium was used to rinse exudate macrophages. Acquired cells were seeded and cultured in complete medium of DMEM with 10% FBS overnight, and then, they were treated with HBV-associated antigens. Mouse macrophage cell lines RAW264.7 were obtained from the American-Type Culture Collection (Manassas, VA, US). The human hepatoma cell lines HepG2 and HepG2.2.15 were obtained from the Shanghai Cell Collection (Chinese Academy of Sciences, Beijing, China). All cells were cultured in DMEM (Gibco-BRL, Grand Island, NY, USA) containing 10% (vol/vol) FBS (Gibco® Sera, AUS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, Grand Island, NY and Scotland, UK) and maintained at 37 °C in a humidified incubator with 5% CO2. HBcAg (ab119441), HBeAg (ab91273), and HBsAg (ab167754) were purchased from Abcam (Abcam, Cambridge, MA, USA). Mouse mAb to GAPDH (60004-1-lg) was purchased from Proteintech Group, Inc. (Rosemont, IL, USA). SOCS-1 (TA321593) was from OriGene (OriGene Technologies, Inc, Rockville, MD, USA). SHIP-1 (ab45142) was purchased from Abcam (Abcam, Cambridge, MA, USA). Antibodies for BCL-6 (#5650), ERK (#4695), p-ERK (#9101), JNK (#9252), p-JNK (#9251), P38 (#9212), p-P38 (#4631), P65 (#4764), p-P65 (#3033), AKT (#2920), and p-AKT (#4060) were obtained from cell signaling (Cell-Signaling Technology, Boston, MA, USA). These corresponding HRP-conjugated secondary antibodies were purchased from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX, USA). PD98059 (ERK inhibitor, HY-12028), SB203580 (P38 inhibitor, HY-10256A), SP600125 (JNK inhibitor, HY-12041), BAY 11-7082 (NF-κB inhibitor, HY-13453), and Wortmannin (PI3K inhibitor, HY-10197) were acquired from MCE (MedChemExpress, Pudong New Area, Shanghai, China).
Co-culture of macrophages and HepG2/HepG2.2.15
Transwell co-culture experiments were performed in 6-, 12-, or 24-well plates with 0.4 μm pore sizes in inner wells (3412, 3401, and 3413, Corning, NY, USA) to physically separate macrophages and HepG2/HepG2.2.15. Approximately 4 × 105/ml macrophages were seeded and adhered to the lower chamber using DMEM with 10% FBS. Next, 3 × 105/ml HepG2/HepG2.2.15 cells were added into the upper chamber in the same medium. The membrane of Boyden Chamber with 0.4 μm pore sizes allowed the interaction of soluble substances between the two cell types. After co-incubation for 12, 24, 36, and 48 h at 37 °C in a humidified incubator with 5% CO2, macrophages were harvested with TRizol for evaluation of cytokine mRNA and miRNA expression levels by q-PCR or with the RIPA lysis buffer supplemented with a protease inhibitor ‘cocktail’ for proteins of signal pathway by Western blot.
q-PCR for mRNA and miRNA quantification
Total RNA was extracted from macrophages with the miRNeasy Mini Kit (217004; Qiagen, Germantown, MD, USA) according to manufacturer instructions. For detection of miR-155 quantification, cDNA was prepared in a reverse-transcription reaction using a miScript II RT kit (218161; Qiagen, Nasdaq, NY, USA). Then, the cDNA templates were analyzed using corresponding primer pairs and miScript SYBR Green PCR kit (218073; Qiagen, Nasdaq, NY, USA) according to manufacturer’s instructions. U6 snRNA was used as an internal control.
For mRNA quantification, cDNA was synthesized using a First-Strand cDNA Synthesis kit (RR047A; TaKaRa, Kusatsu, Shiga, Japan). Expression of TNF-α, IL-6, and IFN-β was determined by quantitative real-time PCR using SYBR Premix Ex Tap™ (TaKaRa, Kusatsu, Shiga, Japan) with GAPDH as an internal normalized reference. Specific primers were used as described previously [15] and indicated as follows: 5′-GCCAC CACGC TCTTC TGTCT-3′ (sense) and 5′-TGAGG GTCTG GGCCA TAGAA C-3′ (antisense) for TNF-α; 5′-ACAAC CACGG CCTTC CCTAC-3′ (sense) and 5′-CATTT CCACG ATTTC CCAGA-3′ (antisense) for IL-6; 5′-AGTTA CACTG CCTTT GCC-3′ (sense) and 5′-GTTGA GGACA TCTCC CAC-3′ (antisense) for IFN-β; and 5′-CAAGG TCATC CATGA CAACT TTG-3′ (sense) and 5′-GTCCA CCACC CTGTT GCTGT AG-3′ (antisense) for GAPDH. Real-time PCR analyses were run on the LightCycler Real-Time PCR System (Roche Diagnostics, USA) as described previously [16].
Enzyme-linked immunosorbent assay (ELISA)
The cell-culture supernatants were collected, and the concentration of IL-6 (KMC0061, Invitrogen, Carlsbad, CA, USA), TNF-α (KMC3011, Invitrogen, Carlsbad, CA, USA), and IFN-β (439407, BioLegend, Inc. San Diego, CA) was measured using a commercially available ELISA kit (Invitrogen, Carlsbad, CA, USA) as described previously [16]. All protocols were conducted according to the manufacturer’s instructions.
Library preparation and sequencing of small RNA
The small RNA library and sequencing were performed by Annoroad Gene Technology (Beijing, China). After RAW264.7 macrophages were stimulated with HBeAg for 0, 24, or 36 h, total RNA was extracted and separated by 15% agarose gels to obtain the small RNA (18–30 nt). Small RNA sample was precipitated by ethanol and centrifuged. In addition, the library was prepared with Small RNA Sample Preparation Kit (Illumina, RS-200-0048, San Diego, CA, USA) according to manufacturer’s instructions. RNA concentration of library was measured using Qubit® RNA Assay Kit (Invitrogen, Carlsbad, CA, USA) in Qubit® 2.0 (Invitrogen, Carlsbad, CA, USA) to preliminary quantify and then diluted to 1 ng/μl. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and after the insert size consistent with expectations, qualified insert size was accurate quantitative using Taqman fluorescence probe of AB Step One Plus Real-Time PCR system (library valid concentration > 2 nM). The qualified libraries were sequenced by an Illumina Hiseq 2500 platform and generated 50 bp single-end reads.
Western blot analysis
The cultured cells were washed with cold phosphate-buffered saline (PBS) and lysed with RIPA lysis buffer (Beyotime Biotechnology, Songjiang District, Shanghai, China) supplemented with a protease inhibitor ‘cocktail’. After the protein concentrations were measured using the bicinchoninic acid assay (Pierce, Rockford, IL, USA), equal amounts of protein lysates were separated by SDS/PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA) for immunoblot analysis as described previously [16, 17]. The membranes were then incubated with the primary antibodies overnight at 4 °C. After washing with TBST, the membranes were hybridized with the corresponding HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Dallas, TX, USA). Finally, an enhanced ECL chemiluminescence reagent kit (Millipore, Billerica, MA, USA) was used to detect the objective protein in accordance with the manufacturer’s protocol.
Application of ago- or antago-miR-155 and RNAi
Ago- or antago-miR-155 and corresponding negative control miRNA were purchased from Genepharma (Pudong new area, Shanghai, China). RNAi of SOCS-1, SHIP-1, and BCL-6 were acquired from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX, USA). These miRNA or/and RNAis were added to cells using the INTERFERin® Transfection Reagents (Polyplus-transfection®, Illkirch, NY, USA) following the manufacturer’s protocol.
Statistical analysis
All data were presented as a result of three or four independent experiments and expressed as the mean ± SD. GraphPad Prism software (GraphPad Software, San Diego, CA, USA) was used for these data analysis. The Student’s t test or one-way analysis of variance (ANOVA) was applied to confirm significant differences between groups. Two-way ANOVA was applied to ensure significant differences between treatments, in different cell cohorts, or at different timepoints. Spearman correlation analysis was performed between the miRNA-155 expression and serum ALT, AST, HBeAg, HBsAg, or HBV-DNA levels. In all cases, Values of p < 0.05 were considered to be statistically significant.
Results
HepG2.2.15, but not HepG2, induced RAW 264.7 macrophages to produce cytokines in the co-culture system
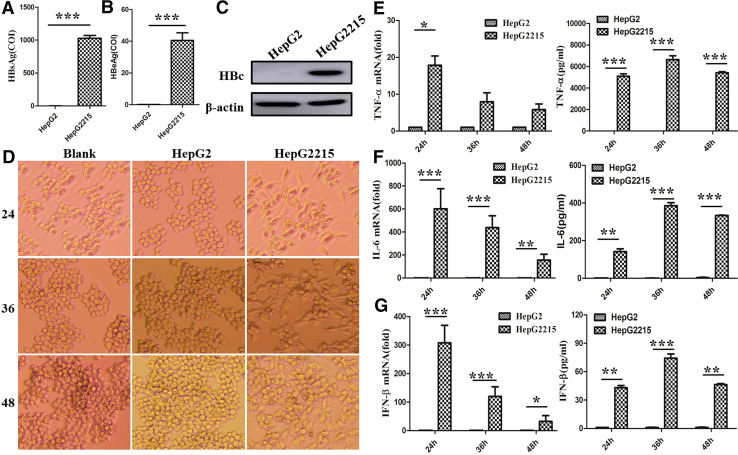
The previous studies from our and other laboratory demonstrated that macrophages play an important role in immune hepatitis, including viral hepatitis and drug-induced hepatitis [6], but the effect and mechanism of HBV-associated antigen for macrophage activation are still not thoroughly understood. Therefore, in this study, we first analyzed the role of HBV-associated antigens, including HBcAg, HBeAg, and HBsAg, in HBV-induced macrophage activation. As shown in Fig. 1a–c, high levels of HBcAg, HBeAg, and HBsAg could be detected in HepG2.2.15 cells, which was integrated HBV genome DNA in HepG2 cells. Second, we examined the role of HBV-associated antigens in macrophage activation using the co-culture system. We found that HepG2.2.15 cells made RAW264.7 macrophages to become more stretched and multilateral (Fig. 1d). However, RAW264.7 cells co-cultured with or without HepG2 cells still kept the round morphology. Previously, we have demonstrated that the change of cellular morphology foreboded macrophage activation upon LPS challenge [16]. Therefore, we further evaluated the status of macrophage activation by detecting the production of inflammatory cytokines. To our surprise, HepG2.2.15 cells increased the expression and secretion of TNF-α, IL-6, and IFN-β of RAW264.7 cells (Fig. 1e–g) compared with HepG2 cells. Taken together, these results imply that HBV antigens might potentiate macrophages activation.
Fig. 1.
HepG2.2.15, but not HepG2, induced RAW 264.7 macrophages cytokine production in the co-culture system. The contents of HBeAg and HBsAg were measured by electrochemiluminescent immunoassay (ECLIA) in cultural supernatant of HepG2 and HepG2.2.15 cells (a, b). The expression of HBcAg was evaluated with western blot assay in HepG2 and HepG2.2.15 cells (c). After HepG2 or HepG2.2.15 and RAW264.7 cells were co-cultured with transwell for 24, 36, or 48 h, the morphology of RAW264.7 macrophages was observed with light microscope (d original magnification, ×100). The expression of TNF-α, IL-6, and IFN-β was tested with q-PCR (the left of e–g), and the secretion of them was analyzed with ELISA (the right of e–g). Data are shown in six independent experiments (mean ± SD of quadruplicates in a, b and triplicates in e–g). *p < 0.05, **p < 0.01, ***p < 0.001
HBeAg played a predominant role in inducing cytokine production of macrophages
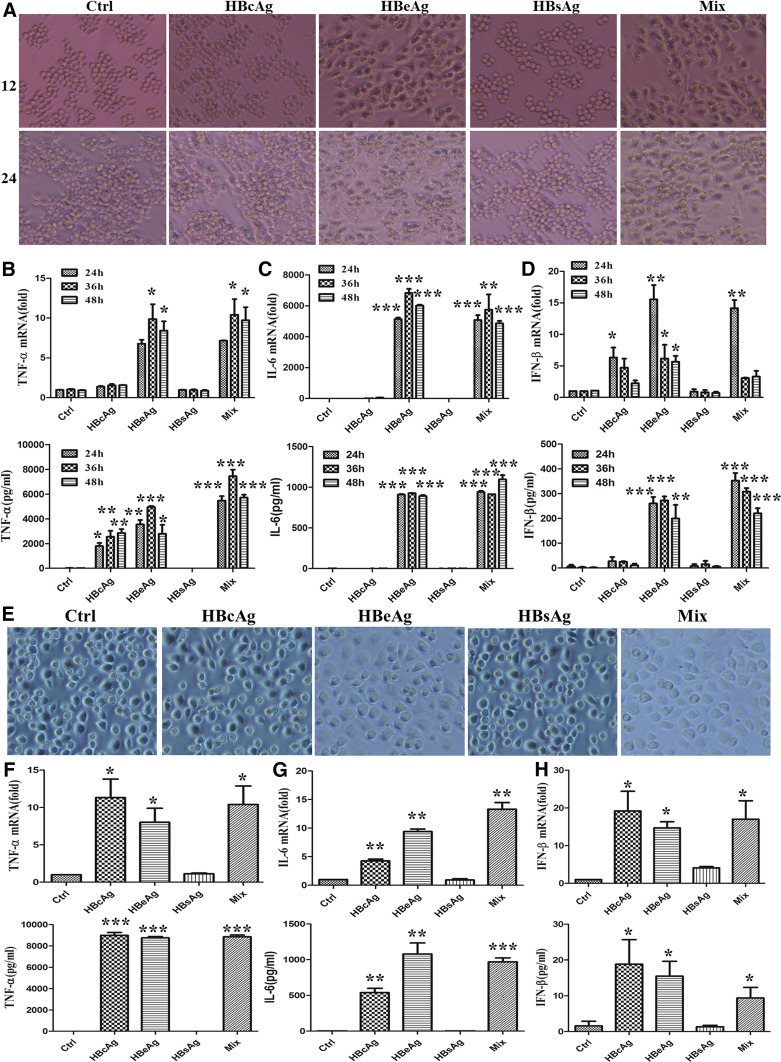
Most generally, except HBV-DNA replication, HBV-associated antigens (including HBcAg, HBeAg, and HBsAg) were also promptly production after HBV infection. To identify which antigens play an essential role in inducing macrophage activation, we stimulated RAW 264.7 macrophages with different antigens (2 μg/ml) or their mixture. As shown in Fig. 2a, cellular morphology of macrophages was greatly changed after stimulation with HBeAg or mixture for 12 and 24 h. Meanwhile, we detected the production of cytokines. Consistently, as shown in Fig. 2b–d, the expression and secretion of TNF-α, IL-6, and IFN-β were remarkably raised in macrophages stimulated with HBeAg or mixture for 24, 36, and 48 h. In addition, we also found that HBcAg played a weak role in the secretion of TNF-α and expression of IFN-β. To further validate the effect of HBV-associated antigens on other macrophages, we confirmed the role of HBeAg with mouse primary peritoneal macrophages. As shown in Fig. 2e, the morphology of peritoneal macrophages was also more flattening and applanate in HBeAg or mixture treated macrophages. At the same time, the production of TNF-α, IL-6, and IFN-β was also increased in these two groups (Fig. 2f–h). It seemed that primary peritoneal macrophages were more sensitive to HBcAg stimulation compared to RAW264.7 macrophage.
Fig. 2.
HBeAg might play a predominant role in macrophage cytokines production. a–d RAW264.7 macrophages were stimulated with HBcAg, HBeAg, HBsAg, or their mixture (2 μg/ml) for 12, 24, 36, or 48 h. The cell morphology was observed with light microscope (a original magnification, ×100). The expression of TNF-α, IL-6, and IFN-β was tested with q-PCR (the top of b–d), and the secretion of them was analyzed with ELISA (the bottom of b–d). e–h Mouse primary peritoneal macrophages were treated with HBcAg, HBeAg, HBsAg, or their mixture (2 µg/ml) for 36 h. The cell morphology was observed with light microscope (e original magnification, ×100). The expression of TNF-α, IL-6 and IFN-β was tested with q-PCR (the top of f–h), and the secretion of them was analyzed with ELISA (the bottom of f–h). Data are representative of three independent experiments (mean ± SD of triplicates in b–d and f–h). *p < 0.05, **p < 0.01, ***p < 0.001
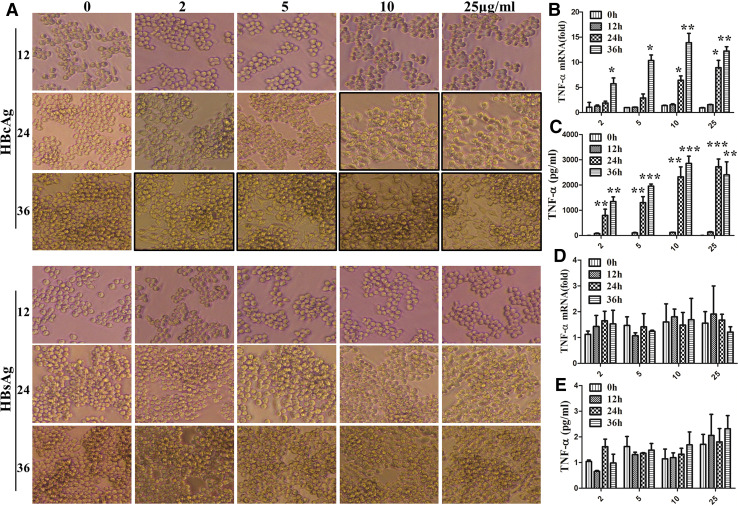
Next, to eliminate the slight role of HBcAg and HBsAg for RAW264.7 macrophages activation not because of dose or timepoints, the RAW264.7 cells were simulated with HBcAg or HBsAg for longer time and larger dose. The results showed that HBcAg indeed altered the morphology of RAW264.7 cells (Fig. 3a, upper) and promoted the expression and secretion of TNF-α (Fig. 3b, c) after incubation with higher dose or longer time, but no significant differences were found for the change of cell morphology (Fig. 3a, lower) and TNF-α production (Fig. 3d, e) of macrophages under stimulation with HBsAg at different time or dose points. All together, these data suggest that HBeAg is a most critical protein in HBV-associated antigen inducing macrophage activation.
Fig. 3.
HBsAg did not affect the production of RAW264.7 macrophage cytokine at any timepoints and concentration. RAW264.7 macrophages were stimulated with HBcAg or HBsAg. The cell morphology was recorded with light microscope (a original magnification, ×100). The expression of TNF-α was tested with q-PCR (b, d), and the secretion of them was analyzed with ELISA (c, e). Data are representative of three independent experiments (mean ± SD of triplicates in b–e). *p < 0.05, **p < 0.01, ***p < 0.001
HBeAg-induced miR-155 expression in macrophages
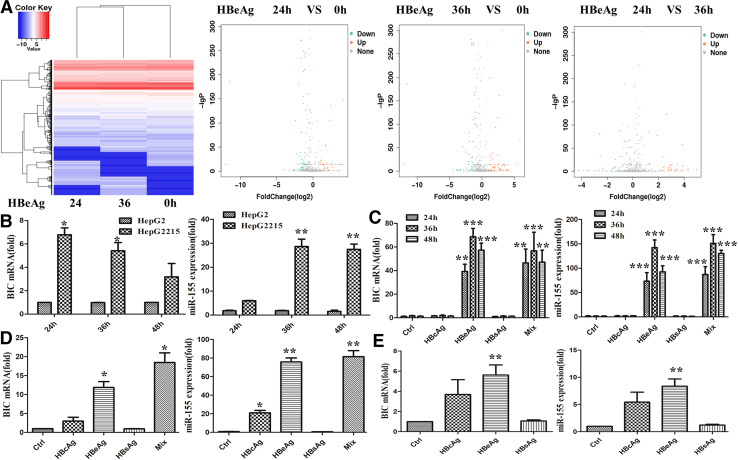
Accumulating data had shown that miRNAs play a critical role in macrophages activation by TLR [9], but the role of HBV-associated antigens in inducing macrophage miRNAs expression has not been clearly identified. Given that the effect of HBeAg on macrophage activation, microRNAs sequencing assay were performed to identify miRNAs involved in HBeAg-induced macrophage activation. The results revealed that miR-155 expression was significantly up-regulated after HBeAg stimulating RAW264.7 macrophage for 24 and 36 h (Fig. 4a). Afterwards, we detected the expression of miR-155 and BIC, the precursor of miR-155, with quantitative real-time PCR in RAW 264.7 cells after co-culture with HepG2 or HepG2.2.15 cells. As shown in Fig. 4b, the expression of BIC and miR-155 were enhanced about 7- and 30-fold. Furthermore, miR-155 expression was induced in a time-dependent manner. To further verify the effect of HBV-associated antigens on miR-155 and BIC expression, we treated RAW264.7 macrophages with different antigens or their mixture for different timepoints. As shown in Fig. 4c, BIC expression was raised approximately 50 fold and miR-155 was up-regulated about more than 100 fold in HBeAg or mixture stimulated macrophages. Similarly, the expression of BIC and miR-155 were also promoted in mouse primary peritoneal macrophages (Fig. 4d) or human peripheral blood monocytes (Fig. 4e) after stimulation with HBeAg and mixture. Notably, miR-155 expression was also increased in peritoneal macrophages after HBcAg stimulation (Fig. 4d). However, no any effect on these two macrophages was found in HBsAg treatment group. This result was in line with the level of cytokines (Figs. 2, 3). Taken together, these results implicated that HBeAg, but not HBcAg and HBsAg, was a pivotal element in inducing miR-155 expression in macrophages.
Fig. 4.
HBeAg-induced miR-155 expression in different kinds of macrophages. RAW264.7 macrophages were stimulated with HBeAg for 24 or 36 h, and microRNA sequencing assay was performed according to the manufacturer’s protocols (a). After HepG2 or HepG2.2.15 and RAW264.7 cells were co-cultured with transwell for 24, 36, or 48 h, the expression of miR-155 and BIC was measured with q-PCR (b). RAW264.7 macrophages were stimulated with HBeAg for 24, 36, or 48 h, and miR-155 and BIC expression was evaluated with q-PCR (c). Mouse primary peritoneal macrophages (d) and peripheral blood monocytes of health controls (e) were treated with HBeAg for 36 h, and miR-155 and BIC expression was evaluated with q-PCR. Data are representative of three independent experiments (mean ± SD of triplicates in b, c and quadruplicates in d, e). *p < 0.05, **p < 0.01, ***p < 0.001
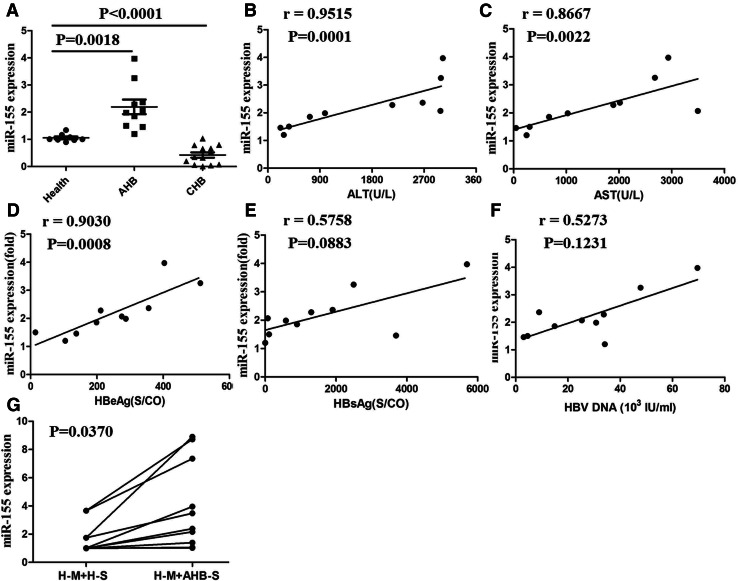
The increased miR-155 in human peripheral blood monocytes of patients with AHB was positively correlated with some clinical parameters
To verify the correlation of miR-155 expression and multiple clinical indexes in patients with HBV infection, we collected peripheral blood monocytes of 8 healthy individuals, 10 patients with AHB and 13 patients with CHB and evaluated miR-155 expression. As shown in Fig. 5a, there was a significant increase of miR-155 expression in peripheral blood monocytes of AHB patients compared with that of healthy controls. Reversely, its expression was decreased in peripheral blood monocytes of CHB patients compared with that of healthy controls. Next, we analyzed the correlation of miR-155 expression and clinical features, such as ALT, AST, HBeAg, HBsAg and HBV-DNA, in AHB patients. As shown in Fig. 5b–d, the expression of miR-155 was positively correlated with serum ALT and AST levels and HBeAg content. Yet, its expression was irrelevant with HBsAg and HBV-DNA (Fig. 5e, f). In addition, peripheral blood monocytes of healthy individuals were stimulated with the serum from AHB patients with higher concentration of HBeAg, and the results showed that miR-155 expression was remarkably induced (Fig. 5g). Taken together, these results indicated that a higher miR-155 expression in peripheral blood monocytes of AHB patients might be closely related to HBeAg contents and contribute to hepatocyte damage. Of course, the exact role of miR-155 in liver injury induced by HBV infection requires further investigation.
Fig. 5.
miR-155 expression of human peripheral blood monocytes was positively correlation with the content of ALT, AST, and HBeAg in serum of patients with AHB. The expression of miR-155 was determined with q-PCR in peripheral blood monocytes of health controls and patients with AHB (a). The correlation between miR-155 expression and the content of ALT (b), AST (c), HBeAg (d), HBcAg (e), or HBV-DNA (f) were analyzed. Peripheral blood monocytes of health controls were collected and cultured with the serum of health controls or patients with AHB (g H-M peripheral blood monocytes of health controls, H-S the serum of health controls, AHB-S the serum of patients with AHB). Data are representative of three independent experiments (mean ± SD of triplicates in a, g)
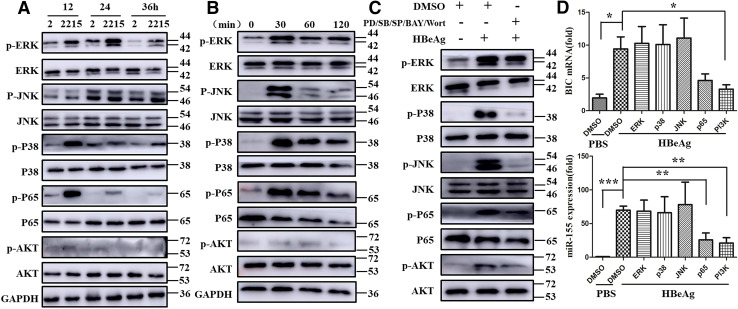
NF-κB and PI3K signaling pathway mediated the up-regulation of miR-155 expression after HBeAg stimulation
The previous studies from our and other laboratory demonstrated that LPS stimulation mainly activate three signal pathway, including MAPK, NF-κB and PI3K [9, 17], and it was also shown that miR-155 expression is regulated by NF-κB activation [18]. To clarify the mechanism of HBeAg regulating miR-155 expression, we tested the activation of above three signal pathways. Similar to the other previous observation, we found MAPK, NF-Κb, and PI3K activation in RAW 264.7 macrophages both in co-culture system and in HBeAg stimulation (Fig. 6a, b). To further reveal the signal pathway responsible for HBeAg-regulated miR-155 expression in macrophage activation, we pre-treated RAW 264.7 cells with the corresponding inhibitors for 30 min and subsequently treated with HBeAg. As shown in Fig. 6c, these inhibitors significantly repressed the phosphorylation of ERK, p38, JNK, p65, and AKT. Furthermore, we found that either inhibition of NF-κB by Bay 11-7082, or PI3K by Wortmannin decreased BIC and miR-155 levels in HBeAg-stimulated macrophages (Fig. 6d), rather than that of MAPK signal pathway. Taken together, these data suggested that HBeAg induced a variety of signal pathway activation, including MAPK, NF-κB, and PI3K. However, it is worth noting that the expression of miR-155 was only modulated through NF-κB or PI3K pathway in HBeAg-stimulated macrophages.
Fig. 6.
NF-κB and PI3K-signaling pathway mediated the up-regulation of miR-155 expression after HBeAg stimulation. The non- and phosphorylation of ERK, JNK, P38, P65, and AKT were examined with western blot in co-culture of HepG2 or HepG2.2.15 and RAW264.7 macrophage (a) and HBeAg treatment experiments (b). RAW264.7 macrophages were pre-treated with the inhibitor of ERK, JNK, P38, P65, and AKT signal pathway (c, d). Western blot analyzed the non- and phosphorylation of ERK, JNK, P38, P65, and AKT (c). The expression of miR-155 and BIC was observed with q-PCR (d). Similar observations were obtained in three independent experiments (mean ± SD of triplicates in d). *p < 0.05, **p < 0.01, ***p < 0.001
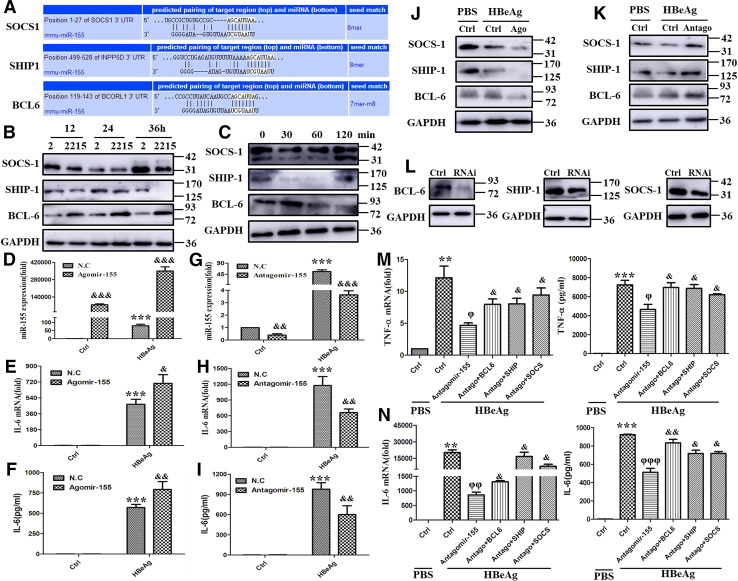
miR-155 promoted HBeAg-induced cytokine production by targeting BCL-6, SHIP-1, and SOCS-1
The previous studies have demonstrated that miR-155-targeted multiple molecules to control the activation and inhibition of different signal pathways [19–21]. Figure 7a shows the binding sites of miR-155, which have been reported, by microRNA prediction programs Targetscan. Subsequently, we tested these three proteins expression in RAW 264.7 macrophages after co-culture with HepG2 or HepG2.2.15 cells or stimulating with HBeAg. As shown in Fig. 7b, c, the expression of SOCS-1 and SHIP-1 was decreased in macrophages after co-culture or HBeAg stimulation. On the contrary, the expression of BCL-6 was increased. To identify the effect of miR-155 on the cytokine production in HBeAg-stimulated macrophages, RAW 264.7 macrophages were transfected with angomir- or antago-mir-155, then stimulated with HBeAg. As shown in Fig. 7d, miR-155 expression was obviously up-regulated in angomir-155 group, especially after HBeAg treatment. Moreover, the expression and secretion of IL-6 were potentiated when miR-155 was overexpressed (Fig. 7e, f). Inversely, the antago-mir-155 transfection significantly down-regulated miR-155 expression, particularly in the HBeAg stimulation group (Fig. 7g). The expression and secretion of IL-6 were also diminished when miR-155 was knocked down (Fig. 7h, i). Meanwhile, we detected the expression of SOCS-1, SHIP-1, and BCL-6, three pivotal targets of miR-155, after transfection with angomir- or antago-mir-155. As shown in Fig. 7j, k, angomir-155 effectively decreased the expression of SOCS-1, SHIP-1, and BCL-6, while antago-mir-155 significantly increased their expression.
Fig. 7.
miR-155 regulated HBeAg inducing cytokines production by targeting BCL6, SHIP1, and SOCS-1. Targetscan of microRNA prediction programs showed the binding sites of miR-155 and BCL6, SHIP1, and SOCS-1 (a). The expression of BCL6, SHIP1, and SOCS-1 was examined with western blot in co-culture of HepG2 or HepG2.2.15 and RAW264.7 macrophage (b) and HBeAg treatment experiments (c). After angomir-155 or negative control miRNA was transfected into RAW264.7 macrophages for 12 h and the cells were treated with HBeAg for 36 h, miR-155 expression (d) and IL-6 production (e, f) were examined with q-PCR and ELISA. After antago-mir-155 or negative control miRNA was transfected into RAW264.7 macrophages for 12 h and the cells were treated with HBeAg for 36 h, miR-155 expression (g) and IL-6 production (h, i) were examined with q-PCR and ELISA. After angomir-155 or negative control miRNA was transfected into RAW264.7 macrophages for 36 h and the cells were treated with HBeAg for 30 min, the expression of BCL6, SHIP1, and SOCS-1 was detected with western blot (j). After antago-mir-155 or negative control miRNA was transfected into RAW264.7 macrophages for 36 h and the cells were treated with HBeAg for 30 min, the expression of BCL6, SHIP1, and SOCS-1 was detected with western blot (k). RNAi of BCL6, SHIP1, or SOCS-1 was transfected into RAW264.7 cells and interference efficiency of them was analyzed with Western blot (l). Both antago-mir-155 or negative control miRNA and RNAi of BCL6, SHIP1, and SOCS-1 or scramble control were transfected into RAW264.7 for 24 h, and the cells were stimulated with HBeAg for 24 h. The expression and secretion of TNF-α and IL-6 were evaluated with q-PCR and ELISA (m, n). Data are representative of four independent experiments (mean ± SD of triplicates in d–n). *p < 0.05, **p < 0.01, ***p < 0.001, &p < 0.05, &&p < 0.01, &&&p < 0.001
Given that SOCS-1, SHIP-1, and BCL-6 were negative regulator in various signal pathways, we simultaneously knocked down the three targets by small RNA interference and miR-155 by antago-mir-155 in RAW 264.7 cells, and HBeAg was used to stimulate these cells to observe cytokine production. As shown in Fig. 7l, these small RNA effectively inhibited their expression in protein level. Besides, they greatly reversed antago-mir-155 caused the decrease of TNF-α and IL-6 expression and secretion (Fig. 7m, n). Taken together, these data supported that the increased miR-155 promoted HBeAg-induced inflammatory cytokine production in macrophages by reducing the expression of negative regulators, such as BCL-6, SHIP-1, and SOCS-1.
Discussion
Hepatitis B virus (HBV) infection remains a major public health problem. In addition, hepatitis B virus invades and replicates within hepatocyte to result in liver acute or chronic inflammatory response. Efficient control of hepatitis B virus requires the coordinate of innate and adaptive immune responses. Innate immunity has been involved in rapidly recognize viral proteins and nucleic acids and tissue damage. It induces inflammatory response through production of chemokines and pro-inflammatory cytokines, diminishes viral infected hepatocytes by NK cells directly mediated killing, and goes along with recruitment and maturation of adaptive immune cells [22].
Macrophages represent a category of heterogeneous population of immune cells with multiple functions in body homeostasis and disease initiation, maintenance, and resolution [16]. They serve as important innate immunocytes and are especially abundant in the liver. Studies indicated that every 100 hepatocytes are along with 20–40 macrophages in healthy rodent livers [23]. Macrophages, named Kupffer cells in the liver, are the first barrier against pathogens accessing into the liver and are activated with PRR, including TLR, Nod-like receptors, RIG-like receptors, C-type lectins, and scavenger receptors [24]. The roles of Kupffer cells are seemingly contradictory during HBV infections. Several studies have revealed that Kupffer cells, exposed upon HBV, are activated to inhibit HBV replication by producing chemokines and pro-inflammatory cytokines in primary hepatocytes [25, 26]. Nevertheless, other studies have supported that HBV decreased the PRR signal pathway and production of pro-inflammatory cytokines by Kupffer cells, and increased the expression of membrane-bound inhibitory ligands and secretion of immune-regulatory cytokines by Kupffer cells [27–29]. These data verify that Kupffer cells are indispensable for the progress of effective anti-viral immunity. In this study, we first observed that HepG2.2.15 cells have been integrated HBV genome and continued to express higher level HBV-associated antigens, including HBcAg, HBeAg, and HBsAg. After co-culture with RAW 264.7 macrophages, HepG2.2.15 cells, but not HepG2 cells, remarkably induced the production of pro-inflammatory cytokines. Similar to the previous research, our data support the conclusion that HBV infection promotes macrophage activation. The difference is that we further evaluated the antigens essential for HBV-inducing macrophage activation. We demonstrated that HBeAg promoted the change of morphology and production of pro-inflammatory cytokines not only in RAW 264.7 macrophages, but also in mouse primary peritoneal macrophages. We have previously reported that cytoskeleton rearrangement facilitated TLR4-inducing macrophage activation [16]. Although HBcAg increased the production of pro-inflammatory cytokines, macrophage morphology was not changed in mouse primary peritoneal macrophages. Therefore, we speculated that HBeAg, but not HBcAg, might affect macrophage activation by regulating cytoskeleton rearrangement, which needs further investigation in the future.
Accumulating data showed that miRNAs, serve as the most widely studied kind of non-coding RNA molecules, play a pivotal role in the regulation of essential biological and pathological processes, such as cell differentiation, proliferation, apoptosis, immunity, autophagy, and metabolism [30–32]. They control gene expression by mediating degradation and/or blocking translation of mRNA. Recently, several miRNAs have been well characterized in the modulation of TLR activation in monocytes or macrophages and have been demonstrated to negatively or positively regulate the activation of MAPK-, NF-κB-, and/or PI3K-signaling pathway and subsequent production and inhibition of pro-inflammatory cytokines [9, 12, 32]. Hence, miRNAs represent a special family to restrict or accelerate inflammatory responses. However, the effect of HBV on miRNAs expression in macrophages remains unknown. In the present study, we have provided evidence to show that the expression of miR-155 is markedly induced by HBV.
Mature miR-155 originates from pri-miR-155 sequence, shortly named BIC gene, and is a sheared product. It, initially identified as a specific miRNA for immune and hematopoietic cells, is among the first miRNAs connected to inflammation and immunity [33, 34]. Actually, miR-155 has been demonstrated to regulate the production of cytokines in multiple immune cells. The expression of miR-155 is induced in various immune cell lineages by treatment with TLR ligands, specific antigens, and inflammatory cytokines [35]. Further studies have suggested that miR-155 also involved in functions outside the immune and hematopoietic systems [36, 37]. Bala et al. demonstrated that miR-155 contributes to alcohol-induced liver injury through increase of TNF-α production in macrophages [18]. In addition, the expression of miR-155 is augmented in plasma and serum in patients with inflammatory and alcoholic liver injuries [38, 39]. These data point out a potential role of miR-155 in liver injury diseases. However, to date, the role of miR-155 in HBV-associated antigens in regulating macrophage activation has not been clearly identified. In this study, we show that the expression of miR-155 is markedly raised by HBeAg, occasionally by HBcAg in different kinds of macrophages. Importantly, we also analyze the correlation of miR-155 expression and several clinical indexes in monocytes of health controls, patients with AHB or CHB. We found that the expression of miR-155 was increased in monocytes of patients with AHB, and decreased in monocytes of patients with CHB, compared with health controls. The reason, that miR-155 expression was conflicting in patients with AHB and CHB, might be the different status of immune response between of them. Moreover, we elucidated that the expression of miR-155 was positively related to ALT, AST, and HBeAg in the serum of patients with AHB.
The previous reports have suggested that miR-155 can be induced by the activation of TLRs and other agonists and subsequently regulate several key targets to feedback corresponding signal pathway in the innate immune response. For example, miR-155 is up-regulated in response to the TLR4 agonist LPS or the TLR3 agonist poly(I:C) during the macrophage inflammatory responses [40, 41]. Next, the increased miR-155 represses the expression of SHIP-1 mRNA and protein and leads to Akt phosphorylation upon LPS stimulation in RAW 264.7 cells [42]. In addition, Fen Du et al.’s study indicated that miR-155 enhanced inflammatory response to LPS through targeting SOCS-1 [43]. Nazari-Jahantigh et al. found that miR-155 directly repressed expression of BCL-6, a transcription factor that attenuates NF-κB signaling, to promote atherosclerosis in macrophages [44]. However, whether miR-155 may also attenuate inflammatory response under other circumstances, including HBV-associated inflammation, has not been delineated. In this study, we found that HBV-related antigens were able to enhance the phosphorylation level of ERK, p38, JNK, p65, and AKT. To further investigate the mechanism of these above signal pathways in regulating miR-155 expression, we pre-treated macrophages with their corresponding inhibitor. We validated that NF-κB and PI3K signal pathway played an important role in HBeAg-induced miR-155 expression in macrophages. More importantly, we indicated that HBeAg-induced miR-155 functions as positive regulator in HBeAg-induced expression of inflammatory cytokines by targeting BCL-6, SHIP-1, and SOCS-1 in macrophages.
In summary, our results have demonstrated that HBV antigens obviously promoted inflammatory cytokine production and miR-155 expression. Moreover, we validated that HBeAg played a pivotal role in macrophage inflammatory cytokine production and miR-155 expression. In addition, miR-155 positively regulated cytokines production in HBeAg-induced macrophage activation by targeting BCL6, SHIP1, and SOCS-1. Importantly, miR-155 expression was enhanced in monocytes of patients with AHB, and positively correlated with ALT, AST, and HBeAg in the serum. Thus, these findings unveil a novel intricate physiological interplay between miR-155 and HBV infection. In other words, miR-155 represents a new positive regulator in macrophage activation in HBV-infection microenvironments, which might be a potential candidate for HBV-induced liver injury.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIFF 17571 kb) Supplementary Fig. 1 RAW264.7 macrophages were stimulated with HBeAg (2000 µg/ml) for 24 h and collected with Trizol. U6, snoRNA202 (another commonly internal control) and miR-155 were reverse transcribed with TapMan-related kit and these microRNA expressions were measured using TaqMan miRNA assays (A–D). Data are representative of three independent experiments (mean ± S.D. of triplicates in A-D)
Supplementary material 2 (TIFF 25690 kb) Supplementary Fig. 2 RAW264.7 macrophages were stimulated with different concentrations HBeAg (including 0, 25, 50, 100, 300, 500, 1000, and 2000 ng/ml) for 24 h and the expression of IL-6, TNF-α, and miR-155 were detected with q-PCR (A, C, E) and the secretion of IL-6 and TNF-α was measured with ELISA (B, D). HBeAg was diluted to different concentrations (0, 25, 50, 100, 300, 500, 1000, and 2000 ng/ml) and detected with chemiluminescence microparticle immunoassay (CMIA) (F). Data are representative of three independent experiments (mean ± S.D. of triplicates in A–E). Data are representative of one experiment (mean ± S.D. of triplicates in F). *p < 0.05, **p < 0.01, ***p < 0.001
Supplementary material 3 (TIFF 54744 kb) Supplementary Fig. 3 RAW264.7 macrophages were stimulated with HBeAg for 24 h and some more magnification photos were recorded with light microscope (original magnification, ×100 or ×200. White box: the specific zoom position)
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81600469, 81472685, and 81772626), the Science and Technology Development Projects of Shandong Province (2017GSF218053 and 2016GSF201126), the Clinical Medical Science and Technology Innovation Program (201704114), the Major Special Plan of Science and Technology of Shandong Province (2015ZDXX0802A01), and the Shandong Province medical and health science and technology development project (2017WS194).
Abbreviations
- HBV
Hepatitis B virus
- HAV
Hepatitis A virus
- HCV
Hepatitis C virus
- HDV
Hepatitis D virus
- HEV
Hepatitis E virus
- HIV
Human immunodeficiency virus
- HBcAg
Hepatitis B core antigen
- HBeAg
Hepatitis B e antigen
- HBsAg
Hepatitis B surface antigen
- IL-6
Interleukin-6
- TNF-α
Tumor necrosis factor-α
- miR-155
MicroRNA-155
- q-PCR
Quantitative real-time polymerase chain reaction
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- AHB
Acute hepatitis B
- CHB
Chronic hepatitis B
- MAPK
Mitogen-activated protein kinase
- BCL-6
B-cell lymphoma 6
- SHIP-1
Src homology-2 domain-containing inositol 5-phosphatase 1
- SOCS-1
Suppressor of cytokine signaling-1
- TLR
Toll-like receptors
- NK
Natural killer
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Wenwen Wang and Hongjun Bian have contributed equally to this work.
References
- 1.WHO. Hepatitis B. http://www.who.int/mediacentre/factsheets/fs204/en/. Accessed July 2017
- 2.GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385(9963):117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hensel KO, Rendon JC, Navas MC, Rots MG, Postberg J. Virus-host interplay in hepatitis B virus infection and epigenetic treatment strategies. FEBS J. 2017;284(21):3550–3572. doi: 10.1111/febs.14094. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Liao Y, Chen J, Cai B, Su Z, Ying B, Lu X, Tao C, Wang L. Epidemiology study of HBV genotypes and antiviral drug resistance in multi-ethnic regions from Western China. Sci Rep. 2015;5:17413. doi: 10.1038/srep17413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhu P, Haque M, Humphries-Bickley T, Ravi S, Song J. Hepatitis B virus immunopathology, model systems, and current therapies. Front Immunol. 2017;8:436. doi: 10.3389/fimmu.2017.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Qi J, Zhao P, Liang X, Ju Y, Liu P, Liu B, Guo C, Zhang L, Ma C, Gao L. T cell immunoglobulin- and mucin-domain-containing molecule-4 attenuates concanavalin A-induced hepatitis by regulating macrophage. J Leukoc Biol. 2010;88(2):329–336. doi: 10.1189/jlb.1209797. [DOI] [PubMed] [Google Scholar]
- 7.Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66(6):1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Tan-Garcia A, Wai LE, Zheng D, Ceccarello E, Jo J, Banu N, Khakpoor A, Chia A, Tham CY, Tan AT, Hong M, Keng CT, Rivino L, Tan KC, Hoe Lee K, Lim SG, Newell EW, Pavelka N, Chen J, Ginhoux F, Chen Q, Bertoletti A, Dutertre CA. Intrahepatic CD206+ macrophages contribute to inflammation in advanced viral-related liver disease. J Hepatol. 2017;67(3):490–500. doi: 10.1016/j.jhep.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Qi J, Qiao Y, Wang P, Li S, Zhao W, Gao C. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 2012;586(8):1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Thounaojam MC, Kundu K, Kaushik DK, Swaroop S, Mahadevan A, Shankar SK, Basu A. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J Virol. 2014;88(9):4798–4810. doi: 10.1128/JVI.02979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. MiR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci USA. 2009;106(37):15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88(2):169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, Wang L, Zhang M, Wang P, Qi J, Zhang L, Gao C. Nuclear to cytoplasmic translocation of heterogeneous nuclear ribonucleoprotein U enhances TLR-induced proinflammatory cytokine production by stabilizing mRNAs in macrophages. J Immunol. 2012;188(7):3179–3187. doi: 10.4049/jimmunol.1101175. [DOI] [PubMed] [Google Scholar]
- 16.Bian H, Li F, Wang W, Zhao Q, Gao S, Ma J, Li X, Ren W, Qin C, Qi J. MAPK/p38 regulation of cytoskeleton rearrangement accelerates induction of macrophage activation by TLR4, but not TLR3. Int J Mol Med. 2017;40(5):1495–1503. doi: 10.3892/ijmm.2017.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W, Qi J, Wang L, Zhang M, Wang P, Gao C. LY294002 inhibits TLR3/4-mediated IFN-β production via inhibition of IRF3 activation with a PI3K-independent mechanism. FEBS Lett. 2012;586(6):705–710. doi: 10.1016/j.febslet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286(2):1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Zhu M, Corbalán-Campos J, Heyll K, Weber C, Schober A. Regulation of Csf1r and Bcl6 in macrophages mediates the stage-specific effects of microRNA-155 on atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35(4):796–803. doi: 10.1161/ATVBAHA.114.304723. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Li F, Jia S, Zhang K, Jiang W, Shang Y, Chang K, Deng S, Chen M. Early secreted antigen ESAT-6 of Mycobacterium Tuberculosis promotes apoptosis of macrophages via targeting the microRNA155–SOCS1 interaction. Cell Physiol Biochem. 2015;35(4):1276–1288. doi: 10.1159/000373950. [DOI] [PubMed] [Google Scholar]
- 22.Bertoletti A, Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Postgrad Med J. 1051;2013(89):294–304. doi: 10.1136/postgradmedj-2011-301073rep. [DOI] [PubMed] [Google Scholar]
- 23.Lopez BG, Tsai MS, Baratta JL, Longmuir KJ, Robertson RT. Characterization of Kupffer cells in livers of developing mice. Comp Hepatol. 2011;10(1):2. doi: 10.1186/1476-5926-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer A, Brown L, Whitehead JP, Prins JB, Fairlie DP. Nutrient and immune sensing are obligate pathways in metabolism, immunity, and disease. FASEB J. 2015;29(9):3612–3625. doi: 10.1096/fj.15-271155. [DOI] [PubMed] [Google Scholar]
- 25.Hosel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, Langenkamp A, Falk C, Büning H, Rose-John S, Protzer U. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50(6):1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 26.Phillips S, Chokshi S, Riva A, Evans A, Williams R, Naoumov NV. CD8(+) T cell control of hepatitis B virus replication: direct comparison between cytolytic and noncytolytic functions. J Immunol. 2010;184(1):287–295. doi: 10.4049/jimmunol.0902761. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Chen Z, Hu C, Qian F, Cheng Y, Wu M, Shi B, Chen J, Hu Y, Yuan Z. Hepatitis B virus surface antigen selectively inhibits TLR2 ligand-induced IL-12 production in monocytes/macrophages by interfering with JNK activation. J Immunol. 2013;190(10):5142–5151. doi: 10.4049/jimmunol.1201625. [DOI] [PubMed] [Google Scholar]
- 28.Bility MT, Cheng L, Zhang Z, Luan Y, Li F, Chi L, Zhang L, Tu Z, Gao Y, Fu Y, Niu J, Wang F, Su L. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog. 2014;10(3):e1004032. doi: 10.1371/journal.ppat.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yong L, Li M, Gao Y, Deng Y, Liu W, Huang D, Ren C, Liu M, Shen J, Hou X. Identification of pro-inflammatory CD205+ macrophages in livers of hepatitis B virus transgenic mice and patients with chronic hepatitis B. Sci Rep. 2017;7:46765. doi: 10.1038/srep46765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 31.Vaporidi K, Vergadi E, Kaniaris E, Hatziapostolou M, Lagoudaki E, Georgopoulos D, Zapol WM, Bloch KD, Iliopoulos D. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;303(3):L199–L207. doi: 10.1152/ajplung.00370.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, Cui K, Liao M, He J, Jiang Y, Yang BB, Xu H, Xu N, Zhang Y. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10(1):70–79. doi: 10.4161/auto.26534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253(1):146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 35.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S. LiewFY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56(5):1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 41.Subedi A, Park PH. Autocrine and paracrine modulation of microRNA-155 expression by globular adiponectin in RAW 264.7 macrophages: involvement of MAPK/NF-κB pathway. Cytokine. 2013;64(3):638–641. doi: 10.1016/j.cyto.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Park EJ, Shen L, Sun D, Pezzuto JM. Inhibitory effect of a callophycin A derivative on iNOS expression via inhibition of Akt in lipopolysaccharide-stimulated RAW 264.7 cells. J Nat Prod. 2014;77(3):527–535. doi: 10.1021/np400800h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du F, Yu F, Wang Y, Hui Y, Carnevale K, Fu M, Lu H, Fan D. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2014;34(4):759–767. doi: 10.1161/ATVBAHA.113.302701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, Weber C, Schober A. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Investig. 2012;122(11):4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (TIFF 17571 kb) Supplementary Fig. 1 RAW264.7 macrophages were stimulated with HBeAg (2000 µg/ml) for 24 h and collected with Trizol. U6, snoRNA202 (another commonly internal control) and miR-155 were reverse transcribed with TapMan-related kit and these microRNA expressions were measured using TaqMan miRNA assays (A–D). Data are representative of three independent experiments (mean ± S.D. of triplicates in A-D)
Supplementary material 2 (TIFF 25690 kb) Supplementary Fig. 2 RAW264.7 macrophages were stimulated with different concentrations HBeAg (including 0, 25, 50, 100, 300, 500, 1000, and 2000 ng/ml) for 24 h and the expression of IL-6, TNF-α, and miR-155 were detected with q-PCR (A, C, E) and the secretion of IL-6 and TNF-α was measured with ELISA (B, D). HBeAg was diluted to different concentrations (0, 25, 50, 100, 300, 500, 1000, and 2000 ng/ml) and detected with chemiluminescence microparticle immunoassay (CMIA) (F). Data are representative of three independent experiments (mean ± S.D. of triplicates in A–E). Data are representative of one experiment (mean ± S.D. of triplicates in F). *p < 0.05, **p < 0.01, ***p < 0.001
Supplementary material 3 (TIFF 54744 kb) Supplementary Fig. 3 RAW264.7 macrophages were stimulated with HBeAg for 24 h and some more magnification photos were recorded with light microscope (original magnification, ×100 or ×200. White box: the specific zoom position)