Abstract
Rab18 is one of the small number of conserved Rab proteins which have been traced to the last eukaryotic common ancestor. It is found in organisms ranging from humans to trypanosomes, and localizes to multiple organelles, including most notably endoplasmic reticulum and lipid droplets. In humans, absence of Rab18 leads to a severe illness known as Warburg-Micro syndrome. Despite this evidence that Rab18 is essential, its role in cells remains mysterious. However, recent studies identifying effectors and interactors of Rab18, are now shedding light on its mechanism of action, suggesting functions related to organelle tethering and to autophagy. In this review, we examine the variety of roles proposed for Rab18 with a focus on new evidence giving insights into the molecular mechanisms it utilizes. Based on this summary of our current understanding, we identify priority areas for further research.
Keywords: Rab18, Warburg-Micro syndrome, Lipid droplets, Secretion, Small GTPases, Tethering, Lipid metabolism
Introduction
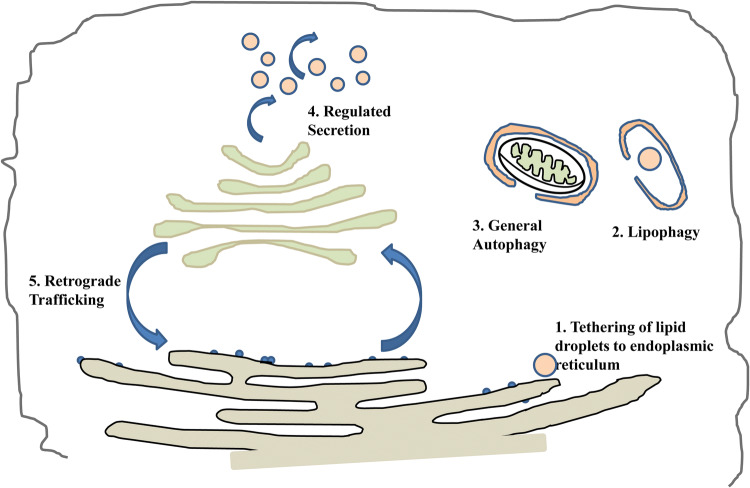
Rab18 was first identified by Chavrier et al. from a partial-length clone [1] and Yu et al. [2] from a full-length clone from a mouse pituitary cDNA library. Yu [2] established that mRNAs were found in a wide range of tissues and cell lines. Rab18 was initially believed to associate with endocytic structures and, in polarized epithelial cells, was localized to membranes near the apical surface [3]. However, subsequent studies suggested roles for Rab18 in various other locations including: the Golgi apparatus [4, 5], the endoplasmic reticulum [5–7] and lipid droplets [6, 8] (Fig. 1). Other studies have provided a confusing abundance of evidence for even more roles for Rab18: in regulated secretion [9], in autophagy [10], and even in pathogen-induced reorganization of the endocytic pathway [11] (Fig. 1).
Fig. 1.
Some reported functions of Rab18. Each of these functions is described at greater length in the main text
Recently, a number of studies have identified potential interaction partners of Rab18 and started to suggest a molecular role for this protein, particularly as related to the endoplasmic reticulum and lipid droplets. While these studies have employed different model systems, and have shown less overlap than could be hoped, for the first time there are concrete clues to the molecular mechanisms by with Rab18 operates. Furthermore, an inherited disease of humans, Warburg-Micro syndrome, characterized by a characteristic set of disorders, including mental retardation and development of cataracts during childhood [12], is caused by inactivation or misregulation of Rab18 [13]. Thus, concrete evidence now exists that Rab18 plays a crucial role in human development, yet the nature of the crucial pathways that fail in the absence of Rab18 are still unclear.
This review will summarize what is currently known regarding the mechanisms by which Rab18 may operate. Substantial progress has been made in identifying its interaction partners and sites of action. However, experiments have been carried out in a variety of cellular systems, and some of the recent literature is contradictory. Thus, there are important unsolved questions requiring further research.
The Rab family of small GTPases
Rab18 is a member of the Rab family of small GTPases within the Ras superfamily. The Rab family consists of 60 + proteins in humans, many of which have been found to localize to specific organelles [14]. Like other Ras-family proteins, Rab proteins are small GTPases, which typically act as molecular switches [14, 15]. When bound to GTP, a “switch” domain is exposed which can bind to interacting proteins referred to as effectors [16]. When GTP is hydrolyzed, the GDP remains bound, but a conformational change hides the switch domain, terminating interaction with effectors. Intrinsic rates of GTP hydrolysis are typically low (hours to days [17, 18]), but GTP hydrolysis is normally triggered by interactions with GTPase activating (GAP) proteins [14]. GTP exchange proteins (GEFs) activate Rabs by causing release of GDP and its replacement by cytoplasmic GTP [14]. An additional important factor in the GTP cycle of Rabs is that it is correlated with membrane association. Most Rabs are prenylated, and have fatty acids covalently attached to their carboxy-terminus (reviewed in [19]). This facilitates membrane association in the GTP state. While in the membrane, Rabs in the GTP form recruit multiple effector proteins (roughly thirty known effectors in the case of Rab5). These effectors can have various functions including: membrane tethering and fusion, modifications of membrane lipids, interactions with microtubule motors, and even recruitment or down-regulation of other Rabs (reviewed in [14]). Rab proteins are inactivated by GAPS, some of which also have effector functions [14]. After GDP hydrolysis, a protein called Rab-GDI extracts Rab-GDP from intracellular membranes [20, 21]. Interaction with GEF proteins is normally required for reinsertion into membranes [14].
While there appears to be only a single Rab-GDI, there are many GEF and GAP proteins with varying degrees of specificities to different Rabs. At least 40 Rab GEFs belonging to several different classes have been identified in human cells (reviewed in [22]), while 44 Tre2-Bub2-Cdc16 (TBC) domain containing proteins are believed to act as GAPs (reviewed in [23]). One GAP not in the TBC family (Rab3GAP) has also been identified ([23, 24]). Rab3GAP acts as a GAP for Rab3, but, importantly, also has GEF activity for Rab18 [25]. Mutation of Rab3GAP, another GAP (TBC1D20) or Rab18 results in Warburg-Micro syndrome [26]. As GEFs have only been identified with specificity for about half of human Rab proteins [22], the existence of a substantial number of unidentified Rab GEFs is likely. There is strong evidence that Rab GEFs play an important role in controlling the specificity of Rab localization to membranes since relocating any of several Rab GEFs to mitochondria resulted in abnormal mitochondrial localization of their substrate Rabs [27].
Warburg-Micro syndrome
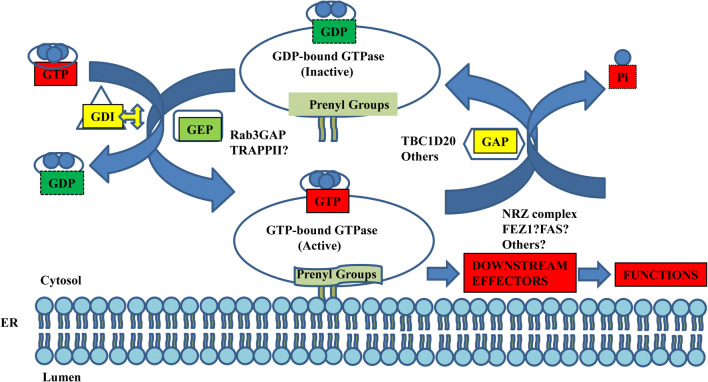
Warburg-Micro syndrome [12] and the similar but less severe Martsolf syndrome [28] are genetic diseases that can be caused by mutations in Rab18 [26], either subunit of Rab3GAP (Rab3GAP1 [29], or Rab3GAP2 [30, 31]) or in another Rab GAP, TBC1D20 [31]. A similar syndrome has been recapitulated in mouse models in which Rab18 [31] or TBC1D20 [31] have been knocked out. Rab3GAP functions as a GEF for Rab18 [25], while TBC1D20 shows GAP activity for Rab18 [13] (Fig. 2). These data strongly suggest that the primary defect in cases of Warburg-Micro syndrome is absence or disregulation of Rab18. Together, these mutations account for about 50% of cases of the disease, suggesting that there may be other undiscovered causative mutations.
Fig. 2.
Schematic showing the major known players in the Rab18 GTP cycle
Symptoms of Warburg-Micro syndrome present at a very early age and include severe mental retardation, absence of the corpus callosum, hypogenitalism and multiple ocular problems, most notably congenital cataract, microcornea and atrophy of the optic nerve [12]. Martsolf’s syndrome is similar, but milder [28]. Current thinking is that the two syndromes represent the ends of a spectrum: Warburg-Micro syndrome results from a severe deficit of Rab18 activity, while Martsolf’s syndrome results from milder or partial deficits.
Part of the collection of disorders reflected in Warburg-Micro syndrome could reflect indirect effects of Rab18, leading to cascading effects on the complex processes involved in development. However, Wu et al. [32] found evidence for a direct role for Rab18 in regulation of neuronal migration. They found that knockdown of Rab18 with shRNAs resulted in impaired radial migration of cortical neurons. In these cells, lysosomal degradation of N-cadherin was accelerated. N-cadherin is essential for development of the cerebral cortex [33, 34]. These results suggest a direct role for Rab18 in brain development, possibly involving regulation of N-cadherin targeting to the cell surface. Other evidence for intracellular roles for Rab18 comes from a mouse model of Warburg-Micro syndrome in which Rab18 was deleted. These cells show abnormal lipid droplet formation, with many small LDs and a small number of “supersized” LDs [35] consistent with results of siRNA knockdowns of Rab18 in cultured cells [36].
It has been noted that one of the cell types most affected in Warburg-Micro syndrome, the cells that form the lens fibers of the eye, are unusual in that they eliminate most of their intracellular organelles as part of their differentiation process. There is evidence that autophagy plays an important role in the development and maintenance of lens tissue and prevention of cataract (reviewed in [37]). The initial removal of organelles still occurred in mice with lens-specific Atg5−/− knockdown, suggesting differentiation of fiber cells did not absolutely depend on autophagy. However, the mice proceeded to develop severe cataracts, suggesting autophagy was important for maintenance of the lens [38]. As Rab18 has been proposed to participate in some autophagic processes, this could suggest that failure or dysregulation of some forms of autophagy could play a role in Warburg-Micro syndrome.
Regulation of the Rab18 GTP cycle
Rab3GAP is the best-established interaction partner of Rab18, with multiple lines of evidence from many independent studies supporting the relationship. Mutations in either subunit of Rab3GAP or of Rab18 produce similar phenotypes in the form of Warburg-Micro syndrome [26, 29, 30]. Further, Rab3GAP has been shown to directly associate with and to function as a GTP exchange factor (GEF) activity for Rab18 [25]. Like Rab18, there is evidence that Rab3GAP plays a role in a diversity of locations, including endoplasmic reticulum [25], the Golgi apparatus [39], secretory vesicles [40], autophagosomes [39] and lipid droplets [10].
Rab3GAP was identified originally as a GAP for Rab3 [24, 41], which plays a role in regulated secretion. It consists of two associated polypeptide chains, the catalytic subunit Rab3GAP1, and the non-catalytic subunit Rab3GAP2. In humans, mutations in either subunit [29, 30] can result in Warburg-Micro syndrome. Gerondopoulos [25] reported that mutations in Rab3GAP (T18P and E24V in Rab3GAP1 and R426C in Rab3GAP2) had no effect on GAP activity for Rab3a or 3b. However, these mutations severely disrupted GEF activity towards Rab18. Phylogenetic analysis suggests that Rab3GAP1 evolved before Rab3. Rab3GAP1 is common to a range of organisms, including vertebrates, C. elegans [39], Drosophila melanogaster [42] and the marine alga Ostreococcus tauri [43]. However, Rab3 is not found in plants, but is specific to metazoans, and may have arisen from a gene duplication of Rab8 during the evolution of neurotransmitter release [44]. Since Rab18 is believed to have been present in the last eukaryotic common ancestor [44], and is also present in a wide range of eukaryotes including vertebrates and C. elegans [10], D. melanogaster [45] and Trypanosoma cruzi [4, 46], it is plausible that Rab3GAP1 evolved initially as an exchange factor for Rab18.
A study by Spang et al. conducted in human primary fibroblasts has suggested that Rab3GAP plays a role in autophagy [39]. They reported that siRNA directed against both subunits of Rab3GAP reduced LC3 cleavage and autophagosome formation after autophagy was induced in human primary fibroblasts by rapamycin treatment. Interestingly, Atg5 punctae were accumulated, suggesting Rab3GAP plays a role in autophagy at an early stage, but subsequent to the recruitment of Atg5 [39]. Inactivation of exchange activity by the mutation R728A, which blocks GAP activity for Rab3 abrogated rescue. However, they reported that simultaneous knockdown of all Rab3 isoforms (Rab3a, 3b, 3c, and 3d) had no effect on autophagy [39], suggesting that Rab3GAP’s role in autophagy was independent of Rab3. Subsequent work by the same group favored the idea that Rab3GAP functioned by regulation of Rab18 instead [10], and they provided evidence that the roles of Rab18 and Rab3GAP in autophagy were interrelated [10].
Independent of its role on secretory granules as a GAP for Rab3, Haines et al. provided evidence that Rab3GAP interacts with the mannose-binding protein ERGIC-53 [47] which is localized to the endoplasmic reticulum, to ER-Golgi intermediate compartment (ERGIC), and to the cis-Golgi network [48, 49]. This interaction between Rab3GAP and ERGIC-53 was dependent on the p97 adaptor protein UBXD1 [47]. The significance of this finding is unknown. Hanes speculated that Rab3GAP and UBXD1 could modulate trafficking of a minor proportion of ERGIC-53 to the cell surface [47]. However, Rab18 is also found on the endoplasmic reticulum [5, 6], and has some role along with Rab3GAP in maintaining ER structure [25]. Martin et al. [6] reported that some Rab18 was on vesiculotubular structures associated with endoplasmic reticulum. They raised the possibility these could be abnormal structures induced by overexpression of Rab18, however association of Rab18 with ERGIC was not ruled out. More detailed studies of the early secretory pathway in the absence of Rab18 could prove insightful.
Li et al. [50] provided evidence that the TRAPPII complex could function as an alternate GEF for Rab18 in HEK293 cells. They reported that activity of TRAPPII was required to recruit Rab18 to LDs, and that Rab3GAP was found on ER, but not on LDs. This contrasts with Spang and Feldmann [10, 39], who reported in two related studies that Rab3GAP both associated with LDs and was required for recruitment of Rab18 to LDs. More research will be required to address this discrepancy, but it is possible that the mechanism of Rab18 recruitment to LDs could depend on cell type or on the metabolic state of the cell. Notably, Spang and Feldmann induced autophagy using starvation and rapamycin [10, 39].
TBC1D20 is the only GAP yet identified for Rab18 although it shows only weak GAP activity for Rab18 in vitro [51], while showing strong GAP activity for Rab1. However, multiple lines of evidence identify it as physiologically relevant. First, mutations in TBC1D20 which impair its GAP activity have been shown to cause Warburg-Micro syndrome in humans [51], and in a mouse model [51]. TBC1D20 participates in processes suspected to involve Rab18 including autophagosome maturation [52] and replication of hepatitis C virus [53]. In another study, Handley et al. found that mutations in TBC1D20 resulted in redistribution of Rab18 from other organelles to ER [13], suggesting that TBC1D20 GAP activity may regulate the intracellular localization of Rab18. It is unknown whether other proteins show GAP activity for Rab18, and whether it might interact with different GAP proteins on different organelles. If so, accumulation of Rab18 on ER could lead to depletion elsewhere, and explain how mutations in a GAP protein could mimic the effect of Rab18 knockdown. More research on the molecular role and intracellular distribution of TBC1D20 will be required.
Association of Rab18 with endoplasmic reticulum and with lipid droplets
Most groups studying Rab18 have identified a proportion of it localized to ER (e.g., [5, 6, 25]). Rab3GAP is also found localized to ER in cells [25], as is TBC1D20 [54], suggesting the presence of some Rab18-related regulatory proteins on ER. Interestingly, an analysis of the Rab proteins present within the last eukaryotic common ancestor (LECA) found 20 Rab proteins in six groups [44]. Rab18 was already found as a distinct Rab in the LECA, suggesting an ancient origin, and the ancestral Rab18 was proposed to originate in a pre-LECA organism, from Rab1 via a gene duplication event. Rab1 has functions associated with ER exit in most eukaryotes [44], and this could suggest an ancestral function for Rab18 related to the endoplasmic reticulum.
In humans, the ancestral Rab18 has undergone additional duplications, generating a family of proteins, including Rab18, and a small family of similar proteins: Rab40a, Rab40b, and Rab40c [44]. The functions of the Rab40 family of proteins are unclear. Rab40b may have a role in the production of invadopodia [55]. Interestingly, Rab40c has been reported to associate both with LDs and with ER-derived structures, similarly to Rab18 [56, 57], and deletion of Rab40c or overexpression of the protein DAB2IP, which shows GAP activity towards Rab40c results in increased LD accumulation [57]. Rab40c has also been reported to mediate clustering of LDs [56]. This suggests that some ancestral functions of Rab18 might be distributed, or potentially redundant with the Rab40 proteins. However, this will only become clear with more research.
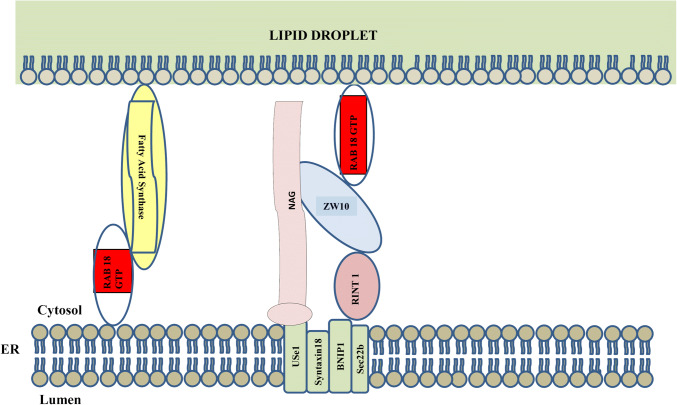
Ozeki et al. proposed, based on electron microscopic evidence, that Rab18 mediated attachment of LDs to endoplasmic reticulum [8]. They also found that Rab18 showed increased association with LDs over time [8], this suggests that Rab18 may mediate attachment of ER to established LDs. Both Ozeki [8] and Martin [6] reported that Rab18 labeled only a subset of LDs. Gillingham [45] identified the NRZ complex as a major effector of Rab18, and proposed it could be involved in tethering of COPI-coated vesicles or of lipid droplets to ER membranes. Xu [36] proposed on the basis of more extensive experiments that Rab18 mediated association of LDs with ER via NRZ tethers and SNARE proteins (Fig. 3). They reported that Rab18 was necessary for the growth of newly formed LDs, suggesting it plays an early role in LD maturation. In their study, Rab18 knockout resulted in accumulation of 100 nm diameter LDs, which formed at a normal rate from endoplasmic reticulum. However, these LDs then failed to expand, suggesting an early role for Rab18 in transferring lipids from the ER.
Fig. 3.
Current model for Rab18/NRZ-mediated tethering of lipid droplets to endoplasmic reticulum. In this model, Rab18 on the LD membrane recruits the NRZ complex consisting of NAG, ZW10 and RINT. Elements of the NRZ complex in turn bind to SNARE proteins (Use1 and BN1 P1) found in ER membranes. Also shown is Rab18 on ER membranes recruiting fatty acid synthase, which could favor enlargement of lipid droplets. Whether Rab18 on ER membranes could also recruit NRZ complex, and whether this complex could participate in tethering interactions, has not been addressed
The discrepancy between [36], which suggests a general role for Rab18 in LD growth and the earlier studies, which only identified Rab18 on a subset of LDs is unlikely to be related to choice of cell lines, as at least some experiments were conducted in 3T3 cells in all of these studies. However, continued interaction of nascent LDs with ER has been described elsewhere [58]. Wilfling et al., using electron microscopy, identified connections between ER and LDs, which they proposed allowed transfer of enzymes in the triglyceride synthesis pathway including GPAT4, and DGAT2, from ER to growing LDs, allowing in situ production of triglycerides on the LD. The molecular mechanisms by which these connections occur are unclear. The same group has proposed that Arf1 and COPI adjusts the phospholipid and surface tension of the LD surface to favor connections [59], but no evidence to date rules out classical Rab-regulated membrane tethering as a component of the process. Potentially, Rab18 could coordinate a variety of interactions between ER and LDs, and some of these interactions might be driven largely by ER-associated Rab18. Thus, the reported lack of Rab18 on some LDs [6, 8] (such as the immature LDs described in [36]) is not inconsistent with a role for Rab18 in regulating association of these LDs with ER. It is also possible that Rab18 remains associated with LDs through part of their life cycle, and is then lost.
Some other studies have proposed a role for Rab18 on ER independent of LDs. Barr et al. [25] reported that both Rab18 and Rab3GAP are required for normal ER structure in a variety of different cell types. They reported that in the absence of either protein, or in primary fibroblasts obtained from Warburg-Micro patients, tubular ER was suppressed, while CLIMP-63 defined ER sheets spread into the cell periphery. Based on that, they propose that Rab18 plays a role in maintaining normal ER structure. This could indicate a distinct function of Rab18 in regulating reticulons or other proteins important for ER morphology. This study did not rule out that inefficient transfer of fatty acid or triglycerides from ER to LDs could lead to toxic effects from accumulation in ER. However, Jayson et al. have claimed that deletion of the gene for Rab18 has no effect on ER morphology in a carcinoma cell line [60].
Secretion
There is evidence for roles for Rab18 in secretion. Vazquez-Martinez et al. [9] reported that endogenous and GFP-tagged Rab18 colocalized with secretory granules in PC12 cells, while GFP-Rab18 S22 N, which does not bind nucleotide, did not. Rab18, in their assays, functioned as a negative regulator of regulated secretion.
Regulated secretion in PC12 cells has some mechanistic similarities to synaptic vesicle release, and is similarly regulated by the small GTPase Rab3a. It is notable that Rab3GAP was identified first as a GAP for Rab3 [24], which plays an important role in regulated secretion. Rab3GAP and Rab18 are ancient proteins conserved over a wide range of eukaryotes and likely originating prior to the last common ancestor of eukaryotes, while Rab3 is phylogenetically more recent, being found only in metazoans [44]. This could suggest that Rab3GAP acquired an interaction with Rab3 as part of the process of the evolution of regulated secretion pathways utilized by multicellular organisms. It could be speculated that Rab18, which Rab3GAP was intrinsically able to recruit, was co-opted to play a role as a negative regulator. Secretory-granule specific interaction partners of Rab18 are not known, and the mechanisms by which Rab18 suppresses secretion are currently unknown. It is interesting, however, that neuronal phenotypes, including altered neurotransmission are found in Warburg-Micro syndrome [61]. This suggests that modulatory effects of Rab18 on regulated secretion are physiologically significant.
Virus assembly
Rab18 has been proposed to play a role in the replication of BK Polyoma virus [62], dengue virus [63], hepatitis B virus [64] and hepatitis C virus [11, 65, 66]. In several of these cases, the virus appears to be targeting Rab18 localization to LDs or what may be roles in lipid metabolism. Some studies give what may be important information about Rab18 interaction partners, or clues about its possible mechanism of action.
Hepatitis C virus has been shown by numerous groups to utilize LD and ER membranes as part of its replication cycle (reviewed in [67]). Salloum et al. [11] reported that the Hepatitis C viral protein NS5A binds to Rab18 on LDs in a GTP-dependent fashion, and that this interaction results in enhanced genome replication. Dansako et al. [66] reported further that the core protein of HCV was recruited to LDs in a Rab18-dependent fashion, and that this recruitment enhanced viral assembly. These findings provide another example of pathogens interacting with Rab proteins to modify cellular functions, and underline that Rab18 plays important roles on ER and LDs.
A whole-genome siRNA screen identified Rab18 together with syntaxin 18 and NRZ complex members RAD50 interactor 1 and ZW10 kinetochore protein, as host factors essential for the replication of BK Polyoma virus [62]. BK Polyoma virus is endocytosed and then goes to the ER by a retrograde transport pathway which shares common elements with the retrograde pathway utilized by ricin and shiga toxins to traffic to the ER via the Golgi apparatus [68, 69]. Elements of the nucleocapsid are then exported to the cytoplasm, and subsequently viral DNA and proteins are imported into the nucleus where they can establish an infection. While the roles played by Rab18, syntaxin 18 or NRZ complex were not established in this study, these proteins were required for BK Polyoma virus to reach the ER, and after knockdown, it accumulated in late endosomes instead [62]. The same NRZ complex, in concert with syntaxin 18, was reported to tether LDs to ER in a Rab18-dependent manner in an independent study [36]. An attractive possibility is that the same NRZ complex involved in tethering of LDs to ER is involved in fusion of retrograde transport intermediates with ER.
Studies of Rab18′s role in virus infection may also reveal candidate Rab18 effectors that have not been uncovered in other studies or to confirm interactions that were previously identified at low confidence. Tang et al. [63] reported that Rab18 recruits the enzyme fatty acid synthase (FAS) to the site of viral replication of Dengue virus. FAS is a cytosolic enzyme complex which synthesizes fatty acid chains with a length of up to 16 carbons, many of which undergo further elongation by other enzymes in the endoplasmic reticulum [70]. FAS was previously identified in a co-immunoprecipitation screen for Rab18 in lysates from 3T3-L1 cells [71], however it was not similarly identified in lysates from HeLa cells in the same screen. Tang [63] also reported that wild-type and constitutively active (Q76L) Rab18 co-precipitated with FAS, but that the Rab18-S22N mutant defective in nucleotide binding failed to co-precipitate with FAS. FAS relocated to the ER and to LDs upon infection with dengue virus. This relocation of FAS could be abrogated with expression of Rab18-S22N [63]. Taken together, these data suggest that FAS could be a physiological effector of Rab18, which is co-opted by the virus in order to modify the lipid composition of intracellular membranes to favor viral replication. Modification of lipid content of intracellular membranes has been reported for other viruses [72]. The association of Rab18 with FAS suggests that Rab18 may be involved in coordinating lipid synthesis pathways with the ER and with LDs. Further research will be required to determine what these roles may be under physiological conditions.
Lipolysis and autophagy
In early work identifying Rab18 on LDs, the Parton group reported that upregulators of lipolysis such as isoproterenol increased association between Rab18 and LDs. This was confirmed in work by the Malagon group [73], which further reported that siRNA knockdown of Rab18 suppressed isoproterenol-induced glycerol release. This could suggest that Rab18 plays a role in regulating at least some forms of lipolysis.
LDs contain primarily triglycerides and cholesterol esters, which are hydrophobic forms in which lipids can be conveniently stored. In most studies, LDs are induced by loading with oleic acid [7], which suggests the primary stored lipids in most experimental systems would be triglycerides. In whole organisms, LDs can contain substantial quantities of cholesterol esters as well. This has physiological relevance, e.g., in the process of conversion of macrophages to foam cells [74], which is believed to be an early step in the development of an atherosclerotic lesion.
Triglycerides can be disassembled, and free fatty acids released from LDs by two primary classes of mechanism. First, cytoplasmic enzymes such as ATGL [75] and HSL [76] can be relocated to the surface of the LD, where they hydrolyze triglycerides to release free fatty acid and glycerol (reviewed in [77]). This mechanism is often regulated by phosphorylation of the enzymes and of surface proteins of the LD [76]. The released fatty acids have various uses, including use as precursors for membrane lipids (primarily in endoplasmic reticulum) and in mitochondria for β-oxidation. LDs can associate specifically with ER and with mitochondria, and this has been proposed to facilitate the transfer of released free fatty acid (reviewed in [78]). As earlier discussed, multiple groups have reported evidence that Rab18 facilitates tight association between LDs and ER. Notably, association of cytoplasmic enzymes with LDs can be modulated and temporally regulated, and does not necessarily lead to the consumption of the entire LD.
Autophagy [79, 80] is the second mechanism by which the contents of LDs can be made accessible to the cell. In autophagy of LDs (lipophagy [81]), a LD is progressively surrounded by a phagophore, which closes to trap the LD in a membrane-bound structure which will then fuse with lysosomes (reviewed in [82]). In the lysosome, lysosomal acid lipase can release free fatty acid from triglyceride and cholesterol esters [82]. In contrast to the action of cytoplasmic enzymes, autophagy is normally thought to irreversibly commit an entire structure to degradation. While the relative importance of autophagy and cytoplasmic enzymes is unclear, it appears to vary between cell types and depending on the mechanism by which degradation of storage forms of lipids was induced. In some circumstances, e.g., in a foam cell model in which LDs were induced in macrophages by heavy loading with lipoprotein particles, autophagy appears to be the primary mechanism by which cholesterol esters are cleaved [74].
Several studies have provided evidence for a link between Rab18, Rab3GAP and autophagy. Makino et al. [83] found that accumulation of free cholesterol leads to Rab18-dependent degradation of perilipin 2, which is a surface protein of LDs. This degradation was inhibited both by ubiquitin inhibitors and by the autophagy inhibitor 3-methyladenine. Other studies provided additional evidence that Rab3GAP [39, 42, 84] and Rab18 [10] play a role in the regulation of autophagy. Some of these studies included data obtained from D. melanogaster [42] and C. elegans [39], suggesting that these roles are conserved over a wide range of organisms. These studies suggested that Rab3GAP and Rab18 favor the lipidation of the Atg8 homologues LC3 and GABARAP, and that Rab3GAP works in part by opposing an anti-autophagy action of FEZ1 and FEZ2. In their hands, knockdown of Rab3GAP could be rescued by a balanced knockdown of FEZ1/2 [39]. In addition to their study, Rab3GAP was identified as an interaction partner of FEZ1 [85]. Since Atg5-positive structures accumulated with Rab3GAP knockdown, they argued that the step regulated by Rab18/Rab3GAP occurred after the recruitment of Atg5 [39]. Notably, while there is evidence that Rab18 induces catabolism of LDs [73] and degradation of some LD proteins, such as PLIN2 [83], the studies by Spang [39] and Feldman [10] were focused on general autophagy, although they did provide evidence that Rab3GAP associated with LDs under pro-autophagy conditions [39]. The roles of Rab18 and Rab3GAP may be cell-type specific as Jayson et al. [60] failed to identify a need for Rab18 in turnover of LDs in human mammary carcinoma cells. Also, unlike deletion of Rab18 or Rab3GAP, deletion of important Atg proteins can lead to embryonic or neonatal lethality in mice (reviewed in [86]). Thus, more research is required to determine the molecular mechanisms involved in Rab18’s regulation of autophagy, and also whether Rab18 plays a prominent role specifically in some cell types or in lipophagy.
It has been reported that Rab18 recruitment onto LDs is driven by Rab3GAP activity [10]. However, Li et al. [50] have argued, based on GAP assays and knockdown experiments that part of the TRAPPII complex, specifically TRAPPC9, functions as the GEF for recruitment of Rab18 to LDs in HEK293T cells. In their hands, knockdown of either TRAPPC9 or COPI led to failure of Rab18 to be recruited to small LDs, and reduced lipolysis, leading to a phenotype of oversized LDs. They reported that Rab3GAP1 was primarily localized to ER, and did not relocalize to LDs under their experimental conditions. The discrepancy between these studies is unclear, and additional work will be required. It cannot be ruled out that there are multiple mechanisms for Rab18 recruitment to LDs related to distinct actions of Rab18. Notably, Li [50] did not distinguish between autophagic lipolysis, and lipolysis resulting from the action of cytoplasmic proteins.
Sidjanin et al. specifically examined the effect of TBC1D20 deletion in lens fiber cells, and argued that inhibited autophagy in these cells led to the loss of lens transparency and cataracts found in Warburg-Micro syndrome [52]. TBC1D20 shows GAP activity for Rab18, as previously discussed, and it is attractive to consider that TBC1D20 could work together with Rab18 and Rab3GAP to regulate autophagy. However, it is also a GAP for Rab1, and in this study the autophagy phenotype could be recapitulated by expression of Rab1b-Q67L, which is defective for GTP hydrolysis, even in the presence of a functional GAP [52]. Given the complexity of autophagy, which is known to involve multiple Rabs and Rab-GAPs, more research will be required.
Conclusions
Rab18 has been implicated in a wide variety of processes including autophagy, secretion and lipid droplet biogenesis. While its mechanistic roles in these processes are still not well characterized, the elucidation of interaction partners has indicated some common threads exist. In particular, Rab3GAP is also implicated at most potential sites of action of Rab18. Multiple lines of evidence suggest that major roles for Rab18 primarily lie in two areas: autophagy and tethering to endoplasmic reticulum. Other roles for Rab18 (e.g. in secretion) may exist. However, their molecular mechanisms are unclear.
A number of recent studies have elucidated important effectors of Rab18 involved in tethering lipid droplets to endoplasmic reticulum. These include members of the NRZ complex, and syntaxin 18. There is evidence that these interactions are involved in the growth of LDs either via direct transfer of triglycerides from ER, or transfer of anabolic enzymes such as DGAT2 via membrane bridges. There is some evidence that effectors of Rab18 such as FAS may play other direct roles in mediating the growth of LDs. While extensive progress has been made, further research will be required to establish the precise roles of Rab18 and interacting proteins in LD–ER interactions. Rab18 could in principle regulate these interactions from either the surface of the LD or from the ER membrane, as it is present in both locations. Further research should include investigation of the geometry of productive interactions of Rab18 with its effectors to clarify whether specific interactions are productive only in cis (within the same membrane) or in trans (between distinct membranes). This may require structural studies to complement the cellular and biochemical methods already in use.
The roles of Rab18 in lipid droplet catabolism and in autophagy are more poorly understood. It is clear that Rab18 plays a role in lipolysis, and also in autophagy, but it is currently unclear whether Rab18’s regulation of lipolysis is via up-regulation of autophagy. There is tantalizing evidence that some of the more striking phenotypes in Warburg-Micro syndrome may relate to defects in autophagy. There is evidence from recent studies that the yeast homologue of syntaxin 18, ufe1, is delivered to sites of autophagosome formation in COPII vesicles [87]. It is possible that Rab18 could be involved in a similar process in higher eukaryotes (Table 1).
Table 1.
Rab18 interactors discussed in this review
| Interactor | Known functions | References |
|---|---|---|
| RAB3GAP1 | GEF for Rab18; GAP for Rab3. Mutated in Warburg-Micro syndrome | [25, 29] |
| RAB3GAP2 | GEF for Rab18; GAP for Rab3. Mutated in Warburg-Micro syndrome | [25, 30, 31] |
| TRAPPC9 | Component of TRAPPII complex. GEF for Rab18 | [50] |
| TBC1D20 | GAP for Rab18. Mutated in Warburg-Micro syndrome | [13, 51] |
| ZW10 | Element of NRZ complex. Proposed Rab18 effector involved in tethering LDs to ER | [36, 62] |
| RINT | Element of NRZ complex. Proposed Rab18 effector involved in tethering LDs to ER | [36, 62] |
| NAG | Element of NRZ complex. Proposed Rab18 effector involved in tethering LDs to ER | [45] |
| Syntaxin 18 | SNARE protein with proposed involvement in tethering LDs to ER via NRZ complex | [36, 62] |
| Fatty acid synthase | Endoplasmic reticulum resident protein involved in fatty acid synthesis | [62] |
| FEZ1/FEZ2 | Involved in various functions. Proposed to have an anti-autophagy action antagonized by Rab18 | [39, 85] |
| NS5A | Hepatitis virus C viral protein involved in viral RNA replication. Rab18 effector | [11] |
Tethering and membrane association could be a common function preserved between distinct cellular processes. Thus, in the absence of other candidate Rab18 effectors, it would be logical to test, e.g., with selective knockdowns, whether syntaxin 18 or NRZ complex members play a role in autophagy, especially in cell systems where a role for Rab18 has been suspected or demonstrated. This would test the attractive hypothesis that many of the seemingly diverse roles of Rab18 in fact take place through a common mechanism.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chavrier P, Parton RG, Hauri H-P, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-P. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Leaf DS, Moore HPH. Gene cloning and characterization of a GTP-binding Rab protein from mouse pituitary AtT-20 cells. Gene. 1993;132:273–278. doi: 10.1016/0378-1119(93)90207-J. [DOI] [PubMed] [Google Scholar]
- 3.Lutcke A, Parton RG, Murphy C, Olkkonen VM, Dupree P, Valencia A, Simons K, Zerial M. Cloning and subcellular localization of novel rab proteins reveals polarized and cell type-specific expression. J Cell Sci. 1994;107:3437–3448. doi: 10.1242/jcs.107.12.3437. [DOI] [PubMed] [Google Scholar]
- 4.Jeffries TR, Morgan GW, Field MC. TbRAB18, a developmentally regulated Golgi GTPase from Trypanosoma brucei. Mol Biochem Parasitol. 2002;121:63–74. doi: 10.1016/S0166-6851(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 5.Dejgaard SY, et al. Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci. 2008;121:2768–2781. doi: 10.1242/jcs.021808. [DOI] [PubMed] [Google Scholar]
- 6.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 7.Martin S, Parton RG. Characterization of Rab18, a lipid droplet-associated small GTPase. Methods Enzymol. 2008;438:109–129. doi: 10.1016/S0076-6879(07)38008-7. [DOI] [PubMed] [Google Scholar]
- 8.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez-Martinez R, Cruz-Garcia D, Duran-Prado M, Peinado JR, Castano JP, Malagon MM. Rab18 inhibits secretory activity in neuroendocrine cells by interacting with secretory granules. Traffic. 2007;8:867–882. doi: 10.1111/j.1600-0854.2007.00570.x. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann A, Bekbulat F, Huesmann H, Ulbrich S, Tatzelt J, Behl C, Kern A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochim Biophys Res Comm. 2017;486:738–743. doi: 10.1016/j.bbrc.2017.03.112. [DOI] [PubMed] [Google Scholar]
- 11.Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013;9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warburg M, Sjo O, Fledelius HC, Pedersen SA. Autosomal recessive microcephaly, microcornea, congenital cataract, mental retardation, optic atrophy, and hypogenitalism: micro syndrome. Am J Dis Child. 1993;147:1309–1312. doi: 10.1001/archpedi.1993.02160360051017. [DOI] [PubMed] [Google Scholar]
- 13.Handley MT, Carpanini SM, Mali GR, Sidjanin DJ, Aligianis IA, Jackson IJ, FitzPatrick DR. Warburg Micro syndrome is caused by Rab18 deficiency or dysregulation. Open Biol. 2015;5:150047. doi: 10.1098/rsob.150047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 15.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 16.Deneka M, Neeft M, van der Sluijs P. Regulation of membrane transport by rab GTPases. Crit Rev Biochem Mol Biol. 2003;38:121–142. doi: 10.1080/713609214. [DOI] [PubMed] [Google Scholar]
- 17.Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–9372. [PubMed] [Google Scholar]
- 18.Shapiro AD, Pfeiffer SR. Quantitative analysis of the interactions between prenyl Rab9, GTP dissociation inhibitor-alpha, and guanine nucleotides. J Biol Chem. 1995;270:11085–11090. doi: 10.1074/jbc.270.19.11085. [DOI] [PubMed] [Google Scholar]
- 19.Pereira-Leal JB, Hume AN, Seabra MC. Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 2001;498:197–200. doi: 10.1016/S0014-5793(01)02483-8. [DOI] [PubMed] [Google Scholar]
- 20.Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 21.Soldati T, Riederer MA, Pfeffer SR. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida M, Oguchi ME, Fukuda M. Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct Funct. 2016;41:61–79. doi: 10.1247/csf.16008. [DOI] [PubMed] [Google Scholar]
- 23.Frasa MAM, Koessmeier KT, Ahmadian MR, Braga VMM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 24.Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- 25.Gerondopoulos A, Bastos RN, Yoshimura S, Anderson R, Carpanini S, Aligianis I, Handley MT, Barr FA. Rab18 and a Rab18 GEF complex are required for normal ER structure. J Cell Biol. 2014;205:707–720. doi: 10.1083/jcb.201403026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bem D, et al. Loss-of-function mutations in Rab18 cause Warburg micro syndrome. Am J Hum Genet. 2011;88:499–507. doi: 10.1016/j.ajhg.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200:287–300. doi: 10.1083/jcb.201209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martsolf JT, Hunter AG, Haworth JC. Severe mental retardation, cataracts, short stature, and primary hypogonadism in two brothers. Am J Med Genet. 1978;1:291–299. doi: 10.1002/ajmg.1320010305. [DOI] [PubMed] [Google Scholar]
- 29.Aligianis I, et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet. 2005;37:221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 30.Borck G, et al. A homozygous RAB3GAP2 mutation causes Warburg Micro syndrome. Hum Genet. 2011;129:45–50. doi: 10.1007/s00439-010-0896-2. [DOI] [PubMed] [Google Scholar]
- 31.Aligianis I, et al. Mutation in Rab3 GTPase-activating protein (RAB3GAP) noncatalytic subunit in a kindred with Martsolf syndrome. Am J Hum Genet. 2006;78:702–707. doi: 10.1086/502681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Q, Sun X, Yue W, Lu T, Ruan Y, Chen T, Zhang D. Rab18, a protein associated with Warburg Micro syndrome controls neuronal migration in the developing cerebral cortex. Mol Brain. 2016;9:19. doi: 10.1186/s13041-016-0198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawauchi T, et al. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010;67:588–602. doi: 10.1016/j.neuron.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Carpanini SM, et al. A novel mouse model of Warburg Micro syndrome reveals roles for RAB18 in eye development and organisation of the neuronal cytoskeleton. Dis Model Mech. 2014;7:711–722. doi: 10.1242/dmm.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, et al. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J Cell Biol. 2018;217:975–995. doi: 10.1083/jcb.201704184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morishita H, Mizushima N. Autophagy in the lens. Exp Eye Res. 2016;144:22–28. doi: 10.1016/j.exer.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Morishita H, Eguchi S, Kimura H, Sasaki J, Sakamaki Y, Robinson ML, Sasaki T, Mizushima N. Deletion of autophagy-related 5 (Atg5) and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation. J Biol Chem. 2013;288:11436–11447. doi: 10.1074/jbc.M112.437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spang N, et al. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-mediated autophagy. Autophagy. 2014;10:2297–2309. doi: 10.4161/15548627.2014.994359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cazares VA, Subramani A, Saldate JJ, Hoerauf W, Stuenkel EL. Distinct actions of Rab3 and Rab27 GTPases on late stages of exocytosis of insulin. Traffic. 2014;15:997–1015. doi: 10.1111/tra.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clabecq A, Henry JP, Darchen F. Biochemical characterization of Rab3-GTPase-activating protein reveals a mechanism similar to that of Ras-GAP. J Biol Chem. 2000;275:31786–31791. doi: 10.1074/jbc.M003705200. [DOI] [PubMed] [Google Scholar]
- 42.Zirin J, Niewuwenhuis J, Samsonova A, Tao R, Perrimon N. Regulators of autophagosome formation in Drosophila muscles. PLoS Genet. 2015;11:e1005006. doi: 10.1371/journal.pgen.1005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanc-Mathieu R, et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014;15:1103. doi: 10.1186/1471-2164-15-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10:1741. doi: 10.1186/1741-7007-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014;31:358–373. doi: 10.1016/j.devcel.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira MG, Visbal G, Costa TFR, Frases S, de Souza W, Atella G, Cunha-E-Silva N. Trypanosoma cruzi epimastigotes store cholesteryl esters in lipid droplets after cholesterol endocytosis Mol. Biochem Parasitol. 2018;224:6–16. doi: 10.1016/j.molbiopara.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Haines DS, et al. Protein interaction profiling of the p97 adaptor UBXD1 points to a role for the complex in modulating ERGIC-53 trafficking. Mol Cell Prot. 2012;11:M111–M016444. doi: 10.1074/mcp.M111.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- 49.Appenzeller C, Andersson H, Kappeler F, Hauri H-P. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Luo X, Zhao S, Siu GKY, Liang Y, Chan HC, Satoh A, Yu SSB. COPI-TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J. 2017;36:441–457. doi: 10.15252/embj.201694866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liegel RP, et al. Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and warburg micro syndrome in humans. Am J Hum Genet. 2013;93:1001–1014. doi: 10.1016/j.ajhg.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidjanin DJ, Park AK, Ronchetti A, Martins J, Jackson WT. TBC1D20 mediates autophagy as a key regulator of autophagosome maturation. Autophagy. 2016;12:1759–1775. doi: 10.1080/15548627.2016.1199300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- 54.Haas AK, Yoshimura S, Stephens DJ, Preisisinger C, Fuchs E, Baar FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 55.Jacob A, Jing J, Lee J, Schedin P, Gilbert SM, Peden AA, Junutula JR, Prekeris R. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J Cell Sci. 2013;126:4647–4658. doi: 10.1242/jcs.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan R, et al. Small GTPase Rab40c associates with lipid droplets and modulates the biogenesis of lipid droplets. PLoS One. 2013;8:e63213. doi: 10.1371/journal.pone.0063213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lui X, Li C, Tan R, Xu X, Wu KKW, Satoh A, Wang T, Yu S. A RasGAP, DAB2IP, regulates lipid droplet homeostasis by serving as GAP toward RAB40c. Oncotarget. 2007;8:85415–85427. doi: 10.18632/oncotarget.19960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilfling F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilfling F, et al. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLIFE. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jayson CBK, Arlt H, Fischer AW, Lai ZW, Farese RV, Walther TC. Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol Biol Cell. 2018;29:2045–2054. doi: 10.1091/mbc.E18-05-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannakopoulou A. Neuronal vacuolation and spinocerebellar degeneration associated with altered neurotransmission. Folia Neuropath. 2017;55:132–145. doi: 10.5114/fn.2017.68580. [DOI] [PubMed] [Google Scholar]
- 62.Zhao L, Imperiale MJ. Identification of Rab18 as an essential host factor for BK polyomavirus infection using a whole-genome RNA interference screen. mSphere. 2017;2:e00291-17. doi: 10.1128/mSphereDirect.00291-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang WC, Lin RJ, Liao CL, Lin YL. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol. 2014;88:6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You X, Liu F, Zhang T, Li Y, Ye L, Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carciogenesis. 2013;34:1644–1652. doi: 10.1093/carcin/bgt089. [DOI] [PubMed] [Google Scholar]
- 65.Chan SC, Lo SY, Liou JW, Lin MC, Syu CL, Lai MJ, Chen YC, Li HC. Visualization of the structure of the hepatitis C virus replication complex. Biochem Biophys Res Commun. 2011;404:574–578. doi: 10.1016/j.bbrc.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 66.Dansako H, Hiramoto H, Ikeda M, Wakita T, Kato N. Rab18 is required for viral assembly of hepatitis C virus through trafficking of the core protein to lipid droplets. Virology. 2014;462–463:166–174. doi: 10.1016/j.virol.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 67.Lavie M, Dubuisson J. Interplay between hepatitis C virus and lipid metabolism during virus entry and assembly. Biochimie. 2017;141:62–69. doi: 10.1016/j.biochi.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 68.Nelson CDS, et al. A retrograde trafficking inhibitor of ricin and shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses. MBio. 2013;4:e00729–e00729. doi: 10.1128/mBio.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandvig K, Skotland T, van Deurs B, Klokk TI. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol. 2013;140:317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 70.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Kistler C (2012) Functional characterisation of Rab18. Doctoral Dissertation, University of Queensland
- 72.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pulido MR, et al. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One. 2011;6:e22931. doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006;7:106–113. doi: 10.1038/sj.embor.7400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clifford GM, Londos C, Kraemer FB, Vernon RG, Yeaman SJ. Translocation of hormone-sensitive lipase and perilipin upon lipolytic stimulation of rat adipocytes. J Biol Chem. 2000;275:5011–5015. doi: 10.1074/jbc.275.7.5011. [DOI] [PubMed] [Google Scholar]
- 77.Wang S, Soni KG, Semache M, Casavant S, Fortier M, Pan L, Mitchell GA. Lipolysis and the integrated physiology of lipid energy metabolism. Mol Genet Metab. 2008;95:117–126. doi: 10.1016/j.ymgme.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 78.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Prespect Biol. 2011;3:1–17. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 81.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 83.Makino A, et al. Acute accumulation of free cholesterol induces the degradation of perilipin 2 and Rab18-dependent fusion of ER and lipid droplets in cultured human hepatocytes. Mol Biol Cell. 2016;27:3293–3304. doi: 10.1091/mbc.E15-10-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Behrends C, Sowa ME, Sygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Assmann EM, Alborghetti MR, Camargo MER, Kobarg J. FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region``. J Biol Chem. 2006;281:9869–9881. doi: 10.1074/jbc.M513280200. [DOI] [PubMed] [Google Scholar]
- 86.Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lemus L, Ribas JL, Sikorska N, Goder V. An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 2016;14:1710–1722. doi: 10.1016/j.celrep.2016.01.047. [DOI] [PubMed] [Google Scholar]