Abstract
Long non-coding RNA (lncRNA) plays an important role in tumor progression and metastasis. Emerging evidence indicates that lncRNA actin filament-associated protein 1-antisense RNA 1 (AFAP1-AS1) is dysregulated in certain tumors. However, the function of AFAP1-AS1 in non-small cell lung cancer (NSCLC) remains elusive. In this study, we conducted global lncRNA profiling and identified that AFAP1-AS1 is significantly upregulated in NSCLC, suggesting that AFAP1-AS1 may be important for lung cancer development. For the first time, the transcription initiation and termination sites of AFAP1-AS1 were identified by rapid amplification of cDNA ends technology, and the sequencing data indicated that AFAP1-AS1 in lung cancer cells is a novel transcript variant. Through gain- and loss-of-function studies, AFAP1-AS1 was demonstrated to promote cell migration and invasion. Mechanistically, AFAP1-AS1 functions through positively regulating the expression of AFAP1 protein. On the other hand, the expression of lncRNA AFAP1-AS1 negatively correlates with CpG methylation status of its gene promoter, identified in both lung cancer cells and patient tissues, and treatment with DNA methyltransferase inhibitor decitabine significantly activates AFAP1-AS1 expression, strongly supporting that AFAP1-AS1 expression is tightly regulated by DNA methylation. Taken together, this study demonstrates that AFAP1-AS1 acts as an oncogene in NSCLC to promote cell migration partly by upregulating AFAP1 expression, while its own expression is controlled by DNA methylation, and highlights its diagnostic and therapeutic values for NSCLC patients.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2923-8) contains supplementary material, which is available to authorized users.
Keywords: Lung cancer, AFAP1-AS1, AFAP1, Cell migration, Cell invasion, DNA methylation
Introduction
Lung cancer is the leading cause of cancer deaths around the world in spite of the improvement in diagnostic and therapeutic techniques. Non-small cell lung cancer (NSCLC) is the most common type and accounts for about 85% of lung cancer. The majority of NSCLC patients are diagnosed at advanced stages, and the prognosis is usually poor [1]. Therefore, it is in urgent need to dissect molecular mechanism of NSCLC development, and explore more accurate biomarkers and therapeutic targets.
Long non-coding RNAs (lncRNAs), a class of transcripts longer than 200 nucleotides in length without protein-encoding capacity, have been initially labeled as genomic “dark matter” [2]. Recently, increasing evidences demonstrate that lncRNAs play an important role in the regulation of gene expression at transcriptional or post-transcriptional levels, and are involved in multiple biological processes, such as cell proliferation, apoptosis, migration and invasion [3, 4]. Thus, dysregulated lncRNA expression may affect the hallmarks of cancers, such as sustaining proliferative signaling, resisting cell death, and activating invasion and metastasis [5, 6]. For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is overexpressed in certain tumors, and functions as an oncogene through controlling alternative splicing process or enhancing the metastasis phenotype of lung cancer cells [7, 8]. LincRNA-p21, a p53-regulated non-coding RNA, affects global gene expression and influences the p53 tumor suppressor pathway by acting in cis as a locus-restricted co-activator for p53-mediated p21 expression [9, 10]. Therefore, lncRNA may be an important target for cancer diagnosis and therapy.
Actin filament-associated protein 1-antisense RNA 1 (AFAP1-AS1) was initially discovered in esophageal adenocarcinoma, which is mapped to the 4p16.1 region of human chromosome 4, and transcribed as lncRNA from the antisense strand of DNA at the AFAP1 protein-coding gene locus [11]. Although the dysregulation of AFAP1-AS1 expression has been reported in esophageal adenocarcinoma [11], nasopharyngeal carcinoma [12], and pancreatic ductal adenocarcinoma [13], the role of AFAP1-AS1 in lung cancer still remains elusive. In this study, we conducted high-throughput RNA-seq to identify the differentially expressed lncRNAs between NSCLC tumors and adjacent normal tissues, and performed rapid amplification of cDNA ends (RACE) to determine the accurate transcription initiation and termination sites of AFAP1-AS1. Moreover, we investigated the role of AFAP1-AS1 in NSCLC through gain- and loss-of-function strategies, and explored the regulation of AFAP1-AS1 expression. Therefore, this study elucidates the potential role of AFAP1-AS1 in NSCLC development and highlights its diagnostic and therapeutic values for NSCLC patients.
Materials and methods
Patient tissue samples
Tumor tissues and their adjacent normal tissues from NSCLC patients were collected from West China Hospital, Sichuan University (China), which was approved by the Ethics Committee of the University Hospital. The patients had no known history of exposure to hypomethylating agents prior to surgery. Written informed consent for research purposes was provided for the patients.
Cell culture and construction of stable cells
The lung adenocarcinoma cell lines H1299, PC9 and H1975 were obtained from American Type Culture Collection (ATCC) and cultured in RPMI-1640 medium (Invitrogen, CA) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin (Sigma, St Louis, MO, USA). All cells were maintained in a humidified 5% CO2 atmosphere at 37 °C.
For lentivirus preparation, 293T cells were transfected using Lipofectamine 2000 with the lentiviral vector of pCDH-CMV-MCS-EF1-copGFP or pLKO.1-derived plasmid, packaging plasmid pCMV-dR8.2 dvpr, and envelope plasmid pCMV-VSVG (System Biosciences), and the resultant lentivirus-infected lung cancer cells. The stable transfectants were sorted by GFP signal, or selected by 1 μg/mL of puromycin.
Plasmid construction and rapid amplification of cDNA ends (RACE)
For AFAP1-AS1 overexpression, the full-length of AFAP1-AS1 was amplified by Phanta Super-fidelity DNA polymerase (Vazyme, China) from cDNAs reverse-transcribed from total RNAs of PC9 cells, and inserted into the vector pCDH-CMV-MCS-EF1-copGFP. For knockdown of human AFAP1-AS1 expression, the annealed oligos designed to target AFAP1-AS1 were inserted into pLKO.1-Puro vector at AgeI and EcoRI sites. For luciferase reporter assay, the promoter region (− 2500 bp to + 100 bp from transcription start site) of AFAP1-AS1 gene was amplified from genomic DNA, and cloned into the pGL3-basic vector (Promega, USA) at KpnI and HindIII sites to get the luciferase reporter plasmids. The transcription initiation and termination sites of AFAP1-AS1 were determined by FirstChoice® RLM-RACE Kit (Ambion, USA) according to the manufacturer’s instructions. The primers and oligonucleotides used in this study are listed in Supplementary Table 1.
RNA isolation and quantitative real-time PCR
Total RNAs were extracted by TRIzol reagent (Life Technologies, Carlsbad, CA, USA), and measured by NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA). For quantitative real-time PCR, cDNAs were generated by M-MLV Reverse Transcriptase Kit (Life Technologies, Carlsbad, CA, USA), and the qPCR was performed with SYBR Green Master Mix using StepOne Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). β-Actin or GAPDH mRNA was used as endogenous control, and the fold changes were calculated using the method. The primers for qPCR are listed in Supplementary Table 1.
Cell migration/invasion assays
Cell migration assays were performed using Transwell chamber (Millipore), and cell invasion assays were done with chambers uniformly covered with Matrigel (BD Biosciences) diluted with RPMI-1640 (1:7). Cells were suspended in RPMI-1640 medium containing 5% BSA and seeded into the top chamber, while RPMI-1640 medium supplemented with 10% FBS was added into the bottom chambers as chemoattractant. After incubation at 37 °C for 20 h, cells that did not migrate or invade through the pores of the Transwell inserts were removed with cotton swab. Cells present at the lower surface of the membrane were fixed by 4% paraformaldehyde for 20 min, stained with 1% crystal violet (Sigma) for 15 min. The cells were counted in at least three randomly selected microscopic fields (× 100) per filter under an inverted phase-contrast microscope. The experiment was repeated in three independent experiments.
Luciferase reporter assay
Using Lipofectamine 2000, cells were co-transfected with luciferase reporter plasmid, Renilla luciferase control vectors (pRL-TK), and AFAP1-AS1 expressing plasmids or empty vector as control. 24 h later, the luciferase activity was measured with Dual Luciferase Reporter Assay System (Promega, USA). The Firefly luciferase signal was normalized to Renilla luciferase signal, and the effect of AFAP1-AS1 on luciferase reporter with AFAP1 promoter region was then normalized with that on luciferase reporter without AFAP1 promoter region.
Fluorescence in situ hybridization (FISH)
The Cy3 fluorescence-labeled probes specifically for AFAP1-AS1 were designed and synthesized by RiboBio Company, and FISH experiments were performed by Ribo™ Fluorescent In Situ Hybridization Kit (RiboBio, China) according to the manufacturer’s instructions. Briefly, cells were fixed in 4% paraformaldehyde for 10 min, exposed to 0.5% Triton X-100 for 5 min, prehybridized at 37 °C for 30 min, and then in situ hybridized at 37 °C overnight with fluorescence-labeled probes. After extensive washing, nuclei were stained with DAPI solution. Imaging was acquired on an Olympus Fluoview laser scanning confocal microscope. The probes to detect 18s rRNA and U6 RNA were used as cytoplasmic and nuclear marker, respectively.
Bisulfite sequencing assay
The methylation status of CpG dinucleotides within two regions (A region: from − 784 to − 537; B region: from + 54 to + 555; relative to the transcription start site of the AFAP1-AS1 gene) was analyzed by bisulfite sequencing assay. Briefly, genomic DNA was isolated by QIAamp DNA Mini Kit (QIAGEN), and subjected to bisulfite conversion using EZ DNA Methylation-Gold Kit (Zymo Research, USA). Then, the bisulfite-modified DNAs were subjected to PCR amplification with TaKaRa EpiTaq HS DNA polymerase (TaKaRa, Japan), and the resultant PCR products were gel purified and cloned into pMD19-T vector using TaKaRa Cloning Kit (TaKaRa, Japan) for sequencing.
Data analysis of RNA-seq
Sequences were aligned to human GRCh38 assembly using HiSat2 v2.0.4 (options --dta -t --rna-strandness RF) [14], and assembled using StringTie v1.3.3b (options --rf) [15]. LncRNAs (long non-coding RNAs) and PCGs (protein-coding genes) were defined by GENCODE (v26) catalogue [16], and separately considered when performing differential expression analysis by edgeR [17]. Specifically, PCGs were filtered with mean TPM (transcripts per million) above 0.3, and identified as significantly differential expression if the fold change ≥ 2 and the adjusted p value < 0.05. LncRNAs were filtered with mean TPM above 0.1, and identified as significantly differential expression if the log2 fold change ≥ 0.75 and the p value < 0.05. Function enrichment analyses were performed for significantly differentially expressed PCG using DAVID [18] and the KOBAS software [19].
Survival analysis
Matched RNA-Seq gene expression and clinical data of 513 lung adenocarcinoma patients and 501 lung squamous cell carcinoma patients were downloaded from TCGA Data portal in October 2017. Expression values (FPKM, fragments per kilobase per million mapped reads) of AFAP1 were log2-transformed. A Cox proportional hazards model was used to determine whether AFAP1 expression was associated with disease-free survival after adjustment for sex, age at diagnosis, tumor histological type, history of other malignancy, history of neoadjuvant treatment and tobacco smoking history. Besides, survival analysis was performed using R package “Survival” from CRAN. The survival curves were constructed according to the Kaplan–Meier method and compared with the log-rank test, using samples with AFAP1 expression above 75th percentile, or below 25th percentile.
Statistical analysis
The experimental data are presented as the mean ± standard deviation (SD). All statistical analyses were performed using Pearson’s correlation coefficient or a two-tailed Student’s t test. The survival curves were calculated using the Kaplan–Meier method and statistically compared using a log-rank test. p < 0.05 was considered statistically significant.
Results
AFAP1-AS1 is upregulated in NSCLC tumors
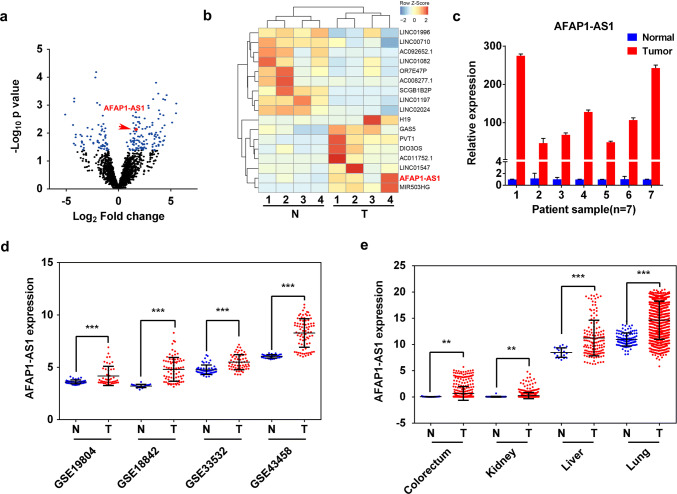
To explore the lncRNAs that have differential expression in NSCLC tissues compared to their paired adjacent normal tissues, we performed high-throughput RNA-Seq analysis (n = 4) and found that 123 and 71 lncRNAs were upregulated and downregulated, respectively, in NSCLC tumors (Fig. 1a). Among them, AFAP1-AS1 expression was significantly higher in NSCLC tumors than that in normal tissues (Fig. 1b), which was further confirmed by quantitative reverse-transcription PCR (qRT-PCR) in another batch of independent NSCLC tumor samples (n = 17) (Fig. 1c and Supplementary Figure 1a), in situ hybridization (n = 10, Supplementary Figure 1b), and the microarray data from Gene Expression Omnibus (GEO) database (Fig. 1d). Interestingly, the higher AFAP1-AS1 expression was also found in other tumors, such as colorectum, kidney and liver tumors (Fig. 1e), implying a universal and important role of AFAP1-AS1 in tumorigenesis. These results prompted us to investigate how AFAP1-AS1 expression is regulated and what’s the role of AFAP1-AS1 in the development of lung cancer.
Fig. 1.
Upregulation of lncRNA AFAP1-AS1 expression in NSCLC tumors. a Volcano plot and b hierarchical cluster plot displaying differentially expressed lncRNAs between NSCLC tumors and adjacent normal tissues. The X-axis represents log2 fold changes and the Y-axis represents log10p values. The blue dots denote the lncRNAs with the significantly differential expression (p < 0.05 and abs (log2 [FC]) > 0.75). AFAP1-AS1, highly expressed in NSCLC tumors, is presented as red dot. c qRT-PCR analysis of AFAP1-AS1 expression in 7 NSCLC tumors and their paired adjacent non-tumor tissues. Comparison of AFAP1-AS1 expression in tumors to normal tissues with the data obtained from GEO database for NSCLC (d) and from TCGA database for colorectum, kidney, liver and lung cancer tumors (e). Data are represented as mean values ± SD. *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t test)
Characterization and cellular distribution of AFAP1-AS1 in NSCLC cells
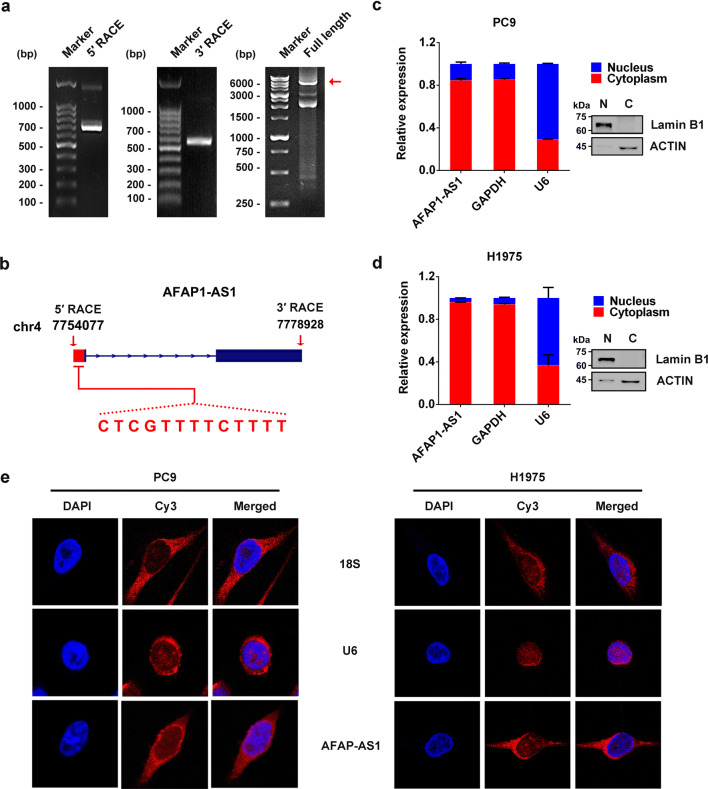
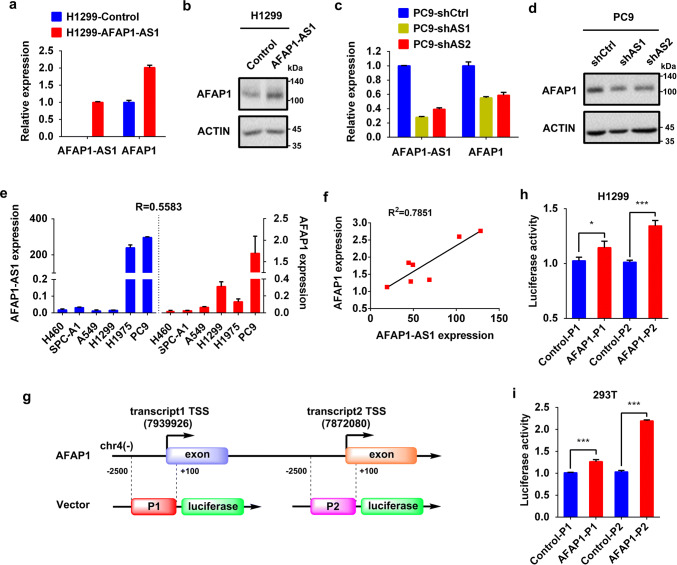
To investigate the cellular function of AFAP1-AS1, we first measured the expression level of AFAP1-AS1 by qRT-PCR in different lung cancer cell lines. As shown in Supplementary Figure 2a, H1975 and PC9 cells exhibited much higher expression of AFAP1-AS1 than other tested cells, such as A549 and H1299 cells. Therefore, PC9 cell was chosen for characterizing AFAP1-AS1, and its total RNAs were extracted and subjected to rapid amplification of cDNA ends (RACE) to identify the transcription initiation and termination sites of AFAP1-AS1. Based on the sequencing results of 5′- and 3′-RACE products, full-length AFAP1-AS1 was amplified and identified by Sanger sequencing to be 6823 nucleotides in length (Fig. 2a). By comparing with the sequence deposited in UCSC Browser database, the full length of AFAP1-AS1 in lung cancer cells we identified also consists of two exons, but has extra 13 nucleotides at its 5′ end (Fig. 2b), indicating it is a novel transcript.
Fig. 2.
Identification of AFAP1-AS1 and its predominant distribution in the cytoplasm of lung cancer cells. a Agarose gel electrophoresis of PCR products generated by 5′-RACE (left panel) and 3′-RACE (middle panel) and of the full-length AFAP1-AS1 (right panel). b Schematic illustration of NR_026892 (dark blue) and AFAP1-AS1 we identified, consisting of two exons. Sequence analysis showed there are 13 extra nucleotides at 5′ end of AFAP1-AS1 in lung cancer cells, which are absent in the NR_026892 sequences. Cell nucleus/cytoplasm fractionation confirmed by Western blots (right panels) and the cellular distribution of AFAP1-AS1 in PC9 cell (c) and H1975 cell (d) detected by qRT-PCR (left panels). Data are shown as the mean ± SD of three independent experiments. e FISH analysis of the cellular distribution of AFAP1-AS1 in PC9 cells (left) and H1975 cells (right)
The cellular distribution of AFAP1-AS1 was examined by qRT-PCR following cytoplasmic/nuclear fractionations, which was confirmed by Western blot analysis (Fig. 2c for PC9 cells and Fig. 2d for H1975 cells) using GAPDH and Lamin B1 as cytoplasmic and nuclear proteins, respectively. The qRT-PCR, adopting GAPDH mRNA and U6 RNA as cytoplasmic and nuclear control, respectively, indicated that AFAP1-AS1 is predominantly located in the cytoplasm of both cells (Fig. 2c, d). It was further validated by RNA fluorescence in situ hybridization (FISH) analyses using Cy3-labeled probes that specifically recognize AFAP1-AS1, because the probe fluorescence signals (red) mostly appeared in the cytoplasm of both PC9 and H1975 cells (Fig. 2e).
AFAP1-AS1 promotes cell migration and invasion in NSCLC cells
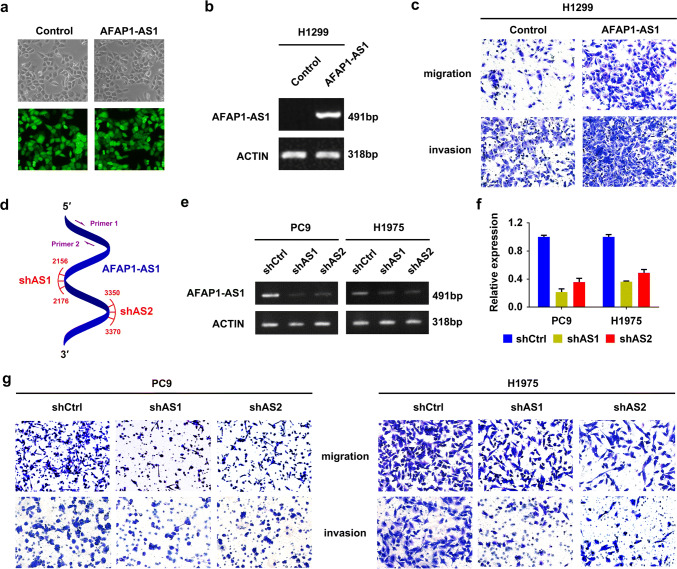
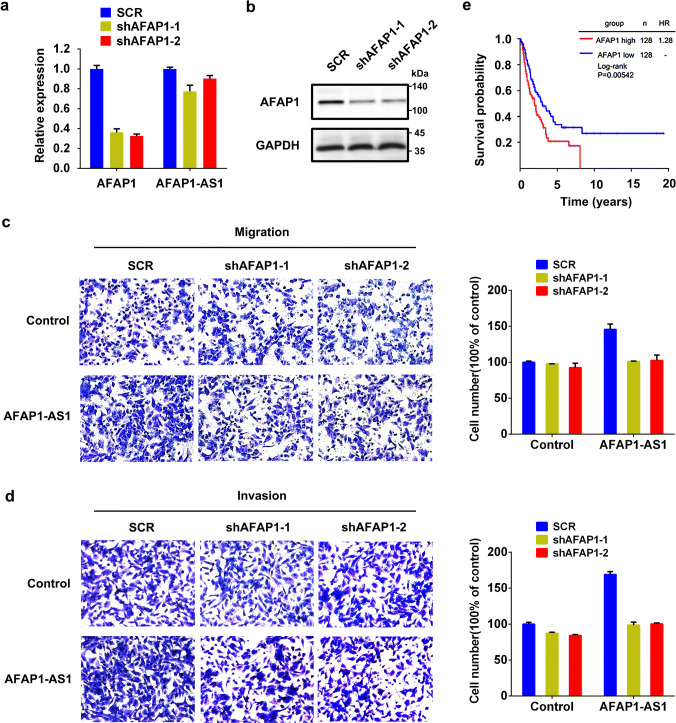
Through gain- or loss-of function experiments, we investigated the biological function of AFAP1-AS1 in NSCLC cells. The stable AFAP1-AS1 overexpressing cell line (H1299-AFAP1-AS1) and its negative control (H1299-control) were established by infection of AFAP1-AS1-containing lentivirus and empty vector, respectively, to H1299 cells, which showed relatively low AFAP1-AS1 expression (Supplementary Figure 2a), and then cell sorting by GFP signal (Fig. 3a). Semi-quantitative RT-PCR confirmed the overexpression of AFAP1-AS1 in H1299-AFAP1-AS1 cells (Fig. 3b). The AFAP1-AS1 overexpression granted the H1299-AFAP1-AS1 cells with significantly increased ability of migration and invasion compared with H1299-control cells (Fig. 3c). However, both MTT assay and colony-formation experiments demonstrated that AFAP1-AS1 overexpression has little effect on cell proliferation and the colony-forming ability (Supplementary Figure 2b and c).
Fig. 3.
AFAP1-AS1 promotes tumor cell migration and invasion in lung cancer cells. a Images of cells with or without AFAP1-AS1 overexpression under fluorescence microscope. b Semi-quantitative RT-PCR analysis of AFAP1-AS1 expression in H1299 cells with or without AFAP1-AS1 overexpression. Actin mRNA was used as control. c Cell migration and invasion assays of H1299-control or H1299-AFAP1-AS1 cells. d Schematic illustration of locations of qRT-PCR primers (primer 1/2) and two independent shRNAs (shAS1 and shAS2) in AFAP1-AS1 gene. Semi-quantitative RT-PCR (e) and qRT-PCR analyses (f) of AFAP1-AS1 expression in the stable transfectants of PC9 and H1975 cells. g Remarkably reduced cell migration and invasion in PC9 cells (left) and H1975 cells (right) following knockdown of AFAP1-AS1 by shRNAs
The loss-of-function assay was conducted through shRNA knockdown strategy to further confirm the function of AFAP1-AS1 in lung cancer cells. For the knockdown, we designed two independent shRNAs (shAS1 and shAS2) specifically targeting AFAP1-AS1 (Fig. 3d and Supplementary Table 1), and prepared the lentivirus particles to infect H1975 and PC9 cells, two cell lines with relatively high expression of AFAP1-AS1 compared to other tested cell lines (Supplementary Figure 2a). After puromycin selection, the stable transfectants expressing AFAS1-AS1 shRNA or scramble shRNA (shCtrl) were established, and the reduction of AFAP1-AS1 expression was validated by both semi-quantitative PCR (Fig. 3e) and qRT-PCR (Fig. 3f). In the following cell migration and invasion assays, it was found that AFAP1-AS1 knockdown was accompanied by significant decrease of cell migration and invasion in both cells (Fig. 3g). Nevertheless, AFAP1-AS1 showed no influence on cell viability and colony formation in these two cell lines (Supplementary Figure 2d–g). Taken together, AFAP1-AS1 plays an oncogenic role in NSCLC cells.
The potential downstream effectors of AFAP1-AS1 involved in metastasis pathway
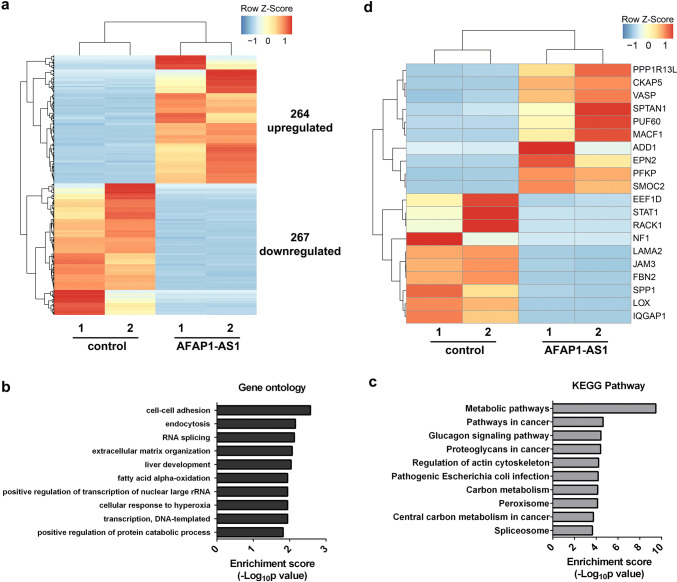
To comprehensively understand the effect of AFAP1-AS1 on NSCLC signalling pathways, we performed RNA-seq to profile differential gene expressions in H1299 cells with AFAP1-AS1 overexpression compared to H1299-control cells. The unbiased genome-scale analysis identified that 531 transcripts displayed differential expression [log2(fold change) > 1 and p < 0.05], with 267 transcripts downregulated and 264 transcripts upregulated (Fig. 4a). Moreover, GO analysis of these differentially expressed genes suggested potential alterations in cell–cell adhesion and extracellular matrix organization as a consequence of AFAP-AS1 overexpression (Fig. 4b), consistent with our observation that AFAP1-AS1 promotes cell migration and invasion. We also performed KEGG analysis and found that regulation of actin cytoskeleton and pathways in cancer are potentially affected by AFAP1-AS1 overexpression (Fig. 4c), further supporting the observed critical role of AFAP1-AS1 in cell migration and invasion. To elucidate the mechanism of AFAP1-AS1 in NSCLC metastasis, the cell migration and invasion-related genes, as displayed in Fig. 4d, were grabbed out of the pool of differentially expressed genes identified by RNA-seq. Among them, PPP1R13L, VASP and SPTAN1, whose high expressions are reported to positively correlate with cell invasion [20–22], are subject to the upregulation by AFAP1-AS1. And STAT1, NF1, and FBN2, whose lower expression is indicated to promote tumor metastasis [23–25], are subject to downregulation by AFAP1-AS1 (Fig. 4d). Thus, our RNA transcriptome profiling data strongly suggested that AFAP1-AS1 promotes NSCLC cell migration and invasion via altering the expression of metastasis-associated genes.
Fig. 4.
Effect of AFAP1-AS1 on gene expression in H1299 cells. a Hierarchical cluster plot of the differentially expressed genes in H1299-AFAP1-AS1 cells compared to H1299-control cells. b GO analyses listing the top ten biological processes regulated by AFAP1-AS1. c KEGG pathway analyses showing the top ten pathways affected by AFAP1-AS1. d Hierarchical cluster plot of the differentially expressed and metastasis-associated genes between H1299-control or H1299-AFAP1-AS1 cells, using a cutoff of fold change > 2 and adjusted p value < 0.05
AFAP1-AS1 upregulates AFAP1 expression in NSCLC cells
The lncRNAs located in the antisense strand of protein-coding genes may function by regulating transcription, splicing, translation, or degradation of their corresponding coding mRNA transcripts [26], which prompted us to investigate the relationship between AFAP1-AS1 and actin filament-associated protein 1 (AFAP1). The qRT-PCR and Western blot analyses revealed that AFAP1-AS1 overexpression in H1299-AFAP1-AS1 cells was accompanied by the increase of AFAP1 at both mRNA (Fig. 5a) and protein levels (Fig. 5b), whereas AFAP1-AS1 has little effect on the half-lives of AFAP1 mRNA and protein (Supplementary Figure 3), suggesting that AFAP1-AS1 promotes AFAP1 gene transcription instead of impacting AFAP1 degradation. The positive correlation of AFAP1 expression with AFAP1-AS1 was also verified in the AFAP1-AS1 knockdown PC9 cell where AFAP1 expression had a marked decrease (Fig. 5c, d), the other tested in NSCLC cell lines (Fig. 5e) and NSCLC patient tissues (Fig. 5f).
Fig. 5.
AFAP1-AS1 positively regulates AFAP1 expression in lung cancer cells. a RNA levels of AFAP1-AS1 and AFAP1 in H1299-control or H1299-AFAP1-AS1 cell, measured by qRT-PCR. b Protein level of AFAP1 in H1299-control or H1299-AFAP1-AS1 cell, examined by Western blot. c RNA levels of AFAP1-AS1 and AFAP1 in PC9 cells with or without AFAP1-AS1 knockdown, determined by qRT-PCR. d Protein levels of AFAP1 in PC9 cells with or without AFAP1-AS1 knockdown, detected by Western blot. e AFAP1-AS1 and AFAP1 mRNA levels in different lung cancer cells, measured by qRT-PCR, and their Pearson’s correlation coefficient (R = 0.5583, p < 0.05) obtained by statistical analysis. f Pearson’s correlation coefficient for correlation between AFAP1-AS1 and AFAP1, calculated based on their respective expression levels in seven NSCLC tumor samples and the paired adjacent normal tissues. g Schematic illustration of constructed luciferase reporter plasmids. Because AFAP1 gene has two transcripts with different transcription start sites, but sharing the same protein-coding sequence, both potential promoters (− 2500 bp to + 100 bp from TSS) were cloned into pGL3-basic plasmid to construct the luciferase reporter plasmids. Dual luciferase reporter assays for AFAP1 promoter activity with AFAP1-AS1 overexpression in H1299 cell (h) and HEK293T cell (i). Renilla luciferase was used as internal control. Statistical analysis was determined by paired Student’s t test. *p < 0.05; ***p < 0.001
To confirm that AFAP1-AS1 affects AFAP1 transcription, we constructed the luciferase reporter plasmids for AFAP1 promoter. Because there are two AFAP1 transcripts sharing the same open-reading frame sequence with different transcription start site (TSS) (Fig. 5g), both promoters were cloned and tested. The dual luciferase report assays in H1299 cells (Fig. 5h) and HEK293T cells (Fig. 5i) co-transfected with firefly luciferase reporter plasmid, Renilla luciferase plasmid as internal control, and AFAP1-AS1 expressing plasmid or empty vector as negative control demonstrated that AFAP-AS1 can promote AFAP1 transcription in both cells.
AFAP1-AS1 enhanced cell migration is mediated by AFAP1
AFAP1 is an adaptor protein of c-Src kinase and it binds to filamentous actin and regulates the activity of c-Src kinase to change the organization of actin cytoskeleton [27]. Combined with the preceding findings that AFAP1-AS1 promotes cell migration and also upregulates AFAP1 expression, we hypothesized that AFAP1-AS1 contributes to cell migration at least in part through AFAP1. To test this hypothesis, two shRNAs specifically targeting different AFAP1 mRNA regions were designed, and the knockdown of AFAP1 expression was validated by qRT-PCR (Fig. 6a) and Western blot (Fig. 6b). With AFAP1 knockdown in H1299-AFAP1-AS1 cells, the migrated and invaded cells significantly decreased (Fig. 6c, d). As a comparison, AFAP1 knockdown in H1299-control cell without AFAP1-AS1 overexpression did not lead to obvious changes in cell migration and invasion (Fig. 6c, d). Therefore, AFAP1-AS1 promoting cell migration is, at least in part, mediated by AFAP1 protein. In addition, Kaplan–Meier curve analyses showed that higher expression of AFAP1 was associated with poorer survival of NSCLC patients (Fig. 6e), regardless of their histotypes (Supplementary Figure 4). Moreover, the cox regression analyses showed that high expression of AFAP1 had a poor effect on disease-free survival in both lung adenocarcinoma patients and squamous cell carcinoma patients (Supplementary Tables 2 and 3).
Fig. 6.
AFAP1 participates in AFAP1-AS1-promoted cell migration and invasion. Knockdown of AFAP1 by shAF1 and shAF2 in H1299 cells, measured by qRT-PCR (a) and Western blot (b). GAPDH was used as control. Dramatically decreased cell migration (c) and invasion (d) in H1299-AFAP1-AS1 cells following AFAP1 knockdown (left panels: images; right panel: number count of migrated and invasive cells). The migrated and invasive cell numbers were counted in at least three randomly selected microscopic fields. The experiment was performed in three independent experiments. e Kaplan–Meier estimates of disease-free survival in NSCLC patients according to AFAP1 expression levels
Promoter CpG hypomethylation leads to increased AFAP1-AS1 expression in NSCLC tumors
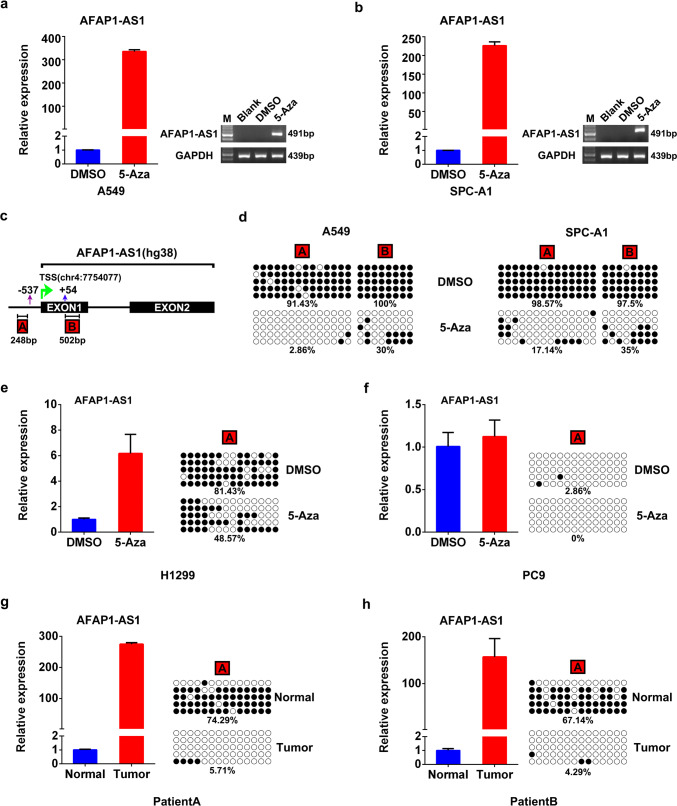
Wu et al. applied high-resolution methylome analysis to identify methylation changes at genomic regions and found that AFAP1-AS1 gene is hypomethylated in esophageal adenocarcinoma [11], so we wondered if the AFAP1-AS1 expression in lung cancer is also regulated by DNA methylation. To test this proposal, both A549 and SPC-A1 cells, which have relatively low level of AFAP1-AS1 among the tested cell lines (Supplementary Figure 2a), were treated with DNA methyltransferase inhibitor decitabine (5-Aza). The significant increase of AFAP1-AS1 expression after the treatment was clearly demonstrated by semi-quantitative RT-PCR and qRT-PCR (Fig. 7a, b), implying that transcription of AFAP1-AS1 gene in A549 and SPC-A1 cells is repressed by hypermethylation of the promoter region. To identify the methylation sites, we dissected the promoter sequence of AFAP1-AS1 gene through bioinformatics analysis. Besides the reported sequence downstream of TSS (B region, 502 bp amplicon) (Fig. 7c) [11], we found another CpG-enriched region starting from 537 bp upstream of TSS (A region, 248 bp amplicon) (Fig. 7c). The bisulfite sequencing analyses showed that 5-Aza treatment of A549 cells dramatically decreased the percentage of CpG methylation in the A region from 91.43 to 2.86% and that in the B region from 100 to 30% (Fig. 7d). The similar reductions of the methylation were also observed in the 5-Aza-treated SPC-A1 cells (Fig. 7d). The data from both cells demonstrated that the A region is more sensitive to 5-Aza treatment than the B region, therefore, the methylation status of only A region was analyzed in the following experiments. As shown in Fig. 7e, in H1299 cells expressing relatively low AFAP1-AS1 (Supplementary Figure 2a), the A region is highly methylated and 5-Aza treatment significantly decreased the CpG methylation, leading to the elevated AFAP1-AS1 expression. However, 5-Aza treatment has little effect on AFAP1-AS1 expression in PC9 cells that has relatively high AFAP1-AS1 expression, which could be due to the existing hypomethylation of AFAP1-AS1 promoter in PC9 cells in the absence of 5-Aza (Fig. 7f). To investigate the clinical relevance, we examined the methylation status of AFAP1-AS1 in NSCLC tumors and their adjacent normal tissues. It was found that the CpGs within the AFAP1-AS1 promoter is highly methylated in the normal tissues, whereas it is hypomethylated in the tumors, corresponding to high expression of AFAP1-AS1 in these tumors and low expression in normal tissues (Fig. 7g, h). Moreover, bioinformatics analysis showed that CpG methylation within AFAP1-AS1 gene promoter is an early event during tumorigenesis, regardless of tumor stages (Supplementary Figure 5). Taken together, our results convincingly support that AFAP1-AS1 expression in lung cells is regulated by DNA methylation.
Fig. 7.
AFAP1-AS1 expression is regulated by DNA methylation in lung cancer cells. AFAP1-AS1 expression after 5-Aza treatment of A549 (a) and SPC-A1 cells (b), measured by qRT-PCR (left panels) and semi-quantitative PCR analyses (right panel). c Schematic diagram of CpG-rich regions within the AFAP1-AS1 gene promoter. The A region starts 54 bp downstream of TSS and the B region starts 537 bp upstream of TSS. Dramatically decreased CpG methylation level of AFAP1-AS1 gene promoter in 5-Aza-treated A549 and SPC-A1 cells (d), detected by bisulfite genomic sequencing. The black solid circles and empty circles represent methylated and unmethylated CpG dinucleotides, respectively. AFAP1-AS1 expression (left panels) and CpG methylation (right panels) status in H1299 cell (e), PC9 cell (f), and patient tissues (g, h), measured by qRT-PCR (left panels) and bisulfite genomic sequencing (right panels), respectively
Discussion
LncRNAs have recently emerged as an important regulator involved in many physiological and pathological processes including tumorigenesis, thus providing a potential target for cancer diagnosis and therapy [4, 6]. Although several studies reported the dysregulation of lncRNA AFAP1-AS1 expression in hepatocellular carcinoma [28], nasopharyngeal carcinoma [12], and gallbladder cancer [29], the role of AFAP1-AS1 in lung cancer remains unclear. In this study, we performed the high-throughput RNA-seq analysis and found that AFAP1-AS1 expression is significantly upregulated in NSCLC tumors compared with their adjacent non-tumor tissues (Fig. 1a, b), which was then validated by qRT-PCR (Fig. 1c) and supported by the bioinformatics analyses of GEO expression profiles (Fig. 1f) and TCGA data (Fig. 1g). Interestingly, based on the TCGA data analysis, the increased expression of AFAP1-AS1 also exists in other types of tumors, such as colorectal cancer, kidney cancer and liver cancer (Fig. 1g), suggesting that AFAP1-AS1 may play universal and important role during tumor progression and metastasis.
The lncRNA AFAP1-AS1 is mapped to the 4p16.1 region of human chromosome 4, and is transcribed from the AFAP1 gene in the antisense direction. Although several papers reported AFAP1-AS1 [11–13, 28–30], its accurate transcription initiation and termination sites have not been identified experimentally. In our study, we first utilized RACE technique to determine the transcription initiation and termination sites (Fig. 2a), and then successfully cloned the full length of AFAP1-AS1 (Fig. 2b). The Sanger sequencing results showed that AFAP1-AS1 in PC9 cell is 6823 nucleotides in length, with extra 13 nucleotides at the 5′ end compared to the deposited NCBI sequence NR_026892 (Fig. 2b), thus AFAP1-AS1 in lung cancer cells is a novel transcript. The following investigation of the cellular distribution revealed that AFAP1-AS1 is predominantly located in the cytoplasm (Fig. 2c–e).
Through gain- and loss-of function experiments, AFAP1-AS1 was identified as an oncogene in NSCLC cells, and it may play a critical role in metastasis. The overexpression of AFAP1-AS1 significantly promoted cell migration and invasion (Fig. 3c), conversely, AFAP1-AS1 knockdown by shRNAs dramatically decreased cell migration and invasion (Fig. 3g). Our observation is consistent with the findings by Han et al. who reported that AFAP1-AS1 knockdown in colorectal cancer cells (CRCs) inhibited the expression of tumor metastasis-associated genes and also suppressed hepatic metastasis of CRC cells in nude mice [31]. In addition, Ma et al. reported that knockdown of lncRNA AFAP1-AS1 in gallbladder cancer cells inhibited epithelial–mesenchymal transition by down-regulating the transcription factor Twist1 and Vimentin and up-regulating the E-cadherin [29]. Therefore, AFAP1-AS1 may act as a universal regulator for metastasis in a wide spectrum of cancers. It was also recognized that in hepatocellular carcinoma [28] and cholangiocarcinoma [32], AFAP1-AS1 not only promotes cell migration but also facilitates cell proliferation, whereas in lung cancer cells AFAP1-AS1 does not facilitate cell proliferation and colony formation (Supplementary Figure 2d–g). Such difference implies that the effect of AFAP1-AS1 on cell growth may be cell specific or tissue specific.
Recently, molecular mechanisms of AFAP1-AS1 in tumorigenesis were investigated in different cancers. Zhang et al. demonstrated that AFAP1-AS may promote HCC development through upregulation of RhoA/Rac2 signaling [28]. In laryngeal carcinoma, AFAP1-AS1 was reported to enhance stemness and chemoresistance by functioning as the sponge of miR-320a, which in turn regulates RBPJ expression [33]. In colorectal cancer, AFAP1-AS1 is associated with the enhancer of zeste homolog 2 (EZH2) to repress the expression of EZH2 target genes [34]. Recent findings have shown that antisense lncRNAs, which are transcribed from the opposite strand of protein002Dcoding genes, can exert their regulatory functions by acting as epigenetic regulators of gene expression and chromatin remodeling [35]. For example, p15 antisense (p15AS) lncRNA suppresses the expression of the cyclin-dependent kinase inhibitor p15 in cis and in trans through heterochromatin formation [26]. From another angle, Carrieri et al. observed that the lncRNA transcribed from the antisense of ubiquitin carboxy-terminal hydrolase L1 (Uchl1) gene can increase UCHL1 protein synthesis [36], hence revealing another layer of gene expression control that occurs at the post-transcriptional level by antisense lncRNAs. Combining with the previous finding that the sense transcript AFAP1, opposite of AFAP1-AS1, is able to change the organization of actin cytoskeleton [37–40], we proposed that AFAP1-AS1 regulates metastasis through controlling AFAP1 expression. And our results showed that AFAP1-AS1 overexpression leads to increased AFAP1 expression in H1299 cells at both mRNA and protein levels (Fig. 5a, b) without changing the half-lives of AFAP1 mRNA and protein (Supplementary Figure 3), while AFAP1-AS1 knockdown markedly decreased AFAP1 expression in PC9 cells (Fig. 5c, d), indicating that AFAP1-AS1 regulates AFAP1 expression at transcriptional level. Moreover, the positive correlation between AFAP1-AS1 and AFAP1 expressions was further confirmed in other tested NSCLC cell lines (Fig. 5e) and patient tissues (Fig. 5f). Through the dual luciferase report assays driven by AFAP1 promoter in the absence or presence of overexpressed AFAP1-AS1, it was confirmed that AFAP1-AS1 indeed enhances the transcription activity of AFAP1 gene promoter (Fig. 5h and i). Next, we looked into the role of AFAP1 in the promotion of cell migration by AFAP1-AS1. And the results showed that AFAP1 knockdown by two independent shRNAs dramatically decreased the migrated and invaded cell number in AFAP1-AS1 overexpressing cell (Fig. 6c, d), indicating that AFAP1-AS1 promotes cell migration is, at least in part, mediated by AFAP1 protein. These findings are consistent with RNA transcriptome sequencing results, GO and KEGG analyses, which showed that AFAP1-AS1 overexpression in H1299 cells significantly changes the expression of genes involved in cell–cell adhesion, extracellular matrix organization, and actin organization (Fig. 4b, c), and is also consistent with the reported role of AFAP1 protein in organization of actin cytoskeleton. In spite of these exciting findings, how AFAP1-AS1 regulates AFAP1 expression still remains obscure. For example, how AFAP1-AS1 interacts and affects AFAP1 expression? Does AFAP1-AS1 directly participate in AFAP1 transcription or via any intermediators? These questions need to be addressed in future investigations.
Similar to protein-coding genes, epigenetic regulation, such as DNA methylation, of lncRNA expression might contribute to carcinogenesis. For example, Heilmann et al. performed a genome-wide screen for differentially methylated lncRNA promoters in tumors versus normal tissues, and identified that lncRNA Esrp2-AS is hypomethylated, leading to the increased expression in human breast cancer [41]. Diaz-Lagares et al. identified that TP53TG1, a p53-induced lncRNA, undergoes cancer-specific promoter hypermethylation-associated silencing, and found that TP53TG1 hypermethylation in primary tumors is associated with poor outcome [42]. We, therefore, hypothesized that AFAP1-AS1 expression in lung cancer cells is possibly regulated by DNA methylation as well. Our results show the negative correlation between AFAP1-AS1 expression and its promoter CpG methylation in both cell lines (Fig. 7a–g) and patient tissues (Fig. 7h, i), indicating the regulation of AFAP1-AS1 expression by DNA methylation in NSCLC tumors. Moreover, treatment with the demethylation reagent 5-Aza significantly increased AFAP1-AS1 expression in the AFAP1-AS1-low-expressing cells (Fig. 7a–g). Therefore, it is clear that AFAP1-AS1 expression in NSCLC is tightly regulated by CpG methylation of its gene promoter, further supporting the idea that epigenetic modification is a common event to cause aberrant expression of lncRNAs in tumors.
In summary, we propose a model to explain how AFAP1-AS1 works in lung cells based on our findings: under normal conditions, the promoter of AFAP1-AS1 gene is highly methylated, leading to low expression in lung epithelial cells. During tumor progression and metastasis, the suppression of AFAP1-AS1 transcription is released by hypomethylation within the promoter and AFAP1-AS1 expression is highly increased, which enhances AFAP1 transcription to promote cell migration and invasion. Taken together, our study elucidates the function and molecular mechanism of AFAP1-AS1 in lung cancer cell migration and highlights its diagnostic and therapeutic values for NSCLC patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by National Key R&D Program of China (2016YFA0502204 and 2017YFA0504304), and National Natural Science Foundation of China (81772960 and 81572739).
Abbreviations
- NSCLC
Non-small cell lung carcinoma
- lncRNA
Long non-coding RNA
- AFAP1-AS1
Actin filament-associated protein 1-antisense RNA 1
- AFAP1
Actin filament-associated protein 1
- RACE
Rapid amplification of cDNA ends
- TSS
Transcription start site
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- FISH
RNA fluorescence in situ hybridization
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Juan He, Ke Wu and Chenglin Guo equally contributed to this work.
References
- 1.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, et al. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Niland CN, Merry CR, Khalil AM. Emerging roles for long non-coding RNAs in cancer and neurological disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144(5):956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137. doi: 10.1186/s12967-015-0490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laska MJ, Lowe SW, Zender L, Hearn S, Vogel U, Jensen UB, et al. Enforced expression of PPP1R13L increases tumorigenesis and invasion through p53-dependent and p53-independent mechanisms. Mol Carcinog. 2009;48(9):832–842. doi: 10.1002/mc.20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Zhang J, Wu J, Luo D, Su K, Shi W, et al. MicroRNA-610 inhibits the migration and invasion of gastric cancer cells by suppressing the expression of vasodilator-stimulated phosphoprotein. Eur J Cancer. 2012;48:1904–1913. doi: 10.1016/j.ejca.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Hinrichsen I, Ernst BP, Nuber F, Passmann S, Schäfer D, Steinke V, et al. Reduced migration of MLH1 deficient colon cancer cells depends on SPTAN1. Mol Cancer. 2014;13:11. doi: 10.1186/1476-4598-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varikuti S, Oghumu S, Elbaz M, Volpedo G, Ahirwar DK, Alarcon PC, et al. STAT1 gene deficient mice develop accelerated breast cancer growth and metastasis which is reduced by IL-17 blockade. Oncoimmunology. 2017;6(11):e1361088. doi: 10.1080/2162402X.2017.1361088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Suzuki M, Nakamura Y, Ohira M, Ando S, Iida T, et al. Aberrant methylation of FBN2 in human non-small cell lung cancer. Lung Cancer. 2005;50:43–49. doi: 10.1016/j.lungcan.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Zhang Y, Li Y, Fan K. Neurofibromatosis type-1 is a prognostic indicator in human gastric carcinoma. Oncotarget. 2017;8:82910–82919. doi: 10.18632/oncotarget.19876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunnick JM, Kim S, Hadsell J, Collins S, Cerra C, Reiser P, et al. Actin filament-associated protein 1 is required for cSrc activity and secretory activation in the lactating mammary gland. Oncogene. 2015;34:2640–2649. doi: 10.1038/onc.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY, et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590–1598. doi: 10.3892/ijo.2016.3385. [DOI] [PubMed] [Google Scholar]
- 29.Ma F, Wang SH, Cai Q, Zhang MD, Yang Y, Ding J. Overexpression of lncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed Pharmacother. 2016;84:1249–1255. doi: 10.1016/j.biopha.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152–159. doi: 10.1016/j.biopha.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49(1):36. doi: 10.1186/s40659-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Zhang H, Wang M, Xu X, Zhao Y, He R, et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394–58404. doi: 10.18632/oncotarget.16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan Z, Xiu C, Song K, Pei R, Miao S, Mao X, et al. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang J, Zhong G, Wu J, Chen H, Jia Y. Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. Am J Cancer Res. 2018;8(5):892–902. [PMC free article] [PubMed] [Google Scholar]
- 35.Magistri M, Faghihi MA, St Laurent G, Wahlestedt G., III Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 37.Flynn DC, Leu TH, Reynolds AB, Parsons JT. Identification and sequence analysis of cDNAs encoding a 110-kilodalton actin filament-associated pp60src substrate. Mol Cell Biol. 1993;13:7892–7900. doi: 10.1128/MCB.13.12.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorfleutner A, Cho Y, Vincent D, Cunnick J, Lin H, Weed SA, et al. Phosphorylation of AFAP-110 affects podosome lifespan in A7r5 cells. J Cell Sci. 2008;121:2394–2405. doi: 10.1242/jcs.026187. [DOI] [PubMed] [Google Scholar]
- 39.Baisden JM, Gatesman AS, Cherezova L, Jiang BH, Flynn DC. The intrinsic ability of AFAP-110 to alter actin filament integrity is linked with its ability to also activate cellular tyrosine kinases. Oncogene. 2001;20:6607–6616. doi: 10.1038/sj.onc.1204802. [DOI] [PubMed] [Google Scholar]
- 40.Qian Y, Baisden JM, Westin EH, Guappone AC, Koay TC, Flynn DC. Src can regulate carboxy terminal interactions with AFAP-110, which influence self-association, cell localization and actin filament integrity. Oncogene. 1998;16:2185–2195. doi: 10.1038/sj.onc.1201753. [DOI] [PubMed] [Google Scholar]
- 41.Heilmann K, Toth R, Bossmann C, Klimo K, Plass C, Gerhauser C. Genome-wide screen for differentially methylated long noncoding RNAs identifies Esrp2 and lncRNA Esrp2-as regulated by enhancer DNA methylation with prognostic relevance for human breast cancer. Oncogene. 2017;36:6446–6461. doi: 10.1038/onc.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, Soler M, Setien F, Goyal A, et al. Epigenetic inactivation of the p53-induced long non-coding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci USA. 2016;113:E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.