Abstract
Embryonic stem cells (ESCs) are immortal stem cells that own multi-lineage differentiation potential. ESCs are commonly derived from the inner cell mass (ICM) of pre-implantation embryos. Due to their tremendous developmental capacity and unlimited self-renewal, ESCs have diverse biomedical applications. Different culture media have been developed to procure and maintain ESCs in a state of naïve pluripotency, and to preserve a stable genome and epigenome during serial passaging. Chromatin modifications such as DNA methylation and histone modifications along with microRNA activity and different signaling pathways dynamically contribute to the regulation of the ESC gene regulatory network (GRN). Such modifications undergo remarkable changes in different ESC media and determine the quality and developmental potential of ESCs. In this review, we discuss the current approaches for derivation and maintenance of ESCs, and examine how differences in culture media impact on the characteristics of pluripotency via modulation of GRN during the course of ICM outgrowth into ESCs. We also summarize the current hypotheses concerning the origin of ESCs and provide a perspective about the relationship of these cells to their in vivo counterparts (early embryonic cells around the time of implantation). Finally, we discuss generation of ESCs from human embryos and domesticated animals, and offer suggestions to further advance this fascinating field.
Keywords: Pluripotent stem cells, Early embryogenesis, Development, Blastocyst, ESC derivation, Gene regulation, Embryo
Introduction
Embryonic stem cells (ESCs) are mostly derived from the inner cell mass (ICM) of blastocyst-stage embryos. Although they possess infinite self-renewal ability, ESCs are able to generate virtually all derivatives of the three embryonic germ layers as well as germ cells during in vitro differentiation and own the potential to return to their original niche in vivo. These unique features make ESCs a convenient candidate for studies in developmental biology, toxicology, disease modeling, and drug testing [1, 2]. However, the ability of ESCs for long-term self-renewal does not recapitulate the limited expansion of the ICM in the early embryo. This issue raises several questions. Do ESCs represent a “locked” state of ICM cells that are captured from a narrow developmental window before implantation or do they acquire specific features during in vitro derivation that distinguishes them from ICM cells? If ESCs acquire distinct properties in vitro, can they still be considered to be natural developmental counterparts of ICM cells? In more practical terms, which criteria should be applied to optimize derivation and maintenance of ESCs for further applications? Is it meaningful to maintain all features of ICM cells during ESC derivation or rather counterproductive? Establishment of ESCs goes along with numerous changes in DNA methylation, histone modifications, gene expression, epithelial/mesenchymal status, and proliferative capacity [3, 4]. Despite these considerable and dynamic changes, the resultant ESCs are highly similar to pre-implantation epiblast cells from which they originated; however, ESCs exhibit certain crucial differences compared to epiblast cells [4, 5]. In this article, we review the emergence of pluripotent cells in the ICM and the continuum of pluripotency around the time of implantation. We discuss the perpetuation of pluripotency in vitro with a focus on culture conditions and extrinsic regulators used to efficiently derive ESCs from the ICM. Finally, we describe the current knowledge about epigenetic control mechanisms and the intrinsic gene regulatory networks (GRNs) that govern maintenance and establishment of different ESCs.
In vivo pluripotency: when to derive ESCs?
During early mammalian development, a totipotent zygote initiates a highly dynamic developmental process to produce a fully functional multicellular organism. The term totipotency has been defined, in its loosest sense, as the ability of a cell to generate cell types from both embryonic (i.e. ectoderm, mesoderm, and endoderm) and extra-embryonic cell lineages (e.g. trophectoderm). However, according to the strictest definition, totipotency is the ability of a cell to give rise to an entire embryo/fetus along with its associated extra-embryonic fetal membranes (a feature typically exhibited by one- to two-cell embryos). In the blastocyst-stage embryo which is the result of the development of a totipotent zygote, the pluripotent ICM and the trophectoderm (TE) cells morphologically separate in the first cell fate bifurcation of the embryo. Global DNA demethylation occurs during the transition from the zygote to the blastocyst. ICM cells undergo genome-wide DNA hypomethylation, with the exception of the parent-specific imprinting control regions (ICRs) that are marked by DNA methylation on one of the two parental alleles [6]. During the second cell fate specification in the embryo, ICM cells segregate into primitive endoderm (PE) and pluripotent epiblast (EPI or primitive ectoderm) lineages [7]. In fact, it has been demonstrated that EPI and PE cells are specified by Nanog and Gata6 expression, respectively; the mutually exclusive expression patterns of Nanog and Gata6 determines the fate of ICM cell segregation into EPI or PE cell lineages, respectively [8]. Despite this association with specific TFs, it was found that modulation of Fgf/Erk signaling could still shift the Nanog- or Gata6-expressing ICM cells towards alternative fate, suggesting that ICM cells (from E3.5 blastocysts) have not yet adopted their final fate and that the activity of Fgf/Erk pathway specifies the final lineage segregation of ICM cells [9]. In the mouse, the first and the second lineage segregations are seen on embryonic days (E)3.5 and E4.5, respectively, when the embryo is preparing for implantation (Fig. 1) but cells maintain the ability to become propagated as pluripotent stem cells (PSCs) in vitro until the late gastrulation stage (E8.25) [10]. Shortly before gastrulation, primordial germ cells (PGCs) arise from the epiblast and are protected from the diverse differentiation stimuli during gastrulation. In contrast to global de novo DNA remethylation in somatic cells of the post-implantation embryo, the DNA of PGCs undergo global demethylation including at ICRs until de novo methylation is initiated in developing gonads, based on the sex of the embryo [11, 12].
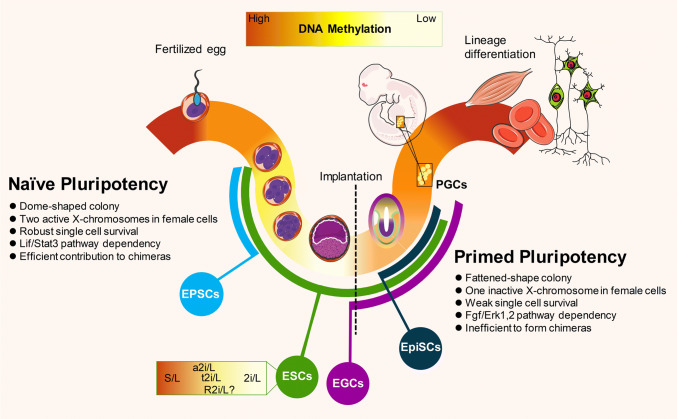
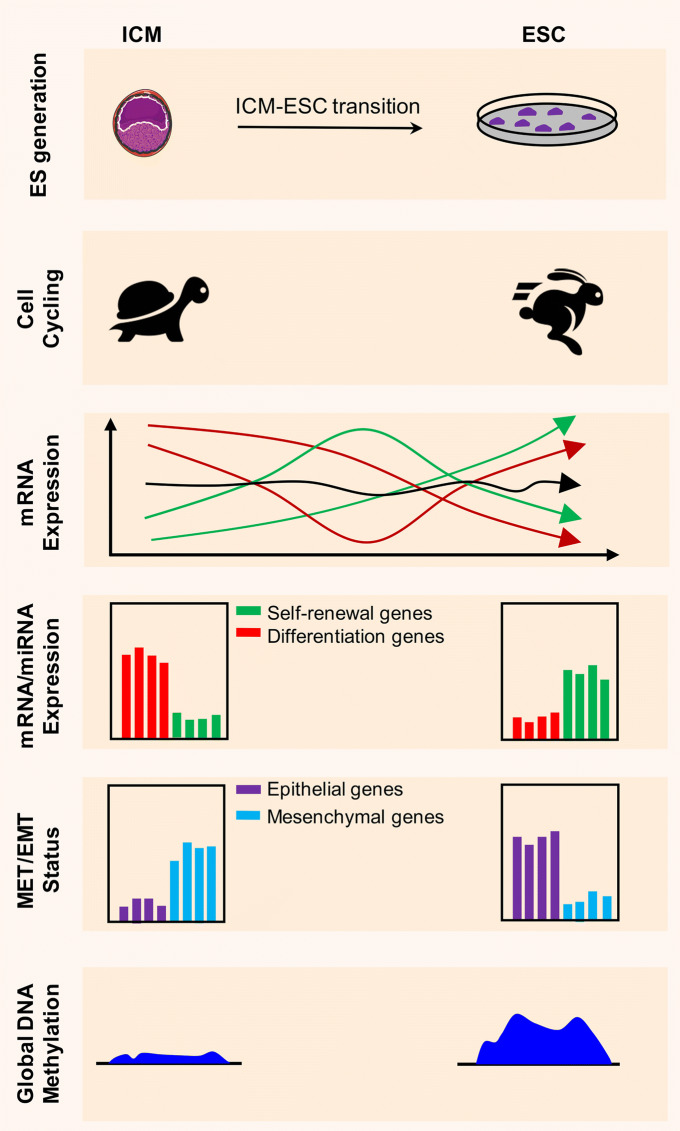
Fig. 1.
Derivation of pluripotent stem cell lines from developing mouse embryos. After sperm and oocyte fertilization, global DNA demethylation occurs in the pre-implantation embryo during the development of a zygote to a blastocyst. After implantation, lineage differentiation is achieved by de novo global methylation. PGCs that emerge from post-implantation epiblast (EPI) are kept away from gastrulation events and undergo global demethylation. Naïve and primed stem cells can be derived from pre- and post-implantation epiblast cells, respectively, which exhibit different molecular and functional properties. Naïve PSC lines are generated in vitro by culturing early embryos at the cleavage to blastocyst stages (ESC lines), from PGCs (EGC lines), and from reprogramming of post-implantation-derived EpiSCs in different culture media, which gives different characteristics to the derived lines. EPSCs derived from single blastomeres in a cocktail of various differentiation chemical inhibitors (see Fig. 2) have the potential to undergo embryonic and extra-embryonic differentiation. The methylation status of ESCs derived and maintained in different culture conditions differ from each other and are not necessarily the same as their in vivo counterpart. EPI epiblasts, ESCs embryonic stem cells, EGCs embryonic germ cells, EPSCs expanded potential stem cells, EpiSCs post-implantation epiblast-derived stem cells, PGCs primordial germ cells
Since pluripotency exists until the blastocyst stage, pluripotent cells are present for approximately 5 days in the mouse embryo, a quarter of the gestation period [13]. The nature of development is the permanent progress; hence, it is likely that a wide range of pluripotent cells with different GRNs and epigenetic signatures might exist during early development. Evidence for the existence of a pluripotency spectrum comes from the derivation and in vitro propagation of two different types of PSC lines, ESCs (which represent naïve pluripotency) and epiblast stem cells (EpiSCs, which represent primed pluripotency) from pre- and post-implantation embryos, respectively [14–16]. The characteristics of these two types of PSC lines have been extensively reviewed [17, 18] and are summarized in Fig. 1. A key point for establishing these two types of PSC lines is the use of different culture conditions for their derivation and long-term maintenance. The growth factors needed to maintain self-renewal in naïve cells are responsible for triggering differentiation in the primed status and vice versa [18]. When somatic cell reprogramming with ectopic expression of the transcription factors (TFs) Oct4, Sox2, Klf4, and c-Myc is performed under culture conditions that favor primed or naïve pluripotency, so-called induced pluripotent stem (iPS) cells will be generated corresponding to the different culture media used [19, 20].
Despite the importance of culture conditions to establish various PSC lines, it seems that the developmental window for acquiring ESCs is broader than for EpiSCs. ESCs can be generated from two- to eight-cell embryos or their isolated single blastomeres, and from the morula to blastocyst-stage embryos [21]. The developmental memories of different embryonic stages from which ESCs are derived appear to become erased with the exception of embryonic germ cells (EGCs) obtained from PGCs in vitro, which retain their epigenetic memory. ICRs that are erased from EGCs are not correctly re-established during in vitro or in vivo differentiation, because parent-specific genomic imprinting occurs only in the developing gonads [22, 23]. Since stability of correct imprinting is necessary for the functionality of PSC lines (discussed later in the manuscript), it is of crucial importance to use culture conditions that support derivation of ESCs with proper ICR methylation allowing unlimited expansion and use for further lineage differentiation applications and other purposes.
In vitro pluripotency: how to derive ESCs?
Historically, the first ESCs were derived either from 129 strain late blastocysts [24] or from blastocysts obtained by mating SWR/J male mice to ICR females [25]. Culture conditions were developed based on media for in vitro cultivation of teratocarcinoma-derived embryonal carcinoma cells, which included fibroblast cells as the feeder layer and fetal calf serum. Further research revealed that feeder cells and serum can be substituted by leukemia inhibitory factor (Lif) [26] and bone morphogenetic protein 4 (Bmp4) [27], respectively. Bmp4 plus Lif (Bmp4/L) or, for economic reasons, serum and Lif (S/L) were established as the standard culture media for cultivation of mouse ESCs. However, neither the culture of the ICM in undefined S/L nor in the defined Bmp4/L conditions promoted successful/efficient generation of ESCs from mouse strains other than 129 strain [28].
In 1997, Brook and Gardner used delayed implantation or diapause blastocysts for highly efficient generation of ESCs from different mouse strains previously considered to be refractory or non-permissive to ESC generation [29]. Diapause is a phenomenon in different species, including many mammals, which occurs in situations considered harmful for further embryo development. In mice, diapause appears naturally in lactating females and can be experimentally induced by ovariectomy, which prevents blastocysts to attach to the uterus for several weeks [30]. Diapause embryos undergo specification of ICM cells into EPI and PE cells. EPI cells from diapause embryos (dia-EPI) have an active pluripotency network while in a state of biosynthetic and proliferative quiescence, which is most likely due to downregulation of c-Myc [31]. Although Lif/Stat3 signal transduction is attributed to the maintenance of dia-EPI cells, it is not involved in the maintenance of ICM or normal EPI cells of E4.5 pre-implantation blastocysts [32]. However, Stat3 signaling has more recently been found to be active and indispensable in four-cell-stage embryos until the blastocyst stage. In four-cell embryos, it is activated by Lif signals, but in blastocysts, Stat3 phosphorylation is induced via autocrine interleukin-6 signaling, which leads to direct Oct4 and Nanog activation by Stat3, and is necessary for maintenance of ICM lineages (but not for the formation of ICM and TE) in vivo [33]. These findings indicate that ESCs are probably derived from EPI progenitors in the ICM or ICM-derived EPI cells, but not from non-segregated ICM cells. The starting point of ESC generation is loosely considered at E3.5 of mouse embryos, when the ICM has not yet segregated into EPI and PE. This time point is well known because of the convenience of flushing blastocysts from the uterus [34]. Consistent with the idea that ESCs are not directly derived from ICM (and, therefore, not from E3.5 blastocysts), the transcriptome and epigenome profiles of the ICM and ESCs show considerable differences. ICM cells express both PE- and EPI cell-specific TFs, whereas ESCs do not express the PE-related genes Pdgfr, Sox17, Gata6, and Gata4 [35, 36]. DNA methylation of ICM cells and S/L-cultured ESCs clearly differs. Global DNA methylation in ICM cells indicates a hypomethylated state with approximately 30% CpG methylation, whereas ESCs cultured in S/L have approximately 80% CpG methylation [37–39] (Fig. 1). Importantly, the highly efficient generation of ESCs from microdissected EPI cells of E4.5 embryos or from dia-EPI strongly suggests that ICM cells first develop into EPI cells in culture prior to ESC generation [28, 29]. The efficiency of ESCs derived from ICM cells is remarkably enhanced with genetic or chemical inhibition of fibroblast growth factor 4 (Fgf4), the main signaling pathway responsible for PE specification/differentiation [40]. The main effects of Lif and Bmp4 signaling in the maintenance of ESCs are mediated by shielding cells against endogenous pro-differentiation pathways in ESCs, such as the autocrine Fgf4 signaling [28, 41]. The high success rate of ESC generation from blastocysts isolated from 129/Sv mice is likely caused by the inherently small number of PE cells in this strain [28]. Notably, it has been shown to be due to augmentation of Jak–Stat3 signaling pathways along with low activity of the mitogen-activated protein kinase (Mapk) pathway in PE cells of 129/Sv strain of mice [42]. Therefore, the mouse strain or developmental stage of the blastocysts determines the efficiency of ESC derivation under S/L conditions. Blastocysts generating EPI cells as the dominant starting cells in the culture show a higher propensity to form ESC lines.
The concept of ground-state pluripotency, which is achieved through efficient inhibition of endogenous pro-differentiation pathways, has facilitated the establishment of ESCs from blastocysts of various refractory and non-permissive mouse strains as well as from rats [41, 43]. According to the ground-state model, ESCs can be successfully and efficiently derived and propagated when ICM cells are insulated from endogenous differentiation stimuli. Application of two small molecule chemicals (named 2 inhibitors or 2i) inhibiting the endogenous pro-differentiation pathways Fgf–Erk and glycogen synthase kinase3β (Gsk3β) has resulted in highly effective ESC production from the ICM, thereby validating the ground-state hypothesis for stem cell generation and maintenance [41]. Of note, Gsk3β inhibition leads to indirect activation of Wnt signaling, which serves as a pro-self-renewal pathway in ESCs. Since Lif upregulates a number of pluripotency-related TFs and promotes clonogenicity, 2i plus Lif (2i/L) was introduced as the optimal setting for establishing ESC cultures under defined serum- and feeder-free conditions [44, 45].
More recently, several other endogenous differentiation pathways such as calcineurin–NFAT [46], Src kinase [47], protein kinase C (PKC) isoforms [48], Tgf-β [21, 49, 50], and the Jnk and p38 branches of Mapk signaling [51, 52] have been described, which tend to promote the exit from pluripotency. Their modulation by various combinations of chemical inhibitors might become useful for efficient derivation and maintenance of ESCs.
Alternative approaches to ESC derivation and maintenance
Cultivation of ESCs in 2i/L medium provides several important advantages over the traditional S/L medium. 2i/L culture markedly enhances ESC derivation from blastocysts, and overcomes the strain type barrier for ESC production in rodents. The 2i/L medium generates ESCs with a high degree of homogeneity and high clonogenicity [44]. However, 2i/L-grown ESCs suffer from extensive global DNA demethylation, even at ICRs, and genetic instability under long-term inhibition of Fgf–Erk signaling, which compromises both the quality and developmental potential of 2i/L-grown ESCs [53, 54]. Inhibition of other differentiation-stimulating pathways and/or modification of the conventional 2i/L cocktail might generate ESCs with superior features compared to 2i/L medium. It has been reported that replacing the small molecule inhibitor of Fgf–Erk signaling either by a Src kinase inhibitor, CGP77675 (also known as alternative 2i or a2i) [47], or reducing the dosage (from 1 to 0.2 μM) of the Fgf–Erk inhibitor (also known as titrated 2i or t2i) gives rise to ESCs with a more stable genetic and epigenetic status [53, 54]. Although the genetic instability in 2i/L cells is attributed to inhibition of Fgf–Erk signaling, surprisingly, dual inhibition of Tgf-β and Fgf–Erk pathways (also known as R2i culture) leads to the generation and maintenance of ESCs with high chromosomal integrity [50]. This finding suggests that inhibition of Fgf–Erk signaling may destabilize the ESC genomic integrity when the Tgf-β pathway is active in the cells. It has been recently reported that addition of the chemicals SB203580 (p38 Mapk inhibitor), JNK inhibitor VIII, XAV939 (Axin stabilizer), A-419259 (inhibitor of Src family kinases) to 2i/L culture results in derivation of so-called expanded potential stem cells (EPSCs) from single blastomeres. EPSCs have a stable genome and enhanced developmental potential giving rise to both embryonic and extra-embryonic lineages [55] (Fig. 2). Since genome stability and a proper epigenomic status are crucial to the developmental capacity of pluripotent cells, it is necessary to use optimized culture media that promote efficient ESC derivation and maintain a stable genome over serial passaging.
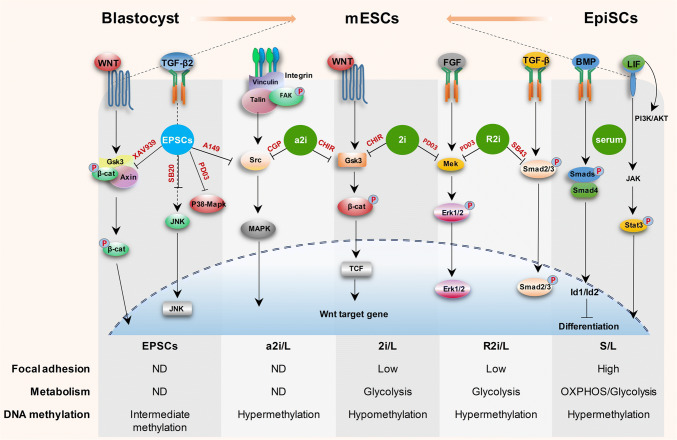
Fig. 2.
Signaling pathways in the naïve mouse pluripotent state. In pre-implantation embryos, ground-state pluripotency is established in the epiblast of the blastocyst and can become ‘primed’ during post-implantation development. These two states of pluripotency not only sustain the ability for self-renewal but also maintain the capacity to differentiate. Naïve pluripotency that can be maintained under serum and LIF (S/L) depends on two extracellular ligands (Lif and Bmp4). Ground-state pluripotency is achieved by the use of two inhibitors, which block Gsk3 and Mapk (2i) plus Lif (2i/L). Alternatively, the ground-state condition is composed of two inhibitors of Tgf-β and Mapk signaling pathways (R2i plus Lif or R2i/L). Inhibition of both Gsk3 and Src kinase (a2i condition) has been found to allow the derivation of ESCs. Block blastomeres’ differentiation by inhibition of p38–Mapk, JNK, Src and the Parp family, and stabilization of Axin create expanded potential stem cells or EPSCs which exhibits embryonic and extra-embryonic lineage differentiation potential. Although the growth factors and pluripotency network genes are important in the determination of a stem cell fate, various metabolic pathways, the degree of cell adhesion to the substrate, and DNA methylation may play a critical role in controlling stem cell fate. Low focal adhesion and the glycolysis pathway in terms of energy metabolism are correlated with pluripotency maintenance of mESCs. PD0325901 inhibitor of Mapk kinase (also known as MEK), SB203580 JNK inhibitor VIII and p38 inhibitor, A149259 Src kinase inhibitor, CGP77675 (CGP) Src inhibitor, XAV939 Parp family members TNKS1/2 inhibitor, AXIN stabilizer and β-catenin and Yap destruction complex, CHIR99021 (CHIR) GSK3 inhibitor, SB431542 (SB43) inhibitor of TGFβ type I receptors (also known as activin receptor-like kinase (ALK)-4, -5, and -7). ND not determined, Lif leukemia inhibitory factor, Bmp4 bone morphogenetic protein 4, mESCs mouse embryonic stem cells, EPSCs expanded potential stem cells
Mechanisms of ESC maintenance
The in vitro derivation and maintenance of ESCs have revealed that modulating different signaling pathways imparts different characteristics to ESCs. In this scenario, while the judge about a signaling pathway alone does not meet our expectation about the actual circumstances present in ESCs, we would address a limited number of molecular pathways, since how different signaling axes cross talk with each other in ESCs has remained poorly understood.
Extrinsic signals that influence ESC behavior
Pluripotency and self-renewal of ESCs are governed by extrinsic signaling pathways, which ripple through intracellular molecular networks, including metabolic processes, TFs’ regulatory circuitry, epigenetic regulators, and regulatory non-coding RNAs (ncRNAs). A number of reports have discussed pluripotency-related signaling pathways [56, 57]. Here, we provide some examples illustrating the impact of individual signaling pathways on others and on the maintenance of pluripotency.
Bmp signaling is among the key signaling axes regulating the undifferentiated state of ESCs. BMPs drive stem cell self-renewal in S/L-based ESC medium. Bmp signaling is highly augmented through the blockade of Tgf-β signaling in R2i/L culture [50]. The Bmp pathway correlates with higher DNA methylation and is less active in 2i/L ESCs [58] where cells exhibit global DNA hypomethylation (Fig. 2). Blockage of Bmp results in massive and rapid R2i/L cell death, whereas 2i/L cells respond less severely to this treatment [50], which highlights a key difference between these two ground-state cultures.
In contrast to pro-self-renewal Bmp4 signaling, Fgf4 enhances differentiation of ESCs via autocrine signaling. Its pharmacological inhibition along with inhibition of Gsk3 (2i/L) or Tgf-β (R2i/L) has been shown to promote self-renewal of ESCs, preventing differentiation [41, 49, 50]. The safety of Fgf4 inhibition has been challenged, since genomic stability is compromised when Fgf4 signaling is blocked for extended periods of time [53], probably because Erk inhibition leads to global DNA demethylation [37], favoring genomic instability and transposition of mobile genetic elements [59]. In light of this finding, the a2i or t2i condition is proposed to be a safer approach to ESC maintenance and generation [53]. We have previously reported the presence of a stable genome in R2i/L ESCs [50], which suggests a protective effect of Tgf-β inhibition on the ESC genome.
Adhesion dynamics and extracellular matrix (ECM) components are important factors that regulate propagation and developmental programming of ESCs. Manipulation of matrix rigidity directs lineage specification in mouse ESCs [60]. Cultured feeder-free, S/L-grown ESCs strongly adhere to gelatinized substrates and grow as a flat layer; however, in ground state supporting media such as 2i/L and R2i/L cultures, ESCs grow as compact colonies on gelatin. Both 2i/L and R2i/L cells express lower levels of integrins compared to S/L cells [61]. Enhanced interaction of ESCs with ECM components, as in S/L medium, negatively affects self-renewal of ESCs via FAK [62] (Fig. 2). Overall, the dynamic activity of diverse signaling routes controls the efficiency by which ESCs are maintained in an undifferentiated state.
Metabolic regulation of ESCs
Metabolic pathways exert important roles in ESCs and during different stages of embryogenesis. In blastocysts, ICM cells are exposed to a hypoxic environment; therefore, they predominantly undergo glycolysis and convert glucose to lactate [63]. Similar to ICM cells, mouse ESCs preferentially use glycolysis, which is necessary for rapid proliferation, although an “on demand” switch from glycolysis to oxidative phosphorylation (OXPHOS) is feasible. A recent study demonstrated that ESCs cultured in ground-state conditions (2i/L and R2i/L) upregulate glycolytic enzymes, thereby proliferating faster than S/L ESCs [64, 65]. Kondoh et al. have reported that inhibition of glycolysis significantly reduces ESC self-renewal [66]. Of note, glycolysis produces less reactive oxygen species (ROS) than OXPHOS, and low levels of ROS support rapid proliferation of PSCs [67]. Metabolic processes are also tightly connected with epigenetic pathways in ESCs. Acetyl-CoA, which is a key metabolic mediator produced via glycolysis, serves as an essential acetyl group donor in histone lysine acetylation reactions [68, 69]. Therefore, metabolic pathways work in concert with other processes to regulate ESC proliferation and developmental potential.
Roles played by transcription factors, non-coding RNAs, and epigenetic factors in the maintenance of ESCs
An integrated regulatory circuitry composed of TFs, epigenetic modifiers, and non-coding RNAs, particularly microRNAs (miRNAs), shapes the behavior of ESCs. Main pluripotency-associated TFs such as Nanog, Oct4, and Sox2 form self-sustaining auto-regulatory as well as feedforward loops which maintain the long-term self-renewal of ESCs [70]. By co-occupying the regulatory regions of thousands of genes in ESCs, these factors regulate the transcription of many genes and serve as main players of a core transcriptional regulatory network which promotes the expression of genes associated with self-renewal and inhibits the transcription of those associated with differentiation lineages [70, 71]. In addition to protein-coding genes, ESC-enriched TFs such as Oct4 and Nanog also bind to the regulatory DNA elements of miRNA genes and promote or inhibit their transcription [72]. The core pluripotency TFs promote the transcription of ESC cycle-regulating (ESCC) miRNAs including miRNAs from miR-290–295 and miR-302–367 clusters which shape the unique cell cycle and other equally important aspects of ESCs [73, 74]. Other sets of miRNAs such as let-7 family members, which are inhibited by pluripotency-associated TFs, tend to destabilize the self-renewal program [75].
In contrast to ground-state ESCs, S/L-grown ESCs express high levels of c-Myc, which not only promotes opening and remodeling of chromatin to drive ESC self-renewal but also inhibits ESC differentiation into primitive endoderm by repressing Gata6 [76]. In S/L ESCs, c-Myc appears to upregulate the miR-290–295 cluster [72, 77]. However, other regulators, such as Gadd45a [78], might activate this miRNA locus in ground-state cells since c-Myc is silenced in ground-state ESCs (similar to diapause embryos) [31, 50, 79]. S/L ESCs also highly express the RNA-binding protein Lin28a which inhibits the maturation of precursor let-7 miRNAs in S/L-grown cells [80]. However, since Lin28a is lowly expressed in ground-state ESCs, some of the let-7 miRNAs are upregulated in ground-state cells. These differentiation-associated miRNAs, which are unexpectedly upregulated in ground-state pluripotency, are reported to promote some features of ground-state pluripotency by targeting c-Myc as well as Lin28a [81, 82]. These findings indicate that ground-state ESCs express a specific repertoire of TFs and other regulatory proteins which are assembled into a specific GRN which is different from that of S/L ESCs, giving rise to different behaviors of these two PSC states.
2i/R2i ESCs not only express different TFs and other regulatory proteins, but also express a distinct set of miRNAs compared to S/L-grown cells. A large miRNA cluster embedded in the 10th intron of the imprinted Sfmbt2 locus is upregulated in S/L-grown ESCs compared to ground-state cells [82, 83]. On the other hand, another large miRNA cluster embedded in the imprinted Dlk1–Dio3 locus is more abundantly expressed in ground-state ESCs than cells grown in S/L [82] (Fig. 3). These data highlight the potential biological importance of genomic imprints in the regulation of different states of naïve pluripotency.
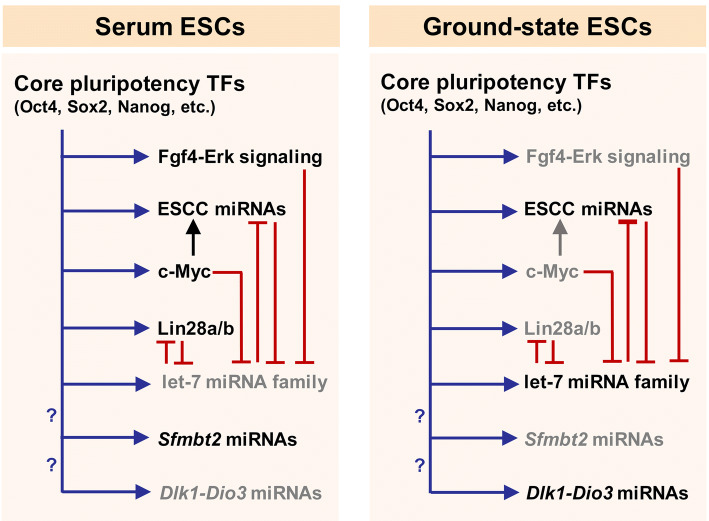
Fig. 3.
miRNA-mediated regulation of GRN in serum and ground-state ESCs. In ESCs, the pluripotency-associated TFs bind to the regulatory regions of their target protein-coding and non-protein-coding genes and promote or repress their expression. In serum ESCs, c-Myc is highly expressed. In cooperation with other TFs, c-Myc promotes the expression of ESCC miRNAs. Lin28 is also highly expressed in serum ESCs and inhibits let-7 maturation. In ground-state ESCs, Lin28 is poorly expressed. This partly explains why let-7 miRNAs are upregulated. Since c-Myc is markedly downregulated in ground-state ESCs, other TFs might promote the high expression level of ESCC miRNAs in these cells. Ground-state ESCs abundantly express the Dlk1-Dio3 locus-embedded miRNAs, whereas serum ESCs highly express a large miRNA cluster located in the 10th intron of the Sfmbt2 gene. It has not been determined which regulator factors control the expression of these two large miRNA clusters. GRN gene regulatory network, miRNA microRNA, TF transcription factor, ESC embryonic stem cell, ESCC miRNAs embryonic stem cell cycle-regulating miRNAs
Contrary to S/L-grown ESCs, 2i/L ESCs display global DNA demethylation [37, 38], probably because de novo DNA methyltransferases (Dnmt3a/3b and Dnmt3l) are downregulated by Prdm14 in 2i/L cells [84–86]. Prdm14 was also reported to inhibit DNA methylation via promotion of active DNA demethylation via ten eleven translocation (Tet) enzymes and stimulation of base-excision repair (BER) [84, 86–88]. However, a recent study shows that neither de novo DNA methyltransferases nor Tet enzymes are involved in global DNA hypomethylation in 2i/L cells; instead, it is shown to be due to the 2i-induced downregulation of Uhrf1, which recruits the maintenance DNA methyltransferase (Dnmt1) to replication foci [89]. Moreover, 2i culture was observed to induce genome-wide loss of H3K9me2 mark, which is required for Uhrf1 recruitment to chromatin in a DNA replication-coupled manner [89]. Prdm14 supports the ground state of pluripotency by diminishing Fgf4–Erk signaling, which is activated by the core pluripotency TFs Oct4, Sox2, and Nanog, and is associated with differentiation leakage observed in S/L-grown ESCs [85] (Fig. 4).
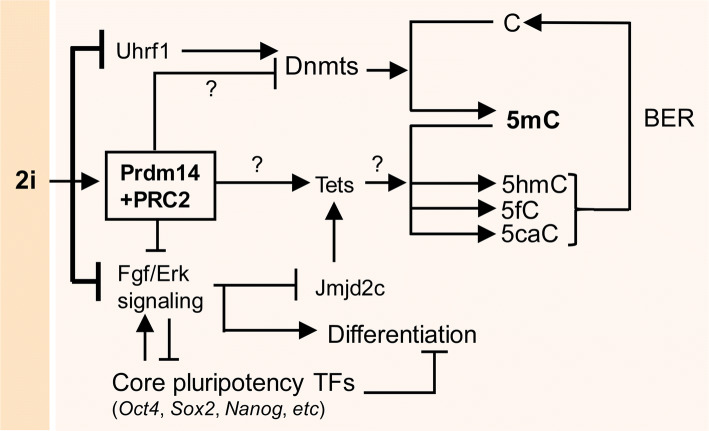
Fig. 4.
How the ESC genome is rapidly and globally demethylated in 2i culture. In 2i ESCs, Prdm14 expression is highly induced and, in conjunction with PRC2, inhibits the expression of Dnmts. The Prdm14–PRC2 complex also activates the expression of Tet enzymes, which catalyze the conversion of 5-methylcytosine (5mC) into 5-hydroxymethyl-cytosine (5hmC), 5-formyl-cytosine (5fC), and 5-carboxyl-cytosine (5caC). Next, the products of Tet activity are converted into an unmodified cytosine through the base-excision repair (BER) pathway. Tet enzymes are reportedly induced by the histone modifier Jmjd2c which is inhibited by Fgf–Erk signaling. In the 2i culture, Fgf–Erk signaling along with the Gsk3 pathway is chemically inhibited, thereby leading to the rapid, genome-wide DNA hypomethylation in 2i ESCs. Of note, it has recently been suggested that Uhrf1 plays a more important role than other factors by recruiting Dnmt1 to replication foci. ESC embryonic stem cell, Tet ten eleven translocation, miRNAs microRNAs, FGF fibroblast growth factor
In 2i/L cells, the genomic imprints are erased and remain unmethylated in somatic cells differentiated from 2i/L cells [54]. Therefore, the 2i/L culture may lead to epigenetic artefacts, which might endanger the safety and quality of 2i/L ESCs. Of note, the R2i/L culture promotes upregulation of Dnmts and preserves a more stable genome compared to the 2i/L culture [37, 87]. Similarly, EPSCs exhibit higher expression of Dnmts and Tet enzymes compared to 2i/L ESCs, a higher level of 5-methylcytosine, and increased numbers of bivalent genomic regions [55]. These results indicate that (1) DNA methylation plays a major role in ESCs exhibiting high or intermediate levels of DNA methylation; (2) global DNA hypomethylation endangers the genomic and epigenomic stability of ESCs (as in 2iL ESCs); and (3) genome-wide DNA demethylation is not a common feature of ground-state pluripotency since alternative culture media which favor ground-state pluripotency exhibit global DNA hypermethylation.
To inhibit differentiation pathways, Prdm14 physically interacts and cooperates with the polycomb group (PcG) proteins to silence target genes, such as Dnmt3a/3b [85]. The simultaneous presence of the repressive H3K27me3 (deposited by PcG proteins) and active H3K4me3 histone modifications (deposited by trithorax group (trxG) proteins) was dubbed “bivalent” and marks genes that are temporarily but nor permanently inactive [90]. ESCs grown in the presence of S/L have a higher number of bivalent chromatin marks than differentiated cells such as mouse embryonic fibroblasts [91]. Genomic regions rich in bivalent chromatin marks or H3K27me3 alone promote long-range promoter–promoter interactions. Such promoter-mediated interactions are absent in 2i/L cells because 2i/L cells contain a significantly smaller number of bivalent marks at their bivalent regions and lower amounts of H3K27me3 marks [92]. In summary, ESCs have an integrated GRN in which various regulatory molecules cooperate with each other to control ESC behavior.
Mechanistic insights into ICM–ESC transition
It is clearly important to decipher the process of how ICM transitions to ESCs with self-renewal capabilities. The relationship of ESC to their in vivo embryonic counterparts would become much clearer, if we understand whether ESCs are captured from a narrow developmental window or undergo different molecular and epigenetic reprogramming events during ICM–ESC transition. This understanding can guide us to choose the optimum culture condition for establishment of ESCs, which in turn has a crucial impact on different characteristics of ESCs. This is not only an academic question but has numerous practical ramifications, since genomic stability and epigenetic configurations of ESC will have lasting effects on ESC-derived cells.
Approaches to study the mechanisms for ICM–ESC transition are based on molecular comparisons between pre-implantation embryonal cells and ESCs or on molecular time-course analysis of ICM outgrowth during establishment of ESCs [3–5, 93]. These studies, although currently incomplete, have revealed highly dynamic changes in the expression and function of key genes and cellular pathways underlying the formation of ESCs from ICM cells of blastocysts. A comprehensive view of temporal changes might clarify the origin of ESCs.
Dynamic gene expression patterns during ESC formation
Single-cell RNA-Seq analyses of ICM cells, ICM outgrowths at days 3 and 5, and established ESCs in S/L culture revealed major changes in the expression of messenger RNA and miRNA transcripts [3]. Many pluripotency-related TFs such as Sox2, Nanog, Esrrb, Cdh1 (E-cadherin), Pecam1, Pim1, Pim3, and Prdm14 are highly expressed in Oct4-expressing cells during the ICM to ESC transition. Other pluripotency-related genes such as Nodal, Eras, Smad1, Zic3, Id1, Id2, Tcf3, and Nr0b1 are upregulated during ICM outgrowth, suggesting potential roles in the immortalization of ESCs. Genes associated with the TE, PE, and different germ layers such as Cdx2, Gata4, Gata6, Hoxd8, Bmp1, Bmp2, Tgfbr2, Tgfbr3, Jak2, Fgf3, Fgf10, Fgfr3, Fgfr4, Sox7, Sox9, and Sox17 are downregulated during generation of ESC in S/L-based medium. The expression profile of a selected set of miRNAs showed highly expressed miRNAs in both ICM and ESCs such as the miR-290–295 cluster; miRNAs that upregulated in ESCs compared to ICM cells, such as miR-302c and miR-367 (miR-302–367 cluster); and miRNAs with reduced expression in ESCs versus ICM that included the let-7 family (associated with differentiation). Interestingly, expression of Lin28, which suppresses maturation of let-7 miRNAs, was positively regulated during ESC formation [3]. These data corresponded to the gradual acquisition of self-renewal ability and repression of differentiation pathways during the course of ESC derivation in S/L-based ESC media.
However, such studies suffer from the use of the S/L condition. The S/L culture exhibits an extremely low efficiency in generating ESCs from most mouse strains as well as from rats. ESCs in S/L culture tend to differentiate spontaneously and show a high degree of heterogeneity regarding expression of important naïve pluripotency TFs such as Nanog [94], Rex1 [95], and Dppa3 (Stella) [96]. These shortcomings might be addressed using culture media that promote highly efficient production of ESCs and acquisition of ground-state pluripotency.
Recently, the process of ESC generation has been carefully analyzed in a time-course transcriptome study using the efficient R2i/L culture system [4]. Previous studies have revealed that the R2i/L culture system more efficiently preserves genomic integrity of PSC lines generated from single blastomeres of pre-implantation and from PGCs of post-implantation embryos compared to the 2i/L condition [21, 50, 97]. Cells were analyzed using high-throughput mRNA profiling at days 0 (ICM), 0.5 (12 h after ICM seeding in R2i/L medium), 1, 2, 3, and 5, as well as established ESC lines at passages 2, 4, and 15. Similar to generation of S/L-based ESCs, it became clear that (1) the number of differentially expressed genes increased considerably in a gradual manner during ground-state ESC derivation; (2) differentiation-associated genes were mostly downregulated; (3) genes associated with self-renewal and immortality were mostly upregulated, although some of the typical pluripotency-associated genes remained unchanged, which was probably due to the pluripotency of both ICM and ESCs; and (4) metabolism and cell adhesion dynamics were strikingly altered. In addition, transcriptional profiles of ICM outgrowths exhibited the most prominent changes at the earliest time points during the course of R2i-based ESC generation. ICM outgrowths at later time points as well as low-passage ESCs showed nearly identical transcriptional profiles compared to E4.5 EPI [4]. These collective findings have indicated that the global gene expression undergoes rapid and substantial changes over the course of ESC formation, suggesting possible functions of genes that exhibit differential expression.
Regulation of epithelial-to-mesenchymal transition (EMT) during ESC generation
Epithelial-to-mesenchymal transition (EMT) and the reverse process, mesenchymal-to-epithelial transition (MET), play critically important roles in tissue remodeling and normal organismal development, as well as in cancer and numerous other diseases [98]. During ESC derivation, this process appears to be tightly regulated and arrest of EMT is necessary for ESC establishment in both 2i/L and R2i/L media. Cellular factors associated with epithelialization, such as E-cadherin and Klf4, as well as some members of the ESCC miRNAs, including miR-302c and miR-367 along with members from the miR-17 family, undergo upregulation during generation of ESCs from ICM [3, 4]. On the other hand, mesenchymal markers such as Snail, Dab2, Eomes, and Tgfbr2, are downregulated during ICM to ESC transition [3, 4], implying that mesenchymal genes might provide a barrier for efficient establishment of ESCs from ICM. In line with these observations, it has been shown that overexpression of typical mesenchymal genes such as Snail or Tgf-β in R2i/L medium (with ~ 100% efficiency of ESC generation) prevents generation of pluripotent ESC lines [4]. Loss of function of Klf4 (as a MET driver) is detrimental to the ICM to ESC transition and significantly reduces the efficiency with which ESCs are obtained [4]. These collective findings highlight the critical significance of EMT inhibition for the successful and efficient generation of ESCs.
Both epithelial-associated factors and miRNAs promote de-differentiation of somatic cells into iPS cells [99–102]. Surprisingly, E-cadherin was reported to replace the master pluripotency TF, Oct4, in somatic cell reprogramming to pluripotency [103], which highlights the importance of epithelialization in cell fate reprogramming to pluripotency. Similar to the requirement of MET for the generation iPS cells and ESCs, epithelialization is also necessary for the successful induction of pluripotency in germ cells [104]. Hence, we reason that epithelialization (MET induction/maintenance or EMT blockade) is a common indispensable requisite for the acquisition of pluripotency from different starting cell types such as the EPI cells of the blastocyst-stage embryos. The finding that epithelialization is crucial for generation of mouse PSC from different sources will have implications for the successful and efficient derivation of ESCs from human blastocysts.
Epigenetic regulations during ESC derivation
Epigenetic regulation contributes substantially to the control of diverse biological processes and developmental pathways. Epigenetic regulators such as histone-modifying enzymes, chromatin-remodeling complexes, and DNA methyltransferases are dynamically regulated during early embryonic development when in vivo pluripotency gradually arises around the time of ICM formation and subsequently disappears when specific embryonic lineages develop around the time of gastrulation [105–110]. In addition, epigenetic regulation modulates key aspects of ESC behavior including survival, cell death pathways, proliferation, and ESC fate decisions [110–112]. Since the process of ICM outgrowth to generate ESCs entails in vitro immortalization of an ephemeral in vivo pluripotent state, epigenetic programs might undergo significant changes and play important functions during this transition. During the course of ESC derivation, epigenetic regulators exhibit highly dynamic patterns of expression [3, 4], which suggests that they may have specific functions in this context. Many of the known epigenetic factors show differential expression during ESC derivation in S/L and/or R2i/L culture conditions; the majority of upregulated epigenetic modifiers (e.g. Dnmts, Suz12, Eed, Mat2b, Mbd2, Mecp2, several Hdac enzymes) are often associated with repressive epigenetic states. In contrast, the majority of downregulated epigenomic factors such as H3K9 demethylases Jmjd2d (also known as Kdm4d) and Jhdm3a (also known as Kdm4a), histone acetyltransferases Ncoa3 and CBP P300, H3K27 demethylase Kdm6b (also known as Jmjd3), and H3K4 methyltransferase Mll3 (also known as Kmt2c) are associated with an active chromatin state [3, 4]. Of note, the DNA demethylases Tet1 and Tet2 are abundantly expressed in both ESCs and ICM cells as well as in Oct4-positive cells in ICM outgrowths [3]. Tet enzymes might reverse methylation at regulatory DNA elements of pluripotency genes introduced by de novo DNA methyltransferases.
Pharmacological inhibition of DNA methyltransferases using RG108 is detrimental to ESC formation in R2i; however, 2i/L cells are not influenced by this treatment [4] probably because they already display highly reduced levels of DNA methylation. It remains to be determined whether ectopically induced hypermethylation by Dnmt overexpression compromises the efficiency of ESC derivation in 2i/L culture. It will be interesting to investigate whether Dnmts and DNA methylation are of similar functional importance for efficient ESC generation in a2i and t2i media, which support proper ICR methylation.
A selected set of analyzed miRNAs is differentially expressed between ICM and ESCs. Pluripotency-associated miRNAs are mostly upregulated, whereas differentiation-associated miRNAs are mostly downregulated during the course of S/L-based ESC derivation [3]. We have recently determined the global expression patterns of miRNAs during the course of ESC generation from ICM in R2i/L culture and observed highly dynamic patterns of miRNA expression (Moradi et al., unpublished data). We identified two major phases of highly differential miRNA expression during this transition. These observations have suggested that miRNAs might play specific roles during ICM–ESC transition. Overall, epigenetic regulators display differential expression patterns and are of crucial importance during the process of ICM outgrowth into immortal ESCs.
The role of sex differences in ESC derivation
ESCs obtained from different sexes show different epigenetic features, as well as other developmental characteristics. Sex differences influence different stages of organismal development. For example, female newborns have a lower mortality rate than males [113]. Importantly, peri-implantation embryogenesis is also affected by sex dimorphism where female embryos develop less rapidly than male embryos [114]. Compared to male ESCs, female ESCs are in a less differentiated state, display lower global DNA methylation levels, accumulate genomic aberrations at imprinted genomic regions over serial passaging, and exit pluripotency less rapidly/efficiently upon differentiation [53, 54, 115]. These features are attributed to the presence of two active X chromosomes (XaXa) in female ESCs. In contrast, XY and XO ESCs are similarly hypermethylated [115, 116]. Sex-specific characteristics of female ESCs have been found to be mainly due to the X chromosome dosage-dependent inhibition of the Fgf–Erk signaling pathway, inhibition of Gsk3β (Wnt signaling activation), and stimulation of Akt signaling, which results in upregulation of key naïve-associated TFs in female ESCs compared to male ESCs [115]. In principle, one or more X-linked genes such as Erk phosphatase Dusp9 [117] along with other X-linked genes and miRNAs [118] downregulate the Fgf–Erk pathway, which leads to repression of Dnmts and global DNA hypomethylation; downregulation of the epigenetic regulator Uhrf1, which is involved in the regulation of maintenance DNA methylation; and delayed differentiation kinetics in female ESCs [53, 54, 115, 116]. Timely X chromosome inactivation is an indispensable prerequisite for the female ESCs to properly exit pluripotency and effective initiation of development of different fetal cell lineages [115]. While female ICM cells retain a XaXa configuration only for a short time period in vivo, female ESCs keep XaXa during the course of prolonged cellular propagation in vitro. Since female and male ICM cells have similarly low global DNA methylation levels, sex-dependent differences in DNA methylation status arise in culture. In male ESCs, the Y chromosome is occasionally lost whereas female ESCs have a high tendency to lose one of their two X chromosomes (or part of it) during cultivation and, therefore, become XO [116], indicating that female ESCs are intrinsically unstable in culture.
Male ESCs cultivated under the hypomethylating culture condition of 2i/L are highly similar to female ESCs cultured in S/L medium in terms of global gene expression, epigenetics, genomic stability, and developmental potential [54]. In 2i/L cultures, the massive erasure of DNA methylation also affects imprinted loci in both male and female ESCs, which leads to biallelic expression of imprinted loci such as Impact [53, 54]. This genome-wide DNA demethylation, which causes aberrant epigenetic changes and defects in genomic imprinting, is more prominent in ‘female’ ESCs cultured in 2i, and renders the cells incompetent for successful contribution to normal full-term embryonic development in somatic cell nuclear transfer and tetraploid complementation experiments, although the cells are able to efficiently contribute to chimeras and two-cell-stage embryos. In contrast to female 2i-grown ESCs, female S/L-grown ESCs as well as female a2i- and t2i-grown ESCs can successfully and efficiently give rise to full-term all-ESC pups and mice. The 2i-grown ESC incompetency has appeared to be due to several chromosomal abnormalities and genomic instabilities, including, but not limited to, trisomy 6 and 8 in injected cells. ESC derivation culture media, which induce global hypermethylation and/or proper ICR imprinting (discussed above), preserve a more stable genome and have a significantly higher success rate in tetraploid embryo complementation experiments [53, 54]. Hence, it seems likely that the erasure of gamete-derived genomic imprints as well as chromosomal instability lead to compromised embryonic and placental development of 2i-derived/grown ESCs. In addition, the histone variant H2AX, which is associated with effective DNA repair and proper embryonic development [119–121], is depleted at developmentally important genes in male 2i-grown ESCs and female S/L-grown ESCs compared to male S/L-grown ESCs. At many genomic loci, aberrant H2AX binding is irreversible [53]. In conclusion, different culture conditions influence ICM–ESC transition in specific ways. The 2i/L culture induces chromosomal and epigenomic aberrations that negatively affect ESC quality, while less potent inhibition of Fgf/Erk in 2i/L culture or dual suppression of Fgf–Erk and Tgf-β signaling (R2i/L culture) gives rise to ESCs with higher genetic and epigenetic integrity.
The origin of ESCs: capturing, reprogramming, or both?
High-throughput profiling and time-resolution analyses have shed light on the relationship of ESCs to early embryonic cells. Understanding the exact embryonic origin of ESCs will help to devise further strategies for obtaining ESCs more efficiently and reproducibly and exploit this knowledge for improving ESC derivation from other sources, such as humans.
A large amount of evidence implies that the starting cells for ESC derivation are EPI cells, which might suggest that EPI cells during in vitro ICM cultivation have been captured to form ESCs. In the S/L condition, however, not all isolated single EPI cells give rise to ESCs, and a maximum of three clones per embryo could be obtained [29, 122]. This low efficiency might be caused by the heterogeneity of EPI cells, variability of undefined culture conditions, or damage to EPI cells during microdissection [122]. On the other hand, expression of key TFs necessary for PGC development in both EPI and ESCs, as well as the high epigenetic similarity which exists between the ESCs and EGCs, suggests the existence of a subpopulation of EPI cells that will give rise to either PGCs or ESC clones in culture. Researchers have tracked the expression of a key gene in PGC development, Blimp1, during transition of the ICM to ESC and determined that ESCs commonly arise from Blimp1-expressing cells in the S/L condition [123]. Sorting of Blimp1-positive cells from ICM outgrowths greatly increased the efficiency of ESC derivation. However, the authors have also observed that Blimp1-negative cells, which constitute a high percentage of EPI cells, can directly give rise to ESCs in 2i/L culture [123]. Later, Blimp1 was shown to be dispensable for long-term maintenance of ESCs under both S/L and 2i/L conditions as well as during EpiSC development and the reprogramming of EpiSCs into ESCs [124] questioning the hypothesis of PGC precursor selection during the ICM to ESC transition.
Expression profiling of 2i-grown ESCs and early mouse embryos from 2-cell through post-implantation embryos (E1.5–E5.5) confirmed that the ability of ICM cells to give rise to ESCs was acquired upon specification of pre-implantation EPI cells [93]. While ICM cells isolated from embryos earlier than E3.75 and older than E5.5 lacked the capacity to become ESCs, all single pre-implantation EPI cells at E4.5 became ESCs under the 2i/L condition. Later, it was shown that 2i-grown ESCs as well as dia-EPI cells possess a nearly identical transcriptional profile as pre-implantation EPI cells. These findings indicate that ESCs are more similar to day 4.5 pre-implantation EPI cells than day 3.5 ICM cells, but did not clarify how early pluripotent embryonal cells acquire long-term self-renewal ability in vitro [93]. Despite the high transcriptional similarity of 2i-grown ESCs with dia-EPI, it needs to be explained how ‘immortal’ ESCs differ from dia-EPI cells, which are in a more silent and dormant condition in terms of cell cycling.
To specify when immortality is acquired during ICM outgrowth into ESCs, a time-resolution global gene expression analysis was conducted during the course of ESC generation in the ground-state R2i/L culture, where ESCs can be derived from various mouse strains with almost 100% efficiency [4, 50]. Interestingly, most of the dynamic transcriptional changes during the ICM–ESC transition occurred as early as 12 h after ICM seeding, while later time points displayed less significant changes compared to the 12-h ICM outgrowth [4].
Functional annotation of up-regulated genes 12 h after ICM culture identified an enrichment of terms related to mitotic cell cycling suggesting that the ability of ESCs for indefinite self-renewal is acquired during the earliest hours of in vitro culture of the ICM. However, the high degree of transcriptional similarity between ESCs at different passages and late ICM outgrowths (days 3 and 5) indicates that ESC identity is gained gradually in vitro, arguing for a reprogramming phenomenon. Yet, the highest degree of similarity of different passage ESCs and the majority of time points of ICM outgrowth is to day 4.5 EPI cells [4] suggesting that ESCs might be obtained through a capturing process. Considering the significant differences of ESCs and EPI cells, particularly in terms of unlimited self-renewal, the enormous transcriptional and epigenetic changes over the course of ESC formation from EPI cells, and phenomena such as EMT blockage, we propose that a combination of both phenomena, i.e, capturing and reprogramming (also called creation), is involved in the generation ESCs from their in vivo counterparts. Figure 5 illustrates the most prominent events occurring during ESC formation from ICM-derived EPI cells.
Fig. 5.
Most notable mechanistic events occurring during ICM-ESC transition. ESCs are generated from the ICM-derived EPI cells through a process which appears to be a combination of reprogramming and capturing. Although ESCs exhibit a transcriptome highly similar to EPI cells, they are highly proliferative as opposed to ICM/EPI cells which are almost in a silent condition in terms of cell cycling. Genes are highly differentially expressed over the course of ESC formation. Some transcripts are either upregulated (green lines in ‘mRNA Expression’ part) or downregulated (red lines in ‘mRNA Expression’ part), whereas others remain unchanged during ICM outgrowth (black line in ‘mRNA Expression’ part). Upregulated transcripts such as, Sox2, Nr0b1, and Id1 appear to be necessary for ESC formation, while downregulated ones such as Cdx2, Tgfbr2, and Gata6 appear to have adverse effects on ICM transition to ESCs. Protein-coding (e.g. Nanog and Eras) and non-coding genes (e.g. miR-302 miRNAs) associated with self-renewal are highly induced, while those associated with differentiation (e.g. Sox17, Hoxd8, and some of let-7 miRNAs) exhibit downregulation. Importantly, epithelial-associated mRNAs and miRNAs show increased expression while those affiliated with mesenchymal phenotype are downregulated during ICM outgrowth into ESCs. Finally, in contrast to ICM cells which exhibit a global DNA hypomethylation, ESCs grown in culture media other than 2i/L mostly show genome-wide hypermethylation of DNA
ESC derivation from domesticated animals
The extensive information about generation and long-term maintenance of mouse ESCs may pave the way for generation of ESC from farmed animals, which are usually refractory to this process. Generation of ESCs from domestic livestock and companion animal species such as cattle, pigs, dogs, chickens, and fish has significant economical values and is of great importance for the production of genetically modified animals as well as biomedical models. Attempts to derive ESCs from domesticated animals are usually based on the outgrowth of ICM or early epiblast, in culture conditions that include a feeder layer and base media supplemented with serum, bFGF, and LIF (reviewed in Ref. [125]). However, the creation of authentic ESC lines with long-term proliferation ability and in vitro or in vivo multi-lineage differentiation potential remains challenging. The few published examples of successful establishment of ESC lines from non-rodent and non-primate species could not be reproduced [126]. Our own attempts to develop stable ESC lines from chick embryos did not meet the acceptable efficiency based on published protocols. Using the R2i/L culture, however, we generated ES-like cells up to four passages from the blastodisc of embryonated eggs [127]. Evidence from our and other studies suggests that ESCs obtained from non-rodent species exhibit a primed morphology similar to mouse EpiSCs. Recently, a study reported generation of stable bovine ESCs [128] using culture conditions, which typically support derivation of a type of mouse EpiSCs similar to the gastrulation epiblast, named “region-selective PSCs” or rsPSCs [129]. The rsPSC culture condition consists of a serum-free base medium supplemented with bFGF and an inhibitor of the canonical Wnt/β-catenin signaling, IWR1. Since most reports dealing with the generation of ESCs from non-rodent and non-primate species could not be reproduced, it is of great importance to optimize culture conditions, allowing reproducible derivation of ESCs. The current state of research suggests that further optimization of culture condition based on the current improvements of primed PSCs’ cultures might enable reproducible derivation and long-term maintenance of valuable domesticated animals.
Derivation of ESCs from humans
Unlimited self-renewing capability and multi-lineage differentiation potential make human ESCs a promising tool for disease modeling and regenerative medicine. Notwithstanding the inefficiency of ESC derivation from non-rodent animals, ESCs can be derived with tolerable efficiency from early human embryos through cleavage stages to late blastocysts [130]. However, because of critical issues such as sensitivity to single cell dissociation, there has been a 17-year delay after establishment of mouse ESC lines before human ESCs were obtained [131]. However, the identification of crucial extrinsic signal molecules including bFGF or ACTIVIN, the use of ROCK inhibitor chemicals as suppressors of single cell-induced apoptosis, and the improvement of defined serum-free media have paved the way for large-scale cultivation of human ESCs [2].
Despite these advances, a limited number of studies have explored the molecular signature during transition of human ICM to ESCs. Human ESCs are morphologically and molecularly similar to mouse EpiSCs [18]. Recently, optimized culture media have been reported to generate human naïve-like ESCs directly from ICM (see below), which might provide the possibility to more accurately investigate the underlying mechanisms of ESC derivation from human blastocysts. Human ESCs originate from a transient epiblast-like structure, designated as post-ICM intermediate (PICMI) [132]. PICMI undergoes X-inactivation in female cells and the transcriptional profile of these structures is close to the human epiblast disc with high expression of NODAL/ACTIVIN signaling, no canonical Wnt/β-catenin signaling, high expression of NANOG, and a mix of early epiblast and late epiblast marker gene expressions [132].
The fact that human ESCs show features of primed ESCs, however, has created some challenges for using human ESCs as an appropriate source for regenerative medicine. In recent years, much effort has been made to overcome primed-associated characteristics using two strategies: (1) by overexpression of naïve-specific genes during conversion of human primed to naïve ESCs and (2) activation of naïve-related signaling pathways by modification of culture conditions during direct derivation of naïve human ESCs from ICM cells (reviewed in [133]) (Fig. 6). However, the molecular and functional properties of naïve human ESCs generated by such protocols differ in terms of differentiation potential, resemblance to pre-implantation human embryos, and the maintenance of genomic integrity [134]. Therefore, the generation of authentic pluripotent human ESCs with naïve-specific features needs further investigation. Mechanistic insights gained from mouse ESC studies should provide a proper platform to produce human ESCs, which exhibit bona fide characteristics of naivety.
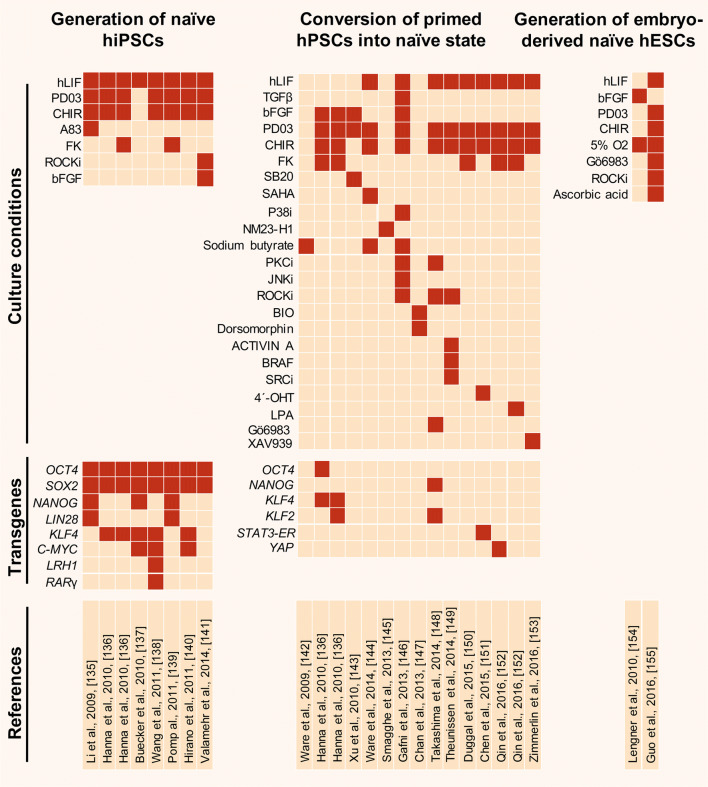
Fig. 6.
Studies to generate naïve human pluripotent stem cells [135–155]. Naïve pluripotency can be established in human cells by direct reprograming of human somatic cells, conversion of conventional primed hESCs/hiPSCs into the naïve pluripotent state, and by direct naïve ESC derivation from pre-implantation embryos via overexpression of pluripotency-related transcription factors or modification of the culture conditions. Current naïve human PSCs require additional efforts in respect of differentiation potential and genome integrity. hiPSCs human induced pluripotent stem cells, hESCs human embryonic stem cells, hPSCs human pluripotent stem cells, hLIF human LIF, PD PD0325901, Mek inhibitor, CHIR CHIR99021, GSK3β inhibitor, A83 A-83-01, ALK4/5/7 inhibitor, FK forskolin, protein kinase A agonist, SB20 SB203580, p38 inhibitor, SAHA suberoylanilide hydroxamic acid or vorinostat, pan-histone deacetylase (HDAC) inhibitor, LPA lysophosphatidic acid, YAP agonist, LRH1 liver receptor homologue 1 or Nr5a2, RARG retinoic acid receptor gamma, XAV939 tankyrase inhibitor
Concluding remarks
The different strategies used to derive and propagate ESCs in culture considerably influence the genomic and epigenetic stabilities of the cells. Culture media promoting DNA hypermethylation and/or proper ICR methylation allow generation of ESCs with high quality and full developmental potential. Although the 2i/L culture induces global hypomethylation similar to ICM methylome in mouse ESCs, the resulting chromosomal instabilities due to extensive DNA demethylation particularly at ICRs pose a potential threat. Global demethylation also negatively affects the developmental potential of ESCs propagated in 2i/L. Since these aberrations are attributed to the potent inhibition of Fgf–Erk signaling, substitution of Fgf–Erk inhibitor by a Src inhibitor or reducing the dosage of the Fgf–Erk inhibitor might prevent defects associated with global DNA demethylation. We have shown that dual inhibition of Fgf–Erk and Tgf-β signaling reverses adverse effects of the inhibition of Fgf–Erk signaling on the ESCs’ (epi)genome and increases DNA methylation, highlighting the protective effect of Tgf-β blockage on the ESC genome. Proper DNA methylation, particularly at ICRs, is of crucial importance for the acquisition of high-quality ESCs with full developmental potential. These insights might also be important for the induction of naivety in human PSCs, where most studies report a global DNA hypomethylation. Because ESCs are transcriptionally highly similar to day 4.5 EPI cells, day 4.5 EPI cells might be the in vivo origin of ESCs, which indicates the existence of a capturing process during ESC derivation. However, key differences between EPI and ESCs, especially the immortality and epithelialization of ESCs compared to their in vivo counterpart as well as dynamic changes in the expression of TFs, epigenetic regulators, and miRNAs suggest reprogramming events. We propose that a combination of both phenomena contributes to the generation of ESCs. Further investigations are needed to clarify how ESCs are related to the embryonic cells from which they are obtained.
Acknowledgements
This work was supported by a grant from Royan Institute, the Iranian Council of Stem Cell Research and Technology, the Iran National Science Foundation (INSF), and Iran Science Elites Federation to H.B.
Footnotes
Seyedeh-Nafiseh Hassani and Sharif Moradi contributed equally to this work.
References
- 1.Pouton CW, Haynes JM. Embryonic stem cells as a source of models for drug discovery. Nat Rev Drug Discov. 2007;6:605–616. doi: 10.1038/nrd2194. [DOI] [PubMed] [Google Scholar]
- 2.Abbasalizadeh S, Baharvand H. Technological progress and challenges towards cGMP manufacturing of human pluripotent stem cells based therapeutic products for allogeneic and autologous cell therapies. Biotechnol Adv. 2013;31:1600–1623. doi: 10.1016/j.biotechadv.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Totonchi M, Hassani SN, Sharifi-Zarchi A, Tapia N, Adachi K, Arand J, Greber B, Sabour D, Arauzo-Bravo MJ, Walter J, Pakzad M, Gourabi H, Scholer HR, Baharvand H. Blockage of the epithelial-to-mesenchymal transition is required for embryonic stem cell derivation. Stem Cell Rep. 2017;9:1275–1290. doi: 10.1016/j.stemcr.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boroviak T, Loos R, Bertone P, Smith A, Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- 7.Posfai E, Tam OH, Rossant J. Mechanisms of pluripotency in vivo and in vitro. Curr Top Dev Biol. 2014;107:1–37. doi: 10.1016/B978-0-12-416022-4.00001-9. [DOI] [PubMed] [Google Scholar]
- 8.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 10.Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, Logan GJ, Teber ET, Tam OH, Stutz MD, Alexander IE, Pickett HA, Tam PP. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yagi M, Yamanaka S, Yamada Y. Epigenetic foundations of pluripotent stem cells that recapitulate in vivo pluripotency. Lab Investig. 2017;97:1133–1141. doi: 10.1038/labinvest.2017.87. [DOI] [PubMed] [Google Scholar]
- 13.Morgani S, Nichols J, Hadjantonakis AK. The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev Biol. 2017;17:7. doi: 10.1186/s12861-017-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, de Sousa Chuva, Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 15.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 16.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 18.Hassani SN, Totonchi M, Gourabi H, Scholer HR, Baharvand H. Signaling roadmap modulating naive and primed pluripotency. Stem Cells Dev. 2014;23:193–208. doi: 10.1089/scd.2013.0368. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Hassani SN, Pakzad M, Asgari B, Taei A, Baharvand H. Suppression of transforming growth factor beta signaling promotes ground state pluripotency from single blastomeres. Hum Reprod. 2014;29:1739–1748. doi: 10.1093/humrep/deu134. [DOI] [PubMed] [Google Scholar]
- 22.Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- 23.Shovlin TC, Durcova-Hills G, Surani A, McLaren A. Heterogeneity in imprinted methylation patterns of pluripotent embryonic germ cells derived from pre-migratory mouse germ cells. Dev Biol. 2008;313:674–681. doi: 10.1016/j.ydbio.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 25.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 27.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 28.Batlle-Morera L, Smith A, Nichols J. Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis. 2008;46:758–767. doi: 10.1002/dvg.20442. [DOI] [PubMed] [Google Scholar]
- 29.Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci U S A. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ptak GE, Tacconi E, Czernik M, Toschi P, Modlinski JA, Loi P. Embryonic diapause is conserved across mammals. PLoS One. 2012;7:e33027. doi: 10.1371/journal.pone.0033027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgartner D, Carnevalli LS, Atzberger A, Haas S, von Paleske L, Boroviak T, Worsdorfer P, Essers MA, Kloz U, Eisenman RN, Edenhofer F, Bertone P, Huber W, van der Hoeven F, Smith A, Trumpp A. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell. 2016;164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- 33.Do DV, Ueda J, Messerschmidt DM, Lorthongpanich C, Zhou Y, Feng B, Guo G, Lin PJ, Hossain MZ, Zhang W, Moh A, Wu Q, Robson P, Ng HH, Poellinger L, Knowles BB, Solter D, Fu XY. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- 35.Santos J, Pereira CF, Di-Gregorio A, Spruce T, Alder O, Rodriguez T, Azuara V, Merkenschlager M, Fisher AG. Differences in the epigenetic and reprogramming properties of pluripotent and extra-embryonic stem cells implicate chromatin remodelling as an important early event in the developing mouse embryo. Epigenetics Chromatin. 2010;3:1. doi: 10.1186/1756-8935-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermitte S, Chazaud C. Primitive endoderm differentiation: from specification to epithelium formation. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130537. doi: 10.1098/rstb.2013.0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, Cook SJ, Andrews S, Branco MR, Reik W. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, Benedetti R, Altucci L, Jansen JH, Walter J, Gut IG, Marks H, Stunnenberg HG. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buehr M, Smith A. Genesis of embryonic stem cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:1397–1402. doi: 10.1098/rstb.2003.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtsuka S, Niwa H. The differential activation of intracellular signaling pathways confers the permissiveness of embryonic stem cell derivation from different mouse strains. Development. 2015;142:431–437. doi: 10.1242/dev.112375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 45.Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Zhu L, Yang A, Lin J, Tang F, Jin S, Wei Z, Li J, Jin Y. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell. 2011;8:46–58. doi: 10.1016/j.stem.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu T, Ueda J, Ho JC, Iwasaki K, Poellinger L, Harada I, Sawada Y. Dual inhibition of Src and GSK3 maintains mouse embryonic stem cells, whose differentiation is mechanically regulated by Src signaling. Stem Cells. 2012;30:1394–1404. doi: 10.1002/stem.1119. [DOI] [PubMed] [Google Scholar]
- 48.Dutta D, Ray S, Home P, Larson M, Wolfe MW, Paul S. Self-renewal versus lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance. Stem Cells. 2011;29:618–628. doi: 10.1002/stem.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hassani SN, Totonchi M, Farrokhi A, Taei A, Larijani MR, Gourabi H, Baharvand H. Simultaneous suppression of TGF-beta and ERK signaling contributes to the highly efficient and reproducible generation of mouse embryonic stem cells from previously considered refractory and non-permissive strains. Stem Cell Rev. 2012;8:472–481. doi: 10.1007/s12015-011-9306-y. [DOI] [PubMed] [Google Scholar]
- 50.Hassani SN, Totonchi M, Sharifi-Zarchi A, Mollamohammadi S, Pakzad M, Moradi S, Samadian A, Masoudi N, Mirshahvaladi S, Farrokhi A, Greber B, Arauzo-Bravo MJ, Sabour D, Sadeghi M, Salekdeh GH, Gourabi H, Scholer HR, Baharvand H. Inhibition of TGFbeta signaling promotes ground state pluripotency. Stem Cell Rev. 2014;10:16–30. doi: 10.1007/s12015-013-9473-0. [DOI] [PubMed] [Google Scholar]
- 51.Xu P, Davis RJ. c-Jun NH2-terminal kinase is required for lineage-specific differentiation but not stem cell self-renewal. Mol Cell Biol. 2010;30:1329–1340. doi: 10.1128/MCB.00795-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan BS, Kwek J, Wong CK, Saner NJ, Yap C, Felquer F, Morris MB, Gardner DK, Rathjen PD, Rathjen J. Src family kinases and p38 mitogen-activated protein kinases regulate pluripotent cell differentiation in culture. PLoS One. 2016;11:e0163244. doi: 10.1371/journal.pone.0163244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J, Huebner AJ, Clement K, Walsh RM, Savol A, Lin K, Gu H, Di Stefano B, Brumbaugh J, Kim SY, Sharif J, Rose CM, Mohammad A, Odajima J, Charron J, Shioda T, Gnirke A, Gygi S, Koseki H, Sadreyev RI, Xiao A, Meissner A, Hochedlinger K. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yagi M, Kishigami S, Tanaka A, Semi K, Mizutani E, Wakayama S, Wakayama T, Yamamoto T, Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Ryan DJ, Wang W, Tsang JC, Lan G, Masaki H, Gao X, Antunes L, Yu Y, Zhu Z, Wang J, Kolodziejczyk AA, Campos LS, Wang C, Yang F, Zhong Z, Fu B, Eckersley-Maslin MA, Woods M, Tanaka Y, Chen X, Wilkinson AC, Bussell J, White J, Ramirez-Solis R, Reik W, Gottgens B, Teichmann SA, Tam PPL, Nakauchi H, Zou X, Lu L, Liu P. Establishment of mouse expanded potential stem cells. Nature. 2017;550:393–397. doi: 10.1038/nature24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menchero S, Rayon T, Andreu MJ, Manzanares M. Signaling pathways in mammalian preimplantation development: linking cellular phenotypes to lineage decisions. Dev Dyn. 2017;246:245–261. doi: 10.1002/dvdy.24471. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H, Jin Y. Signaling networks in the control of pluripotency. Curr Opin Genet Dev. 2017;46:141–148. doi: 10.1016/j.gde.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Gomes Fernandes M, Dries R, Roost MS, Semrau S, de Melo Bernardo A, Davis RP, Ramakrishnan R, Szuhai K, Maas E, Umans L, Abon Escalona V, Salvatori D, Deforce D, Van Criekinge W, Huylebroeck D, Mummery C, Zwijsen A, de Sousa Chuva, Lopes SM. BMP-SMAD signaling regulates lineage priming, but is dispensable for self-renewal in mouse embryonic stem cells. Stem Cell Rep. 2016;6:85–94. doi: 10.1016/j.stemcr.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter M, Teissandier A, Perez-Palacios R, Bourc’his D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife. 2016;5:e11418. doi: 10.7554/eLife.11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taleahmad S, Mirzaei M, Samadian A, Hassani SN, Haynes PA, Salekdeh GH, Baharvand H. Low focal adhesion signaling promotes ground state pluripotency of mouse embryonic stem cells. J Proteome Res. 2017;16:3585–3595. doi: 10.1021/acs.jproteome.7b00322. [DOI] [PubMed] [Google Scholar]
- 62.Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, Fukuhara Y, Abe T, Sato JD, Hata R, Asashima M. Integrins regulate mouse embryonic stem cell self-renewal. Stem Cells. 2007;25:3005–3015. doi: 10.1634/stemcells.2007-0103. [DOI] [PubMed] [Google Scholar]
- 63.Hewitson LC, Leese HJ. Energy metabolism of the trophectoderm and inner cell mass of the mouse blastocyst. J Exp Zool. 1993;267:337–343. doi: 10.1002/jez.1402670310. [DOI] [PubMed] [Google Scholar]
- 64.Taleahmad S, Hassani SN, Hosseini Salekdeh G, Baharvand H. Metabolic signature of pluripotent stem cells. Cell J. 2018;20:388–395. doi: 10.22074/cellj.2018.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taleahmad S, Mirzaei M, Parker LM, Hassani SN, Mollamohammadi S, Sharifi-Zarchi A, Haynes PA, Baharvand H, Salekdeh GH. Proteome analysis of ground state pluripotency. Sci Rep. 2015;5:17985. doi: 10.1038/srep17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 67.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu W, Wang F, Yu Z, Xin F. Epigenetics and cellular metabolism. Genet Epigenetics. 2016;8:43–51. doi: 10.4137/GEG.S32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Melton C, Li YP, Shenoy A, Zhang XX, Subramanyam D, Blelloch R. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep. 2013;4:99–109. doi: 10.1016/j.celrep.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moradi S, Braun T, Baharvand H. miR-302b-3p promotes self-renewal properties in leukemia inhibitory factor-withdrawn embryonic stem cells. Cell J. 2018;20:61–72. doi: 10.22074/cellj.2018.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L, Chen K, Wu Y, Long Q, Zhao D, Ma B, Pei D, Liu X. Gadd45a opens up the promoter regions of miR-295 facilitating pluripotency induction. Cell Death Dis. 2017;8:e3107. doi: 10.1038/cddis.2017.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser A, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ, Collins JJ. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moradi S, Sharifi-Zarchi A, Ahmadi A, Mollamohammadi S, Stubenvoll A, Gunther S, Salekdeh GH, Asgari S, Braun T, Baharvand H. Small RNA sequencing reveals Dlk1-Dio3 locus-embedded microRNAs as major drivers of ground-state pluripotency. Stem Cell Rep. 2017;9:2081–2096. doi: 10.1016/j.stemcr.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan Y, Yang X, Li TT, Gu KL, Hao J, Zhang Q, Wang Y. Significant differences of function and expression of microRNAs between ground state and serum-cultured pluripotent stem cells. J Genet Genom. 2017;44:179–189. doi: 10.1016/j.jgg.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 84.Hackett JA, Dietmann S, Murakami K, Down TA, Leitch HG, Surani MA. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Rep. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 86.Sim YJ, Kim MS, Nayfeh A, Yun YJ, Kim SJ, Park KT, Kim CH, Kim KS. 2i maintains a naive ground state in ESCs through two distinct epigenetic mechanisms. Stem Cell Rep. 2017;8:1312–1328. doi: 10.1016/j.stemcr.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, Hashimoto S, Nakamura T, Sugasawa K, Kojima N, Takada T, Okano M, Seki Y. PRDM14 promotes active DNA demethylation through the ten–eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development. 2014;141:269–280. doi: 10.1242/dev.099622. [DOI] [PubMed] [Google Scholar]
- 88.Okashita N, Sakashita N, Ito K, Mitsuya A, Suwa Y, Seki Y. PRDM14 maintains pluripotency of embryonic stem cells through TET-mediated active DNA demethylation. Biochem Biophys Res Commun. 2015;466:138–145. doi: 10.1016/j.bbrc.2015.08.122. [DOI] [PubMed] [Google Scholar]
- 89.von Meyenn F, Iurlaro M, Habibi E, Liu NQ, Salehzadeh-Yazdi A, Santos F, Petrini E, Milagre I, Yu M, Xie Z, Kroeze LI, Nesterova TB, Jansen JH, Xie H, He C, Reik W, Stunnenberg HG. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol Cell. 2016;62:848–861. doi: 10.1016/j.molcel.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 91.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Joshi O, Wang SY, Kuznetsova T, Atlasi Y, Peng T, Fabre PJ, Habibi E, Shaik J, Saeed S, Handoko L, Richmond T, Spivakov M, Burgess D, Stunnenberg HG. Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell. 2015;17:748–757. doi: 10.1016/j.stem.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 95.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 96.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Attari F, Sepehri H, Ansari H, Hassani SN, Esfandiari F, Asgari B, Shahverdi A, Baharvand H. Efficient induction of pluripotency in primordial germ cells by dual inhibition of TGF-beta and ERK signaling pathways. Stem Cells Dev. 2014;23:1050–1061. doi: 10.1089/scd.2013.0438. [DOI] [PubMed] [Google Scholar]
- 98.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Investig. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 100.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 101.Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, Gao L, Ye D, Zhou Y, Chen J, Wang J, Wu M, Liu H, Kang J. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci USA. 2013;110:2858–2863. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moradi S, Asgari S, Baharvand H. Concise review: harmonies played by microRNAs in cell fate reprogramming. Stem Cells. 2014;32:3–15. doi: 10.1002/stem.1576. [DOI] [PubMed] [Google Scholar]
- 103.Redmer T, Diecke S, Grigoryan T, Quiroga-Negreira A, Birchmeier W, Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.An J, Zheng Y, Dann CT. Mesenchymal to epithelial transition mediated by CDH1 promotes spontaneous reprogramming of male germline stem cells to pluripotency. Stem Cell Rep. 2017;8:446–459. doi: 10.1016/j.stemcr.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol. 2009;7:59. doi: 10.1186/1477-7827-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 108.Marcho C, Bevilacqua A, Tremblay KD, Mager J. Tissue-specific regulation of Igf2r/Airn imprinting during gastrulation. Epigenetics Chromatin. 2015;8:10. doi: 10.1186/s13072-015-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Canovas S, Ross PJ. Epigenetics in preimplantation mammalian development. Theriogenology. 2016;86:69–79. doi: 10.1016/j.theriogenology.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mohammed H, Hernando-Herraez I, Savino A, Scialdone A, Macaulay I, Mulas C, Chandra T, Voet T, Dean W, Nichols J, Marioni JC, Reik W. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 2017;20:1215–1228. doi: 10.1016/j.celrep.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 112.Morey L, Santanach A, Blanco E, Aloia L, Nora EP, Bruneau BG, Di Croce L. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell. 2015;17:300–315. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 113.Migeon BR. Why females are mosaics, X-chromosome inactivation, and sex differences in disease. Gend Med. 2007;4:97–105. doi: 10.1016/S1550-8579(07)80024-6. [DOI] [PubMed] [Google Scholar]
- 114.Seller MJ. Neural tube defects and sex ratios. Am J Med Genet. 1987;26:699–707. doi: 10.1002/ajmg.1320260325. [DOI] [PubMed] [Google Scholar]
- 115.Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Bluthgen N, Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 116.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 117.Choi J, Clement K, Huebner AJ, Webster J, Rose CM, Brumbaugh J, Walsh RM, Lee S, Savol A, Etchegaray JP, Gu H, Boyle P, Elling U, Mostoslavsky R, Sadreyev R, Park PJ, Gygi SP, Meissner A, Hochedlinger K. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell. 2017;20(706–719):e7. doi: 10.1016/j.stem.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/S0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu T, Liu Y, Wen D, Tseng Z, Tahmasian M, Zhong M, Rafii S, Stadtfeld M, Hochedlinger K, Xiao A. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell. 2014;15:281–294. doi: 10.1016/j.stem.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 122.Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 123.Chu LF, Surani MA, Jaenisch R, Zwaka TP. Blimp1 expression predicts embryonic stem cell development in vitro. Curr Biol. 2011;21:1759–1765. doi: 10.1016/j.cub.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bao S, Leitch HG, Gillich A, Nichols J, Tang F, Kim S, Lee C, Zwaka T, Li X, Surani MA. The germ cell determinant blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell. 2012;11:110–117. doi: 10.1016/j.stem.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ezashi T, Yuan Y, Roberts RM. Pluripotent stem cells from domesticated mammals. Annu Rev Anim Biosci. 2016;4:223–253. doi: 10.1146/annurev-animal-021815-111202. [DOI] [PubMed] [Google Scholar]
- 126.Chen X, Ye S, Ying QL. Stem cell maintenance by manipulating signaling pathways: past, current and future. BMB Rep. 2015;48:668–676. doi: 10.5483/BMBRep.2015.48.12.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Farzaneh M, Zare M, Hassani SN, Baharvand H. Effects of various culture conditions on pluripotent stem cell derivation from chick embryos. J Cell Biochem. 2018;119:6325–6336. doi: 10.1002/jcb.26761. [DOI] [PubMed] [Google Scholar]
- 128.Bogliotti YS, Wu J, Vilarino M, Okamura D, Soto DA, Zhong C, Sakurai M, Sampaio RV, Suzuki K, Izpisua Belmonte JC, Ross PJ. Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc Natl Acad Sci USA. 2018;115:2090–2095. doi: 10.1073/pnas.1716161115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu J, Okamura D, Li M, Suzuki K, Luo C, Ma L, He Y, Li Z, Benner C, Tamura I, Krause MN, Nery JR, Du T, Zhang Z, Hishida T, Takahashi Y, Aizawa E, Kim NY, Lajara J, Guillen P, Campistol JM, Esteban CR, Ross PJ, Saghatelian A, Ren B, Ecker JR, Izpisua Belmonte JC. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taei A, Hassani SN, Eftekhari-Yazdi P, Rezazadeh Valojerdi M, Nokhbatolfoghahai M, Masoudi NS, Pakzad M, Gourabi H, Baharvand H. Enhanced generation of human embryonic stem cells from single blastomeres of fair and poor-quality cleavage embryos via inhibition of glycogen synthase kinase beta and Rho-associated kinase signaling. Hum Reprod. 2013;28:2661–2671. doi: 10.1093/humrep/det309. [DOI] [PubMed] [Google Scholar]
- 131.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 132.O’Leary T, Heindryckx B, Lierman S, van Bruggen D, Goeman JJ, Vandewoestyne M, Deforce D, de Sousa Lopes SM, De Sutter P. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat Biotechnol. 2012;30:278–282. doi: 10.1038/nbt.2135. [DOI] [PubMed] [Google Scholar]
- 133.Ware CB. Concise review: lessons from naive human pluripotent cells. Stem Cells. 2017;35:35–41. doi: 10.1002/stem.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu X, Nefzger CM, Rossello FJ, Chen J, Knaupp AS, Firas J, Ford E, Pflueger J, Paynter JM, Chy HS, O’Brien CM, Huang C, Mishra K, Hodgson-Garms M, Jansz N, Williams SM, Blewitt ME, Nilsson SK, Schittenhelm RB, Laslett AL, Lister R, Polo JM. Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat Methods. 2017;14:1055–1062. doi: 10.1038/nmeth.4436. [DOI] [PubMed] [Google Scholar]
- 135.Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 136.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K, Geijsen N. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, Bradley A, Liu P. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 140.Hirano K, Nagata S, Yamaguchi S, Nakagawa M, Okita K, Kotera H, Ainscough J, Tada T. Human and mouse induced pluripotent stem cells are differentially reprogrammed in response to kinase inhibitors. Stem Cells Dev. 2012;21:1287–1298. doi: 10.1089/scd.2011.0283. [DOI] [PubMed] [Google Scholar]
- 141.Valamehr B, Robinson M, Abujarour R, Rezner B, Vranceanu F, Le T, Medcalf A, Lee TT, Fitch M, Robbins D, Flynn P. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Rep. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, Lim R, Tewari M, Gartler SM, Issa JP, Pavlidis P, Duan Z, Blau CA. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, Tesar PJ, Okada J, Margaretha L, Sperber H, Choi M, Blau CA, Treuting PM, Hawkins RD, Cirulli V, Ruohola-Baker H. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Smagghe BJ, Stewart AK, Carter MG, Shelton LM, Bernier KJ, Hartman EJ, Calhoun AK, Hatziioannou VM, Lillacci G, Kirk BA, DiNardo BA, Kosik KS, Bamdad C. MUC1* ligand, NM23-H1, is a novel growth factor that maintains human stem cells in a more naive state. PLoS One. 2013;8:e58601. doi: 10.1371/journal.pone.0058601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 147.Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 148.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty MM, Young RA, Gray NS, Jaenisch R. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Duggal G, Warrier S, Ghimire S, Broekaert D, Van der Jeught M, Lierman S, Deroo T, Peelman L, Van Soom A, Cornelissen R, Menten B, Mestdagh P, Vandesompele J, Roost M, Slieker RC, Heijmans BT, Deforce D, De Sutter P, De Sousa Lopes SC, Heindryckx B. Alternative routes to induce naive pluripotency in human embryonic stem cells. Stem Cells. 2015;33:2686–2698. doi: 10.1002/stem.2071. [DOI] [PubMed] [Google Scholar]
- 151.Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E, Mure M, Afanassieff M, Cleroux E, Guibert S, Chen J, Vallot C, Acloque H, Genthon C, Donnadieu C De, Vos J, Sanlaville D, Guerin JF, Weber M, Stanton LW, Rougeulle C, Pain B, Bourillot PY, Savatier P. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat Commun. 2015;6:7095. doi: 10.1038/ncomms8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qin H, Hejna M, Liu Y, Percharde M, Wossidlo M, Blouin L, Durruthy-Durruthy J, Wong P, Qi Z, Yu J, Qi LS, Sebastiano V, Song JS, Ramalho-Santos M. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zimmerlin L, Park TS, Huo JS, Verma K, Pather SR, Talbot CC, Jr, Agarwal J, Steppan D, Zhang YW, Considine M, Guo H, Zhong X, Gutierrez C, Cope L, Canto-Soler MV, Friedman AD, Baylin SB, Zambidis ET. Tankyrase inhibition promotes a stable human naive pluripotent state with improved functionality. Development. 2016;143:4368–4380. doi: 10.1242/dev.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 155.Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]