Abstract
RNA interference (RNAi) has been widely adopted to repress specific gene expression and is easily achieved by designing small interfering RNAs (siRNAs) with perfect sequence complementarity to the intended target mRNAs. Although siRNAs direct Argonaute (Ago), a core component of the RNA-induced silencing complex (RISC), to recognize and silence target mRNAs, they also inevitably function as microRNAs (miRNAs) and suppress hundreds of off-targets. Such miRNA-like off-target repression is potentially detrimental, resulting in unwanted toxicity and phenotypes. Despite early recognition of the severity of miRNA-like off-target repression, this effect has often been overlooked because of difficulties in recognizing and avoiding off-targets. However, recent advances in genome-wide methods and knowledge of Ago–miRNA target interactions have set the stage for properly evaluating and controlling miRNA-like off-target repression. Here, we describe the intrinsic problems of miRNA-like off-target effects caused by canonical and noncanonical interactions. We particularly focus on various genome-wide approaches and chemical modifications for the evaluation and prevention of off-target repression to facilitate the use of RNAi with secured specificity.
Keywords: Off-target effects, miRNA seed sites, Noncanonical target sites, Ago HITS-CLIP, RNA-Seq, Ribo-Seq, Chemical modification, Abasic pivot, RNAi therapeutics
Introduction
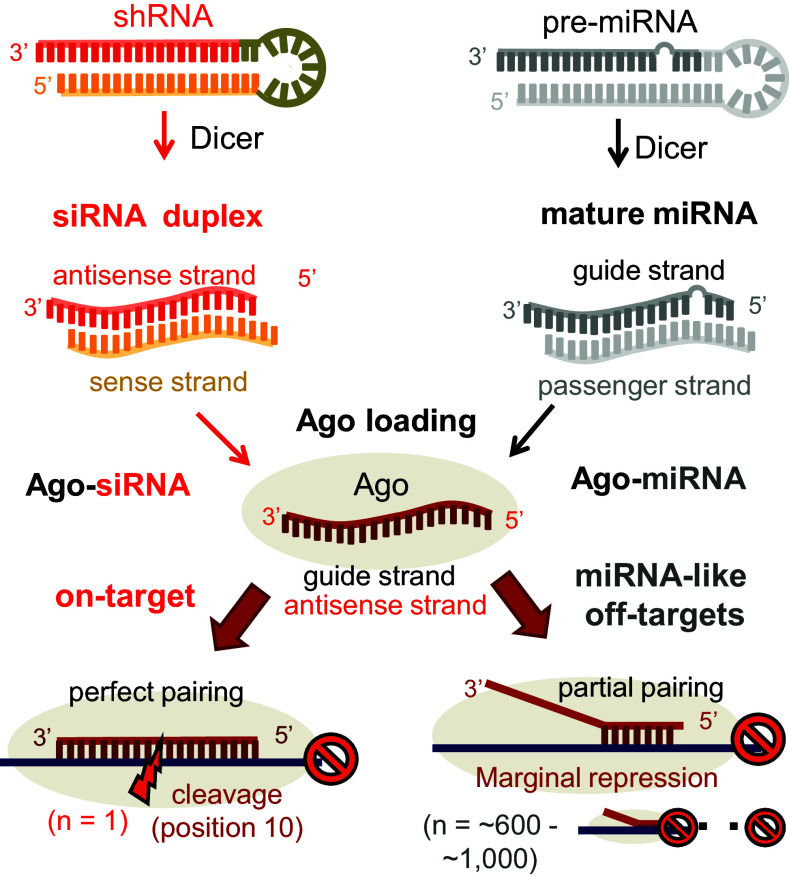
RNA interference (RNAi) is a gene-silencing phenomenon caused by small regulatory RNAs, which allow an RNA-induced silencing complex (RISC) to recognize and regulate target genes in a sequence-specific manner [1, 2]. Such direct modes of action result in effective gene silencing, as compared with the indirect transcriptional inhibition mediated by other antisense RNAs [1]. Small regulatory RNAs can be applied by introducing a small interfering RNA (siRNA), which is a short, ~21-base-pair duplex RNA that contains a perfect sequence match to an intended target transcript and leads to silencing of gene expression [1, 3]. Because of the ease with which siRNAs silence target genes, they have been widely used to study the loss-of-function of various genes and have been developed as promising therapeutics for the treatment of human diseases [4]. Endogenously, small regulatory RNAs are often identified as microRNAs (miRNAs), which suppress hundreds of partially complementary target mRNAs and thereby regulate various biological functions [5]. By sharing the same RNAi effector, small regulatory RNAs loaded onto a RISC always perform the functions of both siRNAs and miRNAs, thus resulting in unintended miRNA-like off-target repression with usage of the siRNA [2, 6] (Fig. 1).
Fig. 1.
Mechanistic overview of miRNA-like repression in siRNAs. Exogenously introduced small RNAs (siRNA or shRNA; indicated in red) and endogenously transcribed miRNAs (indicated in gray) are represented together with functional processing for RNAi-mediated gene silencing. By sharing the same downstream effector protein, Ago, siRNAs intrinsically suppress up to ~1000 miRNA-like off-targets through partial base pairing
In genetic studies, the first identified microRNA (miRNA) was a small non-coding RNA, lin-4, which was found to have a regulatory function in C. elegans development through its mRNA target, lin-14 [7, 8]. miRNAs are abundant in the mammalian genome: currently, more than 2000 human miRNAs have been discovered, according to the miRBase database [9]. To produce miRNAs, primary miRNAs (pri-miRNAs) are initially transcribed in the nucleus by RNA polymerase II [10] or III [11], where regions with ~70-nucleotide (nt) stem-loop structures in pri-miRNAs are subsequently excised by a microprocessor complex containing Drosha [12] and DGCR8 [13]. Such stem-loop miRNA precursors, called pre-miRNAs, are exported to the cytoplasm by exportin-5 [14] and further cleaved by Dicer [15], thereby leading to the production of mature miRNAs. Thus, RNAi can be triggered by introducing small hairpin RNAs (shRNAs), which contain stem-loop structures similar to those in pre-miRNAs [5] (Fig. 1). Mature miRNAs are short dsRNAs (~21 nt) with typical dinucleotide overhangs at the 3′ end, as observed in siRNAs, which are characteristics of products from the class III RNase family [5] (Fig. 1). Dicer also processes long dsRNAs into such siRNA-like structures and directly exhibits an antiviral effect by limiting dsRNA molecules [16–18]. Such RNAi and dsRNA-driven innate immune responses may be beneficial in therapeutic applications of RNAi for virus restriction [18]. Among the double-strands of mature miRNAs or siRNAs, only one strand is incorporated into Argonaute (Ago), a core effector protein in RISC [19], which was initially isolated in an siRNA-associated complex [20]. Because of asymmetric loading [21, 22], the major strand incorporated into Ago is called the “guide strand”, whereas the other strand is called the “passenger strand” (Fig. 1). The guide strand is also called the “antisense strand,” especially in the case of siRNAs, because it is designed to be complementary to the target mRNA (and the passenger strand is the “sense strand” from this perspective).
Although they are not completely specific, siRNAs were initially reported to have selective gene-silencing capability. This conclusion is supported by observations that Ago2 (also known as eIF2C2) cleaves only a perfectly paired target transcript, specifically in cases in which the nucleotide is paired at position 10 from the 5′ end of the guide strand [23–25] (Fig. 1). Otherwise, Ago2 loses cleavage activity for unrelated control sequences, particularly those containing single mismatches in the cleavage sites of the corresponding siRNAs [26, 27]. However, even in the case of partial matches, which are generally observed in miRNA–target interactions in animals (unlike plants, near-perfect matches with targets rarely occur in animals), target genes can be repressed without the catalytic activity of Ago by decreasing mRNA stability and/or inhibiting translation [28]. miRNA-like repression generally has a marginal effect at the individual transcript level and is often mistakenly dismissed as being phenotypically irrelevant [6]. However, given its interrogated effect on global gene expression, miRNA-like repression affects biological functions, thus potentially leading to deleterious phenotypes. In fact, alterations in miRNA regulation have been reported to be responsible for various diseases, such as neurological disorders [29], cardiovascular diseases [30], and various types of cancers [31]. Losses of individual miRNAs have also been observed to cause various biological defects [32]. Notably, all such phenotypic defects are ultimately caused by the dysregulation of miRNA targets. Thus, understanding miRNA-like interactions is important to evaluate phenotypic consequences of miRNA-like repression and to prevent off-target effects.
Canonical miRNA targets: seed-pairing rules
In contrast to siRNAs, which generally have a single perfectly matched target, miRNAs are partially paired with approximately several hundred targets (as estimated by previous studies [33–36], with an average of ~600 targets for mouse brain miRNAs [35]) or ~1000 targets (e.g., 1254 targets identified for miR-124 [35]), thus allowing them to regulate various biological phenomena [37] (Fig. 1). Such incompleteness of sequence matches makes identifying miRNA target sites challenging. Initial studies have attempted to predict miRNA target sites by analyzing several known sites in 3′ untranslated regions (3′UTRs) [38–43]. From these analyses, short local stretches (≥6 nt) of continuous base pairing have been found to be significant for target recognition. The “seed-pairing rule” has been widely used to identify short matches of miRNA target sites, featuring as few as 6-nt pairings in the seed regions (positions 2–8) of miRNAs [40, 44]. In addition, sequence conservation of “A” in the nucleotide opposite to position 1 of the miRNA further improves the prediction of miRNA target sites [45]. Canonical seed matches are defined as 6-mers, 7-mers, and 8-mers [44]. Each type of match is further divided into several subtypes: 6-mer (positions 2–7), offset 6-mer (positions 3–8), A1-7-mer (positions 2–7 with “A” at position 1), m8–7-mer (positions 2–8), and 8-mer (positions 2–8 with “A” at position 1), as illustrated by the examples in Table 1 (6-mer; miR-1 for Hsp 90aa1 [46], offset 6-mer; lin-4 for lin-14 [7, 8], A1-7-mer; miR-375 for Mtpn [47], m8–7-mer; let-7 for hbl-1[48], 8-mer; lin-4 for lin-14 [7, 8]). Notably, a 6-mer match to positions 3–8 is called an “offset 6-mer seed” because of its position and modest effect on repression [49].
Table 1.
Types of miRNA target sites
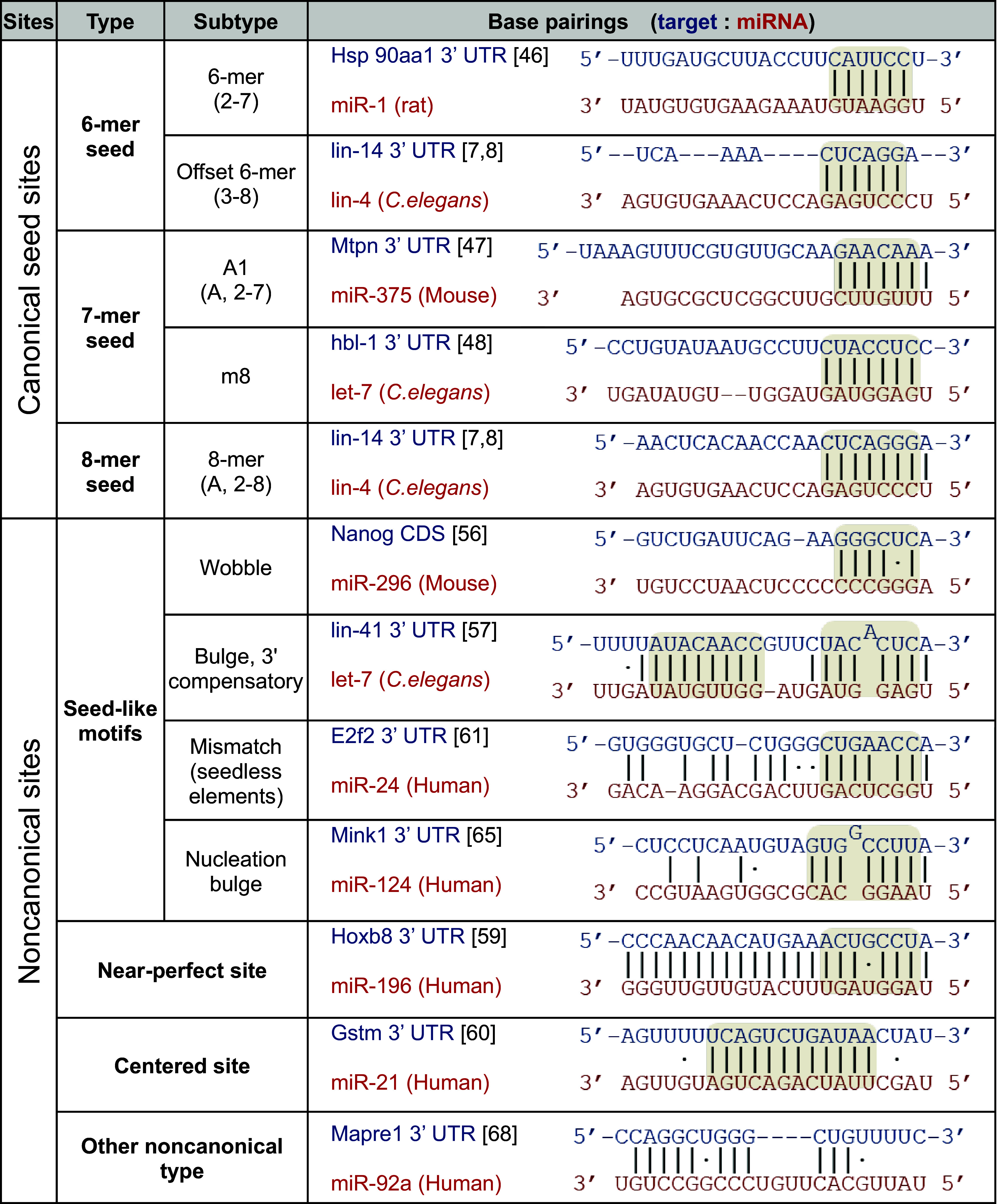
Representative examples of each type are indicated for canonical seed sites and noncanonical sites including their subtypes. Majorly interacting regions between the miRNA and target are highlighted in gray. Solid lines indicate Watson–Crick base pairing and dots indicate G: U wobble pairs
In combination with sequence conservation, which has been observed in 3′UTRs [50], seed-pairing rules enable canonical target sites of miRNAs to be determined [40], and their prediction is further improved by characterizing other features such as the secondary structure [51] and neighboring context information [52]. The seed sites of miRNAs have been validated in many biological studies (Table 1) and have been verified to be functional in miRNA-regulated gene expression [44]. In addition to individually validated target sites, global transcriptome analyses using microarray methods have further confirmed that seed matches are overrepresented in repressed transcripts, depending on miRNA expression [33, 52]. Comparative global analyses of miRNA overexpression or knockdown have shown that seed sites are widespread in miRNA targets [33, 34, 36]. Such unbiased and genome-wide profiling not only validates the “seed-pairing rule” of miRNA–target interactions but also provides evidence that even the marginal repression of most targets may be functional [37]. In support, genetic studies of a single mutation in the miR-96 seed region have shown that alteration of seed pairing leads to the recognition and suppression of different sets of target transcripts, thus causing progressive hearing loss [53, 54].
Noncanonical miRNA targets: widespread seed-like motifs
Although the seed-pairing rules have been informative in identifying miRNA targets, their prevalent use has led to the unintentional bias of studying only canonical seed sites. In fact, perfect seed matches have been continuously observed to be neither sufficient nor necessary for all functional miRNA target sites. For example, a genetic study in C. elegans has verified that lsy-6 miRNA also recognizes noncanonical sites by tolerating imperfect seed matches that form wobble (G:U) pairs with the target mRNA, cog-1 [55]. Such “seed-like motifs” have also been shown to contain mismatches and bulges (Table 1), which play critical roles in miRNA-mediated biological functions [37]. Furthermore, Nanog, Oct4, and Sox2, which are well-known key regulators in induced pluripotent stem cells, have been reported to be regulated by seed-like matches with a wobble (miR-296 for Nanog; Table 1) or bulge for cognate miRNAs (miR-134, miR-296, and miR-470) [56]. In some cases, these elements have been found to be atypically located in their coding sequences (CDSs) [56]. Additionally, seed-like motifs are frequently observed to be accompanied by 3′ complementary pairing (3′ compensatory site), which have been determined to be functional in genetic studies of C. elegans (let-7 for lin-41; Table 1) [57] and Drosophila melanogaster [58]. However, in mammals, such 3′ compensatory sites are rarely observed (less than ~5%) and generally exert modest effects on repression [49, 52]. Exceptionally, a case with near-perfect pairing has been found to effectively induce the cleavage of target mRNA, wherein a wobble in the seed match is compensated by consecutive supplementary pairings in other regions (e.g., miR-196 for Hoxb8; Table 1) [59]. Moreover, a few “centered sites”, which comprise ~11 nts of contiguous base-pairing in the central part of the guide strand of miRNAs, have also been found to induce the cleavage of target transcripts, wherein neither perfect seed matches nor 3′ compensatory pairing has been observed (e.g., miR-21 for Gstm; Table 1) [60]. However, such noncanonical sites of miRNAs are not fully appreciated because of their low occurrence in mammalian target transcripts.
Recent advances in genome-wide methods have revealed that a substantial number of miRNA-dependent transcripts contain sequence elements that deviate from seed matches. Initially, microarray analysis of miR-24 has identified numerous putative target genes that have major functions in the proliferation of leukemia cells, wherein the targets have no seed matches but contain mismatched seed-like motifs named “seedless” recognition elements (e.g., miR-24 for E2f2; Table 1) [61]. Moreover, applying seed-pairing rules to microarray or proteomics results generally yields high false-negatives (~50–70%) in miRNA target identification [34, 36, 62], which can be interpreted to suggest the widespread distribution of noncanonical sites [37]. The prevalent binding of miRNAs with noncanonical sites has been elucidated by the mapping of global miRNA–target interactions through the application of crosslinking and immunoprecipitation (CLIP) methods [63] with high-throughput sequencing (HITS-CLIP, also called CLIP-Seq) [64] to Ago (Ago HITS-CLIP) [35]. The Ago HITS-CLIP results have revealed a noncanonical seed-like motif called a “nucleation bulge” in the Ago–miR-124 complex (miR-124 for Mink1; Table 1) [65]. The nucleation bulge site forms a bulge in the target mRNA between position 5 and 6 of the corresponding miRNA, wherein a nucleotide in the bulge is competent to pair with a nucleotide in the pivot position (position 6), thereby enabling it to be predicted by sequence analysis [65, 66].
In addition, various seed-like motifs have also been revealed by investigating the differences in Ago association, depending on miR-155 expression [67]. Furthermore, seed-like motifs have been directly verified by analysis of chimeras [68–70] that are produced by the ligation process in Ago HITS-CLIP experiments and thus harbor binding sites in conjunction with miRNA sequences. In chimera reads, there are some cases in which miRNAs have limited contacts with their targets through seed-like motifs but do have sequence complementarity to the middle of the miRNA sequences or toward the 3′ end instead (e.g., miR-92 for Mapre1; Table 1) [68]. This finding indicates that more than 15% of miRNA targeting may operate at the 3′ end of miRNAs [68], an abundance sufficient to potentially contribute to Ago target specificity [70] and off-target effects.
Widespread miRNA-like off-target effects
RNAi-mediated gene silencing has been widely applied to various biological studies, including functional genomics and therapeutic target screening [71]. However, increasing data indicate that the initial assumption regarding the specificity of RNAi is relative, but that on-target activity is stronger than off-target repression. The issue of “off-target effects” is intrinsically inevitable because both exogenous siRNAs (including all types of RNAs that eventually produce siRNAs) and endogenous miRNAs share the same downstream effector, Ago, which cannot discriminate between the two types of small RNAs [6, 72] (Fig. 1). siRNAs can function as miRNAs by utilizing the target recognition mechanisms of miRNAs, thus possibly leading to unexpected outcomes, off-target effects. Particularly, when long double-stranded RNA (dsRNA) is used, the effects of unintended gene silencing become more severe. In Drosophila RNAi screens, off-target hits exhibit prevailing short stretches of complementary sequences (of up to 16 nts) to those of siRNAs and promiscuous tandem tri-nucleotide repeats [73]. Furthermore, there are some cases in which phenotypes from siRNA [74–79], dsRNA [73, 80, 81], or shRNA screenings [78] differ for the same gene. Moreover, RNAi-mediated knockdown has been reported to fail to recapitulate the phenotype of mouse gene knockouts because of the off-target effects associated with endogenous miRNA dysregulation [82].
The introduction of synthetic siRNAs or overexpression of shRNAs can compete with endogenous miRNAs for limited amounts of RISC, thus leading to saturation of the RNAi machinery required for the proper endogenous function of RNAi [6]. This saturation often results in cytotoxicity or global perturbation of gene expression as part of the off-target effects, for which the derepression of miRNA targets is caused by inhibiting endogenous miRNA activities [83]. Additionally, short synthetic siRNAs may potentially activate the mammalian innate immune system, thus inducing inflammatory cytokines (interferons, TNFα, IL-6 and others) [84]. Pattern-recognition receptors called Toll-like receptors (TLRs), such as TLR3, TLR7 and TLR8, are known to be responsible for the siRNA-mediated innate immune responses, but the extent of the immunological effects varies with selective expression of TLRs, cell types, oligonucleotide sequences, siRNA structures, and delivery vehicles [6, 84].
In addition to RNA-induced immune responses and/or cytotoxicity, sequence-specific miRNA-like repression is a major cause of off-target effects [6, 74]. Although initial attempts to examine global transcripts supported the effectiveness of siRNA [85, 86], microarray profiling using multiple siRNA sequences subsequently revealed that a large number of off-target transcripts are downregulated depending on specific siRNA sequences, regardless of the target gene [74]. The miRNA-like off-target repression is seed-centric and is mediated primarily by canonical seed sites [76–79, 87, 88], as well as marginally through non-canonical seed-like motifs that tolerate a few mismatches [74–76, 78, 87]. Furthermore, miRNA-like off-target repression is mediated by long matches with noncanonical sites, which can be dictated by as few as 11 contiguous pairings [74] that allow a few mismatches or a G:U wobble [75, 87]. These noncanonical sites are similar to the centered site [60] or the near-perfect site observed in miRNA targets [37, 59].
As observed in miRNA target sites, the efficiency of off-target repression has been reported to be dependent on the types and lengths of seed matches preferentially located in the 3′UTRs [77]. In addition, several features, such as the number of binding sites in a transcript, neighboring context information including GC content and sequence conservation, have been suggested to affect miRNA-like off-target repression [52, 87]. miRNA-like off-target effects often cause more dramatic changes in expected phenotypes than on-target repression in siRNA screens, particularly when large libraries are assessed by using only a single-assay readout, yielding approximately 30% false-positive results [72, 76, 87]. A critical caveat lies in the possibility that miRNA-like off-target effects may confound the proper interpretation of experimental outcomes. As a direct consequence of off-target repression, siRNAs frequently induce unwanted toxic phenotypes [77] or cell growth inhibition [78], independently of on-target silencing effects. In contrast to the evolutionarily conserved phenotypes observed in miRNAs, the off-target effects of siRNAs are generally species-specific; therapeutically potent siRNAs against APOB [89] and PCSK9 [90] have been shown to repress different off-target transcripts and consequently to cause different off-target phenotypes between humans and mice.
Although the siRNA sequences used for functional screening or therapeutic applications have been designed considering the potent sequence homology for an intended target compared with other transcripts, they frequently show significant off-target effects. For example, the development of therapeutic siRNAs for age-related macular edema (AGN-745; Allergan, Bevasiranib; Opko Health Inc.) was discontinued because their effect on the suppression of neovascularization was not caused by on-target silencing of VEGFA A mRNA, but instead resulted from sequence-independent off-target effects that triggered TLR3-mediated immune responses [91]. Moreover, PCS-A2 (Alnylam Pharmaceuticals) [92], an siRNA targeting PCSK9 for treating hypercholesterolemia, has been found to show miRNA-like off-target effects, causing hepatocellular cell death and/or cell cycle arrest [90]. Recently, a trial of ALN-AAT (Alnylam Pharmaceuticals), an siRNA targeting alpha-1 antitrypsin (AAT) for treating AAT deficiency, was halted, and the sequence was replaced with a different nucleotide sequence, presumably because the original sequence was causing microRNA-like off-target effects [93]. More seriously, the recently discontinued revusiran (Alnylam Pharmaceuticals), an siRNA targeting transthyretin for the treatment of amyloidosis with cardiomyopathy, led to the death of 19 patients in a phase 3 trial, a result suspected to be caused by miRNA-like off-target repression [93]. miRNA-like off-target effects in clinical applications can lead to detrimental side effects; hence, ensuring specificity is essential.
General guidelines for evaluating off-target effects
To estimate off-target effects, appropriate controls have been proposed for use in parallel with siRNAs [94]. So-called “unspecific” or “scrambled” sequences are used to evaluate the extent of off-target effects and toxicity caused by RNA delivery and/or innate immune responses. However, such unrelated controls are unable to consider miRNA-like target repression by carrying different seed sequences. To reflect sequence-specific features, siRNAs lacking on-target activity through mismatch mutation of the bases at positions 9–11 can be alternatively used as a control [95] (Table 2). Another method to determine the specificity of siRNAs is to confirm consistent outcomes generated by multiple siRNAs or shRNAs (at least two independent sequences for the same gene) [94, 96]. Failure to observe analogous phenomena may indicate sequence-specific off-target effects. Rescue experiments can also provide proof of specificity and can be used to assess whether the observed phenotype is compensated by expressing the target gene as either cDNA, in cases in which an on-target site is in the 3′UTR, or a functional orthologue that is not silenced because of sequence differences across species [94, 96].
Table 2.
siRNA–target effects and strategies to evaluate and control them
| Off-target effect | Evaluation and control | Advantages and limitations | |
|---|---|---|---|
| miRNA-like off-target repression | General strategies | Use appropriate control sequences (e.g., mismatch mutation in positions 9–11) [94, 95] | Easy but indirect way of evaluating off-target effects, unable to prevent off-targeting, not applicable to therapeutics |
| Rescue using untargeted cDNA or orthologous genes [94, 96] | |||
| Minimizing the concentration of siRNAs [86, 144, 145] | Easy to reduce off-targeting although not complete, on-target activity may not be maintained, pooling may cause more promiscuous off-targeting, not amenable to therapeutic applications | ||
| Use multiple siRNA/shRNA [94, 96] | |||
| Pooling multiple independent siRNAs [146, 147] | |||
| Sequence analysis | Little sequence homology to other mRNAs [140, 141] | Easy to avoid off-targeting although not complete, performance varies depending on bioinformatics methods | |
| GC content and thermodynamic stability in seed regions [142, 143] | |||
| Bioinformatics programs for off-targets in shRNA library screening [99–102] | Developed only for canonical off-targets in shRNA library screening | ||
| Global analysis | Gene expression analysis in the presence of siRNA/shRNA (microarray, RNA-Seq, SILAC, Ribo-Seq) [74, 90] | Unbiased detection of off-target gene regulation, cannot discriminate direct and indirect off-targets | |
| Global siRNA–mRNA interactions (Ago HITS-CLIP, CLASH, CLEAR-CLIP) | Direct detection of siRNA–off-target interactions, experimental procedures are difficult to perform | ||
| Chemical modification | 2′-O-methyl modification (2′-OMe) at position 2 [145] | Selectively reduce off-target effects although not complete, increase metabolic stability, some cases may cause in vivo toxicity, on-target activity may be sacrificed | |
| Locked nucleic acid (LNA) in seed region [155] | |||
| Unlocked nucleic acid (UNA) at position 7 [158] | |||
| DNA substitution in seed region [159] | |||
| Abasic pivot substitution (dSpacer or C2 in position 6) [90] | Complete elimination of seed-mediated miRNA-like repression, superior on-target activity but may not completely maintain on-target activity | ||
| 1-ER triazole I modification at position 1 [174] | Dramatically reduce off-target effects although not complete, superior on-target activity but may not completely maintain on-target activity | ||
| Innate immune response | Evaluate immunological side effects through published data (RNAimmuno database) [103] | Not all pro-inflammatory cases have been investigated | |
| Immune stimulatory sequence motifs (e.g. GU-rich) are excluded [84, 148] | Not all pro-inflammatory sequences have been identified | ||
| Chemical modifications (2′-OMe, LNA, and DNA) [6] | Not all chemical modifications are compatible | ||
| Saturation of RNAi machinery | None yet identified, minimizing the concentration of siRNAs may be tried | Not applicable | |
With the increasing demand for high-throughput screening using RNAi libraries, general guidelines for avoiding false-positive hits have been developed. For instance, the selection of hits has been achieved by using optimal normalization methods with a verified threshold [97] and statistical support to minimize the variation among screens [98, 99]. However, these applications still have the drawbacks of low validation rates and reproducibility among independent screens, possibly because of residual off-target effects. Notably, the frequency of complementary sites to the seed region of the siRNA across the 3′UTRs has been found to correlate with the number of off-targets [79]. This propensity has been used to analyze and reject off-target effects in RNAi screens by applying bioinformatics approaches [71], which have also been integrated into the statistical model to select true positive hits (gespeR) [99]. In addition, RNAi screening produces numerous positive and negative hits with siRNA sequences, and these observations have been utilized to develop interrogative computational tools to define putative off-targets. On the basis of this approach, common seed analysis (CSA) [100] and genome-wide enrichment of seed sequence matches (GESS) [101] methods have been developed and used to deconvolute prominent off-target transcripts in primary screening data. However, analysis of these screening hits on the basis of biological pathways has indicated that the majority of the off-target hits are involved in similar biological processes related to the function of the on-target gene. The Haystack method, which focuses on the analysis of phenotypically related pathways, was developed to identify statistically significant off-targets in functional clusters relevant to a target gene [102]. Nevertheless, all the methods used to identify off-target hits only rely on canonical seed matches, thus limiting the complexity of the miRNA-like off-targets and excluding the possibility of false-positives from noncanonical interactions.
In addition, introduction of the RNA itself can trigger non-specific immune responses activating cellular sensors of foreign RNA (TLRs) and inducing cytokines [84]. Non-specific immunological effects occur regardless of the sequence, but the extent of immune responses varies depending on the cell lines, tissues, model organisms and RNAs of specific types and structures [6, 84]. Manually curated databases such as “RNAimmuno” have gathered the published data for immunological side effects and can be used to evaluate the potential risk of specific RNAi triggers provoking immunological off-target effects [103].
Global analysis of miRNA-like off-targets
Given that miRNAs recognize both canonical and noncanonical sites [37], miRNA-like off-targets have predominant seed matches and seed-like motifs that contain various mismatches, including bulges. Such a promiscuous mode of miRNA-like target recognition makes it difficult to evaluate and identify off-targets, thus ultimately requiring unbiased genome-wide methods to address this issue by detecting changes in global gene expression and miRNA-like target interactions.
Gene expression analysis: miRNA-like target repression
Systematic approaches to detect RNAi-mediated gene silencing began with microarray technologies [85]. The initial use of microarrays was to confirm the cleavage of an on-target mRNA to validate the specificity of the siRNA and to identify targets of an overexpressed plant miRNA, exhibiting near-perfect matches [85, 86]. However, microarray profiling has also indicated some downregulated transcripts other than the intended siRNA target, wherein partial sequence matches have predominantly been observed as miRNAs [74]. Microarray profiling has been widely adopted for analyzing miRNA targets, showing repression of miRNA-dependent target transcripts, among which 3′UTRs most commonly contain seed matches [33]. The microarray analyses have also indicated a tissue-specific shift in transcript profiles similar to that observed in tissues wherein corresponding miRNAs are preferentially expressed (e.g., miR-124 in brain, and miR-1 in muscle) [33]. Moreover, microarray profiling together with bioinformatics analysis has helped to define the characteristics of miRNA-mediated repression [33, 52, 104]. In particular, by comparing the distribution of cumulative fractions according to miRNA-dependent fold changes in gene expression, microarray profiles have successfully illustrated the efficiency of canonical seed sites (6-mer, offset 6-mer, 7-mer-A1, 7-mer-m8, and 8-mer) and their combinatorial effects on miRNA mediated repression, in conjunction with neighboring context information [52, 105]. Microarray analyses have also helped to identify non-canonical miRNA target sites such as centered sites [60] and seedless elements [61]. In addition, for siRNAs, meta-analyses of compiled microarray profiles have revealed that non-canonical nucleation bulge sites as well as seed sites are also widespread in off-targets [90].
Although microarrays provide valuable information, the coverage of detection is restricted by the number of probes and gene annotations [106]. Moreover, a fundamental limitation of microarrays is the high background-to-signal ratio arising from the nature of nucleic acid hybridization, whose strength varies depending on the base composition. Nevertheless, certain limitations have recently been overcome through the adoption of RNA-Seq analysis [107], which was developed to take advantage of the power of high-throughput sequencing methods. Because of the comprehensive and unbiased signature obtained from whole-transcriptome shotgun sequencing, RNA-Seq provides unprecedented accuracy and reproducibility for measuring transcript profiles. RNA-Seq can detect a broad spectrum of gene expression and deduce splice variation, alternative poly-adenylation, and RNA editing. Given that miRNAs can recognize target sites in alternative exons [108], alternative 3′UTRs [109], and non-coding RNAs [110], RNA-Seq analysis expands the repertoire for detecting miRNA-like regulation in whole transcriptomes. In combination with gene ontology analyses, assessing the enrichment of functional terms and annotations can be used to illustrate the biological functions of miRNA-like targets [33]. Although it is not used as frequently for siRNAs than miRNAs, the increasing use of RNA-Seq is expected to resolve the biological consequences of miRNA-like off-target effects. In fact, RNA-Seq analyses have successfully revealed the deleterious side effects caused by the administration of therapeutic siRNAs against PCSK9 [90].
Because miRNAs also regulate the translation of target genes, monitoring protein levels has become required to elucidate miRNA-like off-target effects. In fact, the expression pattern of miRNA regulation is not necessarily correlated with the integrity of the transcripts [28]. To measure changes in protein abundance, stable isotope labeling of amino acids in cell culture (SILAC) has been performed by utilizing mass spectrometry, wherein nonradioactive isotopes (13C or 15N) are used to discriminate mass spectrometry results from different miRNA expression profiles [111]. Although only 12 putative miR-1 targets were initially identified because of the limited coverage of detection (only ~500 proteins), the results monitored in miR-1 versus control transfected HeLa cells have been found to correlate with those obtained from the microarray profiling and computational prediction of seed sites [112]. With the increased efficiency of labeling in SILAC, more comprehensive proteomics results (~5000 proteins) have been achieved for miR-124, miR-1, miR-181 and miR-223 [34]. Most proteins that show marginal changes in expression (~1.5 to 2-fold) predominantly contain seed sites in the 3′UTRs [34]. Similar results have also been obtained by pulsed SILAC, a modified version of the technique for detecting only newly produced proteins depending on miR-1, miR-155, miR-30a, and let-7b expression [36]. SILAC helps to identify miRNA targets in which translation is directly repressed, as demonstrated for miR-29a [113], miR-34a [114], miR-373 [115], miR-143 [116], and miR-21 [117].
Quantitative proteomics has limited potency, owing to the restricted coverage of mass spectrometry, which is less than that of microarray or RNA-Seq analysis. Alternative strategies, which measure translation rates instead of protein levels, have been used to detect actively translated mRNAs in polyribosomes. By combining the purification of polyribosome with microarray analysis, polysome profiling enables the monitoring of the translational states of mRNAs, depending on the miRNA expression [118]. More comprehensively, sequencing-based methods have been developed to quantify ribosomal footprints, mRNA regions that are protected from nuclease treatment, which correlate with the translational efficiency [119]. Ribosomal profiling (Ribo-Seq) provides unprecedented coverage (~90%) of transcripts undergoing translation (translatome) [120] and has been applied to study miRNA-regulated translation [121, 122]. In parallel with transcriptome profiling, Ribo-Seq analyses have revealed properties of miRNA-mediated gene silencing in which mRNA decay is the dominant mechanism underlying substantial repression [121, 122], but translational repression is also effective during the time of miRNA induction [121]. For siRNAs, the same characteristics are expected to be observed for miRNA-like off-target repression. Ribo-Seq analyses are helpful to evaluate miRNA-like off-target repression, especially in cases in which off-targets are regulated only via translational repression. Although the effects of siRNAs at the global protein level have not yet been determined, both quantitative proteomics and translatomics approaches can potentially be used to detect siRNA off-targets at the level of translation.
Global miRNA-like target interactions
Although global gene expression analyses are used to evaluate miRNA-like gene repression, miRNA-dependent changes in gene expression may result from both direct and indirect effects, thus necessitating the development of a method to detect direct binding between miRNAs and targets [37]. Initially, RNA immunoprecipitation (RIP) approaches were developed to biochemically isolate RNAs associated with a specific RNA-binding protein by using cognate antibodies [123–126]. For the RISC complex, the RIP method has been applied to Ago to purify target mRNAs bound by the Ago–miRNA complex, and then the target mRNAs were profiled using microarray (RIP-Chip) [123–126]. RIP-Chip has identified some miRNA targets, which show enrichment of seed sites, but also contain many false-positives, thus raising concern about nonspecific Ago–RNA interactions via in vitro rearrangements [127, 128]. To overcome this issue, the CLIP method [63] was developed by applying ultraviolet (UV) irradiation to covalently crosslink RNA–protein complexes in living cells. This procedure allows for extremely stringent conditions for purification of only the intended RNA–protein complexes, thus minimizing background noise including nonspecific and indirect protein–RNA interactions. Combined with high-throughput sequencing (HITS-CLIP, also called CLIP-Seq) [64] and applied to Ago (Ago HITS-CLIP), the CLIP method has precisely mapped transcriptome-wide miRNA target sites in mouse brain (~93% specificity for seed matches) [35]. For improved resolution, Ago HITS-CLIP results can be analyzed for crosslinking-induced mutation sites (CIMS), for which a deletion occurs at the cross-linked site (~8 to 20%) [129]. Analogously, the photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) method utilizes a uracil (U) analog, 4-thioluridine, to improve the efficiency of UV crosslinking and detects subsequent T/C mutation as cross-linked sites [130]. Moreover, the individual nucleotide resolution of UV crosslinking and immunoprecipitation (iCLIP) method examines truncated cDNAs that indicate the position of the protein–RNA crosslink sites [131]. CLIP-based methods have been widely used for Ago to map miRNA target sites in cultured cells [67, 128, 130, 132–136], tissues [65, 137, 138], and even whole organisms (C. elegans) [69, 139].
With increased specificity and resolution, Ago HITS-CLIP enables defining of canonical seed sites as well as non-canonical seed-like motifs that have been initially elucidated as “nucleation bulges” (≥15% of the total sites) in mouse brain [65]. Moreover, differential Ago HITS-CLIP, devised to accurately identify miR-155 interaction sites using miR-155-deficient T-cells, has also identified seed-like motifs comprising ~20% of Ago–miR-155 binding sites [67]. To further provide direct evidence of miRNA target binding, chimeric reads of such interactions between binding sites and miRNA sequences have been investigated in the Ago HITS-CLIP results, although they are rarely generated [69]. Cross-linking, ligation and sequencing of hybrids (CLASH) [68], modified PAR-CLIP [69], and covalent ligation of endogenous Argonaute-bound RNAs (CLEAR-CLIP) [70] are such methods based on chimeric reads, of which number has been intentionally increased by treating purified Ago–RNA complexes with RNA ligase. These methods have enabled the discovery of widespread non-canonical binding sites consisting mostly of seed-like motifs (~30 to 60%). To understand the functionality of the identified targets sites, Ago CLIP data have been analyzed together with gene expression profiles, and significant miRNA-dependent repression mediated mainly by canonical seed sites and marginally by seed-like motifs has been observed [65, 67–70]. This strategy has also been utilized to examine the efficacy of chemical modifications in preventing off-target effects and functional miRNA-like target interactions [90]. Nevertheless, Ago CLIP-based methods have been frequently used for miRNA studies and not for identifying siRNA off-target sites; however, these methods are expected to be widely adopted to evaluate miRNA-like off-target interactions.
Control of miRNA-like off-target repression
General strategies
Although miRNA-like interactions are inevitable, several strategies have been suggested to minimize the off-target effects. Generally, siRNA or shRNA sequences are designed to exhibit little sequence similarity to other mRNAs (e.g., <11-bp stretch of continuous matches) except their intended on-targets and, particularly, to avoid seed sequences that are frequently observed in 3′UTRs [94, 96]. This criterion can be easily met by using BLAST searches [140] or other suffix array-based searches (RIsearch2) [141] to detect potential off-targets of given siRNA sequences. In addition to sequence similarity, other related features of off-target sites such as GC content and thermodynamic stability in seed regions (positions 2–8) are considered to determine the siRNA sequence [142, 143]. miRNA-like off-target repression is concentration dependent, thus suggesting that minimizing the concentration of siRNAs is critical to improving the target specificity [94, 96]. Thus, it is recommended that siRNA treatment could be titrated down to the level that maintains sufficient on-target activity [86, 144, 145]. Alternatively, pooling of multiple independent siRNAs for the same gene can be used to lower the concentration of each siRNA sequence while preserving on-target activity [146, 147]. However, the pooling approach raises concerns about expanding the number of off-targets with marginal repression, which would presumably result in promiscuous off-target gene regulatory effects. Moreover, introduction of the RNA itself can trigger non-specific immune responses, and thus known stimulatory sequence motifs for innate immune responses (e.g., GU-rich) are generally excluded [84, 148]. Of note, miRNA-like off-target repression can be triggered by the passenger strand, whose sequence should be designed with caution [6]. Although its use has been limited, repeat-targeting siRNAs can be designed to form self-duplexes, in which the sequence of the guide strand can also represent the passenger strand [149]. This strategy has been applied for targeting CAG repeats and selectively inhibiting mutant huntingtin expression without the concern of passenger strand off-targeting [149].
Empirical chemical modifications
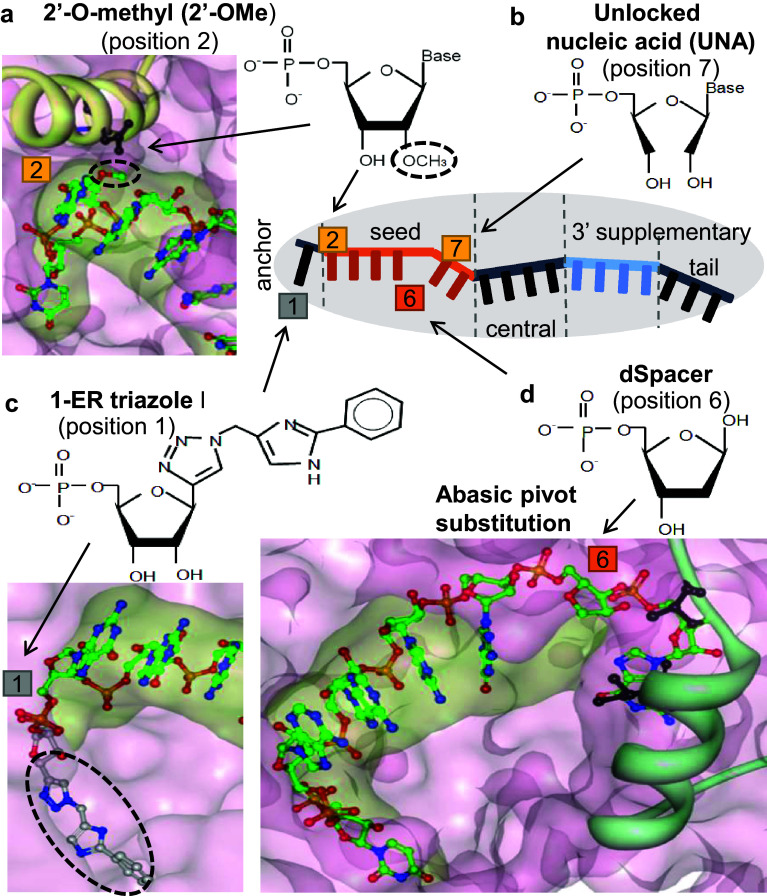
The use of proper siRNA sequences, concentrations, controls, and pooling strategies can circumvent off-target activities, but these are indirect and passive ways to overcome off-target effects. As a direct method, chemical modifications have been used for siRNAs, altering the properties of the nucleotide backbones and bases [6, 72]. Thus, siRNA may preferentially lose miRNA-like target recognition and also exhibit enhanced stability, cellular delivery, and tissue distribution [145, 150–153]. The most conventional 2′-O-methyl modification at position 2 from the 5′ end of the guide strand (2′-OMe) has been empirically found to alleviate off-target repression and subsequent adverse cellular effects (e.g., growth inhibition) [145] (Fig. 2a). The 2′-OMe is generally used in combination with an impaired passenger strand that shows no activity in both on- and off-target repression by introducing the 2′-O-methyl modification at positions 1 and 2 [145]. The 2′-OMe has been found to decrease ~80% of putative off-target transcripts, exerting an ~66% average decrease in off-target repression [145]. Structurally, the nucleotides at positions 1 and 2 of the siRNA or miRNA directly contact the Ago protein. In detail, the 5′ end of the guide strand (position 1) is buried in a deep pocket at the interface of the MID–PIWI domains of Ago, and the 2′-OH at nucleotide position 2 has been implicated in the formation of a hydrogen bond with the asparagine residue of Ago [90, 145, 154] (Fig. 2a). Therefore, the 2′-OMe at these positions may introduce steric constraints and affect Ago-siRNA interactions, thus leading to structural changes and subsequent seed pairing instability for miRNA-like target recognition (Fig. 2a).
Fig. 2.
Structural modes of chemical modifications that prevent miRNA-like off-target repression. Together with various chemical modifications of siRNAs, schematic diagrams of siRNA regions structured by Ago (a gray oval shape) are represented as anchor, seed, central, 3′ supplementary, and tail regions [175]. a 2′-OMe at position 2 is indicated in a structural model of the Ago–siRNA complex (derived from 4F3T[165] by using PyMOL). 2′-OMe is highlighted by a dotted circle. b UNA at position 7. c 1-ER triazole I at position 1 is indicated in a known Ago complex structure [174] (highlighted by a dotted circle). d Abasic pivot substitution. Surface model of dSpacer at position 6 is displayed in an Ago complex [90]
Another chemical modification that is frequently applied to nucleotide backbones of siRNAs is locked nucleic acid (LNA), which links the 2′ oxygen to the 4′ carbon [152, 155]. Such an unusual bond confers resistance to nuclease and structural rigidity, thus enabling thermodynamically stable base-pairing. Generally, LNA modifications of siRNAs improve on-target functionality by enhancing RNA stability and/or strand selectivity over the passenger strand [152, 156]. Intriguingly, despite the increased stability of base pairing, LNA modification of the seed region has been empirically found to attenuate cellular toxicity mediated by miRNA-like off-target repression, which is comparable to that mitigated by 2′-OMe [155]. This result probably reflects the absence of the 2′-OH group in LNA and its inability to participate in a hydrogen bond with Ago, as in the case of 2′-OMe. In contrast to LNA, the unlocked nucleic acid (UNA) modification, which cleaves a covalent bond between the 2′ carbon and 3′ carbon of the nucleotide backbone, is used to increase metabolic stability and is often used in siRNA applications in combination with LNA [157]. Empirically, the UNA modification at position 7 has been found to abrogate miRNA-like repression while preserving the on-target potency of siRNAs [158] (Fig. 2b). The alleviation of miRNA-like activity may be explained by weakened base-pairing of UNA in the seed region caused by the flexible open structure in the nucleotide backbone. In line with the notion that base-pairing of DNA-RNA heteroduplex is weaker than that of the RNA duplex, DNA substitution in the seed region of siRNA has been introduced, thus limiting miRNA-like repression [159].
Individually or in combination, LNA and UNA have been applied to siRNAs for in vivo use and found to exert higher on-target activity along with lower off-target toxicity [156, 157]. However, LNA administration with two trinucleotide motifs (TCC and TGC) has been reported to induce liver toxicity in vivo, revealing a critical limitation to its in vivo application [160]. Notably, chemical modifications applied to 2′-OH (e.g., 2′-OMe, LNA, and DNA) can block RNA-induced innate immune responses [6]. However, any chemical modification that can solve the issue of off-target effects, should be carefully examined for the potential introduction of another issue such as in vivo toxicity. Moreover, all of these above-listed chemical modifications have limited potency to decrease miRNA-like off-target repression, exhibiting generally marginal and varied effects. This is possibly because such modifications are introduced in the nucleotide backbone rather than in the bases, which directly participate in pairing with off-target transcripts.
Mechanism- and structure-guided chemical modifications
Limited potency of the empirical chemical modifications has been attempted to be overcome by rationally designed new strategies. For this, it is particularly important to precisely understand how Ago–miRNA recognizes target mRNAs. Initially, it was postulated that short consecutive matches defined as seed sites might serve as a “nucleus” initiating miRNA–target recognition [161, 162]. In agreement with this concept, seed-to-target binding shows higher affinity in a tethered complex with a PIWI/MID domain protein like Ago than does the short RNA in isolation (up to ~300-fold enhancement) [163]. Similarly, by considering the thermodynamic stability of four-nucleotide pairings in the seed region, the performance of predicting miRNA target sites has been improved, especially when the secondary structure of target sites was also considered [51, 164]. More precisely, a stepwise process comprising nucleation, propagation and cleavage of perfectly matched siRNA–target duplexes has been proposed by structural studies of human Ago–miRNA [165, 166] and Ago–siRNA–target complexes [167]. Here, the miRNA sequences of the seed region in the Ago complex have been found to be geometrically prearranged. Intriguingly, the seed region is structurally disrupted (Fig. 2d)—the miRNA between positions 6 and 7 is interrupted by isoleucine (I365) in an alpha-helical structure of Ago protein, configuring only positions 2–6 into a hybridization-susceptible helical structure [165, 166]. The exposed bases at positions 2–6 have been suggested to play a role in the initial hybridization, subsequently propagating additional base pairs up to position 8 by overcoming the kink at positions 6–7 [167].
Consistent with this structural implication, base pairing at positions 2–6 is termed “transitional nucleation”, as identified in the Ago HITS-CLIP analysis, to explain the nucleation bulge sites as the pattern of noncanonical recognition [65]. In this model, transient 5-nt nucleation (at positions 2–6) is postulated to confer thermodynamic stability and determine the state leading to conformational alterations, wherein the originally matched pivot nucleotide (position 6) bulges out and propagates annealing towards the 3′ end of the miRNA. Recent single-molecule analyses have provided further evidence that Ago2 initially scans target sites by base pairing at positions 2–4 [168] and subsequently mediates rapid and stable binding to the seed region of a miRNA [169, 170], thus serving as a proofreading procedure for target recognition [171]. The transitional nucleation model serves as a general mechanism underlying miRNA-like target recognition [37, 65]. Importantly, base pairing in the pivot position (position 6) has been suggested to play a decisive role in initiating the formation of a functional miRNA–target complex [37, 65].
On the basis of the mechanism- and structure-guided strategies to destabilize transitional nucleation, a base in the pivot of the siRNA is intentionally impaired by substitution with abasic spacers, thus achieving complete elimination of seed-mediated miRNA-like repression [72, 90] (Fig. 2d). Specifically, dSpacer (6pi) substitution, which contains neither a base nor 2′-OH (abasic deoxynucleotide), leads to a complete inability to induce seed-mediated miRNA-like repression (0%, even at the highest applicable concentration; 150 nM siRNA) [90]. The elimination of miRNA-like off-target repression has been further validated at the whole-transcriptome level by using RNA-Seq and Ago HITS-CLIP analyses. Moreover, the incorporation of 6pi into miR-124 abolishes the biological function of inducing neurite outgrowth, whereas other conventional modifications, such as use of 2′-OMe and UNA, have demonstrated limited potency. siRNA with 6pi (siRNA-6pi) also maintains near-perfect on-target activity (~80 to 100%) without altering the slicing activity of Ago2; intact on-target activity (100%) has been observed throughout the concentration ranges routinely used for siRNA experiments (10–75 nM). Superior preservation of on-target activity appears to be caused by decrease steric hindrance, by which 6pi probably provides adequate space to be beneficial for moving the kink outward and widening the central cleft, as envisioned in a ternary human Ago2–miRNA–target structure (Fig. 2d). In support, superior conservation of on-target activity has been observed by using the smallest spacer, C3, which is connected by only three carbons. The abasic pivot substitution is functional in vivo, as validated by its application to therapeutic PCSK9 siRNAs; it efficiently lowers plasma cholesterol in mice and abolishes potentially deleterious off-target phenotypes such as cell death and cell cycle arrest in liver.
To investigate the effects of chemical modifications on the Ago–siRNA structure, the structure of Ago has been examined in the presence of a chemically modified siRNA [172]. Interestingly, the structure of Ago is not changed even though the siRNA is intensively modified with 2′-OMe, 2′-O-(2-methoxyethyl) (2′-OMOE), 2′-fluoro (2′-F) and (E)-5′-vinylphosphate (5′-VP). By contrast, the configuration of the modified siRNA in the 3′ half of the seed region (at positions 5–6) is shifted and becomes disordered (at positions 7–8), probably because it requires plasticity to optimize contacts with Ago or the target transcripts. Such increased flexibility of the modified siRNA can explain the tolerance of Ago exerting conserved siRNA activity. Alternatively, computational screening based on Ago structures have revealed a functional nucleotide analog bearing a triazolyl modification (1-ER triazole I) at the 5′ end of the siRNA [173] (Fig. 2c). Structurally, 1-ER triazole I shows a deep anchorage into the central binding cleft of human Ago2, unlike a natural nucleotide, thus illustrating the possibility that it may modulate base pairing between the siRNA and its corresponding targets [174] (Fig. 2c). Furthermore, 1-ER triazole I attenuates the affinity and repression of seed-mediated off-targeting, but does not affect on-target interaction and activity. Considering the structural properties of 1-ER triazole I, any form of chemical modification that enhances the penetration of the 5′ end is expected to be effective, thus increasing the specificity of siRNAs.
Conclusion
By sharing the same RNAi effectors such as Ago, RNAi-mediated gene silencing is intrinsically concurrent with miRNA-like off-target repression. Although miRNA-like off-target repression has been demonstrated to be deleterious and phenotypically relevant, it has often been overlooked because of difficulties in evaluation and control [6]. In addition to miRNA-like off-target repression, specific solutions to evaluate and mitigate other types of off-target effects have been proposed (Table 2). Several indirect methods, including empirical chemical modifications, have been developed to circumvent the issues of off-target effects, but such conventional methods have limited potency. Nevertheless, recent advances in genome-wide methodologies for profiling global gene expression and miRNA target sites have enabled comprehensive analyses of miRNA-like interactions and repression. Together with increasing knowledge of Ago complex structures, the characteristics of miRNA-like off-target recognition have begun to be elucidated [37].
Such valuable information provides a rational basis for the design of chemical modifications of siRNAs [72] that can effectively eliminate miRNA-like off-target repression [90, 174]. For RNAi-mediated gene silencing, proper evaluation and control of miRNA-like off-target repression are crucial to avoid the misinterpretation of gene silencing results or to prevent adverse effects in clinical applications. However, there are still unknown rules and structural information, particularly for noncanonical modes of miRNA-like target recognition [37, 72], which frequently occur in siRNA off-targets with potentially detrimental effects. Therefore, future studies are required to address the remaining questions and yield further insights regarding new strategies to completely harness the target specificity of RNAi. Moreover, the potential of siRNAs to saturate the endogenous RNAi machinery and to stimulate innate immune responses should also be considered. More work is required to understand the underlying mechanisms and to develop solutions for these issues.
Acknowledgements
We apologize to researchers whose works were not cited in this review because of space limitations. This work was supported by a grant from Korea University to S.W.C, a grant from the Science Research Center Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning to S.W.C (NRF-2015R1A5A1009024), a grant from NRF funded by the Ministry of Science, ICT & Future Planning to H.S (NRF-2017R1D1A1B03030852), and a grant from NRF funded by the Ministry of Education to E.-S. J (NRF-2016R1A6A3A11931948).
Footnotes
Heeyoung Seok and Haejeong Lee have contributed equally.
References
- 1.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czech B, Hannon GJ. Small RNA sorting: matchmaking for argonautes. Nat Rev Genet. 2011;12(1):19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16(9):543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 16.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342(6155):235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342(6155):231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gantier MP. Processing of double-stranded RNA in mammalian cells: a direct antiviral role? J Interferon Cytokine Res. 2014;34(6):469–477. doi: 10.1089/jir.2014.0003. [DOI] [PubMed] [Google Scholar]
- 19.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293(5532):1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 20.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–574. doi: 10.1016/S0092-8674(02)00908-X. [DOI] [PubMed] [Google Scholar]
- 21.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 25.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20(23):6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 27.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31(2):589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32(4):199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Science translational medicine. 2014;6(239):239ps233. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 34.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 37.Seok H, Ham J, Jang ES, Chi SW. MicroRNA target recognition: insights from transcriptome-wide non-canonical interactions. Mol Cells. 2016;39(5):375–381. doi: 10.14348/molcells.2016.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 40.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 41.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1(3):E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila . Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational–experimental approach predicts human microRNA targets. Genes Dev. 2004;18(10):1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 46.Zhu WS, Guo W, Zhu JN, Tang CM, Fu YH, Lin QX, Tan N, Shan ZX. Hsp90aa1: a novel target gene of miR-1 in cardiac ischemia/reperfusion injury. Sci Rep. 2016;6:24498. doi: 10.1038/srep24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 48.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4(5):625–637. doi: 10.1016/S1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 49.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14(4):287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 52.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41(5):614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 55.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat Struct Mol Biol. 2006;13(9):849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 56.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 57.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18(2):132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 60.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38(6):789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mourelatos Z. Small RNAs: the seeds of silence. Nature. 2008;455(7209):44–45. doi: 10.1038/455044a. [DOI] [PubMed] [Google Scholar]
- 63.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 64.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19(3):321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim KK, Ham J, Chi SW. miRTCat: a comprehensive map of human and mouse microRNA target sites including non-canonical nucleation bulges. Bioinformatics. 2013;29(15):1898–1899. doi: 10.1093/bioinformatics/btt296. [DOI] [PubMed] [Google Scholar]
- 67.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48(5):760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell. 2014;54(6):1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014;15(9):591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seok H, Jang ES, Chi SW. Rationally designed siRNAs without miRNA-like off-target repression. BMB Rep. 2016;49(3):135–136. doi: 10.5483/BMBRep.2016.49.3.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443(7109):359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 74.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 75.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278(45):44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 76.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33(14):4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12(7):1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14(5):853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3(10):833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 81.Moffat J, Reiling JH, Sabatini DM. Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol Sci. 2007;28(4):149–151. doi: 10.1016/j.tips.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 82.Baek ST, Kerjan G, Bielas SL, Lee JE, Fenstermaker AG, Novarino G, Gleeson JG. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron. 2014;82(6):1255–1262. doi: 10.1016/j.neuron.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27(6):549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16(9):566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chi JT, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100(11):6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100(11):6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schultz N, Marenstein DR, De Angelis DA, Wang WQ, Nelander S, Jacobsen A, Marks DS, Massague J, Sander C. Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-beta pathway and reveal microRNA regulation of TGFBR2. Silence. 2011;2:3. doi: 10.1186/1758-907X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh S, Wu X, Ljosa V, Bray MA, Piccioni F, Root DE, Doench JG, Boehm JS, Carpenter AE. Morphological profiles of RNAi-induced gene knockdown are highly reproducible but dominated by seed effects. PLoS ONE. 2015;10(7):e0131370. doi: 10.1371/journal.pone.0131370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, Dai H, Sachs AB, Linsley PS. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15(2):308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee HS, Seok H, Lee DH, Ham J, Lee W, Youm EM, Yoo JS, Lee YS, Jang ES, Chi SW. Abasic pivot substitution harnesses target specificity of RNA interference. Nat Commun. 2015;6:10154. doi: 10.1038/ncomms10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452(7187):591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash KN, Maier M, Nechev L, Rajeev KG, Read T, Rohl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, de Fougerolles A, Vornlocher HP, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA. 2008;105(33):11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garber K. Alnylam terminates revusiran program, stock plunges. Nat Biotechnol. 2016;34(12):1213–1214. doi: 10.1038/nbt1216-1213. [DOI] [PubMed] [Google Scholar]
- 94.Cullen BR. Enhancing and confirming the specificity of RNAi experiments. Nat Methods. 2006;3(9):677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- 95.Buehler E, Chen YC, Martin S. C911: a bench-level control for sequence specific siRNA off-target effects. PLoS ONE. 2012;7(12):e51942. doi: 10.1371/journal.pone.0051942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3(10):777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 97.Kaplow IM, Singh R, Friedman A, Bakal C, Perrimon N, Berger B. RNAiCut: automated detection of significant genes from functional genomic screens. Nat Methods. 2009;6(7):476–477. doi: 10.1038/nmeth0709-476. [DOI] [PubMed] [Google Scholar]
- 98.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmich F, Szczurek E, Kreibich S, Dilling S, Andritschke D, Casanova A, Low SH, Eicher S, Muntwiler S, Emmenlauer M, Ramo P, Conde-Alvarez R, von Mering C, Hardt WD, Dehio C, Beerenwinkel N. gespeR: a statistical model for deconvoluting off-target-confounded RNA interference screens. Genome Biol. 2015;16:220. doi: 10.1186/s13059-015-0783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marine S, Bahl A, Ferrer M, Buehler E. Common seed analysis to identify off-target effects in siRNA screens. J Biomol Screen. 2012;17(3):370–378. doi: 10.1177/1087057111427348. [DOI] [PubMed] [Google Scholar]
- 101.Yilmazel B, Hu Y, Sigoillot F, Smith JA, Shamu CE, Perrimon N, Mohr SE. Online GESS: prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinform. 2014;15:192. doi: 10.1186/1471-2105-15-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buehler E, Khan AA, Marine S, Rajaram M, Bahl A, Burchard J, Ferrer M. siRNA off-target effects in genome-wide screens identify signaling pathway members. Sci Rep. 2012;2:428. doi: 10.1038/srep00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olejniczak M, Galka-Marciniak P, Polak K, Fligier A, Krzyzosiak WJ. RNAimmuno: a database of the nonspecific immunological effects of RNA interference and microRNA reagents. RNA. 2012;18(5):930–935. doi: 10.1261/rna.025627.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 105.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11(1):75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53(6):1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3(3):505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 112.Vinther J, Hedegaard MM, Gardner PP, Andersen JS, Arctander P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 2006;34(16):e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bargaje R, Gupta S, Sarkeshik A, Park R, Xu T, Sarkar M, Halimani M, Roy SS, Yates J, Pillai B. Identification of novel targets for miR-29a using miRNA proteomics. PLoS ONE. 2012;7(8):e43243. doi: 10.1371/journal.pone.0043243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, Chung JY, Hewitt SM, Veenstra TD, Khan J. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10(2):479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan GR, Xu SH, Tan ZL, Liu L, He QY. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics. 2011;11(5):912–920. doi: 10.1002/pmic.201000539. [DOI] [PubMed] [Google Scholar]
- 116.Yang Y, Chaerkady R, Kandasamy K, Huang TC, Selvan LD, Dwivedi SB, Kent OA, Mendell JT, Pandey A. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol BioSyst. 2010;6(10):1873–1882. doi: 10.1039/c004401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009;9(5):1374–1384. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7(11):e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335(6068):552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56(1):104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104(49):19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to Argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3(5):e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13(8):1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28(4):598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10(11):1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18(9):1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7):607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8(7):559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 133.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18(2):237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8(8):e1002884. doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim S, Seo D, Kim D, Hong Y, Chang H, Baek D, Kim VN, Lee S, Ahn K. Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe. 2015;17(6):838–851. doi: 10.1016/j.chom.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 136.Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152(1–2):82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boudreau RL, Jiang P, Gilmore BL, Spengler RM, Tirabassi R, Nelson JA, Ross CA, Xing Y, Davidson BL. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81(2):294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, Chen Y, Choi I, Vourekas A, Won KJ, Liu C, Vivek K, Naji A, Friedman JR, Kaestner KH. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19(1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans . Nat Struct Mol Biol. 2010;17(2):173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc. 2007;2(9):2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 141.Alkan F, Wenzel A, Palasca O, Kerpedjiev P, Rudebeck AF, Stadler PF, Hofacker IL, Gorodkin J. RIsearch2: suffix array-based large-scale prediction of RNA–RNA interactions and siRNA off-targets. Nucleic Acids Res. 2017;45(8):e60. doi: 10.1093/nar/gkw1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32((web server issue)):W124–W129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10(1):12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12(7):1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, Wagner J, Rees E, Korn B, Frenzel C, Sachse C, Sonnichsen B, Guo J, Schelter J, Burchard J, Linsley PS, Jackson AL, Habermann B, Buchholz F. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4(4):337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- 147.Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014;42(12):8049–8061. doi: 10.1093/nar/gku480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348(5):1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 149.Fiszer A, Olejniczak M, Galka-Marciniak P, Mykowska A, Krzyzosiak WJ. Self-duplexing CUG repeats selectively inhibit mutant huntingtin expression. Nucleic Acids Res. 2013;41(22):10426–10437. doi: 10.1093/nar/gkt825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37(9):2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33(1):439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 154.Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013;3(6):1893–1900. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Puri N, Wang X, Varma R, Burnett C, Beauchamp L, Batten DM, Young M, Sule V, Latham K, Sendera T, Echeverri C, Sachse C, Magdaleno S. LNA incorporated siRNAs exhibit lower off-target effects compared to 2′-O methoxy in cell phenotypic assays and microarray analysis. Nucleic Acids Sympos Ser. 2008;52:25–26. doi: 10.1093/nass/nrn013. [DOI] [PubMed] [Google Scholar]
- 156.Mook O, Vreijling J, Wengel SL, Wengel J, Zhou C, Chattopadhyaya J, Baas F, Fluiter K. In vivo efficacy and off-target effects of locked nucleic acid (LNA) and unlocked nucleic acid (UNA) modified siRNA and small internally segmented interfering RNA (sisiRNA) in mice bearing human tumor xenografts. Artif DNA PNA XNA. 2010;1(1):36–44. doi: 10.4161/adna.1.1.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Werk D, Wengel J, Wengel SL, Grunert HP, Zeichhardt H, Kurreck J. Application of small interfering RNAs modified by unlocked nucleic acid (UNA) to inhibit the heart-pathogenic coxsackievirus B3. FEBS Lett. 2010;584(3):591–598. doi: 10.1016/j.febslet.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 158.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, Herdewijn P, Wengel J, Kjems J. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38(17):5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36(7):2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Burdick AD, Sciabola S, Mantena SR, Hollingshead BD, Stanton R, Warneke JA, Zeng M, Martsen E, Medvedev A, Makarov SS, Reed LA, Davis JW, 2nd, Whiteley LO. Sequence motifs associated with hepatotoxicity of locked nucleic acid-modified antisense oligonucleotides. Nucleic Acids Res. 2014;42(8):4882–4891. doi: 10.1093/nar/gku142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15(3):331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 162.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 163.Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol Cell. 2009;33(2):204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5(9):813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human Argonaute-2 in complex with miR-20a. Cell. 2012;150(1):100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336(6084):1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346(6209):608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. A dynamic search process underlies microRNA targeting. Cell. 2015;162(1):96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jo MH, Song JJ, Hohng S. Single-molecule fluorescence measurements reveal the reaction mechanisms of the core RISC, composed of human Argonaute2 and a guide RNA. BMB Rep. 2015;48(12):643–644. doi: 10.5483/BMBRep.2015.48.12.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162(1):84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yao C, Sasaki HM, Ueda T, Tomari Y, Tadakuma H. Single-molecule analysis of the target cleavage reaction by the Drosophila RNAi enzyme complex. Mol Cell. 2015;59(1):125–132. doi: 10.1016/j.molcel.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 172.Schirle NT, Kinberger GA, Murray HF, Lima WF, Prakash TP, MacRae IJ. Structural analysis of human Argonaute-2 bound to a modified siRNA guide. J Am Chem Soc. 2016;138(28):8694–8697. doi: 10.1021/jacs.6b04454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Onizuka K, Harrison JG, Ball-Jones AA, Ibarra-Soza JM, Zheng Y, Ly D, Lam W, Mac S, Tantillo DJ, Beal PA. Short interfering RNA guide strand modifiers from computational screening. J Am Chem Soc. 2013;135(45):17069–17077. doi: 10.1021/ja4079754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Suter SR, Sheu-Gruttadauria J, Schirle NT, Valenzuela R, Ball-Jones AA, Onizuka K, MacRae IJ, Beal PA. Structure-guided control of siRNA off-target effects. J Am Chem Soc. 2016;138(28):8667–8669. doi: 10.1021/jacs.6b06137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151(5):1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]