Abstract
Neural stem cells give rise to granule dentate neurons throughout life in the hippocampus. Upon activation, these stem cells generate fast proliferating progenitors that complete several rounds of divisions before differentiating into neurons. Although the mechanisms regulating the activation of stem cells have been intensively studied, little attention has been given so far to the intrinsic machinery allowing the expansion of the progenitor pool. The cell cycle protein Cdk6 positively regulates the proliferation of hippocampal progenitors, but the mechanism involved remains elusive. Whereas Cdk6 functions primarily as a cell cycle kinase, it can also act as transcriptional regulator in cancer cells and hematopoietic stem cells. Using mouse genetics, we show here that the function of Cdk6 in hippocampal neurogenesis relies specifically on its kinase activity. The present study also reveals a specific regulatory mechanism for Cdk6 in hippocampal progenitors. In contrast to the classical model of the cell cycle, we observe that the Cip/Kip family member p27, rather than the Ink4 family, negatively regulates Cdk6 in the adult hippocampus. Altogether, our data uncover a unique, cell type-specific regulatory mechanism controlling the expansion of hippocampal progenitors, where Cdk6 kinase activity is modulated by p27.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2832-x) contains supplementary material, which is available to authorized users.
Keywords: Adult neurogenesis, Dentate gyrus, Cell cycle, Cdks, Cdk inhibitors
Introduction
New neurons are produced throughout life in the dentate gyrus (DG) of the hippocampus. They originate from quiescent neural stem cells (type 1 cells) in the subgranular zone (SGZ) that give rise—after activation—to early progenitors (type 2a cells). Type 2a and their more differentiated progeny type 2b cells rapidly proliferate and expand the pool of newly generated cells, finally giving rise to type 3 cells, which will finally exit the cell cycle and differentiate into granule neurons. In the past 15 years, significant progresses have been made in describing hippocampal neurogenesis at cellular, molecular and functional levels [1]. However, further work remains to be done to elucidate the intrinsic regulatory mechanisms that control and coordinate stem cell activation, progenitor proliferation and cell cycle exit.
In differentiating cells, fate decisions often occur during the G1 phase of the cell cycle [2, 3]. A key step in G1 progression is the sequential phosphorylation of the retinoblastoma protein (Rb) by specific cyclin/cyclin-dependent kinases (Cdks) complexes, namely cyclin E–Cdk2 and cyclin D–Cdk4/6. Rb phosphorylation leads to the accumulation of E2f transcription factors in late G1 and activation of a transcriptional program that permits the entry into S phase. Cdk activity is not only regulated by cyclin binding, but also by interactions with endogenous Cdk inhibitors (CKI) including Ink4 (p15, p16, p18 and p19) and Cip/Kip (p21, p27 and p57) proteins. While Ink4 proteins solely inhibit Cdk4 and Cdk6, the Cip/Kip family can inhibit Cdk1, Cdk2, Cdk4 and Cdk6, but also paradoxically promote the functional assembly of cyclin D–Cdk4/6 complexes [4–7]. This raises the issue of which function of Cip/Kip proteins predominates in vivo, particularly during tissue homeostasis.
In the hippocampus, loss of Cdk6, but not Cdk2 or Cdk4, dramatically reduces the proliferation of progenitors [4, 8], but the cause of this specificity is unknown. Since Cdk4 and Cdk6 are both expressed in hippocampal progenitors, one explanation could be the presence of substrates unique to Cdk6. Alternatively, several studies recently demonstrated that Cdk6 also influences transcription in a kinase-independent fashion [9–11], suggesting that a non-canonical Cdk6 function may participate in the control of hippocampal neurogenesis. A large body of evidence has pointed out an essential role for Cip/Kip inhibitors in restraining proliferation in the hippocampus. Loss of either p21 or p27 causes increased proliferation of progenitors in normal and ischemic conditions [12–14], whereas p27 and p57 promote the quiescence of stem cells [15, 16]. However, there is no evidence yet for a role of Ink4 inhibitors in hippocampal proliferation, questioning which CKI are in charge of modulating Cdk6 activity in these cells.
In the present work, we used mouse genetics to address both the role and the regulation of Cdk6 in hippocampal neurogenesis. We show that mice bearing a kinase-dead allele of Cdk6 exhibit reduced proliferation of progenitors. Surprisingly, we observe a similar phenotype in mice expressing an Ink4-insensitive form of Cdk6, whereas loss of the Ink4 member p18 has no effect on proliferation. Finally, we provide evidence that the anti-proliferative effect of p27 during hippocampal neurogenesis is mediated through Cdk6. Altogether, our results shed new light on the specific interactions between cell cycle regulators underlying the production of new neurons in the hippocampus.
Materials and methods
Animals and administration of BrdU
p18−/− [17], Cdk6−/− [18], Cdk6K43M and Cdk6R31C [19] colonies were maintained on a pure C57BL/6J background. p27−/− [20] and all double transgenic mice were maintained on a C57BL/6J × SV129 background. Genotype was determined by PCR as previously described. Four- to 6-week-old mice were used for immunostaining, western blot and immunoprecipitation experiments, P6–P7 mice were used for western blot experiments and 12-week-old mice were used for the analysis of organ weight. For BrdU assays, BrdU (Sigma-Aldrich) was dissolved in 0.9% NaCl and 0.007 M NaOH and administrated at 50 mg/kg body weight by intraperitoneal (i.p.) injection. Mice were group housed in the animal facility at the University of Liège under standard conditions with food and water ad libitum and were maintained on a 12 h light/dark cycle. All animals were taken care of in accordance with the Declaration of Helsinki and following the guidelines of the Belgian Ministry of Agriculture in agreement with EC laboratory animal care and use regulation (2010/63/UE, 22 September 2010). All experiments were approved by the Animal Care Ethics Committee of the University of Liège. (Protocol no 1078).
Tissue processing and immunostaining
Mice were deeply anesthetized with 1% pentobarbital and perfused with 0.9% NaCl, followed by 4% paraformaldehyde (PFA), except for p18 mice whose brains were directly immersed in 4% PFA. Brains were post-fixed in 4% PFA overnight and then cryoprotected in 20% sucrose. 40-µm-thick free-floating coronal sections were sliced using cryostat (Microm, Prosan, Gent, Belgium) and stored at 4 °C in phosphate-buffered saline (PBS) supplemented with 0.02% sodium azide. For immunostaining, sections underwent antigen retrieval for 30 min at 95 °C (Target Retrieval Solution, Dako) and were incubated overnight at 4 °C with primary antibodies diluted in PBS containing 0.1% Triton, 0.1% Tween 20 and 5% normal donkey serum (blocking solution). The following primary antibodies were used: mouse anti-Ki67 (1:100, BD Biosciences Cat# 550609, RRID:AB_393778), rabbit anti-Ki67 (1:500, Cell Marque Corp Cat# 275R-16, RRID:AB_1158037), rat anti-BrdU (1:500, Bio-Rad/AbD Serotec Cat# OBT0030, RRID:AB_609568), goat anti-Sox2 (1:500, Santa Cruz Biotechnology Cat# sc-17320, RRID:AB_2286684), goat anti-NeuroD1 (1:500, Santa Cruz Biotechnology Cat# sc-1084, RRID:AB_630922), goat anti-Doublecortin (DCX) (1:500, Santa Cruz Biotechnology Cat# sc-8066, RRID:AB_2088494), rabbit anti-Cdk6 (1:1000, Atlas Antibodies Cat# HPA002637, RRID:AB_1233820) and rabbit anti-activated caspase 3 (AC3) (1:500, (Promega Cat# G7481, RRID:AB_430875). After washing in PBS, sections were incubated for 1 h at RT in blocking solution containing the corresponding donkey-raised secondary antibodies conjugated to Alexa Fluor 488, 555 or 647 (Thermo Fisher Scientific). Finally, sections were rinsed in PBS and mounted in VectaShield Hard Set medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). To perform BrdU detection, DNA was denatured with 2 N HCl for 30 min at 37 °C, followed by 0.1 M borate buffer, pH 8.5, for 10 min and immunostaining was performed as described above.
Quantification of cell numbers
All quantifications were performed by an experimenter blind to the experimental conditions. To evaluate cell proliferation and neurogenesis in the DG, we counted the number of Ki67+ and/or BrdU+ cells per section under a 40 × objective (Zeiss Axiovert 10VR microscope) in a sampling of every sixth 40-μm-thick coronal section between − 1.34 and − 3.52 mm from Bregma. DG surface in each section was measured using DAPI. Cell numbers were averaged and expressed as the number of positive cells per section or per mm2 of DG. For counts in the subventricular zone (SVZ), a sampling of every sixth 40-μm-thick coronal section from the most rostral crossing of the corpus callosum (1.10 from Bregma) to the start of the third ventricle (crossing of the anterior commissure, 0.14 from Bregma) was taken and the total number of Ki67+ cells per SVZ was determined using a semiautomatic stereology system (Mercator, Explora Nova). For analysis of double-labeled cells, at least 200 Ki67+ or BrdU+ cells randomly chosen within the DG were analyzed for their coexpression of Sox2/NeuroD1 or Ki67, respectively, using a confocal microscope (Nikon A1 system). For Sox2 or NeuroD1 counts, the percentage of Ki67+ cells that were positive for the marker was multiplied by the number of Ki67+ cells per mm2 of DG of the corresponding animal to obtain the number of double-labeled cells per mm2 of DG.
Western blotting and immunoprecipitation
Adult mice were deeply anesthetized with 1% pentobarbital and killed by cervical dislocation. P6–P7 mice were killed by decapitation. Brains were quickly removed and hippocampi were microdissected under a binocular and lysed on ice in a solution containing 150 mM NaCl, 50 mM Tris–HCl, 60 mM C3H7Na2O6P, 10 mM C6H5Na2O4P·2H2O, 500 mM NaF, 100 mM Na3VO4, 1% NP40 and protease inhibitors (Complete Mini, EDTA-Free, Roche). Protein concentration was determined using Bio-Rad Protein Assay (Bio-Rad). Samples were loaded with equal amounts of proteins (i.e., 30–50 µg) onto a 4–12% Bis–Tris Plus gel (Invitrogen). Following electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membrane (Millipore) before incubation with the following primary antibodies: rabbit anti-Cdk6 (1:500, Atlas Antibodies Cat# HPA002637, RRID:AB_1233820), rabbit anti-phospho-Rb (Ser780) (1:1000, Cell Signaling Technology Cat# 3590S, RRID:AB_2177182), mouse anti-cyclin D2 (1:200, Santa Cruz Biotechnology Cat# sc-53637, RRID:AB_782339) and mouse anti-β-actin horseradish peroxidase conjugated (1:25,000, Sigma-Aldrich Cat# A3854, RRID:AB_262011). Non-conjugated primary antibodies were detected using anti-mouse or anti-rabbit IgG horseradish peroxidase-conjugated antibody, respectively (1:10,000, Abcam). Immunolabeled proteins were revealed by the enhanced chemiluminescent detection system (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific). ImageJ [21] was used for optical density quantification. For immunoprecipitation, hippocampi were incubated in lysis buffer and proteins were extracted as described above. Immunoprecipitation was carried out O/N at 4 °C using the following primary antibodies: rabbit anti-Cdk6 (1:250, Santa Cruz Biotechnology Cat# sc-177, RRID:AB_631225), rabbit anti-p27 (1:250, Santa Cruz Biotechnology Cat# sc-528, RRID:AB_632129), followed by 1 h incubation with protein A/G plus agarose beads (Santa Cruz Biotechnology). Beads were washed six times with the lysis buffer and immunoprecipitates were then subjected to separation using 4–12% Bis–Tris Plus gel for subsequent western blot analysis.
Statistical analysis
Unless otherwise stated, data are reported as mean ± SEM. Statistical analyses were performed using GraphPad Prism (GraphPad). Unpaired two-tailed Student’s t tests were used to compare two groups, and one-way ANOVA followed by a Newman–Keuls post hoc test was used for multiple comparisons. Differences were considered statistically significant at p < 0.05.
Results
Cdk6 kinase activity regulates hippocampal neurogenesis
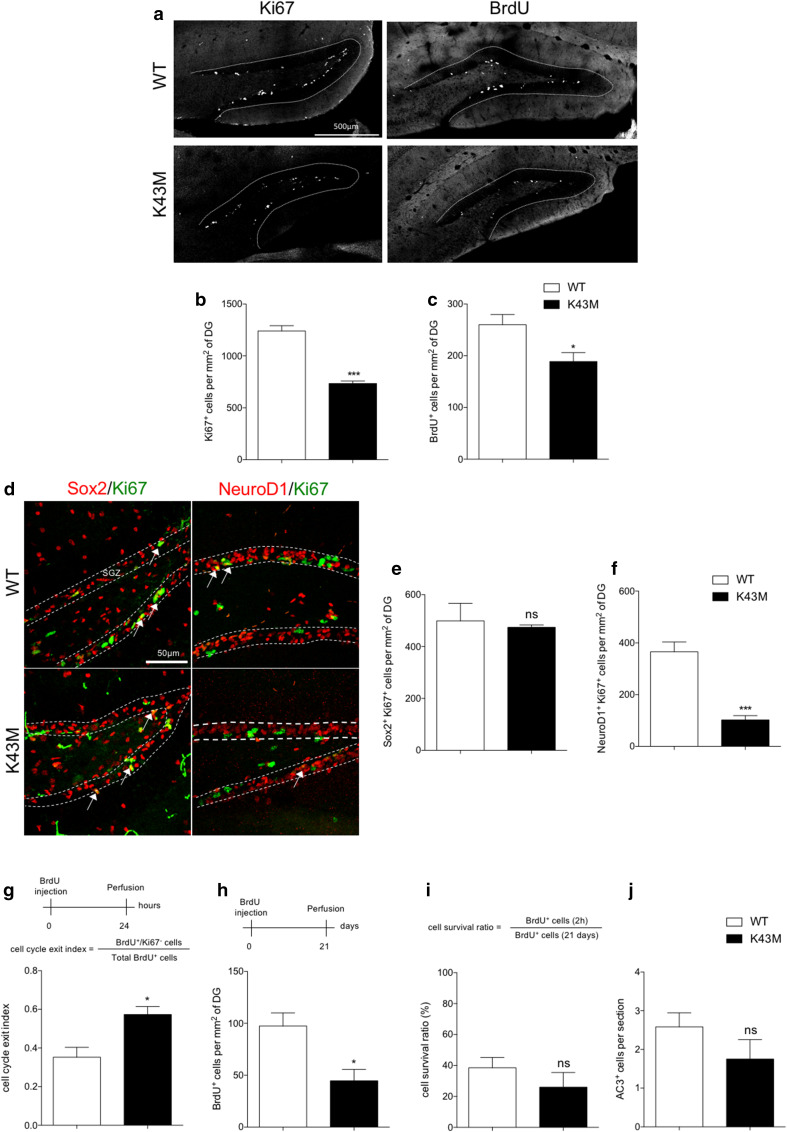
Using a straight knockout model, we previously identified a specific role for Cdk6 in the proliferation of hippocampal progenitors [4]. To determine whether the kinase activity of Cdk6 is required for the regulation of hippocampal proliferation, we analyzed the brain of mice bearing a Cdk6K43M knock-in allele. In these mice, Cdk6 is expressed at normal levels and retains the ability to bind D-type cyclins, Ink4 and Cip/Kip family proteins, but its kinase activity is lost [19]. We first confirmed there was no difference in Cdk6 level between WT and Cdk6K43M/K43M (K43M) hippocampi (Suppl. Fig. 1). We then quantified the number of cells expressing the cell cycle marker Ki67 in the DG of 4- to 6-week-old mice and observed a significant reduction in K43M animals compared to WT (Fig. 1a, b). These results were confirmed by counting the number of bromodeoxyuridine (BrdU)-positive cells 2 h after injection (Fig. 1a, c). While no difference in the number of proliferating type 1/2a (Ki67+/Sox2+) cells was observed between genotypes (Fig. 1d, e), we found a significant decrease in the number of proliferating type 2b/3 (Ki67+/NeuroD1+) cells in K43M animals (Fig. 1d, f). This data suggest the existence of a cell type-specific function for the kinase activity of Cdk6, which mostly controls the proliferation of “late” type 2b/3 hippocampal progenitors as previously shown in mice lacking Cdk6 [4]. Neuronal progenitors undergo a defined number of cell divisions before exiting the cell cycle and differentiating into postmitotic neurons. Hence, we hypothesized that a reduction of the progenitor pool in K43M animals could at least partially result from an enhanced rate of cell cycle exit. To test this hypothesis, WT and K43M mice were injected with BrdU and killed 24 h later to monitor the ratio between BrdU+/Ki67− cells and total BrdU+ cells, corresponding to the fraction of progenitors leaving the cell cycle within 24 h [22, 23]. In agreement with the proliferation data, the cell cycle exit index was increased in K43M animals (Fig. 1g). We next analyzed the consequence of the K43M mutation on the production of newborn DG neurons. For this purpose, WT and K43M mice were injected with BrdU once and killed 3 weeks later, when most of the selection/elimination process of newborn cells was over. We found a significant reduction in the number of BrdU+ cells in K43M animals (Fig. 1h). However, when we compared the number of BrdU+ cells 2 h or 3 weeks after a single BrdU injection, the ratio was not significantly different between WT and K43M animals (Fig. 1i). The number of apoptotic cells expressing activated caspase 3 (AC3) in the DG was also unchanged between genotypes, confirming that K43M mutation does not affect the survival of newborn cells. Overall, our data support a critical role of Cdk6 kinase activity in the proliferation of type 2b/3 hippocampal progenitors. More importantly, the phenotype of K43M mice closely resembles the phenotype described in Cdk6−/− (Cdk6 KO) mice, ruling out a major kinase-independent role for Cdk6 in hippocampal neurogenesis.
Fig. 1.
Cdk6 kinase activity regulates hippocampal neurogenesis. a Representative single-plane confocal images of Ki67 and BrdU staining in the DG of WT and K43M mice. Histograms showing the number of Ki67+ (b) and BrdU+ (c) cells per mm2 of DG in WT and K43M mice. d Representative single-plane confocal images of Sox2/Ki67 and NeuroD1/Ki67 double staining in the DG of WT and K43M mice. White arrows in d point to double-labeled cells. The dotted lines indicate the location of the SGZ. Histograms showing the number of Sox2+Ki67+ (e) and NeuroD1+Ki67+ (f) cells per mm2 of DG in WT and K43M mice. Schematic representation of the experimental design used to determine cell cycle exit (g), neurogenesis (h) and cell survival (i), accompanied by histograms showing the cell cycle exit index (g), the number of BrdU+ cells per mm2 (h), the cell survival ratio (i) and the number of AC3+ cells per section in the DG of WT and K43M mice (j). Data are presented as mean ± SEM and were analyzed by an unpaired two-tailed Student’s t test. N = 4 mice per genotype in b, c, e, f, h and i. N = 3 mice per genotype in j. N = 3 and 4 mice for WT and K43M, respectively in g. *p < 0.05; ***p < 0.001. SGZ subgranular zone, ns not significant
Functional consequences of altering Ink4-mediated inhibition of Cdk6 on hippocampal neurogenesis
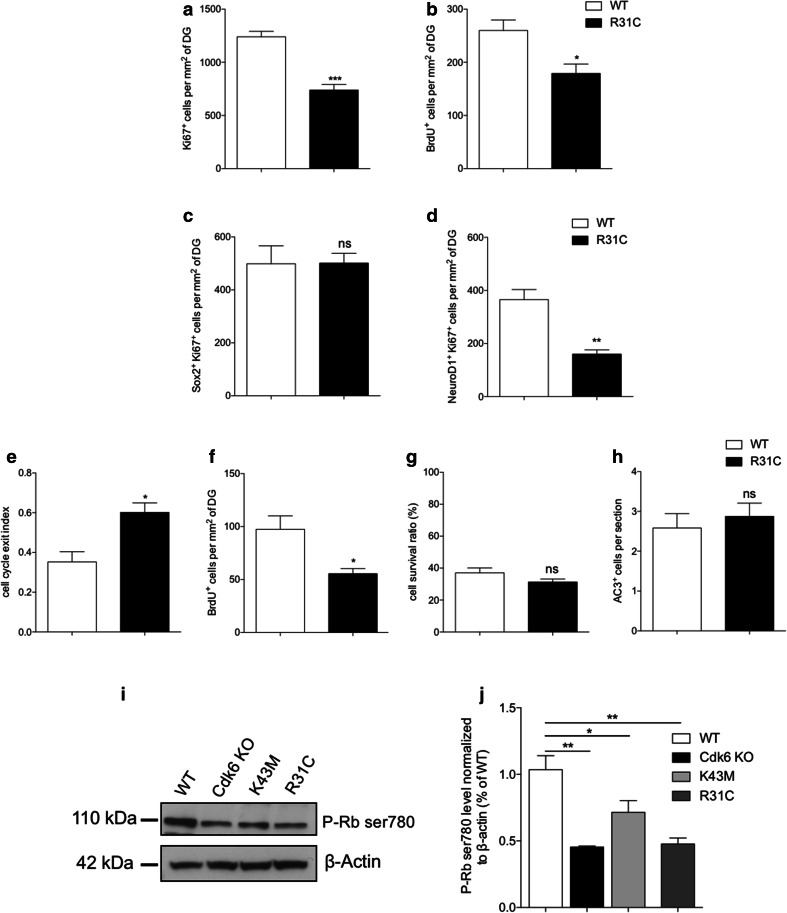
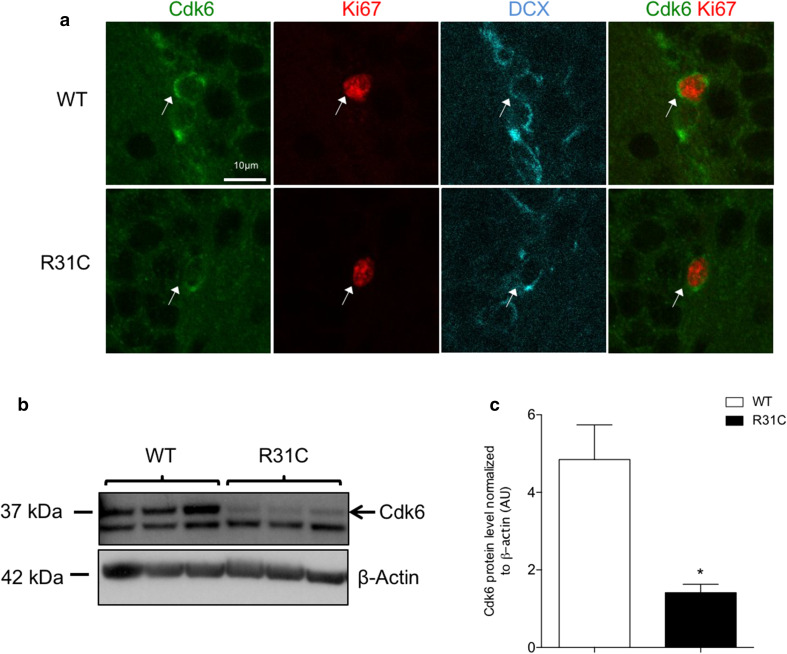
To date, the mechanisms regulating Cdk6 activity during hippocampal neurogenesis remain elusive. Since Ink4 proteins are specific inhibitors of cyclin D–Cdk4/6 complexes, we decided to address their importance in the proliferation of hippocampal progenitors. To that end, we took advantage of a gain-of-function allele, Cdk6R31C, that expresses a form of Cdk6 insensitive to Ink4-mediated inhibition, making it hyperactive [19]. In Cdk6R31C/R31C (R31C) mice, thymocytes—a cell type highly dependent on Cdk6 [18, 24]—display increased proliferation [19]. Unexpectedly, in the adult hippocampus, R31C mice exhibited a phenotype similar to Cdk6 KO and K43M mice. This includes decreased proliferation of type 2b/3 progenitors, increased cell cycle exit, reduced neurogenesis, no difference in AC3+ cells and decreased level of phospho-Rb Ser780, an indicator of Cdk6 activity (Fig. 2a–j). Importantly, we also observed a reduction of proliferation in the subventricular zone (SVZ), the other neurogenic zone, of R31C mice (Suppl. Fig. 2). Such reduction in proliferation might arise from diminished stability of the mutated Cdk6R31C protein. To test this hypothesis, we performed triple immunostaining for Cdk6, Ki67 and DCX to specifically analyze the expression of Cdk6 in proliferating type 2b/3 progenitors. We observed a weaker Cdk6 signal in Ki67+/DCX+ cells in R31C SGZ as compared to WT littermates (Fig. 3a). To confirm these results, we performed western blot analyses to measure Cdk6 protein levels on postnatal day 6 (P6)–P7 hippocampi, a stage corresponding to the peak of proliferation in the DG [25]. Cdk6 protein level was significantly decreased in R31C animals compared to WT (Fig. 3b, c). Altogether, these results indicate that the mutated Cdk6R31C form lacks stability in hippocampal progenitors, unfortunately preventing us from addressing the importance of Ink4 proteins on Cdk6 in these cells. However, these unanticipated data confirm the crucial role of Cdk6 in the proliferation of hippocampal progenitors.
Fig. 2.
Cdk6R31C mutation reduces hippocampal neurogenesis. Histograms showing, respectively, the number of Ki67+ (a), BrdU+ (b), Sox2+Ki67+ (c) and NeuroD1+Ki67+(d) cells per mm2 of DG in WT and R31C mice. Histograms showing, respectively, the cell cycle exit index (e), the number of BrdU+ cells per mm2 (f), the cell survival ratio (g) and the number of AC3+ cells per section in the DG of WT and R31C mice (h). i Representative western blot analysis of phospho-Rb Ser780 level in protein extracts from 4- to 6-week-old WT, Cdk6 KO, K43M and R31C mice. β-actin serves as a loading control. Histogram showing the quantification of the western blots illustrated in j. Data are presented as mean ± SEM and were analyzed by an unpaired two-tailed Student’s t test (a–h) or one-way ANOVA followed by a Newman–Keuls post hoc test (j). N = 4 mice per genotype in a–c, f and g. N = 3 mice per genotype in e, h and j. N = 4 and 3 mice for WT and R31C, respectively, in d. *p < 0.05; **p < 0.01; ***p < 0.001. ns not significant
Fig. 3.
Cdk6R31C mutation decreases Cdk6 protein level in hippocampal progenitors. a Representative single-plane confocal images of Cdk6 (green)/Ki67 (red)/DCX (cyan) triple staining in the SGZ of WT and R31C mice. White arrows point to triple-labeled cells. b Western blot analysis of Cdk6 expression in protein extracts from P6 to P7 WT and R31C hippocampi. Each well corresponds to one independent animal. β-actin serves as a loading control. The black arrow points to the specific Cdk6 band. c Histogram showing the quantification of the western blot depicted in b. Data are presented as mean ± SEM and were analyzed by an unpaired two-tailed Student’s t test. N = 3 mice per genotype. *p < 0.05. AU arbitrary unit
p27 inhibits Cdk6 during hippocampal neurogenesis
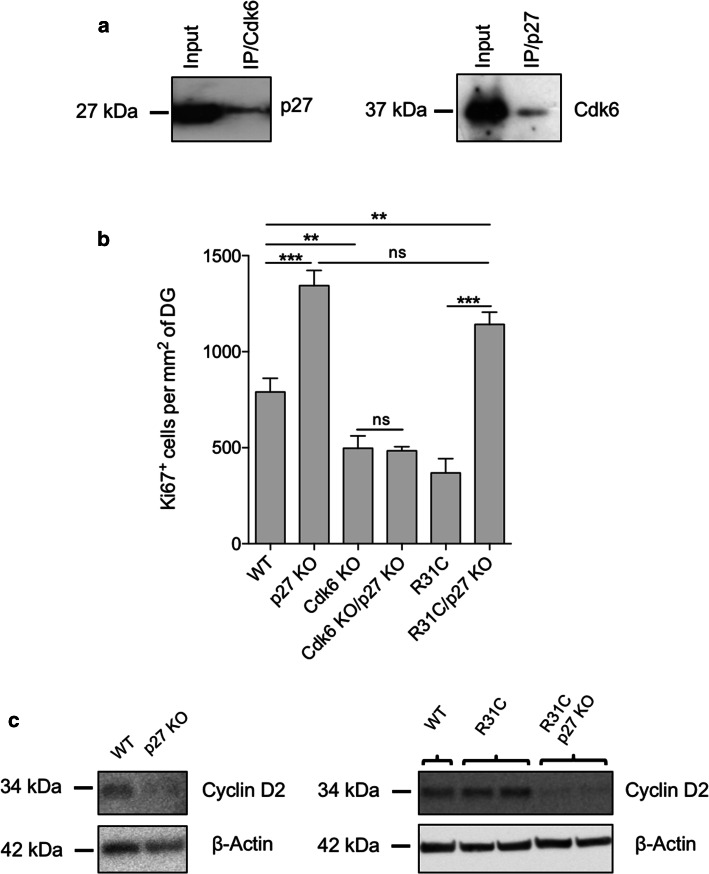
We then explored the regulation of Cdk6 by analyzing KO animals for endogenous Cdk inhibitors. Among the four Ink4 members, p16 is mainly expressed in aging tissues and has no role in hippocampal neurogenesis [26], p19 expression is restricted to postmitotic neurons and p15 is not present in the CNS [27, 28]. Therefore, the best candidate for Cdk6 regulation is p18, which is expressed in the SGZ of the DG [27, 29] and preferentially forms a complex with Cdk6 than Cdk4 [30]. We compared the number of Ki67+ cells between WT and p18−/− (p18 KO) mice and did not observe any difference between groups (Suppl. Fig. 3). This result suggests that p18 does not play an essential role in the proliferation of hippocampal progenitors. Besides Ink4 family members, Cip/Kip proteins, classically considered as Cdk1/2 inhibitors, can also interact with cyclin D–Cdk4/6 complexes. For instance, a report recently demonstrated that loss of p27 in the thymus leads to increased Cdk4/6 kinase activity [31]. In the hippocampus, p27 also hinders the proliferation of type 2/3 cells and induces cell cycle exit [13, 14], making it an excellent candidate to inhibit Cdk6. To establish whether Cdk6 interacts with p27 in the hippocampus, we performed immunoprecipitations and showed indeed the presence of p27/Cdk6 complexes (Fig. 4a). To determine if a link exists between Cdk6 and p27 in hippocampal progenitors in vivo, we then generated mice lacking both proteins. Cdk6−/−; p27−/− (Cdk6 KO/p27 KO) mice develop normally until adulthood. The lack of Cdk6 partially rescues the increased weight of several organs observed in the single p27−/− (p27 KO) mouse, including the spleen, the thymus and the testis (Suppl. Fig. 4). In the brain, as expected, individual loss of Cdk6 and p27 had antagonistic effects on hippocampal proliferation, but surprisingly Cdk6 KO/p27 KO mice exhibit reduced proliferation levels similar to single Cdk6−/− (Cdk6 KO) mice (Fig. 4b). Taken together, these data support the idea that the increased proliferation of type 2b/3 progenitors upon p27 deletion requires Cdk6 activity. Considering that p27 inhibits Cdk6 in hippocampal progenitors, we tested whether the reduced proliferation observed in R31C mice would be rescued by the absence of p27. In sharp contrast to Cdk6 KO/p27 KO mice, Cdk6R31C/R31C; p27−/− (R31C/p27 KO) mice displayed high levels of proliferation similar to those found in p27 KO mice (Fig. 4b). In addition, as previously documented [32, 33], loss of p27 induced a dramatic drop of the level of cyclin D2—an activator of Cdk6 essential for adult neurogenesis—in the hippocampus (Fig. 4c). Finally, the presence of the mutated Cdk6R31C form alone had no effect on cyclin D2 expression, whereas the decrease of cyclin D2 level observed in the absence of p27 was also present in R31C/p27 KO hippocampi (Fig. 4c). In addition, loss of p27 did not rescue Cdk6 decrease in R31C animals (data not shown). These observations further strengthen the inhibitory role of p27 on Cdk6 and also indicate that, in the absence of p27, even a small amount of Cdk6 and cyclin D2 can drive proliferation of hippocampal progenitors.
Fig. 4.
Cdk6 is downstream of p27 in hippocampal progenitors. a Cdk6 or p27 immunoprecipitation was carried out on protein lysates from 4- to 6-week-old mice hippocampi and followed by western blot using p27 or Cdk6 antibody, respectively. Corresponding western blots were performed on crude cell extracts (inputs). b Histogram showing the number of Ki67+ cells per mm2 of DG in WT (n = 5), p27 KO (n = 6), Cdk6 KO (n = 4), Cdk6 KO/p27 KO (n = 5), R31C (n = 3) and R31C/p27 KO (n = 3) mice. Data are presented as mean ± SEM and were analyzed by one-way ANOVA followed by a Newman–Keuls post hoc test. **p < 0.01; ***p < 0.001. c Western blot analysis of cyclin D2 expression in protein extracts from P6 to P7 WT, p27 KO, R31C and R31C/p27 KO hippocampi. Each well corresponds to one independent animal. β-actin serves as a loading control. ns not significant
Discussion
Previous studies have highlighted the essential roles for G1 regulators, such as Cdk6, cyclin D2 and Cip/Kip proteins during hippocampal neurogenesis [4, 5, 12–14, 16]. Although useful, these studies—based on germline knockout strains—cannot discriminate among the roles of specific structural domains for each protein and do not provide information about the interactions existing between these proteins in hippocampal progenitors. Here, we demonstrated that Cdk6 function during hippocampal neurogenesis relies essentially on its kinase activity. Surprisingly, our results also suggest that p27 is the main inhibitor of Cdk6 activity in hippocampal progenitors.
The canonical function of Cdk6 is to phosphorylate Rb to relieve the transcriptional repression of E2F-dependent genes [34]. On the other hand, several reports have identified kinase-independent roles for Cdk6. For example, Cdk6 prevents myeloid differentiation by interfering with the function of the transcription factor Runx1 [35]. In lymphoma cells, Cdk6 is part of a transcription complex that induces the expression of p16 and the pro-angiogenic factor VEGF-A [9]. Using mice expressing a kinase-dead form of Cdk6, Cdk6K43M, we demonstrated that the proliferation defect previously observed in Cdk6 KO hippocampi is mainly due to a lack of Cdk6 kinase activity and not to a transcriptional role of Cdk6. This situation is reminiscent of the thymus, another organ dependent on Cdk6 for its proliferation, since Cdk6 KO and K43M mice exhibit similar thymic hypoplasia [19]. Although Cdk4, a close homolog of Cdk6, also phosphorylates Rb efficiently and is highly expressed in thymocytes and adult hippocampal progenitors, it is not required for their proliferation [4, 36]. Similarly, even if these cell types express at least two types of cyclin D [19, 37, 38], only one of them (i.e., cyclin D2 in the hippocampus and cyclin D3 in the thymus) is essential for proliferation [5, 37]. Collectively, these data indicate that the catalytic activity of cyclin D2–Cdk6 complexes is a major and specific actor of hippocampal progenitor proliferation. Future studies are necessary to identify the major substrate of cyclin D2–Cdk6 complexes in hippocampal progenitors. Indeed, Anders et al. recently identified notable difference of substrate specificity between distinct cyclin D–Cdk4/6 complexes in human cells [39]. One candidate could be Enhancer of zeste homolog 2 (Ezh2), an epigenetic modifier crucial in maintaining high levels of proliferation and neurogenesis in the adult DG [40]. Indeed, Ezh2 is highly phosphorylated by cyclin D3–Cdk6 complexes and much less by cyclin D1–Cdk4.
Although several key cell cycle regulators of hippocampal neurogenesis have been identified, the molecular machinery responsible for fine-tuning their activity remains elusive. Our results shed new light on this regulation. Since (1) we did not observe any increase in proliferation in the hippocampus of p18 KO mice and (2) the Cdk6R31C mutation does not exacerbate the increased proliferation due to p27 loss, it is hence possible that Ink4 binding may not be a relevant mechanism of Cdk6 inhibition during progenitor expansion in the hippocampus. However, it is crucial to emphasize that another Ink4 member could compensate for p18 deficiency and that Cdk6 protein level is decreased in R31C hippocampal progenitors, putting into question some of our findings. Such reduction of Cdk6 level in R31C mice has been documented before [19, 41], but with different outcomes depending on the cellular context. In the thymus, R31C thymocytes exhibit significant increased proliferation as compared to WT [19], whereas Rodríguez-Díez et al. reported a defective progenitor potential in the hematopoietic lineage of R31C mice [41]. Altogether, these results suggest that the regulation, and also the role, of Cdk6 differs among different tissues and cell types. Moreover, given the limitations of the R31C model and the potential redundancies between Ink4 members, the best way to definitively assess the importance of Ink4-mediated inhibition of Cdk6 in hippocampal neurogenesis would be to simultaneously knockout all Ink4 members to prevent any compensatory mechanism.
In contrast to the clear inhibiting action of p27 on cyclin E–Cdk2 complexes, the interaction of p27 with cyclin D–Cdk4/6 complexes is much more complicated. Although p27 inhibits cyclin D–Cdk4/6 in several cellular contexts [42, 43], other studies isolated catalytically active p27–cyclin D–Cdk4/6 complexes and demonstrated that p27 was stabilizing these heterodimers [19, 32, 44], questioning whether the inhibition or activation function of p27 over Cdk4/6 prevails in vivo. Our data show here for the first time that increased organ weight due to p27 loss can be partially compensated by concomitant absence of Cdk6. Moreover, the increased proliferation observed in p27 KO hippocampi is completely abolished in the absence of Cdk6. These results indicate that in hippocampal progenitors, (1) Cdk6 is downstream of p27 and (2) p27 functions mainly as an inhibitor of Cdk6. This partial compensatory effect has also been observed with Cdk4. Indeed, Cdk4 loss partially rescues the increased body weight seen in p27 KO mice [45], whereas Cdk2 loss surprisingly does not [46]. Future work will determine if the inhibition of Cdk4/6 by p27, rather than its activation, is a global regulatory mechanism in every organ. Many studies reported a huge decrease of cyclin D1/D2/D3 levels in p27 KO cells, including the brain [32, 33, 47]. Interestingly, although cyclin D2 KO mice are virtually devoid of cell proliferation in the hippocampus [5], we noted that decreased level of cyclin D2 does not prevent increased proliferation in the absence of p27. Altogether, these observations strengthen the crucial role of cyclin D2–Cdk6 complexes in hippocampal progenitors and reinforce the idea that, in the absence of Cip/Kip inhibition, a small quantity of cyclin D–Cdk4/6 complexes is enough to ensure proliferative activity during tissue homeostasis.
In summary, our results illustrate the specificity of the intrinsic mechanisms driving progenitor proliferation during hippocampal neurogenesis. Tissue-specific cell cycle regulation may find its origin in the distinct expression pattern and function of the G1 regulatory proteins, but also, importantly, in the unique interactions existing between these proteins in a given cell type. Understanding the molecular basis of these differences is a prerequisite to precisely manipulating stem/progenitor cells in regenerative medicine and cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Pierre-Bernard Van Lerberghe for his technical assistance. LN and BM are, respectively, Senior Research Associate and Research Director of the Belgian National Funds for Scientific Research (FRS-FNRS). RV was a postdoctoral researcher of FRS-FNRS. NC and QM were research fellows with the Belgian Fund for Research in Industry and Agriculture (FNRS-FRIA). EG was supported by Marie Curie Action (Cofund) within the EU FP7 program. BM was funded by grants from the FRS-FNRS, the Fonds Léon Fredericq, the Fondation Médicale Reine Elisabeth, Belspo and the Belgian Science Policy (IAP-VII network P7/07).
Abbreviations
- AC3
Activated caspase 3
- DG
Dentate gyrus
- SGZ
Subgranular zone
- Rb
Retinoblastoma protein
- Cdk
Cyclin-dependent kinase
- CKI
Cyclin-dependent kinase inhibitors
- BrdU
Bromodeoxyuridine
- PFA
Paraformaldehyde
- PBS
Phosphate-buffered saline
- DAPI
4′,6-Diamidino-2-phenylindole
- SVZ
Subventricular zone
- PVDF
Polyvinylidene difluoride
Author contributions
NC, EG, RV and BM designed the concept of the experiments. NC, EG, RV, QM, SV, PB, LN and BM collected data and performed data analysis/interpretation. MGH, PH and LM provided study material. RV and BM wrote the manuscript. BM provided financial support.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
Footnotes
Nicolas Caron and Emmanuelle C. Genin: co-first authors.
Renaud Vandenbosch and Brigitte Malgrange: co-last authors.
References
- 1.Kempermann G, Song H, Gage FH. Neurogenesis in the adult hippocampus. Cold Spring Harb Perspect Med. 2015;5:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Dalton S. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25:592–600. doi: 10.1016/j.tcb.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beukelaers P, Vandenbosch R, Caron N, et al. Cdk6-dependent regulation of G(1) length controls adult neurogenesis. Stem Cells. 2011;29:713–724. doi: 10.1002/stem.616. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczyk A, Filipkowski RK, Rylski M, et al. The critical role of cyclin D2 in adult neurogenesis. J Cell Biol. 2004;167:209–213. doi: 10.1083/jcb.200404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Blain SW. Switching cyclin D–Cdk4 kinase activity on and off. Cell Cycle. 2008;7:892–898. doi: 10.4161/cc.7.7.5637. [DOI] [PubMed] [Google Scholar]
- 8.Vandenbosch R, Borgs L, Beukelaers P, et al. CDK2 is dispensable for adult hippocampal neurogenesis. Cell Cycle. 2007;6:3065–3069. doi: 10.4161/cc.6.24.5048. [DOI] [PubMed] [Google Scholar]
- 9.Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheicher R, Hoelbl-Kovacic A, Bellutti F, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125:90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jena N, Sheng J, Hu JK, et al. CDK6-mediated repression of CD25 is required for induction and maintenance of Notch1-induced T-cell acute lymphoblastic leukemia. Leukemia. 2016;30:1033–1043. doi: 10.1038/leu.2015.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pechnick RN, Zonis S, Wawrowsky K, et al. p21Cip1 restricts neuronal proliferation in the subgranular zone of the dentate gyrus of the hippocampus. Proc Natl Acad Sci USA. 2008;105:1358–1363. doi: 10.1073/pnas.0711030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu J, Takagi Y, Harada J, et al. p27Kip1Constrains proliferation of neural progenitor cells in adult brain under homeostatic and ischemic conditions. Stem Cells. 2009;27:920–927. doi: 10.1002/stem.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hörster H, Garthe A, Walker TL, et al. p27kip1 is required for functionally relevant adult hippocampal neurogenesis in mice. Stem Cells (Dayton, Ohio) 2017;35:787–799. doi: 10.1002/stem.2536. [DOI] [PubMed] [Google Scholar]
- 15.Furutachi S, Matsumoto A, Nakayama KI, Gotoh Y. p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 2013;32:970–981. doi: 10.1038/emboj.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreu Z, Khan MA, GonzÁlez-Gómez P, et al. The cyclin-dependent kinase inhibitor p27 kip1 regulates radial stem cell quiescence and neurogenesis in the adult hippocampus. Stem Cells. 2015;33:219–229. doi: 10.1002/stem.1832. [DOI] [PubMed] [Google Scholar]
- 17.Franklin DS, Godfrey VL, Lee H, et al. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 1998;12:2899–2911. doi: 10.1101/gad.12.18.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malumbres M, Sotillo R, Santamaría D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Hu MG, Deshpande A, Schlichting N, et al. CDK6 kinase activity is required for thymocyte development. Blood. 2011;117:6120–6131. doi: 10.1182/blood-2010-08-300517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fero ML, Rivkin M, Tasch M, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/S0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breunig JJ, Silbereis J, Vaccarino FM, et al. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glickstein SB, Monaghan JA, Koeller HB, et al. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29:9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu MG, Deshpande A, Enos M, et al. A requirement for cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res. 2009;69:810–818. doi: 10.1158/0008-5472.CAN-08-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicola Z, Fabel K, Kempermann G. Development of the adult neurogenic niche in the hippocampus of mice. Front Neuroanat. 2015;9:53. doi: 10.3389/fnana.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molofsky AV, Slutsky SG, Joseph NM, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zindy F, Soares H, Herzog KH, et al. Expression of INK4 inhibitors of cyclin D-dependent kinases during mouse brain development. Cell Growth Differ. 1997;8:1139–1150. [PubMed] [Google Scholar]
- 28.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 29.Gong S, Zheng C, Doughty ML, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 30.Noh SJ, Li Y, Xiong Y, Guan KL. Identification of functional elements of p18INK4C essential for binding and inhibition of cyclin-dependent kinase (CDK) 4 and CDK6. Cancer Res. 1999;59:558–564. [PubMed] [Google Scholar]
- 31.Berton S, Pellizzari I, Fabris L, et al. Genetic characterization of p27(kip1) and stathmin in controlling cell proliferation in vivo. Cell Cycle. 2014;13:3100–3111. doi: 10.4161/15384101.2014.949512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng M, Olivier P, Diehl JA, et al. The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryja V, Pacherník J, Faldíková L, et al. The role of p27(Kip1) in maintaining the levels of D-type cyclins in vivo. Biochim Biophys Acta. 2004;1691:105–116. doi: 10.1016/j.bbamcr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto T, Anderson K, Jacobsen SEW, et al. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26:2361–2370. doi: 10.1038/sj.emboj.7601675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow YH, Zhu XD, Liu L, et al. Role of Cdk4 in lymphocyte function and allergen response. Cell Cycle. 2010;9:4922–4930. doi: 10.4161/cc.9.24.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sicinska E, Aifantis I, Le Cam L, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/S1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 38.Glickstein SB, Alexander S, Ross ME. Differences in cyclin D2 and D1 protein expression distinguish forebrain progenitor subsets. Cereb Cortex. 2007;17:632–642. doi: 10.1093/cercor/bhk008. [DOI] [PubMed] [Google Scholar]
- 39.Anders L, Ke N, Hydbring P, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Ji F, Liu Y, et al. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014;34:5184–5199. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez-Díez E, Quereda V, Bellutti F, et al. Cdk4 and Cdk6 cooperate in counteracting the INK4 family of inhibitors during murine leukemogenesis. Blood. 2014;124:2380–2390. doi: 10.1182/blood-2014-02-555292. [DOI] [PubMed] [Google Scholar]
- 42.Bagui TK, Jackson RJ, Agrawal D, Pledger WJ. Analysis of cyclin D3–Cdk4 complexes in fibroblasts expressing and lacking p27(kip1) and p21(cip1) Mol Cell Biol. 2000;20:8748–8757. doi: 10.1128/MCB.20.23.8748-8757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato A, Takahashi H, Takahashi Y, Matsushime H. Inactivation of the cyclin D-dependent kinase in the rat fibroblast cell line, 3Y1, induced by contact inhibition. J Biol Chem. 1997;272:8065–8070. doi: 10.1074/jbc.272.12.8065. [DOI] [PubMed] [Google Scholar]
- 44.Blain SW, Montalvo E, Massagué J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 45.Pei X-H, Bai F, Tsutsui T, et al. Genetic evidence for functional dependency of p18Ink4c on Cdk4. Mol Cell Biol. 2004;24:6653–6664. doi: 10.1128/MCB.24.15.6653-6664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martín A, Odajima J, Hunt SL, et al. Cdk2 is dispensable for cell cycle inhibition and tumor suppression mediated by p27(Kip1) and p21(Cip1) Cancer Cell. 2005;7:591–598. doi: 10.1016/j.ccr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Cerqueira A, Martín A, Symonds CE, et al. Genetic characterization of the role of the Cip/Kip family of proteins as cyclin-dependent kinase inhibitors and assembly factors. Mol Cell Biol. 2014;34:1452–1459. doi: 10.1128/MCB.01163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.