Abstract
Methylation of histone H3 lysine 36 (H3K36) plays crucial roles in the partitioning of chromatin to distinctive domains and the regulation of a wide range of biological processes. Trimethylation of H3K36 (H3K36me3) demarcates body regions of the actively transcribed genes, providing signals for modulating transcription fidelity, mRNA splicing and DNA damage repair; and di-methylation of H3K36 (H3K36me2) spreads out within large intragenic regions, regulating distribution of histone H3 lysine 27 trimethylation (H3K27me3) and possibly DNA methylation. These H3K36 methylation-mediated events are biologically crucial and controlled by different classes of proteins responsible for either ‘writing’, ‘reading’ or ‘erasing’ of H3K36 methylation marks. Deregulation of H3K36 methylation and related regulatory factors leads to pathogenesis of disease such as developmental syndrome and cancer. Additionally, recurrent mutations of H3K36 and surrounding histone residues are detected in human tumors, further highlighting the importance of H3K36 in biology and medicine. This review will elaborate on current advances in understanding H3K36 methylation and related molecular players during various chromatin-templated cellular processes, their crosstalks with other chromatin factors, as well as their deregulations in the diseased contexts.
Keywords: Chromatin, Histone modification, H3K36 methylation, H3K36me2, H3K36me3, H3K27me3, DNA methylation, Epigenetics, Gene transcription, DNA damage repair, Splicing, Cancer, Methyltransferase, Demethylase
Introduction
In eukaryotic cells, the genetic information is stored in the form of chromatin. The basic structural unit of chromatin is the nucleosomal particle that consists of approximately 146 base pairs of DNA wrapped around by a histone octamer composed of two copies of each core histone, H3, H4, and H2A and H2B [1]. Variations in amino acid compositions of the core histones H2A and H3 produce histone variants with distinctive functions. For example, the replication-independent H3 variant termed as H3.3 differs from canonical H3 isoforms, H3.1 and H3.2, by four and five amino acid residues, respectively, and H3.3 is expressed throughout the cell cycle, with its deposition occurring preferentially at active gene bodies [2]. Besides packaging of DNA, the nucleosome is subject to a myriad of dynamic and reversible histone post-translational modifications (PTMs) that occur on both the flexible protruding N-terminal tails and the core globular domains, such as acetylation, methylation, phosphorylation, ubiquitylation and ADP-ribosylation, thereby partitioning chromatin fibers into different domains. It has been increasingly appreciated that histone PTMs can either directly remodel chromatin structure or provide docking platforms for downstream ‘reader’ proteins [1, 3, 4]. Histone PTM represents a fundamental means for regulating virtually all of the chromatin-templated processes.
Histone methylation is mainly attached to the basic side chains of lysine and arginine residues. Various histone lysine methylations impart either the activating or repressive effect on gene transcription, which depends on the site, degree of methylation, genomic location, and the status of other coexisting PTMs. In general, methylations of histone H3 at lysine 4, 36 and 79 (H3K4, H3K36 and H3K79) are linked to the transcriptionally active state, whereas methylations of histone H3 at lysine 9 and 27 (H3K9, H3K27) and histone H4 at lysine 20 (H4K20) are associated with gene silencing [4, 5]. In addition to gene transcription regulation, histone lysine methylations are also found involved in numerous other cellular events including DNA damage repair, DNA replication, and mRNA splicing.
Abundance and genomic distribution of H3K36 methylation
H3K36 methylation exists in three states—mono-, di- and trimethylation (H3K36me1, H3K36me2 and H3K36me3). Mass spectrometry-based measurements of histone modifications have quantified the relative abundance for different H3K36 methylation states in cells [6–8]. Regardless of H3 isoforms (H3.1, H3.2 or H3.3), H3 proteins with either un-methylated or di-methylated H3K36 are the two most abundant forms, each accounting for ~ 20–45% of total H3 in most of the examined mouse tissues, whereas those with H3K36me3 occur in around 5% of total H3 [7]. Similar patterns were found in stem cells and human cancer cells [6, 8, 9]. Thus, methylation of H3K36 appears to be abundant [6–9]. In addition, while H3 proteins with dual H3K36me3 and H3K27me3 are very rare, those with combinatorial low-degree methylations of H3K36 (H3K36me1/2) and H3K27 (H3K27me1/2) are fairly abundant [6, 7, 9, 10], indicating an intimate crosstalk between the two histone PTMs.
Chromatin immunoprecipitation (ChIP) following by sequencing (ChIP-seq) further reveals genomic landscapes of H3K36 methylation. There is a strong correlation of H3K36me3 to gene bodies of the actively transcribed genes [11], and this feature has aided in identification of the previously unknown transcripts such as long non-coding RNAs [12]. H3K36me2, however, shows a very distinctive genomic occupancy pattern. Unlike H3K36me3, H3K36me2 displays a significant enrichment in intergenic regions [13–17] and promoter-associated H3K36me2 was also reported in various cell types [13, 15, 17–19]. Distinctive distributions of H3K36me3 and H3K36me2 are likely due to different enzymes responsible for catalyzing these two methylations, which will be covered in the next sections. While the gene body-associated H3K36me3 is widely perceived as a hallmark of active gene transcription, H3K36me2 remains relatively understudied.
Evidence supporting the functional importance for H3K36 and its methylation
Genetic manipulation of the histone genes represents a powerful tool for assessing functional relevance of an individual histone amino acid that is subject to potential PTMs. While it remains technically infeasible to mutate histone genes in human cells due to existence of a total of 64 histone gene clusters among different chromosomes, such a strategy has been used in model organisms that carry only one H3 gene or one histone gene cluster. In yeast, mutating H3K36 to a non-modifiable residue such as arginine or alanine does not perturb overt growth but leads to a transcription elongation defect as assayed with 6-azauracil [20]. Moreover, yeast cells carrying H3K36A or H3K36R mutations or lacking the H3K36-specific methylase KMT3/Set2 (see also the below section) are sensitive to nutrient stress and exhibit a shortened life span due to loss of H3K36me3 and the resultant upregulation of cryptic transcripts [21, 22]. In Neurospora crassa, a type of filamentous fungi and bread mold, an amino acid substitution of H3K36 to leucine caused various phenotypes including slow growth, poor conidiation and female sterility [23]. A similar developmental requirement for H3K36 was also demonstrated using the fruit fly where mutating H3K36 into arginine led to lethality before completion of pupal development, concurrent increase in acetylation of histone H4, as well as globally dysregulated mRNA expression from the genomic regions that are demarcated by H3K36me3 in wild-type animals [24, 25]. The importance of H3K36 is further strengthened by recent identification of so-called ‘onco’-histones targeting the H3K36 site. ‘Onco’-histone is a term used to describe cancer-associated, recurrent mutations of histones that dominantly drive the malignant development [26]. In this case, the heterozygous mutation of H3K36 to methionine (H3K36M) frequently occurs in specific human cancer types including chondroblastoma [27] and papillomavirus (HPV)-negative head and neck squamous cell carcinomas (HNSCCs) [28], and such an H3K36M mutant was shown to interfere with the global landscape of H3K36 methylation in cancer cells [14, 17, 28–30]. Together, these works support that H3K36 methylation is an evolutionarily conserved event essential for normal development in various eukaryotic organisms.
Signaling of H3K36 methylation via ‘writer’, ‘reader’ and ‘eraser’
Using a variety of model organisms and cell systems, prior studies have identified the molecular players responsible for deposition, recognition or removal of H3K36 methylation, which are often termed as ‘writer’, ‘reader’ and ‘eraser’.
‘Writer’
KMT3/Set2 is the sole H3K36 methyltransferase in yeast that is responsible for production of all three states of H3K36 methylation, whereas there is a labor division and a more complicated landscape of H3K36 methylation in mammalian cells, with at least six different H3K36 ‘writers’ or methyltransferases (Table 1). Specifically, the enzymatic activities of the NSD family proteins (KMT3B/NSD1, KMT3G/NSD2/MMSET/WHSC1, KMT3F/NSD3/WHSC1L1), KMT2H/ASH1L and Metnase/SETMAR are restricted to lower degree methylation of H3K36me2/1 [31–34]; only KMT3A/SETD2, the mammalian homologue of yeast Set2 can further produce H3K36me3, and disrupting SETD2 results in global loss of H3K36me3 but not H3K36me2/1 in mammalian cells [14, 35–37]. Studies also show that NSD1 or NSD2 acts as one of the main ‘writers’ for H3K36me2 in embryonic stem cells (ESCs) [16] and cancer cell lines [13, 15], probably due to its relatively high expression in these cell types. As mentioned above, the epigenomic landscapes of H3K36me3 and H3K36me2 in mammalian cells are quite different, most likely resultant from differential targeting of their corresponding ‘writers’. Thus, H3K36me3 and H3K36me2/1 appear to have distinctive biological functions. Technically, knockout or knockdown of SETD2 [14, 35–37] and NSD1-3 [13, 15] provides a genetic approach for manipulating cellular levels of H3K36me3 and H3K36me2, respectively.
Table 1.
List of H3K36 methylation ‘writers’, ‘readers’ and ‘erasers’
| Domain | Function | References | |
|---|---|---|---|
| Writer | |||
| KMT3/SETD2 | SET | Trimethylation of H3K36 | [31, 33] |
| KMT3B/NSD1 | SET | Mono- and di-methylation of H3K36 | [31, 34] |
| KMT3G/NSD2 | SET | Mono- and di-methylation of H3K36 | [34] |
| KMT3F/NSD3 | SET | Mono- and di-methylation of H3K36 | [31, 34] |
| KMT2H/ASH1L | SET | Mono- and di-methylation of H3K36 | [31] |
| Metnase/SETMAR | SET | Di-methylation of H3K36 during DSB repair | [85] |
| Reader | |||
| MRG15 | Chromo | Reads H3K36me3 and recruits downstream effector proteins such as KDM5B | [66, 67] |
| PHF1; PHF19 | Tudor | Reads H3K36me3 and directs PRC2 complex to active genes | [48, 72, 73] |
| DNMT3B | PWWP | Reads H3K36me3 and guides DNMT3B to catalyze gene-body DNA methylation | [40, 68, 69, 82] |
| DNMT3A | PWWP | Reads H3K36me2/3 in vitro | [40, 81, 82] |
| MSH6 | PWWP | Reads H3K36me3 and recruits the mismatch sensor hMutSa during DNA mismatch repair | [40, 83] |
| LEDGF/PSIP1 | PWWP | Reads H3K36me3 and recruits CtIP to facilitate HR repair of DSB; also reads H3K36me3 and recruits splicing machinery; also reads H3K36me2 and recruits MLL to oncogene promoters in leukemia | [19, 86, 88, 91] |
| ZMYND11/BS69 | PWWP | H3.3K36me3-specific reader | [40, 50, 92] |
| Eraser | |||
| KDM2A/2B | JmjC | Demethylates H3K36me2 | [51, 52] |
| KDM4A/4B/4C | JmjN;JmjC | Demethylates H3K9me3 and H3K36me3 | [53, 55] |
‘Reader’
Following its deposition, H3K36 methylation can serve as docking sites for recruiting the so-called ‘reader’ or effector proteins. Members of ‘Royal family’ protein domains, which include Tudor, PWWP, and chromodomain [38–40], have been documented to engage and ‘read’ different degrees of H3K36 methylation (Table 1; Fig. 1). These ‘reading’ domains establish a ‘cage’-like structure involving a cluster of aromatic residues for engagement of the side chain of the methylated H3K36 (Fig. 1), a phenomenon also seen with ‘reader’ domains specific for other histone methylation as elaborated extensively in previous reviews [39–43]. As the site of H3K36 is in close proximity to DNA in the nucleosomal context, the H3K36 methylation-‘reading’ Royal domains including PWWP [40, 44] and Tudor [45] can synergistically bind H3K36 methylation and DNA. Here, binding to DNA is mediated through a set of electrostatic interactions between the phosphate backbone of DNA and a highly positively charged patch on the surface of Royal domain, therefore, robustly increasing the binding affinity to modified nucleosome without displaying DNA sequence specificity [40, 44, 45].
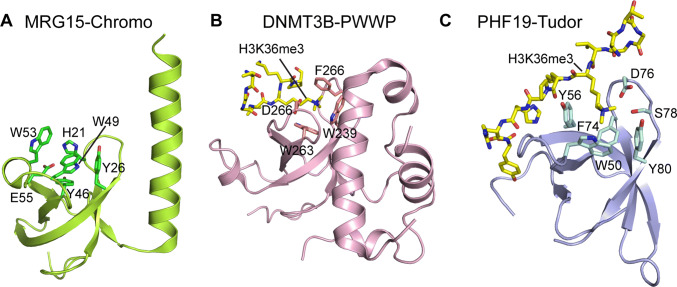
Fig. 1.
Structures of the H3K36 methylation-specific ‘reader’ domains, as exemplified by a the chromodomain of MRG15 (PDB: 2F5K), b the PWWP domain of DNMT3B (PDB: 5CIU) and c the Tudor domain of PHF19 (PDB: 4BD3). The H3K36me3-engaging residues are labeled in each panel, with the histone peptide shown in gold
Studies also reveal distinctive mechanisms by which H3K36 methylation-specific ‘readers’ elicit their respective biological consequences. First, binding to H3K36 methylation by certain ‘readers’ can alter physical properties of nucleosomes, which are again due to proximity of H3K36 to DNA in the nucleosomal context. For instance, it was shown that engagement of H3K36me3 by the Tudor domain of PHF1 enhances accessibility of DNA to regulatory proteins within the H3K36me3-containing nucleosomes [45, 46]. Such a role of PHF1 may be involved in modulation of the methyltransferase activity of Polycomb repressive complex 2 (PRC2), to which PHF1 interacts, or PHF1-mediated DNA damage repair occurs [47, 48]. Additionally, as a more commonly appreciated mode-of-action, H3K36 methylation-specific ‘readers’ can recruit various interacting factors, which carry activities for regulating the chromatin-templated processes. In particular, a list of PWWP domain-containing proteins bind H3K36me3/2 (Table 1) and are known to be involved in gene transcription, chromatin modification and remodeling, mRNA splicing and DNA damage repair, thus linking H3K36me3/2 to modulation of these essential cellular events. Interestingly, NSD2, an H3K36me2 ‘writer’, also harbors a PWWP domain that can bind H3K36me2 and such ‘reading’ of the catalytic products by ‘writer’ suggests a mechanism for genomic spreading and propagation of H3K36me2 [49], a PTM that indeed often displays a pattern of large intergenic domains as shown by ChIP-seq [14, 16]. Furthermore, the H3K36me3-binding PWWP motif of ZMYND11/BS69 is flanked by a bromodomain and an embedded zinc finger, which create a second ‘pocket’ for engaging the H3.3 isoform-unique residue serine 31 (H3.3S31), thus providing a delicate strategy for ‘reading’ H3K36me3 in an H3 isoform-specific manner [50]. Together, recruitment of ‘readers’ represents an important mechanism through which H3K36 methylation elicits its functional consequences. Manipulating the interaction between H3K36 methylation and its specific ‘readers’ provides a means for experimentally dissecting the biology related to this histone PTM.
‘Eraser’
H3K36 methylation is reversible and can be actively removed by certain members of the JmjC signature domain-containing histone lysine demethylases or ‘erasers’ (Table 1), which include the KDM2/JHDM1 and KDM4/JHDM3/JMJD2 family proteins [51–55]. Functional and structural analyses uncovered that the conserved JmjC domain coordinates two cofactors, Fe(II) and α-ketoglutarate, to mediate the hydroxylation and ultimate demethylation of methyl groups. The KDM2/JHDM1 group of ‘erasers’ specifically demethylates H3K36 methylation with a preference for H3K36me2; in contrast, the KDM4/JMJD2/JHDM3 group enzymes exhibit dual-substrate specificity towards H3K9me3 and H3K36me3 [51–55].
Biological processes regulated by H3K36 methylation and associated factors
H3K36me3-mediated silencing of spurious intragenic transcripts
H3K36me3 is a hallmark of active transcriptional elongation. In yeast, Set2 associates with the serine 2 phosphorylated, and to a lesser extent, the serine 5 phosphorylated C-terminal domain (CTD) of RNA polymerase II (RNAPII) through its Set2–Rpb1-interacting (SRI) domain, thereby traveling across the gene body together with RNAPII to lay down H3K36me3 during elongation [20, 56–58] (Fig. 2a). Such transcription-associated H3K36me3 recruits the Rpd3S histone deacetylase complex to the actively transcribed regions, inducing local histone deacetylation to suppress transcription initiation from cryptic alternative start sites within the gene body [59, 60] (Fig. 2a). Mechanistically, recognition of the gene-body-associated H3K36me3 relies on the combined action of the chromodomain of Eaf3, a subunit of Rpd3S, and the plant homeodomain (PHD) of the Rco1 subunit [61–63]. Similarly, SETD2 in mammalian cells interacts with the elongation-competent form of RNAPII to carry out H3K36me3 at the gene bodies of actively transcribed genes [64, 65] (Fig. 2b). Although silencing of SETD2 also leads to aberrant intragenic transcription initiation, no change in histone acetylation levels happens at the affected genes [64], indicating a regulatory mechanism distinctive from what was observed in yeast.
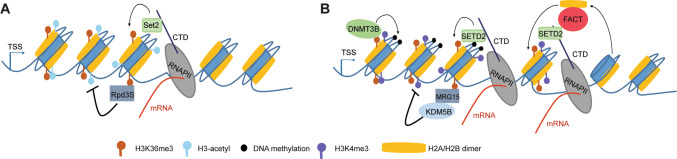
Fig. 2.
Suppression of cryptic intragenic transcripts by H3K36me3. Both the yeast Set2 and the mammalian SETD2 bind the elongating RNAPII and decorate the gene body regions with H3K36me3. In yeast (a), the H3K36me3 recruits the Rpd3S histone deacetylase complex to deacetylate the transcribed body regions to suppress spurious intragenic transcripts. Unlike yeast, the mammalian cells (b) employ different H3K36me3 ‘readers’, a de novo methyltransferase DNMT3B and the MRG15–KDM5B complex, to methylate DNA and remove H3K4me3, respectively, thus leading to inactivation of intragenic promoters; as well, SETD2 recruits the FACT complex to facilitate nucleosome structure recovery after RNAPII passage, thus preventing RNAPII entry into the intragenic promoters
Indeed, recent studies have uncovered multiple SETD2- and H3K36me3-directed signaling pathways for ensuring fidelity of gene transcription. First, similar to its yeast orthologue Eaf3, MRG15 also ‘reads’ H3K36me3 via a conserved chromodomain [66] (Figs. 1a, 2b; Table 1). Here, besides a mammalian Rpd3S-like complex [67], MRG15 also interacts with KDM5B/JARID1B, a histone ‘eraser’ specific to H3K4me3/2. H3K4me3 is a histone PTM correlated with activation of gene promoters. ChIP-seq in mouse ESCs revealed a predominant targeting of KDM5B to intragenic regions in the MRG15-dependent fashion, and depletion of either MRG15 or KDM5B resulted in the increased levels of intragenic H3K4me3 and concurrent cryptic intragenic transcription [67]. This work demonstrated an important requirement of a pathway involving H3K36me3–MRG15–KDM5B for ‘erasing’ H3K4me3 and restricting the usage of alternative intragenic promoters within the gene body, thus safeguarding transcription fidelity during the wake of RNAPII.
Also, recent works found that the SETD2-catalyzed H3K36me3 is recognized by the PWWP domain of DNMT3B (Figs. 1b, 2b; Table 1), which then catalyzes the de novo DNA methylation at the gene body of the actively transcribed gene [68, 69]. In mouse ESCs, SETD2 knockdown resulted in a drastic reduction of H3K36me3, a concomitant loss of DNMT3B binding to gene bodies, and activation of cryptic intragenic transcription initiation. Importantly, loss of Dnmt3b also resulted in an increase of spurious transcripts at the regions affected by SETD2 knockdown and such an abnormal increase in spurious transcripts was suppressed by restoration of a wild-type Dnmt3b but not its enzymatic-dead mutant or a PWWP mutant defective in ‘reading’ H3K36me3. Clearly, there exists an additional signaling pathway in mammalian cells involving SETD2–H3K36me3–DNMT3B, which operates to maintain high degree of DNA methylation at the gene body to protect from spurious RNAPII entry and to ensure transcript fidelity [68, 69].
Further, it is noteworthy that SETD2 per se was also documented to attenuate spurious intragenic transcription through recruiting the FACT chromatin-remodeling complex [64]. Specifically, FACT is recruited in a SETD2-dependent manner to the gene body of actively transcribed genes, where it not only facilitates the RNAPII passage by removing H2A–H2B dimers during the elongation phase but also helps to re-establish nucleosomes after RNAPII has passed through (Fig. 2b). Such activities of the FACT remodeling complex also contribute to prevention of cryptic intragenic promoter activation [64].
Together, to ensure fidelity of gene transcription in the wake of RNAPII, cells employ multiple mechanisms involving Set2/SETD2 and the catalyzed H3K36me3 to suppress spurious transcriptional initiation within the gene bodies.
Shaping chromatin landscapes via crosstalk between H3K36 methylation and PRC2
H3K36 methylation can also mediate its biological outputs through its antagonistic effect on other biologically important histone PTMs. Initially, the in vitro histone methyltransferase assays showed that the H3K27me3-catalyzing activity of PRC2 is impaired in cis by the presence of H3K36me2 or H3K36me3 [70]. A similar observation was seen for H3K4me3 [70]. These in vitro studies indicated a mechanism by which the transcriptionally active chromatin marks counteract the actions of PRC2. Such an antagonism between H3K36me3/2 and PRC2 was substantiated by the in vivo results obtained from various tissue types that include ESCs [16], adipose tissue [71] and cancer cell lines [6, 14, 37]. These findings collectively support that restricting genomic distribution of the PRC2-catalyzed H3K27me3 represents one of the main mechanisms underpinning the biological functions of H3K36me3/2. However, unlike H3K27me3, H3K27me2 often co-exists with H3K36me2/1 on the same H3 tails in cells [6, 7, 9, 10], indicating a more complicated interplay of the two PTMs. In a recent mass spectrometry-based analysis of PRC2-interacting proteins carried out in mouse ESCs, Streubel et al. uncovered Nsd1 [16]. Consistently, ChIP-seq found co-localization of the Nsd1-mediated H3K36me2 with the PRC-catalyzed H3K27me2, and Nsd1 depletion led to genome-wide loss of H3K36me2 and concurrent expansion of H3K27me3 [16] (Fig. 3). Furthermore, in ESCs, PRC2 also associates with the H3K36me3/2 ‘erasers’ [72, 73], as well as the polycomb-like proteins (PHF1 and PH19), which can ‘read’ H3K36me3/2 thereby assisting with PRC2’s intrusion and spreading into the H3K36me3-marked active genes (Figs. 1c, 3; Table 1) [48, 72, 73]. Apparently, biological outcomes of these above complexes oppose each other, thus enabling conversion between different chromatin states, either active, repressed or poised. It remains to be determined how these antagonizing complexes reach a fine balance during dynamic modulation of H3K36me3/2 and H3K27me3/2.
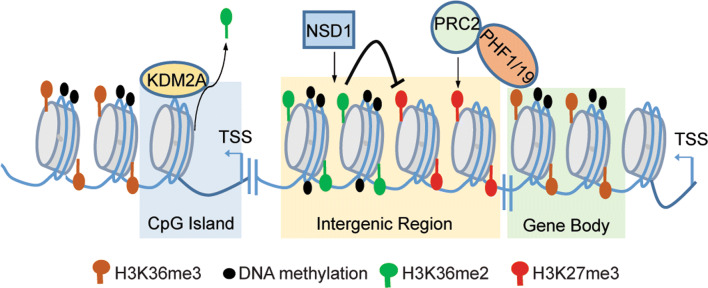
Fig. 3.
Crosstalk of H3K36 methylation with H3K27 methylation and DNA methylation. In the intergenic regions, the NSD1-catalyzed H3K36me2 prevents spreading of the PRC2-catalyzed H3K27me3, and loss of NSD1 causes genome-wide loss of H3K36me2 and DNA methylation, as well as concomitant gain of H3K27me3. Intrusion and spreading of the PRC2-catalyzed H3K27me3 into the active gene bodies are assisted by PHF1 or PHF19, which mediates ‘reading’ of H3K36me3. Furthermore, CpG island elements recruit KDM2A to remove H3K36me2
Shaping epigenomic landscapes via crosstalk between H3K36 methylation and DNA methyltransferases
Growing evidence has linked H3K36 methylation to DNA methylation. First, NSD1 abnormality or mutation is a hallmark for Sotos syndrome, a genetic disorder characterized by facial dysmorphism, macrocephaly, learning disability, and childhood overgrowth [74], and DNA methylome analysis of blood samples from Sotos syndrome patients carrying NSD1 abnormalities has revealed loss of DNA methylation at 7,038 CpG sites, which account for 99.3% of all the differentially methylated CpG sites [75]. Importantly, these hypomethylated genomic sites were found enriched in genes related to neural and cellular development pathways [75]. Remarkably, a comprehensive analysis of genomic datasets from a large set of human tumor samples also found a strong correlation between NSD1 mutation and DNA hypo-methylation [76]. As well, using unsupervised hierarchical clustering of DNA methylation dataset obtained from HPV-negative HNSCCs, Papillon-Cavanagh et al. uncovered a previously unappreciated HNSCC subtype that is featured with either H3K36M substitution or NSD1 mutations, with the most pathogenic missense mutations of NSD1 clustering around its catalytic SET domain [28]. Interestingly, reminiscent to the NSD1-mutated Sotos syndrome, this HNSCC subtype is characterized by global DNA hypo-methylation, particularly at enhancers and intergenic regions rather than promoters [28]. The fact that H3K36me2 reduction is a shared epigenomic alteration in both NSD1-mutated Sotos syndrome and HNSCC patients supports a potential role of H3K36me2 in regulating DNA methylation (Fig. 3). The exact underlying mechanisms remain to be defined. Another evidence highlighting the interplay between H3K36me2 and DNA methylation is from an observation that CpG islands (CGIs) are normally refractory to DNA methylation and also are uniquely devoid of H3K36me2 [77]. Here, un-methylated CGIs serve as the initial recruitment and nucleation sites for the H3K36-specific ‘eraser’ KDM2A through its zinc finger-CxxC domain [77] (Fig. 3). Similarly, KDM2B specifically binds to non-methylated CGIs genome-wide via its zinc finger-CxxC domain. In contrast to KDM2A, KDM2B is preferentially enriched at Polycomb-repressed CGIs where it recruits PRC1 complexes to suppress lineage-specific genes in mouse ESCs [78, 79]. More interestingly, a recent work further demonstrated that H3K36me2 marks are tightly bound by a cysteine-rich domain of Ten-eleven translocation 2 (TET2), a DNA dioxygenase and demethylase, implying that H3K36me2 may also act to fine-tune the dynamic processes of DNA methylation and demethylation [80].
Additionally, as discussed in the above section, the SETD2-mediated H3K36me3 tethers DNMT3B to gene bodies of the active genes via a PWWP ‘reader’ domain (Fig. 1b); consequently, DNA methylation is induced at high levels to prevent cryptic intragenic transcription (Fig. 2b). Notably, various in vitro studies also indicated that the PWWP domain of DNMT3A behaves similarly as that of DNMT3B, with a preferential binding to H3K36me2/3 [81, 82]. However, as revealed by ChIP-seq studies, the DNMT3A’s genomic binding does not follow the transcriptional elongation activity and its binding pattern is not altered following cellular loss of H3K36me3, suggesting a functional difference between DNMT3A and DNMT3B as for their interplay to H3K36 methylation [68, 69].
Overall, accumulating evidence pinpoints the intimate, yet rather complicated crosstalks among H3K36 methylation (either H3K36me3 or H3K36me2), DNMT3B/3A, PRC2 and H3K27me2/3. Further investigation is warranted to delineate the exact regulatory details under different cellular and chromatin contexts.
H3K36 methylation acting to recruit DNA damage repair factors
The DNA mismatch repair (MMR) pathway safeguards the integrity of cellular genomes by correcting replication- and damage-associated base–base mismatches and small insertion–deletion loops. Cells lacking SETD2 displayed an impaired chromatin occupancy of the MMR machinery, resulting in the elevated frequency of microsatellite instability and spontaneous genetic mutation [83]. Mechanistically, H3K36me3 serves as the docking site for recruiting hMutSa (hMSH2–hMSH6), a key mismatch sensor, through an H3K36me3-‘reading’ PWWP domain of the hMSH6 subunit (Fig. 4; Table 1) [83]. The recruited hMSH2–hMSH6 complex then surveys the naked DNA for lesion repair during both DNA replication and gene transcription. Strikingly, the abundance of H3K36me3 fluctuates in response to cell cycle changes, with it being highly enriched in the late G1/early S, the phase when DNA mismatches usually occur, and being largely removed in late S phase and maintained at lower levels throughout the G2/M phase [83]. In support of a critical involvement of H3K36me3 in MMR, the cancer-related mutation of histone H3 Gly34 (H3G34), a histone residue in the vicinity of H3K36, to a bulky side chain residue was shown to suppress MMR and to facilitate genome instability by sterically hindering the interaction of H3K36 to both SETD2 and hMutSα [84].
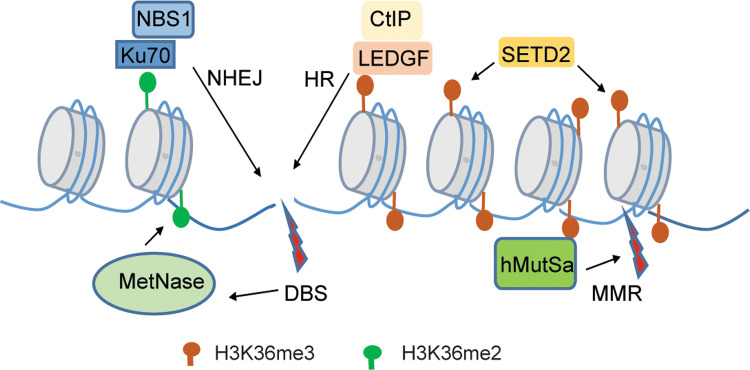
Fig. 4.
H3K36 methylation-mediated promotion of DNA damage repair via both DNA mismatch repair (MMR) and double-strand break (DSB) pathways. Specifically, the SETD2-catalyzed H3K36me3 is recognized by the mismatch sensor hMutSa, thereby promoting MMR during active gene transcription. In response to DSB, Metnase is activated to catalyze H3K36me2 near the DSB site, which in turn recruits the Ku70–NBS1 complex for damage repair via NHEJ. Unlike H3K36me2, the SETD2-catalyzed H3K36me3 also favors HR repair of DSB by orchestrating assembly of an H3K36me3 ‘reader’ complex, LEDGF–CtlP
In addition to MMR, H3K36 methylation is also implicated in the repair of double-strand DNA breaks (DSBs) by non-homologous end joining (NHEJ) and homologous recombination (HR) pathways. Here, Fnu et al. showed that H3K36me2 is markedly induced by Metnase (Table 1), a methylase possessing a SET domain and a transposase nuclease domain, at regions adjacent to the DSB site [85]; such an induced H3K36me2 then enhances recruitment of the NHEJ pathway proteins such as Ku70 and NBS1 for the error-prone DSB repair [85] (Fig. 4). In contrast, during the error-free HR repair of DSB, the constitutive anchoring of LEDGF/PSIP1 to the H3K36me3-marked chromatin through its PWWP ‘reader’ domain can facilitate timely recruitment of CtIP, a DNA damage response factor, to the break sites, thus facilitating the resection events associated with HR [86] (Fig. 4; Table 1). A more recent study also showed that, akin to γ-H2AX, temporal changes of global H3K36me3 levels are induced post-DSB in a SETD2-dependent manner [87]; such an elevation of H3K36me3 peaks 30 min post-exposure to the DSB-inducing agents and drops back to basal levels after 4 h [87]. Moreover, the increased H3K36me3 demarcates the immediate surrounding regions around DSBs and recruits LEDGF, which in turn brings in the histone acetyltransferase KAT5/TIP60 to catalyze acetylation of histone H4 Lys 16 (H4K16ac). H4K16ac and TIP60 are known to mitigate the interaction between the Tudor domain of 53BP1 and H4K20me2, and promote recruitment of BRCA1, thus favoring HR for DSB repair [39, 88]. It is worth noting that H3K36me2 and H3K36me3 appear to exert opposite effect on the choice for DSB repair, either NHEJ or HR. The molecular determinants underlying activation of these distinctive pathways upon DNA damage remain to be defined.
Regulation of mRNA splicing by gene body-associated H3K36me3
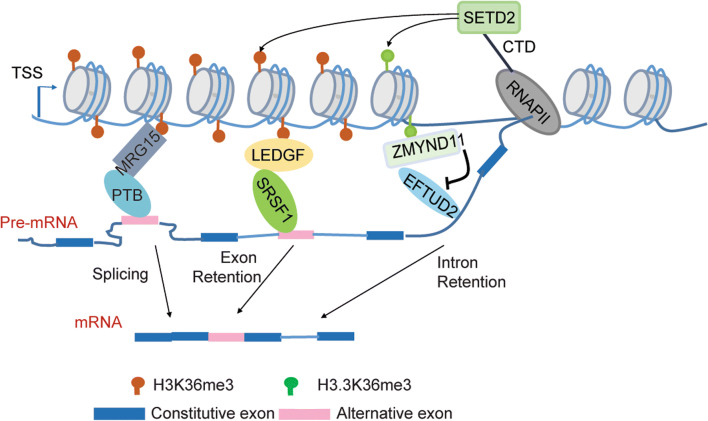
Lines of evidence have highlighted a bidirectional communication between H3K36me3 and co-transcriptional splicing of pre-mRNAs. Such a connection was initially implied by the observation that H3K36me3 is progressively built up in the transcribed gene bodies, with preferential enrichment in exons relative to introns within the same gene [89]. Interestingly, exons included for alternative splicing tend to have higher levels of h3K36me3 than those excluded, and the intron-containing genes are consistently marked by higher H3K36me3 levels than those intronless genes, irrespective of nucleosome occupancy, gene size and transcriptional activity [89]. Subsequent studies then revealed that H3K36me3 plays a crucial role in the recruitment of various splicing-related factors, thus influencing the outcomes of mRNA splicing. For instance, the chromodomain-containing H3K36me3 ‘reader’ protein MRG15 recruits the polypyrimidine tract-binding protein (PTB) to regulate alternative splicing choice [90]; as well, the PWWP domain-containing H3K36me3 ‘reader’ LEDGF contributes to the splicing regulation by acting as an adaptor to couple H3K36me3 with the splicing machinery assembly [91] (Fig. 5; Table 1). Likewise, ZMYND11, an H3.3K36me3-specific ‘reader’, mainly functions to regulate intron retention by antagonizing elongation factor Tu GTP-binding domain containing 2 (EFTUD2), a component of U5 snRNP [92] (Fig. 5; Table 1). Remarkably, splicing can also affect H3K36me3 levels. Here, inhibition of splicing was reported to impair the SETD2 recruitment and reduce the H3K36me3 levels without altering the transcriptional activity, which was demonstrated by the unaffected RNAPII occupancy and transcription elongation rate [89].
Fig. 5.
H3K36me3 modulates alternative splicing of mRNA via its ‘readers’ and recruited splicing machineries. Recognition of H3K36me3 by MRG15 recruits PTB, leading to enhanced splicing of alternative exons, whereas binding of the LEDGF–SRSF1 complex to H3K36me3 facilitates inclusion of alternative exons. Further, ZMYND11 specifically ‘reads’ H3.3K36me3 and opposes the intron-splicing activity of EFTUD2, thus resulting in intron retention
Human pathologies associated with deregulation of H3K36 methylation and related pathways
Because H3K36 methylation is involved in numerous processes critical for cellular functions, its deregulation often results in pathogenesis of various human diseases.
‘Mis-writing’ of H3K36 methylation in disease
NSD1
The t(5;11)(q35;p15.5) chromosomal translocations were reported in ~ 15% of pediatric acute myeloid leukemia (AML) patients and a smaller subset of adult AML cases, resulting in the aberrant chimeric fusion between NSD1 and a nucleoporin-encoding gene, Nup98 [93–97]. Such a NUP98–NSD1 chimeric oncoprotein carries the H3K36 methyltransferase domain of NSD1 and, using murine models, prior studies have shown that NUP98–NSD1 is sufficient in driving AML development, either alone or together with the FLT3 kinase mutation [98, 99]. Studies with the murine AML cells carrying NUP98–NSD1 further demonstrate that it enforces high expression of multiple AML proto-oncogenes such as HOX gene clusters and MEIS1 [98], which are known to arrest hematopoietic differentiation and promote progenitor/stem cell expansion [100–103]. Mechanistically, NUP98–NSD1 directly binds these downstream targets, mediating H3K36 methylation and concurrent histone acetylation to antagonize spreading of the PRC2-catalyzed H3K27me3, a gene-repressive histone PTM known to be associated with terminal differentiation of myeloid cells [98, 104]. Consistently, transcriptome profiling of samples from human AML patients carrying the NUP98–NSD1 translocation also revealed a characteristic HOX-gene expression pattern [93, 95]. In the clinic, NUP98–NSD1 is significantly associated with a poorer prognosis [93, 95], demanding new treatment strategies.
As mentioned above, germ-line haploinsufficiency of NSD1 is a hallmark mutation of Sotos syndrome [74, 105]. Additionally, inactivating mutations of NSD1 were recently found in approximately 10% of HNSCC patients featured by widespread DNA hypo-methylation [28]. A similar NSD1-mutated and DNA-hypomethylated subtype was reported in patients with lung squamous cell carcinoma [106]. Interestingly, HNSCC patients with NSD1 mutations generally display the pronounced survival advantage [107]. Recently, Bui et al. have shown that disruption of NSD1 in the HNSCC cell lines confers a favorable chemotherapeutic sensitivity with a markedly decreased response to cisplatin, presumably due to global DNA hypo-methylation seen in these cells [108]. Moreover, NSD1 inactivation in HNSCC also confers an ‘immune cold’ phenotype, featured by low levels of filtration of anti-tumor immune cells such as the M1 tumor-associated macrophages and CD8+ T cells, as well as low expression of the PD-1 immune checkpoint receptor and its ligands [106]. It has been postulated that NSD1 loss acts as a tumor cell-intrinsic strategy for the production of an ‘immune cold’ phenotype, which potentially affects the efficacy of immunotherapy-based agents [106]. In agreement to its role as a tumor suppressor, NSD1 was found to be epigenetically silenced through promoter DNA hyper-methylation in neuroblastoma, glioma [109] and clear cell renal cell carcinoma (ccRCC) [110], and restoration of the NSD1 expression in these tumor cells compromised their growth [109]. Thus, depending on tissue types, both oncogenic and tumor suppressive roles are reported for NSD1.
NSD2
In about 10–20% of multiple myeloma (MM), a malignancy of antibody-producing plasma cells, NSD2 is amplified and overexpressed due to recurrent t(4;14) chromosomal translocation [111]. Additionally, gain-of-function mutation of NSD2 was reported in a subset of pediatric acute lymphoblastic leukemia [112, 113]. Various studies have shown that the altered NSD2 acts as an oncoprotein, enforcing the gene-expression programs that are crucial for MM tumorigenesis [13, 15, 113–116]. Mechanistically, the pathogenic alterations of NSD2 enhanced genome-wide distribution of H3K36me2 while restricting the PRC2-dependent H3K27me3, both of which are involved in the selection for a gene-expression profile that favors oncogenic transformation of plasma cells [6, 15, 113, 116]. Thus, such the overexpressed and hyper-activated NSD2 enzymes represent an attractive therapeutic target in MM. However, despite recent effort and progress, it remains as a challenge to develop the small-molecule inhibitors of NSD2 that are selective, potent and bioactive. For example, in a recent screening with full-length NSD2 proteins and nucleosome substrates, Coussens et al. identified two out of 16,251 compounds, namely DA3003-1 and PF-03882845, that displayed strong binding affinity to the NSD2’s SET domain in vitro and potent H3K36me2-suppressing effect in U2OS cells [117]; however, multiple targets beyond NSD2 appear to be responsible for the cytotoxic mechanisms [117].
NSD3
Genomic amplification and alterations of NSD3 occur in multiple cancer types, implicating its cancer-promoting role [118–122]. For instance, the fusion between the NUP98 and NSD3 genes was detected in patients with AML or myelodysplastic syndrome [123, 124]. Presumably, the produced NUP98–NSD3 is able to promote hematopoietic transformation in the same fashion as what was already shown for NUP98–NSD1, due to a structural similarity between the two [98]. Recently, Turner-Ivey et al. used a transgenic mouse model to enforce the NSD3 overexpression in the mammary epithelium, and found that a subset of female NSD3-transgenic mice developed frank mammary tumors featured with hyperplasia, ductal dysplasia and carcinoma in situ at the age of 40–50 weeks [120]. It remains unclear, however, as for whether such a cancer-promoting role of NSD3 is dependent on its H3K36 methyltransferase activity.
ASH1L
ASH1L represents another H3K36 methyltransferase that can counteract PRC2- and H3K27me3-mediated gene silencing [19, 125, 126]. And ASH1L was recently shown to be essential for tumorigenicity of the MLL/KMT2-rearranged leukemia [19]. Here, the ASH1L-catalyzed H3K36me2 is recognized by LEDGF, which subsequently recruits the MLL proteins onto the leukemia-related target genes, such as HOXA9 and MEIS1, to enforce their high expression [19]. In addition, this pathway can be suppressed by KDM2A, an H3K36me2 demethylase, and Polycomb group silencing factors [19]. These observations show the importance of well-balanced antagonism between H3K36 and H3K27 methylation for the prevention of malignant development.
SETD2
Loss-of-function mutation or inactivation of SETD2, the non-redundant H3K36me3-inducing methyltransferase, is found to be frequent in a wide range of human cancers, including ccRCC [127], acute leukemia [128, 129], high-grade glioma [130] and enteropathy-associated T cell lymphoma type II [131]. Clearly, SETD2 has generally been proposed to function as a tumor suppressor, a notion recently supported by various SETD2 loss-of-function models [128, 132–134]; evidence also exists, however, showing that SETD2 can be essential for tumor cell viability [135]. In various cancer models, SETD2 inactivation has been shown to perturb a range of critical processes, including gene expression [136], DNA repair [137], genomic instability [138], drug resistance [139], alternative splicing [140, 141] and metabolism [142, 143]. It should be noted that the non-histone substrates of SETD2 such as alpha-tubulin also show perturbed methylation in SETD2-mutated ccRCC cells, contributing to tumor-associated phenotypes [138, 144]. Due to a complicated picture of this topic, readers shall also be referred to comprehensive reviews published recently [33, 145, 146].
‘Onco’-histone: mutation of H3K36 in cancer
Heterozygous mutation of the H3.3 K36M (H3.3K36M) is a frequent event in chondroblastoma, suggesting a dominant-negative or neomorphic nature of the mutation [27]. Also, recurrent mutations of K36M at histone H3.1, H3.2 or H3.3 were detected in HPV-negative HNSCCs [28]. Incorporation of the H3.3K36M ‘onco’-histones into chromatin reduced the genome-wide H3K36me2 and H3K36me3 levels, which is likely due to direct inhibitory effect of H3.3K36M on NSD2 and SETD2, respectively [14, 17]. Fang et al. reported that, in the chondrocytes with the engineered H3.3K36M, the H3K36me2 peaks were drastically depleted from the intergenic regions and became relatively enriched at promoters, gene bodies and transcription end sites, despite the overall reduced levels [17]. Notably, there is a significant correlation between gene expression changes and reduction of H3K36me2 and H3K36me3 at gene bodies, although the underlying mechanism is unclear [17]. Likewise, using the mesenchymal progenitor cells, Lu et al. observed a similar dominant sequestration and repressive effect of H3.3K36M on NSD2 and SETD2, causing global decrease of H3K36me2/3 and arrest of cell differentiation [14]. Additionally, there was concurrent gain of H3K27me3 at intergenic regions, which further induced re-distribution and/or dilution of the H3K27me3-‘reading’ PRC1 complexes, thus providing an explanation for the observed de-repression of polycomb target genes associated with mesenchymal differentiation [14].
Aberrant readout of H3K36 methylation in disease
DNMT3A/3B
Mutations targeting the PWWP domain of DNMT3B were identified from patients with ICF (immunodeficiency, centromeric instability, and facial anomalies) syndrome, a genetic immune disorder featured by defective lymphocytes with pericentromeric DNA hypo-methylation. Such a pathogenic PWWP mutation of DNMT3B (S270P substitution) abrogated the ability of DNMT3B to ‘recognize’ H3K36me3, which is critical for installing proper DNA methylation at the pericentric heterochromatin [82, 147]. Importance of appropriate ‘decoding’ of H3K36 methylation in the diseased setting is further underscored by the recent identification of a heterozygous W330R mutation that ‘hit’ the PWWP domain of DNMT3A in patients of microcephalic dwarfism [148]. Such a W330R mutation disrupted DNMT3A’s binding to H3K36me2/3. However, unlike what was seen with the H3K36me3-binding-defective PWWP mutant of DNMT3B [68, 69], genome-wide profiling of DNA methylation in dwarfism patient-derived cells carrying the PWWP(W330R)-mutated DNMT3A revealed the significantly enhanced DNA methylation at the polycomb-targeted genomic regions, which are enriched with development-related genes and normally show very low degree of DNA methylation [148]. Mechanistically, it was proposed that disrupting the PWWP-mediated binding of H3K36me2/3 might re-shuffle DNMT3A from the H3K36me2/3-covered regions to polycomb-targeted genomic regions, which in return interferes with the PRC2 functions. As a result, hyper-methylation of polycomb targets, as well as concurrent increase in transcriptional activity and gene-active histone PTMs, was seen in cells from dwarfism patients relative to normal cells [148].
ZMYND11
ZMYND11, an H3.3K36me3 ‘reader’, was proposed as a putative tumor suppressor, showing downregulation in various human cancers [149, 150]. Wen et al. showed that, in breast cancer cells, ZMYND11 acts as a transcription co-repressor and blocks the transition of paused RNAPII to the elongation-competent form, and that ZMYND11 represses expression of oncogenes in a mechanism that relies on its capability as an H3.3K36me3-specific ‘reader’ [50]. In addition, chromosomal translocation and missense mutation of ZMYND11 were reported in patients with AML [151–153] and the 10p15.3 microdeletion syndrome [154–156], respectively, but the relationship between these alterations and H3K36me3 is unclear.
‘Mis-erasing’ of H3K36 methylation in disease
KDM2A/2B
KDM2A/2B was shown to have cancer-promoting roles in cancer. KDM2A is significantly amplified in a subset of non-small cell lung cancer (NSCLC), correlating with worse clinical prognosis [157]. Here, KDM2A suppressed the expression of dual-specificity phosphatase 3 (DUSP3) through ‘erasing’ the promoter-associated H3K36me2, an event that then led to ERK1/2 hyper-activation and the enhanced NSCLC cell proliferation and invasion in vivo [157]. Likewise, KDM2B is highly expressed in human AML and is critical for AML initiation and maintenance [158]. Here, the AML-causing leukemia stem cells utilize KDM2B to ‘erase’ H3K36me2 at the tumor suppressor locus p15Ink4b, ensuring its silencing [158]. Furthermore, a previous work demonstrated that the FGF2-triggered transcriptional upregulation of KDM2B in bladder cancer operates in concert with EZH2/KMT6 to repress the EZH2-targeting miR-101, which not only results in EZH2 overexpression but also promotes robust cell proliferation, migration and angiogenesis [159]. Also, in a model of pancreatic ductal adenocarcinoma, the synergistic effect of ectopic expression of KDM2B and KrasG12D is sufficient to drive tumor development via modulating both developmental and metabolic genes [160]. Owing to these works, chemical inhibitors of KDM2A/2B are proposed to be useful for cancer intervention. Recently, compound 9, a cyclopropyl-containing hydroxamate derivative designed based on the crystal structure of KDM7B, was shown to robustly and selectively repress KDM2A, KDM7A, and KDM7B over other KDMs, with IC50 values of 6.8, 0.2, and 1.2 μM, respectively [161]; treatment of cancer cells with compound 9 resulted in arrest of cell cycle progression [161].
KDM4A/B
KDM4A/KDM4B are lysine demethylases with dual specificity for both H3K9me2/3 and H3K36me2/3. In various human cancers, they are found overexpressed due to expression mis-regulation and/or genomic amplification [162, 163]. Prior studies of KDM4A and KDM4B in cancer have largely been focused on their role as the H3K9 demethylase. Recently, Mishra et al. examined a cancer-related phenomenon termed transient site-specific copy gain (TSSG) and reported a correlation of TSSG formation to KDM4B recruitment and H3K36me3 removal [164]. This report supported a role for H3K36me3 ‘mis-erasing’ during oncogenesis [164]. Like KDM2A/2B, the cancer-promoting property of the overexpressed KDM4A/4B makes them valuable targets for anti-cancer agent development. Compound 4 (NSC636819), a selective KDM4A/KDM4B inhibitor designed based on the crystal structure of the KDM4B–pyridine 2,4-dicarboxylic acid–H3K9me3 ternary complex, was identified to block the demethylating activities of KDM4A/4B towards H3K9me3 and H3K36me3 in vitro; however, only the H3K9me3 level was elevated in prostate cancer cells treated with Compound 4 [165].
Conclusions
In summary, H3K36 methylation is a crucial, versatile histone PTM involved in not only the regulation of epigenomic landscapes in cells but also a wide range of biological processes. In a cellular and chromatin context-dependent fashion, H3K36 methylation can recruit different ‘reader’ proteins, which carry out their respective functions for mediating chromatin remodeling, gene transcription regulation, DNA damage repair, or mRNA splicing. Such an important role for H3K36 methylation is reflected by human diseases induced by mis-regulation of various proteins that are responsible for deposition, functional ‘readout’ or removal of H3K36 methylation.
Despite recent advances, questions and challenges remained to be addressed. For example, besides methylation-modulating activities, the regulatory roles of certain H3K36-associated enzymes may rely on their scaffolding functions. An extreme example is a short isoform of NSD3 (NSD3S), which lacks the SET domain and acts mainly as an adaptor protein for bringing together BRD4 and CHD8 to sustain the growth of AML cells [166, 167]. As well, NSD3S was shown to interact with MYC [168]. Association of NSD3 and BRD4 appears to be essential for malignant growth of NUT midline carcinoma (NMC) that is featured with aberrant chromosomal translocation of BRD4–NUT or NSD3–NUT [121]. Also, the bindings of various ‘readers’ to H3K36me3 and H3K36me2 appear to be promiscuous, with a generally low affinity seen for either ligand, as exemplified by the PWWP domain of NSD2, DNMT3A and DNMT3B [19, 49, 81, 82]. Yet, in cells, DNMT3A and DNMT3B display differential patterns of targeting to the H3K36me3 versus H3K36me2-covered genomic regions [68, 69]. How could the promiscuous, low-affinity binding mediate the wanted efficient and selective events in cells? A possibility is that the ‘reader’ modules may simultaneously bind both DNA sequences and H3K36me3/2 as shown for the Tudor domain of PHF1 [45, 46]; alternatively, they may act in concert with other motifs within the same protein or protein complexes to promote a multivalent engagement of histone tails, thus enhancing both selectivity and affinity [40, 42, 44, 45, 50]. Last, unlike potent, selective chemical inhibitors developed for other epigenetic factors such as BRD4 and EZH2 [169–172], those against the H3K36-related regulators (such as the NSD family proteins as discussed above) are currently lacking. Future efforts shall also be directed towards the development of small molecules that specifically target H3K36 methylation-related ‘writers’, ‘erasors’ and/or readers, which could provide attractive therapeutic means for the genetically defined cancer types, for example, those carrying gain-of-function mutations of NSD1 or NSD2.
Acknowledgements
We graciously thank Dr. Jikui Song for assistance with structural illustration. G.G.W. is an American Cancer Society Research Scholar and a Leukemia and Lymphoma Society Scholar. This work was supported by NIH Grants (R01-CA215284, R01-CA218600, and R01-CA211336 to G.G.W.).
Abbreviations
- H3K36
Histone H3 lysine 36
- H3K36me1/2/3
Mono/di/trimethylation of H3K36
- H3K27me3
Trimethylation of histone H3 lysine 27
- PTMs
Post-translational modifications
- ChIP-seq
Chromatin immunoprecipitation (ChIP) following by sequencing
- ESCs
Embryonic stem cells
- PWWP
Pro-Trp-Trp-Pro
- PRC2
Polycomb repressive complex 2
- RNAPII
RNA polymerase II
- CTD
C-terminal domain
- SRI
Set2–Rpb1-interacting domain
- PHD
Plant homeodomain
- TET2
Ten-eleven translocation 2
- MMR
Mismatch repair
- DSB
Double-strand DNA break
- NHEJ
Non-homologous end joining
- HR
Homologous recombination repair
- HPV
Human papillomavirus
- HNSCCs
Head and neck squamous cell carcinomas
- AML
Acute myeloid leukemia
- ccRCC
Clear cell renal cell carcinoma
- MM
Multiple myeloma
- NSCLC
Non-small cell lung cancer
- NMC
NUT midline carcinoma
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S, Smith MM. Histone variants and epigenetics. Cold Spring Harb Perspect Biol. 2015;7:a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159(458–458):e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM, Licht JD, Kelleher NL. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci USA. 2012;109:13549–13554. doi: 10.1073/pnas.1205707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Chen J, Gao Y, Gao J, Liao R, Wang Y, Oyang C, Li E, Zeng C, Zhou S, Yang P, Jin H, Yi W. Quantitative profiling of combinational K27/K36 modifications on histone H3 variants in mouse organs. J Proteome Res. 2016;15:1070–1079. doi: 10.1021/acs.jproteome.5b01164. [DOI] [PubMed] [Google Scholar]
- 8.Bhanu NV, Sidoli S, Garcia BA. Histone modification profiling reveals differential signatures associated with human embryonic stem cell self-renewal and differentiation. Proteomics. 2016;16:448–458. doi: 10.1002/pmic.201500231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S, Liu Y, Li F, Vedadi M, Frye SV, Garcia BA, Zheng D, Jin J, Wang GG. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125:346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, Li W, Gozani O. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, Wunder JS, Dickson BC, Lundgren SM, Jani KS, De Jay N, Papillon-Cavanagh S, Andrulis IL, Sawyer SL, Grynspan D, Turcotte RE, Nadaf J, Fahiminiyah S, Muir TW, Majewski J, Thompson CB, Chi P, Garcia BA, Allis CD, Jabado N, Lewis PW. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Zhang Q, Ezponda T, Shah MY, Zheng Y, Will CM, Small EC, Hua Y, Bulic M, Jiang Y, Carrara M, Calogero RA, Kath WL, Kelleher NL, Wang JP, Elemento O, Licht JD. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10:e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streubel G, Watson A, Jammula SG, Scelfo A, Fitzpatrick DJ, Oliviero G, McCole R, Conway E, Glancy E, Negri GL, Dillon E, Wynne K, Pasini D, Krogan NJ, Bracken AP, Cagney G. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell. 2018;70(371–379):e5. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 17.Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, Zhang H, Yang N, Bradley EW, Ho TH, Rubin BP, Bridge JA, Thibodeau SN, Ordog T, Chen Y, van Wijnen AJ, Oliveira AM, Xu RM, Westendorf JJ, Zhang Z. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science. 2016;352:1344–1348. doi: 10.1126/science.aae0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P, Guo L, Duan ZJ, Tepper CG, Xue L, Chen X, Kung HJ, Gao AC, Zou JX, Chen HW. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32:3121–3131. doi: 10.1128/MCB.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, Ikenouye L, Onishi M, Schneidawind D, Buechele C, Hansen L, Duque-Afonso J, Zhu F, Martin GM, Gozani O, Majeti R, Kutateladze TG, Cleary ML. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016;6:770–783. doi: 10.1158/2159-8290.CD-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniel SL, Hepperla AJ, Huang J, Dronamraju R, Adams AT, Kulkarni VG, Davis IJ, Strahl BD. H3K36 methylation regulates nutrient stress response in Saccharomyces cerevisiae by enforcing transcriptional fidelity. Cell Rep. 2017;19:2371–2382. doi: 10.1016/j.celrep.2017.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen P, Dang W, Donahue G, Dai J, Dorsey J, Cao X, Liu W, Cao K, Perry R, Lee JY, Wasko BM, Carr DT, He C, Robison B, Wagner J, Gregory BD, Kaeberlein M, Kennedy BK, Boeke JD, Berger SL. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adhvaryu KK, Morris SA, Strahl BD, Selker EU. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot Cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meers MP, Henriques T, Lavender CA, McKay DJ, Strahl BD, Duronio RJ, Adelman K, Matera AG. Histone gene replacement reveals a post-transcriptional role for H3K36 in maintaining metazoan transcriptome fidelity. Elife. 2017;6:e23249. doi: 10.7554/eLife.23249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKay DJ, Klusza S, Penke TJ, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, Strahl BD, Duronio RJ, Matera AG. Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell. 2015;32:373–386. doi: 10.1016/j.devcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammad F, Helin K. Oncohistones: drivers of pediatric cancers. Genes Dev. 2017;31:2313–2324. doi: 10.1101/gad.309013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, Nik-Zainal S, Martin S, McLaren S, Goody V, Robinson B, Butler A, Teague JW, Halai D, Khatri B, Myklebost O, Baumhoer D, Jundt G, Hamoudi R, Tirabosco R, Amary MF, Futreal PA, Stratton MR, Campbell PJ, Flanagan AM. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, Ailles L, Karamchandani J, Marchione DM, Garcia BA, Weinreb I, Goldstein D, Lewis PW, Dancu OM, Dhaliwal S, Stecho W, Howlett CJ, Mymryk JS, Barrett JW, Nichols AC, Allis CD, Majewski J, Jabado N. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaran SM, Gozani O. Characterization of H3.3K36M as a tool to study H3K36 methylation in cancer cells. Epigenetics. 2017;12:917–922. doi: 10.1080/15592294.2017.1377870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Ramirez D, Hwang S, Jungbluth A, Frosina D, Ntiamoah P, Healey J, Zhu G, Chen W, Klein M, Hameed M. Histone H3K36M mutation and trimethylation patterns in chondroblastoma. Histopathology. 2019;74:291–299. doi: 10.1111/his.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci. 2017;74:3317–3334. doi: 10.1007/s00018-017-2517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett RL, Swaroop A, Troche C, Licht JD. The role of nuclear receptor-binding set domain family histone lysine methyltransferases in cancer. Cold Spring Harb Perspect Med. 2017;7:a026708. doi: 10.1101/cshperspect.a026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 37.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 39.Lu R, Wang GG. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem Sci. 2013;38:546–555. doi: 10.1016/j.tibs.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin S, Min J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci. 2014;39:536–547. doi: 10.1016/j.tibs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musselman CA, Gibson MD, Hartwick EW, North JA, Gatchalian J, Poirier MG, Kutateladze TG. Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat Commun. 2013;4:2969. doi: 10.1038/ncomms3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson MD, Gatchalian J, Slater A, Kutateladze TG, Poirier MG. PHF1 Tudor and N-terminal domains synergistically target partially unwrapped nucleosomes to increase DNA accessibility. Nucleic acids Res. 2017;45:3767–3776. doi: 10.1093/nar/gkw1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, Kulesza CA, Yasui A, Cote J, Kutateladze TG. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol. 2012;19:1266–1272. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, Strahl BD, Song J, Wang GG. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell. 2013;49:571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sankaran SM, Wilkinson AW, Elias JE, Gozani O. A PWWP domain of histone-lysine N-methyltransferase NSD2 binds to dimethylated Lys-36 of histone H3 and regulates NSD2 function at chromatin. J Biol Chem. 2016;291:8465–8474. doi: 10.1074/jbc.M116.720748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, Li B, Barton MC, Li W, Li H, Shi X. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 52.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 53.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 54.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 55.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 56.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 57.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 59.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 61.McDaniel SL, Fligor JE, Ruan C, Cui H, Bridgers JB, DiFiore JV, Guo AH, Li B, Strahl BD. Combinatorial histone readout by the dual plant homeodomain (PHD) fingers of Rco1 mediates Rpd3S chromatin recruitment and the maintenance of transcriptional fidelity. J Biol Chem. 2016;291:14796–14802. doi: 10.1074/jbc.M116.720193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 64.Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 69.Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 70.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 71.Zhuang L, Jang Y, Park YK, Lee JE, Jain S, Froimchuk E, Broun A, Liu C, Gavrilova O, Ge K. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat Commun. 2018;9:1796. doi: 10.1038/s41467-018-04127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, Lohan AJ, Ferguson N, Shi X, Sinha KM, Loftus BJ, Cagney G, Bracken AP. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 73.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol. 2012;19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faravelli F. NSD1 mutations in Sotos syndrome. Am J Med Genet C Semin Med Genet. 2005;137C:24–31. doi: 10.1002/ajmg.c.30061. [DOI] [PubMed] [Google Scholar]
- 75.Choufani S, Cytrynbaum C, Chung BH, Turinsky AL, Grafodatskaya D, Chen YA, Cohen AS, Dupuis L, Butcher DT, Siu MT, Luk HM, Lo IF, Lam ST, Caluseriu O, Stavropoulos DJ, Reardon W, Mendoza-Londono R, Brudno M, Gibson WT, Chitayat D, Weksberg R. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee ST, Wiemels JL. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res. 2016;44:1105–1117. doi: 10.1093/nar/gkv1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, Koseki H, Brockdorff N, Ponting CP, Kessler BM, Klose RJ. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamagata K, Kobayashi A. The cysteine-rich domain of TET2 binds preferentially to mono- and dimethylated histone H3K36. J Biochem. 2017;161:327–330. doi: 10.1093/jb/mvx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rondelet G, Dal Maso T, Willems L, Wouters J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J Struct Biol. 2016;194:357–367. doi: 10.1016/j.jsb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 83.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fang J, Huang Y, Mao G, Yang S, Rennert G, Gu L, Li H, Li GM. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSalpha interaction. Proc Natl Acad Sci USA. 2018;115:9598–9603. doi: 10.1073/pnas.1806355115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, Humphrey TC. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li L, Wang Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem. 2017;292:11951–11959. doi: 10.1074/jbc.M117.788224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 90.Zhou XL, Zhu RR, Wu X, Xu H, Li YY, Xu QR, Liu S, Huang H, Xu X, Wan L, Wu QC, Liu JC. NSD2 promotes ventricular remodelling mediated by the regulation of H3K36me2. J Cell Mol Med. 2019;23:568–575. doi: 10.1111/jcmm.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, Xu W, Mu S, Wen H, Qiu J, Wang Z, Yang P, Wu F, Hui J, Fu X, Shi X, Shi YG, Xing Y, Lan F, Shi Y. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell. 2014;56:298–310. doi: 10.1016/j.molcel.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ostronoff F, Othus M, Gerbing RB, Loken MR, Raimondi SC, Hirsch BA, Lange BJ, Petersdorf S, Radich J, Appelbaum FR, Gamis AS, Alonzo TA, Meshinchi S. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124:2400–2407. doi: 10.1182/blood-2014-04-570929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thol F, Kolking B, Hollink IH, Damm F, van den Heuvel-Eibrink MM, Michel Zwaan C, Bug G, Ottmann O, Wagner K, Morgan M, Hofmann WK, Gohring G, Schlegelberger B, Krauter J, Ganser A, Heuser M. Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia. 2013;27:750–754. doi: 10.1038/leu.2012.249. [DOI] [PubMed] [Google Scholar]
- 95.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, van Galen JF, Beverloo HB, Sonneveld E, Kaspers GJ, Trka J, Baruchel A, Zimmermann M, Creutzig U, Reinhardt D, Pieters R, Valk PJ, Zwaan CM. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 96.La Starza R, Gorello P, Rosati R, Riezzo A, Veronese A, Ferrazzi E, Martelli MF, Negrini M, Mecucci C. Cryptic insertion producing two NUP98/NSD1 chimeric transcripts in adult refractory anemia with an excess of blasts. Genes Chromosomes Cancer. 2004;41:395–399. doi: 10.1002/gcc.20103. [DOI] [PubMed] [Google Scholar]
- 97.Akiki S, Dyer SA, Grimwade D, Ivey A, Abou-Zeid N, Borrow J, Jeffries S, Caddick J, Newell H, Begum S, Tawana K, Mason J, Velangi M, Griffiths M. NUP98-NSD1 fusion in association with FLT3-ITD mutation identifies a prognostically relevant subgroup of pediatric acute myeloid leukemia patients suitable for monitoring by real time quantitative PCR. Genes Chromosomes Cancer. 2013;52:1053–1064. doi: 10.1002/gcc.22100. [DOI] [PubMed] [Google Scholar]
- 98.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 99.Thanasopoulou A, Tzankov A, Schwaller J. Potent co-operation between the NUP98-NSD1 fusion and the FLT3-ITD mutation in acute myeloid leukemia induction. Haematologica. 2014;99:1465–1471. doi: 10.3324/haematol.2013.100917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, Wang Y, Li S, Huang H, Neilly MB, Wang GG, Jiang X, Liu PP, Jin J, Chen J. PBX3 and MEIS1 cooperate in hematopoietic cells to drive acute myeloid leukemias characterized by a core transcriptome of the MLL-rearranged disease. Cancer Res. 2016;76:619–629. doi: 10.1158/0008-5472.CAN-15-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang GG, Pasillas MP, Kamps MP. Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol. 2006;26:3902–3916. doi: 10.1128/MCB.26.10.3902-3916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, Waggoner D, Turkmen S, Cormier-Daire V, Irrthum A, Rahman N, Childhood Overgrowth C. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brennan K, Shin JH, Tay JK, Prunello M, Gentles AJ, Sunwoo JB, Gevaert O. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep. 2017;7:17064. doi: 10.1038/s41598-017-17298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peri S, Izumchenko E, Schubert AD, Slifker MJ, Ruth K, Serebriiskii IG, Guo T, Burtness BA, Mehra R, Ross EA, Sidransky D, Golemis EA. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun. 2017;8:1772. doi: 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bui N, Huang JK, Bojorquez-Gomez A, Licon K, Sanchez KS, Tang SN, Beckett AN, Wang T, Zhang W, Shen JP, Kreisberg JF, Ideker T. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther. 2018;17:1585–1594. doi: 10.1158/1535-7163.MCT-17-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, Alaminos M, Cheung NK, Rahman N, Esteller M. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su X, Zhang J, Mouawad R, Comperat E, Roupret M, Allanic F, Parra J, Bitker MO, Thompson EJ, Gowrishankar B, Houldsworth J, Weinstein JN, Tost J, Broom BM, Khayat D, Spano JP, Tannir NM, Malouf GG. NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res. 2017;77:4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 112.Oyer JA, Huang X, Zheng Y, Shim J, Ezponda T, Carpenter Z, Allegretta M, Okot-Kotber CI, Patel JP, Melnick A, Levine RL, Ferrando A, Mackerell AD, Jr, Kelleher NL, Licht JD, Popovic R. Point mutation E1099K in MMSET/NSD2 enhances its methyltransferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP, Yan F, Wang Z, Robert McDonald E, 3rd, Wei L, Ma J, Easton J, Yu Z, de Beaumount R, Gibaja V, Venkatesan K, Schlegel R, Sellers WR, Keen N, Liu J, Caponigro G, Barretina J, Cooke VG, Mullighan C, Carr SA, Downing JR, Garraway LA, Stegmeier F. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang Z, Wu H, Chuai S, Xu F, Yan F, Englund N, Wang Z, Zhang H, Fang M, Wang Y, Gu J, Zhang M, Yang T, Zhao K, Yu Y, Dai J, Yi W, Zhou S, Li Q, Wu J, Liu J, Wu X, Chan H, Lu C, Atadja P, Li E, Wang Y, Hu M. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013;73:6277–6288. doi: 10.1158/0008-5472.CAN-13-1000. [DOI] [PubMed] [Google Scholar]
- 116.Swaroop A, Oyer JA, Will CM, Huang X, Yu W, Troche C, Bulic M, Durham BH, Wen QJ, Crispino JD, MacKerell AD, Jr, Bennett RL, Kelleher NL, Licht JD. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene. 2019;38:671–686. doi: 10.1038/s41388-018-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Coussens NP, Kales SC, Henderson MJ, Lee OW, Horiuchi KY, Wang Y, Chen Q, Kuznetsova E, Wu J, Chakka S, Cheff DM, Cheng KC, Shinn P, Brimacombe KR, Shen M, Simeonov A, Lal-Nag M, Ma H, Jadhav A, Hall MD. High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem. 2018;293:13750–13765. doi: 10.1074/jbc.RA118.004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jones DH, Lin DI. Amplification of the NSD3–BRD4–CHD8 pathway in pelvic high-grade serous carcinomas of tubo-ovarian and endometrial origin. Mol Clin Oncol. 2017;7:301–307. doi: 10.3892/mco.2017.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Han X, Piao L, Zhuang Q, Yuan X, Liu Z, He X. The role of histone lysine methyltransferase NSD3 in cancer. Onco Targets Ther. 2018;11:3847–3852. doi: 10.2147/OTT.S166006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turner-Ivey B, Smith EL, Rutkovsky AC, Spruill LS, Mills JN, Ethier SP. Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3. Breast Cancer Res Treat. 2017;164:349–358. doi: 10.1007/s10549-017-4258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.French CA, Rahman S, Walsh EM, Kuhnle S, Grayson AR, Lemieux ME, Grunfeld N, Rubin BP, Antonescu CR, Zhang S, Venkatramani R, Dal Cin P, Howley PM. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4:928–941. doi: 10.1158/2159-8290.CD-14-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 123.Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, Negrini M, Martelli MF, Mecucci C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.V99.10.3857. [DOI] [PubMed] [Google Scholar]
- 124.Taketani T, Taki T, Nakamura H, Taniwaki M, Masuda J, Hayashi Y. NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15) and expression pattern of NSD family genes. Cancer Genet Cytogenet. 2009;190:108–112. doi: 10.1016/j.cancergencyto.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 125.Miyazaki H, Higashimoto K, Yada Y, Endo TA, Sharif J, Komori T, Matsuda M, Koseki Y, Nakayama M, Soejima H, Handa H, Koseki H, Hirose S, Nishioka K. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013;9:e1003897. doi: 10.1371/journal.pgen.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.An S, Yeo KJ, Jeon YH, Song JJ. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem. 2011;286:8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, Yan X, Wei W, Pang Y, Cheng H, Hua C, Zhang Y, Yang X, Lu X, Cao L, Hao L, Dong L, Zou W, Wu J, Li X, Zheng S, Yan J, Zhou J, Zhang L, Mi S, Wang X, Zhang L, Zou Y, Chen Y, Geng Z, Wang J, Zhou J, Liu X, Wang J, Yuan W, Huang G, Cheng T, Wang QF. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46:287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, Kung AL, Lo Nigro L, Ebert BL, Armstrong SA. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. doi: 10.1038/ncomms4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, Fleming A, Hadjadj D, Busche S, Lepage P, Montpetit A, Staffa A, Gerges N, Zakrzewska M, Zakrzewski K, Liberski PP, Hauser P, Garami M, Klekner A, Bognar L, Zadeh G, Faury D, Pfister SM, Jabado N, Majewski J. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Roberti A, Dobay MP, Bisig B, Vallois D, Boechat C, Lanitis E, Bouchindhomme B, Parrens MC, Bossard C, Quintanilla-Martinez L, Missiaglia E, Gaulard P, de Leval L. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat Commun. 2016;7:12602. doi: 10.1038/ncomms12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang YL, Sun JW, Xie YY, Zhou Y, Liu P, Song JC, Xu CH, Wang L, Liu D, Xu AN, Chen Z, Chen SJ, Sun XJ, Huang QH. Setd2 deficiency impairs hematopoietic stem cell self-renewal and causes malignant transformation. Cell Res. 2018;28:476–490. doi: 10.1038/s41422-018-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, Kim-Kiselak C, Cicchini M, Yates TJ, Feldser DM. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77:1719–1729. doi: 10.1158/0008-5472.CAN-16-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moffitt AB, Ondrejka SL, McKinney M, Rempel RE, Goodlad JR, Teh CH, Leppa S, Mannisto S, Kovanen PE, Tse E, Au-Yeung RKH, Kwong YL, Srivastava G, Iqbal J, Yu J, Naresh K, Villa D, Gascoyne RD, Said J, Czader MB, Chadburn A, Richards KL, Rajagopalan D, Davis NS, Smith EC, Palus BC, Tzeng TJ, Healy JA, Lugar PL, Datta J, Love C, Levy S, Dunson DB, Zhuang Y, Hsi ED, Dave SS. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J Exp Med. 2017;214:1371–1386. doi: 10.1084/jem.20160894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Skucha A, Ebner J, Schmollerl J, Roth M, Eder T, Cesar-Razquin A, Stukalov A, Vittori S, Muhar M, Lu B, Aichinger M, Jude J, Muller AC, Gyorffy B, Vakoc CR, Valent P, Bennett KL, Zuber J, Superti-Furga G, Grebien F. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun. 2018;9:1983. doi: 10.1038/s41467-018-04329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bu J, Chen A, Yan X, He F, Dong Y, Zhou Y, He J, Zhan D, Lin P, Hayashi Y, Sun Y, Zhang Y, Xiao Z, Grimes HL, Wang QF, Huang G. SETD2-mediated crosstalk between H3K36me3 and H3K79me2 in MLL-rearranged leukemia. Leukemia. 2018;32:890–899. doi: 10.1038/leu.2017.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kanu N, Gronroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J, Maya-Mendoza A, Mistrik M, Rowan AJ, Patel H, Rabinowitz A, East P, Wilson G, Santos CR, McGranahan N, Gulati S, Gerlinger M, Birkbak NJ, Joshi T, Alexandrov LB, Stratton MR, Powles T, Matthews N, Bates PA, Stewart A, Szallasi Z, Larkin J, Bartek J, Swanton C. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34:5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chiang YC, Park IY, Terzo EA, Tripathi DN, Mason FM, Fahey CC, Karki M, Shuster CB, Sohn BH, Chowdhury P, Powell RT, Ohi R, Tsai YS, de Cubas AA, Khan A, Davis IJ, Strahl BD, Parker JS, Dere R, Walker CL, Rathmell WK. SETD2 haploinsufficiency for microtubule methylation is an early driver of genomic instability in renal cell carcinoma. Cancer Res. 2018;78:3135–3146. doi: 10.1158/0008-5472.CAN-17-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim IK, McCutcheon JN, Rao G, Liu SV, Pommier Y, Skrzypski M, Zhang YW, Giaccone G. Acquired SETD2 mutation and impaired CREB1 activation confer cisplatin resistance in metastatic non-small cell lung cancer. Oncogene. 2018 doi: 10.1038/s41388-018-0429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, Han Y, Wang X, Li Q, Qin J. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Investig. 2017;127:3375–3391. doi: 10.1172/JCI94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, Prins JF, Lieb JD, Rathmell WK, Davis IJ. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24:241–250. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu J, Hanavan PD, Kras K, Ruiz YW, Castle EP, Lake DF, Chen X, O’Brien D, Luo H, Robertson KD, Gu H, Ho TH. Loss of SETD2 induces a metabolic switch in renal cell carcinoma cell lines toward enhanced oxidative phosphorylation. J Proteome Res. 2019;18:331–340. doi: 10.1021/acs.jproteome.8b00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li L, Miao W, Huang M, Williams P, Wang Y. Integrated genomic and proteomic analyses reveal novel mechanisms of the methyltransferase SETD2 in renal cell carcinoma development. Mol Cell Proteomics. 2019;18:437–447. doi: 10.1074/mcp.RA118.000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, Chiang YC, Davis IJ, Fahey CC, Hacker KE, Verhey KJ, Bedford MT, Jonasch E, Rathmell WK, Walker CL. Dual chromatin and cytoskeletal remodeling by SETD2. Cell. 2016;166:950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fahey CC, Davis IJ. SETting the stage for cancer development: SETD2 and the consequences of lost methylation. Cold Spring Harb Perspect Med. 2017;7:a026468. doi: 10.1101/cshperspect.a026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7:50719–50734. doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin CA, Marsh JA, Taglini F, Kilanowski F, Parry DA, Cormier-Daire V, Fong CT, Gibson K, Hwa V, Ibanez L, Robertson SP, Sebastiani G, Rappsilber J, Allshire RC, Reijns MAM, Dauber A, Sproul D, Jackson AP. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet. 2019;51:96–105. doi: 10.1038/s41588-018-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang JP, Yang JK, Li C, Cui ZQ, Liu HJ, Sun XF, Geng SM, Lu SK, Song J, Guo CY, Jiao BH. Downregulation of ZMYND11 induced by miR-196a-5p promotes the progression and growth of GBM. Biochem Biophys Res Commun. 2017;494:674–680. doi: 10.1016/j.bbrc.2017.10.098. [DOI] [PubMed] [Google Scholar]
- 150.Yang H, Zhang C, Zhao X, Wu Q, Fu X, Yu B, Shao Y, Guan M, Zhang W, Wan J, Huang X. Analysis of copy number variations of BS69 in multiple types of hematological malignancies. Ann Hematol. 2010;89:959–964. doi: 10.1007/s00277-010-0966-5. [DOI] [PubMed] [Google Scholar]
- 151.Yamamoto K, Yakushijin K, Ichikawa H, Kakiuchi S, Kawamoto S, Matsumoto H, Nakamachi Y, Saegusa J, Matsuoka H, Minami H. Expression of a novel ZMYND11/MBTD1 fusion transcript in CD7(+)CD56(+) acute myeloid leukemia with t(10;17)(p15;q21) Leuk Lymphoma. 2018 doi: 10.1080/10428194.2018.1464157. [DOI] [PubMed] [Google Scholar]
- 152.de Rooij JD, van den Heuvel-Eibrink MM, Kollen WJ, Sonneveld E, Kaspers GJ, Beverloo HB, Fornerod M, Pieters R, Zwaan CM. Recurrent translocation t(10;17)(p15;q21) in minimally differentiated acute myeloid leukemia results in ZMYND11/MBTD1 fusion. Genes Chromosomes Cancer. 2016;55:237–241. doi: 10.1002/gcc.22326. [DOI] [PubMed] [Google Scholar]
- 153.De Braekeleer E, Auffret R, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, De Braekeleer M. Recurrent translocation (10;17)(p15;q21) in acute poorly differentiated myeloid leukemia likely results in ZMYND11-MBTD1 fusion. Leuk Lymphoma. 2014;55:1189–1190. doi: 10.3109/10428194.2013.820292. [DOI] [PubMed] [Google Scholar]
- 154.Moskowitz AM, Belnap N, Siniard AL, Szelinger S, Claasen AM, Richholt RF, De Both M, Corneveaux JJ, Balak C, Piras IS, Russell M, Courtright AL, Rangasamy S, Ramsey K, Craig DW, Narayanan V, Huentelman MJ, Schrauwen I. A de novo missense mutation in ZMYND11 is associated with global developmental delay, seizures, and hypotonia. Cold Spring Harb Mol Case Stud. 2016;2:a000851. doi: 10.1101/mcs.a000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tumiene B, Ciuladaite Z, Preiksaitiene E, Mameniskiene R, Utkus A, Kucinskas V. Phenotype comparison confirms ZMYND11 as a critical gene for 10p15.3 microdeletion syndrome. J Appl Genet. 2017;58:467–474. doi: 10.1007/s13353-017-0408-3. [DOI] [PubMed] [Google Scholar]
- 156.Cobben JM, Weiss MM, van Dijk FS, De Reuver R, de Kruiff C, Pondaag W, Hennekam RC, Yntema HG. A de novo mutation in ZMYND11, a candidate gene for 10p15.3 deletion syndrome, is associated with syndromic intellectual disability. Eur J Med Genet. 2014;57:636–638. doi: 10.1016/j.ejmg.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 157.Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, Multani AS, Chan CH, Erez B, Saigal B, Chung J, Lin HK, Wu X, Hung MC, Heymach JV, Lee MG. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Investig. 2013;123:5231–5246. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 2011;43:285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park PJ, Bardeesy N. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Investig. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suzuki T, Ozasa H, Itoh Y, Zhan P, Sawada H, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, Tsukada Y, Tumber A, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Schofield CJ, Miyata N. Identification of the KDM2/7 histone lysine demethylase subfamily inhibitor and its antiproliferative activity. J Med Chem. 2013;56:7222–7231. doi: 10.1021/jm400624b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Young LC, Hendzel MJ. The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Biochem Cell Biol. 2013;91:369–377. doi: 10.1139/bcb-2012-0054. [DOI] [PubMed] [Google Scholar]
- 164.Mishra S, Van Rechem C, Pal S, Clarke TL, Chakraborty D, Mahan SD, Black JC, Murphy SE, Lawrence MS, Daniels DL, Whetstine JR. Cross-talk between lysine-modifying enzymes controls site-specific DNA amplifications. Cell. 2018;175:1716. doi: 10.1016/j.cell.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chu CH, Wang LY, Hsu KC, Chen CC, Cheng HH, Wang SM, Wu CM, Chen TJ, Li LT, Liu R, Hung CL, Yang JM, Kung HJ, Wang WC. KDM4B as a target for prostate cancer: structural analysis and selective inhibition by a novel inhibitor. J Med Chem. 2014;57:5975–5985. doi: 10.1021/jm500249n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Q, Zeng L, Shen C, Ju Y, Konuma T, Zhao C, Vakoc CR, Zhou MM. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure. 2016;24:1201–1208. doi: 10.1016/j.str.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xiong J, Pecchi VG, Qui M, Ivanov AA, Mo X, Niu Q, Chen X, Fu H, Du Y. Development of a time-resolved fluorescence resonance energy transfer ultrahigh-throughput screening assay for targeting the NSD3 and MYC interaction. Assay Drug Dev Technol. 2018;16:96–106. doi: 10.1089/adt.2017.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 170.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lu R, Wang GG. Pharmacologic targeting of chromatin modulators as therapeutics of acute myeloid leukemia. Front Oncol. 2017;7:241. doi: 10.3389/fonc.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu B, Konze KD, Jin J, Wang GG. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp Hematol. 2015;43:698–712. doi: 10.1016/j.exphem.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]