Abstract
Misfolded F508del-CFTR, the main molecular cause of the recessive disorder cystic fibrosis, is recognized by the endoplasmic reticulum (ER) quality control (ERQC) resulting in its retention and early degradation. The ERQC mechanisms rely mainly on molecular chaperones and on sorting motifs, whose presence and exposure determine CFTR retention or exit through the secretory pathway. Arginine-framed tripeptides (AFTs) are ER retention motifs shown to modulate CFTR retention. However, the interactions and regulatory pathways involved in this process are still largely unknown. Here, we used proteomic interaction profiling and global bioinformatic analysis to identify factors that interact differentially with F508del-CFTR and F508del-CFTR without AFTs (F508del-4RK-CFTR) as putative regulators of this specific ERQC checkpoint. Using LC–MS/MS, we identified kinesin family member C1 (KIFC1) as a stronger interactor with F508del-CFTR versus F508del-4RK-CFTR. We further validated this interaction showing that decreasing KIFC1 levels or activity stabilizes the immature form of F508del-CFTR by reducing its degradation. We conclude that the current approach is able to identify novel putative therapeutic targets that can be ultimately used to the benefit of CF patients.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2896-7) contains supplementary material, which is available to authorized users.
Keywords: CFTR, Endoplasmic reticulum quality control, Arginine-framed tripeptides, Interactome, Kinesins, Degradation
Introduction
Cystic fibrosis (CF), the most common life-shortening monogenic autosomal recessive disease among Caucasians, is caused by dysfunction of the cystic fibrosis transmembrane conductance regulator (CFTR) protein [1]. This protein functions as a cyclic adenosine monophosphate (cAMP)-regulated chloride (Cl−)/bicarbonate (HCO3−) channel at the apical membrane of epithelial cells. CFTR is a member of the ATP-binding cassette (ABC) transporter superfamily and it is composed of five domains: two membrane-spanning domains (MSD1 and MSD2), each one composed of six transmembrane segments (TM1-6 and TM7-12), that form the pore of the channel allowing Cl−/HCO3− to flow across the membrane, two cytosolic nucleotide-binding domains (NBD1 and NBD2), where ATP is hydrolysed, regulating channel gating and a fifth CFTR-exclusive regulatory domain (RD) that contains multiple phosphorylation sites, relevant for channel activity [2].
The most common CF-causing mutation is the deletion of phenylalanine 508—F508del, that is present in approximately 85% of CF patients in at least one allele. This deletion leads to CFTR misfolding which is recognized by the endoplasmic reticulum (ER) quality control (ERQC) resulting in CFTR retention and targeting for proteasomal degradation. The ERQC consists in a series of checkpoints, two of which include CFTR folding status assessment by the Hsc70 and calnexin chaperones, respectively [3]. Two additional checkpoints were proposed to involve both retention motifs, namely the arginine-framed tripeptides (AFTs) [4, 5] and export motifs, such as the diacidic (DAD) exit code that controls the interaction with the COPII machinery. CFTR possesses four AFTs (which consist in two arginines flanking any amino acid: RXR), namely R29, at the N-terminal tail, R516 and R555, both in NBD1 and R766 in the RD. Disruption of these four AFTs by replacing one arginine in each by a lysine (F508del-4RK-CFTR) allows F508del-CFTR to escape the ERQC and consequently to reach the plasma membrane (PM), partially rescuing F508del-CFTR processing and function without, however, a significant correction in its folding, as shown by single-channel data [6]. These AFTs were proposed to form the third ERQC checkpoint of CFTR—when CFTR achieves a native conformation [wild-type (wt)-CFTR], the four AFTs are buried within the protein and protein exits from the ER [6]. However, in the case of misfolded F508del-CFTR, the AFTs are aberrantly exposed leading to its ER retention and degradation [6]. The cellular machinery required to decode the AFTs is poorly characterized, and not just for CFTR, with still no clear global understanding of the interactors involved in this ERQC checkpoint.
Transcriptomic and proteomic-based approaches showed to be useful to analyse the global gene- and protein-expression patterns which differ in non-disease versus CF disease states, namely for F508del-CFTR [7–9]. Mapping CFTR interactome has also become a key strategy to identify critical interactions that potentially drive the CF phenotype with the ultimate goal of modulating them to rescue misfolded CFTR [10–13].
Here, we used a proteomic interaction profiling approach coupled to global bioinformatic analysis to identify differential protein–protein interactions (PPIs) of F508del-CFTR versus F508del-4RK-CFTR. We found that the F508del-CFTR interactome is enriched in proteins involved in RNA processing and complex organization with a decrease in those related to epithelial integrity when compared to the interactome of F508del-CFTR with abrogated AFT motifs. Among the proteins with increased affinity to F508del-CFTR, we identified kinesin-like protein (KIFC1), a kinesin motor protein involved in important biological processes such as vesicle transport. Our data show that when KIFC1 is downregulated or inhibited, the immature form of F508del-CFTR is stabilized by a decrease in its degradation rate.
We conclude that the approach used here enables further characterization of the molecular mechanisms of CFTR trafficking by identifying novel putative therapeutic targets that can be ultimately used to the benefit of CF patients.
Materials and methods
CFTR constructs and cloning
The F508del-4RK-CFTR cDNA construct (insert) was introduced into the lentiviral expression vector pLVX-Puro by homologous recombination using In-Fusion HD Cloning Kit (Clontech,#121416). First, pLVX-Puro vector was linearized using XhoI. Briefly, a mixture of 20 µl containing 20U XhoI enzyme, 1× Buffer R and pLVX-Puro (2 µg) was incubated at 37 °C for 3 h followed by 20 min at 80 °C. Second, the insert was amplified from the original expression vector by PCR using KOD Hot Start Polymerase (Novagen, #71316) and a set of primers (F:5′-GGACTCAGATCTCGAATGCAGAGGTCGCCTCTGGAAAAG-3′ and R:5′-GAAGCTTGAGCTCGACTAAAGCCTTGTATCTTGCATCTC-3′) according to the manufacturer’s instructions. Linearized pLVX-Puro vector and insert were purified using a 0.5% (w/v) agarose gel using NZYGelpure kit (Nzytech, #MBO1101) according to the manufacturer’s instructions. Homologous recombination was then performed using In-Fusion HD Cloning Kit (Clontech, #121416). Briefly, 20 μl of cloning reaction mixture containing 0.5× In-Fusion HD Enzyme Premix, linearized plasmid (50 ng) and insert (200 ng) was incubated at 50 °C for 15 min and transformed into competent bacteria. Correct insertion was verified by sequencing.
Cell line generation
CFBE41o- cells stably expressing F508del-4RK-CFTR were generated by lentiviral infection. Plasmids with correct sequence were used to calcium phosphate-transfect HEK 293T cell for production of lentiviral particles. Briefly, the transfection reaction mixture of 250 µl included F508del-4RK-CFTR-pLVX-Puro (5 µg), psPAX2 lentiviral packaging plasmid (4 µg) (Addgene, #12260), VSV.G lentiviral packaging plasmid (0.4 µg) (Addgene, #14888), 25 µl CaCl2 (2.5 M) and one-tenth of the volume of TE buffer (1 mM Tris–HCl, 0.1 mM EDTA, pH 7.6). The same volume (250 µl) of 2× HEPES buffered saline (HBS—50 mM HEPES, 280 mM NaCl2 and 1.5 mM Na2HPO4, pH 7.05) was added to the mixture and incubated for 30 min before adding to the cells. Cells were then incubated for 24 h at 37 °C in 5% CO2 before replacing the medium. Lentiviral particles were collected 48 h after transfection and were used to infect CFBE41o- cells. Finally, lentiviral particles were used to transduce parental CFBE41o- cells. Infection mixture of 2 ml containing 50% (v/v) of the collected supernatant with lentiviral particles and 50% (v/v) of DMEM supplemented with 10% (v/v) FBS (GIBCO® Life Technologies, #10270-106) and 8 µg of hexadimethrine bromide (polybrene) (Sigma-Aldrich, #107689-10G) was centrifuged at 10×g for 1 h at 25 °C and then used to infect parental CFBE41o- cells. 24 h post-infection, fresh media containing 40 ng/ml puromycin (50 mg/ml, Sigma-Aldrich, #P8833) was added. 48 h post-infection, puromycin concentration was raised (100 ng/ml).
Cell culture and compound treatment
CFBE41o- cells stably expressing wild-type-CFTR, F508del-CFTR or F508del-4RK-CFTR were grown in EMEM (Lonza, #BE12-611F) supplemented with 10% (v/v) FBS and puromycin (5 µg/ml) (Sigma, #P8833). HEK 293T (Human Embryonic Kidney 293) cells were grown in DMEM (Lonza, #BE12-604F) supplemented with 10% (v/v) FBS. All cell lines were grown at 37 °C in 5% CO2. When applicable, cells were incubated with the corrector VX-809 (3 µM for 24 h). For proteasome inhibiton, cells were treated with MG-132 (25 µM for 2 h). For KIFC1 inhibition, cells were treated with AZ82 (Ak Scientific inc, #SYN5480) (0.4 µM for 24, 48 and 72 h). All treatments used DMSO as vehicle control and were performed in the appropriate media supplemented with 0.1% (v/v) FBS.
CFTR immunoprecipitation
CFTR was immunoprecipitated from whole-cell lysates from CFBE41o- cells stably expressing either F508del- or F508del-4RK-CFTR. Parental CFBE410- cells (not expressing CFTR) were used to define the background using the anti-CFTR monoclonal antibody 596 coupled with rProtein G agarose beads. For the coupling antibody beads, the anti-CFTR 596 was incubated with beads at final concentration of 1 µg/ml at RT for 1 h with rocking. Beads were washed with 10 volumes of sodium borate (0.1 M) pH 9 and resuspended again in 10 volumes of sodium borate (0.1 M) pH 9 with dimethyl pimelimidate.2 HCl (DMP) (Thermo Scientific, #21667) at the final concentration to 20 mM. The DMP and antibody-bound beads were mixed for 30 min at RT. The reaction was terminated by washing the beads once in ethanolamine (0.2 M) pH 8 and twice in PBS. Finally, beads were resuspended in PBS with 0.02% (w/v) sodium azide and stored at 4 °C.
For in vivo cross-linking, immunoprecipitations were performed in the presence of cells incubated with the cleavable chemical cross-linker dithiobis (succinimidylpropinate) (DSP) (Thermo Scientific, #22585) prior to lysis.
For the preparation of cell lysate for CFTR immunoprecipitation, cells were washed in cold PBS supplemented with 0.9 mM CaCl2, 0.5 mM MgCl2, pH 7.2, incubated for 30 min in Triton lysis buffer (TBS) (25 mM Tris–HCl pH 7.4, 150 mM NaCl2, 1% (v/v) Triton-X 100, supplemented with protease inhibitors) at 4 °C and finally scraped and pelleted. For immunoprecipitation, the supernatant from cell lysate was pre-cleared for 1 h at 4 °C with rProtein G agarose beads, followed by incubation with anti-CFTR 596 antibody cross-linked to rProtein G agarose beads overnight at 4 °C. Finally, the sample was washed twice with wash buffer (Tris–HCl 100 mM; NaCl2 300 mM) supplemented with 1% (v/v) Triton-X100 and twice with wash buffer without Triton. Proteins were eluted in DTT (50 mM) for 15 min at RT with rocking, followed by incubation for 5 min at 37 °C with Tris–HCl 90 mM pH 7.2.
To validate the isolated proteins, we specifically assessed if proteins with differential interactions with F508del-CFTR versus F508del-4RK-CFTR also interacted with the AFT motifs. For this, we performed a pull-down of proteins interacting with 10 amino acid long synthetic peptides conjugated with agarose beads and containing the CFTR sequence around each of the AFTs—either the wild-type sequence or the Arg-to-Lys substituted versions. The pull-down was performed using either a mixture of the peptide-conjugated beads corresponding to the non-mutated peptides or to the substituted peptides in lysates from CFBE parental cells.
Sample preparation for LC–MS/MS
Following immunoprecipitation protein, complexes eluted in a solution containing DTT were loaded into filtering columns and washed exhaustively with urea (8 M) in HEPES buffer. After alkylation with iodoacetamide and reduction with DTT, proteins were incubated overnight with trypsin sequencing grade (Promega, Madison, WI).
Collection and analysis of peptide spectra using LC–MS/MS
Peptides generated as described above were desalted and concentrated [14] prior to analysis by nano LC–MS/MS using a Q-Exactive (Thermo, San Jose, CA) mass spectrometer coupled to an EASY-nLC 100 liquid chromatography system (Thermo, San Jose, CA) via a Nanospray Flex Ion Source. Peptides were loaded onto an Acclaim PepMap100 pre-column (75 µm × 2 cm, Thermo, San Jose, CA) connected to an Acclaim PepMap RSLC (50 µm × 15 cm, Thermo, San Jose, CA) analytical column. Peptides were eluted from the column using a linear gradient of 3–30% acetonitrile in 0.1% formic acid at a flow rate of 300 nL/min over 90 min. The mass spectrometer was operated in positive ion mode. Full MS scans were acquired from m/z 350 to 2000 with a resolution of 70.000 at m/z 200. The ten most intense ions were fragmented by higher energy C-trap dissociation with normalized collision energy of 28 and MS/MS spectra were recorded with a resolution of 17.500 at m/z 200. The maximum ion injection time was 120 ms for both survey and MS/MS scans, whereas AGC target values of 3 × 106 and 5 × 105 were used for survey and MS/MS scans, respectively. To avoid repeated sequencing of peptides, dynamic exclusion was applied for 30 s. Singly charged ions or ions with unassigned charge state were also excluded from MS/MS. Data were acquired using Xcalibur software (Thermo, San Jose, CA).
The intensity-based absolute quantification (iBAQ) [15], a label-free proteome quantification method, was used to determine the stoichiometries of the identified proteins. This quantification is based on peak intensity in survey scans, corresponding to the sum of all the peptides intensities divided by the number of observable peptides of a protein and is integrated into the quantitative proteome software package MaxQuant [16, 17]. All data were searched with VEMS [18] and MaxQuant [19]. Mass accuracy was set to 5 ppm for peptides and 10 mDa for peptide fragments. Gaussian weight for fragment ions was set to 5 and the six most intense fragment ions per 100 Da was used for scoring fragment ions. Two missed cleavages were specified and the Human database from UniProtKB (Release 2015_02) was used including permutated protein sequences, leaving Lys and Arg in place, together with common contaminants such as human keratins, bovine serum proteins and proteases. The total number of protein entries searched was 136314. Fixed modification of carbamidomethyl cysteine was included in the search parameters. A list of 8 variable modifications was considered for all data against the full protein database. Protein N-terminal Met-loss is not specified for VEMS searches since VEMS by default checks N-terminal Met-loss. The false discovery rate (FDR) for protein identification was set to 1% for peptide and protein identifications unless otherwise specified. No restriction was applied for minimal peptide length. Identified proteins were divided into evidence groups as defined previously [20]. Statistical analysis and data filtering was performed using R statistical programming language.1
After identification and quantification of the mass spectrometry results, each protein was normalized for the total amount of protein in the respective sample. To filter for unspecific interactions, a combination test was performed between each sample and the background (pull-down from parental CFBE41o- cells) using a R script. For this, the iBAQ values for each protein from each sample replicate in study (F508del-, F508del-4RK-CFTR) were divided by the iBAQ values for the same protein in each replicate of the control (parental cells) resulting in nine combinations. Then, the median for the sample replicates resulting from the combination test was calculated. To find the affinity of a given protein for F508del-4RK-CFTR versus F508del-CFTR, we calculated the log2 of the ratio of the median for each protein in F508del-4RK-CFTR over the median for the same protein in F508del-CFTR. All the pull-downs and MS analyses were performed in triplicate using separate cell cultures.
Pathway, gene ontology and protein–protein interaction analysis
We used available bioinformatics tools to analyse the lists of proteins interacting with the two CFTR variants of interest.
Data were submitted to gene set enrichment analysis (GSEA)2 [21, 22] to assess the distribution of regulated gene sets. GSEA software provides an enrichment score (ES) and p value to define a set of genes differentially expressed among different conditions/phenotypes.
Data were also submitted to The Database for Annotation, Visualization and Integrated Discovery (DAVID)3 [23, 24], to generate a list of the enriched gene ontology (GO) terms in the biological process (BP) and cellular component (CC) domains.
Pathway enrichment analysis was performed using the open source Reactome Pathway database4 [25, 26] accessed using ReactomePA package [27] in R, which provides a gene set enrichment and a p value. PPIs were generated using Agile Protein Interactome DataServed—APID5 [28] accessed and visualized using Cytoscape6 [29] or R.
siRNA transfection
Cells were transfected 24 h after being split. Transfection mixture using Lipofectamine 2000 (1 mg/ml, Invitrogen #11668019) was prepared containing the indicated siRNA at 75 nM (human KIFC1, Dharmacon Cat. No. L-004958-00-0005; human FKBP4, Dharmacon Cat. No. L-006410-00-0005; human YWHAE, Dharmacon Cat. No. L-017302-02-0005; human HNRNPA2B1, Dharmacon Cat. No. D-011690-01; EGFP, Eurofins MWG operon Cat No. 921154) and Lipofectamine (2.5 ng) to a final volume of 500 µL in OptiMEM, according to the manufacturer’s instructions. After transfection, cells were grown in FBS-free media for 24 h before changing the media to EMEM supplemented with 10% (v/v) FBS. 48 Vh post-transfection, cells were harvested and protein and RNA were isolated.
RNA and protein extraction
Cells were washed twice with 1xPBS and lysed with TP + PI buffer (50 mM Tris–HCL pH 7.5, 2 mM MgCl2, 100 mM NaCl2, 10% (v/v) glycerol, 1% NP-40 supplemented with protease inhibitors). The lysate was centrifuged (5000g for 5 min) and 60% of the supernatant was mixed with 15 µl of 5× sample buffer (SB) [156 mM Tris–Cl pH 6.8, 3% SDS, 25% glycerol, 0.05% (w/v) Bromophenol Blue, 249.6 mM DTT] supplemented with 25U Benzonase (Sigma-Aldrich, #E1014-25G) and 3.125 mM MgCl2. The remaining 40% of the supernatant and pellet were used for RNA extraction using the “DNA, RNA and protein purification Kit” (Macherey–Nagel, #740955.250) according to the manufacturer’s instructions.
Reverse transcription-PCR and quantitative real-time PCR
cDNA was synthesized using NZY M-MuLV Reverse Transcriptase (Nzytech, #MBO8301) according to the manufacturer’s instructions. qRT-PCR amplifications were performed in a Cx96 real-time PCR machine using 96 well plates with SsoFastTM Evagreen® supermix reaction mixture (Bio-Rad laboratories, #172-5201) and a set of primers found at Harvard Primerbank7 [30] (human KIFC1, F:5′-GGTGCAACGACCAAAATTACC-3′, R:5′-GGGTCCTGTCTTCTTGGAAAC-3′, PrimerBank ID 167555109c1; human FKBP4, F:5′-GCTGGCTATTAGATGGCACAA-3′, R:5′CCAAGCCTTGATGACCTCCC-3′, PrimerBank ID 206725538c2; human YWHAE, F:5′-ACACCTCATTCCAGCAGCTAA-3′, R:5′-TCTGCCAGATACCTGTGGTAG-3′, PrimerBank ID 195546907c2; human HNRNPA2B1, F:5′-TGGAGGTAGCCCCGGTTATG-3′, R:5’-GGACCGTAGTTAGAAGGTTGCT-3′, PrimerBank ID 156151374c2; and human CAP-1, F:5′-ATGCACCGTGGGTATGCAG-3′, R:5′-AAGCAGCGAGTCAAATGCCT-3′; PrimerBank ID 157649072c1) according to the manufacturer’s instructions. The fold difference in gene expression was calculated by the relative quantification method using the mathematical equation 2−ddCT [7].
Western blotting
Protein extracts from cell lines were separated on 7% (w/v) polyacrylamide gels, transferred to PVDF membranes and analysed by Western blot (WB). Membranes were blocked and probed with 1:5000 mouse anti-CFTR 596 (CFF, Cat No A4), 1:200 rabbit anti-KIFC1 (Proteintech, Cat No 20790-1-AP) or 1:1000 mouse anti-polyubiquitinylated conjugates Mab (FK1) (Enzo Sciences, Cat No BML-PW8805-0500), overnight 4 °C. Membranes were also probed with 1:10,000 mouse anti-alpha-Tubulin (Sigma, Cat No T5168) or 1:3000 mouse anti-calnexin (BD Transduction Laboratories™, Cat No 610523) which were used as loading controls. Then, membranes were washed three times followed by 1 h incubation with 1:3000 secondary antibodies. Antibody dilutions were all made in blocking solution (5% w/v milk/BSA in PBS-T/TBS-T). Chemiluminescent detection was performed using the Clarity™ Western ECL substrate (BioRad, #170-5061) and the Chemidoc™ XRS system (BioRad). The quantification of band intensity was performed using the Image Lab software (BioRad) and normalized to the loading control as appropriate.
Immunoprecipitation of polyubiquitinylated proteins
Polyubiquitinylated proteins were immunoprecipitated from stable CFBE41o-cells expressing F508del-CFTR using the anti-polyubiquitinylated conjugates (FK1) antibody. All the procedures were performed as described above and included an additional conjugation of an anti-IgM with rProtein G agarose beads. Proteins were eluted in 40 µl 1× Laemmli buffer for 30 min at room temperature (RT).
Iodide efflux assay
The CFTR-mediated iodide efflux assay was performed as described [31, 32]. Cells grown in 6-well plates were transfected with either siRNA against EGFP or KIFC1 or treated with VX-809, as given above. 48 h after either transfection or treatment, cells were loaded with iodide in the loading buffer for 30 min at 37 °C, thoroughly washed with iodide-free efflux buffer and equilibrated for 10 min in the same buffer. Cells were then incubated for 5 min either in the presence of iodide-free efflux buffer or in the presence of CFTR stimulators (10 μM Fsk and 50 μM IBMX, Sigma). Cells were lysed and the iodide concentration in each sample was determined using an iodide-sensitive electrode (Orion 96–53; Thermo Scientific, Rockford, IL, USA) with a pH/mV meter and normalized to the amount of protein. In this assay, increased channel activity corresponds to more iodide released from the cells and thus a decreased iodide concentration remaining within the cells.
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). Statistical analysis was performed with GraphPad Prism 7 software. Significance was assessed by the statistical tests indicated in the figure legends.
Results
Mapping the interactome of F508del-CFTR and F508del-4RK-CFTR
To investigate the differential protein interactions between F508del- and F508del-4RK-CFTR, protein complexes were isolated by pulling-down CFTR from CFBE cells expressing these two variants. CFBE cells expressing F508del-4RK-CFTR were produced as described above and characterized by WB (Fig. S1). These cells recapitulate all the characteristics (presence of mature CFTR, additivity with correctors and low temperature) previously reported for this variant when expressed in BHK cells [6, 33]. To discard non-specific interactions, CFBE cells which do not express CFTR protein (parental CFBE cells) were used as a background control.
A total of 834 different proteins (Table S1) were identified: 793 proteins for F508del-4RK-CFTR and 828 proteins for F508del-CFTR group with an overlap of 95% (791 common proteins out of 834 identified proteins). Comparing the different replicates for each cell line, we obtained an overlap of 79% (626 common proteins in all replicates) in F508del-4RK-CFTR and of 82% (675 common proteins in all replicates) in F508del-CFTR, confirming the reproducibility of the approach. Figure 1a summarizes the approach used and the number of proteins obtained and analysed throughout this work.
Fig. 1.
Differential protein interactions for F508del-CFTR versus F508del-4RK-CFTR. a Workflow representing the strategy used for the identification of differential protein–protein interactions. b Log2 plot of the protein abundance ratio for F508del-4RK-CFTR versus F508del-CFTR. Proteins were identified by LC–MS/MS and black dashed lines indicate the threshold (log2 = ±1)
Based on the quantitative profiling of F508del-CFTR versus F508del-4RK-CFTR and using log2 ratio of below − 1 and above 1 as a threshold (see “Material and methods”), we obtained 164 proteins with higher affinity for F508del-4RK-CFTR (above the threshold log2 ratio 1) and 198 proteins with higher affinity for F508del-CFTR (below the threshold log2 ratio − 1) (Fig. 1b and Table S2).
Bioinformatic analysis of the F508del-CFTR and F508del-4RK-CFTR interactomes
Using GSEA tool, we found the following biologic pathways enriched in the F508del-CFTR versus the F508del-4RK-CFTR interactome: epithelial-to-mesenchymal transition (EMT), unfolded protein response (UPR), transcription and cell cycle machineries are enriched among the F508del-CFTR interactors (Fig. S2A, grey dots). On the other hand, mTOR signalling and epithelial integrity (apical junctions) are among the most enriched pathways in the F508del-4RK-CFTR versus the F508del-CFTR interactome (Fig. S2A, black dots).
Using DAVID, we identified the most represented BP and CC GO terms in the two datasets. For the F508del-4RK-CFTR interactome, we found an enrichment in BPs related to cell integrity, such as response to wounding and cell death (Fig. S2B), whereas the F508del-CFTR interactome was enriched in RNA-related processes (Fig. S2B). As to CC, for F508del-4RK-CFTR, we detected an enrichment in proteins associated with vesicles and membrane fraction (Fig. S2C), whereas for F508del-CFTR, the cytoskeleton components and membrane-enclosed lumen seem to be enriched.
Table 1 summarizes the most represented BPs from the bioinformatics analysis performed.
Table 1.
Summary of processes enriched in F508del- and F508del-4RK-CFTR interactomes
| Enrichment | F508del-CFTR | F508del-4RK-CFTR |
|---|---|---|
| Gene set enrichment analysis | EMT | Apical junction |
| UPR | mTOR signalling | |
| Gene ontology (DAVID) | ||
| BP | RNA processing | Response to wounding |
| Response to cell death | ||
| CC | Cytoskeleton | Vesicles |
| Membrane-enclosed lumen | Membrane fraction | |
Generation of protein networks
To further characterize these datasets of F508del-4RK-CFTR and F508del-CFTR interactors, we compared them with previous work identifying the CFTR interactome [12]. For that, we crossed our list of 834 interactors with that of 638 proteins previously described as forming the “core CFTR interactome” (direct and indirect interactors) [12]. We identified a common set of 217 proteins present in both studies, i.e., 26% of our complete list of interactors [12], with an enrichment in upregulated tissue classes “Epithelium” and “Lung” (identified in tissue expression from DAVID).
To find further functional significance for these 834 proteins, we performed a comparison between our work and a genome-wide cell-based secretion screen in which the cell “secretome” was identified [34]. These analyses demonstrated that among the 834 proteins, 143 were previously identified as traffic promoters [34] and 8 as traffic inhibitors [34].
We then focussed on the 198 proteins with higher affinity for F508del-CFTR (over F508del-4RK-CFTR), as these are potential factors involved in its ER retention and/or degradation thus having higher potential to become drug targets. To confirm that this list contains specific interactors, we analysed these proteins with the “Contaminant Repository for Affinity Purification” (CRAPome) database [35]. This analysis revealed that these proteins have a very low average spectral count (SC) in the database (median of 3.4) with only 12% of them being detected with an average SC greater than 10, thus confirming that these are specific interactors. To refine the list, we then searched the ChEMBL database, to find whether inhibitory molecules are available for these 198 interactors [36]. We found 22 proteins for which inhibitors have been reported (Table S3).
To further refine our list, we analysed whether some of these 22 putative hits could also be detected as interactors of the AFT motifs—either wild-type sequence (Arg containing—peptide R) or mutated/abrogated AFTs sequence (Arg-to-Lys substituted versions—peptide K). For this, we used peptides corresponding to these sequences conjugated with agarose beads to pull-down and identified proteins by LC–MS/MS as above. We compared the magnitude of the change in Log2 ratio (F508del-4RK-CFTR/F508del-CFTR) and Log2 ratio (K/R) for these 22 proteins (Fig. S3). These data together with a detailed analysis of the molecules available as well as of the positioning in biological pathways (using the reactome pathway database) and specific protein characteristics (using UniProt) led to the selection of the top 4 proteins—Kinesin Family Member C1 (KIFC1), Heterologous nuclear riboproteins A2/B1 (HNRNPA2B1), 14-3-3 protein epsilon/14-3-3ε (YWHAE) and peptidyl-prolyl cis-transisomerase FK506 binding protein 4 (FKBP4). KIFC1 and HNRNPA2B1 were among the subset of 143 traffic promoters [34] and YWHAE is present in the core CFTR interactome [12].
Using the APID protein–protein interaction database [28], we then created a network (Fig. S4) showing the interactions (represented by straight white lines) between CFTR interactome (depicted in green circle) and the interactomes of these four top hits (depicted in orange circles). The network represents the distribution of the hits relatively to CFTR, comprising proteins which are: (1) in close proximity to CFTR, with just one node distance (edges represented as dashed red lines) and (2) distant from CFTR, with two or more nodes distance (edges represented as full orange lines). Interestingly, FKBP4 presents one node distance to CFTR and the intermediates are proteins previously implicated in ER-associated folding and/or degradation of CFTR, such as Hsp90 (HSP90AA1 and HSP90AB1) and Hsp27 (HSPB1) (Fig. S4). HNRNPA2B1 connects to CFTR through proteasome components (PSMB1 and PSMA3) and appears to be also linked to 14-3-3ε (YWHAE) (Fig. S4). Interestingly, Hsp27 also mediates the interaction between 14-3-3ε and CFTR. Thus, three out of the four top hits fall into different categories in terms of their possible/previous association with CFTR. We then tested the impact of knocking down these three targets by siRNA transfection and assessed F508del-CFTR processing (Fig. S5). Knockdown of all the three targets resulted in a modest, albeit significant, rescue of F508del-CFTR (Fig. S5).
As to KIFC1, no direct association to CFTR was previously reported (Fig. S4) and thus it was selected for a more detailed mechanistic evaluation.
KIFC1 interacts with F508del-CFTR
To confirm the interaction between KIFC1 and F508del-CFTR, we immunoprecipitated F508del-CFTR and detected KIFC1 by WB (Fig. 2a). We then used the reverse approach to that used in LC–MS/MS. We immunoprecipitated KIFC1 from CFBE cells expressing wt-, F508del- or F508del-4RK-CFTR and detected CFTR by WB. CFBE non-CFTR expressing cells (parental) were also used as a control. We were able to detect wt-, F508del- and F508del-4RK-CFTR in the immunoprecipitate of KIFC1, showing that KIFC1 interacts with all these CFTR variants in CFBE cells (Fig. 2b). The absence of signal in the non-CFTR expressing CFBE cells confirmed the specificity of the interaction (Fig. 2b). Interestingly, KIFC1 appears to interact with both immature and mature forms of wt-CFTR, as both bands B and C are detected in the IP (Fig. 2b). Quantification of the interaction (normalization of CFTR detected to the levels of KIFC1 pulled-down) shows a stronger interaction of KIFC1 with F508del-CFTR versus F508del-4RK-CFTR (1.5-fold), which is even more pronounced (threefold) versus wt-CFTR (Fig. 2c). These data confirm the LC–MS/MS results showing that KIFC1 interacts with increased affinity to F508del-CFTR versus F508del-4RK- and also wt-CFTR.
Fig. 2.
Interaction between KIFC1 and CFTR. Immunoprecipitation was performed in CFBE cells expressing wt-CFTR, F508del-CFTR or F508del-4RK-CFTR. CFTR null cells (CFBE parental) were used as a control. a KIFC1 was detected by WB after immunoprecipitation of CFTR with a monoclonal antibody—CFTR Pull-down (top). 20% of the total lysate was analysed as whole-cell lysate (WCL). b CFTR (~ 140 kDa band B and ~ 170 kDa band C was detected by WB after immunoprecipitation of KIFC1 with a specific polyclonal antibody cross-linked to rProtein G agarose beads—KIFC1 Pull-down (top). 20% of the total lysate was analysed as whole-cell lysate (WCL). Equal amount of protein was loaded in each lane, as demonstrated by α-tubulin (~ 50 kDa) loading control. c KIFC1: CFTR interaction was quantified by calculating the CFTR immunoprecipitated normalized to KIFC1 pull-down, total WCL CFTR and loading control. Data are shown as mean ± SEM, n = 3. *p ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test
KIFC1 downregulation promotes stabilization of CFTR immature form
To characterize the role of KIFC1 in its interaction with F508del-CFTR, we assessed how a decrease in KIFC1 total levels affects F508del-CFTR expression and processing by WB. CFBE cells stably expressing F508del-CFTR were thus reverse transfected with a pool of two different siRNAs targeting KIFC1 (siKIFC1). Non-transfected cells and cells transfected with siRNA targeting GFP (siEGFP) were used as negative controls and the same cells incubated for 24 h with the corrector VX-809 (3 µM) were used as a positive control for F508del-CFTR rescue [37].
After transfection with the siRNA, KIFC1 protein and mRNA levels were reduced by ~ 80% (as assessed by WB and qRT-PCR, Fig. 3d, e). This KIFC1 knockdown resulted in increased total levels of immature (band B) F508del-CFTR (1.5-fold) when compared with the control (non-targeting siRNA—siEGFP) (Fig. 3a, b). We also observed that KIFC1 knockdown led to the appearance of mature F508del-CFTR (a 1.5-fold increase when compared to control siRNA) (Fig. 3c). This appearance of mature F508del-CFTR also elicits an increase in CFTR function as assessed by iodide efflux assay (Fig. 3f).
Fig. 3.
Effect of KIFC1 knockdown on F508del-CFTR protein expression and processing. CFBE cells expressing F508del-CFTR were transfected with siRNA pool (four siRNAs) against KIFC1 or EGFP as non-targeting siRNA for 48 h. CT—transfection reagent only was used. Cells were also incubated with VX-809 (3 µM) or DMSO (vehicle control) for 24 h when mentioned. a Detection of F508del-CFTR (~ 140 kDa band B and ~ 170 kDa band C) protein expression (top). Equal amount of protein was loaded in each lane, as demonstrated by calnexin loading control (bottom). b Total CFTR (band B and C) normalized to calnexin and to siEGFP was quantified. c CFTR processing (band C divided by total CFTR) was normalized to siEGFP control. d Fold expression of KIFC1 mRNA levels was obtained by relative quantification (ddCT method) and was normalized to an internal control (CAP-1). e Quantification of KIFC1 protein levels after transfection with siRNA against KIFC1. KIFC1 levels were normalized to calnexin and shown relatively to the siEGFP control. Data are shown as mean ± SEM, n = 3. f Iodide efflux of CFBE-F508del cells either transfected with siRNA against EGFP/KIFC1 or treated with 3 μM VX-809 (or DMSO as control) for 48 h. Fold change of iodide that remained in the cells relative to control condition (siRNA for EGFP or DMSO). Data are shown as mean ± SEM, n = 4. *p ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test
To further confirm the effect of KIFC1 knockdown on F508del-CFTR, we tested a chemical inhibitor of KIFC1 (compound 39 or AZ82) [38] identified using the ChEMBL database (Table S3). AZ82 binds to KIFC1 and inhibits the binding of ATP and release of ADP [38]. This compound is KIFC1-selective, showing no detectable activity against other kinesin motor proteins [38]. CFBE cells expressing F508del-CFTR were incubated with 0.4 µM of AZ82 for 24, 48 and 72 h in parallel with siRNA-KIFC1 for comparison. Treatment with 0.4 µM AZ82 significantly increased the immature form of F508del-CFTR (Band B) at 48 and 72 h, although in lower levels when compared with KIFC1 knockdown (Fig. S6A-B). CFTR band C also increases slightly (Fig. S6C).
KIFC1 expression is increased in cells expressing F508del-CFTR
As KIFC1 has been frequently reported to be overexpressed in many cancers [39, 40], we assessed whether KIFC1 is overexpressed in CFBE cells expressing F508del-CFTR versus wt-CFTR or F508del-4RK-CFTR. WB analysis revealed that KIFC1 protein levels are increased in CFBE cells expressing F508del-CFTR when compared with wt- or F508del-4RK-CFTR expressing cells (Fig. 4a, b). KIFC1 mRNA levels were also quantified by qPCR in CFBE cells expressing the three CFTR variants (Fig. 4c) but also in human lung tissue from either non-CF individuals or CF patients homozygous for F508del (Fig. 4d). Results showed that lung samples from CF patients and F508del-CFTR expressing CFBE cells presented higher levels of KIFC1 mRNA versus their wt- counterparts and in the case of CFBE cells also than cells expressing F508del-4RK-CFTR cells (Fig. 4c, d). Thus, besides a stronger interaction between KIFC1 and F508del-CFTR, there is also a significant increase in the levels of KIFC1 in cells expressing the mutant when compared to ones expressing the wt protein.
Fig. 4.
KIFC1 levels in CFBE cells and human lung tissue. a KIFC1 (~ 74 kDa) protein levels (middle) in CFBE parental cells and CFBE cells expressing wt-, F508del-or F508del-4RK-CFTR, represented as immature form (~ 140 kDa band B) and mature form (~ 170 kDa band C) (top) was determined by Western blotting. Equal amount of protein was loaded in each lane, as demonstrated by α-tubulin (~ 50 kDa) loading control (bottom). b Amount of KIFC1 from experiment A was normalized to loading control and plotted as relative to CFBE parental cells (n = 3). KIFC1 mRNA levels from (C) CFBE cells (n = 3) and d human lung tissue from non-CF individuals (n = 4) and CF patients homozygous for F508del (n = 5) were obtained by relative quantification ddCT method and was normalized to an internal control (CAP-1). Data plotted as mean ± SEM, *p ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test
KIFC1 downregulation prevents misfolded F508del-CFTR from targeting to degradation
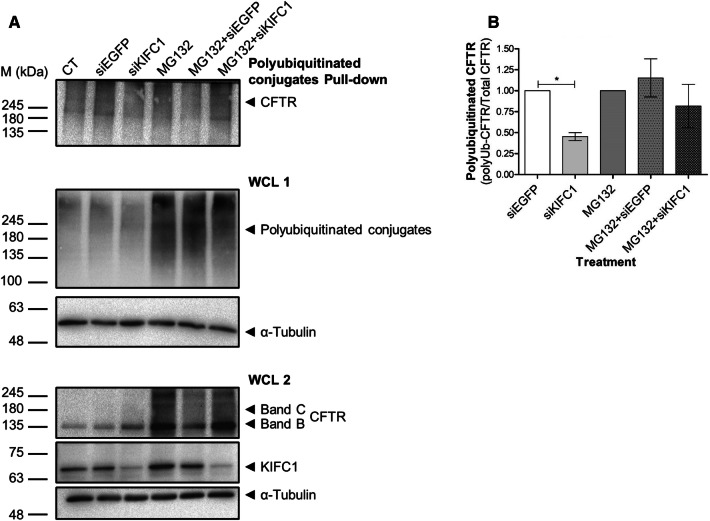
As the decrease in KIFC1 levels leads to an increase in the total amount of F508del-CFTR, next we assessed whether KIFC1 plays a role in CFTR degradation. To this end, we assessed F508del-CFTR ubiquitination under KIFC1 knockdown versus control conditions by pulling-down polyubiquitinated conjugates followed by CFTR detection by WB. Results show that, under KIFC1 knockdown, there is a decrease in the amount of polyubiquitinated F508del-CFTR (Fig. 5a, b). Results also confirm that under this situation there is an increase in both immature and mature form of CFTR in F508del-CFTR CFBE cells (Fig. 5a, b) as above (Fig. 3). When KIFC1 knockdown was combined with treatment with the proteasome inhibitor MG132, we confirmed again the decrease in CFTR polyubiquitination (Fig. 5b) and once again an increase in both immature and mature form of CFTR (Fig. 5a).
Fig. 5.
Polyubiquitination of CFTR under downregulation of KIFC1. F508del-CFTR CFBE cells transfected with siKIFC1 or siEGFP (non-targeting siRNA) were treated for 2 h with 25 µM MG132 compound. CT—transfection reagent only. a CFTR was detected by WB after immunoprecipitation with polyubiquitinated conjugates-specific antibody. 20% of the total lysate was analysed as whole-cell lysate (WCL). Equal amount of protein was loaded in each lane, as demonstrated by α-tubulin (~ 50 kDa) detection. Total polyubiquitinated conjugates are represented in WCL 1 and total CFTR and KIFC1 are represented in WCL 2. b Quantification of polyubiquitinated CFTR, calculated as the ratio of Ub-CFTR to total CFTR. Data shown as the mean ± SEM, n = 3. *p ≤ 0.05 Statistical analysis was performed using two-tailed unpaired Student’s t test
To further understand the mechanism by which KIFC1 regulates CFTR degradation, we used APID accessed in R to analyse the distance of the 198 interactors showing higher affinity to F508del-CFTR (Table S2) to KIFC1 (see Methods). The interactors were subjected to APID1, which accounts for all known interactions, and APID2, which accounts for interactions proved by two or more experiments. Among our list, we did not detect any previously reported direct interactors with KIFC1. However, we found that 97% (APID1) and 68% (APID2) of the interactors are from two to three edges of distance to KIFC1 (Fig. S7). Considering this distance, we found components of the 26S proteasome and small ubiquitin-like modifier 2 (SUMO-2), i.e., proteins involved in degradation-related processes, as possible nodes connecting KIFC1 and CFTR. Interestingly, Hsp90 and FKBP8, proteins involved in the folding process and known CFTR interactors, are also found as possible connectors of KIFC1–CFTR.
Discussion
The most common CF-causing mutation, F508del, leads to CFTR misfolding and thus to its ER retention and early degradation by the proteasome. Here, we focused on one of the mechanisms involved in F508del-CFTR retention—the aberrant exposure of the ER retention motifs (AFTs) which leads to the recognition of F508del-CFTR by the ERQC [5, 6]. In the present study, we mapped the comparative interactome of F508del-CFTR and F508del-4RK-CFTR aiming at identifying biological pathways dictating ER retention and degradation of F508del-CFTR and novel putative therapeutic targets in CF.
Previous studies focused on the identification of the total CFTR interactome. An early sutdy in BHK cells expressing either wt- or F508del-CFTR [10] provided as a major finding that wt- and F508del-CFTR present a differential interaction with Aha1, a Hsp90 cochaperone [10]. Later, a comprehensive interactome study focused on wt- and F508del-CFTR expression in epithelial airway cells (CFBE41o- versus 16HBE14o- cells) and assessed the interactome dynamics upon F508del-CFTR rescue by temperature shift or intervention by histone deacetylase inhibitor (HDACi) [12]. Another study also assessing wt- and F508del-CFTR interactome in CFBE41o- and 16HBE14o- cells revealed mTOR signalling components associated to F508del-CFTR [13]. All these studies identified and characterized interactions which contribute for CFTR folding and trafficking as well as regulatory pathways but did not address particular interactions and regulatory pathways that specifically characterize the ER retention/retrieval processes.
Using a combination of approaches (CFTR immunoprecipitation, mass spectrometry and systems biology-based analysis), we obtained a comprehensive comparative interactomic profiling of F508del-CFTR versus F508del-4RK-CFTR expressed in human respiratory epithelial cells. We identified 834 total proteins of which 791 proteins were detected as interacting with the two variants. Quantitative analysis allowed the identification of two subsets of 198 and 164 proteins showing higher affinity to F508del- or F508del-4RK-CFTR, respectively (Fig. 1b). Comparison of our study with previous ones [12] identified as common proteins known to be involved in CFTR folding and degradation, such as chaperones DnaJ, calreticulin and members of Hsp90 and 70 families, transitional ER ATPase (VCP) or proteasome components [12]. Comparison with functional genomics screens aimed at identifying regulators of CFTR traffic and function also highlighted some common targets, such as the catalytic subunit of casein kinase II CSNK2A1, the assembly factor MNAT1 or the scaffold protein PML [41, 42].
Although similarities shared with other data sets may indicate processes/features with increased significance in CFTR regulation, the novelty of our approach lies in the analysis of the F508del- versus F508del-4RK-CFTR interactomes. Using GSEA, we found an enrichment of EMT and UPR among the F508del-CFTR interactors (Fig. S2A), similarly to what previously described in the context of CF [13] and also in agreement with a previous study assessing the impact of F508del-4RK-CFTR and F508del-CFTR on the whole proteome in BHK cells [11].
GO analysis identified an enrichment in biological processes related to cell integrity, such as response to wounding and cell death (Fig. S2B), among F508del-4RK-CFTR interactors. Such processes are described to be downregulated in CF disease and thus less present among F508del-CFTR interactors. Interestingly, this result is in agreement with previous studies in which a genome-wide microarray study of gene expression in human native nasal epithelial cells from F508del-homozygotic patients versus non-CF controls also identified an enrichment in wound healing in CF cells [7].
Regarding the GO domain CC, we found an enrichment in proteins associated with vesicles and membrane fraction for F508del-4RK-CFTR, which probably relates with its ability to escape the ERQC control thus increasing its interaction with trafficking machinery components (Fig. S2C).
From the diversity of proteins differentially interacting with F508del- versus F508del-4RK-CFTR for which inhibitors are available (Table S3), KIFC1 was selected as a novel interactor showing higher affinity to F508del-CFTR. We focused particularly on this interactor as there were no previous reports of any role in the context of CF (Fig. S4) and also because KIFC1 had also been previously identified as a regulator of secretory traffic [34]. KIFC1 belongs to the kinesin superfamily of proteins (KIFs) which are known to function as motors that move along the microtubules to transport protein complexes and organelles [43] and its excessive activation has been associated with cancer [39, 40].
Validation of the MS results allowed us to detect an increased interaction of KIFC1 with F508del-CFTR over not only F508del-4RK- but also wt-CFTR (Fig. 2). Interestingly, we also validated KIFC1 interaction with CFTR peptides corresponding to the wt AFT sequences but not to the mutated ones (Fig. S3). We then assessed if a decrease in KIFC1 would contribute to promote F508del-CFTR escape from the ER. We found that decreasing KIFC1 levels (with siRNA) or activity (with AZ82) has a positive effect on F508del-CFTR levels and also processing, leading to the appearance of the mature form for the mutant protein (Figs. 3, S6).
Since KIFC1 has a stronger association with F508del-CFTR and stabilizes its immature form, we hypothesized that KIFC1 might be involved in CFTR degradation. Reduced levels of KIFC1 decreased polyubiquitinated F508del-CFTR (Fig. 5), consistent with a role for KIFC1 in promoting F508del-CFTR degradation via polyubiquitination. Using APID, we found that components of the 26S proteasome and SUMO-2 are possible nodes connectors between KIFC1 and CFTR (Fig. S7). Both SUMO modification and the proteasome were reported to mediate F508del-CFTR degradation [44–46]. Thus, our results suggest that KIFC1 might form a link to the degradation machinery promoting F508del-CFTR polyubiquitination and consequently mediating its degradation at the proteasome.
Interestingly, these results lead us to also assess KIFC1 mRNA and protein levels. We detected an overexpression of KIFC1 in cells expressing the CFTR mutant (Fig. 4), a result that was also confirmed in lung samples form F508del homozygous patients (when compared to non-diseased controls) (Fig. 4).
Kinesin family member C1 has been extensively studied as a therapeutic target, since it has an essential role in mitosis and is frequently overexpressed in human cancers [39, 40]. In cancer cells, it functions as the main force to cluster the amplified centrosomes, allowing cells to pass mitosis. In normal cells, without centrosome amplification, KIFC1 appears to be nonessential for cell division, since its knockdown does not lead to a change in the architecture of microtubules and multipolar spindles [47, 48].
Interestingly, upregulation of genes involved in cell proliferation has been identified as a hallmark of CF in a whole-genome microarray study of gene expression comparing human native nasal epithelial cells from F508del-CFTR homozygotes with non-CF controls [7]. In addition, a previous study reported abnormal distribution of CFTR in primary human breast cancer and that reduced levels of CFTR lead to mesenchymal phenotype characterized by loss of E-cadherin (an epithelial cell marker) also observed in EMT process [49]. Thus, our results suggest that KIFC1 upregulation and interaction with F508del-CFTR may be related with this aspect of the CF phenotype and provide a connection to the increased risk of several cancer forms in CF patients [50].
Kinesin family member C1 relevance in F508del-CFTR retention phenotype is also in agreement with its described cellular localization. Besides being present in the nucleus, KIFC1 is also observed to be distributed in the cytoplasm indicating a dynamic binding between KIFC1 and microtubules [51]. This suggests that KIFC1 binding to microtubules may indicate its participation not only in spindle formation but also in protein/vesicle trafficking [51]. In fact, early endosomes require KIFC1 mediating the motility toward the plus end of the microtubules [52]. Interestingly, it was documented that vesicles associated with KIFC1 also contain KIF5B [40], another member of the KIF family also detected in our interactome.
Overall, our results suggest that KIFC1 may be involved not just in CFTR regulation but also in the cancer-propensity reported to occur in CF. In addition, our data supports exploring of KIFC1 as a biomarker of CF lung disease and the negative relationship between elevated KIFC1 levels and CFTR in CF are suggestive of its potential as a drug target.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1 - Characterization of CFBE cells stably expressing F508del-4RK-CFTR. CFBE cells expressing F508del-4RK-CFTR (lane 1-6), wt-CFTR (lane 7) and F508del-CFTR (lane 8) were grown at 37 °C or 26 °C and in some cases incubated with VX-809 (3 µM) for 24 h or DMSO (vehicle compound). Western blot (top) showing both mature form band C (~ 170 kDa) and immature form band B (~ 140 kDa) of CFTR protein. Calnexin (90 kDa) was used as a loading control (bottom). (JPEG 66 kb)
Fig. S2 - Bioinformatics analysis of the proteins differentially interacting with F508del-CFTR and F508del-4RK-CFTR. (A) Hallmark enrichment for F508del-4RK-CFTR and F508del-CFTR using gene set enrichment (GSEA). GSEA was performed for a dataset of 834 proteins. Using GSEA tool, the subset of genes that contribute mostly to the enrichment of each hallmark was identified and plotted according to their affinity to F508del-4RK-CFTR (black dots) versus F508del-CFTR (grey dots) - represented by log2 of the ratio F508del-4RK-CFTR/F508del-CFTR. (B) Biological process represented for F508del-4RK-CFTR and F508del-CFTR using DAVID. From the total of 834 proteins, 198 with more affinity to F508del-CFTR and 164 proteins with more affinity to F508del-4RK-CFTR were used to find the GO terms – biological process enriched in both subsets. p value < 0.05. (C) Cellular component represented for F508del-4RK-CFTR and F508del-CFTR using DAVID. From the total of 834 proteins, 198 with more affinity to F508del-CFTR and 164 proteins with more affinity to F508del-4RK-CFTR were used to find the GO terms – cellular compartment enriched in both subsets. Significant levels are represented by p value < 0.05. (PDF 371 kb)
Fig. S3 - Scatter plot representing the differential protein interactions for the 22 putative hits in F508del-CFTR versus F508del-4RK-CFTR and peptides corresponding to the AFT regions of CFTR. Log2 plot of the protein abundance ratio for F508del-4RK-CFTR versus F508del-CFTR and Log2 plot of the protein abundance ratio for peptide K (corresponding to mutated AFTs with Lys replacement) versus peptide R (Arg containing). Proteins were identified by LC–MS/MS and the threshold (log2 = ±1). (JPEG 113 kb)
Fig. S4 - Force-directed network of CFTR versus top hits interactome. Network comprises the connection of CFTR interactome (depicted in green circle) and the interactome of the selected hits: FKBP4, HNRNPA2B1, KIFC1, YWHAE (depicted in orange circles). All components comprising the interactors are predicted as nodes (yellow circles) and all were identified by LS-MS/MS covered the 834 proteins. Straight white lines illustrate edges that define interactions based on the APID protein interaction database which was accessed using Cytoscape platform. Dashed red lines illustrate the edges in which interactions occur with one node distance and full orange lines the edges in which interactions occur with two or more nodes distance to CFTR. (JPEG 149 kb)
Fig. S5 - Effect of FKBP4, 14-3-3ε and HNRNPA2B1 knockdown on F508del-CFTR protein processing. CFBE cells expressing F508del-CFTR were transfected with siRNA against FKBP4, 14-3-3ε, HNRNPA2B1 or EGFP as non-targeting siRNA for 48 h. CT - transfection reagent only was used. Cells were also incubated with VX-809 (3 µM) or DMSO (vehicle control) are shown. Detection of F508del-CFTR protein expression (top) under (A) FKBP4, (B) 14-3-3ε or (C) HNRNPA2B1 knockdown. Equal amount of protein was loaded in each lane, as demonstrated by calnexin loading control (Bottom). (D) CFTR processing (band C divided by total CFTR) was normalized to siEGFP control. (E) Gene knockdown with siRNA in CFBE cells. Fold expression of FKBP4, 14-3-3ε or HNRNPA2B1 mRNA levels was obtained by relative quantification (ddCT method) and was normalized to an internal control (CAP-1). Data are shown as the mean ± SEM, n = 3. * p ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test. (JPEG 331 kb)
Fig. S6 - Effect of KIFC1 inhibition on F508del-CFTR expression. (A) CFBE cells expressing F508del-CFTR were incubated with 0.4 µM KIFC1 inhibitor (AZ82 compound) or DMSO, vehicle control (non-treated - NT), for 24 h, 48 h or 72 h. As a control for KIFC1 levels, cells were transfected with siRNA pool for KIFC1 or EGFP as non-targeting siRNA for 48 h. Western blot for CFTR (Top). Equal amount of protein was loaded in each lane, as demonstrated by α-tubulin (~ 50 kDa) detection (Bottom). (B) Quantification of CFTR (band B and C) band intensity normalized to loading control and to NT or siEGFP. (C) CFTR processing (band C divided by total CFTR) normalized to NT or siEGFP. Data are shown as mean ± SEM, n = 3. * ρ ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test. (JPEG 144 kb)
Fig. S7 - Distance of F508del-CFTR interactors to KIFC1. The 198 interactors with higher affinity to F508del-CFTR were subjected to APID1 (accounting for all known interactions) and APID2 (accounting for interactions proved by two or more experiments). Each circle represents the distance from the proteins to KIFC1 (from one to five edges). Inside of each circle is represented the number of proteins showing the associated distance. (JPEG 92 kb)
Table S1 - Total interactome identified for F508del-CFTR and F508del-4RK-CFTR. All identified proteins from MS analysis. (XLSX 156 kb)
Table S2 - Protein fold change interaction for F508del-CFTR versus F508del-4RK-CFTR. Fold change interaction was obtained by log2 of the ratio of the amount of the protein detected in association with F508del-CFTR versus F508del-4RK-CFTR. (XLSX 94 kb)
Table S3 - Protein targets and inhibitors listed in ChEMBL database. The 198 proteins with higher affinity for F508del-CFTR (log2 below -1) were searched in ChEMBL. The interactors with available inhibitors are listed along the fold change score (regarding F508del- over F508del-4RK-CFTR interaction). (XLSX 27 kb)
Table S4 - APID interactions linking CFTR and KIFC1. The 198 proteins with stronger affinity with F508del-CFTR were analysed in APID1 and APID2 to identify their possible link to KIFC1, one to five represent the number of edges. (XLSX 30 kb)
Acknowledgements
Work supported by centre Grant UID/MULTI/04046/2013 to BioISI, Romain Pauwels Research Award to C.M.F, and Project MIMED PTDC/EEI-ESS/4923/2014 to A.O.F. SC and JDS are recipient of fellowships from BioSys PhD programme PD/BD/114393/2016 (SFRH/BD/52491/2014) and PD/BD/106084/2015 (SFRH(BD/106084/2015) from FCT, Portugal, respectively. Proteomics Core Facility-SGIKER is part of ProteoRed-ISCIII network and is partially funded by ERDF and ESF.
Abbreviations
- ABC
ATP-binding cassette
- AFTs
Arginine-framed tripeptides
- APID
Agile protein interactome dataserved
- ATP
Adenosine triphosphate
- BHK
Baby hamster kidney
- BP
Biologic process
- cAMP
cyclic adenosine monophosphate
- CC
Cellular compartment
- CFBE41o-
Cystic fibrosis bronchial epithelial cells
- CF
Cystic fibrosis
- CFTR
Cystic fibrosis transmembrane conductance regulator
- DAVID
Database for annotation: visualization and integrated discovery
- DMP
Dimethyl pimelimidate.2 HCl
- DSP
Dithiobis(succinimidylpropinate)
- EMT
Epithelial-to-mesenchymal transition
- ENaC
Epithelial Na+ channel
- ER
Endoplasmic reticulum
- ES
Enrichment score
- ERQC
Endoplasmic reticulum quality control
- GSEA
Gene set enrichment analysis
- GO
Gene ontology
- HDACi
Histone deacetylase inhibitor
- HEK293
Human embryonic kidney 293
- iBAQ
Intensity-based absolute quantification
- MSD
Membrane-spanning domain
- NBD
Nucleotide-binding domain
- NT
Non-treated
- PM
Plasma membrane
- PPIs
Protein–protein interactions
- QC
Quality control
- RD
Regulatory domain
- SB
Sample buffer
- SEM
Standard error of the mean
- SUMO
Small ubiquitin-like modifiers
- TM
Transmembrane segment
- UPR
Unfolded protein response
- WCL
Whole-cell lysate
- Wt
Wild-type
- 16HBE14o-
Human bronchial epithelial cells
Author contributions
SC designed and performed the experiments, analysed data and wrote the manuscript; JDS analysed data; ASC prepared the sample and performed mass spectrometry experiment; KA and RM performed the mass spectrometry experiment. MDA provided advice and comments on the manuscript; AOF guided the experiments design for bioinformatics analysis, secured funding and provided advice; CMF guided the project, secured funding, guided the experiments design and wrote the manuscript. All authors read, revised and approved the manuscript.
Footnotes
R program, https://www.r-project.org/ (Last accessed March 09, 2018).
GSEA database, http://software.broadinstitute.org/gsea/index.jsp (Last accessed February 18, 2017).
DAVID database, https://david.ncifcrf.gov/ (Last accessed March 08, 2018).
Reactome Pathway database, https://reactome.org/ (Last accessed September 30, 2017).
APID database, http://cicblade.dep.usal.es:8080/APID/init.action (Last accessed January 26, 2018).
Cytoscape, http://cytoscape.org (Last accessed January 26, 2018).
Harvard PrimerBank database, https://pga.mgh.harvard.edu/primerbank/ (Last accessed November 06, 2017).
References
- 1.Collins FS. Cystic fibrosis: molecular biology and therapeutic implications. Science. 1992;256:774–779. doi: 10.1126/science.1375392. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Liu F, Chen J. Conformational changes of CFTR upon phosphorylation and ATP binding. Cell. 2017;170(483–491):e8. doi: 10.1016/j.cell.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 3.Farinha CM, Canato S. From the endoplasmic reticulum to the plasma membrane: mechanisms of CFTR folding and trafficking. Cell Mol Life Sci. 2017;74:39–55. doi: 10.1007/s00018-016-2387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron. 1999;22:537–548. doi: 10.1016/S0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 5.Chang XB, Cui L, Hou YX, et al. Removal of multiple arginine-framed trafficking signals overcomes misprocessing of delta F508 CFTR present in most patients with cystic fibrosis. Mol Cell. 1999;4:137–142. doi: 10.1016/S1097-2765(00)80196-3. [DOI] [PubMed] [Google Scholar]
- 6.Roxo-Rosa M, Xu Z, Schmidt A, et al. Revertant mutants G550E and 4RK rescue cystic fibrosis mutants in the first nucleotide-binding domain of CFTR by different mechanisms. Proc Natl Acad Sci USA. 2006;103:17891–17896. doi: 10.1073/pnas.0608312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res. 2013;14:38. doi: 10.1186/1465-9921-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauniyar N, Gupta V, Balch WE, Yates JR. Quantitative proteomic profiling reveals differentially regulated proteins in cystic fibrosis cells. J Proteome Res. 2014;13:4668–4675. doi: 10.1021/pr500370g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roxo-Rosa M, da Costa G, Luider TM, et al. Proteomic analysis of nasal cells from cystic fibrosis patients and non-cystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics. 2006;6:2314–2325. doi: 10.1002/pmic.200500273. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Venable J, LaPointe P, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 11.Gomes-Alves P, Couto F, Pesquita C, et al. Rescue of F508del-CFTR by RXR motif inactivation triggers proteome modulation associated with the unfolded protein response. Biochim Biophys Acta. 2010;1804:856–865. doi: 10.1016/j.bbapap.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Pankow S, Bamberger C, Calzolari D, et al. ∆F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reilly R, Mroz MS, Dempsey E, et al. Targeting the PI3 K/Akt/mTOR signalling pathway in cystic fibrosis. Sci Rep. 2017;7:7642. doi: 10.1038/s41598-017-06588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho AS, Ribeiro H, Voabil P, et al. Global mass spectrometry and transcriptomics array based drug profiling provides novel insight into glucosamine induced endoplasmic reticulum stress. Mol Cell Proteom. 2014;13:3294–3307. doi: 10.1074/mcp.M113.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 16.Arike L, Valgepea K, Peil L, et al. Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteom. 2012;75:5437–5448. doi: 10.1016/j.jprot.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Fabre B, Lambour T, Bouyssié D, et al. Comparison of label-free quantification methods for the determination of protein complexes subunits stoichiometry. EuPA Open Proteom. 2014;4:82–86. doi: 10.1016/j.euprot.2014.06.001. [DOI] [Google Scholar]
- 18.Matthiesen R. Mass spectrometry data analysis in proteomics. Totowa, New Jersey: Humana Press; 2007. [Google Scholar]
- 19.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 20.Matthiesen R, Prieto G, Amorim A, et al. SIR: deterministic protein inference from peptides assigned to MS data. J Proteom. 2012;75:4176–4183. doi: 10.1016/j.jprot.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabregat A, Jupe S, Matthews L, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croft D, Mundo AF, Haw R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, He Q-Y. ReactomePA: an R/bioconductor package for reactome pathway analysis and visualization. Mol BioSyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 28.Alonso-López D, Gutiérrez MA, Lopes KP, et al. APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res. 2016;44:W529–W535. doi: 10.1093/nar/gkw363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendes AI, Matos P, Moniz S, et al. Antagonistic regulation of cystic fibrosis transmembrane conductance regulator cell surface expression by protein kinases WNK4 and spleen tyrosine kinase. Mol Cell Biol. 2011;31:4076–4086. doi: 10.1128/MCB.05152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobo MJ, Amaral MD, Zaccolo M, Farinha CM. EPAC1 activation by cAMP stabilizes CFTR at the membrane by promoting its interaction with NHERF1. J Cell Sci. 2016;129:2599–2612. doi: 10.1242/jcs.185629. [DOI] [PubMed] [Google Scholar]
- 33.Farinha CM, King-Underwood J, Sousa M, et al. Revertants, low temperature, and correctors reveal the mechanism of F508del-CFTR rescue by VX-809 and suggest multiple agents for full correction. Chem Biol. 2013;20:943–955. doi: 10.1016/j.chembiol.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Simpson JC, Joggerst B, Laketa V, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- 35.Mellacheruvu D, Wright Z, Couzens AL, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaulton A, Hersey A, Nowotka M, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Goor F, Hadida S, Grootenhuis PDJ, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Mikule K, Wang W, et al. Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS Chem Biol. 2013;8:2201–2208. doi: 10.1021/cb400186w. [DOI] [PubMed] [Google Scholar]
- 39.Pannu V, Rida PCG, Ogden A, et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget. 2015;6:6076–6091. doi: 10.18632/oncotarget.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal K, Choi DH, Klimov S, et al. A centrosome clustering protein, KIFC1, predicts aggressive disease course in serous ovarian adenocarcinomas. J Ovarian Res. 2016;9:17. doi: 10.1186/s13048-016-0224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomati V, Pesce E, Caci E, et al. High-throughput screening identifies FAU protein as a regulator of mutant cystic fibrosis transmembrane conductance regulator channel. J Biol Chem. 2018;293:1203–1217. doi: 10.1074/jbc.M117.816595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okiyoneda T, Veit G, Sakai R, et al. Chaperone-independent peripheral quality control of CFTR by RFFL E3 ligase. Dev Cell. 2018;44(694–708):e7. doi: 10.1016/j.devcel.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 44.Ahner A, Gong X, Schmidt BZ, et al. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol Biol Cell. 2013;24:74–84. doi: 10.1091/mbc.E12-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen TJ, Loo MA, Pind S, et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 46.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 47.Kwon M, Godinho SA, Chandhok NS, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mountain V, Simerly C, Howard L, et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JT, Jiang XH, Xie C, et al. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochimica et Biophysica Acta (BBA) Mol Cell Res. 2013;1833:2961–2969. doi: 10.1016/j.bbamcr.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 50.Maisonneuve P, Marshall BC, Knapp EA, Lowenfels AB. Cancer risk in cystic fibrosis: a 20-year nationwide study from the United States. J Natl Cancer Inst. 2013;105:122–129. doi: 10.1093/jnci/djs481. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Y-X, Shen H-Q, She Z-Y, et al. C-terminal kinesin motor KIFC1 participates in facilitating proper cell division of human seminoma. Oncotarget. 2017;8:61373–61384. doi: 10.18632/oncotarget.18139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukhopadhyay A, Quiroz JA, Wolkoff AW. Rab1a regulates sorting of early endocytic vesicles. Am J Physiol Gastrointest Liver Physiol. 2014;306:G412–G424. doi: 10.1152/ajpgi.00118.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 - Characterization of CFBE cells stably expressing F508del-4RK-CFTR. CFBE cells expressing F508del-4RK-CFTR (lane 1-6), wt-CFTR (lane 7) and F508del-CFTR (lane 8) were grown at 37 °C or 26 °C and in some cases incubated with VX-809 (3 µM) for 24 h or DMSO (vehicle compound). Western blot (top) showing both mature form band C (~ 170 kDa) and immature form band B (~ 140 kDa) of CFTR protein. Calnexin (90 kDa) was used as a loading control (bottom). (JPEG 66 kb)
Fig. S2 - Bioinformatics analysis of the proteins differentially interacting with F508del-CFTR and F508del-4RK-CFTR. (A) Hallmark enrichment for F508del-4RK-CFTR and F508del-CFTR using gene set enrichment (GSEA). GSEA was performed for a dataset of 834 proteins. Using GSEA tool, the subset of genes that contribute mostly to the enrichment of each hallmark was identified and plotted according to their affinity to F508del-4RK-CFTR (black dots) versus F508del-CFTR (grey dots) - represented by log2 of the ratio F508del-4RK-CFTR/F508del-CFTR. (B) Biological process represented for F508del-4RK-CFTR and F508del-CFTR using DAVID. From the total of 834 proteins, 198 with more affinity to F508del-CFTR and 164 proteins with more affinity to F508del-4RK-CFTR were used to find the GO terms – biological process enriched in both subsets. p value < 0.05. (C) Cellular component represented for F508del-4RK-CFTR and F508del-CFTR using DAVID. From the total of 834 proteins, 198 with more affinity to F508del-CFTR and 164 proteins with more affinity to F508del-4RK-CFTR were used to find the GO terms – cellular compartment enriched in both subsets. Significant levels are represented by p value < 0.05. (PDF 371 kb)
Fig. S3 - Scatter plot representing the differential protein interactions for the 22 putative hits in F508del-CFTR versus F508del-4RK-CFTR and peptides corresponding to the AFT regions of CFTR. Log2 plot of the protein abundance ratio for F508del-4RK-CFTR versus F508del-CFTR and Log2 plot of the protein abundance ratio for peptide K (corresponding to mutated AFTs with Lys replacement) versus peptide R (Arg containing). Proteins were identified by LC–MS/MS and the threshold (log2 = ±1). (JPEG 113 kb)
Fig. S4 - Force-directed network of CFTR versus top hits interactome. Network comprises the connection of CFTR interactome (depicted in green circle) and the interactome of the selected hits: FKBP4, HNRNPA2B1, KIFC1, YWHAE (depicted in orange circles). All components comprising the interactors are predicted as nodes (yellow circles) and all were identified by LS-MS/MS covered the 834 proteins. Straight white lines illustrate edges that define interactions based on the APID protein interaction database which was accessed using Cytoscape platform. Dashed red lines illustrate the edges in which interactions occur with one node distance and full orange lines the edges in which interactions occur with two or more nodes distance to CFTR. (JPEG 149 kb)
Fig. S5 - Effect of FKBP4, 14-3-3ε and HNRNPA2B1 knockdown on F508del-CFTR protein processing. CFBE cells expressing F508del-CFTR were transfected with siRNA against FKBP4, 14-3-3ε, HNRNPA2B1 or EGFP as non-targeting siRNA for 48 h. CT - transfection reagent only was used. Cells were also incubated with VX-809 (3 µM) or DMSO (vehicle control) are shown. Detection of F508del-CFTR protein expression (top) under (A) FKBP4, (B) 14-3-3ε or (C) HNRNPA2B1 knockdown. Equal amount of protein was loaded in each lane, as demonstrated by calnexin loading control (Bottom). (D) CFTR processing (band C divided by total CFTR) was normalized to siEGFP control. (E) Gene knockdown with siRNA in CFBE cells. Fold expression of FKBP4, 14-3-3ε or HNRNPA2B1 mRNA levels was obtained by relative quantification (ddCT method) and was normalized to an internal control (CAP-1). Data are shown as the mean ± SEM, n = 3. * p ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test. (JPEG 331 kb)
Fig. S6 - Effect of KIFC1 inhibition on F508del-CFTR expression. (A) CFBE cells expressing F508del-CFTR were incubated with 0.4 µM KIFC1 inhibitor (AZ82 compound) or DMSO, vehicle control (non-treated - NT), for 24 h, 48 h or 72 h. As a control for KIFC1 levels, cells were transfected with siRNA pool for KIFC1 or EGFP as non-targeting siRNA for 48 h. Western blot for CFTR (Top). Equal amount of protein was loaded in each lane, as demonstrated by α-tubulin (~ 50 kDa) detection (Bottom). (B) Quantification of CFTR (band B and C) band intensity normalized to loading control and to NT or siEGFP. (C) CFTR processing (band C divided by total CFTR) normalized to NT or siEGFP. Data are shown as mean ± SEM, n = 3. * ρ ≤ 0.05. Statistical analysis was performed using two-tailed unpaired Student’s t test. (JPEG 144 kb)
Fig. S7 - Distance of F508del-CFTR interactors to KIFC1. The 198 interactors with higher affinity to F508del-CFTR were subjected to APID1 (accounting for all known interactions) and APID2 (accounting for interactions proved by two or more experiments). Each circle represents the distance from the proteins to KIFC1 (from one to five edges). Inside of each circle is represented the number of proteins showing the associated distance. (JPEG 92 kb)
Table S1 - Total interactome identified for F508del-CFTR and F508del-4RK-CFTR. All identified proteins from MS analysis. (XLSX 156 kb)
Table S2 - Protein fold change interaction for F508del-CFTR versus F508del-4RK-CFTR. Fold change interaction was obtained by log2 of the ratio of the amount of the protein detected in association with F508del-CFTR versus F508del-4RK-CFTR. (XLSX 94 kb)
Table S3 - Protein targets and inhibitors listed in ChEMBL database. The 198 proteins with higher affinity for F508del-CFTR (log2 below -1) were searched in ChEMBL. The interactors with available inhibitors are listed along the fold change score (regarding F508del- over F508del-4RK-CFTR interaction). (XLSX 27 kb)
Table S4 - APID interactions linking CFTR and KIFC1. The 198 proteins with stronger affinity with F508del-CFTR were analysed in APID1 and APID2 to identify their possible link to KIFC1, one to five represent the number of edges. (XLSX 30 kb)