Abstract
Haploid embryonic stem cells (haESCs) contain only one set of genomes inherited from the sperm or egg and are termed AG- or PG-haESCs, respectively. Mammalian haESCs show genome-wide hypomethylation and dysregulated imprinting, whereas they can sustain genome integrity during derivation and long-term propagation. In addition, haESCs exhibit similar pluripotency to traditional diploid ESCs but are unique because they function as gametes and have been used to produce semi-cloned animals. More strikingly, unisexual reproduction has been achieved in mice by using haESCs. In combination with a gene editing or screening system, haESCs represent a powerful tool for studies of underlying gene functions and explorations of mechanisms of genetic and epigenetic regulation not only at the cellular level in vitro but also at the animal level in vivo. More importantly, genetically edited AG-haESC lines may further serve as an ideal candidate for the establishment of a sperm bank, which is a highly cost-effective approach, and a wide range of engineered semi-cloned mice have been produced. Here, we review the historical development, characteristics, advantages and disadvantages of haESCs. Additionally, we present an in-depth discussion of the recent advances in haESCs and their potential applications.
Keywords: Unisexual mice, CRISPR/Cas9, Genetic screen, Self-diploidization, LINE-1, Sperm bank
Introduction
Haploid cells and haploid embryonic stem cells (haESCs)
In nature, haploid animals are very rare, except for the parthenogenetic reproduction of drones [1], wasps [2], male ants [3], mites [4], etc. In mammalian species, only postmeiotic germ cells, including sperm and eggs, are haploid and produce a diploid genome upon fertilization in the mammalian life cycle. In terms of evolution and environmental adaptation, the diploid mammal generated by the fusion between a haploid sperm and egg has several great advantages, which not only effectively maintains the genome stability but also avoids the elimination of organisms caused by unfavorable lethal mutations [5, 6]. In addition, this strategy enhances genetic diversity through hybrid superiority and may generate new species by interspecific crossing or interbreeding. However, studies of recessive gene functions, allele-specific gene functions, imprinted gene regulation and the generation of mutations using diploid cells seems more complex. Moreover, the generation of genetically modified or knockout cell lines with identical alleles is difficult, although gene editing has become more convenient [7–9]. Meanwhile, the production of mouse models with multiple linked gene modifications is more time-consuming because of the requirement for intergenic crossing. In comparison, haploid cells show an obvious advantage in studies of gene function and explorations of epigenomic regulation, gamete development and the generation of complex gene-modified animals.
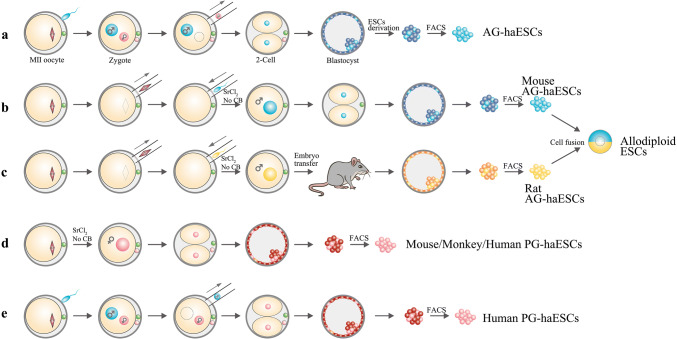
Embryonic stem cells (ESCs) can be derived from the inner cell mass (ICM) of the blastocyst, which exhibit a sustained self-renewal and enormous differentiation potential in vivo and in vitro [10–12]. In addition, these pluripotent stem cells are very powerful tools in both basic research and regenerative medicine. Similar to canonical diploid ESCs (2n ESCs), haploid embryonic stem cells (haESCs) are also derived from the ICM, but from the haploid blastocyst. This artificially generated haploid blastocyst contains only one set of genomes inherited from the sperm or egg (one set of allelic genes of a normal diploid genome). In particular, an androgenetic haploid embryo can be generated by removing the female pronucleus from a fertilized egg or by injecting a sperm head into an enucleated matured oocyte [13, 14] (Fig. 1a, b). Meanwhile, a parthenogenetic haploid embryo can be obtained by artificially activating a mature oocyte [15] (Fig. 1d). Thus, haESCs are divided into androgenetic haESCs (AG-haESCs) and parthenogenetic haESCs (PG-haESCs), according to the origin of the genome.
Fig. 1.
Schematics showing the derivation of mammalian haploid embryonic stem cells. a, b Androgenetic haploid embryos have been generated by removing the maternal pronucleus from the zygote (a) or injecting a sperm head into an enucleated oocyte (b). Then, the mouse AG-haESCs are obtained from the derived AG-ESCs using FACS. c Procedure for producing rat AG-haESCs. Allodiploid ESCs are obtained through the fusion of mouse and rat AG-haESCs. d, e Parthenogenetic haploid embryos are generated from MII oocytes activated by 10 mM SrCl2 in calcium-free CZB medium without CB (d) or through the removal of the male pronucleus from the zygote (e). a–e Blastocysts or ES cells shown in light colors are haploid cells and darker colored cells are diploid cells
The first stable haploid cell line was established in 1970 from an androgenetic embryo of the frog Rana pipiens and was produced by removing the maternal nucleus of the fertilized oocyte with a glass needle (Table 1) [16]. Soon afterwards, haploid cell lines were successfully established from both Drosophila melanogaster and humans (Homo sapiens, near haploid tumor cells KBM7) [17–19]. For decades, many research groups worldwide have been dedicated to generating stable haESC lines. Without exception, the experiments failed because of the frequent self-diploidization of these cells. Not until 2011, certain PG-haESC lines were finally derived from mouse parthenogenetic blastocysts following the development of fluorescence-activated cell sorting (FACS) technology [15, 20–22]. Thereafter, mouse AG-haESCs and rat AG- and PG-haESCs were successfully established through a combination of fine micromanipulation techniques and FACS [13, 14, 23, 24]. Notably, the reconstructed rat androgenetic and parthenogenetic haploid embryos must be first transferred to pseudopregnant rats for in vivo development to the blastocyst stage, and then these haploid blastocysts are recovered from the uterus for the further derivation of AG- or PG-haESCs [23, 24] (Fig. 1c). Meanwhile, monkey and human PG-haESCs were successfully established in 2013 and 2016, respectively [25–27] (Fig. 1d, e), and both lines display great potentials for identifying pathogenic genes and drug screening. Interestingly, a quite unique ESC line called mouse-rat allodiploid ESCs was generated by a cell fusion technique from the haploid ESCs of the two species [28] (Fig. 1b, c). These allodiploid ESCs were reported to be pluripotent and possess a stable diploid genome with a mixture of species-specific allelic genes, which are useful for the identification of genes regulating phenotypic differences between mouse and rat. Additionally, these hybrid cells have proven to be an ideal tool for the study of the molecular mechanisms regulating X chromosome inactivation as well as X-inactivation related escaping genes [28]. Overall, haESCs show an ES-like pluripotency with distinct haploid-related properties, and they further provide a practical system not only for genetic screening but also for generating genetically modified models, which will be carefully discussed below.
Table 1.
The establishment and characteristics of haploid cell lines
| Cell line type | Species | Year | Pluripotency | Genomic ploidy | Semi-cloned animal production | References |
|---|---|---|---|---|---|---|
| Androgenetic haploid ESCs | Frog | 1970 | ND | Stable | ND | [16] |
| Parthenogenetic haploid ESCs | Drosophila melanogaster | 1978 | ND | Stable | ND | [17] |
| Near-haploid tumor cells (KBM7) | Human | 1999 | No | Unstable | No | [19] |
| Parthenogenetic haploid ESCs | Medaka fish | 2009 | Yes | Stable | Yes | [29] |
| Parthenogenetic haploid ESCs | Mouse | 2011,2013,2016 | Yes | Self-diploidization | Yes | [15, 20–22] |
| Androgenetic haploid ESCs | Mouse | 2012 | Yes | Self-diploidization | Yes | [13, 14] |
| Androgenetic haploid ESCs | Macaca fascicularis monkey | 2013 | Yes | Self-diploidization | ND | [25] |
| Androgenetic haploid ESCs | Rat | 2014 | Yes | Self-diploidization | Yes | [23] |
| Parthenogenetic haploid ESCs | Rat | 2017 | Yes | Self-diploidization | ND | [24] |
| Parthenogenetic haploid ESCs | Human | 2016 | Yes | Self-diploidization | ND | [26, 27] |
| Allodiploid ESCs | Mouse and Rat | 2016 | Yes | ND | ND | [28] |
ND not determined
Characteristics of haESCs
Low embryonic development capability and ESC derivation efficiency
A prerequisite for the derivation of AG- and PG-haESCs is the construction of haploid embryos. However, the efficiency of generating mammalian haploid embryos has remained very low for decades. In 1977, Tarkowski reported the bisection of mouse pronuclear stage embryos with a glass needle into halves that each contained a pronucleus, resulting in the production of androgenetic and parthenogenetic haploid embryos from one egg. The average efficiency for producing these haploid blastocysts was 12.8%, and the highest rate did not exceed 29.5%. Additionally, this type of haploid blastocyst contained only half of the number of cells of a normal diploid blastocyst [30, 31], and these haploid embryos developed much more slowly than their diploid counterparts. The underlying mechanisms for this inefficient development of haploid embryos remain unclear. One potential explanation is the increased incidence of apoptosis, as indicated by nuclear condensation and/or DNA fragmentation visualized by a TUNEL assay [31]. With a more comprehensive understanding of the regulation of early embryonic development as well as the roles of signaling pathways and the microenvironment, both human and mouse embryo in vitro culture medium have been continuously modified and improved in recent years [32]. Currently, the development rate of in vitro development of fertilized mouse embryos has reached 90–100% [33]. However, the rate of blastocyst development required to generate mouse PG-haploid embryos is still approximately 10–22% and varies with different genetic backgrounds [15, 22]. In addition, the average efficiency for the establishment of parthenogenetic ESC lines in 2i (GSK3βi; 3 μM CHIR99021 and MEKi; 1 μM PD0325901) medium is approximately 40% [15, 21, 22], which is lower than 2n ESC lines [34, 35]. A more challenging problem is that only 15–30% of these parthenogenetic ESC lines maintain their haploid identities and are able to be called actual PG-haESCs. Interestingly, 129 Sv and B6CBAF1 strains produce a higher percentage (> 60%) of haploid cells in the cell population of the tested mouse strains before sorting, indicating a varying degree of tolerance to haploidy in different genetic backgrounds [15].
Similarly, the in vitro developmental capacity of the AG-haploid embryos (16–23%) and the efficiencies of the derivation of androgenetic ESC lines (6–17%) and actual AG-haESCs (11–26% of AG-ESCs) are even lower than normal in vitro fertilized embryos and 2n ESCs [14, 36]. Regarding the lower blastocyst development rate, AG- and PG-haploid embryos in the morula stage have also been used to establish haESC lines [13, 21]. In addition, the ratio of actual haploid lines among all generated parthenogenetic ESC lines has been slightly improved, potentially due to the shorter time from the 1-cell embryo stage to the first round of sorting of PG-haESCs (from 1-cell to morula stage compared with 1-cell to blastocyst stages). However, the efficiency of generating AG-haESCs (4.4–19% of morulae) by this method still remains low [13, 21]. Impaired proliferation problems with chromosome segregation observed in haploid cells might explain these findings. Even worse, through the activation of a p53-dependent cytotoxic response, many haploid cells die at or shortly after the last mitosis [37–39]. Thus, a P53 deficiency likely helps maintain a higher percentage of haploid cells than wild-type haESCs [37].
Quite unlike the mouse system in which both the AG- and PG-haploid embryos develop to the blastocyst stage in vitro, the reconstructed rat AG- and PG-haploid embryos in the 1-cell stage must be transplanted to recipient pseudopregnant rats for in vivo development [23, 24]. Then, the haploid morulae and blastocysts (4.36%) are collected from the uterus to derive AG-haESC lines. Much higher efficiencies of generating rat androgenetic ESC lines (40% of morulae and blastocysts) and actual rat AG-haESC lines (53% of AG-ESCs) are observed than in the mouse system [23]. Meanwhile, two rat PG-ESC lines (100%) have been established from two parthenogenetic haploid blastocysts, and one PG-haESC line (50% of PG-ESCs) was successfully maintained [24]. However, the developmental potential and derivation efficiency of PG-haESCs are inefficient in both monkey and human systems [25–27] (Table 2).
Table 2.
Haploid embryonic development capability and haESC derivation efficiency
| Cell type | Genetic background | Blastocyst (% of 1-cell embryos) | ES derived (% of blastocysts) | Haploid ES (% of ES cell lines) | References |
|---|---|---|---|---|---|
| PG-haESCs | Mixed strainsa | 181/937 (19.32) | 76/181 (42) | 25/76 (32.9) | [15] |
| PG-haESCs | C57BL/6 | 40/86 (46.5)b | 13/40 (32.5) | 9/13 (69.23) | [21] |
| C57BL/6 × CD1 | 14/30 (46.6)b | 6/14 (42.86) | 2/6 (33.33) | [21] | |
| PG-haESCs | C57BL/6 | 79/590 (13.39) | 38/79 (48.10) | 6/38 (15.79) | [22] |
| AG-haESCs | C57(OG-2) | 194/909 (21.34) | 34/194 (17.53) | 4/34 (11.76) | [14] |
| AG-haESCs | Actin-GFP | 82/490 (16.73) | 5/82 (6.1) | 1/5 (20) | [14] |
| AG-haESCs | 129sv | 46/244 (17.42) | 7/46 (15.22) | 1/7 (14.29) | [36] |
| AG-haESCs | C57(OG-2) | 110/466 (23.60) | 19/110 (17.27) | 5/19 (26.32) | [36] |
| Rat AG-haESCs | DA | 43/986 (4.36)b | 17/43 (40) | 9/17 (53) | [23] |
| Rat AG-haESCs | WDB-Rosa26em1(RT2)Nips | 26/495 (5.25) | 7/26 (26.92) | 1/7 (14.29) | [24] |
| Rat PG-haESCs | WI-Tg (CAG/Venus)Nips | 2/117 (1.71) | 2/2 (100) | 1/2 (50) | [24] |
| Monkey PG-haESCs |
Macaca Fascicularis monkey |
70/181(40) | 10/70 (14) | 2/10 (20) | [25] |
| Human PG-haESCs | ND | ND | 14 | 2/14 (14.28) | [26] |
| Human PG-haESCs | ND | 6/23 (26.08) | 4/6 (66.67) | 2/4 (50) | [27] |
ND not determined
aMixed mouse strains included B6CBAF1, 129B6F1, 129sv, transgenic ROSA26nlsrtTA LC1 Xist2LOX and ROSA26nlsrtTA tetOPXist
bData were obtained from morulae and blastocysts
In summary, both AG- and PG-haploid embryos, as well as haESCs, are more difficult to be generated than their diploid counterparts. Impaired cell proliferation and chromosome segregation, as well as the activation of the apoptotic pathway during development and culture might be the main explanations. Nevertheless, the haploid cells only contain half of the normal diploid genome and show distinct properties with both unique transcriptome and epigenetic networks [14, 36], which will be discussed further.
Intrinsic characteristic of haESCs
Although haploid cells contain half of the DNA content compared to diploid cells [13, 14, 26, 36], the generated mammalian haESCs, including mouse, rat, monkey and human lines, all sustain genome integrity during derivation and propagation without significant copy number variations (CNVs) and show very few single nucleotide polymorphisms (SNPs) in mammals [13, 23, 25, 27, 36]. Currently, most of the mouse PG-haESCs and all the mouse and rat AG-haESCs are derived and cultured on feeder layer. Additionally, the culture medium is supplemented with 2i [13–15, 22, 23, 36, 40], which may help to maintain ground-state pluripotency in rodents [41–43]. As shown in our recent study, 2i is required for the haploidy identity and proper cell proliferation of mouse AG-haESCs [36]. Meanwhile, the withdrawal of 2i may accelerate self-diploidization and slow cell proliferation [36]. Recently, established mouse AG-haESCs and PG-haESCs which could give rise to semi-cloned mice were all derived and maintained in 2i-containing system [13, 14, 22, 36, 40, 44].
Interestingly, several groups have demonstrated that only X chromosome-containing haESC lines that exhibit typical morphologies, such as the 2n ESCs, can be generated and propagated for more than 50 passages without a proliferation crisis in vitro [20, 36]. To date, no haESCs bearing the Y chromosome have been successfully obtained [13, 14, 36]. Additionally, androgenetic embryos carrying the Y chromosome do not survive beyond the 4-cell stage [30]. One potential explanation for these findings is that few genes important for early embryonic development are located on the Y chromosome, and/or the X chromosome is more essential for development. Moreover, a full haploid set of genome with an active X chromosome is the most basic unit needed for preimplantation development [30]. A lack of obvious development defects has been observed in embryos after Y chromosome elimination, and the generated XO mice are even fertile. However, deletion of the X chromosomes leads to serious embryonic lethality [45].
Similar to 2n ESC, the differentiation potential of rodent AG- and PG-haESCs has been examined and verified by chimera experiments as well as teratoma formation assays. In the chimera experiment, haploid cells are injected into the diploid blastocysts and then transferred to pseudopregnant recipients. AG-haploid cells can be detected on E6.5 and E7.5 in mice and rat chimeric embryos, respectively. Meanwhile, PG-haESCs are detected on E7.5. However, no haploid cells can be identified at later stages, which indicates an in vivo self-diploidization of both haESCs during differentiation. Nevertheless, both of these cell types contribute to the chimeric pups recovered on E19.5, as determined by coat color or GFP transgene expression. More interestingly, these self-diploidized haESCs contribute to the development of Oct4-GFP-positive germ cells that have regained their haploid identity. Functionally, the chimeric mice or rats further deliver normal offspring after mating [13, 15, 20, 23, 46].
Both mouse AG- and PG-haESCs, as well as rat AG-haESCs, can differentiate into cells with the lineage of all three germ layers, as evidenced by the results of assays investigating embryoid body (EB) formation in vitro and teratoma formation in vivo [13, 20, 23]. Additionally, haploid epiblast stem cells (haEpiSCs) and somatic cells were differentiated from mouse haESCs [47–49]. However, the haploid cells rapidly undergo self-diploidization upon differentiation [13, 23, 47, 48]. Surprisingly, quite unlike rodent haESCs, human PG-haESCs are compatible with a differentiated somatic fate that maintains the haploid genome, and certain haploid cells have been detected in all three embryonic germ layers following both in vitro and in vivo differentiation [26].
Self-diploidization
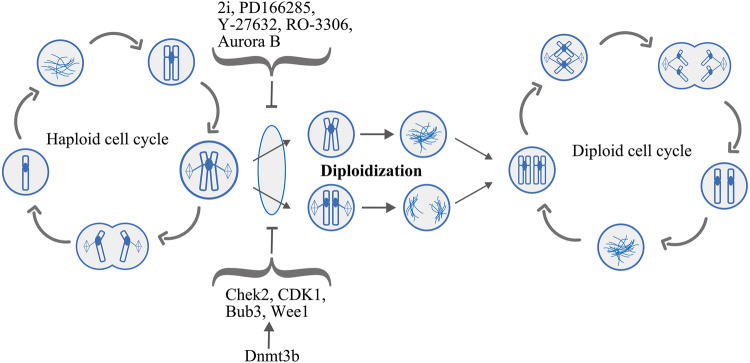
The self-diploidization of haploid cells refers to the process by which haploid cells become diploid due to endogenous replication without cell division [46]. This self-diploidization process occurs rapidly both in haploid embryos and haESCs [15, 20, 46]. As early as 1977, Tarkowski showed that ten of 51 embryos were haploid/diploid mosaics by performing a karyotype analysis of preimplanted mouse haploid embryos, including cleaving eggs, morulae and blastocysts [30]. Then, in 1983, Kaufman reported a chromosome analysis of mouse parthenogenetic morulae revealing that approximately 80% of blastomeres were haploid, whereas the remaining 20% were haploid-diploid and diploid. In addition, a karyotype analysis revealed diploidy with the prototypic 40 chromosomes, even in early passages of all the ESC lines derived from parthenogenetic blastocysts [50]. Similar to mouse haploid embryos, evidence obtained from a time-lapse analysis of images of human haploid parthenogenetic embryos confirmed that approximately 90% of reconstructed embryos undergo self-diploidization at the first cell cycle due to failed cytokinesis and endomitosis [51] (Fig. 2). Therefore, the self-diploidization of mammalian haploid cells begins as early as the embryonic stage, even during the first mitosis stage at the 1-cell stage. In the context of the high self-diploidization ratio, only a small number of haploid cells maintain the haploid property during in vivo or in vitro development to the blastocyst stage. Then, AG- or PG-haESCs may be derived from these haploid cells containing the ICM of the aforementioned blastocysts (Fig. 1).
Fig. 2.
Mechanism of self-diploidization in mouse haESCs. Self-diploidization mainly occurs in metaphase due to mitotic slippage, and mitosis is longer in haESCs. Specifically, the sister chromosomes do not segregate in anaphase or segregate without cytokinesis. Then, these two nuclei duplicate independently and fuse together at metaphase in the next cell cycle. Overexpression of Dnmt3b or the addition of inhibitors to the culture medium reduces the frequency of self-diploidization
However, even if mammalian haESCs are able to be derived from these blastocysts, the true haploid cell proportion among the whole ESC population is ~ 60% in PG-haESCs and even lower in AG-haESCs (~ 5%) during the first few passages [14, 15, 25]. With the assistance of advances in FACS technology accompanied by Hoechst 33342 staining, haESCs are gradually enriched. In addition, the sorting of haploid cells should be performed in the earlier generations at the earliest time point possible. For mouse haESC derivation, haploid sorting should begin as early as passage 3, and these sorted haESCs should be resorted every 3–4 generations. Generally, after 3–5 rounds of sorting, the haploidy of these cells is relatively stabilized and haESC lines can be maintained. Nevertheless, periodic cell sorting is still needed for the long-term maintenance of haploid cells in haESC lines. More simply, two groups have recently reported the isolation of haploid cells by filtration because they are smaller than diploid cells [52, 53]. Thus, this cell strainer-mediated haploid cell isolation method might be a time-saving and cost-effective approach for maintaining haESCs that also better protects the integrity of haploid cells and is more conducive to in vivo analyses of their developmental potential.
Because irreversible self-diploidization may pose substantial challenges for the maintenance and differentiation of haESCs, as well as for genetic analyses by using haESCs, many researchers have focused on identifying the mechanisms and reducing the self-diploidization property of haploid cells. It is indicated that the duration of interphase in haESCs is similar to diploid cells, but a longer mitosis was detected in both human and mouse haploid cells [37, 54], and self-diploidization of haESCs mainly occurs in metaphase due to mitotic slippage [49, 54]. Specifically, the sister chromosomes do not segregate in anaphase of mitosis or segregate without cytokinesis [54] (Fig. 2). The most widely used method to repress self-diploidization is the direct regulation of the cell cycle of haESCs. Hence, the G2/M phase transition is accelerated and re-entry into an extraneous G1/G0 phase is prevented by adding a small molecular inhibitor of Wee1 kinase (PD166285) to the culture medium [55]. Alternatively, mitosis is shortened through the overexpression of Aurora B [54], or mitotic slippage is suppressed by the addition of small molecule inhibitors of CDK1 (CDKi, RO-3306) and ROCK (ROCKi, Y-27632) to the culture medium [49]. These treatments reduce the self-diploidization of haESCs to a certain extent (Fig. 2). In addition, in a previous study, our lab rescued the expression of certain cell cycle-related genes (such as Chek2, Wee1, Cdk1 and Bub3) by overexpressing Dnmt3b in AG-haESCs to alleviate self-diploidization. Moreover, both GSK3βi and MEKi have important roles in stabilizing haploidy [36]. Consistent with these findings, a combination of 2i and PD166285 (Src inhibitor; also inhibits FGFR1, PDGFRβ and Wee1) has been shown to restrain self-diploidization by impeding the exit of haESCs from naïve pluripotency and by shortening the S-G2/M phases [56].
Apoptosis, autophagy and cell death have recently been shown to be involved in the process of self-diploidization [37, 57, 58]. Hence, a depletion of P53 stabilizes haploidy in human HAP1 (generated from KBM7) and mouse haESCs [37]. Meanwhile, mouse PG-haESCs cultured at high passages retain a better haploid state after knockdown of Sirt1, which plays important roles in regulating autophagy [57]. In addition, self-diploidization is also caused by the frequent centrosome loss observed in haploid cells, which further leads to haploid genome instability [58]. Self-diploidization occurs at a rate of 3–9% per day in human PG-haESCs [26]. However, to date, no effective methods for repressing self-diploidization in rat, monkey or human haESCs have been reported.
Notably, the value of haESCs in practical applications is only realized by efficiently inhibiting the frequent self-diploidization. For example, mouse haploid somatic cells of all three germ layers are differentiated from haESCs by repressing the self-diploidization. The haploid differentiated cells were further applied to a genome-wide genetic screen by using piggyBac transposon-based insertional mutagenesis [49]. Based on these findings, many methods have been reported to reduce the incidence of self-diploidization, but the mechanisms that completely stabilize the haploidy and block the occurrence of self-diploidization remain unsolved. Further studies on the mechanisms regulating the cell cycle in haESCs are needed.
DNA methylation of haESCs
DNA cytosine methylation is one of the most important epigenetic modifications in the genome and has been proven to play essential roles in various cellular processes, including genomic imprinting, X chromosome inactivation, retrotransposon silencing and the regulation of gene expression [59–61]. In mammals, the addition of methyl groups to cytosine residues is catalyzed by DNA methyltransferases (DNMTs), including Dnmt1, Dnmt3a and Dnmt3b [62–64]. We recently reported the global hypomethylation profile of AG-haESCs as demonstrated by an UHPLC-MRM-QQQ (ultra-high-performance liquid chromatography-multiple reaction monitoring triple quadrupole) analysis. Specifically, the genomic 5mC content of the AG-haESCs was less than one-fourth of the content in 2n ESCs (XX) and one-sixth of the content in 2n ESCs (XY). This DNA hypomethylation was probably due to the repressed expression of Dnmt3b and aberrant expression of other methylation-related genes, which further affects the methylation level in repetitive sequences, including the retrotransposon long interspersed nuclear element-1 (LINE-1), intracisternal A particles (IAP), major satellite repeats (pericentric repeats), minor satellite repeats (centric repeats) and differentially methylated regions (DMRs) of certain imprinted genes [36]. Consistent with these data, both AG- and PG-haESCs were indicated to be hypomethylated at the global genome level by reduced representation bisulfite sequencing (RRBS). In particular, the reduced methylation level in short interspersed nuclear elements (SINEs), long terminal repeats (LTRs), LINE-1, promoters and enhancers resemble E10.5 primordial germ cells (PGCs). Additionally, the methylation status of imprinted genes is similar to E13.5 PGCs [65]. Interestingly, the hypomethylation of AG-haESCs is not simply caused by the addition of 2i to the culture medium, as these cells still showed hypomethylation after the withdrawal of 2i compared with male ESCs cultured with 2i. Moreover, recovery of DNA methylation only occurred in restricted CpG sites. In addition, DNA methylation is essential for the development of haploid embryos and derivation of haESCs, while no AG-haESCs have been successfully generated in the absence of Dnmt3a or Dnmt3b [36].
Genomic imprinting is mainly achieved by the differential methylation of genes from parents-of-origin of different genders or in different tissue [66]. DMRs and imprinting control regions (ICRs) are DNA sequences that control the expression of related imprinted genes. A dramatic loss of methylation in these regions (both ICRs and DMRs) has frequently been observed in almost all the reported rodent haESCs, particularly in the later passages, which substantially disturbs the regulation of certain imprinted genes that are important for embryogenesis, such as H19, Ifg2 and Gtl2 [13, 14, 23, 40, 67]. Additionally, the expression pattern of these imprinted genes does not show an obvious correlation with their expected parent-of-origin methylation status, whereas only a few imprinted genes maintain their original methylation level [14, 22, 23, 40]. Still quite different from rodent haESCs, human PG-haESCs are able to maintain a typical stable maternal imprinting state in the early and even late passages, which indicates a unique regulation of DNA methylation in human haploid cells [26, 27]. In addition, the culture medium might be one explanation for this difference.
We conclude that the generation of haploid embryos and the derivation and maintenance of haESCs are limited by technical difficulties and imperfect culture systems, which are all inefficient. Nevertheless, haESCs are quite unique because they possess extraordinary genetic and epigenetic regulatory networks, as well as a substantial differentiation potential. Moreover, haESCs can be applied as a useful tool for delineating genome function at both cellular and animal levels. Next, we mainly review the most recent progress in the generation and engineering of semi-cloned animals. In addition, we will discuss several potential applications by using haESCs.
Generation of semi-cloned embryos and animals
Definition and production of semi-cloned animal
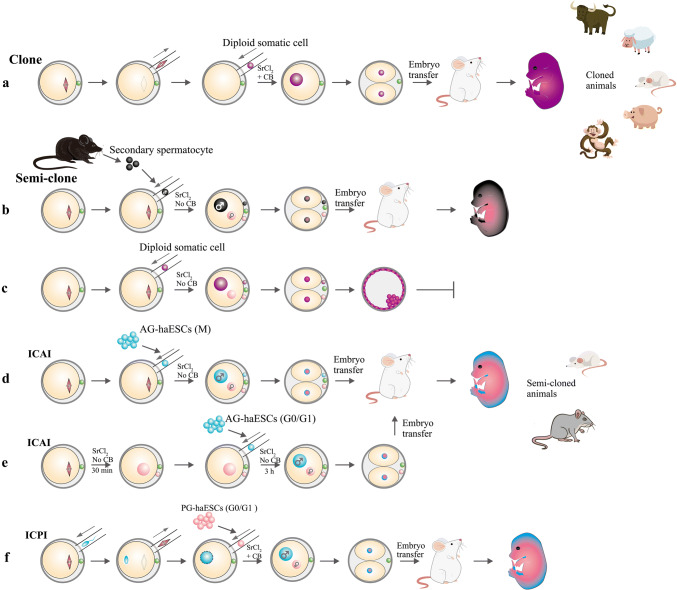
A cloned animal refers to an animal that has developed from a transplanted embryo reconstructed with a nucleus from a donor cell injected into an enucleated matured oocyte [68, 69] (Fig. 3a). Hence, the genetic material, except for the mitochondrial DNA, of a cloned animal almost completely originates from the donor cell instead of the parental oocyte. In recent years, cloned animals of many species have been successfully obtained, including cow [70], mouse [69], goat [71], pig [72], cat [73], rabbit [74], rat [75], dog [76], ferret [77], wolf [78], buffalo [79], camel [80] and even monkey [81] (Fig. 3a). Accordingly, the semi-cloned animal is derived from a semi-cloned embryo, in which half of the genome is inherited from the donor cell and the other half is inherited from the parental oocyte [82]. Currently, semi-cloned embryos are mainly produced via the three approaches described below. First, semi-cloned embryos have been obtained through the fertilization of mature oocytes with secondary spermatocytes (Fig. 3b). Second, a semi-cloned embryo has been generated by transferring the nucleus of a diploid somatic donor cell into an intact mature oocyte, which is slightly different from the nuclear transfer procedure. Then, the resulting embryo is activated without cytochalasin B (CB) to exclude a second polar body and a pseudopolar body (Fig. 3c). Third, a mature oocyte fertilized by an haESC in G0/G1 or M phase has been used generate a semi-cloned embryo (Fig. 3d, e).
Fig. 3.
Schematics showing the procedures used to generate cloned or semi-cloned mammals. a Cloned animals have been produced by injecting a diploid somatic cell into an enucleated oocyte followed by activation with SrCl2 in calcium-free CZB medium supplemented with CB. b Live semi-cloned mice have been produced by injecting a secondary spermatocyte into an intact MII oocyte. c The semi-cloned embryos have been obtained by injecting a diploid somatic cell into an intact MII oocyte followed by activation in media lacking CB. However, no viable animals have been produced using this method. d, e Semi-cloned mice (or rats) have been produced by injecting an M-phase (d) or G0/G1-phase (e) AG-haESC into an intact MII oocyte or a pre-activated MII oocyte, respectively. f The procedure used to generate semi-cloned mice from PG-haESCs in place of a maternal genome. A sperm head is first injected into an oocyte, followed by the removal of the spindle. One hour later, a G0/G1-phase PG-haESC is injected to generate a diploid embryo. Then, semi-cloned embryos and mice can be produced
Semi-cloned embryos and mice generated by using secondary spermatocytes
As early as 1995, Kimura and Yanagimachi described the fertilization of oocytes by secondary spermatocytes isolated from the testis, and 65% of these embryos developed into the blastocyst stage. Additionally, approximately 24% of these artificial fertilized embryos ultimately developed into full-term semi-cloned mice, which all showed normal phenotypes and were fertile [83]. In 2003, the nucleus of a secondary spermatocyte obtained from in vitro cultures of immature testicular tissue was microinjected into an oocyte in MII stage, which further resulted in the generation of a semi-cloned embryo after the extrusion of a pseudopolar body. In addition, these semi-cloned embryos underwent cleavage and developed into the 2-cell stage (74%) and 4-cell stage (18%). However, no viable semi-cloned mice were obtained when these embryos were transplanted into pseudopregnant mice [84]. In conclusion, based on these two initial reports, both in vivo and in vitro secondary spermatocytes can be used to produce semi-cloned embryos, but full-term mice are only obtained by using in vivo developed secondary spermatocytes. This difference is probably due to the inferior in vitro culture system, which may cause improper epigenetic programming of the secondary spermatocytes. Thus, an understanding of the differences between the in vivo and in vitro secondary spermatocytes and subsequent improvements in the culture conditions may facilitate the generation of semi-cloned mice by using in vitro cultured secondary spermatocytes.
Semi-cloned embryos generated using diploid somatic cells
The second approach used to obtain semi-cloned embryos also requires MII oocytes, but with diploid somatic cell nuclei. By applying the haploidization characteristic during fertilization, a diploid somatic cell nucleus can be induced to become haploid. In particular, and quite different from the canonical somatic nuclear transfer procedure, the nucleus of diploid somatic cells (granular cells or muscle fibroblasts) is first injected into a nonenucleated mature oocyte in this procedure. Then, the resulting embryo is activated by SrCl2 and two second polar bodies are extruded into the perivitelline space. Meanwhile, two pronuclei form, with one derived from the oocyte genome and the other from the somatic cell genome. Therefore, the semi-cloned embryos are generated. However, the fertilization rate using the granular cells is quite low (10–29%) and even complicated, which depends on the time of activation and fertilization after HCG injection. Moreover, the blastocyst development rate is only 0–17%, and no full-term mice have been generated [82]. Subsequently, in 2004, Shee-Uan Chen et al. carefully investigated the haploidization of somatic cell nuclei (cumulus cell as donor) in nonenucleated oocytes by examining the microtubular spindle dynamics and developmental potential. The progression to the blastocyst stage occurred at a rate of approximately 15%, and no implantation or live births of semi-cloned pups were obtained from 324 embryos after transfer to ICR surrogate mothers [85]. In addition, chromosomal aberrations were frequently detected in the generated semi-cloned embryos and only 6% of these embryos displayed 40 chromosomes in a cytogenetic analysis [85]. Collectively, although semi-cloned embryos were successfully prepared by this approach with somatic cell nuclei, no live semi-cloned mice have been generated to date (Fig. 3c). Chromosomal abnormalities resulting from segregation errors and potential epigenetic defects are presumed to be the main cause of the low blastocyst rate and in vivo developmental failure of these semi-cloned embryos.
Semi-cloned embryos and mice generated using haploid ESCs
The third approach used to produce semi-cloned embryos is to inject a nucleus of the haESC, which possesses a single set of chromosomes similar to sperm, into an intact mature MII phase oocyte [13, 14] (Fig. 3d, e). Unlike spermatozoa, which have the ability to activate oocytes during fertilization, the reconstructed semi-cloned mouse embryos require artificial activation by SrCl2, but without CB addition. Then, these activated semi-cloned embryos further develop similar to a normal zygote. Notably, the nuclei of haESCs in G0/G1 or M phase can be selected as the donors. In addition, compared with the previous two methods, the haESC-mediated approach is the only method in which cells are both functionally cultured and genetically modified in vitro. Additionally, semi-cloned embryos generated by this approach further support the development of full-term semi-cloned animals. Recently, various strains of semi-cloned mice and rats have been successfully produced by AG-haESCs and/or PG-haESCs [13, 14, 22, 23, 36]. Here, we summarize our current understanding of recently reported engineered semi-cloned animals produced by haESCs.
Semi-cloned mice generated from PG-haESCs
In 2009, the medaka fish PG-haESCs were initially successfully established and used in the procedure designed to generate semi-cloned animals to produce semi-cloned fish. Upon nuclear transfer into unfertilized oocytes, three semi-cloned embryos hatched to produce swimming fry and one finally grew into a fertile female. In addition, these PG-haESCs were genetically modified, and the first engineered semi-cloned fish called ‘Holly’ was obtained. Additionally, germline transmission was also observed in Holly [29]. In mammals, PG-haESCs also functionally replace the maternal genetic material to produce semi-cloned animals. In particular, an intracytoplasmic PG-haESC injection (referred to as ICPI) is performed 1 h after the intracytoplasmic sperm injection (ICSI) with the removal of the maternal spindle (Fig. 3f). In mice, only two full-term pups (0.7%) were obtained after 290 2-cell stage embryos were transferred into pseudopregnant recipients. Meanwhile, only one mouse ultimately survived to adulthood and was fertile [21] (Table 3). The low efficiency of producing semi-cloned mice using PG-haESCs may be attributed to two possible explanations: (1) multiple steps are required for the sophisticated microsurgery of oocytes and are quite difficult to perform, and (2) epigenetic differences exist between the PG-haESC and maternal genome (MII spindle), such as the maintenance of imprinted genes.
Table 3.
Semi-cloned embryo developmental potential
| Species | Gender of ES cell lines | Experimental procedure | Embryos transferred at the 2-cell stage | Full-term pups (% of reconstructed embryos) | Survive to adulthood (% of full-term pups) | Germline transmission ability | References |
|---|---|---|---|---|---|---|---|
| Medaka fish | PG-haESCs | Bimaternal | 667a | 7 (1) | 1 (14) | Yes | [29] |
| Mouse | PG-haESCs | ICPI | 290 | 2 (0.7) | 1 (50) | Yes | [21] |
| Mouse | AG-haESCs | ICAI | 424b | 9 (2.1) | 8 (89) | Yes | [14] |
| Mouse | AG-haESCs | ICAI | 913c | 29 (3) | 12 (41) | Yes | [13] |
| Mouse | DKO AG-haESCs | ICAI | 1993 | 402 (20.2) | NDd | Yes | [40] |
| Mouse | DKO AG-haESCs | ICAI | 400 | 41 (10) | 21 (51) | ND | [36] |
| Mouse | DKO AG-haESCsDnmt3b | ICAI | 305 | 31 (10) | 28 (90.3) | ND | [36] |
| Mouse | DKO PG-haESCs | Bimaternal | 1019 | 158 (15.5) | NDd | Yes | [22] |
| Mouse | PG-haESCsΔIG/Δ5H19/Igf2 | Bimaternal | 450 | 32 (7.1) | 23 (71.9) | Yes | [44] |
| Mouse | PG-haESCsΔIG/Δ5H19 | Bimaternal | 475 | 43 (9) | 31 (72) | Yes | [44] |
| Mouse | TKO PG-haESCs | Bimaternal | 210 | 29 (14) | 27 (93) | Yes | [65] |
| Mouse | 7KO AG-haESCs | Bipaternale | 477 | 12 (2.5) | 0 (0) | / | [65] |
| Rat | AG-haESCs | ICAI | 1142 | 2 (0.2) | 2 (100) | Yes | [23] |
| Rat | Transgenic AG-haESCs | ICAI | 441 | 4 (0.9) | 1 (25) | ND | [23] |
ND not determined
aNumber of reconstructed embryos
bNumber of blastocysts
cNumber of 2-cell embryos and blastocysts from AG-haESCs and gene-modified AG-haESCs
dMost semi-cloned mice grew to adulthood, but specific data are not available
eThese bipaternal mice were produced via a tetraploid embryo complementation assay
Semi-cloned mice generated from AG-haESCs
The AG-haESCs derived from androgenetic blastocysts retain the paternal instead of maternal imprints. Therefore, mouse AG-haESCs have been used to fertilize mature MII oocytes. Two methods have been employed to generate living semi-cloned mice by using AG-haESCs through an intracytoplasmic AG-haESC injection (referred to as ICAI). The first is to inject FACS-selected G0- or G1-phase haploid cells into pre-activated mature oocytes [13]. The other method is to directly inject cultured smaller M phase AG-haESCs into mature oocytes without pre-activation [14, 36]; the oocyte derived from both methods require a subsequent activation step (Fig. 3). Thereafter, these reconstituted embryos form ‘paternal’ pseudo-pronuclei, which will undergo a dynamic demethylation pattern similar to the pattern observed in normal embryos produced by ICSI [13]. After transfer to the recipient mothers in the 2-cell or blastocyst stage, fertile semi-cloned mice have been recovered at E19.5 by cesarean section (Fig. 3d, e). More importantly, as AG-haESCs can be genetically engineered in vitro, AG-haESCs can help to extend genetic screening analyses from the cellular level to the animal level (Table 3). Thus, transgenic semi-cloned mice expressing β-actin-eGFP or Oct4-eGFP have been successfully generated by using transgenic AG-haESCs [13, 14], which also confirms the genetic stability and integrity of haESCs.
In conclusion, AG-haESCs have the ability to act as the sperm to fertilize oocytes and support the development of fertile semi-cloned mice. Additionally, AG-haESCs can feasibly be genetically modified by a gene-editing system and generate transgenic mice with heritable genetic modifications.
Production of unisexual mice
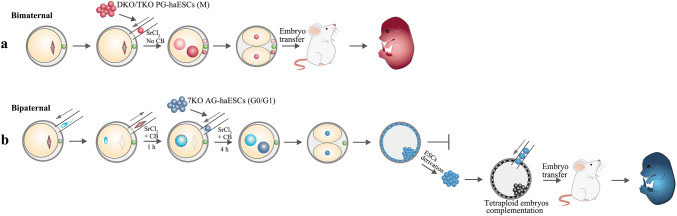
More interestingly, after the removal of H19-DMR and IG-DMR, PG-haESCs have even been used as sperm to fertilize MII oocytes and subsequently produce semi-cloned mice [22, 44] (Fig. 4a). Specially, the H19-DMR and IG-DMR were deleted by the CRISPR/Cas9 system in wild-type PG-haESCs. Among the 91 cell lines, 44 carried both deletions of H19-DMR and IG-DMR (termed DKO PG-haESCs). By applying these DKO PG-haESCs, the efficiency to produce semi-cloned mice can be improved to approximately 15.5%, whereas wild-type PG-haESCs failed to support the full-term development of bimaternal pups when acting as a sperm substitute [22]. Strikingly, 158 relatively normal sized full-term semi-cloned pups were successfully obtained from DKO PG-haESCs (Table 3). Moreover, most of these pups grew to adulthood and delivered offspring after mating with male mice [22]. More recently, a further deletion of the imprinted region in Rasgrf1 restored the growth retardation or other abnormalities observed in the previous DKO PG-haESC-generated semi-cloned mice [65]. As the genetic materials of these semi-cloned mice originate from the oocytes and PG-haESCs, and no paternal genome is involved, this kind of semi-cloned mice is referred to as bimaternal mice.
Fig. 4.
Schematics showing the procedures used to generate unisexual animals. a Bimaternal mice are produced through the injection of M-phase DKO PG-haESCs into intact MII oocytes. b Bipaternal embryos are generated by injecting G0/G1 phase 7KO AG-haES cells into androgenetic haploid embryos that were previously injected with a sperm head, followed by the removal of the maternal spindle. The developmental potential of these bipaternal embryos is limited to E8.5 after embryo transfer. However, bipaternal mice are able to be obtained by an alternative strategy through a tetraploid embryo complementation assay after the derivation of bipaternal diploid ESCs
Researchers have questioned whether AG-haESCs can functionally replace the maternal genome, support fertilization and then generate bipaternal mice. Recently, elegant experiments performed by Hu and colleagues revealed the production of bipaternal mice through a tetraploid complementation assay using androgenetic diploid ESCs derived from androgenetic diploid blastocysts [65]. Specifically, the androgenetic diploid embryo was generated by the co-injection of an AG-haESC in which seven imprinted regions (Nespas, Grb10, Igf2r, Snrpn, Kcnq1, Peg3 and Gnas) were deleted and a sperm into an enucleated oocyte. However, when these androgenetic diploid embryos were transferred to surrogate mothers, their development terminated at E8.5 and none of the placentae further developed beyond E10.5 [65]. As an alternative strategy, androgenetic diploid ESCs from these androgenetic diploid embryos were derived and then injected to tetraploid blastocysts followed by transplantation (Fig. 4b). Finally, 12 full-term, relatively normal bipaternal mice (~ 2.5%) were successfully produced. However, most bipaternal mice died after birth and none survived to adulthood [65] (Table 3).
Taken together, haESCs derived from the paternal or maternal genome inherit the developmental functions of both gametes, even after long-term in vitro culture. After the genetic modification of imprinted regions, both PG-haESCs and AG-haESCs can be separately used in place of the genome from either gamete (sperm or MII spindle) to generate semi-cloned mice. There is reason to believe that with a superior culture system and proper epigenetic regulation, mice will be generated from artificial biparental embryos constructed from AG- and PG-haESCs without either sperm or oocyte genomes in the future.
Strategy used to produce male semi-cloned mice
All the semi-cloned mice that have been produced to date are females, while the Y chromosome of a male semi-cloned mouse is provided by the sperm, but not haESCs. Due to the lack of the Y chromosome in both PG- and AG-haESCs, the production of male semi-cloned mice using these cells as sperm seems impossible. Additionally, an exploration of the functions of the genes located on the Y chromosome and the generation of Y chromosome gene-edited semi-cloned mice are not practicable. However, these concerns may be eliminated through several genetic methods. Conditional overexpression of either Dmrt using the Wt1-BAC transgene system or Sox9 using the Wt1-YAC transgene system in XX mouse fetal gonads causes a female-to-male sex reversal and generates fertile male mice [86, 87]. In contrast, the deletion of a single distal enhancer (Enh13, 557 bp) of Sox9 induces a male-to-female sex reversal [88]. In addition, a minimum Y chromosome (containing Sry, Eif2s3y and Zfy2) knock-in is capable of producing normally functioning sperm in mice lacking the integrated Y chromosome [89]. Thus, we are tempted to speculate that with the help of CRISPR/Cas9-mediated gene knockout, knock-in and gene activation and repression systems, or through the ectopic expression of sex determination-related genes, fertile male semi-cloned mice can be produced when the haESCs are used as sperm.
Generation of semi-cloned rats using AG-haESCs
In addition to mice, semi-cloned rats have also recently been successfully produced via rat AG-haESCs (RAG-haESCs), although at a very low efficiency [23]. Two full-term wild-type (0.2%) and four red fluorescent protein (RFP) transgenic semi-cloned rats (0.9%) were obtained after a rat ICAI procedure (Table 3). Similar to the mouse system, all semi-cloned rats were females, as no Y chromosome-containing RAG-haESC lines have been established or maintained. Of the six rats, three died 1 h after birth because of growth retardation, resulting in a much lower body weight. The remaining three rats survived for more than 1 week, and only one wild-type semi-cloned rat survived to adulthood. After mating with a male SD rat, this rat produced healthy progeny with a normal litter size and coat color separation in accordance with Mendelian law. Although gene-edited RAG-haESCs have been generated by the CRISPR/Cas9 system, only E13.5 genetically modified (Scn4b gene deletion) semi-cloned embryos were detected and no full-term pups were generated [23]. Moreover, no other groups have reported the successful generation of semi-cloned rat, which further indicates the low efficiency of using RAG-haESCs to produce semi-cloned rats [24]. In addition, a semi-cloned rat derived from rat PG-haESCs has not yet been reported. Although one chimeric male rat has been produced by a blastocyst injection of rat PG-haESCs, germline transmission was not examined because female ES cells cannot transmit to offspring from males [24, 90, 91].
Last but not least, although haESCs were successfully established from several other species including monkey and human, semi-cloned organisms of these species have not been produced or tested to date, due to technical limitations, cost or ethical issues.
Applications of haESCs and semi-cloned animals
Advantages of haESCs and semi-cloned animals
Based on the haploid identity and potential of haESCs to generate semi-cloned mice, these cells represent a convenient tool for establishing animal models identifying gene functions. For comparison, traditional mouse models generated with a chimeric system using canonical diploid ESCs take a remarkably long time to produce. Therefore, the advantages of the rapid identification of gene functions by haESCs are described further below. First, the functions and regulatory mechanisms of parental-of-origin imprinted genes can be analyzed in AG- and PG-haESCs in vitro and in vivo [36, 44, 65]. Second, the functions of X-linked genes, pathogenic genes and disease-related recessive genes in self-renewal and in vivo and in vitro differentiation can be explored [28]. Third, gene functions have been analyzed in differentiated haploid somatic cells, which are rarely obtained by any other methods [49]. Fourth, the functions of certain critical paternal genes and dominant lethal genes have been investigated upon fertilization and during early embryonic development and differentiation [40]. Fifth, the phenotype caused by pathogenic genes has been observed [92] and studied more quickly and consistently in animals. Finally, genetic screens combining haESCs and CRISPR/Cas9 are more powerful at both the cellular and animal levels [40, 93].
With the development of gene editing technology, particularly the CRISPR/Cas9 system, the production of animal models has become much simpler, because it eliminates the usage of 2n ESCs and time-consuming genetic background purification from 129 Sv (most 2n ESC lines with germline transmission ability are on the 129 Sv background) to C57BL/6. However, some problems and limitations persist. For example, the injection of constructs into fertilized zygotes by the CRISPR/Cas9 system may frequently result in mosaic founder animals [94]. Additionally, the genotypes of animals in the same litter are quite different, although they may be all identified as complete gene knockout animals. In addition, even in one gene-edited animal, the genotypes of the two alleles are generally different.
In comparison, haESCs have unique characteristics in the production of animal models. First, the editing of genetically linked genes and multiple genes in haESC lines is easy, including not only simple gene knockout but also mutations, tags and labeling, etc. Then, mice containing multiple edited genes are easily and quickly produced, which do not require long-term crossing between single gene-edited mice. Moreover, each mouse produced by one haESC line in the litter possesses the same genotype. Second, complex gene editing of the X chromosome can be first accomplished in haESCs, whereas direct CRISPR/Cas9-mediated injection in zygotes only easily generate X chromosome-related gene knockout mice. Third, complex gene editing of autosomes, including large fragment insertion, deletion, replacement and fusion, as well as conditional knockout (mediated by the Cre-LoxP, Dre-Rox, Flpe-Frt or Nigri-Nox system) in large segment intervals is feasible when producing haESC-mediated semi-cloned mice. However, these gene-edited mice are almost impossible to obtain from direct CRISPR/Cas9-mediated injection in the embryos because of the low efficiency. Additionally, the traditional 2n ESC-mediated procedure requires at least 1 year to produce mice, and the resulting chimeric mice and their offspring may not have a uniform genetic background. Fourth, functional studies of dominantly inherited lethal genes and CRISPR/Cas9-mediated genetic screening during fertilization and early embryonic development are possible using the haESC-mediated semi-cloned mice [40]. Fifth, more interestingly and importantly, the engineered AG-haESCs can serve as a sperm bank for genetically edited mice [40, 92]. In comparison, traditional mouse sperm preservation initially requires first adult male mice, which is a relatively time-consuming and costly process. Besides, these stocked sperm are unable to be further expanded in vitro. In addition, if the modified mice have defects in sperm development, the species is unable to be preserved by this method. Therefore, genetically edited AG-haESCs represent an ideal candidate to establish a sperm bank.
Moreover, as spermatids are unable to be cultured and propagated in vitro, they fail to be epigenetically manipulated followed by selection in the plate. Additionally, sperm and spermatids are impossible to be directly affected or altered in vivo. Besides, sperm-mediated gene transfer (SMGT) generates unpredictable transgenic animals [95–98]. In contrast, AG-haESCs that can be manipulated in vitro have the ability to give rise to complex gene-modified mice, which facilitate explorations of the functions of noncoding RNAs and other epigenetic modifications during reproduction, except for gene knock-out and knock-in mentioned above. With the development of small-scaled ChIP-seq technology, haESCs will serve as a much more powerful tool to study the dynamic genome location of proteins of interest during both preimplantation and postimplantation development after specific genes are genetically tagged. In addition, by taking advantage of the procedure used to produce semi-cloned mice, researchers may be able to precisely explore the paternal metabolic function during fertilization and in offspring by adjusting the composition of the AG-haESC culture medium. Further studies are required to test these possibilities.
Disadvantages of haESCs and semi-cloned animals
Although both mouse AG- and PG-haESCs have the ability to produce full-term semi-cloned pups, a high proportion of these pups display a growth-retarded phenotype and died shortly after birth, which substantially impedes the broad practical applications of haESCs in animal engineering [13, 14, 36]. Aberrant regulation and expression of imprinted genes are thought to be the main causes of this defect [44, 65, 67]. Therefore, after removal of H19 DMR and IG-DMR (DKO) from both AG- and PG-haESCs, the efficiency of generating pups and adulthood survival rate of semi-cloned mice have been substantially improved [22, 40, 44]. Moreover, a further deletion of Rasgrf1 imprinted regions in PG-haESCs can produce much healthier bimaternal mice [40, 65]. However, these semi-cloned mice possess a triple knockout (TKO) genotype. If further gene editing is required with DKO-haESCs or TKO-haESCs, mice with more than three or four modified genes will be produced, which may further complicate the study of gene function. As the extent to which the gene of interest may interact with the noncoding H19 or Gtl2 sequences or protein-coding Rasgrf1 remains uncertain, researchers will experience difficulty in confirming whether the phenotype is truly caused by the loss of the gene of interest. Moreover, although the mixed genotypes can be separated after mating with wild-type mice, this method is not applicable if the gene of interest is located on the same chromosome as H19, Gtl2 or Rasgrf1. Therefore, the possible effects of the deletion of the imprinted region on phenotypes must be carefully considered if genes located on chromosome 7 (H19), 9 (Rasgrf1) or 12 (Gtl2) are planned to be edited. However, with the further development of CRISPR/Cas9-mediated epigenomic modification technology by the dCas9-Dnmt/Tet or dCas9-Suntag system, researchers might be able to epigenetically regulate the dysregulated imprinted genes in haESCs without disrupting the DNA sequences [99, 100].
In addition, the adulthood survival rate of DKO semi-cloned mice is approximately 50%, probably due to the global DNA hypomethylation and dysregulation of certain imprinted genes. Additionally, this genome-wide hypomethylation may further influence the phenotypic analysis of targeted genes at both the cellular and mouse levels. However, the ectopic expression of Dnmt3b in DKO AG-haESCs partially restores the methylation level, enhancing the quality of the cells and further improving the survival rate of DKO semi-cloned mice [36]. Although a further deletion of the Rasgrf1 imprinted region in DKO PG-haESCs ameliorates the defects observed in DKO semi-cloned mice [65], this strategy may not be suitable for AG-haESCs, as Rasgrf1 must be imprinted paternally. Moreover, the maternal deletion of the IG-DMR in the oocyte will lead to postnatal and/or neonatal lethality. As a result, all the full-term F1 pups carrying the IG-DMR deletion from the semi-cloned female mice after mating with wild-type male mice will die shortly after birth [40, 65, 101], whereas the surviving pups have wild-type, H19-DMR or Rasgrf1 imprinted region deletion genotypes [40, 65]. Thus, genes on chromosome 12 are not recommended for engineering in DKO or TKO haESCs.
Recently, the somatic cell cloning efficiency has been substantially improved by removing certain epigenetic barriers, such as H3K9me3, H3K4me3, Xist activation, DNA methylation and remethylation, as well as H3K27me3-loss of imprints [102–104]. Thus, the efficiency of semi-cloning will likely be further improved by regulating the epigenome in haESCs. Meanwhile, the potential applications of haESCs will be further increased.
Whole-genome genetic screens using haESCs
Screening of fish and mouse haESCs
Since haESCs are useful for direct genetic analyses of recessive phenotypes caused by the disruption of a single allele that can induce a loss-of-function phenotype, several vertebrate haESC lines have been used in genetic screens employing various methods. The medaka haESC line represents an ideal tool for the identification of host factors in a genetic screen, as they are susceptible to fish viruses, including Singapore grouper iridovirus (SGIV), spring viremia of carp virus (SVCV) and red-spotted grouper nervous necrosis virus (RGNNV) [105]. Meanwhile, forward and reverse genetic analyses are easily applied to mouse haESCs using the reversible gene trap system, which contains a Cre splice acceptor site and a removable Oct4 binding site [20]. In a single round of retroviral infection in mouse PG-haESCs, 176,178 insertions were precisely identified and approximately 51% of these insertions occurred in promotor regions and intragenic regions, which encompass 8203 individual genes [20]. Additionally, this approach produces homozygous insertions and is feasible for an analysis of the functions of recessive genes. Two clones carrying insertions in the Rarg and Drosha coding regions were established, sequenced and functionally validated. Moreover, a recessive forward genetic screen identified the GPCR Gpr107 (LUSTR1) as a key molecule involved in ricin toxicity for the first time [20].
Screens using RAG-haESCs
Genetic modification and genome-wide screening have also been achieved in rat AG-haESCs through the electroporation of the piggyBac (PB) transposon-based gene-trap system. Using this approach, insertional mutations are efficiently generated throughout the genome. Notably, 51.7% of the insertion sites from randomly selected G418-resistant clones (202 total clones) were located in promoter or intragenic regions and encompassed 132 annotated genes, whereas the remaining insertions occurred within intergenic regions [23]. With an extensive cell population, a RAG-haESC library containing mutations in every gene is theoretically obtainable. In addition, the insertion mutations in trapped genes, including Bcam, Spats2 and Gpb2, were further confirmed to abolish expression [23]. Thus, this study confirmed that a genome-wide genetic screen using rat AG-haESCs is efficient and functional and was able to elucidate the functions of genes involved in a wide variety of biological processes.
Screens using human PG-haESCs
Moreover, the PB transposon-based gene-trap system is also a feasible approach for loss-of-function genetic screens of human PG-haESCs. Using this approach, the disruption of the autosomal gene NUDT5 confers resistance to the purine analogue 6-thioguanine (6-TG) [26]. A genome wide loss-of-function library targeting 18,166 protein-coding genes was further applied in PG-haESCs, and the results showed that transcription factors, cell cycle-related genes and DNA repair-related genes are essential for human pluripotent stem cells (hPSCs). For example, genes in the P53-mTOR pathway were identified as restricting the growth of hPSCs, and their depletion would provide a growth advantage [106].
Screens employing the sgRNA library in vivo
Recently, a CRISPR/Cas9 system consisting of a single guide sgRNA that directs Cas9 nuclease to targeted gene has emerged as a more powerful tool for genome-wide screens compared to other gene manipulation techniques [107–109]. A genetic screen is achieved by combining the (un)controllable Cas9-overexpressing haESCs with a set of particular sgRNA library both in vitro and in vivo for the introduction of organism-wide mutations in mice in a single generation. Four hundred sixty-eight modified semi-cloned mice were obtained using this method via the genetic manipulation of DKO AG-haESCs followed by an ICAI procedure. Among these animals, 230 semi-cloned mice carried monoallelic or biallelic mutations, and most harbored frameshift insertion/deletion (indel) mutations that resulted in a loss-of-function of the targeted allele [40]. For example, all the organs of Scube1 mutant semi-cloned mice carried biallelic mutations and all the full-term mice died 1 h after birth, consistent with a previous report [40, 110]. In addition, as mentioned above, the combination of controllable CRISPR/Cas9 with mouse AG-haESCs is able to deliver uniformly edited mice, avoiding the somatic mosaicism induced by a direct injection of sgRNA and Cas9 (protein or mRNA) into normal fertilized embryos. Thus, this method is a much more convenient and effective tool for producing knockout-based screens in a mouse model in one step.
More recently, a CRISPR/Cas9-mediated base-editing screen was applied in combination with the AG-haESC-mediated semi-cloned mice procedure, which has proven to be an invaluable tool for the in vivo screening of amino acids required for protein function and identification of critical single-nucleotide variations (SNVs) related to diseases. In particular, the base mutant mice are efficiently produced by the injection of AG-haESCs expressing the enhanced third-generation base-editing system and a sgRNA library into mature oocytes. By this approach, mutations that disrupt the stability and protein–protein interactions of DND1 were identified to be critical for the development of PGCs [111].
Screens using haploid somatic cells
Frequent self-diploidization appears to be a challenge for genetic screens by using haESCs. However, upon the inhibition of CDK1 and ROCK, large-scale of genome screening has been extended to haploid somatic cells via a PB transposon-based gene-trap system, thus expanding the application of genetic screening to mammalian haploid cells [49]. A subsequent screen using mouse haploid neural stem-cell-like cells (NSCLCs) identified Park2 as a candidate gene whose depletion induced resistance to Mn2+-induced neurotoxicity. Moreover, this result was validated in a gene-specific knockout [49].
Overall, haESCs and other haploid cells have invaluable potentials in many studies and applications, including essential gene functions [112, 113], multiple biological processes [114–118], disease modeling and mechanisms [119–121], as well as exit from pluripotency [122]. Additionally, other mutation-generating systems, such as N-ethyl-N-nitrosourea (ENU), are also applicable in haploid cells. Taken together, to date, haESCs represent the only system that is useful for both in vitro and in vivo functional screens, and serves as a unique cell resource for quickly generating animal models.
Conclusions
In summary, the establishment of haploid cells, including haESCs, opens an avenue to identify the functions of recessive genes, which were easily cloaked in normal diploid cells in previous studies. Meanwhile, the procedure for generating semi-cloned animals using haESCs has proven to be an effective and economical method for generating a large number of fertile engineered animals. With the combination of a genome-wide scale genetic screening system, haESCs represent as an invaluable platform for studying gene functions and essential biological processes at the cellular and animal levels. Following the improvement and optimization of the derivation efficiency, haploid genome stability and in vivo developmental potential, the applications of haESCs will be further expanded.
Acknowledgements
We would like to thank Mr. Ling Jin for the assistance in the schematics. This study was supported by grants from the National Key R&D Program of China (2016YFA0100400), the National Natural Science Foundation of China (31721003, 31871446 and 31801206), the Shanghai Rising-Star Program (19QA1409600) and China Postdoctoral Science Foundation (2018M640420).
Abbreviations
- AG
Androgenetic
- CB
Cytochalasin B
- CNV
Copy number variation
- CRISPR
Clustered, regularly interspaced short palindromic repeats
- DKO
Double knock-out
- DMR
Differentially methylated region
- DNA
Deoxyribonucleic acid
- DNMT
DNA methyltransferase
- EB
Embryoid body
- ENU
N-ethyl-N-nitrosourea
- ESC
Embryonic stem cell
- FACS
Fluorescence-activated cell sorting
- GFP
Green fluorescent protein
- haESC
Haploid embryonic stem cell
- HCG
Human chorionic gonadotropin
- IAP
Intracisternal A particle
- ICAI
Intracytoplasmic AG-haESC injection
- ICM
Inner cell mass
- ICPI
Intracytoplasmic PG-haESC injection
- ICR
Imprinting control region
- ICSI
Intracytoplasmic sperm injection
- IG DMR
Intergenic germline-derived DMR
- LINE-1
Long interspersed nuclear element-1
- LTR
Long terminal repeat
- PB
PiggyBac
- PG
Parthenogenetic
- PGC
Primordial germ cells
- RGNNV
Red-spotted grouper nervous necrosis virus
- RRBS
Reduced representation bisulfite sequencing
- SGIV
Singapore grouper iridovirus
- SINE
Short interspersed nuclear element
- SMGT
Sperm-mediated gene transfer
- SNPs
Single nucleotide polymorphisms
- SNVs
Single-nucleotide variations
- SVCV
Spring viremia of carp virus
- TET
Tet methylcytosine dioxygenase
- TKO
Triple knock-out
- UHPLC-MRM-QQQ
Ultra-high-performance liquid chromatography-multiple reaction monitoring triple quadrupole
- 2i
GSK3βi and MEKi
- 2n ESC
Diploid ESC
- 5mC
5-Methylcytosine
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenteng He and Jiayu Chen contributed equally to this work.
Contributor Information
Jiayu Chen, Email: chenjiayu@tongji.edu.cn.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
References
- 1.Herrmann M, Trenzcek T, Fahrenhorst H, Engels W. Characters that differ between diploid and haploid honey bee (Apis mellifera) drones. GMR. 2005;4(4):624–641. [PubMed] [Google Scholar]
- 2.Beukeboom LW, Kamping A, Louter M, Pijnacker LP, Katju V, Ferree PM, Werren JH. Haploid females in the parasitic wasp Nasonia vitripennis. Science. 2007;315(5809):206. doi: 10.1126/science.1133388. [DOI] [PubMed] [Google Scholar]
- 3.Glastad KM, Hunt BG, Yi SV, Goodisman MA. Epigenetic inheritance and genome regulation: is DNA methylation linked to ploidy in haplodiploid insects? Proc Biol Sci. 2014;281(1785):20140411. doi: 10.1098/rspb.2014.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weeks AR, Marec F, Breeuwer JA. A mite species that consists entirely of haploid females. Science. 2001;292(5526):2479–2482. doi: 10.1126/science.1060411. [DOI] [PubMed] [Google Scholar]
- 5.Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 6.Otto SP, Goldstein DB. Recombination and the evolution of diploidy. Genetics. 1992;131(3):745–751. doi: 10.1093/genetics/131.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaenisch R, Mintz B. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc Natl Acad Sci USA. 1974;71(4):1250–1254. doi: 10.1073/pnas.71.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans M, Kaufman M. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 11.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H. How is pluripotency determined and maintained? Development. 2007;134(4):635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Shuai L, Wan H, Dong M, Wang M, Sang L, Feng C, Luo GZ, Li T, Li X, Wang L, Zheng QY, Sheng C, Wu HJ, Liu Z, Liu L, Wang XJ, Zhao XY, Zhou Q. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490(7420):407–411. doi: 10.1038/nature11435. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Shi L, Wang B-A, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, Li D, Xu G-L, Li J. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149(3):605–617. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479(7371):131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed JJ, Mezger-Freed L. Stable haploid cultured cell lines from frog embryos. Proc Natl Acad Sci USA. 1970;65(2):337–344. doi: 10.1073/pnas.65.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debec A. Haploid cell cultures of Drosophila melanogaster. Nature. 1978;274(5668):255–256. doi: 10.1038/274255a0. [DOI] [PubMed] [Google Scholar]
- 18.Debec A. Evolution of karyotype in haploid cell lines of Drosophila melanogaster. Exp Cell Res. 1984;151(1):236–246. doi: 10.1016/0014-4827(84)90371-9. [DOI] [PubMed] [Google Scholar]
- 19.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp Cell Res. 1999;252(2):273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 20.Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers S-P, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9(6):563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan H, He Z, Dong M, Gu T, Luo GZ, Teng F, Xia B, Li W, Feng C, Li X, Li T, Shuai L, Fu R, Wang L, Wang XJ, Zhao XY, Zhou Q. Parthenogenetic haploid embryonic stem cells produce fertile mice. Cell Res. 2013;23(11):1330–1333. doi: 10.1038/cr.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong C, Xie Z, Yin Q, Dong R, Yang S, Wu Y, Yang L, Li J. Parthenogenetic haploid embryonic stem cells efficiently support mouse generation by oocyte injection. Cell Res. 2016;26(1):131–134. doi: 10.1038/cr.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Li X, Li T, Jiang MG, Wan H, Luo GZ, Feng C, Cui X, Teng F, Yuan Y, Zhou Q, Gu Q, Shuai L, Sha J, Xiao Y, Wang L, Liu Z, Wang XJ, Zhao XY, Zhou Q. Genetic modification and screening in rat using haploid embryonic stem cells. Cell Stem Cell. 2014;14(3):404–414. doi: 10.1016/j.stem.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Hirabayashi M, Hara H, Goto T, Takizawa A, Dwinell MR, Yamanaka T, Hochi S, Nakauchi H. Haploid embryonic stem cell lines derived from androgenetic and parthenogenetic rat blastocysts. J Reprod Dev. 2017;63(6):611–616. doi: 10.1262/jrd.2017-074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Liu Z, Ma Y, Zhong C, Yin Q, Zhou C, Shi L, Cai Y, Zhao H, Wang H, Tang F, Wang Y, Zhang C, Liu XY, Lai D, Jin Y, Sun Q, Li J. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013;23(10):1187–1200. doi: 10.1038/cr.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagi I, Chia G, Golan-Lev T, Peretz M, Weissbein U, Sui L, Sauer MV, Yanuka O, Egli D, Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532(7597):107–111. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- 27.Zhong C, Zhang M, Yin Q, Zhao H, Wang Y, Huang S, Tao W, Wu K, Chen Z-J, Li J. Generation of human haploid embryonic stem cells from parthenogenetic embryos obtained by microsurgical removal of male pronucleus. Cell Res. 2016;26(6):743–746. doi: 10.1038/cr.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Cui XL, Wang JQ, Wang YK, Li YF, Wang LY, Wan HF, Li TD, Feng GH, Shuai L, Li ZK, Gu Q, Hao J, Wang L, Zhao XY, Liu ZH, Wang XJ, Li W, Zhou Q. Generation and application of mouse-rat allodiploid embryonic stem cells. Cell. 2016;164(1–2):279–292. doi: 10.1016/j.cell.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 29.Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326(5951):430–433. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- 30.Tarkowski AK. In vitro development of haploid mouse embryos produced by bisection of one-cell fertilized eggs. J Embryol Exp Morphol. 1977;38(1):187–202. [PubMed] [Google Scholar]
- 31.Liu L, Trimarchi JR, Keefe DL. Haploidy but not parthenogenetic activation leads to increased incidence of apoptosis in mouse embryos. Biol Reprod. 2002;66(1):204–210. doi: 10.1095/biolreprod66.1.204. [DOI] [PubMed] [Google Scholar]
- 32.Simopoulou M, Sfakianoudis K, Rapani A, Giannelou P, Anifandis G, Bolaris S, Pantou A, Lambropoulou M, Pappas A, Deligeoroglou E, Pantos K, Koutsilieris M. Considerations regarding embryo culture conditions: from media to epigenetics. In Vivo (Athens, Greece) 2018;32(3):451–460. doi: 10.21873/invivo.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, Wang Y, Le R, Wang H, Duan T, Zhang Y, Gao S. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat Cell Biol. 2018;20(5):620–631. doi: 10.1038/s41556-018-0093-4. [DOI] [PubMed] [Google Scholar]
- 34.Yagi M, Kishigami S, Tanaka A, Semi K, Mizutani E, Wakayama S, Wakayama T, Yamamoto T, Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548(7666):224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- 35.Choi J, Huebner AJ, Clement K, Walsh RM, Savol A, Lin K, Gu H, Di Stefano B, Brumbaugh J, Kim SY, Sharif J, Rose CM, Mohammad A, Odajima J, Charron J, Shioda T, Gnirke A, Gygi S, Koseki H, Sadreyev RI, Xiao A, Meissner A, Hochedlinger K. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548(7666):219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He W, Zhang X, Zhang Y, Zheng W, Xiong Z, Hu X, Wang M, Zhang L, Zhao K, Qiao Z, Lai W, Lv C, Kou X, Zhao Y, Yin J, Liu W, Jiang Y, Chen M, Xu R, Le R, Li C, Wang H, Wan X, Wang H, Han Z, Jiang C, Gao S, Chen J. Reduced self-diploidization and improved survival of semi-cloned mice produced from androgenetic haploid embryonic stem cells through overexpression of Dnmt3b. Stem cell reports. 2018;10(2):477–493. doi: 10.1016/j.stemcr.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olbrich T, Mayor-Ruiz C, Vega-Sendino M, Gomez C, Ortega S, Ruiz S, Fernandez-Capetillo O. A p53-dependent response limits the viability of mammalian haploid cells. Proc Natl Acad Sci USA. 2017;114(35):9367–9372. doi: 10.1073/pnas.1705133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131(3):437–440. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16(8):473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 40.Zhong C, Yin Q, Xie Z, Bai M, Dong R, Tang W, Xing YH, Zhang H, Yang S, Chen LL, Bartolomei MS, Ferguson-Smith A, Li D, Yang L, Wu Y, Li J. CRISPR-Cas9-mediated genetic screening in mice with haploid embryonic stem cells carrying a guide RNA library. Cell Stem Cell. 2015;17(2):221–232. doi: 10.1016/j.stem.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15(4):416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Wan H, Feng G, Wang L, He Z, Wang Y, Wang XJ, Li W, Zhou Q, Hu B. Birth of fertile bimaternal offspring following intracytoplasmic injection of parthenogenetic haploid embryonic stem cells. Cell Res. 2016;26(1):135–138. doi: 10.1038/cr.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, He B, Wang X, Shi L, Ping J, Wei Y, Ying W, Wei W, Liu W, Tang C, Li Y, Hu J, Yang H. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017;18(1):224. doi: 10.1186/s13059-017-1354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leeb M, Walker R, Mansfield B, Nichols J, Smith A, Wutz A. Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development. 2012;139(18):3301–3305. doi: 10.1242/dev.083675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuai L, Wang Y, Dong M, Wang X, Sang L, Wang M, Wan H, Luo G, Gu T, Yuan Y, Feng C, Teng F, Li W, Liu X, Li T, Wang L, Wang XJ, Zhao XY, Zhou Q. Durable pluripotency and haploidy in epiblast stem cells derived from haploid embryonic stem cells in vitro. J Mol Cell Biol. 2015;7(4):326–337. doi: 10.1093/jmcb/mjv044. [DOI] [PubMed] [Google Scholar]
- 48.Xu H, Yue C, Zhang T, Li Y, Guo A, Liao J, Pei G, Li J, Jing N. Derivation of haploid neurons from mouse androgenetic haploid embryonic stem cells. Neurosci Bull. 2017;33(3):361–364. doi: 10.1007/s12264-017-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He ZQ, Xia BL, Wang YK, Li J, Feng GH, Zhang LL, Li YH, Wan HF, Li TD, Xu K, Yuan XW, Li YF, Zhang XX, Zhang Y, Wang L, Li W, Zhou Q. Generation of mouse haploid somatic cells by small molecules for genome-wide genetic screening. Cell Rep. 2017;20(9):2227–2237. doi: 10.1016/j.celrep.2017.07.081. [DOI] [PubMed] [Google Scholar]
- 50.Kaufman MH, Robertson EJ, Handyside AH, Evans MJ. Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol. 1983;73(1):249–261. [PubMed] [Google Scholar]
- 51.Leng L, Ouyang Q, Kong X, Gong F, Lu C, Zhao L, Shi Y, Cheng D, Hu L, Lu G, Lin G. Self-diploidization of human haploid parthenogenetic embryos through the Rho pathway regulates endomitosis and failed cytokinesis. Sci Rep. 2017;7(1):4242. doi: 10.1038/s41598-017-04602-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qu C, Yan M, Yang S, Wang L, Yin Q, Liu Y, Chen Y, Li J. Haploid embryonic stem cells can be enriched and maintained by simple filtration. J Biol Chem. 2018;293(14):5230–5235. doi: 10.1074/jbc.RA118.002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freimann R, Wutz A. A fast and efficient size separation method for haploid embryonic stem cells. Biomicrofluidics. 2017;11(5):054117. doi: 10.1063/1.5006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo A, Huang S, Yu J, Wang H, Li H, Pei G, Shen L. Single-cell dynamic analysis of mitosis in haploid embryonic stem cells shows the prolonged metaphase and its association with self-diploidization. Stem Cell Rep. 2017;8(5):1124–1134. doi: 10.1016/j.stemcr.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi S, Lee J, Kohda T, Matsuzawa A, Kawasumi M, Kanai-Azuma M, Kaneko-Ishino T, Ishino F. Induction of the G2/M transition stabilizes haploid embryonic stem cells. Development. 2014;141(20):3842–3847. doi: 10.1242/dev.110726. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Guo A, Xie Z, Tu W, Yu J, Wang H, Zhao J, Zhong C, Kang J, Li J, Huang S, Shen L. Stabilization of mouse haploid embryonic stem cells with combined kinase and signal modulation. Sci Rep. 2017;7(1):13222. doi: 10.1038/s41598-017-13471-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H, Yang L, Cui H. SIRT1 regulates autophagy and diploidization in parthenogenetic haploid embryonic stem cells. Biochem Biophys Res Commun. 2015;464(4):1163–1170. doi: 10.1016/j.bbrc.2015.07.098. [DOI] [PubMed] [Google Scholar]
- 58.Yaguchi K, Yamamoto T, Matsui R, Tsukada Y, Shibanuma A, Kamimura K, Koda T, Uehara R. Uncoordinated centrosome cycle underlies the instability of non-diploid somatic cells in mammals. J Cell Biol. 2018;217(7):2463–2483. doi: 10.1083/jcb.201701151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 60.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev. 2002;118(1–2):187–190. doi: 10.1016/S0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- 62.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 63.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12(7):647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z-K, Wang L-Y, Wang L-B, Feng G-H, Yuan X-W, Liu C, Xu K, Li Y-H, Wan H-F, Zhang Y, Li Y-F, Li X, Li W, Zhou Q, Hu B-Y. Generation of bimaternal and bipaternal mice from hypomethylated haploid ESCs with imprinting region deletions. Cell Stem Cell. 2018 doi: 10.1016/j.stem.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 67.Hu M, Zhao Z, TuanMu LC, Wei H, Gao F, Li L, Ying J, Zhang S. Analysis of imprinted gene expression and implantation in haploid androgenetic mouse embryos. Andrologia. 2015;47(1):102–108. doi: 10.1111/and.12222. [DOI] [PubMed] [Google Scholar]
- 68.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KHS. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 69.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394(6691):369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 70.Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, Doguchi H, Yasue H, Tsunoda Y. Eight calves cloned from somatic cells of a single adult. Science. 1998;282(5396):2095–2098. doi: 10.1126/science.282.5396.2095. [DOI] [PubMed] [Google Scholar]
- 71.Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, Cammuso C, Williams JL, Nims SD, Porter CA, Midura P, Palacios MJ, Ayres SL, Denniston RS, Hayes ML, Ziomek CA, Meade HM, Godke RA, Gavin WG, Overstrom EW, Echelard Y. Production of goats by somatic cell nuclear transfer. Nat Biotechnol. 1999;17(5):456–461. doi: 10.1038/8632. [DOI] [PubMed] [Google Scholar]
- 72.Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000;289(5482):1188–1190. doi: 10.1126/science.289.5482.1188. [DOI] [PubMed] [Google Scholar]
- 73.Shin T, Kraemer D, Pryor J, Liu L, Rugila J, Howe L, Buck S, Murphy K, Lyons L, Westhusin M. A cat cloned by nuclear transplantation. Nature. 2002;415(6874):859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- 74.Chesne P, Adenot PG, Viglietta C, Baratte M, Boulanger L, Renard JP. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat Biotechnol. 2002;20(4):366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- 75.Zhou Q, Renard JP, Le Friec G, Brochard V, Beaujean N, Cherifi Y, Fraichard A, Cozzi J. Generation of fertile cloned rats by regulating oocyte activation. Science. 2003;302(5648):1179. doi: 10.1126/science.1088313. [DOI] [PubMed] [Google Scholar]
- 76.Lee BC, Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ, Hossein MS, Kim JJ, Kang SK, Schatten G, Hwang WS. Dogs cloned from adult somatic cells. Nature. 2005;436(7051):641. doi: 10.1038/436641a. [DOI] [PubMed] [Google Scholar]
- 77.Li Z, Sun X, Chen J, Liu X, Wisely SM, Zhou Q, Renard JP, Leno GH, Engelhardt JF. Cloned ferrets produced by somatic cell nuclear transfer. Dev Biol. 2006;293(2):439–448. doi: 10.1016/j.ydbio.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ, Hwang WS, Hossein MS, Kim JJ, Shin NS, Kang SK, Lee BC. Endangered wolves cloned from adult somatic cells. Cloning Stem Cells. 2007;9(1):130–137. doi: 10.1089/clo.2006.0034. [DOI] [PubMed] [Google Scholar]
- 79.Shi D, Lu F, Wei Y, Cui K, Yang S, Wei J, Liu Q. Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells. Biol Reprod. 2007;77(2):285–291. doi: 10.1095/biolreprod.107.060210. [DOI] [PubMed] [Google Scholar]
- 80.Wani NA, Wernery U, Hassan FA, Wernery R, Skidmore JA. Production of the first cloned camel by somatic cell nuclear transfer. Biol Reprod. 2010;82(2):373–379. doi: 10.1095/biolreprod.109.081083. [DOI] [PubMed] [Google Scholar]
- 81.Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, Sun Q. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 2018 doi: 10.1016/j.cell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 82.Lacham-Kaplan O, Daniels R, Trounson A. Fertilization of mouse oocytes using somatic cells as male germ cells. Reprod Biomed Online. 2001;3(3):205–211. doi: 10.1016/S1472-6483(10)62037-8. [DOI] [PubMed] [Google Scholar]
- 83.Kimura Y, Yanagimachi R. Development of normal mice from oocytes injected with secondary spermatocyte nuclei. Biol Reprod. 1995;53(4):855–862. doi: 10.1095/biolreprod53.4.855. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki S, Sato K. The fertilising ability of spermatogenic cells derived from cultured mouse immature testicular tissue. Zygote. 2003;11(4):307–316. doi: 10.1017/S0967199403002363. [DOI] [PubMed] [Google Scholar]
- 85.Chen SU, Chang CY, Lu CC, Hsieh FJ, Ho HN, Yang YS. Microtubular spindle dynamics and chromosome complements from somatic cell nuclei haploidization in mature mouse oocytes and developmental potential of the derived embryos. Hum Reprod. 2004;19(5):1181–1188. doi: 10.1093/humrep/deh168. [DOI] [PubMed] [Google Scholar]
- 86.Zhao L, Svingen T, Ng ET, Koopman P. Female-to-male sex reversal in mice caused by transgenic overexpression of Dmrt1. Development. 2015;142(6):1083–1088. doi: 10.1242/dev.122184. [DOI] [PubMed] [Google Scholar]
- 87.Vidal VPI, Chaboissier M-C, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat Genet. 2001;28:216. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 88.Gonen N, Futtner CR, Wood S, Garcia-Moreno SA, Salamone IM, Samson SC, Sekido R, Poulat F, Maatouk DM, Lovell-Badge R. Sex reversal following deletion of a single distal enhancer of Sox9. Science. 2018 doi: 10.1126/science.aas9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamauchi Y, Riel JM, Ruthig V, Ward MA. Mouse Y-encoded transcription factor Zfy2 is essential for sperm formation and function in assisted fertilization. PLoS Genet. 2015;11(12):e1005476. doi: 10.1371/journal.pgen.1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagashima H, Giannakis C, Ashman RJ, Nottle MB. Sex differentiation and germ cell production in chimeric pigs produced by inner cell mass injection into blastocysts. Biol Reprod. 2004;70(3):702–707. doi: 10.1095/biolreprod.103.022681. [DOI] [PubMed] [Google Scholar]
- 91.Isotani A, Nakanishi T, Kobayashi S, Lee J, Chuma S, Nakatsuji N, Ishino F, Okabe M. Genomic imprinting of XX spermatogonia and XX oocytes recovered from XX < – > XY chimeric testes. Proc Natl Acad Sci USA. 2005;102(11):4039–4044. doi: 10.1073/pnas.0406769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elling U, Wimmer RA, Leibbrandt A, Burkard T, Michlits G, Leopoldi A, Micheler T, Abdeen D, Zhuk S, Aspalter IM, Handl C, Liebergesell J, Hubmann M, Husa A-M, Kinzer M, Schuller N, Wetzel E, van de Loo N, Martinez JAZ, Estoppey D, Riedl R, Yang F, Fu B, Dechat T, Ivics Z, Agu CA, Bell O, Blaas D, Gerhardt H, Hoepfner D, Stark A, Penninger JM. A reversible haploid mouse embryonic stem cell biobank resource for functional genomics. Nature. 2017;550(7674):114–118. doi: 10.1038/nature24027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forment JV, Herzog M, Coates J, Konopka T, Gapp BV, Nijman SM, Adams DJ, Keane TM, Jackson SP. Genome-wide genetic screening with chemically mutagenized haploid embryonic stem cells. Nat Chem Biol. 2017;13(1):12–14. doi: 10.1038/nchembio.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zuo E, Cai YJ, Li K, Wei Y, Wang BA, Sun Y, Liu Z, Liu J, Hu X, Wei W, Huo X, Shi L, Tang C, Liang D, Wang Y, Nie YH, Zhang CC, Yao X, Wang X, Zhou C, Ying W, Wang Q, Chen RC, Shen Q, Xu GL, Li J, Sun Q, Xiong ZQ, Yang H. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 2017;27(7):933–945. doi: 10.1038/cr.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gandolfi F. Sperm-mediated transgenesis. Theriogenology. 2000;53(1):127–137. doi: 10.1016/S0093-691X(99)00246-0. [DOI] [PubMed] [Google Scholar]
- 96.Spadafora C. Sperm-mediated ‘reverse’ gene transfer: a role of reverse transcriptase in the generation of new genetic information. Hum Reprod. 2008;23(4):735–740. doi: 10.1093/humrep/dem425. [DOI] [PubMed] [Google Scholar]
- 97.Smith K, Spadafora C. Sperm-mediated gene transfer: applications and implications. BioEssays. 2005;27(5):551–562. doi: 10.1002/bies.20211. [DOI] [PubMed] [Google Scholar]
- 98.Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57(5):717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- 99.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233.e217–247.e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pflueger C, Tan D, Swain T, Nguyen T, Pflueger J, Nefzger C, Polo JM, Ford E, Lister R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018;28(8):1193–1206. doi: 10.1101/gr.233049.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35(1):97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 102.Matoba S, Zhang Y. Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell. 2018 doi: 10.1016/j.stem.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W, Wang S, Yin J, Liu W, Cai T, Wang H, Zhang Y, Gao S. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell discovery. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao R, Wang C, Gao Y, Xiu W, Chen J, Kou X, Zhao Y, Liao Y, Bai D, Qiao Z, Yang L, Wang M, Zang R, Liu X, Jia Y, Li Y, Zhang Y, Yin J, Wang H, Wan X, Liu W, Zhang Y, Gao S. Inhibition of aberrant DNA Re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell. 2018;23(3):426.e425–435.e425. doi: 10.1016/j.stem.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 105.Yuan Y, Huang X, Zhang L, Zhu Y, Huang Y, Qin Q, Hong Y. Medaka haploid embryonic stem cells are susceptible to Singapore grouper iridovirus as well as to other viruses of aquaculture fish species. J Gen Virol. 2013;94(Pt 10):2352–2359. doi: 10.1099/vir.0.054460-0. [DOI] [PubMed] [Google Scholar]
- 106.Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N. Defining essential genes for human pluripotent stem cells by CRISPR–Cas9 screening in haploid cells. Nat Cell Biol. 2018 doi: 10.1038/s41556-018-0088-1. [DOI] [PubMed] [Google Scholar]
- 107.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 108.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 110.Tu CF, Yan YT, Wu SY, Djoko B, Tsai MT, Cheng CJ, Yang RB. Domain and functional analysis of a novel platelet-endothelial cell surface protein, SCUBE1. J Biol Chem. 2008;283(18):12478–12488. doi: 10.1074/jbc.M705872200. [DOI] [PubMed] [Google Scholar]
- 111.Li Q, Li Y, Yang S, Huang S, Yan M, Ding Y, Tang W, Lou X, Yin Q, Sun Z, Lu L, Shi H, Wang H, Chen Y, Li J. CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat Cell Biol. 2018 doi: 10.1038/s41556-018-0202-4. [DOI] [PubMed] [Google Scholar]
- 112.Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, Marceau C, Janssen H, Carette JE, Bennett KL, Colinge J, Superti-Furga G, Brummelkamp TR. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350(6264):1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 113.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. Identification of spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015;12(4):554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Winter GE, Radic B, Mayor-Ruiz C, Blomen VA, Trefzer C, Kandasamy RK, Huber KVM, Gridling M, Chen D, Klampfl T, Kralovics R, Kubicek S, Fernandez-Capetillo O, Brummelkamp TR, Superti-Furga G. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014;10(9):768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. An essential receptor for adeno-associated virus infection. Nature. 2016;530(7588):108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Timms RT, Menzies SA, Tchasovnikarova IA, Christensen LC, Williamson JC, Antrobus R, Dougan G, Ellgaard L, Lehner PJ. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat Commun. 2016;7:11786. doi: 10.1038/ncomms11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu J, Ciaudo C. Vector integration sites identification for gene-trap screening in mammalian haploid cells. Sci Rep. 2017;7:44736. doi: 10.1038/srep44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carette JE, Guimaraes CP, Wuethrich I, Blomen VA, Varadarajan M, Sun C, Bell G, Yuan B, Muellner MK, Nijman SM, Ploegh HL, Brummelkamp TR. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat Biotechnol. 2011;29(6):542–546. doi: 10.1038/nbt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jae LT, Raaben M, Riemersma M, van Beusekom E, Blomen VA, Velds A, Kerkhoven RM, Carette JE, Topaloglu H, Meinecke P, Wessels MW, Lefeber DJ, Whelan SP, van Bokhoven H, Brummelkamp TR. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340(6131):479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pettitt SJ, Krastev DB, Pemberton HN, Fontebasso Y, Frankum J, Rehman FL, Brough R, Song F, Bajrami I, Rafiq R, Wallberg F, Kozarewa I, Fenwick K, Armisen-Garrido J, Swain A, Gulati A, Campbell J, Ashworth A, Lord CJ. Genome-wide barcoded transposon screen for cancer drug sensitivity in haploid mouse embryonic stem cells. Sci Data. 2017;4:170020. doi: 10.1038/sdata.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14(3):385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]