Abstract
Rapidly renewing epithelial tissues such as the intestinal epithelium require precise tuning of intercellular adhesion and proliferation to preserve barrier integrity. Here, we provide evidence that desmoglein 2 (Dsg2), an adhesion molecule of desmosomes, controls cell adhesion and proliferation via epidermal growth factor receptor (EGFR) signaling. Dsg2 is required for EGFR localization at intercellular junctions as well as for Src-mediated EGFR activation. Src binds to EGFR and is required for localization of EGFR and Dsg2 to cell–cell contacts. EGFR is critical for cell adhesion and barrier recovery. In line with this, Dsg2-deficient enterocytes display impaired barrier properties and increased cell proliferation. Mechanistically, Dsg2 directly interacts with EGFR and undergoes heterotypic-binding events on the surface of living enterocytes via its extracellular domain as revealed by atomic force microscopy. Thus, our study reveals a new mechanism by which Dsg2 via Src shapes EGFR function towards cell adhesion.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2869-x) contains supplementary material, which is available to authorized users.
Keywords: Desmosomes, Desmosomal cadherins, Intestinal barrier, Cell adhesion
Introduction
Epithelial tissues represent the interface between the organism and the environment, and thus, maintenance of epithelial barrier function is indispensable to prevent entry of pathogens or harmful macromolecules. This is, in particular, crucial for the gastrointestinal epithelium, which comprises a huge surface and faces a plethora of bacteria present in the gut but simultaneously needs to stay permeable to enable the uptake of nutrients and essential macromolecules [1, 2]. Aggravating this situation, the intestinal epithelial tissue shows the highest turnover rate in adult mammals and is exposed to notable mechanical stress resulting from gastrointestinal motility [3, 4]. Hence, precise regulation of intercellular adhesion and proliferation is required to preserve barrier integrity. This function is achieved by a set of adhesion complexes including tight junctions (TJ), adherens junctions (AJ), and desmosomes that tightly connect the polarized enterocytes within the simple columnar epithelium, thereby sealing the paracellular space [5, 6]. Initially, desmosomes were considered to primarily provide the mechanical strength in intercellular cohesion [7]. However, growing evidence suggests that desmosomal cadherins beside their adhesive function actively coordinate signaling pathways, hence mediating proliferation, differentiation, and apoptosis [8–11]. Desmosomal cadherins are transmembrane glycoproteins which interact via their extracellular domains (ED) in a homo- and heterophilic manner, while their tails associate with the plaque proteins plakoglobin (PG), plakophilin (Pkp), and desmoplakin (DP), thereby anchoring the desmosomal complex to the intermediate filament cytoskeleton. Thus, these components constitute the adhesive core of desmosomes [12]. Seven isoforms of desmosomal cadherins are expressed in human epithelial tissues, of which the intestinal epithelium contains Desmoglein 2 (Dsg2) and Desmocollin 2 (Dsc2) only [13–15]. A recent study reports that Dsc2 alone is sufficient to form a functional desmosomal plaque in enterocytes [16]. However, Dsg2 is required for intestinal epithelial barrier properties and abnormal expression of Dsg2 is implicated in colon cancer and inflammatory disorders such as Crohn’s disease [17–20]. In addition, it was shown that Dsg2 besides its role in cellular adhesion has an essential signaling function in regulating p38 mitogen-activated protein kinase (MAPK) signaling in enterocytes [20].
The mechanism by which desmosomal cadherins regulate signaling pathways is an emerging focus of research. Given that the intracellular tail of desmogleins has no enzymatic activity, transduction of signaling requires interaction with signaling components such as kinases. In this context, several molecular mechanisms have been proposed such as matrix metalloprotease-mediated shedding of Dsg2 external domain fragments which may act as ligands for receptor tyrosine kinases (RTK) [21] or displacing kinases from lipid rafts, thereby promoting their activation [22]. The common denominator in both scenarios is the involvement of RTK which are already known to associate with classical cadherins, and thus, both molecules are modulated mutually [23, 24]. Depending on the phosphorylation site, specific signaling molecules can be recruited which, in turn, modulate a variety of downstream signaling cascades [25–29]. Of particular interest to this study is the epidermal growth factor receptor (EGFR) which has been shown to modulate function of desmogleins [30, 31] as well as to be modulated by desmogleins [9, 22, 32]. Furthermore, EGFR is suggested to regulate cell migration, wound healing, and cell extrusion in intestinal epithelial cells [33–35]. Albeit, all these processes require regulation of intercellular junctions; the role of desmosomal cadherins in this context has only been marginally explored.
In this study, we examined how Dsg2 and EGFR cooperate in regulating intestinal epithelial homeostasis to assure barrier integrity. We show that Dsg2 modulates EGFR localization as well as protein level and Src-dependent activation, which both were reduced in Dsg2-deficient DLD1 and Caco2 cells. Here, we demonstrate, for the first time, that Dsg2 and EGFR interact directly using atomic force microscopy (AFM) both on living enterocytes as well as in a cell-free setup. Moreover, these binding events were inhibited by ligand binding to EGFR and depend on EGFR tyrosine kinase activity. In addition, inhibition of EGFR tyrosine kinase activity impaired barrier formation and intercellular adhesion. Furthermore, loss of Dsg2 increased cell proliferation, which was restored after inhibition of EGFR activity. Collectively, our data suggest a new signaling complex consisting of Dsg2 and EGFR which stabilizes the adhesive state of intestinal epithelial cells.
Results
Loss of Dsg2 reduces EGFR levels and prevents its localization to cell borders
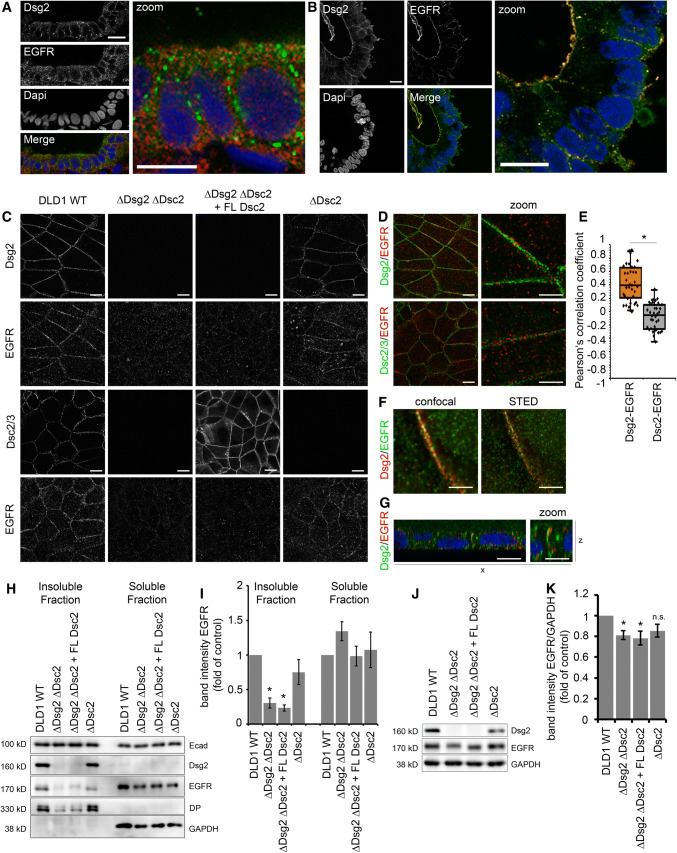
Increasing evidence suggests that desmosomal cadherins beyond their adhesive function are actively involved in regulating signaling cascades. Previously, we demonstrated that Dsg2 regulates barrier integrity in human intestinal cells via p38MAPK signaling [20]. In addition, several studies on keratinocytes linked desmogleins to differentiation and proliferation through regulation of growth factor signaling cascades [8, 9]. We, therefore, asked how desmosomal cadherins influence EGFR signaling in human intestinal cells. First, we performed immunostaining for Dsg2 and EGFR in human colon samples to investigate whether these proteins can be found in close proximity. EGFR is assumed to be located primarily in the basolateral membrane of enterocytes, whereas Dsg2 was shown to be present as part of desmosomes in the basolateral membrane but also outside of desmosomes in the apical membrane, at least in cultured enterocytes [20, 36, 37]. Interestingly, we observed EGFR staining at the basolateral membrane as well as at the apical surface and distinct spots of Dsg2 staining all along the cell borders (Fig. 1a). Besides, a small fraction of apical EGFR staining was overlapping with immunostaining for Dsg2 resulting in yellow spots. Similarly, we observed overlapping staining for EGFR and Dsg2 in human enteroids with an even more pronounced EGFR staining at the apical cell membrane facing the enteroid lumen (Fig. 1b). Using human colon carcinoma DLD1 cells deficient for Dsg2 and/or Dsc2 [16], we next investigated whether localization and protein levels of EGFR are controlled by desmosomal cadherins. DLD1 wild-type (WT) cells in immunostaining displayed linear localization of EGFR along cell borders similar to Dsg2 (Fig. 1c, upper panels) and Dsc2 (Fig. 1c, lower panels). To examine whether desmosomal cadherins affect the localization of EGFR, we immunostained DLD1 cells deficient for Dsg2, Dsc2, or both cadherins. Cells deficient for both, Dsg2 and Dsc2, showed almost complete loss of EGFR staining at cell borders (Fig. 1c, second column). Similar results were obtained using WT and Dsg2-deficient Caco2 cells (Fig. S1A). Reconstitution of Dsc2 was not sufficient to restore this phenotype (Fig. 1c, third column), suggesting that Dsg2 is required for recruiting EGFR to cell borders. In line with this, loss of Dsc2 did not affect EGFR localization (Fig. 1c, fourth column). Moreover, merged images of Dsg2 (green) and EGFR (red) staining revealed co-localization at cell borders with a Pearson correlation coefficient of around 0.4 pointing towards positive correlation of overlapping staining (Fig. 1d, e). In contrast, no correlation between EGFR and Dsc2 staining was observed, as indicated by a correlation coefficient of 0 (Fig. 1d, e). Using STED super resolution microscopy, we analyzed EGFR and Dsg2 localization more closely and found overlapping staining for EGFR and Dsg2 even at high resolution (Fig. 1f) confirming their close co-localization. Furthermore, we observed co-localization all along the cell border but more pronounced at the most apical part of the intercellular cleft (Fig. 1g). In accordance to the immunostaining, triton extraction resulted in a significant reduction of EGFR in the insoluble fraction when Dsg2 was absent (Fig. 1h, i) raising the possibility of a putative signaling complex consisting of Dsg2 and EGFR maybe at sites of desmosomes. Importantly, immunoblotting for Ecad did not result in any changes upon loss of Dsg2 or of Dsc2 (Fig. 1h), suggesting that AJ stay unaffected. Moreover, total protein expression levels of EGFR were reduced significantly in cells deficient for Dsg2 but not Dsc2 (Fig. 1j, k), indicating that Dsg2 might be important for EGFR stability. Similarly, EGFR protein levels were reduced in Dsg2-deficient Caco2 cells (Fig. S1 B–E).
Fig. 1.
Dsg2 knockout results in mislocalization and reduced protein levels of EGFR in intestinal epithelial cells. a Immunostaining for Dsg2 (green) and EGFR (red) in patient tissue sections of the colon revealed co-localization of both proteins. Four independent patient tissue samples were analyzed. Shown is representative image of a section from the colon. Bar 10 µm. b Immunostaining for Dsg2 (green) and EGFR (red) in human enteroids revealed co-localization of both proteins along the cell borders as well as on the surface facing the lumen. Shown is representative image for ten enteroids. Bar 10 µm. c Confluent cell monolayer of DLD1 cells grown on coverslips were immunostained for EGFR, Dsg2, and Dsc2/3. Loss of Dsg2 but not Dsc2/3 results in a major loss of EGFR staining at cell borders. Bar 10 µm. d Merged images of immunofluorescence staining of Dsg2, EGFR, and Dsc2/3 show co-localization of Dsg2 and EGFR but not Dsc2/3 and EGFR. Bar 10 µm (left panel) and 5 µm (right panel). e Evaluation of the Pearson correlation coefficient of EGFR confirms a co-localization of EGFR with Dsg2 but not Dsc2. Shown are boxplots with each point representing one analyzed area along the cell border. *p < 0.05. f STED super resolution microscopy analysis of Dsg2 and EGFR co-immunostaining shows co-localization. Bar 5 µm. g x–z image of Dsg2 and EGFR co-immunostaining shows specific co-localization at the apical site of cell contacts. Bar 10 µm (left panel) and 5 µm (right panel). h EGFR is absent in the Triton X-100 insoluble fraction when Dsg2 is missing in contrast to Ecad that is not affected by Dsg2 knockout. GAPDH served as loading control. i Band intensity of detected EGFR was measured from five independent experiments, showing a significant reduction of EGFR in the insoluble fraction in Dsg2-deficient DLD1 cells. Results are shown as mean ± SE. *p < 0.05. j Total protein level of EGFR in DLD1 cells were assessed by western blotting that revealed reduced EGFR levels in Dsg2-deficient cells. GAPDH served as loading control. k Band intensity of detected EGFR was quantified from at least ten independent experiments, demonstrating a significant reduction of total EGFR protein levels upon Dsg2 knockout. Shown are mean ± SE. *p < 0.05
Dsg2 and EGFR interact directly via their extracellular domains
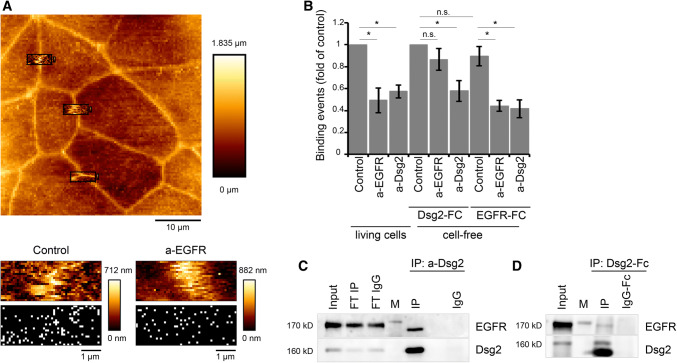
Given the first hints pointing towards a signaling complex consisting of Dsg2 and EGFR, we next investigated whether there is direct interaction. To this end, we conducted AFM on living DLD1 cells using an AFM cantilever coated with recombinant Dsg2-Fc containing the complete extracellular domain of Dsg2 similar as shown recently [20]. First, AFM topography images were created to select specific areas at cell borders for further adhesion measurements (Fig. 2a, upper panel). For interaction studies, the functionalized AFM tip was repetitively lowered on the cell surface, thereby creating an adhesion map consisting of 1000 force–distance curves for each selected area, with each white pixel representing one positive-binding event (Fig. 2a, lower panel). Intriguingly, these binding events were blocked significantly when applying an inhibitory antibody against the extracellular domain of EGFR (Fig. 2a, b), indicating that measured binding events under control conditions partly reflect heterophilic Dsg2–EGFR interaction. To verify that measured binding events were Dsg2-specific, an inhibitory antibody against the extracellular domain of Dsg2 was applied, which reduced the binding events to a similar extent (Fig. 2b), similar as published previously [20]. In addition, we performed cell-free AFM experiments with Dsg2-coated AFM tips on mica sheets functionalized with either recombinant Dsg2-Fc or recombinant EGFR-Fc containing the complete extracellular domain to validate homophilic or heterophilic interactions. Binding frequency was similar when measured Dsg2–Dsg2 or Dsg2–EGFR interaction under the same conditions (Fig. 2b), suggesting that both interactions can occur. Application of the inhibitory EGFR-specific antibody did not reduce homophilic Dsg2 interactions (Fig. 2b), demonstrating that it does not bind un-specifically to Dsg2. In contrast, heterophilic Dsg2–EGFR binding was significantly reduced similar to AFM measurements on living cells (Fig. 2b) underlining the specificity of measured binding events. Bearing in mind that the inhibitory EGFR-specific antibody blocks the ligand-binding site of EGFR, these data further raise the possibility that this binding site might be important for heterophilic EGFR–Dsg2 binding. Application of the inhibitory Dsg2-specific antibody reduced both homophilic Dsg2–Dsg2 as well as heterophilic Dsg2–EGFR interactions (Fig. 2b). Given that the antibody is directed against the extracellular domains 3 and 4 of Dsg2, this result indicates that these subdomains may be required for EGFR–Dsg2 interaction. In addition, we also examined endogenous association between EGFR and Dsg2 in DLD1 cells using co-immunoprecipitation studies. In line with the AFM-based adhesion studies, interaction of EGFR and Dsg2 was detected in DLD1 cells (Fig. 2c). Moreover, we performed a pull-down assay using the recombinant Dsg2-Fc, which also resulted in co-precipitation of EGFR and thus further indicates an interaction of EGFR with the extracellular domain of Dsg2 (Fig. 2d).
Fig. 2.
Dsg2 and EGFR form heterophilic interactions via their extracellular domains. a AFM cell topography image was created to select specific areas along the cell borders for Dsg2 adhesion measurements (upper panel). Adhesion measurements using a Dsg2-coated AFM tip generated adhesion maps with each white pixel representing one binding event (lower panel). b Treatment with an inhibitory EGFR-specific antibody significantly reduced the amount of Dsg2 binding events on living cells similar to an inhibitory anti-Dsg2 antibody. Cell-free AFM measurements using a Dsg2 coated tip and mica sheets coated either with Dsg2 or with EGFR revealed a similar binding frequency of homophilic Dsg2–Dsg2 and heterophilic Dsg2–EGFR interactions, which were significantly blocked by respective antibodies applied to the mica sheet. Graph shows fold-change values from at least three independent experiments ± SE. *p < 0.05. c Co-immunoprecipitation was performed with DLD1 cell lysates and an anti-Dsg2 antibody for immunoprecipitation (IP) of endogenous Dsg2. Immunoblotting for EGFR revealed co-IP of both proteins. FT flow through. d Pull-down assay was performed with DLD1 cell lysates and a Dsg2-Fc fusion protein. Immunoblotting for EGFR revealed co-IP of Dsg2-Fc and EGFR
Ligand binding to EGFR prevents its interaction with Dsg2 at the cell surface
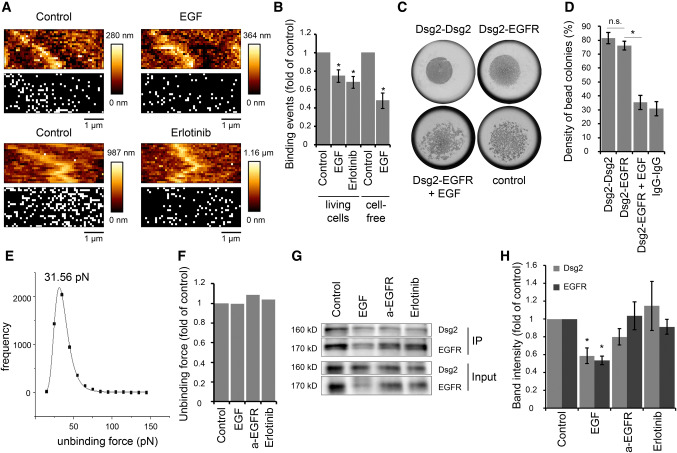
The canonical EGFR signaling pathway involves binding of growth factors to EGFR resulting in receptor dimerization and subsequent cross-phosphorylation of cytoplasmic receptor domains [25, 26, 28, 29]. Therefore, we applied EGF during AFM measurements on living DLD1 cells resulting in a significant reduction of binding frequency (Fig. 3a, b). This prompted us to ask whether EGF-mediated receptor dimerization reduced binding to Dsg2 maybe through masking the binding site. Alternatively, the subsequent auto-phosphorylation might induce decreased Dsg2–EGFR interaction. Hence, we treated the cells with erlotinib, a specific EGFR tyrosine kinase inhibitor to assess whether the tyrosine kinase activity of EGFR affects its binding to Dsg2. Indeed, the inhibitor resulted in reduced binding frequency (Fig. 3a, b), indicating that EGFR kinase activity is important. However, EGF also reduced the binding frequency in cell-free AFM measurements with a Dsg2-coated cantilever on EGFR-coated mica sheets (Fig. 3b), which raises the possibility that Dsg2–EGFR interaction might be also inhibited through steric hindrance upon EGF binding to EGFR. Results from hanging drop bead aggregation assays that revealed a similar inhibitory effect of EGF on interaction of Dsg2- and EGFR-coated beads support this hypothesis (Fig. 3c, d). Next, we analyzed the measured interactions with regard to their unbinding forces to gain insight into the molecular interaction mechanism. Peak fit analysis of measured unbinding force curves under control conditions revealed a distribution peak at 31.56 pN (Fig. 3e) which is in accordance with previously measured unbinding forces using a Dsg2-coated AFM tip on living enterocytes [20]. Interestingly, unbinding forces were similar after application of EGF, the inhibitory EGFR-specific antibody or erlotinib (Fig. 3f). Given that, EGFR is internalized following ligand binding [38, 39], we hypothesized that reduced binding frequency might be due to less available EGFR at the cell surface. Therefore, we performed surface biotinylation assays. As expected, incubation with EGF reduced EGFR levels at the cell surface (IP) as well as in total lysate (Input) (Fig. 3g, h). Interestingly, Dsg2 cell surface levels were also reduced after EGF treatment (Fig. 3g, h), indicating that EGFR and Dsg2 might stabilize one another at the cell surface. In contrast, the inhibitory EGFR-specific antibody as well as Erlotinib had no significant effect on cell surface protein levels of Dsg2 and EGFR (Fig. 3g, h), suggesting that reduced AFM-binding frequency is attributable to inhibition of direct binding. In summary, our data demonstrate that interaction of Dsg2 and EGFR is disrupted upon ligand binding to EGFR as well as after inhibition of the tyrosine kinase activity, suggesting that both direct binding via their extracellular domains as well as ligand-independent activation of EGFR might be involved in this interaction.
Fig. 3.
EGF reduces Dsg2-specific binding events on the cell surface of living DLD1 cells. a Dsg2 adhesion measurements were performed on living DLD1 cells under control conditions and after incubation with EGF or erlotinib. Shown are topography images of selected areas at cell borders and adhesion maps with each white pixel representing one binding event. b Quantification shows a significant reduction of binding events on living DLD1 cells after incubation with both, EGF or erlotinib. Application of EGF also reduced the amount of binding events in the cell-free AFM setup with a Dsg2-coated tip and EGFR-coated mica sheets. Graph bars represent fold-change values ± SE from at least four independent experiments. *p < 0.05. c Peak fit analysis of measured unbinding forces revealed a distribution peak of 31.56 pN under control conditions. d Distribution-peak values of unbinding forces measured on DLD1 cells under control conditions and after incubation with EGF, anti-EGFR antibody, or erlotinib were compared and revealed similar values for all conditions. Graph shows distribution-peak values normalized to control. e, f Hanging drop bead aggregation assay confirms blockade of interaction between Dsg2 and EGFR through EGF. Beads coated with a human IgG Fc fragment served as control. *p < 0.05; n.s. not significant. g Western blot analysis of surface biotinylation assay revealed reduced Dsg2 and EGFR levels on the surface of DLD1 cells after incubation with EGF. h Quantification of Dsg2 and EGFR band intensity from at least seven independent experiments shows a significant reduction of surface protein levels only after incubation with EGF. Graph bars represent fold-change values ± SE. *p < 0.05
Loss of Dsg2 reduces Src-dependent phosphorylation of EGFR
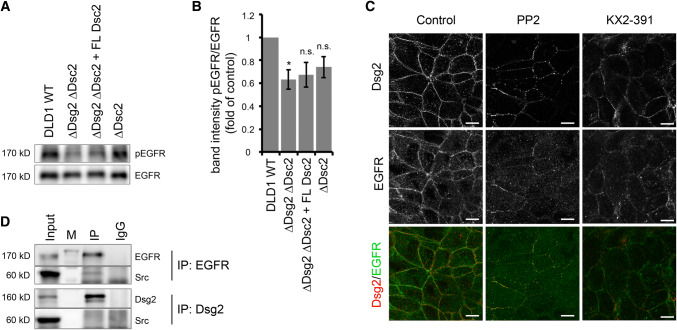
Inhibition of EGFR kinase activity reduced EGFR binding to Dsg2. Hence, we asked whether loss of Dsg2 might reduce phosphorylation of EGFR. As we hypothesized that ligand-independent activation of EGFR is involved in Dsg2–EGFR complex formation and it has been shown that EGFR is phosphorylated on Y845 in an EGF-independent manner [40], we focused on this phosphorylation site. Indeed, DLD1 cells deficient for Dsg2 displayed lower levels of Y845 phosphorylation (Fig. 4a, b). Similarly, loss of Dsg2 in Caco2 cells reduced phosphorylation of EGFR at this site (Fig. S2A and B). Reconstitution of Dsc2 in DLD1 cells deficient for both desmosomal cadherins did not restore the level of phosphorylation. Moreover, loss of Dsc2 alone had no significant effect on phosphorylation levels (Fig. 4a, b), suggesting that Dsg2 and not Dsc2 is required for phosphorylation. Usually, the phosphorylation at Y845 is catalyzed by the tyrosine kinase Src [41–43], which has already been reported to play a role in cell adhesion signaling [40, 44, 45]. Interestingly, Src localization to cell borders remained unchanged upon loss of Dsg2 (Fig. S2C). Furthermore, also protein levels of total and phosphorylated Src as well as distribution in the triton-soluble and -insoluble fractions were not affected (Figs. S2D and S2E). However, inhibition of Src activity using the Src-specific inhibitors PP2 and KX2-391 resulted in reduced and fragmented EGFR and Dsg2 immunostaining at cell borders in DLD1 cells (Fig. 4c). Intriguingly, where remaining both proteins were still co-localizing. Since Src is reported to associate with Dsg3 [45], we next performed immunoprecipitation to investigate whether it also forms a complex with Dsg2 and EGFR. Surprisingly, we found Src only associated with EGFR but not Dsg2 in DLD1 cells (Fig. 4d). Altogether, these data indicate that Src acts upstream of Dsg2 and EGFR; Src activity is required for proper localization of Dsg2 and EGFR to the cell borders and Src-mediated phosphorylation at Y845 stabilizes Dsg2–EGFR interaction.
Fig. 4.
Dsg2 knockout reduces Src-dependent phosphorylation of EGFR. a Level of phosphorylated EGFR at Y845 in DLD1 cells deficient for Dsg2 and Dsc2 was analyzed via Western blot. Loss of Dsg2 resulted in reduced level of pEGFR, which was not rescued by reconstitution of Dsc2. b Intensity of bands detected by a pEGFR-specific antibody was quantified using ImageJ and normalized to total EGFR. Shown are fold-change values ± SE of six independent experiments. *p < 0.05; n.s. not significant. c Immunostaining of Dsg2 and EGFR in DLD1 cells showed reduced and fragmented staining of both proteins at the cell border after application of the Src inhibitors PP2 and KX2-391. Bar 10 µm. d DLD1 cell lysates were used for immunoprecipitation of EGFR (upper panels) or Dsg2 (lower panels) and subjected to western blot analysis for Src, revealing a co-IP of Src with EGFR but not with Dsg2
Dsg2 via EGFR regulates the switch between adhesive and proliferative states in enterocytes
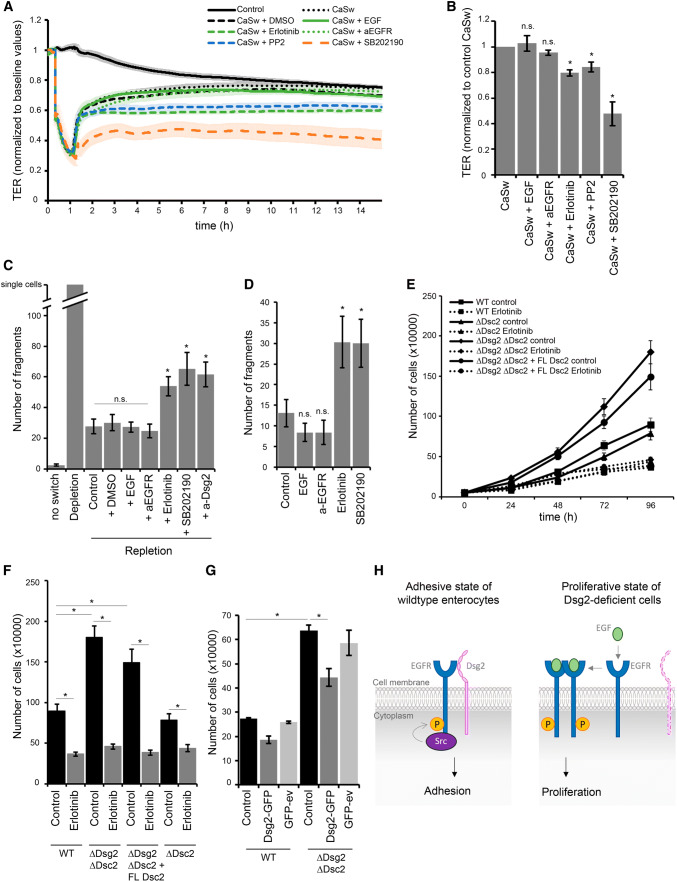
Finally, we were interested in the functional relevance of this new signaling complex. We have previously shown that loss of Dsg2 affects barrier properties via impaired downstream signaling of p38MAPK [20]. Here, we investigated whether EGFR signaling is involved in maintenance of barrier properties in intestinal cells. To explore its role during barrier recovery, we performed Ca2+-switch experiments and applied several EGFR-modulating agents at initiation of repletion. Treatment of DLD1 WT cells with EGF as well as the inhibitory EGFR-specific antibody did not disturb barrier recovery, resulting in similar TER values compared to Ca2+-switch alone (Fig. 5a, b). In contrast, application of erlotinib abolished barrier recovery significantly (Fig. 5a, b). Since we assume that Src activity is required for localization of EGFR to cell borders, we next tested whether inhibition of Src using the inhibitor PP2 affects barrier recovery after Ca2+ switch. Similar to the effect of erlotinib, also Src inhibition prevented barrier reformation significantly (Fig. 5a, b). Furthermore, inhibition of p38MAPK with the inhibitor SB202190 hampered barrier recovery (Fig. 5a, b), similar as published previously [20], which led us to the assumption that p38MAPK might be regulated downstream of the Dsg2–EGFR signaling complex. We obtained similar results using Caco2 cells (Fig. S3A and B). Moreover, Ca2+-switch experiments with DLD1 WT cells grown on filter inserts revealed no difference when applying EGF from apical side to basolateral side with both behaving similar to control conditions (Fig. S3C and D). Treatment with erlotinib in the trans-well system also disturbed barrier under both conditions, but recovery was even more affected by application from basolateral side (Fig. S3C and D). Furthermore, Ca2+-switch experiments with DLD1 cells deficient for both Dsg2 and Dsc2 also revealed significantly impaired barrier recovery after inhibition of EGFR, Src, or p38MAPK, indicating that desmosomal cadherins are not the only target of these signaling pathways in the process of barrier recovery (Fig. S3E and F). Since barrier properties are maintained by TJ, effects on claudins may also be possible. Therefore, we performed immunostaining for Claudin 4 (Cld4) after 2 h repletion together with the respective mediator, which revealed linear staining along cell borders similar to control condition after repletion without any inhibitor or when supplemented with EGF or a-EGFR, whereas staining appeared irregular after repletion in the presence of erlotinib, PP2, or SB202190 (Fig. S3G). Next, we analyzed whether the stability of newly formed cell junctions differs after repletion with EGFR-modulating agents compared to control, using a dispase-based cell dissociation assay. Application of EGF and the inhibitory EGFR-specific antibody had no effect on cell adhesion after Ca2+ switch (Fig. 5c) as well as when applied on a confluent monolayer (Fig. 5d). However, inhibition of EGFR tyrosine kinase activity using erlotinib increased the number of fragments significantly after Ca2+ switch (Fig. 5c) and disrupted cell adhesion of a confluent cell monolayer (Fig. 5d). Similar, inhibition of Dsg2 binding using the Dsg2-specific antibody and inhibition of p38MAPK with SB202190 increased cell monolayer fragmentation after Ca2+ switch as well as under normal conditions (Fig. 5c, d), further indicating that the Dsg2–EGFR complex might regulate a downstream signaling cascade that includes p38MAPK. Several studies implicate a role for Dsg2 in regulating downstream signaling cascades linked to proliferation [8, 22, 32]. In addition, the pivotal function of the EGFR is to drive cell growth and survival [46–48]. Therefore, we investigated whether loss of Dsg2 and, as a consequence, disruption of the Dsg2–EGFR complex affect cell proliferation. Indeed, DLD1 cells deficient for Dsg2 and Dsc2 showed significantly increased cell proliferation compared to WT cells (Fig. 5e, f). Stable reconstitution of Dsc2 in cells deficient for both desmosomal cadherins did not revert proliferation rate and loss of Dsc2 only did not result in increased proliferation compared to WT cells (Fig. 5e, f), suggesting that Dsg2 and not Dsc2 mediates this effect. Treatment with the EGFR tyrosine kinase inhibitor erlotinib diminished cell proliferation under all conditions resulting in values similar to WT cells (Fig. 5e, f). A similar result was obtained using WT and Dsg2-deficient Caco2 cells (Fig. S3 H). These data suggest that increased cell proliferation caused by loss of Dsg2 was mediated through EGFR tyrosine kinase activity. In addition, we performed Dsg2 rescue experiments with transient re-expression of Dsg2–GFP to corroborate the effect on proliferation upon loss of Dsg2. Using confocal microscopy, we confirmed the successful expression of Dsg2–GFP that was found linearly along cell borders in DLD1 double knockout as well as Dsg2-deficient Caco2 cells (Fig. S3I). Analysis of proliferation revealed that transient re-expression of Dsg2 was sufficient to reduce the increased proliferation rate in Dsg2-deficient DLD1 and Caco2 cells (Fig. 5g; Fig. S3J).
Fig. 5.
Dsg2 regulates EGFR activity, thereby suppressing proliferation and supporting an adhesive state in enterocytes. a Barrier recovery of DLD1 WT cells after Ca2+ switch was monitored via TER measurements showing impaired barrier recovery after inhibition of EGFR, Src and p38MAPK activity with respective inhibitors. Inhibitors were applied together with CaCl2 after 1 h depletion with EGTA. b TER values were quantified after 10 h of repletion with respective inhibitors and normalized to control repletion with respective vehicle. Repletion with erlotinib, PP2, and SB202190 resulted in significantly reduced TER values compared to control. Shown are fold-change values ± SE of 4–6 independent experiments. *p < 0.05; n.s. not significant. c Cell adhesion of DLD1 cells was analyzed in a dispase-based dissociation assay after Ca2+ switch and treatment with several inhibitors. Treatment with erlotinib, PP2, and SB202190 during repletion significantly increased monolayer fragmentation compared to control repletion with respective vehicle. Shown is mean ± SE of six independent experiments. *p < 0.05; n.s. not significant. d Confluent DLD1 cell monolayer were treated with EGF, a-EGFR, erlotinib, or SB202190, and analyzed for cell adhesion in a dispase-based cell dissociation assay. Inhibition of EGFR and p38MAPK activity significantly increased number of fragments in contrast to EGF and the EGFR-specific antibody. Shown is mean ± SE of five independent experiments. *p < 0.05; n.s. not significant. e Cell proliferation was determined by cell counting. 50,000 cells were seeded for each cell line with or without erlotinib and cells were counted after 24, 48, 72, and 96 h. f Number of cells after 96 h was quantified. Shown is mean ± SE of at least five independent experiments. *p < 0.05. g Cell proliferation was determined after Dsg2 reconstitution in DLD1 cells deficient for Dsg2 and Dsc2, 48 h after transfection. Shown is mean ± SE of four independent experiments. GFP-ev empty vector, *p < 0.05. h Model of Dsg2-mediated regulation of EGFR. Dsg2 recruits EGFR to cell borders where it is phosphorylated by Src and induces downstream signaling, thereby strengthening adhesion maybe via p38MAPK. Loss of Dsg2 results in activation of the canonical EGFR signaling pathway resulting in proliferation
Altogether, our study has identified a new role for Dsg2 in regulating EGFR activity in intestinal epithelial cells via a novel signaling complex consisting of Dsg2, EGFR, and Src. In this model, EGFR binds to Dsg2 and is phosphorylated by Src, which results in increased adhesion maybe via modulating p38MAPK signaling. In cells deficient for Dsg2, unbound EGFR is activated by ligand binding that leads to dimerization and cross-phosphorylation, thereby inducing pro-proliferative signaling pathways (Fig. 5h).
Discussion
Although many cellular processes regulated by EGFR require a break-up of intercellular junctions and desmosomes are considered to play the leading role in intercellular cohesion, only little is known about the functional interplay of EGFR and desmosomal cadherins. In this study, we propose a new mechanism of EGFR regulation in intestinal epithelial cells via a signaling complex consisting of Dsg2, EGFR, and Src. We demonstrate, for the first time, that Dsg2 and EGFR interact directly via their extracellular domains and that ligand binding as well as inhibition of EGFR tyrosine kinase activity prevent this interaction in AFM measurements. In addition, inhibition of EGFR tyrosine kinase activity significantly reduced cell adhesion, suggesting that the Dsg2–EGFR complex stabilizes desmosomal adhesion. Dsg2 is required for EGFR to localize to cell borders where it binds to Src. In line with this, we detected reduced level of Src-mediated phosphorylation of EGFR at Y845 in Dsg2-deficient cells. Moreover, inhibition of Src activity impaired barrier formation, which was accompanied by fragmented immunostaining of Dsg2 and EGFR at cell borders. Furthermore, loss of Dsg2 enhanced cell proliferation that was restored by inhibition of EGFR kinase activity, indicating that Dsg2 modulates EGFR activity, thereby regulating the switch between adhesive and proliferative states in intestinal epithelial cells.
Our data are in line with the previous studies reporting reduced phosphorylation of EGFR on Y845 upon loss of Dsg2 [22, 32]. EGFR phosphorylation at Y845 is usually catalyzed by Src and regulates a variety of cellular functions through the activation of several downstream events [41–43]. Increased levels of Src and EGFR can be found in various cancer cells, and Y845 phosphorylation-mediated signaling is linked to higher cancer malignancy due to enhanced cell transformation, motility, and invasion [42, 49–52]. Moreover, Src-mediated phosphorylation at Y845 has been reported to promote anti-apoptotic and pro-proliferative cell functions [53–56]. In accordance, recently published data show that reduced level of EGFR Y845 phosphorylation after Dsg2 downregulation suppresses cell proliferation [22, 32]. However, our study provides evidence that loss of Dsg2 and corresponding reduction of EGFR Y845 phosphorylation enhanced cell proliferation of intestinal epithelial DLD1 cells as well as of Caco2 cells. One has to consider that EGFR signaling highly depends on its spatial compartmentalization that determines the biological outcome [57]. After ligand-mediated activation at the cell surface, EGFR molecules are internalized and traverse different routes of the endosome network, while signaling continues [58–61]. EGFR is also known to act as transcription factor in the nucleus [62, 63]. Our data indicate a new mechanism by which EGFR directly interacts with Dsg2 at cell borders, thereby signaling towards adhesion and suppressing proliferation. We observed that this direct interaction is blocked using specific antibodies directed against the extracellular domains of EGFR or Dsg2 in cell-free AFM measurements. A previous study reported that Dsg2 ectodomains, which are capable of binding to the same region as the Dsg2-specific antibody used in this study, inhibit intercellular adhesion and, in addition, increase intestinal epithelial cell proliferation [21], which fits well to our proposed model. Particular attention should be given to the close co-localization of Dsg2 and EGFR which we found not only in cell culture, but similarly in human enteroids as well as in human samples which supports the physiological relevance of direct interaction between Dsg2 and EGFR. Several studies reporting the presence of EGFR in the apical membrane in intestinal cells [64–66] where Dsg2 is located support these observations. Another study suggests EGFR activation through Dsg2-mediated disruption of EGFR association with lipid rafts without direct interaction of EGFR and Dsg2 [22]. Thus, presumably, a different signaling complex is formed, resulting in a different biological function. However, this study was performed in keratinocytes which express more than one desmoglein isoform. This is in contrast to intestinal epithelial cells with only Dsg2 and Dsc2 being expressed [67, 68]. Hence, it is conceivable that desmosomal cadherins exert distinct functions in different tissues. In line with this, in keratinocytes, EGFR was shown to co-localize with Dsg1 that suppressed EGFR activity to promote differentiation [9]. A recent study demonstrated a similar mechanism, by which the classical cadherin Ecad recruits EGFR to AJ and suppresses its activity to foster barrier stability [69]. This is in line with our proposed model, where Dsg2 localizes EGFR to cell borders, thereby suppressing its proliferative activity and strengthening cell adhesion. Similarly, Dsg2 has been reported to support differentiation and thus to act as tumor suppressor gene in gastric cancer [70]. Furthermore, our data indicate that this effect is mediated by increased EGFR phosphorylation at Y845 which appears to be important for the formation of the Dsg2–EGFR signaling complex. Studies from the literature, demonstrating that Y845 phosphorylation induces cell cycle arrest and inhibits growth, also support our hypothesis [71, 72]. Interestingly, another group demonstrated that downregulation of Dsc2 increases proliferation in colon cancer cells, while downregulation of Dsg2 reduces proliferation, which is in contrast to our study [32, 73]. However, this study was conducted in cancer cells harboring a mutation that results in deregulated β-catenin signaling and increased expression of pro-proliferative genes [74, 75]. Given that the EGFR and β-catenin pathways are known to interact at multiple levels ranging from transcriptional to posttranscriptional regulation [76–81], deregulated signaling events downstream of these pathways may account for the different impact on proliferation after Dsg2 downregulation. Similar to our study, EGFR Y845 phosphorylation was reduced upon loss of Dsg2, which points toward comparable EGFR regulation but different downstream signaling. We observed increased proliferation rate in both DLD1 and Caco2 cells upon loss of Dsg2.
Previously, we have shown that Dsg2 regulates intestinal barrier properties via p38MAPK that is activated upon loss of Dsg2 [20]. However, it is still an open question how Dsg2 regulates this signaling pathway. MAPK cascades are known to be downstream targets of the EGFR and increased p38MAPK phosphorylation after EGF stimulation has been reported previously [82, 83]. Furthermore, Src-dependent p38MAPK activation has been shown to modulate the EGF-stimulated response towards migration instead of proliferation during wound closure in intestinal epithelial cells [84]. Hence, a signaling cascade consisting of Src being upstream of the Dsg2–EGFR complex and p38MAPK being regulated downstream of this complex, thereby supporting the adhesive state of intestinal epithelial cells, is conceivable. However, our data do not rule out the possibility that other targets including adhesion molecules as well as other signaling cascades may be coordinated by Dsg2 together with EGFR. For instance, EGFR regulation has recently been linked to AJ [69] and we observed irregular staining of the TJ protein Cld4 after EGFR inhibition. Furthermore, it has been reported that the apical and basolateral fractions of EGFR exert differential signaling functions [85]. Although we observed no difference in outcome when applying EGFR-modulating agents from apical side to basolateral side in this study, future studies may gain more insight into the molecular mechanism underlying the EGFR-mediated regulation of intestinal barrier properties.
Taken together, we propose a new mechanism of EGFR regulation that is important for intestinal epithelial cell homeostasis. Deregulation of any of the involved proteins has potential to induce intestinal disorders such as inflammatory bowel disease (IBD) or cancer. Thus, EGFR signaling has been shown to be reduced in IBD patients, while upregulated levels are often found in various types of cancers, indicating that a precise regulation of EGFR activity is crucial to maintain intestinal barrier integrity [42, 86]. In line with this, growth factors such as EGF promote wound healing in IBD but are also involved in the formation of neoplasia [87–89]. Likewise, Dsg2 is reduced in the mucosa of patients suffering from Crohn’s disease and is implicated in oncogenesis [17, 19]. In addition, tumor necrosis factor α (TNFα), a cytokine implicated in the pathogenesis of Crohn’s disease, has been reported to modulate EGFR activity, as well [90, 91]. Hence, understanding the molecular mechanism of EGFR regulation in intestinal cells might provide the molecular basis for new therapeutic approaches for the treatment of IBD or cancer in the future.
Materials and methods
Cell culture
DLD1 and Caco2 cells were cultured in Dulbecco’s modified Eagle medium (Life Technologies, Carlsbad, CA, USA) supplemented by 10% fetal bovine serum (Biochrom, Berlin, Germany), 50 U/mL penicillin, and 50 U/mL streptomycin (both AppliChem, Darmstadt, Germany), and cultivated in a humidified atmosphere containing 5% CO2 at 37 °C. Experiments were performed 4 days after reaching full confluence. DLD1 knockout cells were generated in the lab of Suzuki (Kwansei Gakuin University, Japan) [16] using the CRISPR/Cas9 system with the pX330 vector (Addgene, Cambridge, MA, USA) according to the method described by Cong et al. [92]. Briefly, human cDNAs for Dsg2 and Dsc2 were obtained by polymerase chain reaction (PCR) with nucleotide sequences that are published in the GenBank, and amplified and cloned into the expression vector pEF1 (Invitrogen, Carlsbad, CA, USA) accompanied with the introduction of an HA tag at the C-terminus. DLD1 cells were co-transfected with the CRISPR/Cas9 construct and the expression vector pCAG-Flag-IRESpuro (Addgene, Cambridge, MA, USA) using the lipofectamine LTX protocol. Puromycin was used for screening, resultant colonies were picked and cultured, and the expression of target protein was analyzed by immunoblotting. In addition, deletion of the target gene was confirmed by amplification of genomic DNA by PCR and subsequent sequencing. For stable expression of full-length Dsc2, cells were transfected with the DNA construct using the Lipofectamine LTX protocol and screened in the presence of G418 (400 µg/mL) for about 2 weeks, followed by immunoblot analysis.
Human tissue samples
Colon samples were obtained from patients who required right or left hemi-colectomy due to colon carcinoma in which the surgical resection routinely involves a part of the healthy small intestine or colon, respectively. All patients gave their written informed consent prior surgery to inclusion in the study and ethical approval was given by the Ethical Board of the University of Würzburg (proposal numbers 113/13, 46/11, 42/16). Tissue samples were collected in a standardized procedure via the Interdisciplinary Bank of Biomaterials and Data Würzburg (IBDW) as described in detail previously [93]. For immunostaining, tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned (1 μm).
Enteroids
Intestinal epithelial cells (IECs) were isolated from healthy human full-wall gut resections, 1 cm2 in size as described previously [94]. Briefly, villi were scraped off the muscle-free mucosa using a sterile glass slide. The remaining tissue was transferred into a 50 mL falcon tube with 20 mL 4 °C cold HBSS (Sigma-Aldrich, St. Louis, MO, USA) and vortexed for 5 s, and the supernatant discarded. This washing step was repeated until the supernatant was completely cleared of cell debris. Afterward, the tissue was incubated in 4 °C cold 2 mM EDTA/HBSS solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 4 °C under gentle rotation on a shaker. Subsequently, the tissue was washed once in 20 mL HBSS by manually inverting the tube five times. The mucosa was transferred in a new tube with 10 mL HBSS and manually shaken five times. This shaking procedure was repeated four times always using a new tube. Each cell fraction was checked for the amount and size of crypts within small drops under the microscope. The supernatants containing the most vital appearing crypts were pooled and centrifuged at 350g for 3 min at room temperature (RT). Pellet was resuspended in 10 mL basal medium, DMEM-F12 Advanced (Invitrogen, Carlsbad, CA, USA) supplemented with N2, B27, Anti–Anti, 10 mM HEPES, 2 mM GlutaMAX-I (all from Invitrogen, Carlsbad, CA, USA), and 1 mM N-acetylcysteine (Sigma-Aldrich, St. Louis, MO, USA), and the crypt number was estimated in a 10 µL drop by microscopy. Crypts were centrifuged in a nonstick 1.5 mL tube at 350g for 3 min at RT and the supernatant was removed. The tube with the cell pellet was placed on ice until further use. The pellet was resuspended in an appropriate amount of cold Matrigel (Corning, Hickory, NC, USA), that is, 5000 crypts/mL. Drops of 50 µL per well were seeded in a 24-well plate and incubated for 10–20 min until the Matrigel was well solidified. The culture medium contained a mixture of 50% fresh basal medium and 50% Wnt3A-conditioned medium.
Furthermore, the following growth factors were added: 500 ng/mL hR-Spondin 1 (PeproTech, Rocky Hill, NY, USA). 100 ng/mL IECs were isolated from human small intestinal tissue and expanded as organoid culture for 3–4 weeks.
Test reagents
Epidermal growth factor receptor activity was inhibited using either a specific mouse anti-EGFR antibody (C225, sodium azide free, Merck Millipore, Darmstadt, Germany) at 0.5 µg/mL for 1 h or the tyrosine kinase activity inhibitor erlotinib (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 2.5 µM for 1 h. For positive stimulation of EGFR activity, the growth factor EGF (Sigma-Aldrich, Munich, Germany) was used at 20 ng/mL for 1 h. Cells were cultured in serum-free medium for 1 h prior to treatment with EGFR-modulating agents. PP2 (Calbiochem, Darmstadt, Germany) and KX2-391 (Biozol, Germany) were used at 10 µM for 1 h to inhibit Src activity and SB202190 (Calbiochem, Darmstadt, Germany) was used at 30 µM for 1 h to inhibit p38 MAPK. Dsg2 binding was inhibited using a specific monoclonal mouse antibody directed against the second and third extracellular repeat domains of Dsg2 (clone 10G11, sodium azide free, Progen, Heidelberg, Germany) applied 1:50. For western blot analysis and immunofluorescence staining, following primary antibodies were used: mouse anti-Dsg2 (clone 10G11) and rabbit anti-Dsg2 (rb5, both Progen, Heidelberg, Germany), mouse anti-Dsc2/3 (clone 7G6) and rabbit anti Claudin-4 (both Life Technologies, Carlsbad, CA, USA), rabbit anti-DP I/II (H-300), rabbit anti-EGFR (clone 1005-G), mouse anti-EGFR (clone A-10) and mouse anti-GAPDH (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-α-tubulin (Abcam, Cambridge, UK), and rabbit anti-phospho-EGFR Tyr845 and rabbit anti-Src (both from Cell Signaling, Danvers, MA, USA). HRP-conjugated goat anti-mouse or goat anti-rabbit (Dianova, Hamburg, Germany) secondary antibodies were used for western blot analysis. Cy3- or Alexa488-labeled goat anti-mouse or goat anti-rabbit antibodies (Dianova, Hamburg, Germany) were used for confocal microscopy, and Star580- and Star635P-labeled antibodies (Abberior) were used for STED. F-actin was visualized using Alexa488-labeled phalloidin (Life Technologies, Carlsbad, CA, USA) and nuclei were counterstained with DAPI (Sigma-Aldrich, Munich, Germany).
Immunofluorescence
Cells were grown on 12 mm glass cover slides, fixed with 2% paraformaldehyde in PBS for 10 min and, subsequently, permeabilized with 0.5% TritonX-100 in PBS containing 0.02% Tween20 (PBS-T) for 10 min following 1 h of blocking in 2% BSA in PBS-T. Human tissue samples embedded in paraffin were sectioned in 1 µm slices and immunostaining was performed after removal of paraffin using 100% Xylol followed by an ethanol series from 100 to 70% and sera dest at the end. Then, samples were permeabilized with 0.1% TritonX-100 in PBS-T for 30 min followed by blocking in 2% BSA in PBS-T. Primary and secondary antibodies were incubated for 1 h at room temperature each. Paraffin sections were incubated with primary antibodies at 4 °C over night. For confocal microscopy, coverslips were placed on glass slides with 60% glycerol in PBS, containing 1.5% N-propyl gallate (Serva, Heidelberg, Germany). Image acquisition was performed using a Leica SP5 confocal microscope with a 63× NA 1.4 PL APO objective (both Leica, Wetzlar, Germany. Co-localization analysis was performed by generating an intensity plot profile for each channel using the ImageJ software and calculating the Pearson’s correlation coefficient between the intensity distributions of two molecules of interest. To this end, 35 cell borders from 7 independent experiments were selected.
Stimulated emission depletion microscopy (STED)
After immunostaining, cells were mounted in 2.5% DABCO in MOWIOL/HEPES (self-made solution). Images were acquired with an Abberior 3D STED confocal microscope. Star580 and Star635P (both from Abberior) were excited at 594 and 638 nm, respectively, using pulsed diode lasers (PDL 594, Abberior Instruments; PiL063X, Advanced Laser Diode Systems). Depletion of fluorescent molecules was conducted at 775 nm with a pulsed fibre laser (PFL-P-30-775B1R, MPB Communications) and emission was detected with an avalanche photodiode detector at 605–625 and 650–720 nm range.
Western blot
Cells were lysed with SDS lysis buffer (25 mM HEPES, 2 mM EDTA, 25 mM NaF, and 1% SDS) supplemented with a protease-inhibitor cocktail (Roche, Mannheim, Germany) followed by sonication and heating to 95 °C for 10 min in Laemmli buffer with 50 mM dithiothreitol (Applichem). Protein amount was determined using a BCA Protein Assay Kit (Pierce/Thermo Scientific, Waltham, MA, USA) and equivalent protein concentrations were resolved by reducing SDS-PAGE. After protein transfer to a nitrocellulose membrane (Life Technologies, Carlsbad, CA, USA) according to the standard protocols, membranes were probed with primary antibodies overnight at 4 °C followed by incubation with secondary antibodies for 2 h at room temperature. Bands were detected with an ECL reaction system (self-made solution) using the Amersham Imager 600 (GE Healthcare Life Sciences, Germany).
TritonX-100 protein extraction
Cells were washed with ice-cold PBS and incubated in a Triton buffer (0.5% Triton X-100, 50 mM MES, 25 mM EGTA, and 5 mM MgCl2) supplemented with 1 mM PMSF (Roth, Germany), Aprotinin, Pepstatin A (both Applichem, Germany), and Leupeptin (VWR, Germany) for 15 min on ice under gentle shaking. Subsequently, cell lysates were centrifuged at 13,000 rpm for 5 min to separate the insoluble from the soluble fraction and pellets were resuspended in SDS lysis buffer followed by sonication. Protein concentration of both fractions was calculated as described above and equivalent amounts of protein were used for western blot analysis.
Cell surface biotinylation
Cells grown in six-well plates were treated with test reagents, washed with ice-cold HBSS, and incubated with 0.25 mM membrane-impermeable EZ-Link Sulfo-NHS-Biotin (Thermo Fischer Scientific, Waltham, USA) for 1 h on ice to prevent internalization. Excess biotin was quenched by washing three times with ice-cold 100 mM Glycin followed by three times washing with ice-cold HBSS. Cells were lysed in lysis buffer (50 mM NaCl, 10 mM PIPES, 3 mM MgCl2, and 1% Triton X-100) for 15 min on ice followed by 5 min centrifugation at 13,000 rpm and measurement of protein concentration of the recovered cell lysate supernatants as described above. For precipitation of biotin-labeled proteins, a protein amount of 500 µg was incubated with NeutrAvidin (High Capacity)-agarose beads (Thermo Fischer Scientific, Waltham, USA) over night at 4 °C. After four times washing with lysis buffer, cell surface proteins were eluted in Laemmli buffer containing 50 mM dithiothreitol at 95 °C and subjected to Western blot analysis.
Co-immunoprecipitation
Cell monolayers were washed with ice-cold HBSS and incubated with RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 0.1% SDS, 1% NP-40, and 0.1 mM EDTA) supplemented with a protease-inhibitor cocktail (Roche, Mannheim, Germany) for 30 min on ice while shaking. Cells were scraped and centrifuged at 13,000 rpm for 5 min at 4 °C, and protein concentration of the lysate was determined as described above. Cell lysates were precleared with protein-G beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at 4 °C, followed by incubation with 1 µg of respective antibody or IgG control for 3 h at 4 °C with gentle rotation. Subsequently, protein-G beads were added and incubated over night at 4 °C. For pull-down assays with Dsg2-Fc, beads were incubated with 0.15 mg/mL Dsg2-Fc in HBSS at 4 °C over night, washed three times with RIPA buffer to remove unbound protein, and incubated with cell lysates at 4 °C again over night. After three times washing with RIPA buffer, immunocomplexes were boiled in 20 µL Laemmli buffer containing 50 mM dithiothreitol at 95 °C for 10 min and subjected to Western blot analysis.
Atomic force microscopy (AFM)
Atomic force microscopy measurements were performed with a Nanowizard III AFM (JPK Instruments, Berlin, Germany) mounted on an optical microscopy (Carl Zeiss, Jena, Germany). The whole setup was placed on a Halcyonics i4 anti-vibration table (Accurion, Goettingen, Germany) in a closed hood to protect measurements from environmental noise. The application of AFM force microscopy on living cells was described in detail before [95]. All measurements were conducted in cell culture medium at 37 °C. For Dsg2 adhesion studies, recombinant Dsg2-Fc (self-made) containing the complete extracellular domain (ED) of Dsg2 was linked to flexible Si3N4 AFM cantilevers (MLCT probes, Bruker, Calle Tecate, CA, USA) via a flexible bifunctional polyethylene glycol linker (Gruber Lab, Institute of Biophysics, Linz, Austria) as described elsewhere [96]. Prior to adhesion measurements on living cells, AFM topography images of 50 × 50 µm and 128 × 128 pixels were created using a force curve-based imaging mode (QI-mode) with a setpoint adjusted to 0.5 nN, a z length of 1500 nm, and a pulling speed of 50 µm/s. Adhesion measurements were performed in the force spectroscopy mode with a relative setpoint of 0.5 nN, a z length of 2 µm, and a pulling speed of 4 µm/s. The same cantilever was used under control conditions and after incubation for 1 h with inhibitory antibodies or EGFR-modulating agents. For each condition, several areas of 2 × 5 µm were selected with 1000 recorded force–distance curves per area. Cell-free AFM measurements were carried out with mica sheets (SPI Supplies, West Chester, USA) instead of cell monolayers, functionalized with either Dsg2-Fc or EGFR-Fc (Sino Biological). To produce comparable results, setpoint was adjusted to the same value as for cell-based experiments. For each condition, several areas of 25 µm × 25 µm with 400 force–distance curves per area were measured using a z length of 0.3 µm and a pulling speed of 1 µm/s. Acquired AFM data were processed using the JPK processing software. Analysis of unbinding force distribution was performed with Origin Pro 2016, 93G (Northampton, MA, USA).
Hanging drop bead aggregation assay
Protein-G-coated polystyrene microbeads (Dynabeads, diameter 2.8 µm; Thermo Fisher Scientific) were coated with recombinant Dsg2- or EGFR-Fc and aggregation assays were carried out as described previously [95]. Briefly, beads were washed with 100 mM sodium phosphate buffer (pH 8.1), blocked with 5% BSA for 1 h at RT, and incubated with 0.15 mg/mL Dsg2-, EGFR-Fc or control-Fc part of human IgGs (Merck Millipore, Darmstadt, Germany) over night at 4 °C under slow overhead rotation. Following washing steps, beads were incubated with 500 mM dimethyl adipimidate·2 HCl (DMA, Thermo Fisher Scientific) for 45 min at RT to covalently cross-link protein-G with Fc parts. After incubation with 200 mM ethanolamine (pH 8.0) for 2 h at RT and several washing steps, about 0.3 µg beads were resuspended in 10 µL HBSS containing 1.8 mM Ca2+. Then, beads were allowed to aggregate in a hanging drop on the underside of a culture dish lid at 37 °C for 1 h, followed by 1 h incubation with EGF. After shaking on an orbital shaker at 1000 rpm for 3 min, a ratio was calculated from of the area covered with beads and the area of total bead extent and expressed as density of bead colonies in percentage.
Calcium switch assay
Cells were incubated with 4 mM EGTA for 1 h, thereby inducing disruption of Ca2+-dependent cell junctions. Reformation of cell junctions was induced by addition of 8 mM CaCl2 supplemented with test reagents or the respective vehicle. Time course of junctional disassembly and reassembly was monitored by measuring the transepithelial resistance (TER) of cells grown on eight-well electrode arrays (Ibidi, 8W10E) with an ECIS model Z theta (Applied Biophysics, Troy, NY, USA) at 800 Hz for DLD1 cells and 400 Hz for Caco2 cells. In addition, cells were grown on filter inserts (Corning, PET membrane, pore size 0.4 µm) and measured with the ECIS 8W Trans-Filter Adapter. Stability of reformed cell junctions was analyzed using a dispase-based cell dissociation assay.
Dispase-based dissociation assay
Dissociation assays were performed in 24-well plates. After treatment with test reagents, cell monolayer were washed with Hank’s buffered saline solution (HBSS; Sigma-Aldrich) and incubated with 150 µL dispase II (2.4 U/mL in HBSS, Sigma-Aldrich) at 37 °C for 25 min. Reaction was stopped by adding 200 µL HBSS and a defined shear stress was applied by pipetting three times with an electrical pipet. Resulting fragments were counted using a binocular microscope (Leica). Every condition was performed in duplicates and each experiment was repeated at least four times.
Proliferation assay
Cellular proliferation was ascertained by counting the number of cells using a Neubauer chamber (Laboroptik, Lancing, UK). 50,000 cells were seeded in 24-well plates in complete DMEM with erlotinib (2.5 µM) or DMSO and counted after 24, 48, 72, and 96 h. Medium was changed every day.
Transfection with Dsg2–GFP
The plasmid encoding for human wild-type Dsg2 with a C-terminal eGFP tag [97] was kindly provided by Katja Gehmlich (University of Oxford, UK). 50,000 cells were seeded and transfected right away with 1 µg endotoxin-free Dsg2–GFP plasmid DNA or the GFP empty vector [98] (pDEST-eGFP-N1, Addgene plasmid # 31796, gift from Robin Shaw, Cedars-Sinai Heart Institute, Los Angeles, USA) using TurboFect™ (Life Technologies) according to the manufacturer’s protocol and incubated for 48 h. Medium was changed 8 h after transfection.
Statistics
All experiments were repeated at least three times. Band intensity was quantified using ImageJ (National Institutes of Health, Bethesda, MD, USA). For statistical analysis, two-tailed Student’s t test was used to analyze two-sample groups and one-way ANOVA followed by Bonferroni correction was used for multiple sample groups. Results are shown as mean ± SE. A p value of < 0.05 was considered significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the DFG priority program SPP 1782. DLD1 cells were a gift from S.T. Suzuki (Kwansei Gakuin University, Japan). STED microscopy was performed in the lab of H. Leonhardt (Ludwig-Maximilians-University, Munich). The authors would like to thank Andreas Meiser and Hartmann Harz for their assistance in STED sample preparation and image acquisition.
Abbreviations
- AFM
Atomic force microscopy
- AJ
Adherens junction
- Cld4
Claudin 4
- Dsc2
Desmocollin 2
- Dsg2
Desmoglein
- DP
Desmoplakin
- Ecad
E-cadherin
- EGFR
Epidermal growth factor receptor
- MAPK
Mitogen-activated protein kinase
- PG
Plakoglobin
- Pkp
Plakophilin
- RTK
Receptor tyrosine kinase
- STED
Stimulated emission depletion microscopy
- TER
Transepithelial resistance
- TJ
Tight junction
- WT
Wild type
Author contributions
HU and JW designed the study. HU performed and analyzed the experiments. VR performed the hanging drop bead aggregation assay. MM obtained and prepared the human tissue samples. CF generated the enteroids. MD generated the Dsg2-deficient Caco2 cell line. All authors interpreted the data. HU wrote the manuscript and prepared the figures. All authors reviewed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 2.Helander HF, Fandriks L. Surface area of the digestive tract—revisited. Scand J Gastroenterol. 2014;49:681–689. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- 3.Gayer CP, Basson MD. The effects of mechanical forces on intestinal physiology and pathology. Cell Signal. 2009;21:1237–1244. doi: 10.1016/j.cellsig.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 5.Capaldo CT, Farkas AE, Nusrat A. Epithelial adhesive junctions. F1000Prime Rep. 2014;6:1. doi: 10.12703/P6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Investig Dermatol. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 8.Brennan D, Hu Y, Joubeh S, Choi YW, Whitaker-Menezes D, O’Brien T, Uitto J, Rodeck U, Mahoney MG. Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. J Cell Sci. 2007;120:758–771. doi: 10.1242/jcs.03392. [DOI] [PubMed] [Google Scholar]
- 9.Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, Cornwell M, Green KJ. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185:1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nava P, Laukoetter MG, Hopkins AM, Laur O, Gerner-Smidt K, Green KJ, Parkos CA, Nusrat A. Desmoglein-2: a novel regulator of apoptosis in the intestinal epithelium. Mol Biol Cell. 2007;18:4565–4578. doi: 10.1091/mbc.e07-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spindler V, Waschke J. Desmosomal cadherins and signaling: lessons from autoimmune disease. Cell Commun Adhes. 2014;21:77–84. doi: 10.3109/15419061.2013.877000. [DOI] [PubMed] [Google Scholar]
- 12.Owen GR, Stokes DL. Exploring the nature of desmosomal cadherin associations in 3D. Dermatol Res Pract. 2010;2010:930401. doi: 10.1155/2010/930401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holthofer B, Windoffer R, Troyanovsky S, Leube RE. Structure and function of desmosomes. Int Rev Cytol. 2007;264:65–163. doi: 10.1016/S0074-7696(07)64003-0. [DOI] [PubMed] [Google Scholar]
- 14.Koch PJ, Goldschmidt MD, Zimbelmann R, Troyanovsky R, Franke WW. Complexity and expression patterns of the desmosomal cadherins. Proc Natl Acad Sci USA. 1992;89:353–357. doi: 10.1073/pnas.89.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waschke J. The desmosome and pemphigus. Histochem Cell Biol. 2008;130:21–54. doi: 10.1007/s00418-008-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara M, Nagatomo A, Tsuda M, Obata S, Sakuma T, Yamamoto T, Suzuki ST. Desmocollin-2 alone forms functional desmosomal plaques, with the plaque formation requiring the juxtamembrane region and plakophilins. J Biochem. 2015;158:339–353. doi: 10.1093/jb/mvv048. [DOI] [PubMed] [Google Scholar]
- 17.Biedermann K, Vogelsang H, Becker I, Plaschke S, Siewert JR, Hofler H, Keller G. Desmoglein 2 is expressed abnormally rather than mutated in familial and sporadic gastric cancer. J Pathol. 2005;207:199–206. doi: 10.1002/path.1821. [DOI] [PubMed] [Google Scholar]
- 18.Schlegel N, Meir M, Heupel WM, Holthofer B, Leube RE, Waschke J. Desmoglein 2-mediated adhesion is required for intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2010;298:G774–G783. doi: 10.1152/ajpgi.00239.2009. [DOI] [PubMed] [Google Scholar]
- 19.Spindler V, Meir M, Vigh B, Flemming S, Hutz K, Germer CT, Waschke J, Schlegel N. Loss of desmoglein 2 contributes to the pathogenesis of Crohn’s disease. Inflamm Bowel Dis. 2015;21:2349–2359. doi: 10.1097/MIB.0000000000000486. [DOI] [PubMed] [Google Scholar]
- 20.Ungewiss H, Vielmuth F, Suzuki ST, Maiser A, Harz H, Leonhardt H, Kugelmann D, Schlegel N, Waschke J. Desmoglein 2 regulates the intestinal epithelial barrier via p38 mitogen-activated protein kinase. Sci Rep. 2017;7:6329. doi: 10.1038/s41598-017-06713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamekura R, Nava P, Feng M, Quiros M, Nishio H, Weber DA, Parkos CA, Nusrat A. Inflammation-induced desmoglein-2 ectodomain shedding compromises the mucosal barrier. Mol Biol Cell. 2015;26:3165–3177. doi: 10.1091/mbc.e15-03-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overmiller AM, McGuinn KP, Roberts BJ, Cooper F, Brennan-Crispi DM, Deguchi T, Peltonen S, Wahl JK, 3rd, Mahoney MG. c-Src/Cav1-dependent activation of the EGFR by Dsg2. Oncotarget. 2016;7:37536–37555. doi: 10.18632/oncotarget.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–235. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- 24.Yap AS, Crampton MS, Hardin J. Making and breaking contacts: the cellular biology of cadherin regulation. Curr Opin Cell Biol. 2007;19:508–514. doi: 10.1016/j.ceb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honegger AM, Kris RM, Ullrich A, Schlessinger J. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proc Natl Acad Sci USA. 1989;86:925–929. doi: 10.1073/pnas.86.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan M, Narasimhan S, de Heus C, Mance D, van Doorn S, Houben K, Popov-Celeketic D, Damman R, Katrukha EA, Jain P, et al. EGFR dynamics change during activation in native membranes as revealed by NMR. Cell. 2016;167(1241–1251):e1211. doi: 10.1016/j.cell.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 27.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/S0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 29.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-K. [DOI] [PubMed] [Google Scholar]
- 30.Klessner JL, Desai BV, Amargo EV, Getsios S, Green KJ. EGFR and ADAMs cooperate to regulate shedding and endocytic trafficking of the desmosomal cadherin desmoglein 2. Mol Biol Cell. 2009;20:328–337. doi: 10.1091/mbc.e08-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorch JH, Klessner J, Park JK, Getsios S, Wu YL, Stack MS, Green KJ. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J Biol Chem. 2004;279:37191–37200. doi: 10.1074/jbc.M405123200. [DOI] [PubMed] [Google Scholar]
- 32.Kamekura R, Kolegraff KN, Nava P, Hilgarth RS, Feng M, Parkos CA, Nusrat A. Loss of the desmosomal cadherin desmoglein-2 suppresses colon cancer cell proliferation through EGFR signaling. Oncogene. 2014;33:4531–4536. doi: 10.1038/onc.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blay J, Brown KD. Epidermal growth factor promotes the chemotactic migration of cultured rat intestinal epithelial cells. J Cell Physiol. 1985;124:107–112. doi: 10.1002/jcp.1041240117. [DOI] [PubMed] [Google Scholar]
- 34.Miguel JC, Maxwell AA, Hsieh JJ, Harnisch LC, Al Alam D, Polk DB, Lien CL, Watson AJ, Frey MR. Epidermal growth factor suppresses intestinal epithelial cell shedding through a MAPK-dependent pathway. J Cell Sci. 2017;130:90–96. doi: 10.1242/jcs.182584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polk DB. Epidermal growth factor receptor-stimulated intestinal epithelial cell migration requires phospholipase C activity. Gastroenterology. 1998;114:493–502. doi: 10.1016/S0016-5085(98)70532-3. [DOI] [PubMed] [Google Scholar]
- 36.Bishop WP, Wen JT. Regulation of Caco-2 cell proliferation by basolateral membrane epidermal growth factor receptors. Am J Physiol. 1994;267:G892–G900. doi: 10.1152/ajpgi.1994.267.5.G892. [DOI] [PubMed] [Google Scholar]
- 37.Scheving LA, Shiurba RA, Nguyen TD, Gray GM. Epidermal growth factor receptor of the intestinal enterocyte. Localization to laterobasal but not brush border membrane. J Biol Chem. 1989;264:1735–1741. [PubMed] [Google Scholar]
- 38.Carpenter G, Cohen S. 125I-labeled human epidermal growth factor. Binding, internalization, and degradation in human fibroblasts. J Cell Biol. 1976;71:159–171. doi: 10.1083/jcb.71.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moro L, Dolce L, Cabodi S, Bergatto E, Boeri Erba E, Smeriglio M, Turco E, Retta SF, Giuffrida MG, Venturino M, et al. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem. 2002;277:9405–9414. doi: 10.1074/jbc.M109101200. [DOI] [PubMed] [Google Scholar]
- 41.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 42.Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc Natl Acad Sci USA. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato K, Sato A, Aoto M, Fukami Y. c-Src phosphorylates epidermal growth factor receptor on tyrosine 845. Biochem Biophys Res Commun. 1995;215:1078–1087. doi: 10.1006/bbrc.1995.2574. [DOI] [PubMed] [Google Scholar]
- 44.Balanis N, Yoshigi M, Wendt MK, Schiemann WP, Carlin CR. beta3 integrin-EGF receptor cross-talk activates p190RhoGAP in mouse mammary gland epithelial cells. Mol Biol Cell. 2011;22:4288–4301. doi: 10.1091/mbc.e10-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rotzer V, Hartlieb E, Vielmuth F, Gliem M, Spindler V, Waschke J. E-cadherin and Src associate with extradesmosomal Dsg3 and modulate desmosome assembly and adhesion. Cell Mol Life Sci. 2015;72:4885–4897. doi: 10.1007/s00018-015-1977-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogdan S, Klambt C. Epidermal growth factor receptor signaling. Curr Biol. 2001;11:R292–R295. doi: 10.1016/S0960-9822(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 47.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 48.Massague J, Pandiella A. Membrane-anchored growth factors. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002503. [DOI] [PubMed] [Google Scholar]
- 49.Chung BM, Dimri M, George M, Reddi AL, Chen G, Band V, Band H. The role of cooperativity with Src in oncogenic transformation mediated by non-small cell lung cancer-associated EGF receptor mutants. Oncogene. 2009;28:1821–1832. doi: 10.1038/onc.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung J, Kim HY, Kim M, Sohn K, Kim M, Lee K. Translationally controlled tumor protein induces human breast epithelial cell transformation through the activation of Src. Oncogene. 2011;30:2264–2274. doi: 10.1038/onc.2010.604. [DOI] [PubMed] [Google Scholar]
- 51.Jung O, Choi YJ, Kwak TK, Kang M, Lee MS, Ryu J, Kim HJ, Lee JW. The COOH-terminus of TM4SF5 in hepatoma cell lines regulates c-Src to form invasive protrusions via EGFR Tyr845 phosphorylation. Biochim Biophys Acta. 2013;1833:629–642. doi: 10.1016/j.bbamcr.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 52.Kannangai R, Sahin F, Torbenson MS. EGFR is phosphorylated at Ty845 in hepatocellular carcinoma. Mod Pathol. 2006;19:1456–1461. doi: 10.1038/modpathol.3800665. [DOI] [PubMed] [Google Scholar]
- 53.Cvrljevic AN, Akhavan D, Wu M, Martinello P, Furnari FB, Johnston AJ, Guo D, Pike L, Cavenee WK, Scott AM, et al. Activation of Src induces mitochondrial localisation of de2-7EGFR (EGFRvIII) in glioma cells: implications for glucose metabolism. J Cell Sci. 2011;124:2938–2950. doi: 10.1242/jcs.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nair VD, Sealfon SC. Agonist-specific transactivation of phosphoinositide 3-kinase signaling pathway mediated by the dopamine D2 receptor. J Biol Chem. 2003;278:47053–47061. doi: 10.1074/jbc.M303364200. [DOI] [PubMed] [Google Scholar]
- 55.Ray RM, Bhattacharya S, Johnson LR. EGFR plays a pivotal role in the regulation of polyamine-dependent apoptosis in intestinal epithelial cells. Cell Signal. 2007;19:2519–2527. doi: 10.1016/j.cellsig.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bakker J, Spits M, Neefjes J, Berlin I. The EGFR odyssey—from activation to destruction in space and time. J Cell Sci. 2017;130:4087–4096. doi: 10.1242/jcs.209197. [DOI] [PubMed] [Google Scholar]
- 58.Francavilla C, Papetti M, Rigbolt KT, Pedersen AK, Sigurdsson JO, Cazzamali G, Karemore G, Blagoev B, Olsen JV. Multilayered proteomics reveals molecular switches dictating ligand-dependent EGFR trafficking. Nat Struct Mol Biol. 2016;23:608–618. doi: 10.1038/nsmb.3218. [DOI] [PubMed] [Google Scholar]
- 59.Haugh JM, Huang AC, Wiley HS, Wells A, Lauffenburger DA. Internalized epidermal growth factor receptors participate in the activation of p21(ras) in fibroblasts. J Biol Chem. 1999;274:34350–34360. doi: 10.1074/jbc.274.48.34350. [DOI] [PubMed] [Google Scholar]
- 60.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 61.Wu P, Wee P, Jiang J, Chen X, Wang Z. Differential regulation of transcription factors by location-specific EGF receptor signaling via a spatio-temporal interplay of ERK activation. PLoS One. 2012;7:e41354. doi: 10.1371/journal.pone.0041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brand TM, Iida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 63.Kamio T, Shigematsu K, Sou H, Kawai K, Tsuchiyama H. Immunohistochemical expression of epidermal growth factor receptors in human adrenocortical carcinoma. Hum Pathol. 1990;21:277–282. doi: 10.1016/0046-8177(90)90227-V. [DOI] [PubMed] [Google Scholar]
- 64.Gonnella PA, Siminoski K, Murphy RA, Neutra MR. Transepithelial transport of epidermal growth factor by absorptive cells of suckling rat ileum. J Clin Investig. 1987;80:22–32. doi: 10.1172/JCI113051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelly D, McFadyen M, King TP, Morgan PJ. Characterization and autoradiographic localization of the epidermal growth factor receptor in the jejunum of neonatal and weaned pigs. Reprod Fertil Dev. 1992;4:183–191. doi: 10.1071/RD9920183. [DOI] [PubMed] [Google Scholar]
- 66.Yoo BK, He P, Lee SJ, Yun CC. Lysophosphatidic acid 5 receptor induces activation of Na(+)/H(+) exchanger 3 via apical epidermal growth factor receptor in intestinal epithelial cells. Am J Physiol Cell Physiol. 2011;301:C1008–C1016. doi: 10.1152/ajpcell.00231.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 68.Mahoney MG, Hu Y, Brennan D, Bazzi H, Christiano AM, Wahl JK., 3rd Delineation of diversified desmoglein distribution in stratified squamous epithelia: implications in diseases. Exp Dermatol. 2006;15:101–109. doi: 10.1111/j.1600-0625.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 69.Rubsam M, Mertz AF, Kubo A, Marg S, Jungst C, Goranci-Buzhala G, Schauss AC, Horsley V, Dufresne ER, Moser M, et al. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat Commun. 2017;8:1250. doi: 10.1038/s41467-017-01170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yashiro M, Nishioka N, Hirakawa K. Decreased expression of the adhesion molecule desmoglein-2 is associated with diffuse-type gastric carcinoma. Eur J Cancer. 2006;42:2397–2403. doi: 10.1016/j.ejca.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 71.Godek J, Sargiannidou I, Patel S, Hurd L, Rothman VL, Tuszynski GP. Angiocidin inhibits breast cancer proliferation through activation of epidermal growth factor receptor and nuclear factor kappa (NF-kB) Exp Mol Pathol. 2011;90:244–251. doi: 10.1016/j.yexmp.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato K, Nagao T, Iwasaki T, Nishihira Y, Fukami Y. Src-dependent phosphorylation of the EGF receptor Tyr-845 mediates Stat-p21waf1 pathway in A431 cells. Genes Cells. 2003;8:995–1003. doi: 10.1046/j.1356-9597.2003.00691.x. [DOI] [PubMed] [Google Scholar]
- 73.Kolegraff K, Nava P, Helms MN, Parkos CA, Nusrat A. Loss of desmocollin-2 confers a tumorigenic phenotype to colonic epithelial cells through activation of Akt/beta-catenin signaling. Mol Biol Cell. 2011;22:1121–1134. doi: 10.1091/mbc.e10-10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. APC mutations in sporadic colorectal tumors: a mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Zhang W, Evans PM, Chen X, He X, Liu C. Adenomatous polyposis coli (APC) differentially regulates beta-catenin phosphorylation and ubiquitination in colon cancer cells. J Biol Chem. 2006;281:17751–17757. doi: 10.1074/jbc.M600831200. [DOI] [PubMed] [Google Scholar]
- 76.Guturi KK, Mandal T, Chatterjee A, Sarkar M, Bhattacharya S, Chatterjee U, Ghosh MK. Mechanism of beta-catenin-mediated transcriptional regulation of epidermal growth factor receptor expression in glycogen synthase kinase 3 beta-inactivated prostate cancer cells. J Biol Chem. 2012;287:18287–18296. doi: 10.1074/jbc.M111.324798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jean C, Blanc A, Prade-Houdellier N, Ysebaert L, Hernandez-Pigeon H, Al Saati T, Haure MJ, Coluccia AM, Charveron M, Delabesse E, et al. Epidermal growth factor receptor/beta-catenin/T cell factor 4/matrix metalloproteinase 1: a new pathway for regulating keratinocyte invasiveness after UVA irradiation. Cancer Res. 2009;69:3291–3299. doi: 10.1158/0008-5472.CAN-08-1909. [DOI] [PubMed] [Google Scholar]
- 78.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/S1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 79.Tan X, Apte U, Micsenyi A, Kotsagrelos E, Luo JH, Ranganathan S, Monga DK, Bell A, Michalopoulos GK, Monga SP. Epidermal growth factor receptor: a novel target of the Wnt/beta-catenin pathway in liver. Gastroenterology. 2005;129:285–302. doi: 10.1053/j.gastro.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, Franken PF, van Gurp L, Meijlink F, van der Valk MA, et al. beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yue X, Lan F, Yang W, Yang Y, Han L, Zhang A, Liu J, Zeng H, Jiang T, Pu P, et al. Interruption of beta-catenin suppresses the EGFR pathway by blocking multiple oncogenic targets in human glioma cells. Brain Res. 2010;1366:27–37. doi: 10.1016/j.brainres.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 82.Vergarajauregui S, San Miguel A, Puertollano R. Activation of p38 mitogen-activated protein kinase promotes epidermal growth factor receptor internalization. Traffic. 2006;7:686–698. doi: 10.1111/j.1600-0854.2006.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frey MR, Dise RS, Edelblum KL, Polk DB. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 2006;25:5683–5692. doi: 10.1038/sj.emboj.7601457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frey MR, Golovin A, Polk DB. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J Biol Chem. 2004;279:44513–44521. doi: 10.1074/jbc.M406253200. [DOI] [PubMed] [Google Scholar]
- 85.Kuwada SK, Lund KA, Li XF, Cliften P, Amsler K, Opresko LK, Wiley HS. Differential signaling and regulation of apical vs. basolateral EGFR in polarized epithelial cells. Am J Physiol. 1998;275:C1419–C1428. doi: 10.1152/ajpcell.1998.275.6.C1419. [DOI] [PubMed] [Google Scholar]
- 86.Alexander RJ, Panja A, Kaplan-Liss E, Mayer L, Raicht RF. Expression of growth factor receptor-encoded mRNA by colonic epithelial cells is altered in inflammatory bowel disease. Dig Dis Sci. 1995;40:485–494. doi: 10.1007/BF02064355. [DOI] [PubMed] [Google Scholar]
- 87.Beck PL, Podolsky DK. Growth factors in inflammatory bowel disease. Inflamm Bowel Dis. 1999;5:44–60. doi: 10.1097/00054725-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 88.Potten CS, Owen G, Hewitt D, Chadwick CA, Hendry H, Lord BI, Woolford LB. Stimulation and inhibition of proliferation in the small intestinal crypts of the mouse after in vivo administration of growth factors. Gut. 1995;36:864–873. doi: 10.1136/gut.36.6.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Janmaat ML, Giaccone G. The epidermal growth factor receptor pathway and its inhibition as anticancer therapy. Drugs Today (Barc) 2003;39(Suppl C):61–80. [PubMed] [Google Scholar]
- 90.Kaiser GC, Polk DB. Tumor necrosis factor alpha regulates proliferation in a mouse intestinal cell line. Gastroenterology. 1997;112:1231–1240. doi: 10.1016/S0016-5085(97)70135-5. [DOI] [PubMed] [Google Scholar]
- 91.McElroy SJ, Frey MR, Yan F, Edelblum KL, Goettel JA, John S, Polk DB. Tumor necrosis factor inhibits ligand-stimulated EGF receptor activation through a TNF receptor 1-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;295:G285–G293. doi: 10.1152/ajpgi.00425.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geiger Joerg BS, Stefan Kircher, Michael Neumann, Andreas Rosenwald, Roland Jahns. Hospital-integrated biobanking as a service—the Interdisciplinary Bank of Biomaterials and Data Wuerzburg (IBDW) Open J Bioresour. 2018;5:6. doi: 10.5334/ojb.38. [DOI] [Google Scholar]
- 94.Schweinlin M, Wilhelm S, Schwedhelm I, Hansmann J, Rietscher R, Jurowich C, Walles H, Metzger M. Development of an advanced primary human in vitro model of the small intestine. Tissue Eng Part C Methods. 2016;22:873–883. doi: 10.1089/ten.tec.2016.0101. [DOI] [PubMed] [Google Scholar]
- 95.Vielmuth F, Hartlieb E, Kugelmann D, Waschke J, Spindler V. Atomic force microscopy identifies regions of distinct desmoglein 3 adhesive properties on living keratinocytes. Nanomedicine. 2015;11:511–520. doi: 10.1016/j.nano.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 96.Andreas E, Linda W, Christian R, Jürgen W, Martin H, Rong Z, Ferry K, Dieter B, et al. A new simple method for linking of antibodies to atomic force microscopy tips. Bioconjugate Chem. 2007;18:1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- 97.Gehmlich K, Asimaki A, Cahill TJ, Ehler E, Syrris P, Zachara E, Re F, Avella A, Monserrat L, Saffitz JE, et al. Novel missense mutations in exon 15 of desmoglein-2: role of the intracellular cadherin segment in arrhythmogenic right ventricular cardiomyopathy? Heart Rhythm. 2010;7:1446–1453. doi: 10.1016/j.hrthm.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hong TT, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, Jensen BC, Colecraft HM, Shaw RM. BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLoS Biol. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.