Abstract
Effects of macromolecular crowding on structural and functional properties of ordered proteins, their folding, interactability, and aggregation are well documented. Much less is known about how macromolecular crowding might affect structural and functional behaviour of intrinsically disordered proteins (IDPs) or intrinsically disordered protein regions (IDPRs). To fill this gap, this review represents a systematic analysis of the available literature data on the behaviour of IDPs/IDPRs in crowded environment. Although it was hypothesized that, due to the excluded-volume effects present in crowded environments, IDPs/IDPRs would invariantly fold in the presence of high concentrations of crowding agents or in the crowded cellular environment, accumulated data indicate that, based on their response to the presence of crowders, IDPs/IDPRs can be grouped into three major categories, foldable, non-foldable, and unfoldable. This is because natural cellular environment is not simply characterized by the presence of high concentration of “inert” macromolecules, but represents an active milieu, components of which are engaged in direct physical interactions and soft interactions with target proteins. Some of these interactions with cellular components can cause (local) unfolding of query proteins. In other words, since crowding can cause both folding and unfolding of an IDP or its regions, the outputs of the placing of a query protein to the crowded environment would depend on the balance between these two processes. As a result, and because of the spatio-temporal heterogeneity in structural organization of IDPs, macromolecular crowding can differently affect structures of different IDPs. Recent studies indicate that some IDPs are able to undergo liquid–liquid-phase transitions leading to the formation of various proteinaceous membrane-less organelles (PMLOs). Although interiors of such PMLOs are self-crowded, being characterized by locally increased concentrations of phase-separating IDPs, these IDPs are minimally foldable or even non-foldable at all (at least within the physiologically safe time-frame of normal PMLO existence).
Keywords: Intrinsically disordered protein, Intrinsically disordered protein region, Macromolecular crowding, Proteinaceous membrane-less organelles, Conformational stability, Induced folding
Introduction
Macromolecular crowding and complexity of the cellular environment
Cell interior is known to be extremely crowded due to the presence of high concentrations (up to 400 mg/ml) of various biological macromolecules, such as carbohydrates, nucleic acids, and proteins [1]. This crowded environment [2, 3] imposes considerable restrictions on the amount of free water [1, 2, 4–7], decreases accessible volume (since macromolecules occupy a significant fraction of the cellular volume (typically 20–30%), the volume occupied by these solutes is unavailable to other molecules, because two molecules cannot be in the same place at the same time) [4, 8], causes confinement [9], as well as affects solvent viscosity, thereby modulating intracellular diffusion [10–12]. Recent study, where the conditions of macromolecular crowding were modelled by solutions containing high concentrations of a model “crowding agent” polyethylene glycol, revealed that the crowder is also able to induce noticeable changes in the solvent properties of water, such as solvent dipolarity/polarizability, hydrogen-bond donor acidity, and hydrogen-bond acceptor basicity [13].
Thermodynamically, the effects of the crowded environment on chemical reactions are attributed to the excluded-volume concept [2–4, 8, 9]. It was pointed out that the crowded environment of biological fluids, and especially related excluded-volume effects, may have significant influence on conformational stability and structure of biological macromolecules [3, 14–17] and alter macromolecular equilibrium, affecting protein folding [18–20], efficiency of various biological reactions [4, 18, 19], binding of small molecules, enzymatic activity, protein–nucleic acid interactions, protein–protein interactions [3, 21], and rates and extent of protein aggregation and amyloid fibril formation [22–24]. Furthermore, the efficiency of excluded-volume effects posed by a crowder is known to be dependent on the relative hydrodynamic dimensions of crowder and target molecule (crowdee), with the strongest effects being ascribed to a situation, where the crowder and the crowdee have comparable hydrodynamic volumes [25–28]. However, when the crowder volume becomes too large, larger “caves” will be formed between the crowder molecules that could accumulate more than one molecule of crowdee. In other words, although globally accessible volume will decrease in the presence of such large crowder, locally accessible volume might increase [25]. Recently, it was pointed out by Kim Sharp that when the steric effects of macromolecular crowders and small molecules, such as ions, are treated on an equal footing, small molecules act as more efficient crowders than the large macromolecules [29]. Although based on these observations, an important conclusion was made that purely excluded-volume effects from macromolecules are more complex than commonly assumed (and potentially are much smaller than generally believed), it was also pointed out that high concentrations of macromolecules in the cell serve as a foundation of the significant non-ideality [29]. Therefore, a significant caution should be used while using high concentrations of “inert” macromolecules (which, in fact, can make specific interactions with proteins and nucleic acids) in experiments in vitro to avoid introduction of unanticipated intersolute interactions [29].
Although, in recent years, the significance of macromolecular crowding as important but mostly neglected variable in biochemical studies is gaining attention [4, 6], the principle shortcoming of a typical test-tube experiment dedicated to the analysis of a biological macromolecule or a biochemical reaction continues to be a failure to acknowledge this phenomenon. In fact, macromolecular crowding is not typically included as a constituent of standard “physiological conditions”, and the vast majority of in vitro experiments are usually performed under the relatively ideal thermodynamic conditions of low protein and moderate salt concentrations. One of the reasons for this negligence is the lack of general understanding of what should be looked at (confinement, excluded volume, changed solvent properties of water, or altered viscosity) while designing the conditions that would appropriately model biological fluids. Traditionally, the effect of excluded volume on the behaviour of query compounds is examined experimentally using concentrated solutions of some crowding agents, such as polymers (e.g., polyethylene glycol, Ucon, polyvinylpyrrolidone, dextran, Ficoll, etc.) or highly soluble “inert” proteins (e.g., lysozyme or serum albumin) [8, 22]. Crowded cellular environment alters molecular diffusion, and the corresponding effects of such environment on the translational mobility can also be modelled by synthetic crowders, concentration, and size of which can modulate probe microviscosity [30]. As far as modelling of the confined intracellular space is concerned, one of the related approaches includes encapsulation of a target protein in silica glass using the sol–gel techniques to create a crowded microenvironment [31, 32]. Although the fraction of the total volume excluded by the silica matrix is lower than the fractional volume occupied by macromolecules in living cell [14, 15], the size of protein-occupied pores in these gels has the same order of magnitude as the diameter of protein [31]. Furthermore, solvents can easily permeate silica matrix due to the porosity of the resulting glass, whereas the encapsulated macromolecules cannot generally escape [14]. This sol–gel glass encapsulation was used for the analysis of several proteins at the variety of solvent conditions [33–36].
One should keep in mind that the potential effects of cellular environment on structural properties, conformational behaviour, and functionality of proteins are much more complex than just the presence of aforementioned effects imposed by macromolecular crowding. For example, in a recent study, it has been pointed out that, in addition to being an energy source for biological reactions, adenosine triphosphate (ATP) might be considered as a biological hydrotrope, i.e., it belongs to a group of small molecules that are able to solubilize hydrophobic molecules in aqueous solutions [37]. In fact, Hymann and co-workers have shown that ATP reduces aggregation and enhances protein solubility. More importantly, ATP, being added at physiologically relevant concentrations (5–10 mM), can inhibit liquid–liquid-phase separation in the solutions of purified intrinsically disordered protein, fused in sarcoma (FUS), leading to the formation of liquid droplets, and efficiently dissolve preformed FUS droplets [37]. Therefore, ATP acts as a crowd controller [38]. Furthermore, other nucleotides, such as guanosine triphosphate (GTP), as well as adenosine diphosphate (ADP) and adenosine monophosphate (AMP) were able to dissolve the FUS droplets (although ADP and AMP did it at higher concentrations than ATP and GTP) [37]. Based on these observations, the authors hypothesized that high intracellular concentrations of ATP, which has long been a puzzle, are needed to keep IDPs soluble and are used as a means that help to regulate proteostasis in vivo [37]. In agreement with these considerations, a more recent work of Weeks and co-workers revealed that, in Xenopus oocyte nucleoli, ATP has a dual role in the maintenance of protein solubility, acting as an endogenous hydrotropic agent and also participating in the energy-dependent destabilization of nucleolar aggregates preceding their hydrotropic solubilisation [39]. Therefore, although the actual mechanisms of such hydrotrope hydrolysis-independent action of ATP (the presence of which is sufficient to solubilize aggregated proteins) are not established and debatable, this study provides interesting insights into certain critical components of the cell apart from just macromolecular crowding.
Overcrowding as a next level of cellular complexity
Although macromolecular crowding clearly complicates the reliable analysis of biological processes, the cellular complexity is further increased by the overcrowding phenomenon, which reflects the fact that the cellular distribution of different biological macromolecules is highly inhomogeneous [40], as they are often assembled into specific intracellular bodies, known as non-membranous cytoplasmic/nucleoplasmic granules, or RNA foci, or proteinaceous membrane-less organelles (PMLOs) [41]. Being abundantly found in the cytoplasm as well as inside the nucleus, mitochondria, and chloroplasts [42, 43], these granules/foci are principally different from the classical membrane-encapsulated organelles (chloroplasts, endoplasmic reticulum, Golgi apparatus, lysosomes, nucleus, mitochondria, and vacuoles) by the fact that their components directly contact with the cytoplasm/nucleoplasm/stroma/matrix because of the lack of the surrounding membrane [44, 45]. PMLOs are many, have specific distribution patterns within the cell, and contain specific proteins and RNAs (and/or DNAs) [42, 43, 46]. Size of these highly dynamic assemblages, whose structural integrity and biogenesis are exclusively defined by protein–protein, protein–DNA, and/or protein–RNA interactions [47, 48], depends on the dimensions of a cell which they are residing in [49]. Because of their liquid-like behaviour, such as the ability to become reversibly deformed when encountering a physical barrier, dripping, fusion, and wetting, PMLOs are considered as a different liquid state of the nucleoplasm, cytoplasm, matrix, or stroma [50–55]. They are formed via highly controlled biological liquid–liquid demixing phase separation (also known as liquid–liquid-phase transition, LLPT) or coacervation [49, 54–60]. This macromolecular condensation defines the overcrowded nature of PMLOs. In fact, concentration of protein(s) that underwent LLPT in these cellular liquid droplets is 10- to 300-fold higher than the content of the same protein(s) in dilute phase [55, 56, 60]. Furthermore, even globally, PMLOs are more crowded than their surrounding cellular fluids. For example, in the Cajal bodies, speckles, and nucleoli of the nucleus of Xenopus oocyte, the total protein concentrations were 136, 162, and 215 mg/mL, respectively, whereas total protein concentration in the surrounding nucleoplasm was 106 mg/mL [61]. Therefore, the interior of PMLOs represents an overcrowded milieu, because the overall protein concentrations found that there noticeably exceeds protein contents of the crowded cytoplasm and nucleoplasm [40].
Major constituents found in all PMLOs are proteins. Although different cellular bodies contain different sets of the resident proteins, many PMLO-associated proteins are intrinsically disordered or hybrid proteins containing ordered domains and intrinsically disordered protein regions [40, 41, 43, 46, 58, 62–69]. These observations suggested that the fluidity and highly dynamic nature of the overcrowded PMLOs, as well as their assembly/disassembly cycles, morphology, and structure are all depend on protein intrinsic disorder.
Intrinsically disordered proteins
Many protein functions are not directly linked to the presence of unique structures, with numerous biologically active proteins either containing intrinsically disordered protein regions (IDPRs) or even being entirely disordered. In fact, all proteomes abundantly contain such structure-less proteins [70–74]. Structurally, these intrinsically disordered proteins (IDPs) and IDPRs are described in terms of highly dynamic ensembles containing rapidly interconverting conformations [73, 75, 76]. Despite of their lack of stable tertiary and/or secondary structure, such IDPs and hybrid proteins possessing ordered domains and functional IDPRs are engaged in extremely diversified biological activities, ranging from signal transduction to recognition, and from regulation of the function of their binding partners and to promotion and guidance of the assembly of supra-molecular complexes [73–106]. Often, IDPRs serve as primary targets for various posttranslational modifications (PTMs) [95, 107–109], and many IDPs/IDPRs are subjected to alternative splicing [110–112]. Functions of IDPs and IDPRs complement functionality of ordered proteins and domains [73, 75, 77, 78, 80, 83, 84, 87, 88, 92, 97, 98, 100, 113–120], and multiple functional advantages are ascribed to IDPs/IDPRs to explain their prevalence in various proteomes and engagement in various biological processes [73, 77, 97, 121–124]. Among these functional advantages are:
large capture radius that is utilized in more efficient search within the interaction space leading to the increased interaction speed;
the ability to be engaged in a wide spectrum of encounter complexes with non-native interactions, but productive for binding;
the ability to form binding interfaces that are large compared to the IDP/IDPR own size;
the ability to overcome steric restrictions;
the presence of energetic frustrations in the binding interfaces that define the IDP/IDPR-binding promiscuity [125];
the ability to be regulated by rapid degradation;
the ability for self-regulation and self-recognition;
the ability to have various entropic chain activities;
the ability of a single IDP/IDPR to interact with multiple structurally diverse partners (one-to-many binding mode);
the ability of many IDPs/IDPRs to interact with a single partner (many-to-one binding mode);
the ability to fold (at least partially) into a specific structure at interaction with a specific partner;
the ability to fold differently according to the template provided by different binding partners;
the ability to preserve noticeable disorder in bound state (i.e., the presence of binding fuzziness);
the ability to form complexes with binding partners using induced folding and/or conformational selection mechanisms;
the ability to be regulated and controlled via multiple posttranslational modifications;
the ability to be regulated by alternative splicing;
the presence of overlapping binding sites;
the ability to masking (or not) of interaction sites;
the ability to escape unwanted interactions via functional misfolding [126];
the ability to be engaged in binding ‘chain reactions’, where interactions with specific partners generate the formation of new binding sites for subsequent interactions with other partners;
the ability to serve as complex interaction centres (scaffolds);
the ability to serve as hubs controlling and regulating sophisticated protein–protein interaction networks;
the ability to act as stochastic machines;
the presence of very different evolutionary rates;
the existence of conditional (cryptic, dormant, or transient) disorder, where functionality of an ordered protein requires its local or even global functional unfolding [127, 128];
the ability to have multiple unrelated functions (moonlighting).
It is recognized now that functional diversity of IDPs/IDPRs can be related to (or originate from) their extreme structural heterogeneity [129–131]. Here, a structure of a protein molecule represents a mosaic of differently (dis)ordered segments, such as foldons (spontaneously foldable regions), non-foldons (regions that do not fold), semi-foldons (semi-folded regions), inducible foldons [regions that can at least partially fold at interaction with binding partner(s)], and unfoldons (regions that need to undergo functional unfolding to make protein active) [129–131]. In addition to such a mosaic structure, where different parts of a protein molecule are (dis)ordered to different degrees, the distribution of foldons, non-foldons, inducible foldons, semi-foldons, and unfoldons is not steady, but constantly changes in time. As a result, protein structure is not crystal-like, but is always morphing over time, with a given protein segment being able to have a different structure at a different time point [129, 130].
An important consideration should be added here related to the complexity of the disorder-based functionality. In their recent work, Naganathan and co-workers emphasized an importance of collapsed states of IDPs in determining protein function [132]. In fact, using the DNA-binding domain of the prokaryotic protein cytidine repressor (CytR) these authors showed that although this protein is substantially disordered in its unbound form, it effectively folds on binding DNA and is also able to undergo a temperature-induced coil-to-globule transition in the absence of DNA [132]. Surprisingly, this temperature-driven formation of the more collapsed state followed the second order-like transition and was accompanied by the disruption of residual structure in CytR [132]. The maximal structural heterogeneity was observed at temperatures ~ 310–313 K, and at 310 K, conformational ensemble of CytR constituted a mixture of forms, such as extended partially helical structures and collapsed conformations with little secondary structure [132]. The authors also showed that this protein preserved capability to interact with DNA at all conditions studied, and that it possessed complex binding isotherms arising from binding heterogeneity [132]. These observations suggest that functionality of IDPs does not always rely on the naturally unfolded configurations or some folded forms induced by binding to specific partners. Instead, non-native collapsed states might be functional too. It is likely that they serve as an alternative functional species whose populations could be tuned by cellular machinery for precise functional control.
Finally, because of their natural abundance, multitude of biological functions, and important regulatory roles of IDPs/IDPRs in various biological processes, misbehaviour of many IDPs/IDPRs is commonly associated with various human maladies [133–139].
IDPs/IDPRs in crowded environment: the good, the bad, and the ugly
We would like to start this section of the review with the important notion that the currently available data on the effects of macromolecular crowding on structural properties of proteins in general (and IDPs in particular) are rather incomplete. In fact, most studies have been focused on the analyses of the effects of crowing agents on conformational stability of proteins in typical unfolding experiments, where proteins that are folded in buffer at room temperature undergo heat denaturation or chemical unfolding in the presence or absence of crowders [18, 19, 140]. On the contrary, the number of studies directly probing the effects of macromolecular crowding on structural properties of IDPs/IDPRs is rather limited. However, even these limited results are sufficient for making some important conclusions on how crowded environment can affect structural behaviour of IDPs/IDPRs.
We will show below that, based on their response to the presence of artificial- and natural-crowding agents, IDPs/IDPRs can be grouped into three major categories, foldable, non-foldable, and unfoldable. Here, foldable IDPs/IDPRs can fold (at least partially) at the artificial crowded environment and inside the living cells. This (partial) folding) is likely to be driven by the crowding-induced formation of a hydrophobic core. Non-foldable IDPs remain mostly unstructured at the crowded conditions. Some of these non-foldable by crowding IDPs may require another protein (or DNA, or RNA, or some other natural-binding partners) to provide a framework for structure formation. Finally, unfoldable IDPs/IDPRs undergo further unfolding being exposed to crowding agents. This is likely due to the interaction of such IDPs/IDPRs with crowding agents.
They should fold: expected outcomes of the macromolecular crowding on IDPs
Despite their lack of stable structure in the unbound state, many IDPs/IDPRs are known to at least partially fold at interaction with their specific partners, such as other proteins, nucleic acids, membranes, or small molecules [73, 77, 78, 80, 82–85, 87–89, 92, 101, 141, 142]. The bound forms of IDPs/IDPRs are either tightly folded or remain substantially disordered [84, 103, 109, 143], and some of them can adopt different structures, being bound to different partners [101, 109, 143–145]. The capability of an IDP/IDPR to functionally fold is related (at least partially) to the presence of noticeable residual structure in their unbound state, which is reflected in their exceptional spatio-temporal heterogeneity [130]. Irrespectively of the scale of their binding-induced folding, it is expected that interaction with specific binding partners would result in the decrease of the hydrodynamic dimensions of IDPs/IDPRs (since this is typically the case for protein-folding process).
Because of all this, and due to the fact that, as a rule, structures of IDPs/IDPRs is extremely responsive to changes in their environment, it was expected that IDPs/IDPRs could be sensitive to the presence of crowding agents, and that their effective folding and compaction would be observed under the conditions of macromolecular crowding. This hypothesis was based on the thermodynamic principles of excluded volume, according to which the presence of a space-filling substance might significantly affect the protein-folding process by favouring a more compact (folded) state over the more extended (unfolded) form [7]. As a matter of fact, at the early stages of the IDP research, when the existence of intrinsic disorder was still questioned by many researchers, this argument (IDP/IDPR has to fold in crowded cellular environment, and, therefore, intrinsic disorder is biologically irrelevant) was frequently brought by many colleagues in private conversations and used by reviewers to illustrate the absurdity and impossibility of protein intrinsic disorder phenomenon.
However, not everything is as simple with macromolecular crowding as it seems. In fact, excluded volume is not the only mechanism that needs to be taken into account while considering the effects of crowded environments on chemical reactions or folding, function, structure, and thermodynamics of a protein molecule [18]. As it was already emphasized, high concentrations of macromolecules change viscosity and solvent properties of crowded milieu. Furthermore, biological macromolecules (and even synthetic polymers used as crowding agents in vitro) are not inert entities, and in addition to perturbing diffusion and creating confined environment, crowders can be engaged in direct physical interactions with the molecule of interest [18]. As a result, globally, macromolecular crowding might affect protein structure, folding, shape, conformational stability, binding of small molecules, enzymatic activity, protein–protein interactions, protein–nucleic acid interactions, and pathological aggregation [19]. Therefore, because of all the different ways of how crowded environment can affect protein structure, the outcomes of the addition of high concentrations of crowding agents are not too transparent.
Curiously, a recent computational analysis, where a coarse-grained model was used to represent proteins with varying sequence contents and investigate changes in the polypeptide chain dimensions caused by the purely repulsive spherical crowders, revealed that the extent of crowding-induced compaction depends on the properties of the protein itself and not only on the size and concentration of the crowder as predicted by the excluded-volume theory [146].
The goal of this review is to analyze available literature describing the effects of crowed environments (both natural, inside the cell, and artificial, modelled by addition of high concentrations of polymers) on structural properties of intrinsically IDPs/IDPRs. Although the amount of currently available information on this subject is rather limited, the analyzed data allow us to make some important conclusions and generalizations. For example, it became clear that, according to their response to macromolecular crowding, IDPs/IDPRs can be grouped into three broad categories: (partially) foldable, non-foldable, and unfoldable. In other words, crowded milieu can cause folding (at least partial) of some IDPs/IDPRs, or shows no noticeable effect on structure of other IDPs/IDPRs, or even results in some further unfolding of IDPs/IDPRs containing residual structure. Therefore, we will talk below about representatives of these three categories, which constitute the good, the bad, and the ugly sides of intrinsic disorder in crowded milieu.
The good: (partially) foldable IDPs/IDPRs
We will start with the presenting examples of IDPs/IDPRs that are able to (partially) fold in the crowded milieu, i.e., “good” IDPs/IDPRs behaving as expected based on the excluded-volume theory. According to the Le Chatelier’s principle, the existence of the volume exclusion should favour compaction as compact state occupies less space [147]. If macromolecular crowding could increase the ratio of folded (compact) to unfolded (less compact) protein by a factor of 100, this would corresponds to the 3 kcal/mol (~ 12.6 kJ/mol) of stabilization at room temperature [148]. However, the energetic contribution of an actual crowding effect was estimated to be in a range of be 1–4 kJ/mol for 100–300 mg/mL of crowding agents [149, 150]. Since essentially larger energy is required to fold a protein (e.g., to change the folded fraction from 10% (ΔGF = 5.3 kJ/mol) to 90% (ΔGF = − 5.3 kJ/mol), a free-energy change of ca. 11 kJ/mol is needed), it would be a mistake to expect to systematically observe the dramatic effects of crowding agents on fractions of folded proteins [151]. Furthermore, even IDPs/IDPRs with “good” behaviour (i.e., those which are able to fold in crowded environment) are not made equal. In fact, due to their highly heterogeneous structural organization, it is obvious that different IDPs can show different responses to the macromolecular crowding that can range from the full-scale folding to rather moderate partial ordering.
IDPs undergoing complete folding in crowded milieu
An illustrative example of the research directly probing the effect of macromolecular crowding on a foldable protein that is unfolded in physiological pH buffers at room temperature is given by the analysis of the conformational behaviour of a destabilized mutant of the immunoglobin G-binding domain of protein L (ProtL) from Streptococcus magnus [151]. The wild type of ProtL is a typical mesophilic protein with the molecular mass of 7 kDa that unfolds in a reversible two-state reaction. The ProtL variant contained seven destabilizing lysine to glutamate substitutions (K×7E) making it an obligate halophile that was preferentially unfolded under “physiological conditions” [152]. In fact, based on the results of the solution NMR analysis in diluted buffer at room temperature, it was concluded that 84% of the K×7E variant molecules populated the unfolded state, whereas only 0.1% wild-type protein molecules were in the unfolded state at the identical conditions [148]. However, at addition of salt, K×7E variant was able to fold to a structure that was indistinguishable from that of the wild-type protein [148, 152, 153]. Combined NMR and circular dichroism (CD) analysis indicated that the addition of 200 mg/mL Dextran-20 to this K×7E variant shifted its conformational equilibrium from preferentially unfolded form toward the folded state, resulting in a structure that matches the structure of the wild-type protein [151]. Careful comparison of the thermodynamic effects of 200 mg/mL of Dextran-20 on the well-folded wild-type ProtL, salt-folded form of its K×7E mutant, and mostly unfolded K×7E mutant in water revealed that the addition of this crowder caused comparable changes in the folding equilibrium constant (ΔΔGU ≈ 2 kJ/mol) for all three forms of this protein [151]. This observation indicated that the scale of the stabilizing effect of macromolecular crowding is independent of the starting stability of the protein. Importantly, this study also clearly indicated that crowding can shift the conformational equilibrium toward the folded form when the polypeptide is in the transition region between folded and unfolded states [151].
In a series of elegant studies, the group of Terrence G. Oas and co-workers conducted a multifactorial analysis of the effect of various factors on the ribonuclease P (RNase P) protein from Bacillus subtilis, which, according to circular dichroism and NMR studies, behaves in 10 mM sodium cacodylate low-ionic strength buffer at neutral pH as a typical extended IDP [154]. The B. subtilis RNase P was shown to gain native α/β structure at addition of various small molecules, such as anions or osmolyte trimethylamine N-oxide (TMAO) [154]. It was pointed out that the tightly coupled folding and binding of this protein in the presence of anions is determined by the preferential binding of anions to the folded state of RNase P (small population of which was hypothesized to be present in the conformational ensemble of this natively unfolded protein at low-ionic strength conditions), whereas TMAO-induced folding represents an independent process that does not interfere with anion-induced folding, and where TMAO molecules do not compete with anions for the two high-affinity anion-binding sites [154]. Subsequent careful thermodynamic analysis confirmed these observations, and indicated structural similarity of the osmolyte- and anion-folded states of RNase P [155]. The existence of a small population of native molecules under the low-ionic strength conditions even in the absence of anions and TMAO was later conformed by NMR-detected amide hydrogen exchange (HX) [156]. Curiously, although, in the original studies, no intermediate was detected in TMAO-induced folding of RNase P [154, 155], subsequent kinetic studies combined with the equilibrium “co-titration” experiments where both TMAO and urea were used to produce a urea–TMAO titration revealed the presence of a significantly populated intermediate state in the RNase P protein-folding process [157]. Complex NMR analysis of this intermediate state suggested that it contained the majority of helix B and the central β-sheet, whereas its N- and C-terminal helical regions were mostly unfolded [158]. It was also pointed out that the pathways of coupled folding and binding of the B. subtilis RNase P can be regulated by the concentration of ligand, where the order of binding and conformational change is ligand concentration-dependent, and where protein conformational dynamics is markedly and variably affected by the ligand [159].
IDPs undergoing significant but partial folding in crowded milieu
As described in the previous section, complete macromolecular crowding-induced folding of the PrtL K×7E mutant represents an exception, and the majority of IDPs/IDPRs undergo only partial folding in crowded milieu. Several illustrative examples of such behaviour, where an IDP folds in the presence of crowders to a molten globule-like conformation, but fails to form a state with the rigid side-chain packing characteristic of the well-folded ordered proteins, are discussed below.
Although, in its heme-bound state, a small globular protein cytochrome c has a well-folded mostly α-helical structure, removal of the heme causes dramatic destabilization of this protein, which is mostly unstructured in aqueous solutions [160], and, at acidic pH, adopts a random coil-like conformation [161]. It was also pointed out that, similar to many other acid-unfolded proteins, at acidic pH, apo-cytochrome c undergoes refolding to a compact conformation with the properties of a molten globule due to the binding of the anion that minimizes the intramolecular charge repulsion causing the initial unfolding [162–164]. Comparable structural changes from mostly unfolded to molten globule-like conformation were induced in apo-cytochrome c by addition of high concentrations of Dextran with the average molecular mass of 35,000 Da [165]. Furthermore, thermodynamic analysis of the Dextran-induced folding of the apo-cytochrome c from the unfolded (U) to molten globule (MG) state suggested that the observed structural transformation was characterized by the ΔG0U-MG value of 10.5 kJ/mol that agreed well with the ΔG0U-MG value of 11 kJ/mol obtained from two-state analysis of salt-induced MG formation in this protein [166]. Formation of MG is independent of rigid side-chain packing needed for the stabilization of ordered forms of globular proteins. The major driving force of anion-induced folding of an acid-unfolded state is the preferential interaction of chloride anions with the positively charged groups of the compact MG state of apo-cytochrome c [162]. On the other hand, since addition of inert Dextran does not affect the charge state of a protein, the formation of the more compact MG state over the more expanded acid-unfolded state of apo-cytochrome c in the presence of this crowder was explained by the ability of dextran to decrease the volume available to a macrosolute, i.e., due to the existence of the excluded volume [165].
Another example of a system capable of rather substantial crowding-induced folding is given the C-terminal domain of the histone H1 [167]. It is known that, structurally, H1 linker histone can be separated into three domains, a central globular domain (~ 80 residues) consisting of a three-α-helix bundle and a β-hairpin, and two disordered tails, a short N-terminal domain (20–35 residues), and a long C-terminal domain (~ 100 residues) [168]. The C-terminal tail is characterized by high content of disorder-promoting residues, containing ~ 40, ~ 12, and ~ 17% of Lys, Pro, and Ala residues. Because of this highly biased amino-acid composition, unbound state of this domain has a little structure in aqueous solution. However, C-tail serves as the primary site of H1 binding to chromatin in vivo [169, 170]. Furthermore, this domain was shown to cooperatively fold on interaction with DNA, with the DNA-bound form of C-tail being characterized by the extremely stable structure that includes α-helix, β-sheet, turns, and loops [171]. Furthermore, in the presence of high concentrations of crowding agents, such as Ficoll 70 and PEG 6000, this functionally important domain was shown to undergo extensive folding, forming the MG-like state [167].
IDPs undergoing limited partial folding in crowded milieu
In addition to utilization of NMR and CD, which are spectroscopic techniques sensitive to crowding-induced changes in protein structure, compaction of several query proteins in crowded milieu was evaluated by small-angle neutron scattering (SANS) and single-molecule fluorescence detection in combination Förster resonance energy transfer (FRET).
For example, the effects of high concentrations of a small globular protein, bovine pancreatic trypsin inhibitor (BPTI), on the hydrodynamic dimensions of an IDP, N protein of bacteriophage λ (the λ N protein) were studied by SANS with D2O-based contrast matching [172]. The λ N protein is a small transcriptional anti-termination factor, which is highly disordered in isolation, as evidenced by NMR and circular dichroism spectroscopy [173]. Although the λ N protein is mostly disordered in isolation, its N-terminal region (the first 22 amino acids) containing the arginine-rich motif is capable of folding to an α-helical conformation at binding to a specific site on the RNA transcript [174], whereas other regions of the λ N protein can interact with RNA polymerase and other components of the transcription complex [175]. It was indicated that although many aspects of SANS are similar to small-angle X-ray scattering, SANS have an important advantage of being scattered by the atomic nuclei. As a result, different isotopes of the same element can have clearly distinguishable scattering properties. The presence of such differential scattering for hydrogen vs. deuterium represents a foundation of the technique of contrast variation (or contrast matching). Here, a contribution of the bystander molecules to the SANS profile can be eliminated by their partial deuteration, because, at the certain ratio of H2O to D2O, known as a match point, the scatter from the partially deuterated molecule will equal that of the solvent, making that molecule invisible for SANS [176]. In the SANS analysis of the effects of high concentrations of BPTI on the λ N protein, the exclusive visualization of the completely deuterated λ N protein in this complex mixture was achieved by eliminating the scattering contrast between the solvent and unlabelled BPTI via adjusting the D2O concentration of the solvent [172]. These experiments showed that the increase in the BPTI concentration was accompanied by detectable decrease in the dimensions of the λ N protein, from Rg = 38 ± 2 Å in diluted solution to Rg = 30 ± 4 Å in the presence of 150 mg/mL BPTI [172]. Importantly, the Rg of 38 Å measured for the λ N protein in the absence of crowding agent corresponded to a monomeric unfolded protein composed of 107 residues [177]. However, the Rg of 30 Å determined for the λ N protein in the presence of high concentrations of BPTI was noticeably larger than the Rg expected for a globular well-folded protein of similar molecular mass (e.g., ribonuclease A, a globular protein of 124 amino-acid residues, has the Rg of 14.8 Å [178, 179]). Comparable SANS-based data were obtained for this uniformly labelled IDP, when high concentrations of equine metmyoglobin were used to create crowded milieu [180]. Therefore, crowding caused only partial collapse/folding of the λ N protein.
An important advantage of single-molecule fluorescence detection in combination with FRET is its ability not only to analyze the effect of macromolecular crowding on structural properties of a query protein, but also to look at the effects of crowded environment on the conformational distributions within the dynamic structural ensemble of an IDP by looking at one protein molecule at a time. Furthermore, single-molecule FRET can be used to study labelled IDPs even in the presence of very large concentrations of unlabelled solutes [181]. Recently, this technique was used in the analysis of the effect of increasing PEG-6000 concentrations on the conformational ensembles of four different IDPs, such as N- and C-terminal segments of human prothymosin-α (ProTα-N and ProTα-C, residues 2–56 and 56–110, respectively), the binding domain of the activator for thyroid hormones and retinoid receptors (ACTR, residues 1–73), and the N-terminal domain of HIV-1 integrase (IN, residues 8–57) [181]. It was pointed out that these three proteins belonged to different IDP classes, where highly charged ProTα cannot be folded under any known conditions, whereas ACTR and IN are able to fold upon binding a protein or a small ligand, respectively [181]. In the related FRET experiments, Alexa Fluor 488 and Alexa Fluor 594 were used as donor and acceptor fluorophores that were introduced to target proteins via cysteine residues incorporated at desired to positions to give sequence separations of 55 (ProTα-N), 54 (ProTα-C), 72 (ACTR), and 49 residues (IN) [181]. Increase in the PEG-6000 concentration from 0 to 40% caused rather different changes in the compaction degree of four IDPRs analyzed in this study. In fact, in the absence of crowding agent, ProTα-C, ProTα-N, ACTR, and IN were characterized by the Rg of 35.4, 30.5, 24.9, and 20.1 Å, respectively [181]. These values reflected that, although behaviour of ACTR and IN was rather close to the behaviour of typical random coil-like polypeptides, ProTα-N and ProTα-C, probably due to their high net charges, were noticeably more expanded. In fact, based on the known scaling law correlating the Rg of unfolded proteins and their length (N) (Rg = (2.08 ± 0.19) N(0.598 ± 0.029)) [177], the random coil-like states of ProTα-C, ProTα-N, ACTR, and IN are expected to have the Rg of 22.8, 22.6, 26.8, and 21.3 Å. In the presence of 35% PEG-6000, the Rg values were decreased to 23.9, 22.6, 20.9, and 18.8 Å, for ProTα-C, ProTα-N, ACTR, and IN, respectively [181]. Although these changes corresponded to the 1.48-, 1.35-, 1.19-, and 1.07-fold increase in the compaction degree of these proteins, collapsed states of all four proteins observed in crowded environment were still rather expanded in comparison with the completely folded conformations of globular proteins of comparable molecular mass (which are expected to have the Rg values in a range of 10–12 Å [182]). Finally, the analysis of the compaction efficiency of PEGs of ten different degrees of polymerization at fixed volume fraction of PEG revealed that the degree of compaction of target proteins was highly dependent on the crowder size, with the increase in the crowder size to about 100 monomers causing the monotonous collapse of IDPs (decrease in their Rg values), which reached a plateau for PEGs of more than ~ 100 monomers [181].
The bad: non-foldable IDPs/IDPRs
A very large part of the currently available literature systematically reports the lack of any noticeable structure-forming effects of high concentrations of crowding agents on IDPs.
For example, Flaugh and Lumb [183] established that molecular crowding modelled by the high concentrations (of up to 250 g/L) of the Dextrans with average molecular weights of 9.5, 37.5, and 77 kDa and Ficoll-70 did not induce a significant folding in two IDPs, FosAD and p27ID. FosAD corresponds to the C-terminal activation domain of human c-Fos (residues 216–310) and is functional for interacting with transcription factors in whole-cell extract [184]. p27ID corresponds to the cyclin-dependent kinase inhibition domain of the cell-cycle inhibitor human p27Kip1 (residues 22–97) and is active as a cyclin A-Cdk2 inhibitor [185]. Both protein domains were shown to by intrinsically disordered as judged by circular dichroism (CD) spectra that were characteristics of the unfolded polypeptide chains, lack of 1H chemical-shift dispersion, and negative 1H–15N nuclear Overhauser effects [184, 185]. In the presence of macromolecular crowding agents, none of these IDPs underwent any significant conformational change reflected in noticeable changes in either circular dichroism or fluorescence spectra. Therefore, molecular crowding effects are not necessarily sufficient to induce ordered structure in IDPs [183].
Calcium-binding protein RCL offers the rare opportunity to study the same polypeptide chain under two drastically distinct folding states: as an extended IDP in its apo-form (RH of 3.2 nm) and as a compact folded structure in the Ca2+-bound form (holo-form, RH of 2.2 nm) [186]. RCL is derived from the RTX-containing domain (Repeat in ToXin) of the adenylate cyclase toxin (CyaA) from Bordetella pertussis. It was recently shown that, although the structural contents of the apo-state and holo-state of RCL were not affected by the crowding agent Ficoll 70, the protein affinity for calcium and thermal stability of both forms were strongly increased by this crowding agent [187].
Cino et al. analyzed the effect of macromolecular crowding on structural properties and conformational dynamics of several IDPs with different extents of residual structures by measuring their NMR spin relaxation parameters in the absence and presence of 160 mg/mL of Ficoll 70 [188]. 1H–15N HSQC spectra of uniformly 15N labelled prothymosin α (ProTα; human isoform 2), thyroid cancer 1 protein (TC-1; human), and α-synuclein (human isoform 1) in dilute and crowded solutions suggested that all three proteins remain mostly disordered under crowded conditions and retained their segmental motions on the nanosecond timescale [188]. These authors also showed that the crowded environment exhibited differential effects on the conformational propensity of distinct regions of an IDP [188].
Similarly, α-synuclein was shown to preserve its mostly unfolded conformation in the presence of several crowding agents [189] and even in the periplasm of the bacterial cells [190]. Recently, these observations were further conformed by the comprehensive solution NMR analysis of α-synuclein in the presence and absence of high concentrations of Ficoll or bovine serum albumin (BSA), which clearly indicated the inability of crowding agents to induce persistent secondary structure in this protein [191].
In addition, the analysis of macromolecular crowding on three dehydrins from Arabidopsis thaliana, Cor47, Lti29, and Lti30 revealed that, on the contrary to the hypothesis these drought-induced IDPs might acquire a biologically active structure upon dehydration, dehydrins were highly resistant to crowding-induced structural changes, being remarkably stable in their disordered state and being only modestly affected by the solvent alterations [192].
One can argue that the lack of structural changes induced in certain IDPs by high concentrations of crowding agents can be attributed to the highly disordered nature in the corresponding query proteins. However, data recently retrieved for the Golgi reassembly and stacking protein (GRASP) from the fungal pathogen Cryptococcus neoformans (CnGRASP) illustrate that this hypothesis is incorrect [193]. GRASPs constitute a family of peripheral membrane proteins that keep the arrangement of the cisternae, are needed for changes in this flattened membrane disk of the endoplasmic reticulum and Golgi apparatus according to the cell needs [194–197], and control organization of the cisternae into stacks and ribbons [198, 199]. In diluted solutions, even in the absence of any denaturing agents, CnGRASP was characterized by structural features typical for the molten globular proteins [200]. Addition of high concentrations of Ficoll-70, PEG-200, and PEG-2000 did not affect the structure of this protein, which retained molten globular conformation even when the crowding mimetic concentrations reached 40% [193].
Between the good and the bad: the two-faced Janus proteins
Because of their complex mosaic and highly dynamic structures containing a set of foldons, non-foldons, inducible foldons, semi-foldons, and unfoldons [129–131], generally, the responses of IDPs/IDPRs to changes in their environment (e.g., to macromolecular crowding) are expected to be highly heterogeneous, with some their parts being able to gain more ordered structure in the presence of crowding agents, and with other parts being insensitive to macromolecular crowding. An illustrative example of such two-faced Janus proteins is given by a 97-residue IDP from Salmonella typhimurium, FlgM, which regulates flagellar synthesis [201] by binding the transcription factor σ28 [202]. Careful multifactorial analysis revealed that the crowded environment enforced only partial folding of FlgM, with approximately half of this IDP-gaining structure inside living Escherichia coli cells and in solutions containing high concentrations (≥ 400 mg/mL) of BSA or ovalbumin in vitro [203]. This ability of crowding-induced folding of the C-terminal half of FlgM was shown to be associated with the functionality of this protein. In fact, in vitro solution NMR analysis revealed that free FlgM was mostly unstructured in the dilute solutions [204]. However, even in the unbound form, the C-terminal half of this protein was characterized by the deviation of Cα chemical shifts from the random coil values, suggesting the presence of a transient secondary structure that included two α-helical regions, residues M60–G73 and A83–A90 [205]. This same C-terminal half of FlgM became structured as a result of specific binding to σ28 at the formation of the FlgM-σ28 complex [204], and was also shown to be partially folded in the crowded environment [203]. On the contrary, the N-terminal half of FlgM retained random coil-like conformation at all the studied conditions [203–205]. Therefore, FlgM represents a case of the two-faced Janus protein, where the first face is exemplified by its C-terminal half that is partially structured in the diluted solutions and folds further into the MG-like conformation in the natural and artificial crowded environment, whereas the second face is exemplified by the N-terminal half of FlgM that remains highly unstructured under all conditions [203].
Recently, two Late Embryogenesis Abundant (LEA) proteins from A. thaliana, AtLEA4–2 and AtLEA4–5, were also shown to possess features of the two-faced Janus proteins [206, 207]. In plants (and some other organisms), LEA proteins accumulate in response to abiotic stress, particularly, dehydration (water limitation) [208–210]. Similar to other hydrophilins, LEA proteins are enriched in hydrophilic and small residues, and the members of the group 4 LEA proteins in plants are characterized by a conserved N-terminal region (about 80 residues long) and a C-terminal region variable in sequence and length. AtLEA4–2 and AtLEA4–5 are characterized by the almost complete lack of ordered structure when fully hydrated [207]. However, under the water-deficit environments, e.g., induced by macromolecular crowding, the conserved N-terminal region of these proteins undergoes transition to α-helical structure, whereas the variable C-terminal region remains mostly disordered [206, 207]. Importantly, similar to FlgM, the conserved N-terminal regions of the group 4 LEA proteins are necessary and sufficient for their chaperone-like activity under water-deficit conditions [206, 207].
The ugly: IDPs/IDPRs that unfold in crowded environment
Since any of the polymers used as model crowding agents can be potentially engaged in non-specific interactions with certain IDPs, one might expect that residual structure of these IDPs can be destabilized in crowded milieu, or, in other words, such IDPs would undergo crowding-induced unfolding. Several examples of this behaviour are listed below.
Amide hydrogen exchange (HX, efficiency of which can be estimated by mass spectrometry) of IDPs in highly concentrated polymer solutions was shown to serve as a useful approach for the evaluation of the IDP transient structure under crowded conditions [211]. Application of this technique to a transiently helical random coil domain of the activator of thyroid and retinoid receptor (ACTR) revealed that, in solutions containing 300 mg/mL of Ficoll, this IDP undergoes noticeable unfolding accompanied by the increase in its HX rate [211]. These data supported the hypothesis that the levels of residual structure in some IDPs can actually decrease in crowded milieu due to the non-specific interaction of said IDPs with natural- and artificial-crowding agents [211].
Although FlgM was described as a Janus protein-containing crowding-foldable and crowding-non-foldable domains [203] (see above), a recent analysis of the effect of polymer and protein crowders on this protein by SANS revealed that, at high concentrations of crowding agents, Rg of FlgM increases [212]. Furthermore, the conformational ensemble of FlgM was shown to include two major populations, collapsed and extended, with compacted conformers being able to fit into the voids between the molecules of crowder, and with extended conformers being able to bind multiple crowders simultaneously, meandering through the crevices of the crowded milieu [212].
IDPs/IDPRs in their natural habitat: effects of cellular crowded environment
It is clear that the in vitro modelling of the complex crowded cellular environment using high concentrations of macromolecular crowders (artificial, e.g., various synthetic polymers; natural, e.g., various inert proteins) is an oversimplification. In fact, natural cellular environment is not only characterized by the presence of high concentrations of macromolecular solutes that can exceed 300 mg/mL and occupy > 30% of the cellular volume, but also affects the diffusion of a target protein, potentially has changed properties of water (in comparison with the “bulk” water in typical in vitro experiments), and might be engaged in interaction (both specific and non-specific) with the target protein, because the intracellular environment comprises biologically active molecules. To address these possibilities, the analysis of protein structure in natural cellular environment should be conducted. Although this seems to be an impossible task, recent advances in the in-cell NMR spectroscopy, which provides structural data at atomic resolution non-invasively, have open new horizons in this field [213–217].
Application of this technique to the analysis of the conformational behaviour of the aforementioned K×7E form of the immunoglobulin G-binding domain of protein L (ProtL) from S. magnus with seven lysine residues replaced by glutamic acids inside the E. coli cells revealed that this ProtL variant failed to fold [148]. This finding indicated that some non-specific interactions between a target protein and cytoplasmic components can overcome the stabilizing excluded-volume effects, which must be present under crowded conditions [148]. Similarly, in-cell NMR analysis of human α-synuclein overexpressed in E. coli clearly showed that this protein is mostly monomeric and disordered in the bacterial cytosol [218]. Using the NMR experiment SOLEXSY (SOLvent EXchange SpectroscopY), to measure the hydrogen–deuterium exchange rates of α-synuclein in buffer and in E. coli revealed that these rates are similar in buffer and cells, further indicating that true disorder can persist inside the crowded cellular interior [219]. This idea of the global insensitivity of the intrinsically disordered structure of human α-synuclein-to-intracellular environment in different mammalian cell types was conformed using a combination of in-cell NMR with electron paramagnetic resonance (EPR) spectroscopy [220]. This study provided atomic-resolution insights into the structure and dynamics of α-synuclein in its natural environment, and showed that the disordered nature of this protein is stably preserved in non-neuronal and neuronal cells [220]. Similarly, in-cell NMR analysis of yeast frataxin showed that the intrinsically disordered N-terminal tail containing the mitochondrial import signal retained its unfolded and highly flexible structure in the cytosol of the E. coli cells [221].
As it was already indicated, the Janus protein FlgM showed very similar structural behaviour in model crowded environment in vitro and inside the cell, with approximately half of this IDP-gaining structure inside the living E. coli cells and in crowded in vitro solutions [203]. The principle similarity of these two structures in natural and artificial crowded environments was further supported by the SOLEXSY analysis, which failed to find a difference between partially folded conformations induced under these conditions [219].
Microinjection of 15N-labelled protein Tau into the Xenopus laevis oocytes followed by the in-cell NMR analysis of intact oocytes revealed that the in-cell NMR spectrum of this protein is similar to the in vitro spectrum of Tau protein bound to the microtubules [222]. Tau is a neuronal 441 residue-long protein that regulates polymerization of tubulin into microtubules (MTs) and plays a role in the pathogenesis of Alzheimer’s disease (AD) [223]. Solution NMR analysis of the isotope-labelled Tau in complex with MTs revealed that it can be considered as another illustration of Janus proteins. In fact, while multiple signals disappeared in the heteronuclear 1H–15N correlation spectrum of its MT-bound state due to direct association of the corresponding residue with the surface of MTs, a significant portion of Tau remained unstructured and projected from the MT surface [224]. In these experiments, the NMR signals were lost for the Tau207–421 fragment corresponding to the proline-rich region and four microtubule-binding repeats reflecting the immobilization of this region upon binding to the MTs [224]. On the other hand, the N-terminal projection domain (residues 1–151) showed heteronuclear NMR spectrum typical for a mostly unfolded polypeptide chain [224]. Very similar NMR spectrum was observed for the 15N-labelled Tau injected into the X. laevis oocytes [222], indicating that although the C-terminal half of Tau protein potentially folds at interaction with cellular MTs, the N-terminal domain preserves it highly disordered state even in the highly crowded cellular environment [222]. Subsequent structural characterization of the MT-bound Tau revealed that interaction with its natural partner converts this predominantly disordered protein into a partially folded state, where the conserved hexapeptides at the beginning of its repeats two and three were converted into a β-hairpin conformation [225].
An application of the IDP-tailored J-modulated proton-less NMR experiment allowing accurate measurement of the backbone one- and two-bond J(15N,13Cα) couplings provided detailed structural description of the intrinsically disordered actin-binding N-terminal domain (residues 2–65) of the Wiskott–Aldrich syndrome protein (WASp)-interacting protein (WIP2-65), which is an important regulator of cytoskeletal changes in various biological systems [226]. In diluted solutions, WIP2-65 conformational ensemble was shown to be resembling the structure assumed by this fragment in its actin-bound form, where residues 28–42 possessed an α-helical tendency, and the remaining parts of the protein were mostly disordered [226]. However, a significant decrease in the structural propensity under the influence of a bacterial cell lysate was observed by the analysis of the J-coupling data, suggesting that cellular crowding caused partial unfolding of the residual structure in this IDP, likely via protein–protein interactions that stabilized the more unfolded state of WIP2-65 [226].
In addition to various NMR techniques, conformational behaviour of IDPs inside the living cells can be analyzed by FRET. Schuler and co-workers showed recently that confocal single-molecule FRET spectroscopy combined with intracellular nanosecond fluorescence correlation spectroscopy (FCS) can provide important information on the dynamics of proteins from the milliseconds to the nanosecond regime in live eukaryotic cells [227]. Here, fluorescently labelled proteins [globular yeast frataxin homologue Yft1 and IgG-binding domain of protein G (GB1), as well as an extended IDP ProTα] were microinjected into cultured eukaryotic cells in a highly controlled manner, providing possibility to precisely control intracellular concentration of labelled protein from the picomolar to nanomolar range [227]. Comparison of the hydrodynamic dimensions and conformational dynamics of ProTα revealed the presence of a remarkable similarity between the intra- and extracellular behaviour of this protein [227], indicating that ProTα belongs to the category of non-foldable by cellular environment IDPs. Furthermore, in line with the well-known dependence of the hydrodynamic volume of ProTα on ionic strength in vitro, increase in the extracellular salt concentration (which cased transient increase in intracellular ionic strength) resulted in noticeable compaction of this protein inside the cell [227].
The Fast Relaxation Imaging (FReI) technique represents a useful approach that can complement in-cell NMR. FReI is pioneered by the group of Martin Gruebele, and represents a smart combination of FRET imaging of biomolecules and biomolecular kinetics induced by the temperature jump relaxation, represents a useful approach that can generate the movies of fast protein dynamics inside living cells [228]. To measure protein dynamics inside the cells, the temperature of the cell is suddenly jumped by a few degrees using an infrared laser, and then, the efficiency of FRET is followed by imaging fluorescence in the donor and acceptor [228]. Although FReI was originally designed as a means for studying folding and stability of globular protein inside the living cell and for looking at the effect of the cell on protein-free-energy landscapes [229–234], this technique is perfectly suited for the analysis of the intracellular conformational behaviour of IDPs [235]. Using this approach, the authors showed that an increase in local temperature from 22.3 to 49 °C resulted in noticeable conformational changes in α-synuclein within the cell. In fact, spatial separation of the FRET labels increased with the temperature, suggesting that α-synuclein adopted more extended conformation at higher temperatures [235]. This is an interesting observation, showing that cellular environment has a pronounced effect on the conformational behaviour of α-synuclein, since, in diluted solutions, α-synuclein was shown to gain more ordered structure in a temperature-dependent manner, being typically more disordered at lower temperatures and more structured at higher temperatures due to the enhancement of hydrophobic interaction at higher temperatures [236].
Data presented in this section indicate that, based on their conformational behaviour in natural cellular environments, IDPs/IDPRs are grouped into the familiar classes (partially) foldable, non-foldable, and unfoldable, which were already described in detail for these proteins/regions in artificial crowded environment generated in vitro by high concentrations of polymers or inert proteins.
IDPs/IDPRs in proteinaceous membrane-less organelles: tenacity of disorder in the overcrowded milieu
We already indicated that eukaryotic cells and bacteria possess various PMLOs, which contain specific proteins and nucleic acids and are specifically distributed within the cell [42, 43, 46]. PMLOs are highly dynamic organelles, whose biogenesis and structural integrity depend on unique protein–protein and protein–nucleic acid interactions [47, 48]. PMLOs are characterized by fluid behaviour [50–55], and because of the lack of membrane encapsulation, the interior of these organelles is freely accessible to the environmental cellular fluids, such as nucleoplasm, cytoplasm, matrix, or stroma. Formation and disintegration of PMLOs represent an illustration of biological liquid–liquid-phase transitions, LLPTs [49, 54–60]. The causative local PMLOs result in the formation of liquid cellular droplets with high protein and nucleic acid concentrations that noticeably exceed content of these biomacromolecules in dilute phase [55, 56, 60, 61]. This biological condensation generates an overcrowded milieu inside the PMLOs [40]. It was indicated that proteomes of PMLOs are enriched in IDPs/IDPRs, indicating that intrinsic disorder plays a crucial role in the fluidity and highly dynamic nature of the PMLOs, and also defines assembly/disassembly cycles, morphology, and structure these overcrowded cellular droplets [40, 41, 43, 46, 58, 62–69]. Since many IDPs serve as “drivers” of the LLPTs leading to the PMLO formation, their concentration inside PMLOs can be high. In other words, these IDPs serve as crowders themselves. Therefore, it is of interest to see how such self-crowding affects the structural properties of IDPs undergoing LLPTs.
Among a multitude of IDPs know to undergo LLPTs leading to the formation of PMLOs or liquid droplets in vitro is a Fused in Sarcoma (FUS) protein, which is an RNA-binding protein, associated with pathogenesis of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) and also related to the chromosomal translocation in certain sarcomas and leukemias. The low-complexity (LC) N-terminally-located domain of FUS (residues 1–163) was shown to undergo phase separation associated with the formation of ribonucleoprotein (RNP) granules [237]. When solution NMR spectroscopy was utilized to directly probe the structural organization FUS liquid-phase-separated assemblies, it was found that the LC domain of FUS (which is a typical IDP in diluted solution) mostly retained disordered structure even in the liquid-phase-separated state [237]. This was demonstrated by an overlay of the 1H–15N and 1H–13C heteronuclear single-quantum coherence (HSQC) spectra measured for the dispersed FUS LC (50 μM) and for the liquid-phase-separated protein (7 mM FUS LC) that showed high similarity [237]. It was pointed out that, since NMR spectra recorded for the phase-separated and dispersed phases of this protein lack large chemical-shift differences, the FUS remains disordered in the phase-separated state and, therefore, the LLPT is not accompanied by significant conformational changes of the LC domain of FUS protein [237].
Elastin-like polypeptides (ELPs) derived from the monomer of elastin, tropoelastin that can undergo LLPT [238–240], serve as a useful proxies for the analysis of the molecular mechanisms underlying phase separation in protein solutions [69, 241–244]. The 1H–15N HSQC analysis of one of such ELPs revealed that this model polypeptide is highly disordered in both the monomeric and phase-separated states [245] This conclusion was supported by the 2D 1H–1H nuclear Overhauser effect spectroscopy (NOESY) experiments reporting the through–space interactions between 1H nuclei, which also indicated that this ELP retained disordered structure in the phase-separated state [245]. Careful analysis of the NOEs arising from short-range interactions between protons in the protein backbone revealed the presence of two distinct type-II β-turns centred on the VPGV and GVGV sequences of both monomeric and phase-separated ELP [245]. Finally, analysis of the 13C and 12C edited 1H–1H NOESY spectra recorded for monomeric and phase-separated samples containing a 1:1 mixture of unlabelled and 13C-labelled ELP revealed the presence of multiple non-specific inter-molecular hydrophobic contacts within the phase-separated ELP [245].
Analysis of the structural properties of the lysine-rich microtubule-binding repeats of Tau protein, which are able to undergo liquid–liquid-phase separation in solution, by solution NMR provided some important insights into this problem [246]. In fact, besides generating very important observations that the microtubule-binding repeats of Tau can form liquid droplets in a phosphorylation-specific manner and that the efficiency of the Tau demixing depends on the number of the microtubule-binding repeats (with three-repeat and four-repeat isoforms of Tau being different in their ability for demixing), this study clearly showed that Tau protein mostly preserves its intrinsically disordered nature inside the Tau-containing membrane-less compartments [246]. In fact, a two-dimensional 1H–15N correlation NMR spectra recorded for the repeat domain of Tau before and after temperature-induced LLPT were characterized by small-signal dispersion typical of the mostly disordered polypeptide chain [246]. However, more detailed analysis of the 1H–13C correlation spectra of Tau protein labelled with the paramagnetic nitroxide tag (1-oxy-2,2,5,5-tetramethyl-d-pyrroline-3-methyl)-methanethiosulfonate (MTSL) attached to the two native cysteines, C291 and C322, revealed that in its droplet state, the aggregation-prone hexapeptides of Tau protein (residues 275VQIINK280 and 306VQIVYK311) are engaged in the intensive intra- and inter-molecular interactions [246]. These observations indicated that, in the phase-separated state, the amyloid hot spots of Tau protein are engaged in the formation of the dynamic molecular mesh, whereas the most parts of this protein retained their intrinsically disordered status [246].
Application of the heteronuclear solution NMR spectroscopy to the analysis of the intrinsically disordered N-terminal domain (residues 1–236) of the germ-granule protein Ddx4 provided further support to the idea that an IDP can continue to be mostly disordered within the highly concentrated phase-separated state [247]. Being a germ cell-specific protein, Ddx4 serves as the major component of the specific PMLOs, nuage/chromatoid bodies, found in the cytoplasm of spermatocytes and spermatids [248]. Ddx4 can be divided into several structural and functional regions, such as a long N-terminal disordered tail (residues 1–260), a catalytic helicase ATP-binding domain (residues 319–502), a helicase C-terminal domain (residues 350–675), and a relatively short disordered C-terminal tail (residues 690–724). The 1H–15N and 1H–13C correlation spectra recorded for the concentrated phase of the Ddx4cond form (which is the phase-separated state of the N-terminal domain of Ddx4 with the protein concentration of 380 mg/mL) were characterized by the sharp resonances, low-signal dispersion, and chemical shifts typical of the intrinsically disordered proteins, indicating mostly disordered state of Ddx4cond in the phase-separated form [247]. The authors also indicated that there was no noticeable line broadening of resonances in the 1H–13C spectra of Ddx4cond relative to spectrum of this protein in diluted form and only minor chemical-shift changes were detected. However, in the phase-separated state, Ddx4 diffused noticeably slower (~ 100-fold) than in its diluted form [247]. This slowed Ddx4 diffusion in the Ddx4cond state was attributed to the presence of a network of interchain interactions, mostly between Phe and Arg residues [247] needed for the establishing both cation–π and π–π interactions that can be crucial for driving the phase separation of Ddx4 and for the Ddx4cond stabilization [60].
NMR-based analysis of the low-complexity (LC) domain of the heterogeneous nuclear ribonuclear protein-2 (hnRNPA2) that serves as a component of the RNA-processing PMLOs and play a role in the mRNA transport and that is able to undergo LLPT [249] revealed that this domain remains predominantly disordered in the phase-separated state [250]. It was emphasized that, similar to Ddx4, the condensed phases of the hnRNPA2 LC domain formed by LLPT are highly concentrated and contain ~440 mg/mL (~ 30 mM) of the protein [250], suggesting that excessive self-crowding has minimal effect of highly dynamic structure of these IDPs.
Data presented in this section suggest that proteins capable of LLPT and used in the PMLO biogenesis are characterized by structures that are minimally affected by the formation of highly concentrated phases. In fact, these proteins are minimally foldable or even non-foldable at all (at least within the limited time period) in the extremely self-crowded milieus of their phase-separated states. It is likely that such insensitivity of disordered structure of these LLPT-driving proteins to self-crowding (once again, within the limited time period) is related to the biogenesis of PMLOs, which are typically present in the cell for some limited time. In fact, some PMLOs are known to be metastable systems that can undergo “maturation”, with their “aging” leading to subsequent transformation of corresponding systems into gels or glass [251]. In other words, biogenesis of PMLOs is a highly dynamic process, and, being formed, many PMLOs exist for the limited time. Within this physiologically safe time-frame of their normal existence, PMLOs are reversible and can undergo disassembly process when not needed. However, under pathological conditions, the biogenesis of PMLOs can be altered, and they could exist for prolonged time, leading to some irreversible changes in the system. In fact, although “freshly” generated droplets formed by the proteins undergoing LLPTs can easily disassemble by changes in their environment, over time, these droplets can mature to more stable states [58, 252], likely due to the formation of amyloid-like fibrils characterized by very high conformational stability. This is because many proteins found in RNP granules contain not only RNA-binding domains but also prion-related low-complexity sequences capable of pathogenic aggregation [58]. In other words, since aggregation and fibrillation are strongly concentration-dependent, dysregulated PMLOs (i.e., PMLOs existing for the period of time exceeding the aforementioned physiologically safe time-frame), with their locally increased concentrations of specific proteins, might inadvertently serve as a kind of amyloid “incubators”.
Conclusions
Because of their mosaic architecture, structures of IDPs or IDPRs can be extremely sensitive to changes in their environment and could be responsive to the presence of crowding agents. Based on the data presented in this review it is clear that the behaviour of IDPs at the conditions of macromolecular crowding is very complex. Despite the fact that, based on the thermodynamic principles of excluded volume, according to which the presence of a space-filling substance would favour a more compact state over the more extended form, it was expected that IDPs/IDPRs should undergo efficient folding and compaction under the conditions of macromolecular crowding, the number of disordered proteins/domains that actually undergo a significant folding in crowded environment is rather limited. In fact, different IDPs/IDPRs show very different response to the presence of artificial- and natural-crowding agents, and can be grouped into three major categories (partially) foldable, non-foldable, and unfoldable by macromolecular crowding. The fact that only some IDPs (partially) fold in crowded environment and that the global structure of many IDPs seems to be insensitive to the presence of crowding agents is not too surprising, since the anticipated overall contribution of the macromolecular crowing to the total free energy of the medium is not too high. Therefore, it is expected that the outputs of the presence of macromolecular crowders in the solution of a query protein will depend on the conformational stability of this protein in terms of its proximity to the transition region describing its potential structural transformation from a highly disordered to ordered state.
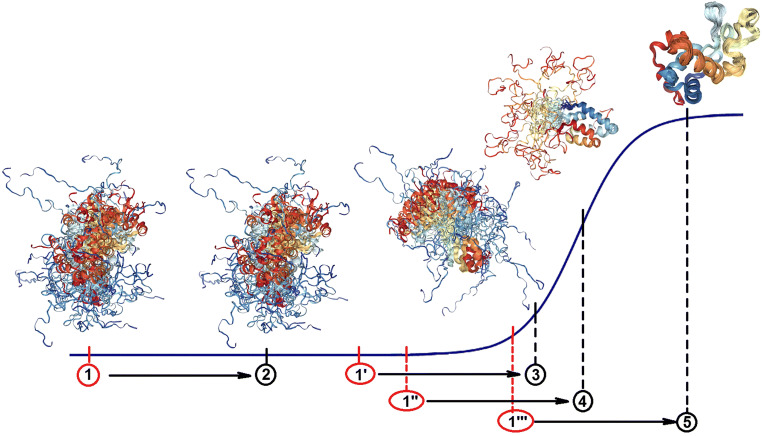
This point is illustrated by Fig. 1, representing an oversimplified model of structural transitions in a disordered protein induced by changes in its environment. It is seen that the end result (2, 3, 4, or 5) associated with the addition of crowder to the solution of an IDP depends on the remoteness of the position of this protein from the transition region in crowder-free environment. In other words, although the distances X1 → X2, X1′ → X3, X1″ → X4, and X1′′′ → X5 are identical, they induce very different changes in the conformational state of a query protein (ΔY1→2, ΔY1′→3, ΔY1″→4, and ΔY1′′′→5). One should also keep in mind that because of the highly heterogeneous spatio-temporal organization of a typical IDP containing regions with the different degrees of (dis)order (foldons, non-foldons, inducible foldons, and semi-foldons, see above), these differently (dis)ordered regions are expected to differently react to the addition of crowders. In other words, in addition to being applicable to the whole protein, a model presented in Fig. 1 can describe crowding-modulated behaviour of its regions that can differently fold in the presence of crowding agents.
Fig. 1.
Oversimplified representation of structural transitions induced in a disordered protein by crowded environment. The overall contribution of macromolecular crowing to the total free energy of a system is the same in all four scenarios. The outputs of the presence of crowders will depend on the conformational stability of a query protein in terms of its proximity to the transition region describing structural transformation of an IDP from highly disordered to ordered state. In this model, the end result (2, 3, 4, or 5) associated with the addition of crowder to the solution of an IDP depends on a remoteness of the position of this protein from the transition region in crowder-free environment. In other words, although the distances X1 → X2, X1′ → X3, X1″ → X4, and X1′′′ → X5 are identical, they induce very different changes (ΔY1→2, ΔY1′→3, ΔY1″→4, and ΔY1′′′→5) in the conformational state of a query protein
It was also pointed out that the biological macromolecules are not inactive entities that serve as inert placeholders that simply restrict cellular volume accessible to a query protein, but instead can be involved in specific and non-specific interactions with this protein. As a result, crowded cellular environment can induce both (partial) excluded-volume-driven folding and (partial) interaction-modulated unfolding of a target protein or its parts (unfoldons). Therefore, since crowding can cause both folding and unfolding of an IDP or its regions, the outputs of the placing of a query protein to the crowded environment would depend on the balance between these two processes. As a result, macromolecular crowding would differently affect the structures of different IDPs.
Finally, a note should be added on the peculiar capability of some IDPs to undergo LLPTs, thereby generating self-crowded PMLOs. An important feature of such IDPs is relative insensitivity of their structure to the self-crowded environment (in fact, they are minimally foldable or even non-foldable at all) and the ability to avoid pathological association and aggregation at least during the limited time of the physiological existence of PMLOs. It is likely that this capability to phase separate but sustain local increase in concentration without undergoing pathological aggregation (at least for a limited time period) represents an important factor driving the evolution of IDPs that contribute to the PMLO biogenesis. One of the features of such IDPs is either their overall high net charge or the presence of highly charged patches. This allows such IDPs to be engaged in the formation of unique complexes, which, despite being characterized by high affinity, show no evidence of folding of the constituents that continue to be highly dynamic and completely disordered in their bound state. The molecular mechanism behind the formation of such tight but highly dynamic complexes is based on strong electrostatic attraction between oppositely charged polypeptides (or oppositely charged regions of a given protein). It is likely that such a kind of global electrostatic attraction does not require defined binding sites or interactions between specific individual residues. Instead, any charged residue of one polypeptide chain (or protein region) can efficiently interact with any oppositely charged reside of another polypeptide chain (or protein region). The possibility of the existence of such kind of complexes, where despite picomolar affinity partners do not fold, being highly dynamic and completely disordered in bound state, was recently demonstrated for a complex between two highly charged and highly disordered proteins, human histone H1 and ProT-α [253].
Compliance with ethical standards
Conflict of interest
There are no conflicts to declare.
Footnotes
Alexander V. Fonin and April L. Darling contributed equally to this work.
References
- 1.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222(3):599–620. doi: 10.1016/0022-2836(91)90499-V. [DOI] [PubMed] [Google Scholar]
- 2.Minton AP. Influence of excluded volume upon macromolecular structure and associations in ‘crowded’ media. Curr Opin Biotechnol. 1997;8(1):65–69. doi: 10.1016/S0958-1669(97)80159-0. [DOI] [PubMed] [Google Scholar]
- 3.Minton AP. Implications of macromolecular crowding for protein assembly. Curr Opin Struct Biol. 2000;10(1):34–39. doi: 10.1016/S0959-440X(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 5.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30(2):345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 6.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26(10):597–604. doi: 10.1016/S0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 7.Minton AP. Protein folding: thickening the broth. Curr Biol. 2000;10(3):R97–99. doi: 10.1016/S0960-9822(00)00301-8. [DOI] [PubMed] [Google Scholar]
- 8.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276(14):10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 9.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalwarczyk T, Ziebacz N, Bielejewska A, Zaboklicka E, Koynov K, Szymanski J, Wilk A, Patkowski A, Gapinski J, Butt HJ, Holyst R. Comparative analysis of viscosity of complex liquids and cytoplasm of mammalian cells at the nanoscale. Nano Lett. 2011;11(5):2157–2163. doi: 10.1021/nl2008218. [DOI] [PubMed] [Google Scholar]
- 11.Jena SS, Bloomfield VA. Probe diffusion in concentrated polyelectrolyte solutions: effect of background interactions on competition between electrostatic and viscous forces. Macromolecules. 2005;38(25):10551–10556. doi: 10.1021/ma0521304. [DOI] [Google Scholar]
- 12.Hou S, Trochimczyk P, Sun L, Wisniewska A, Kalwarczyk T, Zhang X, Wielgus-Kutrowska B, Bzowska A, Holyst R. How can macromolecular crowding inhibit biological reactions? The enhanced formation of DNA nanoparticles. Sci Rep. 2016;6:22033. doi: 10.1038/srep22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira LA, Uversky VN, Zaslavsky BY. Role of solvent properties of water in crowding effects induced by macromolecular agents and osmolytes. Mol BioSyst. 2017;13(12):2551–2563. doi: 10.1039/c7mb00436b. [DOI] [PubMed] [Google Scholar]
- 14.Eggers DK, Valentine JS. Crowding and hydration effects on protein conformation: a study with sol–gel encapsulated proteins. J Mol Biol. 2001;314(4):911–922. doi: 10.1006/jmbi.2001.5166. [DOI] [PubMed] [Google Scholar]
- 15.Eggers DK, Valentine JS. Molecular confinement influences protein structure and enhances thermal protein stability. Protein Sci. 2001;10(2):250–261. doi: 10.1110/ps.36201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bismuto E, Martelli PL, De Maio A, Mita DG, Irace G, Casadio R. Effect of molecular confinement on internal enzyme dynamics: frequency domain fluorometry and molecular dynamics simulation studies. Biopolymers. 2002;67(2):85–95. doi: 10.1002/bip.10058. [DOI] [PubMed] [Google Scholar]
- 17.Minton AP. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: macromolecular crowding and protein stability revisited. Biophys J. 2005;88(2):971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuznetsova IM, Zaslavsky BY, Breydo L, Turoverov KK, Uversky VN. Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Molecules. 2015;20(1):1377–1409. doi: 10.3390/molecules20011377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznetsova IM, Turoverov KK, Uversky VN. What macromolecular crowding can do to a protein. Int J Mol Sci. 2014;15(12):23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepanenko OV, Povarova OI, Sulatskaya AI, Ferreira LA, Zaslavsky BY, Kuznetsova IM, Turoverov KK, Uversky VN. Protein unfolding in crowded milieu: what crowding can do to a protein undergoing unfolding? J Biomol Struct Dyn. 2016;34(10):2155–2170. doi: 10.1080/07391102.2015.1109554. [DOI] [PubMed] [Google Scholar]
- 21.Morar AS, Olteanu A, Young GB, Pielak GJ. Solvent-induced collapse of alpha-synuclein and acid-denatured cytochrome c. Protein Sci. 2001;10(11):2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatters DM, Minton AP, Howlett GJ. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J Biol Chem. 2002;277(10):7824–7830. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Cooper EM, Bower KS, Li J, Fink AL. Accelerated alpha-synuclein fibrillation in crowded milieu. FEBS Lett. 2002;515(1–3):99–103. doi: 10.1016/S0014-5793(02)02446-8. [DOI] [PubMed] [Google Scholar]
- 24.Shtilerman MD, Ding TT, Lansbury PT., Jr Molecular crowding accelerates fibrillization of alpha-synuclein: could an increase in the cytoplasmic protein concentration induce Parkinson’s disease? Biochemistry. 2002;41(12):3855–3860. doi: 10.1021/bi0120906. [DOI] [PubMed] [Google Scholar]
- 25.Zhou HX. Effect of mixed macromolecular crowding agents on protein folding. Proteins. 2008;72(4):1109–1113. doi: 10.1002/prot.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batra J, Xu K, Qin S, Zhou HX. Effect of macromolecular crowding on protein binding stability: modest stabilization and significant biological consequences. Biophys J. 2009;97(3):906–911. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christiansen A, Wang Q, Samiotakis A, Cheung MS, Wittung-Stafshede P. Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry. 2010;49(31):6519–6530. doi: 10.1021/bi100578x. [DOI] [PubMed] [Google Scholar]
- 28.Shahid S, Hassan MI, Islam A. Ahmad F (2017) Size-dependent studies of macromolecular crowding on the thermodynamic stability, structure and functional activity of proteins: in vitro and in silico approaches. Biochim Biophys Acta. 1861;2:178–197. doi: 10.1016/j.bbagen.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Sharp KA. Analysis of the size dependence of macromolecular crowding shows that smaller is better. Proc Natl Acad Sci USA. 2015;112(26):7990–7995. doi: 10.1073/pnas.1505396112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goins AB, Sanabria H, Waxham MN. Macromolecular crowding and size effects on probe microviscosity. Biophys J. 2008;95(11):5362–5373. doi: 10.1529/biophysj.108.131250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan EH, Dave BC, Fukuto JM, Dunn B, Zink JI, Valentine JS. Synthesis of sol–gel encapsulated heme proteins with chemical sensing properties. J Mater Chem. 1999;9:45–53. doi: 10.1039/a805541f. [DOI] [Google Scholar]
- 32.Gottfried DS, Kagan A, Hoffman BM, Friedman JM. Impeded rotation of a protein in sol–gel matrix. J Phys Chem B. 1999;103:2803–2807. doi: 10.1021/jp9840230. [DOI] [Google Scholar]
- 33.Brennan JD. Using fluorescence to investigate proteins entrapped in sol–gel derived materials. Appl Spectrosc. 1999;53:106A–121A. doi: 10.1366/0003702991946514. [DOI] [Google Scholar]
- 34.Dave BC, Dunn B, Valentine JS, Zink JI. Sol–gel encapsulation methods for biosensors. Anal Chem. 1994;66:1120–1127. doi: 10.1021/ac00094a001. [DOI] [Google Scholar]
- 35.Gill I, Ballestros A. Bioencapsulation within synthetic polymers (part 1): sol–gel encapsulated biologicals. Trends Biotechnol. 2000;18:282–296. doi: 10.1016/S0167-7799(00)01457-8. [DOI] [PubMed] [Google Scholar]
- 36.Bismuto E, Irace G. The effect of molecular confinement on the conformational dynamics of the native and partly folded state of apomyoglobin. FEBS Lett. 2001;509:476–480. doi: 10.1016/S0014-5793(01)03087-3. [DOI] [PubMed] [Google Scholar]
- 37.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA. ATP as a biological hydrotrope. Science. 2017;356(6339):753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 38.Rice AM, Rosen MK. ATP controls the crowd. Science. 2017;356(6339):701–702. doi: 10.1126/science.aan4223. [DOI] [PubMed] [Google Scholar]
- 39.Hayes MH, Peuchen EH, Dovichi NJ, Weeks DL. Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. Elife. 2018 doi: 10.7554/eLife.35224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: complex coacervates and membrane-less organelles. Adv Colloid Interface Sci. 2017;239:97–114. doi: 10.1016/j.cis.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Zaslavsky BY, Uversky VN. In aqua veritas: the indispensable yet mostly ignored role of water in phase separation and membrane-less organelles. Biochemistry. 2018 doi: 10.1021/acs.biochem.7b01215. [DOI] [PubMed] [Google Scholar]
- 43.Darling AL, Liu Y, Oldfield CJ, Uversky VN. Intrinsically disordered proteome of human membrane-less organelles. Proteomics. 2018;18(5–6):e1700193. doi: 10.1002/pmic.201700193. [DOI] [PubMed] [Google Scholar]
- 44.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404(6778):604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 45.Pederson T. Protein mobility within the nucleus—what are the right moves? Cell. 2001;104(5):635–638. doi: 10.1016/S0092-8674(01)00258-6. [DOI] [PubMed] [Google Scholar]
- 46.Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589(1):15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 47.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(12):a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27(8):295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203(6):875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 51.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108(11):4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol. 2013;15(10):1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152(4):791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 54.Aggarwal S, Snaidero N, Pahler G, Frey S, Sanchez P, Zweckstetter M, Janshoff A, Schneider A, Weil MT, Schaap IA, Gorlich D, Simons M. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS Biol. 2013;11(6):e1001577. doi: 10.1371/journal.pbio.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holehouse AS, Pappu RV. Protein polymers: encoding phase transitions. Nat Mater. 2015;14(11):1083–1084. doi: 10.1038/nmat4459. [DOI] [PubMed] [Google Scholar]
- 58.Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60(2):208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. doi: 10.1016/j.ceb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57(5):936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16(1):202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38(10):494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206(5):579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA. 2015;112(23):7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng F, Na I, Kurgan L, Uversky VN. Compartmentalization and functionality of nuclear disorder: intrinsic disorder and protein–protein interactions in intra-nuclear compartments. Int J Mol Sci. 2015 doi: 10.3390/ijms17010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, Rosen MK. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63(1):72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215(3):313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, Lopez GP. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat Chem. 2017;9(6):509–515. doi: 10.1038/nchem.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 71.Uversky VN. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol. 2010;2010:568068. doi: 10.1155/2010/568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 73.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 74.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of protein folding. Weinheim: Wiley-VCH; 2005. pp. 271–353. [Google Scholar]
- 76.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60(9):1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 78.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 79.Dunker AK, Obradovic Z. The protein trinity—linking function and disorder. Nat Biotechnol. 2001;19(9):805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 80.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 81.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/S0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 82.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 83.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 84.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269(1):2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 86.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 87.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 88.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 89.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 90.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 91.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30(9):484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 93.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6(5):1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6(5):1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6(5):1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 99.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4(4):229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 100.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genom. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Russell RB, Gibson TJ. A careful disorderliness in the proteome: sites for interaction and targets for future therapies. FEBS Lett. 2008;582(8):1271–1275. doi: 10.1016/j.febslet.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 103.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 104.Uversky VN, Dunker AK. Biochemistry. Controlled chaos. Science. 2008;322(5906):1340–1341. doi: 10.1126/science.1167453. [DOI] [PubMed] [Google Scholar]
- 105.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. BioEssays. 2009;31(3):328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 106.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23(8):1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des. 2013;19:4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 110.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buljan M, Chalancon G, Dunker AK, Bateman A, Balaji S, Fuxreiter M, Babu MM. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23(3):443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 112.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46(6):871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 114.Schulz GE. Nucleotide binding proteins. In: Balaban M, editor. Molecular mechanism of biological recognition. New York: Elsevier/North-Holland Biomedical Press; 1979. pp. 79–94. [Google Scholar]
- 115.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18(5):181–186. doi: 10.1016/0968-0004(93)90111-Y. [DOI] [PubMed] [Google Scholar]
- 116.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 117.Rosenfeld R, Vajda S, DeLisi C. Flexible docking and design. Annu Rev Biophys Biomol Struct. 1995;24:677–700. doi: 10.1146/annurev.bb.24.060195.003333. [DOI] [PubMed] [Google Scholar]
- 118.Plaxco KW, Gross M. Cell biology. The importance of being unfolded. Nature. 1997;386(6626):657–659. doi: 10.1038/386657a0. [DOI] [PubMed] [Google Scholar]
- 119.Lee BM, Xu J, Clarkson BK, Martinez-Yamout MA, Dyson HJ, Case DA, Gottesfeld JM, Wright PE. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J Mol Biol. 2006;357(1):275–291. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 120.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 121.Dunker AK, Obradovic Z, Romero P, Kissinger C, Villafranca E. On the importance of being disordered. PDB Newslett. 1997;81:3–5. [Google Scholar]
- 122.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998;3:473–484. [PubMed] [Google Scholar]
- 123.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins Struct Funct Genet. 2001;42:38–49. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 124.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55(1):104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 125.Jemth P, Mu X, Engstrom A, Dogan J. A frustrated binding interface for intrinsically disordered proteins. J Biol Chem. 2014;289(9):5528–5533. doi: 10.1074/jbc.M113.537068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uversky VN. Intrinsically disordered proteins may escape unwanted interactions via functional misfolding. Biochim Biophys Acta. 2011;1814(5):693–712. doi: 10.1016/j.bbapap.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 127.Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function. Chem Rev. 2014 doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37(12):517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22(6):693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834:932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 131.Uversky VN. Paradoxes and wonders of intrinsic disorder: complexity of simplicity. Intrinsically Disord Proteins. 2016;4(1):e1135015. doi: 10.1080/21690707.2015.1135015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Munshi S, Gopi S, Subramanian S, Campos LA, Naganathan AN. Protein plasticity driven by disorder and collapse governs the heterogeneous binding of CytR to DNA. Nucleic Acids Res. 2018;46(8):4044–4053. doi: 10.1093/nar/gky176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 134.Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114(13):6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept Lett. 2009;16(12):1533–1547. doi: 10.2174/092986609789839377. [DOI] [PubMed] [Google Scholar]
- 136.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 137.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genom. 2009;10(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Uversky VN. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D(2) concept. Expert Rev Proteom. 2010;7(4):543–564. doi: 10.1586/epr.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Uversky VN. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci. 2014;1:6. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Christiansen A, Wang Q, Cheung MS, Wittung-Stafshede P. Effects of macromolecular crowding agents on protein folding in vitro and in silico. Biophys Rev. 2013;5(2):137–145. doi: 10.1007/s12551-013-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 142.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15(1):35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 143.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40(3):1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 144.Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, Romero P, Dunker AK. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012;2012:116–127. [PubMed] [Google Scholar]
- 145.Hsu WL, Oldfield CJ, Xue B, Meng J, Huang F, Romero P, Uversky VN, Dunker AK. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22(3):258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miller CM, Kim YC, Mittal J. Protein composition determines the effect of crowding on the properties of disordered proteins. Biophys J. 2016;111(1):28–37. doi: 10.1016/j.bpj.2016.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci USA. 2005;102(13):4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schlesinger AP, Wang Y, Tadeo X, Millet O, Pielak GJ. Macromolecular crowding fails to fold a globular protein in cells. J Am Chem Soc. 2011;133(21):8082–8085. doi: 10.1021/ja201206t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Christiansen A, Wittung-Stafshede P. Synthetic crowding agent dextran causes excluded volume interactions exclusively to tracer protein apoazurin. FEBS Lett. 2014;588(5):811–814. doi: 10.1016/j.febslet.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 150.Christiansen A, Wittung-Stafshede P. Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin in vitro. Biophys J. 2013;105(7):1689–1699. doi: 10.1016/j.bpj.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aden J, Wittung-Stafshede P. Folding of an unfolded protein by macromolecular crowding in vitro. Biochemistry. 2014;53(14):2271–2277. doi: 10.1021/bi500222g. [DOI] [PubMed] [Google Scholar]
- 152.Tadeo X, Lopez-Mendez B, Trigueros T, Lain A, Castano D, Millet O. Structural basis for the aminoacid composition of proteins from halophilic archea. PLoS Biol. 2009;7(12):e1000257. doi: 10.1371/journal.pbio.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Qvist J, Ortega G, Tadeo X, Millet O, Halle B. Hydration dynamics of a halophilic protein in folded and unfolded states. J Phys Chem B. 2012;116(10):3436–3444. doi: 10.1021/jp3000569. [DOI] [PubMed] [Google Scholar]
- 154.Henkels CH, Kurz JC, Fierke CA, Oas TG. Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein. Biochemistry. 2001;40(9):2777–2789. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]
- 155.Henkels CH, Oas TG. Thermodynamic characterization of the osmolyte- and ligand-folded states of Bacillus subtilis ribonuclease P protein. Biochemistry. 2005;44(39):13014–13026. doi: 10.1021/bi0504613. [DOI] [PubMed] [Google Scholar]
- 156.Henkels CH, Oas TG. Ligation-state hydrogen exchange: coupled binding and folding equilibria in ribonuclease P protein. J Am Chem Soc. 2006;128(24):7772–7781. doi: 10.1021/ja057279+. [DOI] [PubMed] [Google Scholar]
- 157.Chang YC, Oas TG. Osmolyte-induced folding of an intrinsically disordered protein: folding mechanism in the absence of ligand. Biochemistry. 2010;49(25):5086–5096. doi: 10.1021/bi100222h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chang YC, Franch WR, Oas TG. Probing the folding intermediate of Bacillus subtilis RNase P protein by nuclear magnetic resonance. Biochemistry. 2010;49(44):9428–9437. doi: 10.1021/bi100287y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Daniels KG, Tonthat NK, McClure DR, Chang YC, Liu X, Schumacher MA, Fierke CA, Schmidler SC, Oas TG. Ligand concentration regulates the pathways of coupled protein folding and binding. J Am Chem Soc. 2014;136(3):822–825. doi: 10.1021/ja4086726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.de Jongh HH, de Kruijff B. The conformational changes of apocytochrome c upon binding to phospholipid vesicles and micelles of phospholipid based detergents: a circular dichroism study. Biochim Biophys Acta. 1990;1029(1):105–112. doi: 10.1016/0005-2736(90)90442-Q. [DOI] [PubMed] [Google Scholar]
- 161.Damaschun G, Damaschun H, Gast K, Gernat C, Zirwer D. Acid denatured apo-cytochrome c is a random coil: evidence from small-angle X-ray scattering and dynamic light scattering. Biochim Biophys Acta. 1991;1078(2):289–295. doi: 10.1016/0167-4838(91)90571-G. [DOI] [PubMed] [Google Scholar]
- 162.Goto Y, Takahashi N, Fink AL. Mechanism of acid-induced folding of proteins. Biochemistry. 1990;29(14):3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- 163.Goto Y, Calciano LJ, Fink AL. Acid-induced folding of proteins. Proc Natl Acad Sci USA. 1990;87(2):573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hamada D, Hoshino M, Kataoka M, Fink AL, Goto Y. Intermediate conformational states of apocytochrome c. Biochemistry. 1993;32(39):10351–10358. doi: 10.1021/bi00090a010. [DOI] [PubMed] [Google Scholar]
- 165.Sasahara K, McPhie P, Minton AP. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J Mol Biol. 2003;326(4):1227–1237. doi: 10.1016/S0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- 166.Hamada D, Kuroda Y, Kataoka M, Aimoto S, Yoshimura T, Goto Y. Role of heme axial ligands in the conformational stability of the native and molten globule states of horse cytochrome c. J Mol Biol. 1996;256(1):172–186. doi: 10.1006/jmbi.1996.0075. [DOI] [PubMed] [Google Scholar]
- 167.Roque A, Ponte I, Suau P. Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophys J. 2007;93(6):2170–2177. doi: 10.1529/biophysj.107.104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hartman PG, Chapman GE, Moss T, Bradbury EM. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 1977;77(1):45–51. doi: 10.1111/j.1432-1033.1977.tb11639.x. [DOI] [PubMed] [Google Scholar]
- 169.Th’ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem. 2005;280(30):27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 170.Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279(19):20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 171.Roque A, Iloro I, Ponte I, Arrondo JL, Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem. 2005;280(37):32141–32147. doi: 10.1074/jbc.M505636200. [DOI] [PubMed] [Google Scholar]
- 172.Johansen D, Jeffries CM, Hammouda B, Trewhella J, Goldenberg DP. Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys J. 2011;100(4):1120–1128. doi: 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Van Gilst MR, Rees WA, Das A, von Hippel PH. Complexes of N antitermination protein of phage lambda with specific and nonspecific RNA target sites on the nascent transcript. Biochemistry. 1997;36(6):1514–1524. doi: 10.1021/bi961920q. [DOI] [PubMed] [Google Scholar]
- 174.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93(2):289–299. doi: 10.1016/S0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 175.Conant CR, Goodarzi JP, Weitzel SE, von Hippel PH. The antitermination activity of bacteriophage lambda N protein is controlled by the kinetics of an RNA-looping-facilitated interaction with the transcription complex. J Mol Biol. 2008;384(1):87–108. doi: 10.1016/j.jmb.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Heller WT. Small-angle neutron scattering and contrast variation: a powerful combination for studying biological structures. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 11):1213–1217. doi: 10.1107/S0907444910017658. [DOI] [PubMed] [Google Scholar]
- 177.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101(34):12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kumosinski TF, Pessen H. Estimation of sedimentation coefficients of globular proteins: an application of small-angle X-ray scattering. Arch Biochem Biophys. 1982;219(1):89–100. doi: 10.1016/0003-9861(82)90137-0. [DOI] [PubMed] [Google Scholar]
- 179.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78(2):719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goldenberg DP, Argyle B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys J. 2014;106(4):905–914. doi: 10.1016/j.bpj.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Soranno A, Koenig I, Borgia MB, Hofmann H, Zosel F, Nettels D, Schuler B. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc Natl Acad Sci USA. 2014;111(13):4874–4879. doi: 10.1073/pnas.1322611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Smilgies DM, Folta-Stogniew E. Molecular weight-gyration radius relation of globular proteins: a comparison of light scattering, small-angle X-ray scattering and structure-based data. J Appl Crystallogr. 2015;48(Pt 5):1604–1606. doi: 10.1107/S1600576715015551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Flaugh SL, Lumb KJ. Effects of macromolecular crowding on the intrinsically disordered proteins c-Fos and p27(Kip1) Biomacromolecules. 2001;2(2):538–540. doi: 10.1021/bm015502z. [DOI] [PubMed] [Google Scholar]
- 184.Campbell KM, Terrell AR, Laybourn PJ, Lumb KJ. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 2000;39(10):2708–2713. doi: 10.1021/bi9923555. [DOI] [PubMed] [Google Scholar]
- 185.Bienkiewicz EA, Adkins JN, Lumb KJ. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1) Biochemistry. 2002;41(3):752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- 186.Sotomayor-Perez AC, Ladant D, Chenal A. Calcium-induced folding of intrinsically disordered repeat-in-toxin (RTX) motifs via changes of protein charges and oligomerization states. J Biol Chem. 2011;286(19):16997–17004. doi: 10.1074/jbc.M110.210393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sotomayor-Perez AC, Subrini O, Hessel A, Ladant D, Chenal A. Molecular crowding stabilizes both the intrinsically disordered calcium-free state and the folded calcium-bound state of a repeat in toxin (RTX) protein. J Am Chem Soc. 2013;135(32):11929–11934. doi: 10.1021/ja404790f. [DOI] [PubMed] [Google Scholar]
- 188.Cino EA, Karttunen M, Choy WY. Effects of molecular crowding on the dynamics of intrinsically disordered proteins. PLoS One. 2012;7(11):e49876. doi: 10.1371/journal.pone.0049876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Munishkina LA, Cooper EM, Uversky VN, Fink AL. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recognit. 2004;17(5):456–464. doi: 10.1002/jmr.699. [DOI] [PubMed] [Google Scholar]
- 190.McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J Mol Biol. 2006;355(5):893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 191.Bai J, Liu M, Pielak GJ, Li C. Macromolecular and small molecular crowding have similar effects on alpha-synuclein structure. ChemPhysChem. 2017;18(1):55–58. doi: 10.1002/cphc.201601097. [DOI] [PubMed] [Google Scholar]
- 192.Mouillon JM, Eriksson SK, Harryson P. Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 2008;148(4):1925–1937. doi: 10.1104/pp.108.124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mendes LFS, Basso LGM, Kumagai PS, Fonseca-Maldonado R, Costa-Filho AJ. Disorder-to-order transitions in the molten globule-like Golgi reassembly and stacking protein. Biochim Biophys Acta. 2018;1862(4):855–865. doi: 10.1016/j.bbagen.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 194.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17(12):3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91(2):253–262. doi: 10.1016/S0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 196.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18(18):4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010;11(6):827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Veenendaal T, Jarvela T, Grieve AG, van Es JH, Linstedt AD, Rabouille C. GRASP65 controls the cis Golgi integrity in vivo. Biol Open. 2014;3(6):431–443. doi: 10.1242/bio.20147757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jarvela T, Linstedt AD. Isoform-specific tethering links the Golgi ribbon to maintain compartmentalization. Mol Biol Cell. 2014;25(1):133–144. doi: 10.1091/mbc.E13-07-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mendes LF, Garcia AF, Kumagai PS, de Morais FR, Melo FA, Kmetzsch L, Vainstein MH, Rodrigues ML, Costa-Filho AJ. New structural insights into Golgi Reassembly and Stacking Protein (GRASP) in solution. Sci Rep. 2016;6:29976. doi: 10.1038/srep29976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gillen KL, Hughes KT. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173(20):6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Brown KL, Hughes KT. The role of anti-sigma factors in gene regulation. Mol Microbiol. 1995;16(3):397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 203.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99(20):12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4(4):285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 205.Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry. 1998;37(4):1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 206.Cuevas-Velazquez CL, Reyes JL, Covarrubias AA. Group 4 late embryogenesis abundant proteins as a model to study intrinsically disordered proteins in plants. Plant Signal Behav. 2017;12(7):e1343777. doi: 10.1080/15592324.2017.1343777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cuevas-Velazquez CL, Saab-Rincon G, Reyes JL, Covarrubias AA. The unstructured N-terminal region of arabidopsis group 4 late embryogenesis abundant (LEA) proteins is required for folding and for chaperone-like activity under water deficit. J Biol Chem. 2016;291(20):10893–10903. doi: 10.1074/jbc.M116.720318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148(1):6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275(8):5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- 211.Rusinga FI, Weis DD. Soft interactions and volume exclusion by polymeric crowders can stabilize or destabilize transient structure in disordered proteins depending on polymer concentration. Proteins. 2017;85(8):1468–1479. doi: 10.1002/prot.25307. [DOI] [PubMed] [Google Scholar]
- 212.Banks A, Qin S, Weiss KL, Stanley CB, Zhou HX. Intrinsically disordered protein exhibits both compaction and expansion under macromolecular crowding. Biophys J. 2018;114(5):1067–1079. doi: 10.1016/j.bpj.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Selenko P, Wagner G. Looking into live cells with in-cell NMR spectroscopy. J Struct Biol. 2007;158(2):244–253. doi: 10.1016/j.jsb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 214.Ito Y, Selenko P. Cellular structural biology. Curr Opin Struct Biol. 2010;20(5):640–648. doi: 10.1016/j.sbi.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 215.Freedberg DI, Selenko P. Live cell NMR. Annu Rev Biophys. 2014;43:171–192. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- 216.Smith MJ, Marshall CB, Theillet FX, Binolfi A, Selenko P, Ikura M. Real-time NMR monitoring of biological activities in complex physiological environments. Curr Opin Struct Biol. 2015;32:39–47. doi: 10.1016/j.sbi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 217.Plitzko JM, Schuler B, Selenko P. Structural biology outside the box-inside the cell. Curr Opin Struct Biol. 2017;46:110–121. doi: 10.1016/j.sbi.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 218.Binolfi A, Theillet FX, Selenko P. Bacterial in-cell NMR of human alpha-synuclein: a disordered monomer by nature? Biochem Soc Trans. 2012;40(5):950–954. doi: 10.1042/BST20120096. [DOI] [PubMed] [Google Scholar]
- 219.Smith AE, Zhou LZ, Pielak GJ. Hydrogen exchange of disordered proteins in Escherichia coli. Protein Sci. 2015;24(5):706–713. doi: 10.1002/pro.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Theillet FX, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, Selenko P. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature. 2016;530(7588):45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- 221.Popovic M, Sanfelice D, Pastore C, Prischi F, Temussi PA, Pastore A. Selective observation of the disordered import signal of a globular protein by in-cell NMR: the example of frataxins. Protein Sci. 2015;24(6):996–1003. doi: 10.1002/pro.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bodart JF, Wieruszeski JM, Amniai L, Leroy A, Landrieu I, Rousseau-Lescuyer A, Vilain JP, Lippens G. NMR observation of Tau in Xenopus oocytes. J Magn Reson. 2008;192(2):252–257. doi: 10.1016/j.jmr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 223.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33(1):95–130. doi: 10.1016/S0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 224.Sillen A, Barbier P, Landrieu I, Lefebvre S, Wieruszeski JM, Leroy A, Peyrot V, Lippens G. NMR investigation of the interaction between the neuronal protein tau and the microtubules. Biochemistry. 2007;46(11):3055–3064. doi: 10.1021/bi061920i. [DOI] [PubMed] [Google Scholar]
- 225.Kadavath H, Jaremko M, Jaremko L, Biernat J, Mandelkow E, Zweckstetter M. Folding of the Tau protein on microtubules. Angew Chem Int Ed Engl. 2015;54(35):10347–10351. doi: 10.1002/anie.201501714. [DOI] [PubMed] [Google Scholar]
- 226.Rozentur-Shkop E, Goobes G, Chill JH. A J-modulated protonless NMR experiment characterizes the conformational ensemble of the intrinsically disordered protein WIP. J Biomol NMR. 2016;66(4):243–257. doi: 10.1007/s10858-016-0073-6. [DOI] [PubMed] [Google Scholar]
- 227.Konig I, Zarrine-Afsar A, Aznauryan M, Soranno A, Wunderlich B, Dingfelder F, Stuber JC, Pluckthun A, Nettels D, Schuler B. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat Methods. 2015;12(8):773–779. doi: 10.1038/nmeth.3475. [DOI] [PubMed] [Google Scholar]
- 228.Ebbinghaus S, Dhar A, McDonald D, Gruebele M. Protein folding stability and dynamics imaged in a living cell. Nat Methods. 2010;7(4):319–323. doi: 10.1038/Nmeth.1435. [DOI] [PubMed] [Google Scholar]
- 229.Davis CM, Gruebele M. Labeling for quantitative comparison of imaging measurements in vitro and in cells. Biochemistry. 2018;57(13):1929–1938. doi: 10.1021/acs.biochem.8b00141. [DOI] [PubMed] [Google Scholar]
- 230.Gruebele M, Dave K, Sukenik S. Globular protein folding in vitro and in vivo. Annu Rev Biophys. 2016;45:233–251. doi: 10.1146/annurev-biophys-062215-011236. [DOI] [PubMed] [Google Scholar]
- 231.Gelman H, Wirth AJ, Gruebele M. ReAsH as a quantitative probe of in-cell protein dynamics. Biochemistry. 2016;55(13):1968–1976. doi: 10.1021/acs.biochem.5b01336. [DOI] [PubMed] [Google Scholar]
- 232.Guzman I, Gruebele M. Protein folding dynamics in the cell. J Phys Chem B. 2014;118(29):8459–8470. doi: 10.1021/jp501866v. [DOI] [PubMed] [Google Scholar]
- 233.Gelman H, Platkov M, Gruebele M. Rapid perturbation of free-energy landscapes: from in vitro to in vivo. Chemistry. 2012;18(21):6420–6427. doi: 10.1002/chem.201104047. [DOI] [PubMed] [Google Scholar]
- 234.Dhar A, Gruebele M. Fast relaxation imaging in living cells. Curr Protoc Protein Sci Chapter. 2011;28(Unit28):21. doi: 10.1002/0471140864.ps2801s65. [DOI] [PubMed] [Google Scholar]
- 235.Dhar A, Prigozhin M, Gelman H, Gruebele M. Studying IDP stability and dynamics by fast relaxation imaging in living cells. Methods Mol Biol. 2012;895:101–111. doi: 10.1007/978-1-61779-927-3_8. [DOI] [PubMed] [Google Scholar]
- 236.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276(14):10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 237.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60(2):231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Vrhovski B, Jensen S, Weiss AS. Coacervation characteristics of recombinant human tropoelastin. Eur J Biochem. 1997;250(1):92–98. doi: 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- 239.Bellingham CM, Lillie MA, Gosline JM, Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, Keeley FW. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers. 2003;70(4):445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 240.Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Adv Colloid Interface Sci. 2011;167(1–2):94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 241.Mackay JA, Chilkoti A. Temperature sensitive peptides: engineering hyperthermia-directed therapeutics. Int J Hyperthermia. 2008;24(6):483–495. doi: 10.1080/02656730802149570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Li NK, Garcia Quiroz F, Hall CK, Chilkoti A, Yingling YG. Molecular description of the LCST behavior of an elastin-like polypeptide. Biomacromolecules. 2014;15(10):3522–3530. doi: 10.1021/bm500658w. [DOI] [PubMed] [Google Scholar]
- 243.Roberts S, Dzuricky M, Chilkoti A. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 2015;589(19 Pt A):2477–2486. doi: 10.1016/j.febslet.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dzuricky M, Roberts S, Chilkoti A. Convergence of artificial protein polymers and intrinsically disordered proteins. Biochemistry. 2018;57(17):2405–2414. doi: 10.1021/acs.biochem.8b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Reichheld SE, Muiznieks LD, Keeley FW, Sharpe S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc Natl Acad Sci USA. 2017;114(22):E4408–E4415. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8(1):275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, Kay LE. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci USA. 2017;114(39):E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8(1):85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 249.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, Fawzi NL. Mechanistic View of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69(3):465–479 e467. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 252.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, Kragelund BB, Best RB, Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555(7694):61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]