Abstract
Programmed cell death (PCD) is associated with different phases of plant life and provides resistance to different kinds of biotic or abiotic stress. The redox molecule nitric oxide (NO) is usually produced during the stress response and exerts dual effects on PCD regulation. S-nitrosylation, which NO attaches to the cysteine thiol of proteins, is a vital posttranslational modification and is considered as an essential way for NO to regulate cellular redox signaling. In recent years, a great number of proteins have been identified as targets of S-nitrosylation in plants, especially during PCD. S-nitrosylation can directly affect plant PCD positively or negatively, mainly by regulating the activity of cell death-related enzymes or reconstructing the conformation of several functional proteins. Here, we summarized S-nitrosylated proteins that are involved in PCD and provide insight into how S-nitrosylation can regulate plant PCD. In addition, both the importance and challenges of future works on S-nitrosylation in plant PCD are highlighted.
Keywords: Protein S-nitrosylation, Programmed cell death, Plants
Introduction
Programmed cell death (PCD), the process of cellular suicide, is encoded genetically and actively controlled [1]. Although it is not clear how many types of PCD exist in plants, based on a PCD classification criterion, van Doorn classified plant PCD into ‘autolytic’ PCD and ‘non-autolytic’ PCD according to the tonoplast rupture and the cytoplasm subsequent rapid destruction [2]. PCD is involved in plant growth and development such as differentiation of tracheary elements [3] and xylogenesis of pioneer roots [4, 5], development of cereal aleurone cells [6], leaf senescence [7] and floret development [8]. Actually, PCD processes in plants are associated not only with the development of regular specific cell types or tissues but also with their immune responses, which are induced by various biotic stresses, such as plant–pathogen interactions, and abiotic stresses, such as extreme temperature, excessive light and UV radiation, water deprivation, high salinity, high concentrations of heavy metals, and herbicide activity [9–11]. Reactive oxygen species (ROS) and nitrogen species (NOS) are two kinds of redox molecules that are active in plant responses to biotic or abiotic stresses. Among them, nitric oxide (NO) has been reported to have dual effects on PCD during plant stress responses. For example, a negative role of NO in resistance against Cd2+-induced PCD was found in tobacco Bright Yellow-2 (BY-2) cells [12] and yellow lupine plants [13]. Conversely, a positive role of NO in delaying gibberellin-induced PCD [14] and strengthening resistance to the pathogen-induced hypersensitive response (HR) [15] has also been reported.
S-nitrosylation, an important posttranslational modification in which a NO moiety covalently and reversibly binds to a cysteine thiol forms a nitrosothiol, has been investigated in plants over the past few years. S-nitrosylation is thought to account for the influence of NO on cellular signaling via redox-based biochemical regulation of signaling components [16]. S-nitrosylation can regulate the sequences of proteins involved in all major cellular activities [17]. Recently, investigations of S-nitrosylation in plants have focused mainly on the identification of numerous protein candidates via proteomic analyses, and biochemical and computer structural studies have shown that the mechanisms of S-nitrosylation impact the structures of those proteins and thus their function [18, 19]. S-nitrosylation may be responsible for the PCD process in plants by regulating the activities of enzymes that mediate cell death-related signals or by reconstructing the functional domains of some functional proteins in plants, resulting in the PCD process [20, 21]. Reviews about the various test methods of S-nitrosylation in plants and the effects of S-nitrosylation on plant growth, development and resistance to stresses have been published. However, to date, no reviews have highlighted the roles of protein S-nitrosylation in PCD regulation in plants. Thus, in this review, we summarized the S-nitrosylated proteins that are involved in metabolism and photosynthesis, salicylic acid (SA) signaling, ROS-dependent signaling and other pathways in the PCD process, which provides insight into how S-nitrosylation regulates plant PCD.
ROS-related proteins
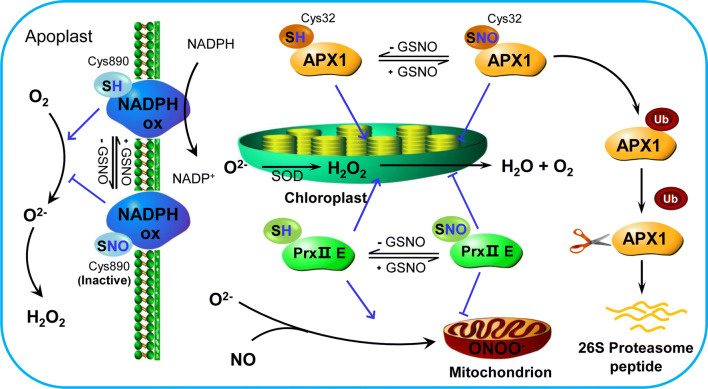
In plants, ROS and NO have been proposed to be the key factors involved in the signaling of PCD induced during the HR, senescence, self-incompatibility, and the response to metal and heat stress (HS) [22–24]. In the PCD process, NO interacts with ROS to influence redox homeostasis. For example, NO regulates ROS levels by inhibiting NADPH oxidase via S-nitrosylation [20]. Similarly, NO affects ROS levels via S-nitrosylation of antioxidant enzymes such as ascorbate peroxidase (APX) and peroxiredoxins (Prxs) in PCD (Fig. 1) and protects plants from various biotic and abiotic stresses [1]. Therefore, S-nitrosylation of ROS-related proteins is one of the important ways by which plants regulate PCD.
Fig. 1.
S-nitrosylation of ROS-related proteins involved in PCD regulation. NADPH oxidase can synthesize ROS, while S-nitrosylation at Cys-890 inactivates NADPH ability, inhibiting ROS production. S-nitrosylation of two antioxidant enzymes, APX1 and PrxII E, also affect ROS accumulation. S-nitrosylation of APX1 at Cys-32 regulates APX1 activity, leading to APX1 ubiquitination and therefore leading to degradation. S-nitrosylation of PrxII E not only inhibited its activity of H2O2-reducing peroxidase but also reduced its ability to detoxify ONOO−, leading to the accumulation of toxic O2− and consequently causing PCD after Pst infection. NADPH ox respiratory burst oxidase homolog, O2 oxygen, O2− superoxide anion, H2O2 hydrogen peroxide, APX1 ascorbate peroxidase 1, SOD superoxide dismutase, PrxII E peroxiredoxin II E, Ub ubiquitination
Ascorbate peroxidase (APX)
APX is one of the key factors in the ascorbate–glutathione (GSH) cycle and regulates the level of cellular hydrogen peroxide (H2O2), which is distributed throughout all cell compartments in higher plants [25]. APX catalyzes the electron transfer from ascorbate to H2O2, which produces dehydroascorbate and water, to scavenge H2O2 [26, 27]. Exogenous NO could inhibit the activities of APX in tobacco suspension cells, although the activity significantly recovered when NO was removed, suggesting that NO might participate in defense responses by affecting APX activity under pathogen attack [28]. Lin et al. also proved that NO and protein S-nitrosylation were integral to H2O2-induced leaf cell death in rice (Oryza sativa) NO excess1 (noe1) plants [29]. In HS- and H2O2-induced PCD of tobacco BY-2 cells, cytosolic ascorbate peroxidase (cAPX) was found to be S-nitrosylated [21]. In vivo and in vitro experiments showed that cAPX S-nitrosylation could be responsible for the rapid decrease in cAPX activity, and S-nitrosylated cAPX1 caused the ubiquitination of cAPX1, resulting in degradation [21] (Fig. 1). Thus, APX S-nitrosylation may be a potential way by which plants regulate PCD.
APX is a potential target of endogenous S-nitrosylation in Arabidopsis [30] (Table 1). The same result in Arabidopsis roots was reported by Correa-Aragunde et al., which was further verified in vitro via an APX recombinant [30]. S-nitrosylated recombinant APX1 caused an increase in its own activity, which contrasts with the results of De Pinto et al., who reported that APX S-nitrosylation inhibited the enzyme’s activity in the PCD process [21]. Despite this contradiction, these two experiments revealed that APX S-nitrosylation could indeed regulate the activity of APX. Furthermore, multiple alignments of the amino acid sequences and the model structure prediction of Arabidopsis APX1 showed that, among the five cysteine residues present in Arabidopsis APX1, Cys-32 and Cys-168, two residues mostly conserved in plants, were candidate sites of S-nitrosylation [31]. Yang et al. reported that Arabidopsis APX1 was S-nitrosylated efficiently in vitro under normal growth conditions [32]. The results of liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analyses and 2,3-diaminonaphthalene (DAN) assays suggested that Cys-32 and Cys-49 were the S-nitrosylated residues in APX1, which was further verified in APX1C32S and APX1C49S mutants. Structure modeling analysis also revealed that both the locations and the surrounding environment of Cys-32 and Cys-49 were largely in line with the consensus motif of S-nitrosylation. However, the authors further confirmed that the enzymatic activity of the Cys-49 recombinant was similar to that of the wild-type; this result differed from that of Cys-32, which increased APX1 activity in the presence of with S-nitrosoglutathione (GSNO), indicating that Cys-49 might not play a role in regulating APX1 activity [32]. Thus, S-nitrosylation of APX1 at Cys-32 can positively regulate APX1 activity, thereby regulating the immune response in plants. With respect to recombinant pea cAPX, APX1 was S-nitrosylated at Cys-32, and APX S-nitrosylation increased under saline conditions [33], suggesting that APX S-nitrosylation could contribute to alleviating oxidative damage induced by salinity stress.
Table 1.
List of S-nitrosylated proteins involved in programmed cell death in plants
| Protein name | Plant species | Tissue types | Effect of S-nitrosylation | References | |
|---|---|---|---|---|---|
| Ascorbate peroxidase 1 | APX1 | N. tabacum | BY-2 cells | Inhibition of activity and ubiquitination | [15] |
| A. thaliana | Seedlings | Activation of activity | [25, 26] | ||
| NADPH oxidases | RBOHD | A. thaliana | Leaves | Inhibition of activity | [14] |
| Peroxiredoxins | PrxII E | A. thaliana | Seedlings | Inhibition of activity | [35] |
| PrxII F | P. sativum L. | Seedlings | Inhibition of activity and conformational changes | [36, 37] | |
| Salicylic acid-binding protein 3 | AtSABP3 | A. thaliana | Seedlings | Inhibition of activity and SA binding | [43] |
| Nonexpresser of Pathogenesis-Related Genes 1 | NPR1 | A. thaliana | Seedlings | Conformational changes | [46] |
| TGACG motif binding factor1 | TGA1 | A. thaliana | Leaves | Conformational and DNA/NPR1-binding behavior changes | [50] |
| Glyceraldehyde 3-phosphate dehydrogenase | GAPDH | A. thaliana | Suspension cells | Inhibition of activity | [59, 61] |
| N. tabacum | BY-2 cells | Inhibition of activity | [56] | ||
| Ribulose-1,5-bisphosphate carboxylase/oxygenase | Rubisco | B. juncea | Seedlings | Inhibition of activity | [63] |
| K. pinnata | Seedlings | Inhibition of activity | [53] | ||
| S-nitrosoglutathione reductase | GSNOR | A. thaliana | Seedlings | Selective autophagy | [73] |
| Metacaspase-9 | AtMC9 | A. thaliana | Seedlings | Inhibition of autoprocessing and activity | [80] |
N.tabacum, Nicotiana tabacum; A.thaliana, Arabidopsis thaliana; P. sativum L., pisum sativum L.; B.juncea, Brassica juncea; K.pinnata, Kalanchoe pinnata
The present study, therefore, provides new insight into the mechanism of the regulation of APX activity via S-nitrosylation mediated by NO-derived molecules. Interestingly, the effects of NO on APX activity are controversial. There are two reasons for this controversy. First, the S-nitrosylation of APX is reversible, whereas the tyrosine nitration of APX is irreversible, both of which lead to an inhibition of APX activity [33]. Second, other APX proteins (with the exception of APX1, which accounts for nearly 70% of APX activity) may either be regulated by NO or not, but this regulation is hard to detect since the altered activity is below the detection limit under assay conditions [32]. Overall, investigations on APX S-nitrosylation in plant immunity are still scarce, and the detailed molecular mechanisms of APX S-nitrosylation need to be further elucidated.
NADPH oxidases (RBOHs)
In plants, extracellular reactive oxygen intermediates (ROIs) can drive PCD and this phenomenon is correlated with stress responses—HR. ROIs derived from the oxidative bust in the HR are produced usually by plasma membrane NADPH oxidases [34]. NADPH oxidases are also considered as RBOHs. As the major producers of ROS, NADPH oxidases transfer electrons from cytoplasmic NADPH or NADH to oxygen (O2) to form superoxide anions (O2−), and O2− is then converted into H2O2 by superoxide dismutase [35] (Fig. 1). ROIs generated by NADPH oxidases can suppress the spread of cell death in Arabidopsis [36]. This suppression might be due to the antagonistic effect of ROIs on SA-dependent death-promoting signals, thereby suppressing cell death in cells surrounding sites of NADPH oxidase activation. S-nitrosylation is another way to regulate RBOHD activity during effector-triggered immunity [37]. High S-nitrosothiol (SNO) concentrations could limit the HR via S-nitrosylation of NADPH oxidase in Arabidopsis [37]. When leaves were exposed to GSNO and S-nitroso-l-cysteine (Cys-NO), the activity of NADPH oxidase was reduced, although this effect was absent in the presence of reduced GSH and dithiothreitol (DTT). NADPH oxidase activity was also significantly reduced in atgsnor1-3 and nox1 plants (atgsnor1-3 and nox1 had high SNO concentrations) that were challenged with the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 (avrB). AtRBOHD is S-nitrosylated in a GSNO or Cys-NO concentration-dependent manner. BST and computer model structure analysis revealed that Cys-890 in the C-terminal portion of AtRBOHD was the site of S-nitrosylation [20] (Fig. 1). AtRBOHD S-nitrosylation at Cys-890 impeded flavin adenine dinucleotide (FAD) binding, thereby inhibiting the activity of AtRBOHD. The biological consequence of AtRBOHD S-nitrosylation was the prevention of ROI synthesis, ultimately inhibiting PCD in response to pathogen induction [20].
As mentioned above, S-nitrosylation of NADPH oxidase might play an important role in stress responses in Arabidopsis. However, to date, the mechanism of NADPH oxidase S-nitrosylation has been reported only in the model plant Arabidopsis. More detailed mechanisms of NADPH oxidase S-nitrosylation in other kinds of plants need studied.
Peroxiredoxins (Prxs)
Prxs are peroxidases in all organisms and exhibit thiol-based catalytic activity. Prxs can be classified into six subfamilies according to their different structures. Among them, the Prx1, Prx5, Prx6, and PrxQ isoforms are present in various organelles in plants [38]. Prxs are distributed in specific subcellular compartments with different specific functions in controlling plant growth and development, cellular metabolism and defense signaling [39]. Prx5, the most diverse and widely distributed Prx subfamily in plants, includes PrxII E and PrxII F, which are located in chloroplasts and mitochondria, respectively [40]. PrxII E and PrxII F can be S-nitrosylated by GSNO under biotic and abiotic stress [41, 42] (Table 1). Romero-Puertas et al. demonstrated that S-nitrosylation of PrxII E could not only inhibit the activity of H2O2-reducing peroxidase but also reduce the enzyme’s ability to detoxify peroxynitrite (ONOO−), leading to the accumulation of toxic O2− and consequently causing PCD after Pst infection [41] (Fig. 1). In pea plants, the activity of PrxII F was similarly reduced because of its S-nitrosylation under salt stress [42, 43]. S-nitrosylation of PrxII F can alter the enzyme’s conformation, which favors the interaction between citrate synthase (CS) and PrxII F and prevents the thermal aggregation of CS, thereby preventing plants from oxidative and nitrosative stress [43] (Table 1). Overall, S-nitrosylation of PrxII E and PrxII F plays an important role in inducing plant PCD and impacting plants’ responses to various stresses.
Both NO and ROS are recognized as mediators or modulators of a wide range of cellular signaling transduction involved in plant PCD. The evidence presented above indicates that S-nitrosylation is a novel way for NO and ROS to regulate PCD. The effect of NO on the crucial components of the antioxidant defense system occurred mainly via the S-nitrosylation of some key ROS-related proteins (Fig. 1). Protein S-nitrosylation could inhibit or accelerate PCD by impacting the activities or interactions the interactions with other proteins (Table 1), thereby affecting plants’ tolerance or resistance to stress.
SA signaling-related proteins
Once challenged by pathogens, plant host cells recognize pathogen effectors, leading to the HR [44]. Localized PCD can induce the accumulation of SA, inducing systemic acquired resistance (SAR) to defend against disease [45]. SA also participates in the signaling in response to abiotic stresses. SNO levels can modulate the accumulation of SA [46]. Salicylic acid-binding protein 3 (SABP3), Nonexpresser of Pathogenesis-Related Genes 1 (NPR1) and TGACG motif binding factor 1 (TGA1) are three important proteins in the SA signaling pathway, and their S-nitrosylation modification also affects the SA signaling pathway during the PCD process (Table 1).
Salicylic acid-binding protein 3 (SABP3)
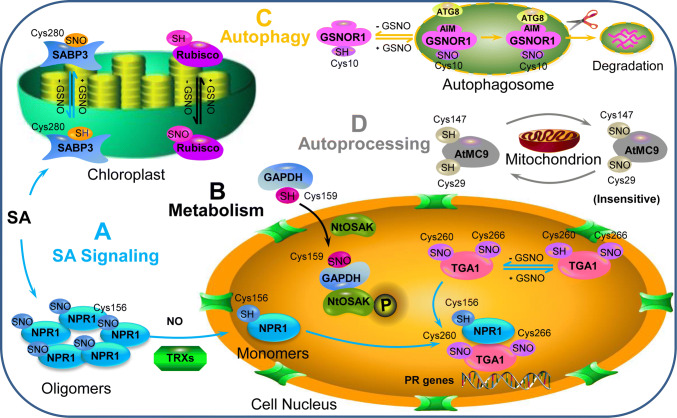
SA is a kind of plant hormone that acts as a key immune activator in defense response systems. SA in plants might bind a wide variety of proteins that are integral to immunity and subsequently modulate their activities [47, 48]. For example, salicylic acid-binding protein 3 (SABP3), a soluble protein molecule with CAT activity, plays a major role in plant immunity. SABP3 exhibited high affinity for SA and presented carbonic anhydrase (CA) activity in the tobacco chloroplast [47]. S-nitrosylation of SABP3 was related to the resistance of pathogen infection in Arabidopsis plants (Table 1). Wang et al. (2009) challenged Arabidopsis plants with Pst, which could be recognized by the R protein RPM1, and reported that AtSABP3 was S-nitrosylated in vivo. The same result was obtained in vitro in AtSABP3 recombinants that were incubated with various concentrations of GSNO [49]. Thus, AtSABP3 was S-nitrosylated in Arabidopsis both in vitro and in vivo. AtSABP3 model structure analysis and LC–MS/MS analysis of AtSABP3 S-biotinylated peptides showed that Cys-280 was the only site of S-nitrosylation of AtSABP3 (Fig. 2). S-nitrosylation of AtSABP3 decreased CA activity during the defense response. Furthermore, [14C]SA binding suggested that SNO-AtSABP3 also significantly reduced SA binding (Table 1). Thus, S-nitrosylation of AtSABP3 at Cys-280 could inhibit the SA-binding ability and CA activity of AtSABP3, thereby negatively modulating plant resistance to stress.
Fig. 2.
S-nitrosylation of SA signaling-related and the metabolism- and photosynthesis -related proteins GSNOR and MC9 involved in PCD regulation. aS-nitrosylation regulates SA signaling (lines in blue): SABP3 can be S-nitrosylated at its Cys-280 during the SA-related defense response. Under normal conditions, S-nitrosylation is beneficial for NPR1 in an oligomeric state in the cytosol. The oligomer can be reduced to monomers by TRXs in a NO-dependent manner and then translocated to the nucleus; S-nitrosylation of TGA1 at two Cys sites results in the formation of the NPR1/TGA system, regulating the expression of downstream PR genes. bS-nitrosylation of two metabolism-related proteins (lines in black): Rubisco and GAPDH can be S-nitrosylated, and S-nitrosylated GAPDH can interact with NtOSAK to respond to stress. c GSNOR S-nitrosylation leads to GSNOR degradation via selective autophagy (lines in orange); dS-nitrosylation of MC9 (lines in gray). SA salicylic acid, SABP3 salicylic acid-binding protein 3, GSNO S-nitrosoglutathione, NPR1 nonexpresser of pathogenesis-related Genes 1, NO nitric oxide, TRXs thioredoxins, TGA1 TGACG motif binding factor1, Rubisco ribulose-1,5-bisphosphate carboxylase/oxygenase, GAPDH glyceraldehyde 3-phosphate dehydrogenase, NtOSAK Nicotiana tabacum osmotic stress-activated protein kinase, GSNOR1 S-nitrosoglutathione reductase 1, ATG8 AUTOPHAGY-RELATED8, AIM ATG8-interacting motif, AtMC9 A. thaliana metacaspase 9
Nonexpresser of Pathogenesis-Related Genes 1 (NPR1)
Nonexpresser of Pathogenesis-Related Genes 1 (NPR1) is another SA-related protein that directly binds SA and activates SA-dependent genes during various immune responses [50]. In unchallenged plants, NPR1 is inactivated and localized to the cytosol as an oligomer formed through intermolecular disulfide bonds (Fig. 2). Once cells were induced by pathogens, the cellular redox state changed, and SA accumulation increased, reducing the oligomeric form of NPR1 to a monomeric form. The monomers then translocated to the nucleus, where they could regulate a group of expressed disease resistance genes [51]. The dynamic equilibrium of NPR1 between oligomers and monomers clearly plays a key role in modulating target gene transcription. S-nitrosylation has been reported to regulate the homeostasis of NPR1 [52] (Table 1). When extracted proteins were pretreated with GSNO, accompanied by an increase in the oligomer, the levels of the monomer decreased, while the total NPR1 levels remained unaffected. This phenomenon indicated that GSNO might impact the conformation of NPR1. BST analysis revealed that GSNO treatment caused NPR1 S-nitrosylation, indicating that it might be S-nitrosylation which affects the NPR1 conformation. In addition, the expression of SA-dependent defense genes was suppressed in atgsnor1-3, suggesting that GSNO could also impact the activity of NPR1 in innate immunity. S-nitrosylation and SA-induced oligomerization were abolished when Cys-156 was mutated [52]. These results were consistent with the results of computational analysis, suggesting that NPR1 is S-nitrosylated at Cys-156. Thus, the S-nitrosylation of NPR1 could facilitate its oligomerization. Moreover, Tada et al. demonstrated that thioredoxins (TRXs) could catalyze the SA-induced NPR1 transformation from an oligomeric to a monomeric form [52] (Fig. 2). Taken together, the above results provide a molecular mechanism of cellular redox changes in NPR1 after pathogen challenge: S-nitrosylation facilitates oligomer formation, while TRXs catalyze monomer release.
TGACG motif binding factor1 (TGA1)
As mentioned above, monomeric NPR1 is translocated from the cytosol to the nucleus and interacts with the reduced form of TGACG motif binding factor1 (TGA1), which increases DNA-binding activity and promotes the expression of PR genes and defense [53, 54] (Fig. 2). Lindermayr et al. used electrophoretic mobility shift assays (EMSAs) to show that GSNO could enhance the DNA-binding activity of TGA1 [55]. A similar result was obtained in the presence of NPR1. In addition, NPR1 and TGA1 were detected to be S-nitrosylated in Arabidopsis. Among them, TGA1 was S-nitrosylated at Cys-260 and Cys-266 in vitro. Thus, S-nitrosylation of TGA and NPR1 might influence DNA binding (Table 1). Interestingly, Lindermayr et al. also reported that the translocation of NPR1 into the nucleus was NO dependent (Fig. 2), which indicated that NO might promote NPR1 translocation [55]. Consequently, it can be speculated that the promotion of GSNO/NO to DNA binding might be due to, on the one hand, GSNO treatment changing the conformation of TGA and/or NPR1, leading to a more effective TGA-NPR1 interaction for DNA binding of TGA; on the other hand, S-nitrosylation of TGA1 might protect TGA1 from the state of disulfide bonds, from which low activity of DNA binding would result [56]. Overall, NO might play an important role in regulating the defense responses of the NPR1/TGA system in plants. However, the contradiction of GSNO/NO with respect to regulating PR gene expression is up-regulated by the NPR1/TGA system but down-regulated when NPR1 oligomers are favored. Neither mechanism has been defined in the physiological context, and the detailed time points of this defense process action have not been pointed out. In this scenario, these conflicting views require further analysis. Wendehenne et al. proposed that NO-induced events might have a temporal hierarchy: NO might not cause NPR1 to be in oligomeric form to inhibit PR genes expressed during the SAR process but rather might regulate the NPR1/TGA cascade at special time points [57]. Under these conditions, S-nitrosylation could be considered a negative feedback loop in controlling SAR.
SABP3, NPR1 and TGA1 are key proteins related to SA, which could accept and translate SA signaling to activate the expression of defense genes. S-nitrosylation of SABP3, NPR1 and TGA1 could change the original ability of these proteins to withstand adversity stress. Among them, S-nitrosylation of SABP3 and NPR1 inhibited the enzymes’ activity and ability, while S-nitrosylation of TGA1 promoted its ability to bind DNA (Table 1). Thus, it is clear that S-nitrosylation is an important way for NO to be involved in hormone signaling in plants.
Metabolism- and photosynthesis-related proteins
The metabolism- and photosynthesis-related enzymes in plants play an important role in responding to biotic and abiotic stresses. Pathogen treatment induced the production of NO and cell death in Arabidopsis and 11 mitochondrial proteins were identified as targets for S-nitrosylation during that process [58]. S-nitrosylation of metabolic and/or photosynthetic proteins in Kalanchoe pinnata [59], Brassica juncea [60], Pisum sativum L. [61] and Nicotiana tabacum [62] have been reported. Until now, proteins Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) have been well studied to be S-nitrosylated.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
GAPDH is one of the five known glycolysis enzymes related to the Calvin–Benson cycle and is sensitive to S-nitrosylation [63]. GAPDH contains a Cys residue in its reactive center that can be inhibited by NO. In Arabidopsis and tobacco, S-nitrosylation of the Cys residues of GAPDH could inhibit the activity of GAPDH [62, 64]. Holtgrefe et al. reported that GAPDH, which has multiple functions as a glycolytic enzyme, could also repair DNA and work as a DNA-binding protein in Arabidopsis [65]. In addition, S-nitrosylation of GAPDH at Cys-159 inactivated GAPDH activity under oxidizing conditions (Table 1), suggesting that the modification of essential Cys residues was a way to transiently protect enzymes involved in metabolism from oxidative damage [65]. In rats, S-nitrosylation of GAPDH could initiate the enzyme’s interaction with the E3 ligase Siah1 and mediate nuclear translocation, thereby resulting in ubiquitin-mediated degradation of nuclear proteins [66]. A similar function of GAPDH has also been reported in plants. In salt-treated cells of tobacco, GAPDH and the osmotic stress-activated protein kinase (NtOSAK) form an immunocomplex, which is related to GAPDH’s ability to translocate to the nucleus and bind DNA (Fig. 2) [62]. GAPDH showed increased and transient S-nitrosylation in response to salt stress, and NtOSAK was activated simultaneously, suggesting that S-nitrosylation of GAPDH might respond to salt-induced osmotic stress and impact NtOSAK [62]. Additionally, the inactivation of GAPDH caused by S-nitrosylation was also discovered in Arabidopsis under H2O2 treatment [67]. This approach represents another way for GAPDH to exercise its function in plants.
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco)
The Cys residue of Rubisco, which plays a central role in photosynthesis, can bind NO, thereby regulating the activity and degradation of the molecule [68] (Fig. 2). This was explained later via both the large and small subunits of Rubisco being identified as S-nitrosylated in Arabidopsis and in Kalanchoe pinnata [59, 69] (Table 1). S-nitrosylation of Rubisco inhibited the enzyme’s activity in a NO-dependent manner [60]. In addition, the action of Rubisco S-nitrosylation upon pathogen infection suggested that there might be an association between Rubisco S-nitrosylation and Rubisco activity during the defense response [70].
S-nitrosoglutathione reductase (GSNOR)
The enzyme GSNOR, which belongs to the class III alcohol dehydrogenase family, is highly conserved and prevalent in Arabidopsis [71], tomato [72], pepper [73], and poplar [74]. GSNOR works as a mobile reservoir of NO to affect GSNO homeostasis between S-nitrosylated proteins and GSNO [75, 76]. During pepper fruit ripening, GSNOR activity diminished, while the content of S-nitrosylated proteins simultaneously increased, suggesting that GSNOR activity was directly correlated with total SNO levels [77]. In addition, GSNOR1 regulated the S-nitrosylation extent of NPR1 and SABP3 in Arabidopsis, thus regulating the disease resistance that depends SA signaling [43, 46]. Therefore, GSNOR plays a critical role in regulating the defense response in biotic stress and abiotic stress by regulating intercellular SNO and NO levels.
GSNOR1 itself could also be S-nitrosylated in a NO-dependent way in plants [78]. S-nitrosylation of GSNOR1 at Cys-10 induced by GSNO could lead to the destabilization of GSNOR1 [72]. GSNOR1, which contains a highly conserved AIM-like motif and has a long half-life, is degraded via autophagy [79]. A key step of phagophore formation is the conjugation of ATG8 with ATG8-interacting proteins [80]. Additionally, specific intermolecular β-sheets, which usually contain an AIM motif, mediate selectivity autophagy [81]. S-nitrosylation of GSNOR1 at Cys-10 increased the exposure of AIM to the surface and allowed the AIM motif to interact with ATG8, thereby facilitating the degradation of GSNOR1 by selective autophagy (Fig. 2). Physiologically, the ability of NO to increase tolerance of Arabidopsis to low-O2 stress during seed germination might be due to the S-nitrosylation of GSNOR1-induced autophagic degradation under hypoxic conditions [79]. Hence, S-nitrosylation of GSNOR1 at Cys-10 plays an important role in facilitating GSNOR1’s selective autophagy (Table 1), thereby increasing the tolerance to low-O2 stress.
Metacaspase 9 (MC9)
Metacaspase, a member of the cysteine protease family, has been demonstrated to be the ancestor of metazoan caspases and plays an important role in plant PCD [82]. Metacaspase, which has specific proteolytic activity, can balance differentiation and cell death during the embryogenesis process in Norway spruce [83]. In animals, caspases inhibit their autoprocessing activity via the S-nitrosylation of cysteine, which is located at the active site under normal conditions [84]. The structure of metacaspase in plants was predicted to be similar to that of caspases in animals, and its acid–base motif favored the S-nitrosylation of its cysteine [85]. These findings prompted Belenghi et al. to investigate the S-nitrosylation of metacaspase and its function in PCD in Arabidopsis (Table 1). In vitro and in vivo experiments showed that A.thaliana metacaspase 9 (AtMC9) was S-nitrosylated at Cys-147, which delayed the autoprocessing of AtMC9 and suppressed proteolytic activity [86] (Fig. 2). However, during the maturation process of Arabidopsis, single-nucleotide polymorphism (SNP) and GSNO treatments caused a twofold decrease in VRPRase activity. Recovery of VRPRase activity after DTT addition indicated that there were other specific cysteine residues within AtMC9. AtMC9 has another highly conserved cysteine residue, Cys-29. This Cys was located inside the catalytic groove and close to Cys-147 but was insensitive to S-nitrosylation (Fig. 2). Single and double Cys mutants of AtMC9 showed that at least one Cys was needed for AtMC9 activity. When treated with SNP, the activity of AtMC929A was highly inhibited [86]. Hence, Cys-29 could act as an alternative nucleophile for catalyzing the proteolytic reaction. Metacaspases in plants remained inactive via S-nitrosylation; moreover, they were insensitive to S-nitrosylation when the genuine catalytic center was replaced with a second Cys during maturation. Collectively, S-nitrosylation plays a crucial role in modifying the structure and function of metacaspases, although some Cys residues are more susceptible to this kind of modification.
Conclusions and future perspectives
PCD is a key event in plant growth and development as well as in stress responses. The PCD process triggered by a series of stimuli is always accompanied by a burst of nitrosation, resulting in a change in S-nitrosylated proteins to respond to stress signaling. Once a plant is subjected to biotic or abiotic stress, the number of S-nitrosylated proteins increases, impacting the PCD process positively or negatively.
In ROS-related proteins, the effects of S-nitrosylation on two antioxidant enzymes, APX1 and Prx, are different. S-nitrosylation of APX1 increases the enzyme’s antioxidative activity, while that of Prx is the opposite, thereby inhibiting or facilitating PCD, respectively. Facilitation of the HR by NO occurs because S-nitrosylation abolishes the ability of NADPH to synthesize ROIs. During SA signaling, S-nitrosylation of SABP3 reduces the activity of CA and the ability of SA binding, negatively regulating resistance to stress. S-nitrosylation of NPR1 sustains the protein in an oligomeric state, which is unfavorable for SA signaling. Conversely, the expression of PR genes, which are mediated by SA signaling, is promoted by S-nitrosylation of TGA1. In addition, S-nitrosylation of some proteins that are involved in metabolism has been reported. Among them, GAPDH S-nitrosylation occurs in response to osmotic stress, while the function of S-nitrosylation of Rubisco and the S-nitrosylation site is still unknown, warranting further attention. Interestingly, metacaspase has important roles in regulating PCD in plants. AtMC9 has two different cysteine residues whose sensitivities to NO differ. S-nitrosylation of Cys-147, which is sensitive to NO, delays the autoprocessing of AtMC9 and suppresses proteolytic activity, while S-nitrosylation at Cys-29 is not sensitive to NO. All S-nitrosylated proteins can be regulated by GSNOR, and GSNOR is the only protein that reportedly can balance the level of SNOs in plants. In fact, S-nitrosylation of GSNOR1 promotes the enzyme’s degradation and increases its ability to defend abiotic stress.
It is clear that S-nitrosylation has important roles in regulating PCD in plants. However, little is known about the potential roles of S-nitrosylation in regulating plant growth and development, aging and death, and resistance to stress. Thus, many studies need to be carried out to explore the S-nitrosylation molecular regulatory mechanism of PCD as well as stress resistance.
Funding
This work was supported by the National Key Research and Development Program (2018YFD1000800); the National Natural Science Foundation of China (nos. 31860568, 31560563 and 31160398); the Research Fund of Higher Education of Gansu, China (no. 2018C-14); the Post-Doctoral Foundation of China (nos. 20100470887 and 2012T50828) and the Natural Science Foundation of Gansu Province, China (nos. 1606RJZA073, 1606RJZA077 and 1606RJYA252).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Locato V, Paradiso A, Sabetta W, De Gara L, de Pinto MC. Advances in botanical research. New York: Academic; 2016. Nitric Oxide-biol Ch and reactive oxygen species in PCD signaling; pp. 165–192. [Google Scholar]
- 2.Van Doorn WG. Classes of programmed cell death in plants, compared to those in animals. J Exp Bot. 2011;62(14):4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- 3.Kwon SI, Cho HJ, Park OK. Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation. Autophagy. 2010;6(8):1187–1189. doi: 10.4161/auto.6.8.13429. [DOI] [PubMed] [Google Scholar]
- 4.Bagniewska-Zadworna A, Byczyk J, Eissenstat DM, Oleksyn J, Zadworny M. Avoiding transport bottlenecks in an expanding root system: xylem vessel development in fibrous and pioneer roots under field conditions. Am J Bot. 2012;99(9):1417–1426. doi: 10.3732/ajb.1100552. [DOI] [PubMed] [Google Scholar]
- 5.Bagniewska-Zadworna A, Arasimowicz-Jelonek M, Smoliński DJ, Stelmasik A. New insights into pioneer root xylem development: evidence obtained from Populus trichocarpa plants grown under field conditions. Ann Bot-London. 2014;113(7):1235–1247. doi: 10.1093/aob/mcu063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fath A, Bethke P, Lonsdale J, Meza-Romero R, Jones R. Programmed cell death in cereal aleurone. Plant Mol Biol. 2000;44(3):255–266. doi: 10.1023/A:1026584207243. [DOI] [PubMed] [Google Scholar]
- 7.Lee IC, Hong SW, Whang SS, Lim PO, Nam HG, Koo JC. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011;52(4):651–662. doi: 10.1093/pcp/pcr026. [DOI] [PubMed] [Google Scholar]
- 8.Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Curá JA, Miralles DJ, Zhu T, Casal JJ. Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 2008;55(6):1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x. [DOI] [PubMed] [Google Scholar]
- 9.Mukhtar MS, McCormack ME, Argueso CT, Pajerowska-Mukhtar KM. Pathogen tactics to manipulate plant cell death. Curr Biol. 2016;26(13):R608–R619. doi: 10.1016/j.cub.2016.02.051. [DOI] [PubMed] [Google Scholar]
- 10.Huysmans M, Lema S, Coll NS, Nowack MK. Dying two deaths-programmed cell death regulation in development and disease. Curr Opin Plant Biol. 2017;35:37–44. doi: 10.1016/j.pbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Bioch. 2010;48(12):909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Ma W, Xu W, Xu H, Chen Y, He Z, Ma M. Nitric oxide modulates cadmium influx during cadmium-induced programmed cell death in tobacco BY-2 cells. Planta. 2010;232(2):325–335. doi: 10.1007/s00425-010-1177-y. [DOI] [PubMed] [Google Scholar]
- 13.Arasimowicz-Jelonek M, Floryszak-Wieczorek J, Deckert J, Rucińska-Sobkowiak R, Gzyl J, Pawlak-Sprada S, Abramowski D, Jelonek T, Gwóźdź EA. Nitric oxide implication in cadmium-induced programmed cell death in roots and signaling response of yellow lupine plants. Plant Physiol Biochem. 2012;58:124–134. doi: 10.1016/j.plaphy.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 2002;129(4):1642–1650. doi: 10.1104/pp.002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaninotto F, La Camera S, Polverari A, Delledonne M. Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 2006;141(2):379–383. doi: 10.1104/pp.106.078857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta KJ. Protein S-nitrosylation in plants: photorespiratory metabolism and NO signaling. Sci Signal. 2011;4(154):jc1. doi: 10.1126/scisignal.2001404. [DOI] [PubMed] [Google Scholar]
- 17.Hess DT, Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem. 2012;287(7):4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astier J, Kulik A, Koen E, Besson-Bard A, Bourque S, Jeandroz S, et al. Protein S-nitrosylation: what’s going on in plants? Free Radic Bio Med. 2012;53(5):1101–1110. doi: 10.1016/j.freeradbiomed.2012.06.032. [DOI] [PubMed] [Google Scholar]
- 19.Wolhuter K, Eaton P. How widespread is stable protein S-nitrosylation as an end-effector of protein regulation? Free Radic Bio MED. 2017;109:156–166. doi: 10.1016/j.freeradbiomed.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478(7368):264. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]
- 21.De Pinto MC, Locato V, Sgobba A, del Carmen Romero-Puertas M, Gadadeta C, Delledonne M, De Gara L. S-Nitrosylation of ascorbate peroxidase is part of the programmed cell death signaling in tobacco By-2 cells. Plant Physiol. 2013;1:113. doi: 10.1104/pp.113.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrano I, Romero-Puertas MC, Sandalio LM, Olmedilla A. The role of reactive oxygen species and Nitric Oxide-biol Ch in programmed cell death associated with self-incompatibility. J Exp Bot. 2015;66(10):2869–2876. doi: 10.1093/jxb/erv083. [DOI] [PubMed] [Google Scholar]
- 23.De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, et al. Nitric Oxide-biol Ch is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 2009;150(1):217–228. doi: 10.1104/pp.108.133397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10(3):310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot. 2002;53(372):1305–1319. doi: 10.1093/jexbot/53.372.1305. [DOI] [PubMed] [Google Scholar]
- 26.Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Biol. 1998;49(1):249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 27.Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, et al. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17(1):268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark D, Durner J, Navarre DA, Klessig DF. Nitric Oxide-biol Ch inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant Microbe In. 2000;13(12):1380–1384. doi: 10.1094/MPMI.2000.13.12.1380. [DOI] [PubMed] [Google Scholar]
- 29.Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, et al. Nitric oxide-biol Ch and protein S-nitrosylation are integral to hydrogen peroxide induced leaf cell death in rice. Plant Physiol. 2011;32:111. doi: 10.1104/pp.111.184531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fares A, Rossignol M, Peltier JB. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Bioph Res Co. 2011;416(3):331–336. doi: 10.1016/j.bbrc.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 31.Correa-Aragunde N, Foresi N, Delledonne M, Lamattina L. Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J Exp Bot. 2013;64(11):3339–3349. doi: 10.1093/jxb/ert172. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Mu J, Chen L, Feng J, Hu J, Li L, et al. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015;1:114. doi: 10.1104/pp.114.255216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, et al. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J Exp Bot. 2013;65(2):527–538. doi: 10.1093/jxb/ert396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant JJ, Loake GJ. Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 2000;124(1):21–30. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino D, Dunand C, Puppo A, Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17(1):9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Torres MA, Jones JD, Dangl JL. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet. 2005;37(10):1130. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- 37.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56(8):1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- 38.Pedrajas JR, Bárcena JA. Antioxidants and antioxidant enzymes in higher plants. Cham: Springer; 2018. Peroxiredoxins: types, characteristics and functions in higher plants; pp. 95–121. [Google Scholar]
- 39.Lee ES, Kang CH, Park JH, Lee SY. Physiological significance of plant peroxiredoxins and the structure-related and multifunctional biochemistry of peroxiredoxin 1. Antioxid Redox Sign. 2018;28(7):625–639. doi: 10.1089/ars.2017.7400. [DOI] [PubMed] [Google Scholar]
- 40.Bréhélin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y. Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol. 2003;132(4):2045–2057. doi: 10.1104/pp.103.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero-Puertas MC, Laxa M, Matte A, Zaninotto F, Finkemeier I, Jones AM, et al. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell. 2007;19(12):4120–4130. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camejo D, del Carmen Romero-Puertas M, Rodríguez-Serrano M, Sandalio LM, Lázaro JJ, Jiménez A, Sevilla F. Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J Proteom. 2013;79:87–99. doi: 10.1016/j.jprot.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Camejo D, Ortiz-Espín A, Lázaro JJ, Romero-Puertas MC, Lázaro-Payo A, Sevilla F, Jiménez A. Functional and structural changes in plant mitochondrial PrxII F caused by NO. J Proteom. 2015;119:112–125. doi: 10.1016/j.jprot.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 44.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 45.Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8(10):1773. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Yun BW, Spoel SH, Loake GJ. A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr Opin Plant Biol. 2012;15(4):424–430. doi: 10.1016/j.pbi.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA. 2002;99(18):11640–11645. doi: 10.1073/pnas.182427699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar D, Klessig DF. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc Natl Acad Sci USA. 2003;100(26):16101–16106. doi: 10.1073/pnas.0307162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, et al. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem. 2009;284(4):2131–2137. doi: 10.1074/jbc.M806782200. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1(6):639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113(7):935–944. doi: 10.1016/S0092-8674(03)00429-X. [DOI] [PubMed] [Google Scholar]
- 52.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, et al. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321(5891):952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe In. 2000;13(2):191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- 54.Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15(9):2181–2191. doi: 10.1105/tpc.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindermayr C, Sell S, Müller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by Nitric Oxide-biol Ch. Plant Cell. 2010;22(8):2894–2897. doi: 10.1105/tpc.109.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, et al. S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci. 2011;181(5):527–533. doi: 10.1016/j.plantsci.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 57.Wendehenne D, Gao QM, Kachroo A, Kachroo P. Free radical-mediated systemic immunity in plants. Curr Opin Plant Biol. 2014;20:127–134. doi: 10.1016/j.pbi.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Palmieri MC, Lindermayr C, Bauwe H, Steinhauser C, Durner J. Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiol. 2010;152(3):1514–1528. doi: 10.1104/pp.109.152579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abat JK, Mattoo AK, Deswal R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata-ribulose-1, 5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008;275(11):2862–2872. doi: 10.1111/j.1742-4658.2008.06425.x. [DOI] [PubMed] [Google Scholar]
- 60.Abat JK, Deswal R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics. 2009;9(18):4368–4380. doi: 10.1002/pmic.200800985. [DOI] [PubMed] [Google Scholar]
- 61.Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC. S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J Exp Botany. 2012;63(5):2089–2103. doi: 10.1093/jxb/err414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wawer I, Bucholc M, Astier J, Anielska-Mazur A, Dahan J, Kulik A, et al. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by Nitric Oxide-biol Ch in response to salinity. Biochem J. 2010;429(1):73–83. doi: 10.1042/BJ20100492. [DOI] [PubMed] [Google Scholar]
- 63.Grennan AK. Protein S-nitrosylation: potential targets and roles in signal transduction. Plant Physiol. 2007;144(3):1237–1239. doi: 10.1104/pp.104.900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005;137(3):921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, et al. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant. 2008;133(2):211–228. doi: 10.1111/j.1399-3054.2008.01066.x. [DOI] [PubMed] [Google Scholar]
- 66.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7(7):665. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 67.Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, et al. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem. 2005;43(9):828–835. doi: 10.1016/j.plaphy.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 68.Marcus Y, Altman-Gueta H, Finkler A, Gurevitz M. Dual role of cysteine 172 in redox regulation of ribulose 1, 5-bisphosphate carboxylase/oxygenase activity and degradation. J Bacteriol. 2003;185(5):1509–1517. doi: 10.1128/JB.185.5.1509-1517.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romero-Puertas MC, Campostrini N, Mattè A, Righetti PG, Perazzolli M, Zolla L, et al. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8(7):1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- 70.Maldonado-Alconada AM, Echevarría-Zomeño S, Lindermayr C, Redondo-López I, Durner J, Jorrín-Novo JV. Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae. Acta Physiol Plant. 2011;33(4):1493–1514. doi: 10.1007/s11738-010-0688-2. [DOI] [Google Scholar]
- 71.Chen R, Sun S, Wang C, Li Y, Liang Y, An F, et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19(12):1377. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 72.Gong B, Wen D, Wang X, Wei M, Yang F, Li Y, Shi Q. S-nitrosoglutathione reductase-modulated redox signaling controls sodic alkaline stress responses in Solanum lycopersicum L. Plant Cell Physiol. 2014;56(4):790–802. doi: 10.1093/pcp/pcv007. [DOI] [PubMed] [Google Scholar]
- 73.Rodríguez-Ruiz M, Mioto P, Palma JM, Corpas FJ. S-nitrosoglutathione reductase (GSNOR) activity is down-regulated during pepper (Capsicum annuum L.) fruit ripening. Nitric Oxide-biol Ch-biol Ch. 2017;68:51–55. doi: 10.1016/j.niox.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 74.Cheng T, Shi J, Dong Y, Ma Y, Peng Y, Hu X, Chen J. Hydrogen sulfide enhances poplar tolerance to high-temperature stress by increasing S-nitrosoglutathione reductase (GSNOR) activity and reducing reactive oxygen/nitrogen damage. Plant Growth Regul. 2018;84(1):11–23. doi: 10.1007/s10725-017-0316-x. [DOI] [Google Scholar]
- 75.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410(6827):490. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 76.Espunya MC, Díaz M, Moreno-Romero J, MartÍnez MC. Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ. 2006;29(5):1002–1011. doi: 10.1111/j.1365-3040.2006.01497.x. [DOI] [PubMed] [Google Scholar]
- 77.Rodríguez-Ruiz M, Mioto P, Palma JM, Corpas FJ. S-nitrosoglutathione reductase (GSNOR) activity is down-regulated during pepper (Capsicum annuum L.) fruit ripening. Nitric Oxide-biol Ch. 2017;68:51–55. doi: 10.1016/j.niox.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 78.He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, et al. Nitric Oxide-biol Ch represses the Arabidopsis floral transition. Science. 2004;305(5692):1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 79.Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, et al. S-Nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell. 2018;71(1):142–154. doi: 10.1016/j.molcel.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 80.Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17(9):526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 81.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584(7):1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 82.Uren AG, O’Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6(4):961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 83.Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Rodriguez-Nieto S, et al. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc Natl Acad Sci USA. 2005;102(40):14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154(6):1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brune B, Mohr S. Protein thiol modification of glyceraldehyde-3-phosphate dehydrogenase and caspase-3 by Nitric Oxide-biol Ch. Curr Protein Pept Sc. 2001;2(1):61–72. doi: 10.2174/1389203013381206. [DOI] [PubMed] [Google Scholar]
- 86.Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inzé D, Delledonne M, Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem. 2007;282(2):1352–1358. doi: 10.1074/jbc.M608931200. [DOI] [PubMed] [Google Scholar]