Abstract
Protein kinase C ε (PKCε) has emerged as an oncogenic protein kinase and plays important roles in cancer cell survival, proliferation, and invasion. It is, however, still unknown whether PKCε affects cell proliferation via glucose metabolism in cancer cells. Here we report a novel function of PKCε that provides growth advantages for cancer cells by enhancing tumor cells glycolysis. We found that either PKCε or Smad2/3 promoted aerobic glycolysis, expression of the glycolytic genes encoding HIF-1α, HKII, PFKP and MCT4, and tumor cell proliferation, while overexpression of PKCε or Smad3 enhanced aerobic glycolysis and cell proliferation in a protein kinase D- or TGF-β-independent manner in PC-3M and DU145 prostate cancer cells. The effects of PKCε silencing were reversed by ectopic expression of Smad3. PKCε or Smad3 ectopic expression-induced increase in cell growth was antagonized by inhibition of lactate transportation. Furthermore, interaction of endogenous PKCε with Smad2/3 was primarily responsible for phosphorylation of Ser213 in the Samd3 linker region, and resulted in Smad3 binding to the promoter of the glycolytic genes, thereby promoting cell proliferation. Forced expression of mutant Smad3 (S213A) attenuated PKCε-stimulated protein overexpression of the glycolytic genes. Thus, our results demonstrate a novel PKCε function that promotes cell growth in prostate cancer cells by increasing aerobic glycolysis through crosstalk between PKCε and Smad2/3.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2914-9) contains supplementary material, which is available to authorized users.
Keywords: Protein kinase C, TGF-β signaling, Warburg effect, Tumor growth
Introduction
Many tumor cells adopt a metabolic phenotype characterized by high rates of glucose uptake and lactate production regardless of oxygen concentration, a phenomenon commonly referred to as the Warburg effect [1, 2]. Several lines of evidence demonstrated that oncogene activation or loss of tumor suppressor genes, such as mutations in Ras [3, 4], AKT [5], Myc [6], and p53 [7, 8], increase glucose uptake and lactate production, which provides several advantages for tumor cells, such as lower production of reactive oxygen species, protection from apoptosis and exertion of tumor drug resistance [9]. However, the underlying molecular mechanisms that connect oncogenes to metabolic pathways and tumor cell growth remain poorly defined. A better understanding of the interactions of oncogenes with glucose metabolism may shed light on new therapeutic strategies for tumor metabolic therapy.
Protein kinase C (PKC) belongs to the family of serine/threonine kinases that regulate an adverse set of cellular processes including proliferation, apoptosis, cell survival and migration, and there is a substantial amount of evidence linking PKC to tumorigenesis [10]. There are at least eleven different isoforms of this family that are classified into three sub-families (classical, novel, and atypical). PKCε, an isoform belonging to the novel PKC sub-family, has been identified as a transforming oncogene; and regulates tumor occurrence [11–13], invasion [14], metastasis [15], proliferation [16], and survival [17, 18]. Moreover, PKCε is overexpressed in numerous cancers including colon, breast, stomach, prostate, thyroid and lung cancers, and considered as an important marker of negative disease outcome and a therapeutic target of cancer [10, 19, 20].
Mounting evidences have highlighted the engagement of PKCε with the progression of prostate cancer. PKCε is highly expressed in prostate cancer and in recurrent disease, whereas it is barely detectable in normal or benign human prostatic epithelium [15]. Ectopic expression of PKCε in androgen-dependent prostate cancer cells contributes to the acquisition of androgen independence [12, 21]. In transgenic mouse model, overexpression of PKCε in prostate leads to the formation of preneoplastic lesions [12, 22]. In contrast, genetic ablation of PKCε inhibits the development of prostate cancer and bone metastasis [15, 22]. However, the mechanisms underlying contribution of PKCε to tumor progression and metastasis remain unclear.
Members of the TGF-β family regulate a wide range of biological processes including cell proliferation, migration, differentiation, apoptosis, and extracellular matrix deposition [23]. Ligand binding to TGF-β receptors (TGF-βRI and TGF-βRII) initiates the formation of a Smad2/3/4 complex and its translocation to the nucleus (Smad pathway), and then regulates the transcription of target genes. The Smad pathway is essential for TGF-β-induced tumor suppression in epithelium and at the early stage of tumor progression. On the other hand, TGF-β elicits epithelial-to-mesenchymal transition (EMT), cell invasion and metastasis by interplay with other signaling pathways via both Smad-dependent and Smad-independent mechanisms [24, 25]. However, the mechanisms underlying cross-talk between the core TGF-β/Smad pathway and other signal cascades in promoting tumor progression remain largely obscure.
In this paper, we investigated impact of PKCε and Smad2/3 signaling on aerobic glycolysis and cell proliferation in prostate cancer cells. Our study reveals a novel molecular mechanism of PKCε in promoting cell growth by enhancing glycolysis through its crosstalk to Smad3 in the prostate cancer cells.
Materials and methods
Chemicals and reagents
PMA, EGF and reagents were provided by Promega (Madison, WI, USA) and Sigma (St. Louis, MO, USA), respectively. Lipofectamine3000 for siRNA transfection (Invitrogen), HilyMax for plasmid transfection (Dojindo, Kumamoto, Japan). All-in-One First-Strand cDNA Synthesis Kit, All-in-One qPCR Mix (GeneCopoeia, MD, USA), and the ChIP-IT Express Enzymatic Chromatin Immunoprecipitation Kit (Active Motif, CA, USA) were obtained commercially. The glycolysis sampler kit (Cell Signaling Technology, MA, USA), PKCε and Smad2/3 primary antibodies (Santa Cruz Biotechnology, CA, USA), Alexa 488- and 594-conjugated secondary antibodies (Molecular Probes, Invitrogen) and the antibodies targeting phosphorylated Smad3 at Ser213, Flag, α-tubulin along with all unconjugated secondary antibodies were also purchased commercially.
Cell culture and siRNA transfection
The prostate cancer cell lines PC-3M, PC-3 and DU145 were from American Type Culture Collection (ATCC) and cultured according to the manufacturer’s recommendations. The siRNAs and plasmids were transfected into cells using Lipofectamine3000 and Hilymax, respectively, following the manufacturer’s instructions.
Plasmid construction
Plasmid Flag-Smad3 and constitutively active form of PKCε (PKCε*) were provided generously by Prof. Mitsuyasu Katoa and Prof. Q. Jane Wang, respectively. The mutant Smad3-S213A vector was constructed by PCR mutagenesis using the following primers: 5′ CCGAATCCGATGGCCCCAGCACATAATAACTTGGACCT 3′ and 5′ AGGTCCAAGTTATTATGTGCTGGGGCCATCGGATTCGG 3′. One nucleotide of Smad3 was mutated from T to G for the Flag-Smad3 vector. Successful mutation was confirmed by DNA sequencing.
RNA oligonucleotides
si-PKCε, si-PKD1, si-PKD2, si-PKD3 si-Smad2, si-Smad3, si-HIF-1a and negative control (siCTL) were purchased from GenePharma (GenePharma, Suzhou, China), The siRNA sequences in the present study are shown in Supplementary Table S1.
Infection of lentivirus and generation of stable cell lines
Lentivirus vectors Lv-GFP (GFP) and Lv-GFP-PKCε (PKCε) were purchased from Genepharma (GenePharma, Suzhou, China) and used to infect into PC-3 cells, respectively. After 3 days infection, puromycin obtained from Sigma (St. Louis, MO, USA) was used to select stably infected cells for 7 days, PKCε expression at protein expression level was verified by Western blotting.
Lactate and glucose measurements
Glucose and lactate concentrations in culture media were determined as described previously [26]. Briefly, fresh medium was added to a 12-well plate of sub-confluent cells, and lactate and glucose concentrations in the media were measured after 30–60 min (Lactate Reagent Kit) or 6–24 h (BioProfile Analyzer) and the results were normalized to the number of cells each well.
Real-time quantitative PCR amplification
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, RT-qPCR was carried out using All-in-One First-Strand cDNA Synthesis Kit and All-in-One qPCR Mix (GeneCopoeia) according to the manufacturer’s protocol, respectively. The sequences of primers are listed in Supplementary Table S2.
Co-immunoprecipitation (Co-IP) and immunoblotting
Immunoprecipitation and immunoblotting were performed as described in our previous studies [27]. After washing with cold PBS twice, cells were lysed with IP lysis buffer [50 mM Tris–HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA-2Na; 10% TritonX-100; 0.5 mM Na4P2O7·10H2O; 1 mM C3H7Na2O6P·5(H2O); 1 mM Na3VO4] for 10 min on ice, subsequently, harvested and incubated with primary antibodies as indicated overnight at 4 °C, protein A/G was added into cell lysis for 1 h to precipitate the antibody combined with proteins. The complex was analyzed by Western blotting. Total protein was further subjected to SDS-PAGE, transferred to nitrocellulose filter membrane (NC), and probed with the corresponding antibodies. Overnight after incubation with primary antibodies at 4 °C, the membranes were washed with PBST (0.05%Tween-20) and incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. Proteins were detected by enhanced chemiluminescence substrates (Perkin Elmer).
Chromatin immunoprecipitation (ChIP)
ChIP assay were performed using the ChIP-IT Express Enzymatic Chromatin Immunoprecipitation Kit (Active Motif) according to the manufacturer’s protocol. RT-qPCR of co-immunoprecipitated target genomic DNA fragments was performed with promoter-specific primers listed in Supplementary Table S3. Smad7, (F: TAGAAACCCGATCTGTTGTTTGCG; R: CCTCTGCTCGGCTGGTTCCACTGC), was used as a positive control [28], cycling parameters for reactions were at 95 °C for 10 min, followed by 40 cycles at 95 °C for 20 s; 60 °C, 30 s; and at 72 °C for 30 s, fold enrichment in the bound fractions relative to input was calculated as described previously [27].
Immunofluorescence microscopy
PC-3M cells were incubated in 4% formaldehyde for 15 min at room temperature. For immunofluorescence, the primary antibodies were anti-PKCε and anti-Smad2/3. To detect nuclei, cells were co-stained with 4ʹ-6-diamidino-2-phenylindole (DAPI, Invitrogen). Fluorescence images were collected with a FluoView FV1000 confocal microscopy (Olympus).
Statistics
All statistical analyses were conducted using GraphPad Prism V software. A p value of less than 0.05 was considered statistically significant. Statistical differences between two groups with stimulation were determined by two-way ANOVA. One sample t test was performed to determine statistical differences between two groups in RT-qPCR analysis.
Results
Depletion of PKCε inhibits aerobic glycolysis and cell proliferation in prostate cancer cells
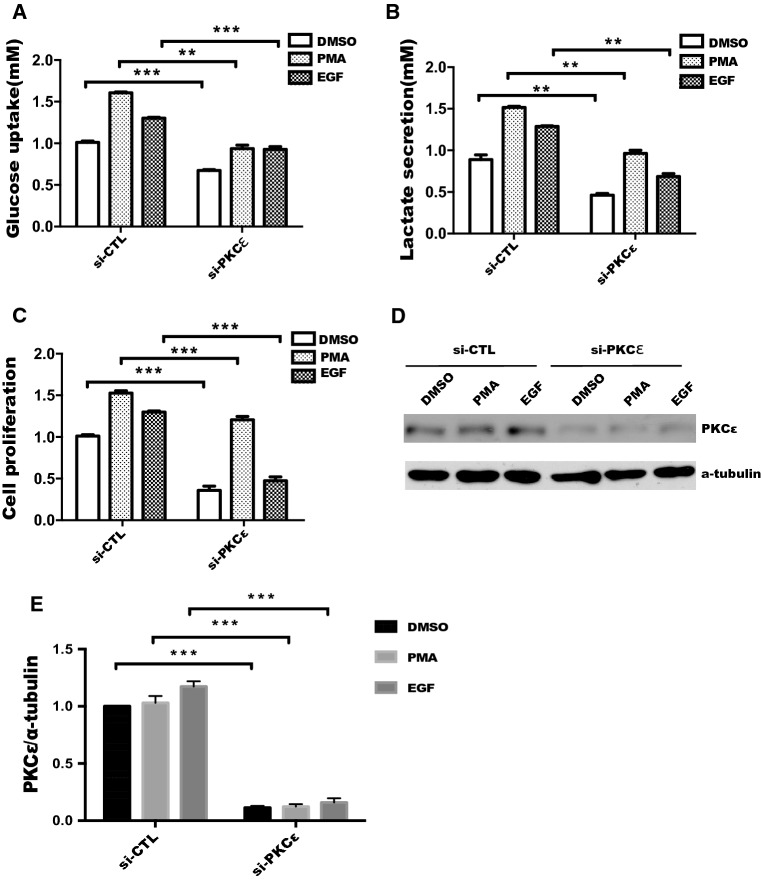
PKCε has been implicated in the development and progression of numerous cancers including prostate cancer, and plays an important role in tumor growth, invasion and metastasis [15, 29]. However, the contribution of PKCε to glycolysis in cancer cells remains unknown. In this context, we first measured the glucose consumption and lactate production in prostate cancer cells treated with PMA or EGF, known agonists of PKCε [30–32]. As shown in Fig. S1, the glucose consumption and lactate production in prostate cancer cells were significantly increased by PMA or EGF treatment, suggesting that activation of PKCε regulates glycolysis in prostate cancer cells. To evaluate the specific effect of PKCε on glycolysis, PKCε siRNA (si-PKCε) was transfected into DU145 cells. The results showed that glucose uptake and lactate production were significantly inhibited by PKCε silencing with or without PMA and EGF treatment compared with those in the siRNA control group (Fig. 1 a, b). Meanwhile, depletion of PKCε inhibited proliferation with or without PMA and EGF treatment (Fig. 1c). In addition, knockdown of endogenous PKCε was verified with or without PMA and EGF treatment (Fig. 1 d, e). These results suggested that activation of PKCε promotes cell proliferation by increasing aerobic glycolysis in prostate cancer cells.
Fig. 1.
Silencing of PKCε represses aerobic glycolysis and cell proliferation in prostate cancer cells. DU145 cells were transfected with siRNA control (siCTL) or siRNA-PKCε (si-PKCε) for 24 h, cells were starved in serum-free medium for an additional 18 h, followed by treatment with or without 100 nM PMA or 50 ng/mL EGF for 24 h, level of glucose uptake (a) and lactate production (b) were measured as described in “Materials and methods”. c Cell proliferation was performed by the CCK-8 assay after transfection and treated as described in a. Data represent the mean ± SEM of three independent experiments, and were analyzed by two-way ANOVA with multiple comparisons, followed by Bonferroni post hoc test for significance. Statistical significance, ***p < 0.001 and **p < 0.01. d Sufficient silencing of PKCε in DU145 cells was validated by Western blotting and quantified in e
PKCε upregulates glycolytic genes expression in a PKD-independent manner
To determine the effect of PKCε on glycolytic gene expression in prostate cancer, we transiently transfected DU145 and PC-3M cells with constitutive active mutant of PKCε (PKCε*). As shown in Fig. 2a, overexpression of PKCε* increased expression of glycolytic genes, including HIF-1α, HKII, PFKP and MCT4. In contrast, depletion of endogenous PKCε decreased the expression of the genes (Fig. 2b). As HIF-1α is involved in promoting vessel formation and enhancing tumor growth through the glycolytic pathway [33, 34], we further tested whether HIF-1α mediated PKCε-induced upregulation of glycolytic genes. As shown in Fig. 2c, HIF-1α silencing antagonized PKCε*-triggered expression of glycolytic genes. These results imply that PKCε promotes aerobic glycolysis through HIF-1α-mediated activation of glycolytic genes in prostate cancer cells.
Fig. 2.
PKCε promotes aerobic glycolysis and glycolytic proteins expression in a PKD-independent manner. DU145 and PC-3M cells were transiently transfected with constitutive active mutant of PKCε (PKCε*) plasmid (a) or with siRNA-PKCε (b), expression of glycolytic-related proteins as indicated was measured by Western blotting. c DU145 cells were transfected with pcDNA3.1, PKCε* plasmid or PKCε* plus siRNA-HIF-1α, and the total protein was detected by Western blotting. d DU145 cells were starved with serum-free medium for 18 h, then treated with the PKC inhibitor GÖ6983 or PKD inhibitor GÖ6976 for an additional 1 h, followed by incubation with or without 100 nM PMA, level of lactate secretion was analyzed as described in “Materials and methods”. e Lactate production (left) and glucose consumption (right) were analyzed in DU145 cells transfected by siCTL, si-PKD1, si-PKD2, and si-PKD3 with or without PMA treatment, as described in “Materials and methods”. f DU145 cells were transfected with siRNA targeting PKD1, PKD2 and PKD3. Western blotting was used to detect indicated protein expression. Data represent the mean ± SEM of three independent experiments and were analyzed by two-way ANOVA with multiple comparisons (PKD knockdown effect and PMA treatment effect), followed by Bonferroni post hoc test for significance. ***p < 0.001, **p < 0.01
Given that PKD is activated at the activation loop by PKCε and acts as an important downstream target in intact cells and cancer cells [29], we evaluated whether PKDs were involved in PKCε-mediated glycolysis in response to PMA treatment in prostate cancer cells. We found that GÖ6983, a PKC inhibitor, rather than GÖ6976, a PKD inhibitor, blocked lactate production induced by PMA treatment in DU145 (Fig. 2d) or PC-3M cells (Fig. S2). To further confirm that the specific PKD isoforms were not involved in PKCε-mediated tumor glycolysis, PKDs silencing (si-PKD1, si-PKD2, si-PKD3) were shown to have no effect on glucose uptake and lactate production with or without PMA treatment in DU145 cells (Fig. 2e). Similarly, expression of the glycolytic genes was not affected by knockdown of PKDs (Fig. 2f). Thus, these results suggest PKCε contributes to glycolysis in a PKD-independent manner in prostate cancer cells.
Smad2 and Smad3 promote aerobic glycolysis in a TGF-β independent manner
Mounting evidence has demonstrated aberrant TGF-β signaling pathway in human cancer. Prior to tumor initiation and early progression, TGF-β acts as a tumor suppressor; however, at later stages, it is often a tumor promoter [35]. Interestingly, it has been shown that Smad3 is overexpressed in surgical specimens of human prostate cancer, which correlates with expression of proliferating cell nuclear antigen (PCNA) and Gleason scores. In contrast, overexpression of dominant-negative Smad3 (Smad3D) does not alter tumorigenicity, but it significantly reduced the rate of tumor growth [36]. To determine its potential role in metabolism of prostate cancer, we first transiently transfected Smad3 (Flag-Smad3) or a control pcDNA3.1-flag (Flag) plasmid into DU145 cells, then treated the cells with TGF-β1. As shown in Fig. 3a, overexpression of Smad3 significantly increased glucose uptake (left panel) and lactate production (right panel) compared with control plasmid with or without TGF-β1 treatment, whereas silencing of endogenous Smad2 or Smad3 remarkably reduced glucose uptake (left panel) and lactate secretion (right panel) (Fig. 3b). Similar results were observed in PC-3M cells transfected with siRNA of Smad3 with or without TGF-β1 treatment (Fig. 3c). Meanwhile, upregulation of glucose uptake and lactate secretion induced by PMA treatment was not changed by SB431542 inhibitor of TGF-β1RII in DU145 cells (Fig. S3). Similarly, glucose uptake and lactate secretion were also not induced by TGF-β1 treatment in three kind of colorectal cancer cells (Fig. S4). In addition, silencing of Smad2 and Smad3 inhibited cells proliferation in DU145 cells (Fig. 3d). Thus, these data suggest that Smad2 or Smad3 promotes aerobic glycolysis and cell proliferation in a TGF-β1-independent way in prostate cancer cells.
Fig. 3.
Increased glycolysis induced by Smad2 or Smad3 was independent of TGF-β1 stimulation in prostate cancer cells. a DU145 cells were transfected with Flag-pcDNA3.1 (Flag) control or Flag-Smad3 plasmid. After transfection for 24 h, cells were starved for 16 h and then treated with 50 ng/mL TGF-β1 for 18 h. Glucose consumption (left panel) and lactate production (right panel) were measured as described in “Materials and methods”. b DU145 cells were transiently transfected with control siRNA (siCTL), si-Smad2 or si-Smad3, after transfection for 48 h, glucose consumption (left panel) and lactate production (right panel) were measured as described in a. c PC-3M cells were transfected with siCTL or si-Smad3. At 24-h post-transfection, cells were treated and measured as described in a. Data represent the mean ± SEM of three independent experiments, and were analyzed by two-way ANOVA with multiple comparisons, followed by Bonferroni post hoc test for significance versus siCTL. ***p < 0.001 and *p < 0.05. d DU145 cells were transiently transfected with siCTL, si-Smad2, si-Smad3 or combined with siRNA of Smad2 and Smad3. Cell proliferation was determined by the CCK-8 assay after transfection for 48 h. Data represent the mean ± SEM of three independent experiments and were analyzed by one-way ANOVA with multiple comparisons, followed by Dunnett’s post hoc test for significance versus siCTL. ***p < 0.001
Smad2 and Smad3 are key regulators in PMA- and EGF-mediated glycolysis in prostate cancer cells
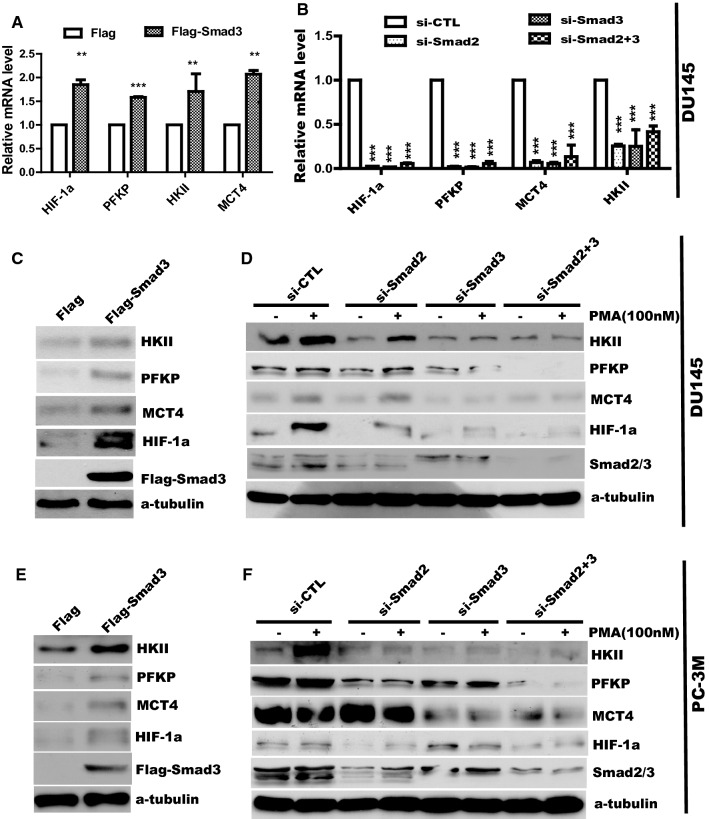
To identify the target of the Smad2- and Smad3-mediated aerobic glycolysis, RT-qPCR was performed to determine expression of glycolytic genes in DU145 cells transfected with Smad3 or control plasmid. As shown in Fig. 4a, overexpression of Smad3 (Flag-Smad3) significantly increased expression of HIF-1α, HKII, PFKP and MCT4 transcripts. Conversely, depletion of Smad2 (si-Smad2), Smad3 (si-Smad3), or both Smad2 and Smad3 (si-Smad2+3) led to downregulation of the genes compared with non-targeting siRNA control (siCTL) (Fig. 4b). Similar results were found in PC-3M cells transfected with siRNAs of Smad2 or Smad3 (Fig. S5).
Fig. 4.
Smad2 and Smad3 promote the expression of mRNA and proteins involved in glycolysis in prostate cancer cells. DU145 cells were transfected with Flag and Flag-Smad3 plasmid (a) or with siRNA of Smad2 and Smad3 (b) as indicated. After 48-h transfection, total RNA was extracted and performed to detect HIF-1α, PFKP, HKII and MCT4 by real-time PCR. Data represent the mean ± SEM of three independent experiments and were analyzed by one-way ANOVA with multiple comparisons, followed by Dunnett’s post hoc test for significance versus siCTL. ***p < 0.001, **p < 0.01, *p < 0.05. DU145 cells (c, d) and PC-3M cells (e, f) were transfected with plasmid or siRNA as indicated, the total protein was collected to detect related glycolytic protein by Western blotting
We further evaluated the effect of Smad2/3 on the expression of these targets at the protein level. In agreement with the PCR results, protein levels of HIF-1α, HKII, PFKP and MCT4 were enhanced in DU145 (Fig. 4c) and PC-3M (Fig. 4e) cells after ectopic expression of Smad3. Moreover, expression of the genes substantially declined at the baseline and after PMA stimulation in the cells transfected with siRNA of Smad2 and Smad3 (Fig. 4d, f).
PKCε interacts with Smad2/3 and phosphorylates Ser213 in the Smad3 linker region
Given that PKC-dependent phosphorylation of Smad3 leads to downregulation of the growth inhibitory and apoptotic action of TGF-β [37]. We evaluated whether PKCε interacts with Smad2 and Smad3 in prostate cancer cells. Immunofluorescence assay showed that endogenous PKCε co-localized with Smad3 in the nucleus of PC-3M prostate cancer cells (Fig. 5a). Furthermore, co-immunoprecipitation demonstrated that endogenous PKCε interacted with Smad2/3 both in DU145 and in PC-3M cells (Fig. 5b, c). Similar result was observed in HEK293 cells transfected with Flag-Smad3 plasmid with or without PMA treatment (Fig. S6).
Fig. 5.
PKCε interacts with Smad2/3 and is responsible for the Smad3 linker region phosphorylation. a Co-localization of PKCε and Smad2/3 by immunofluorescence in PC-3M cells. b, c DU145 or PC-3M cells were grown to 70–80% confluence, whole cell lysates were immunoprecipitated (IP) with antibodies against endogenous Smad2/3 or PKCε and co-precipitating PKCε or Smad2/3 were detected by immunoblotting. d DU145 cells were transiently transfected with pcDNA3.1 or the constitutively active mutant of PKCε (PKCε*) for 48 h, cell lysates were separated by SDS-PAGE and assayed with the antibodies against the indicated proteins; e DU145 (left panel) or PC-3M (right panel) cells were transfected with control siRNA (siCTL) or si-PKCε, cell lysates were separated by SDS-PAGE and assayed with the antibodies against the indicated proteins
Current research found that linker region of Smad3 is a critical prognostic indicator in the progression of human cancer, including prostate cancer and colorectal cancer [36, 38–40]. We further explored whether PKCε promotes phosphorylation of Smad3 at the linker region. As shown in Fig. 5d, overexpression of constitutive active mutant of PKCε (PKCε*) dramatically enhanced phosphorylation of Ser213 at the linker region of Smad3. Silencing of PKCε in DU145 (left panel) and PC-3M (right panel) cells attenuated phosphorylation of Ser213 at the Smad3 linker region (Fig. 5e).
PKCε activation is required for binding of Smad3 to the promoter of glycolytic genes
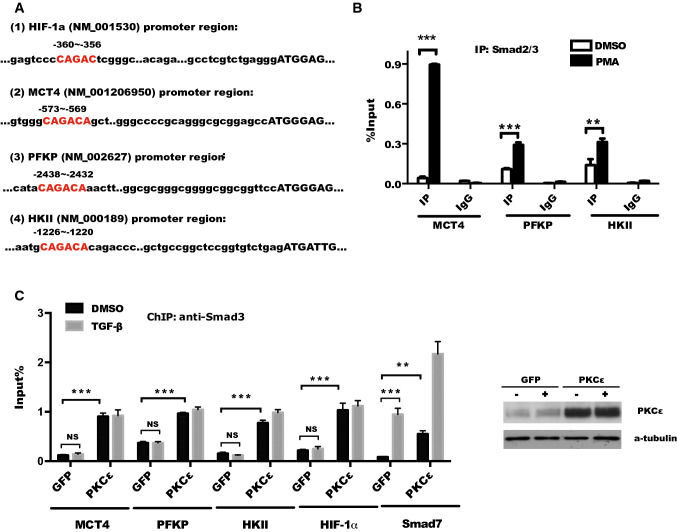
Since PKC directly phosphorylates receptor-regulated Smad proteins and results in inhibition of direct DNA binding and gene activation by Smad3 but not Smad2 [37, 41–43]. To further investigate whether phosphorylation of Smad3 induced by PKCε promotes the binding of Smad3 to the promoter of glycolytic genes, we used UCSC online software to identify a putative consensus sequence CAGACA of Smad3 bind element (SBE) [44] in the promoter of the target gene (HIF-1α, MCT4, PFKP, HKII) (Fig. 6a). The possibility of Smad3 binding to the promoter of MCT4, PFKP, and HKII was verified using Chromatin Immunoprecipitation (ChIP) followed by RT-qPCR. As shown in Fig. 6b, the binding of Smad2/3 to the promoter of MCT4, PFKP, HKII, and HIF-1α genes was markedly increased in response to PMA stimulation in DU145 cells.
Fig. 6.
PKCε is required for the binding of Smad3 to the promoter of glycolytic genes in prostate cancer cells. a The putative consensus sequence CAGACA of Smad3 binding element (SBE) and the number of the bases from transcriptional start codon (ATG, + 1) within promoter of the indicated target gene were analyzed by UCSC online software. b Smad3 binding to the promoter of glycolytic genes was determined in DU145 cells with or without PMA (100 nM) treatment using CHIP followed by RT-qPCR assay. c Smad3 binding to the promoter of glycolytic genes and Smad7 (positive control) were analyzed by CHIP assay in PC-3 cells infected with lentivirus GFP or PKCε in response to TGF-β1 (50 ng/mL) treatment. The ectopic expression of PKCε in PC-3 cells was confirmed by Western blotting (right panel). Data represent the mean ± SEM of three independent experiments and were analyzed by two sample tests for significance versus siCTL. ***p < 0.001; **p < 0.01, and *p < 0.05
Given that Smad2 differs from Smad3 mainly in the N-terminal MH1 domain where Smad2 contains two additional stretches of amino acids that lack in Smad3, leading to failure of Smad2 to activate transcription through the same CAGA DNA-binding elements, compared to Smad3 [45]. Thus, we explored whether PKCε-mediated tumor glycolytic genes transactivation is dependent on TGF-β-Smad3 signaling pathway in response to TGF-β treatment. CHIP assay and RT-qPCR showed that overexpression of PKCε significantly increased Smad3 binding to the promoter of MCT4, PFKP, HKII, and HIF-1α with or without TGF-β treatment. However, as a positive control, Smad3 binding to the promoter of Smad7, a specific target of TGF-β/Smad3 pathway [42], was remarkably enhanced with TGF-β stimulation, although its binding to the promoter of Smad7 was also slightly increased by overexpression of PKCε in PC-3 cells (Fig. 6c). These data suggest that PKCε regulates glycolysis via Smad3 binding to the promoter of glycolytic genes but not in TGF-β-dependent manner in prostate cancer cells.
Smad3 and lactate transporter mediate PKCε-induced glycolysis and cell proliferation in prostate cancer cells
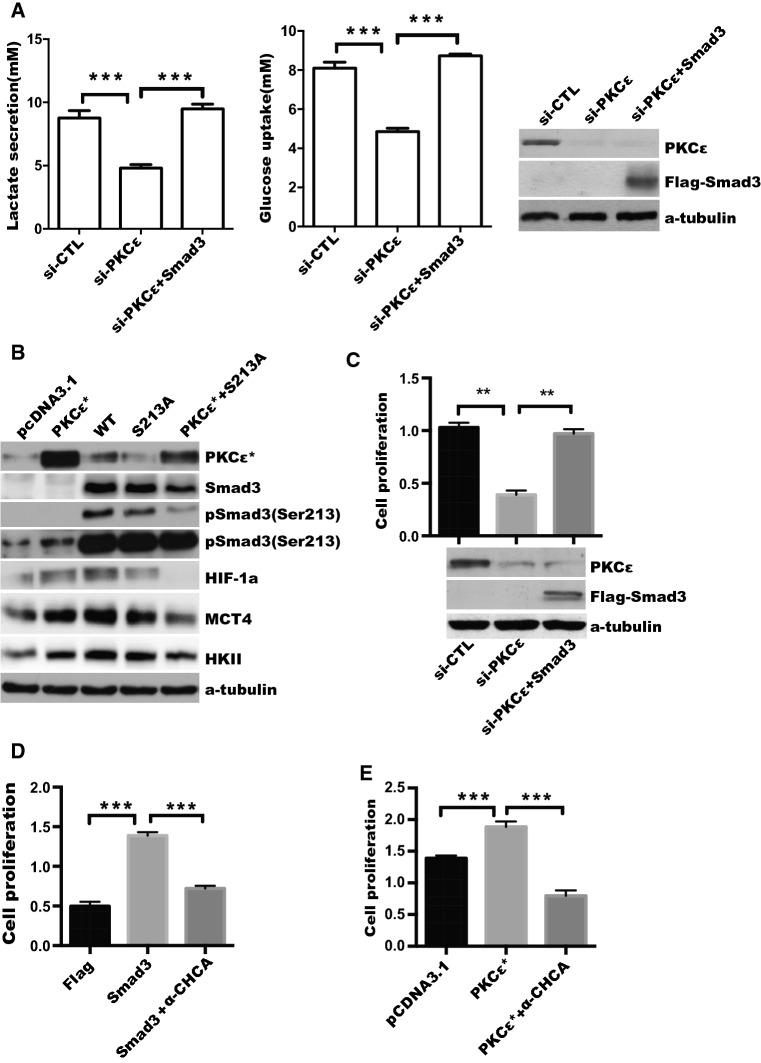
To test the hypothesis that PKCε-mediated Smad3 activity plays a critical role in regulation of tumor glycolysis and tumor growth, we examined whether the effect of overexpression of Smad3 on the glycolysis and PKCε depletion-induced tumor cell proliferation. SiRNA of PKCε and Flag-Smad3 plasmid were co-transfected into DU145 cells, and glucose uptake and lactate production, as well as tumor cell proliferation were analyzed. As shown in Fig. 7a, overexpression of Smad3 reversed PKCε silencing-induced downregulation of glucose uptake (left panel) and lactate production (middle panel). To further confirm the expression of glycolytic genes is induced by PKCε-mediated phosphorylation of Smad3 (Ser213), we transfected or co-transfected constitutive active mutant of PKCε and S213A mutant of Smad3 (S213A) into DU145 cells. We found that expression of mutant Smad3 (S213A) attenuated the effect of PKCε on protein expression of glycolytic genes, especially HIF-1a (Fig. 7b). Furthermore, forced Smad3 expression reversed inhibition of cell proliferation caused by PKCε depletion (Fig. 7c). In addition, attribution of PKCε and Smad3 expression-triggered enhancement of cell proliferation to the lactate secretion was evaluated. As expected, the a-CHCA inhibitor of MCT4 lactate transporter [46], significantly blocked tumor cell proliferation caused by ectopic expression of PKCε or Smad3 (Fig. 7d, e).
Fig. 7.
PKCε upregulates aerobic glycolysis and cell proliferation via Smad3 and lactate transporter in prostate cancer cells. a DU145 cells were co-transfected with si-PKCε and Flag-pcDNA3.1 or Flag-Smad3 (Smad3) plasmid, after 24-h transfection, cells were starved with serum-free medium for another 24 h, level of glucose and lactate production were measured as described in “Materials and methods”. Silencing of PKCε and overexpression of Smad3 were confirmed by Western blotting (right). b DU145 cells were transfected with pcDNA3.1, constitutive active mutant of PKCε (PKCε*), WT, S213A mutant of Smad3 (S213A) or PKCε* plus S213A mutant plasmids, respectively, as indicated and relative glycolytic proteins were detected by Western blotting. c DU145 cells were transfected with siCTL, si-PKCε, or si-PKCε plus Flag-Smad3, respectively. Cell proliferation was determined by CCK8 assay. Silencing of PKCε and overexpression of Smad3 were confirmed by Western blotting (bottom). d DU145 cells were transfected with Flag-pcDNA3.1 (Flag) or Flag-Smad3 (Smad3) for 18 h, and then treated with DMSO or MCT4 inhibitor (α-CHCA, 5 μg/mL) for another 24 h, the CCK-8 assay was used to measure the cell viability at OD value of 450 nm. e DU145 cells were transfected with pcDNA3.1 or the constitutively active mutant of PKCε, and then treated with the MCT4 inhibitor (α-CHCA). The CCK-8 assay as described in d. Data represent the mean ± SEM of three independent experiments and were analyzed by one-way ANOVA with multiple comparisons, followed by Dunnett’s post hoc test for significance. ***p < 0.001, **p < 0.01, and *p < 0.05
Discussion
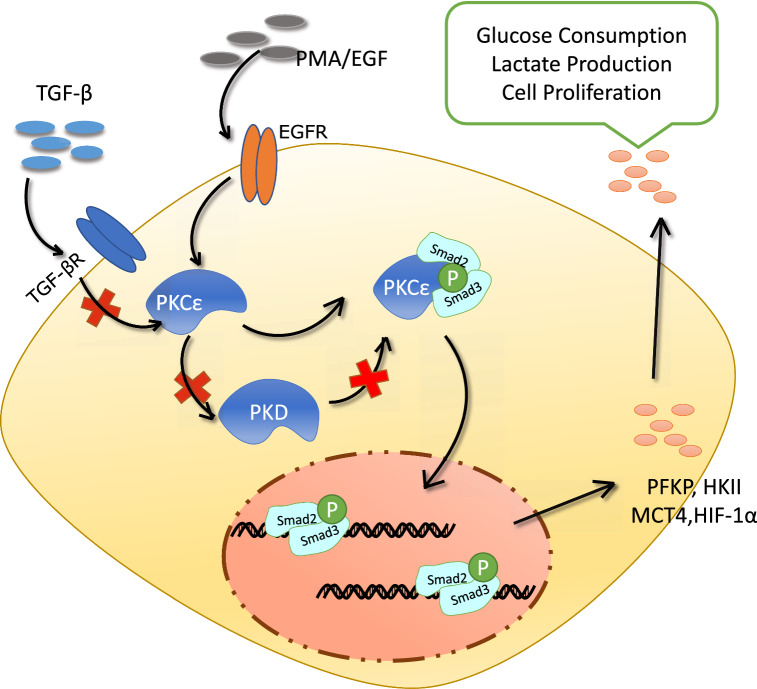
PKCε plays a critical role in upregulating pyruvate kinase M2 (PKM2) expression and promotes tumorigenesis [32]. Nevertheless, the contribution of PKCε to glycolysis during tumor progression remains largely unclear. Although the mechanisms and the role of TGF-β/Smad3 activation during tumor cell proliferation and metastasis have been extensively explored [47–51], it remains elusive how Smad3 is regulated in response to PKCε stimulation in the cancer cell growth and metabolism [37]. In this study, we demonstrate the cross-talk of PKCε with Smad2/3 cascade promoted cell proliferation by upregulation of tumor glycolysis in prostate cancer cells. Activation of PKCε enhanced phosphorylation of Smad3 (Ser213) in the linker region and increased its binding to the promoter of key glycolytic genes, thereby resulting in tumor Aerobic glycolysis and tumor cell proliferation (Fig. 8).
Fig. 8.
Schematic illustration for interplay of PKCε with Smad3 signaling in regulation of glycolysis and cell proliferation via induction of glycolytic genes expression in prostate cancer cells. PKCε interacts with and phosphorylates the linker region of Smad3 (Ser213), promoting its binding to the glycolytic gene promoters to upregulate the glycolytic gene expression in a TGF-β- and PKD-independent manner. Inhibition of PKCε blocks tumor cell proliferation and reduces glucose uptake and lactate production, and ectopic expression of Smad3 reverses this process, and consequently increases the aerobic glycolysis and proliferation. Red X represents the indicated pathway does not work in this model
High expression of PKCε has been regarded as the hallmark of prostate cancer development [11]. Mounting evidence has demonstrated that ectopic expression of PKCε is oncogenic and involved in prostate cancer development, aggressiveness, invasion, proliferation, and survival [52]. Although a number of targets of PKCε, such as caveolin-1, ILK, Akt, Bax, Stat3, and uPA, have been identified [13, 27, 29, 52], the immediate signaling pathways through which PKCε regulates tumor metabolism are largely unknown. The PKC family, particularly the novel PKCs, directly phosphorylate and activate downstream of PKD [53]. The DAG/PKC/PKD canonical pathway has been shown to mediate the mitogenic signaling of GPCR agonists to promote prostate cancer cell proliferation and invasion [54]. It is, however, still unclear whether PKC/PKD axis is involved in the tumor metabolism. Our findings revealed that PKCε depletion reduced tumor glucose uptake and lactate production, which is attributed to a decrease in MCT4, PFKP, HKII, and HIF-1α expression in prostate cancer cells. Consistent with our findings, Yang et al. demonstrated that ectopic expression of PKCε upregulated PKM2 expression, resulting in alteration of cell metabolism and promotion of tumorigenesis [32]. In line with this finding, we demonstrated that alterations in aerobic glycolysis of prostate cancer cells were caused by PKC, but not PKD using PKD inhibitor or PKDs silencing. Thus, these data strongly suggest that the contribution of PKC to tumor glycolysis is a PKD-independent in prostate cancer cells.
The involvement of aberrantly high activity of PKCε in regulating NF-κB and Stat3 is well established in both prostate adenocarcinoma of transgenic mouse model and human tumor tissues [11]. It is of interest that PKC-dependent phosphorylation of Smad3 has been found to be a key event in the PMA-dependent inactivation of TGF-β-induced growth inhibition and cell death [37]. Our findings showed, for the first time, that activation of Smad2/3 by PKCε promotes tumor glycolysis in a TGF-β-independent manner in prostate cancer cells. Depletion of Smad2 and Smad3 reduced protein expression of the glycolytic genes in prostate cancer cells. PKCε interacts with Smad2/3 and is primarily responsible for phosphorylation of Ser213 in the Smad3 linker region, which mediates the malignant signaling that allows human metastatic cancer to adopt more invasive and proliferative properties required for progression [55]. Nevertheless, there are no consensus PKC substrate phosphorylation sites (K/RXSXK/R) in the amino acid sequences of Smad2 and Smad3 [56], suggesting that the Ser213 of Smad3 may not be directly phosphorylated by PKCε.
It has been shown that loss of expression of TGF-β receptors occurs frequently in lung, gastric, prostate, and bladder cancers, due to frame shift, missense mutations, and methylation of the TGF-βRI promoter, which preferentially disables the tumor-suppressive action of TGF-β by attenuating the tumor-suppressive arm of the signaling pathway [57–60]. Accordingly, a specific inhibitor of TGF-β receptor and TGF-β1 stimulation had no impact in tumor glycolysis, which is possibility attributed to the decrease in expression of TGF-βRI or TGF-βRII. Meanwhile, PKCε-Smad pathways regulate glycolysis in a TGF-β/Smad signaling-independent responses; the precise mechanism which cross-talk of Smad3 and PKCε-mediated tumor Aerobic glycolysis and cell growth remains to be determined. In contrast, transforming growth factor alpha (TGF-α), a member of the epidermal growth factor family, is elevated and its potential use as a prognostic biomarker in various tumors, like gastric carcinoma [61] or melanoma [62]. Interestingly, TGF-α stimulates activation of ERK1/2 and DNA synthesis in independent of PKC activity in human pancreatic cancer cells [63]. On the other hand, in colon cancer cells, TGF-α regulates cell adhesion function through PKC-mediated specific phosphorylation sites of S6K [64]. However, the cross-talk between TGF-α and PKC, especially PKCε in prostate cancer is not clear.
The current data showed that Smad2 contains two additional stretches of amino acids in the N-terminal MH1 domain that are lacking in Smad3, which leads to its inability to activate transcription of target genes via the same CAGA DNA-binding elements for Smad3 binding [45]. Recently, a report from Wang et al. showed that apelin, a bioactive peptide, activated PKCε in tubular epithelial cells, which in turn decreased phosphorylation of Smad3 in the COOH-terminal region and increased Smad7 levels [65]. Linker phosphorylation of Smad3 indirectly inhibited its COOH-terminal phosphorylation, and the proliferative effect mediated by RTK-dependent pSmad3L pathway antagonized TGF-β signaling through the cytostatic pSmad3C pathway in normal epithelial cells [55]. By studying the impact of PKCε perturbations on the transcriptional regulation of glycolytic genes by Smad, we also demonstrate that PKCε-stimulated glycolysis in cancer cells by regulation of phosphorylation of Smad3 linker region. ChIP and RT-qPCR analyses in the PC-3 cells overexpressing PKCε with or without stimulation of TGF-β showed that Smad3 binding to the promoter of HKII, PFKP, MCT4, and HIF-1α was increased by ectopic expression of PKCε, and TGF-β has no significant impact, supporting the conclusion that PKCε activation results in Smad3 binding to the promoter of the glycolytic genes and promotes the expression of these target genes, which further contributes to cell proliferation and metabolism in prostate cancer cells. Further work on the interplay between PKCε and Smad3 in regulating metabolism, including lipid metabolism, is guaranteed.
In conclusion, our results demonstrate that the activation of PKCε enhances Smad2/3-mediated glycolysis, thereby promoting cell proliferation in prostate cancer cells. Our findings reveal a novel function of PKCε that promotes cell proliferation by enhancing aerobic glycolysis through its crosstalk to Smad2/3 in prostate cancer cells. The interplay of the PKCε with Smad2/3 pathways in regulating glucose metabolism may have important implications in prostate cancer development and potentially provide novel molecular targets for prevention of and treatment of the cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported financially by the National Natural Science Foundation of China (Grant nos. 81472407, 81672540, 81772761, 81272852); Oversea Hong Kong & Macao Scholars Collaborative Research Fund of NSFC in China (Grant no. 81328020); Science and Technology Foundation of Guangzhou in China (Grant no. 201607010351, 210707010303); the National Institutes of Health (Grant R01CA142580 to Wang), and Department of Defense award (PC150190 to Wang).
Abbreviations
- PKC ε
Protein kinase C epsilon
- Smad
Contraction of Sma and Mad (mothers against decapentaplegic)
- HKII
Hexokinase 2
- PFKP
Phosphofructokinase, platelet
- HIF-1α
Hypoxia inducible factor 1 alpha
- MCT4
Monocarboxylate transporter 4
- PKD
Protein kinase D
- TGF-β1
Transforming growth factor-β
- PMA
Phorbol-12-myristate-13-acetate
Author contributions
FD and WFX conceived and designed the experiments, WFX, FYZ, SYL, GHL, and XJL performed experiments and analyzed data, WFX and FD wrote the manuscript, QJW and FD revised manuscript.
Compliance with ethical standards
Conflict of interest
The authors declared that they have no competing interests.
Footnotes
Wanfu Xu, Fangyin Zeng and Songyu Li contributed equally to this work.
References
- 1.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 3.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26(9):877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Garcia S, Lopez-Gonzalez JS, Baez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H. Tumor cell metabolism: an integral view. Cancer Biol Ther. 2011;12(11):939–948. doi: 10.4161/cbt.12.11.18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science (New York, NY) 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Wang L, Wang Z, Yang Y, Tian J, Liu G, Guan D, Cao X, Zhang Y, Hao A. GRIM-19 opposes reprogramming of glioblastoma cell metabolism via HIF1alpha destabilization. Carcinogenesis. 2013;34(8):1728–1736. doi: 10.1093/carcin/bgt125. [DOI] [PubMed] [Google Scholar]
- 10.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7(4):281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 11.Hafeez BB, Zhong W, Weichert J, Dreckschmidt NE, Jamal MS, Verma AK. Genetic ablation of PKC epsilon inhibits prostate cancer development and metastasis in transgenic mouse model of prostate adenocarcinoma. Can Res. 2011;71(6):2318–2327. doi: 10.1158/0008-5472.can-10-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benavides F, Blando J, Perez CJ, Garg R, Conti CJ, DiGiovanni J, Kazanietz MG. Transgenic overexpression of PKCepsilon in the mouse prostate induces preneoplastic lesions. Cell Cycle (Georgetown, Tex) 2011;10(2):268–277. doi: 10.4161/cc.10.2.14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafeez BB, Zhong W, Mustafa A, Fischer JW, Witkowsky O, Verma AK. Plumbagin inhibits prostate cancer development in TRAMP mice via targeting PKCepsilon, Stat3 and neuroendocrine markers. Carcinogenesis. 2012;33(12):2586–2592. doi: 10.1093/carcin/bgs291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu CT, Cheng CC, Wu JR, Pan SM, Wu WS. PKCepsilon-mediated c-Met endosomal processing directs fluctuant c-Met-JNK-paxillin signaling for tumor progression of HepG2. Cell Signal. 2015;27(7):1544–1555. doi: 10.1016/j.cellsig.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez-Uzquiza A, Lopez-Haber C, Jernigan DL, Fatatis A, Kazanietz MG. PKCepsilon is an essential mediator of prostate cancer bone metastasis. Mol Cancer Res. 2015;13(9):1336–1346. doi: 10.1158/1541-7786.mcr-15-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Li Z, Zhang C, Zhang S, Ji Y, Chen F. Knockdown of PKCepsilon expression inhibits growth, induces apoptosis and decreases invasiveness of human glioma cells partially through Stat3. J Mol Neurosci. 2015;55(1):21–31. doi: 10.1007/s12031-014-0341-4. [DOI] [PubMed] [Google Scholar]
- 17.Gurbuz N, Park MA, Dent P, Abdel Mageed AB, Sikka SC, Baykal A. Cystine dimethyl ester induces apoptosis through regulation of PKC-delta and PKC-epsilon in prostate cancer cells. Anticancer Agents Med Chem. 2015;15(2):217–227. doi: 10.2174/1871520614666141120121901. [DOI] [PubMed] [Google Scholar]
- 18.Sarveswaran S, Thamilselvan V, Brodie C, Ghosh J. Inhibition of 5-lipoxygenase triggers apoptosis in prostate cancer cells via down-regulation of protein kinase C-epsilon. Biochim Biophys Acta 1813. 2011;12:2108–2117. doi: 10.1016/j.bbamcr.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11(12):937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toton E, Ignatowicz E, Skrzeczkowska K, Rybczynska M. Protein kinase Cepsilon as a cancer marker and target for anticancer therapy. Pharmacol Rep. 2011;63(1):19–29. doi: 10.1016/S1734-1140(11)70395-4. [DOI] [PubMed] [Google Scholar]
- 21.Wu D, Foreman TL, Gregory CW, McJilton MA, Wescott GG, Ford OH, Alvey RF, Mohler JL, Terrian DM. Protein kinase cepsilon has the potential to advance the recurrence of human prostate cancer. Can Res. 2002;62(8):2423–2429. [PubMed] [Google Scholar]
- 22.Garg R, Blando J, Perez CJ, Wang H, Benavides FJ, Kazanietz MG. Activation of nuclear factor kappaB (NF-kappaB) in prostate cancer is mediated by protein kinase C epsilon (PKCepsilon) J Biol Chem. 2012;287(44):37570–37582. doi: 10.1074/jbc.M112.398925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamato D, Burch ML, Piva TJ, Rezaei HB, Rostam MA, Xu S, Zheng W, Little PJ, Osman N. Transforming growth factor-beta signalling: role and consequences of Smad linker region phosphorylation. Cell Signal. 2013;25(10):2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Ten Dijke P, Egorova AD, Goumans MJ, Poelmann RE, Hierck BP. TGF-beta signaling in endothelial-to-mesenchymal transition: the role of shear stress and primary cilia. Sci Signal. 2012;5(212):2. doi: 10.1126/scisignal.2002722. [DOI] [PubMed] [Google Scholar]
- 25.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10(6):415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 26.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Z, Zeng F, Xu W, Wang C, Ke Z, Wang QJ, Deng F. PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci. 2012;125(Pt 20):4800–4811. doi: 10.1242/jcs.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okita Y, Kamoshida A, Suzuki H, Itoh K, Motohashi H, Igarashi K, Yamamoto M, Ogami T, Koinuma D, Kato M. Transforming growth factor-beta induces transcription factors MafK and Bach1 to suppress expression of the heme oxygenase-1 gene. J Biol Chem. 2013;288(28):20658–20667. doi: 10.1074/jbc.M113.450478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Can Res. 2008;68(10):3844–3853. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 30.Ren B, Li X, Zhang J, Fan J, Duan J, Chen Y. PDLIM5 mediates PKCepsilon translocation in PMA-induced growth cone collapse. Cell Signal. 2015;27(3):424–435. doi: 10.1016/j.cellsig.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Petiti JP, Gutierrez S, Mukdsi JH, De Paul AL, Torres AI. Specific subcellular targeting of PKCalpha and PKCepsilon in normal and tumoral lactotroph cells by PMA-mitogenic stimulus. J Mol Histol. 2009;40(5–6):417–425. doi: 10.1007/s10735-010-9255-9. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, You MJ, Koh MY, Cote G, Aldape K, Li Y, Verma IM, Chiao PJ, Lu Z. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48(5):771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Investig. 2013;123(9):3664–3671. doi: 10.1172/jci67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang Y, Han J, Kim SJ, Kim J, Lee MJ, Jeong S, Ryu MJ, Seo KS, Choi SY, Shong M, Lim K, Heo JY, Kweon GR. Suppression of mitochondrial respiration with auraptene inhibits the progression of renal cell carcinoma: involvement of HIF-1alpha degradation. Oncotarget. 2015;6(35):38127–38138. doi: 10.18632/oncotarget.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drabsch Y, ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31(3–4):553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- 36.Lu S, Lee J, Revelo M, Wang X, Lu S, Dong Z. Smad3 is overexpressed in advanced human prostate cancer and necessary for progressive growth of prostate cancer cells in nude mice. Clin Cancer Res. 2007;13(19):5692–5702. doi: 10.1158/1078-0432.CCR-07-1078. [DOI] [PubMed] [Google Scholar]
- 37.Yakymovych I, Ten Dijke P, Heldin CH, Souchelnytskyi S. Regulation of Smad signaling by protein kinase C. FASEB J. 2001;15(3):553–555. doi: 10.1096/fj.00-0474fje. [DOI] [PubMed] [Google Scholar]
- 38.Kim SH, Kim KH, Ahn S, Hyeon J, Park CK. Smad3 and Smad3 phosphoisoforms are prognostic markers of gastric carcinoma. Dig Dis Sci. 2013;58(4):989–997. doi: 10.1007/s10620-012-2470-3. [DOI] [PubMed] [Google Scholar]
- 39.Yao B, Zhao J, Li Y, Li H, Hu Z, Pan P, Zhang Y, Du E, Liu R, Xu Y. Elf5 inhibits TGF-beta-driven epithelial-mesenchymal transition in prostate cancer by repressing SMAD3 activation. Prostate. 2015;75(8):872–882. doi: 10.1002/pros.22970. [DOI] [PubMed] [Google Scholar]
- 40.Kang HY, Lin HK, Hu YC, Yeh S, Huang KE, Chang C. From transforming growth factor-beta signaling to androgen action: identification of Smad3 as an androgen receptor coregulator in prostate cancer cells. Proc Natl Acad Sci USA. 2001;98(6):3018–3023. doi: 10.1073/pnas.061305498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17(11):3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger EP. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor beta. J Biol Chem. 2000;275(15):11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 43.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1(4):611–617. doi: 10.1016/S1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Zhang Z, Zou K, Cheng Y, Yang M, Chen H, Wang H, Zhao J, Chen P, He L, Chen X, Geng L, Gong S. MiR-1 suppresses tumor cell proliferation in colorectal cancer by inhibition of Smad3-mediated tumor glycolysis. Cell Death Dis. 2017;8(5):e2761. doi: 10.1038/cddis.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennler S, Huet S, Gauthier JM. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18(8):1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- 46.Kikutani Y, Kobayashi M, Konishi T, Sasaki S, Narumi K, Furugen A, Takahashi N, Iseki K. Involvement of monocarboxylate transporter 4 expression in statin-induced cytotoxicity. J Pharm Sci. 2016;105(4):1544–1549. doi: 10.1016/j.xphs.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Ali A, Zhang P, Liangfang Y, Wenshe S, Wang H, Lin X, Dai Y, Feng XH, Moses R, Wang D, Li X, Xiao J. KLF17 empowers TGF-beta/Smad signaling by targeting Smad3-dependent pathway to suppress tumor growth and metastasis during cancer progression. Cell Death Dis. 2015;6:e1681. doi: 10.1038/cddis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Acharya S, Sahin O, Zhang Q, Saito Y, Yao J, Wang H, Li P, Zhang L, Lowery FJ, Kuo WL, Xiao Y, Ensor J, Sahin AA, Zhang XH, Hung MC, Zhang JD, Yu D. 14-3-3zeta turns TGF-beta’s function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell. 2015;27(2):177–192. doi: 10.1016/j.ccell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu JR, Tai Y, Jin Y, Hammell MC, Wilkinson JE, Roe JS, Vakoc CR, Van Aelst L. TGF-beta/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 2015;29(3):250–261. doi: 10.1101/gad.248963.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G, Qin L, Ye Q, Dong Q, Ren N, Jia H. Organ microenvironment affects growth and metastasis of hepatocellular carcinoma via the TGF-beta/Smad pathway in mice. Exp Ther Med. 2013;5(1):133–137. doi: 10.3892/etm.2012.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A. TGF-beta/SMAD/GLI2 signaling axis in cancer progression and metastasis. Can Res. 2011;71(17):5606–5610. doi: 10.1158/0008-5472.CAN-11-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, Wilding G, Verma AK. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Can Res. 2007;67(18):8828–8838. doi: 10.1158/0008-5472.can-07-1604. [DOI] [PubMed] [Google Scholar]
- 53.Tinsley JH, Teasdale NR, Yuan SY. Involvement of PKCdelta and PKD in pulmonary microvascular endothelial cell hyperpermeability. Am J Physiol Cell Physiol. 2004;286(1):C105–C111. doi: 10.1152/ajpcell.00340.2003. [DOI] [PubMed] [Google Scholar]
- 54.Xu X, Jin T. The novel functions of the PLC/PKC/PKD signaling axis in G protein-coupled receptor-mediated chemotaxis of neutrophils. J Immunol Res. 2015;2015:817604. doi: 10.1155/2015/817604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuzaki K, Kitano C, Murata M, Sekimoto G, Yoshida K, Uemura Y, Seki T, Taketani S, Fujisawa J, Okazaki K. Smad2 and Smad3 phosphorylated at both linker and COOH-terminal regions transmit malignant TGF-beta signal in later stages of human colorectal cancer. Cancer Res. 2009;69(13):5321–5330. doi: 10.1158/0008-5472.CAN-08-4203. [DOI] [PubMed] [Google Scholar]
- 56.Kang JH, Toita R, Kim CW, Katayama Y. Protein kinase C (PKC) isozyme-specific substrates and their design. Biotechnol Adv. 2012;30(6):1662–1672. doi: 10.1016/j.biotechadv.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 58.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, Byrom D, Riera A, Rossell D, Mangues R, Massague J, Sancho E, Batlle E. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 60.Hata A, Shi Y, Massague J. TGF-beta signaling and cancer: structural and functional consequences of mutations in Smads. Mol Med Today. 1998;4(6):257–262. doi: 10.1016/S1357-4310(98)01247-7. [DOI] [PubMed] [Google Scholar]
- 61.Fanelli MF, Chinen LT, Begnami MD, Costa WL, Jr, Fregnami JH, Soares FA, Montagnini AL. The influence of transforming growth factor-alpha, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial–mesenchymal transition on overall survival of patients with gastric cancer. Histopathology. 2012;61(2):153–161. doi: 10.1111/j.1365-2559.2011.04139.x. [DOI] [PubMed] [Google Scholar]
- 62.Tarhini AA, Lin Y, Yeku O, LaFramboise WA, Ashraf M, Sander C, Lee S, Kirkwood JM. A four-marker signature of TNF-RII, TGF-alpha, TIMP-1 and CRP is prognostic of worse survival in high-risk surgically resected melanoma. J Transl Med. 2014;12:19. doi: 10.1186/1479-5876-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seufferlein T, Van Lint J, Liptay S, Adler G, Schmid RM. Transforming growth factor alpha activates Ha-Ras in human pancreatic cancer cells with Ki-ras mutations. Gastroenterology. 1999;116(6):1441–1452. doi: 10.1016/S0016-5085(99)70509-3. [DOI] [PubMed] [Google Scholar]
- 64.Sawhney RS, Cookson MM, Sharma B, Hauser J, Brattain MG. Autocrine transforming growth factor alpha regulates cell adhesion by multiple signaling via specific phosphorylation sites of p70S6 kinase in colon cancer cells. J Biol Chem. 2004;279(45):47379–47390. doi: 10.1074/jbc.M402031200. [DOI] [PubMed] [Google Scholar]
- 65.Wang LY, Diao ZL, Zheng JF, Wu YR, Zhang QD, Liu WH. Apelin attenuates TGF-beta1-induced epithelial to mesenchymal transition via activation of PKC-epsilon in human renal tubular epithelial cells. Peptides. 2017;96:44–52. doi: 10.1016/j.peptides.2017.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.