Abstract
During the past decade, we have learnt that the most common DNA modification, 5-methylcytosine (5mC), playing crucial roles in development and disease, is not stable but can be actively reversed to its unmodified form via enzymatic catalysis involving the TET enzymes. These ground-breaking discoveries have been achieved thanks to technological advances in the detection of the oxidized forms of 5mC and to the boldness of individual scientists. The TET enzymes require molecular oxygen for their catalysis, making them important targets for hypoxia research. They also require special cofactors which enable additional levels of regulation. Moreover, mutations and other genetic alterations in TETs are found, especially in myeloid malignances. This review focuses on the kinetic and inhibitory properties of the TET enzymes and the role of TETs in cellular differentiation and transformation and in cancer.
Keywords: Cancer, DNA methylation, EMT, Gene regulation, 5hmC, Hypoxia
Introduction
The enzyme family of 2-oxoglutarate-dependent dioxygenases (2-OGDDs) gained a new family member in 2009 when the conversion of 5mC to 5-hydroxymethyl cytosine (5hmC) in DNA was found to be associated with catalysis by TET1 [1]. TET1 in 10q22 had actually been cloned a few years earlier as a leukemia-associated protein with a CXXC domain (LCX) as a fusion partner of mixed-lineage leukemia (MLL) in 11q23, and was suggested as playing a role in the pathogenesis of 11q23-associated leukemia [2]. The first acute myeloid leukemia (AML) patient with this TET1 translocation was subsequently characterized and the name Ten-Eleven Translocation was suggested [3]. Based on sequence homology, the existence of three isoenzymes in human and mouse was recognized, although no function could yet be associated with these proteins [3]. An attempt to identify mammalian enzymes that would modify 5mC by means of structurally informed iterative sequence profile searches using the oxygenase domains of Trypanosomal JBP1 and JBP2, which are 2-OGDDs and catalyze the first step in β-d-glucosyl hydroxymethyluracil (base J) synthesis, revealed homology with the TETs [1]. Further experimental data then showed that TET1 overexpression resulted in a reduction in 5mC levels in genomic DNA and the appearance of a novel species, identified as 5hmC [1]. Human embryonic stem cells (ES) and Purkinje cells were the first cell types reported to contain 5hmC [1, 4]. Subsequently two isoenzymes, TET2 and TET3, were shown to possess similar catalytic activity [5].
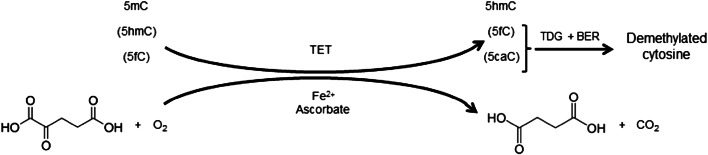
The 2-OGDD family has ~ 70 members in mammals. The others in addition to the TETs are, for example, numerous histone lysine demethylases (KDMs), the prolyl 4-hydroxylases that modify the hypoxia-inducible factor (HIF-P4Hs) or collagens (collagen P4Hs), the hypoxia-inducible factor asparagine hydroxylase FIH and the obesity-associated FTO, the first identified RNA demethylase (for a complete list see [6]). The 2-OGDDs share the same reaction mechanism and cofactors, but their substrates vary from DNA to RNA, proteins and fatty acids. 2-OGDDs require Fe2+, 2-oxoglutarate (2-OG/α-ketoglutarate), molecular oxygen [6] and many require a reducing agent, typically vitamin C (ascorbate) to support the reaction [6, 7]. The cofactors are coordinated at the active site by conserved residues, iron by two histidines and an aspartate and 2-oxoglutarate by a positively charged residue, an arginine or a lysine. In TETs this latter is an arginine. The catalytic domains possess a double-stranded β-helix (DSBH) structure known as a jelly roll. Following cofactor and substrate binding, the molecular oxygen oxidizes Fe2+, inducing substrate oxidation and decarboxylation of 2-OG to succinate and CO2 (Fig. 1). In the case of TETs, hydroxylation of the 5mC substrate in the DNA CpG dinucleotides to 5hmC can be followed by further oxidation of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) catalyzed by the TETs themselves (Fig. 1) [5, 8].
Fig. 1.
Schematic representation of the enzymatic reactions catalyzed by the TET enzymes resulting in DNA demethylation. TDG thymine DNA glycosylase, BER base excision repair
The human TETs are large proteins, full-length TET1 being composed of 2039 amino acids (aa), TET2 of 1921 aa and TET3 of 1803 aa [5], although shorter variants of TET1 and TET3 are also found [9–11]. The aminoterminal parts of TET1 and TET3 contain a DNA-binding CXXC domain, whereas this is lacking in TET2 but is coded by its neighboring gene IDAX [12]. The catalytic domains are situated in the C-termini and contain a cysteine-rich domain and a DSBH domain. A large low-complexity insert is found within the DSHB domain that may have regulatory roles via post-translational modifications [13, 14]. According to mRNA and protein expression databases, the human TETs are widely expressed (https://www.ebi.ac.uk/gxa/home, http://www.proteinatlas.org/), while experimental data suggest that Tet1 is preferentially expressed in ES cells (ESCs) whereas Tet2 and Tet3 are expressed in many tissues and have overlapping expression profiles [5].
The catalytic activity of the TETs is strongly dependent on Fe2+ and 2-OG [1, 5, 15], whereas the omission of ascorbate did not significantly reduce TET1 catalytic activity [1]. The reported K m values for Fe2+ and 2-OG of TET1 and TET2 are similar to those for the collagen P4H-I (Table 1). Mutations of the iron-binding residues of TET1 and TET2 impair their catalytic activity and increase the K m value for Fe2+ up to ~ 60-fold while reducing the V max values by half [1, 15]. Divalent metals other than Fe2+ can typically act as competitive inhibitors for 2-OGDDs [16, 17]. The reported K m values of TET1 and TET2 for molecular oxygen are 0.3–30 µM [15, 18] (Table 1), suggesting that they can retain their catalytic activity under low oxygen tension and do not act as cellular oxygen sensors, unlike the HIF-P4Hs, with K m values of ≥ 70 µM for oxygen (Table 1). In vitro, TETs possess a nanomolar affinity for a substrate with 5mC (Table 1), but their affinity for substrates containing 5hmC and 5fC appears to be weaker than that for 5mC [5, 19]. However, in a situation of high TET catalytic activity or overexpression, the levels of both the 5mC substrate and 5hmC are likely to reduce because of substrate exhaustion [20, 21].
Table 1.
K m values of TETs and several other 2-OGDDs for cosubstrates and substrates
| Fe2+ (µM) | 2-Oxoglutarate (µM) | Oxygen (µM) | Substrate (µM) | |
|---|---|---|---|---|
| TET1 | 5a | 55a | 0.3–30a,b | 0.075a |
| TET2 | 4a | 60a | 0.5–30a,b | 0.125a |
| HIF-P4H-1 | 0.05c | 2d | 230e | 0.01–0.02f |
| HIF-P4H-2 | 0.05c | 1d | 67–250e,f,g | 0.14f |
| HIF-P4H-3 | 0.1c | 12d | 230e | 0.07f |
| Collagen P4H-I | 2h | 20h | 40h | 0.2i |
| FIH | 0.5j | 25j | 90–240j,k | 100–220i,j,k |
| KDM4A | ND | 23l | 173l | 23–656l,m |
| ABH2 | ND | 4n | ND | 0.08–4n,o |
Diverse roles of oxidized 5mC bases
DNA methyltransferases (DNMTs) convert unmethylated cytosine to 5mC, which has a well-established role as a transcriptional repressor and therefore a regulator of gene expression [22]. The methylation of cytosine was for a long time assumed to be permanent, but the discovery of the ability of the TET enzymes to convert 5mC to 5hmC, and further to 5fC and 5caC has suggested a new mechanism by which DNA can be demethylated [1, 23]. In 2011, He et al. [8] showed how 5mC converted to 5caC by the TET enzymes can be demethylated via excision by thymine DNA glycosylase (TDG) and base excision repair (BER), demonstrating a role for the TET enzymes in DNA demethylation (Fig. 1) [8]. Nevertheless, it has also been suggested recently that the oxidized forms of 5mC have their own functions independent to that of DNA demethylation. Many endogenous proteins have been identified as potential reader proteins for 5hmC, 5fC and 5caC [24], and 5fC, for example, has been associated with tissue development [25].
5hmC is the first and most abundant form of the oxidized 5mCs (Fig. 1) [22] and the one that has been studied the most. 5hmC has many important functions in embryonic development, hematopoiesis, hematological malignancies and other cancers, and high levels are found in many types of stem cells and also in neural cell lines. Since TET enzymes are also highly expressed in these cell types [1], it has been suggested that the TETs and 5hmC both control pluripotency and cell differentiation, and lack of 5hmC is also associated with the malignant progression of cancer cells [26, 27].
CpG islands are short regions rich of CpG dinucleotides usually found in promoter regions of many genes. In non-cancer cells these regions are mostly nonmethylated [28, 29]. Methylated status of CpG islands near promoter regions is associated with transcriptional repression [28, 29]. Aberrant methylation of CpG islands near promoter regions of tumor suppressor genes has been a major focus of DNA methylation research since discovery of CpG island methylator phenotype (CIMP) in colorectal cancer [30]. Since TET enzymes convert 5mC to 5hmC, reduced 5hmC levels resulting from abnormal TET function, can be associated with dysregulated DNA demethylation which can result in increased 5mC levels, CIMP and hypermethylator phenotype [18, 31]. Reduced global 5hmC levels, possibly associated with a hypermethylator phenotype of chromatin or CpG islands, are found in hematological malignancies as well as in several solid tumors [9, 26, 32–36]. However, in contrast to CIMP and the hypermethylator phenotype, recent findings suggest that in human seminomas TET1 is overexpressed and both 5mC and 5hmC levels are decreased probably reflecting the germ cell origin of these tumors [20, 21]. Due to reduced 5hmC levels being an epigenetic hallmark of a number of cancers, it has been suggested that TETs and 5hmC levels may control cell differentiation and epithelial-to-mesenchymal transition (EMT) [27, 37], and low 5hmC has been associated with a poorer prognosis [33].
TET mutants in cancers
TET2 appears to be the most frequently mutated of the TET1-3 genes, its mutations being found in ~ 20% cases of myelodysplastic syndrome (MDS), ~ 20% myeloproliferative neoplasms (MPN), ~ 20% AML and ~ 45% chronic myelomonocytic leukemia (CMML) [38–40]. TET2 mutations have also been found in ~ 15% T cell lymphomas, often appearing in these together with a DNMT3A mutation [41]. The majority of the mutations are somatic heterozygous missense mutations that do not cluster to a certain site in the TET2 gene [38–40]. The mutations residing in the C-terminal Cys-rich domain or the DSBH domain have been shown to impair hydroxylation of 5mC [42]. Some mutations that target the Fe2+-binding histidine or the 2-OG-coordinating arginine have been shown to increase the K m value for these cofactors by at least 50- to 80-fold and reduce the V max value, indicating significant loss of function [15]. As the TET2 mutants studied in detail do not show complete loss of function, increasing the local concentration of iron or 2-OG in the bone marrow might, for example, be a mechanism for restoring its catalytic activity and reversing the oncogenic properties of TET2 mutants [15, 42]. Altogether, these data suggest that TET2 is a critical tumor suppressor, especially in myeloid tissue.
TET1 has frequently been found in the MLL-containing chromosome translocations in AML, T-cell lymphoma and B-cell acute lymphoblastic leukemia (B-ALL) [3, 43]. In AML, it is significantly upregulated, being a direct target gene of the MLL fusion proteins and resulting in increased 5hmC levels [43]. Somatic splice site, missense or nonsense/frameshift mutations in TET1 have been found at low frequencies (~ 1%) in AML and in ~ 6% of T-cell acute lymphoblastic leukemia (T-ALL) [44]. No mutations in TET3 have been characterized, suggesting that they are not tolerated, which is supported by the lethal nature of its knockout in mouse embryos [45] whereas Tet1 −/− and Tet2 −/− mice largely develop normally, although TET2 deficiency leads to myeloid malignances later [46–49].
TETs and hypoxia
As the TETs require oxygen for their catalysis, it is of interest to study how hypoxia affects the catalytic activity of TETs and global 5hmC levels. As many tumors are known to be hypoxic [50], a connection between TETs and tumorigenesis has been hypothesized. Moreover, the effects of hypoxia on the expression of TET mRNAs and transcription of the genes regulated by TETs have been examined. The response of TETs to hypoxia appears to be cell-type specific. In neuroblastoma cells, hypoxia increases the global 5hmC levels accumulating in canonical hypoxia response genes and induces TET1 mRNA levels via hypoxia-inducible factor (HIF)1 [51]. The full induction of the hypoxia response in these cells is nevertheless dependent on TET1 [51]. Interestingly, in the neuroblastoma cells hypoxia-inducible genes appear to be regulated in a multilayered manner including HIF stabilization and epigenetic regulation via TETs and 5hmC level, and not all HIF target genes are regulated equally via TETs [15]. In human retinal pigment epithelial (RPE) cells, chemical hypoxia induced by cobalt chloride increased TET1 and TET2 mRNA levels but reduced the methylation status of the promoter regions of the corresponding genes [52], while in several cancer cells and murine ES cells hypoxia specifically reduces 5hmC levels in gene promoters and causes either a modest or no effect on TETs 1-3 mRNA levels [18]. In hypermethylated cancer patient samples, a reduction in 5hmC levels was seen especially in tumor-suppressor gene promoters [18]. Data have been presented to suggest that these reductions are specifically due to severe pathophysiological tumor hypoxia that impairs the TET catalytic activity [18]. Modest hypoxia (2–5% O2) did not inactivate the TETs [18], an observation which is in line with the reported K m values for TET1 and TET2 (Table 1).
TETs and 2-OG analogues
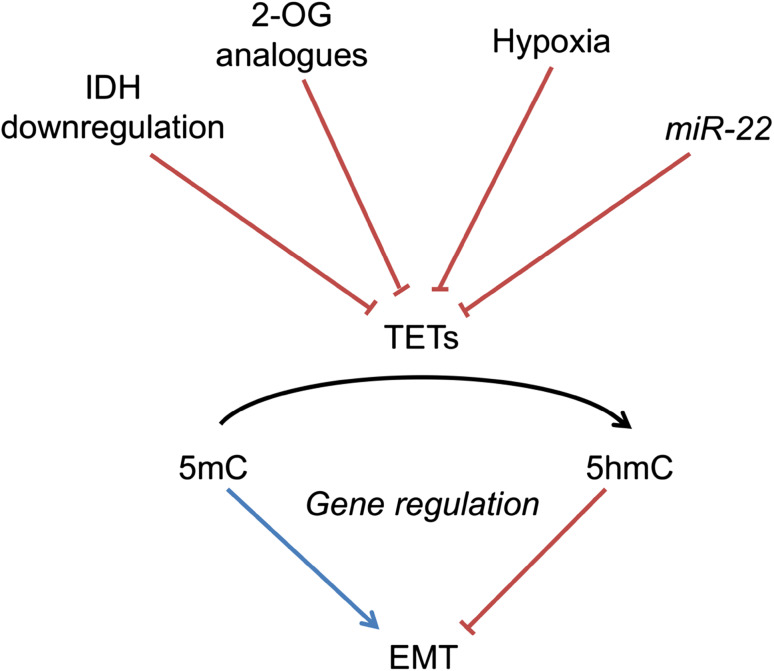
The catalytic activity of the TET enzymes can be inhibited by natural or synthetic 2-OG analogues that act as competitive inhibitors with respect to the cofactor (Figs. 1, 2). Of the Krebs cycle intermediates, succinate and fumarate, which accumulate in succinate dehydrogenase (SDH) and fumarate hydratase (FH) mutant tumors, respectively [53], have been shown to impair TET catalytic activity (Fig. 2) [15, 54]. The IC50 values of TET1 and TET2 for fumarate and succinate were about 400–500 µM, and they reduced global 5hmC levels in neuroblastoma cells (Table 2) [15]. As these oncometabolites can accumulate to high millimolar levels in FH- and SDH-mutant tumor samples [53], inhibition of TETs is likely to be involved in their pathogenesis, although fumarate and succinate are also potent inhibitors of the HIF-P4Hs and collagen P4H-I, and succinate additionally of KDM4E histone lysine demethylase (Table 2). Further support for the role of TET inhibition by succinate in tumorigenesis is provided by the evidence on SDH-mutant paragangliomas, which present a hypermethylator phenotype [55]. In mouse ES cells, a high 2-OG/succinate ratio is found to contribute to pluripotency-associated gene expression via TETs [56]. Dimethylfumarate (DMF), a cell-permeable form of fumarate, has been used for a few years now as an efficient immunomodulatory agent in the treatment of relapsing–remitting multiple sclerosis [57]. Though not studied in detail, treatment with DMF is likely to signal via several 2-OGDDs, including the TETs (Table 2). R-2-hydroxyglutarate (R-2HG), generated by isocitrate dehydrogenase (IDH) mutations in glioblastoma, AML, cholangiocarcinoma or chondrosarcoma, impairs TET activity [58] even though the IC50 values of TET1 and TET2 for R-2HG are at a high millimolar level (Table 2). Within the 2-OGDD family, several KDMs, FIH, the alkylation damage-correcting DNA demethylase ABH2 and collagen P4H-I are more sensitive than TETs to inhibition by R-2HG, whereas this does not efficiently inhibit the HIF-P4Hs, but rather can support their activity (Table 2) [58–60]. However, as R-2HG can accumulate to 5–35 mM levels in IDH-mutant tumors [61] inhibition of TET catalytic activity is also likely to occur in them. S-2-hydroxyglutarate (S-2HG), an enantiomer of R-2HG that has been found to accumulate in a rare metabolic disease caused by a defect in S-2HG dehydrogenase [62, 63], inhibits TET1 and TET2 with about fourfold lower IC50 values than R-2HG (Table 2) [15, 58]. Citrate, a key regulator of lipid metabolism and inhibitor of HIF-P4H-3, FIH and collagen P4H-I, was not an effective inhibitor of TET1 or TET2 (Table 2). Like other 2-OGDDs, TETs can be targeted with synthetic 2-OG analogue inhibitors such as dimethyloxalyl glycine (DMOG) [64] that have shown a potential for treating ischemic, anemic or inflammatory conditions by being efficient inhibitors of the HIF-P4Hs [65–68]. However, in the case of the TETs such inhibitors of catalytic activity may only possess therapeutic potential for the treatment of MLL-fusion AMLs, in which TET1 overexpression is involved in the disease mechanism [43].
Fig. 2.
Factors influencing gene regulation via TET enzymes
Table 2.
IC50 values of TETs and several other 2-OGDDs for 2-oxoglutarate analogues
| Fumarate (µM) | Succinate (µM) | R-2-hydroxyglutarate (µM) | S-2-hydroxyglutarate (µM) | Citrate (µM) | |
|---|---|---|---|---|---|
| TET1 | 390a | 540a | 4000b | 1000b | > 5000a |
| TET2 | 400a | 570a | 5000b | 1600b | > 5000a |
| HIF-P4H-1 | 120c | 830c | Not an inhibitorb | 630b,d | 6300c |
| HIF-P4H-2 | 80c | 510c | Not an inhibitorb/7300e | 1150b,d | 4800c |
| HIF-P4H-3 | 60c | 570c | Not an inhibitorb | 90b,d | 550c |
| Collagen P4H-I | 190c,d | 400d,f | 1800b | 310b | 450d,g |
| FIH | > 10,000c | > 10,000c | 1100–1500b,e | 190–300b,e | 850c |
| KDM4A | 1500h | 800h | 20e | 30e | ND |
| KDM4C | ND | ND | 80e | 100e | ND |
| KDM4E | 2300i | 320i | ND | ND | ND |
| ABH2 | ND | ND | 420e | 150e | ND |
TET enzymes contribute to cell differentiation and transformation
Altered activity of the TET enzymes and the concomitant changes in 5hmC levels and DNA methylation have recently been associated with pathological changes in cell differentiation and transformation. First, reduced global 5hmC levels were found in many human cancers, e.g. melanoma, glioblastoma, seminoma, breast, prostate, urothelial, gastric and renal cancers [20, 21, 26, 27, 32, 35, 69–72], while later, loss of 5hmC was found to be an epigenetic hallmark of many cancers and silencing of the TET enzymes was found to be a key mediator not only of reduced 5hmC levels but also of EMT in cancer cells [26, 27].
Loss of 5hmC has been shown to be an epigenetic hallmark of human melanoma [26, 73]. Low 5hmC levels have been shown to be a marker of melanoma progression and to correlate negatively with Breslow scores and mitotic rate, well-established markers used for melanoma staging. Loss of 5hmC is present in both primary melanomas and metastases and appears to be mediated by downregulation of IDH2 and TET expression [26]. The most dramatic decrease was seen in the TET2 mRNA levels and, interestingly, modeling the overexpression of IDH2 or TET2 increased the previously lost 5hmC levels and suppressed tumor invasion and growth. Later article paper by Gong et al. shed light on the mechanism linking loss of 5hmC to melanoma progression. It was reported that TGF-β1-induced downregulation of TET2 and TET3 was responsible for EMT in melanoma cells [74]. TGF-β1 increased the methylation of TET2 and TET3 promoters through recruitment of DNMT3A methyltransferase, and a compound inhibiting gene methylation, 5-aza-2′-deoxycytidine, was able to reverse the downregulation of TET2 and TET3 and furthermore inhibit EMT [74]. Also, overexpression of TET2 was able to inhibit the EMT process and suppress tumor growth and metastasis. In addition to melanoma, TET1 expression is downregulated in non-Hodgkin B-cell lymphoma and acute B-lymphocytic leukemia [75], and also in prostate cancer [71]. TET1 downregulation was also linked to lower expression of tissue inhibitors of metalloproteinase (TIMPs), which in turn contributes to tumor invasion [71].
Interestingly, oncogenic micro-RNAs have also been shown to downregulate TETs, and TETs themselves have been shown to regulate the expression of tumor-suppressor micro-RNAs [27]. It was found during an attempt to explain the connection between TETs and EMT that TETs and TDG-mediated DNA demethylation are essential for mesenchymal epithelial transition and somatic cell differentiation [37]. TET-mediated demethylation of a tumor-suppressor micro-RNA family called miR-200, which is responsible for modulating the expression of EMT transcription factors such as Zeb1 and Zeb2, was reported to be essential for cell differentiation [37]. Song et al. [27] in turn showed that oncogenic miR-22 is able to downregulate TETs and, therefore, inhibit expression of the tumor-suppressive miR-200 family.
Since the cancer-associated 2-OG analogues such as fumarate, succinate, R-2HG and S-2HG have been shown to inhibit the TETs (Table 2) [15, 54], it is intriguing to hypothesize that, in addition to downregulation of TETs, inhibition of the catalytic activity of TETs by these compounds could be responsible for the low 5hmC levels and EMT in cancers with IDH1 or 2, FH or SDH loss-of-function mutations [53, 55, 58]. Interestingly, it has recently been shown that fumarate is also able to promote EMT in FH −/− cells by inhibiting the TET-mediated demethylation of the antimetastatic miRNA cluster mir-200ba429 [76].
Taken together, the TET enzymes have been consistently shown to be key mediators in cell differentiation and transformation, most notably in EMT. The DNA demethylation controlled by TETs appears to be a dynamic process that can control the state of cell differentiation. In cancers, pathological states resulting in hypoxia, the accumulation of 2-OG analogues, a scarcity of 2-OG, e.g. via IDH downregulation, or micro-RNAs, such as miR22, can affect the activity or expression of the TET enzymes and, therefore, result in pathological cell differentiation and transition, leading to increased tumor aggressiveness and invasion (Fig. 2).
The diagnostic and prognostic value of 5hmC levels in cancer
As low 5hmC levels have been found in many cancers, several evaluations of its value as a diagnostic or prognostic marker have been published. Promising suggestions that 5hmC levels could be used as a diagnostic or prognostic tool in many cancers have emerged.
Since Lian et al. [26] showed that loss of 5hmC is an epigenetic hallmark of melanoma, it has been reported that 5hmC expression is not only associated with the prognosis for melanoma but could also be used for microstaging the disease and more notably for differentiation between benign congenital nevi and malignant melanoma [73, 77]. In these studies, malignant melanomas showed an almost complete loss of 5hmC, whereas benign proliferative nodules displayed high 5hmC expression [77]. Low 5hmC levels were also associated with a high mitotic rate and high Breslow scores [73]. Besides melanoma, reduced 5hmC levels are found in laryngeal and esophageal squamous cell carcinomas and are associated with a higher TNM score and lower overall survival [32, 78, 79]. In non-small cell lung cancer, 5hmC expression correlated negatively with a higher tumor stage, lymph node metastasis and primary tumor size [34]. Orr et al. [35] associated low 5hmC levels in cases of malignant human glioma with the neural progenitor phenotype and shorter survival. In additional, low 5hmC levels have been associated with increased aggressiveness and a poorer prognosis in adult T-cell leukemia and chronic lymphocytic leukemia [36, 80]. Interestingly, low 5hmC levels in these leukemias were linked to low levels of TET2 and IDH expression, which was also correlated significantly with the prognosis, underlying the fact that TETs mediate these effects in cancers.
A recent meta-analysis combining data from numerous cancers of various types reports that decreased 5hmC levels correlate with overall cancer progression and poor survival [33]. In general, these new studies show that determining the 5hmC levels in various tumors and cancers could be a valuable diagnostic tool and an aid to cancer staging and prediction of the prognosis for the disease. In addition to the use of 5hmC as a diagnostic and prognostic tool, cancers with low 5hmC levels might be more susceptible to treatment with hypomethylating agents. One promising aspect is that previous studies have shown AML and MDS patients with TET2 mutations and low 5hmC levels to have a higher response rate to these agents [81, 82].
Conclusions
The TET enzymes play central roles in regulating gene expression via DNA demethylation in myeloid tissue, ES cells and several other tissues. Dysregulation of TET function and imbalance in genomic 5mC/5hmC levels are associated with oncogenic transformation. Although kinetic analyses suggest that the TET enzymes do not act as cellular oxygen sensors, pathological tumor hypoxia is thought to impair their catalytic activity. The 2-OG analogue oncometabolites such as fumarate, succinate, R-2HG and S-2HG have been shown to signal via TETs. However, we do not currently have any precise knowledge of their potency, or that of oxygen deprivation, for inhibiting all other 2-OGDDs, including several KDMs that can be considered equally credible to the TETs as potential regulators of gene expression and oncogenic transformation. The overall contribution of TET inhibition to carcinogenesis, therefore, remains to be resolved.
Acknowledgements
This work was supported to P.K. by the Academy of Finland Grant 218129, the S. Jusélius Foundation, the Emil Aaltonen Foundation, Finnish Cancer Organizations and the Jane and Aatos Erkko Foundation and to T.L. by the Emil Aaltonen Foundation and the Finnish Medical Foundation.
References
- 1.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 3.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40:4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 7.Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
- 8.He Y, Li B, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song C, Zhang K, He C, Xu G. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Xia W, Wang Q, Towers AJ, Chen J, Gao R, Zhang Y, Yen CA, Lee AY, Li Y, Zhou C, Liu K, Zhang J, Gu TP, Chen X, Chang Z, Leung D, Gao S, Jiang YH, Xie W. Isoform switch of TET1 regulates DNA demethylation and mouse development. Mol Cell. 2016;64:1062–1073. doi: 10.1016/j.molcel.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Liu N, Wang M, Deng W, Schmidt CS, Qin W, Leonhardt H, Spada F. Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules. PLoS One. 2013;8:e62755. doi: 10.1371/journal.pone.0062755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, Li Z, Liu J, Szabo PE, Lu Q, Xu GL, Song J, Pfeifer GP. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016;14:493–505. doi: 10.1016/j.celrep.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, Hogan PG, Aravind L, Rao A. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer C, Gobel K, Nagaraj N, Colantuoni C, Wang M, Muller U, Kremmer E, Rottach A, Leonhardt H. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT) J Biol Chem. 2015;290:4801–4812. doi: 10.1074/jbc.M114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laukka T, Mariani CJ, Ihantola T, Cao JZ, Hokkanen J, Kaelin WG, Jr, Godley LA, Koivunen P. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J Biol Chem. 2016;291:4256–4265. doi: 10.1074/jbc.M115.688762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 17.Hirsila M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 2005;19:1308–1310. doi: 10.1096/fj.04-3399fje. [DOI] [PubMed] [Google Scholar]
- 18.Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, Ghesquiere B, Van Dyck L, Boeckx B, Schoonjans L, Hermans E, Amant F, Kristensen VN, Peng Koh K, Mazzone M, Coleman M, Carell T, Carmeliet P, Lambrechts D. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, Lou Z, Zhang L, Li W, Gong W, Liu M, Sun C, Yin X, Li J, Tan X, Wang P, Wang Y, Fang D, Cui Q, Yang P, He C, Jiang H, Luo C, Xu Y. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527:118–122. doi: 10.1038/nature15713. [DOI] [PubMed] [Google Scholar]
- 20.Nettersheim D, Heukamp LC, Fronhoffs F, Grewe MJ, Haas N, Waha A, Honecker F, Waha A, Kristiansen G, Schorle H. Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PLoS One. 2013;8:e82881. doi: 10.1371/journal.pone.0082881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benesova M, Trejbalova K, Kucerova D, Vernerova Z, Hron T, Szabo A, Amouroux R, Klezl P, Hajkova P, Hejnar J. Overexpression of TET dioxygenases in seminomas associates with low levels of DNA methylation and hydroxymethylation. Mol Carcinog. 2017;56:1837–1850. doi: 10.1002/mc.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasanthakumar A, Godley LA. 5-Hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 2015;208:167–177. doi: 10.1016/j.cancergen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Ito Shinsuke, Shen Li, Dai Qing, Wu Susan C, Collins Leonard B, Swenberg James A, He Chuan, Zhang Yi. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iurlaro M, Ficz G, Oxley D, Raiber E, Bachman M, Booth MJ, Andrews S, Balasubramanian S, Reik W. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iurlaro M, McInroy GR, Burgess HE, Dean W, Raiber E, Bachman M, Beraldi D, Balasubramanian S, Reik W. In vivo genome-wide profiling reveals a tissue-specific role for 5-formylcytosine. Genome Biol. 2016;17:141. doi: 10.1186/s13059-016-1001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, Richardson AL, Pandolfi PP. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med. 2017;49:e323. doi: 10.1038/emm.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Yu Y, Luo M, Zhang Z, Shi S, Feng X, Chen Z, He J. Loss of 5-hydroxymethylcytosine is an independent unfavorable prognostic factor for esophageal squamous cell carcinoma. PLoS One. 2016;11:e0153100. doi: 10.1371/journal.pone.0153100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Shi X, Guo L, Li Y, Luo M, He J. Decreased 5-hydroxymethylcytosine levels correlate with cancer progression and poor survival: a systematic review and meta-analysis. Oncotarget. 2017;8:1944–1952. doi: 10.18632/oncotarget.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y, Gu J, Wu Y, Long X, Ge DI, Xu J, Ding J. Low level of 5-hydroxymethylcytosine predicts poor prognosis in non-small cell lung cancer. Oncol Lett. 2016;11:3753–3760. doi: 10.3892/ol.2016.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Damme M, Crompot E, Meuleman N, Maerevoet M, Mineur P, Bron D, Lagneaux L, Stamatopoulos B. Characterization of TET and IDH gene expression in chronic lymphocytic leukemia: comparison with normal B cells and prognostic significance. Clin Epigenet. 2016;8:132. doi: 10.1186/s13148-016-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, Xu GF, Dai HQ, Shi YG, Li X, Hu B, Tang F, Pei D, Xu GL. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 40.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–496. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 41.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 42.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Jiang X, Li Z, Li Y, Song CX, He C, Sun M, Chen P, Gurbuxani S, Wang J, Hong GM, Elkahloun AG, Arnovitz S, Wang J, Szulwach K, Lin L, Street C, Wunderlich M, Dawlaty M, Neilly MB, Jaenisch R, Yang FC, Mulloy JC, Jin P, Liu PP, Rowley JD, Xu M, He C, Chen J. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, Lahortiga I, Luca R, Yan J, Hulselmans G, Vranckx H, Vandepoel R, Sweron B, Jacobs K, Mentens N, Wlodarska I, Cauwelier B, Cloos J, Soulier J, Uyttebroeck A, Bagni C, Hassan BA, Vandenberghe P, Johnson AW, Aerts S, Cools J. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 46.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Mariani CJ, Vasanthakumar A, Madzo J, Yesilkanal A, Bhagat T, Yu Y, Bhattacharyya S, Wenger RH, Cohn SL, Nanduri J, Verma A, Prabhakar NR, Godley LA. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014;7:1343–1352. doi: 10.1016/j.celrep.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alivand MR, Soheili ZS, Pornour M, Solali S, Sabouni F. Novel epigenetic controlling of hypoxia pathway related to overexpression and promoter hypomethylation of TET1 and TET2 in RPE cells. J Cell Biochem. 2017;118:3193–3204. doi: 10.1002/jcb.25965. [DOI] [PubMed] [Google Scholar]
- 53.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, Hargreaves IP, Heales SJ, Chung YL, Griffiths JR, Dalgleish A, McGrath JA, Gleeson MJ, Hodgson SV, Poulsom R, Rustin P, Tomlinson IP. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 54.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, Buffet A, Marcaillou C, Bertherat J, Amar L, Rustin P, De Reynies A, Gimenez-Roqueplo AP, Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 56.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dargahi N, Katsara M, Tselios T, Androutsou ME, de Courten M, Matsoukas J, Apostolopoulos V. Multiple sclerosis: immunopathology and treatment update. Brain Sci. 2017 doi: 10.3390/brainsci7070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG., Jr Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rzem R, Veiga-da-Cunha M, Noel G, Goffette S, Nassogne MC, Tabarki B, Scholler C, Marquardt T, Vikkula M, Van Schaftingen E. A gene encoding a putative FAD-dependent l-2-hydroxyglutarate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proc Natl Acad Sci USA. 2004;101:16849–16854. doi: 10.1073/pnas.0404840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Topcu M, Jobard F, Halliez S, Coskun T, Yalcinkayal C, Gerceker FO, Wanders RJ, Prud’homme JF, Lathrop M, Ozguc M, Fischer J. l-2-Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum Mol Genet. 2004;13:2803–2811. doi: 10.1093/hmg/ddh300. [DOI] [PubMed] [Google Scholar]
- 64.Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D’Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 66.Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG, Taylor CT. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology. 2008;134:156–165. doi: 10.1053/j.gastro.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 67.Ockaili R, Natarajan R, Salloum F, Fisher BJ, Jones D, Fowler AA, 3rd, Kukreja RC. HIF-1 activation attenuates postischemic myocardial injury: role for heme oxygenase-1 in modulating microvascular chemokine generation. Am J Physiol Heart Circ Physiol. 2005;289:542. doi: 10.1152/ajpheart.00089.2005. [DOI] [PubMed] [Google Scholar]
- 68.Hill P, Shukla D, Tran MG, Aragones J, Cook HT, Carmeliet P, Maxwell PH. Inhibition of hypoxia inducible factor hydroxylases protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2008;19:39–46. doi: 10.1681/ASN.2006090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munari E, Chaux A, Vaghasia AM, Taheri D, Karram S, Bezerra SM, Gonzalez Roibon N, Nelson WG, Yegnasubramanian S, Netto GJ, Haffner MC. Global 5-hydroxymethylcytosine levels are profoundly reduced in multiple genitourinary malignancies. PLoS One. 2016;11:e0146302. doi: 10.1371/journal.pone.0146302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, Netto GJ, De Marzo AM, Yegnasubramanian S. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, Huang HD, Lu YY, Teng YC, Lin ST, Lin RK, Tang FM, Lee SB, Hsu HM, Yu JC, Hsiao PW, Juan LJ. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 72.Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X, Li X, Yuan B, Ke Y, He C, Zhou L, Liu J, Ci W. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26:103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JJ, Cook M, Mihm MC, Xu S, Zhan Q, Wang TJ, Murphy GF, Lian CG. Loss of the epigenetic mark, 5-hydroxymethylcytosine, correlates with small cell/nevoid subpopulations and assists in microstaging of human melanoma. Oncotarget. 2015;6:37995. doi: 10.18632/oncotarget.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gong F, Guo Y, Niu Y, Jin J, Zhang X, Shi X, Zhang L, Li R, Chen L, Ma RZ. Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma. Oncotarget. 2017;8:315–328. doi: 10.18632/oncotarget.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Z, Chen L, Dawlaty MM, Pan F, Weeks O, Zhou Y, Cao Z, Shi H, Wang J, Lin L, Chen S, Yuan W, Qin Z, Ni H, Nimer SD, Yang FC, Jaenisch R, Jin P, Xu M. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep. 2015;13:1692–1704. doi: 10.1016/j.celrep.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa Ana, Henriques Sofia, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MGB, Rajeeve V, Cardaci S, Foster S, Yun H, Cutillas P, Warren A, Gnanapragasam V, Gottlieb E, Franze K, Huntly B, Maher ER, Maxwell PH, Saez-Rodriguez J, Frezza C. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavlova O, Fraitag S, Hohl D. 5-Hydroxymethylcytosine expression in proliferative nodules arising within congenital nevi allows differentiation from malignant melanoma. J Investig Dermatol. 2016;136:2453–2461. doi: 10.1016/j.jid.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 78.Murata A, Baba Y, Ishimoto T, Miyake K, Kosumi K, Harada K, Kurashige J, Iwagami S, Sakamoto Y, Miyamoto Y, Yoshida N, Yamamoto M, Oda S, Watanabe M, Nakao M, Baba H. TET family proteins and 5-hydroxymethylcytosine in esophageal squamous cell carcinoma. Oncotarget. 2015;6:23372–23382. doi: 10.18632/oncotarget.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Wu K, Shao Y, Sui F, Yang Q, Shi B, Hou P, Ji M. Decreased 5-Hydroxymethylcytosine (5-hmC) predicts poor prognosis in early-stage laryngeal squamous cell carcinoma. Am J Cancer Res. 2016;6:1089–1098. [PMC free article] [PubMed] [Google Scholar]
- 80.Marçais A, Waast L, Bruneau J, Hanssens K, Asnafi V, Gaulard P, Suarez F, Dubreuil P, Gessain A, Hermine O, Pique C. Adult T cell leukemia aggressivenness correlates with loss of both 5-hydroxymethylcytosine and TET2 expression. Oncotarget. 2016;8:52256–52268. doi: 10.18632/oncotarget.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, Quesnel B, Vey N, Gelsi-Boyer V, Raynaud S, Preudhomme C, Ades L, Fenaux P, Fontenay M, Groupe Francophone des Myelodysplasies, (GFM) Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 82.Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, Wang H, Caughey B, Stojanov P, Getz G, Garcia-Manero G, Kantarjian H, Chen R, Stone RM, Neuberg D, Steensma DP, Ebert BL. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J Biol Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 84.Koivunen P, Hirsila M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J Biol Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- 85.Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Myllyharju J, Kivirikko KI. Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berg RA, Kishida Y, Sakakibara S, Prockop DJ. Hydroxylation of (Pro-Pro-Gly)5 and (Pro-Pro-Gly)10 by prolyl hydroxylase. Evidence for an asymmetric active site in the enzyme. Biochemistry. 1977;16:1615–1621. doi: 10.1021/bi00627a014. [DOI] [PubMed] [Google Scholar]
- 88.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 89.Hancock RL, Masson N, Dunne K, Flashman E, Kawamura A. The activity of JmjC histone lysine demethylase KDM4A is highly sensitive to oxygen concentrations. ACS Chem Biol. 2017;12:1011–1019. doi: 10.1021/acschembio.6b00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams ST, Walport LJ, Hopkinson RJ, Madden SK, Chowdhury R, Schofield CJ, Kawamura A. Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics. 2014;9:1596–1603. doi: 10.4161/15592294.2014.983381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O’Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 92.Majamaa K, Hanauske-Abel HM, Gunzler V, Kivirikko KI. The 2-oxoglutarate binding site of prolyl 4-hydroxylase. Identification of distinct subsites and evidence for 2-oxoglutarate decarboxylation in a ligand reaction at the enzyme-bound ferrous ion. Eur J Biochem. 1984;138:239–245. doi: 10.1111/j.1432-1033.1984.tb07907.x. [DOI] [PubMed] [Google Scholar]
- 93.Tuderman L, Myllyla R, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 1. Role of co-substrates. Eur J Biochem. 1977;80:341–348. doi: 10.1111/j.1432-1033.1977.tb11888.x. [DOI] [PubMed] [Google Scholar]
- 94.Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J Med Chem. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]