Abstract
Rab proteins are master regulators of intracellular membrane trafficking, but they also contribute to cell division, signaling, polarization, and migration. The majority of the works describing the mechanisms used by Rab proteins to regulate cell motility involve intracellular transport of key molecules important for migration. Interestingly, a few studies indicate that Rabs can modulate the activity of Rho GTPases, important regulators for the cytoskeleton rearrangements, but the mechanisms behind this crosstalk are still poorly understood. In this work, we identify Rab6 as a negative regulator of cell migration in vitro and in vivo. We show that the loss of Rab6 promotes formation of actin protrusions and influences actomyosin dynamics by upregulating Cdc42 activity and downregulating myosin II phosphorylation. We further provide the molecular mechanism behind this regulation demonstrating that Rab6 interacts with both Cdc42 and Trio, a GEF for Cdc42. In sum, our results uncover a mechanism used by Rab proteins to ensure spatial regulation of Rho GTPase activity for coordination of cytoskeleton rearrangements required in migrating cells.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03057-w) contains supplementary material, which is available to authorized users.
Keywords: Rab proteins, Rab6, small GTPases, cell migration
Introduction
Rab proteins constitute the largest family of the Ras superfamily of small GTPases, with more than 60 members in humans. The role of Rab proteins in vesicle transport was identified for the first time in yeast already in the 1980s, and since then, more and more studies have established them as the master regulators of intracellular membrane traffic [1–4].
To perform their tasks each Rab protein cycles between membrane and cytosol, switching between a GTP-bound and a GDP-bound conformation. Upon membrane recruitment, Rab proteins in their GTP-bound state can bind a variety of different effector molecules, including sorting adaptors, tethering factors, fusion regulators, kinases, phosphatases, and motor proteins [4, 5].
Lately, it has been demonstrated that in addition to regulating intracellular traffic, Rab proteins take part in several other cellular processes. Indeed, these small GTPases are also important for regulation of mitotic spindle positioning and abscission during cell division, apical lumen formation and polarization of epithelial cells, and nutrient sensing and signaling [6–9].
Interestingly, an increasing amount of evidence demonstrates that Rab proteins also have a role in cell migration [10–12]. So far, regulation of cytoskeleton dynamics involved in cell shape and motility has been mainly attributed to another family of small GTPases, namely, the Rho proteins, and thus, the contribution by the Rab family to this process remains much less characterized [13, 14]. We recently demonstrated that Rab7b can affect cell migration through regulation of the actin cytoskeleton [10]. By directly interacting with the actin motor myosin II, Rab7b is able to modulate the activity of RhoA and thereby the phosphorylation of myosin light chain (MLC). In this way, Rab7b influences actin cytoskeleton dynamics, including the formation of stress fibers, cell adhesions, and thus cell migration [10].
Intriguingly, also Rab6 is known to interact directly with myosin II in a GTP-dependent manner [15]. Rab6 is an evolutionary conserved and ubiquitously expressed Rab protein [16, 17]. It locates to the Golgi apparatus and Golgi-derived vesicles and regulates many trafficking routes, both anterograde and retrograde, between the Golgi apparatus, endoplasmatic reticulum (ER), plasma membrane (PM), and endosomes [17–21]. In addition, Rab6 is involved in cell division and phagosome maturation [22–24].
To execute all these functions, Rab6 interacts with many different effector molecules, including motor proteins and their interactors such as KIF1C, KIF5B, KIF20A (also known as Rabkinesin-6), the dynein–dynactin complex through Bicaudal D, myosin II, and myosin VA [15, 18, 25–29].
In this study, we investigated whether Rab6, similar to Rab7b, can regulate cell migration through its interaction with myosin II. Our results show that Rab6 knockdown indeed affects cell migration by increasing cell speed and, therefore, wound closure. We also demonstrate that Rab6 depletion influences the actomyosin system by upregulation of Cdc42 activity and downregulation of MLC phosphorylation. Intriguingly, we discover that Rab6 interacts with both the Rho GTPase Cdc42 and Trio, a GEF for Cdc42 [30]. By modulating Cdc42 activity, Rab6 regulates the formation and dynamics of actin-dependent protrusions such as filopodia. We finally confirm that Rab6 is a negative regulator of cell migration using xenotransplantation of human cancer cells into zebrafish embryos. In sum, our results support a novel emerging mechanism for Rab proteins in the regulation of actin cytoskeleton dynamics and cell migration by crosstalk with Rho GTPases.
Materials and methods
Cell culture
U2OS, HeLa, and H1299 cells were grown in Dulbecco´s modified Eagle´s medium (DMEM; Lonza, BioWhittaker). RPE-1 cells were grown DMEM F-12 (Lonza, BioWhittaker). Both DMEM and DMEM F-12 were supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Constructs and antibodies
pEGFP-C1 was purchased from BD Biosciences, Clontech. pEGFP-C1 Rab6A wt, pEGFP-C1 Rab6A Q72L, and pEGFP-C1 Rab6A T27 N were a gift from Marci Scidmore (Addgene plasmid #49,469, #49,483, and #49,484, respectively) [31]. pcDNA3-EGFP-Cdc42-Q61L and pcDNA3-EGFP-Cdc42-T17 N were a gift from Gary Bokoch (Addgene plasmid #12,986 and #12,976, respectively) [32]. pTriEx- mCherry-cdc42 Q61L and the biosensor constructs, pTriEX Cdc42 wt and pTriEX Cdc42 G12 V, were a gift from Luis Hodgson (Albert Einstein College of Medicine, NY, USA). mCherry-Cdc42-C-10 was a gift from Michael Davidson (Addgene plasmid # 55,014). pCMV-LifeAct-RFP was purchased from Ibidi. pEGFP-C3 Cdc42 wt was a gift from Keith Burridge, University of North Carolina, Chapel Hill, USA. pcDNA 3.1-HA Rab6A, pcDNA 3.1-HA Rab6A Q72L, and pcDNA 3.1-HA Rab6A T27 N were purchased from Genscript. pEGFPC1-Trio was a kind gift from Jaap D. Van Buul (University of Amsterdam, The Netherlands) [33].
Primary antibodies used in this study were: anti-giantin (Abcam, ab24586, 1:1000), anti-tubulin (Life Technologies, # 13-8000, 1:12,000), anti-phospho-myosin light chain 2 (Ser19) (Cell Signaling Technology, # 3671, 1:300), anti-myosin light chain (Sigma-Aldrich, # M4401, 1:50), anti-GFP (Abcam, ab6556, 1:3000), anti-HA (Abcam, ab9110, 1:1000), anti-RhoA (Cytoskeleton Inc., ARH04, 1:500), anti-Rac1 (Cytoskeleton Inc., ARC03, 1:500), anti-Cdc42 (Cytoskeleton Inc., ACD03, 1:250), anti-Cdc42 (Abcam, ab155940, 1:200), anti-Trio (Abnova, H00007204-A01), anti-Rab6A (Abcam, ab95954, 1:200), anti-actin (Cytoskeleton Inc., AAN01, 1:500), anti-N-WASP (Cell Signaling Technology, # 30D10, 1:300), anti-IQGAP1 (BD Laboratories, #610,611, 1:300), anti-DOCK10 (Abcam, ab75258, 1:1000), and mouse IgG1 (× 0931, Dako, 1:50). Rhodamine-conjugated phalloidin was purchased from Invitrogen (R415). The SiR-actin kit was purchased from Cytoskeleton, Inc. and used according to manufacturer’s protocol (Cytoskeleton, Inc./Spirochrome #CY-SC001). For immunofluorescence experiments, Alexa Fluor secondary antibodies (Invitrogen) were used at dilution 1:200. Secondary antibodies conjugated to horseradish peroxidase for immunoblotting studies (GE Healthcare) were diluted 1:5000. Hoechst (Life Technologies, H3569) was used at 0.2 µg/ml and DAPI (Sigma-Aldrich, D9542) was used at 0.1 µg/ml.
Transfection and RNA interference
U2OS cells were transiently transfected using Lipofectamine 2000 (Life Technologies), while HeLa cells were transfected using FuGENE 6 (ProMega), following the producer’s protocol. Cells were transfected at approximately 50–70% confluency for 24 h prior to further execution of experiments. U2OS, RPE-1, and H1299 cells were transfected with siRNA using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) according the manufacturer’s instructions. The cells were transfected either the day after plating or the same day by reverse transfection, and analyzed after 72 h. siRNA transfections in HeLa cells were performed using Oligofectamine (Invitrogen) as described previously [34]. In short, cells were plated in 6-cm dishes 1 day prior to transfection (~ 4 × 105 cells/dish), and replated 72 h after transfection. Experiments were performed after 48 h.
Nontargeting control siRNA (sense sequence 5′-ACUUCGAGCGUGCAUGGCUTT-3′ and antisense 5′-AGCCAUGCACGCUCGAAGUTT-3′) was purchased from MWG-Biotech (Ebersberg, Germany).
siRNAs against Rab6 (siRab6 #1, J-008975-08; siRab6 #2. J-008975-09; siRab6 #3, J-008975-07; siRab6 #4, J-008975-10). Trio (siTrio, J-005047-05) and DOCK 10 (siDOCK10, J-023079-05) were purchased from Dharmacon™.
For rescue experiments, cells were first transfected with siRNA using RNAiMax (for U2OS cells) or Oligofectamine (for HeLa cells). After 48 h, the cells were transfected with pEGFP-Rab6 using Lipofectamine 2000 (for U2OS cells) or FuGENE 6 (for HeLa cells) according to the manufacturers’ instructions. The experiments were performed 72 h after siRNA transfection.
Cell migration assays
Cells were grown to form confluent monolayers in IncuCyte ImageLock 96-well plates (Essen Bioscience) and scratched with IncuCyte® WoundMaker (Essen Bioscience). Cell migration was monitored by time-lapse imaging using an IncuCyte® ZOOM (10× objective, Essen Bioscience). Relative wound density (percent) was calculated using the IncuCyte® ZOOM software analysis program. Cell tracking and quantification of velocity and directionality was done using Fiji/ImageJ manual tracking plugin and Ibidi Chemotaxis software.
Particle image velocimetry (PIV) analysis was done using OpenPIV and OpenPIV spatial analysis toolbox in MATLAB R2015b. The interrogation windows were set to 32 × 32-pixels with 50% overlap.
Cell proliferation and cell death assays
Cell proliferation and cell death assays were performed on cells subjected to wound healing assays as described above. For the cell proliferation assays, the Click-iT™ Plus EdU Alexa Fluor™ 488 Imaging Kit (Molecular Probes) was used according to the manufacturer’s instructions. Briefly, medium containing 10 µM EdU was added to the cells before imaging, and after 24 h, the cells were fixed using 3% paraformaldehyde and permabilized with 0.5% Triton® X-100. Incorporated Edu was detected by adding the Click-iT® Plus reaction cocktail as described by the manufacturer, and DNA was stained with 5 µg/ml Hoechst® 33342 (Molecular Probes). Cells were imaged using an Andor Dragonfly microscope with a 10× objective and quantification of the percentage of Edu-positive cells was done using the Fiji/ImageJ analysis software.
For the cell death assays, medium containing 250 nM IncuCyte® Cytotox Reagent (Essen Bioscience) was added to label dying cells green. The number of green objects per image was calculated using the IncuCyte® ZOOM software analysis program. Cell death was quantified by dividing the number of green objects at each timepoint with the number of green objects at time 0.
Golgi reorientation measurements
Cells were grown to form a confluent monolayer on glass slides, scratched with a pipette tip, and incubated for 2 h at 37 °C and 5% CO2. Subsequently, the cells were fixed and stained with anti-giantin, rhodamine-conjugated phalloidin and Hoechst to visualize the Golgi complex, actin cytoskeleton, and nuclei, respectively. Golgi reorientation was measured by dividing the cells on the wound edge into three equal sectors and calculating the percentage of cells having their Golgi apparatus in the front sector (facing in the direction of migration) as previously described [10].
Cell-spreading assay
Cells were seeded onto fibronectin-coated coverslips (10 µg/ml) and incubated for 1 h at 37 °C and 5% CO2 before fixation and staining with rhodamine-conjugated phalloidin and DAPI. Quantification of the cell area was done using the Fiji/ImageJ analysis software.
Micropatterns
96-well CYTOO plates (CYTOO, 20-900-00) containing L-shaped micropattern (1100 µm2) were coated with 20 µg/ml sterile fibronectin (Sigma F2006) in PBS for 2 h at room temperature. 3 × 103 cells were then added to each well and kept at room temperature for 30 min and subsequently at 37 °C for 3 h. Cells were then fixed and stained with rhodamine-conjugated phalloidin and Hoechst.
Analysis of actin distribution in cells seeded on the L-shaped micropatterns was performed in ImageJ, using the CYTOOL-IP Reference Cell macro (RefCell) that provides a statistical representation of the spatial distribution of intracellular compartments, in our case the actin fibers. Using the Reference Cell macro, a normalized mean cell was obtained by alignment and overlay of 40 cells per experiment and averaging the signals for each condition. A Reference Cell was constructed by making a projection of the filtered and aligned images from a stack [35]. The rigidity/collapse of the actin fibers along the hypotenuse was quantified using the Hypotenuse Macro [35]. Briefly, thresholded images of the empty l-micropatterns were used to define the theoretical hypotenuse of the triangle shape. Next, thresholding of the actin stained images was performed to define the actual cell shape. The hypotenuse macro was modified by shifting the theoretical hypotenuse of 2.4 µm to be able to recognize smaller difference in the collapse of the actin fibers. The difference in the areas between the theoretical hypotenuse and the actual cell border was then measured and represented as percentage of collapsed cells (curved when the area difference resulted in a positive value) and non-collapsed cells (straight when no differences in the areas between the theoretical hypotenuse and the actual cell border were detected).
Immunofluorescence and live-cell microscopy
Cells were grown on coverslips, fixed with 3% paraformaldehyde, and quenched using 50 mM NH4Cl. Afterwards, 0.25% saponin in PBS 1× was used for permeabilization and 5% FCS in PBS 1× for blocking. The cells were incubated with primary antibodies at room temperature for 20 min, washed in PBS/saponin, incubated with secondary antibodies in darkness at room temperature for 20 min, washed again in PBS/saponin, and finally mounted with Mowiol.
Fixed cells were imaged with an Olympus FluoView 1000 IX81 confocal laser scanning microscope (inverted) using a 60× PlanApo NA 1.35 objective, or with an Olympus FluoView 1000 BX61WI confocal laser scanning microscope (upright) using a 60× PlanApo 1.4 objective and FV1000 software.
For live-cell imaging, cells were seeded on MatTek glass-bottom dishes. Before imaging, the culture medium was replaced to phenol red-free DMEM. For live-cell image acquisition the cells were kept in an incubation chamber (Solent Scientific) at 37 °C and 5% CO2 and imaged using a Yokagawa CSU22 spinning-disk confocal unit with an Andor Ixon EMCCD camera and a 60× NA 1.42 objective provided with the Andor iQ1.8 software.
Total internal reflection fluorescence (TIRF) microscopy acquisition was done on an Andor Dragonfly microscope using a 100× objective with NA 1.45 and the Fusion software.
To measure Cdc42 activity in live cells, a biosensor for Cdc42 based on Förster resonance energy transfer (FRET) was used [49]. This single-chain biosensor incorporates the donor/acceptor FRET pair of the monomeric Cerulean (mCer) and monomeric Venus (mVen) fluorescent proteins. When Cdc42 is in an inactive GDP-bound state, the mCer and mVen are at a distance from each other, only given off a weak FRET signal when excited. However, when Cdc42 upon binding to GTP changes its conformation, the mCer and mVen fluorescent proteins are brought into closer proximity and therefore the FRET signal increases. For excitation of the biosensor (excitation of mCer), the 405 nm laser was used to reduce the amount of mVen cross excitation to a minimum. The emission was measured using the Zeiss LSM880 microscope equipped with a 32 array spectral GaAsP detector using a 63× oil C Plan Apo objective with NA 1.4 or a 40× oil Plan Apo objective with NA 1.3 and the ZEN Black software. To measure the activity of Cdc42, the emission values were recorded at 477 nm, which corresponds to the maximum emission peak of mCer, and at 530 nm, which corresponds to the maximum emission peak for mVen. Since the 405 nm laser was used for excitation, which does not excite mVen, the observed emission at 530 nm of mVen corresponds to the FRET in the sample. To make our measurements comparable between cells, we divided the intensities at 530 nm by the intensities at 477 nm to obtain the FRET/mCer ratio. Image acquisition was performed on a focal plane close to the cell surface/dish.
Optical tweezers
HeLa cells were seeded on MatTek glass-bottom dishes at low confluency to assure a good access for the optical tweezers. Before mounting the dishes on the optical tweezers setup, the medium was replaced by phenol red-free DMEM containing 1 µm carboxylated latex beads (Bangs Laboratories). The experiments were performed on an optical tweezers setup (NanoTracker2, JPK, Germany) equipped with a confocal unit (C2, Nikon, Japan) and a temperature-controlled stage at 37 °C. Before every experiment, the optical trap and the QPD detection system were calibrated using the implemented calibration function based on thermal fluctuations [36]. To investigate the filopodial properties, a trapped bead was moved close to the filopodial tip. After binding of the bead to the filopodium, the filopodium usually starts to retract [37, 38]. The bead’s position was tracked during the entire retraction process of each filopodium. Hereby, a force clamp was used, where the set point has been changed every 10 s to test the reaction on different forces. The trajectory segments were analyzed by a linear fit of the projection on the main retraction velocity, which reveals a velocity for the respective counteracting force during acquisition of the segment. Velocities below a threshold of 0.5 nm/s were considered as a stall event.
Immunoblotting
Cell lysates were subjected to SDS-PAGE and blotted onto polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were incubated with primary antibodies diluted in 2% blotting grade non-fat dry milk (BioRad), followed by secondary antibodies conjugated to horseradish peroxidase (HRP) (GE Healthcare). Either the ECL Prime Western Blotting Detection (GE Healthcare) or the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) were used to detect the chemiluminescent signals. Quantification of band intensity was done by densitrometry analysis with the ImageQuant TL software (GE Healthcare) or with the Carestream software.
RhoA, Rac1, and Cdc42 activation assay
RhoA, Rac1, and Cdc42 activation was assessed using the Activation Assay Biochem Kit (Cytoskeleton, Inc.) according to manufacturer’s protocol. Briefly, GTP-bound RhoA was immunoprecipitated from cell lysates with beads coupled to the Rho-binding domain (RBD) of the Rho effector protein rhotekin, while Rac1 and Cdc42 were immunoprecipitated using beads coupled to the Interactive Binding (CRIB/PDB) region of the Cdc42/Rac1 effector protein p21 activated kinase I (PAK1). Immunoprecipitated samples and total lysates were analyzed by immunoblotting.
F- and G-actin quantification
F- and G-actin extractions were performed with the G-Actin/F-actin In Vivo Assay Biochem Kit (Cytoskeleton, Inc.) according to the manufacturer’s instructions. Briefly, U2OS cells were lysed in a buffer that stabilized and maintained the globular and filamentous forms of actin, incubated for 10 min at 37 °C, and subjected to ultracentrifugation for 2 h at 100,000×g at room temperature. The supernatants, containing the G-actin fractions, and the re-suspended pellets, containing F-actin, were subjected to Western Blot analysis. The amounts of F-actin and G-actin relative to the total actin were quantified by detecting the intensity of the bands using the program ImageQuant (Amersham Biosciences). To control that the F-actin was efficiently pelleted during centrifugation, 100× phalloidin (AE01, Cytoskeleton inc.) was added at 1× final concentration to an additional control sample directly after lysis.
Co-immunoprecipitation
Co-immunoprecipitation experiments were done using the GFP-Trap®_MA (Chromotek) according to the producer’s protocol. In short, lysates from cells transfected with GFP-fusion proteins were incubated for 1 h at 4 °C with magnetic beads coupled to anti-GFP antibodies for co-immunoprecipitation. Immunoprecipitated samples and total lysates were subsequently loaded on SDS-PAGE gels and subjected to western blotting analysis.
Dynabead protein G (Life Technologies) was used according to the manufacturer’s protocol for IP of HA-Rab6. Briefly, Dynabeads (0.6 µg) were washed in RIPA buffer and incubated with either IgG isotype control or with the antibody against HA, for 60 min with end-over-end-rotation at room temperature. Precleared cell lysates were thereafter incubated with the antibody-coupled beads for 90 min with end-over-end-rotation at room temperature. Immunoprecipitated samples were loaded on SDS-PAGE and analyzed by western blotting.
Zebrafish xenotransplants
Wild-type zebrafish embryos were kept at 28.5 °C in standard embryo medium [39] containing 0.003% phenylthiourea (Aldrich). The experiments were conducted in agreement with the provisions enforced by the Norwegian national animal research authority (NARA).
Embryos were decorionated 24 h post-fertilization (hpf). At day 2 post-fertilization, H1299 were injected in the otic vesicle. For this, larvae were first sedated in a 230 µg/ml tricaine bath before being placed on a dish containing 2% of hardened agarose gel in water.
Before the injection, 105 H1299 cells were transfected for 16–20 h with either a control siRNA or a siRNA against Rab6. For rescue experiments, cells were electroporated with GFP-Rab6 (Amaxa nucleofector, Lonza) the day after silencing. 6–7 h after electroporation the cells were injected in the otic vesicle of zebrafish embryos. Before the injection, cells were stained with Q-tracker 655 cell-labelling kit (Life Technologies) for 45 min at 37 °C following the manufacturer’s instructions. After staining, cells were trypsinized, re-suspended in culture medium, and centrifuged for 8 min at 400×g. The supernatant was discarded and 1 ml PBS −/− Ca2+/Mg2+ was added to each cell pellet. Cells were re-suspended and centrifuged again for 8 min, at 400×g. The PBS was removed until the pellet was only partially covered and re-suspended in the small remaining volume of PBS. The cell suspension was then loaded onto a glass capillary connected to a pump (Eppendorf Femtojet Express) and controlled using a micromanipulator (MN153, Narishige). Prior to injection, the number of cells was regulated by varying the applied pressure of the pump. 2–3 nl of cell suspension was injected in the otic vesicle of each animal that were subsequently placed at 35 °C to facilitate cell growth. No adverse effects on embryo development were observed. Images were acquired directly after the injection (time zero) and at 24 and 48 h post-injection using a Leica DFC365FX stereomicroscope with a 1.0× PlanApo lens. For the quantification of cell migration, the distance of the cells from the injection site was measured as the area obtained by drawing a line along the cells outside of the injection site at 24 and 48 h after the injection. The quantification was performed using the software ImageJ.
Image processing and analysis
Image analysis and processing was performed using ImageJ (National Institutes of Health) and Adobe Photoshop (Adobe Systems). Analysis of the total filopodia number per cell was done using FiloQuant plugin for ImageJ, kindly provided by Guillaume Jacquemet, University of Turku, Finland.
For the analysis of filopodia at the leading edge of migrating cells, the FiloQuant plugin was used to measure the cell border, and the filopodia were manually counted to include only protrusions extending from the membrane facing the wound.
For the quantification of Cdc42-positive vesicles in the cell front of migrating cells, the total number of Cdc42-positive vesicles was calculated using ImageJ. A line parallel to the migration front was drawn over the perinuclear region, half-way between the nucleus and the cell border and the number of vesicles above the line in the cell front was counted and expressed as percentage compared to the total number of vesicles per cell.
Statistical analysis
Assessment of statistical differences was done by two-tailed paired Student’s t test using the Excel software, or ANOVA followed by post hoc tests in the GraphPad Prism 8 software as indicated in the figure legends. In the figures, statistical significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001. The statistical analysis of the filopodial retraction force and velocity was done in Matlab using a two-sample Student’s t test and a two-sample Kolmogorov–Smirnov test.
Results
Rab6 depletion increases cell migration
Rab proteins are master regulators of intracellular membrane transport. However, increasing evidence shows that Rabs are also involved in the process of cell migration [10–12]. Rab7b interferes with cell migration by modulating actomyosin dynamics through direct interaction with the actin motor myosin II [10]. Since also Rab6 interacts directly with myosin II [15], we investigated whether Rab6, similar to Rab7b, is important for the process of cell migration.
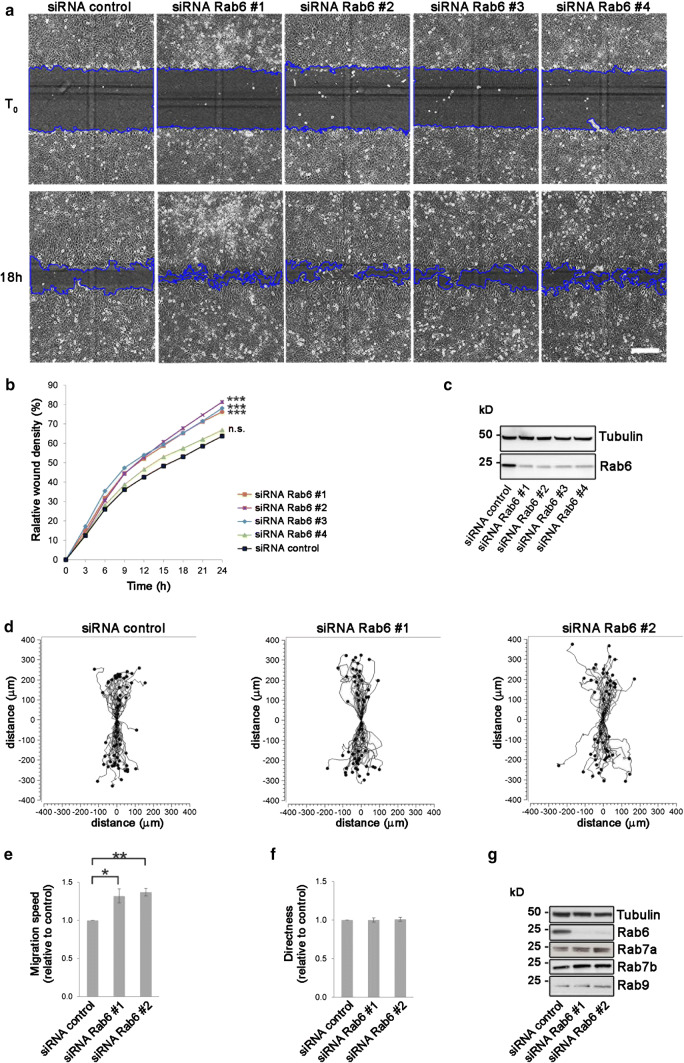
To test our hypothesis, we performed a wound healing assay using U2OS cells transfected with an siRNA control or with four different siRNAs against Rab6 (siRNA Rab6 #1, siRNARab6 #2, siRNA Rab6 #3, and siRNA Rab6 #4). The quantification of the relative wound density showed that Rab6 knockdown does indeed affect cell migration by increasing the rate of wound closure (Fig. 1a–c). Importantly, all the four different siRNAs used to knockdown Rab6 gave similar results, with an increase in wound density of approximately 20% 24 h after wounding (except siRNA Rab6 #4 that resulted in only 5% increase), indicating that Rab6 specifically influences cell migration and that the result obtained was unlikely caused by off-target effects.
Fig. 1.
Rab6 silencing promotes cell migration. a U2OS cells transfected with control siRNA or 4 different siRNAs against Rab6 (#1, #2, #3, #4) were scratch-wounded and imaged every 3rd hour for 24 h. Representative images of (T0) and 18 h after scratching are shown. Scale bar: 300 µm. b Quantification of the relative wound density (%) as function of time for control cells and Rab6-depleted cells. Data represents the mean of three independent experiments. ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t = 24 h). c Cell lysates from each sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). d Representative track plots of the single-cell distances of migration for cells transfected with siRNA control, siRNA Rab6#1 or siRNA Rab6#2. Individual tracks are shown so that each starts at the origin (distance 0). Quantification of the mean ± SEM of speed (e) and directness (f) relative to the control. n > 150 cells from at least five independent experiments. *P < 0.05; **P < 0.01 (paired Student’s t test). g Lysates from cells transfected with either siRNA control or siRNA against Rab6 (#1 and #2) were subjected to Western blot analysis with the indicated antibodies
We further analyzed in more detail the effect of Rab6 depletion on migrating cells. For this purpose, we chose the two siRNAs that gave the strongest effect in the wound healing assay (siRNA Rab6 #1 and siRNA Rab6 #2) and measured single-cell speed and directionality in both U2OS (cancer cell line) and RPE-1 cells (epithelial cell line). The single-cell analysis revealed that the depletion of Rab6 increased cell speed by about 30% compared to control cells in both U2OS and RPE-1 cells, while directionality was only modestly affected (Fig. 1d–g; Suppl. Fig. 1a–f). The movement of continuous sheets of epithelial RPE-1 cells was also visualized by velocity fields, showing that the increased migration speed upon Rab6 depletion can be observed throughout the cell monolayer (Suppl. Fig. 1g).
The re-expression of Rab6 in U2OS cells depleted for this small GTPase, using either oligos, rescued the migration at levels similar to the control (Suppl. Fig. 2a, b), thus excluding that this effect was a consequence of increased proliferation or reduced cell death (Suppl. Fig. 2c, d). This further validates the specificity of the Rab6 siRNAs and of the migration phenotype upon Rab6 knockdown.
In addition to U2OS and RPE-1 cells, we also measured the effect on cell migration upon Rab6 silencing on HeLa cells. Similar to the previously tested cell lines, also migration in HeLa cells was increased in cells silenced with either oligos (Suppl. Fig. 2e, g).
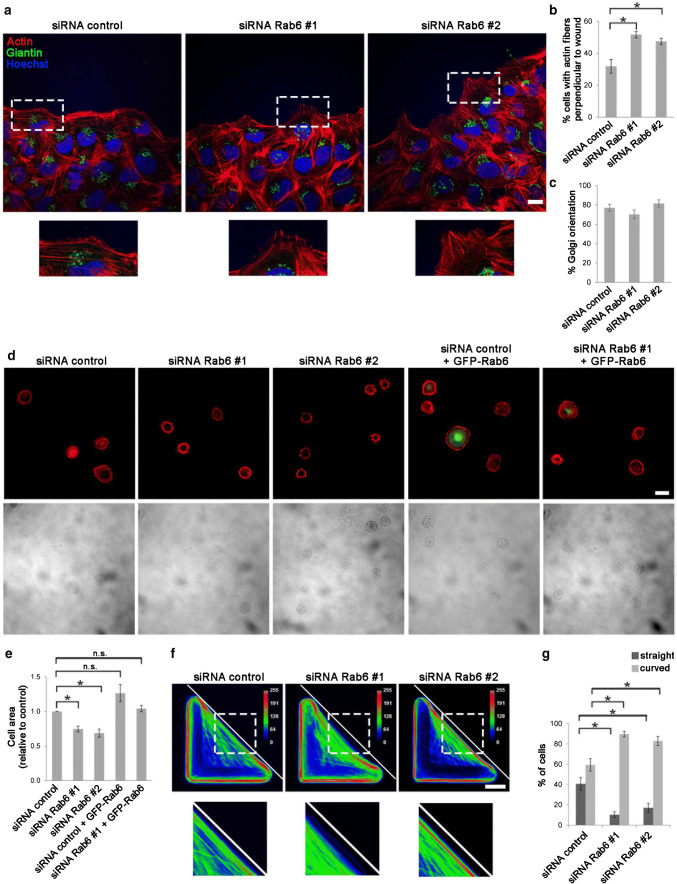
Having discovered that depletion of Rab6 increases cell migration, and knowing that Rab6 directly interacts with myosin II [15], we next investigated whether the effect of Rab6 on cell migration is a consequence of an effect on the actomyosin cytoskeleton. U2OS cells transfected with siRNA control or siRNAs against Rab6 were subjected to wound healing assays and the arrangement of the actin cytoskeleton was analyzed by confocal microscopy. The results showed a clear difference in the formation of actin protrusions at the leading edge of the migrating cells. In about 50% of the cells silenced for Rab6 the actin fibers were oriented perpendicular to the wound in contrast to only 30% for control cells (Fig. 2a, b). This result suggests that Rab6 influences actin dynamics.
Fig. 2.
Rab6 influences actin dynamics and cell spreading. a U2OS cells treated with siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratched with a pipet tip and fixed after 2 h. Cells were immunostained with an antibody against giantin. Actin was labeled with rhodamine-conjugated phalloidin and nuclei with Hoechst. The lower insets show magnifications of the boxed areas. Scale bar: 20 µm. b Quantification of the percentage of cells with actin fibers perpendicular to the wound is represented as mean ± SEM; n > 150 cells from four independent experiments. *P < 0.05 (paired Student’s t test). c Quantification of the percentage of cells having Golgi located between the nucleus and the leading edge. The graph shows the mean ± SEM; n > 120 cells from four independent experiments. d U2OS cells transfected with siRNA control, siRNA Rab6 #1, siRNA Rab6 #2, or silenced with siRNA control or siRNA Rab6 #1 and subsequently transfected with GFP-Rab6 were plated on fibronectin-coated coverslips and left to adhere for 1 h before fixation and staining with Hoechst and rhodamine-conjugated phalloidin. Lower panel shows transmission images for comparison. Scale bar: 20 µm. e Quantification of the average area. The graph represents the mean ± SEM normalized to the control; n > 90 cells from at least three independent experiments. *P < 0.05 (one-way ANOVA followed by Tukey’s post test). f U2OS cells treated with siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were plated onto dishes with fibronectin-coated L-shaped micropattern and left to adhere for 3.5 h before fixation and staining with rhodamine-conjugated phalloidin. Color-coded frequency map of averaged Z-projected images from one representative experiment are shown. Scale bar: 10 µm. g The graph shows the quantification for each condition of the percentage of cells with straight or curved hypotenuse (mean ± SEM) normalized to the control. n > 60 cells from three independent experiments. *P < 0.05; **P < 0.01 (paired Student’s t test)
We also checked whether Rab6 influences cell polarization in migrating cells. It is known that the Golgi apparatus reorients in migrating cells from a random perinuclear position to the area between the nucleus and the leading edge [40, 41]. However, Golgi reorientation was unaffected after Rab6 depletion (Fig. 2a–c).
All together, these data indicate that Rab6 depletion promote cell migration and actin fiber reorientation.
Rab6 is required for cell spreading
Having observed that Rab6 affects cell motility, we further investigated whether Rab6 is required for cell spreading. U2OS cells transfected with siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were trypsinized and plated on fibronectin-coated cover slips, fixed and finally stained with rhodamine–phalloidin. Quantification of the cell area clearly shows that knockdown of Rab6 decreases cell spreading in U2OS cells by circa 30% for both the tested oligos (Fig. 2d, e).
In line with this, overexpression of Rab6 leads to an increased cell area by almost 30% (Fig. 2d, e), indicating that Rab6 is a modulator of cell spreading. Moreover, in samples knocked down using siRNA Rab6 #1, transfection of GFP-Rab6 rescued the spreading defect by restoring the normal cellular area (Fig. 2d, e).
To shed more light on how Rab6 influences cell migration and cell spreading, we took advantage of L-shaped adhesive micropatterns. Cells plated on regular culture dishes can take on a variety of different shapes and are constantly changing as their cytoskeleton reorganizes. Having all cells confined to specific and identical shapes makes it easier to compare them and identify possible effects on cytoskeleton architecture after Rab6 knockdown. On L-shaped fibronectin-coated micropatterns, cells are forced to assume a right-angled triangular shape with the adhesive micropattern on two sides and a long non-adhesive stretch along the hypotenuse (Fig. 2f).
U2OS cells were seeded on fibronectin-coated L-shaped micropatterns for 3.5 h before fixation and staining with rhodamine–phalloidin to visualize the actin network. Interestingly, more than 80% of the cells silenced for Rab6 displayed an actin network architecture that appeared less straight and more collapsed along the non-adhesive side resulting in a hypotenuse with higher curvature, compared to 60% of the control cells (Fig. 2f, g). To overcome the absence of the underlying adhesive substrate, cells are dependent on actomyosin contraction. Indeed, upon myosin inhibition, the non-adhesive stretch takes on a relaxed (curved) instead of straight cell border [42]. Therefore, our results suggest that the increased curvature after depletion of Rab6 could be a consequence of reduced actomyosin contractility.
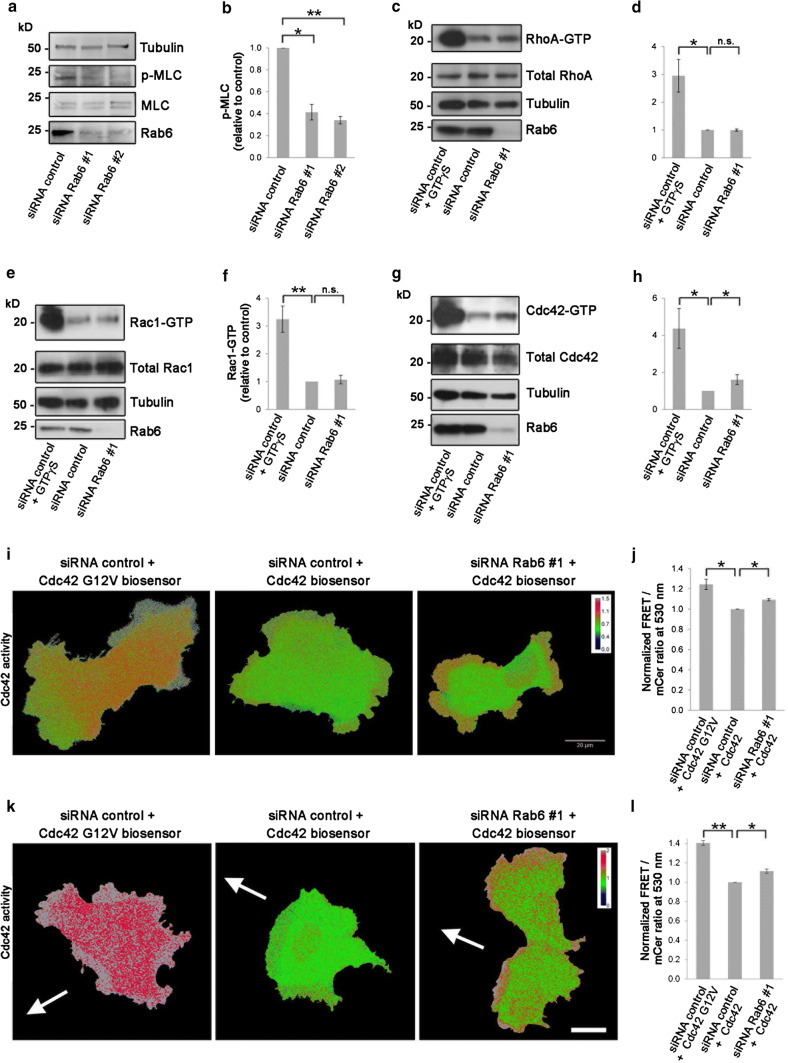
To test this hypothesis, we next checked if Rab6 knockdown alter the phosphorylation status of MLC, as phosphorylation of MLC is known to regulate myosin II assembly and contraction [43]. As Ser19 is the primary phosphorylation site of MLC [44–46], we investigated whether MLC phosphorylation on Ser19 was affected upon Rab6 silencing. In line with our hypothesis, MLC phosphorylation was strongly downregulated after knockdown of Rab6 (Fig. 3a, b).
Fig. 3.
Rab6 regulates MLC phosphorylation and Cdc42 activity. a Lysates from U2OS cells transfected with control siRNA, siRNA Rab6 #1 or siRNA Rab6 #2 were subjected to western blot analysis using antibodies against phosphorylated myosin light chain (p-MLC), total MLC and tubulin (as a loading control). b Quantification of the amount of p-MLC normalized to the total amount of MLC for each of the indicated sample. The data represent the mean ± SEM relative to the siRNA control sample of three independent experiments. c–h Lysates from U2OS cells treated with control siRNA or siRNA Rab6 #1 were mixed with beads coupled to either GST–Rhotekin–RBD to pull down the active form (GTP-bound) of RhoA (c), or to GST–PAK–PBD to pull down the active forms of Rac1 (e) or Cdc42 (g) and analyzed by western blot. As a positive control, cells were loaded with GTPγS. The levels of active RhoA (d), Rac1 (f), and Cdc42 (h) were normalized to the amount of tubulin and plotted relative to the intensities of GTP-bound Rho proteins in the control sample. The graphs represent the mean ± SEM normalized to the control of at least three independent experiments. i U2OS cells treated with siRNA control or siRNA Rab6 #1 and transfected with either Cdc42 G12 V biosensor or Cdc42 wt biosensor as indicated, were imaged live with spectral imaging. Scale bar: 20 µm. j Graph shows the mean ± SEM of the normalized FRET/mCerulean ratios for the indicated samples; n > 70 cells from three independent experiments. k U2OS cells treated with siRNA control or siRNA Rab6 #1 and transfected with either Cdc42 G12 V biosensor or Cdc42 wt biosensor as indicated, were scratched and let migrate for 4 h before live spectral imaging. The white arrows indicate the direction of migration. Scale bar: 20 µm. l Graph shows the mean ± SEM of the normalized FRET/mCerulean ratios for the indicated samples; n > 12 cells from four independent experiments. *P < 0.05; **P < 0.01 (paired Student’s t test)
Taken together, these results suggest that Rab6 is important for regulation of actin dynamics and actomyosin contractility by regulating the activity of myosin II.
Cdc42 activity is regulated by Rab6
The phosphorylation status of MLC is tightly regulated by a myosin specific phosphatase and several different kinases working at distinct places in the cell to locally control myosin activity [45, 47–52]. The upstream control of these kinases is orchestrated by members of the Rho GTPase family that are master regulators of cytoskeleton rearrangements [47, 53, 54]. We, therefore, hypothesized that Rab6 influences myosin phosphorylation through the regulation of Rho GTPase family members. To test this, we investigated if Rab6 knockdown affected the activity of the most extensively characterized Rho GTPases, namely, RhoA, Rac1, or Cdc42 [53, 54], by pull-down assays.
GST-tagged Rho-binding domain (RBD) of the RhoA effector Rhotekin was used to pull down the active form of RhoA, while GST-tagged p21 Binding Domain (PBD) of the Cdc42 and Rac1 effector protein p21 activated kinase I (PAK) was used to pull down the active forms of Rac1 and Cdc42 in U2OS cell lysates treated with siRNA control or siRNA Rab6 #1. As shown in Fig. 3c–f, Rab6 depletion did not affect RhoA or Rac1 activity. However, Cdc42 activity was increased more than 1.6-fold compared to control cells, indicating that Rab6 regulates actin dynamics through the modulation of Cdc42 activity (Fig. 3g, h).
To further detect the localized Cdc42 activation in live cells, we next took advantage of a fluorescence resonance energy transfer (FRET)-based biosensor for Cdc42 [55]. U2OS cells treated with siRNA control or siRNA Rab6 #1 were transfected with either wild-type Cdc42 biosensor or Cdc42 G12 V constitutively active mutant biosensor as control, and imaged for FRET analysis. We measured Cdc42 activity in regions at the cell periphery as we had previously detected differences in actin arrangement in cell protrusions after Rab6 depletion (Fig. 2a, b). Spectral imaging and measurement of the FRET/Cerulean ratio at 530 nm confirmed a significant increase in Cdc42 activity in cells depleted for Rab6 compared to the control, thus verifying the results from the pull-down assay and further demonstrating a localized activation of Cdc42 at the cell periphery (Fig. 3i, j). A similar increase in Cdc42 activity in cells depleted for Rab6 was measured also in migrating U2OS cells (Fig. 3k, l). All together, these results support a model in which Rab6 knockdown induces the activation of Cdc42 at the cell periphery.
Cdc42 and its GEF Trio interact with Rab6
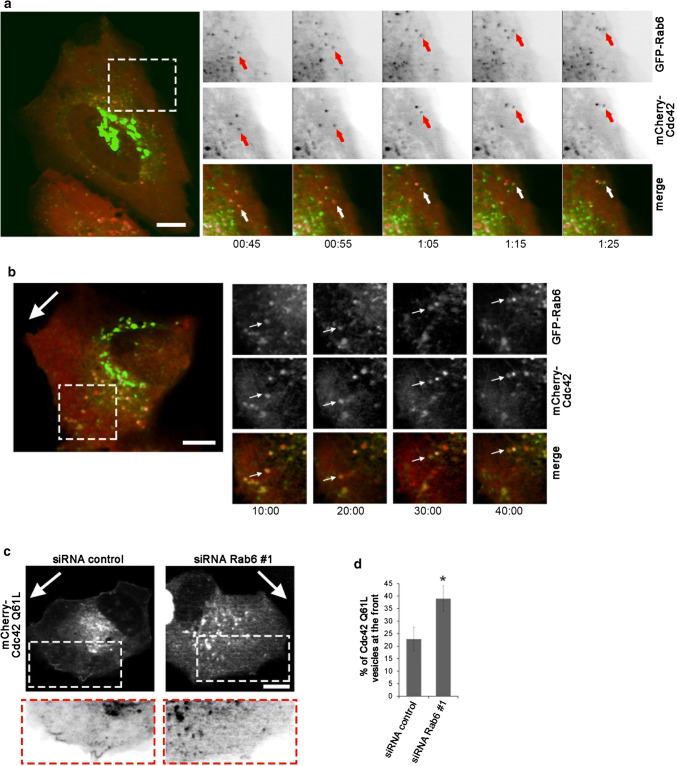
To understand how Rab6 affects Cdc42 activity, we next looked at the intracellular localization and dynamics of these molecules by live-cell confocal microscopy. U2OS cells were co-transfected with GFP-Rab6 and mCherry-Cdc42 before time-lapse image acquisition. As shown in Fig. 4a, GFP-Rab6 was present in the perinuclear/Golgi region from where Rab6-positive vesicles emerged and moved towards the cell periphery as previously reported [18, 56]. Some of these vesicles were also positive for mCherry-Cdc42, although the percentage of mCherry-Cdc42-positive vesicles that co-localized with GFP-Rab6 varied considerably between cells. Interestingly, time-lapse imaging revealed that these vesicles moved from the perinuclear region towards the cell periphery (Fig. 4a, Movie 1). Similar dynamics were also observed in migrating cells, where vesicles positive for both Rab6 and Cdc42 were observed to move towards the cell periphery at the leading edge, but also often return back towards the cell centre (Fig. 4b).
Fig. 4.
Cdc42 is transported towards the cell periphery in Rab6-positive vesicles. a U2OS cells co-transfected with GFP-Rab6 and mCherry-Cdc42 were imaged using a spinning-disk confocal microscope for the indicated time points. The arrows indicate a vesicle positive for both Rab6 and Cdc42 moving towards the cell periphery. Scale bar: 10 µm. b U2OS cells co-transfected with GFP-Rab6 and mCherry-Cdc42 were scratch-wounded with a pipet tip and imaged 3 h later by using a spinning-disk confocal microscope for the indicated time points. The image represents maximum-intensity projections of z-stacks. The big white arrow indicates the direction of migration. Magnifications of the boxed area are shown in the insets. The arrows in the insets indicate a vesicle positive for both Rab6 and Cdc42 moving both towards the cell periphery and back. Scale bar: 10 µm. c U2OS cells treated with siRNA control or siRNA Rab6 #1 and transfected with pTriEx-mCherry-Cdc42 Q61L were scratched with a pipet tip and imaged 3 h later. The images represent maximum-intensity projections of z-stacks. The white arrows indicate the direction of migration. Magnifications of the boxed areas are shown in the insets. Scale bar: 10 µm. d Quantification of the percentage of Cdc42 Q61L-positive vesicle in the cell front compared to the total number of Cdc42 Q61L-positive vesicles within the cell. The graph shows the mean ± SEM from two independent experiments (n = 8). *P < 0.05 (paired Student’s t test)
As Rab6 depletion promotes the activation of Cdc42, we then studied whether it also influences the localization of GTP-bound Cdc42. For this, we transfected control cells and cells silenced for Rab6 with the constitutively active mutant of Cdc42. We left the cells to migrate for 3 h after a scratch was made with a pipette tip in the confluent cell monolayer, and then, we imaged the cells using time-lapse video microscopy. Interestingly, the percentage of Cdc42-positive vesicles present in the front of the migrating cells was higher in cells silenced for Rab6, suggesting that Rab6 might have a role in balancing the transport of active Cdc42 to the cell periphery (Fig. 4c, d).
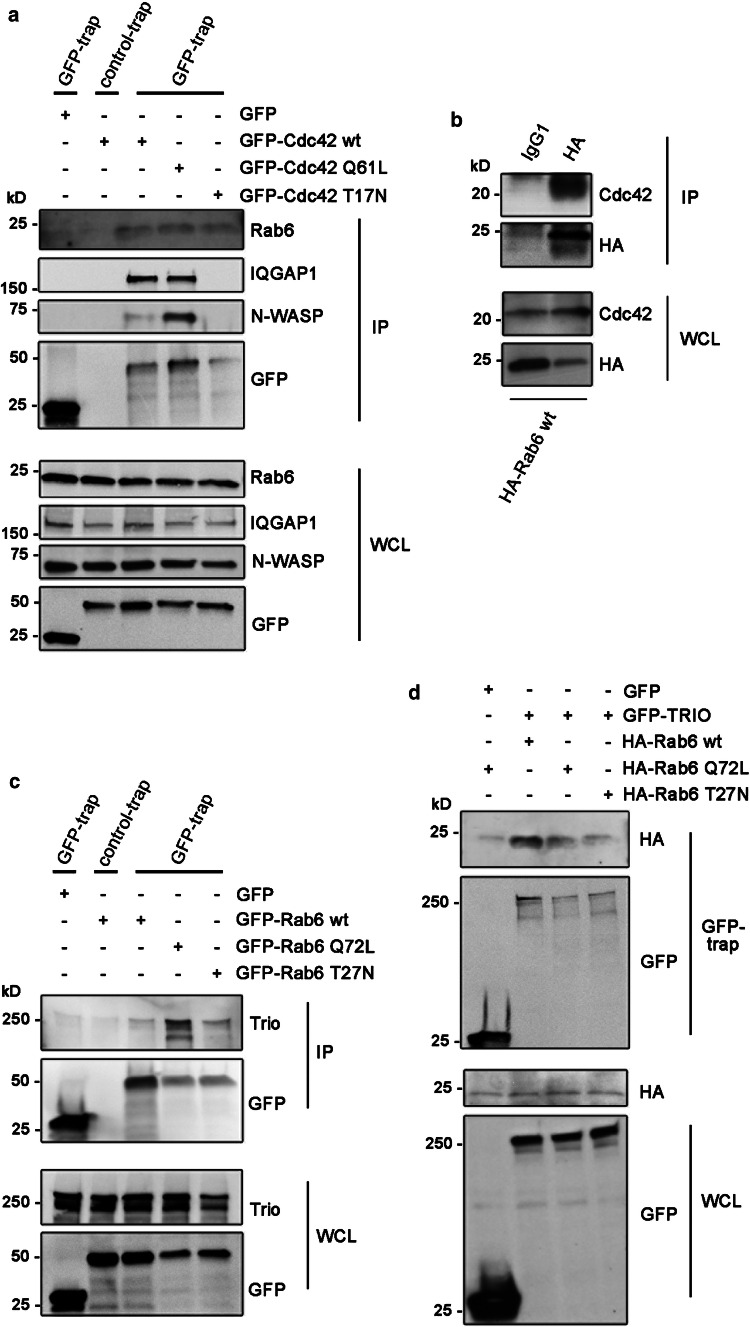
Having observed that Cdc42 is present on Rab6-positive vesicles, we next investigated if these two small GTPases also interact. To test this, we performed co-immunoprecipitation (co-IP) experiments in U2OS cells transiently transfected with either GFP, GFP-Cdc42 wt, GFP-Cdc42 Q61L (constitutively active mutant), or GFP-Cdc42 T17 N (dominant negative mutant). The results in Fig. 5a show that Rab6 interacts with all forms of Cdc42, and indicate that this interaction does not depend on the nucleotide binding state of Cdc42. As controls, both N-WASP and IQGAP1, known interactors of Cdc42, were immunoprecipitated by Cdc42 wt and its constitutively active mutant, but not by the dominant negative mutant (Fig. 5a), in agreement with the previous reports [57–59]. This further supports the specificity of the interaction of Rab6 with all forms of Cdc42. We additionally performed a reverse co-IP, where U2OS cells were transiently transfected with HA-Rab6. HA-Rab6 was able to immunoprecipitate endogenous Cdc42, again confirming the interaction between Rab6 and Cdc42 (Fig. 5b).
Fig. 5.
Cdc42 and its GEF Trio interact with Rab6. a Lysates from U2OS cells transiently transfected with either GFP, GFP-Cdc42 wt, GFP-Cdc42-Q61L or GFP-Cdc42-T17 N were subjected to immunoprecipitation with GFP-Trap or control-Trap magnetic agarose beads. Whole-cell lysates (WCL) and immunoprecipitates (IP) were subjected to western blot analysis with the indicated antibodies. b Lysates from U2OS cells transiently transfected with HA-Rab6 wt, were subjected to immunoprecipitation with an antibody against HA or an isotype control (IgG1). WCL and IP were subjected to western blot analysis using antibodies against HA or Cdc42. c HeLa cells were transiently transfected with GFP, GFP-Rab6 wt, GFP-Rab6 Q72L, or GFP-Rab6 T27 N, lysed and subjected to immunoprecipitation with GFP-Trap or control-Trap magnetic agarose beads. WCL and immunoprecipitates IP were subjected to western blot analysis using the indicated antibodies. d U2OS cells transiently transfected with either GFP or GFP-TRIO, together with HA-Rab6 wt, HA-Rab6 Q72L or HA-Rab6 T27 N, were subjected to immunoprecipitation with GFP-Trap magnetic agarose beads. WCL and IP were subjected to western blot analysis with the indicated antibodies
Cdc42 is a small GTPase, whose activity is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) that activate and inactivate it, respectively. Intriguingly, in a study from 2014, a GEF for the Rho family GTPases, Trio, was identified as a putative Rab6-binding protein [5]. Importantly, while Trio has been considered a GEF for RhoG and Rac1, a recent work has challenged this view by demonstrating that Trio, although with a lower exchange rate than RhoG and Rac1, potently activates both RhoA and Cdc42 [30].
We, therefore, investigated whether Rab6 modulates Cdc42 activity by regulating the recruitment of a Cdc42 GEF, and verified the putative interaction between Rab6 and Trio by co-IP in HeLa cells as U2OS cells express lower levels of endogenous Trio. Cells were transiently transfected with GFP, GFP-Rab6 wt, GFP-Rab6 Q72L (constitutively active mutant), or GFP-Rab6 T27 N (dominant negative mutant) and the immunoprecipitation was performed using GFP-Trap magnetic agarose beads. Immunoblotting using an antibody against Trio indeed demonstrated that GTP-bound Rab6 and endogenous Trio interact (Fig. 5c). We also performed a reverse co-IP in U2OS cells by transiently transfecting cells with either GFP or GFP-Trio, as well as HA-tagged Rab6 wt, HA-Rab6 Q72L, or HA-Rab6 T27 N, and verified the interaction between Rab6 and Trio (Fig. 5d). Taken together, these findings indicate that Rab6 is able to recruit both Cdc42 and the GEF Trio and thus suggest a possible mechanism for Rab6 to modulate Cdc42 activity.
To further investigate whether the enhanced activation of Cdc42 upon Rab6 knockdown depends on Trio, we silenced both Rab6 and Trio and measured Cdc42 activity. As shown in Suppl. Fig. 3a, b, Trio depletion alone inhibits Cdc42 activation down to levels comparable to another well-known GEF for Cdc42, DOCK10. The activity of Cdc42 in Rab6 and Trio double-knockdown cells was also significantly reduced, indicating that the effect on Cdc42 activation upon of Rab6 knockdown depends, at least to a certain extent, on Trio.
We next studied the contribution of Trio on cell migration. In line with previous work [60], silencing of Trio considerably inhibited cell migration in a wound healing assay (Suppl. Fig. 3c). Overexpression of Rab6 wt did not further influence the effect of Trio depletion, while double-knockdown of Trio and Rab6 reduced the Rab6-dependent increase in cell migration to the level of control cells. Taken together, these results suggest that the increased migration upon Rab6 depletion may be at least in part dependent on Trio-mediated activation of Cdc42.
Rab6 depletion influences F/G-actin ratio and filopodia formation and dynamics
Cdc42 is a regulator of actin cytoskeleton dynamics that promotes formation of filopodia [61]. Since our results indicate that Rab6 influences Cdc42 activity at the cell periphery, we next checked whether actin polymerization and filopodia formation were altered after Rab6 knockdown.
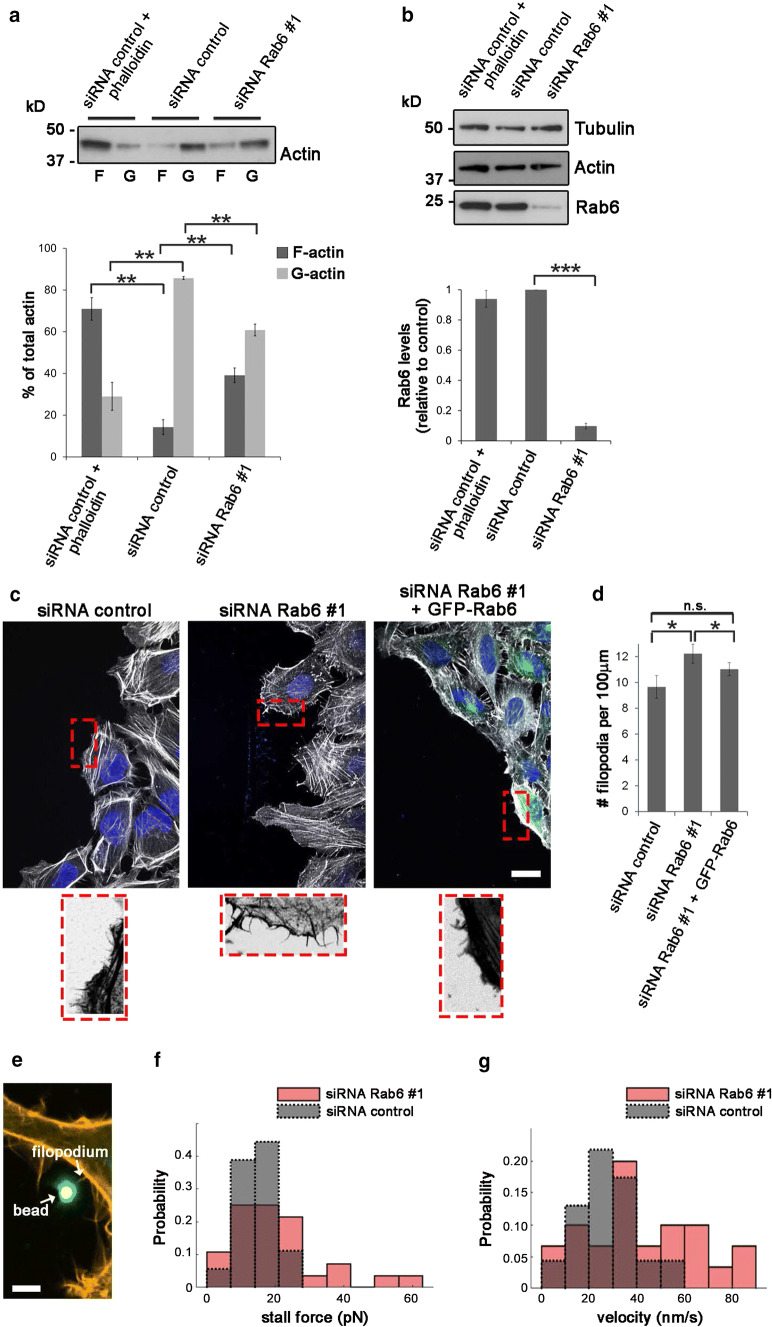
To determine if Rab6 influences actin polymerization, we quantified the relative amounts of cellular G-actin and F-actin in control cells and cells depleted for Rab6. Cell lysates were subjected to high-speed centrifugation to separate the G- and F-actin pools, followed by quantitative Western blot analysis to assess the ratio of F-to-G-actin. As expected, pretreatment with phalloidin, which stabilizes F-actin, increased the F/G-actin ratio with almost 2.5-fold compared to control cells (Fig. 6a, b). Importantly, Rab6 knockdown also increased the ratio between F/G-actin of 0.6-fold compared to control cells (Fig. 6a, b).
Fig. 6.
Filopodia formation and dynamics are affected by Rab6 depletion. a F-actin (F) and G-actin (G) pools were separated from lysates of U2OS cells transfected with siRNA control or siRNA Rab6 #1 by ultracentrifugation. Phalloidin was added to a control sample to verify that the F- and G-actin pools were successfully separated. Samples were loaded on SDS-PAGE and subjected to Western blot analysis using an antibody against actin. The graph shows the percentage of F-actin and G-actin relative to the total amount of actin. Data represents the average of four independent experiments. **P < 0.01 (paired Student’s t test). b Cell lysates from each of the indicated sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). The intensities of the bands were quantified using densitometry, normalized against the amount of tubulin, and plotted relative to the intensities obtained in cells transfected with siRNA control. The values represent the mean ± SEM for four independent experiments. ***P < 0.001 (paired Student’s t test). c HeLa cells treated with siRNA control or siRNA Rab6 #1, or silenced for Rab6 and subsequently transfected with GFP-Rab6, were scratched and let migrate for 3 h before fixation and staining with DAPI and rhodamine-conjugated phalloidin. The lower insets show magnifications of the boxed areas. Scale bar: 20 µm. d Quantification of the number of filopodia per 100 µm of cell membrane facing the wound. The graph represents the mean ± SEM; n > 50 cells from three independent experiments. P < 0.05 (one-way ANOVA followed by Fisher’s LSD test). e Confocal image of HeLa cells transfected with LifeAct-RFP showing an optically trapped bead attached to a filopodium. Scale bar: 2 µm. f Distribution of the mean filopodial retraction velocity from 8 independent experiments (n = 30 for siRNA Rab6 #1 and n = 23 for siRNA control). Average values: 41.2 nm/s for siRNA Rab6 #1 and 28.5 nm/s for siRNA control. The measured velocities reject the null hypothesis that they originate from the same sample using a two-sample Kolmogorov–Smirnov test and a two-sample t test at a 5% significance level. g Distribution of measured stall forces from 8 independent experiments (n = 27 for siRNA Rab6 #1 and n = 15 for siRNA control). Mean stall force for siRNA Rab6 #1 = 12.6 pN and for siRNA control = 10.6 pN. Neither a two-sample Kolmogorov–Smirnov test nor a two-sample t test of the data does allow to reject the null hypothesis that they originate from the same sample
As this result is in line with increased actin polymerization, we next investigated whether the augmented pool of F-actin in cells knocked down for Rab6 could correlate with an increase in filopodia formation. We silenced HeLa cells for Rab6 as filopodia are easier to detect in this cell line than in U2Os cells, before fixation and staining of actin filaments with rhodamine–phalloidin. Consistent with the augmented Cdc42 activity after Rab6 knockdown, quantification revealed an increase in filopodia number by 17% in cells depleted for Rab7b compared to control cells (Suppl. Fig. 4a, b). Interestingly, the number of filopodia at the leading edge of migrating cells was even higher (27% more than in control cells) upon Rab6 depletion and the re-expression of GFP-Rab6 in these cells was able to reduce the filopodia number (Fig. 6c, d). Since Rab6 is able to affect filopodia formation, it is reasonable to expect that Rab6 is localized to filopodia. To confirm this, U2OS cells were transiently transfected with GFP-Rab6 and imaged using TIRF microscopy (TIRFM). TIRFM showed that Rab6-positive vesicles were transported towards the cell surface and often localized to filopodia (Suppl. Fig. 4c, Movie 2).
Filopodia are dynamic structures consisting of F-actin bundles. Their growth and retraction is dependent on actin polymerization rate at the tip of the filopodia and on the actin retrograde flow [38]. To further investigate whether Rab6 in addition to filopodia formation also influences filopodia dynamics, we measured the retraction velocity and the force exerted by filopodia on optically trapped beads in control cells and in cells silenced for Rab6 (Fig. 6e). As shown in Fig. 6g, the velocity of filopodia retraction increased in cells knocked down for Rab6 as the mean retraction velocity of filopodia resulted in 41.2 ± 23.9 nm/s compared to 28.5 ± 12.7 nm/s in control cells. In addition, the mean stall force of the filopodia was quantified. The stall force is evaluated by the considering both the highest counteracting force at which still a retraction could be determined and the lowest counteracting force which led to a stall of the retraction process. The mean stall force of the filopodia in cells depleted for Rab6 (15.7 ± 11 pN) was slightly higher than the one of the control cells (13.5 ± 5.9pN) (Fig. 6f). In sum, our results show that the enhanced activation of Cdc42 induced by Rab6 silencing results in an increase in filopodia formation and influences filopodia dynamics.
Rab6 knockdown promotes spread of cancer cells in vivo
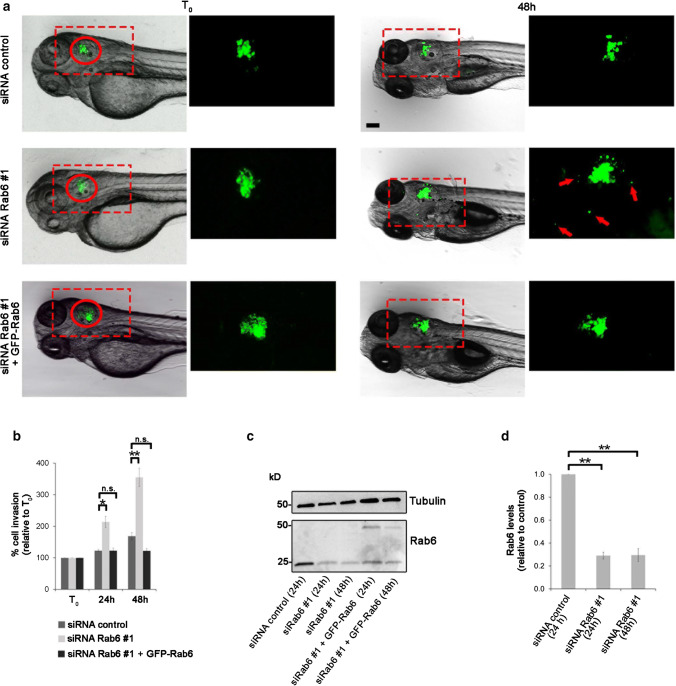
In this study, we have shown using in vitro systems that Rab6 depletion leads to increased cell migration by inhibiting myosin II phosphorylation and promoting Cdc42 activation. This results in increased actin polymerization that in turn leads to an enhanced formation of cell protrusions. We next investigated whether Rab6 depletion also promotes cell migration in vivo. To test this, we examined if Rab6 silencing influences the dissemination of human cancer cells with known invasion/metastasis potential, such as H1299 [62, 63] in zebrafish embryos. 20 h after silencing, control cells, or cells silenced for Rab6 were labeled with fluorescent quantum dots (QDs) and injected into the otic vesicle of zebrafish larvae at 48 h post-fertilization. Analysis was performed at 24 and 48 h post-injection by acquiring images using a stereomicroscope, as the spread of cancer cells could be seen in the body of the fish. The invasion potential of these cells was assessed by calculating the ability of the cells to migrate out from the otic vesicle. The results show that, compared to the control, cells silenced for Rab6 were able to migrate from the otic vesicle already at 24 h post-injection and even more at 48 h post injections (Fig. 7). Moreover, transfection of GFP-Rab6 in cells knocked down using siRNA Rab6 #1 rescued the increased invasive ability induced by Rab6 depletion (Fig. 7a, c). The effect of Rab6 depletion on in vivo cell migration was additionally confirmed by using a second siRNA targeting Rab6. Similar to the results obtained with siRNA #1, cells silenced with Rab6 #2 were able to migrate in zebrafish embryos from the otic vesicle at 24 h post-injection and at 48 h post injections more than control cells (suppl. Fig. 5). In conclusion, using a zebrafish xenograft model, we demonstrated that silencing of Rab6 potentiates invasion of H1299 cells in vivo, further confirming the role of Rab6 in cell migration.
Fig. 7.
Rab6 knockdown promotes spread of cancer cells in zebrafish embryos following xenotransplantation. a H1299 cells transfected with control siRNA, siRNA Rab6 #1, or silenced with siRNA Rab6 #1 and subsequently transfected with GFP-Rab6, were stained with QDs Q-Tracker 655 and injected into the otic vesicle (red circle) of 2-dpf zebrafish wild-type embryos. Representative images of (T0) and 48 h after injection are shown. In contrast to the control cells (top panels), and to cells silenced with siRNA Rab6 #1 and transfected with GFP-Rab6 (lower panels), that remained confined to the injection area, cell silenced for Rab6 (middle panels) showed higher ability to migrate outside from the otic vesicle at 48 h (arrows). Magnification of the boxed areas are shown in the inset on the right of each panel. Scale bar: 100 µm. b Quantification of cell migration was performed by measuring the area occupied by the cells at 24 h and 48 h after injection and was normalized to the area occupied by the cells in the otic vesicle at T0. The graph represents the mean ± SEM from n > 10 embryos. c H1299 cells transfected with control siRNA or siRNA Rab6 #1 for 24 or 48 h, or silenced with siRNA Rab6 #1 and subsequently transfected with GFP-Rab6 for 24 or 48 h, were subjected to western blot analysis using antibodies against Rab6 and tubulin (as a loading control). *P < 0.05; **P < 0.01 (one-way ANOVA followed by Fisher’s LSD test). d Quantification of Rab6 levels normalized to the amount of tubulin, and plotted relative to the intensities obtained in cells transfected with siRNA control for each of the indicated sample. The data represent the mean ± SEM relative to the siRNA control sample of three independent experiments. **P < 0.01 (paired Student’s t test)
Discussion
Rab6 is known to regulate the transport between the endoplasmatic reticulum, Golgi apparatus, plasma membrane, and endosomes [17–21]. In this study, we reveal a novel function of this GTPase as a negative regulator of cell migration by interacting with both a Rho family member and its GEF and influencing actin cytoskeleton organization.
Rab proteins are known to interact with different motor proteins to facilitate distinct processes in intracellular trafficking [64–67]. Between them, only Rab6 and Rab7b have so far been shown to directly interact with the non-processive motor protein myosin II [10, 15]. Interestingly, while Rab6 is reported to mediate the fission of vesicles from the Golgi apparatus by recruiting myosin II [15], the interaction between Rab7b and myosin II has proven to be important not only for vesicular transport but also for proper cytoskeleton dynamics required in cell migration [10].
In the present study, we reveal a new function for Rab6 in the regulation of cytoskeleton organization and cell migration by showing that Rab6 modulates myosin II activity. Indeed, depletion of Rab6 decreases myosin II phosphorylation on Ser 19 (Fig. 3a, b). These results are further supported by the decreased ability of cells knocked down for Rab6 to generate enough actomyosin contractility to support full extension of their free edge when grown on L-shaped micropatterns (Fig. 2f, g).
Myosin II knockdown or inhibition by drugs has been generally shown to increase the speed of migrating cells in many different cell types [68–71]. These studies support our finding that increased cell migration after Rab6 knockdown is a consequence of a decrease in myosin II phosphorylation. Even though there is no general consensus on the role of myosin II in cell spreading and on the effects of its inhibition [72–77], Nisenholz et al. recently found that the balance of the forces exerted by myosin II between the periphery and the center of the cell is important for the regulation of cell spreading [78]. They demonstrated that the forces exerted by myosin II in the periphery facilitated cell spreading, while those exerted more centrally opposed spreading. Based on this model, the decrease in cell spreading we measured after Rab6 knockdown could be a consequence of less myosin-dependent forces exerted towards the cell periphery due to the decreased myosin II phosphorylation.
Intriguingly, our results also show that depletion of Rab6 increases Cdc42 activity (Fig. 3g, h). In addition, using a Cdc42 FRET biosensor construct, we demonstrate that the Cdc42 activity is increased at the cell periphery in both migrating and non-migrating cells (Fig. 3i, l). It is well established that the activation of the small GTPase Cdc42 promotes Arp2/3-dependent actin nucleation and polymerization important for initiation and dynamics of filopodia [53, 79–82]. In agreement with this, we found an increase not only in the amount of F-actin after depletion of Rab6, but also in cell protrusions and filopodia formation (Figs. 2a, b, 6a–d, Suppl. Fig. 4a, b).
It is tempting to hypothesize that the increased activation of Cdc42 after Rab6 knockdown could be the cause for the reduced phosphorylation of myosin II. Indeed, phosphorylation of myosin II on Ser 19 is tightly controlled by myosin specific phosphatase and kinases, some of which are regulated by Cdc42 [44–46, 83, 84]. In line with this hypothesis, studies on force generation of cortical actin and cell protrusions have shown that less myosin II contractility in the cortical actin can lead to more protrusions [85, 86]. This is again consistent with the increased formation of protrusions and filopodia observed in cells silenced for Rab6. In the light of this, our results are compatible with a model, where Rab6 modulates Cdc42 activity at the cell periphery, thereby regulating actin cytoskeleton dynamics and myosin II activity, which results in decreased spreading and increased filopodia formation and protruding potential. Indeed, we demonstrate that Cdc42 is present on Rab6-positive vesicles directed towards the cell periphery that afterwards can also move back towards the center of the cell (Fig. 4a, b). Most interestingly, depletion of Rab6 redistributes active Cdc42 towards the leading edge of migrating cells (Fig. 4c, d). This suggests that Rab6 may modulate either how much Cdc42 should be transported to the cell periphery or its time of residence at the periphery of the cells, and that upon Rab6 depletion this control is lost.
How then, does Rab6 regulate Cdc42 activity? Evidence of crosstalk between the Rab and Rho family of small GTPases in the regulation of cytoskeleton dynamics is starting to emerge as some Rab proteins have been shown to regulate the activity of Rho family members [10, 12, 67, 87–90]. However, a general mechanism behind this regulation is not well characterized. Our results show that Rab6 and Cdc42 are not only present on the same transport vesicle, but can also interact. This interaction is independent on the nucleotide binding state of Cdc42 (Fig. 5a, b), suggesting that Rab6 may recruit another factor for the direct modulation of Cdc42 activity. As we found that GTP-bound Rab6 binds to the Rho family GEF Trio (Fig. 5c, d), it is tempting to speculate that the recruitment of Trio by active Rab6 may prevent the activation of Cdc42.
Trio is a member of the Dbl family of GEFs whose activity is inhibited by direct interaction with phosphorylated myosin II [91]. A reduction in myosin II phosphorylation decreases the Dbl GEF–myosin II interaction, resulting in increased GEF activity [91]. In light of this, the reduced amount of phosphorylated myosin II we measured after knockdown of Rab6 could trigger the increase in Trio GEF activity and thereby promote Cdc42 activation. As both Trio and Rab6 are known to bind to myosin II [15, 91], an intriguing scenario is, therefore, that Rab6, Trio and myosin II may form a complex for the specific regulation of Cdc42 activity. The formation of such complex could explain how Rab6 recruits Trio and prevents Cdc42 activation: active Rab6 binds to both myosin II and Trio, sequestering this GEF as the binding of myosin II to Trio inhibits its GEF activity [91].
In line with the involvement of Trio in the Rab6-dependent modulation of Cdc42 activity, double knockdown of Rab6 and Trio-reduced Cdc42 activation and to some extent also the Rab6-dependent increase in cell migration (Suppl. Fig. 3). However, as both the activation of Cdc42 and the migration in the double-knockdown cells were not decreased to the same levels as Trio silencing alone, this suggests that the increased migration caused by Rab6 depletion is only in part dependent on Trio-mediated activation of Cdc42. Therefore, additional mechanisms contribute to Cdc42 activation or to the increased migration, and based on our results, one of these is likely to involve myosin II phosphorylation.
In conclusion, in this study, we have revealed a new role for Rab6 in cell migration. We have also elucidated the underlying molecular mechanisms involved by showing that Rab6 can influence filopodia formation through the modulation of Cdc42 activity. Intriguingly, we further demonstrated that Rab6 knockdown promotes cancer cell spreading after xenotransplantation in zebrafish embryos (Fig. 7, Suppl. Fig. 5), in line with the evidence that an increased protrusion potential and filopodia formation contribute to cancer cell invasion [92, 93], thus establishing Rab6 as a negative regulator of cell migration.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Movie 1: Cdc42 is transported towards the cell periphery in Rab6-positive vesicles. U2OS cells co-transfected with GFP-Rab6 and mCherry-Cdc42 were imaged using a spinning-disk confocal microscope at 1 second intervals. A vesicle positive for both GFP-Rab6 and mCherry-Cdc42 moving towards the cell periphery is tracked over time. Scale bar: 10 µm (AVI 2233 kb)
Movie 2: Rab6-positive vesicles are transported towards filopodia. U2OS cells transfected with GFP-Rab6 and labeled with SiR-actin, were imaged with a Total Internal Reflection fluorescent (TIRF) microscope. (AVI 188 kb)
Supplementary figure 1: Rab6 silencing promotes cell migration in RPE-1 cells. (a) RPE-1 cells treated with siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and imaged every 15th minute for 24 h. Representative images of (T0) and 24 h after scratching are shown. Scale bar: 300 µm. Quantification of single-cell speed (b) and directness (c) is represented as mean ± s.e.m.; n>230 cells from five independent experiments. (d) Cell lysates from each of the indicated samples were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (e) The intensities of the bands were quantified using densitometry, normalized against the amount of tubulin, and plotted relative to the intensities obtained in cells transfected with siRNA control. The values represent the mean ± s.e.m. for five independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (paired Student’s t test). (f) Representative track plots of the single-cell migration distances. Individual tracks are shown so that each starts at the origin (distance 0). (g) Velocity vector fields of particle image velocimetry analysis (PIV). Scale bar: 100 µm (TIFF 1719 kb)
Supplementary figure 2: Rab6 depletion promotes cell migration independently from cell proliferation. (a) U2OS cells treated with either siRNA control, siRNA Rab6 #1, siRNA Rab6 #2, or treated with the same siRNAs and afterwards transfected with GFP-Rab6 were scratch-wounded and imaged for 24 h. The graph represents the mean from three independent experiments of the relative wound density (%) as function of time. ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=24h). (b) Cell lysates from each sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (c) U2OS cells treated with either siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and imaged in medium containing Cytotox Green reagent to label dying cells green. Cell death during migration was quantified by dividing the number of green objects at each timepoint with the number of green objects at time 0. The graph represents the mean from three independent experiments. (d) U2OS cells treated with either siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and incubated in medium (with or without serum as indicated) containing the modified thymidine analogue Edu. The graph shows the percentage of Edu-positive cells after 24 h, relative to Edu-positive control cells in medium containing serum. The data represents the mean ± s.e.m from three independent experiments. (e) HeLa cells treated with either siRNA control, siRNA Rab6 #1, or siRNA Rab6 #2 were scratch-wounded and imaged for 24 h. Representative images of (T0) and 24 h after scratching are shown. Scale bar: 300 µm. (f) Quantification of the relative wound density (%) as function of time for control cells and Rab6-depleted cells. Data represents the mean of three (siRNA #1) or two (siRNA #2) independent experiments. *P < 0.05; ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=24h). (g) Cell lysates from each sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (TIFF 809 kb)
Supplementary figure 3: Role of Trio in Rab6-dependent Cdc42 activation and cell migration. (a) Lysates from U2OS cells transfected with control siRNA, siRNA Rab6 #1, siRNA Trio, siRNA Rab6 #1 and siRNA Trio, or siRNA DOCK10 were mixed with beads coupled to GST–PAK–PBD to pull down the active forms of Cdc42 and analyzed by western blot. (b) Quantification of the levels of active Cdc42 were normalized to the amount of tubulin and plotted relative to the intensities of GTP-bound Cdc42 in the control sample. The graphs represent the mean ± s.e.m. normalized to the control of at least three independent experiments. *P < 0.05; **P < 0.01 (paired Student’s t test). (c) U2OS cells treated with either siRNA control, siRNA Rab6 #1, siRNA Trio, siRNA Rab6 #1 and siRNA Trio, or treated with siRNA Trio and afterwards transfected with GFP-Rab6, were scratch-wounded and imaged for 24 h. The graph represents the mean from at least three independent experiments of the relative wound density (%) as function of time. ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=21h). (TIFF 309 kb)
Supplementary figure 4: Rab6 and filopodia. (a) HeLa cells treated with siRNA control or siRNA Rab6 #1 were fixed and stained with DAPI and rhodamine-conjugated phalloidin. The lower insets show magnifications of the boxed areas. Scale bar: 20 µm. (b) Quantification of the number of filopodia per 100 µm cell membrane length. The graph represents the mean ± s.e.m.; n>100 cells from four independent experiments. *P < 0.05 (paired Student’s t test). (c) U2OS cells transfected with GFP-Rab6 and labeled with SiR-actin, were imaged with a Total Internal Reflection fluorescent (TIRF) microscope. Right panels: arrows in the insets point to Rab6-positive vesicles moving towards filopodia. Scale bar: 10 µm. (TIFF 698 kb)
Supplementary figure 5: Rab6 knockdown promotes spread of cancer cells in zebrafish embryos following xenotransplantation. (a) H1299 cells transfected with control siRNA or siRNA Rab6 #2 were stained with QDs Q-Tracker 655 and injected into the otic vesicle (red circle) of 2-dpf zebrafish wild-type embryos. Images were taken soon after the injection (T0) as well as 24 and 48 h later. In contrast to the control cells (left panels) that remained confined to the injection area, cell silenced for Rab6 (right panels) showed higher ability to migrate outside from the otic vesicle, both at 24 h and at even more at 48 h (arrows). Magnification of the boxed areas are shown in the inset on the right of each panel. Scale bar: 100 µm. (b) Quantification of cell migration was performed by measuring the area occupied by the cells at 24 h and 48 h after injection and was normalized to the area occupied by the cells in the otic vesicle at T0. The graph represents the mean ± s.e.m. from n>15 embryos. *P < 0.05 (paired Student’s t test). (c) H1299 cells transfected with control siRNA or siRNA Rab6 #2 for 24 or 48 h were subjected to western blot analysis using antibodies against Rab6 and tubulin (as a loading control). (TIFF 477 kb)
Acknowledgements
We acknowledge the NorMIC Oslo imaging platform (Department of Biosciences, University of Oslo), Catherine Anne Heyward, and Frode Skjeldal for technical assistance, Hesso Farhan and Salim Ghannoum (Institute of Basic Medical Sciences, University of Oslo) for assistance and access to the IncuCyte ZOOM instrument. We thank Luis Hodgson (Albert Einstein College of Medicine, USA), Keith Burridge (University of North Carolina, USA), and Jaap D. Van Buul (University of Amsterdam, The Netherlands), for the kind gift of the pTriEX Cdc42 constructs, pEGFP-C3 Cdc42 plasmids, and pEGFPC1-Trio respectively. We thank Guillaume Jacquemet (University of Turku, Finland) for providing the FiloQuant plugin for Fiji/ImageJ, Giorgio Scita (IFOM, Milan, Italy) and Fabio Giavazzi (University of Milan, Italy) for advice on PIV analysis. We are grateful to Bruno Goud (Institut Curie, Paris, France) for critically reading the manuscript. The financial support of the Norwegian Cancer Society [Grants 5760850 to C.P. and 4604944 to O.B.], the Research Council of Norway [grants 239903 to C.P., 230779 to O.B., and through its Centre of Excellence funding scheme, Project Number 179573], and a Mayent-Rothschild- Institut Curie Award to OB is gratefully acknowledged.
Author contributions
KV, IK, NAG, MBD, FK, and CP performed the experiments and analyzed data. FF helped with setting up the Zebrafish xenotransplantation model. CP conceived and supervised the project. IK and CP wrote the manuscript with input from all the authors. CP and OB procured funding.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Katharina Vestre and Ingrid Kjos contributed equally to this work.
References
- 1.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 2.Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49(4):527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 3.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62(2):317–329. doi: 10.1016/0092-8674(90)90369-P. [DOI] [PubMed] [Google Scholar]
- 4.Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128(17):3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
- 5.Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014;31(3):358–373. doi: 10.1016/j.devcel.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12(11):1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibieza P, Prekeris R (2017) Rab GTPases and cell division. Small GTPases. 10.1080/21541248.2017.1313182 [DOI] [PMC free article] [PubMed]
- 8.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16(17):1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Thomas JD, Zhang YJ, Wei YH, Cho JH, Morris LE, Wang HY, Zheng XF. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26(5):754–769. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borg M, Bakke O, Progida C. A novel interaction between Rab7b and actomyosin reveals a dual role in intracellular transport and cell migration. J Cell Sci. 2014;127(22):4927–4939. doi: 10.1242/jcs.155861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linford A, Yoshimura S, Nunes Bastos R, Langemeyer L, Gerondopoulos A, Rigden DJ, Barr FA. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22(5):952–966. doi: 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134(1):135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 13.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 14.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17(8):496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 15.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12(7):645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 16.Goud B, Zahraoui A, Tavitian A, Saraste J. Small GTP-binding protein associated with Golgi cisternae. Nature. 1990;345(6275):553–556. doi: 10.1038/345553a0. [DOI] [PubMed] [Google Scholar]
- 17.Antony C, Cibert C, Geraud G, Santa Maria A, Maro B, Mayau V, Goud B. The small GTP-binding protein rab6p is distributed from medial Golgi to the trans-Golgi network as determined by a confocal microscopic approach. J Cell Sci. 1992;103(Pt 3):785–796. doi: 10.1242/jcs.103.3.785. [DOI] [PubMed] [Google Scholar]
- 18.Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, Akhmanova A. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13(2):305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 19.White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, Stelzer EH. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147(4):743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monier S, Jollivet F, Janoueix-Lerosey I, Johannes L, Goud B. Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic. 2002;3(4):289–297. doi: 10.1034/j.1600-0854.2002.030406.x. [DOI] [PubMed] [Google Scholar]
- 21.Patwardhan A, Bardin S, Miserey-Lenkei S, Larue L, Goud B, Raposo G, Delevoye C. Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes. Nat Commun. 2017;8:15835. doi: 10.1038/ncomms15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 2000;19(21):5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miserey-Lenkei S, Waharte F, Boulet A, Cuif MH, Tenza D, El Marjou A, Raposo G, Salamero J, Heliot L, Goud B, Monier S. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic. 2007;8(10):1385–1403. doi: 10.1111/j.1600-0854.2007.00612.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Jiang C, Jin M, Gong Y, Zhang X. The role of Rab6 GTPase in the maturation of phagosome against Staphylococcus aureus. Int J Biochem Cell Biol. 2015;61:35–44. doi: 10.1016/j.biocel.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Echard A, Jollivet F, Martinez O, Lacapere JJ, Rousselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279(5350):580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 26.Short B, Preisinger C, Schaletzky J, Kopajtich R, Barr FA. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr Biol. 2002;12(20):1792–1795. doi: 10.1016/S0960-9822(02)01221-6. [DOI] [PubMed] [Google Scholar]
- 27.Lee PL, Ohlson MB, Pfeffer SR (2015) Rab6 regulation of the kinesin family KIF1C motor domain contributes to Golgi tethering. Elife. 10.7554/elife.06029 [DOI] [PMC free article] [PubMed]
- 28.Miserey-Lenkei S, Bousquet H, Pylypenko O, Bardin S, Dimitrov A, Bressanelli G, Bonifay R, Fraisier V, Guillou C, Bougeret C, Houdusse A, Echard A, Goud B. Coupling fission and exit of RAB6 vesicles at Golgi hotspots through kinesin-myosin interactions. Nat Commun. 2017;8(1):1254. doi: 10.1038/s41467-017-01266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsay AJ, Jollivet F, Horgan CP, Khan AR, Raposo G, McCaffrey MW, Goud B. Identification and characterization of multiple novel Rab-myosin Va interactions. Mol Biol Cell. 2013;24(21):3420–3434. doi: 10.1091/mbc.E13-05-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peurois F, Veyron S, Ferrandez Y, Ladid I, Benabdi S, Zeghouf M, Peyroche G, Cherfils J. Characterization of the activation of small GTPases by their GEFs on membranes using artificial membrane tethering. Biochem J. 2017;474(7):1259–1272. doi: 10.1042/BCJ20170015. [DOI] [PubMed] [Google Scholar]
- 31.Moorhead AR, Rzomp KA, Scidmore MA. The Rab6 effector Bicaudal D1 associates with Chlamydia trachomatis inclusions in a biovar-specific manner. Infect Immun. 2007;75(2):781–791. doi: 10.1128/IAI.01447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subauste MC, Von Herrath M, Benard V, Chamberlain CE, Chuang TH, Chu K, Bokoch GM, Hahn KM. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J Biol Chem. 2000;275(13):9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- 33.van Rijssel J, Hoogenboezem M, Wester L, Hordijk PL, Van Buul JD. The N-terminal DH-PH domain of Trio induces cell spreading and migration by regulating lamellipodia dynamics in a Rac1-dependent fashion. PLoS One. 2012;7(1):e29912. doi: 10.1371/journal.pone.0029912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Progida C, Malerod L, Stuffers S, Brech A, Bucci C, Stenmark H. RILP is required for the proper morphology and function of late endosomes. J Cell Sci. 2007;120(Pt 21):3729–3737. doi: 10.1242/jcs.017301. [DOI] [PubMed] [Google Scholar]
- 35.Degot S, Auzan M, Chapuis V, Beghin A, Chadeyras A, Nelep C, Calvo-Munoz ML, Young J, Chatelain F, Fuchs A (2010) Improved visualization and quantitative analysis of drug effects using micropatterned cells. J Vis Exp. 10.3791/2514 [DOI] [PMC free article] [PubMed]
- 36.Gittes F, Schmidt CF. Signals and noise in micromechanical measurements. Methods Cell Biol. 1998;55:129–156. doi: 10.1016/S0091-679X(08)60406-9. [DOI] [PubMed] [Google Scholar]
- 37.Kress H, Stelzer EH, Holzer D, Buss F, Griffiths G, Rohrbach A. Filopodia act as phagocytic tentacles and pull with discrete steps and a load-dependent velocity. Proc Natl Acad Sci USA. 2007;104(28):11633–11638. doi: 10.1073/pnas.0702449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bornschlogl T, Romero S, Vestergaard CL, Joanny JF, Van Nhieu GT, Bassereau P. Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc Natl Acad Sci USA. 2013;110(47):18928–18933. doi: 10.1073/pnas.1316572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westerfield M. The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio) 4. Eugene: Univ. of Oregon Press; 2000. [Google Scholar]
- 40.Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci USA. 1982;79(8):2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, Yoshimura S, Nakamura N, Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182(5):837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thery M, Pepin A, Dressaire E, Chen Y, Bornens M. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Motil Cytoskeleton. 2006;63(6):341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe T, Hosoya H, Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell. 2007;18(2):605–616. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betapudi V. Life without double-headed non-muscle myosin II motor proteins. Front Chem. 2014;2:45. doi: 10.3389/fchem.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikebe M, Hartshorne DJ. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 1985;260(18):10027–10031. [PubMed] [Google Scholar]
- 47.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271(34):20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K, Kano Y, Amano M, Onishi H, Kaibuchi K, Fujiwara K. Rho-kinase–mediated contraction of isolated stress fibers. J Cell Biol. 2001;153(3):569–584. doi: 10.1083/jcb.153.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem. 2004;259(1–2):197–209. doi: 10.1023/B:MCBI.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- 50.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, Behrouz B, Melrose HL, Hentati E, Puschmann A, Evans DM, Conibear E, Wasserman WW, Aasly JO, Burkhard PR, Djaldetti R, Ghika J, Hentati F, Krygowska-Wajs A, Lynch T, Melamed E, Rajput A, Rajput AH, Solida A, Wu RM, Uitti RJ, Wszolek ZK, Vingerhoets F, Farrer MJ. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniel JL, Adelstein RS. Isolation and properties of platelet myosin light chain kinase. Biochemistry. 1976;15(11):2370–2377. doi: 10.1021/bi00656a019. [DOI] [PubMed] [Google Scholar]
- 52.Singh TJ, Akatsuka A, Huang KP. Phosphorylation of smooth muscle myosin light chain by five different kinases. FEBS Lett. 1983;159(1–2):217–220. doi: 10.1016/0014-5793(83)80449-9. [DOI] [PubMed] [Google Scholar]
- 53.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11(12):471–477. doi: 10.1016/S0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- 55.Hanna S, Miskolci V, Cox D, Hodgson L. A new genetically encoded single-chain biosensor for Cdc42 based on FRET, useful for live-cell imaging. PLoS One. 2014;9(5):e96469. doi: 10.1371/journal.pone.0096469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grigoriev I, Yu KL, Martinez-Sanchez E, Serra-Marques A, Smal I, Meijering E, Demmers J, Peranen J, Pasterkamp RJ, van der Sluijs P, Hoogenraad CC, Akhmanova A. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr Biol. 2011;21(11):967–974. doi: 10.1016/j.cub.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 57.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391(6662):93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 58.Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15(12):2997–3005. doi: 10.1002/j.1460-2075.1996.tb00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz J, Proff J, Havemeier A, Ladwein M, Rottner K, Barlag B, Pich A, Tatge H, Just I, Gerhard R. Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS One. 2012;7(9):e44358. doi: 10.1371/journal.pone.0044358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou C, Zhuang Z, Deng X, Xu Y, Zhang P, Zhu L. Knockdown of Trio by CRISPR/Cas9 suppresses migration and invasion of cervical cancer cells. Oncol Rep. 2018;39(2):795–801. doi: 10.3892/or.2017.6117. [DOI] [PubMed] [Google Scholar]
- 61.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15(4):1942–1952. doi: 10.1128/MCB.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Yang J, Li J, Shen X, Le Y, Zhou C, Wang S, Zhang S, Xu D, Gong Z. MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Sci Rep. 2015;5:13677. doi: 10.1038/srep13677. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Moshal KS, Ferri-Lagneau KF, Haider J, Pardhanani P, Leung T. Discriminating different cancer cells using a zebrafish in vivo assay. Cancers (Basel) 2011;3(4):4102–4113. doi: 10.3390/cancers3044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seabra MC, Coudrier E. Rab GTPases and myosin motors in organelle motility. Traffic. 2004;5(6):393–399. doi: 10.1111/j.1398-9219.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- 65.Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol. 2010;20(6):329–336. doi: 10.1016/j.tcb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 66.Horgan CP, McCaffrey MW. Rab GTPases and microtubule motors. Biochem Soc Trans. 2011;39(5):1202–1206. doi: 10.1042/BST0391202. [DOI] [PubMed] [Google Scholar]
- 67.Kjos I, Vestre K, Guadagno NA, Borg Distefano M. Progida C (2018) Rab and Arf proteins at the crossroad between membrane transport and cytoskeleton dynamics. Biochim Biophys Acta. 1865;10:1397–1409. doi: 10.1016/j.bbamcr.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 68.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration–myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125(Pt 9):2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9(3):299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 70.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281(47):35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 71.Jorrisch MH, Shih W, Yamada S. Myosin IIA deficient cells migrate efficiently despite reduced traction forces at cell periphery. Biol Open. 2013;2(4):368–372. doi: 10.1242/bio.20133707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116(3):431–443. doi: 10.1016/S0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 73.Qiu Y, Brown AC, Myers DR, Sakurai Y, Mannino RG, Tran R, Ahn B, Hardy ET, Kee MF, Kumar S, Bao G, Barker TH, Lam WA. Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc Natl Acad Sci USA. 2014;111(40):14430–14435. doi: 10.1073/pnas.1322917111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai Y, Rossier O, Gauthier NC, Biais N, Fardin MA, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123(Pt 3):413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakatsuki T, Wysolmerski RB, Elson EL. Mechanics of cell spreading: role of myosin II. J Cell Sci. 2003;116(Pt 8):1617–1625. doi: 10.1242/jcs.00340. [DOI] [PubMed] [Google Scholar]
- 76.Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J Cell Sci. 2012;125(Pt 24):5974–5983. doi: 10.1242/jcs.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66(9):4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 78.Nisenholz N, Paknikar A, Koster S, Zemel A. Contribution of myosin II activity to cell spreading dynamics. Soft Matter. 2016;12(2):500–507. doi: 10.1039/c5sm01733e. [DOI] [PubMed] [Google Scholar]
- 79.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 80.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/S0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 81.Carlier MF, Ducruix A, Pantaloni D. Signalling to actin: the Cdc42-N-WASP-Arp2/3 connection. Chem Biol. 1999;6(9):R235–R240. doi: 10.1016/S1074-5521(99)80107-0. [DOI] [PubMed] [Google Scholar]
- 82.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160(3):409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367(6458):40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 84.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1(5):253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 85.van Leeuwen FN, van Delft S, Kain HE, van der Kammen RA, Collard JG. Rac regulates phosphorylation of the myosin-II heavy chain, actinomyosin disassembly and cell spreading. Nat Cell Biol. 1999;1(4):242–248. doi: 10.1038/12068. [DOI] [PubMed] [Google Scholar]
- 86.Ridley AJ. Rho GTPases and cell migration. J Cell Sci. 2001;114(Pt 15):2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 87.Jian Q, Miao Y, Tang L, Huang M, Yang Y, Ba W, Liu Y, Chi S, Li C. Rab23 promotes squamous cell carcinoma cell migration and invasion via integrin beta1/Rac1 pathway. Oncotarget. 2016;7(5):5342–5352. doi: 10.18632/oncotarget.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Margiotta A, Progida C, Bakke O. Bucci C (2017) Rab7a regulates cell migration through Rac1 and vimentin. Biochim Biophys Acta. 1864;2:367–381. doi: 10.1016/j.bbamcr.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 89.Chevallier J, Koop C, Srivastava A, Petrie RJ, Lamarche-Vane N, Presley JF. Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 2009;583(7):1096–1101. doi: 10.1016/j.febslet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 90.Bravo-Cordero JJ, Cordani M, Soriano SF, Diez B, Munoz-Agudo C, Casanova-Acebes M, Boullosa C, Guadamillas MC, Ezkurdia I, Gonzalez-Pisano D, Del Pozo MA, Montoya MC. A novel high-content analysis tool reveals Rab8-driven cytoskeletal reorganization through Rho GTPases, calpain and MT1-MMP. J Cell Sci. 2016;129(8):1734–1749. doi: 10.1242/jcs.174920. [DOI] [PubMed] [Google Scholar]
- 91.Lee CS, Choi CK, Shin EY, Schwartz MA, Kim EG. Myosin II directly binds and inhibits Dbl family guanine nucleotide exchange factors: a possible link to Rho family GTPases. J Cell Biol. 2010;190(4):663–674. doi: 10.1083/jcb.201003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacquemet G, Hamidi H, Ivaska J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr Opin Cell Biol. 2015;36:23–31. doi: 10.1016/j.ceb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 93.Jacquemet G, Paatero I, Carisey AF, Padzik A, Orange JS, Hamidi H, Ivaska J. FiloQuant reveals increased filopodia density during breast cancer progression. J Cell Biol. 2017;216(10):3387–3403. doi: 10.1083/jcb.201704045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1: Cdc42 is transported towards the cell periphery in Rab6-positive vesicles. U2OS cells co-transfected with GFP-Rab6 and mCherry-Cdc42 were imaged using a spinning-disk confocal microscope at 1 second intervals. A vesicle positive for both GFP-Rab6 and mCherry-Cdc42 moving towards the cell periphery is tracked over time. Scale bar: 10 µm (AVI 2233 kb)
Movie 2: Rab6-positive vesicles are transported towards filopodia. U2OS cells transfected with GFP-Rab6 and labeled with SiR-actin, were imaged with a Total Internal Reflection fluorescent (TIRF) microscope. (AVI 188 kb)
Supplementary figure 1: Rab6 silencing promotes cell migration in RPE-1 cells. (a) RPE-1 cells treated with siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and imaged every 15th minute for 24 h. Representative images of (T0) and 24 h after scratching are shown. Scale bar: 300 µm. Quantification of single-cell speed (b) and directness (c) is represented as mean ± s.e.m.; n>230 cells from five independent experiments. (d) Cell lysates from each of the indicated samples were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (e) The intensities of the bands were quantified using densitometry, normalized against the amount of tubulin, and plotted relative to the intensities obtained in cells transfected with siRNA control. The values represent the mean ± s.e.m. for five independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (paired Student’s t test). (f) Representative track plots of the single-cell migration distances. Individual tracks are shown so that each starts at the origin (distance 0). (g) Velocity vector fields of particle image velocimetry analysis (PIV). Scale bar: 100 µm (TIFF 1719 kb)
Supplementary figure 2: Rab6 depletion promotes cell migration independently from cell proliferation. (a) U2OS cells treated with either siRNA control, siRNA Rab6 #1, siRNA Rab6 #2, or treated with the same siRNAs and afterwards transfected with GFP-Rab6 were scratch-wounded and imaged for 24 h. The graph represents the mean from three independent experiments of the relative wound density (%) as function of time. ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=24h). (b) Cell lysates from each sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (c) U2OS cells treated with either siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and imaged in medium containing Cytotox Green reagent to label dying cells green. Cell death during migration was quantified by dividing the number of green objects at each timepoint with the number of green objects at time 0. The graph represents the mean from three independent experiments. (d) U2OS cells treated with either siRNA control, siRNA Rab6 #1 or siRNA Rab6 #2 were scratch-wounded and incubated in medium (with or without serum as indicated) containing the modified thymidine analogue Edu. The graph shows the percentage of Edu-positive cells after 24 h, relative to Edu-positive control cells in medium containing serum. The data represents the mean ± s.e.m from three independent experiments. (e) HeLa cells treated with either siRNA control, siRNA Rab6 #1, or siRNA Rab6 #2 were scratch-wounded and imaged for 24 h. Representative images of (T0) and 24 h after scratching are shown. Scale bar: 300 µm. (f) Quantification of the relative wound density (%) as function of time for control cells and Rab6-depleted cells. Data represents the mean of three (siRNA #1) or two (siRNA #2) independent experiments. *P < 0.05; ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=24h). (g) Cell lysates from each sample were subjected to Western blot analysis with antibodies against Rab6 and tubulin (as a loading control). (TIFF 809 kb)
Supplementary figure 3: Role of Trio in Rab6-dependent Cdc42 activation and cell migration. (a) Lysates from U2OS cells transfected with control siRNA, siRNA Rab6 #1, siRNA Trio, siRNA Rab6 #1 and siRNA Trio, or siRNA DOCK10 were mixed with beads coupled to GST–PAK–PBD to pull down the active forms of Cdc42 and analyzed by western blot. (b) Quantification of the levels of active Cdc42 were normalized to the amount of tubulin and plotted relative to the intensities of GTP-bound Cdc42 in the control sample. The graphs represent the mean ± s.e.m. normalized to the control of at least three independent experiments. *P < 0.05; **P < 0.01 (paired Student’s t test). (c) U2OS cells treated with either siRNA control, siRNA Rab6 #1, siRNA Trio, siRNA Rab6 #1 and siRNA Trio, or treated with siRNA Trio and afterwards transfected with GFP-Rab6, were scratch-wounded and imaged for 24 h. The graph represents the mean from at least three independent experiments of the relative wound density (%) as function of time. ***P < 0.001 compared to control (two-way repeated measures ANOVA followed by Tukey’s post test for t=21h). (TIFF 309 kb)
Supplementary figure 4: Rab6 and filopodia. (a) HeLa cells treated with siRNA control or siRNA Rab6 #1 were fixed and stained with DAPI and rhodamine-conjugated phalloidin. The lower insets show magnifications of the boxed areas. Scale bar: 20 µm. (b) Quantification of the number of filopodia per 100 µm cell membrane length. The graph represents the mean ± s.e.m.; n>100 cells from four independent experiments. *P < 0.05 (paired Student’s t test). (c) U2OS cells transfected with GFP-Rab6 and labeled with SiR-actin, were imaged with a Total Internal Reflection fluorescent (TIRF) microscope. Right panels: arrows in the insets point to Rab6-positive vesicles moving towards filopodia. Scale bar: 10 µm. (TIFF 698 kb)
Supplementary figure 5: Rab6 knockdown promotes spread of cancer cells in zebrafish embryos following xenotransplantation. (a) H1299 cells transfected with control siRNA or siRNA Rab6 #2 were stained with QDs Q-Tracker 655 and injected into the otic vesicle (red circle) of 2-dpf zebrafish wild-type embryos. Images were taken soon after the injection (T0) as well as 24 and 48 h later. In contrast to the control cells (left panels) that remained confined to the injection area, cell silenced for Rab6 (right panels) showed higher ability to migrate outside from the otic vesicle, both at 24 h and at even more at 48 h (arrows). Magnification of the boxed areas are shown in the inset on the right of each panel. Scale bar: 100 µm. (b) Quantification of cell migration was performed by measuring the area occupied by the cells at 24 h and 48 h after injection and was normalized to the area occupied by the cells in the otic vesicle at T0. The graph represents the mean ± s.e.m. from n>15 embryos. *P < 0.05 (paired Student’s t test). (c) H1299 cells transfected with control siRNA or siRNA Rab6 #2 for 24 or 48 h were subjected to western blot analysis using antibodies against Rab6 and tubulin (as a loading control). (TIFF 477 kb)