Abstract
Lysophosphatidylcholine (LysoPC) has been shown to induce the expression of inflammatory proteins, including cyclooxygenase-2 (COX-2) and interleukin-6 (IL-6), associated with cardiac fibrosis. Here, we demonstrated that LysoPC-induced COX-2 and IL-6 expression was inhibited by silencing NADPH oxidase 1, 2, 4, 5; p65; and FoxO1 in human cardiac fibroblasts (HCFs). LysoPC-induced IL-6 expression was attenuated by a COX-2 inhibitor. LysoPC-induced responses were mediated via the NADPH oxidase-derived reactive oxygen species-dependent JNK1/2 phosphorylation pathway, leading to NF-κB and FoxO1 activation. In addition, we demonstrated that both FoxO1 and p65 regulated COX-2 promoter activity stimulated by LysoPC. Overexpression of wild-type FoxO1 and S256D FoxO1 enhanced COX-2 promoter activity and protein expression in HCFs. These results were confirmed by ex vivo studies, where LysoPC-induced COX-2 and IL-6 expression was attenuated by the inhibitors of NADPH oxidase, NF-κB, and FoxO1. Our findings demonstrate that LysoPC-induced COX-2 expression is mediated via NADPH oxidase-derived reactive oxygen species generation linked to the JNK1/2-dependent pathway leading to FoxO1 and NF-κB activation in HCFs. LysoPC-induced COX-2-dependent IL-6 expression provided novel insights into the therapeutic targets of the cardiac fibrotic responses.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2916-7) contains supplementary material, which is available to authorized users.
Keywords: COX-2, Lysophosphatidylcholine, IL-6, NOX, ROS, NF-κB, FoxO1
Introduction
Heart diseases involve the pathological development of several functional and structural defects in the heart [1]. Cardiac fibroblasts (CFs) play a major role in the repair and remodeling of the heart during myocardial injury [2]. However, over-secretion of cytokines from CFs, such as interleukin (IL)-1β, tumor necrosis factor (TNF)-α, IL-6, and transforming growth factor-β (TGF-β) [3], amplifies inflammatory cascades, contributing to cardiac fibrosis and hypertrophy [4]. Lysophosphatidylcholine (LysoPC) is produced by phospholipase A2 (PLA2) via the hydrolysis of cell membrane-derived phosphatidylcholine (PC). It regulates several cellular events including oxidative stress, differentiation, and inflammation [5–7]. The levels of LysoPC are also increased in ischemic and insulin-resistant hearts [8, 9], suggesting its association with cardiomyocyte apoptosis in fibrosis [8, 10], providing important evidence for an association between LysoPC and pathogenesis of various cardiovascular diseases [11]. Previous reports have indicated that IL-6 is a key mediator in tissue remodeling that promotes acute inflammation to chronic fibrotic state and induces recurrent inflammatory cascades [4]. Induction of IL-6 in rat CFs is associated with fibroblast proliferation and differentiation via a TGF-β-mediated pathway [12–14]. LysoPC is a potential inducer to stimulating the release of pro-inflammatory cytokines including IL-6 [6]. Nevertheless, the pathological relationship between LysoPC-induced inflammatory cytokines and cardiac fibrosis remains unclear.
LysoPC has been shown to induce intracellular generation of reactive oxygen species (ROS) in various cell types [15, 16]. ROS is a group of highly reactive molecules, including hydrogen peroxide (H2O2), hydroxyl radical (OH·), and superoxide anion (O·−2). NAPDH oxidase (NOX) is a major source of ROS generation such as H2O2 and O·−2 via an NADPH-dependent one- or two-electron reduction of oxygen [17]. Activation of NOX is hypothesized to increase oxidative stress, contributing to pathogenesis of cardiac remodeling and heart failure [18, 19]. NOX-derived ROS are recognized as major second messengers involved in activation of various signaling components leading to COX-2 expression via NF-κB [20]. Upregulation of COX-2 is linked to the development of heart failure involving macrophage infiltration and fibroblast proliferation [21, 22]. Brkic et al. indicated that LysoPC induced COX-2 expression in endothelial cells [23]. However, whether COX-2 further induced IL-6 secretion in human cardiac fibroblasts (HCFs) is still unknown.
Forkhead transcription factors of the O class 1 (FoxO1) have been shown to be involved in COX-2 expression induced by IL-1β [24], whereas FoxO1 negatively regulates COX-2 expression by sphingosine-1-phosphate [25]. FoxO1 is a critical player in the development of physiological heart and cardiac remodeling during heart failure [26, 27]. FoxO1 also regulates cellular energy metabolism, cell survival, and autophagy in the heart [28–30]. Moreover, accumulation of nuclear FoxO1 increases the expression of pro-inflammatory cytokines, including IL-6 [31, 32]. Based on these findings, the functional roles of FoxO1 in physiological and pathological processes are still controversial. Therefore, we aimed to investigate the mechanisms by which LysoPC primed NOX/ROS-JNK1/2-dependent NF-κB and FoxO1 activation, leading to COX-2-induced IL-6 expression in HCFs.
Methods
Antibodies and reagents
CM-H2DCFDA and M-MLV Reverse Transcriptase kit were obtained from Invitrogen (Carlsbad, CA, USA). Human cytokine antibody array C3 was obtained from Ray Biotech (Norcross, GA, USA). Anti-phospho-FoxO1 (Ser256) (#9461) was obtained from Cell signaling (Danvers, MA, USA). Anti-COX-2 (ab62331), anti-FoxO1 (ab52857), and anti-phospho-p65 (ab86299) were obtained from Abcam (Cambridge, UK). Anti-FoxO1 (sc-11350), anti-p65 (sc-398442), anti-lamin A (sc-20680), and β-actin (sc-47778) antibodies were obtained from Santa Cruz (Santa Cruz, CA, USA). Anti-GAPDH (#MCA-1D4) was obtained from EnCor Biotechnology (Gainesville, FL, USA). LysoPC (L-0906) and TRIzol were obtained from Sigma-Aldrich (St. Louis, MO, USA). LysoPC was dissolved in 50% ethanol and filtered through a 0.22 μm syringe filter. A final concentration of 0.25% ethanol was used for cell culture treatment. Celecoxib and diphenyleneiodonium chloride (DPI) were obtained from Biomol (Plymouth Meeting, PA, USA). AS1842856 was obtained from EMD Millipore (Billerica, MA, USA). Helenalin was obtained from Cayman Chemicals (Ann Arbor, MI, USA). Kapa Probe Fast qPCR Kit was obtained from KAPA Biosystems (Wilmington, MA, USA). GenMute™ siRNA Transfection Reagent was obtained from SignaGen Lab (Gaithersburg, MD, USA). X-tremeGENE™ HP DNA Transfection Reagent and β-gal reporter assay kit were obtained from Roche Applied Sciences (Indianapolis, IN, USA). SDS-PAGE supplies and BSA were obtained from MDBio Inc (Taipei, Taiwan). DMEM/F-12 medium was obtained from Corning (Manassas, VA, USA). FBS was obtained from Gibco (Grand Island, NY, USA).
Ethics of animal experimentation
Male ICR mice (25–30 g, 8 weeks old) were purchased from the National Laboratory Animal Centre (Taipei, Taiwan). All animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines and approved by the IACUC committee of Chang Gung University (CGU 16-046), and were carried out in accordance with the guidelines of the National Institute of Heath for the Care and Use of Laboratory Animal.
Collection and incubation of ex vivo tissues
ICR mice were anesthetized with one injection of Zoletil (40 mg/kg i.p.) and xylazine (10 mg/kg i.p.). After anesthesia, mice were withdrawn with lined forceps on the paws, and then the chests were opened and the hearts were quickly removed for experiments. The slices of cardiac apexes were pretreated with the respective inhibitors for 1 h and then exposed to LysoPC for 6 h while in Krebs solution (119 mM NaCl, 25 mM NaHCO3, 4.7 mM KCl, 11 mM glucose, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, pH 7.4) bubbled with 95% O2 and 5% CO2 at 37 °C. The homogenates of cardiac apexes were prepared and lysed in a lysis buffer (25 mM Tris–HCl (pH 7.4), 25 mM NaCl, 25 mM NaF, 25 mM sodium pyrophosphate, 1 mM sodium vanadate, 2.5 mM EDTA, 2.5 mM EGTA, 0.05% (w/v) Triton X-100, 0.5% (w/v) SDS, 0.5% (w/v) deoxycholate, 0.5% (w/v) NP-40, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM PMSF) and subjected to western blot analysis and RT/qPCR.
Cell culture
Human cardiac fibroblasts were purchased from ScienCell Research Laboratories (San Diego, CA, USA) and maintained in DMEM/F-12 medium supplemented with 10% FBS, as previously described [33].
Preparation of samples and western blot analysis
Growth-arrested HCFs were incubated without or with different concentrations of LysoPC at 37 °C for the indicated time intervals. When pharmacological inhibitors were used, they were added 1 h prior to the exposure of LysoPC. After incubation, the cells were rapidly washed with ice-cold PBS and lysed with a sample buffer containing 125 mM Tris–HCl, 1.25% SDS, 6.25% glycerol, 3.2% β-metacaptoethanol, and 7.5 nM bromophenol blue with pH 6.8. Samples were denatured, subjected to SDS-PAGE using a 10% (w/v) running gel, and transferred to nitrocellulose membrane (BioTrace™ NT membrane, Pall Life Sciences, Ann Arbor, MI, USA). The membranes were immunoblotted with one of the primary antibodies (1:1000 dilution) overnight at 4 °C, followed by incubation with a peroxidase-conjugated secondary antibody at room temperature for 2 h. The immunoreactive bands were visualized by enhanced chemiluminescence reagent (Western Lighting Plus; Perkin Elmer, Waltham, MA, USA). The images of the immunoblots were acquired by using a UVP BioSpectrum 500 imaging system (Upland, CA, USA) and densitometry analysis was conducted using UN-SCAN-IT gel software (Orem, UT, USA).
cDNA microarray analysis
Growth-arrested HCFs were incubated with LysoPC for either 6 h or 12 h. Total RNA was extracted with TRIzol (Sigma-Aldrich, St. Louis, MO, USA) which was used to prepare aRNA amplification (Amino Allyl MessageAmp II aRNA Amplification Kit; Ambion) and labeled with Cy5. The labeled aRNAs were hybridized to the Human Whole Genome OneArray™ (HOA 6.2; Phalanx Biotech Group, Hsinchu, Taiwan) according to Phalanx OneArray™ Plus protocol and examined using Agilent 0.1 XDR protocol (Phalanx Biotech Group, Hsinchu, Taiwan). The raw fluorescence intensities of each spot were analyzed by GenePix software and subsequently loaded into Rosetta Resolver System (version 7.2, Rosetta Biosoftware, Seattle, WA, USA) to process analyzed data. Normalized data are expressed as log2 of LysoPC-treatment relative to the control group.
Reverse-transcription PCR and qPCR analyses
Total RNA was extracted with TRIzol reagent according to the protocol of the manufacturer.
First-strand cDNA synthesis was performed with 5 μg of total RNA using Oligo(dT)15 as primers in a final volume of 20 μl [25 ng/μl Oligo(dT)15, 0.5 mM dNTPs, 10 mM DTT, 2 units/μl RNase inhibitor, and 10 unit/μl of superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA)]. The synthesized cDNAs were used as templates for PCR reaction using Q-Amp™ 2× screeningFire Taq master mix (Bio-Genesis Technologies, Taipei, Taiwan) and primers for the target genes. qPCR was performed by using Kapa Probe Fast qPCR Kit Master Mix Universal (KAPA Biosystems, Wilmington, MA, USA) on a StepOnePlus™ real-time PCR system (ThermoScientific-Applied Biosystems). The relative amount of the target gene was calculated using (Ct = threshold cycle). The primer sequences are given in Supplementary Tables 1 and 2.
Determination of NADPH oxidase activity by chemiluminescence assay
Human cardiac fibroblasts were seeded in six-well plates, reaching about 90% confluence, and then transferred to fresh DMEM/F-12 medium for 24 h. Cells were then exposed to LysoPC for the indicated time intervals, gently scraped, and then centrifuged at 8000×g for 12 min at 4 °C. The cell pellet was resuspended in 100 μl ice-cold PBS containing 150 mM sucrose and 1 mM EGTA. To a final 200 μl volume of pre-warmed (37 °C) PBS containing either NAD(P)H (0.1 mM) or lucigenin (0.2 mM), 10 μl of cell suspension was added to initiate the reaction followed by immediate measurement of chemiluminescence using a luminometer (Synergy H1 Hybird Reader, BioTek) in an out-of-coincidence mode. Appropriate blanks and controls were established, and chemiluminescence was recorded. Neither NAD(P)H nor NADH enhanced the background chemiluminescence of lucigenin alone (30–40 counts/min). Chemiluminescence was continuously measured for 12 min, and the activity of NADPH oxidase was expressed as counts per million cells.
Evaluation of ROS generation
Human cardiac fibroblasts were stimulated with LysoPC and then incubated in DMEM/F-12 medium containing 5 μM CM-H2DCFDA. The fluorescent intensity was detected by using a fluorescence microplate reader (Synergy H1 Hybird Reader, BioTek) with EX495 nm and EM529 nm.
Transient transfection with siRNAs
Human cardiac fibroblasts were plated in 12-well, 6-well plates or 10-cm dish, reaching about 90% confluence, and transferred to fresh DMEM/F-12 medium before transfection. p65 siRNA (HSS109161) was obtained from Invitrogen Life Technologies (Carlsbad, CA, USA), and siRNAs of NOX1 (SASI_Hs02_00342845), NOX2 (SASI_Hs01_00086110, NOX4 (SASI_Hs02_00349918), FoxO1 (SASI_Hs01_00076732), and scramble siRNA were obtained from Sigma-Aldrich (St. Louis, MO, USA). NOX5 siRNA (1070614_8103) was obtained from MDBio Inc (Taipei, Taiwan) [34]. The siRNA sequences are shown in Supplementary Table 3. Transient transfection of siRNA was carried out using GenMute™ siRNA Transfection Reagent according to the instructions of the manufacturer (SignaGen Lab. Gaithersburg, MD, USA). The siRNA (100 nM) was added to each well and then incubated at 37 °C for 6 h. The cells were transferred to DMEM/F-12 medium containing 10% FBS for an additional 6 h, washed twice with PBS, and then maintained in serum-free DMEM/F-12 medium for 24 h before treatment with LysoPC.
Construction of FoxO1 plasmid DNA
For the construction of pCMV-Tag2B, the FoxO1 protein-encoding sequence was amplified by PCR using the following primer sequences: sense 5′-GGGGA TATCATGGCCGAGGCGCCTCAGGTGGTGGAGA-3′ and antisense 5′-GGGAAGCTTTCAGCCTGACACCCAGCTATGTG-3′. Ser256-to-Ala256 FoxO1 mutant (S256A FoxO1), Ser256-to-Asp256 FoxO1 mutant (S256D FoxO1), Ser319-to-Ala319 FoxO1 mutant (S319A FoxO1), and Ser319-to-Asp319 FoxO1 mutant (S319D FoxO1) were cloned. The Ser256 and Ser319 residues of FoxO1 were replaced with an alanine and aspartic acid residue, respectively, by priming with oligonucleotide 5′-CTCCTAGGAG AAGAGCTGCATGCAATGGACAACAACAGTAAATTTGCT-3′ (S256A), 5′-CTCCT AGGAGAAGAGCTGCATGACATGGACAACAACAGTAAATTTGCT-3′ (S256D), 5′-TACTATTAGTGGGAGACTCGCACCCATTATGACCGAACAG-3′ (S319A), and 5′-TACTATTAGTGGGAGACTCGACCCCATTATGACCGAACAG-3′ (S319D). The PCR products were cloned into the EcoRV-HindIII site of the pCMV-Tag2B vector.
Transient transfection of plasmid DNA
Human cardiac fibroblasts were seeded in six-well plates or 10-cm dishes, reaching 90% confluence, transferred to serum-free DMEM/F-12 medium, and transiently transfected with plasmid DNA using an X-tremeGENE™ HP DNA Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA). Briefly, 3 μg of plasmid DNA (pCMV-Tag2B-FoxO1, -FoxO1 mutants, or pCMV-Tag2B) into six-well or 15 μg of plasmid DNA into 10-cm dish was diluted with Opti-MEM (200µl/well or 500µl/dish) (Gibco, Life Technologies, Gaithersburg, MD, USA) and incubated at 25 °C for 15 min. The X-tremeGENE™ HP DNA Transfection Reagent was added to the DNA diluents [3:1 ratio of reagent (μl) to DNA (μg)] and incubated at 25 °C for 20 min. Subsequently, the transfection complex was added into each well and incubated at 37 °C in a humidified 5% CO2 atmosphere. After 48 h of transfection, the cells were incubated with LysoPC and harvested.
Measurement of COX-2 promoter activity
For construction of the COX-2-luc plasmid, human COX-2 promoter, a region spanning from − 484 to + 37 was cloned into pGL3-basic vector, as previously described [35]. HCFs were co-transfected with pGL3b-cox-2, pCMV-Tag2B-FoxO1, pCMV-Tag2B (as a control group), or pCMV-β-gal plasmid (as an internal control). Promoter activities of COX-2 were determined using a luciferase-e assay HIT kit (BioThema, Handen, Sweden) and normalized with β-Gal reporter gene.
Assessment of cytokine secretion
Human cardiac fibroblasts were starved and exposed to LysoPC in the presence or absence of pharmacological inhibitors for the indicated time durations. The media were collected and the levels of cytokines and chemokines were determined using a human cytokine antibody array (Ray Biotech Inc., Norcross, GA, USA). The dot intensities of different cytokines were semi-quantified using UN-SCAN-IT and normalized to the intensity of internal positive control.
Immunofluorescence staining
Human cardiac fibroblasts were seeded on coverslips in six-well culture plates, reaching 90% confluence, transferred to serum-free DMEM/F-12 medium overnight, and then stimulated with 40 μM LysoPC for the indicated time durations. After washing twice with ice-cold PBS, the cells were fixed with 4% (w/v) paraformaldehyde in PBS for 30 min, and then permeabilized with 0.1 % Triton X-100 in PBS for 15 min. The staining was performed by incubating with 5% BSA for 2 h at 37 °C, followed by incubation with a primary anti-phospho-FoxO1S256 polyclonal antibody (1:100 dilution) overnight in PBS with 1% BSA. The cells were washed thrice with PBS and incubated for 2 h with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody (1:100 dilution; Jackson Immunoresearch) in PBS with 1% BSA. Finally, cells were washed thrice with PBS and then mounted with aqueous mounting medium containing DAPI (H1200; Vector Lab, Burlingame, CA, USA). Images were captured with a fluorescence microscope (Axiovert 200 M; Carl Zeiss, Thornwood, NY, USA).
Chromatin immunoprecipitation assay
Human cardiac fibroblasts were transfected with FoxO1 plasmid or incubated with LysoPC. Samples were prepared and chromatin immunoprecipitation (ChIP) assay was performed. Protein–DNA complexes were fixed by 1% formaldehyde in DMEM/F-12 medium and the reaction was stopped by 0.125 M glycine. The fixed cells were washed and lysed in a lysis buffer (1% SDS, 10 mM EDTA, 1 mM PMSF, 50 mM Tris–HCl, pH 8.1). The cell lysates were sonicated using a Misonix Sonicator S-4000 (pulse on for 20 s and off for 15 s at amplitude 20 for 90 cycles; Farmingdale, NY, USA) at 4 °C until the chromosome DNA was broken to approximately 200–500 base pairs. The samples were centrifuged, and the soluble chromatin was pre-cleared by incubation with salmon sperm DNA–protein agarose A slurry (Upstate, Billerica, MA, USA) for 30 min at 4 °C with rotation. The samples were then centrifuged at 4000×g for 2 min and the supernatant was transferred to a new tube. The samples were quantified and adjusted to the same protein concentrations. One portion of the sample was used as DNA input control. Soluble chromatin was immunoprecipitated either without (control) or with an anti-FoxO1, anti-p65, or anti-Flag M2 antibody for 3 h at 4 °C, and then incubated with salmon sperm DNA–protein agarose A slurry overnight at 4 °C with rotation. The samples were successively washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1), high-salt buffer (same as low-salt buffer but with 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1), and Tris–EDTA (10 mM Tris–HCl and 1 mM EDTA, pH 8.0), prior to elution (1% SDS, 100 mM NaHCO3). The cross-linked protein–DNA complexes were reversed by incubation at 65 °C overnight. DNA fragments were purified by using an EasyPure PCR/Gel Extraction kit (Bioman, Taiwan). The purified DNA was subjected to PCR amplification. The primer sequences were as follows—FoxO1: forward primer 5′-AAGACATCTGGCGGAAACC-3′, reverse primer 5′-ACAATTGGTCGCTAACCGAG-3′ (− 299 to + 7); proximal NF-κB: forward primer 5′-GGCAAAGACTGCGAAGAAGA-3′, reverse primer 5′-AAAATCGGAAACCCAGGAAG-3′; and distal NF-κB: forward primer 5′-CCTCGACCCTCTAAAGACGTA-3′, reverse primer 5′-AGCCAGTTCTGGACTGATCG-3′, which were specifically designed from the COX-2 promoter region (− 320 to − 171) and (− 499 to − 335), respectively. PCR fragments were analyzed on 3% agarose in 1× TAE gel containing ethidium bromide. In addition, qPCR was performed with KAPA SYBR FAST qPCR Kit Master Mix Universal (KAPA Biosystems, Wilmington, MA, USA) on a StepOnePlus™ real-time PCR system.
Isolation of subcellular fractions
Human cardiac fibroblasts were seeded in 10-cm dishes, reaching 90% confluence, transferred to serum-free DMEM/F-12 medium for 24 h, and then incubated with LysoPC for the indicated time durations. The subcellular fractions were prepared by using a NE-PER nuclear and cytoplasmic extraction kit according to the instructions of the manufacturer (Thermo Scientific, Rockford, IL, USA). Cells were gently scraped and then centrifuged at 2000×g for 12 min at 4 °C. The pellet was suspended in 300 μl CRE I, and then vortexed for 15 s. After incubation for 10 min, 25 μl CRE II was added to the sample, sonicated for 10 s at level of amplitude 10 (Misonix Sonicator S-4000), and then incubated for 30 min. The samples were centrifuged at 16,000×g for 5 min. The supernatants (cytoplasmic extracts) were transferred to new tubes. The pellets (nuclear extracts) were lysed with 150 μl NER and sonicated for 10 s at level of amplitude 10. All the reagents contained protease inhibitor cocktails (1 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) and the procedures were carried out at 4 °C. Protein concentration was determined by a BCA assay, and 20 μg of protein from each sample was analyzed by western blot analysis.
Co‐immunoprecipitation assay
Human cardiac fibroblasts were seeded in 10-cm dishes, reaching 90% confluence, transferred to serum-free DMEM/F-12 medium for 24 h, and then incubated with LysoPC for the indicated time durations. The nuclear extracts were isolated and lysed in a RIPA buffer (20 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% NP40, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM PMSF, pH 8.0). The supernatants containing 1 mg of protein were incubated without (input) or with anti-FoxO1 or anti-p65 antibody under gentle rocking at 4 °C for 2 h, and after addition of 50 μl of 50% protein A‐agarose beads tumbled overnight at 4 °C. The agarose beads were extensively washed with a lysis buffer without Triton X‐100 and NP40. The samples were added to 2× western blot loading buffer and the western blot analysis was performed.
Data and statistical analysis
All the data were estimated using a GraphPad Prizm Program (GraphPad, San Diego, CA, USA). Quantitative data were expressed as the mean ± SEM of at least three individual experiments (n ≥ 3), and analyzed with a one-way ANOVA followed by Tukey’s post hoc test at a *p < 0.01 or #p < 0.05 level of significance. Error bars were omitted when they fell within the dimensions of the symbols.
Results
LysoPC enhances COX-2 expression and promoter activity
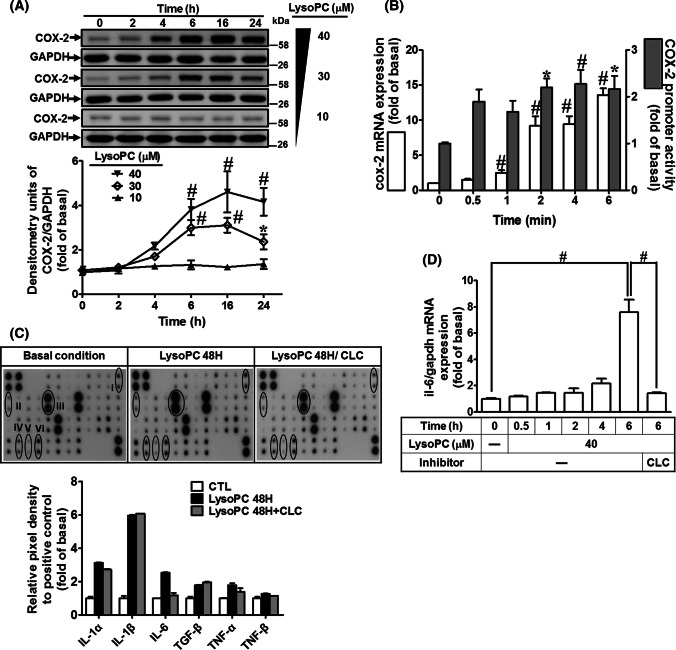
To determine the effect of LysoPC on COX-2 expression, as shown in Fig. 1a, LysoPC-induced COX-2 protein expression in a time- and concentration-dependent manner was studied and a maximal response was observed within 16 h. Next, the results of RT/qPCR analysis showed that LysoPC induced an approximate 13.6-fold increase in cox-2 mRNA expression within 6 h (Fig. 1b). To explore the LysoPC-regulated COX-2 transcription, LysoPC induction was studied and an approximate 2.3-fold increase in COX-2 promoter activity was observed within 4 h (Fig. 1b). These data suggested that LysoPC regulates COX-2 expression at the transcriptional level in HCFs.
Fig. 1.
LysoPC induces COX-2-mediated IL-6 expression. a HCFs were treated with various concentrations of LysoPC for the indicated time intervals. The levels of COX-2 and GAPDH proteins were determined by western blot (n = 6). b HCFs were incubated with LysoPC (40 μM) for the indicated time points. The levels of COX-2 and GAPDH mRNA were determined by RT/qPCR (open bars; n = 7). Cells were co-transfected with a pGL3b-COX-2-luc and pCMV-β-gal plasmids with LysoPC (40 μM) for the indicated time intervals (black bars; n = 5). c HCFs were pretreated with celecoxib (10 μM) for 1 h and then incubated with or without LysoPC (40 μM) for 48 h. The conditioned media were analyzed by cytokine antibody arrays. (1) IL-1α, (2) IL-1β, (3) IL-6, (4) TGF-β, (5) TNF-α, (6) TNF-β. d HCFs were incubated with LysoPC for the indicated time intervals in the absence or presence of celecoxib (10 μM). The levels of IL-6 and GAPDH mRNA expression were determined by RT/qPCR (n = 6). Data are presented with mean ± SEM, and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
LysoPC induces COX-2-mediated IL-6 expression
The expression of potential fibrotic genes in HCFs induced by LysoPC was determined by cDNA microarray analysis. The data showed that the pro-fibrotic cytokine genes (il1a, il1b, il6, il33, and TNF-α) and PTGS2 (encoding COX-2 protein) were upregulated in HCFs treated with LysoPC (Supplementary Table 4). To confirm the effects of LysoPC on the profile of cytokines, the conditioned media were subjected to a human inflammatory cytokine array. The levels of IL-1α, IL-β, and IL-6 were predominately increased in the LysoPC-treated HCFs (Fig. 1c). Our previous study demonstrated that sphingosine-1-phosphate induces COX-2 expression, contributing to IL-6 production via a PGE2/EP autocrine pathway [35]. Thus, we determined whether LysoPC-induced IL-6 secretion shared a similar mechanism in HCFs. We found that pretreatment with celecoxib (a selective COX-2 inhibitor) attenuated LysoPC-induced IL-6 secretion (Fig. 1c) and mRNA expression (Fig. 1d). These results suggested that LysoPC-induced IL-6 secretion is mediated via a COX-2-dependent mechanism in HCFs.
NADPH oxidase/ROS is involved in LysoPC-induced COX-2 expression
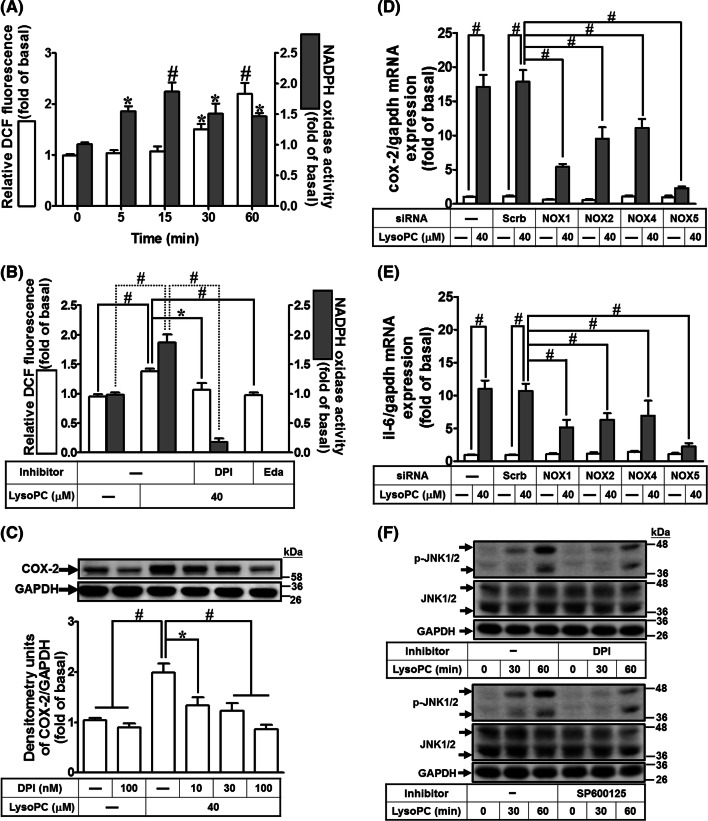
Previous studies have demonstrated that LysoPC increased intracellular ROS production in various cell types [5, 10, 15]. Thus, we explored whether LysoPC induced intracellular ROS generation in HCFs. Our results showed that the levels of ROS generation in cells induced by LysoPC were elevated in a time-dependent manner ranging from 30 to 60 min (Fig. 2a). We further determined whether the ROS generation was due to promotion of NOX activity by LysoPC. As shown in Fig. 2a, LysoPC induced NOX activity in a time-dependent manner with a maximal response within 5–60 min. Moreover, pretreatment with either edaravone (a scavenger of ROS) or DPI (a NOX inhibitor) inhibited the LysoPC-induced ROS generation as well as NOX activation (Fig. 2b), suggesting that LysoPC primes NOX activation leading to ROS generation. The family of NOX comprises seven members: NOX1–5 and DUOX1–2, although, DUOX1/2 is not expressed in the cardiovascular system [36]. Previous studies have demonstrated that ROS are involved in COX-2 expression via NOX2 activation [20]. Our results also demonstrated that pretreatment with DPI attenuated LysoPC-induced COX-2 protein expression in a concentration-dependent manner (Fig. 2c). Moreover, we identified the expression of NOX isoforms in HCFs using RT/PCR. The isoforms of NOX that expressed in HCFs were NOX1, 2, 4, and 5 (Supplementary Fig. 1A). To ensure the roles of NOX isoforms in COX-2 expression, specific NOX siRNAs were used to knock down their respective mRNA targets (Supplementary Fig. 1B–E). Transfection with NOX1, NOX2, NOX4, or NOX5 reduced LysoPC-induced COX-2 expression in HCFs (Fig. 2d). Although NOX/ROS-dependent IL-6 expression has been confirmed in human vascular smooth muscle cells [25], the role of NOX/ROS in COX-2-mediated IL-6 expression was not well defined for HCFs. RT/qPCR analysis showed that IL-6 mRNA expression was downregulated after transfection with NOX1, NOX2, NOX4, or NOX5 siRNA (Fig. 2e). These results suggested that LysoPC-induced IL-6 expression is mediated via a NOX/ROS-dependent cascade in HCFs.
Fig. 2.
NADPH oxidase/ROS are involved in LysoPC-induced COX-2 and IL-6 expression. a HCFs were treated with LysoPC (40 μM) for the indicated time intervals. The generation of ROS and activity of NADPH oxidase were determined using a CM-H2DCFDA probe (open bars; n = 10) and lucigenin chemiluminescence analysis (black bars; n = 10), respectively. b HCFs were pretreated with either DPI (100 nM) or edaravone (Eda; 100 nM) for 1 h and then incubated with LysoPC for 30 min. The cell lysates were subjected to determine the ROS generation (open bars; n = 5) and NADPH oxidase activity (black bars; n = 7). c HCFs were pretreated with DPI for 1 h and then incubated with LysoPC for 6 h. The levels of COX-2 and GAPDH protein were determined by western blot (n = 6). d, e HCFs were transfected with siRNA of scramble, NOX1, NOX2, NOX4, or NOX5 and then incubated with LysoPC for 6 h. The levels of COX-2, IL-6, and GAPDH mRNA were determined by RT/qPCR (d, n = 5; e, n = 5). f HCFs were pretreated with either DPI (100 nM) or SP600125 (1 μM) for 1 h and then treated with LysoPC for the indicated time intervals. The levels of JNK1/2, phospho-JNK1/2, and GAPDH protein were determined by western blot (n = 5). The densitometry measurements are presented in Supplementary Fig. 2B and C. Data are presented with mean ± SEM and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
LysoPC has been shown to stimulate phosphorylation of MAPKs, including JNK1/2 [10, 37]. Therefore, we tested the role of JNK1/2 in COX-2 expression using the inhibitor of JNK1/2 (SP600125) which attenuated LysoPC-induced COX-2 expression in a concentration-dependent manner (Supplementary Fig. 2A). We further examined whether NOX/ROS played a role in LysoPC-mediated JNK1/2 activation. The results showed that pretreatment with either DPI or SP600125 attenuated JNK1/2 phosphorylation stimulated by LysoPC (Fig. 2f; Supplementary Fig. 2B and C), suggesting that LysoPC-induced COX-2 expression is mediated via activation of NOX/ROS-dependent JNK1/2 pathway in HCFs.
NF-κB is involved in LysoPC-mediated COX-2 and IL-6 expression
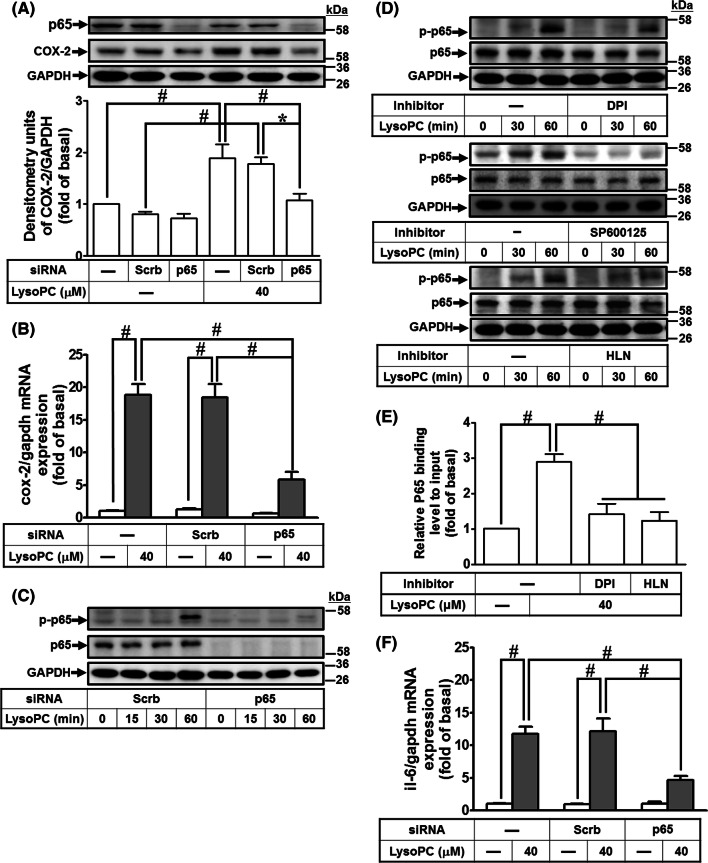
Previous studies have demonstrated that activation of NF-κB leads to expression of pro-inflammatory cytokines and COX-2 induced by LysoPC [23, 38]. To investigate the role of NF-κB in LysoPC-mediated COX-2 expression, the HCFs were transfected with p65 siRNA, which knocked down p65 protein and attenuated LysoPC-induced COX-2 protein and mRNA expression (Fig. 3a, b). In addition, LysoPC-stimulated p65 phosphorylation, with a maximal response at 60 min, was reduced after p65 siRNA transfection (Fig. 3c; Supplementary Fig. 3A). While investigating NOX/ROS-mediated p65 activation, our data indicated that LysoPC-stimulated p65 phosphorylation was attenuated by DPI, SP600125, or helenalin (an inhibitor of NF-κB) (Fig. 3d; Supplementary Fig. 3B–D), suggesting that LysoPC-induced COX-2 expression is mediated via NOX/ROS-JNK1/2-dependent NF-κB p65 activation in HCFs.
Fig. 3.
NF-κB is involved in LysoPC-induced COX-2 and IL-6 expression. a, b HCFs were transfected with siRNA of scramble or p65 and then treated with LysoPC for 6 h. a The levels of p65, COX-2, and GAPDH protein were determined by western blot (n = 6). b The levels of COX-2 and GAPDH mRNA were determined by RT/qPCR (n = 6). c HCFs were transfected with siRNA of scramble or p65, and then treated with LysoPC for the indicated time intervals. The levels of p65, phospho-p65, and GAPDH protein were determined by western blot (n = 6). The densitometry measurements of phospho-p65 are presented in Supplementary Fig. 3A. d HCFs were pretreated with DPI (100 nM; n = 7), SP600125 (1 μM; n = 5), or helenalin (HLN, 1 μM; n = 7) for 1 h, and then treated with LysoPC for the indicated time interval. The levels of p65, phospho-p65, and GAPDH protein were determined by western blot. The densitometry measurements of phospho-p65 are presented in Supplementary Fig. 3B–D. e HCFs were pretreated with DPI (100 nM) or helenalin (1 μM) for 1 h, and then incubated with LysoPC for 1 h. The DNA binding activity of NF-κB was determined by a ChIP assay. Quantification of p65 immunoprecipitated DNA was performed by an SYBR system for qPCR, and the results are shown as the fold change normalized to input control (n = 4). f HCFs were transfected with siRNA of scramble or p65, and then treated with LysoPC for 6 h. The levels of IL-6 and GAPDH mRNA were determined by RT/qPCR (n = 6). Data are presented as mean ± SEM, and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
COX-2 contains two NF-κB binding elements in its promoter regions, specifically at − 449 to − 437 and − 225 to − 214 [39]. Thus, we determined which NF-κB binding elements of the COX-2 promoter was manipulated by LysoPC, which led to COX-2 expression. The results of ChIP revealed that the binding activity of p65 was increased after flanking of proximal NF-κB elements using an anti-p65 or anti-phospho-p65 antibody (Supplementary Fig. 4B and C). Moreover, we investigated the interaction between activated NF-κB and COX-2 promoter stimulated by LysoPC via NOX/ROS pathways. Our results showed that LysoPC-stimulated binding of p65 to the proximal NF-κB-binding element on COX-2 promoters was attenuated by either DPI or helenalin (Fig. 3e). These results suggested that NOX/ROS-stimulated phosphorylated p65 binds to a proximal NF-κB-binding element, leading to COX-2 expression in HCFs. While determining whether LysoPC-activated p65 affected IL-6 expression in HCFs, we found that transfection with p65 siRNA knocked down p65 protein expression and markedly suppressed LysoPC-induced IL-6 gene expression (Fig. 3f). Together, these results suggested that LysoPC-primed NOX/ROS signaling regulates NF-κB activation, leading to COX-2-dependent IL-6 expression in HCFs.
FoxO1 is involved in LysoPC-mediated COX-2 and IL-6 expression
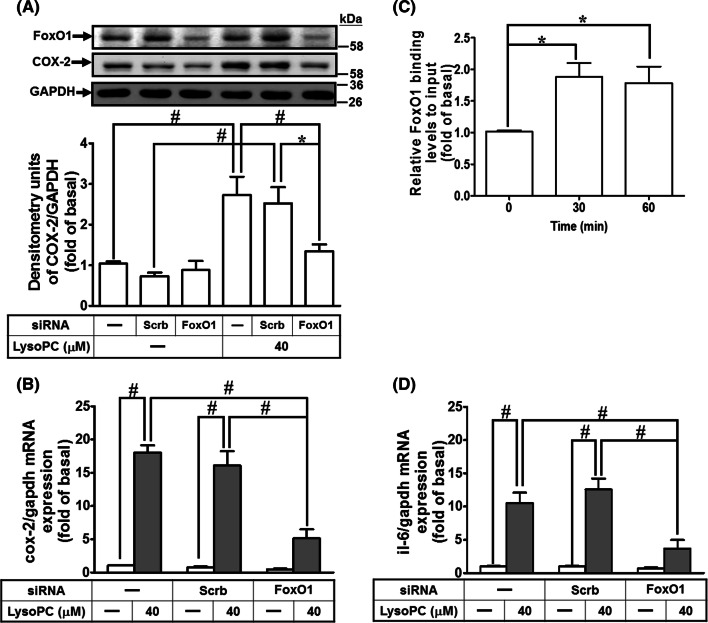
While determining the role of FoxO1 in the LysoPC-induced COX-2 expression, we found that knockdown of FoxO1 protein after transfection with FoxO1 siRNA reduced LysoPC-induced COX-2 protein and mRNA expression in HCFs (Fig. 4a, b). Next, we determined whether FoxO1 bound to COX-2 promoter in HCFs treated with LysoPC. ChIP analysis showed that LysoPC stimulated the binding of FoxO1 to the COX-2 promoter in a time-dependent manner, with a maximal response within 30–60 min (Fig. 4c). Furthermore, transfection with FoxO1 siRNA attenuated the LysoPC-induced IL-6 mRNA expression (Fig. 4d). These results suggested that LysoPC-mediated FoxO1-dependent COX-2 induction leads to IL-6 expression in HCFs.
Fig. 4.
FoxO1 is involved in LysoPC-mediated COX-2 expression and IL-6 expression. a, b HCFs were transfected with siRNA of scramble or FoxO1 and then treated with LysoPC for 6 h. a The levels of COX-2, FoxO1 and GAPDH protein were determined by western blot (n = 6). b The levels of COX-2 and GAPDH mRNA expression were determined by RT/qPCR (n = 7). c HCFs were treated with LysoPC for the indicated time intervals. DNA binding activity of FoxO1 was determined by a ChIP assay. Quantification of data was performed by an SYBR system for qPCR, and the results are shown as the fold change normalized to input control (n = 5). d HCFs were transfected with siRNA of scramble or FoxO1 and then treated with LysoPC for 6 h. The levels of IL-6 and GAPDH mRNA were determined by RT/qPCR (n = 6). Data are presented as mean ± SEM, and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
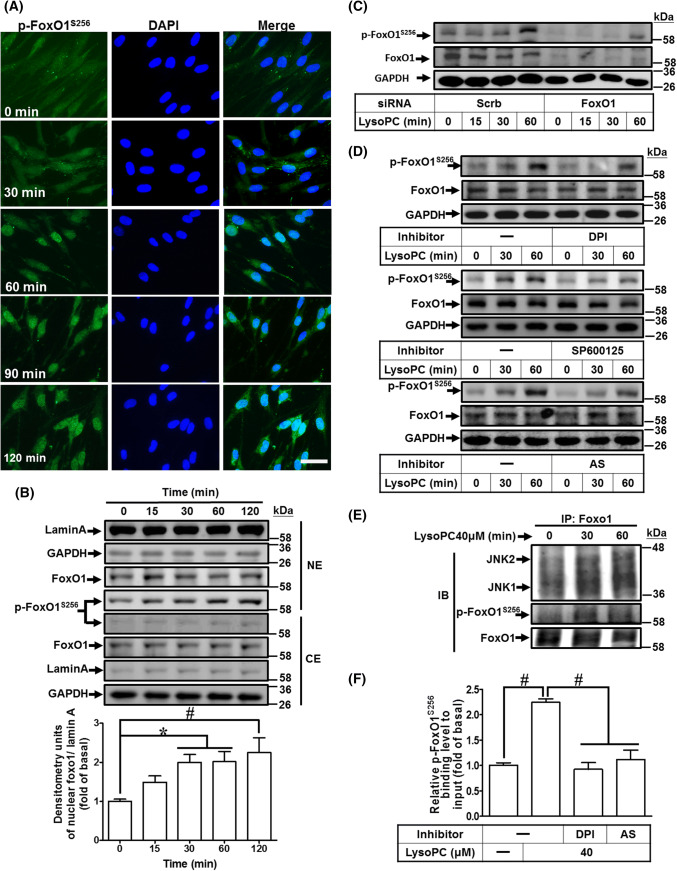
LysoPC-mediated phosphorylation of FoxO1 at Ser256 enhances nuclear localization and binding activity with COX-2 promoter
Phosphorylation at Ser256 of human FoxO1 results in differential transcriptional activities [40], including, increase in transcriptional activity of nuclear FoxO1, leading to expression of pro-inflammatory genes [31, 32, 41]. While investigating whether LysoPC stimulated phosphorylation at Ser256 of FoxO1, the immunofluorescence images showed that LysoPC induced FoxO1 phosphorylation at Ser256 residue and its nuclear accumulation (Fig. 5a). The nuclear accumulation of phosphorylated FoxO1S256 was also confirmed by western blot (Fig. 5b), and was attenuated after transfection with FoxO1 siRNA (Fig. 5c; Supplementary Fig. 5A). Indeed, nuclear accumulation of FoxO1 increases transcriptional activity in response to oxidative stresses [42]. Thus, we investigated whether LysoPC-stimulated phosphorylation of FoxO1S256 was mediated through NOX/ROS signaling. LysoPC-stimulated phosphorylation of FoxO1S256 was attenuated by DPI, SP600125, or AS1842856 (an inhibitor of FoxO1) (Fig. 5d; Supplementary Fig. 5B–D). To further validate the binding between JNK1/2 and FoxO1, the cell lysates were immunoprecipitated with an anti-FoxO1 antibody after LysoPC stimulation and then analyzed by western blot using an antibody as indicated. The results showed that the levels of JNK1/2 and phosphorylation of FoxO1S256 were increased after treatment with LysoPC within 30–60 min (Fig. 5e), suggesting that phosphorylation of FoxO1 is regulated by a NOX/ROS-JNK1/2 pathway. Moreover, we investigated whether phosphorylation of FoxO1S256 was associated with LysoPC-induced COX-2 gene expression. The interaction between FoxO1 and COX-2 promoter was determined by a ChIP analysis. As shown in Fig. 5f, LysoPC-induced phosphorylation of FoxO1S256 enhanced the binding activity of FoxO1 with COX-2 promoter, which was attenuated by either DPI or AS1842856. These results suggested that LysoPC-stimulated phosphorylated FoxO1S256 interacts with COX-2 promoter, contributing to increase in the transcriptional activity via NOX/ROS-JNK1/2 pathways in HCFs.
Fig. 5.
LysoPC-mediated phosphorylation at Ser256 of FoxO1 increases nuclear localization and DNA binding activity on COX-2 promoter. a, b HCFs were incubated with LysoPC (40 μM) for the indicated time points. a Immunofluorescence staining was performed with an anti-phospho-FoxO1S256 antibody, labeled with FITC (green) and DAPI (blue), and observed by using a fluorescence microscope (scale bar, 100 μm). b The cytosolic and nuclear fractions were prepared and subjected to western blot analysis. Lamin A and GAPDH were used as a marker protein for the nuclear and cytosolic fractions, respectively. Quantification data of nuclear phospho-FoxO1S256 data is presented in the bottom panel (n = 6). c HCFs were transfected with siRNA of scramble or FoxO1 and then treated with LysoPC for the indicated time intervals. The levels of FoxO1, phospho-FoxO1S256, and GAPDH protein were determined by western blot (n = 7). The densitometry measurements of phospho-FoxO1S256 are presented in Supplementary Fig. 5A. d HCFs were pretreated with DPI (100 nM; n = 7), SP600125 (1 μM; n = 5), or AS1842856 (AS, 100 nM; n = 7) for 1 h, and then treated with LysoPC for the indicated time intervals. The levels of phospho-FoxO1S256, FoxO1, and GAPDH protein were determined by western blot. The densitometry measurements of phospho-FoxO1S256 are presented in Supplementary Fig. 5B–D. e HCFs were treated with LysoPC for the indicated time intervals and subjected to immunoprecipitation assay using an anti-FoxO1 antibody. The immunoprecipitates were analyzed by western blot using an anti-JNK1/2, anti-phospho-FoxO1S256, or anti-FoxO1 (as an internal control) antibody. Data are representative of three independent experiments (n = 3). f HCFs were pretreated with DPI (100 nM) or AS1842856 (100 nM) for 1 h and then incubated with LysoPC for 1 h. The DNA binding activity of phospho-FoxO1S256 was determined by a ChIP assay. Quantification of data was performed by an SYBR system for qPCR, and the results are shown as the fold change normalized to input control (n = 5). Data are presented as mean ± SEM, and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01. NE nuclear extract, CE cytosolic extract
FoxO1 coordinates with NF-κB in regulating COX-2 promoter activity
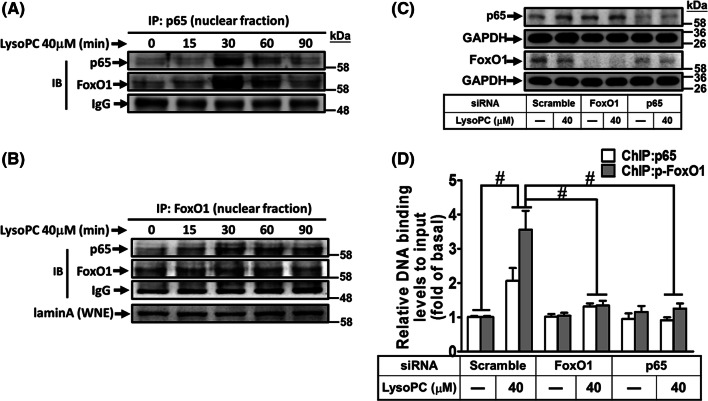
While investigating the association of FoxO1 with p65 in response to LysoPC in HCFs, the results of co-immunoprecipitation showed that LysoPC enhanced p65 translocation and its interaction with FoxO1 (Fig. 6a). Similarly, the nuclear FoxO1 enhanced p65 activity upon LysoPC stimulation in a time-dependent manner (Fig. 6b). Next, we examined whether activation of FoxO1 enhanced p65 binding to COX-2 promoters. The ChIP results showed that knockdown of either FoxO1 or p65 expression by FoxO1 or p65 siRNA, respectively (Fig. 6c), attenuated immunoprecipitated DNA with an anti-p65 or anti-phospho-FoxO1S256 antibody (Fig. 6d). These findings suggested that LysoPC-induced FoxO1 interacts with p65 and coordinately enhances its binding to COX-2 promoters, leading to COX-2 gene expression in HCFs.
Fig. 6.
FoxO1 coordinates with NF-κB in regulating COX-2 promoter activity. a, b HCFs were treated with LysoPC for the indicated time intervals, and then the nuclear fraction was prepared. Nuclear fractions were subjected to immunoprecipitation assay using the indicated antibodies. Immunoprecipitates were analyzed by western blot using an anti-p65, anti-FoxO1, or anti-laminA (as an internal control) antibody. IP immunoprecipitation, IB immunoblotting, WNE whole nuclear extract. Data are representative of three independent experiments. c, d HCFs were transfected with siRNA of scramble, FoxO1, or p65 and then treated with LysoPC for 1 h. The cells lysates were subjected to western blot analysis (c) and a ChIP analysis (d). The DNA binding activity of FoxO1 and NF-κB was determined by a ChIP assay, using an anti-phospho-FoxO1S256 or anti-p65 antibody. Quantification of data is shown as fold change after normalizing to input control (n = 5). Data are presented as mean ± SEM and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
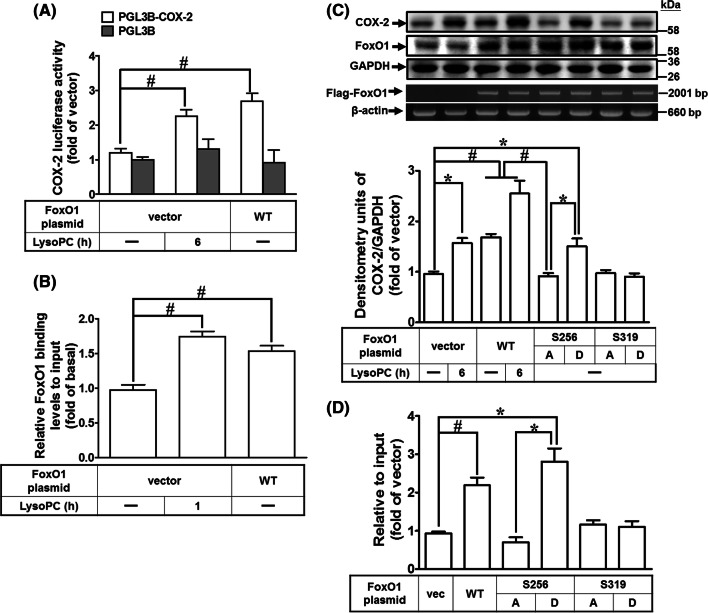
Overexpression of wild-type or S256D FoxO1 enhances COX-2 promoter activity
To corroborate our findings above, we explored the effects of overexpression of FoxO1 on COX-2 expression. Exposure of HCFs to LysoPC induced approximately 2.3-fold increase in the COX-2 promoter activity (Fig. 7a). Co-transfection of pCMV Tag-2B FoxO1 (wild-type) further increased the COX-2 promoter activity by approximately 2.7-fold as compared to that of empty vector alone (Fig. 7a). The ChIP results showed that overexpression of FoxO1 or treatment with LysoPC significantly increased the binding activity of FoxO1 on COX-2 promoters (Fig. 7b). FoxO1 is also phosphorylated by Akt at Tyr24, Ser256, or Ser319, while post-transcriptional modification independently regulates FoxO1 transcriptional activity [27, 40, 43]. To directly assess the impact of FoxO1 phosphorylation on COX-2 expression, we modified the Flag-tagged FoxO1 to encode phospho-silencing mutants (S256A and S319A FoxO1) and phospho-mimic mutants (S256D and S319D FoxO1). Overexpression of wild-type FoxO1 and S256D FoxO1 was associated with a 1.6- and 1.7-fold increase in COX-2 expression, respectively (Fig. 7c). The levels of chimeric Flag-FoxO1 were expressed as the levels of mRNA, determined by RT/PCR analysis. However, overexpression of S256D FoxO1 significantly enhanced COX-2 expression as compared with that of S256A FoxO1. In contrast, neither S319A nor S319D FoxO1 mutant affected COX-2 expression. Immunoprecipitation of Flag was used to determine the DNA binding activity of FoxO1 mutants on COX-2 promoter. Consistently, ChIP analysis demonstrated that the interaction of FoxO1 with the COX-2 promoter was enhanced in HCFs overexpressing either wild-type FoxO1 or S256D FoxO1 (Fig. 7d). These results suggested that phosphorylation at Ser256 residue of FoxO1 is sufficient to upregulate COX-2 gene expression in HCFs.
Fig. 7.
Overexpression of wild-type FoxO1 and S256D FoxO1 enhances DNA binding activity on COX-2 promoter. a HCFs were co-transfected with pGL3b, pGL3b-cox-2-luc, pCMV-β-gal, pCMV-Tag2B, or pCMV-Tag2B-FoxO1 (WT) plasmid for 48 h and treated with LysoPC (40 μM) for 6 h. The luciferase activity was determined and normalized to the respective β-gal activity (n = 4). b HCFs were transfected with pCMV-Tag2B or pCMV-Tag2B-FoxO1 (WT) for 48 h and then treated with LysoPC (40 μM) for 1 h. Quantification of data were determined by an SYBR system for qPCR, and the results are shown as the fold change normalized to input control (n = 5). c, d HCFs were transfected with pCMV-Tag2B-FoxO1, S256A FoxO1, S256D FoxO1, S319A FoxO1 or S319D FoxO1 for 48 h and then treated with LysoPC (40 μM) for 6 h. c The levels of COX-2, FoxO1 and GAPDH protein were analyzed by western blot. Expressions of chimeric Flag-FoxO1 and β-actin were determined by RT/PCR. Quantification of data is shown in the bottom panel (n = 7). d DNA binding activity of FoxO1 constructs on COX-2 promoter was determined by a ChIP assay using an anti-Flag M2 antibody and analyzed by an SYBR system for qPCR (n = 7). Data are presented as mean ± SEM and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
LysoPC induces COX-2 and IL-6 expression in ex vivo mouse heart tissue
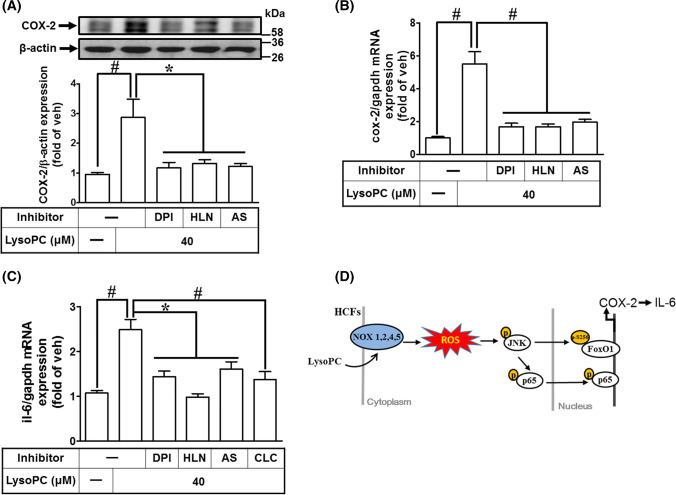
We investigated whether LysoPC induced COX-2 expression in ex vivo mouse heart tissues. LysoPC-induced COX-2 protein and mRNA expressions were reduced by DPI, helenalin, or AS1842856 (Fig. 8a, b). Moreover, our results demonstrated that LysoPC-induced IL-6 expression was inhibited by DPI, helenalin, AS1842856, or celecoxib, as determined by RT/qPCR analysis (Fig. 8c). These results suggested that induction of COX-2 by LysoPC is associated with IL-6 expression in ex vivo mouse heart tissues and is mediated through NOX/ROS, FoxO1, and NF-κB in the heart.
Fig. 8.
LysoPC induces COX-2 and IL-6 expression in ex vivo mouse heart tissue. a–c Mouse heart segments were pretreated with DPI (100 nM), AS1842856 (100 nM), helenalin (1 μM), or celecoxib (10 μM) for 1 h and then exposed to LysoPC (40 μM) for 6 h while in Krebs solution bubbled with 95% O2 and 5% CO2 at 37 °C. The heart tissues were homogenized and subjected to western blot analysis or RT/qPCR to determine the levels of COX-2 protein (a; n = 6) and mRNA (b; n = 7) or the levels of IL-6 mRNA (c; n = 7). d Schematic diagram illustrates that LysoPC activated FoxO1 and NF-κB mediated a COX-2-dependent IL-6 expression via NOX/ROS-JNK1/2-dependent pathway in HCFs. Data are presented as mean ± SEM and analyzed by one-way ANOVA with Tukey’s post hoc tests. *p < 0.05; #p < 0.01
Discussion
LysoPC induced COX-2-dependent IL-6 expression in HCFs, which may be associated with pro-fibrotic responses. Our findings showed that LysoPC-induced COX-2-dependent IL-6 expression is mediated through NOX/ROS/JNK1/2 leading to FoxO1S256 and NF-κB activation (Fig. 8d). These results were confirmed by ex vivo studies, indicating the involvement of NOX, NF-κB, or FoxO1 in LysoPC-mediated responses. We found that nuclear accumulation of phospho-FoxO1S256 was significantly increased and it was bound to the COX-2 promoter. In contrast with previous reports, the phosphorylation of FoxO1 at Ser256 strongly promoted nuclear exclusion [43]. Moreover, overexpression of S256D FoxO1, but not of S256A FoxO1, promoted DNA binding activity and turned on COX-2 expression in HCFs. These data suggested that interaction between FoxO1 and p65 may coordinately regulate COX-2 gene expression in HCFs. Our findings provided new insights into the mechanisms underlying LysoPC-induced COX-2-dependent IL-6 expression through phosphorylated p65 and FoxO1S256-mediated transcriptional activation in HCFs.
Induction of COX-2 could enhance macrophage infiltration and fibroblast activation during myocardial infarction in patients and experimental animals [21, 22]. COX-2 catalyzes the conversion of arachidonic acid into PGs, which activate their specific receptors. For example, PGE2 enhances IL-6 production through EP1, EP2, or EP4 in various cell types [44, 45]. Our previous study also demonstrated that S1P induces COX-2 expression associated with PGE2/IL-6 production in human tracheal smooth muscle cells [25, 35]. In this study, we found that inhibition of COX-2 activity attenuated LysoPC-induced IL-6 expression in HCFs and ex vivo mouse heart tissues. IL-6 has been shown to either enhance collagen synthesis leading to cardiac fibrosis via activation of CFs [13] or induce TGF-β expression leading to tissue remodeling via TGF-β/smad3 activation [12, 14]. Therefore, we speculated that activation of the COX-2/PG axis promotes IL-6 production and further triggers pro-fibrotic responses in CFs, which is an important issue to be considered for future studies.
LysoPC stimulates NOX-derived ROS generation in various types of cells [15, 16]. Consistent with these findings, LysoPC stimulated NOX activation and increased ROS generation in HCFs. ROS are believed to activate various signaling molecules, leading to COX-2 expression [20, 25]. Our findings confirmed that LysoPC-induced COX-2 expression was dependent on NOX/ROS generation. NOX1, 2, 4, and 5 were involved in LysoPC-mediated responses as confirmed after transfection of cells with their respective siRNAs. In mouse heart, knockout of either NOX2 or NOX4 reduces ROS production, hypertrophy, interstitial fibrosis, and apoptosis [18, 19]. Moreover, JNK1/2 can be activated by ROS signaling [46]. Our results supported the NOX-derived ROS generation mediated JNK1/2 activation, contributing to LysoPC-induced COX-2 expression in HCFs.
NF-κB is a key player in the regulation of COX-2 transcription [39]. LysoPC has been shown to mediate phosphorylation and degradation of IκB, resulting in the nuclear translocation of NF-κB p65 in endothelial cells [38]. LysoPC-activated NF-κB is also involved in the expression of endothelial adhesion molecules and COX-2 expression [5, 23]. Consistent with these findings, our results supported that LysoPC-induced COX-2 and IL-6 expressions were mediated through NF-κB activation. κB sites are highly conserved among species [39]. The human COX-2 promoter contains two NF-κB binding elements, which are different from those of rodents [39]. The rodent COX-2 promoter lacks the proximal NF-κB binding element, whereas the proximal NF-κB-binding element of the human COX-2 promoter is a major regulatory element, contributing to COX-2 promoter activation upon treatment with LysoPC in HCFs. Nguyen et al. mentioned that the COX-2 promoter is differentially regulated between mice and humans [39]. We found that LysoPC regulated COX-2-dependent IL-6 expression via the proximal NF-κB-binding element in the COX-2 promoter. Redox signaling has been shown to activate NF-κB through various signaling pathways and thereby enhance the expression of its targeted proteins [5]. Our previous results demonstrated that ATPγS-induced COX-2 expression is mediated via NOX/ROS generation-dependent NF-κB activation in human pulmonary alveolar epithelial cells [20]. Here, we demonstrated that activated p65 interacted with the COX-2 promoter and initiated its gene transcription, which was inhibited by DPI, or helenalin in HCFs. In addition, increase in NF-κB activity is mediated via JNK1/2-dependent phosphorylation at Ser176 of IKKα, contributing to IκB degradation and subsequent NF-κB activation [47]. Consistent with previous reports, inhibition of JNK1/2 by SP600125 attenuated LysoPC-stimulated p65 phosphorylation. Thus, we concluded that NOX-derived ROS resulted in an increase of NF-κB activity and upregulation of COX-2 and IL-6 gene expression via a NOX/ROS-JNK1/2-dependent pathway in HCFs.
FoxO1 is a multifunctional transcription factor [24, 25, 31, 32]. Knockdown of FoxO1 attenuates the IL-1β-induced COX-2 expression in primary human myometrial cells [24]. In contrast, IL-1β-induced COX-2 expression is not altered by knockdown of either FoxO1 or FoxO3 in human chondrocytes [48]. However, regulation of FoxOs’ downstream targets is highly cell lineage dependent in response to oxidative stress [49]. In this study, we found the involvement of FoxO1 in COX-2 expression by transfection of cells with FoxO1 siRNA. The interaction between FoxO1 and COX-2 promoter was confirmed using a ChIP assay, demonstrating that induction of COX-2 by LysoPC was mediated via an increase in the binding activity of FoxO1 on the COX-2 promoters. In particular, overexpression of FoxO1 enhanced either COX-2 promoter activity or FoxO1 DNA association. We found that the FoxO1 binding element of the COX-2 promoter was located between positions − 299 and + 7. FoxO1 has been shown to be involved in IL-6 expression induced by IL-1β, LPS, and TNF-α [24, 31, 32]. In this study, knockdown of FoxO1 attenuated LysoPC-induced COX-2-dependent IL-6 expression in HCFs. These results suggested that FoxO1 is involved in LysoPC-induced COX-2-dependent IL-6 expression associated with cardiac inflammation.
On the basis of previous studies, cooperative regulation of inflammatory genes by NF-κB and FoxO1 is controversial. Several genes such as the IL-1β promoter contain both FoxO1 and NF-κB response elements [31, 50]. Fan et al. mentioned that NF-κB potentially interacted with FoxO1 because response motifs of NF-κB were significantly enriched across the FoxO1 cistrome [31]. For the chemotactic ligand CCL20, overexpression of FoxO1 increased the binding of the active NF-κB dimer, whereas FoxO1 silencing decreased NF-κB binding to its response element. Since FoxO1 does not directly bind to the CCL20 promoter, FoxO1 may serve as a coactivator of NF-κB in the nucleus to amplify NF-κB signaling [51]. Thus, in some cases such as IL-1β, FoxO1 binds to a response element nearby the NF-κB binding element to enhance transcription, whereas, in CCL20, FoxO1 is speculated to physically interact with NF-κB and enhance NF-κB-mediated CCL20 transcription. Our data revealed that FoxO1 can associate with nuclear p65, a component of NF-κB, and knockdown of FoxO1 attenuated p65 binding to the proximal NF-κB binding element of the COX-2 promoter in HCFs. We speculated that the spatial conformation of proximal NF-κB binding element is close to the FoxO1 binding element, inferring that p65 and FoxO1 coordinately regulated COX-2 gene expression in HCFs.
The transcriptional activity of FoxO1 is dependent on its phosphorylation status and subcellular localization. Akt-mediated FoxO1 activation has been shown to promote nuclear exclusion of FoxO1 and negatively regulate IRS-dependent gene expression in response to IGF signaling [43]. Therefore, nuclear accumulation of FoxO1 increases DNA binding activity and results in CCL20 induction in keratinocytes and mouse models [41]. Kinase-dead mutation of Akt blocks FoxO1 phosphorylation at Tyr24, Ser256, and Ser319, which restricts FoxO1 expression in the cytoplasm [27] and enhances the expression of pro-inflammatory genes including TNF-α, IL-1β, and IL-6 [31, 32]. Our novel findings indicated that LysoPC-stimulated FoxO1S256 phosphorylation increased the nuclear accumulation of FoxO1 and its binding activity on COX-2 promoter. Previous reports have indicated that under oxidative stress, phosphorylated JNK1/2 accelerates FoxO1 nuclear localization, leading to its target gene expression [42]. Here, we demonstrated that blocking of NOXs and JNK1/2 activity attenuated LysoPC-stimulated phosphorylation of FoxO1S256. In addition, our data also demonstrated that overexpression of S256D FoxO1 enhanced COX-2 induction, whereas S256A, S319D, and S319A FoxO failed to alter COX-2 gene expression. These results suggested that phosphorylation of FoxO1 at Ser256 is required for COX-2 expression in HCFs, leading to cardiac inflammation.
With respect to the limitations of the present study, we recognized the inhibition of COX-2 activity as a target may also incur side effects on the cardiovascular system. Increase in COX-2 activation leads to metabolism and production of various prostanoids by cell-dependent isomerases of different tissues, which may also be involved in pathological and physiological regulation. It is crucial to further investigate the modulation of these prostanoids and their receptors in response to LysoPC. Nevertheless, our results provide new insights into COX-2 gene expression, involving a synergistic regulation of FoxO1 and p65 in HCFs. In fact, our previous data demonstrated FoxO1 as a transcription repressor for COX-2 gene expression in human tracheal smooth muscle cells [25]. We suggest that FoxO1-mediated COX-2 regulation is a key mechanism and represents different patterns in various tissues exposed to different stimuli.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 The expression of NOXs isoforms in HCFs (A) Total RNA was extracted from growth-arrested HCFs. The levels of NOX1-5 mRNA were determined by RT/PCR analysis. β-actin was used as an internal control. The star symbol indicates each replicon. (B-E) HCFs were transfected with Scramble, NOX1, NOX2, NOX4 or NOX5 siRNA for 36 h, and then incubated with LysoPC for 6 h. The levels of NOXs mRNA expression were determined by RT/qPCR analysis (n = 4). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 799 kb)
Supplementary Fig. 2 (A) HCFs were pretreated with SP600125 (0.3, 1, 3 μM) for 1 h and then incubated with LysoPC for 6 h. The levels of COX-2 and GAPDH protein were determined by western blot analysis (n = 5). (B-C) Representative data of western blotting are presented in Fig. 2E (DPI: n = 5; SP600125: n = 5). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 436 kb)
Supplementary Fig. 3 (A-D) Representative data of western blotting are presented in Fig. 3C (n = 6) and 3D (DPI: n = 7; SP600125: n = 5; Helenalin: n = 7). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 407 kb)
Supplementary Fig. 4 (A) Schematic diagram of positions of NF-κB binding elements and ChIP primer pairs on COX-2 promoter. (B-C) HCFs were treated with LysoPC for the indicated time intervals. The DNA binding activity of NF-κB was determined by ChIP assays using an anti-p65 or anti-phosph-p65 antibody. The immunoprecipitated DNA and inputs were analyzed by an SYBR system for qPCR using a pair of primers flanking the NF-κB binding site within the human COX-2 promoter (proximal NF-κB: -320 ~ -171; distal NF-κB: -499 ~ -335). Representative gel (upper panels) and quantification of data are shown as the fold change normalized to input control (n = 5; bottom panels). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01 (TIFF 498 kb)
Supplementary Fig. 5 (A-D) Representative data of western blotting are presented in Fig. 5C (n = 7) and 5D (DPI: n = 7; SP600125: n = 5; AS1842856: n = 7). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01 (TIFF 410 kb)
Acknowledgements
We thank Ms. Yu-Wen Chen for her technical assistance. This work was supported by the Ministry of Education, Taiwan, Grant numbers: EMRPD1H032 and EMRPD1H055; the Ministry of Science and Technology, Taiwan, Grant numbers: MOST104-2320-B-182A-003-MY3, MOST105-2320-B-182-005-MY3 MOST107-2320-B-182A-011, and MOST107-2320-B-182-020-MY2; Chang Gung Medical Research Foundation, Taiwan, Grant numbers: CMRPD1F0022, CMRPD1F0023, CMRPD1F0551, CMRPD1F0552, CMRPG3E2232, CMRPG3F1532 CMRPG3F1533, CMRPG3H0061, and CMRPG5F0202.
Abbreviations
- COX-2
Cyclooxygenase-2
- DPI
Diphenyleneiodonium chloride
- DUOX
Dual oxidase
- EP
Prostaglandin E2 receptor
- FoxO
FoxO Forkhead box protein O
- HCFs
Human cardiac fibroblasts
- IL
Interleukin
- LysoPC
Lysophosphatidylcholine
- NOX
NADPH oxidase
- PC
Phosphatidylcholine
- PGE2
Prostaglandin E2
- PLA2
Phospholipase A2
- ROS
Reactive oxygen species
- TGF-β
Transforming growth factor-β
- TNF
Tumor necrosis factor
References
- 1.Metra M, Teerlink JR. Heart failure. Lancet. 2017;390:1981–1995. doi: 10.1016/S0140-6736(17)31071-1. [DOI] [PubMed] [Google Scholar]
- 2.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sziksz E, Pap D, Lippai R, Beres NJ, Fekete A, Szabo AJ, Vannay A. Fibrosis related inflammatory mediators: role of the IL-10 cytokine family. Mediators Inflamm. 2015;2015:764641. doi: 10.1155/2015/764641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belperio J, Horwich T, Abraham WT, Fonarow GC, Gorcsan J, 3rd, Bersohn MM, Singh JP, Sonel A, Lee LY, Halilovic J, Kadish A, Shalaby AA. Inflammatory mediators and clinical outcome in patients with advanced heart failure receiving cardiac resynchronization therapy. Am J Cardiol. 2016;117:617–625. doi: 10.1016/j.amjcard.2015.11.049. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Fang P, Li Y, Kuo YM, Andrews AJ, Nanayakkara G, Johnson C, Fu H, Shan H, Du F, Hoffman NE, Yu D, Eguchi S, Madesh M, Koch WJ, Sun J, Jiang X, Wang H, Yang X. Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler Thromb Vasc Biol. 2016;36:1090–1100. doi: 10.1161/ATVBAHA.115.306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin X, Qiu C, Zhao L. Lysophosphatidylcholine perpetuates macrophage polarization toward classically activated phenotype in inflammation. Cell Immunol. 2014;289:185–190. doi: 10.1016/j.cellimm.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Scholz H, Eder C. Lysophosphatidylcholine activates caspase-1 in microglia via a novel pathway involving two inflammasomes. J Neuroimmunol. 2017;310:107–110. doi: 10.1016/j.jneuroim.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Huang JP, Cheng ML, Wang CH, Shiao MS, Chen JK, Hung LM. High-fructose and high-fat feeding correspondingly lead to the development of lysoPC-associated apoptotic cardiomyopathy and adrenergic signaling-related cardiac hypertrophy. Int J Cardiol. 2016;215:65–76. doi: 10.1016/j.ijcard.2016.03.239. [DOI] [PubMed] [Google Scholar]
- 9.Nam M, Jung Y, Ryu DH, Hwang GS. A metabolomics-driven approach reveals metabolic responses and mechanisms in the rat heart following myocardial infarction. Int J Cardiol. 2017;227:239–246. doi: 10.1016/j.ijcard.2016.11.127. [DOI] [PubMed] [Google Scholar]
- 10.Chen HM, Hsu JH, Liou SF, Chen TJ, Chen LY, Chiu CC, Yeh JL. Baicalein, an active component of Scutellaria baicalensis Georgi, prevents lysophosphatidylcholine-induced cardiac injury by reducing reactive oxygen species production, calcium overload and apoptosis via MAPK pathways. BMC Complement Altern Med. 2014;14:233. doi: 10.1186/1472-6882-14-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncalves I, Edsfeldt A, Ko NY, Grufman H, Berg K, Bjorkbacka H, Nitulescu M, Persson A, Nilsson M, Prehn C, Adamski J, Nilsson J. Evidence supporting a key role of Lp-PLA2-generated lysophosphatidylcholine in human atherosclerotic plaque inflammation. Arterioscler Thromb Vasc Biol. 2012;32:1505–1512. doi: 10.1161/ATVBAHA.112.249854. [DOI] [PubMed] [Google Scholar]
- 12.Ma F, Li Y, Jia L, Han Y, Cheng J, Li H, Qi Y, Du J. Macrophage-stimulated cardiac fibroblast production of IL-6 is essential for TGF beta/Smad activation and cardiac fibrosis induced by angiotensin II. PLoS One. 2012;7:e35144. doi: 10.1371/journal.pone.0035144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melendez GC, McLarty JL, Levick SP, Du Y, Janicki JS, Brower GL. Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension. 2010;56:225–231. doi: 10.1161/HYPERTENSIONAHA.109.148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JH, Zhao L, Pan X, Chen NN, Chen J, Gong QL, Su F, Yan J, Zhang Y, Zhang SH. Hypoxia-stimulated cardiac fibroblast production of IL-6 promotes myocardial fibrosis via the TGF-beta1 signaling pathway. Lab Investig. 2016;96:1035. doi: 10.1038/labinvest.2016.84. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhuri P, Rosenbaum MA, Birnbaumer L, Graham LM. Integration of TRPC6 and NADPH oxidase activation in lysophosphatidylcholine-induced TRPC5 externalization. Am J Physiol Cell Physiol. 2017;313:C541–C555. doi: 10.1152/ajpcell.00028.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelher MR, McLaughlin NJ, Banerjee A, Elzi DJ, Gamboni F, Khan SY, Meng X, Mitra S, Silliman CC. LysoPCs induce Hck- and PKCdelta-mediated activation of PKCgamma causing p47phox phosphorylation and membrane translocation in neutrophils. J Leukoc Biol. 2017;101:261–273. doi: 10.1189/jlb.3A0813-420RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teixeira G, Szyndralewiez C, Molango S, Carnesecchi S, Heitz F, Wiesel P, Wood JM. Therapeutic potential of NADPH oxidase 1/4 inhibitors. Br J Pharmacol. 2017;174:1647–1669. doi: 10.1111/bph.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao QD, Viswanadhapalli S, Williams P, Shi Q, Tan C, Yi X, Bhandari B, Abboud HE. NADPH oxidase 4 induces cardiac fibrosis and hypertrophy through activating Akt/mTOR and NFkappaB signaling pathways. Circulation. 2015;131:643–655. doi: 10.1161/CIRCULATIONAHA.114.011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parajuli N, Patel VB, Wang W, Basu R, Oudit GY. Loss of NOX2 (gp91phox) prevents oxidative stress and progression to advanced heart failure. Clin Sci (Lond) 2014;127:331–340. doi: 10.1042/CS20130787. [DOI] [PubMed] [Google Scholar]
- 20.Lin CC, Lee IT, Wu WL, Lin WN, Yang CM. Adenosine triphosphate regulates NADPH oxidase activity leading to hydrogen peroxide production and COX-2/PGE2 expression in A549 cells. Am J Physiol Lung Cell Mol Physiol. 2012;303:L401–L412. doi: 10.1152/ajplung.00090.2012. [DOI] [PubMed] [Google Scholar]
- 21.Wong SC, Fukuchi M, Melnyk P, Rodger I, Giaid A. Induction of cyclooxygenase-2 and activation of nuclear factor-kappaB in myocardium of patients with congestive heart failure. Circulation. 1998;98:100–103. doi: 10.1161/01.CIR.98.2.100. [DOI] [PubMed] [Google Scholar]
- 22.Scheuren N, Jacobs M, Ertl G, Schorb W. Cyclooxygenase-2 in myocardium stimulation by angiotensin-II in cultured cardiac fibroblasts and role at acute myocardial infarction. J Mol Cell Cardiol. 2002;34:29–37. doi: 10.1006/jmcc.2001.1484. [DOI] [PubMed] [Google Scholar]
- 23.Brkic L, Riederer M, Graier WF, Malli R, Frank S. Acyl chain-dependent effect of lysophosphatidylcholine on cyclooxygenase (COX)-2 expression in endothelial cells. Atherosclerosis. 2012;224:348–354. doi: 10.1016/j.atherosclerosis.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lappas M. Forkhead box O1 (FOXO1) in pregnant human myometrial cells: a role as a pro-inflammatory mediator in human parturition. J Reprod Immunol. 2013;99:24–32. doi: 10.1016/j.jri.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Hsu CK, Lin CC, Hsiao LD, Yang CM. Mevastatin ameliorates sphingosine 1-phosphate-induced COX-2/PGE2-dependent cell migration via FoxO1 and CREB phosphorylation and translocation. Br J Pharmacol. 2015;172:5360–5376. doi: 10.1111/bph.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappel BA, Stohr R, De Angelis L, Mavilio M, Menghini R, Federici M. Posttranslational modulation of FoxO1 contributes to cardiac remodeling in post-ischemic heart failure. Atherosclerosis. 2016;249:148–156. doi: 10.1016/j.atherosclerosis.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Wilhelm K, Happel K, Eelen G, Schoors S, Oellerich MF, Lim R, Zimmermann B, Aspalter IM, Franco CA, Boettger T, Braun T, Fruttiger M, Rajewsky K, Keller C, Bruning JC, Gerhardt H, Carmeliet P, Potente M. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature. 2016;529:216–220. doi: 10.1038/nature16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hariharan N, Ikeda Y, Hong C, Alcendor RR, Usui S, Gao S, Maejima Y, Sadoshima J. Autophagy plays an essential role in mediating regression of hypertrophy during unloading of the heart. PLoS One. 2013;8:e51632. doi: 10.1371/journal.pone.0051632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puthanveetil P, Wan A, Rodrigues B. FoxO1 is crucial for sustaining cardiomyocyte metabolism and cell survival. Cardiovasc Res. 2013;97:393–403. doi: 10.1093/cvr/cvs426. [DOI] [PubMed] [Google Scholar]
- 30.Gopal K, Saleme B, Al Batran R, Aburasayn H, Eshreif A, Ho KL, Ma WK, Almutairi M, Eaton F, Gandhi M, Park EA, Sutendra G, Ussher JR. FoxO1 regulates myocardial glucose oxidation rates via transcriptional control of pyruvate dehydrogenase kinase 4 expression. Am J Physiol Heart Circ Physiol. 2017;313:H479–H490. doi: 10.1152/ajpheart.00191.2017. [DOI] [PubMed] [Google Scholar]
- 31.Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S, Glass CK, Olefsky JM. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown J, Wang H, Suttles J, Graves DT, Martin M. Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem. 2011;286:44295–44305. doi: 10.1074/jbc.M111.258053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CC, Yang CC, Wang CY, Tseng HC, Pan CS, Hsiao LD, Yang CM. NADPH oxidase/ROS-dependent VCAM-1 induction on TNF-alpha-challenged human cardiac fibroblasts enhances monocyte adhesion. Front Pharmacol. 2015;6:310. doi: 10.3389/fphar.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizvi F, Heimann T, O’Brien WJ. Expression of NADPH oxidase (NOX) 5 in rabbit corneal stromal cells. PLoS One. 2012;7:e34440. doi: 10.1371/journal.pone.0034440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu CK, Lee IT, Lin CC, Hsiao LD, Yang CM. Sphingosine-1-phosphate mediates COX-2 expression and PGE2/IL-6 secretion via c-Src-dependent AP-1 activation. J Cell Physiol. 2015;230:702–715. doi: 10.1002/jcp.24795. [DOI] [PubMed] [Google Scholar]
- 36.Griendling KK. Novel NAD(P)H oxidases in the cardiovascular system. Heart. 2004;90:491–493. doi: 10.1136/hrt.2003.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe N, Zmijewski JW, Takabe W, Umezu-Goto M, Le Goffe C, Sekine A, Landar A, Watanabe A, Aoki J, Arai H, Kodama T, Murphy MP, Kalyanaraman R, Darley-Usmar VM, Noguchi N. Activation of mitogen-activated protein kinases by lysophosphatidylcholine-induced mitochondrial reactive oxygen species generation in endothelial cells. Am J Pathol. 2006;168:1737–1748. doi: 10.2353/ajpath.2006.050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bi X, Song J, Gao J, Zhao J, Wang M, Scipione CA, Koschinsky ML, Wang ZV, Xu S, Fu G. Activation of liver X receptor attenuates lysophosphatidylcholine-induced IL-8 expression in endothelial cells via the NF-kappaB pathway and SUMOylation. J Cell Mol Med. 2016;20:2249–2258. doi: 10.1111/jcmm.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen LK, Cavadas MA, Kholodenko BN, Frank TD, Cheong A. Species differential regulation of COX2 can be described by an NFkappaB-dependent logic AND gate. Cell Mol Life Sci. 2015;72:2431–2443. doi: 10.1007/s00018-015-1850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langlet F, Haeusler RA, Linden D, Ericson E, Norris T, Johansson A, Cook JR, Aizawa K, Wang L, Buettner C, Accili D. Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell. 2017;171(824–835):e818. doi: 10.1016/j.cell.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Ponugoti B, Tian C, Xu F, Tarapore R, Batres A, Alsadun S, Lim J, Dong G, Graves DT. FOXO1 differentially regulates both normal and diabetic wound healing. J Cell Biol. 2015;209:289–303. doi: 10.1083/jcb.201409032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamori D, Kaneto H, Nakatani Y, Matsuoka TA, Matsuhisa M, Hori M, Yamasaki Y. The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J Biol Chem. 2006;281:1091–1098. doi: 10.1074/jbc.M508510200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Gan L, Pan H, Guo S, He X, Olson ST, Mesecar A, Adam S, Unterman TG. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277:45276–45284. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 44.Gomi K, Zhu FG, Marshall JS. Prostaglandin E2 selectively enhances the IgE-mediated production of IL-6 and granulocyte-macrophage colony-stimulating factor by mast cells through an EP1/EP3-dependent mechanism. J Immunol. 2000;165:6545–6552. doi: 10.4049/jimmunol.165.11.6545. [DOI] [PubMed] [Google Scholar]
- 45.Inoue H, Takamori M, Shimoyama Y, Ishibashi H, Yamamoto S, Koshihara Y. Regulation by PGE2 of the production of interleukin-6, macrophage colony stimulating factor, and vascular endothelial growth factor in human synovial fibroblasts. Br J Pharmacol. 2002;136:287–295. doi: 10.1038/sj.bjp.0704705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son Y, Kim S, Chung HT, Pae HO. Reactive oxygen species in the activation of MAP kinases. Methods Enzymol. 2013;528:27–48. doi: 10.1016/B978-0-12-405881-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 47.Tsai KH, Wang WJ, Lin CW, Pai P, Lai TY, Tsai CY, Kuo WW. NADPH oxidase-derived superoxide anion-induced apoptosis is mediated via the JNK-dependent activation of NF-kappaB in cardiomyocytes exposed to high glucose. J Cell Physiol. 2012;227:1347–1357. doi: 10.1002/jcp.22847. [DOI] [PubMed] [Google Scholar]
- 48.Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349–3358. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harant H, Eldershaw SA, Lindley IJ. Human macrophage inflammatory protein-3alpha/CCL20/LARC/Exodus/SCYA20 is transcriptionally upregulated by tumor necrosis factor-alpha via a non-standard NF-kappaB site. FEBS Lett. 2001;509:439–445. doi: 10.1016/S0014-5793(01)03138-6. [DOI] [PubMed] [Google Scholar]
- 51.Miao H, Zhang Y, Lu Z, Yu L, Gan L. FOXO1 increases CCL20 to promote NF-kappaB-dependent lymphocyte chemotaxis. Mol Endocrinol. 2012;26:423–437. doi: 10.1210/me.2011-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 The expression of NOXs isoforms in HCFs (A) Total RNA was extracted from growth-arrested HCFs. The levels of NOX1-5 mRNA were determined by RT/PCR analysis. β-actin was used as an internal control. The star symbol indicates each replicon. (B-E) HCFs were transfected with Scramble, NOX1, NOX2, NOX4 or NOX5 siRNA for 36 h, and then incubated with LysoPC for 6 h. The levels of NOXs mRNA expression were determined by RT/qPCR analysis (n = 4). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 799 kb)
Supplementary Fig. 2 (A) HCFs were pretreated with SP600125 (0.3, 1, 3 μM) for 1 h and then incubated with LysoPC for 6 h. The levels of COX-2 and GAPDH protein were determined by western blot analysis (n = 5). (B-C) Representative data of western blotting are presented in Fig. 2E (DPI: n = 5; SP600125: n = 5). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 436 kb)
Supplementary Fig. 3 (A-D) Representative data of western blotting are presented in Fig. 3C (n = 6) and 3D (DPI: n = 7; SP600125: n = 5; Helenalin: n = 7). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01. (TIFF 407 kb)
Supplementary Fig. 4 (A) Schematic diagram of positions of NF-κB binding elements and ChIP primer pairs on COX-2 promoter. (B-C) HCFs were treated with LysoPC for the indicated time intervals. The DNA binding activity of NF-κB was determined by ChIP assays using an anti-p65 or anti-phosph-p65 antibody. The immunoprecipitated DNA and inputs were analyzed by an SYBR system for qPCR using a pair of primers flanking the NF-κB binding site within the human COX-2 promoter (proximal NF-κB: -320 ~ -171; distal NF-κB: -499 ~ -335). Representative gel (upper panels) and quantification of data are shown as the fold change normalized to input control (n = 5; bottom panels). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01 (TIFF 498 kb)
Supplementary Fig. 5 (A-D) Representative data of western blotting are presented in Fig. 5C (n = 7) and 5D (DPI: n = 7; SP600125: n = 5; AS1842856: n = 7). Data are expressed as the mean ± SEM, and analyzed by a one-way ANOVA with Tukey’s post hoc tests. *, p < 0.05. #, p < 0.01 (TIFF 410 kb)