Abstract
Extracellular vesicles (EVs) are released by cells to the extracellular environment to mediate inter-cellular communication. Proteins, lipids, nucleic acids and metabolites shuttled in these vesicles modulate specific functions in recipient cells. The enrichment of selected sets of proteins in EVs compared with global cellular levels suggests the existence of specific sorting mechanisms to specify EV loading. Diverse post-translational modifications (PTMs) of proteins participate in the loading of specific elements into EVs. In this review, we offer a perspective on PTMs found in EVs and discuss the specific role of some PTMs, specifically Ubiquitin and Ubiquitin-like modifiers, in exosomal sorting of protein components. The understanding of these mechanisms will provide new strategies for biomedical applications. Examples include the presence of defined PTM marks on EVs as novel biomarkers for the diagnosis and prognosis of certain diseases, or the specific import of immunogenic components into EVs for vaccine generation.
Keywords: Extracellular vesicles, Exosome, Post-translational modifications, Exosomal sorting, Ubiquitin
Introduction
Extracellular vesicles
Extracellular vesicles (EVs) are 50–300 nm-sized nanovesicles secreted to the extracellular milieu by a vast variety of cell types to promote inter-cellular communication on cells that capture them. Such vesicle exchange is involved in many physio-pathological situations such as tumour progression, metastasis, immunomodulation and the spreading of infections [1]. EVs have a heterogeneous nature, thus they are classified according to their origin into three groups: exosomes, shedding vesicles and apoptotic bodies [2]. Exosomes are of endosomal origin. Cells release them by fusing multivesicular bodies (MVBs)-containing intraluminal vesicles (ILVs) with the plasma membrane. Shedding vesicles form directly at the plasma membrane. Finally, apoptotic bodies are vesicles that result from apoptosis and contain cellular waste [3, 4]. EVs are membrane-covered vesicles that contain proteins, nucleic acids (DNA, mRNA and miRNAs), lipids and metabolites [5]. Proteins that are enriched in exosomes include tetraspanins (CD9, CD63, CD81), cytoskeleton components (Syntenin, Drebrin, ERMs), Endosomal Sorting Complexes Required for Transport (ESCRT complex) (TSG101, ALIX), Heat-Shock proteins (HSP70 and 90), Annexins, Rab proteins, among others [6]. Exosome membranes are also different from the membrane of their producer cells, mainly enriched in ceramide, sphingolipids, glycerophospholipids and cholesterol [7, 8].
Almost every cell, prokaryotic and eukaryotic, can secrete EVs. These particles appear in many fluids, including urine, amniotic fluid, blood, bile, semen, cerebrospinal fluid, breast milk, ascites and cell cultures (reviewed in [2]). Moreover, EVs contain specific external and internal markers, which vary depending on their origin. For instance, CD24 is a marker for urine and amniotic fluid exosomes [9]. In addition, a single cell type can simultaneously secrete different populations of exosomes [10, 11]. For example, polarized cells can deliver different populations of exosomes through the apical and basolateral poles [12].
Regarding cargo sorting, EVs receive cargoes through both ESCRT-dependent and independent mechanisms [13, 14]. The existence of heterogeneous MVBs populations becomes apparent when different components of endosomal trafficking or the ESCRT pathway are silenced or overexpressed, and when exosomal secretion is disturbed and not all exosomal markers are affected to the same extent [10, 15]. This enables distinguishing between global markers, e.g. TSG101, ALIX or HSP70; and cell-specific markers, e.g. class II Major Histocompatibility Complex (MHC-II) [3]. EV cargo also evolves with global changes to the whole cells. For example, oxidative stress modulates MVB biogenesis, endocytosis and consequently EVs composition [12].
Post-translational modifications
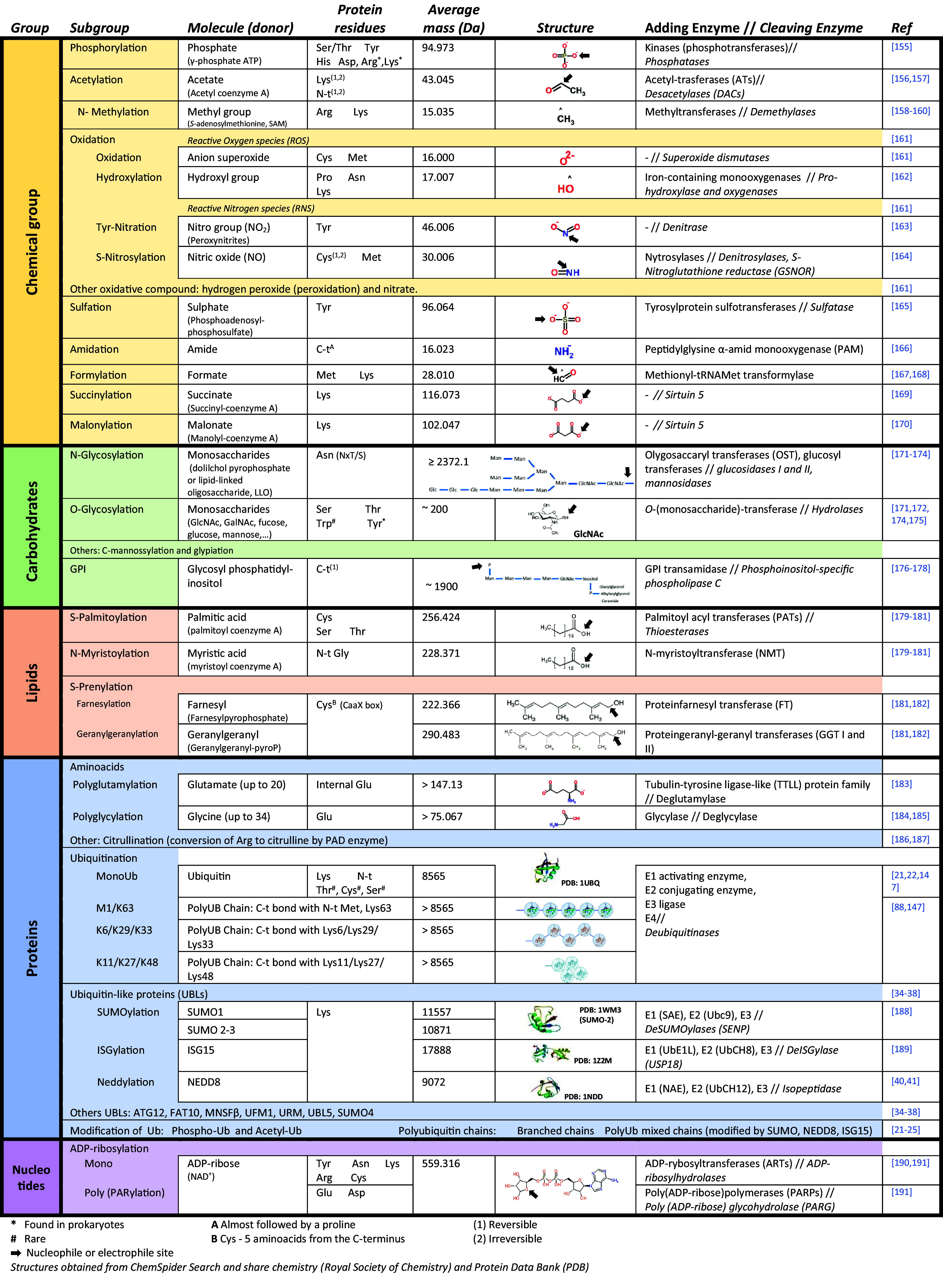
Post-translational modifications (PTMs) include a collection of add-ons that provide variability to a defined set of proteins that all cells use to enable most biological processes, adapting the functionality of those proteins by varying their interactive capability, resistance to degradation, etc. In this sense, PTMs exponentially increase the level of the proteome complexity [16]. These add-ons are molecules of diverse nature that attach to specific amino acids. Multiple amino acids can receive varied PTMs. The types of added molecule include chemical groups, carbohydrates, lipids, aminoacids, proteins and nucleotides. This process is controlled by specialized enzymes responsible for the addition or removal of each modification [17]. PTMs and their regulation constitute a cellular communication code to respond in a fast, versatile and accurate mode. The fulcrum of this review is that PTMs can direct exosome loading. Exosomes do not mirror the protein composition of their parental cells, and some proteins appear enriched in exosomes by selective mechanisms of protein cargo sorting controlled by specific PTMs [18]. The main PTMs are summarized in Table 1. These include addition of chemical groups (phosphorylation, acetylation, methylation, oxidation, hydroxylation, S-nitrosylation, sulfation or sulfonation, and others), carbohydrates [N-glycosylation, O-glycosylation, glycosylphosphatidylinositol (GPI)-anchoring], lipids (myristoylation, palmitoylation, prenylation) and nucleotides (ADP-rybosylation, PARylation). To make this large problem tractable, we focus in Ubiquitin and Ubiquitin-like modifiers (UBLs) as major controllers of EV protein loading.
Table 1.
Post-translational modifications of proteins
Ubiquitination and Ubiquitin-like modifiers (UBLs)
This group of PTMs represents one of the most important regulatory systems of protein location, stability and function. The C-terminal (C-t) Gly of ubiquitin and most UBLs attaches covalently mainly to the ε-amino group of a Lys (K) residue of the substrate protein, forming an isopeptide bond. The conjugation of ubiquitin and UBLs requires the consecutive action of three specific enzymes: Ubl-activating enzymes (E1s), Ubl-conjugating enzymes (E2s) and Ubl-protein ligases (E3s). Specific proteases reverse this process, removing ubiquitin from target proteins [19, 20]. Ubiquitin is a highly conserved, small protein of 8.5 kDa involved in diverse functions, such as protein degradation, endocytic trafficking, signal transduction and DNA repair, among others. Mono-ubiquitination is the modification of a specific Lys in a target protein. The same protein can bear multiple mono-ubiquitinated residues simultaneously. Besides, the ubiquitin protein itself contains seven Lys (K6, 11, 27, 29, 33, 48 and 63) in its sequence. These Lys can bind to other molecules of ubiquitin, generating seven types of polymeric ubiquitin chains, constituting the so called “ubiquitin code”. Moreover, M1 chains are polymeric chains formed by ubiquitin N-terminal (N-t) Met. All these chains vary in length, ranging from two to more than ten residues. They can be homotypic if the same residue is modified during elongation as in M1, K11, 48 or 63-linked chains, or heterotypic if the same chain displays different linkages [21, 22]. Moreover, a single ubiquitin can be modified with multiple chains or UBLs (NEDD8, SUMO, ISG15), generating branched chains or mixed chains, respectively. Finally, ubiquitin can be acetylated or phosphorylated [21–25]. K48-linked ubiquitin was the first described poly-ubiquitin chain related to degradation [21, 26]. On the other hand, K63-linked chains relate to many biological processes, and K63-modified proteins play important roles in diverse cellular signalling pathways, e.g. Toll-like receptor (TLR) cascade and endosomal transport [21, 27–30]. However, the specific functions of other mixed, branched or heterogeneous chains remain almost unknown [28] (Table 1). The consensus is that proteins modified with K29 and K48-linked chains are proteasome-bound for degradation. Nevertheless, M1, K6, K11, K27 and K63 chains and mono-ubiquitination regulate different cellular processes such as DNA repair, translation, inflammation and endocytic trafficking [21, 27–33] (Table 1).
UBLs share many features with ubiquitin, including their sequence and three-dimensional structure, which is a compact β-grasp fold. The main members of this group are SUMO1, SUMO2-3, SUMO4, ISG15, NEDD8, ATG12, FAT10, MNSFβ, UFM1, URM and UBL5 (reviewed in [34–38]). Neural precursor cells-Expressed Developmentally Down-regulated protein 8 (NEDD8) is the closest UBL to ubiquitin. NEDD8 mainly modifies proteins from the Cullin RING Ligases (CRLs) superfamily. RING E3 Ligases transfer ubiquitin directly from E2 enzymes to the substrate protein forming a complex scaffolding by Cullin. CRLs play an important role in protein ubiquitination for proteasomal degradation under the control of NEDD8 [39, 40]. NEDD8 also forms chains through one of its six Lys [41]. Small ubiquitin-related modifier (SUMO) is an UBL involved in nuclear transport and organization, DNA repair, transcription, chromatin remodelling and ribosome biogenesis. It is an essential modification to maintain cell homeostasis in stress responses and has become relevant in many diseases [42]. Interferon-stimulated gene 15 (ISG15) is induced by type I interferons (IFN-I), exposure to bacterial lipopolysaccharide (LPS), viral double-stranded RNA (dsRNA), ischemia and aging [43]. It is an essential part of the innate immune response to fight against bacterial and virus infections [44]. Besides, ISG15 is also overrepresented in various types of tumours [45]. Interestingly, ISG15 can also function as a free unconjugated form with an immunomodulatory role [46].
Exploring the sorting of modified proteins into EVs
Sorting mechanisms to MVBs
Exosomes originate in the endocytic pathway, whereas shedding vesicles form from direct budding of plasma membrane. Therefore, the mechanisms involved in protein secretion are different [18]. Here, we will discuss how PTMs could be involved in the sorting of proteins into EVs.
ESCRT-dependent and independent sorting mechanisms into ILVs
Presently, mechanisms of protein incorporation into the ILVs of MVBs for exosomal secretion or lysosomal degradation remain unclear. One key part of the machinery that controls the incorporation of ubiquitinated proteins into ILVs is the ESCRT complex [47, 48]. This complex includes four sub-complexes that sequentially recognize protein cargos and include them into ILVs. ESCRT-0 comprises HRS and STAM1/2 subunits, which are proteins with several distinct ubiquitin-binding domains (UBDs) that form a cargo-recognition module. HRS recognizes ubiquitinated proteins through a FYVE domain necessary to interact with phosphatidyl inositol 3-phosphate (PI3-P) molecules of endosomal membranes. HRS also displays PSAP domains that connect with the ESCRT-I subunit TSG101, which also binds ubiquitinated proteins. Moreover, the PSAP domains of small integral membrane protein of the lysosome/late endosome (SIMPLE) also bind TSG101. SIMPLE has a PPxY motif that interacts with NEDD4 to be ubiquitinated, and di-Leu and YKRL motives, which are signatures of endocytic functions [14, 49]. Mutations in these motifs impair the secretion of SIMPLE to exosomes, altering MVBs biogenesis [49]. Moreover, Hrs conditional knock-out (KO) in dendritic cells (DC) significantly reduces exosome release, pointing to an important role for ESCRT components in exosome secretion [50].
Deubiquitination is mandatory prior to packaging cargo into ILVs. To remove ubiquitin, ESCRT-III recruits deubiquitinating enzymes. ESCRT-III subunits CHMP2 A, B, CHMP3, CHMP4 A, B, C and CHMP6 bind to lipid membranes through their positively charged N-t domains, allowing high order oligomerization that results in structures over 600 kDa. Yeast ESCRT-III Snf7 (CHMP4 in humans), is not only implicated in neck-shaped oligomeric assembly for vesicle budding, but also in its scission. In this regard, the necessary energy to dissociate budding vesicles from membrane is provided by the ATPase activity of the Vacuolar protein 4 (VPS4) Complex (SKD1, CHMP5, LIP5) (reviewed in [47, 48]).
Ubiquitination of cargo proteins is necessary for recognition by ESCRT-0 components, whereas deubiquitination is a crucial step for sorting them into ILVs. However, whether ubiquitination is mandatory for driving proteins into exosomes is currently controversial. The sorting of some proteins seems independent of ubiquitination, e.g. MHCII loading into DC exosomes after T cell activation, which is associated to tetraspanin-enriched microdomains (TEMs) formed by CD9 [13, 51]. Furthermore, CD81 is important for the sorting of its associated proteins into exosomes [42]. Other studies support that the ubiquitination drives MHCII-containing MVBs to degradation upon fusion with the lysosomal system [52] (Fig. 1).
Fig. 1.

Schematic representation of some PTMs found in EVs. Endocytosis of plasma membrane receptors (MHC-II, LMP2A and EGFR) transfer them into the endocytic pathway. Depending on the added PTMs, these receptors can have different fates. Ubiquitinated-MHC-II is preferentially directed to degradative MVBs for degradation in lysosomes, whereas, Ub-LMP2A and ph-EGFR are usually incorporated into ILVs of exocytic MVBs, which are fused with plasma membrane to be secreted as EVs [52, 77, 116]. In the case of acetylated-GRP78 and ISGylated-TSG101, these PTMs act as cellular retention tags, promoting their degradation in lysosomes [128, 129]. However, Ub-HSP70, SUMOylated-hnRNPA2B1, SUMOylated-α-synuclein, ph-AKT, oxidized and phosphorylated γ-synuclein and citrullinated vimentin are loaded into ILVs and driven to EVs [80, 104, 106, 108, 118, 154]. Besides, unmodified TSG101 and GRP78 proteins can also be present in EVs [128, 129]. Figure key is shown at the right side. The images in the figures are not scaled
In addition, ESCRT components can be ubiquitinated themselves, for example HRS. HRS mono-ubiquitination prevents binding of ubiquitinated cargo, regulating the flow of specific proteins into MVBs [47]. The E3 ubiquitin ligase implicated in ubiquitin turnover can thus control the destiny of MVBs. For example, the activity of Mahogunin on TSG101 disrupts MVB lysosomal trafficking, whereas another E3 ubiquitin ligase called Tal abrogates MVB biogenesis [53, 54]. Associated molecule with the SH3 domain of STAM (AMSH) ubiquitin isopeptidase and ubiquitin isopeptidase Y (UBPY) removes mono-ubiquitin from HRS to recycle ESCRT components, preventing their degradation. Moreover, AMSH deubiquitinates cargoes modified with K63-linked chains, whereas UBPY recognizes both K63 and K48-linked chains. The action of these enzymes indicated a deubiquitination step that is required to sort cargoes into ILVs, and hence, into exosomes [55, 56].
However, mono and poly-ubiquitinated proteins can be found in exosomes, but not associated to the exosomal membrane [57]. Cargo deubiquitination before its packaging into ILVs seems essential, but how some ubiquitinated proteins end up in exosomes remains controversial. These contradictory results suggest that ubiquitinated proteins can evade deubiquitination; or finish up in exosomes through ESCRT-independent mechanisms. Such mechanisms do exist. In these, sorting depends on tetraspanins, sphingolipids and ceramide [58–60]. For example, LYPX(n)L motifs in the protein ALIX are implicated in its interaction with CD63 tetraspanin, and together with transmembrane proteins syndecans and syntenin induce budding and membrane abscission of ILVs [61]. In fact, an extensive study of ESCRT components downregulation revealed that of 23 ESCRT proteins studied, not all of them interfered with exosome secretion in the same way [11]. When HRS, TSG101, VPS22 and VPS24 are simultaneously silenced, MVBs are still produced and only the formation of Epidermal Growth Factor (EGF)+-MVBs is affected, highlighting a possible mechanism of substrate selectivity [62]. Proteolipid (PLP)+-exosomes localize together with flotillin and GPI in endosomal compartments, but not with HRS. These vesicles were enriched in cholesterol and ceramide, and their secretion depended on ceramide production by the activity of sphingomyelinase 2 (nSMase2) [63]. The enzyme sphingomyelin synthase SMS2 regulates the secretion of amyloid-β (Aβ)-peptide+-exosomes. In addition, the transfer of CD63+-exosomes from T cells to APCs during immune synapse is ceramide-dependent [59, 60]. This is likely related to the fact that ceramide creates spontaneous curvature of endosomal membranes, triggering ILVs budding. Besides, other lipids metabolites such as sphingosine-1-phosphate (S1P), diacylglycerol (DAG), cholesterol, phosphatidyl choline, or lysobisphosphatidic acid (LBPA) are involved in the MVB biogenesis and exosome secretion, although how protein cargoes are selectively transported into this kind of vesicles remains unknown [64–71].
Ubiquitinated proteins in EVs
Ubiquitin was first observed in human urine exosomes showing a wide range of molecular weight complexes by Western blot, which suggested the presence of ubiquitinated proteins in exosomes [72]. Specific antibodies for ubiquitin and poly-ubiquitin chains allowed to identify mono-, oligo- and poly-ubiquitinated proteins in exosomes [57]. Various studies and proteomic analysis identified specific ubiquitinated proteins (reviewed in [73]) (Table 2 and Fig. 1). For example, myeloid-derived suppressor cells (MDSC) secrete exosomes enriched in ubiquitinated proteins, including endosomal trafficking proteins and histones [74]. Moreover, high molecular weight complexes of non-classical human leukocyte antigen-G (HLA-G) are ubiquitinated, but not glycosylated, in exosomes obtained from ascitic and pleural exudates of patients [75]. Shedding vesicles also contain modified proteins such as arrestin domain-containing protein 1 (ARRDC1), which is ubiquitinated by the E3 ubiquitin ligase WW domain-containing protein 2 (WWP2). An arrestin domain in ARRDC1 anchors it to the plasma membrane, enabling its interaction with the PSAP domain of TSG101 to generate direct membrane budding [76]. In the case of ubiquitinated Epstein–Bar virus latent membrane protein 2A (LMP2A), exosome sorting depends on cholesterol, whereas the phosphorylated form was not detected in exosomes [77] (Table 2 and Fig. 1).
Table 2.
Post-translational modified proteins found in EVs
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is ubiquitinated on Lys13 by the association and regulation of Nedd4 family-interacting protein 1 (Ndfip1), an adaptor protein of the Nedd4 family E3 ubiquitin ligases, appearing in this form in exosomes, although modified-Ndfip1 does not. However, its presence is necessary for the import of Nedd4, Nedd4-2 and ITCH to exosomes [14, 78]. Alanine replacement on di-Leu (LL) and YKRL motifs of SIMPLE reduces it secretion into exosomes, as well as a mutation on its PTAP motif, which controls SIMPLE binding to TSG101 for incorporation into ILVs. However, point mutations of the two PPxY motifs of SIMPLE, a binding domain for Nedd4 type E3 ubiquitin ligases, such as Nedd4 and ITCH, increase exosomal incorporation; hence SIMPLE ubiquitination does not direct its own sorting [49]. In other cases, such as Fas Ligand (FasL), ubiquitination of the proline-rich domain (PRD)-flanking Lys and phosphorylation of multiple tyrosines promote its internalization in MVBs and its secretion into EVs [79]. The COP9 signalosome (CSN)-associated protein CSN5 mediates ubiquitin isopeptidase activity. Deletion of its JAB1/MPN/Mov34 metalloenzyme (JAMM) domain increases the sorting of ubiquitinated HSP70 and HIV Gag proteins into exosomes; whereas a mutation of its associated deubiquitin activity domain promotes the enrichment of non-modified proteins in EVs [80]. These results highlight that CSN5 participates in ubiquitin-dependent and independent protein vectorization into exosomes [80] (Table 2). MARCH1, an E3 ubiquitin ligase for MHCII and CD86, is incorporated into exosomes dependent on its C-t tyrosin-based motifs [81]. This endosome sorting motif is also important for sorting of transferrin receptor into exosomes due to its interaction with ALIX [82]. Nevertheless, whether MARCH1 is ubiquitinated or not in exosomes remains unclear, although this region is important for MARCH1 auto-ubiquitination [83].
Human urine exosomes contain approximately 15% of ubiquitinated proteins. Of these, 21% are transmembrane proteins, underscoring that deubiquitination is not mandatory for the incorporation of protein cargo into MVBs and exosomes [84]. Although, ubiquitination-specific motifs remain unidentified, this PTM has preference for basic amino acids and positively charged residues upstream of the ubiquitination site, hydrophobic residues adjacent to it and additional Lys 6 amino acids upstream [84]. Besides, poly-ubiquitin chains appear following this order of enrichment: K63 > K48 > K11 > K6 > K29 > K33 > K27. This shows that K63, K48 and K11-linked chains are the most representative poly-ubiquitin topologies in exosomes [84]. Furthermore, platelet exosomes contain poly-ubiquitin chains and in higher numbers compared to platelets microparticles [85] (Table 2).
Deciphering the ubiquitin code in EVs
The ubiquitin code comprises the combination of mono-, poly- and branched-ubiquitin chains and even other PTMs, which allow a wide range of possibilities for the regulation of proteins. K48-linked chains require at least four ubiquitin units to attach to the target protein to enable recognition by the 26S proteasome [86, 87]. In particular, the proteasome subunit S5a/Rpn10 contains an ubiquitin-interacting motif (UIM) in the hydrophobic patch of its C-t region, which enables recognition of this specific poly-ubiquitin chain. Inside the proteasome, ubiquitin molecules are removed from target proteins for recycling, whereas the rest of the proteins are cleaved into small peptides (3–25 amino acids of length). There is some evidence suggesting that other chains, e.g. M1, K6, K11, K27 and K29 could be involved in proteasomal degradation of target proteins [21, 30, 33, 86]. Moreover, a new conjugation factor, named E4, participates in the generation of efficient poly-ubiquitinated-linked chains under stress conditions to direct modified proteins to the proteasome, demonstrating the existence of an E4-dependent multi-ubiquitination pathway for degradation [88] (Fig. 2). K63-linked chains do not end up in the proteasome as they bind specifically to the ESCRT-0 complex, specifically to UIMs found in HRS and STAM1 components [27, 89]. These interactions condense proteins modified with K63-linked poly-ubiquitin chains into endosomes, promoting their import into ILVs of MVBs and possibly, their subsequent delivery into exosomes [90]. In fact, K63-linked chain is the most representative ubiquitin topology found in EVs [84] (Fig. 2).
Fig. 2.

The ubiquitin code in cellular functions. MHC-I and II and TCR are preferentially retained in endosomes by the action of specific E3-Ub-ligases MARCH-IV, I and Cbl, respectively. Whereas Ub-MHC-II is mainly degraded by lysosomes, Ub-MHC-I (HLA-G) can be sorted into EVs. Signalling proteins downstream of TCR, TNFR or TLR can be modified by K63-linked chains to mediate signal transduction or by K48-linked ones, such as IκBα to be degraded in the proteasome. M1-linked chain is added to NF-κB to allow its nuclear translocation to start gene transcription. K27-linked chains are related with DNA repair processes, while K6, K11 and K63 chains can modify mitochondrial proteins to initiate mitophagy of damaged mitochondria by the E3-Ub-ligase Parkin. Proteins with K11 and K48-chains are principally transported to the proteasome to be degraded. Most of the proteins modified with K63-linked chains interact with ESCRT-0 and are vectorised in ILVs of exocytic MVBs to be delivered into EVs. Although Ub-MHC-II is mainly degraded, unmodified MHC-II and Ub-MHC-I (HLA-G) are usually incorporated in exosome membrane. Besides, K11, K48 and K63-linked chains can be found in EVs. Figure key is shown at the right side. The images in the figures are not scaled
How ubiquitin-binding proteins detect unequivocally a specific conformation of poly-ubiquitin chains may explain the selective targeting of a given protein within the cell. For example, M1 and K63-linked chains display a fairly linear or open space conformation, whereas K6, K11, and K48-linked chains create closed structures [21, 33, 91, 92] (Table 1). Open-conformation chains only limitation is that imposed by isopeptide bonds that connect them [21, 33]. However, closed conformation chains have multiple steric hindrances, which alters ubiquitin space structure by exposing and concealing different parts of ubiquitin residues, creating diverse topologies intrinsic to a specific type of linkage [21, 91]. In the cell, specific proteins bind to a unique type of ubiquitin linkage topology [91]. Because distances between ubiquitin units within a chain are different in K63 or K48-linked chains, UBDs of cellular machinery display small spaces between UIMs to specifically recognize K48-chains, whereas larger spacers are used for K63 binding [91, 92]. The use of a variety of UBDs structures not only allows discriminating between open and close conformations, it also enables distinguishing among different open structures such as K63- and M1-linked chains [91, 92]. All these possibilities of ubiquitin topologies and UBDs compose a fine and selective language among ubiquitinated-target proteins and ubiquitin-interacting proteins.
In a similar way, depending on the different distances between UIMs inside UBDs, certain E3-ubiquitin ligases elongate specific types of K-linked chains associated to target proteins. Besides, the wide variety of E3-ubiquitin ligases described in eukaryotic cells and their tissue restrictions amplify the code of ubiquitin chains. For example, the Cbl family of E3-ubiquitin ligases are highly expressed in hematopoietic cells and are involved in the downregulation of many signalling pathways controlling T and B cell receptors and integrins [93, 94]. However, MARCH1 is represented in antigen presenting cells (APCs), modifying class II MHC molecules, downregulating their expression in the plasma membrane, whereas MARCH-IV regulates class I MHC molecules using a similar mechanism [95, 96]. In this regard, a possible explanation regarding how K48-linked poly-ubiquitin chains evade proteasomal degradation and travel into exosome involves the collaboration of certain ubiquitin-binding proteins located in endosomal pathway (Fig. 2).
Branched poly-ubiquitin chains display the particular signalling properties of their K48 or K63 components, although they are recognized by multiple linkage-specific enzymes [97]. This underscores the predominance of certain chain topologies within a mixed-linkage poly-ubiquitin chain. The sorting of ubiquitin chains other than K63-linked requires mechanisms different from those dependent on ESCRT components. The incorporation of SUMO-2/3 following ubiquitination creates heterologous SUMO-2/3-Ubiquitin chains responsible for enhancing the degradation of IκBα by the proteasome after the activation of the Tumour Necrosis α (TNFα) signalling pathway [23]. In stress situations, NEDD8 can also modify ubiquitin Lys48 [25]. A recent study described how ISGylation of ubiquitin Lys29 negatively controls the turnover of ubiquitinated proteins [24].
Interestingly, ubiquitin can be phosphorylated on Ser65 by PTEN induced putative kinase 1 (PINK1), enhancing its interaction with the mitochondrial E3 ubiquitin ligase parkin [98]. This phosphorylation-dependent mechanism increases the enzymatic activity of parkin, decreasing the mitochondrial membrane potential [98]. Moreover, ubiquitin becomes acetylated in Lys6 and Lys48 without affecting substrate mono-ubiquitination of histone H2B [99]. However, both acetylations inhibit K11, K48 and K63-linked poly-ubiquitin chain elongation, providing a novel regulatory step of control of mono- and poly-ubiquitination [99].
Because many PTMs can modify proteins in the same residue, competition and crosstalk between different PTMs depending on different cell factors can change protein function and localization [100]. In summary, deciphering the sophisticated ubiquitin code is necessary to unveil specific functions of every chain topology in the selective import of proteins into exosomes.
Other PTMs in EVs
Proteomic studies have contributed extensively to define the protein composition of EVs [72, 84, 101–103]. Heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) is present in exosomes in its sumoylated form, and that is necessary for the sorting of EXOmotif-containing microRNAs into exosomes [104]. On the other hand, a study suggested that SUMO-1 could modulate the toxicity of α-synuclein in neurodegenerative disorders such as Parkinson’s disease (PD) [105]. Besides, EVs can contribute to disseminate neurotoxic proteins, which favours the development of PD pathogenesis. Hence, sumoylated α-synuclein is incorporated into exosomes by a mechanism dependent on the ESCRT complex and phospholipids [106] (Table 2 and Fig. 1).
Nevertheless, ubiquitin and UBLs are not the only PTMs detected in EVs [73, 107]. For example, many phosphorylated proteins appear in exosomes, including the calcium-dependent phospholipid-binding protein Annexin A2, which is enriched in cholesterol-rich lipid raft micro-domains of exosome membranes [101, 108, 109]. The localization of this protein depends on the phosphorylation of Tyr23, which prevents its endosomal degradation and allows its incorporation into exosomal membranes [109]. Other phosphorylated proteins found in exosomes include Tau, type 1 insulin receptor substrate (IRS-1), Crystalline alphaB (CryAB), Epidermal Growth Factor Receptor (EGFR), Na+-Cl− co-transporter (NCC), Aquaporin 2, FasL and proteins implicated in cellular processes such as apoptosis, survival and metabolism [79, 101, 108, 110–117] (Table 2 and Fig. 1).
γ-Synuclein acquires features of prion-like proteins by oxidation, which triggers the aggregation of other proteins, e.g. α-synuclein [118]. The most frequent oxidized residues of γ-synuclein are Met38 and Tyr39. Oxidized γ-synuclein is released into exosomes and incorporated in neighbouring cells, amplifying the toxic protein-aggregation cascade that finally causes a degenerative synucleinopathy [118]. Besides, other oxidized proteins appear in EVs as described in other studies (reviewed in [107]) (Table 2 and Fig. 1). Moreover, several studies have revealed the presence of glycosylated proteins in EVs (reviewed in [73, 107]). Notably, EVs contain mainly mannose or sialic acid enriched-glycans and N-linked glycoproteins. In particular, exosomes contain large amounts of the sialoglycoprotein galectin-3-binding protein (LGALS3BP) [119–122] (Table 2).
Additionally, plasma membrane anchors induce vesicle budding directly from the plasma membrane of highly oligomeric proteins, such as the HIV Gag protein and the yeast cytoplasmic protein Tya [123, 124]. An evaluation of different membrane anchors for Tya demonstrated that myristoylation was the most effective enhancer of the formation of shedding vesicles, whereas other types such as, phosphatidylinositol-(4,5)-bisphosphate and phosphatidylinositol-(3,4,5)-trisphosphate-binding domains, prenylation/palmitoylation tag or the type-1 plasma membrane protein CD43 did not cause this effect [124]. This illustrates the role of protein anchors for the induction of shedding vesicles, which is similar to retrovirus budding [124] (Table 2). Besides, CD55 or decay-accelerating factor (DAF) anchored to a GPI molecules is selectively secreted to EVs [125].
The role of UBL and other PTMs in the sorting of proteins to EVs
The function of PTMs such as oxidation, phosphorylation, glycosylation and citrullination in the incorporation of proteins into EVs, has been recently reviewed in [107]. Briefly, these PTMs can regulate EVs release and uptake, acting as a disposal mechanism for cellular harmful components or as a repair mechanism in physio-pathological conditions [107]. Similar to ubiquitin, the UBL SUMO also plays an important role in protein sorting to EVs. For example, SUMOylation is necessary for α-synuclein sorting into exosomes in an ESCRT-dependent manner [106] (Table 2). A recent study showed that other UBL called autophagy-related protein 12 (ATG12) and its E3 ligase ATG3 interact with ALIX, regulating diverse functions carried out by ALIX, for example, viral budding and MVBs biogenesis [126]. This observation suggests that ATG12 could also participate in exosome biogenesis. Besides, isoprenylation and palmitoylation may also participate in protein delivery into exosomes, as in the case of the CINCCKVL palmitoylation and isoprenylation motif, which is present in human Ras Homolog Family Member B (RhoB), determines its incorporation into ILVs in a cholesterol-dependent way and consequently in exosomes [127]. Furthermore, the expression of reporter proteins containing this palmitoylation/isoprenylation motif is enough to define a late-endosomal fate for these proteins [127].
On the contrary, ISG15, which is recognized as an immune alarm signal by cells, has an opposite role in protein sorting to exosomes. A recent study showed that ISGylation tags proteins for degradation, thereby avoiding exosome secretion and spreading [43, 128] (Fig. 1). Besides, acetylation can abrogate the sorting of proteins into exosomes. High glucose-regulated protein 78 (GRP78) is expressed and secreted into exosomes by colon cancer cells to prepare the tumor microenvironment [129]. Its secretion in exosomes is impaired by chemical inhibitors of histone deacetylases (HDACs) and silencing of HDAC6 protein, which causes the dissociation of GRP78 from HDAC6 and promotes GRP78 aggregation and interaction with VPS34 complex [129]. The use of an acetylation mimetic mutant of GRP78 (K633Q) also impairs exosome sorting and blocks tumour cell growth in vivo [129] (Fig. 1).
On the other side, the role of the other PTMs, gathered in Table 1, remains largely unexplored. This could suggest that a role for these PTMs is unlikely, or that they can work as a cellular retaining signal for some proteins, directing them to other cellular processes such as the autophagy-lysosome degradation pathway.
Targeting post-translationally modified proteins into EVs for biomedical applications
Deciphering the role of PTMs for protein sorting into exosomes could be useful for medical purposes, such as association of certain modified proteins with concrete physio-pathological situations, highlighting PTMs of exosomal proteins as biomarkers for prognosis, diagnosis and treatment of diseases.
PTM marks in EVs as biomarkers for diagnosis and prognosis
EVs have potential as non-invasive biomarkers for several pathological conditions, particularly due to their accessibility from biological fluids, facilitating the detection of certain pathologies [130–133]. A specific PTM of a protein in EVs in particular diseases could constitute a potential biomarker (Fig. 3). For example, the secretion of modified Tau protein aggravates development and progression of neurodegenerative processes. The majority of modified (phospho-Thr-181) Tau detected in human cerebrospinal fluid samples is secreted via exosomes, constituting a well-characterized biomarker for Alzheimer’s disease (AD) [115]. In fact, ph-Tau in combination with Aβ activate diverse signalling pathways, increasing the production of hyperphosphorylated Tau oligomers, which are then secreted by exosomes within the brain, accelerating neurodegeneration [110]. Moreover, some AD-related proteins such as ph-Tau, Aβ1-42, neurogranin (NRGN) and the repressor element 1-silencing transcription factor (REST) contained in neuronal-derived exosomes appear in plasma [117]. The transference of these proteins contained in exosomes isolated from AD patients’ plasma into the central nervous system of a normal mouse causes Tau aggregation and induces AD-like neuropathology [117]. Other studies carried out with Tau-transgenic mice (rTg4510) revealed a high grade of pathology and higher accumulation of Tau in their exosomes compared to control ones [113].
Fig. 3.
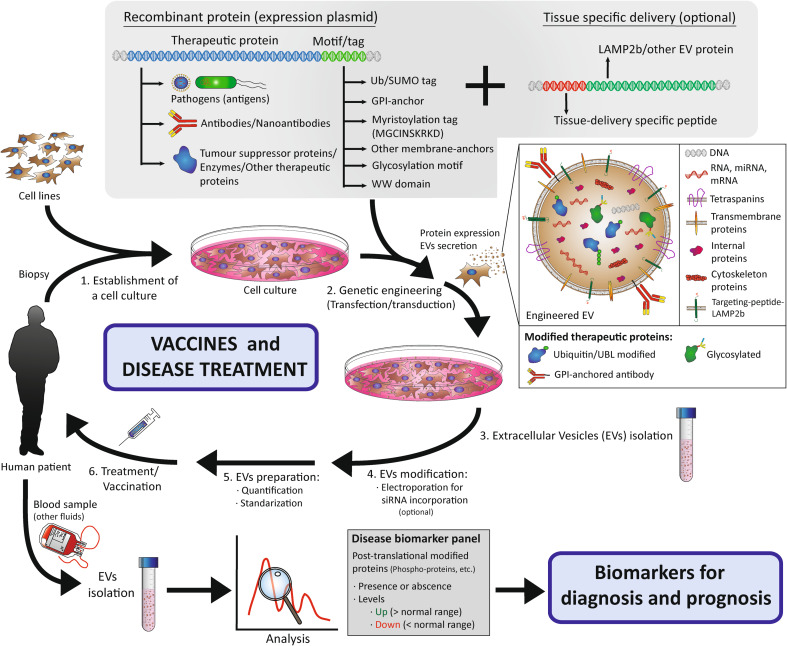
Targeting post-translationally modified proteins into EVs for biomedical applications. The process for exosome engineering to use as a vaccine or treatment comprises various steps: (1) EVs can be isolated from different cell cultures, either cell lines or cells obtained from human biopsies. (2) These cells can be transfected or transduced with specific constructs coding for engineered therapeutic proteins, that will be targeted to EVs by the addition of a specific PTM motif or tag. Therapeutic proteins include: pathogen antigens for the generation of vaccines; antibodies or nanoantibodies for activation or blocking of signalling pathways, or any kind of protein with a therapeutic potential. Besides, the expression of LAMP2b fused to a tissue-delivery targeting-peptide defines a specific fate for these EVs once in the blood stream. (3) The EVs can be isolated by diverse procedures. (4) Additionally, an extra step can be performed to incorporate inside EVs therapeutic small interference RNAs by electroporation. (5, 6) Finally, EVs preparations, once standardized and quantified, could be administered intravenously for disease treatment or vaccination. On the other hand, EVs obtained from blood samples (or other fluids) can be analysed to identify certain condition biomarkers (post-translational modified protein) for disease prognosis and diagnosis. The images in the figures are not scaled
On the other hand, several phosphorylations of IRS-1 in different residues associate to insulin resistance syndrome in diabetes mellitus as well as to degenerative diseases such as AD and fronto-temporal dementia [111]. Exosomes released by retinal pigment epithelial (RPE) cells into vitreous humour contain a vast subset of phosphorylated proteins in age-related macular degeneration (AMD) patients [108]. Oxidative stressed RPE cells modify the phosphorylation profile of exosomal proteins compared to control cells, thus their detection could represent a source of biomarkers for diagnosis and prognosis of eye-degenerative diseases [108].
Besides, detection of modified exosomal proteins bears potential as a biomarker of treatment progression. For instance, cetuximab works as a therapeutic antibody to block EGFR activation in tumour cells [116]. EVs coming from cetuximab-treated cells represent well their originating cells, containing lower level of phosphorylated EGFR after treatment [116]. Monitoring total and phosphorylated forms of EGFR present in EVs from patient blood could reflect the efficacy of anti-tumour therapy [116].
Moreover, detection of some exosomal biomarkers can reveal the existence of side effects during treatment. For example, a fraction of kidney transplant patients treated with tacrolimus as part of the immunosuppressive therapy developed hypertension 6 months after surgery [114]. Urinary exosomes of hypertensive patients are enriched in the phosphorylated form of thiazide-sensitive NCC compared to non-hypertensive patients [114]. Hence, the measurement of total and phosphorylated NCC in urinary exosomes can be used as a biomarker of hypertension [114].
Vaccines
EVs also represent a promising tool for other therapeutic applications, such as generation of vaccines. EVs and many viruses share common mechanisms for protein sorting and particle release [134]. The study of EVs physiology can help us to understand virus infections, but also to generate virus-like particles suitable for vaccination exploiting the exosome pathway by sorting virus proteins into exosomes. For instance, exosomes isolated from infected cells can prime mice and protect them from a following lethal infection [135]. In the case of macrophages infected with the intracellular bacteria Mycobacterium tuberculosis, more than 40 mycobacterial components appear in exosomes [136] (Table 2). In addition, the administration of soluble mycobacterial proteins in human epithelial and mouse macrophage cell lines promotes sorting of similar bacterial proteins into exosomes [136]. Ubiquitination of mycobacterial proteins such as HspX and GroES acts as a signal for their incorporation into ILVs and secretion as EVs [136]. Moreover, vaccination using these exosomes in mice protects them against subsequent aerosolized infection with the whole infective bacteria [136]. This is just an example of how vaccination with a PTM-protein is a new strategy to initiate a protective immune response against a specific pathogen. An option for heterologous protein sorting into exosomes, which can be used as a vaccine, consists of protein overexpression with the addition of an ubiquitin tag, avoiding strategies based on inactivated or non-replicative pathogens [128, 137] (Fig. 3). This approach enables the exosomal delivery of proteins not usually found in exosomes, e.g. GFP. For instance, the use of exosomes produced by a stable cell line and containing recombinant fusion proteins from M. tuberculosis evoked T cell immune responses [137]. However, although exosome-based vaccines could trigger antigen-specific immune responses, they also contain other proteins that could trigger side- or off-target effects. In addition to ubiquitin, GFP SUMOylation drives it into exosomes, suggesting that SUMO could work as a specific sorting tool for antigen incorporation into exosomes for vaccination schemes [106] (Fig. 3).
Disease treatment
EVs are a promising tool to design new strategies for disease treatment. The size of these small particles make them suitable to carry proteins, lipids, RNA, DNA or small chemical drugs, protecting them from external factors in the blood stream [138, 139]. They can be easily isolated in high amounts from diverse culture cells types. Furthermore, the target specificity of EVs prevents some secondary effects observed in traditional therapies [138]. One of the most potent advantages of exosomes is their capability of crossing the brain blood barrier (BBB), which opens a wide variety of possibilities for diagnosis and treatment of neurodegenerative diseases [140]. In fact, exosomes carrying a specific siRNA and a brain-delivery peptide signal could even silence the expression of the targeted gene [141]. Generating exosomes targeted to a certain type of tissue can be accomplished by the expression of specific tissue-delivery peptide fused to the N-t of lysosome-associated membrane glycoprotein 2 (LAMP2), a transmembrane protein which causes their incorporation into EVs [141]. Engineered targeting peptides-LAMP2 sometimes undergo degradation even when localized to exosomes, reducing their potential as therapeutic tools. A recent study has discovered that the incorporation of glycosylating motifs enhances their delivery and protects them from damage [142].
The use of engineered EVs based on biologically active proteins could be used to treat some diseases. The addition of a WW tag (a domain-containing PPxY motif) to a certain protein determines its incorporation into exosomes [139]. The l-domains of Ndfip1 recognize this particular mark, targeting this protein to exosomes [14]. In the same way, we propose the use of known PTMs motifs, such as the myristoylation sequence MGCINSKRKD, in the N-t of the protein of interest to drive it specifically into exosomes, as other authors have described for Tya [124]. Moreover, addition of a C-t ubiquitin or SUMO tag could deliver proteins into exosomes, including targeting-peptides-LAMP2 with glycosylated motifs for specific tissue delivery [106, 137, 142]. Besides, protein glycosylation has improved the efficacy of therapeutic proteins [143]. Thus, addition of glycosylating motifs to the protein of interest could also mediate its incorporation to exosomes, although protein glycosylation is still technically complex for industrial production [73, 107] (Fig. 3).
PTM-dependent sorting approaches could contribute to halting tumour progression and development. PTEN, a tumour suppressor phosphatase, is ubiquitinated in exosomes and is active in recipient cells, promoting the dephosphorylation of AKT [78]. Hence, the uptake of exosomes containing the C-t domain of PTEN by cancer cells reduces proliferation, migration and metastasis, postulating such a strategy as an anti-tumour treatment [144]. On the other hand, GPI modification of blocking-nanoantibodies that recognize receptors present in tumour cells, favours their attachment to exosomes. Exosome-attached EGFR-nanoantibodies approach can confer a specific selectivity to EVs against tumour cells, decreasing their EGFR-proliferative signalling pathways [145]. Hence, GPI anchors can be used to target particular proteins to EVs for biomedical applications [146] (Fig. 3). To sum up, all these strategies remark the potential application of EVs-containing post-translational modified proteins for selective delivery of therapeutic components into specific tissues.
Concluding remarks
Although the mechanisms that define the sorting of proteins into EVs are not fully understood, PTMs are likely to play an important role. Many post-translationally modified proteins have been detected in EVs [73, 107], whereas others that have not been identified in EVs could have a role in protein delivery, participating in the incorporation of other proteins, or being removed before secretion. Ubiquitination or SUMOylation of some proteins have a clear role in their incorporation into exosomes, whereas others such as ISGylation or acetylation oppose such mechanisms, driving the modified protein to degradation [106, 128, 137]. Moreover, exosome secretion can be mediated by diverse mechanisms, such as the ESCRT complex, lipids and tetraspanins, increasing the complexity and diversity of ILV-protein sorting and exosome release [18]. In this regard, EVs populations are heterogeneous as each vesicle contains a specific repertoire of proteins potentially different from its neighbours [10, 11]. In addition, there are other factors that increase the complexity of this system, for example the fact that almost every cell can secrete EVs containing common and cell-type-specific markers. Then, the final destination of each population of EVs can be different, working as an autocrine or paracrine signal or even travelling throughout the blood stream to other tissues.
Regarding ubiquitination, its own structure and functional complexity increases the number of factors to be taken into consideration. Compartimentalization and specificity of the different E3 ligases participate in protein localization [93, 94]. In addition, competition or crosstalk with other PTMs such as SUMOylation or Neddylation alters the fate of the modified protein [100]. Also, the complexity of the ubiquitin code includes homotypic, heterotypic, mixed or branched chains that distribute protein populations among many cellular processes such as proteasome degradation or endocytic trafficking, forming a ubiquitin-based protein ZIP code that has yet to be revealed [20–25, 33, 147]. Finally, the study of ubiquitinated proteins or even UBLs is difficult technically. Proteomics approaches to find ubiquitinated proteins in whole proteome typically used trypsin digestion of the sample. Nowadays, the discovery of new UBLs highlights that the sole use of this enzyme may mask the appearance of additional variations due to the same di-Gly residue shared by all these kind of modifications [148]. Hence, this information can only be used to localize specific sites on proteins susceptible to be modified by these kind of PTMs [148]. Nevertheless, some approaches tried to discern among these PTMs by terminal mutation and protein overexpression [149]. Besides, these proteomic approaches could also help to discover or define any possible consensus sequence for protein delivery into EVs, either protein mono-ubiquitination and selective chain formation or other PTMs [102, 150, 151].
In addition, EVs represent a novel non-invasive and selective therapeutic tool to be developed for prognosis, diagnosis or treatment of diverse diseases, with very promising preliminary results [138, 139, 152, 153]. PTMs confer specific properties to proteins, playing a relevant role in EVs protein sorting. Hence, the use of PTMs represents a useful tool for the generation of therapeutically engineered EVs.
Acknowledgements
The authors thank Dr. Miguel Vicente-Manzanares and Dr. Manuel Gómez-Gutiérrez for assistance with English editing. This work was supported by Grants to Francisco Sánchez-Madrid, SAF2014-55579-R; INDISNET-S2011/BMD-2332; PIE13/00041; H2020 European Research Council ( ERC-2011-AdG 294340-GENTRIS); and was co-funded by Fondo Europeo de Desarrollo Regional (FEDER). Irene Fernández-Delgado and Olga Moreno-Gonzalo are supported by Formación de Profesorado Universitario (FPU) program from the Spanish Ministry of Education, Culture and Sports. The CNIC is supported by the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) and by the Pro CNIC Foundation.
Footnotes
Olga Moreno-Gonzalo and Irene Fernandez-Delgado contributed equally to this work.
References
- 1.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 4.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Vallabhaneni KC, Penfornis P, Dhule S, Guillonneau F, Adams KV, Mo YY, Xu R, Liu Y, Watabe K, Vemuri MC, Pochampally R. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget. 2015;6(7):4953–4967. doi: 10.18632/oncotarget.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 7.Llorente A, Skotland T, Sylvanne T, Kauhanen D, Rog T, Orlowski A, Vattulainen I, Ekroos K, Sandvig K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831(7):1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72(9):1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 10.Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012 doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 12.Sreekumar PG, Kannan R, Kitamura M, Spee C, Barron E, Ryan SJ, Hinton DR. AlphaB crystallin is apically secreted within exosomes by polarized human retinal pigment epithelium and provides neuroprotection to adjacent cells. PLoS One. 2010;5(10):e12578. doi: 10.1371/journal.pone.0012578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschow SI, Nolte-’t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, Wauben MH, Stoorvogel W. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10(10):1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 14.Putz U, Howitt J, Lackovic J, Foot N, Kumar S, Silke J, Tan SS. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J Biol Chem. 2008;283(47):32621–32627. doi: 10.1074/jbc.M804120200. [DOI] [PubMed] [Google Scholar]
- 15.van der Vlist EJ, Arkesteijn GJ, van de Lest CH, Stoorvogel W, Nolte-’t Hoen EN, Wauben MH. CD4(+) T cell activation promotes the differential release of distinct populations of nanosized vesicles. J Extracell Vesicles. 2012 doi: 10.3402/jev.v1i0.18364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011 doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knorre DG, Kudryashova NV, Godovikova TS. Chemical and functional aspects of posttranslational modification of proteins. Acta Nat. 2009;1(3):29–51. [PMC free article] [PubMed] [Google Scholar]
- 18.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 20.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 22.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aillet F, Lopitz-Otsoa F, Egana I, Hjerpe R, Fraser P, Hay RT, Rodriguez MS, Lang V. Heterologous SUMO-2/3-ubiquitin chains optimize IkappaBalpha degradation and NF-kappaB activity. PLoS One. 2012;7(12):e51672. doi: 10.1371/journal.pone.0051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan JB, Arimoto K, Motamedchaboki K, Yan M, Wolf DA, Zhang DE. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci Rep. 2015;5:12704. doi: 10.1038/srep12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjerpe R, Thomas Y, Kurz T. NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J Mol Biol. 2012;421(1):27–29. doi: 10.1016/j.jmb.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270(29):17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 27.Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem. 2003;278(14):12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: beyond the Usual Suspects’ review series. EMBO Rep. 2008;9(6):536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 30.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278(38):35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 31.Hammond-Martel I, Yu H, el Affar B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24(2):410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009;8(4):544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Michel MA, Elliott PR, Swatek KN, Simicek M, Pruneda JN, Wagstaff JL, Freund SM, Komander D. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol Cell. 2015;58(1):95–109. doi: 10.1016/j.molcel.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 36.Taherbhoy AM, Schulman BA, Kaiser SE. Ubiquitin-like modifiers. Essays Biochem. 2012;52:51–63. doi: 10.1042/bse0520051. [DOI] [PubMed] [Google Scholar]
- 37.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 38.Vierstra RD. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012;160(1):2–14. doi: 10.1104/pp.112.200667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16(1):30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 41.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7(3):1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 43.Villarroya-Beltri C, Guerra S, Sanchez-Madrid F. ISGylation—a key to lock the cell gates for preventing the spread of threats. J Cell Sci. 2017 doi: 10.1242/jcs.205468. [DOI] [PubMed] [Google Scholar]
- 44.Morales DJ, Lenschow DJ. The antiviral activities of ISG15. J Mol Biol. 2013;425(24):4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai SD. ISG15: a double edged sword in cancer. Oncoimmunology. 2015;4(12):e1052935. doi: 10.1080/2162402X.2015.1052935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell JA, Lenschow DJ. Emerging roles for immunomodulatory functions of free ISG15. J Interferon Cytokine Res. 2013;33(12):728–738. doi: 10.1089/jir.2013.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H, Guariglia S, Yu RY, Li W, Brancho D, Peinado H, Lyden D, Salzer J, Bennett C, Chow CW. Mutation of SIMPLE in Charcot-Marie-Tooth 1C alters production of exosomes. Mol Biol Cell. 2013;24(11):1619–1637. doi: 10.1091/mbc.E12-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399(3):384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Hernandez D, Gutierrez-Vazquez C, Jorge I, Lopez-Martin S, Ursa A, Sanchez-Madrid F, Vazquez J, Yanez-Mo M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288(17):11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gauvreau ME, Cote MH, Bourgeois-Daigneault MC, Rivard LD, Xiu F, Brunet A, Shaw A, Steimle V, Thibodeau J. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic. 2009;10(10):1518–1527. doi: 10.1111/j.1600-0854.2009.00948.x. [DOI] [PubMed] [Google Scholar]
- 53.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18(4):1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol Biol Cell. 2008;19(2):754–763. doi: 10.1091/mbc.E07-09-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16(2):160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 56.Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281(18):12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 57.Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood Cells Mol Dis. 2005;35(3):398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Mazurov D, Barbashova L, Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013;280(5):1200–1213. doi: 10.1111/febs.12110. [DOI] [PubMed] [Google Scholar]
- 59.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuyama K, Sun H, Mitsutake S, Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J Biol Chem. 2012;287(14):10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 62.Stuffers S, Sem Wegner C, Stenmark H, Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10(7):925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 63.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 64.Alonso R, Rodriguez MC, Pindado J, Merino E, Merida I, Izquierdo M. Diacylglycerol kinase alpha regulates the secretion of lethal exosomes bearing Fas ligand during activation-induced cell death of T lymphocytes. J Biol Chem. 2005;280(31):28439–28450. doi: 10.1074/jbc.M501112200. [DOI] [PubMed] [Google Scholar]
- 65.Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, Weinhage T, Menck K, Hupfeld T, Koch R, Trumper L, Wulf GG. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci USA. 2011;108(37):15336–15341. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bollag WB, Xie D, Zheng X, Zhong X. A potential role for the phospholipase D2-aquaporin-3 signaling module in early keratinocyte differentiation: production of a phosphatidylglycerol signaling lipid. J Investig Dermatol. 2007;127(12):2823–2831. doi: 10.1038/sj.jid.5700921. [DOI] [PubMed] [Google Scholar]
- 67.Kajimoto T, Okada T, Miya S, Zhang L, Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat Commun. 2013;4:2712. doi: 10.1038/ncomms3712. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392(6672):193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 69.Laulagnier K, Grand D, Dujardin A, Hamdi S, Vincent-Schneider H, Lankar D, Salles JP, Bonnerot C, Perret B, Record M. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004;572(1–3):11–14. doi: 10.1016/j.febslet.2004.06.082. [DOI] [PubMed] [Google Scholar]
- 70.Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, Parton RG, Vilbois F, Gruenberg J. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303(5657):531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 71.Strauss K, Goebel C, Runz H, Mobius W, Weiss S, Feussner I, Simons M, Schneider A. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J Biol Chem. 2010;285(34):26279–26288. doi: 10.1074/jbc.M110.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno-Gonzalo O, Villarroya-Beltri C, Sanchez-Madrid F. Post-translational modifications of exosomal proteins. Front Immunol. 2014;5:383. doi: 10.3389/fimmu.2014.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burke MC, Oei MS, Edwards NJ, Ostrand-Rosenberg S, Fenselau C. Ubiquitinated proteins in exosomes secreted by myeloid-derived suppressor cells. J Proteome Res. 2014;13(12):5965–5972. doi: 10.1021/pr500854x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alegre E, Rebmann V, Lemaoult J, Rodriguez C, Horn PA, Diaz-Lagares A, Echeveste JI, Gonzalez A. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur J Immunol. 2013;43(7):1933–1939. doi: 10.1002/eji.201343318. [DOI] [PubMed] [Google Scholar]
- 76.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109(11):4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ikeda M, Longnecker R. Cholesterol is critical for Epstein-Barr virus latent membrane protein 2A trafficking and protein stability. Virology. 2007;360(2):461–468. doi: 10.1016/j.virol.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Putz U, Howitt J, Doan A, Goh CP, Low LH, Silke J, Tan SS. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal. 2012;5(243):ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 79.Zuccato E, Blott EJ, Holt O, Sigismund S, Shaw M, Bossi G, Griffiths GM. Sorting of Fas ligand to secretory lysosomes is regulated by mono-ubiquitylation and phosphorylation. J Cell Sci. 2007;120(Pt 1):191–199. doi: 10.1242/jcs.03315. [DOI] [PubMed] [Google Scholar]
- 80.Liu Y, Shah SV, Xiang X, Wang J, Deng ZB, Liu C, Zhang L, Wu J, Edmonds T, Jambor C, Kappes JC, Zhang HG. COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting. Am J Pathol. 2009;174(4):1415–1425. doi: 10.2353/ajpath.2009.080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bourgeois-Daigneault MC, Thibodeau J. Identification of a novel motif that affects the conformation and activity of the MARCH1 E3 ubiquitin ligase. J Cell Sci. 2013;126(Pt 4):989–998. doi: 10.1242/jcs.117804. [DOI] [PubMed] [Google Scholar]
- 82.Geminard C, De Gassart A, Blanc L, Vidal M. Degradation of AP2 during reticulocyte maturation enhances binding of hsc70 and Alix to a common site on TFR for sorting into exosomes. Traffic. 2004;5(3):181–193. doi: 10.1111/j.1600-0854.2004.0167.x. [DOI] [PubMed] [Google Scholar]
- 83.Bourgeois-Daigneault MC, Thibodeau J. Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J Immunol. 2012;188(10):4959–4970. doi: 10.4049/jimmunol.1102708. [DOI] [PubMed] [Google Scholar]
- 84.Huebner AR, Cheng L, Somparn P, Knepper MA, Fenton RA, Pisitkun T. Deubiquitylation of protein cargo is not an essential step in exosome formation. Mol Cell Proteom. 2016;15(5):1556–1571. doi: 10.1074/mcp.M115.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srikanthan S, Li W, Silverstein RL, McIntyre TM. Exosome poly-ubiquitin inhibits platelet activation, downregulates CD36 and inhibits pro-atherothombotic cellular functions. J Thromb Haemost. 2014;12(11):1906–1917. doi: 10.1111/jth.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125(Pt 3):539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 87.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 88.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 89.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32(4):552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Leon S, Raposo G, Reggiori F, Haguenauer-Tsapis R. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol Biol Cell. 2012;23(11):2170–2183. doi: 10.1091/mbc.E11-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6(8):610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 92.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 93.Huang F, Gu H. Negative regulation of lymphocyte development and function by the Cbl family of proteins. Immunol Rev. 2008;224:229–238. doi: 10.1111/j.1600-065X.2008.00655.x. [DOI] [PubMed] [Google Scholar]
- 94.Thien CB, Langdon WY. c-Cbl: a regulator of T cell receptor-mediated signalling. Immunol Cell Biol. 1998;76(5):473–482. doi: 10.1046/j.1440-1711.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 95.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78(3):1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jabbour M, Campbell EM, Fares H, Lybarger L. Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J Immunol. 2009;183(10):6500–6512. doi: 10.4049/jimmunol.0901521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21(5):727–740. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 99.Ohtake F, Saeki Y, Sakamoto K, Ohtake K, Nishikawa H, Tsuchiya H, Ohta T, Tanaka K, Kanno J. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 2015;16(2):192–201. doi: 10.15252/embr.201439152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14(4–5):513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 101.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20(2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schey KL, Luther JM, Rose KL. Proteomics characterization of exosome cargo. Methods. 2015;87:75–82. doi: 10.1016/j.ymeth.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marimpietri D, Petretto A, Raffaghello L, Pezzolo A, Gagliani C, Tacchetti C, Mauri P, Melioli G, Pistoia V. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS One. 2013;8(9):e75054. doi: 10.1371/journal.pone.0075054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vijayakumaran S, Wong MB, Antony H, Pountney DL. Direct and/or indirect roles for SUMO in modulating alpha-synuclein toxicity. Biomolecules. 2015;5(3):1697–1716. doi: 10.3390/biom5031697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kunadt M, Eckermann K, Stuendl A, Gong J, Russo B, Strauss K, Rai S, Kugler S, Falomir Lockhart L, Schwalbe M, Krumova P, Oliveira LM, Bahr M, Mobius W, Levin J, Giese A, Kruse N, Mollenhauer B, Geiss-Friedlander R, Ludolph AC, Freischmidt A, Feiler MS, Danzer KM, Zweckstetter M, Jovin TM, Simons M, Weishaupt JH, Schneider A. Extracellular vesicle sorting of alpha-synuclein is regulated by sumoylation. Acta Neuropathol. 2015;129(5):695–713. doi: 10.1007/s00401-015-1408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Szabo-Taylor K, Ryan B, Osteikoetxea X, Szabo TG, Sodar B, Holub M, Nemeth A, Paloczi K, Pallinger E, Winyard P, Buzas EI. Oxidative and other posttranslational modifications in extracellular vesicle biology. Semin Cell Dev Biol. 2015;40:8–16. doi: 10.1016/j.semcdb.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 108.Biasutto L, Chiechi A, Couch R, Liotta LA, Espina V. Retinal pigment epithelium (RPE) exosomes contain signaling phosphoproteins affected by oxidative stress. Exp Cell Res. 2013;319(13):2113–2123. doi: 10.1016/j.yexcr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valapala M, Vishwanatha JK. Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J Biol Chem. 2011;286(35):30911–30925. doi: 10.1074/jbc.M111.271155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chiarini A, Armato U, Gardenal E, Gui L, Dal Pra I. Amyloid beta-exposed human astrocytes overproduce phospho-tau and overrelease it within exosomes, effects suppressed by calcilytic NPS 2143-further implications for Alzheimer’s therapy. Front Neurosci. 2017;11:217. doi: 10.3389/fnins.2017.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kapogiannis D, Boxer A, Schwartz JB, Abner EL, Biragyn A, Masharani U, Frassetto L, Petersen RC, Miller BL, Goetzl EJ. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer’s disease. FASEB J. 2015;29(2):589–596. doi: 10.1096/fj.14-262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kore RA, Abraham EC. Phosphorylation negatively regulates exosome mediated secretion of cryAB in glioma cells. Biochim Biophys Acta. 2016;1863(2):368–377. doi: 10.1016/j.bbamcr.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Polanco JC, Scicluna BJ, Hill AF, Gotz J. Extracellular vesicles isolated from the brains of rTg4510 mice seed tau protein aggregation in a threshold-dependent manner. J Biol Chem. 2016;291(24):12445–12466. doi: 10.1074/jbc.M115.709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rojas-Vega L, Jimenez-Vega AR, Bazua-Valenti S, Arroyo-Garza I, Jimenez JV, Gomez-Ocadiz R, Carrillo-Perez DL, Moreno E, Morales-Buenrostro LE, Alberu J, Gamba G. Increased phosphorylation of the renal Na+-Cl− cotransporter in male kidney transplant recipient patients with hypertension: a prospective cohort. Am J Physiol Renal Physiol. 2015;309(10):F836–F842. doi: 10.1152/ajprenal.00326.2015. [DOI] [PubMed] [Google Scholar]
- 115.Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NC, Hall GF. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287(6):3842–3849. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Dommelen SM, van der Meel R, van Solinge WW, Coimbra M, Vader P, Schiffelers RM. Cetuximab treatment alters the content of extracellular vesicles released from tumor cells. Nanomedicine (Lond) 2016;11(8):881–890. doi: 10.2217/nnm-2015-0009. [DOI] [PubMed] [Google Scholar]
- 117.Winston CN, Goetzl EJ, Akers JC, Carter BS, Rockenstein EM, Galasko D, Masliah E, Rissman RA. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement (Amst) 2016;3:63–72. doi: 10.1016/j.dadm.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Surgucheva I, Sharov VS, Surguchov A. Gamma-synuclein: seeding of alpha-synuclein aggregation and transmission between cells. Biochemistry. 2012;51(23):4743–4754. doi: 10.1021/bi300478w. [DOI] [PubMed] [Google Scholar]
- 119.Batista BS, Eng WS, Pilobello KT, Hendricks-Munoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. J Proteome Res. 2011;10(10):4624–4633. doi: 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Escrevente C, Grammel N, Kandzia S, Zeiser J, Tranfield EM, Conradt HS, Costa J. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS One. 2013;8(10):e78631. doi: 10.1371/journal.pone.0078631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krishnamoorthy L, Bess JW, Jr, Preston AB, Nagashima K, Mahal LK. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat Chem Biol. 2009;5(4):244–250. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Palmisano G, Jensen SS, Le Bihan MC, Laine J, McGuire JN, Pociot F, Larsen MR. Characterization of membrane-shed microvesicles from cytokine-stimulated beta-cells using proteomics strategies. Mol Cell Proteom. 2012;11(8):230–243. doi: 10.1074/mcp.M111.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5(6):e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286(16):14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rabesandratana H, Toutant JP, Reggio H, Vidal M. Decay-accelerating factor (CD55) and membrane inhibitor of reactive lysis (CD59) are released within exosomes during in vitro maturation of reticulocytes. Blood. 1998;91(7):2573–2580. [PubMed] [Google Scholar]
- 126.Murrow L, Malhotra R, Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol. 2015;17(3):300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oeste CL, Pinar M, Schink KO, Martinez-Turrion J, Stenmark H, Penalva MA, Perez-Sala D. An isoprenylation and palmitoylation motif promotes intraluminal vesicle delivery of proteins in cells from distant species. PLoS One. 2014;9(9):e107190. doi: 10.1371/journal.pone.0107190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sanchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Z, Zhuang M, Zhang L, Zheng X, Yang P, Li Z. Acetylation modification regulates GRP78 secretion in colon cancer cells. Sci Rep. 2016;6:30406. doi: 10.1038/srep30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gasecka A, Boing AN, Filipiak KJ, Nieuwland R. Platelet extracellular vesicles as biomarkers for arterial thrombosis. Platelets. 2017;28(3):228–234. doi: 10.1080/09537104.2016.1254174. [DOI] [PubMed] [Google Scholar]
- 131.Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekstrom K, Wang X, Principe S, Shah N, Ashraf NM, Fatima F, Neder L, Kislinger T. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11(12):688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 132.Perez-Hernandez J, Cortes R. Extracellular vesicles as biomarkers of systemic lupus erythematosus. Dis Markers. 2015;2015:613536. doi: 10.1155/2015/613536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sadovska L, Eglitis J, Line A. Extracellular vesicles as biomarkers and therapeutic targets in breast cancer. Anticancer Res. 2015;35(12):6379–6390. [PubMed] [Google Scholar]
- 134.Nolte-’t Hoen E, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci USA. 2016;113(33):9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martin-Jaular L, Nakayasu ES, Ferrer M, Almeida IC, Del Portillo HA. Exosomes from Plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PLoS One. 2011;6(10):e26588. doi: 10.1371/journal.pone.0026588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smith VL, Jackson L, Schorey JS. Ubiquitination as a mechanism to transport soluble mycobacterial and eukaryotic proteins to exosomes. J Immunol. 2015;195(6):2722–2730. doi: 10.4049/jimmunol.1403186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cheng Y, Schorey JS. Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol Bioeng. 2016;113(6):1315–1324. doi: 10.1002/bit.25884. [DOI] [PubMed] [Google Scholar]
- 138.Gyorgy B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25(6):1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Soria FN, Pampliega O, Bourdenx M, Meissner WG, Bezard E, Dehay B. Exosomes, an unmasked culprit in neurodegenerative diseases. Front Neurosci. 2017;11:26. doi: 10.3389/fnins.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 142.Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166–8172. doi: 10.1074/jbc.M114.621383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sola RJ, Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs. 2010;24(1):9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ahmed SF, Das N, Sarkar M, Chatterjee U, Chatterjee S, Ghosh MK. Exosome-mediated delivery of the intrinsic C-terminus domain of PTEN protects it from proteasomal degradation and ablates tumorigenesis. Mol Ther. 2015;23(2):255–269. doi: 10.1038/mt.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles. 2016;5:31053. doi: 10.3402/jev.v5.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heider S, Dangerfield JA, Metzner C. Biomedical applications of glycosylphosphatidylinositol-anchored proteins. J Lipid Res. 2016;57(10):1778–1788. doi: 10.1194/jlr.R070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37(Pt 5):937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 148.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 149.Hendriks IA, Vertegaal AC. A high-yield double-purification proteomics strategy for the identification of SUMO sites. Nat Protoc. 2016;11(9):1630–1649. doi: 10.1038/nprot.2016.082. [DOI] [PubMed] [Google Scholar]
- 150.Pocsfalvi G, Stanly C, Fiume I, Vekey K. Chromatography and its hyphenation to mass spectrometry for extracellular vesicle analysis. J Chromatogr A. 2016;1439:26–41. doi: 10.1016/j.chroma.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 151.Pocsfalvi G, Stanly C, Vilasi A, Fiume I, Capasso G, Turiak L, Buzas EI, Vekey K. Mass spectrometry of extracellular vesicles. Mass Spectrom Rev. 2016;35(1):3–21. doi: 10.1002/mas.21457. [DOI] [PubMed] [Google Scholar]
- 152.Dorayappan KD, Wallbillich JJ, Cohn DE, Selvendiran K. The biological significance and clinical applications of exosomes in ovarian cancer. Gynecol Oncol. 2016;142(1):199–205. doi: 10.1016/j.ygyno.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Santangelo L, Battistelli C, Montaldo C, Citarella F, Strippoli R, Cicchini C. Functional roles and therapeutic applications of exosomes in hepatocellular carcinoma. Biomed Res Int. 2017;2017:2931813. doi: 10.1155/2017/2931813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cloutier N, Tan S, Boudreau LH, Cramb C, Subbaiah R, Lahey L, Albert A, Shnayder R, Gobezie R, Nigrovic PA, Farndale RW, Robinson WH, Brisson A, Lee DM, Boilard E. The exposure of autoantigens by microparticles underlies the formation of potent inflammatory components: the microparticle-associated immune complexes. EMBO Mol Med. 2013;5(2):235–249. doi: 10.1002/emmm.201201846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 156.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 157.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864(10):1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 158.Bedford MT. Methylation of proteins. Encyclopedic reference of genomics and proteomics in molecular medicine. Berlin: Springer; 2006. [Google Scholar]
- 159.Lanouette S, Mongeon V, Figeys D, Couture JF. The functional diversity of protein lysine methylation. Mol Syst Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28(6):329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schöneich C, Barrón LB. Posttranslational oxidative modifications of proteins. Encycl Anal Chem. 2006 [Google Scholar]
- 162.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58(5–6):737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem Res Toxicol. 1996;9(5):836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 164.Nakamura T, Lipton SA. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011;18(9):1478–1486. doi: 10.1038/cdd.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang YS, Wang CC, Chen BH, Hou YH, Hung KS, Mao YC. Tyrosine sulfation as a protein post-translational modification. Molecules. 2015;20(2):2138–2164. doi: 10.3390/molecules20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 167.Clark BF, Marcker KA. The role of N-formyl-methionyl-sRNA in protein biosynthesis. J Mol Biol. 1966;17(2):394–406. doi: 10.1016/S0022-2836(66)80150-X. [DOI] [PubMed] [Google Scholar]
- 168.Wisniewski JR, Zougman A, Mann M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36(2):570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7(1):58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteom. 2011;10(12):M111 012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8(11):765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 173.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21(5):576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 174.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43R–56R. doi: 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- 175.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33(3):151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 176.Galian C, Bjorkholm P, Bulleid N, von Heijne G. Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J Biol Chem. 2012;287(20):16399–16409. doi: 10.1074/jbc.M112.350009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol Pharm Bull. 2002;25(4):409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- 178.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5(2):110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- 179.Nadolski MJ, Linder ME. Protein lipidation. FEBS J. 2007;274(20):5202–5210. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- 180.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451(1):1–16. doi: 10.1016/S0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 181.Triola G. The protein lipidation and its analysis. J Glycom Lipidom S. 2011;2:001. [Google Scholar]
- 182.Roskoski R., Jr Protein prenylation: a pivotal posttranslational process. Biochem Biophys Res Commun. 2003;303(1):1–7. doi: 10.1016/S0006-291X(03)00323-1. [DOI] [PubMed] [Google Scholar]
- 183.van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283(7):3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- 184.Bre MH, Redeker V, Vinh J, Rossier J, Levilliers N. Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in paramecium. Mol Biol Cell. 1998;9(9):2655–2665. doi: 10.1091/mbc.9.9.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bre MH, Levilliers N, Gorovsky MA, Gaertig J. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila . J Cell Biol. 2000;149(5):1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Baka Z, Gyorgy B, Geher P, Buzas EI, Falus A, Nagy G. Citrullination under physiological and pathological conditions. Jt Bone Spine. 2012;79(5):431–436. doi: 10.1016/j.jbspin.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 187.Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI. Citrullination: a posttranslational modification in health and disease. Int J Biochem Cell Biol. 2006;38(10):1662–1677. doi: 10.1016/j.biocel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 188.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 189.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31(1):119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22(9):1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu C, Yu X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr Protein Pept Sci. 2015;16(6):491–501. doi: 10.2174/1389203716666150504122435. [DOI] [PMC free article] [PubMed] [Google Scholar]