Abstract
Cell stress such as hypoxia elicits adaptive responses, also on the level of mitochondria, and in part is mediated by the hypoxia-inducible factor (HIF) 1α. Adaptation of mitochondria towards acute hypoxic conditions is reasonably well understood, while regulatory mechanisms, especially of respiratory chain assembly factors, under chronic hypoxia remains elusive. One of these assembly factors is transmembrane protein 126B (TMEM126B). This protein is part of the mitochondrial complex I assembly machinery. We identified changes in complex I abundance under chronic hypoxia, in association with impaired substrate-specific mitochondrial respiration. Complexome profiling of isolated mitochondria of the human leukemia monocytic cell line THP-1 revealed HIF-1α-dependent deficits in complex I assembly and mitochondrial complex I assembly complex (MCIA) abundance. Of all mitochondrial MCIA members, we proved a selective HIF-1-dependent decrease of TMEM126B under chronic hypoxia. Mechanistically, HIF-1α induces the E3-ubiquitin ligase F-box/WD repeat-containing protein 1A (β-TrCP1), which in turn facilitates the proteolytic degradation of TMEM126B. Attenuating a functional complex I assembly appears critical for cellular adaptation towards chronic hypoxia and is linked to destruction of the mitochondrial assembly factor TMEM126B.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2779-y) contains supplementary material, which is available to authorized users.
Keywords: Hypoxia-inducible factor (HIF), Complexome profiling, AKT, β-TrCP, MCIA, Mitochondrial respiration
Introduction
Diabetes, autoimmune disorders, or cancer are characterized by hypoxic episodes imposing a stress signal to cells [1–4]. Hypoxia arises when the oxygen demand exceeds oxygen supply, with the severity of hypoxia ranging from acute to chronic states [5]. In contrast to acute hypoxia, the term chronic hypoxia is not well defined towards underlying molecular or biochemical alterations [6]. Previously, we defined chronic hypoxia as a situation when hypoxia-inducible factor (HIF)-1α and HIF-2α levels decrease from initial induction to levels slightly above normoxia [7]. HIF consists of a regulated alpha subunit and a constitutively expressed beta subunit. Under hypoxia, the alpha subunits escape proteasomal degradation, translocate to the nucleus, and dimerize with the beta subunit to enhance transcription of target genes [8, 9]. Under ambient oxygen level, the alpha subunits get hydroxylated by prolyl hydroxylase (PHD) enzymes, which bind the von Hippel–Lindau protein, followed by their degradation, preventing target gene induction [10, 11]. Thus, decreased HIF-1α accumulation under chronic vs. acute hypoxia can partially be explained by feedback regulatory mechanisms comprising PHD2 abundance and activity [12, 13]. Moreover, under chronic hypoxia secondary, HIF-unrelated pathways are operating that affect mRNA and/or protein stability [14].
At the cellular level, mitochondria are central to several anabolic and catabolic pathways, in part linked to their function in generating ATP by oxidative phosphorylation (OXPHOS). The ATP synthase uses the mitochondrial membrane potential to generate ATP. Electrons from NADH enter the respiratory chain at complex I (NADH:quinone oxidoreductase) and are passed step-wise by mobile redox carriers to complex III (ubiquinol-cytochrome c oxidoreductase), complex IV (cytochrome c oxidase), and molecular oxygen [15, 16]. Thus, hypoxic adaptation of these organelles appears rational. Hypoxia reduces the mitochondrial mass but also alters protein composition of respiratory chain complexes, presumably to avoid a potential hazardous formation of superoxide anions [7, 17–20]. Composition of complex IV is adjusted by an HIF-1-dependent increase of the cytochrome c oxidase subunit (COX) 4-2, which optimizes complex IV activity for hypoxic conditions, and degradation of COX4-1 [21]. In contrast, complex I activity is reduced by an HIF-1-driven induction of NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4-like 2 (NDUFA4L2) [22]. Interestingly, NDUFA4L2 was also shown to reduce oxidative stress and protect against ischemia–reperfusion injury [23–25]. In addition, metabolic changes like HIF-1-dependent upregulation of pyruvate dehydrogenase kinase 1 followed by a decreased citric acid cycle activity were shown to reduce mitochondrial respiration after 24 h of hypoxic incubation [26]. In many organisms, complexes I, III, and IV are organized in catalytically active stoichiometric associations called supercomplexes that stabilize individual complexes, exhibit narrow substrate flow, and are suggested to prevent superoxide radical formation [27–31]. Complex I as a crucial electron acceptor requires a precise and coordinated assembly of all its 45 proteins to a functional complex [16, 32–34]. Coordinated by assembly factors, the generation of complex I starts with formation of protein subcomplexes, which are step-wise recruited, resulting in a mature complex I [17, 33, 35, 36]. A defect in one of the assembly factors results in accumulation of intermediates of preassembled modules: (i) the N-module (NADH:dehydrogenase module) that binds NADH and transfers electrons via proteins that contain FMN and FeS clusters, (ii) the Q module that contains iron sulfur proteins to transfer electrons to ubiquinone, and (iii) the P module as a membrane embedded part that facilitates proton pumping [37]. The assembly factors acyl-CoA dehydrogenase family member 9 (ACAD9), evolutionarily conserved signaling intermediate in toll pathway (ECSIT), complex I intermediate-associated protein 30 (NDUFAF1, CIA30), and transmembrane protein 126B (complex I assembly factor TMEM126B, TMEM126B) themselves compose the mitochondrial complex I assembly complex (MCIA) that facilitates membrane arm assembly; and together with translocase of inner mitochondrial membrane domain-containing protein 1 (complex I assembly factor TIMMDC1, TIMMDC1), the connection with the intermediate of the Q module that contains iron sulfur proteins [38–40].
Recent work with complex I defects increased our understanding of the complex I assembly sequence and factors involved in this procedure [33]. Regulation of complex I assembly in response to variable physiologic conditions such as substrate or oxygen supply has not been studied in detail. This also includes the challenge to control expression and stability of nuclear and mitochondrial encoded complex I subunits and assembly factors.
Here, we show that HIF-1 reduces complex I assembly and function by upregulating F-box/WD repeat-containing protein 1A (β-TrCP1), which degrades complex I assembly factor TMEM126B under chronic hypoxia and consequently prevents formation of the mitochondrial complex I assembly complex (MCIA).
Results
Decreased mitochondrial respiration under chronic hypoxia
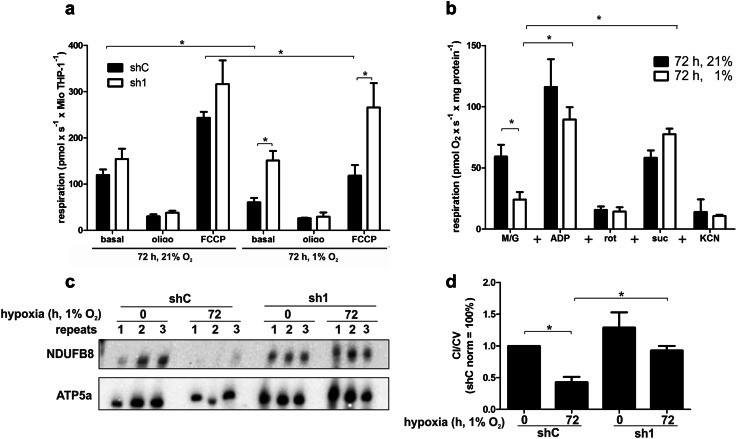
While the decrease of mitochondrial mass under chronic hypoxia is well established, the previous studies did not focus on mitochondrial activity. Therefore, we monitored oxygen consumption in the human leukemia monocytic cell line THP-1 cultured under normoxia vs. hypoxia. A chronic hypoxic episode was established by cultivating cells for 72 h at 1% oxygen [7]. To explore the role of HIF-1 and HIF-2, experiments were performed in cells with a knockdown of HIF-1α (sh1) and HIF-2α (sh2) compared to control cells (shC) (Online Resource 1, Fig. S1a). Under normoxia, basal respiration was comparable in shC and sh1 (Fig. 1a). Inhibition of ATP synthase with oligomycin (oligo) decreased oxygen consumption in both clones, while the uncoupler carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), added to determine the maximal oxygen consumption capacity, increased cellular respiration with no significant difference between control and knockdown cells. However, incubating THP-1 cells for 72 h under hypoxia (1% O2) significantly decreased basal oxygen consumption in shC, but not in sh1 cells. Respiration of sh1 cells was still comparable to normoxia. In contrast, sh2 cells behaved similar to shC except for a higher respiratory rate after FCCP treatment under normoxic conditions (Online Resource 1, Fig. S1b). Under chronic hypoxia oligomycin also reduced respiration, while FCCP enhanced oxygen consumption in shC cells to rates observed under basal, but not FCCP-treated, normoxic conditions (Fig. 1a). Interestingly, FCCP significantly increased respiration in sh1 cells, making it comparable to maximal rates observed under normoxia. These results indicate that the function of the respiratory chain is subjected to regulation by HIF-1α under hypoxia.
Fig. 1.
HIF-1 regulates complex I function and abundance under chronic hypoxia. a High-resolution respirometry of THP-1 control (shC) or HIF-1α knockdown (sh1) cells cultured for 72 h at 21 or 1% oxygen. 2 µg/ml oligomycin (oligo) and 1 µM FCCP were added to determine fractions of ATP producing (state 3 respiration) and maximal oxygen consumption rates (n = 4). b THP-1 cells were cultured for 72 h at 21 or 1% oxygen. Mitochondria were isolated and analyzed by respirometry (n = 3). Mitochondria were subsequently supplemented with malate/glutamate (M/G, 5 mM), ADP (2 mM), rotenone (rot, 5 µM), succinate (suc, 5 mM), and potassium cyanide (KCN, 2 mM). c Mitochondria from shC to sh1 cells grown under normoxic and hypoxic conditions were solubilized with dodecylmaltoside and resolved by blue native electrophoresis (BNE). BNE-immunoblots for complex I (anti NDUFB8) and V (anti ATP5a) were performed. d Blots from c were quantified (n = 6). All data are shown as mean values ± SEM, *p < 0.05
Complex I accounts for decreased oxygen consumption under hypoxia
To determine how HIF-1α reduced mitochondrial oxygen consumption under chronic hypoxia, we asked which of the respiratory chain complexes might be affected. Mitochondria were isolated from normoxic and hypoxic THP-1 cells, to analyze their substrate-specific respiration. The basal complex I-dependent respiratory activity, determined in the presence of malate and glutamate (M/G), revealed reduced oxygen consumption in mitochondria isolated from hypoxic vs. normoxic cells (Fig. 1b). The subsequent addition of ADP (state 3 respiration) enhanced oxygen consumption in mitochondria from normoxic to hypoxic cells, with more pronounced effects in normoxic mitochondria, while inhibition by rotenone (rot) shut down complex I-dependent respiration to similar levels in both conditions. However, further addition of succinate (suc) restored respiration by feeding electrons into the respiratory chain via complex II, indicating that the respiratory chain downstream of complex I was not affected by hypoxia. Interestingly, hypoxic mitochondria supplemented with suc had a higher respiratory rate compared to normoxic mitochondria (p = 0.0635), suggesting an enhanced complex II activity as a compensatory mechanism for decreased complex I levels under hypoxia. Finally, the complex IV inhibitor potassium cyanide (KCN) was added, which reduced respiration to levels comparable to rot treated mitochondria. We next asked if altered complex I abundance or stability contributes to the decrease of complex I-dependent respiration under hypoxic conditions. Isolated mitochondria were solubilized with dodecylmaltoside and individual OXPHOS complexes were separated by blue native (BN) gel electrophoresis (BNE) (Fig. 1c, for a complete time course, see Online Resource 1, Fig. S1c). Western analysis of BN gels was performed with antibodies against NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit (NDUFB) 8 (complex I) and ATP synthase subunit alpha (ATP5a) (complex V). Subsequent densitometry demonstrated a significant reduction of total complex I in control cells (shC) under chronic hypoxic conditions (Fig. 1d). Interestingly, attenuated expression of complex I was absent in sh1. Since respirometry and BNE indicate that complex I was the limiting factor for oxygen consumption under chronic hypoxia, it appeared imperative to obtain detailed information about complex I composition.
HIF-1α disturbs complex I assembly under hypoxia
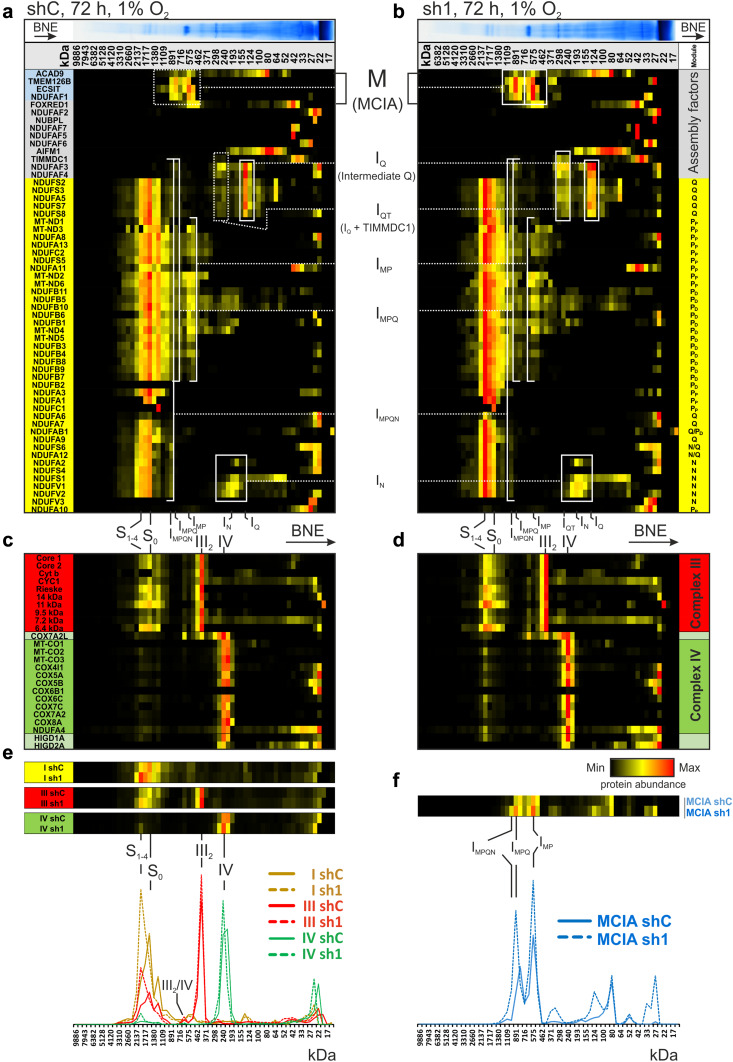
After identifying complex I as a target of HIF-1α, regulating mitochondrial respiration under chronic hypoxia, we studied details on complex I assembly and stability by complexome profiling (Fig. 2, Online Resource 2) [38]. Lower levels of complex I in shC cells (Fig. 2a) compared to HIF-1α knockdown sh1 cells (Fig. 2b) were confirmed by reference profiles of the average of all identified complex I subunits (Fig. 2e).
Fig. 2.
Complexome profiling of chronic hypoxic shC and sh1 cells. a, b Abundance of mitochondrial protein complexes from chronic hypoxic shC and sh1 THP-1 cells was normalized to maximum appearance and depicted in a heatmap, showing one representative experiment. Abundance profiles of subunits of complex I (yellow) from a shC and b sh1 cells were placed to visualize assembly intermediates. Components of MCIA complex (blue) and other assembly factors of complex I (grey) were placed on top. c, d Protein abundance profiles of complex III (red), and complex IV (green) from c shC and d sh1 mitochondria. e Reference profiles (average of subunits) of complex I (yellow), III (red), and IV (green) from shC (continues line) and sh1 mitochondria (dashed line) were plotted to demonstrate supercomplex abundance. f Reference profiles of MCIA from shC (continues line) and sh1 (dashed line). Assignments of complexes: S0, supercomplex containing complex I and III; S1–4, supercomplex containing S0 and 1–4 copies of complex IV; III2, dimer of complex III; IV, complex IV; III2/IV, supercomplex containing complex III and complex IV; M, MCIA or mitochondrial complex I assembly factor complex; IQ, intermediate of Q module; IQT, IQ with bound TIMMDC1; IMP, intermediate containing subunits of P module and MCIA; IMPQ, connection of IMP and IQT; IN, intermediate of N-module; IMPQN, connection of all intermediates still containing assembly factors. Complex I subunit location to P-, Q-, and N-module according to [63]
Interestingly, supercomplexes containing complex I, a dimer of complex III, and 0–4 copies of complex IV (S0, S1–4) are formed in mitochondria under chronic hypoxic condition (Fig. 2a–e). The heatmap clearly demonstrates that in hypoxia, sh1 cells assemble these supercomplexes in a higher abundance compared to cells with normal HIF-1α expression in chronic hypoxia (Fig. 2a–e). Individual complexes III and IV appear at comparable levels (Fig. 2c–e). Interestingly, hypoxia-inducible gene (HIGD) 1A and HIGD2A cluster together with complex IV and confirm recent publications, claiming, and interaction of HIG proteins with complex IV [41–43]. These results confirm an HIF-1α-dependent decrease of complex I.
An intermediate (IQ) containing NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit (NDUFA) 5 and iron–sulfur proteins NADH dehydrogenase [ubiquinone] iron–sulfur protein (NDUFS) 2, NDUFS3, NDUFS7, and NDUFS8 accumulate together with the assembly factors NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor (NDUFAF) 3 and NDUFAF4 to a higher level in shC, indicating that in the presence of HIF-1α, the formation of complex I stalled at an early point in the assembly sequence. Moreover, in sh1 cells, we identified a larger assembly intermediate IQT of the Q module that appears in a mass range between 260 and 300 kDa containing IQ and at least the recently characterized assembly factor TIMMDC1 [40]. The identification of these Q module intermediates suggests that the assembly sequence either is interrupted at this stage in shC, while it is processed further in sh1 cells, or that complex I disintegrates to smaller complexes.
Intermediates of the membrane arm of complex I or P module were identified in both cells. NDUFB1, NDUFB5, NDUFB10, and the mitochondrial encoded subunits NADH-ubiquinone oxidoreductase chain (ND) 4 and ND5 of the distal membrane part co-migrate first as small subcomplexes at ~ 200 kDa and later in intermediate IMP together with additional subunits and the MCIA complex. The largest intermediates IMP, IMPQ, and IMPQN co-migrate with the MCIA complex and exhibit maxima in the native mass region 575, 890, and 990 kDa, respectively (Fig. 2f). A very prominent difference between control and HIF-1α knockdown cells was the abundance of MCIA complex containing ACAD9, NDUFAF1, ECSIT, and TMEM126B (Fig. 2a, b). The MCIA complex, which is essential to connect the preformed IQT and the membrane arm, appears in higher abundance in sh1 cells, suggesting that HIF-1α controls mid-stage complex I assembly (Fig. 2f). For validation, 2D-BNE coupled with Western blotting was performed for the IQ protein NDUFS2 and for NDUFB6 (Online Resource 1, Fig. S2). While NDUFB6 only showed marginal differences between shC and sh1 cells, NDUFS2 accumulated in lower molecular complexes in sh1 cells like seen in the complexome within IQ.
HIF-1α controls complex I formation in chronic hypoxia at the level of MCIA
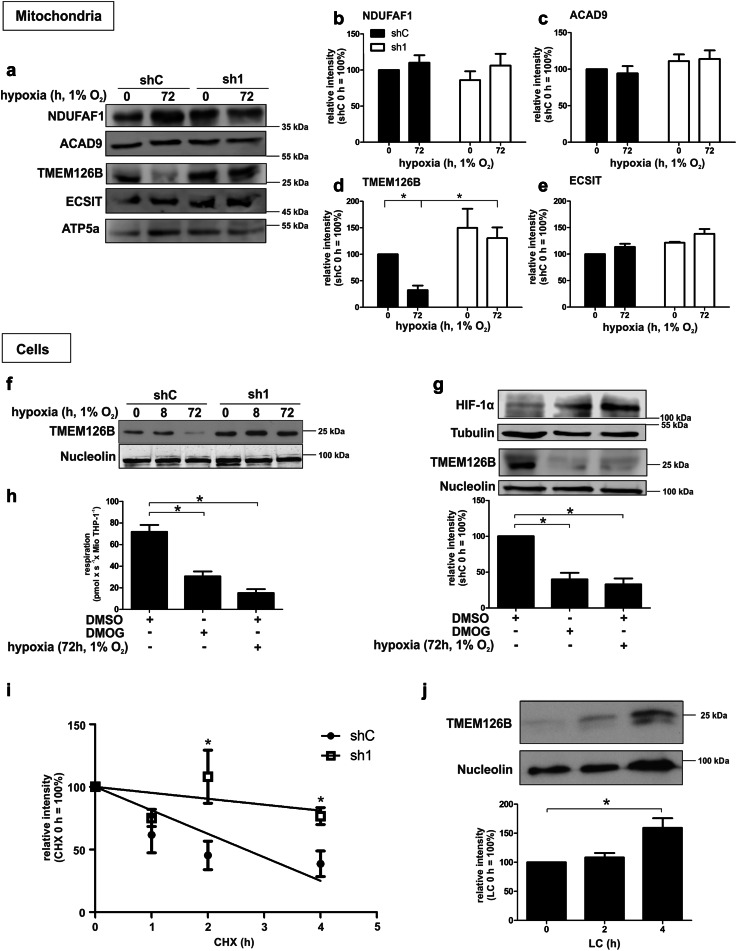
Complexome profiling revealed a decrease of MCIA and especially its component TMEM126B in shC compared to sh1 cells under chronic hypoxic conditions (Online Resource 1, Fig. S3a). Since MCIA is responsible for a substantial part of complex I assembly, we analyzed its members NDUFAF1, ACAD9, ECSIT, and TMEM126B in isolated mitochondria by Western analysis (Fig. 3a), followed by quantification (Fig. 3b–e). In line with complexome data, only TMEM126B showed significant changes in abundance compared to the other MCIA members. The protein decreased in control but not in sh1 cells after 72 h of hypoxia pointed to an HIF-1 dependence. To elucidate TMEM126B regulation at the protein level in total cell lysate under chronic hypoxic conditions, we performed Western analysis in shC vs. sh1 cells, incubated under acute (8 h) and chronic (72 h) hypoxia (Fig. 3f, for a complete time course, see Online Resource 1, Fig. S3b). In shC cells, TMEM126B expression decreased under chronic hypoxic conditions (72 h). However, the decrease was not evident in the absence of HIF-1α, i.e., sh1 cells. To validate the role of hydroxylases and thus HIF in affecting TMEM126B abundance, we treated cells with the PHD inhibitor dimethyloxalylglycine (DMOG), and dimethylsulfoxide (DMSO) as a solvent control (Fig. 3g). DMOG, used to stabilize and activate HIF-1, decreased TMEM126B expression as effectively as incubating cells for 72 h under hypoxia. In line, respirometry showed decreased oxygen consumption in DMOG-treated and hypoxic cells, validating that respiration under hypoxia is reduced in a hydroxylase- and likely HIF-dependent manner (Fig. 3h). Interestingly, mRNA expression of TMEM126B analyzed by qPCR revealed comparable values under acute and chronic hypoxia in shC or sh1 cells, excluding a transcriptional regulatory component (Online Resource 1, Fig. S3c). Therefore, we suggest that HIF-1α affects either TMEM126B translation or protein stability.
Fig. 3.
Regulation of MCIA and TMEM126B under chronic hypoxia. a Western analysis of MCIA components of isolated mitochondria. b–e Quantification of data from A (n = 3). f Western analysis of TMEM126B in shC and sh1 cells cultured under acute and chronic hypoxia. g Western analysis and quantification of TMEM126B and HIF-1α after exposing THP-1 cells to dimethyloxalylglycine (DMOG, 1 µM) or hypoxia for 72 h (n = 3). h Respirometry of THP-1 cells treated for 72 h with dimethyl sulfoxide (DMSO), dimethyloxalylglycine (DMOG, 1 µM), or 1% oxygen (n = 3). i Half-life determination of TMEM126B in THP-1 cells, incubated for 72 h under hypoxia and treated with cycloheximide (CHX, 10 µg/ml) for indicated times (n = 7). j Western analysis and quantification of TMEM126B in shC THP-1 cells incubated for 72 h under hypoxia and subsequently treated with lactacystin (LC, 10 µM) for indicated times (n = 7). All data are expressed as mean values ± SEM, *p < 0.05
Chronic hypoxia destabilizes TMEM126B
Following the idea that TMEM126B is destabilized in shC cells, we incubated cells for 72 h under hypoxia and added cycloheximide (CHX) for the last 1–4 h to block translation (Fig. 3i). As indicated, the TMEM126B protein half-life is higher in sh1 (t1/2 of 23.1 h) compared to shC cells (t1/2 of 2.5 h), with significant differences after 2 and 4 h of CHX-treatment, pointing to an increased TMEM126B protein stability in sh1 cells. To explore molecular mechanisms of TMEM126B degradation, THP-1 cells were incubated for 72 h under hypoxia, prior to adding the proteasome inhibitor lactacystin (LC) for the last 2 or 4 h. With the proteasome being blocked, TMEM126B protein started to accumulate at 2 h and reached highest levels after 4 h (Fig. 3j). Thus, destabilization of TMEM126B under chronic hypoxia is HIF-1α mediated and facilitated by the proteasome.
β‐TrCP destabilizes TMEM126B
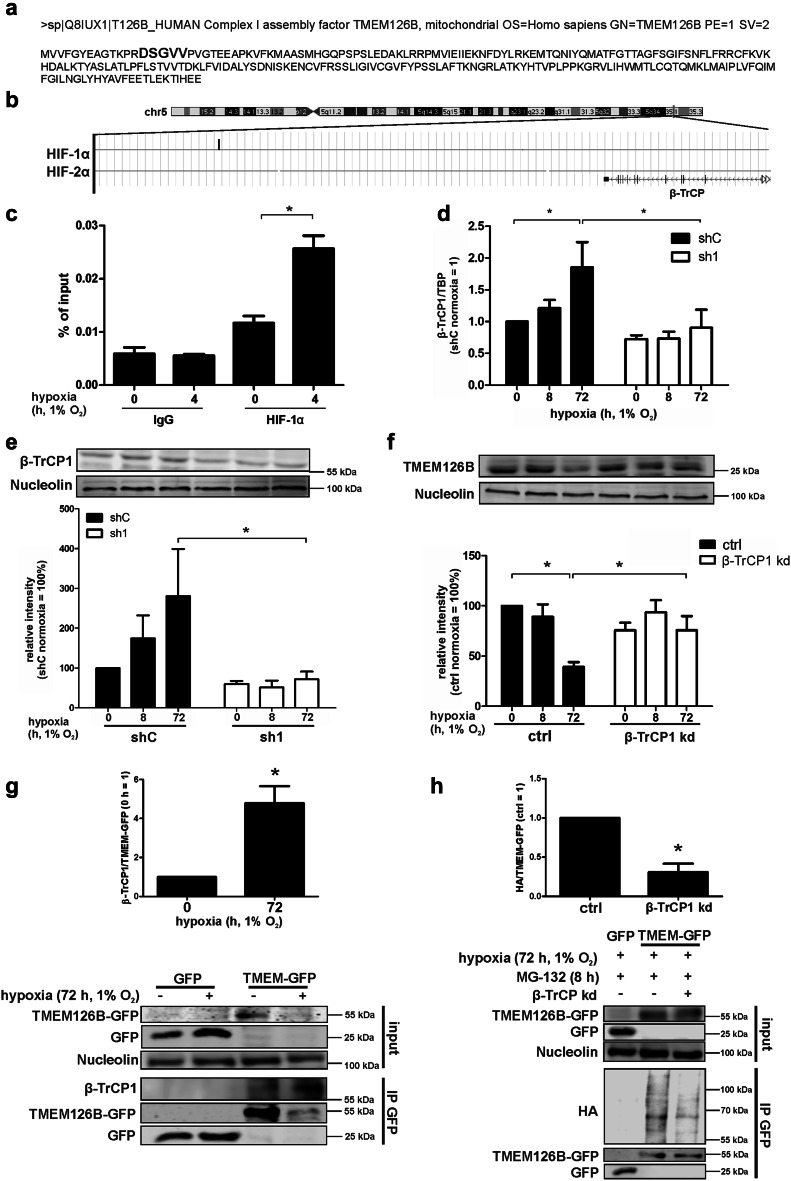
Considering proteasomal degradation of TMEM126B under chronic hypoxia, we searched for detailed regulatory mechanisms. Inspecting the amino acid sequence of TMEM126B, we noticed a motif similar to DpSGXXpS, the binding motif for F-box/WD repeat-containing protein 1A (β‐TrCP1) (Fig. 4a, bolt), pointing to an E3 ubiquitin ligase complex-mediated TMEM126B regulation. Western analysis of the components belonging to the E3-ligase system, cullin4A, DNA damage‐binding protein 1, S-phase kinase-associated protein 1, and β‐TrCP1, showed an increase of cullin4A in sh1 cells, while β‐TrCP1 expression under hypoxia was clearly HIF-1-dependent (for Western analysis of all E3-ligase components, see Online Resource 1, Fig. S4a). Off note, chromatin immune precipitation (ChIP) coupled with deep-sequencing in human macrophages revealed binding of HIF-1α, but not HIF-2α, to the gene coding for β‐TrCP (Fig. 4b) [44]. To validate in silico analysis, we incubated human macrophages for 4 h under hypoxia to allow HIF-binding to the DNA and performed ChIP as described by Tausendschön et al. for HIF-1α, HIF-2α, and IgG as a negative control (Online Resource 1, Fig. S4b). HIF-1 binding was significantly enhanced compared to HIF-2, thus, confirming in silico data. In addition, we performed ChIP analysis for HIF-1α and IgG under normoxic vs. hypoxic conditions in THP-1 and MDA-MB-231 cells, to validate a binding of HIF-1 to β-TrCP1 under hypoxic conditions only (Fig. 4c, Online Resource 1, Fig. S4c).
Fig. 4.
β-TrCP degrades TMEM126B. a Amino acid sequence of TMEM126B, showing the predicted β-TrCP binding motif in bolt. b ChIP-sequencing data showing HIF-1α binding in a region near the gene encoding β-TrCP as visualized by the genome browser. c Validation of HIF-binding to a region near the gene encoding β-TrCP in THP-1 cells after 4 h hypoxia to the region identified in b by ChIP. d qPCR analysis of β-TrCP1 (n = 5). e Western analysis and quantification of β-TrCP1 in control (shC) or HIF-1α knockdown (sh1) cells under normoxia as well as 8 or 72 h hypoxia (n = 4). f For a knockdown of β-TrCP1, MDA-MB-231 cells were treated with IPTG (1 mM) for 3 days and subsequently incubated under normoxia/hypoxia for times indicated to analyze protein TMEM126B (n = 3). g MDA-MB-231 cells expressing GFP or GFP-tagged TMEM126B were incubated for 72 h in hypoxia. GFP was immunoprecipitated and Western analysis was performed for GFP and nucleolin (input) or β-TrCP1 and GFP (IP). Afterwards, the ratio of β-TrCP1 to TMEM126B was calculated (n = 3). h MDA-MB-231 cells with an inducible knockdown of β-TrCP1 and overexpressing GFP or GFP-tagged TMEM126B were transfected with a construct expressing HA-tagged ubiquitin. For a knockdown of β-TrCP1, cells were treated with IPTG (1 mM) for 3 days. Cells were incubated for 72 h in hypoxia and treated with MG-132 8 h prior harvest. GFP was immunoprecipitated and Western analysis were performed for GFP, β-TrCP1 and nucleolin (input) or HA and GFP (IP). Afterwards, the ratio of HA to TMEM126B was calculated (n = 4). All data are expressed as mean values ± SEM, *p < 0.05
Next, we determined a β-TrCP1 mRNA increase after 72 h of hypoxia, which was completely absent in sh1 cells (Fig. 4d). Based on similar mRNA expression in shC and sh1 cells, β-TrCP2 unlikely accounts for HIF-dependent TMEM126B degradation under chronic hypoxia (Online Resource 1, Fig. S4d). Since HIF-1α is predominantly stabilized under acute hypoxia, we postulate secondary HIF-1α-mediated effects, transmitting signals under chronic hypoxic episodes (Online Resource 1, Fig. S1a) [7, 14]. The regulatory feature of β‐TrCP1 became also evident at the protein level (Fig. 4e). Its expression slightly increased under 8 h hypoxia and became significant at 72 h hypoxia in shC cells, while β-TrCP1 abundance remained low in sh1 cells under otherwise identical experimental conditions. Since the impact of hypoxia on TMEM126B expression was conserved in different cell lines, i.e., MCF-7, Hep3B, and MDA-MB-231, we proceeded using MDA-MB-231 cells for mechanistic studies, as these are easier to transfect compared to macrophages (Online Resource 1, Fig. S4e). To analyze the role of β‐TrCP1 for TMEM126B degradation, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible knockdown was created in MDA-MB-231 cells and an efficient knockdown was confirmed by Western analysis (Online Resource 1, Fig. S4f). We then followed TMEM126B expression at 0, 8, and 72 h of hypoxia comparing β‐TrCP knockdown and control cells (Fig. 4f). TMEM126B expression decreased in control cells at 72 h, while its expression remained comparable to values seen under normoxia in β-TrCP1 knockdown cells. These data suggest that β‐TrCP1 regulates TMEM126B expression under hypoxia and, moreover, link this to HIF-1α. A perquisite for β‐TrCP1 to facilitate ubiquitination and subsequent protein degradation is the interaction to its target protein. To verify the interaction between β‐TrCP1 and TMEM126B, we performed immunoprecipitation (IP). Since endogenous TMEM126B levels are low, especially under chronic hypoxia, and do not allow IP, MDA-MB-231 cells were stably transduced to express GFP-tagged TMEM126B (TMEM-GFP) or GFP (Online Resource 1, Fig. S5a, b and c). Western analysis of the cell lysate (input) confirmed degradation of TMEM-GFP under chronic hypoxia (Fig. 4g). Next, an IP of GFP was performed. Corresponding Western analysis confirmed the decrease of TMEM-GFP under hypoxia, as seen in the input samples. Importantly, β‐TrCP1 was found associated with TMEM-GFP but not GFP. Supporting our hypothesis, more β-TrCP1 was pulled down together with TMEM-GFP when cells were incubated under hypoxia compared to normoxia. Normalizing the amount of β‐TrCP1 to immunoprecipitated TMEM126B-GFP revealed a significantly enhanced binding of β‐TrCP1 to TMEM126B under chronic hypoxia (Fig. 4g). To further prove a β‐TrCP1-dependent ubiquitination of TMEM126B, TMEM-GFP overexpressing β‐TrCP1 knockdown cells were transiently transfected with a plasmid expressing HA-tagged ubiquitin (Fig. 4h). Cells were incubated for 72 h under hypoxia and treated with the proteasome inhibitor MG-132 (10 µM) for the last 8 h. The knockdown and an equal input was validated by Western analysis of GFP and β‐TrCP1. Western analysis using an HA antibody revealed increased accumulation of HA-tagged ubiquitin in control cells compared to the β‐TrCP1 knockdown, while no HA-signal was detected in GFP transduced cells. Lanes corresponding to a molecular weight of 55–100 kDa were quantified to detect changes in TMEM126B ubiquitination (Fig. 4h). Since TMEM126B is nuclear encoded and β‐TrCP1-dependent degradation is supposed to occur in the cytoplasm, we fractionated normoxic and chronic hypoxic MDA-MB-231 cells, followed by Western analysis for β‐TrCP1 in the cytosolic (C) vs. mitochondrial (M) fraction (Online Resource 1, Fig. S5d). As to our expectations, β‐TrCP1 predominantly resides in the cytosol. We used ATP5a, respectively, tubulin, to confirm purity of the mitochondrial vs. cytosolic fractions. Taking into consideration that binding of β‐TrCP1 to its potential target sequence requires serine phosphorylation within or in close proximity to the binding motive we asked, which kinases are involved.
TMEM126B is phosphorylated by Akt to facilitate β‐TrCP1 binding
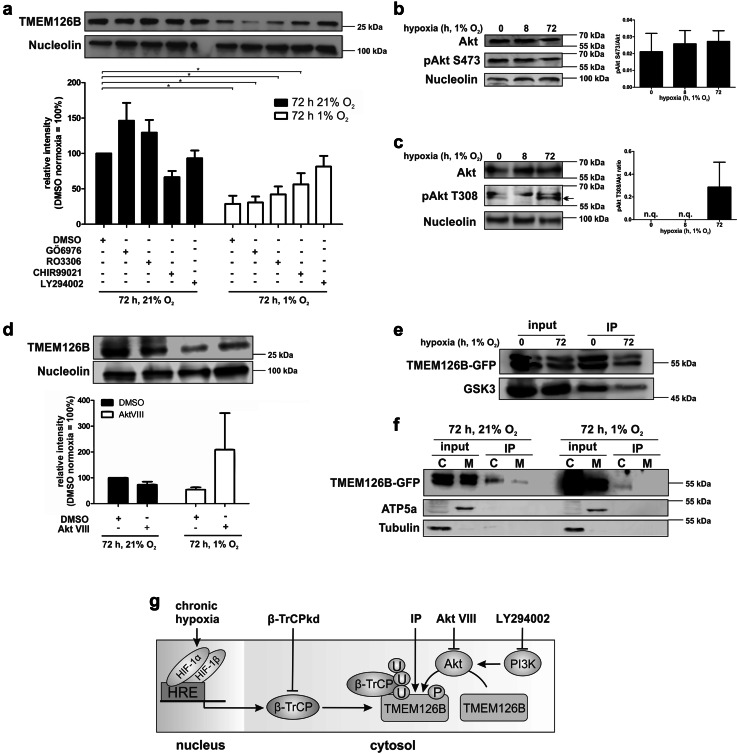
To identify potential kinases responsible for TMEM126B phosphorylation, we blocked protein kinase C with GÖ6976, cyclin-dependent kinase 1 with RO3306, glycogen synthase kinase-3 (GSK3) with CHIR99021, and Phosphatidylinositol 3-kinase (PI3K) with LY294002, while DMSO served as a vehicle control (Fig. 5a).
Fig. 5.
Akt phosphorylates TMEM126B. a MDA-MB-231 cells were incubated with GÖ6976 (10 µM), RO3306 (5 µM), CHIR99021 (1 µM), or LY294002 (15 µM) under normoxic or hypoxic conditions. Western analysis and quantification of TMEM126B normalized to nucleolin (n = 6). b Western analysis of Akt and phospho-Akt pS473 (pAkt S473) under hypoxia and quantification (n = 5). c Western analysis of Akt and phospho-Akt pT308 (pAkt T308) under hypoxia and quantification (n = 3, n.q. not quantifiable). d Western analysis and quantification of TMEM126B in cells treated with Akt VIII (1 µM) and incubated under normoxia or chronic hypoxia (72 h) (n = 4). e Phospho-Akt substrates were immunoprecipitated and analyzed for TMEM126B and GSK3 by Western analysis (n = 3). f Phospho-Akt substrates were immunoprecipitated from mitochondrial (M) and cytosolic (C) fractions and analyzed for TMEM126B, ATP5A, and tubulin. g Proposed pathways indicating TMEM126B degradation, sites of intervention, and methodological aspects. All data are expressed as mean values ± SEM, *p < 0.05
Blocking these kinases under normoxia left TMEM126B protein expression unaltered, with the exception of GSK3 inhibition, which decreased its abundance. Hypoxia decreased TMEM126B expression in DMSO-, GÖ6976-, and RO3306-treated cells. Inhibition of GSK3 increased TMEM126B expression under hypoxia to some extent, an effect that became more pronounced using the PI3K inhibitor. Analyzing the amino acid sequence of TMEM126B using NetPhos 2.0, we predicted PI3K to target RAC-alpha serine/threonine-protein kinase (Akt) as a candidate causing TMEM126B phosphorylation. To ensure that Akt is phosphorylated and thus activated, we analyzed its phosphorylated and total protein amount in MDA-MB-231 cells, revealing a constitutively phosphorylated Akt on serine 473 (Fig. 5b). These results were validated by calculating the pAkt-to-Akt ratio. Interestingly, threonine 308 appears to be phosphorylated under chronic hypoxia and indicates an increased activity of Akt (Fig. 5c) [45]. To estimate the influence of Akt on TMEM126B abundance, we used Akt VIII to block Akt under normoxia and hypoxia (Fig. 5d). Under normoxia Akt VIII slightly decreased TMEM126B expression compared to DMSO controls. After 72 h of hypoxia, TMEM126B expression decreased and inhibition of Akt largely reversed this effect, suggesting that Akt facilitates TMEM126B phosphorylation/degradation. To verify TMEM126B as an Akt substrate, we used antibodies recognizing the pAkt motif to pull down potential pAkt substrates (Fig. 5e, Online Resource 1, S5e). As described, an IP was performed using cells overexpressing GFP-tagged TMEM126B. Following IP of pAkt substrates, TMEM126B and GSK3, as known Akt and β-TrCP1 target, were verified by Western analysis (Fig. 5e). Both proteins were detected in the input control. The IP showed a strong signal under normoxic conditions for both, TMEM126B as well as GSK3, implying Akt activity under control conditions. Compared to normoxia, we detected a weaker signal for both proteins under hypoxia, suggesting not only phosphorylation but also degradation of TMEM126B and GSK3 (Fig. 5e). Considering that TMEM126B is degraded in the cytosol, we should be able to detect TMEM126B in the cytosol, where it is likely phosphorylated and, thus detectable by the Akt substrate antibody. We fractionated cells as described above using TMEM126-GFP expressing cells, followed by an IP for Akt substrates (Fig. 5f). TMEM126B was detected in the input of both normoxic and chronic hypoxic cells. Following an IP, TMEM126B was clearly abundant in the cytosolic fraction (C), with only some protein being detected in the mitochondrial fraction (M) under normoxia. Taking the large amount of input into account, only a weak TMEM126B band was detected in the cytosol under chronic hypoxia, probably due to its degradation. The fractions were checked for purity as outlined above. These data suggest an involvement of Akt, respectively, the PI3K–Akt–GSK3 axis in TMEM126B degradation. Although the precise mechanism of phosphorylation needs further investigation, we elucidated a HIF-1-dependent induction of β-TrCP1 under chronic hypoxia, its binding to TMEM126B, leading to TMEM126B ubiquitination and degradation in the cytosol (Fig. 5g).
Discussion
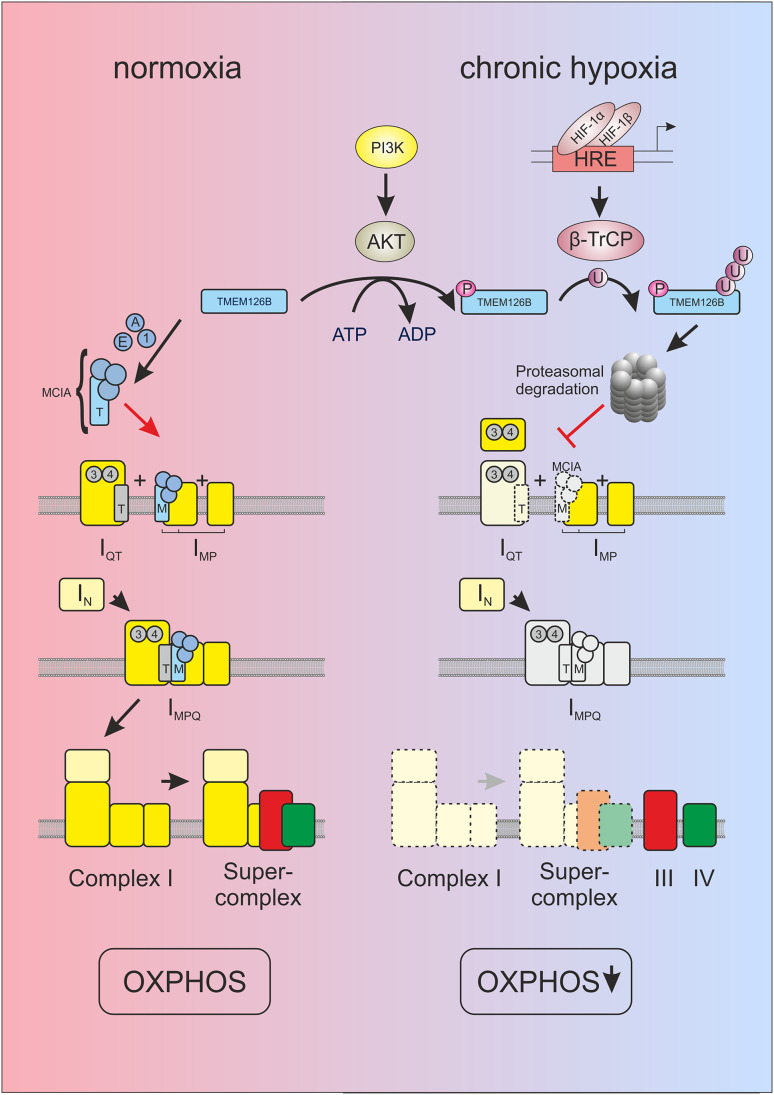
Complexome profiling of isolated mitochondria coupled with functional high-resolution respirometry identified a deficient complex I assembly under chronic hypoxia. Moreover, complexome profiling allows the analysis of multiprotein assemblies in the mitochondrial compartment and provides insights into the size and composition of complexes under normoxia or chronic hypoxia. Whether distinct changes result from assembly deficiencies or complex/supercomplex disintegration has to be elucidated in further studies. We identified defects in the formation of MCIA, a complex required for the correct assembly of complex I. TMEM126B was recently identified as a member of MCIA and its shRNA-mediated knockdown almost completely abolished complex I assembly together with severely impaired mitochondrial respiration [38]. We now provide evidence that the assembly factor TMEM126B, and thus MCIA, is regulated with consequences for complex I formation and function under chronic hypoxia (Fig. 6).
Fig. 6.
Scheme of HIF-1α-dependent complex I regulation under chronic hypoxia. In line with the current model of complex I assembly [33, 39, 40] under normoxic conditions (left panel), TMEM126B enters mitochondria and gathers with ACAD9 (A), ECSIT (E) and NDUFAF1 (1) to build the MCIA complex (M) to assemble the membrane arm (IMP). Subunits of the Q modules assemble with the factors NDUFAF3 (3) and NDUFAF4 (4), and are recruited to the membrane by assembly factor TIMMDC1 (T) (IQT). MCIA and TIMMDC1 connect IQT and IMP to a large intermediate (IMPQ). Connection of the preassembled N-module (IN) completes assembly of complex I. Supercomplexes are formed and contribute to OXPHOS. Recent work suggested supercomplex formation coupled with late stage complex I assembly [64]. In hypoxia (right panel), HIF-1α is stabilized and induces β-TrCP. TMEM126B is encoded by a nuclear gene and is imported into mitochondria. Upon phosphorylation by AKT, TMEM126B is polyubiquitinated by β-TrCP and degraded by the proteasome. Consequently, not only formation of MCIA fails but also the subsequent connection of assembly intermediates of complex I and supercomplex assemblies. Complex III and IV are present as individual complexes that may contribute to the formation of important anaplerotic metabolites and supporting allowing complex II respiration
MCIA was well assembled in sh1 cells, containing TMEM126B together with ACAD9, NDUFAF1, and ECSIT. TMEM126B may recruit the hydrophilic assembly factors NDUFAF1, ECSIT, and ACAD9 to the membrane arm intermediates of complex I connecting them with the preformed Q module under assistance of TIMMDC1 [39, 40]. Hindering complex I assembly in shC cells in chronic hypoxia goes along with the enhanced abundance of subcomplexes of the Q module. An interference with these early assembly steps was already noticed in TMEM126B knockdown cells [38]. Other MCIA members such as NDUFAF1 or ACAD9 also affect correct complex I formation and mutations in the corresponding genes were found in patients with complex I deficiency [36, 46–48]. These findings support the assumption that the altered expression of a single assembly factor can affect complex I formation and function.
We discovered a unique molecular mechanism showing that degradation of TMEM126B and the subsequent complex I assembly defect under chronic hypoxia is a consequence of HIF-1-dependent induction of the E3-ubiquitin ligase β-TrCP. It is known that HIF-1 promotes anaerobic respiration under conditions when molecular oxygen is low, but several studies report a role of HIF-1 in aerobic respiration, as well [49, 50]. Fukuda and coworker expanded the shift from oxidative phosphorylation towards anaerobic respiration by showing changes in the composition of complex IV under acute hypoxia by HIF-1-dependent regulation of COX4-2 and activation of the LON protease that degrades COX4-1 [51]. Swapping complex IV subunits enhanced mitochondrial respiration under reduced oxygen availability. Concerning complex I, an earlier study reported the induction of NDUFA4L2 in a HIF-1-dependent manner after 6 h of hypoxia, thus reducing complex I activity [22]. While the work of Fukuda and Tello reveals short-term adaptive responses, which alter mitochondrial respiration under acute, i.e., 24 h of hypoxia, our data provide evidence that complex I is attenuated under chronic hypoxia, while electron flow via complex II remains intact.
The biological consequence of specifically interfering with complex I assembly under chronic hypoxia warrants further studies. Hypothetically, a role of complex I in modulating the formation of reactive oxygen species and/or affecting the NADH/NAD+ ratio may be linked to chronic hypoxic periods under inflammatory conditions, diabetes, or cancer. One can speculate that complex I and, thus, oxidative phosphorylation can easily be restored upon reoxygenation. Studies in the future need to analyze how fast TMEM126B is incorporated into MCIA and how this affects the assembly and function of complex I. It will also be of interest to identify the site of β-TrCP-mediated TMEM126B ubiquitination, to mutate this site and to study mitochondrial activity with this non-degradable TMEM126B mutant in vitro and in tumor cells in vivo. Interestingly, we found Akt to be phosphorylated at threonine 308 under chronic hypoxia. This goes in line with a study by Guo et al., showing that under normoxia Akt gets prolyl-hydroxylated by PHD2, is bound by the von Hippel–Lindau protein, and, consequently, is inactivated [45]. Under hypoxia, prolyl-hydroxylases are inhibited and Akt gains function by threonine 308 phosphorylation. It is also possible that the remaining electron transfer activity by FAD-dependent dehydrogenases, e.g., succinate dehydrogenase, electron-transferring-flavoprotein (ETF), and glycerol-3-phosphate dehydrogenase (GPDH) to ubiquinone and subsequently to complex III, IV, and molecular oxygen preserves the mitochondrial membrane potential, enables upstream anaplerotic reactions, and guarantees signaling qualities of these organelles [52]. This is supported by an enhanced respiration of hypoxic compared to normoxic mitochondria supplied with succinate. Interestingly, mitochondrial activity in general and respiration in particular affects macrophage polarization and, thus, their ability to produce cytokines but also touches on cancer cells, thereby supporting cancer progression [53–57]. It was shown that macrophages of mice lacking a functional complex I enhance cytokine production, pointing to an influence of complex I in macrophage polarization [58].
While adaption of cells to acute hypoxia is reasonably understood, mechanisms and physiological consequences to handle chronic hypoxia are less explored. After 8–24 h, hypoxia respiratory chain complexes are regulated by substrate availability or altered subunit composition. In contrast, under chronic hypoxia, complex assembly is affected. Complexome profiling uncovered a deficient complex I assembly under chronic hypoxia. Changes result from HIF-1-dependent expression of the E3-ligase β-TrCP1 and subsequent proteasomal degradation of TMEM126B. This disturbed complex I assembly and in turn decreased oxygen consumption. To encompass the molecular mechanism is a first step to understand the role of complex I inactivation under chronic hypoxia and in the future allows addressing its impact on cellular metabolism and biological function.
Methods
Cell culture
THP-1 (derived from a male donor) and MDA-MB-231 (derived from a female donor) cells were incubated at 37 °C with 5% CO2 in DMEM medium (GE Healthcare, Munich, Germany) with 10% FCS and 1% penicillin/streptomycin (PAA Laboratories, Cölbe, Germany). THP-1 cells were stably transduced with a lentiviral shRNA (Mission shRNA) against HIF-1α (sh1: TRCN0000003810) or HIF-2α (sh2: TRCN0000003804) and selected using puromycin. Controls (shC) are THP-1 cells transduced with a pLKO.1-puro vector. Monocytes were differentiated to macrophages with 10 nM TPA for 5 days followed by 1 day incubation without TPA. MDA-MB-231 cells were transduced with a lentiviral pGIPZshRNAmir TMEM126B set (shTMEM: RHS4531, Thermo Scientific, Karlsruhe, Germany) or as control with pGIPZ lentiviral shRNAmir control (RHS4346). The knockdown of β-TrCP was created by stably transducing MDA-MB-231 cells with a PLKO-PURO-IPTG-3XLACOIXM12-8083378BTRCP1B vector (Sigma, Munich, Germany), selected with puromycin and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). To create MDA-MB-231 cells overexpressing transcript variant 1 of TMEM126B, the variant was cloned into a pSEW vector (pSEW_TMEM). For IP, these cells were transduced with the pSEW_TMEM vector or a control vector expressing GFP. Afterwards cells were seeded and incubated 24 h before hypoxic incubation. To detect ubiquitination, a HA-ubiquitin expressing plasmid was transfected into the cells using JetPRIME (Polyplus transfection, Illkirch, France) according to the manufacturer’s instructions.
Treatments and hypoxic incubations
Cells were incubated at 1% O2 in a hypoxic incubator (Invivo2 400, Baker Ruskinn, Leeds, UK). Fresh, hypoxic medium was provided to cells after 24 and 48 h without reoxygenation. Proteins were harvested under hypoxic conditions, RNA directly after removing cells from the incubator. To inhibit kinase activity, cells were treated with GÖ6976 (10 µM), RO3306 (5 µM), CHIR99021 (1 µM), LY294002 (15 µM), or Akt VIII (1 µM) during hypoxic incubation. Inhibitors were added freshly when changing medium. Dimethyloxalylglycine (DMOG, 1 µM), cycloheximide (CHX, 10 µg/ml), or lactacystin (LC, 10 µM) was provided at different time points prior to harvesting cells, always guaranteeing total incubation times of 72 h.
Western analysis
Cells were lysed in buffer containing 4% SDS, 150 mM NaCl, and 100 mM Tris/HCl, pH 7.4, and sonicated. Protein content was determined by a protein assay kit (Bio-Rad, Munich, Germany) and 50 µg protein were loaded on a 10% SDS gel. Gels were blotted using a Trans Blot Turbo blotting system (Bio-Rad). Membranes were blocked in 5% milk in TBS-T for nucleolin (Santa Cruz, Heidelberg, Germany) and tubulin (Sigma) or 5% BSA in TBS-T for TMEM126B (Atlas Antibodies), β-TrCP (Cell Signaling, Danvers, USA), ACAD9 (Atlas Antibodies), ECSIT (Atlas Antibodies), NDUFAF1 (Atlas Antibodies), Akt and pAkt (Cell Signaling), GSK3 (Cell Signaling), NDUFA8 (Sigma), antiserum against bovine subunit α and β of ATP synthase raised in rabbits, and NDUFB6 (Atlas Antibodies). Enhanced chemiluminescence on a C-DIGIT scanner (Licor, Lincoln, USA) or fluorescence on an Odyssey scanner (Licor) was quantified with Image Studio Digits 5.0 (Licor).
Immunoprecipitation
ChIP was performed according to Tausendschön et al. [44]. Phospho-Akt substrate IP was performed as follows. MDA-MB-231 overexpressing TMEM126B were lysed in IP lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3Vo4, and 1 µg/ml leupeptin) and sonicated. 10 µl of phospho-Akt substrate antibody coupled to Sepharose beads (Cell Signaling) were added and incubated overnight on a rotary shaker, centrifuged, washed three times with IP lysis buffer, and prepared for Western blotting. To coIP β-TrCP, TMEM-GFP overexpressing MDA-MB-132 cells were lysed in coIP lysis buffer I (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP40, 1 mM DTT), supplemented with 25 µl GFP-Trap_A beads (ChromoTek, Munich, Germany), incubated on a rotary shaker overnight, centrifuged, washed in wash buffer I (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% NP40), and prepared for Western blotting. For detection of HA-ubiquitin, GFP IP was performed using MDA-MB-231 cells expressing GFP-coupled TMEM126B, HA-tagged ubiquitin, followed by treatment with MG-132 (10 µM) for 8 h prior harvest. Cells were lysed in coIP lysis buffer II (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, and 0.5% NP-40), incubated for 30 min on ice, and centrifuged. The supernatant was diluted with wash buffer II (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA), supplemented with 25 µl GFP-Trap_A beads (ChromoTek), incubated on a rotary shaker overnight, centrifuged, washed in wash buffer, and prepared for Western blotting.
RNA analysis
RNA was isolated using peqGold (Peqlab, Erlangen, Germany) and measured using a Nanodrop ND-1000 spectrophotometer (Peqlab). Reverse transcription was performed with the Maxima First Strand cDNA Synthesis Kit for RT-PCR (Thermo Scientific). RNA expression of TMEM126B (fwd: 5’-GGTGGTGTTCGGGTATGAGG-3’, rev: 5’-TCTTGAAAACCTTGGGCGCT-3’), β-TrCP (mRNA: fwd: 5’-AACAGCTGTGCCAGACTCTGCTTA-3’, rev: 5’-GCTTGCTGAGAGTTTCCGTTGCTT-3’, ChIP: fwd: 5’-CAGCCACCGCAGTTCTTTAT-3’, rev: 5’-TGCCCTATGGAGAGAACCAC-3’) was analyzed using SYBR green fluorescent mix (Thermo Scientific) on a CFX96 Real Time PCR Detection System (Bio-Rad) and normalized to TBP (fwd: 5’-GGGCCGCCGGCTGTTTAACT-3’, rev: 5’-GGGCCGCCGGCTGTTTAACT-3’).
Isolation of mitochondria and fractionation
THP-1 cells were washed, carefully harvested in PBS, centrifugated (10 min, 1000×g, 4 °C), and resuspended in CCM I buffer (250 mM sucrose, 1 mM EDTA, 20 mM Tris/HCl, pH 7.4) containing protease-inhibitor (Roche, Grenzach-Wyhlen, Germany). The suspension was pressed ten times through a 25G needle, followed by a centrifugation step (10 min, 1000×g, 4 °C). The nuclear fraction was stored and supernatant was taken and centrifuged again (10 min, 6000×g, 4 °C). The pellet was resuspended in CCM I. For IP, the supernatant was stored as cytosolic fraction. The protein concentration was measured by a Lowry protein assay kit (Bio-Rad).
Respiration measurements
The oxygen consumption of trypsinized cells and isolated mitochondria was determined by high-resolution respirometry as described [38, 59]. Briefly, for the determination of cellular oxygen consumption, 0.5–1 × 106 cells were resuspended in 2 ml medium and transferred into an Oxygraph-2k (Oroboros, Innsbruck, Austria). After recording, basal respiration at 37 °C, 2 µg/ml oligomycin (Sigma) was added to inhibit ATP synthesis. Subsequently, the uncoupler carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) (Sigma) was added in 1 µM steps to assess maximal capacity of the electron-transferring chain. Finally, 2 mM KCN was added to determine the mitochondria-independent oxygen consumption of cells. For respirometry of isolated mitochondria, mitochondria (0.192–2.332 mg protein) were resuspended in 2 ml buffer (200 mM sucrose, 10 mM KH2PO4, 10 mM Tris/HCl, 10 mM MgSO4, 2 mM EDTA, and pH 7.0) and fueled by the NADH + H+-generating substrates malate/glutamate (5 mM each) at 37 °C. State 3 respiration was subsequently induced by adding 2 mM ADP. 5 µM rotenone (Sigma) and 5 mM of the complex II substrate succinate were added. Finally, 2 mM KCN was applied to determine mitochondria-independent oxygen consumption rates.
Isolation of macromolecular complexes by blue native gels
Isolated mitochondria were aliquoted into portions of 400 µg protein content and sedimented to obtain pellets. Mitochondria were resuspended in 40 µl buffer A (50 mM NaCl, 50 mM imidazole pH 7, 1 mM EDTA, and 2 mM aminocaproic acid) and solubilized with 16 µl 20% digitonin (w/v in water) to obtain a detergent/protein ratio of 8 g/g. Samples were centrifuged for 10 min at 22,000g and protein content of the supernatant was determined. 50 µg total protein was loaded onto 3–16% gradient gels following blue native electrophoresis (BNE) [59]. For complexome profiling, BN gels were stained with Coomassie. Following separation of mitochondrial OXPHOS complexes by BNE, subunits of protein complexes were separated by 2D BN/SDS-PAGE. 2D gels were stained with silver. Gels were scanned by an office scanner (Epson perfection 2400 PHOTO) for documentation.
Sample preparation for complexome profiling
Blue native gels were fixed in 50% (v/v) methanol, 10% (v/v) acetic acid, and 10 mM ammonium acetate for 30 min, and stained with Coomassie [0.025% Serva Blue G, 10% (v/v) acetic acid] [59]. Each lane was cut into 60 equal fractions and collected in 96 filter well plates (30–40 µm PP/PE, Pall Corporation). Pieces were destained in 60% methanol and 50 mM ammonium bicarbonate (ABC). Solutions were removed by centrifugation for 2 min at 1500 rpm. Proteins were reduced in 10 mM DTT, 50 mM ABC for 1 h at 56 °C and alkylated for 45 min in 30 mM iodoacetamide. Samples were digested for 16 h with trypsin (sequencing grade, Promega) at 37 °C in 50 mM ABC, 0.01% Protease Max (Promega) and 1 mM CaCl2. Peptides were eluted in 30% acetonitrile and 3% formic acid, centrifuged into a fresh 96 well plate, dried in a speed vac, and resolved in 1% acetonitrile and 0.5% formic acid (S2).
Mass spectrometry
Liquid chromatography/mass spectrometry (LC/MS) was performed on a Thermo Scientific™ Q ExactivePlus mass spectrometer coupled to an ultra-high-performance liquid chromatography unit (Thermo Scientific Dionex Ultimate 3000) via a Nanospray Flex Ion-Source (Thermo Scientific). Peptides were loaded on a C18 reversed-phase precolumn (Zorbax 300SB-C18, Agilent Technologies) followed by separation on in-house packed 2.4 µm ReprosilC18 resin (Dr. Maisch GmbH) picotip emitter tip (diameter 100 µm, 15 cm long, New Objectives) using a gradient from mobile phase A (4% acetonitrile, 0.1% formic acid) to 50% mobile phase B (80% acetonitrile, 0.1% formic acid) for 30 min. Each run was finished by washout with 99% B for 5 min and column equilibration for 13 min with 99% B. Mass spectrometry (MS) data were recorded by data-dependent Top10 acquisition [selecting the ten most abundant precursor ions in positive mode for high energy collision dissociation fragmentation (HCD)]. The full MS scan range was 300–2000 m/z with resolution of 70,000 at m/z 200, and an automatic gain control (AGC) value of 3 × 106 total ion counts with a maximal ion injection time of 160 ms. Only higher charged ions (2 +) were selected for MS/MS scans with a resolution of 17,500, an isolation window of 2 m/z and an automatic gain control value set to 1 × 105 ions with a maximal ion injection time of 150 ms. Following the fragmentation event, all selected ions were excluded in a time frame of 30 s. Data were acquired in profile mode by Xcalibur software. The LC Unit was controlled by Chromeleon Xpress software. The performance of both units LC and MS was integrated by DCMSLink.
Mass spectrometry and data analysis for complexome profiling
MS data were analyzed by MaxQuant (1.5.3.30) [60]. Proteins were identified using human proteome database UniProtKB with 68,506 entries, released in 4/2015. Variable modifications were N-terminalacetylation, oxidation of methionine, deamidation on asparagine, and glutamine, and fixed modification was carbamidomethylation on cysteines. False discovery rate (FDR) was 1%. Intensity-based absolute quantification (IBAQ) values were recorded and used to inspect migration profiles with NOVA software (0.5.7) [61, 62]. IBAQ values were normalized to median appearance of proteins from hypoxic control cells (shC). Subunits of complexes I, III, IV, and complex I assembly factors were selected and normalized to maximum of the lanes from hypoxic samples to visualize them in a heatmap. Slice number of the maximum appearance of mitochondrial complex III dimer (483,695 Da), complex IV (220,156 Da), complex V (618,824 Da), and respiratory supercomplex containing complex I, III dimer, and one copy of complex IV (1,663,827 Da) was used for native mass calibration.
Statistics
Data are expressed as mean values ± SEM. Statistically significant differences were calculated after analysis of variance (ANOVA) and Bonferroni’s test or Student’s t test; p < 0.05 was considered as significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Tanja Keppler for excellent technical assistance, Jana Meisterknecht for competent support with BNE and 2D-BNE/SDS gels, and Ilka Siebels for technical assistance in respiration measurements.
Abbreviations
- ACAD9
Acyl-CoA dehydrogenase family member 9
- ADP
Adenosine diphosphate
- Akt
RAC-alpha serine/threonine-protein kinase
- ATP
Adenosine triphosphate
- ATP5a
ATP synthase subunit alpha
- β-TrCP
F-box/WD repeat-containing protein 1A
- BNE
Blue native electrophoresis
- ChIP
Chromatin immunoprecipitation
- CHX
Cycloheximide
- COX
Cytochrome c oxidase subunit
- DMOG
Dimethyloxalylglycine
- DMSO
Dimethyl sulfoxide
- ECSIT
Evolutionarily conserved signaling intermediate in toll pathway
- ETF
Electron transfer flavoprotein
- FAD
Flavin adenine dinucleotide
- FCCP
Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- GFP
Green fluorescent protein
- GPDH
Glycerol-3-phosphate dehydrogenase
- GSK3
Glycogen synthase kinase-3
- HIF
Hypoxia-inducible factor
- HIGD
Hypoxia-inducible gene
- IP
Immunoprecipitation
- IPTG
Isopropyl-β-d-thiogalactopyranoside
- KCN
Potassium cyanide
- LC
Lactacystin
- MCIA
Mitochondrial complex I assembly complex
- NAD
Nicotinamide adenine dinucleotide
- ND
NADH-ubiquinone oxidoreductase chain
- NDUFA
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit
- NDUFAF1
Complex I intermediate-associated protein 30
- NDUFA4L2
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4-like 2
- NDUFB
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit
- NDUFS
NADH dehydrogenase [ubiquinone] iron–sulfur protein
- OXPHOS
Oxidative phosphorylation
- PHD
Prolyl hydroxylase
- PI3K
Phosphatidylinositol 3-kinase
- rot
Rotenone
- shC
Cells transduced with control short hairpin RNA
- sh1
Cells transduced with short hairpin RNA against HIF-1α
- sh2
Cells transduced with short hairpin RNA against HIF-2α
- TIMMDC1
Translocase of inner mitochondrial membrane domain-containing protein 1
- TMEM126B
Transmembrane protein 126B
Author contributions
DF performed and planed the experiments and wrote the paper. IW performed complexome profiling and participated on paper writing. SD performed respiratory measurements. TS gave his expertise concerning β-TrCP experiments ND discussed data and literature. BB performed data interpretation, wrote the paper, and designed the project.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft [SFB 815, project Z1 (I.W.) and project A8 (B.B.)].
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Scholz CC, Taylor CT. Targeting the HIF pathway in inflammation and immunity. Curr Opin Pharmacol. 2013;13(4):646–653. doi: 10.1016/j.coph.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Palazon A, Goldrath AW, Nizet V, et al. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41(4):518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mucaj V, Shay JE, Simon MC. Effects of hypoxia and HIFs on cancer metabolism. Int J Hematol. 2012;95(5):464–470. doi: 10.1007/s12185-012-1070-5. [DOI] [PubMed] [Google Scholar]
- 4.Catrina S-B, Okamoto K, Pereira T, et al. Hyperglycemia regulates hypoxia-inducible factor-1 protein stability and function. Diabetes. 2004;53(12):3226–3232. doi: 10.2337/diabetes.53.12.3226. [DOI] [PubMed] [Google Scholar]
- 5.Bayer C, Shi K, Astner ST, et al. Acute versus chronic hypoxia. Why a simplified classification is simply not enough. Int J Radiat Oncol Biol Phys. 2011;80(4):965–968. doi: 10.1016/j.ijrobp.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 6.Bayer C, Vaupel P. Acute versus chronic hypoxia in tumors. Controversial data concerning time frames and biological consequences. Strahlenther Onkol. 2012;188(7):616–627. doi: 10.1007/s00066-012-0085-4. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrmann DC, Wittig I, Heide H, et al. Chronic hypoxia alters mitochondrial composition in human macrophages. Biochim Biophys Acta. 1834;12:2750–2760. doi: 10.1016/j.bbapap.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Huang LE, Gu J, Schau M, et al. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95(14):7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh CW, O’Rourke JF, Nagao M, et al. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J Biol Chem. 1997;272(17):11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 10.Appelhoff RJ, Tian YM, Raval RR, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279(37):38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 11.Jiang BH, Zheng JZ, Leung SW, et al. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272(31):19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 12.Berchner-Pfannschmidt U, Tug S, Kirsch M, et al. Oxygen-sensing under the influence of nitric oxide. Cell Signal. 2010;22(3):349–356. doi: 10.1016/j.cellsig.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Kapitsinou PP, Rajendran G, Astleford L, et al. The endothelial PHD2/HIF-2 axis regulates pulmonary artery pressure in mice. Mol Cell Biol. 2016 doi: 10.1128/MCB.01055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrmann DC, Tausendschon M, Wittig I, et al. Inactivation of tristetraprolin in chronic hypoxia provokes the expression of cathepsin B. Mol Cell Biol. 2015;35(3):619–630. doi: 10.1128/MCB.01034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 16.Brandt U. Energy converting NADH. Quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 17.Janssen RJ, Nijtmans LG, van den Heuvel LP, et al. Mitochondrial complex I. Structure, function and pathology. J Inherit Metab Dis. 2006;29(4):499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- 18.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 19.Lee I, Bender E, Kadenbach B. Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol Cell Biochem. 2002;234–235(1–2):63–70. doi: 10.1023/A:1015921513720. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrmann DC, Brune B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017;12:208–215. doi: 10.1016/j.redox.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405(1):1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 22.Tello D, Balsa E, Acosta-Iborra B, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting complex I activity. Cell Metab. 2011;14(6):768–779. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Lai RK, Xu IM, Chiu DK, et al. NDUFA4L2 fine-tunes oxidative stress in hepatocellular carcinoma. Clin Cancer Res. 2016;22(12):3105–3117. doi: 10.1158/1078-0432.CCR-15-1987. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Bai C, Guo J, et al. NDUFA4L2 protects against ischaemia/reperfusion-induced cardiomyocyte apoptosis and mitochondrial dysfunction by inhibiting complex I. Clin Exp Pharmacol Physiol. 2017;44(7):779–786. doi: 10.1111/1440-1681.12768. [DOI] [PubMed] [Google Scholar]
- 25.Hernansanz-Agustín P, Ramos E, Navarro E, et al. Mitochondrial complex I deactivation is related to superoxide production in acute hypoxia. Redox Biol. 2017;12:1040–1051. doi: 10.1016/j.redox.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papandreou I, Cairns RA, Fontana L, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Schagger H, de Coo R, Bauer MF, et al. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem. 2004;279(35):36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- 28.Acin-Perez R, Fernandez-Silva P, Peleato ML, et al. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32(4):529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Maranzana E, Barbero G, Falasca AI, et al. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal. 2013;19(13):1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19(8):1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;4:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Dieteren CE, Willems PH, Vogel RO, et al. Subunits of mitochondrial complex I exist as part of matrix- and membrane-associated subcomplexes in living cells. J Biol Chem. 2008;283(50):34753–34761. doi: 10.1074/jbc.M807323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62(7):497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 34.Guerrero-Castillo S, Baertling F, Kownatzki D, et al. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 2017;25(1):128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Vogel RO, Janssen RJ, Ugalde C, et al. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 2005;272(20):5317–5326. doi: 10.1111/j.1742-4658.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- 36.Nouws J, Nijtmans L, Houten SM, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12(3):283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Lazarou M, Thorburn DR, Ryan MT, et al. Assembly of mitochondrial complex I and defects in disease. Biochim Biophys Acta. 2009;1793(1):78–88. doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Heide H, Bleier L, Steger M, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16(4):538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Andrews B, Carroll J, Ding S, et al. Assembly factors for the membrane arm of human complex I. Proc Natl Acad Sci USA. 2013;110(47):18934–18939. doi: 10.1073/pnas.1319247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guarani V, Paulo J, Zhai B, et al. TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex. Mol Cell Biol. 2014;34(5):847–861. doi: 10.1128/MCB.01551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayashi T, Asano Y, Shintani Y, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112(5):1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidoni S, Harbour ME, Guerrero-Castillo S, et al. MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 2017;18(7):1727–1738. doi: 10.1016/j.celrep.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 43.Vukotic M, Oeljeklaus S, Wiese S, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15(3):336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 44.Tausendschon M, Rehli M, Dehne N, et al. Genome-wide identification of hypoxia-inducible factor-1 and -2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochim Biophys Acta. 1849;1:10–22. doi: 10.1016/j.bbagrm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Guo J, Chakraborty A, Liu P, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353(6302):929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haack TB, Danhauser K, Haberberger B, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42(12):1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 47.Dunning CJ, McKenzie M, Sugiana C, et al. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26(13):3227–3237. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nouws J, Nijtmans LG, Smeitink JA, et al. Assembly factors as a new class of disease genes for mitochondrial complex I deficiency. Cause, pathology and treatment options. Brain. 2012;135(Pt 1):12–22. doi: 10.1093/brain/awr261. [DOI] [PubMed] [Google Scholar]
- 49.Taylor CT. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem J. 2008;409(1):19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 50.Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist. 2004;9(Suppl 5):10–17. doi: 10.1634/theoncologist.9-90005-10. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda R, Zhang H, Kim JW, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 52.Chandel NS. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22(2):204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 53.El Kasmi KC, Stenmark KR. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin Immunol. 2015;27(4):267–275. doi: 10.1016/j.smim.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan Z, Xie N, Cui H, et al. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. 2015;194(12):6082–6089. doi: 10.4049/jimmunol.1402469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tannahill GM, Iraci N, Gaude E, et al. Metabolic reprogramming of mononuclear phagocytes in progressive multiple sclerosis. Front Immunol. 2015;6:106. doi: 10.3389/fimmu.2015.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biswas SK. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43(3):435–449. doi: 10.1016/j.immuni.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin Z, Wei W, Yang M, et al. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014;20(3):483–498. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 61.Giese H, Ackermann J, Heide H, et al. NOVA. A software to analyze complexome profiling data. Bioinformatics. 2015;31(3):440–441. doi: 10.1093/bioinformatics/btu623. [DOI] [PubMed] [Google Scholar]
- 62.Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 63.Wirth C, Brandt U, Hunte C, et al. Structure and function of mitochondrial complex I. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 64.Moreno-Lastres D, Fontanesi F, Garcia-Consuegra I, et al. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15(3):324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.