Abstract
Polycystin-1 (PC1) and polycystin-2 (PC2) are transmembrane proteins encoded by the Pkd1 and Pkd2 genes, respectively. Mutations in these genes are causative for the development of autosomal-dominant polycystic kidney disease. A prominent feature of this disease is an unbalanced cell proliferation. PC1 and PC2 physically interact to form a complex, which localizes to the primary cilia of renal epithelial cells. Recently, PC1 and PC2 have also been described to be present in primary cilia of radial glial cells (RGCs) and to contribute to the planar cell polarity of late RGCs and E1 ependymal cells. As neural progenitor cells (NPCs), early RGCs have to balance proliferation for expansion, or for self-renewal and differentiation to generate neurons. It is not known whether the polycystins play a role in this process. Here, we show that PC1 and PC2 are expressed in RGCs of the developing mouse cerebral cortex during neurogenesis. Loss-of-function analysis and cell-based assays reveal that a reduction of PC1 or PC2 expression leads to increased NPC proliferation, while the differentiation to neurons becomes impaired. The increased NPC proliferation is preceded by enhanced Notch signaling and accompanied by a rise in the number of symmetric cell divisions. The transcription factor STAT3 seems to be mechanistically important for polycystin signaling in NPCs as either STAT3 knockdown or inhibition of STAT3 function abrogates the increased proliferation driven by reduced polycystin expression. Our findings indicate that PC1 and PC2 are critical for maintaining a balance between proliferation and differentiation of NPCs.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03072-x) contains supplementary material, which is available to authorized users.
Keywords: Nestin, Ventricular zone, BrdU labeling, MAP2, CBFRE sensor
Introduction
Mutations in the genes encoding polycystin-1 (PC1) and polycystin-2 (PC2) account for almost all cases of autosomal-dominant polycystic kidney disease (ADPKD), one of the most common genetic diseases [1–7]. Cyst formation in ADPKD is associated with a dysregulation of renal epithelial cell proliferation. Renal cyst growth can be caused by eliminating or even only reducing the expression of PC1 or PC2, but also by expression of mutated polycystin proteins, indicating that the levels of functionally active PC1 and PC2 are critical for normal function [8–11]. Several signaling pathways have been connected to the dysregulation of renal epithelial cell proliferation including the JAK/STAT signaling pathway [3–6, 12–14]. In particular the activity of the transcription factor STAT3 has been found to be regulated by PC1, and STAT3 has been shown to be strongly activated in cyst-lining cells. PC1 is a large integral membrane protein structurally resembling a receptor or adhesion molecule comprising a very large extracellular region, 11 transmembrane domains, and a relatively short carboxy-terminal cytoplasmic tail [4, 6]. PC2, also called TRPP2, is a member of the family of transient receptor potential (TRP) cation channels [15]. PC1 and PC2 form a receptor-ion channel complex by interacting via the coiled-coil domains present in each of these proteins [16–18]. This heteromeric molecular complex localizes to the plasma membrane and to primary cilia. The solitary primary cilium protruding from the apical surface of most kidney tubule cells functions as a specialized cellular compartment for signal integration of sensory input [3, 6, 7]. Here, PC1 and PC2 are thought to form a mechanosensory complex that translates fluid flow into second messenger signals including PC2-mediated Ca2+ signaling [3, 15, 19, 20].
During cerebral cortex development in the brain, all neurons develop from neural progenitor cells (NPCs). In the ventricular zone, neuroepithelial cells (NECs) are the primary, and radial glial cells (RGCs) the subsequently emerging NPCs, both of which are immunopositive for the marker nestin [21–26]. During the cell cycle in interphase, NECs and RGCs can generate primary cilia, which protrude into the lateral ventricles, where the cerebrospinal fluid (CSF) circulates [25, 27]. Multiciliated ependymal (E1) cells, which in the adult brain line the walls of the ventricles and propel the CSF, are derived from RGCs. They display location-specific planar cell polarity (PCP) in the orientation and positioning of their motile cilia [28]. The development of PCP in late RGCs set in around embryonic day 16.5 (E16.5) [27]. Recently, it has been demonstrated that PC1 and PC2 co-localize to the primary cilia of RGCs. Further, it has been shown that genetic ablation of PC1 or PC2 affects PCP in RGCs and E1 cells [28]. However, expression analysis by in situ hybridization studies has revealed a remarkably early expression of PC1 in the developing brain, considerably before PCP in RGCs cells is going to be established. These data suggest additional functions of PC1 or the complex of PC1 and PC2 in NECs and RGCs [29, 30].
To examine putative further roles of PC1 and PC2 in NPC development, this study initially investigated the expression and localization of PC1 and PC2 proteins in the embryonic cerebral cortex. Actually, PC1 and PC2 were expressed in nestin-positive NPCs in the ventricular zone with a prominent localization at the apical site of these cells facing the lumen of the lateral ventricle. Functional analysis in primary cell culture showed that RNA interference (RNAi)-mediated knockdown of PC1 or PC2 expression increased NPC proliferation and decreased differentiation to microtubule-associated protein 2 (MAP2)-positive neurons. Additionally, reducing PC1 or PC2 expression stimulated Notch signaling, which is thought to be essential for preserving NPC character and inhibiting neurogenesis. This study further shows that lowering the expression of the Pkd1 or Pkd2 gene products raised the number of cell clusters comprising exclusively cells, which are all immunopositive for the NPC marker nestin. Consistently, the number of mixed cell clusters, containing both nestin-positive and also nestin-negative cells, were reduced. These results suggest that PC1 and PC2 may be involved in the regulation of differentiative versus proliferative NPC divisions. Mechanistically, the transcription factor STAT3 seems to antagonize the role of PC1 and PC2 in regulating NPC differentiation. Together, our findings uncover a role of PC1 and PC2 in maintaining a balance between proliferation and differentiation of nestin-positive NPCs.
Materials and methods
Cell culture and transfection
Primary cells were prepared from the cerebral cortices of C57BL/6J mouse embryos at embryonic day 13.5 (E13.5) as described [31]. This cell culture comprised predominantly NPCs and a small amount of neurons, and is denoted NPC culture in the following. The cells were cultured in Neurobasal A medium containing the B27 supplement, 0.5 mM l-glutamine and penicillin/streptomycin (all Invitrogen). To promote the proliferation of NPCs, recombinant bFGF was added (Peprotech 100-18B, 10 ng/ml f. c.). For all experiments, the cells were transfected 1 day after plating at days in vitro 1 (DIV1). The transfections were performed with Effectene (Qiagen) or Lipofectamine Stem (Thermo Fisher Scientific) according to the manufacturer’s instructions. For the analysis of NPC proliferation and differentiation, 50% of the culture medium was exchanged at DIV3, and fresh bFGF was added. For proliferation analysis, the cells were labeled at DIV4 for 2.5 h with BrdU (Sigma-Aldrich, 5 μg/ml f. c.), fixed with 4% paraformaldehyde (Carl Roth), and processed for BrdU staining. The STAT3 inhibitor S3I-201 (Sigma-Aldrich) was added to the NPC culture at DIV2 (10 μM f. c.). For the analysis of NPC apoptosis, the cells were fixed at DIV4 and stained for cleaved caspase 3. To evaluate the differentiation potential of the NPC culture, the cells were fixed at DIV4 and processed for MAP2 staining, or assessment of the fluorescence derived from the Tα1p-DsRed2 sensor construct. For cell cluster analysis, the NPC culture was fixed at DIV4 and then processed for nestin staining. The analysis of BrdU-positive cell pairs with regard to Notch signaling was performed following 6 h of BrdU labeling at DIV3, cell fixation at DIV4, and triple staining for RFP (driven by the polycystin shRNA constructs), GFP (driven by the CBFRE-EGFP sensor construct) and BrdU. For the analysis of Notch signaling, the fluorescence derived from the CBFRE-EGFP sensor construct was assessed after fixation and GFP staining of the NPC culture at DIV3. For the experiments with Bis(2-aminophenoxy)ethane tetraacetic acid acetoxymethyl ester (BAPTA-AM, Thermo Fisher Scientific), following transfection the NPC culture was incubated with 3 μM BAPTA-AM for 45 min in Hank’s balanced salt solution (HBSS) w/o Ca2+/Mg2+. After two washes with phosphate-buffered saline (PBS) the cells were cultured as described above.
DNA constructs
The target sequences for the shRNA constructs Pkd1–kd1, Pkd1–kd2, Pkd2–kd1, and Pkd2–kd2 on the backbones of the pCGLH or pCRLH vectors were 5′-GTCTGCTTGTCCAGTTTGA-3′, 5′-GCACATCTGGCTCTCCATA-3′, 5′-TGATGAGCTCCAATGTGTA-3′, and 5′-TTTGTGTCTGTCAAAGACA-3′. The constructs Pkd1–kd1, Pkd1–kd2, Pkd2–kd1, and Pkd2–kd2 correspond to the siPKD1/12632, siPKD1/10071, siRNA1, and siRNA4 sequences, respectively, and have been specified and validated elsewhere [32, Fig. 1E and Suppl. Fig. 3C]; [33, Fig. 2a, b, and Suppl. Table 1]. The target sequences for the shRNA constructs kdcontrol and STAT3–kd1/2 were 5′-AACCCCAACCCCAGCCTTGAT-3′, 5′-TGCAGGATCTGAATGGAAA-3′, and 5′-TGCAGGATCTAGAACAGAA-3′, respectively, and have been described earlier [31, 34, 35].
Fig. 1.
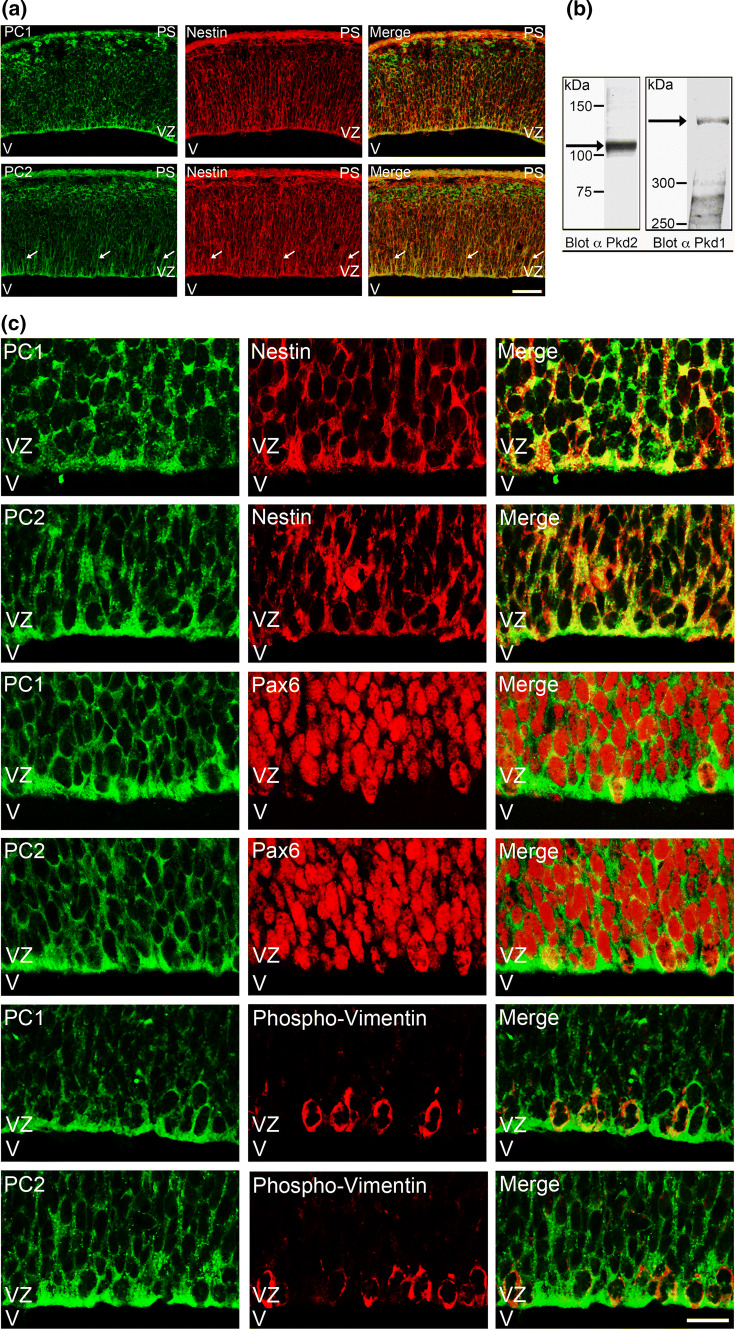
PC1 and PC2 are expressed in nestin-positive NPCs during neurogenesis. a Coronal sections of E12.5 embryonic mouse brains were analyzed by indirect immunofluorescence staining with polyclonal antibodies to PC1 or PC2, respectively, followed by a monoclonal antibody to nestin. PC1 and PC2 (green) were expressed in the ventricular zone (VZ) of E12.5 brains, labeled by the neuroepithelial and radial glia marker nestin (red). PC1 and PC2 expression clearly overlapped with that of nestin (merge), most prominently at the apical site of the ventricular zone, near the ventricle (V). Especially PC2 could also be detected in nestin-positive radial glial fibers (arrows). Scale bar, 50 μm. b The expression of PC1 and PC2 (arrows) in crude membrane fractions of E14 embryonic mouse brains as assessed by Western blotting with polyclonal antibodies to PC1 or PC2, respectively. c Coronal sections of E14.5 embryonic mouse brains were analyzed by indirect immunofluorescence staining with polyclonal antibodies to PC1 or PC2, respectively, followed by monoclonal antibodies to nestin, Pax6, or phospho-vimentin, respectively. Detail pictures of the VZ are shown. PC1 and PC2 were expressed in nestin-positive cells. In addition, both polycystins were expressed in cells labeled by Pax6, a transcription factor that is upregulated in RGCs. PC1 and PC2 were also detected in cells labeled by phospho-vimentin, a cytoskeletal marker of mitotic radial glial cells (RGCs) in the VZ. Scale bar, 15 μm. PS pial surface, V ventricle, VZ ventricular zone
Fig. 2.
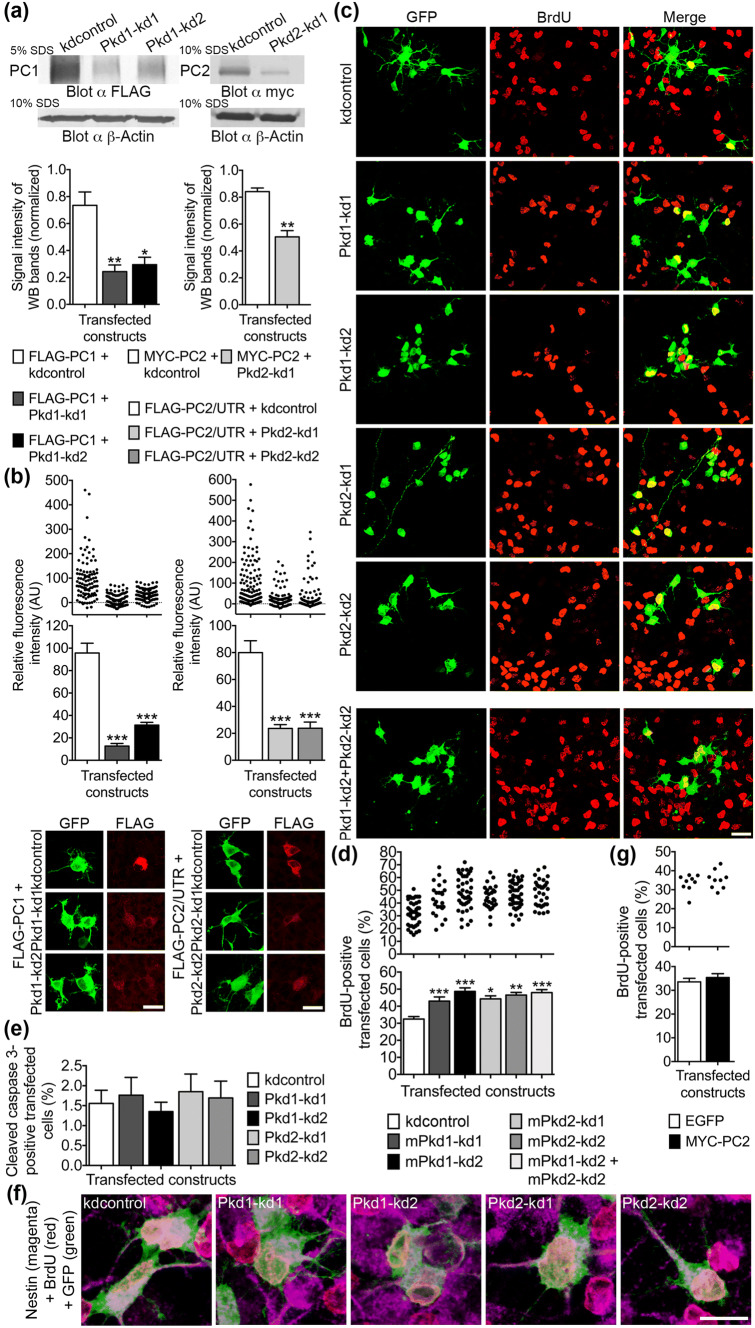
Reduced PC1 or PC2 expression leads to an increase in the number of proliferating NPCs. a Recombinant FLAG-tagged PC1 was co-transfected with the shRNA knockdown constructs Pkd1–kd1 or Pkd1–kd2, both specific for PC1, or the control shRNA construct kdcontrol into HEK293 cells. Whole cell lysates were prepared 32 h after transfection and subjected to SDS-PAGE and Western blot (WB). Following protein quantification, equal amounts of total lysate proteins were loaded for each experimental condition. A monoclonal anti-FLAG antibody was used for the detection of FLAG-tagged PC1 (FLAG-PC1). Due to the very large size of PC1 (~ 450 kDa), the SDS-PAGE was performed with 5% acrylamide (5% SDS). The control SDS-PAGE (loading control) for the detection of β-Actin (~ 40 kDa) contained 10% acrylamide (10% SDS). Exactly the same volumes were loaded onto this 10% gel as was done for the 5% gel. The intensities of six WB bands specific for FLAG-PC1 taken from three independent experiments were measured and normalized to the intensities of the corresponding β-actin bands. Data are presented as mean ± SEM in a histogram (**p < 0.01, *p < 0.05; Dunn’s multiple comparison post hoc test following Kruskal–Wallis). Both shRNA knockdown constructs, Pkd1–kd1 and Pkd1–kd2, reduced the amount of overexpressed FLAG-PC1 (to 32% and 41% of control) when compared to kdcontrol. The evaluation of the knockdown efficiency of the shRNA construct Pkd2–kd1 specific for PC2 was performed in a similar manner as for the PC1-specific shRNA constructs. After being separated on a 10% SDS-PAGE, MYC-PC2 and β-actin were detected on the same WB membrane. Pkd2–kd1 reduced the amount of overexpressed MYC-PC2 to 60% when compared to kdcontrol. As the binding site for the PC2-specific knockdown construct Pkd2–kd2 is in the 3′-UTR of PC2, we had to analyze an recombinant PC2 expression construct including the 3′-UTR (FLAG-PC2/UTR). Unfortunately, this construct could not be expressed in a stable manner in HEK293 cells preventing an analysis of the knockdown efficiency of Pkd2–kd2 with this technique. The key of a does also apply to b. b FLAG-PC1 was co-expressed with either kdcontrol, Pkd1–kd1, or Pkd1–kd2, respectively, in cultured E13.5 neocortical cells. Similarly, FLAG-PC2/UTR was co-expressed with either kdcontrol, Pkd2–kd1, or Pkd2–kd2 in NPCs. An indirect immunostaining was performed 4 days after transfection to detect the FLAG epitopes of the overexpression constructs in transfected cells. Only a small number of transfected cells (expressing the shRNA constructs and labeled green) co-expressed FLAG-PC1 or FLAG-PC2/UTR (labeled red), respectively. The relative fluorescence intensities elicited by anti-FLAG immunostaining (red) of all transfected cells (green) were measured and corrected for background. Data are presented as mean ± SEM in histograms (***p < 0.005; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in scatter plots (one dot represents the red fluorescence intensity of one analyzed cell). Examples of cells co-expressing either FLAG-PC1 or FLAG-PC2/UTR (FLAG is labeled red) together with the corresponding knockdown or control constructs (these shRNA constructs drive GFP expression and are labeled green) are shown. Both shRNA constructs specific for PC1 strongly reduced the expression of FLAG-PC1 (to 13% and 33% of control, respectively). Both shRNA constructs specific for PC2 strongly reduced the expression of FLAG-PC2/UTR (to 30% of control, respectively). Scale bars, 10 μm. c Primary cells prepared from the neocortices of E13.5 mice were cultured in the presence of 20 ng/ml bFGF, transfected on days in vitro 1 (DIV1) with the indicated shRNA constructs (in the vector pCGLH driving GFP expression; to better compare the single knockdowns with the double knockdown in terms of the amount of transfected plasmid DNA, all single knockdown constructs were co-transfected with the kdcontrol construct), and subjected to BrdU labeling 3 days later (DIV4). Indirect immunofluorescence co-stainings were performed to mark GFP-positive transfected cells (green) and BrdU-positive proliferating cells (red). Overlay pictures are presented to illustrate the BrdU-positive transfected cells (merge). Scale bar, 15 μm. d The percentage of BrdU-positive transfected cells was assessed. Reducing PC1 expression by the shRNA-mediated knockdown with either the construct Pkd1–kd1 or Pkd1–kd2 significantly increased the percentage of BrdU-positive proliferating cells (to 132% and 150%, respectively) when compared to the control (kdcontrol). Similarly, Pkd2–kd1 or Pkd2–kd2 increased the percentage of BrdU-positive proliferating cells (to 143% and 148%, respectively) when compared to the control. Data are presented as mean ± SEM in a histogram (***p < 0.005, **p < 0.01, *p < 0.05; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the percentage of BrdU-positive transfected cells of a group of 100 analyzed transfected cells). e Primary neocortical cells were prepared and cultured as described above. Without preceding BrdU labeling, the cells were fixed and co-immunostained for GFP and cleaved caspase 3 as an indicator of apoptotic cells. Neither knockdown of PC1 nor knockdown of PC2 significantly altered the percentage of apoptotic transfected cells when compared to the control (Kruskal–Wallis). Data are presented as mean ± SEM. f Triple immunostainings were performed to demonstrate that the BrdU-positive proliferating cells expressing kdcontrol or the polycystin-specific knockdown constructs co-express the RGC marker nestin. All BrdU-positive (labeled red) cells (100 cells for each shRNA construct analyzed), which expressed the different shRNA constructs (labeled green), co-expressed nestin (labeled magenta). Examples of triple stained NPCs are shown for each shRNA construct. Scale bar, 10 μm. g The overexpression of MYC-PC2 did not affect the number of BrdU-positive proliferating cells when compared to cells overexpressing EGFP. Data are presented as mean ± SEM in a histogram (Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the percentage of BrdU-positive transfected cells of a group of 300 analyzed transfected cells). The effect of overexpression of FLAG-PC1 on NPC proliferation could not be assessed as the expression rate of this construct was too low to allow the gathering of a sufficient number of cells for analysis
The constructs CBFRE-EGFP (Addgene plasmid #17705) and Tα1p-DsRed2 (Addgene plasmid #17707) were gifts from Nicholas Gaiano [36]. The construct pcDNA4/TO 3xFlag-m-PKD1-2xMyc/His, in the following designated FLAG-PC1, was a gift from Thomas Weimbs (Addgene plasmid #83452) [37]. The construct M-PKD2 (OF2-3), in the following designated MYC-PC2, was a gift from Gregory Germino (Addgene plasmid #21370) [38]. The construct Stat3 FLAG pRc/CMV, in the following designated STAT3-FLAG, was a gift from Jim Darnell (Addgene plasmid #8707) [39]. The construct FLAG-PC2/UTR corresponds to the coding region and the 3′-UTR of mouse Pkd2 according to NM_008861.3, cloned into the vector p3xFLAG-CMV-10 (Merck-Sigma).
Biochemical procedures
E14 mouse brains were homogenized in ice-cold phosphate-buffered saline containing a protease inhibitor cocktail (Roche, cOmplete). After sedimentation, the pellet was denoted crude membrane fraction, and was further extracted with detergent according to [31]. The SDS-PAGE was performed with 5% acrylamide under reducing conditions. A subsequent Western blot was analyzed with polyclonal antibodies to PC1 (Bioss, bs-2157R) or PC2 (Santa Cruz Biotechnology, sc-28331), respectively. The lysates of the HEK293 cells for the evaluation of the knockdown constructs were performed according to [31]. Monoclonal antibodies for Western blot detection were mouse anti-FLAG (Merck-Sigma, M2) or mouse anti-MYC (Roche, 9E10).
Immunohistochemistry and immunocytochemistry
Embryonic mouse brains of the indicated ages were fixed with 4% paraformaldehyde. Cryostat sections (14 μm thick) were prepared, permeabilized with PBS supplemented with 5% fetal calf serum and 0.2% Triton X-100, and then forwarded to indirect immunostainings. For detection, polyclonal antibodies to PC1 (Bioss, bs-2157R), PC2 (Santa Cruz Biotechnology, sc-25749), Pax6 (abcam, ab5790), Tbr2 (abcam, ab23345), and monoclonal antibodies to PC1 (Santa Cruz Biotechnology, sc-130554), PC2 (Santa Cruz Biotechnology, sc-28331), nestin (BD Biosciences, clone Rat-401), and phospho-vimentin (pS55, Enzo Life Sciences, clone 4A4) were applied. After fixation and permeabilization, the NPC cultures were indirectly stained with polyclonal antibodies to GFP (abcam, ab6556 and ab13970), DsRed (Clontech, #632496), cleaved caspase 3 (Chemicon, AB3623; Cell Signaling Technology, #9661), GFAP (Diagnostic BioSystems, RP014), and with monoclonal antibodies to BrdU (Sigma-Aldrich, clone BU-33), MAP2 (Sigma-Aldrich, clone HM-2), nestin (BD Biosciences, clone Rat-401), GFAP (Chemicon, MAB360), and GFP (Invitrogen, clone 3E6). The secondary antibodies Alexa Fluor 488 or 568-conjugated goat anti-rabbit, Alexa Fluor 488-conjugated goat anti-chicken, and Alexa Fluor 488, 568 or 647-conjugated goat anti-mouse (Molecular Probes) were used. All shRNA constructs applied in this study either drive GFP or RFP expression (GFP, green fluorescent protein and RFP, red fluorescent protein). Sections and cells were imaged with a fluorescence microscope (Leica, DM 6000 B) equipped with a Cool SNAP ES II digital camera.
Image acquisition and data analysis
Images were captured on a Leica TCS SP5 II scanning confocal microscope with 20 × HCX PL APO, 0.7 NA (dry) and 40 × HCX PL APO, 1.25 NA (oil) objectives. Z-stacks were collected at 1.0-μm steps for immunohistochemical analysis of E12.5 cortical slices, and at 0.8-μm steps for immunocytochemical analysis (10–15 sections). For higher magnification images of E14.5 cortical slices, Z-stacks were collected at 0.5-μm steps (three sections). For cell proliferation and apoptosis analysis, the percentage of BrdU or cleaved caspase 3-positive transfected cells, respectively, was determined. To assess the activities of the tubulin α1 promoter or the C promoter-binding factor 1 (CBF1)-responsive element (CBFRE), the DsRed2 or GFP fluorescence driven by the corresponding sensor constructs Tα1p-DsRed2 or CBFRE-EGFP, respectively, was measured. The quantification of the fluorescence intensities was performed after indirect immunostainings with the corresponding antibodies as described [31]. Net fluorescence was quantified. The measurements were obtained from regions of interest (ROIs). These ROIs represented the areas of the cell bodies of the analyzed cells. The areas of the cell bodies were defined by the fluorescence of the “transfection control channel” (e.g. if the Tα sensor fluorescence should be measured on the red channel, then the cell area would have been defined on the green channel, because the knockdown constructs in the pCGLH vector drive GFP expression). The expression level of the knockdown constructs did not differ significantly.
The Leica filter cubes (GFP, with BP470/40 for excitation and BP525/50 for suppression; TX2ET, with BP560/40 for excitation and BP 645/75 for suppression) were chosen such that a bleed through of fluorescence was minimized. The measurements represented an averaged intensity. The same acquisition parameters for each set of control and experimental samples were used. All the imaging was done in the same session. Data acquisition was performed with wide-field epifluorescence microscopy using the equipment described above and MetaMorph image analysis software (Visitron Systems).
To obtain the percentage of cell clusters either containing solely nestin-positive transfected cells, or comprising nestin-positive and nestin-negative transfected cells (mixed clusters), all cell clusters were counted that include at least two transfected cells spaced at a distance of not more than two cell diameters. For monitoring Notch signaling in sibling cell pairs originating from a common mother cell, the cell pairs chosen for the analysis of the GFP fluorescence derived from the CBFRE-EGFP sensor construct had to meet the following criteria: (1) each cell of a chosen cell pair had to be co-transfected with the respective polycystin knockdown construct and the CBFRE-EGFP sensor construct monitoring Notch signaling; (2) each cell of a chosen cell pair had to be BrdU positive; (3) the chromatin pattern (revealed by the BrdU staining) of both cells of a chosen cell pair had to be similar; and (4) the two cells of a chosen cell pair should be spaced at a distance of not more than one cell diameter.
Acquisition and analysis of data were performed by investigators blind to the experimental conditions.
Statistical analysis
Statistical analysis was done with Excel 2011 (Microsoft) and Prism 6 (GraphPad) software. Significance between groups was assessed by the Kruskal–Wallis test followed by post hoc analysis (Dunn’s multiple comparison test) or the Mann–Whitney test. The number of replicates for each experiment, the number of cell counts, the statistics including mean ± SEM for each group, and the related statistical test are presented in the Supplementary Table 1. Values of p < 0.05 were considered significant.
Results
PC1 and PC2 are expressed in nestin-positive NPCs during neurogenesis
In situ hybridization studies have suggested an early onset of PC1 expression in the developing brain [29, 30]. To examine, whether the polycystin proteins are expressed in the developing neocortex when neurogenesis is ongoing, we first performed immunostainings of coronal sections of E12.5 mouse brain (Fig. 1a). We employed polyclonal antibodies to the Pkd1 and Pkd2 gene products. These antibodies recognized PC1 or PC2, respectively, in crude membrane fractions of E14 brain extracts in the Western blot (Fig. 1b). The immunostainings revealed a prominent expression of PC1 and PC2 in nestin-positive cells in the ventricular zone (VZ) of E12.5 mouse neocortex (Fig. 1a). From the morphological appearance (radial glial cells extend characteristic nestin-positive processes, called radial glial fibers, towards the pial surface), these cells most likely represent RGCs. The co-expression of PC1 or PC2 with nestin was most prominent at the apical site of the cells in the VZ facing the lateral ventricle (V). Nestin is a marker for NPCs and at the apical site of the VZ predominantly labels NECs and RGCs [22, 25, 26]. We next conducted immunostainings of coronal sections of E14.5 mouse brain (Fig. 1c). Higher magnification images confirmed that PC1 and PC2 are co-expressed with nestin in the VZ. Pax6 is a transcription factor that is upregulated in RGCs. Co-immunostainings showed that PC1 and PC2 are expressed in Pax6-positive cells in the VZ. Phospho-vimentin was used as a marker for mitotically active RGCs in the VZ [40]. Co-immunostainings revealed that PC1 and PC2 are not only expressed in resting RGCs but also expressed in phospho-vimentin-positive mitotically active RGCs in the VZ. Tbr2 is a transcription factor which becomes active in basal progenitors (BPs, also called intermediate neural progenitors, INPs), a NPC type that is generated from NEC or RGCs [41]. However, most BPs leave the VZ to migrate to the subventricular zone. When performing co-immunostainings with a monoclonal anti-PC2 antibody and a polyclonal Tbr2 antibody, a weak expression of PC2 in Tbr2-positive cells in the transition from the VZ to the subventricular zone was detected (Suppl. Fig. 1).
Taken together, our results show that during active neurogenesis PC1 and PC2 are expressed most prominently in nestin-positive NPCs at the VZ, most likely RGCs.
The proliferation of FGF2-sensitive NPCs is controlled by PC1 and PC2
Nestin-positive NPCs have proliferative potential and are endowed with the ability for self-renewal [21, 24, 25]. Therefore, in a first step to figure out the functions of PC1 and PC2 in NPCs, we set out to examine the proliferative capacity of NPCs in cultures prepared from E13.5 mouse neocortex. In these cultures, we reduced the expression of PC1 by RNA interference employing the shRNA constructs Pkd1–kd1 and Pkd1–kd2 specific for the Pkd1 gene product [32]. The shRNA constructs Pkd2–kd1 and Pkd2–kd2 were applied to specifically reduce the expression of PC2 [33]. To prove the knockdown efficiency of the PC1-specific shRNA constructs, we co-expressed recombinant FLAG-PC1 with either kdcontrol, a shRNA construct which does not bind any specific target sequence, Pkd1–kd1 or Pkd1–kd2, respectively, in HEK293 cells. We found that the knockdown constructs Pkd1–kd1 and Pkd1–kd2 were able to significantly reduce the amount of overexpressed FLAG-PC1 (Fig. 2a, left panel), when analyzed in the Western blot. Additionally, FLAG-PC1 was co-expressed with these shRNA constructs in cultured E13.5 neocortical cells. Relatively few cells co-expressed FLAG-PC1 with the shRNA constructs, most likely because of a low transfection efficiency caused by the size of the plasmid (~ 18,000 bp). Nevertheless, a sufficient number of co-expressing cells could be collected. After conducting immunostainings to detect the FLAG epitope, we quantified the immunofluorescence of transfected cells, and affirmed that also in this assay both PC1-specific knockdown constructs strongly reduced the expression of FLAG-PC1 (Fig. 2b, left panel). Similarly, we substantiated the knockdown efficiency of the PC2-specific shRNA constructs Pkd2–kd1 and Pkd2–kd2 (Fig. 2a, b, right panels). However, the target sequence of the shRNA construct Pkd2–kd2 is not situated in the coding region of Pkd2 (encoding PC2) but in the 3′-UTR. Unfortunately, a recombinant FLAG-PC2/UTR construct comprising the coding region and the 3′-UTR of Pkd2 could not be expressed in HEK293 cells in a stable manner. A small but sufficient number of cultured E13.5 neocortical cells co-expressed FLAG-PC2/UTR together with the shRNA constructs. The bad expression of the FLAG-PC2/UTR construct may be due to the fact that the 3′-UTR of Pkd2 harbors a strong mir-17-5-p microRNA-binding site (http://www.targetscan.org). Both knockdown constructs specific for PC2 significantly reduced the amount of FLAG-PC2/UTR expressed by E13.5 neocortical cells (Fig. 2b, right panel). The knockdown construct Pkd2–kd1 was further evaluated in HEK293 cells. A recombinant MYC-PC2 construct without the 3′-UTR was co-expressed with either kdcontrol or Pkd2–kd1. The expression analysis via Western blot confirmed that Pkd2–kd1 significantly reduced the expression of overexpressed MYC-PC2 (Fig. 2a, right panel).
By means of the knockdown constructs directed to PC1 or PC2, respectively, the proliferative capacity of NPCs in cultures prepared from E13.5 mouse neocortex was analyzed by determining the number of 5-bromo-2′-deoxyuridine (BrdU)-positive cells (Fig. 2c). We found that both shRNA constructs directed to PC1 increased the number of BrdU-positive transfected cells (Fig. 2c, d). The same result was obtained when reducing the expression of PC2 (Fig. 2c, d). The effects of reducing PC1 and PC2 expression on the proliferation of NPCs were not additive as assessed by a double knockdown using the constructs Pkd1–kd2 and Pkd2–kd2 (Fig. 2d). None of the PC1- or PC2-specific knockdown constructs applied did significantly influence the cell survival of the transfected cells (Fig. 2e). A triple immunostaining to detect GFP+/BrdU+/nestin+-positive cells served as an example to show that essentially all proliferating cells analyzed for each shRNA construct were nestin-positive NPCs (Fig. 2f). Overexpression of MYC-PC2 did not significantly change the proliferation of NPCs when compared to EGFP (Fig. 2g). We observed, however, that the number of cortical cells elaborating neurites increased by the overexpression of MYC-PC2 (data not shown). The effect of PC1 overexpression on NPC proliferation could not be evaluated because the number of cells expressing the FLAG-PC1 construct was not high enough to allow an analysis (see above). In conclusion, PC1 and PC2 control the proliferation of NPCs in culture.
PC1 and PC2 promote the differentiation of NPCs
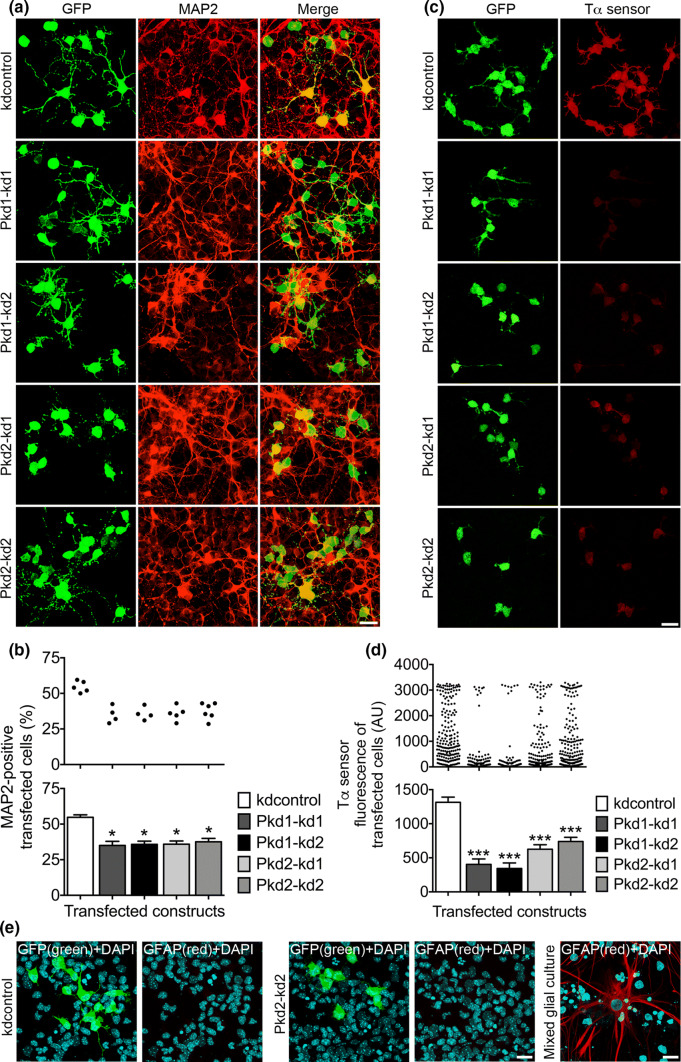
The results of the proliferation studies described above suggest that the polycystin proteins would promote the differentiation of NPCs. To verify this inference, the expression of PC1 or PC2 in E13.5 NPC culture was reduced by transfection of the shRNA constructs directed to PC1 or PC2, respectively. Then, we analyzed the differentiation of NPCs by determining the number of neurons emerging from the transfected cells. MAP2 was used as a neuronal marker. We found that either knockdown of PC1 or knockdown of PC2 expression reduced the number of MAP2-positive cells 3 days after transfection (Fig. 3a, b). To further prove the impact of PC1 and PC2 in driving the differentiation of NPCs towards a neuronal cell fate, we employed the sensor construct Tα1p-DsRed2 reporting the expression driven by the tubulin α1 promoter (Tα1p) [36]. The activity of this promoter has been shown to increase in neurogenic INPs, and in young neurons. Very few RGCs have been found to drive expression from this promoter [36]. E13.5 NPCs in culture were co-transfected with polycystin knockdown constructs and the Tα1p-DsRed2 sensor construct. Three days after transfection we measured the DsRed fluorescence intensity as an indicator of the Tα1p activity of the co-transfected cells. The data obtained from these experiments demonstrated that a reduction of either PC1 or PC2 expression resulted in a strong decrease of Tα1p activity (Fig. 3c, d). Interestingly, the data values of DsRed fluorescence in the case of kdcontrol covered the whole range of low fluorescence to high fluorescence. In contrast, a small gap in the middle range of the Tα sensor fluorescences could be observed in case of PC2 knockdown, and a large gap was evident in case of PC1 knockdown. From the morphological appearance (these cells extended neurites), the cells presenting high DsRed fluorescence were most likely young neurons. These results suggest that knockdown of PC2 and especially of PC1 forces the NPCs to divide symmetrically proliferative, generating two new NPCs with low Tα1p activity.
Fig. 3.
The expression of PC1 and PC2 is vital for the neurogenic differentiation of NPCs. a Primary mouse E13.5 neocortical cells were cultured in the presence of 20 ng/ml bFGF, transfected on DIV1 with the indicated constructs, and analyzed 3 days later following fixation. Double staining immunocytochemistry was performed to label GFP-positive transfected cells (green) and MAP2-positive differentiating cells (red). Overlay pictures are presented to illustrate the MAP2-positive transfected cells (merge). Scale bar, 15 μm. b The percentage of MAP2-positive transfected cells was ascertained. The percentage of MAP2-positive transfected cells was reduced by knockdown of either PC1 (to ~ 64% for both Pkd1 knockdown constructs) or PC2 (to ~ 65% for Pkd2–kd1 and ~ 69% for Pkd2–kd2), when compared to the control (kdcontrol). Data are given in a histogram as mean ± SEM (*p < 0.05; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the percentage of MAP2-positive transfected cells of a group of 600 analyzed transfected cells). c Primary neocortical cells were prepared and cultured as described above. However, on DIV1 the cells were co-transfected with the indicated shRNA constructs and the Tα1p-DsRed2 construct to report the expression from the Tα1p. The cells were fixed on DIV4. The transfected cells were co-immunostained for GFP (green) and DsRed (Tα sensor; red) to quantify the activity of the Tα1p. Scale bar, 15 μm. d Knockdown of either PC1 (to ~ 31% for Pkd1–kd1 and ~ 26% for Pkd1–kd2) or PC2 (to ~ 48% for Pkd2–kd1 and ~ 56% for Pkd2–kd2) led to a strong decrease of the expression from the Tα1p, when compared to the control (kdcontrol). Data are presented in a histogram as mean ± SEM (***p < 0.005; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the red fluorescence intensity of one analyzed cell). e Primary neocortical cells were prepared and cultured as described in a. The cells were co-stained for DAPI (to label all cell nuclei) and GFP (the transfected shRNA constructs drive GFP expression), or DAPI and glial fibrillary acidic protein (GFAP; to label astrocytes). No astrocytes could be detected in our NPC culture. A mixed glial culture prepared from E19 cortices served as positive control for the GFAP staining of astrocytes. Scale bars, 10 μm
Finally we examined, to which extent a differentiation to GFAP-positive astrocytes occured in our NPC culture (Fig. 3e). We did not observe any GFAP-positive astrocytes in our cultures, regardless which shRNA constructs were expressed. This result is in accordance with published data revealing that early cortical progenitor cells do not undergo astrocytic differentiation [42]. Together, these findings reveal that PC1 and PC2 promote the differentiation of NPCs towards a neuronal cell fate.
PC1 and PC2 restrict Notch signaling in NPCs
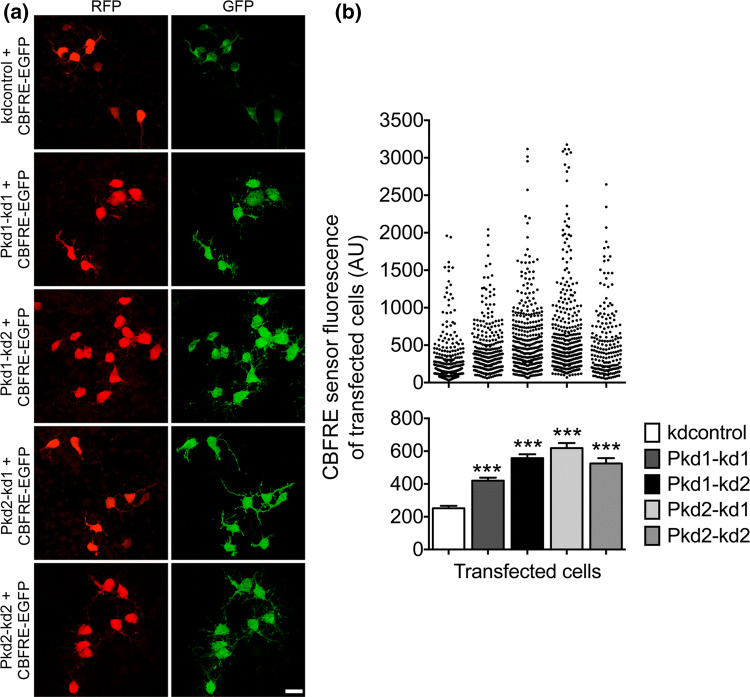
Active Notch signaling has been documented to be fundamental for maintaining the early progenitor cell identity and for inhibiting neuronal differentiation [36, 43–45]. Our data show that reducing PC1 or PC2 expression restrains the differentiation of NPCs into neurons. This result suggests that a decrease of PC1 or PC2 expression could lead to an enhanced Notch signaling in NPCs. To test this assumption directly, we made use of a sensor construct known to report canonical Notch signaling. On activation of Notch, the intracellular domain of this protein (NICD) is cleaved and translocates into the nucleus to interact with the transcriptional regulator CBF1 (also called CSL or RBP-J). The NICD-CBF1 complex then activates Notch target genes such as Hes1 and Hes5. We applied a CBFRE sensor construct containing a CBF1-responsive element upstream of EGFP [36]. This construct, CBFRE-EGFP, was co-transfected with the different polycystin knockdown constructs into E13.5 NPCs in culture. Two days after transfection the GFP fluorescence intensities of the co-transfected cells were measured as an indicator of the strength of Notch signaling. We found that knockdown of PC1 or PC2 expression considerably increased the number of cells with higher CBFRE sensor fluorescence, indicative of enhanced Notch signaling (Fig. 4a, b). Cells with low CBFRE sensor activity in the mouse neocortical VZ have been characterized as neurogenic INPs [36]. Collectively, our results are in line with the idea that PC1 and PC2 restrict Notch signaling in NPCs to promote the neuronal differentiation pathway of these cells.
Fig. 4.
Notch signaling is activated when PC1 or PC2 expression is lowered. a Primary cells prepared from the neocortices of E13.5 mice were cultured in the presence of 20 ng/ml bFGF, co-transfected on DIV1, and analyzed 2 days later following fixation. The co-transfection was carried out with the indicated shRNA constructs (in the vector pCRLH driving RFP expression) and the CBFRE-EGFP construct which allowed us to report the CBF1 activity. A co-immunostaining for RFP (red) and GFP (CBFRE sensor; green) was performed to quantify the activity from the CBF1-responsive element (CBFRE) of transfected cells. Scale bar, 15 μm. b A strong increase of CBF1 activity reported from the CBFRE was detected in the case of PC1 knockdown (to ~ 167% for Pkd1–kd1 and ~ 222% for Pkd1–kd2) or PC2 knockdown (to ~ 247% for Pkd2–kd1 and ~ 209% for Pkd2–kd2) when compared to the control (kdcontrol). Data are presented in a histogram as mean ± SEM (***p < 0.005; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the green fluorescence intensity of one analyzed cell)
PC1 and PC2 drive asymmetric divisions of NPCs
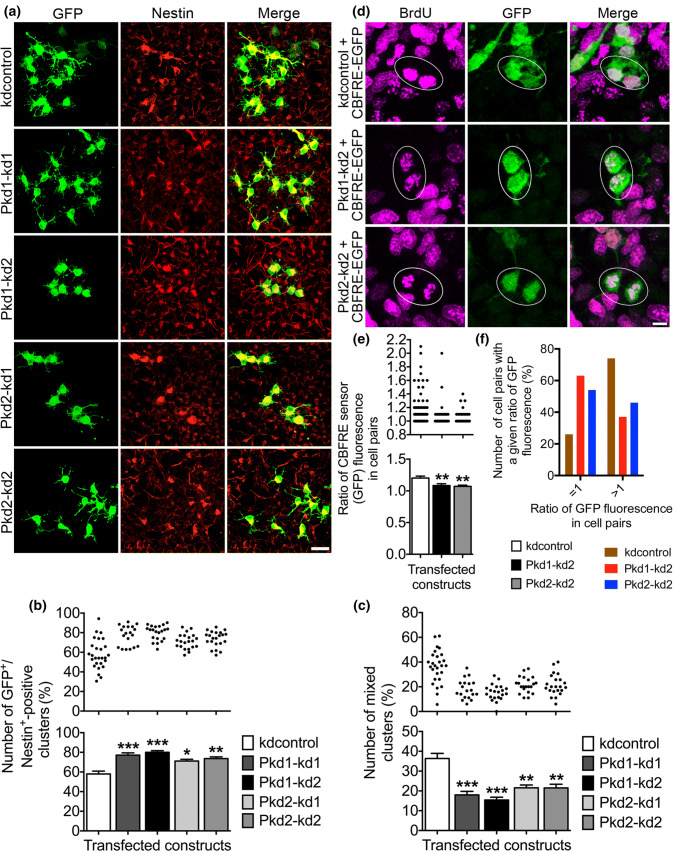
In our NPC culture, there are three modes of cell division that are most likely: (1) in a proliferative division, a nestin-positive NPC divides symmetrically into two new nestin-positive NPCs thus expanding the progenitor pool; (2) in a consumptive neurogenic division, a nestin-positive NPC divides symmetrically into two nestin-negative cells (most likely neurons, or possibly nestin-negative late NPCs); and (3) in a differentiative neurogenic division, a nestin-positive NPC divides asymmetrically into one nestin-positive NPC and one nestin-negative cell (most likely a neuron, or possibly a nestin-negative late NPC), this way maintaining the progenitor pool but also promoting neuronal differentiation [22–26, 35, 46, 47]. As PC1 and PC2 restricted Notch signaling in NPCs, and Notch signaling has been implicated in maintaining NPC identity, we asked whether PC1 and PC2 could promote neuronal differentiation by regulating the mode of NPC division. In an initial experiment to answer this question, we transfected E13.5 NPCs in culture with PC1-specific or PC2-specific shRNA constructs driving GFP expression. After allowing several rounds of cell division, a cell cluster analysis was performed by determining the percentage of cell clusters that encompassed solely GFP and nestin-positive (GFP+/nestin+) transfected cells (Fig. 5a, b). These cell clusters represented cell division mode (1). Then we counted the number of mixed cell clusters comprising GFP-positive but nestin-negative (GFP+/nestin−) cells in addition to GFP+/nestin+ transfected cells (Fig. 5a, c). These cell clusters most likely arose due to cell division mode (3); however, a combination of cell division modes (1) and (2) could also be conceivable.
Fig. 5.
Reduced PC1 or PC2 expression promotes symmetric divisions of NPCs. a Primary cells prepared from the neocortices of E13.5 mice were cultured in the presence of 20 ng/ml bFGF, transfected on DIV1 with the indicated constructs, and analyzed 3 days later following fixation. Immunofluorescence co-stainings were performed to label GFP-positive transfected cells (green) and nestin-positive progenitor cells (red). Overlay pictures are presented to illustrate the nestin-positive transfected cells in clusters (merge). Scale bar, 15 μm. b, c Knockdown of either PC1 (to ~ 133% for Pkd1–kd1 and ~ 138% for Pkd1–kd2) or PC2 (to ~ 122% for Pkd2–kd1 and ~ 127% for Pkd2–kd2) led to a rise in the number of cell clusters containing only nestin-positive transfected cells, when compared to the control (kdcontrol). Additionally, in the case of knockdown of PC1 (to ~ 49% for Pkd1–kd1 and ~ 42% for Pkd1–kd2) or PC2 (to ~ 59% for Pkd2–kd1 and Pkd2–kd2), the number of mixed clusters was decreased, when compared to the control (kdcontrol). Mixed clusters comprised not only GFP+/nestin+ double-positive cells, but also GFP-positive but nestin-negative cells (GFP+/nestin−). Data are presented in a histogram as mean ± SEM (***p < 0.005, **p < 0.01, *p < 0.05; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the percentage of GFP+/nestin+ or GFP+/nestin− clusters, respectively, in a group of 35 analyzed cell clusters containing transfected cells). d Primary cells prepared from neocortices of E13.5 mice were cultured in the presence of 20 ng/ml bFGF, co-transfected on DIV1 with the indicated constructs, and subjected to 6 h of BrdU labeling on DIV3. The cells were fixed 18 h after the BrdU labeling. An indirect triple immunostaining for RFP (red, label of transfected cells; not shown), BrdU (magenta), and GFP (green) was performed to determine the ratio of the CBFRE sensor fluorescence of transfected BrdU-positive cell pairs (marked by oval windows). Scale bar, 5 μm. e Knockdown of either PC1 or PC2 resulted in a decrease of the ratio of the CBFRE sensor fluorescence of transfected BrdU-positive cell pairs (to ~ 90% of control). Data are presented in a histogram as mean ± SEM (**p < 0.01; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the ratio of the CBFRE sensor fluorescences of both BrdU-positive sibling cells of one cell pair). f Deduced from the data shown in e, the percentage of BrdU-positive cell pairs with a ratio of GFP fluorescence that is equal to 1 (meaning equal CBFRE sensor fluorescences of both BrdU-positive sibling cells) or greater than 1 (meaning unequal CBFRE sensor fluorescences of both BrdU-positive sibling cells) is presented
We found that knockdown of PC1 or PC2 expression increased the number of cell clusters containing only GFP+/nestin+ double-positive cells, but decreased the number of mixed cell clusters. This result suggests that knockdown of PC1 or PC2 expression promotes proliferative symmetric cell divisions of nestin-positive NPCs, but reduced the number of asymmetric neurogenic cell divisions according to cell division mode (3). For additional proof of the hypothesis, that PC1 and PC2 promote asymmetric cell divisions, we again took advantage of the CBFRE sensor construct monitoring Notch signaling. We performed BrdU labeling of NPCs in culture suffering from reduced PC1 or PC2 expression, respectively. Next, BrdU-positive pairs of cells (siblings, originating from a common ancestor mother cell) co-transfected with the CBFRE sensor construct and the respective polycystin knockdown constructs were selected (Fig. 5d). Then, the GFP fluorescence intensities (originating from the CBFRE sensor) of both cells of a chosen cell pair were measured. In the end, the ratio of these fluorescence intensities was determined to figure out whether both cells of a BrdU-positive cell pair do present equal or unequal levels of Notch signaling. The data showed that knockdown of PC1 or PC2 expression led to an increased number of BrdU-positive cell pairs comprising cells with equal levels of Notch signaling (the ratio of GFP fluorescence in these cell pairs is 1; Fig. 5f), probably preferring the proliferative symmetric mode of cell division (Fig. 5e, f). Taken together, the results of these experiments support the view that PC1 and PC2 drive asymmetric neurogenic divisions of NPCs.
STAT3 antagonizes PC1 and PC2 in controlling NPC proliferation
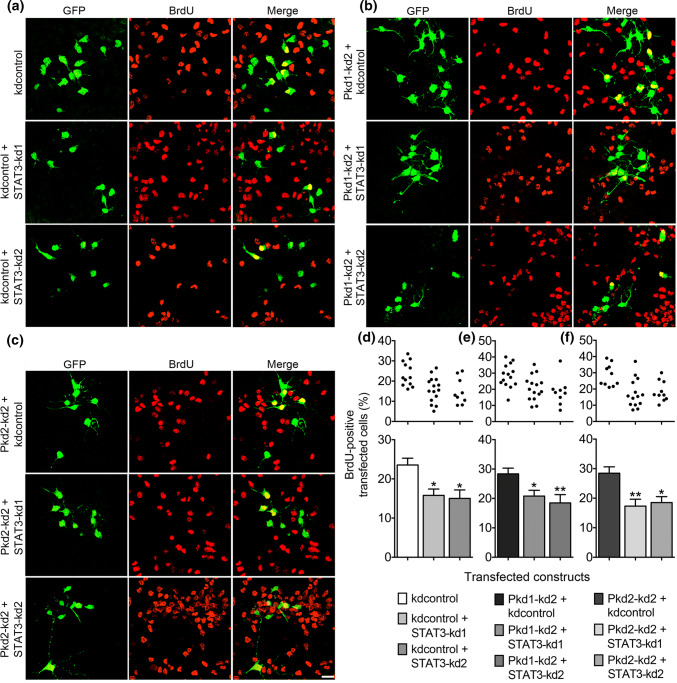
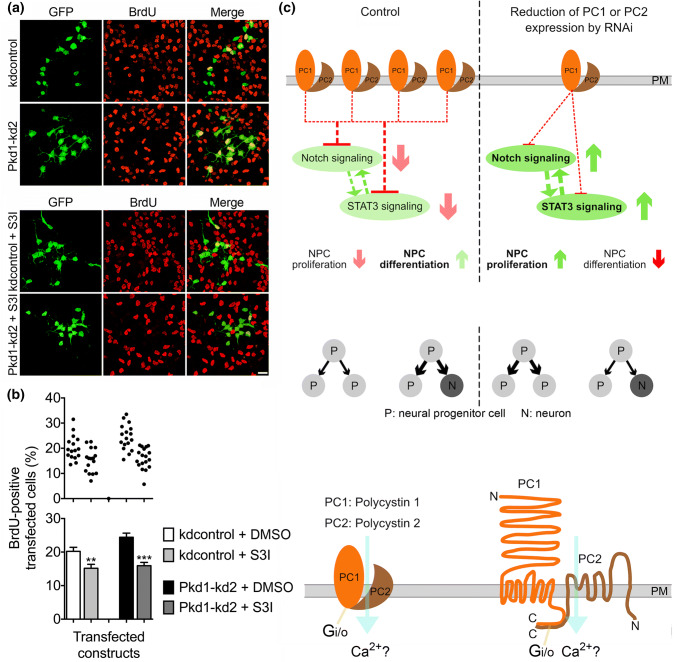
The data obtained so far in this study suggest a role of the polycystin proteins in restricting the proliferative expansion of the NPC pool while promoting neuronal differentiation. The transcription factor STAT3 has been described to preserve a pool of RGCs by driving symmetric divisions of RGCs, thereby maintaining the stemness of these NPCs [35]. Furthermore, reduced STAT3 expression in the cortex has led to a decrease in the number of proliferating RGCs and to a rise in the number of dividing INPs. As in the cortex, STAT3 has also been shown to inhibit the neuronal differentiation of NPCs in cell culture [35]. Hence, STAT3 and the polycystin proteins seem to operate in an opposite manner with respect to the proliferation and fate determination of NPCs. Thus, we asked whether reducing STAT3 expression could inhibit the increased proliferation of NPCs in culture elicited by knockdown of PC1 or PC2 expression. Again, the proliferative capacity was assessed by determining the number of BrdU-positive cells. Two independent STAT3 knockdown constructs were applied to reduce STAT3 expression [35]. According to published data, both STAT3 knockdown constructs reduced the number of BrdU-positive transfected cells (Fig. 6a, d). STAT3 overexpression stimulated proliferation by increasing the percentage of BrdU-positive transfected cells (Suppl. Fig. S2). Furthermore, scaling down STAT3 expression abrogated the increase in NPC proliferation elicited by knockdown of PC1 (Fig. 6b, e) or PC2 expression (Fig. 6c, f). Finally, the STAT3 inhibitor S3I-201 was applied to decrease NPC proliferation [48]. This STAT3 inhibitor not only decreased NPC proliferation but also abolished the rise in NPC proliferation evoked by knockdown of PC1 expression (Fig. 7a, b). Together, these data suggest that STAT3 antagonizes PC1 and PC2 in controlling NPC proliferation.
Fig. 6.
Knockdown of PC1 or PC2 expression increases the proliferation of NPCs in a STAT3-dependent manner. a–c E13.5 neocortical cells were treated and stained as described in Fig. 2. GFP-positive transfected cells are labeled green and BrdU-positive proliferating cells are shown in red. Overlay pictures are presented to illustrate the BrdU-positive transfected cells (merge). Scale bar, 15 μm. d The proliferation of NPCs decreased when reducing STAT3 expression by the shRNA constructs STAT3–kd1 (to ~ 67%) or STAT3–kd2 (to ~ 64%), respectively, compared to the control (kdcontrol). e, f Reducing STAT3 expression abrogated the increased proliferation of NPCs evoked by knockdown of PC1 (to ~ 73% for STAT3–kd1 and to ~ 65% for STAT3–kd2) or PC2 (to ~ 61% for STAT3–kd1 and to ~ 65% for STAT3–kd2). Data are presented as mean ± SEM in a histogram (**p < 0.01, *p < 0.05; Dunn’s multiple comparison post hoc test following Kruskal–Wallis) and in a scatter plot (one dot represents the percentage of BrdU-positive transfected cells of a group of 300 analyzed transfected cells)
Fig. 7.
Inhibiting STAT3 function abrogates the effect of PC1 knockdown on NPC proliferation. a Cells were treated as described in Fig. 2. S3I-201 (S3I), a cell-permeable STAT3 inhibitor, was added 24 h after transfection, where indicated. Scale bar, 15 μm. b S3I-201 reduced cell proliferation under control conditions (kdcontrol + S3I versus kdcontrol) to ~ 75%. Inhibition of STAT3 by S3I-201 abrogated the increased proliferation of NPCs evoked by knockdown of PC1 (to ~ 66%). Data are presented as mean ± SEM in a histogram (***p < 0.005, **p < 0.01; Mann–Whitney test) and in a scatter plot (one dot represents the percentage of BrdU-positive transfected cells of a group of 300 analyzed transfected cells). c Schematic model of the proposed function of PC1 and PC2 in NPC differentiation. PC1 and PC2 are likely to operate as a complex, because either a reduction of PC1 or PC2 expression results in the same functional and cellular phenotypes. A reduction of polycystin expression in NPCs functionally leads to increased CBF1-dependent Notch signaling and to enhanced STAT3 signaling. The reflections on the cellular level are increased NPC proliferation and decreased NPC differentiation. A symmetric division mode generating two new NPCs is preferred. How Notch signaling and STAT3 signaling are mechanistically connected is an open issue. Deduced from these results, we suggest that the PC1/PC2 complex directly or indirectly inhibits or modifies CBF1-dependent Notch signaling and STAT3 signaling, thus promoting NPC differentiation at the expense of NPC proliferation. An asymmetric division mode generating one NPC and one neuron predominates. Arrows with strong color stand for experimental results, arrows with faint color and dashed lines symbolize suggested functions. Published data have shown that PC1 and PC2 are transmembrane proteins, which span the plasma membrane (PM) several times, and which physically interact with their C-terminal domains to form a complex. PC2 can operate as a calcium-permeable cation channel, and PC1 can activate G -proteins as Gi/o [4, 6, 15–18, 38, 58]. C C-terminus, N N-terminus
Discussion
This study identifies PC1 and PC2 as critically involved in the neuronal differentiation of NPCs.
PC1 and PC2 were shown to be prominently expressed in nestin and Pax6-positive cells in the VZ during times of active neurogenesis. These cells are likely RGCs, which represents a prominent type of NPCs. Knockdown of PC1 or PC2 expression increased the NPC proliferation and reduced the differentiation of NPCs to MAP2-positive neurons in primary NPC culture. Furthermore, knockdown of PC1 or PC2 expression stimulated CBF1-dependent Notch signaling and promoted symmetric divisions of NPCs. The transcription factor STAT3 was shown to be essential for the increase in NPC proliferation driven by knockdown of PC1 or PC2 expression (Fig. 7c).
PC1 and PC2 have been extensively analyzed in the context of ADPKD [1–7]. These proteins physically interact to form a complex with a prominent, but not exclusive, localization in the primary cilia of kidney tubule cells. Increased cell proliferation and the growth of epithelial-lined cysts, which in the end destroy the normal renal parenchyma, have been widely observed in human ADPKD kidneys and in mouse models for this disease. Intriguingly, both overexpression of mutated PC1 or PC2, and also a reduction of PC1 or PC2 expression is sufficient to initiate cystogenesis [8–11]. Recently, a complex of PC1 and PC2 has been described in the primary cilia of P0 RGCs, and their genetic ablations affect planar cell polarity in RGCs and E1 ependymal cells [27, 28]. We observed PC1 and PC2 expression in apical RGCs in the VZ of E12.5 and E14.5 mouse neocortex. During this time, RGCs are actively involved in neurogenesis. Like NECs, apical RGCs can pass through symmetric proliferative cell divisions, thereby expanding the progenitor pool. However, as neurogenesis progresses, apical RGCs increasingly switch to an asymmetric differentiative, but self-renewing mode of cell division, which preserves one RGC per cell division [22–26]. We found that a reduction of PC1 or PC2 expression in NPCs, like in kidney epithelial cells, led to increased cell proliferation. Concomitantly, the neuronal differentiation was reduced. Our data provide strong evidence that a reduction of PC1 or PC2 expression promotes symmetric proliferative cell divisions. Together, these data suggest that PC1 and PC2 drive asymmetric differentiative cell divisions of NPCs. In our cell culture model, the PC1- and PC2-mediated asymmetric differentiative cell divisions were most likely neurogenic, as knockdown of PC1 or PC2 expression reduced the activity of the Tα1p-DsRed2 sensor and decreased the number of MAP2-positive neurons.
Active Notch signaling has been implicated in the maintenance of RGC identity in the developing mouse neocortex [43–45]. Additionally, CBF1-dependent Notch signaling has been shown to promote RGC character in the VZ, while loss of CBF1 signaling has been demonstrated to stimulate neurogenesis [36]. The results from this study demonstrated that a reduction of PC1 or PC2 expression enhances CBF1-dependent Notch signaling, strengthening the view that PC1 and PC2 are vital for advancing NPC differentiation. Furthermore, knockdown of PC1 or PC2 expression led to more progeny with an equal distribution of CBF1 activity of both daughter cells originating from one mother cell during cell division. This result suggests that PC1 and PC2 raise the percentage of cell divisions generating daughter cells with an unequal distribution of CBF1 activity implying an asymmetry in Notch signaling in paired daughter cells. An asymmetry of Notch signaling in daughter cells of asymmetric cell divisions resulting in one differentiating sibling (with low Notch signaling) and one self-renewing sibling (with high Notch signaling) has also been described in the zebrafish brain [49]. The transcription factor STAT3 has emerged as an important propelling factor for renal epithelial cell proliferation during development and during cystic growth [5, 13]. PC1 in complex with PC2 has been found to regulate STAT3, which is aberrantly activated in the kidneys of ADPKD patients and in ADPKD mouse models. In the developing mouse neocortex, STAT3 has been shown to be expressed in nestin-positive NPCs in the VZ, most of which have been specified as RGCs [35, 50]. Here, STAT3 signaling has increased the proportion of dividing RGCs and has maintained the stemness of RGCs at mid-neurogenesis, whereas knockdown of STAT3 expression has led to premature neurogenesis. Additionally, elevated STAT3 activity has been correlated with symmetric divisions of RGCs. Our findings revealed that either STAT3 knockdown or inhibition of STAT3 function inhibited the enhanced NPC proliferation driven by reduced polycystin expression. These results suggest that PC1 and PC2 reduce, inhibit, or modify elevated STAT3 signaling in NPCs leading to enhanced NPC differentiation.
Intriguingly, PC1 and PC1-regulated STAT3 have been described in different physiological settings both as a stimulator of cell proliferation, but also as an inducer of growth arrest and cell differentiation. For example, PC1 has recently been shown to promote osteoblastic differentiation by upregulating Runx2 expression via JAK2/STAT3 signaling [51]. Hence, an arising concern is how STAT3 can be a stimulator of cell proliferation, but also an inducer of cell differentiation. Probably, the regulation of STAT3 activity by post-translational modifications could be important in this context. Stimulated by seminal work that had elaborated important cytokine signaling pathways, many studies analyzing STAT3 activity have focused on the phosphorylation of the tyrosine residue 705 (pY705) [52]. Tyrosine-phosphorylated STAT3 has been shown to drive differentiation towards an astroglial cell fate in later cortical development (starting around E15), but has not been detected during NPC self-renewal [53]. However, the phosphorylation of the serine residue 727 (pS727) of STAT3 might also be important for the context-dependent regulation of STAT3 activity [53]. For instance, the non-phosphorylatable STAT3 mutant STAT3-S727A has been shown to considerably reduce the number of nestin-positive NPCs developing from mouse embryonic stem cells [54]. Phosphorylation of STAT3-S727 has been found in NPCs derived from embryonic stem cells by expression of the homeobox protein Hoxb1, which also maintained active Notch signaling [55]. The growth factors FGF2 and EGF have been demonstrated to increase the number of NPCs which are immunopositive for STAT3-pS727 staining [56]. In our experimental system, knockdown of PC1 or PC2 expression in NPCs subjected to growth stimulation by FGF2 also led to an increase in the number of STAT3-pS727-positive cells (data not shown) suggesting that the phosphorylation of STAT3-S727 could contribute to the maintenance of NPC identity driven by a reduction of polycystin expression.
Our data show that a reduction of PC1 or PC2 expression stimulated the proliferation of NPCs in a STAT3-dependent manner and enhanced CBF1-dependent Notch signaling. Notch-dependent growth of NPCs has been functionally associated with Notch-dependent phosphorylation of STAT3-S727 [57]. On the other hand, STAT3 has been described to increase the expression of the Notch ligand DLL1 thus stimulating Notch signaling and leading to the maintenance of NPCs [50]. How the STAT3 signaling and Notch signaling modulated by PC1/PC2 are mechanistically interconnected is challenging for future studies.
This study additionally raises the question why knockdown of either PC1 or PC2 induces a similar effect on proliferation and differentiation. Since the double knockdown of PC1 and PC2 did not evoke a stronger effect on NPC proliferation compared to the single knockdowns (Fig. 2d), and the overexpression of MYC-PC2 did not decrease NPC proliferation (Fig. 2g), a likely explanation would be that PC1 and PC2 have to operate as a heteromeric complex to induce the differentiation of NPCs. A known function of a co-assembled PC1/PC2 complex is that PC1 can activate the PC2 ion channel function, eventually allowing the influx of Ca2+ ions [18, 38]. In preliminary experiments, we inhibited Ca2+-signaling by applying the cell-permeant Ca2+-chelator BAPTA-AM. In our hands, the NPCs tolerate a single load of BAPTA-AM up to a concentration of 5–10 μM. When NPCs expressing kdcontrol were passively loaded with 3 μM BAPTA-AM after the transfection, an increase of NPC proliferation could be observed 3 days later. The stimulation of NPC proliferation by knockdown of PC1 was not affected by a single application of 3 μM BAPTA-AM. Interestingly, however, the stimulation of NPC proliferation by knockdown of PC2 was reduced by a single application of this Ca2+-chelator (data not shown). Although BAPTA-AM induced NPC proliferation in the control situation, it decreased proliferation in the case of PC2 knockdown. A conceivable explanation of this result could be that knockdown of PC2 enables PC1 to operate independently of PC2 leading to increased NPC proliferation. This function of PC1 is proposed to depend on Ca2+-signaling. If the Ca2+-signaling is inhibited by BAPTA-AM, PC1 would no longer be able to stimulate NPC proliferation. Although only a preliminary hypothesis, it would be in accordance to published data demonstrating that PC1 can activate Gi/o-type G-proteins (Fig. 7c) to modulate the activity of Ca2+ and K+ channels, an effect that is abrogated by co-expressed PC2 [58].
In conclusion, the present findings show that PC1 and PC2 are expressed in the mouse neocortex to the right time at the right place to be involved in RGC differentiation. All data obtained in primary NPC culture strengthen the view that PC1 and PC2 are involved in regulating CBF1-dependent Notch signaling and in controlling the mode of NPC division (symmetric or asymmetric). This way, PC1 and PC2 take part in governing the decision of NPCs to proliferate for expansion, or for self-renewal and differentiation to generate neurons.
Our results implicate STAT3 signaling as functionally connected to PC1 and PC2 in maintaining a balance between proliferation and differentiation of NPCs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Anne Lehner and Stefanie Raith for excellent technical assistance.
Abbreviations
- ADPKD
Autosomal-dominant polycystic kidney disease
- BAPTA-AM
Bis(2-aminophenoxy)ethane tetraacetic acid acetoxymethyl ester
- BP
Basal progenitor
- BrdU
5-Bromo-2′-deoxyuridine
- CBF1
C promoter-binding factor 1
- DIV
Days in vitro
- E
Embryonic
- INP
Intermediate neural progenitor
- MAP2
Microtubule-associated protein 2
- NEC
Neuroepithelial cell
- NPC
Neural progenitor cell
- PC1
Polycystin-1
- PC2
Polycystin-2
- P
Postnatal
- RGC
Radial glial cell
- RNAi
RNA interference
- STAT3
Signal transducer and activator of transcription 3
- Tα1p
Tubulin α1 promoter
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Simons M, Walz G. Polycystic kidney disease: cell division without a c(l)ue? Kidney Int. 2006;70(5):854–864. doi: 10.1038/sj.ki.5001534. [DOI] [PubMed] [Google Scholar]
- 2.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J. Polycystins and primary cilia: primers for cell-cycle progression. Annu Rev Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 4.Chapin HC, Caplan MJ. The cell biology of polycystic kidney disease. J Cell Biol. 2010;191(4):701–710. doi: 10.1083/JCB.201006173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weimbs T, Talbot JJ. STAT3 signaling in polycystic kidney disease. Drug Discov Today Dis Mech. 2013;10(3–4):e113–e118. doi: 10.1016/j.ddmec.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong AC, Harris PC. A polycystin-centric view of cyst formation and disease: the polycystins revisited. Kidney Int. 2015;88(4):699–710. doi: 10.1038/ki.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma M, Gallagher AR, Somlo S. Ciliary mechanisms of cyst formation in polycystic kidney disease. Cold Spring Harb Perspect Biol. 2017;9(11):1–16. doi: 10.1101/cshperspect.a028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris PC. What is the role of somatic mutation in autosomal dominant polycystic kidney disease? J Am Soc Nephrol. 2010;21(7):1073–1076. doi: 10.1681/ASN.2010030328. [DOI] [PubMed] [Google Scholar]
- 9.Lantinga-van Leeuwen IS, Dauwerse JG, Baelde HJ, Leonhard WN, van de Wal A, Ward CJ, Verbeek S, Deruiter MC, Breuning MH, de Heer E, Peters DJ. Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum Mol Genet. 2004;13(24):3069–3077. doi: 10.1093/hmg/ddh336. [DOI] [PubMed] [Google Scholar]
- 10.Chang MY, Parker E, Ibrahim S, Shortland JR, Nahas ME, Haylor JL, Ong AC. Haploinsufficiency of Pkd2 is associated with increased tubular cell proliferation and interstitial fibrosis in two murine Pkd2 models. Nephrol Dial Transplant. 2006;21(8):2078–2084. doi: 10.1093/ndt/gfl150. [DOI] [PubMed] [Google Scholar]
- 11.Jiang ST, Chiou YY, Wang E, Lin HK, Lin YT, Chi YC, Wang CK, Tang MJ, Li H. Defining a link with autosomal-dominant polycystic kidney disease in mice with congenitally low expression of Pkd1. Am J Pathol. 2006;168(1):205–220. doi: 10.2353/ajpath.2006.050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhunia AK, Piontek K, Boletta A, Liu L, Qian F, Xu PN, Germino FJ, Germino GG. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109(2):157–168. doi: 10.1016/S0092-8674(02)00716-X. [DOI] [PubMed] [Google Scholar]
- 13.Talbot JJ, Shillingford JM, Vasanth S, Doerr N, Mukherjee S, Kinter MT, Watnick T, Weimbs T. Polycystin-1 regulates STAT activity by a dual mechanism. Proc Natl Acad Sci USA. 2011;108(19):7985–7990. doi: 10.1073/pnas.1103816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick D, Bertuccio CA, Chapin HC, Lal M, Chauvet V, Caplan MJ. Polycystin-1 cleavage and the regulation of transcriptional pathways. Pediatr Nephrol. 2014;29(4):505–511. doi: 10.1007/s00467-013-2548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch T, Köttgen M, Hofherr A. TRPP2 ion channels: critical regulators of organ morphogenesis in health and disease. Cell Calcium. 2017;66:25–32. doi: 10.1016/j.ceca.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci USA. 1997;94(13):6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet. 1997;16(2):179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 18.Giamarchi A, Feng S, Rodat-Despoix L, Xu Y, Bubenshchikova E, Newby LJ, Hao J, Gaudioso C, Crest M, Lupas AN, Honoré E, Williamson MP, Obara T, Ong AC, Delmas P. A polycystin-2 (TRPP2) dimerization domain essential for the function of heteromeric polycystin complexes. EMBO J. 2010;29(7):1176–1191. doi: 10.1038/emboj.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalagiorgou G, Basdra EK, Papavassiliou AG. Polycystin-1: function as a mechano-sensor. Int J Biochem Cell Biol. 2010;42(10):1610–1613. doi: 10.1016/j.biocel.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Lemos FO, Ehrlich BE. Polycystin and calcium signaling in cell death and survival. Cell Calcium. 2018;69:37–45. doi: 10.1016/j.ceca.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127(24):5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 22.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franco SJ, Müller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77(1):19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141(11):2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 25.Laguesse S, Peyre E, Nguyen L. Progenitor genealogy in the developing cerebral cortex. Cell Tissue Res. 2015;359(1):17–32. doi: 10.1007/s00441-014-1979-5. [DOI] [PubMed] [Google Scholar]
- 26.Popovitchenko T, Rasin MR. Transcriptional and post-transcriptional mechanisms of the development of neocortical lamination. Front Neuroanat. 2017;11:102–119. doi: 10.3389/fnana.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohata S, Álvarez-Buylla A. Planar organization of multiciliated ependymal (E1) cells in the brain ventricular epithelium. Trends Neurosci. 2016;39(8):543–551. doi: 10.1016/j.tins.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohata S, Herranz-Pérez V, Nakatani J, Boletta A, García-Verdugo JM, Álvarez-Buylla A. Mechanosensory genes Pkd1 and Pkd2 contribute to the planar polarization of brain ventricular epithelium. J Neurosci. 2015;35(31):11153–11168. doi: 10.1523/JNEUROSCI.0686-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillaume R, D’Agati V, Daoust M, Trudel M. Murine Pkd1 is a developmentally regulated gene from morula to adulthood: role in tissue condensation and patterning. Dev Dyn. 1999;214(4):337–348. doi: 10.1002/(SICI)1097-0177(199904)214:4<337::AID-AJA6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Guillaume R, Trudel M. Distinct and common developmental expression patterns of the murine Pkd2 and Pkd1 genes. Mech Dev. 2000;93(1–2):179–183. doi: 10.1016/S0925-4773(00)00257-4. [DOI] [PubMed] [Google Scholar]
- 31.Brandt N, Franke K, Rašin MR, Baumgart J, Vogt J, Khrulev S, Hassel B, Pohl EE, Šestan N, Nitsch R, Schumacher S. The neural EGF family member CALEB/NGC mediates dendritic tree and spine complexity. EMBO J. 2007;26(9):2371–2386. doi: 10.1038/sj.emboj.7601680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battini L, Macip S, Fedorova E, Dikman S, Somlo S, Montagna C, Gusella GL. Loss of polycystin-1 causes centrosome amplification and genomic instability. Hum Mol Genet. 2008;17(18):2819–2833. doi: 10.1093/hmg/ddn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.AbouAlaiwi WA, Takahashi M, Mell BR, Jones TJ, Ratnam S, Kolb RJ, Nauli SM. Ciliary polycystin-2 is a mechanosensitive calcium channel involved in nitric oxide signaling cascades. Circ Res. 2009;104(7):860–869. doi: 10.1161/CIRCRESAHA.108.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz J, Franke K, Frick M, Schumacher S. Different roles of the small GTPases Rac1, Cdc42, and RhoG in CALEB/NGC-induced dendritic tree complexity. J Neurochem. 2016;139(1):26–39. doi: 10.1111/jnc.13735. [DOI] [PubMed] [Google Scholar]
- 35.Hong S, Song MR. Signal transducer and activator of transcription-3 maintains the stemness of radial glia at mid-neurogenesis. J Neurosci. 2015;35(3):1011–1023. doi: 10.1523/JNEUROSCI.2119-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449(7160):351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 37.Doerr N, Wang Y, Kipp KR, Liu G, Benza JJ, Pletnev V, Pavlov TS, Staruschenko A, Mohieldin AM, Takahashi M, Nauli SM, Weimbs T. Regulation of polycystin-1 function by calmodulin binding. PLoS One. 2016;11(8):e0161525. doi: 10.1371/journal.pone.0161525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanaoka K, Qian F, Boletta A, Bhunia AK, Piontek K, Tsiokas L, Sukhatme VP, Guggino WB, Germino GG. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature. 2000;408(6815):990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- 39.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9(8):984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 40.Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10(7):819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- 41.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molné M, Studer L, Tabar V, Ting YT, Eiden MV, McKay RD. Early cortical precursors do not undergo LIF-mediated astrocytic differentiation. J Neurosci Res. 2000;59(3):301–311. doi: 10.1002/(SICI)1097-4547(20000201)59:3<301::AID-JNR3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 43.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26(2):395–404. doi: 10.1016/S0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 44.Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr Opin Cell Biol. 2009;21(6):733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Berstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16(7):846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101(9):3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 48.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, Sebti SM, Turkson J. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces anti-tumor activity. Proc Natl Acad Sci USA. 2007;104(18):7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong Z, Yang N, Yeo SY, Chitnis A, Guo S. Intralineage directional Notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron. 2012;74(1):65–78. doi: 10.1016/j.neuron.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimatsu T, Kawaguchi D, Oishi K, Tekeda K, Akira S, Masuyama N, Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133(13):2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- 51.Dalagiorgou G, Piperi C, Adamopoulos C, Georgopoulou U, Gargalionis AN, Spyropoulou A, Zoi I, Nokhbehsaim M, Damanaki A, Deschner J, Basdra EK, Papavassiliou AG. Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK/STAT3 signaling axis. Cell Mol Life Sci. 2017;74(5):921–936. doi: 10.1007/s00018-016-2394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 53.Poser SW, Park DM, Androutsellis-Theotokis A. The STAT3-Ser/Hes3 signaling axis: an emerging regulator of endogenous regeneration and cancer growth. Front Physiol. 2013;4:273–279. doi: 10.3389/fphys.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32(5):1149–1160. doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gouti M, Gavalas A. Hoxb1 controls cell fate specification and proliferative capacity of neural stem and progenitor cells. Stem Cells. 2008;26(8):1985–1997. doi: 10.1634/stemcells.2008-0182. [DOI] [PubMed] [Google Scholar]
- 56.Nagao M, Sugimori M, Nakafuku M. Cross talk between Notch and growth factor/cytokine signaling pathways in neural stem cells. Mol Cell Biol. 2007;27:3982–3994. doi: 10.1128/MCB.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signaling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442(7104):823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 58.Delmas P, Nomura H, Li X, Lakkis M, Luo Y, Segal Y, Fernández-Fernández JM, Harris P, Frischauf AM, Brown DA, Zhou J. Constitutive activation of G-proteins by polycystin-1 is antagonized by polycystin-2. J Biol Chem. 2002;277(13):11276–11283. doi: 10.1074/jbc.M110483200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.