Abstract
With dual capacities for unlimited self-renewal and pluripotent differentiation, pluripotent stem cells (PSCs) give rise to many cell types in our body and PSC culture systems provide an unparalleled opportunity to study early human development and disease. Accumulating evidence indicates that the molecular mechanisms underlying pluripotency maintenance in PSCs involve many factors. Among these regulators, recent studies have shown that long non-coding RNAs (lncRNAs) can affect the pluripotency circuitry by cooperating with master pluripotency-associated factors. Additionally, trans-spliced RNAs, which are generated by combining two or more pre-mRNA transcripts to produce a chimeric RNA, have been identified as regulators of various biological processes, including human pluripotency. In this review, we summarize and discuss current knowledge about the roles of lncRNAs, including trans-spliced lncRNAs, in controlling pluripotency.
Keywords: Trans-spliced RNA, Pluripotency, Human embryonic stem cells, Long non-coding RNAs, Induced pluripotent stem cells
Introduction
Pluripotent stem cells (PSCs) categorically include both embryonic stem cells (ESCs) and induced PSCs (iPSCs). Both these subcategories of human PSCs (hPSCs) are able to give rise to many cell types from our body [1, 2] and this broad differentiation potential, known as pluripotency, makes hPSCs a valuable platform for studying early events in human embryonic development. Moreover, the capacity for unlimited self-renewal makes hPSCs a feasible cell source for use in regenerative medicine. The molecular pluripotency circuit in hPSCs is composed of and maintained by multilayered, coordinated gene expression networks. Thus, the core components of this circuitry are transcription factors, especially including NANOG, OCT4 and SOX2. Collectively, these transcription factors provide a point of integration for extracellular signals and orchestrate with epigenetic modifications on histones or chromatin to maintain hESCs and iPSCs in a long-term, proliferative state while suppressing differentiation. Recent studies have shown that, in addition to protein-coding genes, lncRNAs (> 200 nucleotides) are vital to various regulatory mechanisms of pluripotency maintenance and reprogramming. As such, the expression of lncRNAs has been shown to affect promoter-driven gene transcription [3, 4], epigenetic modification of histones and chromatin [5, 6], post-transcriptional regulation [7], miRNA availability through sponge function [8, 9], and imprinting [10, 11]. Several previous reviews have comprehensively discussed the expression and function of lncRNAs in pluripotency maintenance, reprogramming and lineage differentiation, including the mechanisms by which lncRNA regulates these biological events [12–14].
Both protein-coding mRNAs and lncRNAs are spliced and poly-adenylated to generate mature transcripts. However, splicing can occur either in cis or in trans [15, 16]. Cis-splicing joins exons from a single precursor mRNA (pre-mRNA), while trans-splicing joins exons from two or more distinct pre-mRNAs [17–20]. The best-characterized example of trans-splicing involves spliced-leader (SL) RNA in trypanosomes and nematodes. In SL trans-splicing, an identical 5′ capping exon is spliced onto various pre-mRNAs by a process that is dependent on canonical spliceosome function and the secondary structure of the SL RNA. This form of trans-splicing plays a pivotal role in processing the polycistronic transcriptional units of trypanosomes [18, 21] and is involved in the growth recovery of nematodes [22]. However, trans-splicing events in higher eukaryotes are not SL-type, and the functions and mechanisms of trans-splicing in these organisms are still unclear. In higher eukaryotes, trans-spliced RNAs have been shown to regulate apoptosis and axon guidance decisions in Drosophila [23, 24] and are associated with cancer development in humans [25]. With recent advances in next-generation sequencing technology, a continuously increasing number of trans-splicing events have been discovered in human cells or tissues. These events have many functions and recently a functional role for trans-spliced RNAs in human pluripotent stem cells has been described [5].
In this review, we summarily describe the molecular circuity underlying pluripotency maintenance in pluripotent stem cells and then highlight new discoveries that demonstrate a critical role for lncRNA in regulating pluripotency. Furthermore, we provide a summary of known trans-splicing events and describe how these events function in various biological process. Finally, we discuss how trans-spliced and non-trans-spliced lncRNAs are coordinated with transcription factors, epigenetic modification complexes and signaling molecules to constitute a molecular pluripotency network in hPSCs, focusing on pluripotency maintenance in ESCs and the establishment of pluripotency through reprogramming in iPSCs.
Human pluripotent stem cells (hPSCs): hESCs and iPSCs
During preimplantation stages, a fertilized egg undergoes a series of cleavage divisions and forms a compact embryonic sphere, known as a morula, which consists of blastomeres. The human morula further develops into a blastocyst, which contains an inner cell mass (ICM) and a fluid-filled cavity surrounded by a thin layer of trophectodermal epithelium. The ICM eventually gives rise to the embryo and the trophoblasts contribute to placenta formation. At this preimplantation stage, hESCs may be derived from the ICM and these cells will retain the pluripotency that is characteristic of the blastocyst. The successful derivation of hESCs has facilitated studies of regulatory mechanisms that operate during early embryo development and has also benefited studies on regenerative medicine. However, the generation of hESCs critically compromises the integrity of human embryos, thus raising the ethical question of whether it is acceptable to destroy human embryos for the purpose of hESC derivation. Fortunately, in 2007, Shinya Yamanaka’s group first demonstrated that iPSCs can be reprogrammed from human somatic cells by the expression of four transcription factors, OCT4, SOX2, KLF4 and c-MYC [26]. These iPSCs possess functional characteristics that are similar to hESCs, but unlike hESCs, the derivation of human iPSCs does not involve human embryos. Therefore, the use of iPSCs avoids the complex ethical issues surrounding the derivation and use of hESCs, while providing a viable source of hPSCs for experimentation and potential clinical application. Various in vitro and in vivo studies have shown that iPSCs are able to give rise to cell types representing all three embryonic germ layers [27–29], including endodermal hepatocytes [30], mesodermal cardiomyocytes [31], and various neuronal subtypes found in brain [32, 33] or spinal cord [27] (ectoderm). As such, iPSC technology provides an easy and efficient means to generate hPSCs from individuals with familial or sporadic forms of disease, as well as unlimited numbers of specific types of human cells that are not otherwise accessible for disease modeling and drug discovery. Moreover, iPSCs can be derived from any individual, providing immune-compatible, person-specific iPSCs for medical use. The iPSC reprogramming process also offers an opportunity to explore how the molecular circuity of pluripotency is established and maintained [34]. Overall, hESCs and iPSCs represent two valuable, but distinct, platforms for regenerative medicine and basic research on pluripotency circuitry.
The transcriptional regulatory network in pluripotency maintenance
OCT4/Oct4 (homeodomain transcription factor), SOX2/Sox2 (HMG-box transcription factor) and NANOG/Nanog (homeodomain transcription factor), which are all highly expressed in the ICM, the epiblast and undifferentiated PSCs, are the core molecules of a transcriptional regulatory network that controls pluripotency maintenance [35–38]. The functional roles of OCT4/Oct4, SOX2/Sox2 and NANOG/Nanog in pluripotency were first revealed in vivo by creating null mutations in mouse embryos. Knockout of Oct4 in mice prevented the formation of a pluripotent ICM population within blastocysts [39], and knockout of Sox2 expression caused a similar phenotype [38]. Ablation of Nanog is lethal in early embryonic stages, suggesting that its role is also critical in early development [36, 40]. In agreement with these findings, in vitro studies of mouse ESCs (mESCs) showed that disruption of Oct4 or Sox2 expression results in the loss of pluripotency and promotes trophoblastic differentiation [39, 41]. On the other hand, overexpression of Oct4 or Sox2 in mESCs can perturb the pluripotent state and, respectively, promote differentiation toward primitive endoderm or neuroectoderm [42, 43]. Perturbing Nanog expression in mESCs causes the loss of pluripotency and promotes in vitro differentiation toward an extraembryonic endoderm lineage [36]. Similar to the mouse studies, knockdown of OCT4 expression in hESCs induces trophectoderm differentiation [44, 45]. Additionally, overexpression of OCT4 in hESCs does not necessarily induce autonomous differentiation or an exit from pluripotency, but under certain conditions may promote in vitro differentiation toward an endodermal lineage [46]. Knockdown of NANOG expression induces neuroectodermal subsets of genes with anterior–posterior identities, while overexpression does not induce differentiation, but instead completely prevents neuroectodermal differentiation [47]. Functional studies in both mESCs and hESCs have revealed that OCT4/Oct4, SOX2/Sox2 and NANOG/Nanog can bind their own promoters/enhancers and the promoters of other genes encoding pluripotency-associated factors to activate expression [37, 41, 48–52]. Further, these three transcription factors are known to occupy the promoters of genes that specify differentiation of extra-embryonic, endodermal, mesodermal and ectodermal lineages, and inactivate expression [37].
Collectively, the previously mentioned studies have revealed a mechanism in which OCT4/Oct4, SOX2/Sox2 and NANOG/Nanog can bind together at their own promoters and promoters of genes encoding other core pluripotency-associated transcription factors, forming an interconnected auto-regulatory loop to activate downstream pluripotency-associated effectors and repress lineage-associated gene expression.
Signaling pathways that regulate pluripotency in hPSCs
The convergence of extracellular signaling events acting on intrinsic core pluripotency-associated transcription factors, OCT4, SOX2 and NANOG, to regulate gene expression at promoters and enhancers is a common theme in the literature describing pluripotency maintenance [53–55]. Interestingly, while hPSCs and mESCs share the same core pluripotency-associated transcription factors, the supporting signaling pathways have proven to be quite different. In mESCs, it known that activation of LIF and BMP signaling pathways is essential for pluripotency maintenance [53, 54]. However, LIF and BMP do not promote human pluripotency, and, alternatively, activation of Activin/Nodal, FGF, and WNT signaling pathways is critical regulators [56–59]. These findings have led to the discovery of two distinct pluripotency states, namely naïve and primed pluripotency, which describe ICM-derived mESCs and epiblast-derived epiblast stem cells (EpiSCs), respectively [60, 61]. Since mouse EpiSCs and hPSCs have similar growth requirements, as well as similar cellular and molecular characteristics, it is believed that hPSCs are in a primed pluripotency state and, thus, are likely to originate from human epiblasts [59].
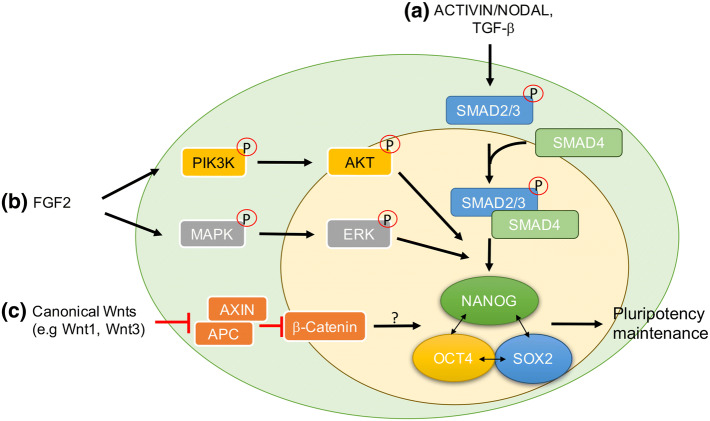
The roles of Activin/Nodal and FGF signaling pathways in maintaining pluripotency of hPSCs were first described by Vallier et al. and Ludwig et al., who demonstrated that inhibition of Activin/Nodal signaling pathway in hESCs promotes in vitro differentiation [55, 62]. Further studies showed that activation of the Activin/Nodal signaling mediator, SMAD2/3, promotes NANOG expression [56, 57] and inhibits differentiation-related BMP signaling by competing with the common mediator, SMAD4 [63] (Fig. 1a). However, neither Activin nor Nodal alone is sufficient to sustain hESCs in an undifferentiated state for an extended period. FGF2 can contribute to pluripotency maintenance in hESCs by activating the MAPK/ERK signaling cascade as a competence factor, which is necessary to cooperate with Activin/Nodal signaling [55] (Fig. 1b). FGF2 may also act to stimulate PI3K/AKT signaling and enhance SMAD2/3 occupancy on the promoters of pluripotency-associated genes [64, 65]. In addition to facilitating pluripotency-related gene transcription, both FGF-activated MAPK/ERK and PI3K/AKT signaling cascades can repress differentiation-related BMP signaling pathways to further promote pluripotency [57, 66, 67] (Fig. 1b).
Fig. 1.
Signaling pathways in human pluripotency maintenance. In hPSCs: a Activin/Nodal signaling molecules induce SMAD2/3 phosphorylation and interaction with SMAD4. The SMAD2/3–SMAD4 complex then translocates to the nucleus, where it promotes NANOG expression. NANOG cooperates with OCT4 and SOX2 to maintain pluripotency in hPSCs. b FGF2 activates phosphorylation of PI3K/AKT or MAPK/ERK signaling cascades, which cooperate with SMAD2/3–SMAD4 complexes to promote pluripotency in hPSCs. c Canonical Wnts inhibit the activity of the APC/AXIN/GSK-3β complex, by which β-catenin is protected from degradation. β-Catenin translocates to the nucleus, where it cooperates with TCF/LEF family members to regulate expression of target genes. Whether the canonical Wnt/β-catenin cascade maintains pluripotency or promotes differentiation of hESCs remains controversial
Wnt ligands are secreted glycoproteins that are involved in regulating diverse biological processes, such as cell proliferation, differentiation, migration, and asymmetric cell division. It is established that Wnt ligands exert their functions through the canonical Wnt/β-catenin cascade, or non-canonical Wnt/JNK and Wnt/Calcium-related pathways [68]. The activation of canonical Wnt/β-catenin signaling protects β-catenin from degradation by inhibiting the APC/AXIN/GSK-3β complex (Fig. 1c). Subsequently, stabilized β-catenin can translocate to the nucleus, where it interacts with TCF/LEF family transcription factors to regulate gene expression. Activation of canonical Wnt/β-catenin signaling maintains the naïve pluripotency state of mESCs [69], but the role of canonical Wnt/β-catenin signaling is less clear in hPSCs. Sato et al. reported that activation of Wnt/β-catenin signaling by either WNT3A or GSK-3β inhibitor, BIO, can maintain pluripotency in hESCs [70]. In contrast, Bone et al. and Nakanishi et al. demonstrated that WNT3A or BIO induces primitive and definitive endoderm differentiation of hESCs [71, 72]. Other studies further showed that the activation of canonical Wnt/β-catenin signaling may maintain the expression of pluripotency markers but does not support the long-term maintenance of hESCs [73–75]. Therefore, the role of canonical Wnt/β-catenin signaling in the maintenance of pluripotency or promotion of differentiation in hESCs will require further study.
Unlike canonical Wnt/β-catenin signaling, non-canonical Wnt signaling is usually recognized as a differentiation-related pathway. For example, the non-canonical Wnt/JNK cascade is known to regulate cell polarity during morphogenesis processes, such as gastrulation and neural tube closure during early embryonic development [76–78]. On the other hand, non-canonical Wnt/Calcium-related cascades control differentiation to specific cell lineages, as well as migration. This non-canonical pathway operates via activation of heterotrimeric G proteins to promote phospholipase C-mediated release of calcium from intracellular stores, which in turn activates downstream effectors such as PKC, CamKII and NFAT [79, 80].
LncRNA regulation of pluripotency circuitry
lncRNA, which refers to non-coding RNA transcripts longer than 200 nucleotides, is an emerging category of molecules that function in a variety of biological processes, including X-chromosome inactivation [81], imprinting [82], epigenetic modifications [5, 83], transcriptional regulation [84, 85], miRNA sequestration [8], nuclear trafficking [86] and nuclear body formation [2, 87]. The development of platforms for screening lncRNAs, such as targeted microarrays or next generation sequencing, has accelerated the discovery of lncRNA functions in various cell types. Accumulating evidence shows that lncRNA also functions in pluripotent stem cells. Dinger et al. were the first to show that the expression of a set of lncRNAs is controlled by NANOG, OCT4 and SOX2 during mESC differentiation and directly confirmed the regulatory role of NANOG and OCT4 on lncRNA promoters by chromatin immunoprecipitation (ChIP) [88]. Sheik Mohamed et al. demonstrated that modulation of lncRNA-AK141205 expression induced the loss of pluripotency and promoted differentiation of mESCs, suggesting a functional role for lncRNA in pluripotency maintenance [89].
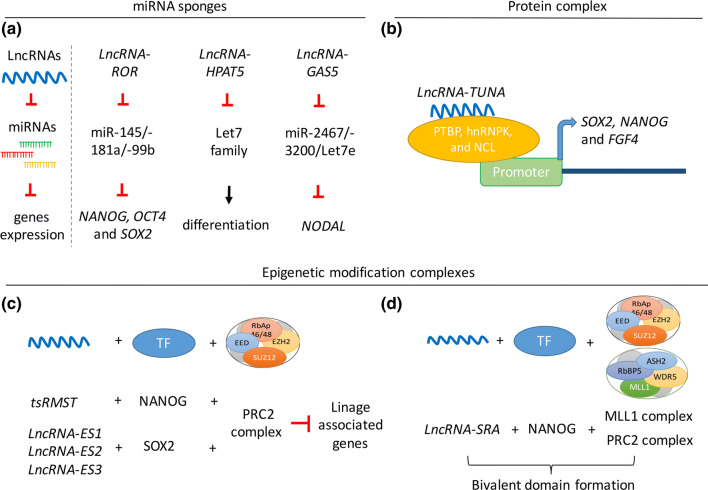
The roles of lncRNAs in hPSCs have also been studied. Loewer et al. first discovered over 100 lncRNAs with significantly higher expression levels in hPSCs, as compared to fibroblasts [9]. Among these hPSC-enriched lncRNAs, lncRNA-ROR was shown to modulate NANOG, OCT4 and SOX2 expression in hESCs by competing with miR-145/181a/99b [8, 9], while lncRNA-HPAT5 counteracted let-7 activity to protect hESCs from differentiation [90]. Furthermore, Xu et al. demonstrated that the lncRNA, GAS5, can act as an miR-2467/3200/let7e sponge to regulate NODAL signaling and maintain hESC pluripotency [91]. These findings are summarized in Fig. 2a.
Fig. 2.
The roles of lncRNAs in regulating hPSCs pluripotency. a lncRNAs (lncRNA-ROR, lncRNA-HPAT5 and lncRNA-GAS5) act as miRNA sponges to inhibit miRNA activity, promoting pluripotency maintenance of hPSCs. b lncRNA-TUNA interacts with PTBP1, hnRNPK, and NCL1 to regulate NANOG and SOX2 expression through promoter occupancy. c lncRNAs (tsRMST and lncRNA-ES1/ES2/ES3) interact with the PRC2 complex and pluripotency-associated factors (NANOG or SOX2) to inhibit expression of linage-associated genes. d lncRNA-SRA interacts with MLL1 complex, PRC2 complex and the pluripotency-associated factor, NANOG, to promote bivalent domain formation
In addition to functioning as a miRNA sponge, lncRNAs can also form complexes with proteins to regulate pluripotency maintenance. For example, Rana’s group demonstrated that lncRNA-TUNA forms a complex with RNA-binding proteins, PTBP1, hnRNPK and NCL, to regulate NANOG, SOX2 and FGF4 expression through promoter occupancy [92] (Fig. 2b). Ng et al. identified 36 lncRNAs with expression profiles that mirrored expression of OCT4 and NANOG during hESC differentiation. The authors further demonstrated that impaired expression of three of the identified hESC-enriched lncRNAs (lncRNA-ES1, lncRNA-ES2 and lncRNA-ES3) in hESCs resulted in upregulation of early lineage-associated genes and downregulation of pluripotency-associated genes by a process dependent on interaction with SUZ12, a PRC2 repressive complex component, and SOX2 [83]. Yu et al. reported that a trans-spliced lncRNA, lncRNA-tsRMST interacts with NANOG and PRC2 complex to repress differentiation-associated genes and signaling pathways to promote pluripotency maintenance [5, 93] (Fig. 2c). Further, a recent report showed that lncRNA-SRA can interact with both the MLL1/SET1 activating complex and the PRC2 repressive complex, suggesting that lncRNA may also be involved in bivalent domain formation, which is functionally important in the balance of pluripotency and differentiation [94] (Fig. 2d). Finally, lncRNA has also been shown to contribute to pluripotency reprogramming. For instance, disruption of lncRNA-SRA was found to decrease the number of reprogrammed iPSC colonies [95], and lncRNA-ROR and lncRNA-HPAT5 act as miRNA sponge to promote pluripotency reprogramming and iPSC generation [9, 90]. In contrast, lncRNA-p21 hampers pluripotency reprogramming of iPSCs by interacting with SETDB1 and DNMT1 to sustain H3K9me3 modifications and CpG methylation [96]. Based on this mounting evidence, lncRNA can be considered as an emerging player in pluripotency maintenance and reprogramming. For further detailed examples regarding the expression, function roles and the mechanism by which lncRNA regulates pluripotency and reprogramming, readers are referred to the review by Ghosal et al. [14] and Chakrabarti et al. [97].
Trans-spliced RNA (tsRNA)
RNA splicing removes introns from pre-mRNA and joins exons to generate mature and functional mRNA. Cis-splicing combines exons from the same pre-mRNA transcript, whereas trans-splicing uses two or more pre-mRNA transcripts to form chimeric, non-colinear transcripts, which may either encode new protein products or serve as regulatory non-coding RNAs. Trans-splicing can occur within a single gene or between different genes [15, 16] and was first discovered in Trypanosoma brucei and other trypanosomes, where the process entails a short-SL RNA being spliced onto the 5′ termini of all mRNAs [98–100]. SL-type trans-splicing was later found to occur in nematodes, such as Caenorhabditis elegans and Panagrellus redivivus [20, 101]. Intriguingly, SL-type trans-splicing can be processed by HeLa cell extracts, suggesting that the machinery is broadly conserved in eukaryotes [20, 102–104]. However, no observations of SL-type splicing events have been reported in higher eukaryotes. In higher eukaryotes, the most well-known trans-spliced RNAs are Mod4, which regulates apoptosis, and Lola, which regulates axon guidance, in Drosophila [23, 24, 105–107]. In a global exploration of trans-splicing events in insect linages by Kong et al., Mod4 trans-splicing was found to be conserved in two Diptera and two Lepidoptera species [108]. Interestingly, Kong et al. also showed that 146 trans-spliced RNAs resemble cognate genes in different species, suggesting that trans-splicing may function as a buffer system to preserve the function of genes that have undergone “breakup” during the evolution of insect linages [108].
In contrast with the numerous trans-splicing events that have been identified in insects or lower eukaryotes, only around 20 have been identified in humans to date. Moreover, more than half of the known human trans-splicing events were identified in cancer cell lines or tissues, and the functions of most human trans-splicing events are still not well characterized (Table 1). The most prominent examples of known functional trans-splicing in normal human cells are JAZF1-JJAZ1 and tsRMST. JAZF1-JJAZ1 is translated into a chimeric protein with anti-apoptotic function and is believed to be a prerequisite for chromosomal exchange [16, 109, 110], while tsRMST recruits the PRC2 complex to repress differentiation-associated genes in hESCs [5].
Table 1.
Summary of trans-splicing events identified in human
| Trans-splicing | Biological function | Cell types | Validation | Read through | References | |||
|---|---|---|---|---|---|---|---|---|
| RT-PCR | Sequencing | Northern, RPA Western or FISH | Genome rearrangement | |||||
| TMEM79-SMG5 | Cancer marker |
LNCaP cells Cancer tissue |
Yes | Yes | No | No | No | [111] |
| TSNAX-DISC1 | Cell growth | Endometrial carcinomas | Yes | Yes | Western | No | Unknown | [112] |
| hER-α | May modulate hER-αbinding | MCF7, T47D, ZR75, cells, mammary gland, ovary, liver, endometrium | Yes | Yes | RPA | No | – | [113] |
| SP1 | May modulate the activity of SP1 | Hep2 cells | Yes | Yes | RPA | No | – | [114] |
| RGS12TS | Induction of nuclear abnormalities | COS-7, HEK293T cells | Yes | Yes | Western | No | – | [115] |
| PAX3-FOXO1 | Activates MYOD and MYOG | Rhabdomyosarcoma | Yes | Yes | FISH, Western | No | No | [116, 117] |
| PJA2-FER | Cancer maker | Non-small cell lung cancer | Yes | Yes | No | No | No | [118] |
| JAZF1-JJAZ1 | Anti-apoptotic | Endometrial stromal cells | Yes | Yes | FISH | No | No | [25] |
| ZC3HAV1l-CHMP1A | Onset of chromosomal translocation | Mammary epithelial cells | Yes | Yes | No | No | No | [119] |
| AF4, AF9, ELL, ENL, MLL | Onset of chromosomal translocation | PBMCs | Yes | Yes | No | No | No | [120] |
| CYP3A43-3A4 CYP3A43-3A5 | May alter cellular location | HepG2 cells and normal Liver tissue. | Yes | Yes | RPA | No | No | [121] |
| CYCLIND1-TROP2 | Cell growth | OVCA-432, MCF7 cells | Yes | Yes | Northern, RPA | No | No | [122] |
| CoAA-RBM4 | Regulates CoAA activity | Wild range of cell lines and normal tissues | Yes | Yes | Western | Unknown | Unknown | [123] |
| CDC2L2 | Unknown | Testis | Yes | No | FISH | Unknown | – | [124] |
| CAMK2G-SRP72 | Unknown | Islet cells | Yes | Yes | Western | Unknown | No | [125] |
| ACAT1-Amp | Unknown | THP-1, HEK293 cells | Yes | Yes | Northern | Unknown | No | [126–128] |
| SLC45A3-ELK4 | Cancer maker | (Benign) prostate cancer tissues and LNCaP cells | Yes | Yes | FISH | No | No | [124] |
| tsRMST | Pluripotency maintenance | hESCs | Yes | Yes | RPA | No | No | [5] |
RPA RNase protection assay
The function of tsRMST in human pluripotency
RMST (ENSG00000255794.6) is located on the q arm of chromosome 12 (chr12:97431653-97565015). The entire linear RMST transcript contains 13 exons and is ~ 2.6 kb in length. RMST was first identified as a cancer marker in alveolar-subtype rhabdomyosarcoma [129], and a subsequent study in mice showed that Rmst is also expressed in the ventral mesencephalic floor plate and anterior dorsal midline cells [130]. The transcript was then identified as a neurogenic lncRNA that is important for neural differentiation. This discovery was based on genome-wide profiling and functional screening of lncRNAs with differential expression during in vitro differentiation of hESCs [83]. Importantly, a further mechanismic study showed that RMST expression is important for the binding of SOX2 to neurogenic genes, such as ASCL1, NEUROG2, HEY2, and DLX1 [131].
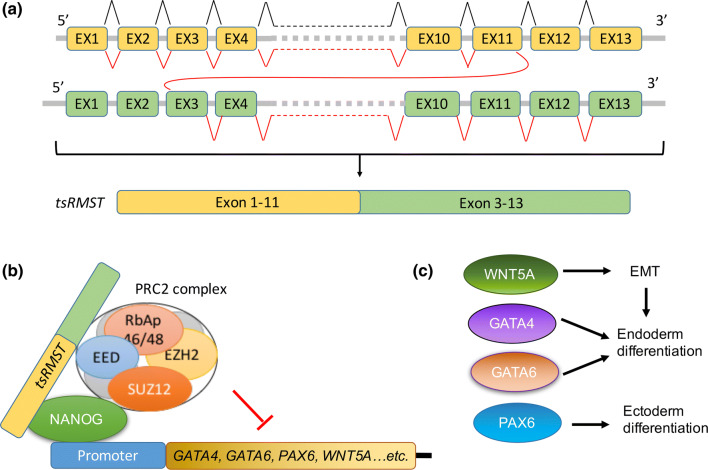
tsRMST is the trans-spliced isoform of RMST, in which the 3′ terminus of RMST exon 11 is joined to the 5′ terminus of exon 3, forming a linear RNA with scrambled exon order (Fig. 3a). The enrichment of tsRMST in the oligo-dT purified mRNA fraction indicated that tsRMST is polyadenylated. Furthermore, tsRMST was found to be degraded by RNaseR treatment, which confirmed that tsRMST is not a circular RNA (circRNA) with scrambled exon order [5]. When the tsRMST sequence was examined by a coding potential calculator [132], it showed low protein coding potential, suggesting that tsRMST is likely to be a trans-spliced lncRNA. Furthermore, the conserved exon–intron boundaries in both RMST and tsRMST suggest that conventional splicing machinery is involved in the trans-splicing process, and the downregulation of tsRMST in differentiated hESCs suggests that trans-splicing is tightly regulated. It has been demonstrated that splicing factors and complementary sequences in flanking introns regulate the biogenesis of circRNAs, another type of alternatively spliced RNA with scrambled exon order. Therefore, it will be interesting to explore whether trans-splicing events are regulated through similar mechanisms [133–135].
Fig. 3.
tsRMST promotes pluripotency maintenance in hESCs. a The exon order of tsRMST is shown. EX exon. b In hPSCs, interaction of tsRMST with NANOG and PRC2 complex promotes occupancy on inactive genes, such as GATA4, GATA6, PAX6 and WNT5A. c The expression of GATA4 and GATA6 promote endoderm differentiation of hPSCs, while PAX6 promotes ectoderm differentiation. The expression WNT5A further activates epithelial–mesenchymal transition (EMT) to promote endoderm differentiation
tsRMST is highly expressed in hPSCs, including hESCs and human iPSCs, as compared to differentiated somatic cells. The disruption of tsRMST expression in hESCs hampers pluripotency-associated gene expression, suggesting a role in pluripotency maintenance. Mechanistically, tsRMST interacts with pluripotency factor, NANOG, and PRC2 complex component, SUZ12, which acts to suppress lineage differentiation and promote pluripotency. These interactions suggest that tsRMST may act as a co-repressor of NANOG and the PRC2 complex to maintain pluripotency. Interestingly, a study by our group found that tsRMST does not interact with SOX2; however, an interaction between SOX2 and the colinear RMST transcript was shown by Ng et al. [83]. Therefore, trans-splicing may modify the protein interactome of RMST or other lncRNAs. Considering that tsRMST and RMST contain highly similar nucleotide sequences with the only difference being in the trans-splice junction, it is reasonable to suspect that trans-splicing may alter the RNA structure to modulate the protein interactome of lncRNAs in humans. ChIP-Seq-based global analysis of gene occupancy by SUZ12 and NANOG indicated that NANOG and the PRC2 complex co-occupy inactive genes, such as early lineage-associated transcription factors PAX6, GATA4 and GATA6, as well as the signaling ligand, WNT5A [5, 93]. In hESCs with impaired tsRMST expression, NANOG and the PRC2 complex are not associated with PAX6, GATA4, GATA6 and WNT5A promoters, suggesting that tsRMST promotes NANOG and PRC2 complex occupancy on inactive genes in hESCs (Fig. 3b). With these results in mind, we proposed a model for regulation of hESCs, wherein tsRMST interacts with NANOG and the PRC2 complex to repress expression of signaling ligands and early lineage-associated transcription factors to promote pluripotency. Along these lines, we observed that downregulation of tsRMST disrupted the repressive complex and activated GATA4, GATA6, PAX6 and WNT5A expression during in vitro differentiation of hESCs. Moreover, the expression of WNT5A in differentiated hESCs further activated epithelial–mesenchymal transition to promote endoderm differentiation (Fig. 3c). Thus, tsRMST blocks differentiation by affecting multiple layers of the pluripotency maintenance machinery, specifically repressing both core transcription factors and signaling ligands.
Conclusions and perspectives
In hPSCs, the pluripotency circuitry is tightly regulated at multiple layers to orchestrate self-renewal, cell differentiation and pluripotency. Master pluripotency-associated transcription factors (i.e., NANOG, OCT4, SOX2) form the core network that promotes the pluripotency program and suppresses differentiation programs. Meanwhile, an additional layer of regulation centers around signaling molecules, such as FGF2, NODAL/ACTIVIN and WNTs, which support the pluripotency core network by activating kinase cascades. The role of lncRNA as an additional regulatory layer in pluripotency circuitry is an emerging concept and is supported by recent findings that individual lncRNAs can inhibit differentiation-associated miRNAs, such as miR-145 and miR-34a, to promote the expression of core transcription factors.
Our exploration of the functions of trans-splicing in humans has only just begun and many unanswered questions remain. How many trans-splicing products can be found in human transcriptome? Are there unknown functions for tsRMST and other trans-splicing events? How are trans-splicing events regulated? What splicing machinery is utilized in human trans-splicing and does it resemble the canonical splicing machinery? With so many central questions remaining, it will be interesting and fruitful to continue exploring the unknown field of trans-splicing in humans.
Acknowledgements
This work was supported by Grants from Academia Sinica (Thematic Project, AS-104-TP-B09 and AS-103-TP-B10) and the Ministry of Science and Technology (MOST104-0210-01-09-02/105-0210-01-13-01/106-0210-01-15-02/106-2321-B-001-049-MY3) and National Health Research Institutes (HRI-EX104-10320SI).
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, Lu Z, Li H, Na J, Tang F, Wang J, Zhang YE, Shen X. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18:637–652. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu CS, Yu CY, Chuang CY, Hsiao M, Kao CF, Kuo HC, Chuang TJ. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24:25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Bluthgen N, Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 16.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings KE. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Nilsen TW. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet. 2001;17:678–680. doi: 10.1016/S0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 20.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 22.Zaslaver A, Baugh LR, Sternberg PW. Metazoan operons accelerate recovery from growth-arrested states. Cell. 2011;145:981–992. doi: 10.1016/j.cell.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc Natl Acad Sci USA. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiuchi T, Giniger E, Aigaki T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 2003;17:2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 29.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 30.Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- 31.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.RES.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 32.Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, Zhan S, Kronenberg MS, Lichtler A, Liu HX, Chen FP, Yue L, Li XJ, Xu RH. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xi J, Liu Y, Liu H, Chen H, Emborg ME, Zhang SC. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30:1655–1663. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David L, Polo JM. Phases of reprogramming. Stem Cell Res. 2014;12:754–761. doi: 10.1016/j.scr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 37.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 40.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 42.Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 44.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 45.Zafarana G, Avery SR, Avery K, Moore HD, Andrews PW. Specific knockdown of OCT4 in human embryonic stem cells by inducible short hairpin RNA interference. Stem Cells. 2009;27:776–782. doi: 10.1002/stem.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Leng Ng CK, Herath W, Lili S, Hutchins AP, Robson P, Kolatkar PR, Stanton LW. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32:938–953. doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 48.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 49.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 50.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 55.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 56.Shin M, Alev C, Wu Y, Nagai H, Sheng G. Activin/TGF-beta signaling regulates Nanog expression in the epiblast during gastrulation. Mech Dev. 2011;128:268–278. doi: 10.1016/j.mod.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 60.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 61.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, de Sousa Chuva, Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 62.Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, Llanas RA, Thomson JA. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 63.Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 64.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McLean AB, D’Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 66.Avery S, Zafarana G, Gokhale PJ, Andrews PW. The role of SMAD4 in human embryonic stem cell self-renewal and stem cell fate. Stem Cells. 2010;28:863–873. doi: 10.1002/stem.409. [DOI] [PubMed] [Google Scholar]
- 67.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 68.Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci. 2015;72:4157–4172. doi: 10.1007/s00018-015-2028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 71.Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakanishi M, Kurisaki A, Hayashi Y, Warashina M, Ishiura S, Kusuda-Furue M, Asashima M. Directed induction of anterior and posterior primitive streak by Wnt from embryonic stem cells cultured in a chemically defined serum-free medium. FASEB J. 2009;23:114–122. doi: 10.1096/fj.08-111203. [DOI] [PubMed] [Google Scholar]
- 73.Ullmann U, Gilles C, De Rycke M, Van de Velde H, Sermon K, Liebaers I. GSK-3-specific inhibitor-supplemented hESC medium prevents the epithelial-mesenchymal transition process and the up-regulation of matrix metalloproteinases in hESCs cultured in feeder-free conditions. Mol Hum Reprod. 2008;14:169–179. doi: 10.1093/molehr/gan001. [DOI] [PubMed] [Google Scholar]
- 74.Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 75.Cai L, Ye Z, Zhou BY, Mali P, Zhou C, Cheng L. Promoting human embryonic stem cell renewal or differentiation by modulating Wnt signal and culture conditions. Cell Res. 2007;17:62–72. doi: 10.1038/sj.cr.7310138. [DOI] [PubMed] [Google Scholar]
- 76.Axelrod JD. Progress and challenges in understanding planar cell polarity signaling. Semin Cell Dev Biol. 2009;20:964–971. doi: 10.1016/j.semcdb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 77.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. New insights into the mechanism of Wnt signaling pathway activation. Int Rev Cell Mol Biol. 2011;291:21–71. doi: 10.1016/B978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 78.Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 80.De A. Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin (Shanghai) 2011;43:745–756. doi: 10.1093/abbs/gmr079. [DOI] [PubMed] [Google Scholar]
- 81.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 82.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 83.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 86.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 87.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Durruthy-Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, Davila J, Mall M, Wong WH, Wysocka J, Au KF, Reijo Pera RA. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet. 2016;48:44–52. doi: 10.1038/ng.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu C, Zhang Y, Wang Q, Xu Z, Jiang J, Gao Y, Gao M, Kang J, Wu M, Xiong J, Ji K, Yuan W, Wang Y, Liu H. Long non-coding RNA GAS5 controls human embryonic stem cell self-renewal by maintaining NODAL signalling. Nat Commun. 2016;7:13287. doi: 10.1038/ncomms13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu CY, Kuo HC. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells. 2016;34:2052–2062. doi: 10.1002/stem.2386. [DOI] [PubMed] [Google Scholar]
- 94.Wongtrakoongate P, Riddick G, Fucharoen S, Felsenfeld G. Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet. 2015;11:e1005615. doi: 10.1371/journal.pgen.1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deng C, Li Y, Zhou L, Cho J, Patel B, Terada N, Li Y, Bungert J, Qiu Y, Huang S. HoxBlinc RNA recruits Set1/MLL complexes to activate hox gene expression patterns and mesoderm lineage development. Cell Rep. 2016;14:103–114. doi: 10.1016/j.celrep.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bao X, Wu H, Zhu X, Guo X, Hutchins AP, Luo Z, Song H, Chen Y, Lai K, Yin M, Xu L, Zhou L, Chen J, Wang D, Qin B, Frampton J, Tse HF, Pei D, Wang H, Zhang B, Esteban MA. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boothroyd JC, Cross GA. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 99.De Lange T, Berkvens TM, Veerman HJ, Frasch AC, Barry JD, Borst P. Comparison of the genes coding for the common 5′ terminal sequence of messenger RNAs in three trypanosome species. Nucleic Acids Res. 1984;12:4431–4443. doi: 10.1093/nar/12.11.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parsons M, Nelson RG, Watkins KP, Agabian N. Trypanosome mRNAs share a common 5′ spliced leader sequence. Cell. 1984;38:309–316. doi: 10.1016/0092-8674(84)90552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bektesh S, Van Doren K, Hirsh D. Presence of the Caenorhabditis elegans spliced leader on different mRNAs and in different genera of nematodes. Genes Dev. 1988;2:1277–1283. doi: 10.1101/gad.2.10.1277. [DOI] [PubMed] [Google Scholar]
- 102.Van Doren K, Hirsh D. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature. 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- 103.Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–164. doi: 10.1016/S0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- 104.Konarska MM, Padgett RA, Sharp PA. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/S0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 105.Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dorn R, Krauss V. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica. 2003;117:165–177. doi: 10.1023/A:1022983810016. [DOI] [PubMed] [Google Scholar]
- 107.Goeke S, Greene EA, Grant PK, Gates MA, Crowner D, Aigaki T, Giniger E. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat Neurosci. 2003;6:917–924. doi: 10.1038/nn1105. [DOI] [PubMed] [Google Scholar]
- 108.Kong Y, Zhou H, Yu Y, Chen L, Hao P, Li X. The evolutionary landscape of intergenic trans-splicing events in insects. Nat Commun. 2015;6:8734. doi: 10.1038/ncomms9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:5. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 110.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 111.Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li N, Zheng J, Li H, Deng J, Hu M, Wu H, Li W, Li F, Lan X, Lu J, Zhou Y. Identification of chimeric TSNAX–DISC1 resulting from intergenic splicing in endometrial carcinoma through high-throughput RNA sequencing. Carcinogenesis. 2014;35:2687–2697. doi: 10.1093/carcin/bgu201. [DOI] [PubMed] [Google Scholar]
- 113.Flouriot G, Brand H, Seraphin B, Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J Biol Chem. 2002;277:26244–26251. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- 114.Takahara T, Kanazu SI, Yanagisawa S, Akanuma H. Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J Biol Chem. 2000;275:38067–38072. doi: 10.1074/jbc.M002010200. [DOI] [PubMed] [Google Scholar]
- 115.Chatterjee TK, Fisher RA. Novel alternative splicing and nuclear localization of human RGS12 gene products. J Biol Chem. 2000;275:29660–29671. doi: 10.1074/jbc.M000330200. [DOI] [PubMed] [Google Scholar]
- 116.Linardic CM. PAX3–FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuan H, Qin F, Movassagh M, Park H, Golden W, Xie Z, Zhang P, Sklar J, Li H. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 118.Kawakami M, Ishikawa R, Amano Y, Sunohara M, Watanabe K, Ohishi N, Yatomi Y, Nakajima J, Fukayama M, Nagase T, Takai D. Detection of novel paraja ring finger 2-fer tyrosine kinase mRNA chimeras is associated with poor postoperative prognosis in non-small cell lung cancer. Cancer Sci. 2013;104:1447–1454. doi: 10.1111/cas.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fang W, Wei Y, Kang Y, Landweber LF. Detection of a common chimeric transcript between human chromosomes 7 and 16. Biol Direct. 2012;7:49. doi: 10.1186/1745-6150-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kowarz E, Merkens J, Karas M, Dingermann T, Marschalek R. Premature transcript termination, trans-splicing and DNA repair: a vicious path to cancer. Am J Blood Res. 2011;1:1–12. [PMC free article] [PubMed] [Google Scholar]
- 121.Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 122.Guerra E, Trerotola M, Dell’ Arciprete R, Bonasera V, Palombo B, El-Sewedy T, Ciccimarra T, Crescenzi C, Lorenzini F, Rossi C, Vacca G, Lattanzio R, Piantelli M, Alberti S. A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- 123.Brooks YS, Wang G, Yang Z, Smith KK, Bieberich E, Ko L. Functional pre-mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem. 2009;284:18033–18046. doi: 10.1074/jbc.M109.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jehan Z, Vallinayagam S, Tiwari S, Pradhan S, Singh L, Suresh A, Reddy HM, Ahuja YR, Jesudasan RA. Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res. 2007;17:433–440. doi: 10.1101/gr.5155706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Breen MA, Ashcroft SJ. A truncated isoform of Ca2+/calmodulin-dependent protein kinase II expressed in human islets of Langerhans may result from trans-splicing. FEBS Lett. 1997;409:375–379. doi: 10.1016/S0014-5793(97)00555-3. [DOI] [PubMed] [Google Scholar]
- 126.Hu GJ, Chen J, Zhao XN, Xu JJ, Guo DQ, Lu M, Zhu M, Xiong Y, Li Q, Chang CC, Song BL, Chang TY, Li BL. Production of ACAT1 56-kDa isoform in human cells via trans-splicing involving the ampicillin resistance gene. Cell Res. 2013;23:1007–1024. doi: 10.1038/cr.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu C, Chen J, Lin S, Liu J, Chang CC, Chang TY. Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. J Biol Chem. 1999;274:36139–36145. doi: 10.1074/jbc.274.51.36139. [DOI] [PubMed] [Google Scholar]
- 128.Yang L, Lee O, Chen J, Chen J, Chang CC, Zhou P, Wang ZZ, Ma HH, Sha HF, Feng JX, Wang Y, Yang XY, Wang L, Dong R, Ornvold K, Li BL, Chang TY. Human acyl-coenzyme A:cholesterol acyltransferase 1 (acat1) sequences located in two different chromosomes (7 and 1) are required to produce a novel ACAT1 isoenzyme with additional sequence at the N terminus. J Biol Chem. 2004;279:46253–46262. doi: 10.1074/jbc.M408155200. [DOI] [PubMed] [Google Scholar]
- 129.Chan AS, Thorner PS, Squire JA, Zielenska M. Identification of a novel gene NCRMS on chromosome 12q21 with differential expression between rhabdomyosarcoma subtypes. Oncogene. 2002;21:3029–3037. doi: 10.1038/sj.onc.1205460. [DOI] [PubMed] [Google Scholar]
- 130.Uhde CW, Vives J, Jaeger I, Li M. Rmst is a novel marker for the mouse ventral mesencephalic floor plate and the anterior dorsal midline cells. PLoS One. 2010;5:e8641. doi: 10.1371/journal.pone.0008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 132.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 134.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 135.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]