Abstract
MD2, a 160-residue accessory glycoprotein, is responsible for the recognition and binding of Gram-negative bacterial membrane component, lipopolysaccharide (LPS). Internalization of pathogen inside the mononuclear phagocytes has also been attributed to MD2 which leads to the clearance of pathogens from the host. However, not much is known about the segments in MD2 that are responsible for LPS interaction or internalization of pathogen inside the defense cells. A 16-residue stretch (MD54) from MD2 protein has been identified that possesses a short heptad repeat sequence and four cationic residues enabling it to participate in both hydrophobic and electrostatic interactions with LPS. An MD54 analog of the same size was also designed in which a leucine residue at a heptadic position was replaced with an alanine residue. MD54 but not its analog, MMD54 induced aggregation of LPS and aided in its internalization within THP-1 monocytes. Furthermore, MD54 inhibited LPS-induced nuclear translocation of NF-κB in PMA-treated THP-1 and TLR4/MD2/CD14-transfected HEK-293T cells and the production of pro-inflammatory cytokines. In addition, in in vivo experiments, MD54 showed marked protection and survival of mice against LPS-induced inflammation and death. Overall, we have identified a short peptide with heptad repeat sequence from MD2 that can cause aggregation of LPS and abet in its internalization within THP-1 cells, resulting in attenuation of LPS-induced pro-inflammatory responses in vitro and in vivo.
Electronic supplementary material
The online version of this article (10.1007/s00018-017-2735-2) contains supplementary material, which is available to authorized users.
Keywords: MD2, Lipopolysaccharide (LPS) aggregation, Sepsis, Attenuation of LPS-induced pro-inflammatory responses, Heptad repeat, Synthetic peptides
Introduction
Sepsis is a clinical symptom which can lead to the failure of circulation, metabolism, renal, hepatic, and neuro-endocrine functions. It is initiated through an attack of bacteria, fungi, viruses, and parasites on the blood, urinary tract, lungs, skin or, other tissues [1]. These microbes contain molecular signatures on their surfaces that are recognized by pattern recognition receptors (PRRs) present in the host defense cells, including monocytes, macrophages, dendritic cells, etc. For example, lipopolysaccharide (LPS), an outer membrane component of Gram-negative bacteria [2–4], is a pathogen-associated molecular pattern (PAMP) which is recognized by PRRs [1, 5]. LPS in the aggregated form first binds to a serum protein, namely, lipopolysaccharide-binding protein (LBP) which is facilitated by albumin [6]. This binding brings about a drastic change in LPS resulting in its conversion from aggregated to monomeric form [7]. The LBP-extracted LPS monomer is then transferred to CD14 in an albumin-dependent manner which shields the hydrophobic lipid A portion of LPS when it moves through a hydrophilic environment that in turn efficiently shuttles the LPS to MD2 in the TLR4/MD2 complex [8–10]. MD2 after coming in contact with LPS initiates a series of key events that involve the dimerization of TLR4 causing TLR4, MD2, and LPS to form an active complex [11]. The formation of this dimeric active complex at extracellular level transmits the signal in the cytoplasm, probably through the transmembrane domain of TLR4 to activate a cascade of complex, but regulated events that include nuclear translocation of nuclear factor-κB (NF-κB) and production of pro-inflammatory cytokines [12, 13]. Production of these pro-inflammatory cytokines as a result of microbial invasion is a natural response of the host innate immune system, the purpose of which is to activate mononuclear phagocytes (monocytes and macrophages) [1] at the site of invasion for eliminating the pathogen by phagocytosis [14]. However, when the remains of microorganisms such as LPS from dead or live bacteria spread in the host circulatory system, often, an uncontrolled activation of host immune response occurs, causing sepsis. Sepsis is characterized by symptoms such as fever, endothelial damage, capillary leakage, peripheral vascular dilatation, and coagulation disorders, which frequently leads to multi-organ failure and even death of the patients [1, 14].
In recent years, sepsis has been a major cause of ICU deaths. No proper preventive measure is yet available to the patients of sepsis [15]. Numerous LPS-like TLR4/MD2 antagonists, such as fatty acid chain-containing CRX-526, E5531, and E5564 (eritoran), targeting MD2 protein directly have been evaluated in clinical and pre-clinical studies, but they failed at later stages of trials. In addition, small natural and synthetic chemicals such as JSH, curcumin, xanthohumol, and CAPE behave as MD2 inhibitors and block the LPS to bind the TLR4/MD2 complex, causing inflammatory signaling inactivated, thereby preventing sepsis, but their specificities are not yet defined [16]. Therefore, there is an urgent demand for chemical entities that inhibit the LPS-induced cytokine productions in host cells. Often, the undesired toxicity of the molecules with anti-endotoxin property hampers the prospects of drug development against sepsis. Besides, many a times alterations in such molecules for reducing their toxicity compromise with their efficacy to inhibit LPS-induced cytokine production [17–19]. Thus, the design of an anti-LPS (anti-endotoxin) molecule with negligible toxicity is a primary step towards the development of an anti-sepsis agent.
Short peptides from both human and mouse MD2 protein have been derived that possess features of LPS-binding proteins. These peptides have been further investigated for their anti-endotoxin property. For example, various peptides derived from different amino acid regions of human MD2 showed in vitro and in vivo LPS neutralization activity [20, 21]. Furthermore, the peptide derived from 119 to 132 region of the mouse MD2 protein attenuated LPS-induced pro-inflammatory responses in vitro as well as in vivo [22]. These results further indicate that MD2 protein sequence can further be explored for identifying peptides with anti-LPS property.
Crystal structure of the dimeric complex comprising MD2, TLR4, and LPS [11] presents a detailed picture of atomic-level interactions among these molecules and thus provides a significant opportunity to identify and characterize anti-endotoxin peptides either from TLR4 or from MD2. A segment encompassing the amino acid region, 54–69 of human MD2, was selected that comprises a heptad repeat sequence and contains amino acids that are involved with hydrophobic and electrostatic interactions with LPS as per the crystal structure of TLR4–MD2–LPS [11]. Therefore, in the quest of anti-endotoxin peptide, we felt that it is worth to investigate the ability of this peptide to neutralize LPS-induced pro-inflammatory cytokine production as well as its probable mode of action. The results presented here reveal the identification and characterization of an MD2-derived short anti-endotoxin peptide that can induce aggregation in LPS, and facilitate its internalization and neutralization in human monocytes, thus demonstrating its potential as an anti-sepsis agent.
Materials and methods
Materials
Rink amide MBHA resin (loading capacity 0.4–0.8 mmol/g), N-α-F-moc, and necessary side-chain-protected amino acids were purchased from Novabiochem. Coupling reagents for peptide synthesis that include 1-hydroxybenzotriazole (HOBt), 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), and N, N-diisopropylethylamine (DIPEA) were purchased from Sigma, USA. Dichloromethane (DCM), N, N-dimethylformamide (DMF), and piperidine were of standard grades and were procured from reputed local companies. Acetonitrile (HPLC grade) was procured from Merck, India. Trifluoroacetic acid (TFA), sodium dodecyl sulfate (SDS), E. coli 0111:B4 lipopolysaccharide (LPS), FITC–LPS E. coli 0111:B4, and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma. For cell culture, RPMI 1640 and fetal bovine serum were purchased from sera laboratories West Sussex, UK. The GIBCO 100X antibiotic–antimycotic, lipofectamine 2000 reagent, and trypsin (EDTA) were purchased from Invitrogen corporations. 5(6)-Carboxytetramethyl rhodamine N-succinimidyl ester was purchased from Molecular Probes (Eugene, OR, USA). Dual-luciferase® reporter assay kit was procured from Promega Corporation. Rests of the reagents were of analytical grade and procured locally, and buffers were prepared in Milli Q water.
Cytokine estimation kits
Pro-inflammatory cytokine estimation kits and reagents that include Human TNF-α ELISA Set, Human IL-1β ELISA Set II, Human IL-8 ELISA Set, Mouse TNF-α ELISA Set II, Mouse IL-6 ELISA Set, and TMB substrate reagent set were purchased from BD Biosciences.
Antibodies
Human anti-NF-κB p105/p50, Human anti-NF-κB p65, Anti-rabbit IgG HRP, Alexa flour 488 Secondary IgG, and proliferating cell nuclear antigen (PCNA) antibodies were purchased from Cell Signaling Technology (CST), whereas anti-MD2 and anti-TLR4 antibody were procured from Santa Cruz Biotechnology.
Plasmids
Expression plasmids for Flag–pCMV TLR4, pEF-Bos–HA MD2, and pcDNA3-His CD14 were kind gifts from Dr. Douglous Golenbock (Medicine Department, University of Massachusetts Medical School, Worcester, MA, USA) whereas NF-κB-Luc, pRL–TK control, and pcDNA3 empty vector were kindly provided by Dr. Anila Dwivedi (Endocrinology Div. CSIR-CDRI, Lucknow, India).
Cell lines and animals
THP-1 (A human monocytic cell line derived from an acute monocytic leukemia patient), HEK-293T (human embryonic kidney cell line), and murine 3T3 (murine fibroblast cells) cell lines were obtained from CSIR-Central Drug Research Institute, Lucknow cell line repository. The cell lines were maintained by usual protocol in an Innova CO2 incubator. Animals for experiments were provided by National Laboratory Animal Center, CSIR-CDRI (Lucknow, India). All animal procedures were carried out according to the protocols approved by the Institutional Animal Ethics Committee (approval no. IAEC/2015/90). Animals were properly anesthetized before experiments, and care was taken in all the animal experiments to minimize the sufferings to the animals. Our animal protocols adhered to the guidelines of CPCSEA (Committee for the Purpose of Control and Supervision of Experiments on Animals), Govt. of India.
Peptide synthesis, fluorescent labeling, and purification
An MD2 protein-derived peptide, MD54 and its analog, and MMD54 were synthesized by employing a peptide synthesizer (model: PS3, Protein Technologies, Inc.) utilizing solid-phase method on rink amide MBHA resin using F-moc chemistry [23–25]. Labeling at the N-termini of peptides with a fluorescent probe (rhodamine), cleavage of the peptides from the resins, and their HPLC purifications were achieved by the standard procedures [24, 26–31]. Experimental molecular masses of the peptides were detected by MALDI–TOF mass spectra. The purified peptides were ~ 95% homogeneous as determined by analytical reverse phase chromatography.
Assay of hemolytic activity of the peptides
Hemolytic activity assay of the peptides against human red blood cells (hRBCs) in PBS was performed to examine the ability of the peptides to lyse the hRBCs as previously described [17]. For this purpose, fresh human blood was collected in the presence of an anticoagulant from a healthy volunteer. The collected blood was washed three times with PBS and hemolytic activity of the peptides was determined as reported previously [17].
Cell culture
Murine 3T3 and human embryonic kidney 293T (HEK-293T) were grown in DMEM media, while THP-1 (human monocytic cell line derived from an acute monocytic leukemia patient) cells were grown in suspension in RPMI 1640 medium, supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% antibiotic (Gibco anti-anti) at 37 °C in a humidified 5% CO2 (v/v) incubator. THP-1 cells at a density of 1 × 106 cells/ml were treated with 0.3 mg/ml PMA for 24 h [32] to induce plastic-adherent properties in the cells. They were further rested in complete RPMI 1640 medium for an additional 24 h before stimulations with various treatments. Cells were counted with the help of an Olympus inverted microscope for the experiments.
Cell viability assay
The viability of the cells was determined to check the toxic activity of peptides against murine 3T3 cells by the standard procedures as described earlier [30, 33].
Measurement of cytokine expression levels in supernatant
Enzyme-linked immunosorbent assays (ELISA) were performed to estimate the secreted TNF-α and IL-1β after incubation of 4 h and IL-8 after incubation of 6 h in LPS and peptide-treated THP-1 cells in RPMI 1640 supplemented with 10% FBS. Media and only LPS-treated cells served as negative and positive controls, respectively. The experiments were performed according to manufacturer’s protocol. Briefly, plates were coated with respective capture antibody overnight at 4 °C followed by blocking (10% FBS in PBS) and incubation of samples for 2 h at room temperature (RT). Samples were washed five times with PBST and incubated with detection antibody with HRP-conjugate for 1 h at RT. The samples were washed again and added TMB substrate solution was added for 30 min at RT for color development. The reaction was stopped by adding stop solution and the reading was taken at 450 nm and corrected at 570 nm in BIOTEK microtiter plate reader.
Preparation of nuclear fractions
It has been described in the Online Resource.
SDS-PAGE and western blotting
It has been described in the Online Resource.
Co-immunoprecipitation
MD2 was co-precipitated with TLR4 to detect the effect of MD54 on the formation of TLR4–MD2–LPS complex as previously reported [16]. THP-1 cells were seeded in six well plates and after 24 h cells were pre-treated with the peptide MD54 and MMD54 (for 1 h and then LPS was added for a further 30 min). Cells were then lysed in radioimmunoprecipitation assay buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml aprotinin, leupeptin, pepstatin, 1 mM Na3VO4, and 1 mM NaF) after washing with cold 1× phosphate-buffered saline. The lysates were spun down at 15,000g for 15 min after 10 min incubation with lysis buffer on ice. A total of 500 μg of cell lysate was used for immunoprecipitation by incubation with 20 μl Protein A/G Sepharose beads (Sigma) and 10 μg primary antibody TLR4 for 2 h at 4 °C under constant shaking according to the manufacturer’s instructions. The immunoprecipitates were washed 3–4 times with a cold PBS. Supernatants were separated and subjected to immunoblotting. Immunoprecipitates were separated on a 10% SDS-PAGE followed by immunoblotting with MD2 primary antibody.
Plasmid isolation
Plasmids from respective constructs were isolated using Macherey–Nagel plasmid isolation kit according to the manufacturer’s protocol.
Luciferase reporter assay of NF-κB activity
One day before transfection, HEK-293T cells were plated in either 24-well plate or 6-well plate, so that the cells are 50–80% confluent at the time of transfection. At least 3 h before transfection, complete DMEM was removed and serum-free DMEM was added to the cells without antibiotics. Expression plasmids FLAG–CMV1–TLR4 (5 ng/well), pcDNA3–CD14 (15 ng/well), pEFBos–HA–MD2 (5 ng/well) together with NF-κB-luc (100 ng/well), and pRL-luc (10 ng/well) for 24 well [34], were transiently transfected with lipofectamine 2000 reagent according to the manufacturer’s protocol. After 4 h of transfection, complete medium (DMEM supplemented with 10% FBS and 1% antibiotic) was added to the cells. After 24 h of transfection, cells were treated with peptide and LPS with respective concentrations for 16–24 h prior to lysate preparation. Dual-luciferase reporter assay was performed according to the manufacturer’s protocol. Briefly, the cells were lysed in passive lysis buffer (Promega) and luciferase activity was measured in a luminometer (GLOMAX).
Human ethics statement
All standard procedures duly approved by the ethical committee of the Institute (CSIR-Central Drug Research Institute, Lucknow, India) were taken care during the collection of blood from a healthy volunteer. The study bears the Institutional Ethics Committee Approval No. CDRI/IEC/2014/A5.
Circular dichroism spectroscopy
Circular dichroism (CD) [17, 25] spectra of these peptides were recorded on a JASCO J-815 spectropolarimeter in phosphate-buffered saline (PBS, pH 7.4) and in the presence of LPS in PBS. The spectropolarimeter was calibrated routinely with 10-camphor sulphonic acid. The samples were scanned at room temperature (~ 25 °C) with the help of capped quartz cuvettes of 0.2 cm path length at a wavelength range of 250–200 nm. An average of three scans was taken for each sample with a scan speed of 50 nm/min and data interval of 0.2 nm. The JASCO Spectra Manager software was used to collect the spectra and the spectra analysis software was used to calculate the helicity. Spectra were baseline corrected by subtracting a blank spectrum containing only buffer. The spectra were plotted as CD [millidegree (mdeg) vs. wavelength].
Binding of FITC–LPS onto THP-1 cells in the presence of the peptides
FITC-labeled LPS (10 μg/ml) was incubated with either MD54 or MMD54 in the equimolar ratio at different concentrations for 1 h at 37 °C in RPMI 1640. Afterwards, ~ 105 THP-1 cells were treated with LPS–peptide mixture for 30 min at room temperature with gentle shaking. The cells were washed with ice chilled PBS (pH 7.4) extensively to remove the unbound LPS from the samples. Binding of FITC–LPS-to-THP-1 cells in the presence of MD54 and its analog was monitored by measuring the mean fluorescence intensity of 10,000 cells for each sample in terms of log FL1 height with a BD FACS Caliber flow cytometer. Fluorescence of untreated cells was measured to determine the auto-fluorescence of these cells.
Effect of the peptides on aggregation of FITC–LPS
Aggregation of FITC–LPS in the presence of these peptides was examined in aqueous buffer (PBS) as described previously [35] by fluorescence studies. FITC–LPS (0.5 µg/ml) was treated with increasing concentrations of a particular peptide. The changes in the emission of FITC in PBS (7.4) were monitored by a Perkin Elmer LS55 spectrofluorimeter with excitation and emission wavelengths at 488 and 515 nm, respectively, and excitation and emission slits set at 8 and 6 nm, respectively.
Aggregation of FITC–LPS in the presence of a peptide could be reflected in the decrease in its fluorescence as a result of its quenching [17, 36]. Further proteinase K, a de-quencher, was added to FITC–LPS alone or in the presence of the peptides to observe any de-quenching of FITC fluorescence as a result of dissociation of FITC–LPS aggregates.
Dynamic light-scattering study
Dynamic light-scattering (DLS) experiments were carried out on a Malvern Zeta Sizer Nano ZS (Malvern, UK) with a backscattering detection of 173°, equipped with an He–Ne laser (l = 632.8 nm) at 37 °C. A set of 12 measurements (3 runs each) for LPS, MD54–LPS, and MMD54–LPS suspensions (in PBS) were run and the size distributions and mean count rate of LPS and peptide–LPS aggregates were determined. All measurements were carried out using dispersant RI = 1.33 and disposable polystyrene cells.
In vivo studies, treatment of mice
Female Balb/c mice (average weight ~ 22 g) (National laboratory animal center, CSIR-Central Drug Research Institute, Lucknow, India) were given a standard laboratory diet and water ad libitum and were housed under controlled environmental conditions. All the mice were divided into six experimental groups with five animals in each group for the administration of PBS, MD54, MMD54, LPS, and LPS along with MD54 and MMD54, respectively. Mice were treated with 10 mg/kg of E. coli 0111:B4 LPS in the presence and the absence of MD54 and its analog in different experimental groups. The mice treated with PBS only, or MD54 or its analog in the absence of LPS was considered as experimental controls to monitor the self-level of pro-inflammatory markers in mice when non-stimulated with LPS. To study the in vivo efficacy of these peptides in inhibiting LPS toxicity, the mice were simultaneously treated with MD54 or its analog along with LPS and kept under observation for 120 h. All the experimental sets of peptides/LPS were administered by intraperitoneal injections (i.p). Survival of mice was monitored up to 120 h after LPS and peptide administrations. All animal procedures were in accordance with the standards set forth in the guidelines for the IAEC and CSIR-Central Drug Research Institute animal house facilities.
For serum TNF-α and IL-6 measurement, mice were anesthetized with ether and blood was collected after 4 h either by tail vein or retro-orbital sinus aseptically. Serum levels of TNF-α and IL-6 were determined by an ELISA kit for TNF-α and IL-6. Plasma samples were diluted with assay diluent (10% FBS in PBS).
Statistical analysis
Each data point is an average of three independent experiments and results are given as standard error of mean (± SEM), Student’s t test and one-way analysis of variance using Turkey’s test were performed using Graph Pad Prism Version 5.00. p values of ≤ 0.05, ≤ 0.01, and ≤ 0.001 between different groups were considered as significant, highly significant, and very highly significant and marked with *, **, and ***, respectively, vs. LPS.
Results
Identification of heptad repeat in MD2 protein
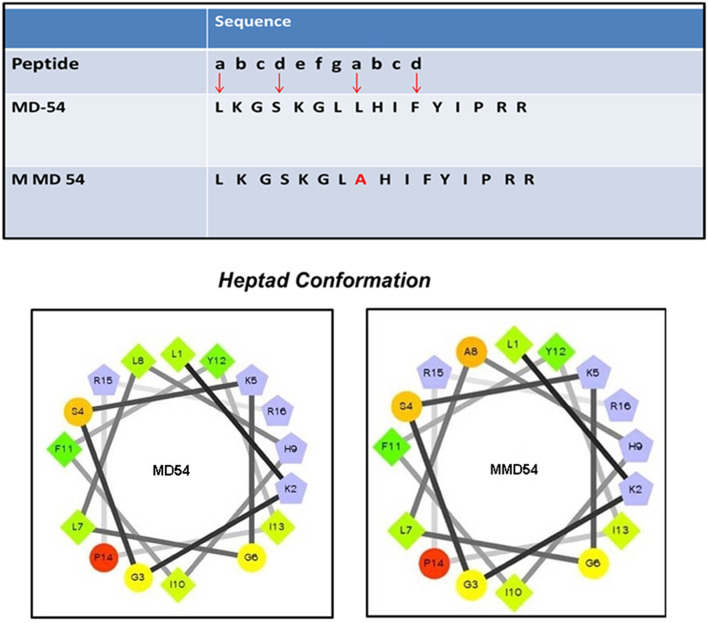
Heptad repeat is an important structural element that participates in many molecular interactions [37]. It is the repetition of seven amino acids (a–g) in which each ‘a’ and ‘d’ positions are occupied by hydrophobic residues such as leucine, isoleucine, or phenylalanine [17]. The amino acid (a.a.) segment 54–69 contains a small stretch of heptad repeat which is located in the a.a. region 54–64 (Fig. 1). Two ‘a’ positions of this heptad repeat are occupied by two leucine residues, while two ‘d’ positions are occupied by a serine and a phenylalanine residue. Crystal structure of TLR4/MD2/LPS complex shows that leucine 54 and lysine 58 are involved in hydrophobic and electrostatic interactions with LPS [11], respectively. Moreover, for anti-microbial peptide melittin, it was demonstrated that substitutions of leucine residues with alanine residues at ‘a’ positions of its leucine zipper sequence impaired the peptide–LPS interaction and anti-LPS (anti-endotoxin) property of the peptide, implicating the role of such structural motif in interaction with LPS [17]. Furthermore, besides lysine residues at the 55th and 58th positions towards its N-terminus, this stretch also contains two cationic arginine residues (positions, 68 and 69) at its C-terminus. Thus altogether, the presence of four positively charged amino acids could facilitate the interaction of negatively charged LPS with the peptide (MD54) corresponding to a.a. 54–69. Considering the composition and the amino acid sequence features of MD54 as mentioned above, it was of interest to investigate the interaction of this peptide with LPS and examine its anti-LPS (anti-endotoxin) property. Along with the wild-type peptide MD54, an analog (MMD54) of the same length was designed, by replacing the leucine residue at position 61, corresponding to ‘a’ position of the heptad repeat with an alanine residue.
Fig. 1.
Heptad conformation and the helical wheel presentation of MD54 and MMD54
Table 1 shows the amino acid sequences of MD54 and its analog with their observed and calculated molecular masses and HPLC retention times. Helical wheels of the peptides show appreciable segregation of polar and non-polar amino acid residues, further indicating amphipathic nature of these peptides (Fig. 1).
Table 1.
Sequence of the peptide and it analog with observed and calculated mass with HPLC retention time
| Peptide | Sequence | Observed mass | Calculated mass | Retention time (min) |
|---|---|---|---|---|
| MD54 | LKGSKGLLHIFYIPRR | 1898.50 | 1897.2 | ~ 13–14 |
| MMD54 | LKGSKGLAHIFYIPRR | 1856.42 | 1855.14 | ~ 12–13 |
In MMD54, the alanine residue, placed in place of the leucine residue, has been marked in bold
In vitro toxic property of the peptides
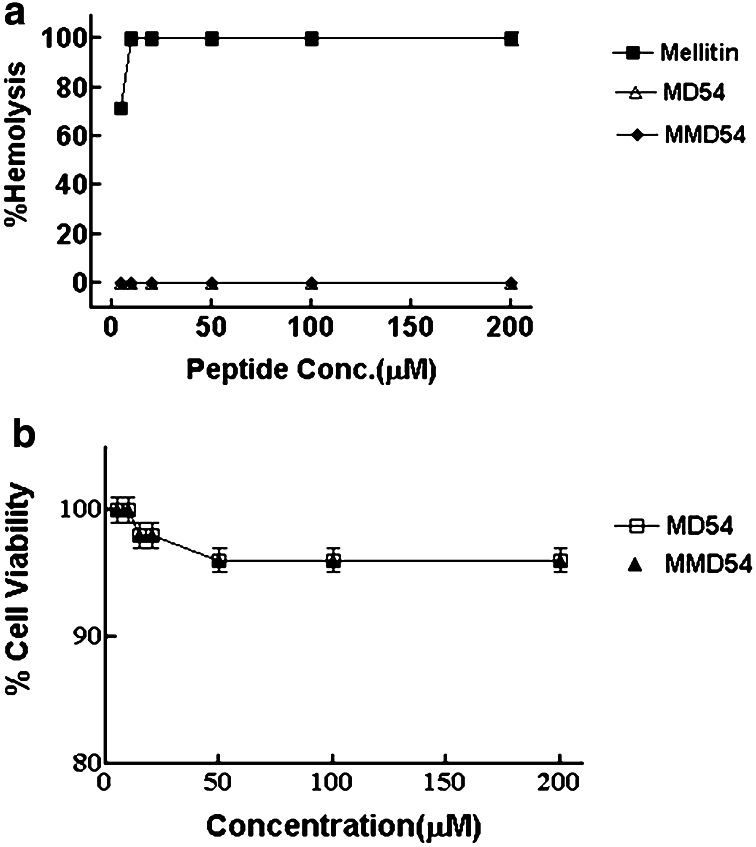
Both the wild-type peptide (MD54) and its analog (MMD54) were examined for their hemolytic activities against hRBCs and for their cytotoxic properties against murine 3T3 cells. MD54 and MMD54 did not show any significant hemolytic activity up to 200 μM peptide concentrations. MTT assay also showed ~ 90% viability of the 3T3 cells at such high peptide concentrations (Fig. 2a, b). Thus, the data suggested significant non-toxic nature of both the MD2-derived peptides.
Fig. 2.
Determination of toxicity of MD54 and MMD54. a, b % hemolysis of hRBCs and viability of murine 3T3 cells, respectively, in the presence of MD54 and its MMD54. Each data point is an average of three independent experiments and results are given as standard error of mean (± SEM)
MD54 interacts with LPS
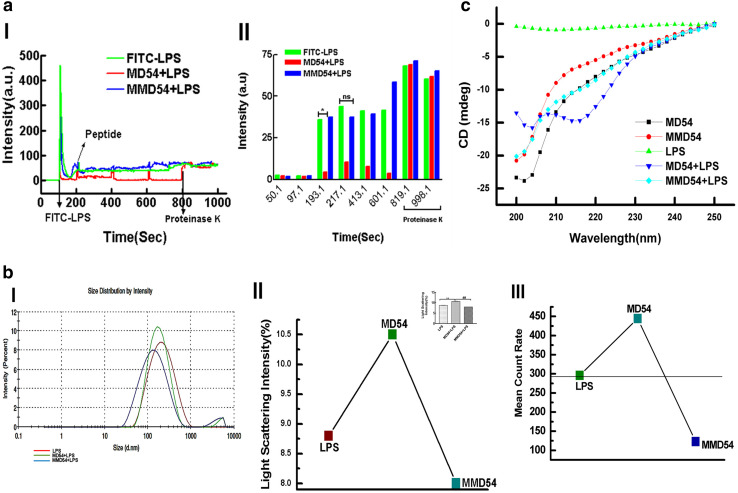
Interaction of MD54 with LPS was probed by fluorescence, DLS, and CD studies. Fluorescence studies were performed by employing FITC–LPS. Fluorescence signal of FITC–LPS was recorded in the absence and the presence of either MD54 or MMD54, followed by the addition of proteinase K to these individual samples. Results indicate that after the addition of MD54 (15, 30, and 50 μM) at three different timepoints (~ 200, 400, and 600 s), the overall fluorescence of FITC–LPS decreased slightly (Fig. 3a) which could be indicative of its more aggregation in the presence of MD54. However, when MMD54 was added to FITC–LPS instead of MD54 in a similar experiment, its fluorescence increased to some extent, which could indicate its dissociation in the presence of this peptide. Whereas, fluorescence of FITC–LPS alone remained almost the same. Interestingly, when proteinase K (1 mg/ml), a de-quencher which cleaves peptide bonds and releases FITC–LPS free, was added to the combination of FITC–LPS and MD54 in PBS, a significant increase in FITC fluorescence was observed. This increase in fluorescence was due to de-quenching of FITC fluorescence resulting from the dissociation of FITC–LPS from FITC–LPS–MD54 aggregates by proteinase K [38]. When proteinase K was added to MMD54-treated FITC–LPS in PBS, increase in FITC fluorescence was much lesser as compared to that of MD54-treated FITC–LPS (Fig. 3a). Fluorescence of FITC–LPS alone increased to some extent [Fig. 3a(I, II)] in the presence of proteinase K, but it was much less as compared to that observed when proteinase K was added to FITC–LPS that was pre-treated with MD54. It is to be noticed that after proteinase K treatment, fluorescence of FITC–LPS alone and FITC–LPS treated with either MD54 or MMD54 showed very similar levels. Overall, the results indicated that MD54 but not its alanine-substituted analog, caused aggregation of LPS.
Fig. 3.
LPS aggregation and dissociation in the presence of MD54 and MMD54. a I FITC–LPS (0.5 µg/ml) was treated with varying concentrations of peptides (10, 30, and 50 μM) to detect the aggregation of LPS as evident by the quenching of its fluorescence. FITC–LPS was added at around 100 s and peptide was added initially to FITC–LPS at 200 s after every 200 s until 600 s. Proteinase K, a de-quencher of FITC, was added after 800 s, the fluorescence profile was recorded up to 1000 s. II, and bar graphs indicate the changes in the fluorescence of FITC–LPS without and with additions of MD54, MMD54, and proteinase K at different timepoints. *p < 0.05, Student t test was performed using Graph Pad Prism Version 5.00 vs. LPS. b I, Size distribution of LPS (12.5 μg/ml) and LPS + MD54, MMD54 (12.5 + 12.5 μg/ml), II, light-scattering intensity, inset shows a statistical graph **,##p < 0.01, One-way analysis of variance using Turkey’s test was performed using Graph Pad Prism Version 5.00 vs. LPS and MMD54, respectively, and III, Mean Count Rate [KCPS (kilo counts per second)] of LPS alone and in the presence of peptides. c CD spectra of peptides (100 μg/ml) recorded at LPS concentration (50 μg/ml) in PBS (pH 7.4)
DLS studies of LPS in the presence of MD54 and MMD54 were performed, to get further evidence on its aggregation in the presence of any of these peptides. The intensity is certainly weighted according to the scattering intensity of each particle fraction which changes as the density or number of particles change. As a result of interaction of MD54 with LPS, the density or the number of particles increased. Light-scattering intensity of LPS (%) in the presence of MD54 increased to 10.5% as compared to LPS alone (8.8%), while in the presence of MMD54, it was (8%), lower than the LPS level. Thus, the data suggest that the density or the aggregation of LPS increases due to its interaction with MD54 [Fig. 3b(II)]. A statistical graph (p = 0.0016) in the inset demonstrates a significant difference between LPS and LPS in combination with MD54 [Fig. 3b(II)]. Second, the mean count rate (KPS) which is the number of photons detected during the interaction per second, defining the stability as a function of time, was also measured. We observed that the count rate increased following the incubation of LPS with MD54, while it decreased in the presence of MMD54. Thus, it was inferred that only MD54 induced a significant aggregation in LPS [Fig. 3b(III)].
Interaction of MD2-derived peptides with LPS was further examined by CD studies. Significant changes in the shape of the CD spectrum of MD54 was observed when LPS was added to it (Fig. 3c). Analysis of the CD spectra was performed with the secondary structure estimation software, supplied with the spectropolarimeter JASCO-1500. Secondary structural data in Table 2 indicate the presence of appreciable amounts of β-sheet, turn, and random coil structures in both MD54 and its analog, MMD54, in PBS. However, when LPS was added, there was significant induction of more β-sheet characteristics in both the peptides at the cost of turn structures. Yet, in the presence of LPS, MD54 adopted more β-sheet and lesser random coil structure characteristics than MMD54 under the same condition. Overall, the CD data indicated interaction of both the peptides with LPS, but the wild-type peptide MD54 was more structured in the presence of LPS than its analog MMD54.
Table 2.
Secondary structure prediction of MD54 and MMD54
| Secondary structure | MD54 only | MMD54 only | MD54 + LPS | MMD54 + LPS |
|---|---|---|---|---|
| α-Helix | 1 | 0 | 0.0 | 0.0 |
| β-Sheet | 37 | 31.3 | 47.7 | 43.8 |
| Turn | 17.8 | 20.8 | 2.4 | 0 |
| Random | 44.2 | 48 | 49.9 | 56.2 |
MD54 promotes significant binding of FITC–LPS onto THP-1 cells
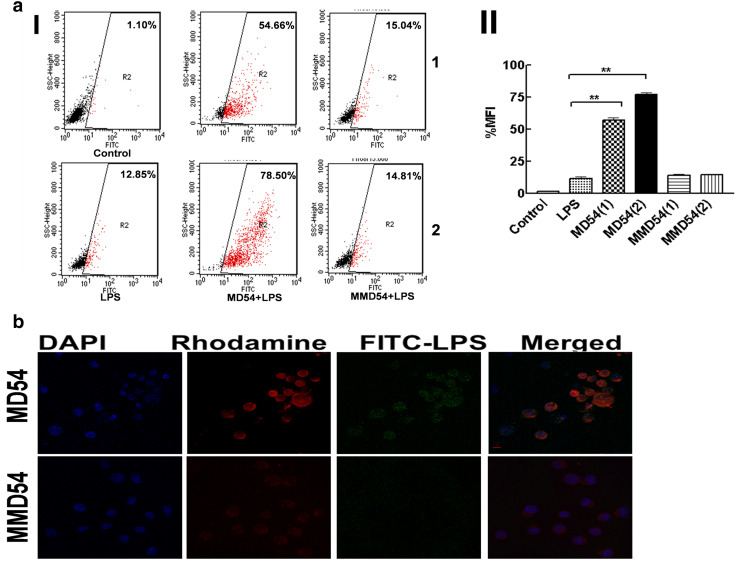
Influence of MD54 on the binding of LPS-to-THP-1 cells was studied by employing FITC–LPS and a flow cytometry approach. Binding of FITC–LPS-to-THP-1 cells was evidenced by an appreciable increase in fluorescence of these cells [mean fluorescence intensity (MFI), 1.10–12.85%]. However, when MD54 was added to THP-1 cells in combination with FITC–LPS, a dramatic enhancement in fluorescence of the cells was observed. MFI of THP-1 cells increased from 12.85% in the absence of any peptide to 54.66 and 78.50% at 80 and 100 µM concentrations of MD54, respectively. Remarkably, when the same experiment was performed with MD54 analog, MMD54, very little changes in fluorescence of the cells were observed and the MFI of the cells was very close to that observed for FITC–LPS only (MFI, 15.04 and 14.81%). The results indicated that MD54 but not its analog, MMD54 promoted the binding of FITC–LPS-to-THP-1 cells. The enhancement of fluorescence in THP-1 cells as a result of binding of FITC–LPS in the presence of MD54 did not depend on the order of addition of peptide to cells, i.e., when MD54 was pre-incubated with THP-1 cells, and then, FITC–LPS was added to the incubated cells or MD54 was added after the addition of FITC–LPS onto these cells or MD54 and FITC–LPS were added simultaneously to THP-1 cells (data not shown). THP-1 cells naturally possess the receptors for LPS. However, the results indicated that MD54 very significantly enhanced the fluorescence of THP-1 cells following their binding to FITC–LPS. The results [Fig. 4a(I)] probably suggested that MD54 but not MMD54 induced alteration in conformation/assembly of FITC–LPS creating a situation conducive to its stronger binding to THP-1 cells. Moreover, a very significant enhancement in fluorescence of THP-1 cells after binding to FITC–LPS in the presence of MD54 probably reflected that FITC–LPS was localized in a more hydrophobic environment of these cells in the presence of this peptide. A bar graph with statistical evaluation of these results is shown in Fig. 4a(II).
Fig. 4.
Binding of FITC–LPS in the presence of increasing concentrations of MD54 and MMD54. Effect of MD54 and MMD54 on the binding of LPS-to-THP-1 cells as studied by flow cytometry and confocal microscopy. a FITC–LPS (0.5 µg/ml) was incubated with two concentrations [80 and 100 µM concentrations for upper (1) and lower (2) panels, respectively] of individual peptides for 1 h in RPMI 1640, and then, macrophage cells were treated with each LPS–peptide mixture. [I] Fluorescence of the cells for each set of experiments was recorded with a BD FACS Caliber flow cytometer. Fluorescence of the cells treated with only FITC–LPS but not incubated with any of the peptides was also recorded. Control fluorescence of the cells was recorded without treating them with FITC–LPS or any of the peptides. 10,000 events were counted for each experiment. [II] A statistical graph of the mean fluorescence intensity (MFI) of the cell bound to FITC–LPS. Graph with lowest MFI denotes auto-fluorescence of cells. **p < 0.01, one-way analysis of variance using Turkey’s test was performed using Graph Pad Prism Version 5.00 vs. LPS. b Confocal laser-scanning microscopy images: THP-1 cells were treated with a mixture of FITC–LPS (100 μg/ml) and rhodamine-labeled peptides (0.1 μM). The first column shows the DAPI fluorescence signal, the second column shows the rhodamine-labeled peptide signal, the third column shows FITC–LPS fluorescence signal and the fourth column shows the overlay (merged) of the two probes enabling one to visualize co-localization (reflected in yellowish red), and arrows in red indicate the merged color. Scale bars, 10 μm
Furthermore, we performed confocal microscopic experiments to ascertain the localization of FITC–LPS in THP-1 cells in the presence of MD54. To visualize MD54 in these cells, it was fluorescently labeled with rhodamine (Rho). Significant green fluorescence was observed onto the cells that were treated with FITC–LPS in the presence of Rho-labeled MD54 (Fig. 4b, upper panel). Whereas green fluorescence was almost invisible when in the same experiment, Rho-MD54 was replaced with Rho-MMD54 (Fig. 4b, lower panel). The results indicate that MD54 but not its analog, MMD54, promotes the binding and localization of FITC–LPS inside the THP-1 cells. Looking into the FACS data (Fig. 4a) of significant enhancement of fluorescence of THP-1 cells following their binding to FITC–LPS, pre-treated with MD54 and the localization of FITC–LPS throughout the cytoplasm of these cells in the presence of MD54 in the confocal images (Fig. 4b), probably one can speculate that fluorescent probe attached to LPS could be located in the hydrophobic membrane environment of some organelles in THP-1 cells which, however, cannot be ascertained from the present study. The presence of appreciable (yellowish red) color (as indicated by red colored arrows) in the merged fluorescence image suggests co-localization of FITC–LPS and Rho-MD54 which provide further evidence that MD54 facilitates the internalization of FITC–LPS inside the THP-1 cells (Fig. 4b).
Moreover, to look into the specificity of MD54 towards LPS, FACS experiments were performed to study the binding of a non-specific fluid phase marker FITC–dextran onto THP-1 cells in the presence of MD54. Unlike in case of FITC–LPS, MD54 did not promote the binding of FITC–dextran-to-THP-1 cells, since no significant increase in fluorescence of THP-1 cells was observed when they were incubated with FITC–dextran and MD54. Like MD54, its analog, MMD54 too, did not facilitate the binding of FITC–dextran-to-THP-1 cells (Fig. S1). Overall, the results indicate that the MD54- and MD2 protein-derived peptide having a short heptad repeat sequence specifically facilitated the internalization of FITC–LPS in THP-1 cells.
MD54 but not its analog showed appreciable anti-endotoxin property
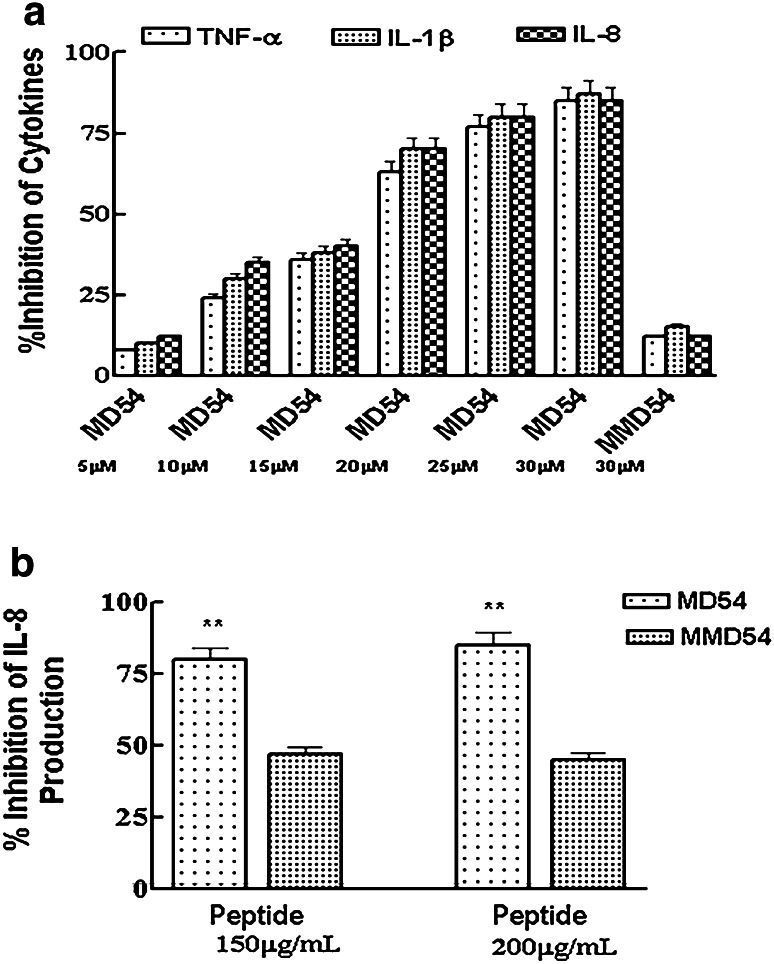
Though a majority of AMPs with the ability to inhibit LPS-induced pro-inflammatory cytokine production induce dissociation of LPS aggregates [17, 35], the MD2-derived peptide, MD54 induced aggregation in LPS. Moreover, as discussed in the previous section, MD54 significantly induced the internalization of FITC–LPS in THP-1 cells. In other words, MD54 appeared to induce significant alterations in FITC–LPS when it is in aqueous buffer or onto THP-1 cells. Therefore, it was of interest to examine if MD54 could inhibit LPS-induced pro-inflammatory responses in THP-1 cells. Indeed, MD54 inhibited LPS-induced production of TNF-α, IL-β, and IL-8 in THP-1 cells in a dose-dependent manner. It inhibited ~ 50% LPS-stimulated pro-inflammatory cytokine production at ~ 15 μM concentration, while at ~ 30 μM concentration, MD54 showed ~ 90% inhibition. However, the analog MMD54 with just a single amino acid substitution in MD54 was almost inactive in inhibiting LPS-induced pro-inflammatory cytokine responses (Fig. 5a).
Fig. 5.
Effect of MD54 and MMD54 onto the secretion of cytokines in LPS-stimulated Cells. a Percentage inhibition of levels of LPS-induced secretions of TNF- α, IL-1β, and IL-8 in THP-1 cell lines, respectively. b Percentage inhibition of levels of LPS-induced secretions of IL-8 in HEK-293T transiently transfected with TLR4/MD2/CD14 in the presence of MD54 and MMD54 by ELISA experiments. Each data point is an average of three independent experiments and results are given as the standard error of mean (± SEM); **p < 0.01, Student t test was performed using Graph Pad Prism Version 5.00 vs. LPS
Furthermore, this effect was also studied in a non-TLR4 and MD2-expressing HEK-293T cells that were transiently transfected with TLR4/CD14/MD2. Since IL-8 is the principal cytokine released by the HEK-293T TLR4-transfected cells [34], it was measured at different concentrations of these peptides. The results showed that MD54 inhibited IL-8 production in these transfected cells more significantly than MMD54 (Fig. 5b).
MD54 inhibits NF-κB activation
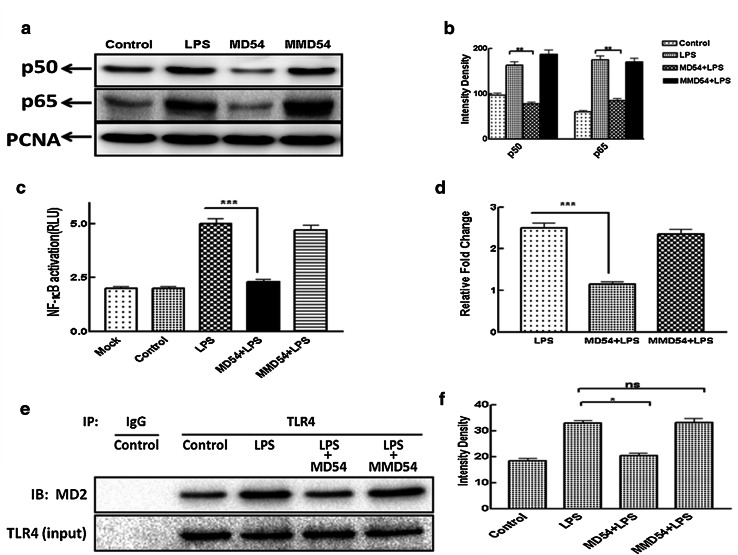
In LPS-induced pro-inflammatory cytokine production, one of the key steps is the translocation of NF-κB into the nucleus of macrophages/monocytes. Molecules that inhibit LPS-stimulated pro-inflammatory cytokine production also inhibit LPS-induced nuclear translocation of NF-κB [21]. Therefore, LPS-induced nuclear translocation of p65/p50 subunits of NF-κB was investigated in the absence and the presence of MD54 and MMD54 in PMA-stimulated THP-1 cells. The results showed that LPS-induced the translocation of p65/p50 into the nucleus while MD54 significantly inhibited its nuclear translocation. Furthermore, MMD54 did not show significant inhibition of nuclear translocation of NF-κB p65/p50 subunit which was consistent with its low anti-LPS (anti-endotoxin) property (Fig. 6a, b).
Fig. 6.
Study of effect of MD54 and MMD54 on the translocation of NF-κB into the nucleus in LPS-stimulated THP-1 and HEK-293T cells and MD54-induced attenuation of LPS-induced interaction between MD2 and TLR4. a Western blot of the nuclear fraction of LPS-stimulated THP-1 cells for p65/p50 subunit of NF-κB in the absence and the presence of MD54 and MMD54. b Intensity graph. **p < 0.01, one-way analysis of variance using Turkey’s test was performed using Graph Pad Prism Version 5.00 vs. LPS, c NF-κB activation in HEK-293T transfected cells. d Relative fold change in NF-κB activation in HEK-293T after treatment of LPS and peptides (RLU control = 1). Treatment of the peptides is as marked in the figure, whereas control and LPS represent untreated and LPS-treated THP-1 and HEK-293T without the addition of peptides (Mock represent un-transfected cell treated with peptides). ***p < 0.001, Student t test was performed using Graph Pad Prism Version 5.00 vs. LPS. e Immunoprecipitation assay showed that MD54 pretreatment (10 μM) significantly reduced the formation of the TLR4–MD2 complex induced by 100 ng ml−1 LPS. f Densitometric analysis was done using the ImageJ software. Data represented are WB analyses of at least two independent experiments (*p < 0.05)
Furthermore, LPS-induced nuclear translocation of NF-κB in the presence of these peptides was also studied in HEK-293T cells, transiently transfected with MD2/TLR4/CD14 by Luciferase assay. The results of Luciferase assay indicated the inhibition of NF-κB activation in the presence of MD54, suggesting its anti-LPS (anti-endotoxin) property (Fig. 6c, d).
MD54 attenuates LPS-induced interaction between MD2 and TLR4
A major upstream event leading to NF-κB activation following the LPS treatment in macrophages or monocytes is the formation of the TLR4–MD2 complex. Therefore, we further examined the effect of MD54 and MMD54 on LPS-induced MD2/TLR4 association in THP-1 monocytes by co-immunoprecipitation assay. Briefly, THP-1 cell lysate was immunoprecipitated with an anti-TLR4 antibody, followed by immunoblot detection of MD2 in the immunoprecipitates. As shown in Fig. 6e, LPS stimulation significantly increased the association and complex formation between MD2 and TLR4, which was significantly reduced in the presence of MD54, whereas MMD54 did not show any significant inhibition of LPS-stimulated complex formation between TLR4 and MD2. As a control, the immunoprecipitation by IgG did not result in precipitation of MD2. A statistical graph (Fig. 6f) shows the significant difference in LPS-stimulated TLR4/MD2 complex formation in the presence of the MD2-derived peptide, MD54 but not its analog, MMD54.
MD54 protects mice from LPS challenge
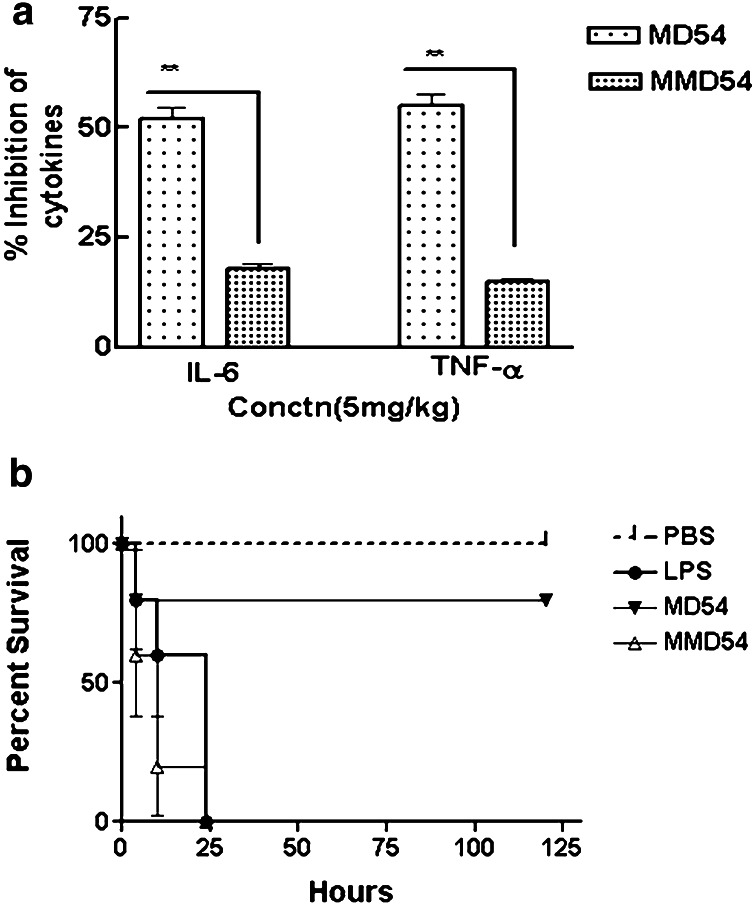
In vivo efficacy of MD54 was investigated in mice as this peptide was significantly non-toxic and also showed appreciable in vitro anti-LPS property. When MD54 was administered alone in mice (5 mg/kg body weight), no mortality of animal was observed indicating its non-toxic nature in vivo (data not shown). Subsequent to LPS challenge (10 mg/kg body weight) by i.p. injection, MD54 at a single dose of 5 mg/kg body weight showed 55 and 52% inhibition of LPS-induced production of TNF-α and IL-6, respectively, in comparison with LPS alone, while MMD54 did not show any significant inhibition of production of these cytokines in mice. MD54 (5 mg/kg) also significantly improved the survival of LPS (10 mg/kg) challenged mice. Mortality was reduced from 100 to 20% (for n = 5) when MD54 (5 mg/kg) was administered with LPS (10 mg/kg) in mice in a 5 day experiment (p < 0.05). Whereas the mice group administered with only LPS (10 mg/kg) or LPS and MMD54 (5 mg/kg) together died within 24 h (Fig. 7a, b) [39, 40].
Fig. 7.
Peptide protects mice from LPS challenge. a In vivo estimation of percentage inhibition of TNF-α and IL-6 was done after administration of 10 mg/kg LPS with 5 mg/kg of peptide. Student t test was performed using Graph Pad Prism Version 5.00 vs. MMD54 (**p < 0.01, p = 0.0033). b Survival analysis of the LPS challenged peptide was observed for 5 days. Female Balb/c mice (6–8 week; n = 5) were subjected to LPS challenge with peptide, and survival was determined every 12 h for 120 h after LPS challenge. Survival in MD54 (80%) as compared to LPS and MMD54 (0%) was statistically significant [*p < 0.05, log-rank test (p = 0.0419)]
Discussion
The key finding of this study is the identification of a 16-residue MD2 segment (a.a. 54–69), namely, MD54 which promotes aggregation of LPS, facilitates its internalization in THP-1 monocytes, and thereby leads to neutralization/detoxification of LPS-induced pro-inflammatory responses in these cells.
MD54 is an amphipathic peptide, comprising of almost an equal number of hydrophobic and hydrophilic amino acid residues including four cationic charges (Fig. 1; Table 1). Moreover, this stretch possesses a heptad repeat sequence which has been implicated in the interaction of melittin with LPS [17]. CD studies (Fig. 3c) demonstrated that interaction of MD54 with LPS leads to significant conformational changes particularly with induction of more β-sheet structure. Secondary structural analyses suggest that MD54 was more structured both in PBS and in the presence of LPS (Fig. 3c; Table 2) than its analog, MMD54, which has a leucine residue substituted with an alanine residue at an ‘a’ position of the heptad repeat. The results are supportive of an earlier notion of a possible role of this motif in interaction with LPS [17].
The adoption of β-sheet structure by MD54 peptide in LPS matches with the crystal structure of the corresponding segment of MD2 in the presence of LPS and TLR4. The data indicated that MD54 segment (a.a. 54–69) comprises an important stretch that could contribute to structural property of the whole MD2 protein in the presence of its interacting molecules. MD2 plays a crucial role in the defense mechanism of the host by recognizing and binding to LPS and further facilitating the downstream signaling events through TLR4. Therefore, further studies were carried out to look into the interaction of MD54 with LPS. In the presence of MD54, fluorescence of FITC–LPS decreased slightly (Fig. 3a). Furthermore, the addition of proteinase K significantly increased the fluorescence of FITC–LPS that was pre-treated with MD54 (Fig. 3a). This increase in fluorescence was due to de-quenching of FITC fluorescence resulting from the dissociation of the aggregated FITC–LPS–MD54 complex. Taken together, these results indicated that MD54 induced aggregation in FITC–LPS. On the other hand, when FITC–LPS was pre-treated with the MD54 analog, MMD54, and then, proteinase K was added, a very weak increase in FITC fluorescence observed (Fig. 3a). The data further suggested a sequence specific interaction of MD54 with FITC–LPS.
Further evidence on the aggregation of FITC–LPS in the presence of MD54 was accomplished by the light-scattering experiment. Studies with Zeta sizer indicated an increase in the intensity of scattered light, suggesting the formation of the more aggregated state of LPS in the presence of MD54. While in the presence of the analog MMD54, a significant decrease in light scattering was observed indicating that a single leucine to alanine substitution impaired the ability of MD54 to promote aggregation in LPS (Fig. 3b).
MD54 altered the binding of FITC–LPS-to-THP-1 cells very significantly. A dramatic increase in fluorescence of THP-1 cells in the presence of MD54 by FACS studies (Fig. 4a) indicated a possible internalization of FITC–LPS in these cells as reported previously [41, 42]. The FACS results were supported by confocal microscopic images (Fig. 4b) and showed the localization of FITC–LPS inside the THP-1 cells in the presence of MD54. MMD54 weakly facilitated the internalization of FITC–LPS into these cells (Fig. 4b). FACS studies also showed that MD54 is specific for LPS. It did not facilitate the internalization of a non-specific fluid phase marker, FITC–dextran (Fig. S1) into THP-1 cells.
The mode of action of MD54 is contrasting to the antimicrobial peptides (AMPs) that usually interact with LPS and disintegrate its aggregation, thereby not allowing it to activate downstream signaling events required for the production of pro-inflammatory responses [17]. However, there are certain proteins such as rBPI, ECP, Limulus, and anti-LPS factor which help in the aggregation of LPS and disturb its hydrophobic core, thereby neutralizing its toxicity [43–45]. Recently, based on this model, Uppu et al. [46] designed a synthetic amphiphilic polymeric compound which formed pseudo-aggregates with LPS and thus not allowing it to bind its receptor and hence inhibited LPS-induced inflammatory responses. In the present study, we observed that MD54 not only induced aggregation in LPS, but also promoted its internalization in THP-1 cell, leading to its reduced toxicity and neutralization (Fig. 5). Furthermore, translocation of NF-κB subunits to the nucleus is a key event that occurs following the TLR4–MD2–LPS complex formation. MD54 significantly inhibited LPS-induced nuclear translocation of NF-κB subunits, p60 and p65 (Fig. 6a). Moreover, it also inhibited LPS-induced production of pro-inflammatory cytokines, TNF-α, IL-1β, and IL-8 (Fig. 5a) in PMA-stimulated THP-1 cells and THP-1 cells, respectively, that could be the possible implications of inhibition of LPS–MD2–TL4 complex formation in the presence of this peptide. The study to look into the effect of MD54 on LPS and its interacting proteins was extended in HEK-293T cells that naturally do not express proteins associated with LPS-induced signaling events. Our results based on the transient transfection delineated the requirement of LPS-interacting protein machinery for LPS-induced activation of NF-κB which was efficiently inhibited by MD54 (Fig. 6b, c). In addition to that, MD54 significantly inhibited LPS-induced IL-8 production in these transfected cells (Fig. 5a). Moreover, to examine the underlying mechanisms of LPS inhibition, immunoprecipitation experiments were performed and co-precipitation of TLR4/MD2 significantly increased in the LPS alone sample, while MD54 treatment significantly inhibited the association between MD-2 and TLR4 following LPS stimulation in THP-1 cells (Fig. 6e, f). Further studies indicated that MD54 inhibited LPS-induced production of pro-inflammatory cytokines in mice in vivo (Fig. 7a) and provided significant survival to mice against LPS challenge (Fig. 7b).
Thus, the results revealed the identification of a short, amphipathic peptide (MD54) with a heptad repeat sequence from MD2 that interacted with LPS, induced aggregation and facilitated its internalization in THP-1 cells further inhibiting its ability to produce pro-inflammatory responses. Formation of TLR4–MD2–LPS complex is a crucial event in the host defense mechanism which starts with the interaction of MD2 and LPS [47]. Considering the ability of MD54 to interact with LPS and induce its aggregation, probably, one can speculate that MD54 peptide possesses crucial amino acid sequences that could participate in the MD2–LPS–TLR-4 complex formation which is also supported by the immunoprecipitation experiments. Furthermore, this study may aid in the design of novel anti-endotoxin peptides with similar a mode of action.
Conclusion
The present study demonstrated that a 16-residue peptide, MD54 derived from extracellular adopter protein, MD2 induces aggregation in endotoxin (LPS) and facilitates THP-1 monocytes to uptake the aggregated LPS complex and neutralize it, thereby resulting in the inhibition of production of pro-inflammatory cytokines both in vitro in monocytes and in vivo in mice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The CSIR-CDRI communication number for this article is 9609. This work was supported by a Council of Scientific and Industrial Research (CSIR) network project BioDiscovery (BSC0120). We are thankful to Dr. Douglous Golenbock (Medicine Department, University of Massachusetts Medical School, Worcester MA) and Dr. Anila Dwivedi (Endocrinology div. CSIR-CDRI, Lucknow.) for providing us with the plasmid constructs. The authors are extremely thankful to Prof. Surajit Bhattacharjya, School of Biological Sciences, Nanyang Technological University, Singapore for editing the revised version of the manuscript. The authors thankfully acknowledge the anonymous reviewers for their constructive criticism and comments in improving the quality of the manuscript. Garima Pant of Sophisticated Analytical Instrumentation Facility (SAIF) and Rima Roy from Molecular and Structural Biology Division, CSIR-CDRI are acknowledged for assistance in using the Confocal Microscope. A. L. Vishwakarma and R. K Purshottam from SAIF and CSIR-CDRI are acknowledged for assistance in using FACS and HPLC facility, respectively. We are thankful to Pharmaceutics Department for providing us the Zeta sizer instrument accessibility. We also thankfully acknowledge National Laboratory Animal Centre, CSIR-Central Drug Research Institute, Lucknow, India for providing us the animals.
Abbreviations
- CD14
Cluster of differentiation 14
- HEK-293T
Human embryonic kidney cells 293
- LBP
Lipopolysaccharide-binding protein
- LPS
Lipopolysaccharide
- MD2
Lymphocyte antigen 96
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- PAMPs
Pathogen-associated molecular pattern
- PRRs
Pattern recognition receptor
- TLR4
Toll-like receptor 4
- PCNA
Proliferating cell nuclear antigen
- ICU
Intensive Care Unit
- AMP
Anti-microbial peptide
Author contributions
JKG and AT conceived the idea and designed the experiments. AT did the major part of the experiments. MKH and NI helped in western and in vivo experiments. AKT and SS assisted us in the peptide synthesis and biophysical experiments, respectively. JKG and AT analyzed the results and wrote the manuscript. All the authors were consulted on the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
References
- 1.Van Amersfoort ES, Van Berkel TJ, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16(3):379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365(9453):63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Scott MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA. 2000;97(16):8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenfeld Y, Shai Y. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta. 2006;1758(9):1513–1522. doi: 10.1016/j.bbamem.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Fenton MJ, Golenbock DT. LPS-binding proteins and receptors. J Leukoc Biol. 1998;64(1):25–32. doi: 10.1002/jlb.64.1.25. [DOI] [PubMed] [Google Scholar]
- 6.Gioannini TL, Zhang D, Teghanemt A, Weiss JP. An essential role for albumin in the interaction of endotoxin with lipopolysaccharide-binding protein and sCD14 and resultant cell activation. J Biol Chem. 2002;277(49):47818–47825. doi: 10.1074/jbc.M206404200. [DOI] [PubMed] [Google Scholar]
- 7.Gioannini TL, Teghanemt A, Zhang D, Levis EN, Weiss JP. Monomeric endotoxin:protein complexes are essential for TLR4-dependent cell activation. J Endotoxin Res. 2005;11(2):117–123. doi: 10.1177/09680519050110020801. [DOI] [PubMed] [Google Scholar]
- 8.Plociennikowska A, Hromada-Judycka A, Borzecka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. 2015;72(3):557–581. doi: 10.1007/s00018-014-1762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274(16):10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, Rowe DC, Golenbock DT. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. 2004;6(15):1361–1367. doi: 10.1016/j.micinf.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 12.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300(5625):1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 13.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16(1):3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Ulevitch RJ, Tobias PS. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr Opin Immunol. 1999;11(1):19–22. doi: 10.1016/S0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 15.Yang QW, Mou L, Lv FL, Zhu PF, Wang ZG, Jiang JX, Wang JZ. Novel TLR4-antagonizing peptides inhibit LPS-induced release of inflammatory mediators by monocytes. Biochem Biophys Res Commun. 2005;329(3):846–854. doi: 10.1016/j.bbrc.2005.01.162. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Shan X, Chen G, Jiang L, Wang Z, Fang Q, Liu X, Wang J, Zhang Y, Wu W, Liang G. MD-2 as the target of a novel small molecule, L6H21, in the attenuation of LPS-induced inflammatory response and sepsis. Br J Pharmacol. 2015;172(17):4391–4405. doi: 10.1111/bph.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava RM, Srivastava S, Singh M, Bajpai VK, Ghosh JK. Consequences of alteration in leucine zipper sequence of melittin in its neutralization of lipopolysaccharide-induced proinflammatory response in macrophage cells and interaction with lipopolysaccharide. J Biol Chem. 2012;287(3):1980–1995. doi: 10.1074/jbc.M111.302893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tack BF, Sawai MV, Kearney WR, Robertson AD, Sherman MA, Wang W, Hong T, Boo LM, Wu H, Waring AJ, Lehrer RI. SMAP-29 has two LPS-binding sites and a central hinge. Eur J Biochem. 2002;269(4):1181–1189. doi: 10.1046/j.0014-2956.2002.02751.x. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Y, Dai H, Bommineni YR, Soulages JL, Gong YX, Prakash O, Zhang G. Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken. FEBS J. 2006;273(12):2581–2593. doi: 10.1111/j.1742-4658.2006.05261.x. [DOI] [PubMed] [Google Scholar]
- 20.Mancek M, Pristovsek P, Jerala R. Identification of LPS-binding peptide fragment of MD-2, a toll-receptor accessory protein. Biochem Biophys Res Commun. 2002;292(4):880–885. doi: 10.1006/bbrc.2002.6748. [DOI] [PubMed] [Google Scholar]
- 21.Yan H, Gu CG, Xu FL, Wu XH, Yin HH, Hu CX, Zhu XD, Liu F, Ge HJ, Chen LY, Zhang XY, Wang ZG, Xing Z, Li L. Identification of synthetic peptides that inhibit lipopolysaccharide (LPS) binding to myeloid differentiation protein-2 (MD-2) J Immunother. 2013;36(3):197–207. doi: 10.1097/CJI.0b013e31828eed62. [DOI] [PubMed] [Google Scholar]
- 22.Duan GJ, Zhu J, Wan JY, Li X, Ge XD, Liu LM, Liu YS. A synthetic MD-2 mimetic peptide attenuates lipopolysaccharide-induced inflammatory responses in vivo and in vitro. Int Immunopharmacol. 2014;10(9):1091–1100. doi: 10.1016/j.intimp.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Bouchayer E, Stassinopoulou CI, Tzougraki C, Marion D, Gans P. NMR and CD conformational studies of the C-terminal 16-peptides of Pseudomonas aeruginosa c551 and Hydrogenobacter thermophilus c552 cytochromes. J Pept Res. 2001;57(1):39–47. doi: 10.1034/j.1399-3011.2001.00792.x. [DOI] [PubMed] [Google Scholar]
- 24.Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35(3):161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 25.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89(21):10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oren Z, Shai Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: structure-function study. Biochemistry. 1997;36(7):1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad A, Yadav SP, Asthana N, Mitra K, Srivastava SP, Ghosh JK. Utilization of an amphipathic leucine zipper sequence to design antibacterial peptides with simultaneous modulation of toxic activity against human red blood cells. J Biol Chem. 2006;281(31):22029–22038. doi: 10.1074/jbc.M602378200. [DOI] [PubMed] [Google Scholar]
- 28.Asthana N, Yadav SP, Ghosh JK. Dissection of antibacterial and toxic activity of melittin: a leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J Biol Chem. 2004;279(53):55042–55050. doi: 10.1074/jbc.M408881200. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh JK, Ovadia M, Shai Y. A leucine zipper motif in the ectodomain of Sendai virus fusion protein assembles in solution and in membranes and specifically binds biologically-active peptides and the virus. Biochemistry. 1997;36(49):15451–15462. doi: 10.1021/bi971152i. [DOI] [PubMed] [Google Scholar]
- 30.Pandey BK, Srivastava S, Singh M, Ghosh JK. Inducing toxicity by introducing a leucine-zipper-like motif in frog antimicrobial peptide, magainin 2. Biochem J. 2011;436(3):609–620. doi: 10.1042/BJ20110056. [DOI] [PubMed] [Google Scholar]
- 31.Yadav SP, Kundu B, Ghosh JK. Identification and characterization of an amphipathic leucine zipper-like motif in Escherichia coli toxin hemolysin E. Plausible role in the assembly and membrane destabilization. J Biol Chem. 2003;278(51):51023–51034. doi: 10.1074/jbc.M310052200. [DOI] [PubMed] [Google Scholar]
- 32.Mookherjee N, Brown KL, Bowdish DM, Doria S, Falsafi R, Hokamp K, Roche FM, Mu R, Doho GH, Pistolic J, Powers JP, Bryan J, Brinkman FS, Hancock RE. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J Immunol. 2006;176(4):2455–2464. doi: 10.4049/jimmunol.176.4.2455. [DOI] [PubMed] [Google Scholar]
- 33.Nordahl EA, Rydengard V, Morgelin M, Schmidtchen A. Domain 5 of high molecular weight kininogen is antibacterial. J Biol Chem. 2005;280(41):34832–34839. doi: 10.1074/jbc.M507249200. [DOI] [PubMed] [Google Scholar]
- 34.Rallabhandi P. Measuring TLR function in transfectants. Curr Protoc Immunol. 2010;Chapter 14:Unit 14.16. doi: 10.1002/0471142735.im1416s91. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J Biol Chem. 2006;281(3):1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 36.de Haas CJ, van Leeuwen HJ, Verhoef J, van Kessel KP, van Strijp JA. Analysis of lipopolysaccharide (LPS)-binding characteristics of serum components using gel filtration of FITC-labeled LPS. J Immunol Methods. 2000;242(1–2):79–89. doi: 10.1016/S0022-1759(00)00207-6. [DOI] [PubMed] [Google Scholar]
- 37.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132(17):6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SC, Kim JY, Jeong C, Yoo S, Hahm KS, Park Y. A plausible mode of action of pseudin-2, an antimicrobial peptide from Pseudis paradoxa. Biochim Biophys Acta. 2011;1808(1):171–182. doi: 10.1016/j.bbamem.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Xu R, Feng L, Guo W, Cao N, Qian C, Teng P, Wang L, Wu X, Sun Y, Li J, Shen Y, Xu Q. A novel chromone derivative with anti-inflammatory property via inhibition of ROS-dependent activation of TRAF6–ASK1–p38 pathway. PLoS One. 2012;7(8):e37168. doi: 10.1371/journal.pone.0037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas RC, Bath MF, Stover CM, Lambert DG, Thompson JP. Exploring LPS-induced sepsis in rats and mice as a model to study potential protective effects of the nociceptin/orphanin FQ system. Peptides. 2014;61:56–60. doi: 10.1016/j.peptides.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Fermino ML, Polli CD, Toledo KA, Liu FT, Hsu DK, Roque-Barreira MC, Pereira-da-Silva G, Bernardes ES, Halbwachs-Mecarelli L. LPS-induced galectin-3 oligomerization results in enhancement of neutrophil activation. PLoS One. 2011;6(10):e26004. doi: 10.1371/journal.pone.0026004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitchens RL, Munford RS. CD14-dependent internalization of bacterial lipopolysaccharide (LPS) is strongly influenced by LPS aggregation but not by cellular responses to LPS. J Immunol. 1998;160(4):1920–1928. [PubMed] [Google Scholar]
- 43.Kaconis Y, Kowalski I, Howe J, Brauser A, Richter W, Razquin-Olazaran I, Inigo-Pestana M, Garidel P, Rossle M, Martinez de Tejada G, Gutsmann T, Brandenburg K. Biophysical mechanisms of endotoxin neutralization by cationic amphiphilic peptides. Biophys J. 2011;100(11):2652–2661. doi: 10.1016/j.bpj.2011.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andra J, Koch MH, Bartels R, Brandenburg K. Biophysical characterization of endotoxin inactivation by NK-2, an antimicrobial peptide derived from mammalian NK-lysin. Antimicrob Agents Chemother. 2004;48(5):1593–1599. doi: 10.1128/AAC.48.5.1593-1599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulido D, Nogues MV, Boix E, Torrent M. Lipopolysaccharide neutralization by antimicrobial peptides: a gambit in the innate host defense strategy. J Innate Immun. 2012;4(4):327–336. doi: 10.1159/000336713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uppu DS, Haldar J. Lipopolysaccharide neutralization by cationic-amphiphilic polymers through pseudoaggregate formation. Biomacromolecules. 2016;17(3):862–873. doi: 10.1021/acs.biomac.5b01567. [DOI] [PubMed] [Google Scholar]
- 47.Gangloff M, Gay NJ. MD-2: the Toll ‘gatekeeper’ in endotoxin signalling. Trends Biochem Sci. 2004;29(6):294–300. doi: 10.1016/j.tibs.2004.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.