Abstract
Saturated fatty acids, such as palmitate, lead to circadian disruption in cell culture. Moreover, information regarding the effects of unsaturated fatty acids on circadian parameters is scarce. We aimed at studying the effects of low doses of saturated as well as unsaturated fatty acids on circadian metabolism in vivo and at deciphering the mechanism by which fatty acids convey their effect. Mice were fed non-obesogenic doses of palm or olive oil and hepatocytes were treated with palmitate and oleate. Mice fed non-obesogenic doses of palm oil showed increased signaling towards fatty acid synthesis, while olive oil increased signaling towards fatty acid oxidation. Low doses of palmitate and oleate were sufficient to alter circadian rhythms, due to changes in the expression and/or activity of key metabolic proteins. Palmitate, but not oleate, counteracted the reduction in lipid accumulation and BMAL1-induced expression of mitochondrial genes involved in fatty acid oxidation. Palmitate was also found to interfere with the transcriptional activity of CLOCK:BMAL1 by preventing BMAL1 deacetylation and activation. In addition, palmitate, but not oleate, reduced PER2-mediated transcriptional activation and increased REV-ERBα-mediated transcriptional inhibition of Bmal1. The inhibition of PER2-mediated transcriptional activation by palmitate was achieved by interfering with PER2 nuclear translocation. Indeed, PER2 reduced fat accumulation in hepatocytes and this reduction was prevented by palmitate. Herein, we show that the detrimental metabolic alteration seen with high doses of palmitate manifests itself early on even with non-obesogenic levels. This is achieved by modulating BMAL1 at several levels abrogating its activity and expression.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03023-6) contains supplementary material, which is available to authorized users.
Keywords: Palmitate, Oleate, Metabolism, Liver, Hepatocyte, Clock, Circadian
Introduction
Mammals have developed an endogenous circadian clock located in the brain suprachiasmatic nuclei (SCN) of the anterior hypothalamus that responds to the environmental light–dark cycle. The SCN receives light information from the retina and transmits synchronization cues to peripheral clocks, e.g., in the liver, heart or adipose tissue, regulating cellular and physiological functions [1–3]. The clock mechanism in both SCN neurons and peripheral tissues consists of CLOCK and BMAL1 (brain–muscle-Arnt-like 1) that heterodimerize and bind to E-box sequences to mediate transcription of a large number of genes, including Periods (Per1, Per2, Per3) and Cryptochromes (Cry1, Cry2). PERs and CRYs constitute a part of the negative feedback loop and when they are produced in the cytoplasm, they oligomerize and translocate to the nucleus to inhibit CLOCK:BMAL1-mediated transcription [1, 4].
The rhythmic expression and activity of metabolic pathways are mainly attributed to the robust and coordinated expression of clock genes in tissues, such as liver, adipose tissue and muscle [5, 6]. In addition, clock proteins have been shown to link the core clock mechanism with metabolic pathways: (1) REV-ERBα, the negative regulator of Bmal1 expression [7] is induced during normal adipogenesis [8]. (2) RORα, the positive regulator of Bmal1 expression [9] has been shown to regulate lipogenesis and lipid storage in skeletal muscle. (3) CLOCK:BMAL1 heterodimer regulates the expression of Rev-erbα and Rorα [7, 9, 10]. (4) Peroxisome proliferator-activated receptor α (PPARα), involved in lipid and lipoprotein metabolism, binds directly to the Bmal1 promoter [11], and, in turn, the CLOCK:BMAL1 heterodimer regulates PPARα expression [11–13]. (5) PGC1α, a PPARγ transcriptional co-activator that regulates energy metabolism, stimulates the expression of the clock genes, Bmal1 and Rev-erbα, through co-activation of the ROR family of orphan nuclear receptors [14]. (6) Adenosine monophosphate-activated protein kinase (AMPK), a sensitive sensor of low energy and nutrient state in the cell, phosphorylates and, as a result, activates casein kinase I epsilon (CKIε), which, phosphorylates the PER proteins and, thereby, enhances their instability and degradation [15, 16]. (7) AMPK also phosphorylates the CRY proteins and enhances their degradation [17]. (8) One of the key factors in the mammalian target of rapamycin (mTOR) pathway, protein 70 S6 kinase (P70S6K), rhythmically phosphorylates BMAL1 allowing it to both associate with the translational machinery and stimulate circadian oscillations of protein synthesis [18]. (9) Sirtuin 1 (SIRT1), a key factor involved in metabolism, interacts directly with CLOCK and deacetylates BMAL1 and PER2 [19–21].
High-fat content in food has been shown to influence clock oscillation and function in various animal studies [22–25]. These findings hint towards the possible involvement of lipids in circadian control [26]. It was recently shown that 3 days of high-fat diet (HFD) were sufficient to impose reprogramming of the circadian clock, including loss of oscillation of a large number of normally oscillating genes, a phase advance of an additional subset of oscillating transcripts and induction of de novo oscillating transcripts [27]. The rapid influence of the diet on the clock (3 days only) suggests that the nutritional challenge, and possibly lipids themselves, and not merely the development of obesity, are sufficient to alter clock function.
The long-chain saturated fatty acid palmitate (C16:0) is particularly detrimental to hepatocytes by inducing lipotoxicity, such as insulin resistance, oxidative stress and cell death at high dose and for prolonged treatment [28]. On the other hand, the monounsaturated fatty acid oleate (C18:1) has been shown to prevent or alleviate the toxic effect of saturated free fatty acids and to be protective against insulin resistance and metabolic disorders [29]. Palmitate, the most abundant saturated fatty acid in the circulation of obese animals, has been demonstrated to alter the expression of clock genes in cell lines [30–32]. However, the effect of palmitate on the clock has never been tested in vivo. In cell culture, palmitate was found to destabilize protein–protein interaction between CLOCK:BMAL1 in a dose and time-dependent manner. Furthermore, SIRT1 activators reversed the inhibitory action of palmitate on CLOCK:BMAL1 interaction [31]. In addition, although docosahexaenoic acid (DHA) altered the circadian profile of Bmal1, the disruptive effect of palmitate in the presence of DHA on Bmal1 was less pronounced, suggesting a protective role for DHA [33, 34]. Thus, saturated fatty acids, such as palmitate, have been implicated in circadian disruption in cell culture, whereas little information exists regarding the effects of unsaturated fatty acids on circadian parameters. Therefore, our goal was to study in vivo effects of low doses of palmitate and oleate on circadian metabolism and use cell culture models to study the mechanism by which fatty acids convey this effect.
Materials and methods
Animals, treatments and tissues
Ten-week-old C57BL/6 male mice (Harlan Laboratories, Jerusalem, Israel) were housed in a temperature- and humidity-controlled facility (23–24 °C, 60% humidity). Mice were entrained to 12 h light and 12 h darkness for 2 weeks with food available ad libitum and then were randomly assigned to either olive oil (n = 24) or palm oil diet (n = 24) for 3 weeks. Control diet contained cornstarch 52.7% (w/w), casein 20% (w/w), sucrose 10% (w/w), soybean oil 7% (w/w), cellulose 5% (w/w), mineral mix 4% (w/w), vitamin mix 1% (w/w), methionine 0.3% (w/w). In palm and olive oil diet groups, soybean oil was replaced with olive oil (fatty acid composition: C:14 ˂ 0.03, C:16 18.4%, C:16.1 2.01%, C:18 3.75%, C:18-1 68.2%, C:18-2 7.6%) or palm oil (fatty acid composition: C:12 0.2%, C:14 1.1%, C:16 44.3%, C:18 4.6%, C:18-1 39%, C:18-2 10%). Body weight was recorded weekly and at the end of the experiment. Daily food intake was monitored every 2 days. Mice were placed on an overnight fast (12 h) before anesthesia and blood and liver were removed every 4 h around the circadian cycle in total darkness under a dim red light to avoid the masking effects of light. Tissues were immediately frozen in liquid nitrogen and stored at − 80 °C. Mice were humanely killed at the end of the experiment. Animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985). The joint ethics committee of the Hebrew University and Hadassah Medical Center approved this study.
Enzymatic colorimetric tests
Total cholesterol levels were determined using the Cobas kit (Roche Diagnostics, Burgess Hill, UK) and analyzed in a Roche/Hitachi analyzer (Roche Diagnostics). Assays were performed according to the manufacturer’s instructions.
Triglyceride and free fatty acid measurement
Triglyceride levels in liver tissue were determined using a Triglyceride Quantification Kit (Abcam, Cambridge, UK)), according to the manufacturer’s instructions. Free fatty acid levels in liver tissue were determined using a quantification Kit (Abcam), according to the manufacturer’s instructions.
Cell culture experiments
Mouse hepatocyte AML-12 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, Rehovot, Israel) supplemented with 10% fetal calf serum (FCS), 100 mg/l l-glutamine and 100 mg/l penicillin at 37 °C in 5% CO2. For circadian gene and protein expression, cells were synchronized with a 1-h pulse of 1 mM dexamethasone (Sigma). After 1 h, medium was replaced with medium supplemented with palmitate or oleate mixed in 1% bovine serum albumin to a final concentration of 50 μM. Following 6 h of incubation, cells were harvested in triplicates per treatment per time-point every 6 h for 24 h. Three independent experiments were performed.
Fluorometric detection of fat content in intact cells by Nile red staining
The lipid content in cultured cells was determined fluorometrically using Nile red [35]. Cells were washed with phosphate-buffered saline (PBS), pH 7.4, and then incubated for 10 min at 37 °C with 1 μg/ml Nile red in PBS. Subsequently, cells were washed with PBS and measured by flow cytometry with excitation at 488 nm and emission at 575 nm (FACS Calibur, Becton–Dickinson, CA, USA). Intracellular fat accumulation was viewed using light/fluorescence eclipse E400 Nikon microscope with 40 × 0.75 objective or the 100 × 1.3 oil objective, using light or the fluorescence filters for Tetramethylrhodamine (TRITC) or 4, 6-diamidino-2-phenylindole (DAPI). Pictures were taken with DP71 camera (Olympus, Airport City, Israel), controlled by CellSens software (Olympus).
RNA extraction and quantitative real-time PCR
RNA was extracted from cells using TRI Reagent (Sigma). Total RNA was DNase I-treated using RQ1 DNase (Promega, Madison, WI, USA) for 2 h at 37 °C, as was previously described [36]. 2 μg of DNase I-treated RNA were reverse transcribed using MMuLV reverse-transcriptase and random hexamers (Promega). One twentieth of the reaction was then subjected to quantitative real-time PCR using SYBR Green Supermix (Quanta Biosciences, Beverly, MA, USA) and primers (Table S1) spanning exon–exon boundaries and the ABI Prism 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Gene expressing was normalized to actin. Reaction conditions were as follows: 3 min at 95 °C, 10 s at 95 °C, 45 s at 60 °C. The fold change in target gene expression was calculated by the 2−ΔΔCt relative quantification method (Applied Biosystems).
Western blot analyses
Liver tissue samples and cells were homogenized in lysis buffer as was previously described [37]. Nuclear extracts were prepared as was previously described [38]. In brief, treated cells were disrupted in cold hypotonic buffer (20 mM Hepes, pH 7.0, 10 mM KCl, 1 mM MgCl2, 2 mM dithiothreitol (DTT), 0.1% Triton X-100, 20% glycerol, 2 mM phenylmethylsulfonyl fluoride (PMSF), 1% protease inhibitor cocktail). Nuclei were pelleted by centrifugation at 3000 rpm for 5 min. The pellets were resuspended in cold extraction buffer (20 mM HEPES, pH 7.0, 10 mM KCl, 1 mM MgCl2, 2 mM DTT, 0.1% Triton X-100, 20% glycerol, 2 mM PMSF, 1% protease inhibitor cocktail, 420 mM NaCl). The samples were rotated for 30 min at 4 °C and then centrifuged at 14,000 rpm for 1 h. The supernatant was collected and protein concentration was measured using the Bradford reagent. Samples were kept frozen at − 80 °C. Samples were run onto a 10% SDS–polyacrylamide gel. After electrophoresis, proteins were semi-dry-transferred onto nitrocellulose membranes. Blots were incubated with antibodies against AMPK and its phosphorylated form (pAMPK), acetyl CoA carboxylase (ACC) and its phosphorylated form (pACC), P70S6K and its phosphorylated form (pP70S6K) (Cell Signaling Technology, Beverly, MA, USA), SIRT1, BMAL1 and PER2 (Abcam), Ac-BMAL1 (Merck, Darmstadt, Germany) and after several washes, with horseradish peroxidase conjugated secondary antibody (Pierce, Rockford, IL, USA). Anti-mouse antibody (MP Biomedicals, Solon, OH, USA) was used to detect actin, the loading control. The immune reaction was detected by enhanced chemiluminescence (Santa Cruz Biotechnologies, Santa Cruz, CA, USA). Finally, bands were quantified by scanning and densitometry and expressed as arbitrary units. For calculation of average levels, all samples were run separately several times and were normalized to the same sample in all blots. Densitometry after normalization to the normalizer was performed and averaged for each treatment.
Transfections and luciferase reporter assays
AML-12 cells were transfected using lipofectamin 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. Five hours after transfection, cells were incubated overnight in DMEM supplemented with 10% FCS and antibiotics in the presence or absences of fatty acids and then cells were harvested. For luciferase reporter assays, 48 h after transfection, cells were washed with PBS and harvested in Reporter Lysis Buffer (Promega). Luciferase activities were assayed with the Dual-Luciferase Reporter Assay System (Promega) using a Tristar LB-941 (Berthold Technologies, Bad Wildbad, Germany) luminometer. β-galactosidase or Renilla were co-transfected and used as normalizers. The amount of DNA per well was adjusted with the pcDNA3.1 empty vector.
Lipid extraction
Five micrograms of heptadecanoic acid (C17:0) was added to 0.5 ml sample. Subsequently, 10 ml of methanol–chloroform solution (2:1, v/v) was added and samples were incubated at 4 °C for 14 h. The upper phase was discarded and the lower phase containing total lipids was evaporated (Sheldon’s vacuum oven, Cornelius, OR, USA) at 65 °C. For gas chromatography analyses, fatty acid methylesters were generated by incubation of the total lipid extract with 2 ml of 5% (v/v) methanolic H2SO4 at 65 °C for 1 h in sealed vials. Samples were then cooled to room temperature and 1.5 ml of petroleum ether and 3 ml of double-distilled water were added. The upper phase was collected and evaporated in a vacuum oven at 65 °C. Dried samples were dissolved in 30 μl of petroleum ether.
Gas chromatography (GC) analysis
Chromatographic analysis was performed with a 6890 N gas chromatograph (Agilent Technologies, Wilmington, DE, USA) equipped with a fused-silica (60 m × 0.25 mm ID, 0.25 μm film) capillary column (DB-23, Agilent Technologies). The following conditions were applied: oven temperature was programmed from 130 to 170 °C at a rate of 27 °C/min; 170 to 215 °C at a rate of 2 °C/min, held at 215 °C for 8 min; 215 to 250 °C at a rate of 40 °C/min and held at 250 °C for 5 min. Run time was 37.9 min. Helium was used as the carrier gas at a flow rate of 2.21 ml/min. Flame ionization detector temperature was 280 °C, and injector temperature was 270 °C. Air and hydrogen flows were adjusted to 40 ml/min H2 and 450 ml/min air to give maximal detector response. The split ratio was set at 10:1 and 1 μl sample was injected. Peak identification was based on relative retention times of 2 external standards. The area of each fatty acid peak was recorded using ChemStation software (Agilent Technologies), and fatty acid molecular weights were calculated using the internal standard’s area under the peak. The amount of product synthesized after BMAL1 overexpression (de novo) was divided by the total amount of the substrate in the culture system.
Statistical analyses
All results are expressed as mean ± SE. Tukey HSD was performed for the evaluation of significant differences in average daily expression levels of metabolic genes. One-way ANOVA (time of day) test was performed to analyze the circadian pattern of clock and metabolic genes with several time-points. For all analyses, the significance level was set at p < 0.05. Statistical analysis was performed with JMP software (version 7) (SAS Institute Inc., Cary, NC, USA). Further analysis of circadian rhythmicity was performed using Circwave software (version 1.4) (Circadian Rhythm Laboratory, University of Groningen, Groningen, Holland).
Results
Effect of palm oil/palmitate and olive oil/oleate on liver/hepatocyte metabolism
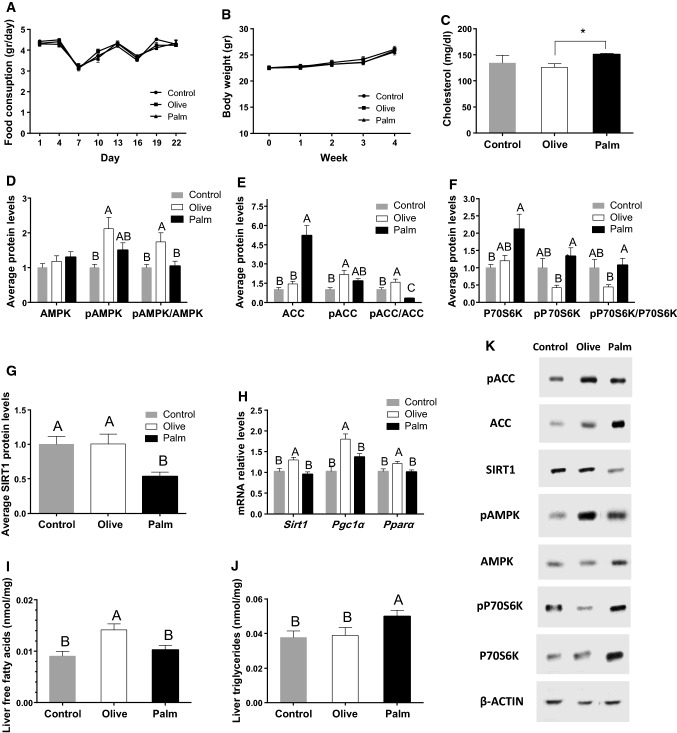
The short-term (3 weeks) effects of palm and olive oil treatment on metabolism were first investigated in mice and compared to a control group on chow diet that contains soybean oil. Although all groups consumed the same amount of fat and calories (Fig. 1a) and showed no statistical difference in body weight (Fig. 1b), palm oil diet increased serum total cholesterol levels compared to olive oil diet (p = 0.01, Student’s t test) (Fig. 1c). To accurately assess the effect of the oils on metabolism, 6 time-points throughout the circadian cycle were used to measure the daily average expression and/or activation levels of the key metabolic regulators AMPK, ACC, P70S6K and SIRT1. These metabolic factors showed circadian rhythmicity (data not shown). The olive oil diet showed higher ratio of the activated phosphorylated form of AMPK (pAMPK) to AMPK compared to control and palm oil diet (p = 0.01, Tukey’s HSD) (Fig. 1d, k). In parallel, olive oil diet led to a significantly higher ratio of the inactive phosphorylated ACC (pACC) to ACC (p = 0.03, Tukey’s HSD), whereas palm oil diet led to a significantly lower levels of pACC/ACC (p = 0.008, Tukey’s HSD) compared to the control diet (Fig. 1e, k). Olive oil diet also led to a significantly lower ratio of the active phosphorylated P70S6K (pP70S6K) to P70S6K, the downstream target of mTOR compared to palm oil diet (p = 0.04, Tukey’s HSD) (Fig. 1f, k). Furthermore, the palm oil diet showed significantly lower daily SIRT1 protein levels (p = 0.04, Tukey’s HSD) (Fig. 1g, k) and olive oil diet showed higher mRNA levels of SIRT1 (p = 0.01, Tukey’s HSD) and higher mRNA levels of its downstream targets Pgc1α and Pparα (p < 0.001 and p = 0.02, respectively, Tukey’s HSD) (Fig. 1h). Olive oil diet showed higher concentrations of liver free fatty acids (p = 0.004, Tukey’s HSD), whereas palm oil diet also led to significantly higher levels of liver triglycerides (p = 0.03, Tukey’s HSD) (Fig. 1i, j). The triglycerides showed differences between the light phase and the dark (data not shown).
Fig. 1.
Effect of palm and olive oil on liver metabolism. a Daily food consumption. b Weekly body weight. c Total plasma cholesterol. d Daily average protein levels of AMPK, pAMPK and pAMPK/AMPK ratio in mouse liver. e Daily average protein levels of ACC, pACC and pACC/ACC ratio in mouse. f Daily average protein levels of P70S6K, pP70S6K and pP70S6K/P70S6K ratio in mouse liver. g Daily average protein levels of SIRT1 in mouse liver. h Daily average mRNA levels of Sirt1, Pgc1α, Pparα in mouse liver. i Liver free fatty acids. j Liver triglycerides. k Representative protein bands. Mice were fed palm or olive oil diet for 3 weeks. Liver and plasma samples were collected every 4 h for 24 h. Proteins were analyzed by Western blotting. mRNA was determined by real-time PCR. Data are presented as mean ± SE; n = 24/group, n = 4/time-point. Asterisks denote significant differences (p < 0.0 5, Student’s t test)
To test whether the metabolic effect was due to palmitate and oleate, the predominant fatty acids in palm and olive oil, respectively, we treated AML-12 hepatocytes with low concentrations of these fatty acids. Whereas neither oleate nor palmitate resulted in fat accumulation (Fig. S1A), cells treated with oleate showed significantly increased and palmitate-decreased pAMPK levels compared to control (p < 0.005, Tukey’s HSD) (Fig. S1B, F). Oleate treatment also increased pACC levels compared to control (p = 0.01, Tukey’s HSD) with no effect of palmitate (p > 0.05, Tukey’s HSD) (Fig. S1C, F). Palmitate treatment increased pP70S6K (p < 0.0001, Tukey’s HSD) and decreased P70S6K (p < 0.0001, Tukey’s HSD) levels resulting in significantly increased pP70S6K/P70S6K ratio (p < 0.0001, Tukey’s HSD) with no effect of oleate (Fig. S1D, F). Oleate treatment showed significantly higher SIRT1 protein (Fig. S1E, F) and mRNA daily levels (p = 0.02 and p = 0.0002, respectively, Tukey’s HSD) and the mRNA of downstream targets Pgc1α (p = 0.03, Tukey’s HSD) and Pparα (p < 0.001, Tukey’s HSD) with no effect of palmitate (Fig. S1G). Taken together, in the liver/hepatocytes, olive oil/oleate activate the AMPK–SIRT1 signaling pathway which may lead to inhibition of fatty acid synthesis and increased fatty acid oxidation, whereas palm oil/palmitate activate mTOR signaling which may lead to increased fatty acid synthesis and inhibition of fatty acid oxidation.
Effect of palm oil/palmitate and olive oil/oleate on circadian rhythms
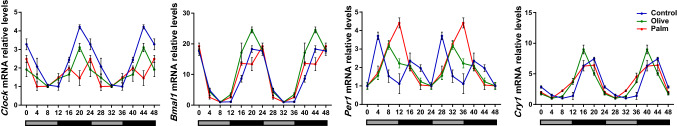
As the aforementioned key metabolic factors, i.e., AMPK, mTOR, and SIRT1, have intricate relationships with the core clock mechanism [39] and their modulation by the oils may lead to changes in the phase of gene expression, we analyzed clock gene expression at 6 time-points around the circadian cycle in mouse liver (Fig. 2). Clock genes oscillated robustly in liver and cell culture (p < 0.05, 1-way ANOVA, CircWave). Bmal1, Clock, Per1, and Cry1 showed advanced rhythms in both olive oil and palm oil groups compared to the control group, but with a larger advance in the olive oil group (Fig. 2, Table S2). Similarly, treatment of AML-12 hepatocytes with oleate resulted in phase advances in Bmal1, Clock, Per1, and Cry1, while treatment with palmitate resulted in phase delays in Bmal1, Per1, and Cry1 (Fig. S2, Table S3). Cry1 expression dampened as a result of oleate treatment. Taken together, these results show that low doses of fatty acids are sufficient to alter clock gene expression in liver.
Fig. 2.
Effect of palm oil and olive oil diet on circadian rhythms. Clock gene oscillation in liver. Mice were fed palm or olive oil diet for 3 weeks. Liver samples were collected every 4 h for 24 h. mRNA was determined by real-time PCR. Data are mean ± SE; n = 24/group, n = 4/time-point
Palmitate counteracts BMAL1 activity in reducing fat accumulation
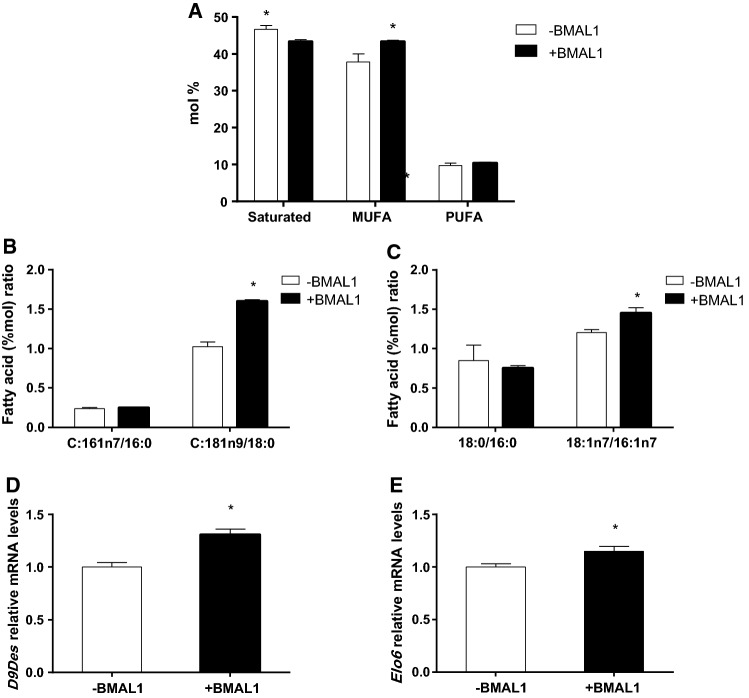
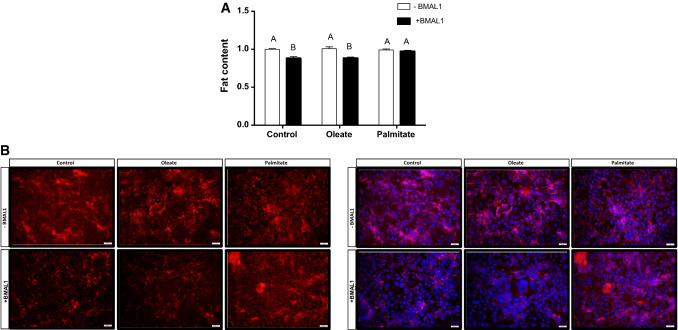
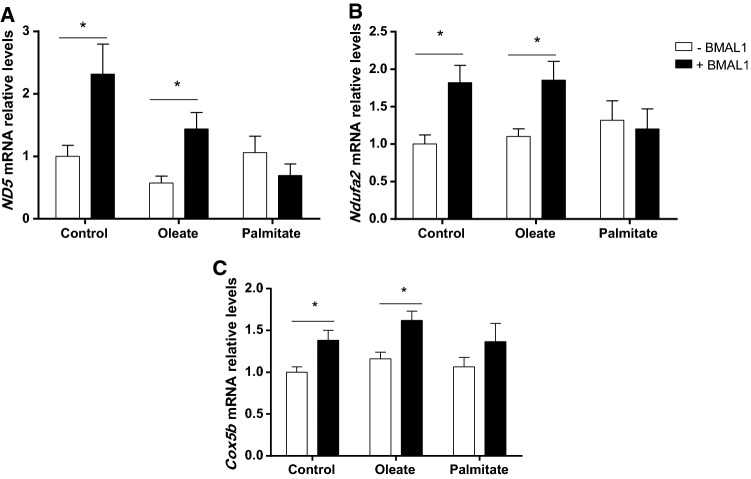
As BMAL1 has been reported to be involved in lipid accumulation in adipose tissue [39], we next studied the effect of fatty acids on BMAL1 activity in AML-12 hepatocytes. In the absence of fatty acids, BMAL1 overexpression modified cellular fatty acid composition leading to 5.4% elevation in monounsaturated fatty acids (p = 0.04, Student’s t test) and 3.3% reduction in saturated fatty acids (p = 0.02, Student’s t test) (Fig. 3a). The fatty acid that was mostly elevated following BMAL1 overexpression was oleic acid (c18:1n9), by 2.6% (p = 0.002, Student’s t test) (Table S4). Other monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) were also significantly increased following BMAL1 overexpression (Table S4), however, in total, PUFA did not change. We next analyzed the effect of BMAL1 overexpression on fatty acid desaturation and elongation indices. Suggested by the higher C18:1n9/C18:0 ratio, but not C16:1n7/C16:0 ratio (Fig. 3b), BMAL1 increased Δ9 desaturase activity index (p < 0.0001, Student’s t test). BMAL1 also increased elongase 6 activity (p < 0.03, Student’s t test) suggested by the higher C18:1n7/C16:1n7, but not C18:0/C16:0 ratio (Fig. 3c). Indeed, Δ9 desaturase (p < 0.001, Student’s t test) and elongase 6 (p = 0.01, Student’s t test) mRNA expression were increased following BMAL1 overexpression (Fig. 3d, e). When measured by flow cytometry, BMAL1 overexpression led to reduced lipid accumulation in AML-12 hepatocytes (p < 0.0001, Student’s t test) (Fig. 4). This result was also found when BMAL1-overexpressing hepatocytes were treated with oleate (p = 0.0002, Student’s t test). In contrast, when BMAL1-overexpressing hepatocytes were treated with palmitate this reduction in fat accumulation was abolished (p > 0.05, Student’s t test) (Fig. 4). These results are further supported by the finding that palmitate, but not oleate, counteracted the increase in the expression of mitochondrial genes whose protein products participate in fatty acid oxidation in cells overexpressing BMAL1, such as NADH-ubiquinone oxidoreductase chain 5 (ND-5), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 2 (NDUFA2), Cytochrome c oxidase subunit 5B (COX5b) (Fig. 5). Taken together, our results show that BMAL1 overexpression leads to reduced fat accumulation in hepatocytes and this reduction is counteracted by palmitate.
Fig. 3.
BMAL1 overexpression regulates fatty acid composition. a Fatty acid composition. b Δ9 desaturase activity. c Elongase 6 activity. d Δ9 desaturase mRNA in AML-12 hepatocytes and AML-12 hepatocytes overexpressing BMAL1. e Elongase 6 mRNA in AML-12 hepatocytes and AML-12 hepatocytes overexpressing BMAL1. AML-12 hepatocytes were transfected with Bmal1 and cell lipid extracts were methylated and fatty acid amounts were analyzed by GC. mRNA was determined by real-time PCR. Values are mean ± SE (n = 4). Asterisks denote significant difference (p < 0.05, Student’s t test)
Fig. 4.
Palmitate counteracts BMAL1 activity in reducing fat accumulation. a Intracellular lipid content. Values are mean ± SE (n = 12). Asterisks denote significant difference (p < 0.05, Student’s t test). b Fluorescence microscopy images showing lipid content stained by Nile red (left panel) and Nile red merged with DAPI staining (right panel). AML-12 hepatocytes were transfected with Bmal1 and treated with 50 μM palmitate, oleate or BSA (control)
Fig. 5.
Palmitate counteracts BMAL1-induced expression of mitochondrial genes involved in fatty acid oxidation. a ND-5 mRNA. b NDUFA2 mRNA. c COX5b mRNA. AML-12 hepatocytes were transfected with Bmal1 and treated with 50 μM palmitate, oleate or BSA (control). mRNA was determined by real-time PCR. Values are mean ± SE (n = 9). Asterisks denote significant difference (p < 0.05, Student’s t test)
Palmitate inhibits CLOCK:BMAL1-mediated expression
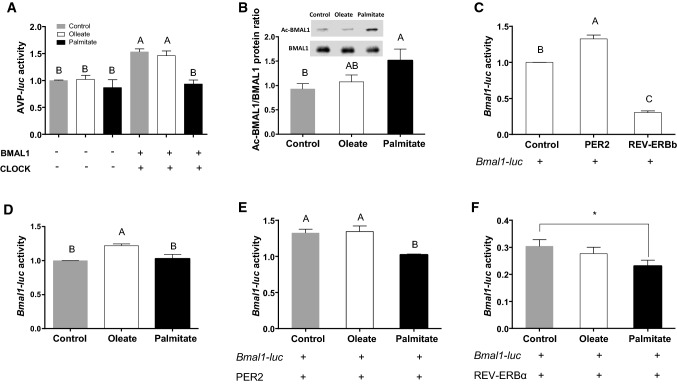
To study whether palmitate may interfere with BMAL1 transcriptional activity by affecting its function in CLOCK:BMAL1-mediated gene expression, we examined the effect of palmitate and oleate treatment on the activity of the clock-controlled promoter, arginine vasopressin (AVP) promoter-driven luciferase reporter (AVP-luc) in AML-12 hepatocytes. CLOCK and BMAL1 overexpression induced luciferase activity (p ˂ 0.0001, Tukey’s HSD) (Fig. 6a). This luciferase induction was abolished by palmitate, but not oleate (p ˂ 0.0001, Tukey’s HSD) (Fig. 6a). To study how palmitate may interfere with CLOCK:BMAL1 activity, we measured endogenous BMAL1 acetylation (AcBMAL1) level, as BMAL1 requires deacetylation to become active [40]. Six hours after exposure of hepatocytes to fatty acids, the ratio of AcBMAL1/BMAL1 increased (p = 0.04, Tukey’s HSD) with palmitate, but not oleate (Fig. 6b). Taken together, these results demonstrate that palmitate interferes with BMAL1 activity by preventing its deacetylation.
Fig. 6.
Palmitate modulates BMAL1 activity and expression. a Luciferase activity. AVP-luc was co-transfected with Bmal1 and Clock and AML-12 hepatocytes were treated with palmitate, oleate or BSA (control). Luciferase activity was normalized to β–galactosidase activity (n = 12). b AcBMAL1/BMAL1 protein ratio. AML-12 hepatocytes were treated with palmitate, oleate or BSA (control) (n = 6). c Luciferase activity. Bmal1-luc was co-transfected with Per2 or Rev-erbα. d Luciferase activity. Bmal1-luc transfected cells were treated with palmitate, oleate or BSA (control) (n = 12). e Luciferase activity. Bmal1-luc was co-transfected with Per2 and cells were treated with palmitate, oleate or BSA (control) (n = 12). f Luciferase activity. Bmal1-luc was co-transfected with Rev-erbα and cells were treated with palmitate, oleate or BSA (control) (n = 9). Different letters denote significant difference (p < 0.05, Tukey’s HSD); asterisks denote significant difference (p < 0.05, Student’s t test)
Palmitate modulates BMAL1 expression
Our results in vivo showed no significant change in average daily levels of Bmal1 mRNA or BMAL1 protein under olive or palm oil diet (Fig. S3). To study whether palmitate and oleate can acutely affect BMAL1 expression in cell culture, we transfected AML-12 hepatocytes with a luciferase reporter whose expression is driven by the Bmal1 promoter (Bmal1-luc) [41]. The Bmal1 regulators, PER2 and REV-ERBα [42], were also co-transfected. While REV-ERBα reduced Bmal1-luc activity (p ˂ 0.0001, Tukey’s HSD), PER2 induced Bmal1-luc activity (p ˂ 0.0001, Tukey’s HSD) (Fig. 6c). Luciferase levels significantly increased after oleate treatment (p < 0.05, Tukey’s HSD), but not palmitate (Fig. 6d). Similar induction was achieved in the presence of oleate (p = 0.04, Student’s t test), suggesting that it does not affect PER2-mediated BMAL1 expression (Fig. 6e). In contrast, palmitate prevented PER2-mediated luciferase activity (p > 0.05, Student’s t test) (Fig. 6e), suggesting that it interferes with PER2-mediated BMAL1 expression. When REV-ERBα was co-transfected with Bmal1-luc, luciferase activity was reduced in the presence of oleate to the same extent as in the absence of oleate (Fig. 6f). However, palmitate led to a much greater reduction in luciferase activity (p < 0.05, Student’s t test) (Fig. 6f), suggesting that it amplified the inhibitory effect REV-ERBα has on BMAL1 expression. Taken together, these results suggest that acute treatment of palmitate reduces Bmal1 transcription levels by reducing its PER2-mediated induction and increasing its REV-ERBα-mediated inhibition.
Palmitate affects PER2 nuclear localization
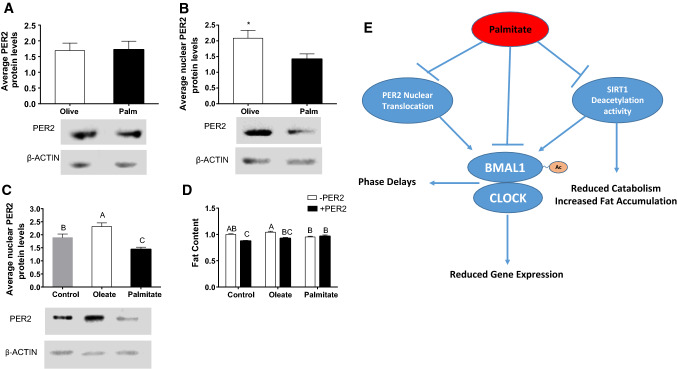
To investigate why PER2-mediated BMAL1 induction is abrogated, we measured PER2 protein nuclear levels. Oleate or palmitate treatment did not affect whole cell levels of PER2 (Fig. 7a). While oleate treatment increased nuclear PER2 levels (p = 0.04, Tukey’s HSD), palmitate decreased nuclear PER2 levels (p = 0.02, Tukey’s HSD) in AML-12 hepatocytes (Fig. 7b). In parallel, mice fed palm oil diet had significantly lower levels of nuclear PER2 in the liver compared to mice fed olive oil diet (p = 0.02, Student’s t test) (Fig. 7c). Next, we wanted to test whether PER2 overexpression by itself affects fat accumulation in the presence or absence of fatty acids. Interestingly, PER2 overexpression by itself significantly reduced fat content in AML-12 hepatocytes (p ˂ 0.0001, Student’s t test). Cells overexpressing PER2 treated with oleate showed similar reduction in fat content (p < 0.001, Student’s t test) (Fig. 7d). In contrast, cells overexpressing PER2 treated with palmitate did not show the reduction in fat content (p > 0.05, Student’s t test). Taken together, these results indicate that PER2 reduces fat accumulation in AML-12 hepatocytes and that this reduction is prevented by palmitate.
Fig. 7.
Palmitate abrogates PER2 nuclear localization. a Daily PER2 average whole-cell protein levels in mouse liver. b Daily PER2 average nuclear protein levels in mouse liver. Mice were fed with palm or olive oil diet for 3 weeks. c Daily PER2 average nuclear protein levels in AML-12 treated with fatty acids (n = 45). d Fat content of AML-12 hepatocytes transfected with Per2 and treated with palmitate, oleate or BSA (control). Intracellular lipid levels were evaluated using Nile red staining and flow cytometry (n = 12). Different letters denote significant difference (p < 0.05, Tukey’s HSD); asterisks denote significant difference (p < 0.05, Student’s t test). e A model delineating how palmitate modulates clock and metabolism. Palmitate regulates clock functionality by inhibiting CLOCK:BMAL1-mediated transcriptional activation, by attenuating deacetylation of BMAL1 via SIRT1 inhibition and by inhibiting BMAL1 expression by interfering with PER2 nuclear translocation. All these activities of palmitate lead to attenuated and delayed circadian gene expression, reduced fatty acid oxidation, and increased fat accumulation
Discussion
In this study, we show that non-obesogenic doses of palmitate increase signaling towards fatty acid synthesis, while oleate increases signaling towards fatty acid oxidation. Our findings with non-obesogenic doses of palmitate are congruent with the reported detrimental effects of high doses and prolonged treatment of palmitate on hepatocytes, inducing insulin resistance, oxidative stress and cell death [28]. Similarly, our results with low doses of oleate are compatible with the previous reports that oleate alleviates the toxic effect of saturated free fatty acids and is protective against insulin resistance and metabolic disorders [29]. Thus, the detrimental metabolic alteration seen with high doses of palmitate manifests itself early on even with non-obesogenic levels.
These low doses of palmitate and oleate were sufficient to alter circadian rhythms. Circadian alteration is most likely achieved due to changes in the key metabolic factors AMPK, mTOR and SIRT1, that have been shown to have intricate relationships with the core clock mechanism [39] and their modulation leads to changes in the phase of gene expression. We have previously shown that activation of the AMPK–SIRT1 axis leads to phase advances [36, 43], as was seen here for oleate. So far, palmitate has been demonstrated to alter clock gene expression only in cell culture and at high doses [30–32]. However, herein we show that palmitate affects circadian rhythms even at low doses in vivo as well as in cell culture. In addition, low doses of oleate also showed altered circadian rhythms.
Supplementation of low-doses of palmitate did not lead to significant changes in Bmal1 gene and BMAL1 protein expression in vivo. These results are similar to other reports in which low-doses of palmitate were used [31, 33]. A decrease in Bmal1 oscillatory amplitude has been reported but with a sixfold higher palmitate concentration [30, 34]. Although it was previously shown that DHA altered the circadian profile of Bmal1, the disruptive effect of palmitate in the presence of DHA on Bmal1 was less pronounced, suggesting a protective role for DHA [33, 34]. Thus, although unsaturated fatty acids alter circadian rhythms, saturated fatty acids play a more disruptive role.
Although BMAL1 has been reported to regulate lipid accumulation in adipose tissue [39], its role in the liver is less clear. Bmal1 loss-of-function causes swollen mitochondria incapable of adapting to different nutrient conditions accompanied by diminished respiration and elevated oxidative stress. Consequently, liver-specific Bmal1 knockout mice accumulate oxidative damage and develop hepatic insulin resistance [44]. We found that in the absence of fatty acids, BMAL1 overexpression led to an overall reduction in lipid accumulation and increased ratio of unsaturated/saturated fatty acids by increasing the expression and activity of desaturases and elongases. In the presence of oleate, BMAL1 still had the same effect, but palmitate abrogated the reduction in fat accumulation, suggesting interference with BMAL1 ability to induce the expression of enzymes involved in fatty acid oxidation, such as ND5, NDUFA2, COX5b. Indeed, we show herein that palmitate interferes with the transcriptional activity of the CLOCK:BMAL1 heterodimer. Moreover, palmitate was reported to destabilize the interaction between CLOCK and BMAL1 in a dose and time-dependent manner [31]. In addition, we found that palmitate, but not oleate, interfered with the BMAL1 deacetylation preventing its activation. This is achieved by down-regulation of SIRT1 protein levels in mouse liver. Indeed, SIRT1 activators that induce BMAL1 deacetylation reversed the inhibitory action of palmitate on CLOCK:BMAL1 interaction [31].
In addition to the role of palmitate in interfering with BMAL1 deacetylation and transcriptional activity, we also found that, unlike oleate, palmitate reduced PER2-mediated transcriptional activation and increased REV-ERBα-mediated transcriptional inhibition of Bmal1. Indeed, PER2 has been documented to have a positive role in BMAL1 expression [42, 45, 46]. The inhibition of PER2-mediated transcriptional activation by palmitate was achieved by interfering with PER2 nuclear translocation. The role of PER2 in BMAL1 expression is further demonstrated by the finding that similarly to BMAL1, PER2 overexpression reduced fat accumulation in hepatocytes. However, this reduction was prevented by palmitate. Although in mouse liver overall daily levels did not change with non-obesogenic concentrations of the fatty acids, however, oleate was shown to have a positive acute effect on Bmal1 transcription in cell culture.
In summary, in hepatocytes, oleate activates the AMPK–SIRT1 signaling pathway leading to inhibition of fatty acid synthesis and increased fatty acid oxidation, whereas palmitate activates mTOR signaling leading to increased fatty acid synthesis. This is achieved by modulating BMAL1 at several levels abrogating its activity and expression (Fig. 7 ). The intricate relationship between core clock proteins and fatty acids warrants further study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Prof. Gad Asher, the Weizmann Institute of Science, for his kind contribution of plasmids harboring Per2, Rev-erbα and the Bmal1-luc.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 3.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 4.Froy O, Chang DC, Reppert SM. Redox potential: differential roles in dCRY and mCRY1 functions. Curr Biol. 2002;12:147–152. doi: 10.1016/S0960-9822(01)00656-X. [DOI] [PubMed] [Google Scholar]
- 5.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 6.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 7.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 8.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- 9.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 11.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 12.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, Katayama S. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb. 2005;12:169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 15.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPER2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 17.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161:1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 20.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD + salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh M, Yang S, Tseng H, Hwang L-L, Chen C, Shieh K. Abnormal expressions of circadian-clock and circadian clock-controlled genes in the livers and kidneys of long-term, high-fat-diet-treated mice. Int J Obes. 2010;34:227–239. doi: 10.1038/ijo.2009.228. [DOI] [PubMed] [Google Scholar]
- 23.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Yanagihara H, Ando H, Hayashi Y, Obi Y, Fujimura A. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int. 2006;23:905–914. doi: 10.1080/07420520600827103. [DOI] [PubMed] [Google Scholar]
- 25.Barnea M, Madar Z, Froy O. High-fat diet followed by fasting disrupts circadian expression of adiponectin signaling pathway in muscle and adipose tissue. Obesity (Silver Spring) 2010;18:230–238. doi: 10.1038/oby.2009.276. [DOI] [PubMed] [Google Scholar]
- 26.Adamovich Y, Aviram R, Asher G. The emerging roles of lipids in circadian control. BBA Mol Cell Biol Lipids. 2015;1851:1017–1025. doi: 10.1016/j.bbalip.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim SH, Akazawa Y, Cazanave SC, Bronk SF, Elmi NA, Werneburg NW, Billadeau DD, Gores GJ. Glycogen synthase kinase-3 (GSK-3) inhibition attenuates hepatocyte lipoapoptosis. J Hepatol. 2011;54:765–772. doi: 10.1016/j.jhep.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fick LJ, Fick GH, Belsham DD. Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem Biophys Res Commun. 2011;413:414–419. doi: 10.1016/j.bbrc.2011.08.103. [DOI] [PubMed] [Google Scholar]
- 31.Tong X, Zhang D, Arthurs B, Li P, Durudogan L, Gupta N, Yin L. Palmitate inhibits SIRT1-dependent BMAL1/CLOCK interaction and disrupts circadian gene oscillations in hepatocytes. PLoS ONE. 2015;10:e0130047. doi: 10.1371/journal.pone.0130047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil-Lozano M, Wu WK, Martchenko A, Brubaker PL. High-fat diet and palmitate alter the rhythmic secretion of glucagon-like peptide-1 by the rodent L-cell. Endocrinology. 2016;157:586–599. doi: 10.1210/en.2015-1732. [DOI] [PubMed] [Google Scholar]
- 33.Greco JA, Oosterman JE, Belsham DD. Differential effects of omega-3 fatty acid docosahexaenoic acid and palmitate on the circadian transcriptional profile of clock genes in immortalized hypothalamic neurons. Am J Physiol Regul Integr Comp Physiol. 2014;307:R1049–R1060. doi: 10.1152/ajpregu.00100.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SM, Neuendorff N, Chapkin RS, Earnest DJ. Role of inflammatory signaling in the differential effects of saturated and poly-unsaturated fatty acids on peripheral circadian clocks. EBioMedicine. 2016;7:100–111. doi: 10.1016/j.ebiom.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman H, Frumin I, Gutman R, Chapnik N, Lorentz A, Meylan J, le Coutre J, Froy O. Long-term restricted feeding alters circadian expression and reduces the level of inflammatory and disease markers. J Cell Mol Med. 2011;15:2745–2759. doi: 10.1111/j.1582-4934.2010.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman H, Froy O. Expression of human β-defensin 1 is regulated via c-myc and the biological clock. Mol Immunol. 2008;45:3163–3167. doi: 10.1016/j.molimm.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Aharoni-Simon M, Reifen R, Tirosh O. ROS-production-mediated activation of AP-1 but not NFκB inhibits glutamate-induced HT4 neuronal cell death. Antioxid Redox Signal. 2006;8:1339–1349. doi: 10.1089/ars.2006.8.1339. [DOI] [PubMed] [Google Scholar]
- 39.Froy O, Garaulet M. The circadian clock in white and brown adipose tissue: mechanistic, endocrine, and clinical aspects. Endocr Rev. 2018;39:261–273. doi: 10.1210/er.2017-00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 41.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnea M, Madar Z, Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology. 2009;150:161–168. doi: 10.1210/en.2008-0944. [DOI] [PubMed] [Google Scholar]
- 44.Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, Alexander RK, Unluturk U, Li X, Kong X, Hyde AL, Gangl MR, Mair WB, Lee CH. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22:709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akashi M, Okamoto A, Tsuchiya Y, Todo T, Nishida E, Node K. A positive role for PERIOD in mammalian circadian gene expression. Cell Rep. 2014;7:1056–1064. doi: 10.1016/j.celrep.2014.03.072. [DOI] [PubMed] [Google Scholar]
- 46.Wang M, Zhong Z, Zhong Y, Zhang W, Wang H. The zebrafish Period2 protein positively regulates the circadian clock through mediation of RAR-related orphan receptor alpha (Rorα) JBC. 2014;M114:605022. doi: 10.1074/jbc.M114.605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.