Abstract
Rab44 is an atypical Rab GTPase that contains some additional domains such as the EF-hand and coiled-coil domains as well as Rab-GTPase domain. Although Rab44 genes have been found in mammalian genomes, no studies concerning Rab44 have been reported yet. Here, we identified Rab44 as an upregulated protein during osteoclast differentiation. Knockdown of Rab44 by small interfering RNA promotes RANKL-induced osteoclast differentiation of the murine monocytic cell line, RAW-D or of bone marrow-derived macrophages (BMMs). In contrast, overexpression of Rab44 prevents osteoclast differentiation. Rab44 was localized in the Golgi complex and lysosomes, and Rab44 overexpression caused an enlargement of early endosomes. A series of deletion mutant studies of Rab44 showed that the coiled-coil domain and lipidation sites of Rab44 is important for regulation of osteoclast differentiation. Mechanistically, Rab44 affects nuclear factor of activated T-cells c1 (NFATc1) signaling in RANKL-stimulated macrophages. Moreover, Rab44 depletion caused an elevation in intracellular Ca2+ transients upon RANKL stimulation, and particularly regulated lysosomal Ca2+ influx. Taken together, these results suggest that Rab44 negatively regulates osteoclast differentiation by modulating intracellular Ca2+ levels followed by NFATc1 activation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-017-2607-9) contains supplementary material, which is available to authorized users.
Keywords: Rab44, Rab GTPase, Osteoclast, Intracellular Ca2+ levels, NFATc1
Introduction
The Rab GTPase family controls intracellular membrane trafficking by switching between active GTP-bound and inactive GDP-bound forms [1]. Comprehensive genomics and phylogenetic analyses have revealed that 247 eukaryotic genomes contain approximately 8000 Rabs, and that the human genome encodes 66 Rabs [2]. At present, the Rab database is publicly available as “Rabifier” at http://www.RabDB.org [3, 4]. Among the Rab GTPase family members, Rab 1–43 are the conventional small GTPase Rab proteins as with a molecular weight of about 20–30 kDa. By contrast, Rab44 and Rab45 are atypical Rab-GTPases that contain some additional domains such as the EF-hand and coiled-coil domains as well as Rab-GTPase domain [5]. Several studies have shown that Rab45 is a self-interacting protein localized in the perinuclear region of the cells [6], and that it is implicated in p38-mediated apoptosis of chronic myeloid leukemia progenitor cells [7]. Rab44 is a “large Rab GTPase” that is evolutionarily conserved from the basal metazoan to the mammalian [2]. The deduced amino acid sequences of Rab44 cDNA show that human Rab44 encodes an N-terminal EF-hand domain, a mid-region containing coiled-coil domain, and a C-terminal Rab-GTPase domain, while mouse Rab44 lacks an N-terminal EF-hand domain. Although Rab44 genes have been found in mammalian genomes, studies concerning Rab44 have not yet been reported. Here, we discovered that the expression of Rab44 is upregulated during osteoclast differentiation, as deduced by DNA microarray analysis.
Osteoclasts are bone-resorbing multinucleated cells formed by the fusion of mononuclear progenitors of the monocyte/macrophage lineage [8, 9]. Osteoclast differentiation of macrophages is mediated by mainly receptor activator of nuclear factor κ-B ligand (RANKL). RANKL stimulation activates the following six essential signaling pathways: nuclear factor of activated T cells cytoplasmic-1 (NFATc1); nuclear factor kappa B (NF-κB); phosphatidylinositol 3-kinase (PI3K)/Akt; Jun N-terminal kinase (JNK); extracellular signal-regulated kinase (ERK), and p38 mitogen-activated protein kinase (MAPK) [10–12]. Upon bone resorption, osteoclasts configure a lysosome-related organelle, termed as the ruffled border, which constructs an acidic extracellular microenvironment, and simultaneously releases many acid hydrolases, including cathepsin K (CTSK), tartrate-resistant acid phosphatase (TRAP, also known as ACP5), and matrix metalloproteinase (MMP)-9 [13, 14]. Recently, our research group has shown that Rab27A regulates transport of cell surface receptors, thereby modulating multinucleation and lysosome-related organelles in the osteoclasts [15]. Although it is speculated that osteoclasts have a specific system for the regulation of the unique organelle, their regulatory mechanisms remain poorly understood.
In this study, we have shown by knockdown using small interfering RNA (siRNA) and overexpression systems that Rab44 negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation.
Materials and methods
Antibodies and reagents
Recombinant RANKL was prepared as described previously [16]. Rat monoclonal anti-LAMP1 (Cat. no. 1921-01) was purchased from Southern Biotechnology Inc, (Birmingham, AL, USA). Rabbit polyclonal anti-c-fms (Cat. no. sc-692) and mouse monoclonal anti-NFATc1 (Cat. no. sc-7294) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit polyclonal anti-GFP (Cat. no. 598) from MBL (Nagoya, Japan). Rabbit polyclonal anti-RANK (Cat. no. 4845S), rabbit monoclonal anti-phospho-p38 (Cat. no. 4511S, Thr180/Tyr182), rabbit polyclonal anti-p38 MAPK (Cat. no. 9212S), rabbit monoclonal anti-phospho-IκBα (Cat. no. 2859S, Ser32), rabbit monoclonal anti-IκBα (Cat. No. 4812S), rabbit polyclonal anti-phospho-JNK (Cat. no. 9251S, Thr183/Tyr185), rabbit polyclonal anti-JNK (Cat. No. 9252S), rabbit monoclonal anti-phospho-Akt (protein kinase B) (Cat. no. 4060S, Ser473), rabbit monoclonal anti-Akt (Cat. no. 4691S), rabbit polyclonal anti-phospho-Erk1/2 (Cat. no. 9101S, Thr202/Tyr204), rabbit polyclonal anti-Erk1/2 (Cat. no. 9102S), rabbit monoclonal anti-GAPDH (Cat. no. 2118S), Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rat IgG were from Cell Signaling Technology (Danvers, MA, USA). Rat monoclonal anti-LAMP1 (Cat. no. 553792), mouse monoclonal anti-EEA1 (Cat. no. 610457), mouse monoclonal anti-GM130 (Cat. no. 610823) were from BD Biosciences (Franklin Lakes, NJ, USA). Mouse monoclonal anti-KDEL (Cat. no. ADI-SPA-827) was from Enzo Life Sciences (Farmingdale, NY, USA). Alexa Fluor 546 goat anti-mouse was from Molecular Probes/Invitrogen (Eugene, OR, USA). Reagents were purchased as follows: Fluo 4-AM was purchased from Dojindo (Tokyo, Japan). Fura Red AM, Vibrant Dil Cell-Labeling Solution, 4′,6-diamidino-2-phenylindole (DAPI) and LysoSensor Yellow/Blue dextran were from Thermo Fisher (Waltham, MA, USA). Ionomycin, and ML-SA1 were from WAKO (Osaka, Japan). Bafilomycin A1 was from BioViotica (Liestal, Switzerland). Other reagents, including the protease inhibitor cocktail, were from Sigma-Aldrich (Tokyo, Japan). The Osteo Assay Stripwell Plate was from Corning, Inc. (Corning, NY, USA).
Cell culture
Osteoclasts were differentiated from RAW-D cells, a subclone of the RAW264.7 murine macrophage cell line, in α-minimum essential medium (MEM) containing 10% fetal bovine serum with RANKL (100 ng/mL) at 37 °C in 5% CO2 for the indicated time periods. RAW-D cells were kindly provided by Prof. Toshio Kukita (Kyushu University, Japan). Isolation of BMMs was carried out according to a previously described method [16]. Briefly, marrow cells from the femurs and tibias of mice were cultured overnight in α-MEM containing 10% FBS in the presence of M-CSF (50 ng/mL) at 37 °C in 5% CO2. Non-adherent cells were harvested to stroma-free bone marrow cell culture system containing 50 ng/mL M-CSF. After 3 days, the adherent cells were harvested as bone marrow macrophages (BMMs). BMMs were replated and further cultured in the presence of M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 72 h.
Immunofluorescence microscopy
Cells were cultured on cover glasses and fixed with 4.0% paraformaldehyde in phosphate buffered saline (PBS) for 30 min at 25 °C. The fixed cells were then washed with PBS for 5 min twice, and permeabilized with 0.2% Triton X-100 in PBS for 15 min. The cells were blocked with 5% normal goat serum (NGS) in PBS for 1 h, and subsequently incubated overnight at 4 °C with primary antibodies. Following this, the cells were washed with PBS three times, and then stained by second antibodies such as Alexa Fluor 488 goat anti-rabbit IgG or Alexa Fluor 488 goat anti-rat IgG, or Alexa Fluor 546 goat anti-mouse IgG, or nuclear staining with DAPI was then performed. The samples were subjected to microscopy using a laser-scanning confocal imaging system (LSM800; Carl Zeiss, AG, Jena, Germany).
Quantitative real-time polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed using oligo(dT) 15 primer (Promega, Madison, WI, USA) and Revertra Ace (Toyobo, Osaka, Japan). Quantitative RT-PCR was performed using a LightCycler 480 (Roche Diagnostics, Mannheim, Germany). cDNA was amplified using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technology, La Jolla, CA, USA). The following primer sets were used:
GAPDH, forward: AAATGGTGAAGGTCGGTGTG and reverse: TGAAGGGGTCGTTGATGG;
Rab44, forward: AGAGACCACACACACTCTC and reverse: CTCCTGTAAGTCTGTTCTTG;
NFATc1, forward: TCATCCTGTCCAACACCAAA and reverse: TCACCCTGGTGTTCTTCCTC;
RANK, forward: CTTGGACACCTGGAATGAAGAAG and reverse: AGGGCCTTGCCTGCATC;
c-fms, forward: TTGGACTGGCTAGGGACATC and reverse: GGTTCAGACCAAGCGAGAAG;
DC-STAMP, forward: CTAGCTGGCTGGACTTCATCC and reverse: TCATGCTGTCTAGGAGACCTC;
OC-STAMP, forward: TGGGCCTCCATATGACCTCGAGTAG and reverse: TCAAAGGCTTGTAAATTGGAGGAGT;
CTSK, forward: CAGCTTCCCCAAGATGTGAT and reverse: AGCACCAACGAGAGGAGAAA;
Integrin β3, forward: TGTGTGCCTGGTGCTCAGA and reverse: AGCAGGTTCTCCTTCAGGTTACA;
Src, forward: AGAGTGCTGAGCGACCTGTGT and reverse: GCAGAGATGCTGCCTTGGTT;
CTR, forward: CGCATCCGCTTGAATGTG and reverse: TCTGTCTTTCCCCAGGAAATGA;
MMP9, forward: TATTTTTGTGTGGCGTCTGAGAA and reverse: GAGGTGGTTTAGCCGGTGAA;
c-Fos, forward: CCAGTCAAGAGCATCAGCAA and reverse: AAGTAGTGCAGCCCGGAGTA.
Western blot analysis
Cells were rinsed twice with ice-cold PBS, and lysed in a cell lysis buffer [50 mM Tris–HCl (pH 8.0), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM PMSF, and proteinase inhibitor cocktail]. For the phosphorylated protein, phosphatase inhibitors (20 mM sodium fluoride and 2 mM orthovanadate) were added into the cell lysis buffer. The protein concentration was measured using BCA Protein Assay Reagent (Thermo Pierce, Rockford, IL, USA). An equal amount of protein (5 μg) was applied to each lane. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were electroblotted onto a polyvinylidene difluoride membrane. The blots were blocked with 5% nonfat milk solution containing Tris-buffered saline (TBS)/0.1% Tween 20 for 1 h at 25 °C, probed with various antibodies overnight at 4 °C, washed, incubated with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA), and finally detected with ECL-Prime (GE Healthcare Life Sciences, Tokyo, Japan). The immunoreactive bands were analyzed by LAS4000-mini (Fuji Photo Film, Tokyo, Japan).
Small interfering RNA (siRNA)
siRNA experiments in RANKL-stimulated RAW-D cells or BMMs were performed as described previously [15]. The target sequence of murine Rab44 siRNA was: UCUAAGGACUGUUUGGCUGGAUCGG. RAW-D cells or BMMs plated on dishes or plates were cultured in the presence of 500 ng/mL RANKL (for RAW-D cells) or 500 ng/mL RANKL and 30 ng/mL M-CSF (for BMMs) in antibiotic-free media for 1 day. The next day, the siRNA was transfected into RAW-D cells or BMMs using Lipofectamine RNAiMAX™ transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cells were incubated with 10 pmol of siRNA for 24 h. For TRAP staining, the cells were incubated for the indicated days.
TRAP staining
Cells were fixed with 4% paraformaldehyde at 4 °C for 60 min and then treated with 0.2% Triton X-100 in PBS at room temperature for 5 min. Finally, cells were incubated with 0.01% naphthol AS-MX phosphate (Sigma-Aldrich) and 0.05% fast red violet LB salt (Sigma-Aldrich, Tokyo, Japan) in the presence of 50 mM sodium tartrate and 90 mM sodium acetate (pH 5.0) for TRAP activity. TRAP-positive red-colored cells with three or more nuclei were considered as mature osteoclasts.
Bone resorption assay
Bone resorption assay was performed by the method of as described previously [17]. Briefly, RAW-D cells were seeded onto Osteo Assay Stripwell Plates coated with thin calcium phosphate films (Corning, New York, USA), and incubated with RANKL (500 ng/mL) for 1 day. BMMs were plated on dentin slices and cultured for a day with 30 ng/mL M-CSF and 500 ng/mL RANKL. Consequently, cells were incubated with siRNA (control or Rab44-specific), and then cultured for 6 days until multinucleated osteoclasts were formed in the presence of 500 ng/mL RANKL (RAW-D cells) or 30 ng/mL M-CSF and 500 ng/mL RANKL (BMMs). After incubation, osteoclasts from RAW-D cells were dissolved in 5% sodium hypochlorite. Dentin slices were sonicated in 70% isopropanol to remove cells. Resorption pits on dentin slices were visualized using 1% toluidine blue staining. Images of the resorption pit were taken with a reverse phase microscope (Olympus, Tokyo, Japan). The ratios of the resorbed areas to the total areas were calculated using the Image J software (http://rsbweb.nih.gov/ij/) as described previously [16, 18].
Single-cell Ca2+ oscillations measurements
Ca2+ oscillations were measured by a method described previously [19, 20] with some modifications. Cells were stimulated with RANKL (100 ng/mL) for 24 h, and subsequently transfected with either control siRNA or Rab44-specific siRNA for 24 h in the presence of RANKL. For measurements, cells were washed with α-MEM twice and then loaded with 3 μM Fluo 4-AM, 3 μM Fura Red-AM for 1 h in serum-free α-MEM. Cells were washed twice with Hank’s balanced salt solution (HBSS) and subjected to confocal laser-scanning microscopy (Nikon A1R; Nikon, Japan). An excitation wavelength of 488 nm was used. The emission wavelengths of 500–530 and 600–680 nm were used for Fluo 4-AM and Fura Red AM, respectively. Fluorescence intensities were acquired simultaneously at 5 s intervals. To measure single-cell Ca2+ oscillations, the ratio of the fluorescence intensity of Fluo 4-AM to Fura Red AM was calculated. The ratio of basal level was divided by the maximum ratio acquired at adding 10 μM ionomycin, 20 μM ML-SA1, and was indicated as the percent of maximum ratio, respectively.
Retrovirus construction and expression of mouse Rab44 in RAW-D cells
The full-length cDNA of mouse Rab44 was generated by PCR using cDNA derived from bone marrow macrophages incubated with M-CSF and RANKL for 48 h. The Rab44 mutant cDNAs were produced by PCR. The primers were used for EGFP (for in-fusion) forward: AATTAGATCTCTCGAGATGGTGAGCAAGGGCGAGGAGCT and reverse: GTGAGCACTGAGGGCCTTGTACAGCTCGTCCATGCCGAG;
EGFP for Δcoiled-coil reverse: GTGTCCTCTTGTCACCTTGTACAGCTCGTCCATGCC GAG;
Rab44 WT forward: GACGAGCTGTACAAGGCCCTCAGTGCTCACATGCAGGA and reverse: ATTCGTTAACCTCGAGTCAGTGGCAGCAGCCAGCTCTCTT;
Δcoiled-coil forward: GACGAGCTGTACAAGGTGACAAGAGGACACTTGGATACT GC and reverse: ATTCGTTAACCTCGAGTCAGTGGCAGCAGCCAGCTCTCTT;
ΔRab forward: GACGAGCTGTACAAGGCCCTCAGTGCTCACATGCAGGA and reverse: ATTCGTTAACCTCGAGCAAGAA GACGACATGGTAGAGGTAGTC;
C723A forward: GACGAGCTGTACAAGGCCCTCAGTGCTCACATGCAGGA and reverse: ATTCGTTAACCTCGAGTCAGTGGCAGGCGCCAGCTCTCTT;
C723A/C724A forward: GACGAGCTGTACAAGGCCCTCAGTGCTCACATGCAGGA and reverse: ATTCGTTAACCTCGAGTCAGTGGGCGGCGCCAGCTCTCTTG. The cDNAs were amplified by PCR using PrimeSTAR GXL DNA polymerase (Takara, Tokyo) with 40 cycles of denaturation at 94 °C for 10 s, annealing at 60 °C except for EGFP and WT (62 °C) for 30 s, and extension at 72 °C for 3 min. To express EGFP-Rab44 fusion protein, the amplified fragments were fused with a linearized pMSCVpuro-EGFP, which was kindly provided by Prof Kosei Ito (Nagasaki University, Japan), using In-Fusion cloning kit (Clontech, Mountain View, CA, USA). A control vector was composed by only EGFP cDNA. All vectors placed EGFP at its N terminus. Vectors were transfected into HEK293T cells by using the Lipofectamine 2000 kit (Life Technologies, Gaithersburg, MD, USA), according to the manufacturer’s instructions. After incubation at 37 °C in 5% CO2 for 48 h, the virus-containing supernatants were collected and used to infect to RAW-D cells. Rab44 overexpressing cells were selected by puromycin (3 μg/mL) in α-MEM and every 3 days the medium was changed with new one. About 2 weeks later, several cloned cells were obtained.
Lysosomal pH measurement
The endolysosomal pH in macrophages was determined using our previous method of Yanagawa et al. [21] with some modifications. Briefly, cells were stimulated with RANKL (100 ng/mL) for 24 h, and subsequently transfected with either a control siRNA or a Rab44-specific siRNA, and concomitantly incubated with LysoSensor Yellow/Blue dextran (250 μg/mL) for 24 h in the presence of RANKL. For calibration curve, RAW-D cells were incubated for 1.5 h with bafilomycin A1 (1 μM) in MES buffer (5 mM NaCl, 115 mM KCl, 1.2 mM MgSO4, 25 mM MES) of pH 4.5, 5.0, 5.5, 6.0, and 7.0, respectively. Then cells were washed with HBSS and the fluorescence intensities were acquired by ARVO (PerkinElmer, MA, USA). The excitation wavelengths of 355 nm and the emission wavelengths of 460 or 535 nm were used. The ratio of the fluorescence of 535–460 nm and the calibration curve were used for calculating lysosomal pH.
Statistical analysis
Quantitative data are presented as mean ± standard deviation (SD). The Tukey–Kramer method was used to identify differences between concentrations when a significant difference (*P < 0.05 or **P < 0.01) was determined by analysis of variance (ANOVA).
Results
Rab44 is upregulated during osteoclast differentiation
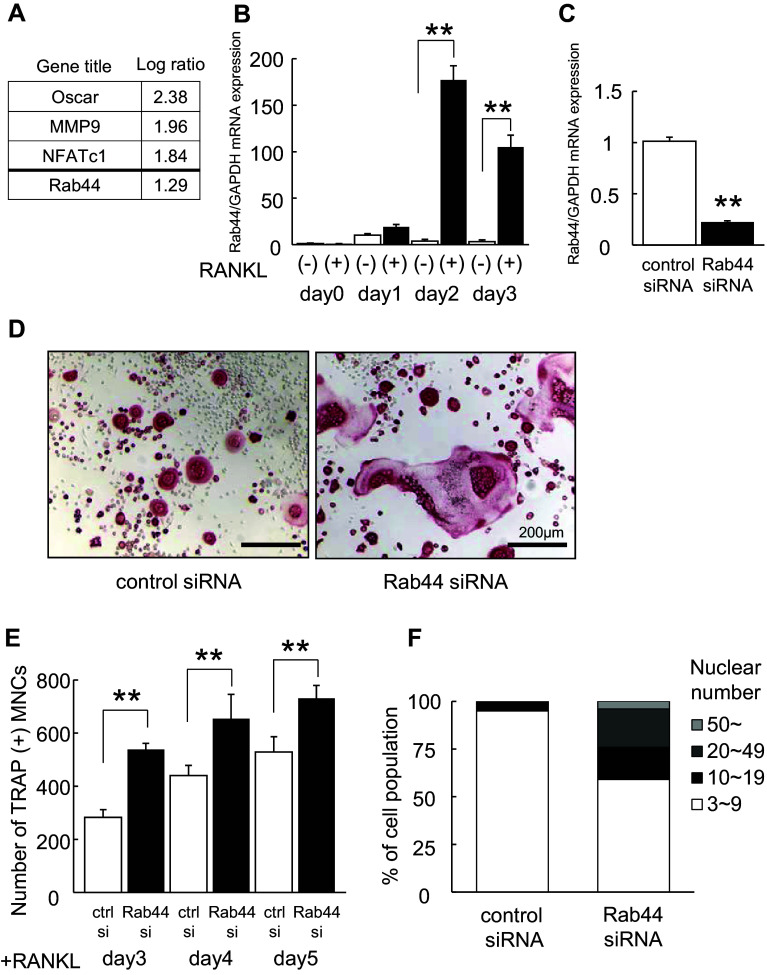
Our previous study indicated that when bone marrow-derived macrophages (BMMs) are cultured with macrophage colony-stimulating factor (M-CSF) and RANKL for 3 days on a plastic surface or a dentin slice, the macrophages cultured on the plastic surface were rapidly differentiated into osteoclasts compared to these on the dentin slice [15]. Comparing the mRNA levels of osteoclasts cultured under the two different conditions, DNA microarray analysis showed that 1363 genes were upregulated and 881 genes were downregulated among a total of 40,130 genes. Among several upregulated genes, including osteoclast marker genes osteoclast-associated receptor (Oscar), MMP9, and NFATc1, we identified Rab44, which has never been reported earlier (Fig. 1a).
Fig. 1.
Upregulation of Rab44 during osteoclastogenesis and knockdown of Rab44 in RAW-D-derived osteoclasts. a List of upregulated transcripts in osteoclasts cultured under rapid differentiation conditions compared to those under slow differentiation conditions. Bone marrow-macrophages were cultured on plastic plate (rapid differentiation conditions) and dentin (slow differentiation conditions) for 72 h in the presence of M-CSF (30 ng/mL) and RANKL (50 ng/mL). Total RNA from these cells was analyzed by Affymetrix Microarray system. b Quantitative RT-PCR analysis of Rab44mRNA expression levels in RANKL-stimulated RAW-D cells. The data are represented as mean ± SD of values from three independent experiments. **P < 0.01; compared with the control cells. c Knockdown efficacy was evaluated by measuring the Rab44 mRNA levels. After incubation with RANKL (100 ng/mL) for 24 h, cells were transfected with control or Rab44-specific siRNA (10 pmol) for an additional 24 h in the presence of RANKL. **P < 0.01; compared with the control cells. d RAW-D cells were transfected with either control or Rab44-specific siRNA. Following stimulation with RANKL (100 ng/mL) for 72 h, osteoclasts were analyzed by TRAP-staining. Bars 200 μm. e The number of TRAP-positive multinucleated osteoclasts per viewing field was counted. **P < 0.01; compared with the control cells. f Total nucleus number of TRAP positive multinucleated osteoclasts, but not TRAP-negative mononucleated cells following a 72 h culture, was counted and classified per viewing field
To confirm whether Rab44 is up-regulated during osteoclast differentiation, we measured the mRNA level of Rab44 in the murine monocytic cell line, RAW-D, by quantitative real-time polymerase chain reaction (RT-PCR) analysis. Two days after stimulation with RANKL, the Rab44 level in RANKL-stimulated RAW-D cells was 170-fold higher than that in unstimulated cells, and it remained at more than a 100-fold higher even after 3 days of stimulation (Fig. 1b). Similar results were observed in the native bone marrow-derived macrophages (BMMs). The expression levels of Rab44 gradually increased in the RANKL-stimulated BMMs (Fig S1A). These results indicate that Rab44 expression significantly increases during the differentiation of macrophages into osteoclasts.
Rab44 knockdown promotes osteoclast differentiation
To examine the biological function of Rab44 during osteoclast differentiation, we performed knockdown experiments siRNA transfection. Three days after treatment with RANKL, the knockdown efficacy in RANKL-stimulated RAW-D cells was determined (Fig. 1c). Depletion of Rab44 by siRNA reduced its expression level by approximately 80% compared with control siRNA (Fig. 1c). TRAP staining revealed that, upon stimulation with RANKL for 3 days, Rab44 knockdown enhanced multinucleated cell (MNC) formation in osteoclasts compared with the control (Fig. 1d). The number of TRAP-positive and MNCs was significantly increased in Rab44-knockdown osteoclasts compared with that in control cells (Fig. 1e). On counting the nuclear number of control and Rab44-knockdown osteoclasts, Rab44-knockdown osteoclasts containing more than ten nuclei accounted for approximately 40% of the total number, whereas control osteoclasts accounted for 3% only (Fig. 1f). Similar results were found in BMMs wherein the knockdown of Rab44 by siRNA decreased its expression by approximately 60% compared with the control siRNA (Fig. S1B). TRAP staining showed that Rab44-knockdown osteoclasts derived from BMMs were bigger in size compared to the control osteoclasts (Fig. S1C). The number of TRAP-positive multinucleated cells was higher in Rab44-depleted BMMs than that of the control cells at 3 days (Fig. S1D).
To test whether Rab44-depleted osteoclasts show enhanced maturation, we analyzed the mRNA levels of some osteoclast marker genes in control and Rab44-knockdown osteoclasts. Quantitative RT-PCR analysis of RANKL-stimulated RAW-D cells showed that mRNA levels of transmembrane 7 superfamily member 4 (DC-STAMP), osteoclast stimulatory transmembrane protein (OC-STAMP), CTSK, calcitonin receptor (CTR), and integrin β3 were significantly higher in Rab44-depleted osteoclasts than in control cells (Fig. S2A). However, the levels of Src and c-Fos were indistinguishable between the control and Rab44-knockdown cells (Fig. S2A). Similarly, in the BMM-derived osteoclasts, increased mRNA levels of DC-STAMP, OC-STAMP, CTR, CTSK, and integrin β3 were observed. However, c-Fos levels were significantly lower in the Rab44-depleted osteoclasts than in the control cells (Fig. S2B). These results indicate that Rab44-knockdown osteoclasts exhibit enhanced maturation with occasional exceptions.
We further analyzed bone resorption activity of control and Rab44-depleted osteoclasts by pit formation assay. Upon comparing the resorption pit areas, Rab44-knockdown osteoclasts displayed a marked increased resorbing activity, while the control osteoclasts showed a moderate resorbing activity (Fig. S3A). The calculated resorption area resorbed by Rab44-knockdown osteoclasts was significantly larger than that by the control osteoclasts (Fig. S3B). In the BMM-derived osteoclasts, bone resorption assay using dentin slices revealed that the Rab44-depleted osteoclasts displayed a markedly increased resorbing activity compared with the control osteoclasts (Fig. S3C). The resorption area of Rab44-knockdown osteoclasts was markedly larger than that of the control osteoclasts (Fig. S3D). These results indicate that Rab44-knockdown leads to an increased resorbing activity. Considering that Rab44 knockdown causes multinucleation and an enhanced marker gene expression, and bone-resorbing activity, we concluded that Rab44 deficiency promotes osteoclast differentiation and function.
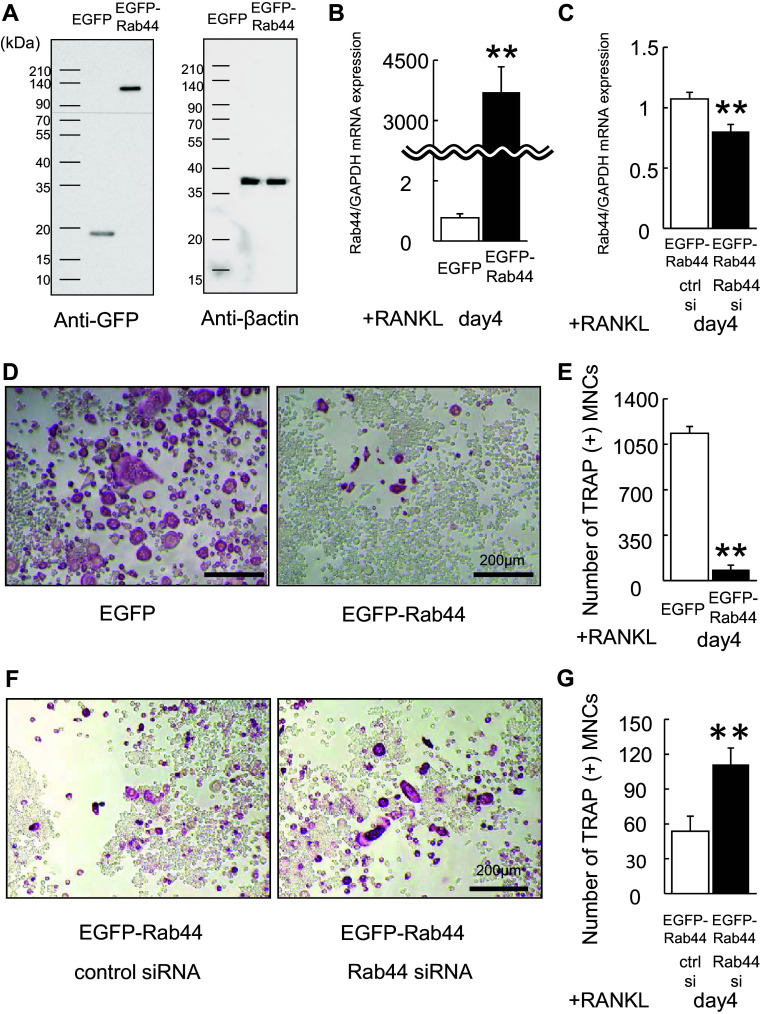
Rab44 overexpression prevents osteoclast differentiation
To assess whether Rab44 overexpression inhibits osteoclast differentiation, we transfected RAW-D cells with a vector encoding Rab44-enhanced green fluorescent protein (EGFP) or EGFP alone (control). Western blot analysis using anti-GFP antibody revealed that EGFP-Rab44 was a major band with a molecular mass of approximately 120 kDa (Fig. 2a). The mRNA expression levels of Rab44 in EGFP-Rab44-expressing RAW-D cells were markedly higher than those in control cells (Fig. 2b). Upon RANKL stimulation, TRAP staining showed that Rab44-overexpressing osteoclasts were of markedly smaller size compared to control osteoclasts (Fig. 2d). The number of TRAP-positive MNCs derived from Rab44-overexpressing macrophages was markedly lower than that in control cells (Fig. 2e). To exclude the possibility of off-target effects of siRNA, we further performed knockdown rescue experiments using Rab44-expressing RAW-D cells. Upon transfection with siRNA, the mRNA expression level of Rab44 in Rab44-siRNA and EGFP-Rab44 overexpressing cells was significantly lower than that in control siRNA-and EGFP-Rab44-overexpressing cells (Fig. 2c). Furthermore, we confirmed that Rab44 depletion partially recovered MNC formation in Rab44-overexpressing cells compared to the control, which continued to display reduced MNC formation in Rab44-expressing cells (Fig. 2f, g). Upon measuring the bone resorption activity by pit formation assay, Rab44-overexpressing osteoclasts had a markedly reduced activity, whereas the control-expressing osteoclasts exhibited an optimal activity (Fig. S2E, F).
Fig. 2.
Overexpression of Rab44 in RAW-D cells. a RAW-D cells were transduced with either retrovirus vector containing EGFP-tagged Rab44 or only EGFP (control). The cultured cells were harvested at day 3 and lysates were subjected to western blot analysis with anti-GFP antibody or anti-β actin antibody as a control. b Quantitative RT-PCR analysis of the Rab44 mRNA expression levels in RAW-D cells expressing EGFP or EGFP-Rab44 after 4 days stimulation with RANKL. The data are represented as mean ± SD of values from five independent experiments. **P < 0.01; compared with the control cells. c Knockdown efficacy was evaluated by measuring by Rab44mRNA levels in EGFP-Rab44-expressing RAW-D cells transfected together with control or Rab44 siRNA after 4 days RANKL stimulation. **P < 0.01; compared with the control cells. d The EGFP and EGFP-Rab44-overexpressing RAW-D cells were cultured with RANKL (100 ng/mL) for the 4 days. The cells were fixed and stained for TRAP. Bars 200 μm. e The number of TRAP-positive MNCs in control and Rab44-overexpressing cells was counted at indicated day. **P < 0.01; compared with the control cells. F Rab44-overexpressing RAW-D cells were transfected with control or Rab44-specific siRNA for 24 h, and stimulated with RANKL (100 ng/mL) for the 4 days. The cells were fixed and stained for TRAP. Bars 200 μm. G The number of TRAP-positive MNCs in Rab44-overexpressing cells transfected with control or Rab44 siRNA was counted at the indicated day. **P < 0.01; compared with the control cells
To examine whether Rab44-overexpressing osteoclasts exhibit impaired maturation, we examined the mRNA levels of several osteoclast marker genes in control and Rab44-overexpressing osteoclasts (Fig. S4). Quantitative RT-PCR analysis revealed that mRNA levels of all marker genes, including DC-STAMP, OC-STAMP, CTSK, MMP9, receptor activator of nuclear factor κ-B (RANK), and colony stimulating factor 1 receptor (c-fms) were significantly lower in Rab44-overexpressing osteoclasts compared to control cells (Fig. S4). These results imply that Rab44 expression induces to prevent all marker gene expression in Rab44-expressing osteoclasts compared to the control osteoclasts. Taken together, these data indicate that Rab44 overexpression inhibits osteoclast differentiation.
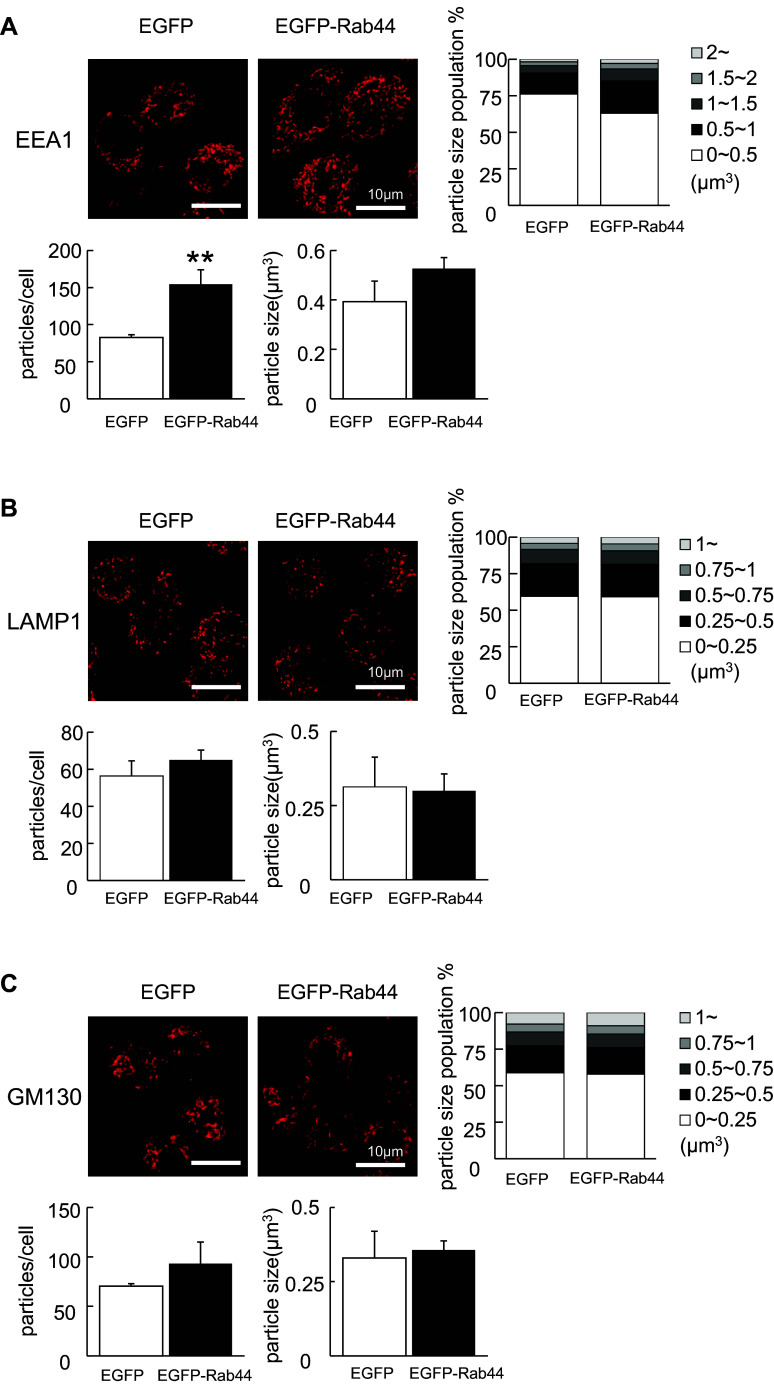
EGFP-Rab44 is localized in the Golgi complex and lysosomes, and Rab44 overexpression causes an enlargement of early endosomes
To explore the subcellular localization of Rab44 in RAW-D cells, we compared the localization of EGFP-Rab44 against several organelle marker proteins. EGFP-Rab44 was partially co-localized with GM130 (Golgi) and LAMP1 (late endosomes/lysosomes), but not with KDEL (endoplasmic reticulum; ER) and EEA1 (early endosomes) (Fig. 3). These data indicate that Rab44 is localized in the Golgi complex, late endosomes and lysosomes in RAW-D cells.
Fig. 3.
Subcellular localization of EGFP-Rab44-overexpressing RAW-D cells. The cells on cover glasses were fixed, permeabilized with 0.2% Triton X-100 in PBS, and then allowed to react with antibodies against a KDEL (marker for ER), b GM130 (marker for the Golgi complex), c LAMP1 (marker for late endosomes/lysosomes), and d EEA1 (marker for early endosomes). After washing, the samples were incubated with a fluorescence-labeled secondary antibody and then were visualized by confocal laser microscopy. Bar 10 μm
During morphological observation, we noticed that Rab44 overexpression causes enlargement of EEA1-positive early endosomes. Confocal microscopic analysis using 3D-image software revealed that the number of EEA1-positive compartments was significantly increased following overexpression of Rab44 compared with control cells (Fig. 4a). Moreover, the size of EEA1-positive early endosomes in Rab44-overexpressing RAW-D cells was slightly larger than that in control EGFP-expressing cells, although there was no statistically significant difference between them (Fig. 4a). However, such morphological alterations were not observed in LAMP-1-positive late endosomes/lysosomes or GM130-positive Golgi complex between control and Rab44-overexpressing cells (Fig. 4b, c).
Fig. 4.
Comparison of organelle size between EGFP-Rab44-overexpressing and control RAW-D cells. The cells on cover glasses were fixed, permeabilized with 0.2% Triton X-100 in PBS, and then allowed to react with antibodies against a EEA1 b LAMP1, and c GM130 on the cover glasses. After washing, the samples were incubated with a fluorescence-labeled secondary antibody and then were visualized by confocal laser microscopy. The data are representative of five independent experiments. Bar 10 μm. The number of these organelles as particles per a cell was quantified from images obtained by confocal microscopy using IMARIS 6.0 software (Bitplane, Zurich, Switzerland). Data are the means and standard deviation from three independent experiments. The particle size was measured by (μm3) using IMARIS and classified. **P < 0.01; compared with the control cells
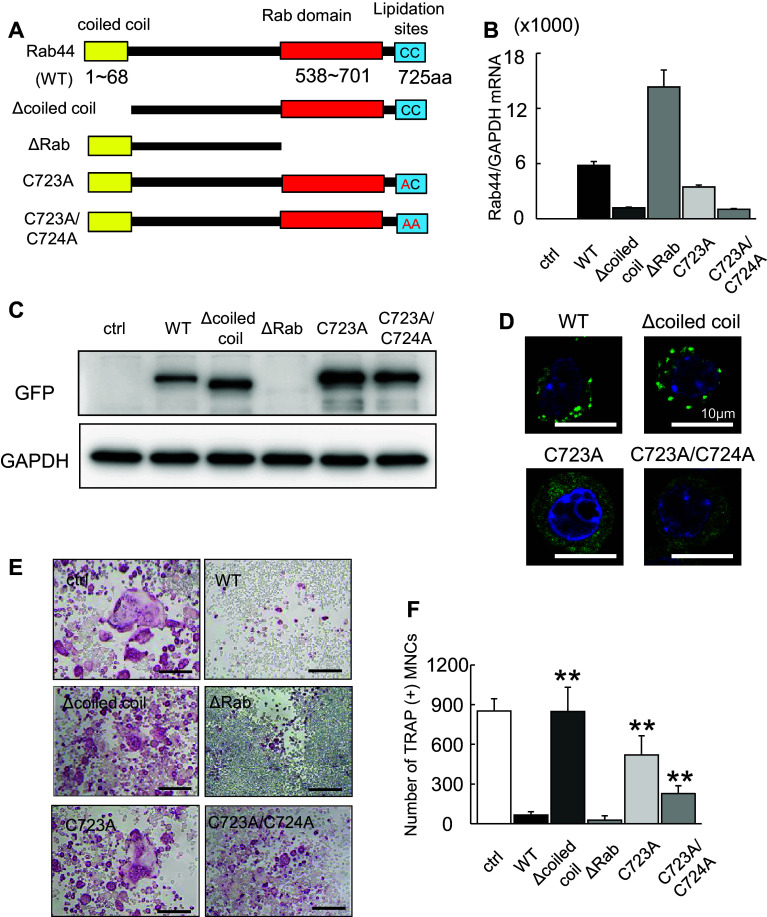
The coiled-coil domain and lipidation sites of Rab44 is important for regulation of osteoclast differentiation
The deduced amino acid sequences of Rab44 cDNA show that human Rab44 encodes an N-terminal EF-hand domain, a mid-region coiled-coil domain, and a C-terminal Rab-GTPase domain, while mouse Rab44 lacks the N-terminal EF-hand domain. At the C terminus, Rab44 contains possible lipidation sites at 723 and 724 residues. We, therefore, constructed some mutants, such as coiled-coil domain deletion (Δcoiled-coil), Rab domain deletion (ΔRab), and point mutants (C723A, C723A/C724A) as well as the wild-type (WT) (Fig. 5a). The mRNA levels of the transfected mutants in RAW-D cells were remarkably different (Fig. 5b). When we confirmed the protein levels of these mutants, we found that the ΔRab protein was undetected, but its mRNA was detectable, suggesting that this mutant was degraded as a misfolded protein (Fig. 5c). The localization of the point mutant C723A or C723A/C724A was dispersed throughout the cytoplasm, while that of WT or Δcoiled-coil cells was detected in punctate structures in the cells (Fig. 5d). Upon RANKL stimulation, TRAP staining revealed that WT-overexpressing osteoclasts were of markedly smaller size compared to control (EGFP) osteoclasts (Fig. 5e). The Δcoiled-coil mutant-overexpressing osteoclasts displayed a similar to that of control osteoclasts (Fig. 5e). The mutants of C723A, C723A/C724A expressing cells were of moderately smaller size (Fig. 5e). Indeed, the number of TRAP-positive MNCs expressing Δcoiled-coil mutant was markedly increased than that in WT-overexpressing osteoclasts (Fig. 5f). However, the ΔRab had inhibitory effects on osteoclast differentiation, although the detailed mechanisms are currently unknown (Fig. 5e). These results indicate that the lipidation sites at 723 and 724 are important, and the coiled-coil domain of Rab44 is essential for regulation of osteoclast differentiation, and that the Rab domain is required for protein expression.
Fig. 5.
Effects of expression of Rab44 mutants on osteoclast differentiation. a Schematic representation of mouse Rab44 and its mutants used in this study. Rab44 consists of a coiled-coil domain, and a Rab domain. Lipidation sites are at amino acid residues of 723 and 724. b Quantitative RT-PCR analysis of Rab44 mRNA expression levels of osteoclasts expressing EGFP-tagged Rab44 mutants. c Cell lysates (same protein amounts) were subjected to SDS–PAGE followed by western blotting with antibodies against GFP or GAPDH (control). d Typical EGFP fluorescence images of RAW-D cells expressing EGFP-Rab44 mutants and WT. The cells on cover glasses were fixed, and then allowed to DAPI staining. Bars 10 μm. e Typical TRAP-staining images of osteoclasts expressing EGFP-Rab44 mutants, WT and EGFP alone as a control. RAW-D cells expressing the control or Rab44 were cultured with RANKL (100 ng/mL) for 4 days. TRAP staining was performed. Bars 200 μm. f The number of TRAP-positive MNCs per viewing field was counted. **P < 0.01; compared with the WT cells
Rab44 modulates NFATc1 signaling in RANKL-stimulated macrophages
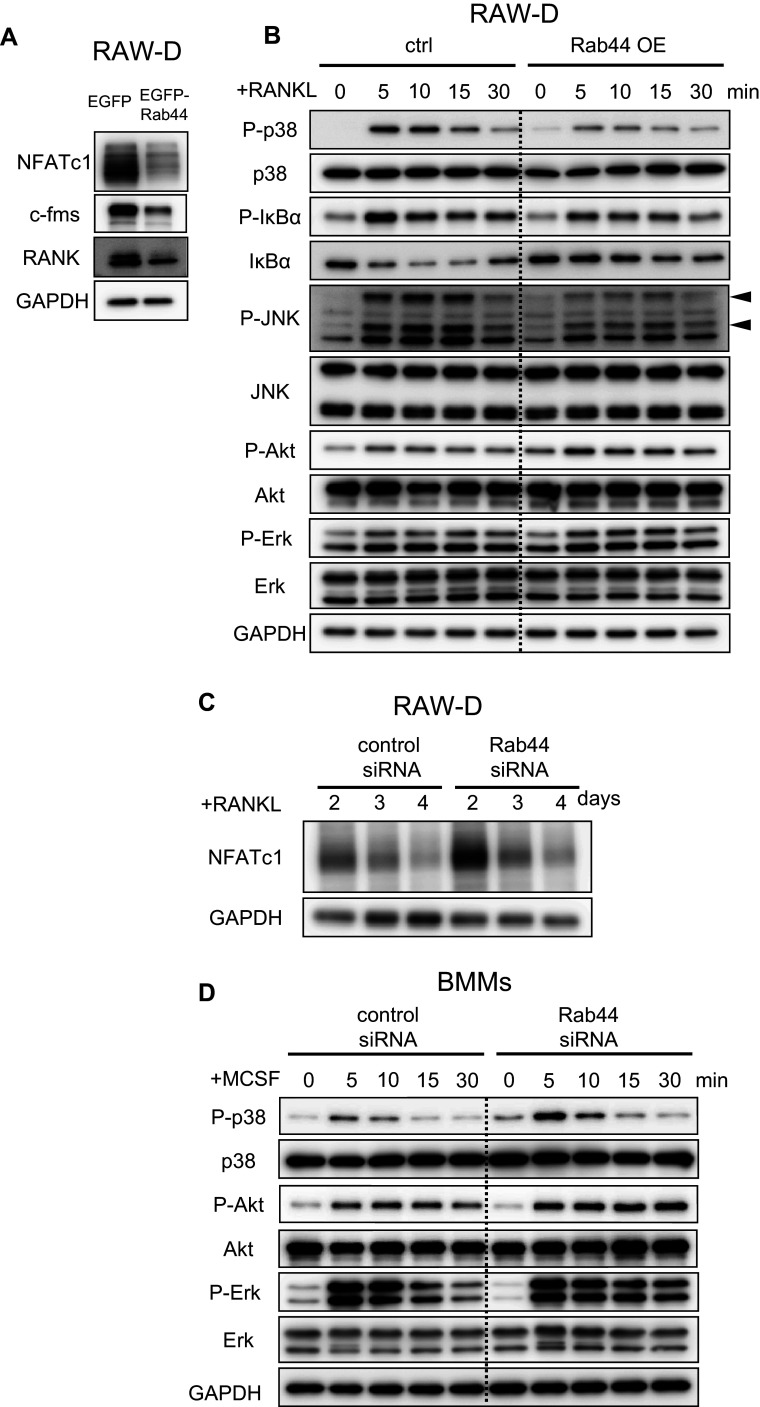
To analyze molecular mechanisms regulating activation of Rab44-overexpressing osteoclasts, we investigated signaling of control and Rab44-overexpressing RAW-D macrophages following stimulation with RANKL (Fig. 6). Importantly, western blot analysis revealed that NFATc1 expression level was markedly decreased in Rab44-overexpressing RAW-D cells compared with cells expressing the control (EGFP) (Fig. 6a). In addition, the protein levels of c-fms, M-CSF receptor, and RANK, RANKL receptor, in Rab44-expressing RAW-D cells were lower than in control cells (Fig. 6a). The phosphorylation levels of p-38, IκBα and JNK were slightly decreased in Rab44-expressing RAW-D cells compared to the control cells, although the phosphorylation levels of Akt and Erk were indistinguishable between control and Rab44-expressing RAW-D macrophages (Fig. 6b). Conversely, upon transfection with siRNA for 2 days, the NFATc1 expression level in Rab44-knockdown osteoclasts was apparently higher than that in control cells (Fig. 6c). Moreover, we examined the signaling pathways of the control and Rab44-knockdown BMMs following stimulation with M-CSF (Fig. 6d). The phosphorylation levels of p-38 were slightly increased in Rab44-depleted BMMs at 5 or 10 min compared to the control cells. Conversely, the phosphorylation levels of Akt and Erk were slightly increased in Rab44-depleted BMMs (Fig. 6d). These results indicate that overexpression of Rab44 predominantly affects the NFATc1 expression levels; however, other signaling pathways were also slightly affected.
Fig. 6.
Effects of Rab44 on NFATc1 signaling in RANKL-stimulated macrophages. a Control or Rab44-expressing RAW-D cells were cultured with RANKL (100 ng/mL) for 1 day. Cell lysates (same protein amounts) were subjected to SDS-PAGE followed by western blotting with antibodies against NFATc1, c-fms, RANK, and GAPDH (control). b Control or Rab44-expressing RAW-D cells were pre-incubated for 2 h in serum-free media in the absence of RANKL. After adding RANKL, the cells were incubated for the indicated times, consequently harvested. Cell lysates (same protein amounts) were subjected to SDS-PAGE followed by western blotting with antibodies to p-p-38, p-38, p-IκBα, IκBα, p-JNK, JNK, p-Akt, Akt, p-Erk, Erk, and GAPDH (control). c The protein levels of NFATc1 in the control and Rab44-knockdown cells after RANKL stimulation. Control or Rab44-knockdown cells were cultured with RANKL (100 ng/mL) for 2–4 days. Cell lysates (same protein amounts) were subjected to SDS-PAGE followed by western blotting with antibodies against NFATc1, and GAPDH (control). d The control or Rab44-knockdown bone marrow-macrophages (BMMs) were pre-incubated for 2 h in serum free media in the absence of M-CSF. After adding M-CSF (50 ng/mL), the cells were incubated for the indicated time periods, and subsequently harvested. Cell lysates (equivalent protein amounts) were subjected to SDS-PAGE followed by western blotting with antibodies to p-p-38, p-38, p-Akt, Akt, p-Erk, Erk, and GAPDH (control)
Rab44 regulates lysosomal Ca2+ influx and acidic pH in the endosomes/lysosomes
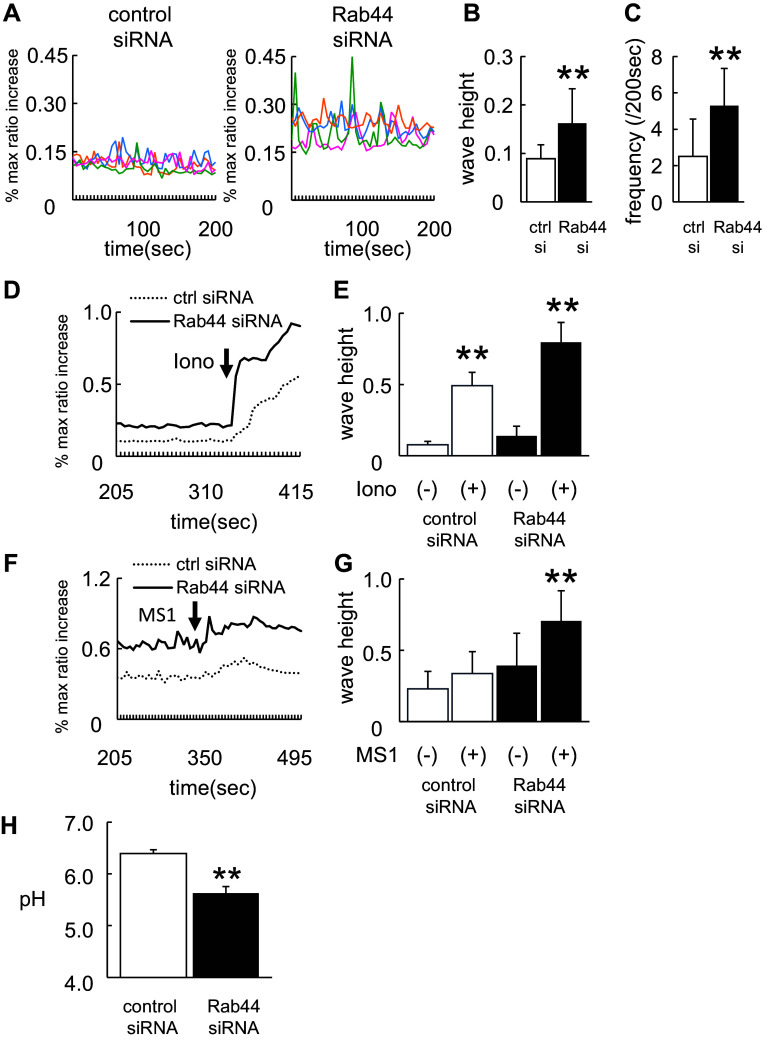
Because NFATc1 is amplified by transient intracellular Ca2+ oscillations, we assumed that Rab44-knockdown RAW-D cells exhibit elevated intracellular Ca2+ fluxes following stimulation with RANKL. When we measured the intracellular Ca2+ levels in macrophages treated with RANKL, confocal laser-scanning microscopic analysis revealed that the Ca2+ levels were significantly higher in Rab44-knockdown cells than in control cells (Fig. 7a–c). To examine the effects of compounds other than RANKL on Ca2+ fluxes, we tested ionomycin, a selective Ca2+ ionophore (Fig. 7d, e). Treatment with ionomycin enhanced the intracellular Ca2+ influx in Rab44-knockdown cells compared with control cells (Fig. 7d, e). Considering that Rab44 is localized in the Golgi complex and late endosomes/lysosomes, we evaluated calcium release from the lysosome by using mucolipin synthetic agonist 1 (ML-SA1), which is a specific agonist for the lysosomal calcium channels including transient receptor potential channel mucolipins (TRPML1-3) [22, 23]. Treatment of Rab44-depleted cells with ML-SA1 significantly increased Ca2+ fluxes compared to control group (Fig. 7f, g). Since the lysosomal Ca2+ levels are regulated by acidic pH, we measured the endosomal/lysosomal pH within the cells by incubation with LysoSensor. Rab44-knockdown cells showed a lower endosomal/lysosomal pH than control cells (Fig. 7h). These results indicate that Rab44 knockdown enhanced lower pH in the endosomes and lysosomes. Taken together, these findings indicate that Rab44 knockdown enhances the Ca2+-NFATc1 signaling pathway in osteoclasts and increases Ca2+ oscillations by mainly regulating lysosome-operated Ca2+-influx.
Fig. 7.
Effects of Rab44 on lysosomal Ca2+ influx and acidic pH in the endosomes/lysosomes. a Ca2+ oscillations in control and Rab44-knockdown RAW-D cells after RANKL treatment. Cells were stimulated with RANKL (100 ng/mL) for 24 h, and then transfected with control or Rab44 siRNA for further 24 h. The cells were washed and then loaded with 3 μM Fluo 4-AM, or 3 μM Fura Red AM for 1 h in serum-free α-MEM. The cells were then washed and analyzed by confocal laser-scanning microscopy. Data show the results obtained from four cells. b Mean wave height of Ca2+ oscillations in control and Rab44-knockdown RAW-D cells after RANKL-treatment. Each column indicates mean ± SD. Results are representative of ten cells. **P < 0.01; compared with the control cells. c Mean frequency of Ca2+ oscillations in the control and Rab44-knockdown cells after RANKL-treatment. Each wave height 0.05 or more (ratio) were counted. Each column indicates mean ± SD. Results are representative of 10 cells. **P < 0.01. d, f Effects of ionomycin or ML-SA1 on the RANKL-induced Ca2+ levels. Cells were stimulated with RANKL for 24 h, and then was transfected with control or Rab44 siRNA for 24 h. Subsequently, cells were loaded with 3 μM Fluo 4-AM, or 3 μM Fura Red AM for 1 h in serum-free α-MEM. Cells were stimulated with 10 μM ionomycin (d) or 20 μM ML-SA1 (f), and consequently analyzed by confocal laser-scanning microscopy. Results are representative of 30 cells, and are expressed as mean ± SD for five independent experiments. Arrow indicates the point of addition of 10 μM ionomycin (d) or 20 μM ML-SA1 (f). e, g Mean wave height of Ca2+ oscillations in the control and Rab44-knockdown cells after RANKL-treatment in the absence (−) or presence (+) of 10 μM ionomycin (e) or 20 μM ML-SA1 (g). Each column indicates mean ± SD. Number of cells studied was 30 cells. **P < 0.01. Results are representative of five independent experiments. h Measurement of lysosomal pH in control and Rab44-knockdown cells. Cells were stimulated with RANKL for 24 h, and then were transfected with control or Rab44 siRNA, and concomitantly incubated with LysoSensor Yellow/Blue dextran in control or Rab44-knockdown cells for 24 h. After washing with PBS, the fluorescence in each cell type was measured by the emission intensity ratio at 430 and 535 nm using an excitation at 340 nm. **P < 0.01; compared with the control cells
Discussion
In this study, we have demonstrated the increase in Rab44 expression during osteoclast differentiation. Rab44-knockdown osteoclasts showed multinucleated and giant cells, increased expression of osteoclast differentiation marker genes, and increased bone resorbing activity. Conversely, Rab44 overexpression prevented osteoclast differentiation with decreased expressions of osteoclast differentiation marker genes. The EGFP-Rab44 protein was localized in the Golgi complex and lysosomes, and Rab44 overexpression caused an enlargement of early endosomes. Among several mutants that were constructed, osteoclasts expressing Δcoiled-coil mutant displayed a phenotype similar to that of the control osteoclasts indicating that the coiled-coil domain of Rab44 is essential for regulation of osteoclast differentiation. Among the six major signaling pathways involved in the osteoclast differentiation, Rab44 mostly affected NFATc1 signaling in RANKL-stimulated macrophages. Upon treatment with RANKL, the intracellular Ca2+ levels were significantly higher in Rab44-depleted cells than those in control cells, and particularly regulated lysosomal Ca2+ influx. Thus, it is likely that Rab44 negatively regulates osteoclast differentiation and function by modulating intracellular Ca2+ influx followed by NFATc1 activation.
Although Rab44 was mainly localized in the Golgi complex and lysosomes, Rab44 overexpression caused an enlargement of early endosomes. Similar results in macrophages have been observed with Rab20, which is localized in many components of the endocytic pathway, including the ER, Golgi apparatus, and phagosomes [24]. Rab20 overexpression induces a dramatic enlargement of early and late endosomes, suggesting that this enlargement might be a result of homotypic fusion promoted by Rab20 [24]. In addition to morphological changes, Rab44 functionally affected the acidic pH in the endosomes/lysosomes. These results indicate that Rab44 probably modulates lysosomal functions. Considering that Rab44 overexpression causes an enlargement of early endosomes, it is likely that Rab44 regulates the endocytic pathways among the Golgi–endosomes–lysosomes.
Several lines of evidence indicate that lysosomal Ca2+ levels depend on lysosomal pH [25]. Regulation of lysosomal Ca2+ levels is governed by TRPMLs, which are permeable to monovalent cations and to Ca2+ among other divalent cations [26, 27]. Importantly, their current is regulated by luminal pH. Our results show that Rab44 affects lysosomal pH and Ca2+ influx. Consistent with our findings, a recent study has shown that lysosomal Ca2+ influx is essential for osteoclast differentiation [23]. Deficiency of TRPML1 in mice or pharmacological inhibition of lysosomal Ca2+ influx by ML-SA1 markedly reduced osteoclast differentiation owing to lack of RANKL-induced cytosolic Ca2+ oscillations [23]. Thus, it is possible that Rab44 directly regulates lysosomal Ca2+ influx during osteoclastogenesis.
The finding that Rab44 regulates intracellular Ca2+ levels may be explained by their genetic structure. In general, Rab44 encodes EF-hand motifs, a coiled-coil domain, and a Rab GTPase domain. Rab44 containing the EF-hand motifs are conserved in vertebrates, including human, rat, etc. However, because mouse Rab44 lacking the EF-hand motif functions in the regulation of intracellular Ca2+ levels, the other domain(s) still function in regulation of intracellular Ca2+ levels. This notion is consistent with that of the Δcoiled-coil mutant. Therefore, the observation that the Δcoiled-coil mutant lacks the inhibitory effects on osteoclast differentiation implies that coiled-coil domain may be involved in the regulation of intracellular Ca2+ levels.
So far, several large Rab-GTPases have been identified, including Rab 44, Rab45 and CRACR2A (Ca2+ release-activated Ca2+ channel regulator 2A) [5]. Among these, the gene structure and function of Rab44 are reminiscent of those of CRACR2A. CRACR2A is a lymphocyte-specific large Rab GTPase that contains multiple functional domains, including EF-hand motifs, a coiled-coil domain, and a Rab-GTPase domain [28]. Moreover, studies using gene silencing in cells and knockout mice indicate that CRACR2A deficiency impairs the Ca2+-NFAT signaling pathway in mouse T cells [28, 29]. CRACR2A has been shown to be directly associated with Orai1 channels and stromal interaction molecule 1 (STIM1) [30, 31]. Several lines of evidence indicate that when the Ca2+ sensor STIM1 recognizes depletion of ER-stored Ca2+ levels, it translocates to the ER–plasma membrane junctions, inducing plasma membrane Ca2+ entry through the Orai1 channels [32–35]. Therefore, Rab44 may interact with a Ca2+-regulating protein(s). Given that Rab44 and its binding proteins regulates lysosomal Ca2+ entry, it is of importance to determine the similarities or differences between Rab44 and CRACR2A, and to further investigate the Rab44 binding protein(s) like Orai1 and STIM1.
In conclusion, this study shows that Rab44 negatively regulates osteoclast differentiation by modulating intracellular Ca2+ levels followed by NFATc1 activation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1- Effects of Rab44 on osteoclastogenesis of bone marrow-derived macrophages (BMMs). A Quantitative RT-PCR analysis for Rab44 mRNA expression levels in the bone marrow-macrophages (BMMs) stimulated with M-CSF (30 ng/mL) and RANKL (100 ng/mL) at 0–3 days. The data are represented as mean ± SD of values from three independent experiments. **P < 0.01; compared with the control cells. B Knockdown efficacy of BMM-derived osteoclasts was evaluated by measuring the Rab44 mRNA levels. After incubation with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 24 h, the cells were transfected with control or Rab44-specific siRNA (10 pmol) for an additional 24 h in the presence of M-CSF and RANKL. **P < 0.01; compared with the control cells. C BMMs were transfected with either control or Rab44-specific siRNA. Following stimulation with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 72 h, osteoclasts were analyzed by TRAP-staining. Bars, 200 μm. D The number of TRAP-positive multinucleated osteoclasts per viewing field was counted. **P < 0.01; compared with the control cells. Figure S2- Comparison of mRNA levels of various osteoclast marker proteins in Rab44-knockdown and control cells. A, RAW-D cells were cultured with RANKL (100 ng/mL) for 2 days. After mRNA isolation from these cells, RT-PCR was performed. **P < 0.01, compared with the control cells. B, Bone marrow-derived macrophages (BMMs) were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 4 days. RT-PCR was performed on the mRNA isolated from these cells. **P < 0.01; compared with the control cells. Figure S3- Effects of Rab44 on the bone resorbing activity of osteoclasts. A, E RAW-D cells were seeded onto Osteo Assay Stripwell Plates with RANKL (500 ng/mL) for 7 days. Photographs of the bone resorbing activity of each osteoclast. Bars, 400 μm. B, F The resorption area was determined using Image J software. (significance compared with control or transfected cells. **P < 0.01). C Bone marrow-derived macrophages (BMMs) were seeded onto dentin slices with M-CSF (30 ng/mL) and RANKL (500 ng/mL) for 7 days. Images of the bone resorbing activity of each osteoclast. Bars, 100 μm. D The resorption area was determined using Image J software. (significance compared with the control transfected cells, **P < 0.01). Figure S4- Comparison of mRNA levels of various osteoclast marker proteins in Rab44-overexpressing and control cells. RAW-D cells were cultured with RANKL (100 ng/ml) for 3 days. After isolation of mRNA in these cells, RT-PCR was performed. **P < 0.01, compared with the control cells. (PDF 260 kb)
Acknowledgements
We thank Dr, Kazuhisa Nishishita for providing recombinant RANKL. This work was supported by JSPS KAKENHI Grant numbers 15H05298, 16K15790, and for Research Fellow of Japan Society for the Promotion of Science Grant number to 16J03008.
Abbreviations
- NFATc1
Nuclear factor of activated T-cells c1
- RANKL
Nuclear factor κ-B ligand
- NF-κB
Nuclear factor kappa B
- PI3K/Akt
Phosphatidylinositol 3-kinase
- JNK
Jun N-terminal kinase
- ERK
Extracellular signal-regulated kinase
- MAPK
Mitogen-activated protein kinase
- CTSK
Cathepsin K
- TRAP
Tartrate-resistant acid phosphatase
- MMP
Matrix metalloproteinase
- siRNA
Small interfering RNA
- LAMP1
Lysosomal associated membrane protein 1
- NGS
Normal goat serum
- MEM
Minimum essential medium
- DAPI
4′,6-Diamidino-2-phenylindole
- PBS
Phosphate buffered saline
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- M-CSF
Macrophage colony-stimulating factor
- Oscar
Osteoclast-associated receptor
- RT-PCR
Real-time polymerase chain reaction
- MNC
Multinucleated cell
- DC-STAMP
Transmembrane 7 superfamily member 4
- OC-STAMP
Osteoclast stimulatory transmembrane protein
- CTR
Calcitonin receptor
- EGFP
Enhanced green fluorescent protein
- TRPML
Transient receptor potential channel mucolipin
- CRACR2A
Ca2+ release-activated Ca2+ channel regulator 2A
- STIM1
Stromal interaction molecule 1
- BMMs
Bone marrow-derived macrophages
References
- 1.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 2.Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10:71. doi: 10.1186/1741-7007-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, Pereira-Leal JB. Thousands of rab GTPases for the cell biologist. PLoS Comput Biol. 2011;7(10):e1002217. doi: 10.1371/journal.pcbi.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surkont J, Diekmann Y, Pereira-Leal JB. Rabifier2: an improved bioinformatic classifier of Rab GTPases. Bioinformatics (Oxford, England) 2016 doi: 10.1093/bioinformatics/btw654. [DOI] [PubMed] [Google Scholar]
- 5.Srikanth S, Woo JS, Gwack Y. A large Rab GTPase family in a small GTPase world. Small GTPases. 2016 doi: 10.1080/21541248.2016.1192921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shintani M, Tada M, Kobayashi T, Kajiho H, Kontani K, Katada T. Characterization of Rab45/RASEF containing EF-hand domain and a coiled-coil motif as a self-associating GTPase. Biochem Biophys Res Commun. 2007;357(3):661–667. doi: 10.1016/j.bbrc.2007.03.206. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura S, Takemura T, Tan L, Nagata Y, Yokota D, Hirano I, Shigeno K, Shibata K, Fujie M, Fujisawa S, Ohnishi K. Small GTPase RAB45-mediated p38 activation in apoptosis of chronic myeloid leukemia progenitor cells. Carcinogenesis. 2011;32(12):1758–1772. doi: 10.1093/carcin/bgr205. [DOI] [PubMed] [Google Scholar]
- 8.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 9.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 10.Darnay BG, Ni J, Moore PA, Aggarwal BB. Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif. J Biol Chem. 1999;274(12):7724–7731. doi: 10.1074/jbc.274.12.7724. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto M, Sudo T, Saito T, Osada H, Tsujimoto M. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL) J Biol Chem. 2000;275(40):31155–31161. doi: 10.1074/jbc.M001229200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, Abu-Amer Y. Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. J Biol Chem. 2001;276(1):563–568. doi: 10.1074/jbc.M008198200. [DOI] [PubMed] [Google Scholar]
- 13.Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genom Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacombe J, Karsenty G, Ferron M. Regulation of lysosome biogenesis and functions in osteoclasts. Cell Cycle. 2013;12:2744–2752. doi: 10.4161/cc.25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada-Sugawara M, Sakai E, Okamoto K, Fukuda M, Izumi T, Yoshida N, Tsukuba T. Rab27A regulates transport of cell surface receptors modulating multinucleation and lysosome-related organelles in osteoclasts. Sci Rep. 2015;5:9620. doi: 10.1038/srep09620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai E, Shimada-Sugawara M, Nishishita K, Fukuma Y, Naito M, Okamoto K, Nakayama K, Tsukuba T. Suppression of RANKL-dependent heme oxygenase-1 is required for high mobility group box 1 release and osteoclastogenesis. J Cell Biochem. 2012;113(2):486–498. doi: 10.1002/jcb.23372. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi Y, Sakai E, Sakamoto H, Fumimoto R, Fukuma Y, Nishishita K, Okamoto K, Tsukuba T. Inhibitory effects of tert-butylhydroquinone on osteoclast differentiation via up-regulation of heme oxygenase-1 and down-regulation of HMGB1 release and NFATc1 expression. J Appl Toxicol. 2014;34(1):49–56. doi: 10.1002/jat.2827. [DOI] [PubMed] [Google Scholar]
- 18.Sakai E, Shimada-Sugawara M, Yamaguchi Y, Sakamoto H, Fumimoto R, Fukuma Y, Nishishita K, Okamoto K, Tsukuba T. Fisetin inhibits osteoclastogenesis through prevention of RANKL-induced ROS production by Nrf2-mediated up-regulation of phase II antioxidant enzymes. J Pharmacol Sci. 2013;121(4):288–298. doi: 10.1254/jphs.12243FP. [DOI] [PubMed] [Google Scholar]
- 19.Kajiya H, Okamoto F, Nemoto T, Kimachi K, Toh-Goto K, Nakayana S, Okabe K. RANKL-induced TRPV2 expression regulates osteoclastogenesis via calcium oscillations. Cell Calcium. 2010;48(5):260–269. doi: 10.1016/j.ceca.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 21.Yanagawa M, Tsukuba T, Nishioku T, Okamoto Y, Okamoto K, Takii R, Terada Y, Nakayama KI, Kadowaki T, Yamamoto K. Cathepsin E deficiency induces a novel form of lysosomal storage disorder showing the accumulation of lysosomal membrane sialoglycoproteins and the elevation of lysosomal pH in macrophages. J Biol Chem. 2007;282(3):1851–1862. doi: 10.1074/jbc.M604143200. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Xiong J, Lu Y, Xia X, Zhu MX. Differential mechanisms of action of the mucolipin synthetic agonist, ML-SA1, on insect TRPML and mammalian TRPML1. Cell Calcium. 2014;56(6):446–456. doi: 10.1016/j.ceca.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkhembaatar M, Gu DR, Lee SH, Yang YM, Park S, Muallem S, Shin DM, Kim MS. Lysosomal Ca2+ signaling is essential for osteoclastogenesis and bone remodeling. J Bone Miner Res. 2016 doi: 10.1002/jbmr.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei G, Schnettger L, Bronietzki M, Repnik U, Griffiths G, Gutierrez MG. Interferon-gamma-inducible Rab20 regulates endosomal morphology and EGFR degradation in macrophages. Mol Biol Cell. 2015;26(17):3061–3070. doi: 10.1091/mbc.E14-11-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen KA, Myers JT, Swanson JA. pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci. 2002;115(Pt 3):599–607. doi: 10.1242/jcs.115.3.599. [DOI] [PubMed] [Google Scholar]
- 26.Kiselyov K, Colletti GA, Terwilliger A, Ketchum K, Lyons CW, Quinn J, Muallem S. TRPML: transporters of metals in lysosomes essential for cell survival? Cell Calcium. 2011;50(3):288–294. doi: 10.1016/j.ceca.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatachalam K, Wong CO, Zhu MX. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 2015;58(1):48–56. doi: 10.1016/j.ceca.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, Gwack Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2 + sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12(5):436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srikanth S, Kim KD, Gao Y, Woo JS, Ghosh S, Calmettes G, Paz A, Abramson J, Jiang M, Gwack Y. A large Rab GTPase encoded by CRACR2A is a component of subsynaptic vesicles that transmit T cell activation signals. Sci Signal. 2016;9(420):ra31. doi: 10.1126/scisignal.aac9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frischauf I, Fahrner M, Jardin I, Romanin C. The STIM1: orai interaction. Adv Exp Med Biol. 2016;898:25–46. doi: 10.1007/978-3-319-26974-0_2. [DOI] [PubMed] [Google Scholar]
- 31.Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4:2963. doi: 10.1038/ncomms3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15(13):1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169(3):435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437(7060):902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454(7203):538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1- Effects of Rab44 on osteoclastogenesis of bone marrow-derived macrophages (BMMs). A Quantitative RT-PCR analysis for Rab44 mRNA expression levels in the bone marrow-macrophages (BMMs) stimulated with M-CSF (30 ng/mL) and RANKL (100 ng/mL) at 0–3 days. The data are represented as mean ± SD of values from three independent experiments. **P < 0.01; compared with the control cells. B Knockdown efficacy of BMM-derived osteoclasts was evaluated by measuring the Rab44 mRNA levels. After incubation with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 24 h, the cells were transfected with control or Rab44-specific siRNA (10 pmol) for an additional 24 h in the presence of M-CSF and RANKL. **P < 0.01; compared with the control cells. C BMMs were transfected with either control or Rab44-specific siRNA. Following stimulation with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 72 h, osteoclasts were analyzed by TRAP-staining. Bars, 200 μm. D The number of TRAP-positive multinucleated osteoclasts per viewing field was counted. **P < 0.01; compared with the control cells. Figure S2- Comparison of mRNA levels of various osteoclast marker proteins in Rab44-knockdown and control cells. A, RAW-D cells were cultured with RANKL (100 ng/mL) for 2 days. After mRNA isolation from these cells, RT-PCR was performed. **P < 0.01, compared with the control cells. B, Bone marrow-derived macrophages (BMMs) were cultured with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 4 days. RT-PCR was performed on the mRNA isolated from these cells. **P < 0.01; compared with the control cells. Figure S3- Effects of Rab44 on the bone resorbing activity of osteoclasts. A, E RAW-D cells were seeded onto Osteo Assay Stripwell Plates with RANKL (500 ng/mL) for 7 days. Photographs of the bone resorbing activity of each osteoclast. Bars, 400 μm. B, F The resorption area was determined using Image J software. (significance compared with control or transfected cells. **P < 0.01). C Bone marrow-derived macrophages (BMMs) were seeded onto dentin slices with M-CSF (30 ng/mL) and RANKL (500 ng/mL) for 7 days. Images of the bone resorbing activity of each osteoclast. Bars, 100 μm. D The resorption area was determined using Image J software. (significance compared with the control transfected cells, **P < 0.01). Figure S4- Comparison of mRNA levels of various osteoclast marker proteins in Rab44-overexpressing and control cells. RAW-D cells were cultured with RANKL (100 ng/ml) for 3 days. After isolation of mRNA in these cells, RT-PCR was performed. **P < 0.01, compared with the control cells. (PDF 260 kb)