Abstract
Type 2 diabetes is a complex metabolic disorder characterized by insulin resistance and pancreatic β-cell dysfunction. Deregulated glucose and lipid metabolism are the primary underlying manifestations associated with this disease and its complications. Long non-coding RNAs (lncRNAs) are a novel class of functional RNAs that regulate a variety of biological processes by a diverse interplay of mechanisms including recruitment of epigenetic modifiers, transcriptional and post-transcriptional regulation, control of mRNA decay, and sequestration of transcription factors. Although the underlying causes that define the diabetic phenotype are extremely intricate, most of the studies in the last decades were mostly centered on protein-coding genes. However, current opinion in the recent past has authenticated the contributions of diverse lncRNAs as critical regulatory players during the manifestation of diabetes. The current review will highlight the importance of lncRNAs in regulating cellular processes that govern metabolic homeostasis in key metabolic tissues. A more in-depth understanding of lncRNAs may enable their exploitation as biomarkers or for therapeutic applications during diabetes and its associated complications.
Keywords: Liver, Muscle, Gene regulation, Diabetic complications, Biomarkers, Metabolic diseases
Introduction
The central dogma of molecular biology was until the last few years, thought to revolve around protein-coding genes via flow of information from genes (carriers of genetic information) to RNA which acts as a mediator and then finally to proteins that play important functional roles, essential for all aspects of life. Technical advances in the human genome and evidence from various high-throughput genomic platforms have revealed that the human genome is more complex with only < 2% of the genome coding for protein-coding genes and the major fraction being assigned to non-coding RNAs (ncRNAs) [1]. The increasing complexity of the mammalian genome is mainly due to the expanding world of these ncRNAs [2]. High-throughput studies have highlighted tens of thousands of transcripts with meagre protein-coding potential; most of them being ascribed to miRNAs which are a small subclass of ncRNAs involved in transcriptional and post-transcriptional gene silencing. Gradually, emerging evidences unearthed another prevalent and functionally diverse class of ncRNAs known as long non-coding RNAs (lncRNAs) that are defined as transcripts greater than 200 nucleotides in length, mostly transcribed by RNA polymerase II (RNA pol II) and primarily thought to be without any coding potential. However, a few reports show that lncRNAs can translate into short peptides, although this event seems to be rare in occurrence [3, 4]. These peptides are believed to play roles in the regulation and function of other proteins within the cell [5, 6]. Similar to mRNAs, lncRNAs are 5′ capped, 3′ polyadenylated, frequently spliced, and multi-exonic [7, 8]. Although lncRNAs are studied in diverse species, including plants [9], yeast [10], viruses [11], prokaryotes [12], and eukaryotes [13], they are poorly conserved among different species as compared to mRNAs [14]. They are generally more cell type-specific and are usually less abundantly expressed [8, 14] and, therefore, were earlier considered as transcriptional noise and non-functional [15]. LncRNAs have been observed in the nucleus, cytosol, or both [16], consistent with their important roles in many biological processes including transcriptional and post-transcriptional gene regulation, genome packing, cellular structure integrity, chromatin organization, genomic imprinting, protein localization, cell cycle, and apoptosis [8, 17, 18]. The recent transcriptome annotation of the human genome [14] and that of other vertebrate model organisms including mouse [8], rat [19], and zebrafish [20] have uncovered a comprehensive list of such lncRNAs.
Classification
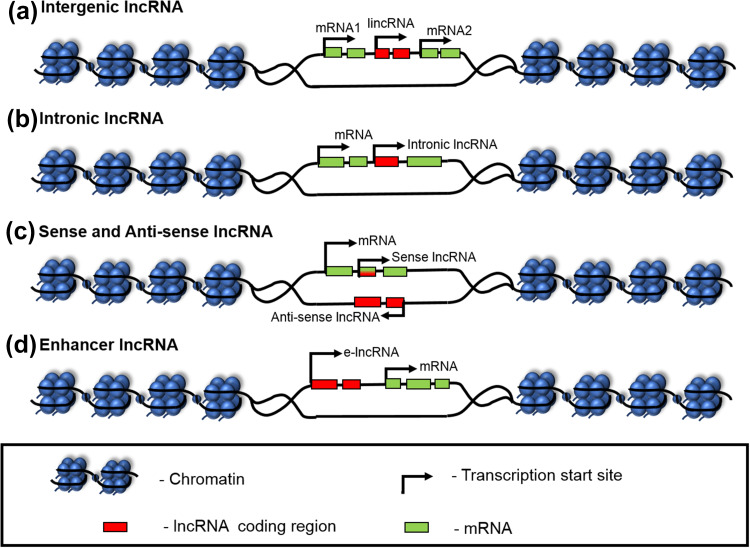
LncRNAs are classified on the basis of their function and genomic location relative to protein-coding genes. These include (1) Intergenic lncRNAs (lincRNAs)—also known as long intergenic non-coding RNAs (lincRNAs)—are transcribed from genomic loci between the protein-coding genes or intergenic regions (Fig. 1a). They exhibit many features similar to protein-coding genes like transcriptional activation and methylation marks [21] and are implicated in diverse cellular processes, embryonic stem cell pluripotency, and cell proliferation [22, 23]. (2) Intronic lncRNAs—are transcribed from the introns of the protein-coding genes in either the sense or antisense direction (Fig. 1b). These are less stable and poorly conserved than intergenic lncRNAs [21]. (3) Sense lncRNAs—are transcribed from the exonic regions of the sense strand of protein-coding genes. As shown in Fig. 1c, they can cover the entire protein-coding gene or may overlap with a part of it. This is a much less explored class as compared to the linc and antisense RNAs, although a few studies suggest both RNA and protein-coding functions of sense lncRNAs [24]. (4) Antisense lncRNAs—are transcribed from the antisense strand of a protein-coding gene [14] and they tend to (a) overlap an exon of a sense gene or (b) occur as transcripts from the intronic region of a sense gene that does not have any exon–exon overlap or (c) cover the entire sequence of a sense gene through its intron (Fig. 1c). Around 32% lncRNAs are antisense to coding transcripts in the human genome [14] and approximately 87% coding transcripts have been shown to have antisense partners in the mouse genome [25]. (5) Enhancer lncRNAs (e-lncRNAs)—are transcribed either mono or bidirectionally from the enhancer regions of protein-coding genes and have H3K4me1 marks on their promoter regions (Fig. 1d) [26, 27]. These are usually < 2 kb in length with sparse proof of being spliced or polyadenylated. They function to regulate mRNA transcription and contribute to enhancer-mediated transcriptional activation of several coding genes.
Fig. 1.
Location of lncRNAs with respect to protein-coding genes. a Intergenic lncRNAs: transcribed from genomic regions between two protein-coding genes, b intronic lncRNAs: transcribed from intronic regions within protein-coding genes, c sense and antisense lncRNAs: sense lncRNAs transcribed from exonic regions of sense strands of protein-coding genes, while antisense lncRNAs transcribed from antisense strands of protein-coding genes, and d enhancer lncRNAs: transcribed from enhancer regions of protein-coding genes
Mechanism of action
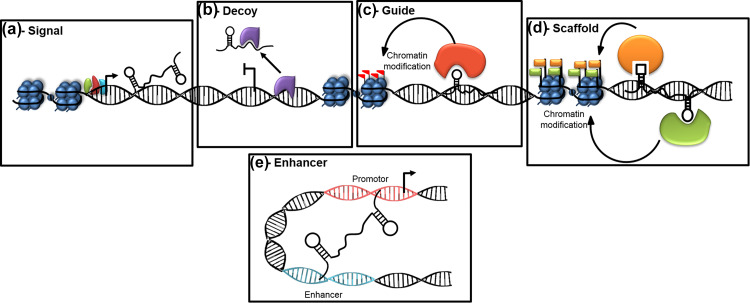
LncRNAs are involved in a wide variety of cellular and molecular functions. Based on their targeting mechanisms (Fig. 2), lncRNAs can be classified as: (1) signals where they show cell type-specific expression to integrate developmental cues, interpret cellular context, and respond to diverse stimuli [28], (2) decoys where they bind and titrate away protein targets [28], (3) guides where they bind and direct localization of ribonucleoprotein complexes (transcriptional and epigenetic regulatory factors) to specific targets [28], (4) scaffolds where they act as platforms to bind to multiple proteins to form a ribonucleoprotein complex and by doing so, bring the effectors together in both, time and space [28] and (5) enhancers where they increase the association between promoters and enhancer regions by chromosomal looping [29]. Mechanistically, lncRNAs participate in epigenetic reprogramming, in transcriptional or post-transcriptional regulation (Fig. 3). LncRNAs have been shown to recruit chromatin-modifying complexes to specific genomic loci and thereby regulate cell cycle, apoptosis, differentiation, cell adhesion, and DNA repair (Fig. 3). For example, the lncRNA, HOTAIR that is over expressed in many cancers like colorectal cancer, breast cancer, and laryngeal squamous cell carcinoma [30, 31] or Xist that recruits the chromatin-modifying complex, PRC2 to the target site, modify the epigenetic program of the cell. Tsix is an overlapping antisense lncRNA to Xist, which negatively regulates Xist expression to inactivate one X chromosome [32]. In addition, lncRNAs may interact with different proteins, RNA and DNA to regulate gene expression at different levels (Fig. 3). A large number of lncRNAs are known to regulate gene expression through transcriptional interference. For example, the, lncRNA HSR1 (heat shock RNA-1) binds with the trimeric HSF1 (heat shock factor 1) and induces transcription of genes (heat shock induced genes) by binding to their promoters, thereby acting as a thermo-sensing device as in some bacteria [33]. LncRNAs also regulate subcellular localization or cellular trafficking of transcription factors. For example, the lncRNA, NRON (non-coding repressor of NFAT) controls the localization of NFAT (nuclear factor of activated T cells) and consequently regulates transcription. LncRNAs are also believed to regulate splicing and post-transcriptional events. For example, the lncRNA Zeb2NAT (Zeb2 natural antisense transcript) overlaps with the 5′ splice site in the intron of the Zeb2 gene and inhibits splicing of the intron to allow translation of the Zeb2 mRNA [34] which consequently downregulates E-cadherin mRNA and protein levels during epithelial–mesenchymal transition. Some lncRNAs also act as precursors for siRNAs or interact directly or indirectly with miRNAs that in turn regulates gene expression. Thus, lncRNAs are now identified to interact with DNA, proteins, mRNAs, miRNAs, siRNAs, and other antisense RNAs, and consequently influence their function and status.
Fig. 2.
Molecular interactions during lncRNA action. a As a signal: lncRNA action reflects the combinatorial actions of transcription factors (shown in green, red, and blue) or signaling pathways which dictate gene regulation in space and time. b As a decoy: lncRNAs titrate away the transcription factors or other proteins (shown in purple) from the genomic locus. c As a guide: lncRNAs can recruit chromatin-modifying complexes (shown in red) to target genes. d As a scaffold: lncRNAs can bring together multiple proteins (shown in green and orange) to form a ribonucleoprotein complex which then acts on the chromatin. e As an enhancer: lncRNAs can cause chromosomal looping
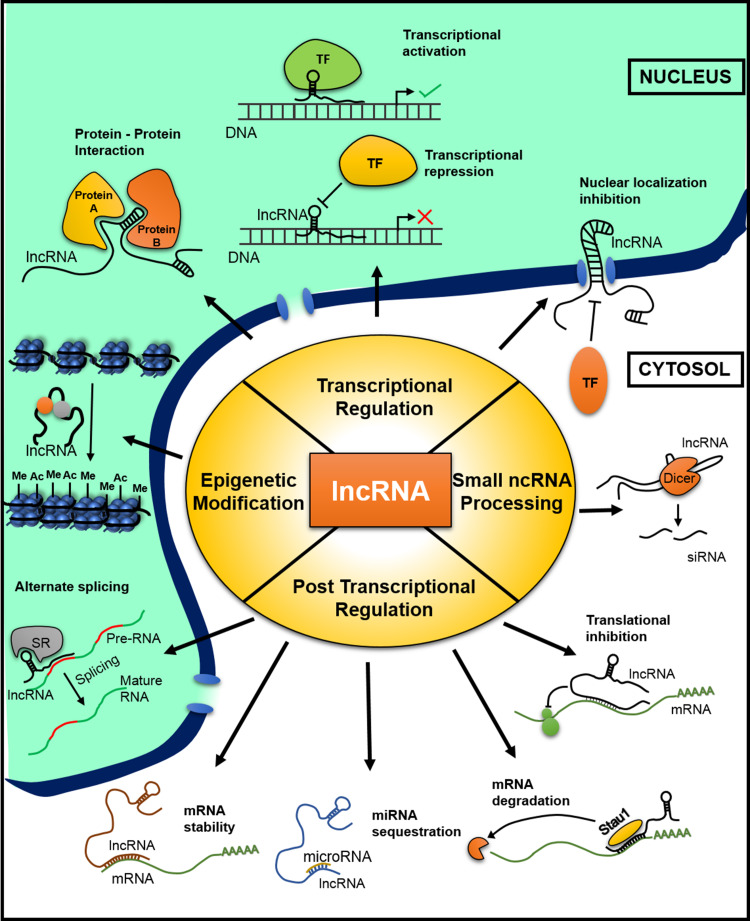
Fig. 3.
Cellular events that are modulated by lncRNAs. LncRNAs regulate cellular mechanisms by epigenetic modification, transcription regulation, small ncRNA processing, and post-transcriptional regulation. Epigenetic modifications include DNA methylation and histone modifications. Transcriptional regulation includes protein–protein interactions that might affect transcription, transcriptional activation/inhibition, and nuclear localization of transcription factors. Small ncRNA processing, additionally, also involves splicing of lncRNA into siRNA by DICER. Post-transcriptional regulation includes alternate splicing, mRNA stability, miRNA sequestration, mRNA degradation, and translational inhibition
Role of lncRNAs in complex diseases
Deregulation of lncRNAs expression has been observed in several diseases [35–37]. Specifically, altered levels of the lncRNA, H19 have been strongly demonstrated in diverse types of cancer [38–40]. In gastric cancer, H19 expression is unregulated as compared to normal and this leads to increased cell proliferation, whereas treatment with H19 siRNA promotes cell apoptosis [41]. H19 interacts with ISM1 and induces metastasis in this cancer [42]. Various studies indicate that growth arrest-specific transcript 5 (GAS5) is a tumor suppressor lncRNA and is downregulated in human HCC, and this correlation associates with advanced tumor growth [43]. The lncRNA, maternally expressed gene 3 (MEG3), is downregulated in human colorectal cancer (CRC), and its overexpression in cancer tissues suppresses cell proliferation and promotes cell cycle arrest and apoptosis [44]. The lncRNA, DANCR (antidifferentiation non-coding RNA), is elevated in colorectal cancer, and this is related to shorter survival rates of patients [45]. From genome-wide analysis of circulatory lncRNAs, the mitochondrial lncRNA, LIPCAR, was identified as a novel biomarker for cardiac remodeling and its higher expression correlates with mortality in heart failure patients [46]. LncRNAs like NORN, AK087060, FRM4, UBE3A-AS, and IPW are also implicated in several neuronal disorders, with altered expression in Down syndrome, Rett syndrome, fragile X syndrome, Angelman syndrome, and Prader–Willi syndrome, respectively [35]. These suggest that lncRNAs are significant in various biological processes and affect diverse cellular events that subsequently manifest as complex disease phenotypes. The following section will discuss the roles of lncRNAs in metabolic tissues and deregulation of which are implicated in varied metabolic phenotypes associated with diabetes.
Long non-coding RNAs in metabolism and metabolic diseases
LncRNAs are emerging as important regulatory elements that control metabolic tissues’ development and function [47]. Regulation of metabolism and glucose homeostasis are fundamental biochemical processes, orchestrated, and fine-tuned by reciprocal signaling between different tissues/organs that serve regulatory functions, including absorption of sugar from the intestine, secretion of insulin from the pancreas, glucose production in the liver, and uptake of glucose by fat, muscle, and other tissues. Pancreatic islets have a critical role in regulating systemic glucose metabolism through the secretion of key endocrine hormones, insulin and glucagon, where insulin is the anabolic master regulator which controls peripheral as well as central nervous system-related aspects of metabolism. Resistance towards insulin action is a key step in the development of metabolic disorders. Dysregulation of any of these processes underlies the pathogenesis of major metabolic disorders including obesity, Type 2 Diabetes (T2D), dyslipidemia, and non-alcoholic fatty liver disease (NAFLD). The discovery of lncRNAs has added a new layer of complexity to the regulatory network that impinges on metabolic homeostasis and disease [48]. Interestingly, several GWAS studies have reported lncRNAs to be mapped to diabetic susceptible loci [49–52], all suggesting towards critical roles of lncRNAs in insulin resistance, diabetes, and its associated complications.
LncRNAs as regulators of islet function
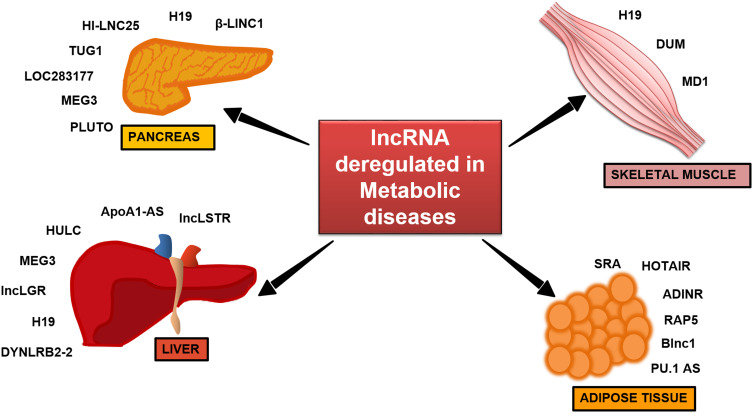
The pancreatic islet is an important central node to researchers to understand the pathophysiology of diabetes [53]. The possible regulation of islet development and function by lncRNAs was first demonstrated by Ding et al., where the lncRNA, H19 (Fig. 4), was shown to be involved in transgenerational transmission of gestational diabetes mellitus which leads to impaired islet structure and function [54]. To understand the roles of lncRNAs in regulating pancreatic function, several research groups have profiled lncRNA expression in mouse and human pancreatic islets [55, 56]. Transcriptome analysis in pancreatic β-cells of type 2 diabetes patients identified tissue-specific and dynamically regulated abnormally expressed lncRNAs. These lncRNAs are often located near islet-specific chromatin domains containing islet-specific coding genes or mapped to diabetes susceptible genetic loci. Knockdown of HI-LNC25, a β cell-specific lncRNA conserved between mouse and human resulted in decreased GLIS3, an important islet transcription factor, thereby suggesting its functional importance in pancreatic β cells [56] (Fig. 4). A coexpression analysis has identified that the lncRNA, LOC283177, correlates with the expression of insulin synthesis and secretion [51] (Fig. 4). Yin et al. demonstrated that silencing of the lncRNA, TUG1 in vivo increased apoptosis in pancreatic β cells and decreased insulin secretion leading to elevated fasting glucose levels (Fig. 4). Expression of TUG1 is decreased in a non-obese diabetic (NOD) mouse and is suppressed by glucose treatment in pancreatic Nit-1 cells, indicating its association with diabetes [57]. Another lncRNA, MEG3 was reported to be downregulated in the pancreatic tissue of Type 1 Diabetic (T1D) and T2D mice models and its expression was dynamically modulated by glucose in Min6 and primary mouse islet cells (Fig. 4). In vivo silencing of MEG3 led to impaired glucose tolerance and decreased insulin secretion, as also evident by the reduced insulin-positive cells. There was a significant decrease in the Pdx-1 and MafA levels indicating MEG3 as a novel β-cell regulator [58]. Deletion of a conserved lncRNA, βlinc1 (β-cell long intergenic non-coding RNA 1) in adult mice results in defective islet development and disruption of glucose homeostasis [59] (Fig. 4). Decreased levels of the lncRNA, PLUTO (Fig. 4) in islets of T2D or impaired glucose tolerant subjects affect the 3D chromatin structure and transcription of Pdx-1, a key β cell transcription factor implicating its role in insulin synthesis and β cell-specific regulatory network [60]. In spite of these reports, the elucidation of lncRNA-mediated molecular mechanisms in pancreatic biology still awaits further detailed investigations.
Fig. 4.
LncRNAs in metabolism and different metabolic diseases. Deregulation of different lncRNAs as reported in different metabolic tissues, like liver, skeletal muscle, pancreas, and adipose tissue, leads to various metabolic diseases and complications therein
Role of lncRNAs in hepatic metabolism
Liver is a unique organ which plays a central role in maintaining whole body glucose and lipid homeostasis. It is capable of both, utilizing and producing glucose. So far, very few studies have demonstrated the importance of lncRNAs in regulating hepatic metabolism. A study by Li et al. in 2015 identified a liver enriched lncRNA, lncLSTR (Fig. 4), decreased levels of which are associated with enhanced triglycerides (TG) clearance in mice. It regulates systemic lipid homeostasis by regulating the TDP-43/FXR/apoC2-pathway where lncLSTR forms a complex with TDP-43 to regulate Cyp8b1 expression. This is a key enzyme in the bile acid synthesis pathway, which through FXR induces apoC2 expression and, hence, reduces plasma TG levels. Interestingly, depletion of lncLSTR in mice results in decreased blood glucose levels [61]. An antisense lncRNA transcript of ApoA1 (ApoA1-AS) (Fig. 4) negatively regulates ApoA1 expression in the liver by epigenetically regulating the APO gene cluster [62]. Certain lncRNAs induced by hormones and lipoproteins are believed to regulate lipid metabolism. For example, oxidized LDL (Ox-LDL) induces the expression of the lncRNA, lincRNA-DYNLRB2-2 (Fig. 4), which results in upregulation of GPR119 and ABCA1 levels and increases APOA1-mediated cholesterol efflux from the liver [63]. An abnormally overexpressed lncRNA HULC (Fig. 4) in hepatocellular carcinoma (HCC) activates PPARα and ACSL1 in hepatoma cells, and increases triglyceride and cholesterol levels in these cells. Furthermore, increased cholesterol through a positive feedback loop upregulates HULC expression through the retinoid receptor, RXRA [64]. Increased hepatic lncRNA MEG3 levels in high-fat diet (HFD) mice, ob/ob mice, and on treatment of palmitate, oleate, or linoleate in primary hepatocytes are associated with increased G6pc, Pepck, and FoxO1 levels (Fig. 4). This leads to increased hepatic gluconeogenesis and impaired insulin-stimulated glycogen accumulation in hepatocytes which manifests as hepatic insulin resistance. In addition, MEG3 interference in high-fat diet and ob/ob mice significantly attenuates impaired glucose tolerance [65]. A fasting induced lncRNA, liver glucokinase repressor (lncLGR) (Fig. 4), suppresses glucokinase activity by interacting with hnRNPL and represses glycogen storage in the mouse liver [66]. A novel human-specific lncRNA, lncHR1 (Fig. 4), was recently identified as a negative regulator of SREBP-1c expression and to regulate lipid metabolism in both cultured hepatoma cells and in transgenic mice [67]. A recent study from our laboratory has identified decreased H19 (Fig. 4) levels in the db/db mice liver which regulates hepatic gluconeogenesis. Silencing H19 levels in human hepatoma and primary hepatocytes increased the levels of gluconeogenic genes and hepatic glucose output [68]. In addition, H19 is also implicated in liver development and in progression of other liver diseases like steatosis, fibrosis, cirrhosis, and hepatocellular carcinoma [69].
Roles of lncRNAs in the skeletal muscle tissue
Gao et al. in 2014, have shown decreased H19 lncRNA (Fig. 4) levels in the skeletal muscle of diabetic human subjects and mice and its role in impairing insulin signaling and decreasing glucose uptake. H19 directly binds to the let-7 miRNA, which results in decreased bioavailability of let-7 and consequently affects the expression of let-7 target genes, namely the insulin receptor (Insr) and lipoprotein lipase (lpl), thereby impairing insulin signaling. Insulin treatment in vivo and in vitro directly suppresses H19 expression via PI3K/AKT-dependent phosphorylation of KSRP (miRNA processing factor), which promotes biogenesis of let-7 and H19 destabilization. Thus, the H19/let-7 double-negative feedback loop participates in impaired glucose metabolism in muscle cells [70]. In addition, H19 depletion in mouse myogenic C2C12 cell line is known to accelerate muscle differentiation [71]. Interestingly, H19 also generates the miRNA, miR-675 which regulates Smad expression during myogenesis [72]. Several studies have also reported lncRNAs as regulators of muscle differentiation and regeneration [73]. LncRNA Dum (Fig. 4) (Dppa2 Upstream Binding Muscle) silences its neighboring gene, Dppa2 gene by recruiting DNMTs, and this regulates myogenesis [74]. Lnc-MD1 (Fig. 4) is a muscle differentiation specific lncRNA and acts as a microRNA sponge for miR-133 and miR-135 that are critical for myogenesis and muscle differentiation [75]. Since muscle acquires its metabolic and regulatory function during differentiation, deregulation of this event may lead to metabolic abnormalities.
LncRNAs in regulating glucose metabolism and differentiation in the adipose tissue
A few reports have demonstrated the role of lncRNAs in regulating glucose metabolism in the adipose tissue with some lncRNAs shown to regulate adipocyte differentiation [73, 76–78] (Fig. 4). A steroid receptor RNA activator (SRA) lncRNA (Fig. 4), highly expressed in the adipose tissue, was earlier known to coactivate steroid receptor-dependent reporter gene expression and promote adipogenesis [79]. Mouse knockout models of the Sra1 gene (SRA−/−) display improved insulin sensitivity and are resistant to diet induced obesity with improved glucose tolerance. Knockdown of another lncRNA, ADINR (Fig. 4), results in reduced expression of key adipogenic regulators, namely C/EBPα, PPARγ, lipoprotein lipase, and fatty acid-binding protein 4. ADINR binds to PA1 and recruits the histone methyl transferase complex at the C/EBPα locus to transcriptionally regulate C/EBPα levels and, hence, adipogenesis [80]. An antisense lncRNA, PU.1 AS transcript of PU.1 mRNA, promotes adipogenesis by preventing the translation of PU.1 mRNA by forming an mRNA-AS lncRNA duplex where PU.1 protein reduces PPARγ levels [81] (Fig. 4). As compared to abdominal adipocytes, the lncRNA, HOTAIR, is differentially expressed in gluteal adipocytes and plays a role in adipocyte differentiation [76].
Taken together, these studies highlight the importance of lncRNAs in the development and function of metabolic tissues, and therefore, their altered levels are closely associated with the onset and progression of insulin resistance and diabetes.
Roles of lncRNAs in diabetic complications
Apart from being involved in major metabolic tissues during diabetes as discussed above, lncRNAs are implicated in complications associated with diabetes. Diabetic retinopathy is one of the common complications in diabetic patients, which leads to impaired or loss of vision. Altered expression of lncRNAs, namely MALAT1 [82, 83] and MEG3 [84], are reported to be associated with diabetic retinopathy. In STZ-induced diabetic rats, the expression of MALAT1 is elevated in the endothelial cells of the retina and knockdown of MALAT1 ameliorates retinopathy in STZ-induced rats [82]. The lncRNA, MEG3, was also found to be downregulated in the retina of STZ-induced diabetic mice and its in vitro knockdown in retinal endothelial cells was found to regulate cell proliferation, viability, and migration [84]. Hyperglycemia as in diabetes causes upregulation of ANRIL levels in endothelial cells [85, 86], and this elevates the levels of the PRC2 subunit, EZH2 that consequently promotes the expression of VEGF, a key promoter of angiogenesis [85]. Another major complication associated with diabetes is diabetic nephropathy, and this is considered a major cause of end-stage renal disease and disability in diabetic patients [87]. Recent studies show that lncRNAs play important roles in the development of diabetic nephropathy and accumulation of extracellular matrix (ECM) proteins. There is higher expression of the lncRNA, PVT1, during diabetic nephropathy, and this increase leads to increased fibrosis due to accumulation of ECM proteins in renal cells [88]; downregulation of PVT1 reduces ECM accumulation [88]. LncRNA PVT1 is also a host to miR-1207-5p and this miRNA is shown to regulate the expression of fibronectin1 (FN1), plasminogen activator inhibitor-1 (PAI1), and transforming growth factor beta 1 (TGFβ1) [89]. In renal tube injury during diabetes, the lncRNA, MIAT, is under-expressed, and this negatively correlates with creatinine and BUN levels in the serum of these subjects. It has been shown to regulate cell viability of proximal convoluted renal tubules [90]. In diabetic nephropathic mice, the lncRNA, MGC, is increased in renal mesangial cells. Interestingly, this lncRNA harbours a cluster of approximately 40 miRNAs, and is regulated by the ER stress marker C/EBP homologous protein (CHOP) [91]. In CHOP-deficient mice, there is decreased expression of the lncRNA, MGC, and the clustered miRNAs, and these mice have shown an improvement in diabetic nephropathy [91]. Diabetic nephropathy is also associated with increased levels of lincRNA, Gm4419, and this exerts its action by interacting with NF-κβ. Knockdown of this lincRNA in renal mesangial cells lowers cellular proliferation and inhibits expression of NF-κβ in hyperglycemic states [92]. The lncRNA, TUG1, that is upregulated in diabetic nephropathy acts as sponge for miR-377 and regulates PPAR-γ expression which further modulates the expression of FN1, collagen type IV alpha 1 chain (COL4A1), PAI1, and TGFβ1 in renal mesangial cells [93]. Diabetic cardiomyopathy is a critical end-stage complication associated with diabetes. Several such cardiovascular complications and myocardial dysfunction in diabetic patients lead to heart failure [94]. Differential expression analysis in cardiac tissue from normal and diabetic rats shows that the lncRNA, MALAT1, is upregulated during cardiomyopathy and knockdown of this lncRNA improves left ventricular systolic function by reducing myocardial inflammation in diabetic rats [95, 96]. Decreased expression of the lncRNA, H19, is also reported during diabetes [68, 70], and this often results in decreased expression of the exonic miRNA, miR-675 [97, 98]. mir-675 directly targets the voltage-dependent anion channel 1 (VDAC1) which is involved in mitochondria-mediated apoptosis in the cardiac tissue during diabetes. H19 overexpression in diabetic rats reduces oxidative stress, apoptosis, and inflammation, and improves ventricle function [98]. LncRNAs NONRATT021972 and uc.48+ are reported to be associated with diabetic neuropathic pain [99, 100], and inhibition of both have been shown to alleviate such neuropathic pain by activating the P2X3 receptor. Impaired wound closure is a notable complication associated with diabetes and a recent report shows decreased levels of the lncRNA, Lethe in such impaired dorsal wounds of diabetic mice. This was demonstrated to be associated with increased ROS production, possibly through regulation of NOX2 expression [101].
All these suggest towards important roles of various lncRNAs in complications associated with diabetes and, therefore, assume importance to be studied in detail.
Future perspectives
Metabolic abnormalities associated with diabetes are diversely complex, and these are determined by intricate networks and cross-talks between several cellular entities. LncRNAs play major regulatory roles in cellular metabolism and their discovery has opened up new domains in the understanding of the mechanisms of deregulated cellular physiology during diabetes. LncRNAs are now increasingly being identified in many body fluids like whole blood, plasma, serum, urine, saliva, gastric juice, etc., and this projects them to be exploited as potential biomarkers in several diseases. Their high stability in body fluids, especially when included in exosomes or apoptotic bodies, and their patterns of expression that mimics the progress of a disease, make them ideal biomarker candidates [102]. Such lncRNAs are capable of resisting the action of abundant quantities of ribonucleases in different body fluids [103] and, therefore, offers promise to be used as an effective and convenient biomarker. In fact, a few lncRNAs have been considered as biomarkers for human cancers. For example, the lncRNA, PCA3 for prostate cancer [104], H19 for gastric cancer [105], HULC in hepatocellular carcinoma [106], UCA1 in bladder carcinoma [107], etc., are being increasingly considered. These, together with the fact that several lncRNAs are increasingly being implicated in diverse aspects of cell metabolism, indicate the immense possibility of lncRNAs being exploited as possible biomarkers to follow the onset and progression of diabetes in future. In addition, as compared to protein-coding genes, lncRNAs are better considered as ideal candidates for therapeutic intervention. Studies show that normalizing lncRNA levels are beneficial in several diseased states. Inhibition of LncRNA Z38 suppresses cell proliferation and tumorigenesis [108], and that of the lncRNA, linc00974 in hepatocellular carcinoma promotes apoptosis and cell cycle arrest [109]. Therapeutically regulating H19 levels has shown promise in pancreatic and bladder cancer [110, 111]. Several lncRNA-based therapeutic approaches for cancer are in different stages of clinical trials [112], suggesting that lncRNAs can be exploited for therapeutic application in other diseases, as well. These approaches primarily silence lncRNAs using antisense oligonucleotides (ASOs), locked nucleic acids, siRNAs or use ribozymes, aptamers, small molecule inhibitors, or synthetic lncRNA mimics. H19 overexpression using specific constructs has also been shown to reduce the size of tumors in human trials of several carcinomas [112, 113]. Use of nanoparticles or extracellular vehicles for targeted delivery of such moieties to ensure specificity is also being investigated. With the steady progressing pace of research on lncRNAs, it appears that soon they will be considered as major contributors of diseases and emerge as better and effective cellular entities for therapeutic interventions. Since lncRNAs can interact with DNA, mRNA, other ncRNAs, and proteins, they seem to be more potent and viable in terms of applicability and usefulness.
Conclusions
To conclude, it would be apt to state that lncRNAs are widely implicated in diverse domains of cell metabolism and their altered expression is associated with diabetes and its complications. Although originally thought to be non-functional, lncRNA genes transcribe into lncRNAs that exert important and specific functions in regulating cellular pathways. Due to this specificity, lncRNAs are considered better therapeutic targets. In addition, their expression patterns in tissues quite follow the progress of diseases and this get reflected in diverse body fluids which, therefore, make them suitable to be exploited as better biomarkers. Although still in the nascent stage, these properties of lncRNAs are beginning to be exploited and there are a few instances of them being used as biomarkers in diverse forms of cancer. Therefore, the use of lncRNAs both as biomarkers and therapeutic targets for diabetes and its associated complications is highly envisaged in future. However, this would require further investigations, both in vivo and in vitro and critical networking among researchers, clinicians, and patients. Nevertheless, the implications of lncRNAs in diverse facets of insulin resistance and diabetes are indicative of their roles in the diagnosis, prognosis, and therapy of this disease in future.
Acknowledgements
This work was supported by funding from the Council of Scientific and Industrial Research (CSIR), New Delhi, India (BSC0123). NG and DK acknowledge CSIR, New Delhi, India for their fellowship.
Compliance with ethical standards
Conflict of interest
None to declare.
References
- 1.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 3.Bánfai B, Jia H, Khatun J, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tupy JL, Bailey AM, Dailey G, et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo T, Plaza S, Zanet J, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 7.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 9.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 10.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein HD, Zopf D, Freymann DM, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci USA. 1993;90:5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brosius J. Waste not, want not-transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 18.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y, Fuscoe JC, Zhao C, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaushik K, Leonard VE, Kv S, et al. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One. 2013;8:e83616. doi: 10.1371/journal.pone.0083616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:924–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 26.Duvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mousavi K, Zare H, Dell’orso S, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Feng J, Wu T, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 32.Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 33.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 34.Beltran M, Puig I, Pena C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Vondervoort II, Gordebeke PM, Khoshab N, et al. Long non-coding RNAs in neurodevelopmental disorders. Front Mol Neurosci. 2013;6:53. doi: 10.3389/fnmol.2013.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233–1239. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013;14:18790–18808. doi: 10.3390/ijms140918790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matouk IJ, DeGroot N, Mezan S, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 40.Barsyte-Lovejoy D, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 41.Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 44.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851–4859. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 46.Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 47.Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanson RL, Craig DW, Millis MP, et al. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes. 2007;56:975–983. doi: 10.2337/db06-1072. [DOI] [PubMed] [Google Scholar]
- 50.Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fadista J, Vikman P, Laakso EO, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motterle A, Sanchez-Parra C, Regazzi R. Role of long non-coding RNAs in the determination of beta-cell identity. Diabetes Obes Metab. 2016;18(Suppl 1):41–50. doi: 10.1111/dom.12714. [DOI] [PubMed] [Google Scholar]
- 53.Eliasson L, Esguerra JL. Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol (Oxf) 2014;211:273–284. doi: 10.1111/apha.12285. [DOI] [PubMed] [Google Scholar]
- 54.Ding GL, Wang FF, Shu J, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku GM, Kim H, Vaughn IW, et al. Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol Endocrinol. 2012;26:1783–1792. doi: 10.1210/me.2012-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moran I, Akerman I, van de Bunt M, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin DD, Zhang EB, You LH, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol Biochem. 2015;35:1892–1904. doi: 10.1159/000373999. [DOI] [PubMed] [Google Scholar]
- 58.You L, Wang N, Yin D, et al. Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J Cell Physiol. 2016;231:852–862. doi: 10.1002/jcp.25175. [DOI] [PubMed] [Google Scholar]
- 59.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes Dev. 2016;30:502–507. doi: 10.1101/gad.273821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akerman I, Tu Z, Beucher A, et al. Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab. 2017;25:400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li P, Ruan X, Yang L, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu YW, Yang JY, Ma X, et al. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J Lipid Res. 2014;55:681–697. doi: 10.1194/jlr.M044669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui M, Xiao Z, Wang Y, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 65.Zhu X, Wu YB, Zhou J, Kang DM. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun. 2016;469:319–325. doi: 10.1016/j.bbrc.2015.11.048. [DOI] [PubMed] [Google Scholar]
- 66.Ruan X, Li P, Cangelosi A, Yang L, Cao H. A long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep. 2016;14:1867–1875. doi: 10.1016/j.celrep.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Cheng M, Niu Y, et al. Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int J Biol Sci. 2017;13:349–357. doi: 10.7150/ijbs.16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goyal N, Sivadas A, Shamsudheen K, et al. RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci Rep. 2017;7:8312. doi: 10.1038/s41598-017-08281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pope C, Mishra S, Russell J, Zhou Q, Zhong X-B. Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases. 2017;5:11. doi: 10.3390/diseases5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao Y, Wu F, Zhou J, et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Zhao Y, Bao X, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Divoux A, Karastergiou K, Xie H, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring) 2014;22:1781–1785. doi: 10.1002/oby.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei S, Du M, Jiang Z, Hausman GJ, Zhang L, Dodson MV. Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell Mol Life Sci. 2016;73:2079–2087. doi: 10.1007/s00018-016-2169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu B, Gerin I, Miao H, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao T, Liu L, Li H, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Reports. 2015;5:856–865. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang F, Tong Q. Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol. 2008;295:C213–C220. doi: 10.1152/ajpcell.00422.2007. [DOI] [PubMed] [Google Scholar]
- 82.Liu J, Yao J, Li X, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan B, Tao Z-F, Li X-M, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy aberrant expression of lncRNAs in early DR. Investig Ophthalmol Vis Sci. 2014;55:941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 84.Qiu G-Z, Tian W, Fu H-T, Li C-P, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun. 2016;471:135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 85.Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy role of ANRIL in diabetic retinopathy. Investig Ophthalmol Vis Sci. 2017;58:470–480. doi: 10.1167/iovs.16-20569. [DOI] [PubMed] [Google Scholar]
- 86.Siddiqui A, Hussain S, Azam A, et al. ANRIL polymorphism rs1333049, a novel genetic predictor for diabetic retinopathy complication. Meta Gene. 2017;14:33–37. doi: 10.1016/j.mgene.2017.07.011. [DOI] [Google Scholar]
- 87.Fox CS, Larson MG, Leip EP, Meigs JB, Wilson PW, Levy D. Glycemic status and development of kidney disease. Diabetes Care. 2005;28:2436–2440. doi: 10.2337/diacare.28.10.2436. [DOI] [PubMed] [Google Scholar]
- 88.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One. 2013;8:e77468. doi: 10.1371/journal.pone.0077468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun. 2015;468:726–732. doi: 10.1016/j.bbrc.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 91.Kato M, Wang M, Chen Z, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yi H, Peng R, L-y Zhang, et al. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8:e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duan L-J, Ding M, Hou L-J, Cui Y-T, Li C-J, Yu D-M. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 94.Aneja A, Tang WW, Bansilal S, Garcia MJ, Farkouh ME. Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am J Med. 2008;121:748–757. doi: 10.1016/j.amjmed.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 95.Zhang M, Gu H, Chen J, Zhou X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int J Cardiol. 2016;202:753–755. doi: 10.1016/j.ijcard.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 96.Zhang M, Gu H, Xu W, Zhou X. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol. 2016;203:214–216. doi: 10.1016/j.ijcard.2015.10.136. [DOI] [PubMed] [Google Scholar]
- 97.Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X, Wang H, Yao B, Xu W, Chen J, Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 2016;6:36340. doi: 10.1038/srep36340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peng H, Zou L, Xie J, et al. lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Mol Neurobiol. 2017;54:511–523. doi: 10.1007/s12035-015-9632-1. [DOI] [PubMed] [Google Scholar]
- 100.Wang S, Xu H, Zou L, et al. LncRNA uc. 48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal. 2016;12:139–148. doi: 10.1007/s11302-015-9488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zgheib C, Hodges MM, Hu J, Liechty KW, Xu J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS One. 2017;12:e0177453. doi: 10.1371/journal.pone.0177453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 105.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 107.Wang X-S, Zhang Z, Wang H-C, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 108.Deng R, Liu B, Wang Y, et al. High expression of the newly found long noncoding RNA Z38 promotes cell proliferation and oncogenic activity in breast cancer. J Cancer. 2016;7:576–586. doi: 10.7150/jca.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang J, Zhuo H, Zhang X, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanna N, Ohana P, Konikoff FM, et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. 2012;19:374–381. doi: 10.1038/cgt.2012.10. [DOI] [PubMed] [Google Scholar]
- 111.Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med. 2010;8:134. doi: 10.1186/1479-5876-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mizrahi A, Czerniak A, Levy T, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]