Abstract
Background
Liver fibrosis is the common sequel of chronic liver diseases. Recent studies have identified hepatic stellate cells as the primary cell type mediating hepatic fibrogenesis. It has been demonstrated that hepatic stellate cells undergo a process of activation during the development of liver fibrosis. During the activation process, hepatic stellate cells acquire myofibroblast-like phenotype featuring the expression of smooth muscle alpha actin. Interferons have been employed for the treatment of viral hepatitis. However, it is unclear what is the effect of interferons on the prevention and treatment of liver fibrosis. Moreover, it is not clear whether there are any differences among interferon alpha, interferon beta, and interferon gamma in the treatment of liver fibrosis. Therefore, our objective in current study is to investigate the effects of rat interferon-α, interferon-β, and interferon-γ on the proliferation and activation of rat hepatic stellate cells.
Results
Rat interferon-β and interferon-γ significantly inhibited rat hepatic stellate cell proliferation while rat interferon-α did not affect the cell proliferation under the same culture condition. Inhibition of cell proliferation was confirmed by both WST-1 cell proliferation assay and 5-bromo-2'-deoxy-uridine incorporation assay. Similar results were observed regarding interferons regulation of hepatic stellate cell activation. Both rat interferon-β and interferon-γ reduced smooth muscle α-actin abundance after 6 days treatment, but rat interferon-α did not alter smooth muscle α-actin level.
Conclusions
Our results indicate that rat interferon-α and interferon-β have different biological effects on rat hepatic stellate cells and suggest that there are different signaling events between interferon-α and interferon-β in hepatic stellate cells.
Background
Liver fibrosis is the common sequel of chronic liver injury of variable origin (viral infection, metabolic diseases and toxin). Recent studies have identified hepatic stellate cells (HSCs) as the primary cell type mediating hepatic fibrosis [1,2]. In normal liver, HSCs are the site of storage and metabolism of vitamin A [3,4]. During hepatic fibrogenesis, HSCs proliferate and undergo a process of activation, developing a myofibroblast-like appearance. Activated HSCs appear to lose lipid droplet, increase rough endoplasmic reticulum, and express smooth muscle alpha actin (SMA) [5-7]. Activated HSCs also increase the synthesis of extracellular matrix components that form the major part of the fibrotic liver [8,9].
Interferons were first discovered as anti-viral soluble protein and at present, interferon-α is the main medication for the treatment of viral hepatitis [10,11]. Interferons consist of type 1 interferon, which includes interferon-α, -β and -ω, and type II interferon, which is interferon γ. Several subtypes are observed in human interferon. Interferon-α has at least 14 subtypes, interferon-β has 3 subtypes and interferon-γ has 5 subtypes [12,13]. Biological activities of interferons are initiated by the interaction of interferons with cell surface type 1 and/or type II interferon receptors. This interaction brings together two receptor subunits. These two receptor subunits are not pre-associated on the cell surface but rather are induced to be associated in the presence of ligand [14]. The formation of the heteromeric receptor results in the formation of a functionally active receptor that leads to the activation of cytoplasmic proteins and interferon signaling [15]. IFN-α and IFN-β appear to utilize type 1 IFN receptor complex and activate similar intracellular signaling pathways [16,17]. IFN-γ seems to activate type II IFN receptor. Ligand-induced association of both types of IFN receptor results in the phosphorylation of the receptors by Janus kinases [18-20], which subsequently results in the activation of STATs (signal transducers and activators of transcription) proteins by additional phosphorylation events [21,22]. Such events lead to the formation of IFN-inducible transcription factors that bind to IFN response elements presented in IFN-inducible gene [23].
Although it is generally considered that IFN-α and IFN-β utilize a common receptor complex, a number of observations suggest that differences may occur in the abilities of IFN-α and/or IFN-β to induce certain biological effects. These include the preferential induction of an IFN-specific gene [24,25], different growth inhibitory effects [26], and erythropoietic effects [27]. One possible explanation for the different signaling events between IFN-α and IFN-β would be the existence of IFN-β specific receptor associated phosphoprotein (BRAP), which appears to be tyrosine phosphorylated and to be associated with IFNAR1 (interferon alpha-receptor 1) [28,29].
Results
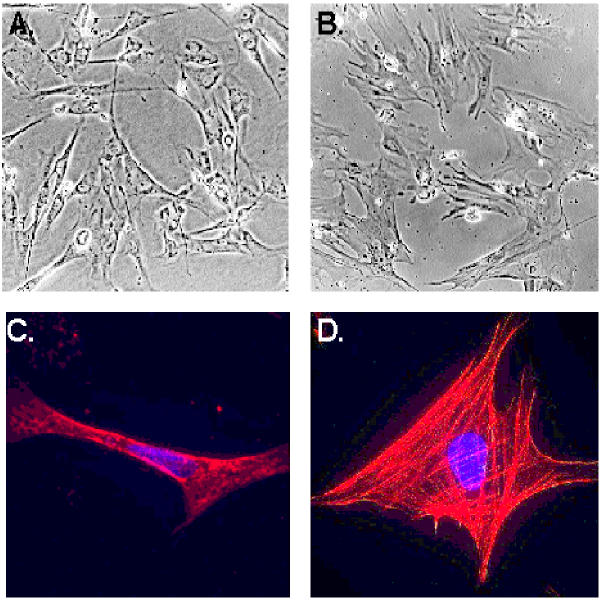
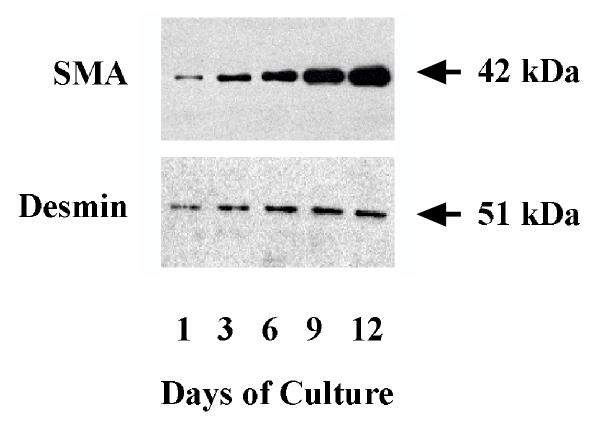
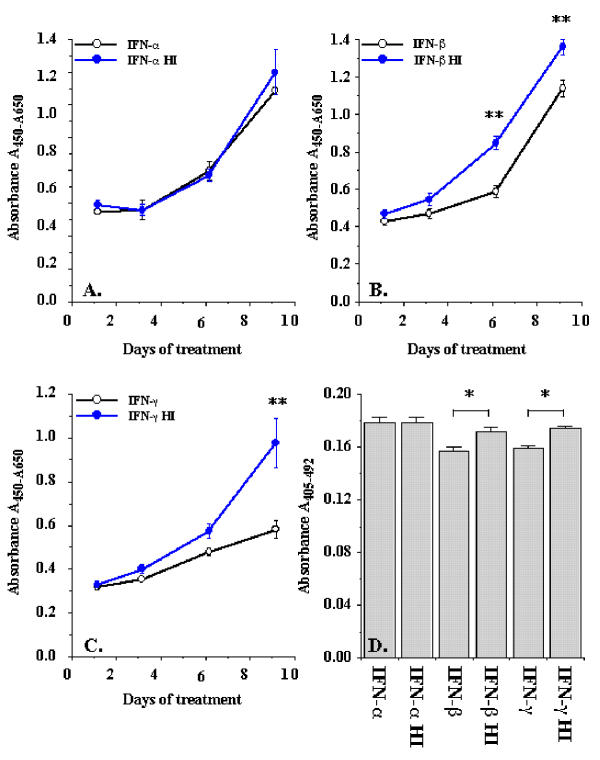
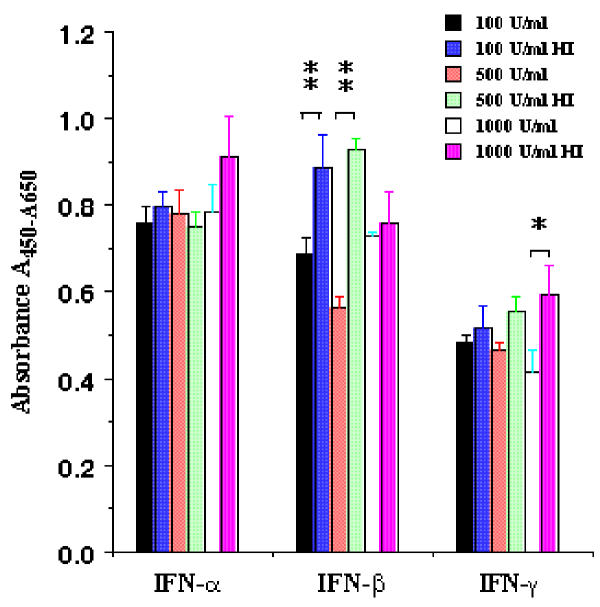
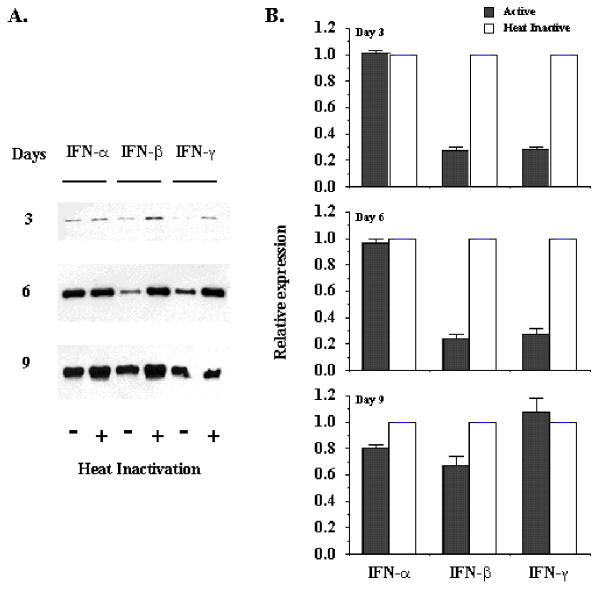
Primary rat hepatic stellate cells were isolated and cultured for 9 days before the cells were detached and sub-cultured for the study of interferon regulation of proliferation and activation. Figure 1 shows the morphology of sub-cultured HSCs at days 3 (Figure 1A) and 12 (Figure 1B) under phase contract microscope. The fluorescent images of SMA expression in rat HSCs were presented in Figure 1C and 1D for sub-cultured 3-day and 12-day HSCs respectively. At day 12, all HSCs displayed myofibroblast phenotype and expressed SMA. The expression of SMA and desmin proteins at different times (1, 3, 6, 9, and 12 days) of the sub-cultured HSCs was shown in Figure 2. SMA gradually increased from day 1 to day 12 and reached a maximum level at day 12 while desmin abundance did not elevate as much as observed on the SMA. The result indicated that HSCs fully differentiated into myofibroblast-like phenotype after 12 days sub-culture in vitro. We then examined the effect of IFN-α, IFN-β and IFN-γ on cell proliferation and DNA synthesis in sub-cultured HSCs. Figure 3 showed the effect of IFN-α, IFN-β and IFN-γ on proliferation of subcultured HSCs by WST-1 cell proliferation assay. As shown in Figure 3A, IFN-α did not affect sub-cultured HSC proliferation and it also did not affect Bromo-2'-deoxy-urindne (BrdU) incorporation in HSCs (Figure 3D). In contrast, both IFN-β and IFN-γ significantly inhibited subcultured HSC proliferation and BrdU incorporation. IFN-β inhibition of HSC proliferation was observed at day 6 (32% decrease relative to control) and still could be observed at day 9 (18% decrease relative to control) (Figure 3B). IFN-γ also inhibited HSC proliferation at day 9 (40% decrease relative to control) (Figure 3C). Both IFN-β and IFN-γ significantly inhibited BrdU incorporation in HSCs (about 10 % decrease relative to control for both IFN-β and IFN-γ) (Figure 3D). Moreover, both IFN-β and IFN-γ reduced the cell number after 7 days treatment (500 U/ml IFB-β = 4.5 × 105 ± 0.10 × 105 cells/ml vs. control = 5.2 × 105 ± 0.12 × 105 cells/ml, p < 0.01; 500 U/ml IFN-γ = 4.8 × 105 ± 0.1 × 105 cells/ml vs. control = 6.0 × 105 ± 0.15 × 105 cells/ml, p < 0.01) The inhibitory effect of IFN-β on HSC proliferation was not dose dependent (Figure 4) while the effect of INF-γ on HSC proliferation seemed to correlate with the dose. However, the statistically significant difference of inhibition was only observed at the highest concentration of INF-γ employed.
Figure 1.
Morphology of sub-cultured rat HSCs and expression of SMA in HSCs. HSC morphology under phase-contrast microscope at days 3 and 12 was shown at panels A and B respectively. The expression of SMA in HSCs at the same days was presented in panels C and D.
Figure 2.
Expression of SMA and desmin in sub-cultured HSCs. Hepatic stellate cells were isolated from Sprague-Dawley rat and cultured for 9 days. Cells were then incubated with trypsin and sub-cultured for days indicated. The upper panel represents abundance of SMA while the lower panel represents abundance of desmin.
Figure 3.
Interferons regulation of rat hepatic stellate cell proliferation. Sub-cultured hepatic stellate cells were incubated with 500 U/ml of interferons for the days indicated. Cell proliferation was performed by WST-1 cell proliferation and BrdU incorporation assays. Panel A exhibits the effect of IFN-α on cell proliferation. Panel B indicates the effect of IFN-β on cell proliferation. Panel C displays the effect of IFN-γ on cell proliferation. Panel D exhibits the IFNs effect on BrdU incorporation in rat HSCs. The data represent mean ± SEM. HI represents heat-inactivation. * indicates p < 0.05 and ** indicates p < 0.01.
Figure 4.
Dose dependent effect of IFNs on rat HSC proliferation. Sub-cultured HSCs were incubated with three concentrations of IFNs (100, 500 and 1000 U/ml) for 6 days. WST-1 cell proliferation reagents assayed cell proliferation. The data represent mean ± SEM. HI represents heat-inactivation. * presents p < 0.05 and ** indicated p < 0.01
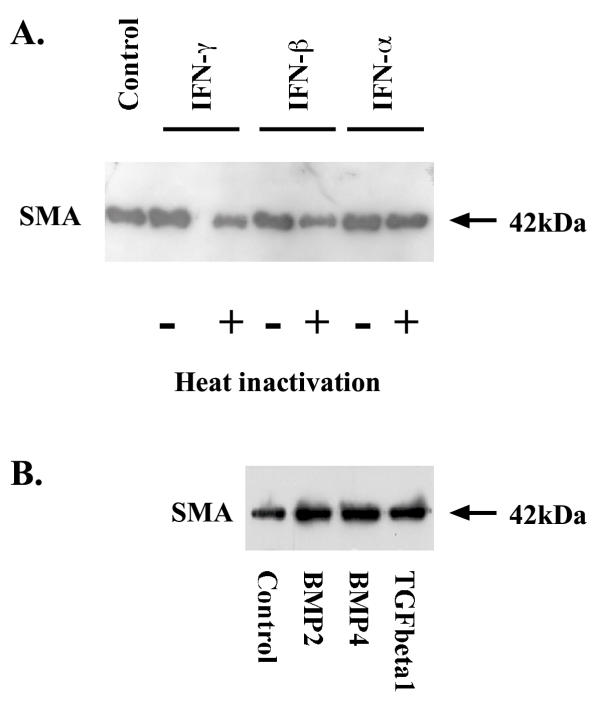
We then examined the effect of IFNs on the expression of SMA, which is a phenotypic marker of activated HSCs. Western blot analysis indicated that both IFN-β and IFN-γ decreased SMA expression in sub-cultured HSCs when they were exposed to the cytokines for 3 or 6 days (Figure 5A), but IFN-α did not affect SMA abundance after the same length of treatment. However, when sub-cultured HSCs were exposed to the cytokines for 9 days, both IFN-α and IFN-β reduced SMA level in a small attitude while IFN-γ did not alter the abundance of SMA. Analyses of the blots by densitometric scanning revealed that IFN-β and IFN-γ reduced the SMA level by 76% ± 3% and 73% ± 7% respectively after HSCs were incubated with 500 U/ml IFN-β and IFN-γ for six days (Figure 5B). In addition, we also observed that water alone did not affect the expression of SMA in 6-day rat HSCs (Figure 6A) as well as transforming growth factor beta1, bone morphogenetic protein 2 and bone morphogenetic protein 4 promoted the expression of SMA in rat HSCs (Figure 6B).
Figure 5.
Effect of interferons on SMA abundance in rat HSCs. Sub-cultured HSCs were incubated with 500 U/ml of IFN-α, IFN-β and IFN-γ for 3, 6, or 9 days. Panel A exhibits the typical Western blot of SMA expression. Panel B represents densitometric data of Western blot analysis. Active form of IFNs was compared with heat-inactivated (HI) form of IFNs, which were arbitrarily set at 1. The data represent mean ± SEM from four experiments.
Figure 6.
Effect of interferons, transforming growth factor beta1, bone morphogenetic protein 2 and bone morphogenetic protein 4 on SMA abundance in rat HSCs. Sub-cultured HSCs were incubated with water only, 500 U/ml of different IFNs for 6 days and the abundance of SMA in HSCs was documented in panel A, while sub-cultured HSCs were treated with 5 ng/ml of transforming growth factor beta1, 5 ng/ml of bone morphogenetic protein 2 and 5 ng/ml of bone morphogenetic protein 4 for 4 days and the abundance of SMA in HSCs was presented in panel B. HI indicates heat-inactivation.
Discussion
The present study investigated the effect of IFN-α, IFN-β, and IFN-γ on the proliferation and SMA expression of rat hepatic stellate cells cultured on uncoated plastic dish. Rat hepatic stellate cells have been documented to exhibit proliferation and morphological change in experimental hepatic fibrosis [30] or in human liver specimens obtained from patients with fibrotic liver disease [31,32]. IFN-α is an effective drug for the treatment of patients with hepatitis B virus or C virus infection. The antiviral efficiency of IFN-α is almost the same as IFN-γ; however, IFN-α has fewer side effects than IFN-γ. In the other aspect, IFN-γ has anti-fibrogenic effect as it is documented that IFN-γ inhibits collagen synthesis in several cell types [33-36]. Moreover, IFN-γ delays phenotypic trans-differentiation of rat hepatic stellate cells in vitro [37]. Furthermore, IFN-γ reduces SMA expression in human HSCs, arterial smooth muscle cells and dermal myofibroblasts [38,39]. Our results with IFN-γ were consistent with these reports. We also observed that IFN-γ reduced SMA expression in cultured rat HSCs. Several clinical studies have suggested that IFN-α has anti-fibrogenic activity. Most of the studies were conducted in patients with chronic hepatitis C. This suggests that IFN-α may reduce the histological fibrosis index of the Knodell score in responders, which may be consequent to the antiviral properties of the drug [10,11,40]. Furthermore, some studies point to a direct anti-fibrogenic effect of IFN-α, independent of its antiviral property [41]. In these studies, IFN-α decreased collagen concentration and SMA index in not only responders but also non-responders or relapsers. An in vitro study confirmed these results by documenting that IFN-α-2c inhibits human hepatic stellate cell proliferation and collagen product at the concentration (10000 U/ml) higher than a single therapeutic dose [42] (peak plasma concentrations range between 40 U/ml to 150 U/ml [43,44]). In our current study, we did not observe that rat IFN-α inhibited rat hepatic stellate cell proliferation and SMA expression at the concentrations from 100 to 1000 U/ml. In contrast to IFN-α, we showed that rat IFN-β significantly inhibited rat hepatic stellate cell proliferation and SMA expression at the concentration of 500 U/ml. Although the concentration was higher than the single therapeutic dose of IFN-α used in clinical treatment, it is within the range of drug accumulation after repeated administration, which is by the factor of 2 to 5 of the single administration [45]. Moreover, the difference between this study and others regarding the effect of IFN-α on HSCs could also be due to the different species employed in studies. In this study, rats were employed while in the other study [42] human HSCs were used. In addition, more than 14 subtypes of human IFN-α have been identified and each subtype of IFN-α has different binding affinity to type 1 IFN receptor [46]. At present, only one subtype of rat IFN-α has been identified.
The effect of IFNs on rat HSC proliferation in this study was low and marginal especially at early time points. Since rat HSCs were cultured in 10% serum condition, proliferation of rat HSCs has been stimulated by the other factors in serum. In other studies relating to HSC proliferation, serum-reducing condition has been employed which utilized 0.1%-1% serum [47]. Dramatic differences of cell proliferation and activation of HSCs were observed in this culture condition. Recently we also observed that when rat HSCs were cultured in 5% serum condition or serum-free condition for a short period, IFNs and other cytokines could dramatically affect cell proliferation and expression of SMA in rat HSCs (data not shown).
Our results also showed the first time that IFN-α and IFN-β induced different biological effects on rat hepatic stellate cells. The different biological effects of IFN-α and IFN-β have been documented in human glioma cells [26] and in the selected Tyk2-deficient cell lines [48,49]. In these cells, growth inhibition and gene expression are responsive to IFN-β but not to IFN-α. The mechanism of the differences between IFN-α and IFN-β biological effects is still unknown. However, it is known that type 1 IFNs bind to un-associated type 1 IFN receptor and assemble two chains of IFNAR1 and IFNAR2. The assembled type 1 IFN receptor will be phosphorylated by associated kinases, which would lead to intracellular signaling events. One difference between IFN-α and IFN-β at the receptor level is that there is a phosphoprotein selectively involved in IFN-β signaling [24,25]. Two different research groups demonstrated this tyrosine-phosphorylated protein to be IFNAR2.2 [28,29]. By employing specific antibody against IFNAR2.2, they documented that IFNAR2.2 is present in Daudi cells as a cell surface protein approximately 90–100 kDa, which is tyrosine-phosphorylated and associated with IFNAR1 upon stimulation of cells with IFN-β but not IFN-α. Their studies suggest that there are some differences in receptor interaction between IFN-α and IFN-β in HSCs. However, it is still unclear why this phosphoprotein is not related with IFN-α and what IFN-β specific responses are associated with the IFN-β induced phosphoprotein. Our results suggested that HSCs might serve as the cell type to investigate the different responses of IFN-α and IFN-β.
Conclusions
Rat interferon-α and interferon-β have different biological effects on rat hepatic stellate cells and different signaling events might exist between interferon-α and interferon-β in hepatic stellate cells.
Materials and methods
Materials
Rat interferon α (specific activity = 1 × 108 units/mg), rat interferon β (specific activity = 3 × 107 units/mg), rat interferon γ (specific activity = 4.6 × 106 units/mg) were purchased from PBL Biomedical Laboratories (New Brunswick, NJ). To remove biological activity of rat interferons, the stock solution containing rat interferons was boiled in water for 10 minutes to denature the proteins (heat inactivation – HI). Cell proliferation reagent WST-1, and 5-Bromo-2'-deoxy-uridine (BrdU) Labeling and Detection Kit II, collagenase D, pronase, DNase 1, and the antibody against smooth muscle α-actin (SMA) were purchased from Roche Molecular Biochemicals (Laval, QC). Anti-mouse IgG immunoglobulin conjugated to horseradish peroxidase and Enhanced Chemiluminescence Detection Kit were purchased from Amersham Pharmacia Biotech, Inc. (Baie d'Urfe, QC). All buffers and reagents were purchased from Sigma (St. Louis, MO).
Rat hepatic stellate cells
Male Sprague-Dawley rats (450–550 gram body weight) were provided by Central Animal Care of the University of Manitoba and maintained under 12-hour light/dark cycles with food and water ad libitum. In conducting the research described in this report, all animals received humane care in compliance with the Institution's guidelines (Animal Protocol No. 98-053), which is in accordance with criteria set by the Canadian Council on Animal Care. Hepatic stellate cells were isolated by two steps of collagenase and pronase methods [50]. Briefly, rat liver was perfused with 0.125 mg/ml collagenase D, 0.5 mg/ml pronase and 15 μg/ml DNase 1 in Hank balanced salt solution (HBSS) supplemented with 10 mM HEPES, 4.2 mM sodium bicarbonate for 20 minutes and incubated with 0.125 mg/ml collagenase D, 0.5 mg/ml pronase for another 15 minutes with constant low speed stirring at 37°C. After removing hepatocytes, HSCs were separated from other non-parenchymal cells by centrifugation on 11.3% Nycodenz with sodium chloride. HSCs were harvested from the interface between suspension buffer and 11.3% Nycodenz solution, washed and plated on uncoated plastic tissue culture dish (Costar) at a density of 25,000 cells/cm2. A good separation of rat HSC in our laboratory could reach 2 × 107 cells per liver. Purity of HSC preparation was assessed by their typical light microscopic appearance, vitamin A specific autofluorescence. Purity of HSCs was about 97%. Cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine at 37°C in a humidified atmosphere of 5% CO2 and 95% air. The first change of medium was made 24 hours after seeding and the second change of medium was about 20 hours later. Sub-cultured HSCs (the first passage of primary HSCs) were obtained from 9 days old primary culture of HSCs. Microphotography and fluorescent microphotography: Sub-cultured HSCs were imaged and photographed on an Olympus inverted-phase microscope (CK-40) using a mounted Olympus 35-mm camera (Carsen Group Inc. Markham, ON) and TMAX 400 Kodak black-and-white film (Eastern Kodak Co., Rochester, NY). For fluorescent microphotography, HSCs grown on uncoated Nunc 8-well glass-slide dishes were rinsed with phosphate-buffered-saline, fixed in ice-cold paraformaldehyde, rinsed and stained with HOECHST mix for nuclei (5 mg/ml HOECHST 33258 and 0.5% Saponin in culture medium containing 10% fetal bovine serum) and antibody against SMA at dilution of 1:100, followed by Cy3-conjugated rabbit anti-mouse IgG at dilution of 1:250. A fluorescence image was obtained by Olympus True Research Microscope (Olympus AX70) with 60X oil-objective and ImagePro software (Carsen Group Inc. Markham, ON).
Cell proliferation assay
Cell proliferation was determined using both cell proliferation reagents WST-1 [51] and 5-Bromo-2'-deoxy-uridine (BrdU) Labeling and Detection Kit II methods [52]. The first passage HSCs (5 × 103) in 100 μl culture medium was seeded into 96 well plates in complete culture medium. After one day of culture in a 37°C-humidified incubator, the medium was carefully removed, and 100 μl of fresh medium containing different concentrations of rat IFN-α, rat IFN-β and rat IFN-γ were added into the wells. The cells were treated continuously with IFNs for the days indicated and the medium containing IFNs were changed every other day. Cell proliferation was documented after 1, 3, 6 and 9 days of IFNs treatment. At the end of experiments, cells were incubated with 10 μl of the cell proliferation reagent WST-1 for 2 hours or 1 μl BrdU labeling reagent for 4 hours. The absorbance of the treated samples against a blank control was measured using a THERMOmax microplate reader (ELISA) (Molecular Devices Co., Menlo Park, CA). The wavelength for measuring absorbance of the WST-1 product was 420 nm and reference wavelength was 650 nm while measuring wavelength for BrdU was 405 nm and reference wavelength for BrdU was 492 nm according to the filters available in the ELISA reader. Cell proliferation was performed in eight wells and each experiment was completely repeated on three occasions.
Western blot analyses of SMA
sub-culture HSCs on different days was lysed in 100 μl of protein extract solution (1 mM Tris-HCl pH7.5, 1 mM EDTA pH 8.0, 10 mM NaCl, 1% sodium dodecyl sulfate (SDS), 1 mM PMSF and 0.25 M sucrose) [53]. The cell membrane was broken by sonicating the cells for 1 minute with Sonicator (Vibra Cell, Sonics and Material Inc. Danbury, CT) and cell debris was pelleted by centrifugation at 14000 rpm at 4°C for 5 minutes. The protein content of cellular lysates was calculated by the Lowry method [54]. Twenty μg of protein was boiled for 5 minutes, separated on 12% sodium dodecyl sulfate-polyacrylamide (SDS-polyacrylamide) gel electrophoresis under reducing conditions and transferred to Nitroplus-2000 membrane (Micron Separations Inc. Westborough, MA). Non-specific antibody binding was blocked by pre-incubation of the membranes with 5% skim milk in 1 x Tris-buffered-saline (TBS) for one hour at room temperature. Membranes were then incubated overnight at 4°C with primary antibodies against SMA at dilution of 1:1000 in 1 x TBS containing 2% skim milk. After washing, they were incubated with sheep anti-mouse IgG at 1:1000 dilutions for 1 hour at room temperature. Bands were visualized by employing the enhanced chemiluminescence kit per the manufacturer's instruction.
Statistical analyses
To analyze differences in the treatment groups, we performed the ANOVA and Fisher's PLSD test as Post hoc test using StatView (version 5.0) software (SAS Institute Inc. Cary, NC). Differences with p values below 0.05 were judged to be significant.
Authors' contributions
Dr. Shen H carried out the isolation of hepatic stellate cells, Western blot analysis and cell proliferation assay. Mrs. Zhang MN participated in the cell imaging. Dr. Minuk GY participated in the design of the study. Dr. Gong Y conceived of the study and participated in its design and coordination.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by a grant from Serono Laboratory Inc (Norwell MA). We sincerely thank Dr. Birgit J. Maschek for her contribution and coordination to this project.
Contributor Information
Hong Shen, Email: hongshen2000@yahoo.com.
Manna Zhang, Email: mazhang@cc.umanitoba.ca.
Gerald Y Minuk, Email: gminuk@cc.umanitoba.ca.
Yuewen Gong, Email: ygong@ms.umanitoba.ca.
References
- Friedman SL. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med. 1993;328:1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- Gressner AM. Transdifferentiation of hepatic stellate cells (Ito cells) to myofibroblasts: a key event in hepatic fibrogenesis. Kidney Int Suppl. 1996;54:S39–45. [PubMed] [Google Scholar]
- Hendriks HF, Verhoofstad WA, Brouwer A, de Leeuw AM, Knook DL. Perisinusoidal fat-storing cells are the main vitamin A storage sites in rat liver. Exp Cell Res. 1985;160:138–149. doi: 10.1016/0014-4827(85)90243-5. [DOI] [PubMed] [Google Scholar]
- Blomhoff R, Wake K. Perisinusoidal stellate cells of the liver: important roles in retinol metabolism and fibrosis. Faseb J. 1991;5:271–277. doi: 10.1096/fasebj.5.3.2001786. [DOI] [PubMed] [Google Scholar]
- Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15:234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Boyles JK, Gabbiani G, Friedman SL. Rat hepatic lipocytes express smooth muscle actin upon activation in vivo and in culture. J Submicrosc Cytol Pathol. 1992;24:193–203. [PubMed] [Google Scholar]
- Burt AD, Le Bail B, Balabaud C, Bioulac-Sage P. Morphologic investigation of sinusoidal cells. Semin Liver Dis. 1993;13:21–38. doi: 10.1055/s-2007-1007335. [DOI] [PubMed] [Google Scholar]
- Seyer JM, Hutcheson ET, Kang AH. Collagen polymorphism in normal and cirrhotic human liver. J Clin Invest. 1977;59:241–248. doi: 10.1172/JCI108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent G, Gay S, Inouye T, Bahu R, Minick OT, Popper H. Vitamin A-containing lipocytes and formation of type III collagen in liver injury. Proc Natl Acad Sci U S A. 1976;73:3719–3722. doi: 10.1073/pnas.73.10.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Castilla A, Ruiz J, Civeira MP, Prieto J. Randomised trial of lymphoblastoid alpha-interferon in chronic hepatitis C. Effects on inflammation, fibrogenesis and viremia. J Hepatol. 1993;17:390–396. doi: 10.1016/s0168-8278(05)80223-5. [DOI] [PubMed] [Google Scholar]
- Schvarcz R, Glaumann H, Weiland O, Norkrans G, Wejstal R, Fryden A. Histological outcome in interferon alpha-2b treated patients with chronic posttransfusion non-A, non-B hepatitis. Liver. 1991;11:30–38. doi: 10.1111/j.1600-0676.1991.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Pestka S, Meager A. Interferon standardization and designations. J Interferon Cytokine Res. 1997;17 Suppl 1:S9–14. [PubMed] [Google Scholar]
- Pestka S. The human interferon-alpha species and hybrid proteins. Semin Oncol. 1997;24:S9–S17. [PubMed] [Google Scholar]
- Cohen B, Novick D, Barak S, Rubinstein M. Ligand-induced association of the type 1 interferon receptor components. Mol Cell Biol. 1995;15:4208–4214. doi: 10.1128/mcb.15.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.biochem.56.1.727. [DOI] [PubMed] [Google Scholar]
- Uze G, Lutfalla G, Mogensen KE. Alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Croze E, Wang C, Murti A, Basu L, Mullersman JE, Pfeffer LM. Role of interferon alpha/beta receptor chain 1 in the structure and transmembrane signaling of the interferon alpha/beta receptor complex. Proc Natl Acad Sci U S A. 1994;91:9602–9606. doi: 10.1073/pnas.91.20.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici O, Yan H, Domanski P, Handa R, Smalley D, Mullersman J, Witte M, Krishnan K, Krolewski J. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type 1 interferon receptor by p135tyk2 tyrosine kinase. Mol Cell Biol. 1994;14:8133–8142. doi: 10.1128/mcb.14.12.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick D, Cohen B, Tal N, Rubinstein M. Soluble and membrane-anchored forms of the human IFN-alpha/beta receptor. J Leukoc Biol. 1995;57:712–718. doi: 10.1002/jlb.57.5.712. [DOI] [PubMed] [Google Scholar]
- Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/S0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- Haque SJ, Williams BR. Signal transduction in the interferon system. Semin Oncol. 1998;25:14–22. [PubMed] [Google Scholar]
- Pestka S. The interferon receptors. Semin Oncol. 1997;24:S9–18. [PubMed] [Google Scholar]
- Rani MRS, Foster GR, Leung S, Leaman D, Stark GR, Ransohoff RM. Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J Biol Chem. 1996;271:22878–22884. doi: 10.1074/jbc.271.37.22878. [DOI] [PubMed] [Google Scholar]
- Platanias LC, Uddin S, Colamonici OR. Tyrosine phosphorylation of the alpha and beta subunits of the type 1 interferon receptor. Interferon-beta selectively induces tyrosine phosphorylation of an alpha subunit-associated protein. J Biol Chem. 1994;269:17761–17764. [PubMed] [Google Scholar]
- Rosenblum MG, Yung WK, Kelleher PJ, Ruzicka F, Steck PA, Borden EC. Growth inhibitory effects of interferon-beta but not interferon-alpha on human glioma cells: correlation of receptor binding, 2',5'-oligoadenylate synthetase and protein kinase activity. J Interferon Res. 1990;10:141–151. doi: 10.1089/jir.1990.10.141. [DOI] [PubMed] [Google Scholar]
- Means RT, Jr, Krantz SB. Inhibition of human erythroid colony-forming units by interferons alpha and beta: differing mechanisms despite shared receptor. Exp Hematol. 1996;24:204–208. [PubMed] [Google Scholar]
- Croze E, Russell-Harde D, Wagner TC, Pu H, Pfeffer LM, Perez HD. The human type 1 interferon receptor. Identification of the interferon beta-specific receptor-associated phosphoprotein. J Biol Chem. 1996;271:33165–33168. doi: 10.1074/jbc.271.52.33165. [DOI] [PubMed] [Google Scholar]
- Platanias LC, Uddin S, Domanski P, Colamonici OR. Differences in interferon alpha and beta signaling. Interferon beta selectively induces the interaction of the alpha and betaL subunits of the type 1 interferon receptor. J Biol Chem. 1996;271:23630–23633. doi: 10.1074/jbc.271.39.23630. [DOI] [PubMed] [Google Scholar]
- Mak KM, Leo MA, Lieber CS. Alcoholic liver injury in baboons: transformation of lipocytes to transitional cells. Gastroenterology. 1984;87:188–200. [PubMed] [Google Scholar]
- Mak KM, Lieber CS. Lipocytes and transitional cells in alcoholic liver disease: a morphometric study. Hepatology. 1988;8:1027–1033. doi: 10.1002/hep.1840080508. [DOI] [PubMed] [Google Scholar]
- Schmitt-Graff A, Kruger S, Bochard F, Gabbiani G, Denk H. Modulation of alpha smooth muscle actin and desmin expression in perisinusoidal cells of normal and diseased human livers. Am J Pathol. 1991;138:1233–1242. [PMC free article] [PubMed] [Google Scholar]
- Amento EP, Bhan AK, McCullagh KG, Krane SM. Influences of gamma interferon on synovial fibroblast-like cells. Ia induction and inhibition of collagen synthesis. J Clin Invest. 1985;76:837–848. doi: 10.1172/JCI112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB, Sandell LJ, Stephenson ML, Krane SM. Immune interferon suppresses levels of procollagen mRNA and type II collagen synthesis in cultured human articular and costal chondrocytes. J Biol Chem. 1986;261:9049–9055. [PubMed] [Google Scholar]
- Reginato AM, Sanz-Rodriguez C, Diaz A, Dharmavaram RM, Jimenez SA. Transcriptional modulation of cartilage-specific collagen gene expression by interferon gamma and tumour necrosis factor alpha in cultured human chondrocytes. Biochem J. 1993;294:761–769. doi: 10.1042/bj2940761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom J, Feldman G, Freundlich B, Jimenez SA. Inhibition of excessive scleroderma fibroblast collagen production by recombinant gamma-interferon. Association with a coordinate decrease in types I and III procollagen messenger RNA levels. Arthritis Rheum. 1986;29:851–856. doi: 10.1002/art.1780290706. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Maher JJ, Jarnagin WR, Gabbiani G, Friedman SL. Inhibition of rat hepatic lipocyte activation in culture by interferon-gamma. Hepatology. 1992;16:776–784. doi: 10.1002/hep.1840160325. [DOI] [PubMed] [Google Scholar]
- Hansson GK, Hellstrand M, Rymo L, Rubbia L, Gabbiani G. Interferon gamma inhibits both proliferation and expression of differentiation-specific alpha-smooth muscle actin in arterial smooth muscle cells. J Exp Med. 1989;170:1595–1608. doi: 10.1084/jem.170.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmouliere A, Rubbia-Brandt L, Abdiu A, Walz T, Macieira-Coelho A, Gabbiani G. Alpha-smooth muscle actin is expressed in a subpopulation of cultured and cloned fibroblasts and is modulated by gamma-interferon. Exp Cell Res. 1992;201:64–73. doi: 10.1016/0014-4827(92)90348-c. [DOI] [PubMed] [Google Scholar]
- Castilla A, Prieto J, Fausto N. Transforming growth factors beta 1 and alpha in chronic liver disease. Effects of interferon alfa therapy [see comments]. N Engl J Med. 1991;324:933–940. doi: 10.1056/NEJM199104043241401. [DOI] [PubMed] [Google Scholar]
- Manabe N, Chevallier M, Chossegros P, Causse X, Guerret S, Trepo C, Grimaud JA. Interferon-alpha 2b therapy reduces liver fibrosis in chronic non-A, non-B hepatitis: a quantitative histological evaluation. Hepatology. 1993;18:1344–1349. [PubMed] [Google Scholar]
- Mallat A, Preaux AM, Blazejewski S, Rosenbaum J, Dhumeaux D, Mavier P. Interferon alfa and gamma inhibit proliferation and collagen synthesis of human Ito cells in culture. Hepatology. 1995;21:1003–1010. [PubMed] [Google Scholar]
- Radwanski E, Perentesis G, Jacobs S, Oden E, Affrime M, Symchowicz S, Zampaglione N. Pharmacokinetics of interferon alpha-2b in healthy volunteers. J Clin Pharmacol. 1987;27:432–435. doi: 10.1002/j.1552-4604.1987.tb03044.x. [DOI] [PubMed] [Google Scholar]
- Witter F, Barouki F, Griffin D, Nadler P, Woods A, Wood D, Lietman P. Biologic response (antiviral) to recombinant human interferon alpha 2a as a function of dose and route of administration in healthy volunteers. Clin Pharmacol Ther. 1987;42:567–575. doi: 10.1038/clpt.1987.198. [DOI] [PubMed] [Google Scholar]
- Smith CI, Weissberg J, Bernhardt L, Gregory PB, Robinson WS, Merigan TC. Acute Dane particle suppression with recombinant leukocyte A interferon in chronic hepatitis B virus infection. J Infect Dis. 1983;148:907–913. doi: 10.1093/infdis/148.5.907. [DOI] [PubMed] [Google Scholar]
- Pestka S. The human interferon alpha species and receptors. Biopolymers. 2000;55:254–287. doi: 10.1002/1097-0282(2000)55:4<254::AID-BIP1001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Dooley S, Delvoux B, Lahme B, Mangasser-Stephan K, Gressner AM. Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology. 2000;31:1094–1106. doi: 10.1053/he.2000.6126. [DOI] [PubMed] [Google Scholar]
- Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- Gong W, Roth S, Michel K, Gressner AM. Isoforms and splice variant of transforming growth factor beta-binding protein in rat hepatic stellate cells. Gastroenterology. 1998;114:352–363. doi: 10.1016/s0016-5085(98)70488-3. [DOI] [PubMed] [Google Scholar]
- Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, Boyd MR. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- Magaud JP, Sargent I, Mason DY. Detection of human white cell proliferative responses by immunoenzymatic measurement of bromodeoxyuridine uptake. J Immunol Methods. 1988;106:95–100. doi: 10.1016/0022-1759(88)90276-1. [DOI] [PubMed] [Google Scholar]
- Gong Y, Zhang M, Minuk GY. Regulation of transforming growth factor-beta1 gene expression and cell proliferation in human hepatocellular carcinoma cells (PLC/PRF/5) by tamoxifen. J Lab Clin Med. 1999;134:90–95. doi: 10.1016/s0022-2143(99)90058-6. [DOI] [PubMed] [Google Scholar]
- Fryer HJ, Davis GE, Manthorpe M, Varon S. Lowry protein assay using an automatic microtiter plate spectrophotometer. Anal Biochem. 1986;153:262–266. doi: 10.1016/0003-2697(86)90090-4. [DOI] [PubMed] [Google Scholar]