Abstract
Src homology 2 domain-containing phosphatase (SHP2) is a phosphatase that mediates signaling downstream of multiple receptor tyrosine kinases (RTK) and is required for full activation of the MAPK pathway. SHP2 inhibition has demonstrated tumor growth inhibition in RTK-activated cancers in preclinical studies. The long-term effectiveness of tyrosine kinase inhibitors (TKI) such as the EGFR inhibitor osimertinib in non-small cell lung cancer (NSCLC) is limited by acquired resistance. Multiple clinically identified mechanisms underlie resistance to osimertinib, including mutations in EGFR that preclude drug binding as well as EGFR-independent activation of the MAPK pathway through alternate RTK (RTK-bypass). It has also been noted that frequently a tumor from a single patient harbors more than one resistance mechanism and the plasticity between multiple resistance mechanisms could restrict the effectiveness of therapies targeting a single node of the oncogenic signaling network. Here we report the discovery of IACS-13909, a specific and potent allosteric inhibitor of SHP2 that suppresses signaling through the MAPK pathway. IACS-13909 potently impeded proliferation of tumors harboring a broad spectrum of activated RTK as the oncogenic driver. In EGFRmut osimertinib-resistant NSCLC models with EGFR-dependent and EGFR-independent resistance mechanisms, IACS-13909, administered as a single agent or in combination with osimertinib, potently suppressed tumor cell proliferation in vitro and caused tumor regression in vivo. Together, our findings provide preclinical evidence for using a SHP2 inhibitor as a therapeutic strategy in acquired EGFR inhibitor-resistant NSCLC.
Keywords: SHP2, allosteric inhibitor, osimertinib resistance, receptor tyrosine kinase, EGFR, NSCLC
Introduction
The Src homology 2 domain containing phosphatase 2 (SHP2, encoded by PTPN11) is a critical regulator of oncogenic MAPK signaling. The SHP2 protein contains N-terminal and C-terminal Src homology 2 (SH2) domains, a protein tyrosine phosphatase (PTP) domain and a C-terminal tail. The inactive SHP2 protein is maintained in a closed state by interdomain interactions between the N-terminal SH2 and PTP domains, preventing substrate access to the active site. SHP2 mutations at the interface of the SH2 and PTP domains, such as the somatic mutations found in juvenile myelomonocytic leukemia (JMML) and the germline mutations found in Noonan’s Syndrome, open and activate the protein (1) (2).
In addition to being an oncogenic driver in rare malignancies, SHP2 critically mediates MAPK pathway signaling downstream of a broad spectrum of receptor tyrosine kinases (RTK), including EGFR, HER2, MET, PDGFR (3–7). The intracellular phospho-tyrosine residues on activated RTKs interact with the SH2 domains on wildtype SHP2, resulting in an open, active conformation of SHP2 and subsequent activation of the downstream MAPK signaling cascade. SHP2 also sits upstream of RAS in the MAPK pathway and full activation of RAS requires input from SHP2, particularly the nucleotide cycling mutant of RAS (i.e., G12C) (8–11).
The development of multiple generations of tyrosine kinase inhibitors (TKI) has transformed the clinical landscape of non-small lung cancer (NSCLC), yet acquired resistance remains as a major challenge. Osimertinib is a mutant-selective third-generation EGFR inhibitor (EGFRi) that targets both EGFR activating mutations (e.g., exon 19 deletion, L858R) and EGFR-dependent on-target resistance mutation towards the first-generation EGFRi (i.e., T790M) (12). It is currently a front-line therapy for EGFRmut NSCLC, with average progression free survival of ~19 months in previously untreated patients (13). Clinical and preclinical studies have revealed numerous resistance mechanisms. Among these, EGFR-dependent mechanisms such as resistance mutations in EGFR (e.g., C797S, reversal to wildtype EGFR) occur in 20~50% of relapsed patients. Other clinically observed resistance mechanisms which also apply to earlier generations of EGFRi include activation of alternate RTKs (e.g., MET, FGFR, HER2, IGF1R), PIK3CA mutations, and mutations in RAS/RAF pathway that maintain downstream ERK activation (14–16). In addition, non-signaling mechanisms such as epithelial-mesenchymal transition (EMT), acquisition of stem-like properties, and metabolic rewiring have also been reported in preclinical models (17,18). Importantly, it has been noted that tumor from a single patient may harbor more than one resistance mechanism (16,19), suggesting that with oncogenic shock from EGFRi, the tumor may switch to multiple alternate drivers. For example, EGFR C797S mutation and MET amplification have been reported to coexist in the same tumor sample from a patient who relapsed on osimertinib treatment (16). The heterogeneity and plasticity in resistance mechanisms makes it challenging to treat patients with a therapy targeting a single RTK.
Since SHP2 critically mediates the signaling of multiple RTKs, and several resistance mechanisms towards osimertinib are through RTK signaling, we hypothesize that a SHP2 inhibitor may be effective in addressing the heterogeneous mechanisms of osimertinib resistance. In this study, we report the discovery of IACS-13909, a novel, potent, and selective allosteric inhibitor of SHP2 that suppresses signaling through the MAPK pathway and inhibits proliferation of RTK-activated tumors in vitro and in vivo. Importantly, we provide preclinical data showing IACS-13909, either administered as a single agent or in combination with osimertinib, potently suppresses tumor cell proliferation in vitro and causes tumor regression in vivo in tumors with EGFR-dependent and EGFR-independent resistance mechanisms.
Materials and Methods
Additional/detailed methods are provided in supplementary information.
In vitro enzymatic assay
Phosphatase activity of full-length SHP2 or SHP2 phosphatase domain was measured using fluorogenic 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP; Molecular Probes) as the substrate. Detailed method is in supplementary information.
X-ray crystallography
Co-crystals of SHP2:IACS-13909 were generated and a 2.4 Å structure solved by X-ray crystallography. Details for crystal generation, structure determination and data analysis are in supplementary information.
Cell culture and generation of engineered lines
All cell lines unless specified were obtained from an internal cell bank, which conducted STR finger printing and PCR-based mycoplasma test on all cryopreserved batches. STR finger printing was also conducted with all engineered cell lines and derivatives. Unless specified, experiments were conducted with cells that were <6 weeks after thawing.
All cells were cultured at 37 °C with 5% CO2. KYSE-520, NCI-H1975, NCI-H1975 CS, LS411N, HCC827, HCC4006 cells were cultured in RPMI1640 (Thermo Fisher) +10% FBS (Sigma). MIA PaCa-2, MV-4–11 and 293T cells were cultured in high glucose DMEM (Thermo Fisher) +10% FBS. MV-4–11-Luc cells were from the Experimental Therapeutics Core at Dana-Farber Cancer Institute, and cultured in high glucose DMEM +10% FBS+1 μg/mL puromycin.
The HCC827-ER1 cells that harbors MET amplification (Crown bioscience UK) were cultured in RPMI1640+10%FBS+42 μM erlotinib (20). The cell line was derived from HCC827 (ATCC) at Crown bioscience, by culturing the cells in the presence of escalated concentrations of erlotinib. The HCC4006-OsiR cells were generated by culturing HCC4006 cells in the presence of 1 μM osimertinib for ~3 months, and were maintained in RPMI1640+10%FBS+1 μM osimertinib.
NCI-H1975 CS cells that harbor EGFRL858R/T790M/C797S mutation were generated through CRISPR Cas9 mediated point mutation at EGFR C797 site, by Synthego Corporation. To generate these cells, ribonucleoproteins containing the Cas9 protein and synthetic chemically modified sgRNA were electroporated into the cells using Synthego’s optimized protocol. Editing efficiency was assessed upon recovery, 48 hours post electroporation. Genomic DNA was extracted from a portion of the cells, PCR amplified and sequenced using Sanger sequencing. The resulting chromatograms are processed using Synthego Inference of CRISPR edits software (ice.synthego.com).
KYSE-520 cells stably overexpressing SHP2 WT or SHP2 P491Q were generated by infecting the parental cells with concentrated lentiviruses, and cultured in the media of the parental cells with 1 μg/ml puromycin (Thermo Fisher). The cells were split whenever needed, cultured for ~2 weeks, and frozen down for future experiments.
Cell proliferation assays
In vitro clonogenic assays were conducted with adherent lines plated in 12-well or 24-well plates, treated for 2 weeks, and stained with crystal violet. Ex vivo spheroid proliferation assay was conducted with cells freshly isolated from PDX, plated in U-bottom ultra-low attachment 96-well plates (Corning) without matrix, and treated for 6 days. Detailed procedures are described in supplementary information.
RNA sequencing (QuantSeq) and data analysis
RNA libraries were prepared with the QuantSeq 3’ mRNA-Seq FWD Kit (Lexogen), following the vendor’s standard protocols. Briefly, libraries were generated with 500 ng total RNA input and 11 cycles of PCR amplification of the cDNA. Batches of up to 40 samples were multiplexed and each batch was run on NextSeq 500 (Illumina) using the High Output Kit v2 (Illumina).
Sample analyses were conducted using R Bioconductor. Transcript compatibility counts were obtained with kallisto (v0.44.0) (21) running the pseudo mode with GENCODE 23 transcript annotations (22). Gene counts were obtained by summing all reads that uniquely mapped, and differential expression analysis was carried out using DESeq2’s (23) default settings. Heatmaps were generated in GraphPad Prism 8.0.
Mouse studies
All in vivo work was either approved by the Institutional Animal Care and Use Committee (IACUC) of MD Anderson Cancer Center or by the relevant committee of the testing facility. Female mice were used, and body weight was typically 20~28 g when treatment started.
All subcutaneous models were implanted with 50% Matrigel (Corning). Cell numbers and mouse strains are: KYSE-520, 3 million in NSG (Jackson); NCI-H1975, 1 million in CD-1 nude (Charles River); NCI-H1975 CS, 3 million in NSG; HCC827 and HCC827-ER1, 5 million in Athymic nude (Envigo). Tumor size was measured with caliper and calculated using a standard formula: length × width2/2. Dosing volume was 10 mL/kg/day. IACS-13909 was formulated in 0.5% methylcellulose, and osimertinib/erlotinib in 0.5% HPMC. For the combination studies, IACS-13909 was dosed in the morning, and osimertinib was dosed in the afternoon, with a 6-hour interval in between. In order to pool tumor measurements from independent experiments, biweekly measurements differ by one day across studies are considered as at the same timepoint.
For studies with the MV-4–11 orthotopic model (Experimental therapeutics core, Dana-Farber Cancer Institute), NSG mice were implanted with 2 million MV-4–11-Luc cells (250 μL) intravenously. Mice were enrolled into treatment groups using total flux bioluminescence value, two days post-implantation. After dosing ended, all animals were monitored for survival, and euthanized once morbidity and/or stage 3 paralysis was observed.
Data plotting and statistical analysis
Unless specified, data plotting and statistical analysis was conducted using GraphPad Prism 8.0. Graph with error bars represent mean ± SEM.
Results
IACS-13909 is a potent and selective allosteric inhibitor of SHP2
In order to discover novel SHP2 inhibitors with drug-like properties, we utilized structure-based design principles starting from known SHP2 allosteric inhibitors, and identified IACS-13909 (Figure 1A). In an in vitro enzymatic assay, IACS-13909 potently suppressed the phosphatase activity of purified full-length, recombinant human SHP2 protein, with an IC50 of ~15.7 nM (Figure 1B). In comparison, in a similar assay using the SHP2 phosphatase domain, IACS-13909 did not suppress phosphatase activity at concentrations up to 50,000 nM, the highest concentration tested (Figure 1C), suggesting IACS-13909 acts outside the phosphatase domain. The Kd of IACS-13909 binding to SHP2 is ~ 32 nM, as determined by isothermal titration calorimetry analysis (Figure S1A). IACS-13909 is highly selective for SHP2. When tested at 10 μM against a panel of 22 phosphatases, the compound only showed significant inhibition of SHP2 (>50% inhibition) (Table S1). It is notable that IACS-13909 demonstrated no inhibition of full-length SHP1, the phosphatase that is structurally most similar to SHP2.
Figure 1. IACS-13909 is a potent and selective allosteric inhibitor of SHP2.
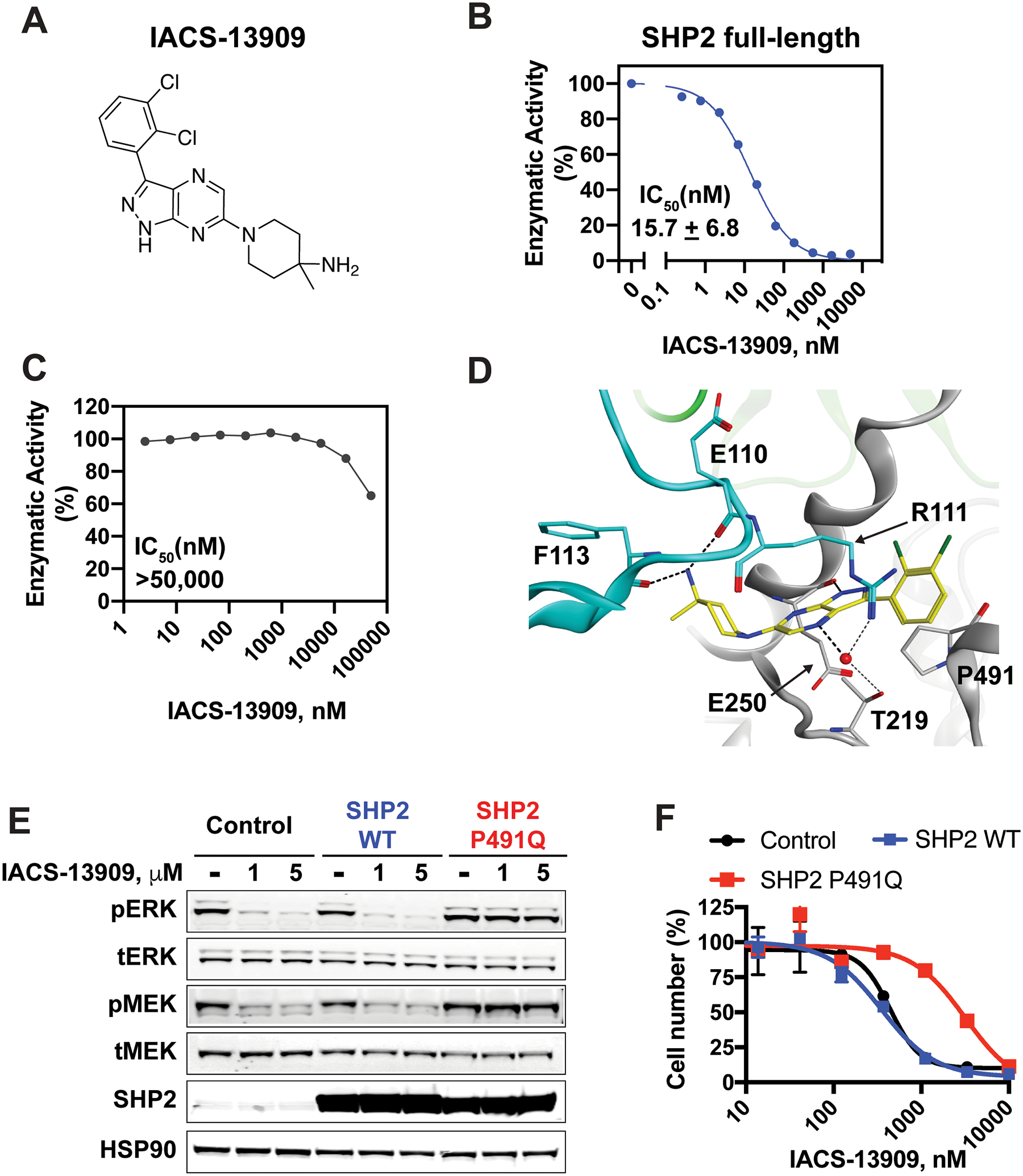
(A) The structure of IACS-13909. (B) Dose response of IACS-13909 in an in vitro enzymatic assay with purified full-length human SHP2, in the presence of 1 μM bistyrosylphorphorylated peptide. The dose response curve is from a single representative experiment. The IC50 is calculated from 59 independent tests. (C) The effect of IACS-13909 in an in vitro enzymatic assay with purified, human SHP2 phosphatase domain. N=10. Since 50% inhibition was not achieved, IC50 was defined as above the top tested concentration. (D) Crystal structure of IACS-13909 with purified human SHP2 protein, at 2.4 A°, determined by X-ray crystallography. PDB=6WU8. The phosphatase domain is highlighted in grey, C-SH2 in cyan and N-SH2 in green. (E) The impact of IACS-13909 on pERKT202/Y204 and pMEK1/2S217/221 levels in KYSE-520 cells overexpressing dsRED (control), SHP2 WT or SHP2 P491Q mutant. Cells were treated with IACS-13909 for 2 hours and processed for Western blotting. (F) The in vitro anti-proliferative effect of IACS-13909 on cells used in E, determined by a 14-day clonogenic assay. N=2 from the same experiment. This experiment has been repeated with another allosteric SHP2 inhibitor with similar observation.
To elucidate where IACS-13909 interacts with SHP2 protein, we solved the crystal structure of SHP21–530 with IACS-13909 at 2.40 Å resolution with Rfree of 0.270 (PDB=6WU8) (Figure 1D and Table S2). The refined structure contains two protomer chains of SHP21–530 and two molecules of IACS-13909 in the asymmetric unit. The crystal structure confirmed that the compound binds outside the active site, at the interface between the phosphatase domain (grey) and the C-terminal SH2 domain (cyan), a key allosteric pocket of the protein (24) and stabilizes the inactive state of the enzyme. Key hydrogen bond interactions were observed between the backbone carbonyls of Glu110 and Phe113 and the basic amine group of IACS-13909 as well as between the backbone carbonyl of Glu250 and the pyrazol N-H of the compound. A water molecule bridges between the sidechains of Thr219 and Arg111 and the pyrazine core of the compound and we observe cation-∏ stacking interactions between the Arg111 sidechain and the dichlorobenzene of IACS-13909. Together, these data confirm that IACS-13909 is a direct allosteric inhibitor of SHP2.
IACS-13909 inhibits the proliferation and MAPK pathway signaling of tumor cell lines driven by a broad spectrum of RTKs in vitro
Since SHP2 is a critical mediator of oncogenic signaling, a SHP2 inhibitor might be useful as an anti-cancer agent (2). We evaluated the in vitro anti-proliferative effect of IACS-13909 in a panel of 283 cancer cell lines with diverse genomic drivers, using a 10-day two-dimensional proliferation assay. Among the exceptional responder lines (with GI50 ≤ 1 μM), many harbored genetic alterations of RTK or were RTK-addicted (sensitive to TKI or RTK shRNA according to DRIVE) (25). Particularly, all six cell lines with GI50 ≤ 100 nM harbored RTK alterations - DK-MG (EGFR vIII+), BV-173 (BCR-ABL), KG-1 (OP2-FGFR1), KU812 (BCR-ABL), SW-13 (ERBB4-IKZF2) (26) and MV-4–11 (FLT3-ITD) (Figure S1B). In addition, BRAF V600 mutation appears to be a predictor of IACS-13909 resistance, with 19 out of 23 BRAF V600 mutated cell lines having GI50 > 5 μM. Consistent with the proliferation data, IACS-13909 suppressed pERK in RTK-dependent lines, such as KYSE-520 (EGFRamp) (Figure S1C), MV-4–11 (FLT3-ITD) (Figure S1D), but did not suppress pERK or pMEK in LS411N cells harboring BRAFV600E (Figure S1E). It is noteworthy that majority of KRASmut cell lines in this analysis were resistant to IACS-13909 (Figure S1B), likely due to low coverage of cell lines expressing the nucleotide-cycling KRAS G12C mutant in this panel and limitations of the two-dimensional culture system in evaluating KRASmut cancers. Together, these data demonstrate the anti-tumor activity of IACS-13909 in cancer cell lines harboring a broad spectrum of activated RTKs, consistent with literature (24).
IACS-13909 inhibits the proliferation and MAPK pathway signaling in RTK-activated cancer cells in vitro due to on-target SHP2 inhibition
To ensure the anti-proliferative effect of IACS-13909 in RTK-activated cancer cell lines in vitro was due to inhibition of SHP2, we leveraged the SHP2 Proline 491 to Glutamine (P491Q) mutant. Based on the X-ray crystal structure, Pro491 lines the allosteric binding pocket of SHP2 adjacent to the pyrazolopyrazine ring of IACS-13909. Sequence and structural alignment with SHP1 (PDB=3PS5) showed Glutamine 485 in this position in SHP1 and suggested that a P491Q mutation will abolish IACS-13909 binding to SHP2 due to steric clashes with the glutamine side chain (Figure 1D) but should still yield a catalytically competent protein (8). Therefore, we stably overexpressed dsRed (control), SHP2 WT or SHP2 P491Q in the KYSE-520 cells, an EGFRamp esophageal cancer cell line. Western blotting showed that exogenous SHP2 was expressed at a much higher level than endogenous SHP2 (Figure 1E). In control cells or cells overexpressing SHP2 WT, IACS-13909 potently suppressed pERK and pMEK levels, but not in cells overexpressing SHP2 P491Q. Similarly, in an in vitro clonogenic assay, whereas IACS-13909 potently suppressed the proliferation of control cells and cells overexpressing SHP2 WT (GI50 <1 μM), overexpression of SHP2 P491Q significantly reduced sensitivity of IACS-13909 (with 7.8-fold shift in IACS-13909 GI50) (Figure 1F). The rescue is specific to SHP2 inhibitor, because EGFR inhibitor erlotinib demonstrated identical sensitivity in control, SHP2 WT and SHP2 P491Q overexpressing cells (Figure S1F and S1G). It is noteworthy that SHP2 P491Q overexpression did not confer complete resistance of KYSE-520 cells to IACS-13909; this is likely because the presence of endogenous SHP2 in the mutant overexpressing KYSE-520 cells that confers some signaling through the MAPK pathway. Together, these data suggest that IACS-13909 suppresses cell proliferation and signaling through the MAPK pathway in RTK-dependent tumor cells due to its inhibitory effect on SHP2.
In vivo anti-tumor activity of IACS-13909 in models driven by a broad spectrum of RTKs
In order to determine the activity of IACS-13909 in vivo, we first evaluated the pharmacokinetic (PK) properties of IACS-13909 in mice, rats and dogs. IACS-13909 demonstrated >70% bioavailability (%F), low clearance rate (Cl), and half-lives of ≥7 hours across species, suggesting that the compound is suitable for once per day (QD) oral dosing (Table S3).
We selected two RTK-dependent cell lines for in vivo evaluation – the EGFRamp esophageal cancer cell line KYSE-520 as a representative solid tumor cell line, and the FLT3-ITD+ acute myeloid leukemia (AML) cell line MV-4–11 as a representative blood cancer cell line. In mice with established subcutaneous KYSE-520 tumors, IACS-13909 dosed orally at 70 mg/kg QD potently suppressed tumor growth, with 100% tumor growth inhibition (TGI) observed following 21 days of dosing (Figure 2A). Importantly, the treatment was well-tolerated, with body weight maintained throughout the study (Figure 2B). A higher dose of IACS-13909, such as 100 mg/kg QD was not tolerated in mice, suggesting 70 mg/kg QD is approximately the maximally tolerated dose of IACS-13909 in mice.
Figure 2. IACS-13909 suppresses proliferation and MAPK pathway signaling of RTK-activated tumors in vivo.
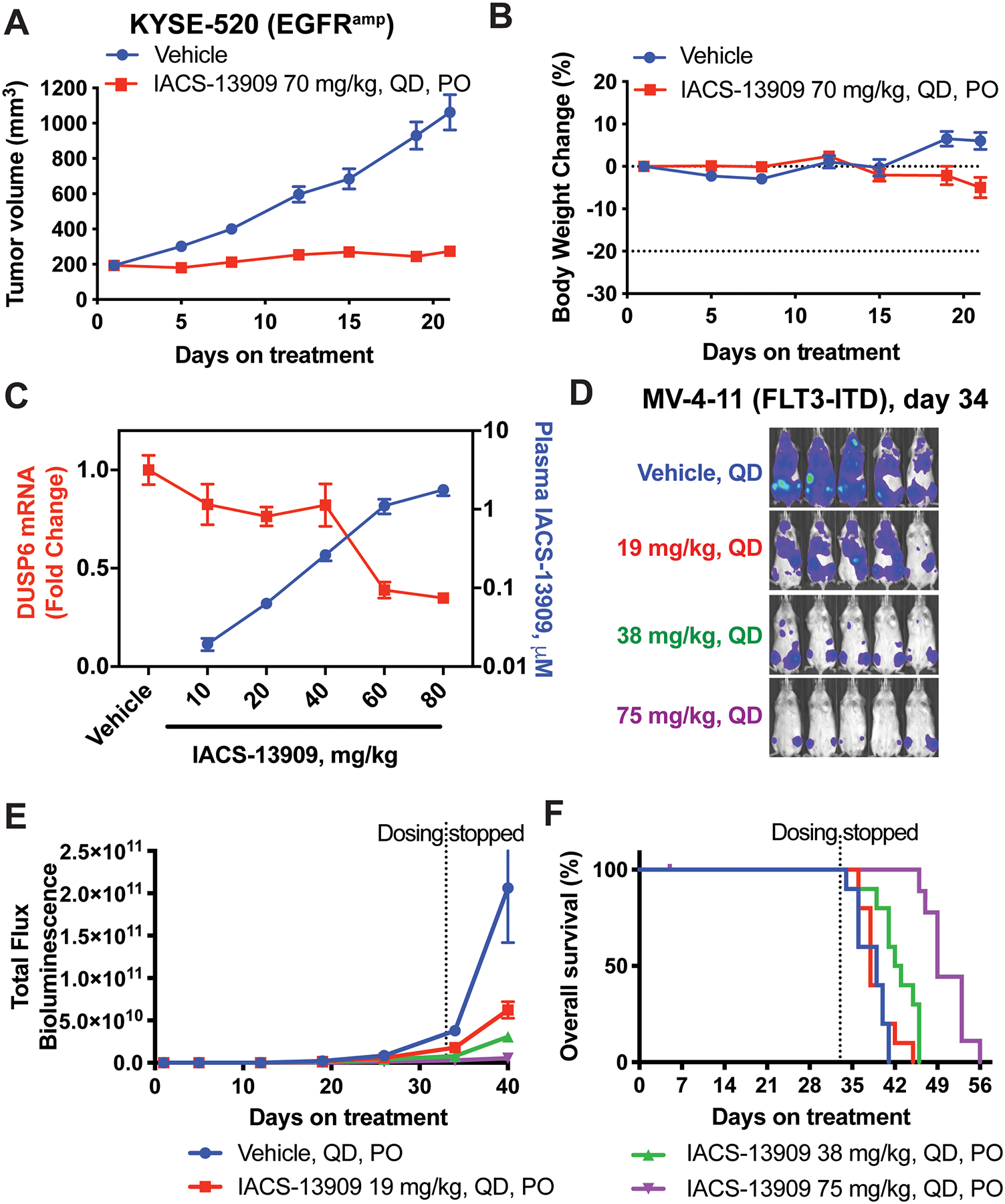
Tumor growth curve (A) and mouse body weight change (B) of the KYSE-520 subcutaneous xenograft model in mice, when treated with either vehicle (0.5% methylcellulose) or IACS-13909 at 70 mg/kg QD orally for 21 days. N=9 mice per group. (C) Plasma concentration of IACS-13909 (blue curve) and DUSP6 mRNA level in KYSE-520 subcutaneous tumor samples (red curve) from mice treated with vehicle or IACS-13909. Plasma and tumor samples were harvested 24 hours after a single dose treatment. N=3 mice/group/timepoint. (D-F) Anti-tumor efficacy of IACS-13909 on MV-4–11 orthotopic mouse model. Mice were injected with MV-4–11-Luc cells through tail vein, and treated with different doses of IACS-13909 QD orally. N=10 mice/group. (D) Representative mouse images from bioluminescence imaging indicating tumor volume on day 34. (E) Quantitated tumor volume determined by bioluminescence imaging. (F) Kaplan-Meier curve showing the overall survival of the mice with or without IACS-13909 treatment. The dotted vertical line indicates when dosing stopped.
To confirm that the in vivo anti-tumor efficacy by IACS-13909 was due to SHP2 inhibition, we analyzed KYSE-520 tumors and blood from mice treated with different dosing levels of IACS-13909. The mRNA levels of DUSP6, an ERK-dependent gene, was used as a readout of SHP2 activity and MAPK pathway signaling in tumors (27). IACS-13909 achieved dose-dependent plasma exposure at 24 hours after a single dose treatment, and demonstrated dose-dependent suppression of DUSP6 transcript levels in KYSE-520 tumors (Figure 2C). An inverse correlation between tumor DUSP6 mRNA level and plasma concentration was observed. Among the doses tested, IACS-13909 at 60 mg/kg or 80 mg/kg that span the dose that results in tumor stasis in this model (Figure 2A), maintained DUSP6 mRNA suppression at >50% throughout the 24-hour dosing interval. These data demonstrate that IACS-13909 potently suppresses MAPK pathway signaling and inhibits growth of an RTK-dependent subcutaneous solid tumor model in vivo.
We further tested the in vivo anti-tumor efficacy of IACS-13909 in the FLT3-ITD+ MV-4–11 leukemia orthotopic model. Mice were implanted with MV-4–11 cells expressing luciferase through tail vein injection, and systemic tumor growth was rapidly established. Mice were randomized based on tumor luminescence levels, and then treated with IACS-13909 at different dosing levels for five weeks. Dose-dependent suppression of systemic tumor burden was observed (Figures 2D and 2E), with IACS-13909 75 mg/kg QD causing nearly 100% TGI. Importantly, consistent with the suppression of tumor burden, IACS-13909 extended the overall survival of the mice in a dose-dependent manner (Figure 2F). These data demonstrate the dose-dependent anti-tumor efficacy of IACS-13909 in an RTK-dependent disseminated leukemia model.
IACS-13909 demonstrates anti-tumor activity in tumors harboring EGFR-dependent resistance mutation in vitro and in vivo
Multiple EGFR TKIs (e.g., erlotinib, gefitinib, osimertinib) are currently approved in the US for the front-line treatment of patients with EGFR-activated metastatic NSCLC (13). Most patients on EGFRi treatment will ultimately experience disease progression, with acquired resistance being a major clinical challenge. EGFR mutations in the proximity of the compound binding site (e.g., T790M for erlotinib/gefitinib, C797S or L792H for osimertinib) that preclude drug binding are clinically observed resistance mechanisms (14,15). Considering that tumors with an EGFR resistance mutation still depend on EGFR and also that SHP2 is a critical mediator of EGFR signaling, we hypothesized that IACS-13909 might have activities in these tumors.
To evaluate the effect of SHP2 inhibition on cancer cells harboring EGFR resistance mutations, we used NSCLC NCI-H1975 cells that harbor both an EGFR activating mutation (L858R) and resistance mutation (T790M). The NCI-H1975 cells are resistant to erlotinib and sensitive to osimertinib (12). In addition, we generated the NCI-H1975 CS cells in which EGFR C797S mutation was introduced through the CRISPR-cas9 technology (Figure S2). The NCI-H1975 CS cells demonstrated significantly reduced sensitivity towards osimertinib in an in vitro clonogenic assay, compared with the parental cells (Figure 3A). Importantly, IACS-13909 potently suppressed the proliferation of both the parental cells and NCI-H1975 CS cells in a dose-dependent manner, with similar potency (GI50 ~ 1 μM; Figure 3B). Consistent with the proliferation data, osimertinib at up to 300 nM failed to suppress the levels of pERK or pEGFR in NCI-H1975 CS cells (Figure 3C), although in the parental cells osimertinib 10 nM is sufficient for potent suppression of pERK and pEGFR (12). Unlike osimertinib, IACS-13909 suppressed pERK in NCI-H1975 CS cells in a dose-dependent manner. As expected, treatment with IACS-13909 did not reduce pEGFR since SHP2 is downstream of EGFR (Figure 3C). Together, these data demonstrate that IACS-13909 suppresses the proliferation and MAPK pathway signaling in osimertinib-resistant cells harboring an EGFR-dependent resistance mutation in vitro.
Figure 3. IACS-13909 suppresses the proliferation and MAPK pathway signaling of EGFR TKI-resistant EGFRmut NSCLC models harboring an EGFR-dependent resistance mutation.
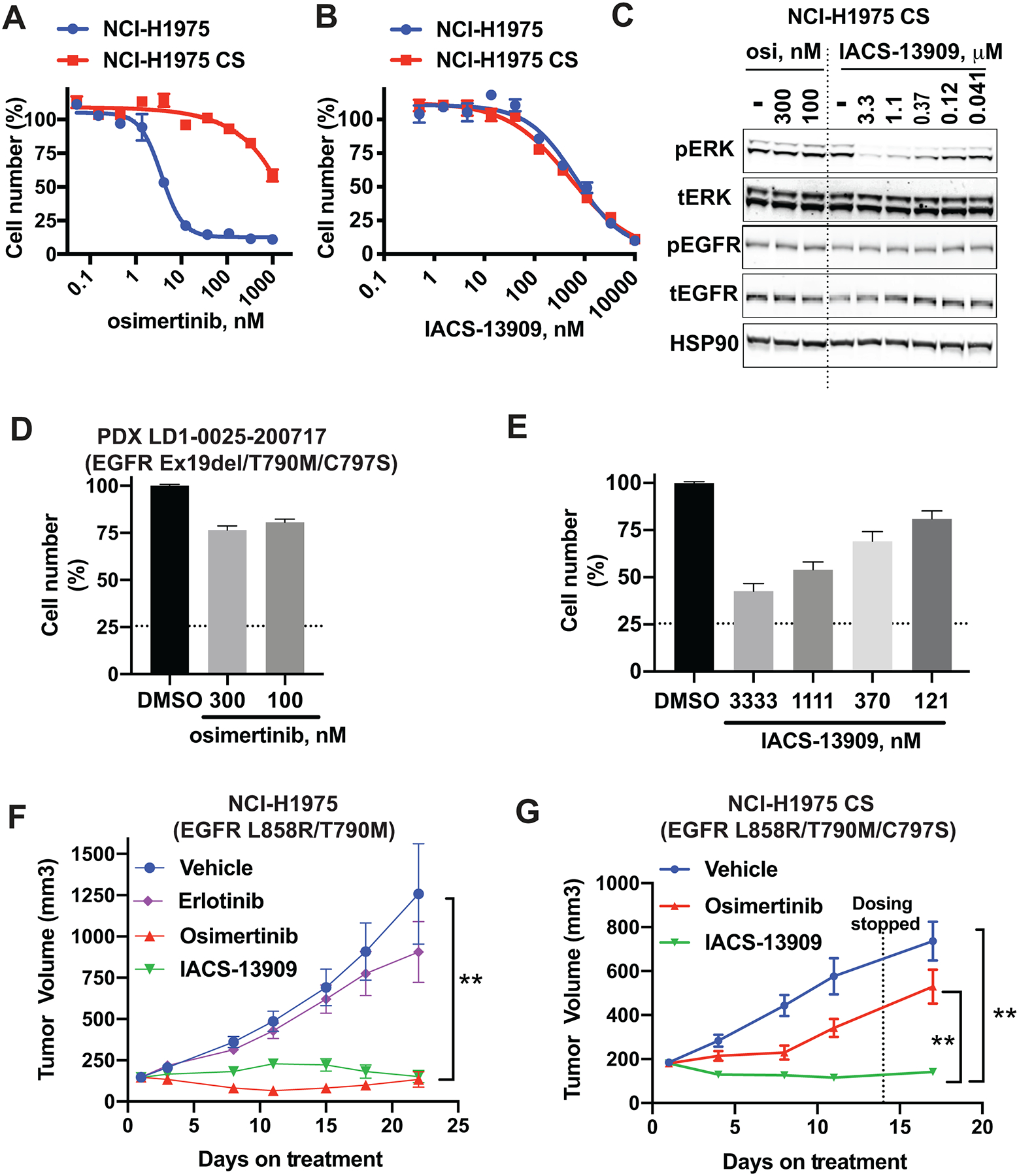
Anti-proliferative activity of osimertinib (A) and IACS-13909 (B) in NCI-H1975 parental and NCI-H1975 CS cells, determined by a 14-day clonogenic assay. The NCI-H1975 parental cells harbor EGFRL858R/T790M, and the NCI-H1975 CS cells harbor EGFRL858R/T790M/C797S, where the C797S mutation on EGFR was introduced through CRISPR. N=3. Confirmation of the C797S mutation was provided in Figure S2. (C) The impact of osimertinib or IACS-13909 on pERK1/2 T202/Y204 and pEGFRY1068 levels in NCI-H1975 CS cells in vitro. Cells were treated with IACS-13909 or osimertinib for two hours and processed for Western blotting. (D-E) Anti-proliferative activity of osimertinib (D) and IACS-13909 (E) on primary cells isolated from NSCLC PDX LD1-0025-200717 harboring EGFRex19del/T790M/C797S, determined by a 6-day ex vivo spheroid assay. The dotted horizontal line indicates the relative viable cell number when compound was added. N=4. (F) Tumor growth curve of the NCI-H1975 parental subcutaneous xenograft model in mice, when treated with either vehicle, erlotinib 10 mg/kg QD, osimertinib 5 mg/kg QD or IACS-13909 at 70 mg/kg QD orally, for 21 days. N=10 mice per group. 2-way ANOVA was used to compare the growth curve of IACS-13909-treated tumors vs vehicle treated tumors. **, p<0.01. (G) Tumor growth curve of the NCI-H1975 CS subcutaneous xenograft model in mice, when treated with either vehicle, osimertinib 5 mg/kg QD or IACS-13909 at 70 mg/kg QD orally, for 12 days. The dotted vertical line denotes the final dose. N=10 mice per group. 2-way ANOVA was used to compare the growth curve of IACS-13909-treated tumors vs osimertinib-treated tumors and vehicle-treated tumors. **, p<0.01.
To confirm the activity of IACS-13909 in human primary cancer cells, we used the LD1-0025-200717 model, which is a patient derived xenograft model (PDX) established from the hydrothorax of a NSCLC patient who progressed on treatment with osimertinib. The tumor model harbors EGFRex19del/T790M/C797S, and is resistant to erlotinib, osimertinib and the combination of the two agents in vivo (28). To rapidly assess the anti-proliferative effect of IACS-13909 in this model, we conducted an ex vivo spheroid proliferation assay with cells freshly isolated from tumors grown in mice. As expected, osimertinib treatment at 300 nM, a concentration that is approximately 40-fold higher than the GI50 of osimertinib in NCI-H1975 cells that harbor EGFRL858R/T790M (Figure 3A), had little impact on proliferation of the LD1-0025-200717 spheroids ex vivo (Figure 3D), confirming that this model is resistant to osimertinib. In contrast, IACS-13909 demonstrated dose-dependent suppression of proliferation of the LD1-0025-200717 spheroids ex vivo, with GI50 ~ 1 μM (Figure 3E), similar to the GI50 of IACS-13909 in the NCI-H1975 cells. These data demonstrate the activity of IACS-13909 in primary cells derived from an osimertinib-resistant EGFRex19del/T790M/C797S PDX ex vivo.
To determine the in vivo activity of IACS-13909 in tumors harboring an EGFR-dependent resistance mutation, we tested IACS-13909 in the NCI-H1975 parental and NCI-H1975 CS subcutaneous xenograft models in mice. As expected, in the NCI-H1975 parental tumor harboring EGFRL858R/T790M, erlotinib treatment at 10 mg/kg QD delivered orally failed to suppress tumor proliferation, and treatment with osimertinib at 5 mg/kg QD caused regression of the established tumor. Treatment with IACS-13909 at 70 mg/kg QD demonstrated robust anti-tumor efficacy, with tumor regression observed (Figure 3F). In mice bearing the NCI-H1975 CS tumors harboring EGFRL858R/T790M/C797S (Figure 3G), treatment with osimertinib demonstrated little anti-tumor efficacy, which was distinct from the response observed in the parental tumors, confirming that this model is resistant to osimertinib in vivo. Importantly, treatment with IACS-13909 at 70 mg/kg QD also demonstrated robust anti-tumor efficacy in the NCI-H1975 CS model, with tumor regression observed. Together, these data demonstrate robust activity of IACS-13909 in osimertinib-resistant tumors harboring an EGFR-dependent resistance mutation in vivo.
In cells harboring EGFR-independent resistance mechanisms, IACS-13909 single agent or in combination with osimertinib suppresses proliferation and MAPK pathway signaling in vitro
Beyond EGFR-specific mutations, a major resistance mechanism observed with multiple generations of EGFR inhibitors is RTK-bypass, i.e., the compensatory activation of alternate RTKs that maintains downstream activation of the MAPK pathway with EGFR inhibited (14,29–31). Prompted by the anti-proliferative effect of IACS-13909 in many RTK-activated human cancer cell lines, we tested the activity of IACS-13909 in EGFRmut NSCLC cells with RTK-bypass. We generated the HCC4006-osimertinib resistant (OsiR) model by culturing HCC4006 cells harboring EGFRex19del in the presence of 1 μM osimertinib for an extended period of time (~3 months), and confirmed the reduced osimertinib sensitivity in the OsiR derivative (Figure 4A). We further demonstrated that the HCC4006-OsiR cells did not harbor resistance mutations in EGFR (i.e., T790M or C797S etc) and had decreased pEGFR compared with the parental cells, suggesting that these cells were switching to other oncogenic drivers (Figure 4B). Indeed, the HCC4006-OsiR cells demonstrated increased expression of multiple RTKs, including FGFR1, Axl, PDGFR, and IGF1Rβ. The OsiR cells had also undergone EMT, with decreased expression of epithelial marker E-cadherin and increased expression of mesenchymal markers vimentin and Zeb1 (Figure 4B).
Figure 4. IACS-13909 suppresses the proliferation and MAPK pathway signaling of osimertinib-resistant EGFRmut NSCLC cells harboring RTK-bypass in vitro.
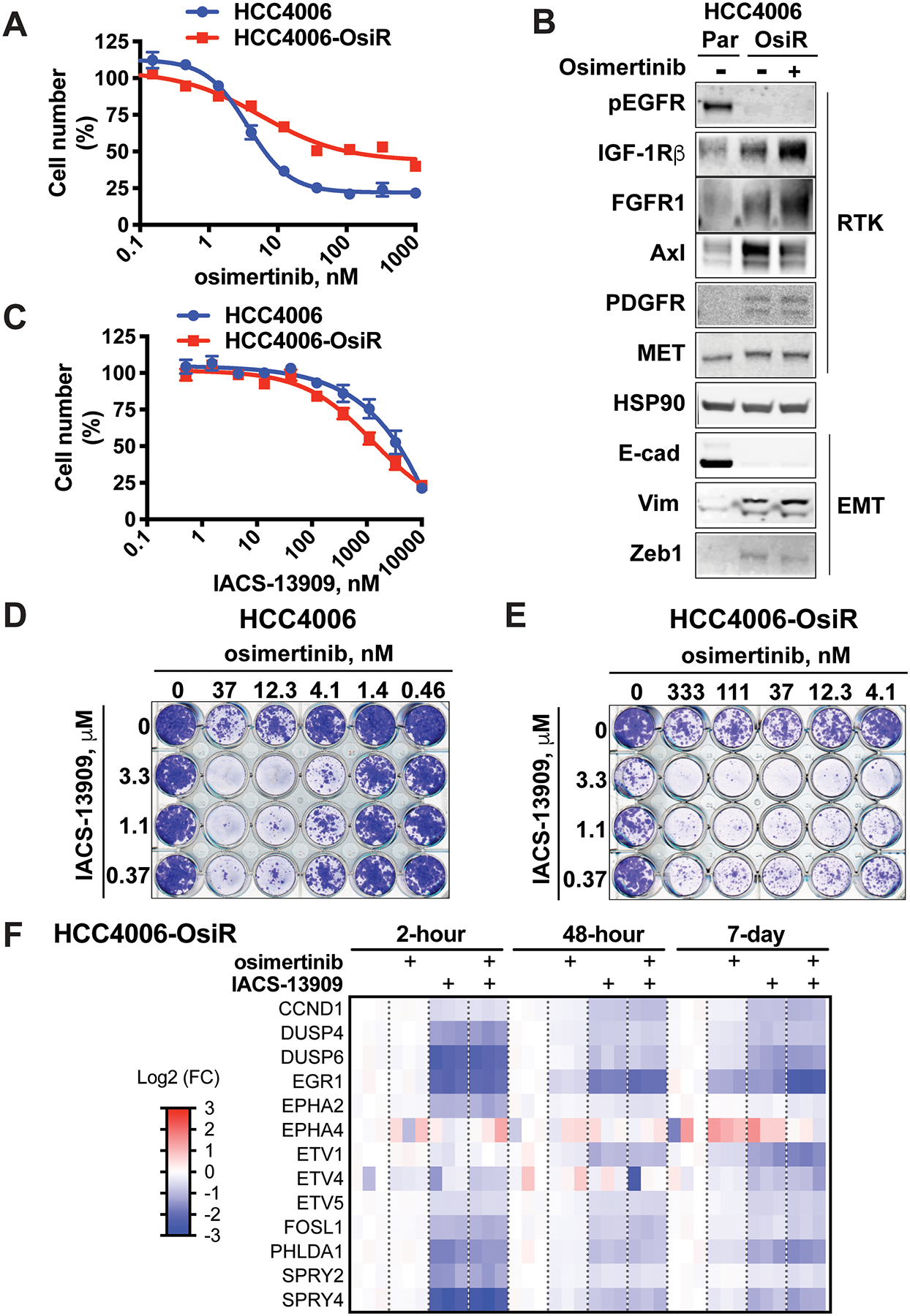
(A) Activity of osimertinib in EGFRmut HCC4006 and HCC4006-OsiR models, determined by clonogenic assays. N=3. (B) The level of various RTKs and EMT markers in HCC4006 parental (Par) and OsiR models. The OsiR models were maintained in the presence of 1 μM osimertinib. To generate samples for signaling analysis, cells were cultured in the absence of osimertinib for one day, then treated with DMSO or 1 μM osimertinib for 2 hours before being harvested and processed for Western blotting. (C) Activity of IACS-13909 in EGFRmut HCC4006 and HCC4006-OsiR models, determined by in vitro clonogenic assays. N=4~8. (D-E) Anti-proliferative activity of IACS-13909 in combination with osimertinib in HCC4006 (D) and HCC4006-OsiR (E) cell lines, determined by in vitro clonogenic assays. Percent of inhibition, calculated from quantitated cell number is provided in Figures S3A and S3B. Bliss score calculation is provided in Figure S3C and S3D. N=4. (F) Heatmap from gene expression analysis showing the impact of osimertinib (100 nM) and IACS-13909 (3 μM) either as single agent or in combination on a MAPK signature (MPAS-plus signature) in HCC4006-OsiR cells over a timecourse in vitro. N=3.
We conducted in vitro proliferation assays with HCC4006 parental and HCC4006-OsiR cells, which were treated with IACS-13909 either as a single agent or in combination with osimertinib. Despite the reduced osimertinib sensitivity observed in the OsiR cells, IACS-13909 showed comparable single agent anti-proliferative effect in the parental and OsiR cells in clonogenic assays (Figure 4C). Importantly, treatment with the combination of IACS-13909 and osimertinib resulted in a synergistic anti-proliferative effect in both models (Figures 4D, 4E, S3A and S3B), with positive bliss scores in the majority of the concentrations tested (Figures S3C and S3D).
To understand the mechanism of action underlying the anti-proliferative effect, signaling analysis was conducted in HCC4006-OsiR cells treated with osimertinib and/or IACS-13909 in vitro. The MPAS (MAPK pathway activity score) signature is composed of 10 genes that reflects MAPK pathway activity (27). Based on the MPAS signature, a 13-gene signature (“MPAS-plus”) was developed, which includes three additional MAPK-targeted genes (ETV1, EGR1 and FOSL1) (32,33). Osimertinib alone failed to potently suppress MAPK pathway signaling in the HCC4006-OsiR cells, as demonstrated by lack of suppression of DUSP6 mRNA levels and other MPAS-plus genes. In contrast, IACS-13909 potently suppressed MAPK pathway signaling, both as a single agent and in combination with osimertinib (Figure 4F). The suppression was achieved with 2-hour, 48-hour and 7-day treatment of IACS-13909, suggesting sustained suppression of MAPK pathway, despite the observed partial adaptation (less suppression with prolonged treatment compared with acute treatment). This is consistent with the notion that SHP2 inhibition suppresses the signaling downstream of multiple RTKs, therefore delaying the multi-RTK mediated rapid adaptation that is commonly observed towards MAPK pathway inhibitors (34–36). It is also noteworthy that treatment with the combination of IACS-13909 and osimertinib did not cause further suppression of the MAPK pathway compared with IACS-13909 single agent in vitro, suggesting potential additional non-MAPK mediated mechanisms for the synergistic anti-proliferative effect between osimertinib and IACS-13909 in the in vitro setting.
Combination of IACS-13909 and osimertinib extends the durability of osimertinib response in osimertinib-sensitive tumors, and causes tumor regression in osimertinib-resistant tumors with RTK-bypass
Our in vitro data with IACS-13909 either as single agent or in combination with osimertinib in the HCC4006-OsiR model that harbors EGFR-independent resistance mechanisms prompted us to conduct further evaluation in vivo. The osimertinib-resistant EGFRmut NSCLC HCC827-ER1 cells were generated by exposing HCC827 cells, which harbor an EGFR activating mutation (EGFRex19del), to erlotinib in culture (20). The HCC827-ER1 cells do not harbor EGFR-dependent resistance mutations, but do have amplified c-MET, a genetic alteration observed in tumors from patients who have relapsed on erlotinib and osimertinib (14,29). The HCC827-ER1 cells are resistant to erlotinib and also to osimertinib (20).
In the osimertinib-sensitive HCC827 xenograft model (Figure 5A), IACS-13909 70 mg/kg dosed daily as a single agent potently suppressed tumor growth, leading to tumor stasis, and osimertinib dosed as a single agent at 5 mg/kg QD caused robust tumor regression. As expected, treatment with the combination of IACS-13909 and osimertinib yielded tumor regression, similar to that observed with osimertinib alone, during the period of compound administration. However, following cessation of dosing, tumors in mice treated with the combination did not grow whereas those treated with osimertinib showed significant growth beginning ~30 days after the final dose. Importantly, the combination treatment in HCC827 xenograft model was tolerated, as shown by the maintenance of body weight during the study (<10% average body weight loss; Figure S4A). Thus, treatment with the combination of IACS-13909 and osimertinib resulted in a more durable anti-tumor response (Figure 5A), consistent with the in vitro observation in the HCC4006 cells (Figure 4D).
Figure 5. Anti-tumor efficacy of treatment with IACS-13909 and osimertinib, alone and in combination, in a MET-amplified EGFRi acquired resistant model in vivo.
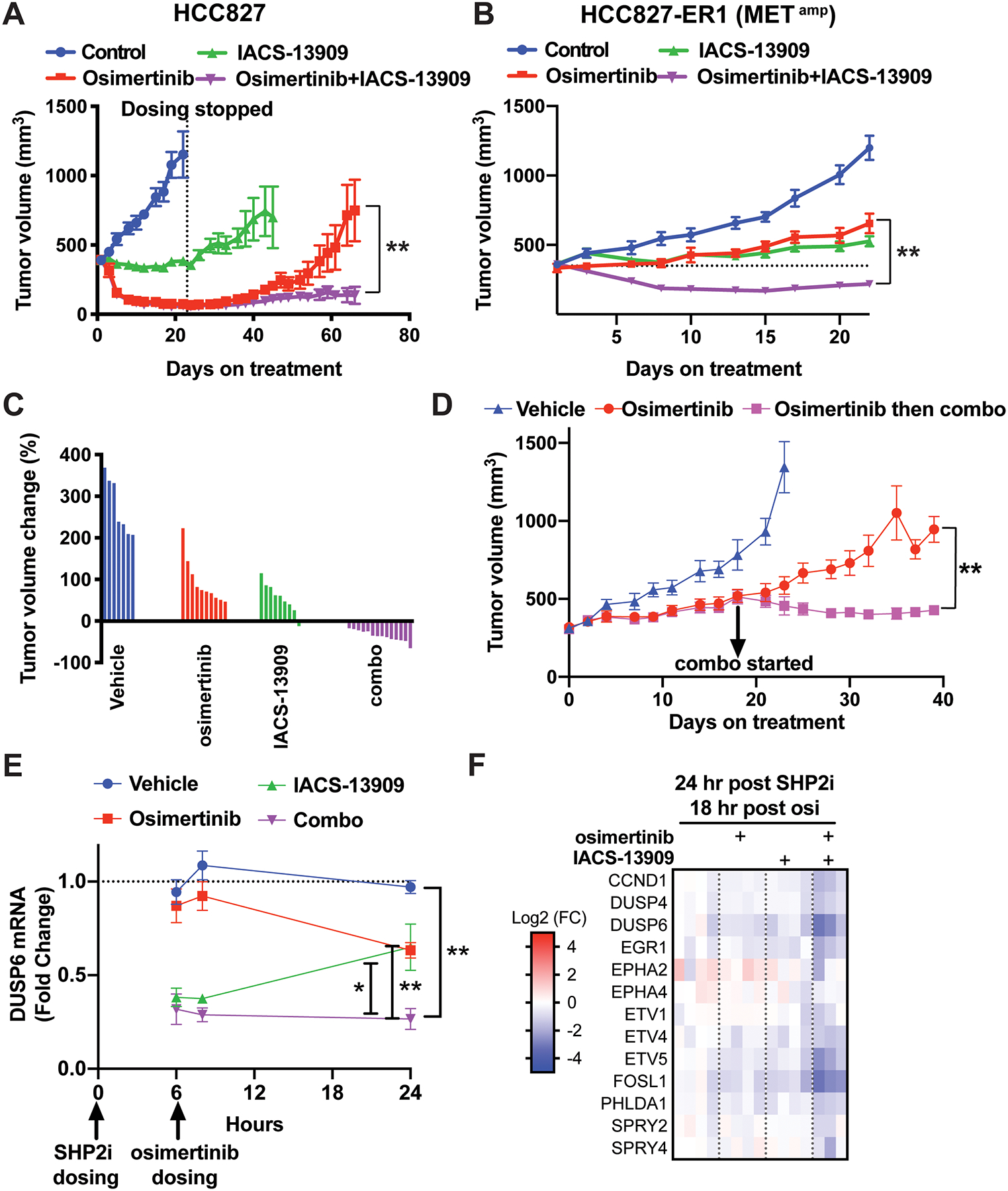
Tumor growth curves of EGFRmut HCC827 (A) and HCC827-ER1(B) xenograft models treated with vehicle (0.5% methylcellulose, QD+0.5% HPMC, QD), osimertinib (0.5% methylcellulose, QD + osimertinib 5 mg/kg, QD), IACS-13909 (IACS-13909 70 or 80 mg/kg, QD+0.5% HPMC, QD), or the combination (IACS-13909 70 or 80 mg/kg, QD + osimertinib 5 mg/kg, QD). In A, n=10 mice per group. In B, n≥10 mice for all groups. In both A and B, the graphs represent pooled data from two independent experiments. In one experiment, IACS-13909 was used at 70 mg/kg, and in the other IACS-13909 was used at 80 mg/kg. 2-way ANOVA was used to compare the tumor growth curves of osimertinib single agent group vs the combination group. **, p<0.01. (C) Relative tumor volume change on day 22 from data shown in B, when dosing ended. The tumor volume of each mouse was normalized to the tumor volume when dosing started. 0 indicates tumor stasis, <0 indicates tumor regression. (D) Anti-tumor efficacy of treatment with the combination of IACS-13909+osimertinib on HCC827-ER1 tumors that outgrew on osimertinib treatment. When average tumor volume reached 300 mm3, mice bearing HCC827-ER1 tumors were treated with vehicle (n=5), or osimertinib (5 mg/kg, QD, n=10). When tumors progressed on osimertinib treatment and reached 500 mm3, the treated tumors were re-randomized and subjected to treatment with osimertinib (5 mg/kg, QD; n=5) or the combination (osimertinib 5 mg/kg, QD + IACS-13909 70 mg/kg, QD; n=5). 2-way ANOVA was used to compare osimertinib single agent group vs combination group. **, p<0.01. (E) Modulation of DUSP6 mRNA levels in HCC827-ER1 subcutaneous tumors during a 24-hour time period following one day treatment, as conducted in B. Tumor samples were harvested at hour 6, 8 and 24. DUSP6 mRNA levels were determined by qRT-PCR. N=4 mice per group for each timepoint. 2-tail t-test was conducted to compare the combination group vs vehicle or single agent groups. *, p<0.05; **, p<0.01. (F) Gene expression analysis was conducted with tumor samples harvested at the 24-hour post SHP2i/18-hour post osimertinib timepoint in E. Modulation of a MAPK-pathway signature (MPAS-plus) by osimertinib, IACS-13909 and the combination is shown.
In the osimertinib-resistant HCC827-ER1 model (Figure 5B), tumors in mice treated with IACS-13909 70 mg/kg QD demonstrated tumor stasis, similar to the response in the HCC827 parental model. However, tumors in mice treated with osimertinib continued to grow on treatment, indicating reduced osimertinib sensitivity in HCC827-ER1 as compared to the HCC827 model. Importantly, the combination treatment of IACS-13909 and osimertinib caused robust regression of the HCC827-ER1 tumor, similar to the single agent effect of osimertinib in the parental model. Waterfall plot of tumor volume demonstrated that almost all mice treated with the combination for three weeks had tumor regression (Figure 5C). The combination of osimertinib and IACS-13909 in the HCC827-ER1 xenograft model was also tolerated, as shown by the maintenance of body weight during the study (≤5% body weight loss; Figure S4B). This result demonstrates that addition of the SHP2 inhibitor IACS-13909 leads to re-sensitization of the osimertinib-resistant HCC827-ER1 model harboring RTK-bypass to treatment with osimertinib.
We further determined if combination of osimertinib and IACS-13909 can inhibit the growth of HCC827-ER1 tumors that progressed on osimertinib treatment. We started osimertinib single agent treatment when average tumor volume was ~300 mm3; the tumors progressed and average tumor volume reached ~500 mm3 (67% increase in tumor volume) within three weeks. At this time, the osimertinib-treated mice were re-randomized and enrolled into treatment with osimertinib alone or with the combination of osimertinib and IACS-13909. Whereas tumors in mice treated with osimertinib alone continued to grow, tumors in mice treated with the combination of osimertinib and IACS-13909 demonstrated significantly inhibited tumor growth, with tumor regression (Figure 5D).
To gain insight into the mechanism of the combination treatment, we analyzed HCC827-ER1 tumors from mice treated with osimertinib, IACS-13909 or the combination. Mice were dosed following the same schedule as in the efficacy study, with a 6-hour interval between IACS-13909 dosing and osimertinib dosing (Figure 5E), and samples were harvested at three time points during the 24-hour dosing cycle. In the HCC827-ER1 tumors, osimertinib treatment modestly suppressed DUSP6 mRNA levels. IACS-13909 potently suppressed DUSP6 mRNA levels at 6-hour and 8-hour after treatment, with the extent of suppression decreased afterwards. Importantly, the combined treatment with the two compounds maintained the potent suppression of DUSP6 mRNA levels throughout the 24-hour dosing cycle (Figure 5E). To further confirm the combinational effect on MAPK pathway signaling, we evaluated the MPAS-plus panel (Figure 5F). At close to dosing trough, while each monotherapy had little effect on the mRNA levels of the genes on the panel, the combination of IACS-13909 and osimertinib more potently suppressed the level of most of the MPAS-plus genes. These data suggest that combined treatment with osimertinib and IACS-13909 potently suppresses MAPK pathway signaling, to a larger extent than either single agent, in an osimertinib-resistant model with RTK-bypass, consistent with the efficacy data.
Discussion
In this study, we report the discovery of IACS-13909, a potent and selective allosteric SHP2 inhibitor. Our in vitro and in vivo data demonstrate that IACS-13909 has anti-tumor activity and suppresses MAPK pathway signaling in RTK-dependent cancers. Importantly, IACS-13909 exhibits anti-tumor efficacy in osimertinib-resistant models that harbor clinically relevant resistance mechanisms. In osimertinib-resistant tumors with EGFR-dependent resistance mutations, such as the C797S mutation in the NCI-H1975 CS cells, EGFR remains the primary oncogenic driver and signals through SHP2. Thus, although osimertinib is not able to potently suppress EGFR here, inhibition of SHP2, which lies downstream of EGFR, blocks signaling through the MAPK pathway (Figure 6A). In osimertinib-resistant tumors in which the MAPK pathway is activated due to activation of an alternate RTK, such as MET in the HCC827-ER1 cells, the alternate RTK signals through SHP2 to maintain the MAPK pathway activity (Figure 6B). Together, our preclinical data demonstrate that the SHP2 inhibitor IACS-13909 is effective in overcoming both EGFR-dependent and EGFR-independent resistance mechanisms towards osimertinib. Importantly, the ability of SHP2 inhibition in targeting multiple resistance mechanisms is anticipated to address the heterogeneity and plasticity of osimertinib resistance.
Figure 6. Proposed model for IACS-13909 in overcoming both EGFR-dependent and EGFR-independent osimertinib resistance mechanisms.
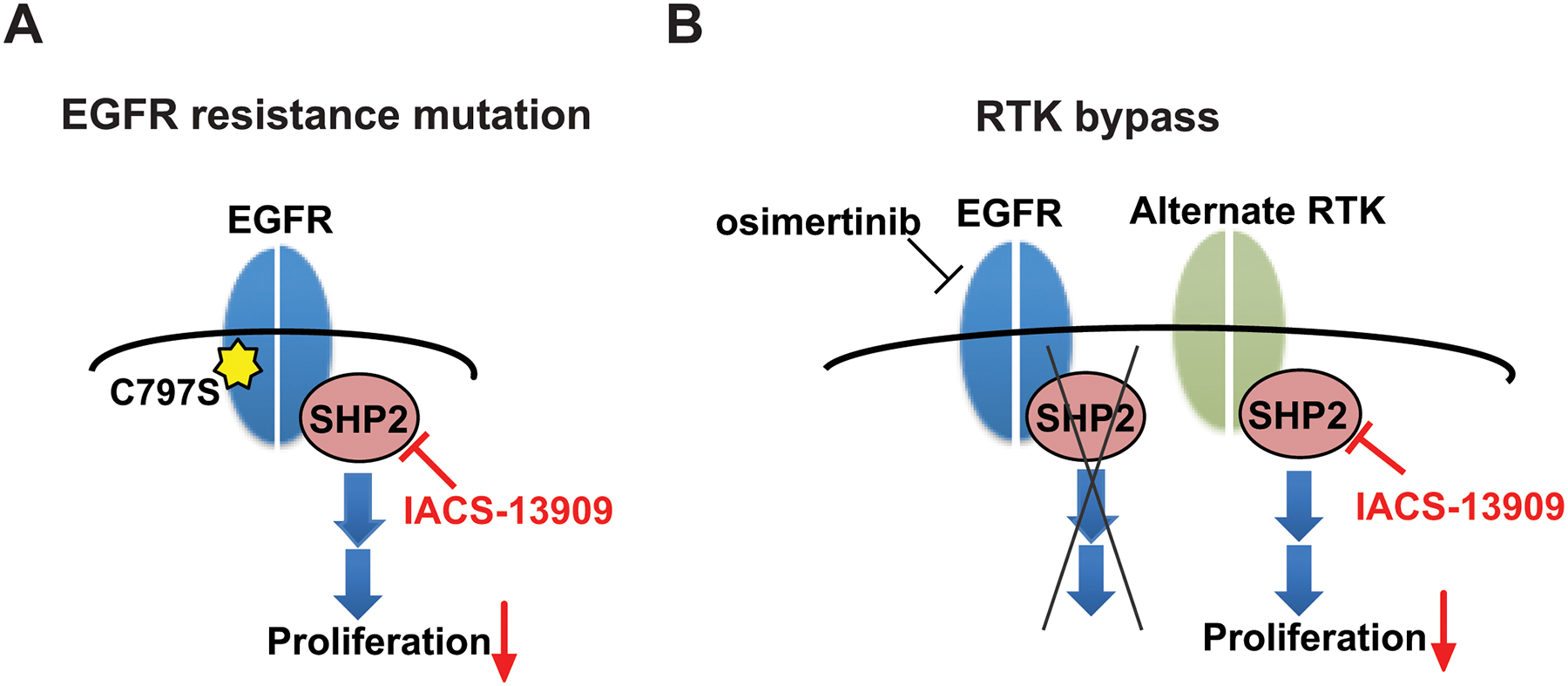
(A) In tumors harboring an EGFR mutation that confers resistance to osimertinib (e.g., C797S), EGFR remains as the primary oncogenic driver and signals through SHP2. SHP2 inhibition by an allosteric SHP2 inhibitor such as IACS-13909 is effective in inhibiting proliferation of the tumor. (B) In tumors where inhibition of EGFR results in compensatory activation of one or more RTKs (“RTK-bypass”), a SHP2 inhibitor can inhibit tumor cell proliferation by blocking signaling downstream of the activated RTKs.
A major challenge in targeting the RTK/MAPK pathway is acquired resistance, whereby a tumor initially responds to treatment but regrows on continued treatment. This can be attributed to both the adaptability of the cancer cells and the heterogeneity of the primary tumors. Cancer cells very often harbor a primary oncogenic driver. When the primary driver is blocked, other oncogenic drivers either within the same cells or from a different clone emerge as the alternate driving force for tumor growth. Combining one drug targeting the primary oncogenic driver and a second drug suppressing multiple potential alternative drivers is an attractive strategy. To improve the therapeutic index, ideally the first drug is mutant selective; to ensure broad targeting of potential secondary drivers, the second drug should target wildtype protein as well. Here we propose combining EGFR mutant-selective inhibitor osimertinib (12) and SHP2 allosteric inhibitor IACS-13909 that is not mutant selective. At tolerated doses, such combination achieves more durable response compared with osimertinib single agent in osimertinib-sensitive EGFRmut NSCLC tumors, and causes tumor regression in osimertinib-resistant EGFRmut NSCLC xenograft tumors in mice.
Our data demonstrate that IACS-13909 has anti-tumor activity in cancers with a broad range of RTKs as the oncogenic driver. While we provide data showing SHP2 inhibition can overcome both EGFR-dependent and EGFR-independent osimertinib resistance in EGFRmut NSCLC, a SHP2 inhibitor can be used more broadly. Several additional combination strategies with an allosteric SHP2 inhibitor have been proposed in overcoming resistance to targeted agents. First of all, in ALK inhibitor resistant preclinical models with RTK-bypass as a resistance mechanism, in vivo efficacy for the treatment with combination of ALK inhibitor ceritinib and SHP099 has been reported (37). Secondly, the clinical response to MEK inhibitors is limited by adaptive feedback activation through multiple RTKs (35,38), therefore combination of SHP2i and MEKi have demonstrated anti-tumor efficacy in mice (8,36). In addition to MEKi, preclinical data for combining with ERKi have been reported (39). A major challenge with combining SHP2i and MEKi (or ERKi) when both molecules target wildtype enzymes is the therapeutic index. SHP2 inhibitors were used at reduced doses or dosing frequencies in both combination strategies in mice (36,39). It is speculated that sustained shutdown of the MAPK pathway in normal tissue may not be tolerated, therefore reduced dose or dosing frequency that leads to pulsatile shutdown of the MAPK pathway had to be performed. Most recently, multiple approaches have identified combining a KRASG12C mutant-specific inhibitor and SHP2 inhibitor as a strategy for achieving more robust and durable response (40,41). Identifying the most optimal combination strategy for a SHP2 allosteric inhibitor requires additional preclinical work and, most importantly, clinical trials. Currently, several SHP2 allosteric inhibitors (TNO155, RMC-4630, JAB-3068, JAB-3312 and RLY-1971) are under early phase clinical development. An advanced derivative of IACS-13909 will enter Phase 1 clinical trial in later 2020.
Supplementary Material
Statement of Significance.
Findings highlight the discovery of IACS-13909 as a potent, selective inhibitor of SHP2 with drug-like properties, and targeting SHP2 may serve as a therapeutic strategy to overcome tumor resistance to osimertinib.
Acknowledgements
The authors thank DV-MS at MDACC for mouse husbandry and care; Research Histology, Pathology & Imaging Core at MDACC Science Park for histology service; and all members at TRACTION and the Institute for Applied Cancer Science at MDACC for discussions. The authors also thank ChemPartners, Eurofins, Crown Bioscience, LIDE biotech, and the experimental therapeutics core at the Dana-Farber Cancer Institute for service.
Conflicts of interest
The authors declare the following potential competing interests. This work was funded by Navire Pharma Inc., a BridgeBio company. The University of Texas MD Anderson Cancer Center and Navire Pharma, Inc. are parties to a Collaboration and License pursuant to which MD Anderson and Navire will collaborate on the conduct of research and development of products. Under this Agreement, the Board of Regents of The University of Texas System, on behalf of MD Anderson, received equity, milestone payments and royalties in Navire. Proceeds may be distributed based on UT System Intellectual Property Policy. N.E.K. is a consultant to The University of Texas MD Anderson Cancer Center and Navire Pharma, Inc. All other authors are employees of The University of Texas MD Anderson Cancer Center. G.F.D. reports personal fees from and stock ownership in Frontier Medicines, Forma Therapeutics, Metabomed, BiovelocITA, Nurix and Orionis Biosciences; and personal fees from The FIRC Institute of Molecular Oncology, Blueprint Medicines, Taiho Pharmaceuticals. T.P.H. reports personal fees and stock ownership from Cullgen Inc.
References
- 1.Chang TY, Dvorak CC, Loh ML. Bedside to bench in juvenile myelomonocytic leukemia: insights into leukemogenesis from a rare pediatric leukemia. Blood 2014;124:2487–97 [DOI] [PubMed] [Google Scholar]
- 2.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev 2007;17:23–30 [DOI] [PubMed] [Google Scholar]
- 3.Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 2003;23:7875–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batth TS, Papetti M, Pfeiffer A, Tollenaere MAX, Francavilla C, Olsen JV. Large-Scale Phosphoproteomics Reveals Shp-2 Phosphatase-Dependent Regulators of Pdgf Receptor Signaling. Cell Rep 2018;22:2784–96 [DOI] [PubMed] [Google Scholar]
- 5.Maroun CR, Naujokas MA, Holgado-Madruga M, Wong AJ, Park M. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol 2000;20:8513–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Agazie YM. Molecular mechanism for SHP2 in promoting HER2-induced signaling and transformation. J Biol Chem 2009;284:12226–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu S, Sayad A, Chan G, Yang W, Lu Z, Virtanen C, et al. SHP2 is required for BCR-ABL1-induced hematologic neoplasia. Leukemia 2018;32:203–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, et al. SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov 2018;8:1237–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med 2018;24:961–7 [DOI] [PubMed] [Google Scholar]
- 10.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 2018;20:1064–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24:954–60 [DOI] [PubMed] [Google Scholar]
- 12.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113–25 [DOI] [PubMed] [Google Scholar]
- 14.Le X, Puri S, Negrao MV, Nilsson MB, Robichaux J, Boyle T, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murtuza A, Bulbul A, Shen JP, Keshavarzian P, Woodward BD, Lopez-Diaz FJ, et al. Novel Third-Generation EGFR Tyrosine Kinase Inhibitors and Strategies to Overcome Therapeutic Resistance in Lung Cancer. Cancer Res 2019;79:689–98 [DOI] [PubMed] [Google Scholar]
- 16.Ortiz-Cuaran S, Scheffler M, Plenker D, Dahmen L, Scheel AH, Fernandez-Cuesta L, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res 2016;22:4837–47 [DOI] [PubMed] [Google Scholar]
- 17.Martin MJ, Eberlein C, Taylor M, Ashton S, Robinson D, Cross D. Inhibition of oxidative phosphorylation suppresses the development of osimertinib resistance in a preclinical model of EGFR-driven lung adenocarcinoma. Oncotarget 2016;7:86313–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab 2014;2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moores SL, Chiu ML, Bushey BS, Chevalier K, Luistro L, Dorn K, et al. A Novel Bispecific Antibody Targeting EGFR and cMet Is Effective against EGFR Inhibitor-Resistant Lung Tumors. Cancer Res 2016;76:3942–53 [DOI] [PubMed] [Google Scholar]
- 21.Yi L, Pimentel H, Bray NL, Pachter L. Gene-level differential analysis at transcript-level resolution. Genome Biol 2018;19:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 2016;535:148–52 [DOI] [PubMed] [Google Scholar]
- 25.McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 2017;170:577–92 e10 [DOI] [PubMed] [Google Scholar]
- 26.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagle MC, Kirouac D, Klijn C, Liu B, Mahajan S, Junttila M, et al. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis Oncol 2018;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feifei Zhang ZZ, Yuan Long, Wenhua Xu, Shizhu Zhao, Yang Yang, Hui Liu, Jijun Cheng, Shun Lu, Danyi Wen. PDX model of pleural effusion of lung cancer patient for testing on targeted drug sensitivity and resistance (abstract). 2018 2018 Apr 14–18; Chicago, IL. Philadelphia (PA): AACR; Cancer Res 2018;78(13 Suppl):Abstract nr 1164. [Google Scholar]
- 29.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal 2013;6:re6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, et al. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia 2009;11:448–58, 2 p following 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A 2009;106:4519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long JE, Wongchenko MJ, Nickles D, Chung WJ, Wang BE, Riegler J, et al. Therapeutic resistance and susceptibility is shaped by cooperative multi-compartment tumor adaptation. Cell Death Differ 2019;26:2416–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed TA, Adamopoulos C, Karoulia Z, Wu X, Sachidanandam R, Aaronson SA, et al. SHP2 Drives Adaptive Resistance to ERK Signaling Inhibition in Molecularly Defined Subsets of ERK-Dependent Tumors. Cell Rep 2019;26:65–78 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016;534:647–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H, Liu C, Velazquez R, Wang H, Dunkl LM, Kazic-Legueux M, et al. SHP2 Inhibition Overcomes RTK-Mediated Pathway Reactivation in KRAS-Mutant Tumors Treated with MEK Inhibitors. Mol Cancer Ther 2019;18:1323–34 [DOI] [PubMed] [Google Scholar]
- 37.Dardaei L, Wang HQ, Singh M, Fordjour P, Shaw KX, Yoda S, et al. SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med 2018;24:512–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 2012;149:307–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.NICHOLS RJ, GOLDSMITH MA, SCHULZE C, SMITH J, WILDES DE, KELSEY S, et al. ; REVOLUTION MEDICINES, INC., assignee. SHP2 INHIBITOR COMPOSITIONS AND METHODS FOR TREATING CANCER. USA: 2019. [Google Scholar]
- 40.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


