Abstract
In 2000, the US National Cancer Institute established the Early Detection Research Network (EDRN) to identify, develop and validate biomarkers to improve the detection of early stage cancers and risk assessment. This consortium of more than 300 investigators at academic institutions and in the private sector are working collaboratively to bring biomarkers and imaging methods to clinical fruition. Although significant roadblocks have hindered the field of biomarker discovery and validation, the EDRN has helped overcome many of them by setting well-defined strategies and milestones focused on solving defined unmet clinical needs. The EDRN has implemented measures to improve biomarker discovery and validation, such as data sharing, use of common data elements, generating multi-disciplinary and multi-institutional collaborations within a cohesive and productive team environment, and putting emphasis on quality control and data replication for all candidate biomarkers for reaching a “go” or “no go” decision. A measure of the success of the EDRN are the number of biomarkers tests or devices approved by the Food and Drug Administration to which EDRN investigators have made significant contributions and the number of biomarkers tests developed by EDRN investigators that are available in Clinical Laboratory Improvement Amendments (CLIA) laboratories.
Introduction
The mission of the US National Cancer Institute’s Early Detection Research Network (EDRN) is to discover, develop and validate biomarkers and imaging methods to detect early stage cancers and for risk assessment and to translate these biomarkers and imaging methods into clinical tests. The EDRN is a ‘Team of Investigators’ that, for the past 20 years, has dedicated their cooperative and collaborative efforts to the discovery and validation of biomarkers to detect cancers at early stages. These investigators, drawn from individual laboratories and institutions where they have been highly-successful as individuals, have been transformed into a group where success is measured by the collective success of the group. Together, this group of investigators, led by National Cancer Institute Program Officers, an Executive Committee and Working Groups, has discovered and validated biomarkers that have changed the management of cancer.
Translational biomarker research bridges the gap between basic research that identifies biomolecules involved in or the result from carcinogenesis and the clinical application of these discoveries. Cultural separations between scientific disciplines often create barriers to the development of the multidisciplinary teams required for translational research. Biomarker research requires extensive interactions among academic researchers, industry, and clinicians to shape research strategies (1).
At a superficial level, cancer biomarker discovery and validation appear to be a trivial process. A PubMed search for “cancer” and “biomarkers” gives more than 24,000 publications in 2018; 2200 when limited to “cancer biomarkers” and “early detection.” The ‘Valley of Death’ of biomarkers is often harsher than that for new pharmaceuticals, illustrated by the fact that fewer than 30 cancer biomarkers have been approved by the Food and Drug Administration (FDA), and most of these are for monitoring response to therapy. Barriers to success include the performance characteristics needed to a make a biomarker clinically useful, especially for early detection or screening, tumor heterogeneity, a discovery process with many hazards and biases (many of which have been identified by the EDRN), a validation process that is cumbersome and expensive, and regulatory requirements. The team approach taken by the EDRN is designed to address these barriers. Prior to the EDRN and indeed persistently outside the EDRN, biomarker science is ‘stuck’ in the discovery step with very little interest or ability to formally validate a marker and achieve FDA approval.
The EDRN has been successful by developing an interactive group of institutions and processes with a singular, common goal to discover, validate, and establish clinical applications for new cancer biomarkers. The organization is almost uniquely porous; collaborations with industry and non-EDRN academic institutions are actively sought to collaborate through informal and formal mechanisms, which includes the EDRN Associate Membership Program (https://edrn.nci.nih.gov/funding-opportunities). Extensively characterized and curated biospecimens, often from rigorously collected ‘reference sets’ and prospective cohorts, are made available to both EDRN and external investigators whose applications have been deemed meritorious based on rigorous and defined criteria, i.e. Will the investigation advance the science of cancer detection and prognosis and ultimately, will it benefit the patient?
History of the EDRN
In 1998, the US National Cancer Institute convened a panel of outside experts, the Early Detection Implementation Working Group, to give advice on early cancer detection and to address the major recommendations of the Cancer Prevention Program Group (EDRN Initial Report, October, 2000 https://edrn.nci.nih.gov/docs).
In 1999, the NCI’s Board of Scientific Advisors reviewed the recommendations of the Early Detection Implementation Working Group and recommended the formation of the EDRN. The objectives of the Network will include:
development and testing of biomarkers and technologies for early cancer detection and to obtain preliminary data on the biomarker’s performance that will be used to guide further testing;
conduct early phase evaluations of promising, analytically proven biomarkers or technologies, which will include measures of diagnostic accuracy, sensitivity, and specificity;
based on these early phase evaluations, perform subsequent large definitive validation studies in the field of cancer detection and screening;
encourage collaboration and rapid dissemination of information among awardees to ensure progress and avoid fragmentation of effort.
The EDRN was formally established in 2000 (2). At that time, a veritable explosion in new biomarkers was underway. Many of these biomarkers had made their way into clinical practice without rigorous evaluation of their performance and their impact on clinical outcomes. Few had achieved or even sought FDA approval. Historically, this was because too few biomarker studies used appropriate study designs, employed technologies with too low a sensitivity, too few biospecimens, or used biospecimens that were not appropriate for the clinical question being asked. To address these deficiencies, the EDRN took the lead in developing a systematic strategy to implement rigorous criteria for biomarker discovery and validation.
The EDRN has since been renewed by the US National Cancer Institute in 2005, 2010, and 2016. The process for renewal involves a number of reviews and evaluations by a Network Consulting Team, NCI leadership, subcommittees of the NCI Board of Scientific Advisors, and the whole of the Board of Scientific Advisors. While the basic structure of the EDRN has remained the same, there have been modifications in the EDRN’s goals to reflect progress in the field and recommendations of the Network Consulting Team and the Board of Scientific Advisors, e.g. increased emphasis on biomarker validation, inclusion of imaging methods, and development of biomarkers to distinguish aggressive early stage cancers from indolent cancers to reduce overtreatment. These renewals were open competitions, and applications were evaluated by independent review panels convened by the National Cancer Institute. There has been approximately 40% turnover of the laboratories and centers each cycle, which is indicative of the open and competitive nature of the reviews.
EDRN Organizational Structure
The four major components of the EDRN (Biomarker Developmental Laboratories, Biomarker Reference Laboratories, Clinical Validation Centers, and Data Management and Coordinating Center ) have distinct but complementary roles and work synergistically to facilitate the discovery, development, and validation of cancer biomarkers (Figure 1).
Figure 1: EDRN Organizational Structure:
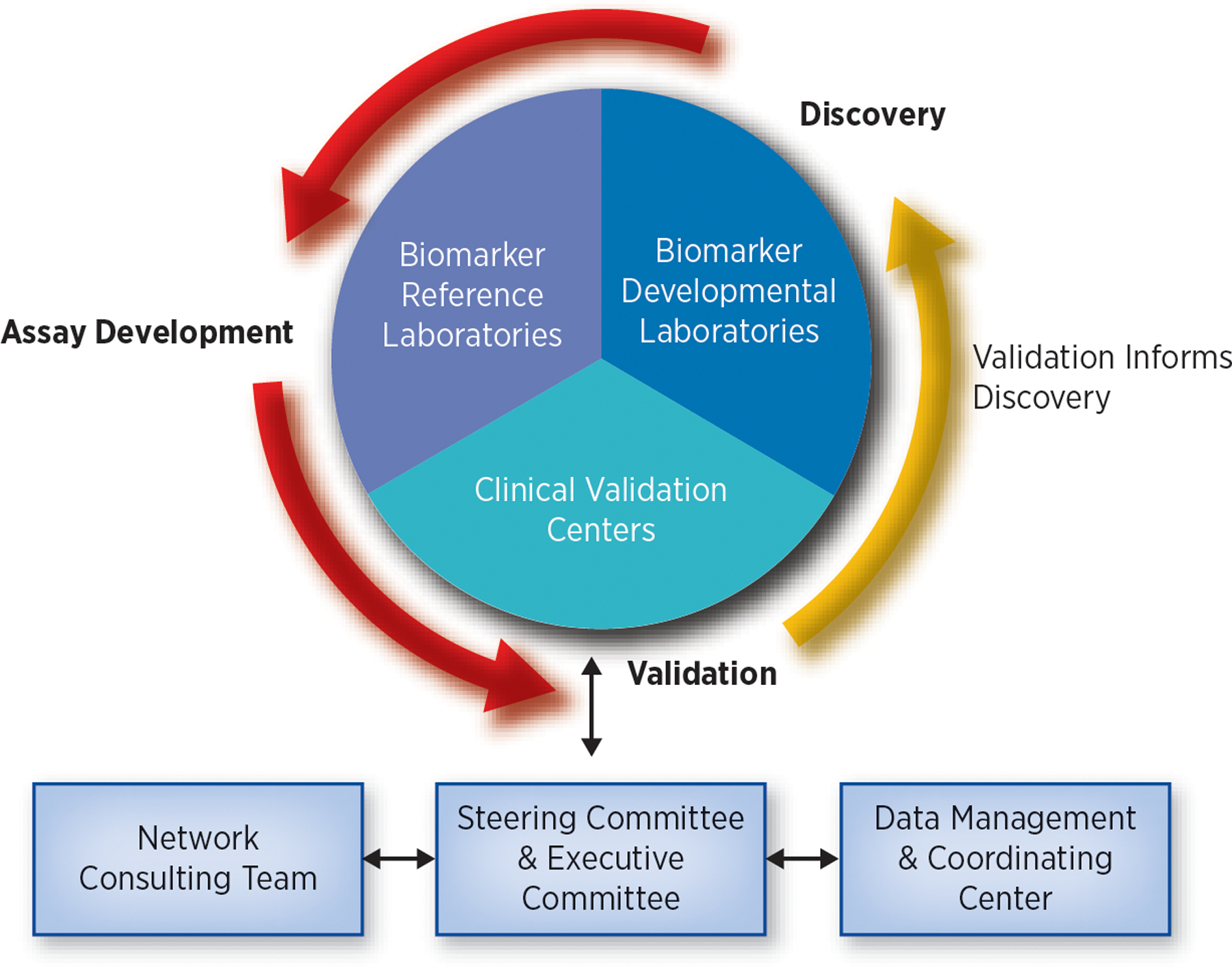
The four structural components of the EDRN have distinct but complementary roles. Biomarker Developmental Laboratories discover, develop, and characterize new biomarkers or refine existing biomarkers. Biomarker Reference Laboratories conduct assays for EDRN validation trials. Clinical Validation Centers conduct validation trials on biomarkers developed by both EDRN and non-EDRN investigators. Data Management and Coordinating Center assists with protocol design, works with the Clinical Validation Centers to conduct biomarker validation trials and maintains the data and biospecimen tracking systems. EDRN Steering Committee is composed of all the EDRN Principal Investigators and is responsible for overseeing the activities of EDRN and setting priorities. Network Consulting Team is composed of independent scientists and clinicians. They review and evaluate the research conducted by the EDRN.
Modified from Srivastava, S. The Early Detection Research Network: 10-Year Outlook. Clinical Chemistry, 2013;59 (1):60–67.
Biomarker Developmental Laboratories (BDLs):
BDLs discover, develop, and characterize new biomarkers or refine existing biomarkers. Within the EDRN, BDLs are the primary source of new biomarkers or panels of biomarkers on which the EDRN conducts validation trials. They also develop new assays to detect candidate biomarkers.
Biomarker Reference Laboratories (BRLs):
The primary role of the BRLs is to conduct assays for EDRN validation trials. The assays are performed on blinded biospecimens to minimize bias in the analysis and independently verify the assay performance.
Clinical Validation Centers (CVCs):
The primary role of the CVCs is to conduct validation trials on biomarkers. CVCs also provide high-quality, well-annotated biospecimens to the BDLs for biomarker development studies. The use of biospecimens collected using rigorous standard operating procedures helps minimize false discoveries.
Data Management and Coordinating Center (DMCC):
One of the major roles of the DMCC is to work with the CVCs to conduct biomarker validation trials. The DMCC assists with protocol design, monitors the trial, and maintains the data and biospecimen tracking system. The DMCC is responsible for analyzing the results of the trials, thereby reducing bias as it is independent from the laboratories that discovered the biomarkers. The DMCC provides statistical advice to the BDLs, develops theoretical and applied approaches and collaborates with the EDRN Informatics Center.
EDRN Steering Committee:
The EDRN Steering Committee is composed of all the EDRN Principal Investigators (PIs) and is responsible for overseeing the activities of the EDRN and setting priorities. The Steering Committee meets in person twice a year. The Executive Committee, which consists of the EDRN Chair and Co-Chair and the elected Chairs of the Collaborative Groups, have monthly conference calls.
Collaborative Groups:
Within the EDRN there are four organ-specific Collaborative Groups — Breast and Gynecologic Cancers (current focus is on breast and ovarian cancers), Colorectal and Other Gastrointestinal Cancers (current focus is on colorectal, esophageal and pancreatic cancers), Lung and Upper Aerodigestive Cancers (current focus is on lung cancer and mesothelioma), and Prostate and Other Urologic Cancers (current focus is on prostate cancer). All EDRN PIs, co-investigators and many Associate Members are members of one or more of these collaborative groups. They have monthly conference calls and meet twice a year in person to update each other on their progress and to develop collaborative projects that use the resources of all the investigators. These projects frequently involve comparing the performance of biomarkers from different laboratories in a common set of biospecimens and when appropriate combining these biomarkers to create a panel. These projects are supported by the grantees’ set-aside funds, which are restricted for this purpose.
Interagency Agreements:
The EDRN collaborates with four other Federal agencies: the National Aeronautics and Space Administration’s Jet Propulsion Laboratory (JPL), which supports EDRN informatics; Pacific Northwest National Laboratory (PNNL), which supports the development of proteomic-based assays; the Department of Defense Center for Prostate Disease Research (CPDR), which provides valuable biospecimens collected from subjects with prostatic diseases with high representation of African Americans; and the National Institute of Standards and Technology (NIST), which assists in the development of standards and references.
Associate Member Program:
The EDRN Associate Member Program is a mechanism through which non-EDRN investigators can work collaboratively with the EDRN. Type A Associate Members are investigators who have promising biomarkers that apply to the EDRN for resources to verify or validate their biomarker’s performance. Category B Associate Members are investigators who participate in EDRN validation studies and often help with patient accrual. Category C Associate Members are non-EDRN investigators, industrial partners and cancer advocates who participate in EDRN activities and conferences. There are currently more than 300 Associate Members. Details on the Associate Member Program and how to apply can be found on the EDRN website at (https://edrn.nci.nih.gov/funding-opportunities).
EDRN Collaborative Environment
This integrated structure is a unique and important aspect of the EDRN. The bringing together of investigators with expertise in cancer biology and biomarker development with clinicians having expertise in cancer screening and early diagnosis ensures that the discovery efforts are performed with specific diagnostic requirements in mind – in what patient population and for what purpose will the biomarkers or imaging modality be used and what performance (sensitivity and specificity) and accuracy are required to make them clinically useful? Too often, discovery efforts by non-EDRN investigators are not closely aligned with potential clinical applications, e.g. use only late stage cancers or only healthy controls with no confounding conditions. It is notable that more than 50% of the biomarkers or panels being validated by the EDRN CVCs were developed by EDRN BDLs.
The success of the EDRN has been due, in part, to a tremendous commitment of its members. A highly democratic, yet mission-focused, agenda has been adopted that seeks out the best science from within the EDRN and outside the EDRN, identifies and prioritizes opportunities for new applications, and oversees the conduct and reporting of pivotal translational studies. Throughout the process, quality control measures are implemented to minimize the risk of false discovery or bias.
Collaborative Funds:
The EDRN has developed an innovative funding mechanism to support collaboration within the EDRN and with investigators outside of the EDRN. These funds reside at the DMCC and are used to support post-award projects. Requests for the release of these funds are reviewed by the EDRN Steering Committee and NCI. They are used primarily to support large multi-center biomarker validation trials that involve patient accrual, biospecimen collections, assays of defined biomarkers and the Associate Members.
Foundations and International Partnerships:
The EDRN has strategic alliances with non-profit foundations, such as the Canary Foundation and the Lustgarten Foundation, Federal agencies, such as NIST and the FDA, and professional organizations, such as the American Society of Clinical Oncology (ASCO). The EDRN and Japan Agency for Medical Research and Development (AMED) hold joint meetings yearly to discuss ongoing projects and establish collaborative projects on cancer early detection.
This collaborative environment facilitates high quality, productive biomarker research and well-designed biomarker validation studies. This achievement has been widely lauded by a number of peer-reviewed journals, such as Nature (3), Science and JNCI (4), and professional associations, such as Institute of Medicine, which cited EDRN as a model for conducting big science (5). Indeed, China and the United Kingdom have each developed collaborations with the EDRN because of an interest of those countries in setting up a similar infrastructure.
Novel Approaches
Guidelines
A landmark achievement of the EDRN was the development of a five-phase approach for biomarker development (6). This provides a systematic approach that identifies the most promising biomarkers and is used to determine which biomarkers are moved forward to validation (Figure 2).
Figure 2: Phases of Biomarker Discovery and Validation:
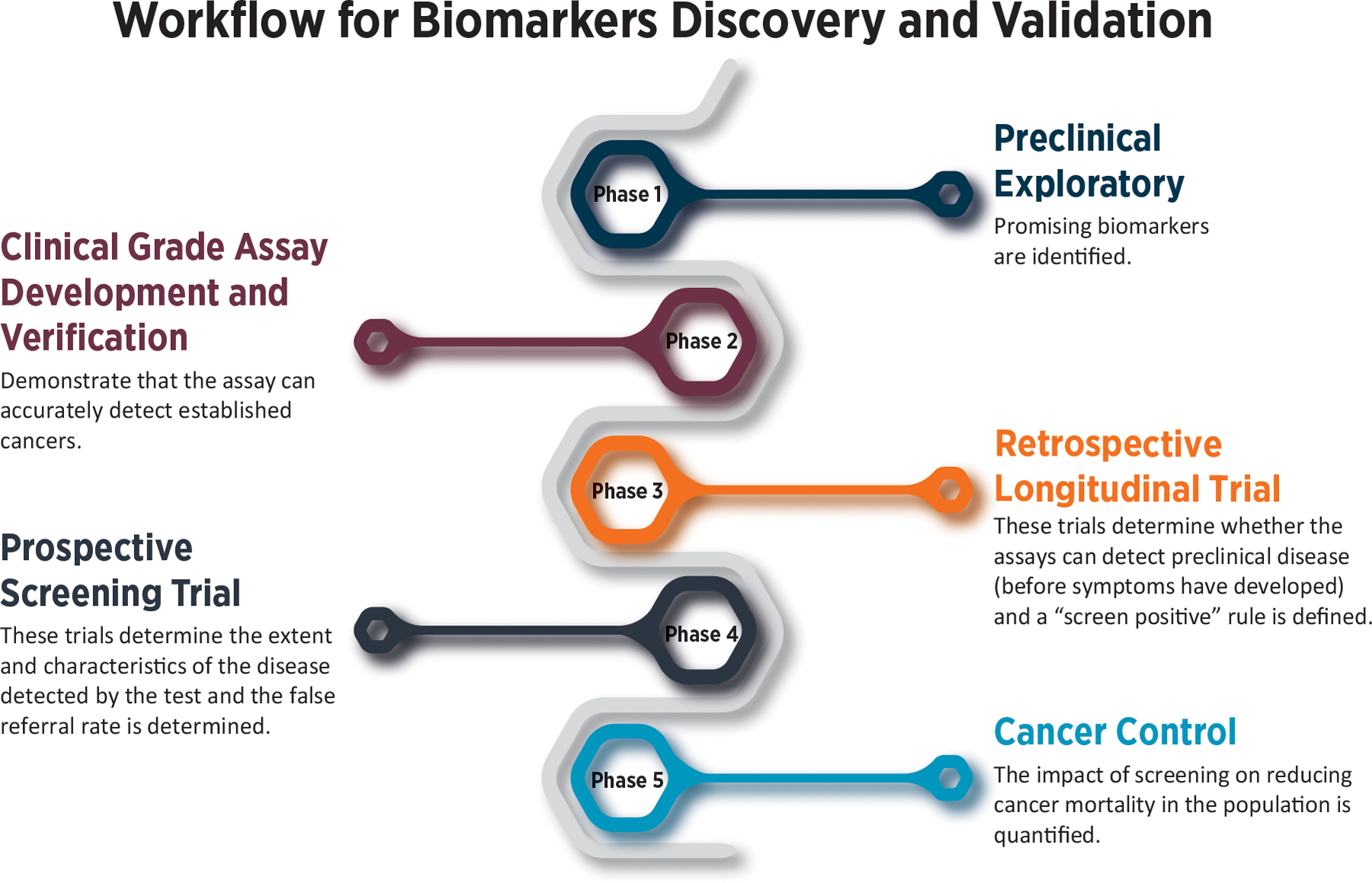
The EDRN has developed a five-phase approach for biomarker development and validation. The 5-step approach to biomarker validation includes Phase 1 – Preclinical Exploratory; Phase 2 – Clinical Grade Assay Development and Verification; Phase 3 – Retrospective Longitudinal Trial, Phase 4 – Prospective Screening Trial, and Phase 5 – Cancer Control.
Adapted from Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. Journal of the National Cancer Institute. 2001;93(14):1054–61.
This rigorous, systematic method for biomarker discovery and validation is used throughout the EDRN process and requires a highly collaborative organization that includes a broad range of expertise in which each collaborator has an essential role. The organization has been compared by some to the Mission Control Room of NASA: if a single component in a pre-launch sequence is not performing at 100%, the launch is ‘held’ until the issue is resolved. Similarly, because of the essential nature of each of the components in the process of biomarker validation, all components are regarded as equals. If there are issues, for example with potential biases related to specimen processing or in ensuring generalizability of the samples, these are resolved prior to initiation of the scientific process. As a result, the broad range of individuals involved, from biostatisticians, to clinicians, to scientists, to technology experts are equally valued and respected within the organization. In this context, it has been the Team that ensures and celebrates the success, not an individual.
In addition to these 5-phase guidelines, the EDRN has developed and adopted the PRoBe study design, a rigorous approach to reduce bias during all phases of biomarker discovery and validation (7). PRoBe stands for prospective-specimen-collection, retrospective-blinded-evaluation. It is “a nest-case control study design that involves prospective collection of biospecimens before outcome ascertainment from a study cohort that is relevant to the clinical applications.” Critical features of this design are that patients are enrolled prior to diagnosis, cases and controls are enrolled under the same conditions, and samples are collected and processed identically. For a screening application, this design may require large numbers of study participants because the incidence of most cancers is low. For instance, as the incidence of colon cancer in a screening population is about 0.5%, 20,000 patients would need to be enrolled to obtain 100 cancers.
EDRN Specimen Reference Sets - Advancing Research Through Better Biospecimen Practices
The EDRN is a pioneer in the development of ‘biospecimen reference sets’ for most of the major epithelial cancers, including breast, lung, colon, prostate, pancreas and liver. This concept was established in partnership with NIST, which develops standards for a variety of industrial and technology evaluations.
Reference samples are sets of samples with cases and controls statistically powered for a specific intended clinical application. Multiple EDRN sites contribute specimens to these sets; this is critical as single sites rarely have sufficient numbers of early stage cancers (8). These sets of biospecimens are collected using standard operating procedures with associated EDRN-established common data elements (8, 9). These reference sets are collected prospectively in an appropriate setting to answer pressing clinical questions. They allow the direct comparison and assessment of the performance characteristics of different platforms as well as the performance characteristics of individual candidate biomarkers using the same specimens in a blinded fashion.
Historically, biomarker discovery has been pursued using lower-value biospecimens collected from ‘sets of convenience.’ As a result, many biomarkers that appeared to be highly-promising were found to not validate when tested using more appropriate, higher quality specimens. In contrast, EDRN investigators use high-quality, prospectively collected biospecimens for discovery. In so doing, investigators have found false-discovery rates to be substantially reduced, allowing resources to be committed to biomarkers that are most likely to succeed. EDRN investigators have demonstrated that this process of identifying biomarkers early on that will not be clinically useful. This prevents waste of precious resources (time, specimens) and prevents biomarkers making their way into the clinic where there is a potential for patient harm.
Nine EDRN biospecimen reference sets are stored at NCI Frederick, which ensures their availability to the entire research community (Table 1) https://edrn.nci.nih.gov/resources/sample-reference-sets. For example, the EDRN pancreatic cancer reference set is comprised of serum/plasma samples from subjects with pancreatic cancer (n=60 early stage and 40 late stage cancers), chronic pancreatitis (n=63), acute benign biliary obstruction (n=31), and healthy controls (n=61) (10). Also, in most instances when the EDRN undertakes a biomarker validation trial, sufficient aliquots of specimens are collected to allow for the validation of future biomarkers. For example, biospecimens from a hepatocellular carcinoma biomarker validation trial were used to create a biospecimen validation set that contains specimens from 400 patients with HCC and 400 cirrhosis controls (11).
Table 1:
EDRN Biospecimen Reference Sets
| Reference Set | Type of Specimens | Participants # |
Participant Groups |
|---|---|---|---|
| Bladder Cancer | Serum, whole urine, DNA from blood | 497 | Bladder cancer cases Healthy controls High Risk controls |
| Breast Cancer | Serum, plasma, buffy coat | 832 | Pre-diagnosis specimens DCIS cases Invasive cancer cases LCIS cases Benignlater cancer cases Normallater cancer cases Benign Disease Atypia controls Benign Disease non-Atypia controls Normal controls |
| Cancers in Women Endometrium, Ovary, Breast | Serum, plasma | 536 | Cases (pooled) Controls (pooled) |
| Colon Cancer | Serum, plasma, whole urine | 150 | Cases Adenoma controls Normal controls |
| Liver Cancer | Serum, plasma | 871 | Cases Controls |
| Lung Cancer | Serum, plasma | 1,205 | Cases Controls High risk controls with CT nodule High risk controls with no CT nodule |
| Pancreatic Cancer | Serum, plasma | 255 | Cases Controls |
| Prostate Cancer | Serum, plasma, buffy coat, RNA, supernatant fluid, whole urine | 900 | Initial Biopsy w/ Cancer cases Repeat Biopsy w/Cancer cases Confirmed but no biopsy controls Initial Biopsy w/o Cancer controls Repeat Biopsy w/o Cancer controls |
| Prostate Cancer (retrospective) | Serum | 663 | Cases Controls |
These reference sets have been supplied to more than 70 investigators (from academia, industry and foreign laboratories), most of whom are not members of the EDRN. Equally important are the other biologic samples that have been provided by individual EDRN sites to other EDRN and non-EDRN investigators through a merit-based system. More than 200 Materials Transfer Agreements (MTAs) have been executed for sample transfer.
Scientific Advances
Biomarker Development
EDRN investigators have developed many novel technologies to enhance the discovery and development of promising biomarkers to enrich the pipeline for future validation trials. New genomic and proteomic technologies are being used to identify candidate biomarkers for early detection. The multiple disciplines represented within the EDRN (i.e., clinical and basic science, biostatistics and bioinformatics) have enabled the successful implementation of these technologies. Descriptions of many of the biomarkers discovered by EDRN investigators are described in the EDRN 5th Report (https://edrn.nci.nih.gov/docs/EDRN5.pdf).
EDRN investigators have published more than 2,500 peer-reviewed articles; more than 20% are in high impact journals (IF>10). EDRN investigators currently have more than 64 patents and more than 12 licenses which is indicative of the practical applications sought within the Network. The EDRN initiated over 60 network collaborative projects, and over 30 collaborations have been formed between EDRN laboratories and biotechnology or diagnostic companies. More than 1,000 biomarkers have been discovered, developed or evaluated by EDRN investigators (Biomarker Database; http://cancer.jpl.nasa. gov/documents/applications/biomarker-database), and approximately 300 of these were found to have sufficient accuracy to be moved forward for consideration in pre-validation studies. Equally important is that hundreds of these biomarkers have been discarded using the biomarker triage system developed by the EDRN (12).
An essential role of the EDRN is to conduct validation trials on biomarkers with sufficient preliminary data demonstrating that they likely have the accuracy (sensitivity and specificity) to be clinically useful. The primary role of the EDRN CVCs is to conduct these trials. These trials typically recruit thousands of patients from multiple sites, the assays are performed on blinded specimens by an independent laboratory, such as an EDRN BRL, and the data analyzed by the EDRN DMCC.
Because clinical biomarker validation studies are time consuming and expensive, it is imperative that only the most promising biomarkers with good preliminary data on their diagnostic accuracy be moved forward into a validation trial. The EDRN has developed a systematic approach to rapidly triage candidate biomarkers with sequential go or no go decision steps (Figure 3). The ultimate prioritization of the most promising candidates is testing in the EDRN standard reference sets. Only when biomarkers successfully pass this rigorous process of prioritization and verification is a large clinical validation study undertaken.
Figure 3: EDRN Biomarker Triage System:
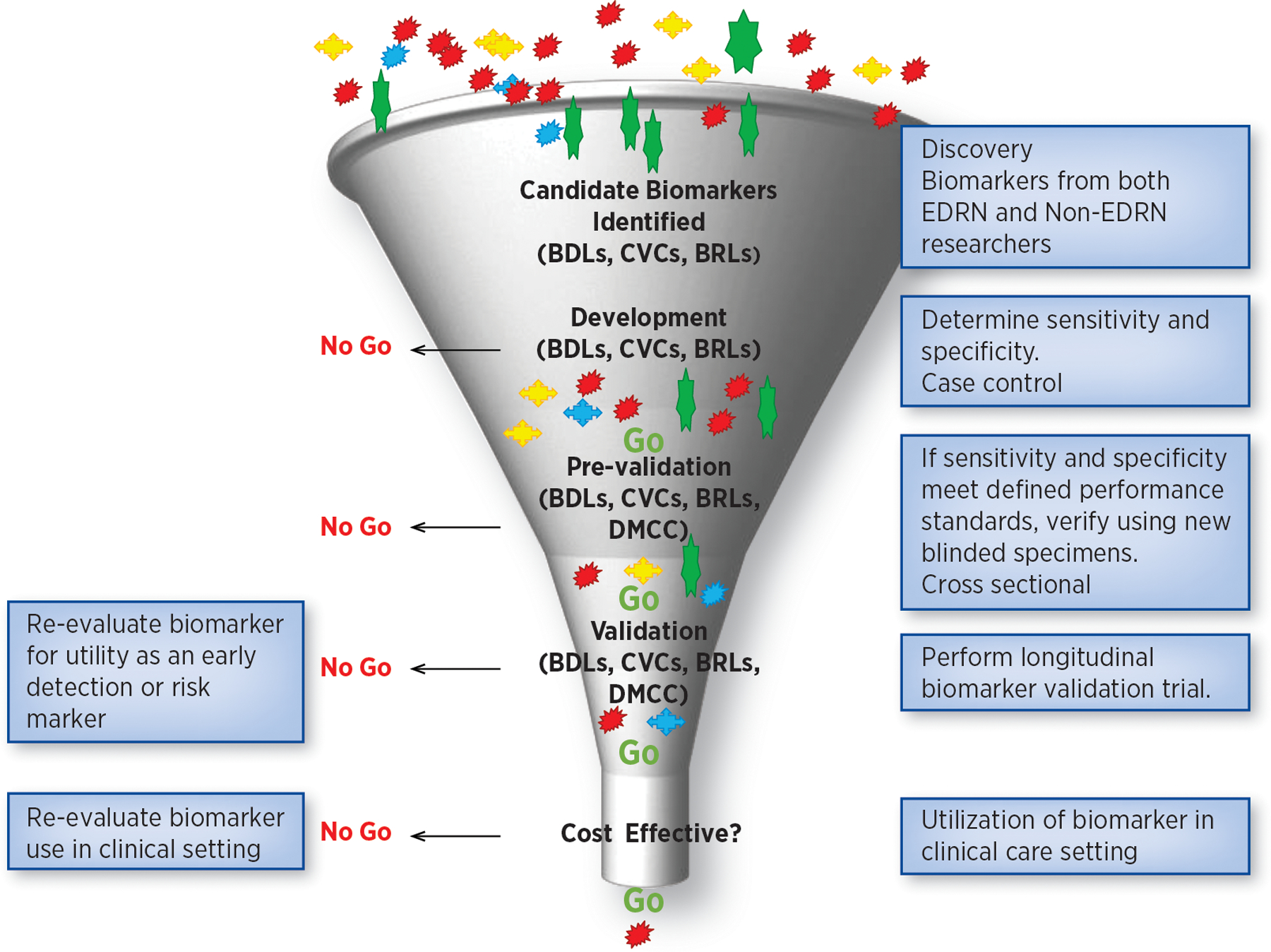
EDRN has developed a triage system with a series of “go or no-go” decisions. Only when biomarkers have passed this rigorous process of verification, is a large clinical validation study undertaken.
This “go/no go” decision schema determines which biomarkers can progress to verification, and then to validation (12). The EDRN has established clear milestones and performance criteria for reaching a “go” or “no-go” decision. These evaluation criteria include 1) does the biomarker accurately distinguish early stage cancers from healthy and diseased controls, i.e., pancreatic cancer from pancreatitis, 2) can the results be verified in an independent cohort, 3) are the reported sensitivity and specificity statistically valid, 4) does the biomarker address a defined clinical need, and 5) does the biomarker outperform currently used markers, or add significant value to them. Using these criteria, the EDRN Steering Committee evaluates newly discovered biomarkers and makes go no go recommendations. This allows the EDRN to focus its resources on those biomarkers most likely to have an impact on clinical practice.
Landmark Successes: FDA Approved Biomarkers and Tests Available in CLIA Laboratories
Given the ‘Valley of Death’ between biomarker discovery and validation, it is notable that 8 diagnostic tests or devices supported by the EDRN have been approved by the FDA for clinical use and that 18 EDRN developed biomarker tests are available in Clinical Laboratory Improvement Amendments (CLIA) approved laboratories (Table 2). Details on the EDRN-supported biomarkers that received FDA approval are listed below.
Table 2:
Tests in Clinical Laboratory Improvement Amendments (CLIA) Laboratories
| Biomarker Assay | Purpose | EDRN Principal Investigator CLIA Laboratory |
|---|---|---|
| MiCheck (Glypican-1 protein and related signalling molecules) | Differentiate aggressive prostate cancer from non-aggressive cancer and no cancer | Daniel Chan, Ph.D. Minomic, Inc |
| Videssa (a multi-protein biomarker blood test) | Distinguish benign from malignant breast lesions | Joshua LaBaer, M.D., and Karen Anderson M.D. Provista |
| DetermaVu | Facilitate clinical decision making in lung cancer | Louise Showe, Ph.D. OncoCyte |
| Precepta (23-gene expression panel) | Detection of lung cancer | Avrum Spira, M.D. Veracyte Inc. |
| Esoguard (methylated vimentin and cyclin A1) | Detection of Barrett’s esophagus | Sandford Markowitz, M.D. PAVmed |
| Decipher Prostate Cancer Classifier Test (SChLAP1 and other lncRNAs) | Determination of prostate cancer aggressiveness | Arul Chinnaiyan, M.D., Ph.D. GenomeDx |
| Protein panel (TIMP1, LRG1 and CA19–9) | Detection of pancreatic cancer | Samir Hanash M.D., Ph.D. Cosmos Wisdom |
| Mucin panel (MUC4, MUC5AC, MUC16 and MUC 17) | Detection of pancreatic cancer | Surinder Batra, Ph.D. Sanguine Diagnostic and Therapeutics |
| MiPS (Mi Prostate Score Urine test), Multiplex analysis of TMPRSS2:ERG gene fusion, PCA3 and serum PSA | Detection of prostate cancer | Arul Chinnaiyan, M.D., Ph.D. Gen-Probe |
| IHC and FISH for TMPRSS2:ERG fusion | Detection of prostate cancer | Arul Chinnaiyan, M.D., Ph.D. Roche |
| GSTP1 methylation | Decision making regarding repeat biopsies in prostate cancer | David Sidransky, M.D. OncoMethylome |
| Mitochondrial deletion | Detection of prostate cancer | National Institute of Standards and Technology (NIST) Mitomics |
| Proteomic panel | Detection of lung cancer | William Rom, M.D., M.P.H. Celera |
| Aptamer-based markers | Detection of lung cancer | William Rom, M.D., M.P.H. Somalogic |
| 80-gene panel** **(This panel has been refined; Percepta®, a 23-gene classifier, is now available through Veracyte) |
Detection of lung cancer | Avrum Spira, M.D., M.Sc. Allegro/Veracyte |
| Vimentin methylation in stool | Detection of colon cancer | Sanford Markowitz, M.D., Ph.D. LabCorp |
| Galectin-3 ligand | Detection of advanced adenomas and colon cancer | Robert Bresalier, M.D. BG Medicine |
| GP73 | Risk of hepatocellular carcinoma | Timothy Block, Ph.D. Beckman Coulter |
| 8-gene Panel for Barrett’s Esophagus (BE) | Progression Prediction of BE | Stephen Meltzer, M.D. Diagnovus |
[−2]proPSA: The FDA approved this test to help determine if a man with a total prostate specific antigen (PSA) between 4 –10 ng/mL is likely to have cancer and for whom biopsy is appropriate. This test may reduce the number of unnecessary biopsies in men unlikely to have cancer. (http://healthworkscollective.com/barbaraduck/34852/fda-approves-new-blood-test-prostate-cancer-men-psa-levels-just-above-normal-range )
PCA3: Overexpression of the PCA3 gene, measured in urine, is highly specific to prostate cancer. When evaluated with other risk factors, the PROGENSA PCA3 assay helps physicians identify which men should undergo a repeat prostate biopsy. (http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm294907.htm)
OVA1: This test helps detect ovarian cancer in women with a pelvic mass. It identifies those who may benefit from referral to a gynecological oncologist for surgery. (http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm182057.htm)
ROMA Algorithm: This Algorithm uses the CA-125 and HE4 blood markers to determine the likelihood that an ovarian pelvic mass is malignant (13).
AFPL3% and DCP: These markers were approved by the FDA in 2011 to help determine which patients with chronic liver diseases are at high risk for the development of hepatocellular carcinoma in (Wako Diagnostics). The EDRN performed an independent validation of both AFPL3% and DCP (11), and subsequently provided Wako blinded specimens to test their new, more sensitive instrument.
Overa: This test is designed to help determine which women are at high risk for having a malignancy and, therefore, would benefit from care with a gynecologic oncologist (14). Overa was approved by the FDA in 2016, and Vermillion, Inc. offers the test.
CancerSEEK: Five EDRN investigators participated in the development and testing of CancerSEEK, which is a multianalyte test that simultaneously determines the levels of eight proteins and the presence of cancer gene mutations in circulating DNA (15). In 2019, Thrive Earlier Detection Corp. received the FDA’s Breakthrough Device designation and has launched an effort to further investigate and commercialize this technology.
EsoCheck: This device is designed to allow patients to undergo a non-invasive five-minute office-based procedure to detect Barrett’s Esophagus (BE) without the need for endoscopy. EsoCheck has the potential to save lives through the early detection of esophageal abnormalities (16). This device received FDA approval in 2019, and PAVmed markets this device.
Robust Informatics Infrastructure Supporting Collaboration and Communications
In collaboration with the Jet Propulsion Laboratory (JPL), the EDRN has developed a number of informatic tools and databases to support biomarker development and validation, including tools to support the processing, capture, curation and sharing of data before publication, a national biomarker knowledge system, a biomarker data infrastructure consisting of 1000 biomarkers, 200 protocols, 2500 publications, and 100 terabytes of data, and pilot imaging projects. These tools and databases are accessible at https://edrn.nci.nih.gov/informatics and link EDRN research institutions together into a virtual knowledge system (17).
Early on the EDRN recognized the need for an effective knowledge system and made a significant investment in bioinformatics to support biospecimen collections and distribution, collection of study specific data, and storage of data on the performance of biomarkers developed by EDRN investigators. These data can be accessed and shared via a transparent, grid-type architecture. The EDRN has focused on five critical informatics goals: (1) define an information model that will address EDRN research and clinical needs; (2) a system to distribute all components of the knowledge system to users within and outside the EDRN; (3) software interfaces to capture and access data resources across the EDRN; (4) provide a secure transfer and distribution infrastructure that meets United States federal regulations for data sharing; and (5) provide an integrated portal to allow access to the various EDRN databases and informatics tools.
This infrastructure captures and links data from across the EDRN using nearly a thousand annotations of cancer biomarkers to terabytes of analysis results in the EDRN data commons, known as LabCAS. LabCAS provides support to capture data from validation studies linking data from laboratory tracking tools at the DMCC to the analysis of data captured in the EDRN. Capabilities are also in place to run repeatable analysis pipelines for genomics, image analysis, and other complex data types. These can be customized for each study. Several validation study teams are working with JPL to capture their data. Similar capabilities are being developed for alignment of massive imaging data for pancreatic cancer. As EDRN is amassing more data, there is greater opportunity for applying advanced data mining within and across different repositories of data. Integrated tools provide access to and visualization of the data in the data commons. Several of the tools are open source tools and are developed through collaborations with NCI’s Information Technology for Cancer Research (ITCR) program. The entire knowledge environment is integrated with the EDRN portal, providing secure, multi-layer access to data for EDRN, NCI, research, and public communities. Approximately 2500 unique visitors come to the portal each month.
EDRN BIG Data and Data Science
The EDRN is active in the research on next generation capabilities for crowd sourcing, machine learning, computational analysis, and visualization. JPL and Caltech have been working to bring in tools, such as Zoonverse, which have been successfully used in fields such as astronomy of crowdsourcing. These tools have been useful for generating large labeled sets required to train machine learning algorithms in the classification of features in images. JPL and Caltech have been working with different PIs, in particular with lung imaging, to explore the use of these tools to improve the capture, annotation and construction of databases using crowdsourcing and collaborative methods in data analysis with a goal of looking at how data-driven approaches (e.g. deep learning) can be applied. The capture of EDRN data in LabCAS provides a foundation for opening up new possibilities in these areas and enabling new analysis approaches for consortiums like the EDRN which are highly distributed and diverse. Additionally, Caltech, JPL, and NCI have explored the use of nascent virtual reality (VR) capabilities for analysis of multi-dimensional data. A recent prototype exploring lung cancer was presented at SigGraph 2019, one of the foremost research conferences in visualization and graphics. The VR prototype demonstrated multidimensional 3D data visualization of EDRN’s radiology data, integrating with LabCAS. The prototype demonstrated opportunities for entirely new approaches for data exploration as data increases in size and complexity.
The following provides an overview of EDRN’s progress in data science:
Standards and a process for capturing highly annotated cancer biomarker data.
Development of the LabCAS data commons infrastructure enabling EDRN to capture data, run pipelines, and link analytics including 77 collections and 23,200 files.
Deployment and instantiation of LabCAS on Amazon Web Services to support massive scalability and computation as well as collaborative analysis tools.
Integration of analytical tools including OHIF, caMicroscope, QuPath, 3D Slicer.
Development of cancer biomarker ontology and common data elements for annotating data in the data commons, and sharing of these standards with the research community by deposition in caDSR.
- Integration of EDRN’s data commons LabCAS into validation studies/reference sets including:
- Breast Reference Set
- Prostate MRI
Development of a genomics pipeline
Development of a Secretome tool/pipeline
Development of a pipeline for miRNA Measurements
- Capture and curation of 940 EDRN biomarkers in the EDRN biomarker database
- Linking of EDRN biomarker studies to researched and discovered biomarkers
- Linking of publications and data
- Linking of external data sources
Development and collaboration on OncoMX Portal through ITCR with George Washington University and integration with EDRN to link biomarkers and gene mutations.
Development of crowd sourcing techniques based on Zoonverse for analysis of lung imaging data with UCLA and Moffitt.
- Development of the EDRN Portal to integrate sites, protocols, biomarkers, and data into a searchable knowledge environment
- Collaboration and coordination with the DMCC to maintain portal information
- Daily integration of databases to support an integrate biomarker data environment
- Collaboration with NCI’s Center for Bioinformatics and Information Technology to provide operational support for running the portal.
Research and development of image alignment and an automated data pipeline to support automated alignment of 3D imaging for biomarker discovery in pancreatic cancer.
Presentations to the HTAN on the biomarker knowledge environment and model-drive architecture for large scale data integration, sharing and analytics.
Pilot an experimental VR platform, spun out from Caltech, for exploring lung images as a future concept for multi-dimension data analysis.
Conclusions
The EDRN structure has fostered a culture of Team Science. Most EDRN biomarker developmental and validation research is supported by the individual PI’s cooperative agreement, peer-reviewed award, and EDRN PIs have the primary responsibility for the conduct of this research. However, the advantage of a network like the EDRN and its Team Science approach becomes apparent when the investigators meet to discuss their work and establish collaborative projects. Presentation of their recent research results to other EDRN PIs during Collaborative Group and Steering Committee meetings provides them feedback on their ongoing research, allowing adjustment in light of peer input and progress being made by other EDRN PIs. EDRN awards include ‘set-aside’ funds that can only be used for new team and other collaborative projects that take advantage of the expertise, resources, and platforms of several different PIs.
While the organization of the EDRN provides a sound foundation for this high degree of collaboration, its success would not have been possible without the enlightened leadership of NCI program staff, the Chairs and Co-Chairs of the EDRN Executive Committee, and the members of working groups and consulting teams that have provided advice to the EDRN over the years. The vision of NCI leadership to establish such an organization should be congratulated, and this type of structure should be seriously considered for future large-scale enterprises that require a host of specialties, organizations, and institutions to achieve ‘Big Science’ discoveries. The AACR-NCI Think Tank Charting the Future of Cancer Prevention noted in 2008 that the “EDRN should be tapped as a potential partner in the effort to develop response biomarkers. Efficient translational research in prevention requires that trials enroll primarily individuals at high risk. This is another way to take advantage of the EDRN success. The goal of better risk assessment entails research on targets and pathways of the early stages of pre-cancer. Promising agents tested in high-risk individuals for short periods of time can build momentum for some of the needed reforms.” The Committee further stated that “Without the EDRN, research into new biomarkers of early cancer detection and risk would have remained on the periphery of research with a strong, but fragmented laboratory presence and little translational interest in the academic scientific community. But with the Network, a new translational paradigm has defined the organization, approaches, and standards by which biomarkers are being developed and assessed. The Network created major focus, energy and new research in the field of early detection. The Network’s publications, meetings, funding opportunities and infrastructure have fashioned a new environment for cancer prevention research.”
Footnotes
Statement of Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- 1.Wagner PD, Srivastava S. New paradigms in translational science research in cancer biomarkers. Translational research: The journal of laboratory and clinical medicine 2012;159(4):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava S, Kramer BS. Early detection cancer research network. Laboratory investigation; a journal of technical methods and pathology 2000;80(8):1147–8. [DOI] [PubMed] [Google Scholar]
- 3.Gewin V Missing the mark. Nature 2007;449(7164):770–1. [DOI] [PubMed] [Google Scholar]
- 4.Schully SD, Carrick DM, Mechanic LE, Srivastava S, Anderson GL, Baron JA, et al. Leveraging Biospecimen Resources for Discovery or Validation of Markers for Early Cancer Detection. JNCI: Journal of the National Cancer Institute 2015;107(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Io Medicine, Council NR. Large-Scale Biomedical Science: Exploring Strategies for Future Research Nass SJ, Stillman BW, editors. Washington, DC: The National Academies Press; 2003. 296 p. [Google Scholar]
- 6.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. Journal of the National Cancer Institute 2001;93(14):1054–61. [DOI] [PubMed] [Google Scholar]
- 7.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. Journal of the National Cancer Institute 2008;100(20):1432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Kagan J, Pepe M, Thornquist M, Ann Rinaudo J, Dahlgren J, et al. The Early Detection Research Network’s Specimen reference sets: paving the way for rapid evaluation of potential biomarkers. Clinical chemistry 2013;59(1):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winget MD, Baron JA, Spitz MR, Brenner DE, Warzel D, Kincaid H, et al. Development of common data elements: the experience of and recommendations from the early detection research network. International journal of medical informatics 2003;70(1):41–8. [DOI] [PubMed] [Google Scholar]
- 10.Haab BB, Huang Y, Balasenthil S, Partyka K, Tang H, Anderson M, et al. Definitive Characterization of CA 19–9 in Resectable Pancreatic Cancer Using a Reference Set of Serum and Plasma Specimens. PloS one 2015;10(10):e0139049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137(1):110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava S The early detection research network: 10-year outlook. Clinical chemistry 2013;59(1):60–7. [DOI] [PubMed] [Google Scholar]
- 13.Moore RG, Jabre-Raughley M, Brown AK, Robison KM, Miller MC, Allard WJ, et al. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. American journal of obstetrics and gynecology 2010;203(3):228.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman RL, Herzog TJ, Chan DW, Munroe DG, Pappas TC, Smith A, et al. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. American journal of obstetrics and gynecology 2016;215(1):82.e1-.e11. [DOI] [PubMed] [Google Scholar]
- 15.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science (New York, NY) 2018;359(6378):926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moinova HR, LaFramboise T, Lutterbaugh JD, Chandar AK, Dumot J, Faulx A, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Science translational medicine 2018;10(424). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crichton DJ, Mattmann CA, Thornquist M, Anton K, Hughes JS. Bioinformatics: biomarkers of early detection. Cancer biomarkers : section A of Disease markers 2010;9(1–6):511–30. [DOI] [PubMed] [Google Scholar]


