Abstract
The skeletal muscle is the largest organ in mammals and is the primary motor function organ of the body. Our previous research has shown that long non-coding RNAs (lncRNAs) are significant in the epigenetic control of skeletal muscle development. Here, we observed progressive upregulation of lncRNA 4930581F22Rik expression during skeletal muscle differentiation. Knockdown of lncRNA 4930581F22Rik hindered skeletal muscle differentiation and resulted in the inhibition of the myogenic markers MyHC and MEF2C. Furthermore, we found that lncRNA 4930581F22Rik regulates myogenesis via the ERK/MAPK signaling pathway, and this effect could be attenuated by the ERK-specific inhibitor PD0325901. Additionally, in vivo mice injury model results revealed that lncRNA 4930581F22Rik is involved in skeletal muscle regeneration. These results establish a theoretical basis for understanding the contribution of lncRNAs in skeletal muscle development and regeneration.
Keywords: lncRNA 4930581F22Rik, Skeletal muscle, Differentiation, Regeneration, ERK/MAPK signaling
Highlights
-
•
First discovery of lncRNA 4930581F22Rik is upregulated during skeletal muscle development and promotes myogenesis.
-
•
LncRNA 4930581F22Rik regulates skeletal muscle differentiation via ERK/MAPK pathway.
-
•
LncRNA 4930581F22Rik is involved in skeletal muscle regeneration.
1. Introducion
The skeletal muscle, constituting approximately 45 % of the total body weight, is the largest tissue in the human body [1]. They play crucial roles in the regulation of posture, autonomous movement, respiration, metabolism, and other fundamental aspects of human existence [2]. Regenerative repair disorders following skeletal muscle injury present a significant challenge in the field of regenerative medicine. Diseased skeletal muscles can seriously impair motor function and jeopardize life [3]. During normal muscle regeneration, damaged muscle fibers are first cleared by inflammatory cells and then replaced by tissue-resident muscle stem cells (MuSCs), known as satellite cells (SCs) [4]. Under the regulation of myogenic regulatory factors (MRFs), the activated satellite cells undergo further differentiation and fusion to form multinuclear muscle tubes, ultimately giving rise to contractile myofibers. Studying myoblast differentiation in vitro provides a robust model system to investigate the essential signaling mechanisms that regulate the genetic network throughout myogenesis [5]. Therefore, understanding the differentiation and developmental functions of skeletal muscles is essential for the advancement of human medicine.
The process of skeletal muscle regeneration depends on the highly coordinated expression of several genes. The dysregulation of genes associated with regeneration may impede the regenerative capacity of muscle, thereby compromising its functionality [6,7]. These regulatory factors do not act independently, they form a complex regulatory network and signaling pathway to ensure the orderly regulation of skeletal muscle development. Moreover, the MRFs and Myocyte Enhancer Factor 2 (MEF2) families have been studied previously [8]. Myosin heavy chain (MyHC), a well-demonstrated marker of muscle type, is not expressed in myoblasts, but is only expressed in differentiated myotubes. Myogenic differentiation 1 (MyoD) is a determinant of myogenesis initiation in the muscles [9,10]. Myocyte enhancer factor 2A and 2C (Mef2A, Mef2C) are core transcription factors that have an important impact on skeletal muscle fibers [11]. Collectively, these factors cooperatively orchestrates the balance between proliferation and differentiation.
The mitogen-activated protein kinase (MAPK) signaling pathway is a phosphorylated kinase signaling cascade that plays a pivotal role in the long-term effective response of cells and regulates numerous cellular processes, including cell division, differentiation, and release of inflammatory mediators [12,13]. Multiple genes participate in the signal transduction of the MAPK cascade [14,15]. Our previous studies showed that the branching MAPK signaling pathways, c-Jun N-terminal Kinase (JNK)/MAPK, P38 mitogen-activated protein kinase (P38)/MAPK, and Extracellular Regulated protein Kinase (ERK)/MAPK pathways are implicated during myogenic development and muscle regeneration processes [[16], [17], [18]]. It has been established that inhibition of JNK/MAPK signaling promotes the differentiation of C2C12 cells, while the inhibition of P38/MAPK signaling has the opposite effect [16]. ERK/MAPK signaling is inhibited during skeletal muscle development, and inhibition of ERK1/2 promotes myogenesis [17]. Moreover, the method by which these intricate signaling mechanisms collectively regulate the same biological processes need to be further investigated.
Previous research has shown the vital role of non-coding RNAs, such as microRNAs (miRNAs) and lncRNAs, in the development of skeletal muscles [19]. LncRNAs play important roles in various biological functions including epigenetic modification [20,21] and mRNA transcription, splicing, stability, and translation [[22], [23], [24]]. Functionally, lncRNAs can regulate the expression of nearby genes in cis or the expression of distant genes in trans [25]. Recent studies have revealed the role of lncRNAs in maintaining muscle homeostasis [19]. For instance, as one of the earliest identified lncRNAs, the regulation of H19 in skeletal muscle and myocardial development in mammals have received considerable attention. H19 promotes skeletal muscle satellite cells by cis regulation, myogenic differentiation, and muscle formation [[26], [27], [28], [29]]. In our previous studies, the expression levels of lncRNA 4930581F22Rik, lncRNA Sap30bpos, lncRNA Gm41609 and lncRNA Gm26737 have significantly changed during skeletal muscle differentiation. Notably, the lncRNA 4930581F22Rik is consistently highly expressed during skeletal muscle differentiation [30]. However, the precise function of the lncRNAs remains unclear. The expression of 4930581F22Rik has rarely been reported and was recently found to be significantly downregulated in liver tissues of mice infected with C. sinensis [31]. The method by which these intricate signaling mechanisms collectively regulate the same biological processes need to be further investigated.
In the present study, we characterized the expression patterns and potential functions of lncRNA 4930581F22Rik. A gradual increase in lncRNA 4930581F22Rik expression during skeletal muscle differentiation was observed. Knockdown of lncRNA 4930581F22Rik significantly inhibited muscle differentiation and the expression of myogenic markers. It was found that lncRNA 4930581F22Rik modulates the differentiation program by regulating the ERK/MAPK signaling pathway, which can be neutralized by the ERK-specific inhibitor PD0325901. Additionally, in vivo mice injury model result revealed that lncRNA 4930581F22Rik is involved in skeletal muscle regeneration. These results establish a theoretical basis for comprehending the role of lncRNAs in the development and regeneration of skeletal muscles.
2. Materials and methods
2.1. Cell culture
The C2C12 mouse skeletal myoblast cell line was acquired from the Cellular Library of the National Collection of Authenticated Cell Cultures (Shanghai, China). Cells were grown in growth medium (GM) containing Dulbecco's Modified Eagle's Medium (DMEM, Gibco), 100 U/ml penicillin, 10 % fetal bovine serum (FBS, Gibco), and 100 μg/ml streptomycin (1 × penicillin–streptomycin) at 37 °C and 5 % carbon dioxide in an incubator. C2C12 cells were cultured in 6-well plates at a concentration of 5 × 10^4 cells/ml per well. Once the C2C12 cells reached about 90 % confluence, GM was then switched to the differentiation medium (DM) consisting of DMEM with 2 % horse serum (HyClone, Australia). The cells were initially cultured for fewer than five passages.
2.2. Animals
C57BL/6J mice were obtained from GemPharmatech Co., Ltd (Nanjing, China) and were kept in pathogen-free cages at the animal facility with controlled temperature, humidity, and ventilation. All animals handling and procedures were approved by the Animal Care and Use Committee of Jennio Biotech Co., Ltd. (No. 2023-A055). For conducting the mouse muscle injury and regeneration experiment, 25 μl 10 μM of cardiotoxin (CTX, Merck Micropore, 217503) was injected into 6-week-old male tibialis anterior muscle (TA), with 0.9 % saline as a control solution. The regenerating muscles were harvested three days post-injection. The collected muscles were used for hematoxylin and eosin (H&E) staining, or were frozen in liquid nitrogen for subsequent protein or RNA extraction.
2.3. RNA transfection
We acquired small interfering RNA (siRNA) directed against lncRNA 4930581F22Rik and the respective negative control siRNA (si-NC) from GenePharma Co., Ltd (Suzhou, China). Following the manufacturer's instructions, the cells were subjected to reverse transfection using Lipofectamine 2000 (Invitrogen). Briefly, the transfection complex containing siRNA was added to 12-well plates, followed by seeding C2C12 cells at a density of 50,000 cells/well. The final concentration of siRNA for each transfection was 100 nM. After transfection, C2C12 cells were incubated in growth medium for two days until they reached 90 % confluence, and then switched to differentiation medium for three days. Subsequently, the cells were harvested for further analysis. The sequences of the siRNAs are detailed in Table 1.
Table 1.
Sequences of siRNAs.
| sense | antisense | |
|---|---|---|
| si-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| si-4930581F22Rik-#1 | GGAAUAGAAGUCCAUCCUUTT | AAGGAUGGACUUCUAUUCCTT |
| si-4930581F22Rik-#2 | GGGACUCUCCAAUUCACAATT | UUGUGAAUUGGAGAGUCCCTT |
| si-4930581F22Rik-#3 | GCAUAGGUGGAAGCUCUUUTT | AAAGAGCUUCCACCUAUGCTT |
To suppress the ERK signaling pathway's activity, 6 h after siRNA transfection, C2C12 cells were treated with 2 μM or 10 μM PD0325901 (PD, Sigma-Aldrich), and the DMSO (Sigma-Aldrich) treated sample serving as the internal control.
2.4. RNA extraction and qRT-PCR
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen, #15596018). Subsequently, first-strand cDNA was synthesized for PCR analysis utilizing HiScript III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, R323-01). The quantitative real-time PCR was carried out employing ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711-02), with the GAPDH gene serving as the internal control. Relative gene expression levels were determined by the 2−ΔΔCt method. Comparisons between the two groups were performed by using the unpaired t-test. Primers synthesis for qRT-PCR in this study was provided by Shengon Biotech (Shanghai, China). The sequences of the qPCR were detailed in Table 2.
Table 2.
qPCR primers.
| Gene Name | primer | sequence (5'→3′) |
|---|---|---|
| MHC | MHC-RT-qF | CGCAAGAATGTTCTCAGGCT |
| MHC-RT-qR | GCCAGGTTGACATTGGATTG | |
| Mef2C | Mef2c-RT-qF | ATCCCGATGCAGACGATTCAG |
| Mef2c-RT-qR | AACAGCACACAATCTTTGCCT | |
| MyoD | MyoD-RT-qF | CTACCCAAGGTGGAGATCCTG |
| MyoD-RT-qR | CACTGTAGTAGGCGGTGTCGT | |
| MyoG | MyoG-RT-qF | ATCCAGTACATTGAGCGCCTAC |
| MyoG-RT-qR | GACGTAAGGGAGTGCAGATTGT | |
| Ki67 | Ki67-RT-qF | GAGGAGAAACGCCAACCAAGAG |
| Ki67-RT-qR | TTTGTCCTCGGTGGCGTTATCC | |
| 4930581F22Rik | RT-4930581F22Rik-F | GTGCAGAGACCACAGGACAA |
| RT-4930581F22Rik-R | TTACAGGCGTGTACCACCAC | |
| Sap30bpos | RT-Sap30bpos-F | ACATTGCCACTGTGCTCTGT |
| RT-Sap30bpos-R | GCAACTGCAGTGCATCCATT | |
| Gm41609 | RT-Gm41609-F | TGGTCATGATGTTTTGTGCAGG |
| RT-Gm41609-R | GGCCCTTCTAGCCCAAAGTT | |
| Gm29773 | RT-Gm29773-F | CTGTGTAGACCTGGCTGTCC |
| RT-Gm29773-R | GTACACGCCTTCAATCGCAG | |
| Gm41556 | RT-Gm41556-F | TCTGAGCATGAGACTCCTGTC |
| RT-Gm41556-R | GAGGTGTCTCCATGCCTCAG | |
| Gm26737 | RT-Gm26737-F | TTGTGGCGAGATCGGAGTG |
| RT-Gm26737-R | TACCGCTCCAGGCTCCTAAC | |
| GAPDH | GAPDH-RT-qF | CGTCCCGTAGACAAAATGGT |
| GAPDH-RT-qR | TCAATGAAGGGGTCGTTGAT |
2.5. Western blot
The cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Shanghai Wanhaotian Biotechnology) containing a cocktail of phosphatase and protease inhibitors (Roche). Equal amounts of protein extract (20 μg) were separated using SDS-PAGE, which were performed with a 10 % acrylamide separating gel and 4 % acrylamide stacking gel containing 10 % SDS. And then transferred onto a nitrocellulose filter membrane. The cell membranes were blocked with 5 % skim milk for 1 h at room temperature and incubated overnight at 4 °C in the corresponding primary antibodies (diluted to 1:2000 using primary antibody dilution (Beyotime)). The membranes were incubated with a secondary antibody (diluted 1:5000) for 1 h at room temperature for imaging. In the current study, the following antibodies were used: antibodies for MyHC (R&D, MAB4470) were sourced from R&D, antibodies for GAPDH (#2118), MEF2A (#9736), MEF2C (#5030), p-ERK (#4370S), ERK (#4695S), p-JNK (#4671S), JNK (#9258S), p-P38 (#4511S), P38 (#9212S), and c-Raf (#9422) were purchased by Cell Signaling Technology. MyoG (sc-12732) and MyoD (sc-760) were obtained from Santa Cruz. The protein ladder (molecular weight marker, MK) used in this study was purchased from Thermo Fisher Scientific (#26619). Secondary antibodies against mouse (#7076s) and rabbit (#7074s) proteins were purchased from Cell Signaling Technology. All proteins were detected using a chemiluminescent horseradish peroxidase (HRP) substrate (microwell, WBKLS0500), and the chemiluminescence of the membrane was captured using a Bio-Rad luminescence imaging system. The gray value of each image was quantified using an ImageJ analyzer. Results are presented as mean ± SEM. Comparisons between the two groups were performed using the unpaired t-test.
2.6. Luciferase reporter assays
C2C12 cells were plated in 96-well plates at a concentration of 2000 cells per well to investigate the impact of ERK signaling pathway activity on lncRNA 4930581F22Rik knockdown cells. In the next day, the cells were transfected with siRNAs (si-NC, si- # 1, si- # 2, and si- # 3), together with the reporter plasmid (ERK pathway reporter or control plasmid) using Lipofectamine 2000 reagent (Invitrogen). Following this, the cells were cultured in growth medium for two days before switching to differentiation medium for the subsequent three days. Subsequently, cell lysis was performed using passive lysis buffer, and luciferase activity was quantified utilizing a Dual Luciferase Assay Kit (Promega). The transfection of siRNA and pathway reporter constructs was used in 96-well plates at concentrations of 100 nM/well and 50 ng/well, respectively. All luciferase assays were conducted a minimum of three times.
2.7. Data collation software, databases, and statistical methods
The experimental data underwent statistical analysis using Graphpad Prism (version 9). Charts were generated using Adobe Photoshop CS5 and Adobe Illustrator. All quantitative results were presented as mean ± SEM. The statistical significance of the differences between two groups was determined through an unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. ns: No significant difference.
3. Results
3.1. LncRNA 4930581F22Rik is gradually elevated and expression during skeletal muscle differentiation
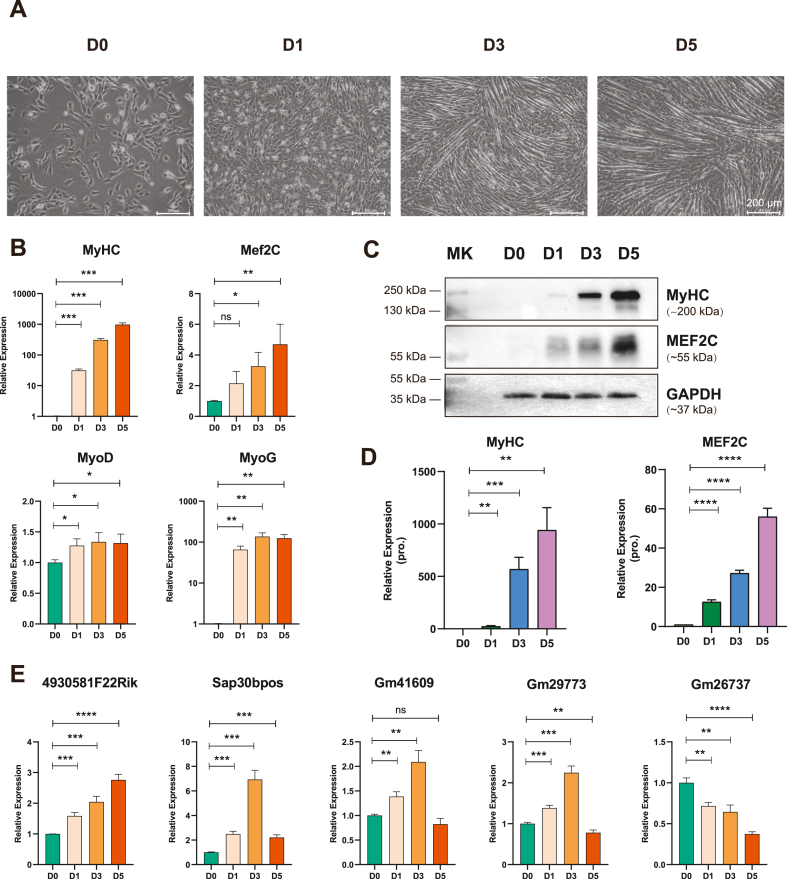
LncRNA 4930581F22Rik is widely expressed in a variety of organs, localizes to chromosome 9 with a full length of 2402 nt, and consists of six exons [32]. The expression of 4930581F22Rik is widely observed in various organs, particularly in the adult liver and kidneys (Supplementary Fig. 1). To explore the expression pattern of lncRNA 4930581F22Rik during the process of skeletal muscle differentiation, we utilized mouse C2C12 myoblasts as a model system to mimic skeletal muscle differentiation. C2C12 myoblasts expanded in the presence of 10 % fetal bovine serum and started differentiated by withdrawal from 2 % horse serum. As previously reported, significant changes in the cell phenotype were observed [30,33]. After three days of differentiation (D3), the cells were fused. After five days of differentiation (D5), prominent myotube formation was evident (Fig. 1A). The RNA expression levels of key differentiation markers such as MyHC, Mef2c, MyoD, and MyoG, exhibited signature profiles throughout the differentiation of C2C12 myoblasts (Fig. 1B). In line with the RNA findings, a significant rise in MyHC and MEF2C protein expression was observed during this progression (Fig. 1C and D, and Supplementary Fig. 2).
Fig. 1.
Expression pattern of lncRNA 4930581F22Rik during skeletal muscle differentiation. (A) Phase-contrast microscopy of differentiating C2C12 myoblasts in growth medium or 1, 3 or 5 days in the differentiation medium. (B) Quantitative real-time PCR analysis of skeletal muscle differentiation markers MyHC, Mef2C, MyoD and MyoG at indicated differentiating time points during C2C12 myoblasts differentiation. (C) Western blot analysis of skeletal muscle differentiation markers MyHC and MEF2C at indicated differentiating time points differentiation. GAPDH was used as a loading control. The molecular weight of the targeting protein is indicated as the marker (MK) showed. (D) Quantification of protein expression levels for skeletal muscle differentiation markers MyHC and MEF2C at indicated differentiating time points during C2C12 myoblasts differentiation. (E) Quantitative real-time PCR analysis of the concerned lncRNAs at indicated differentiating time points during C2C12 myoblasts differentiation. ns: no significant difference. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
Subsequently, we examined the expression patterns of several lncRNAs, including 4930581F22Rik. The upregulation of lncRNA 4930581F22Rik, Sap30bpos, Gm41609, and Gm29773 during C2C12 differentiation was consistent with the findings of our previous study. In particular, expression of lncRNA 4930581F22Rik gradually increased throughout the differentiation process, which prompted us to further investigate its functional role (Fig. 1E).
3.2. Inhibition of lncRNA 4930581F22Rik impedes the process of skeletal muscle differentiation
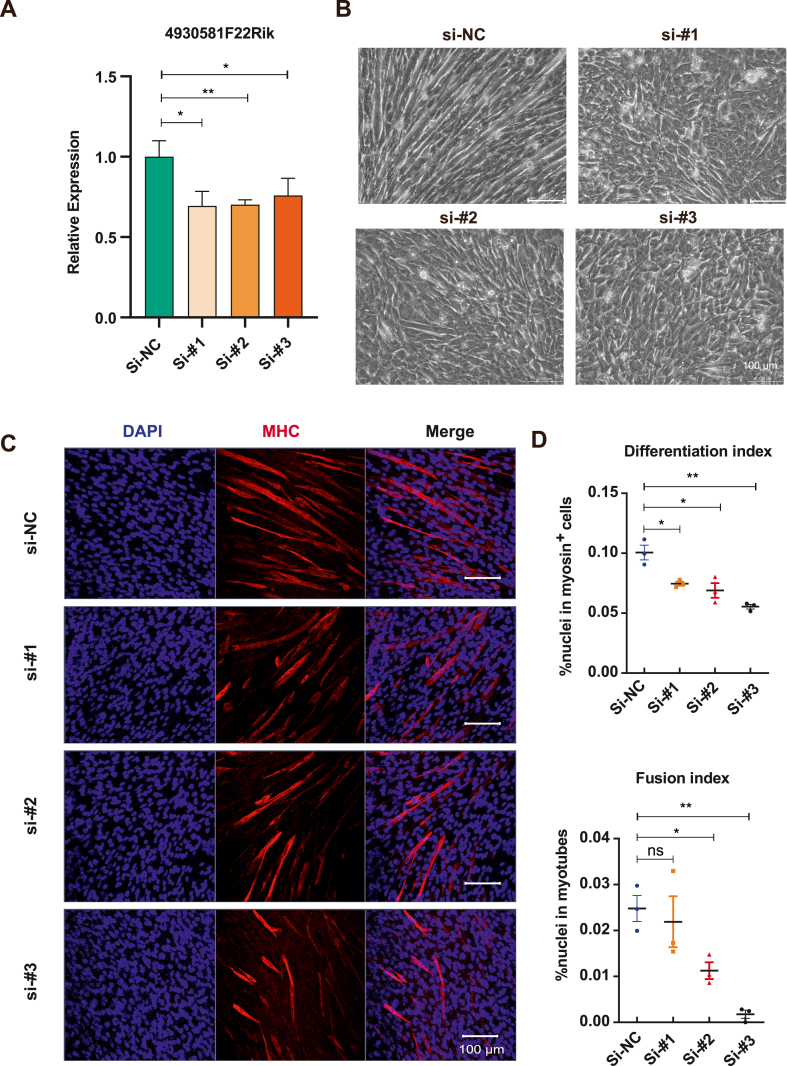
To examine the effect of lncRNA 4930581F22Rik on skeletal muscle differentiation, we used three distinct siRNAs to knock down lncRNA 4930581F22Rik. Following transfection, the cells were cultured in growth medium for two days and subsequently switched to differentiation medium for another three days before being harvested for RNA extraction. RT-PCR analysis showed that all three siRNAs successfully knocked down the lncRNA 4930581F22Rik (Fig. 2A). Morphological examination revealed that, compared with the negative control (si-NC) cells, all three siRNAs effectively impeded muscle cell fusion (Fig. 2B). Immunofluorescence staining of MyHC also showed that myofilament morphology was not only shorter and tapered, but also significantly decreased after lncRNA 4930581F22Rik downregulation (Fig. 2C). Statistical analysis also indicated that lncRNA 4930581F22Rik downregulation not only inhibited myoblast differentiation, but also inhibited the fusion ability of myoblasts (Fig. 2D). These results indicated that lncRNA 4930581F22Rik promotes skeletal muscle differentiation.
Fig. 2.
Knockdown of lncRNA 4930581F22Rik impedes skeletal muscle differentiation. (A) Quantitative real-time PCR analysis evaluated the knockdown efficiency of lncRNA4930581F22Rik with three different siRNAs. (B) The morphology changes of myoblasts that transfected with siRNAs. (C) Effects of 4930581F22Rik knockdown on MyHC expression and myotube formation. Scale bar: 100 μm. (D) Effect on differentiation index and fusion index after knockdown of 4930581F22Rik. ns: no significant difference. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
3.3. Knockdown of lncRNA 4930581F22Rik impedes myogenic markers expression
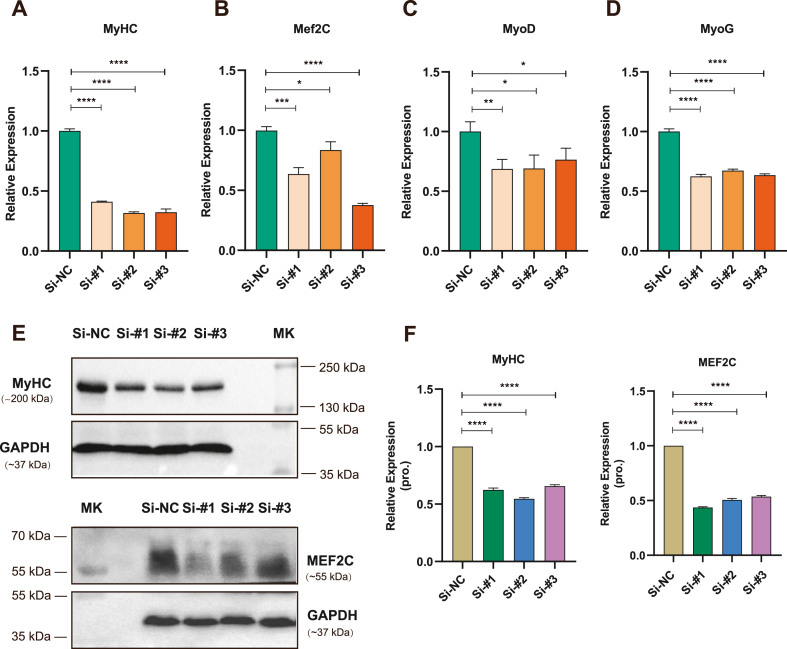
To investigate the impact of lncRNA 4930581F22Rik on skeletal muscle differentiation, lncRNA 4930581F22Rik was knocked down by three individual siRNAs. Following transfection, the cells were then collected to analyze RNA and protein expression levels, evaluating the presence of myogenic markers MyHC, Mef2C, MyoD, and MyoG. Remarkably reduced RNA expression levels were observed for each myogenic marker, particularly MyHC, which exhibited greater than 60 % decrease (Fig. 3A–D). Moreover, western blot analysis confirmed a significant decrease in the protein expression levels of MyHC and MEF2C, consistent with the reduction RNA levels. (Fig. 3E and F, and Supplementary Fig. 3). Thus, experimental results showed that the inhibition of lncRNA 4930581F22Rik led to impaired skeletal muscle differentiation accompanied by decreased expression of key myogenic markers.
Fig. 3.
Knockdown of lncRNA 4930581F22Rik impairs the expression of myogenic markers. (A–D) Quantitative real-time PCR analysis was performed to assess the expression levels of skeletal muscle differentiation markers MyHC, Mef2C, MyoD, and MyoG in myoblasts transfected with siRNAs. (E) Western blot analysis was conducted to examine the protein levels of skeletal muscle differentiation markers MyHC and MEF2C in myoblasts transfected with siRNAs. (F) Quantification of protein expression for skeletal muscle differentiation markers MyHC and MEF2C in myoblasts transfected with siRNAs. GAPDH was used as a loading control. The molecular weight of the targeting protein is indicated as the marker (MK) showed. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
3.4. LncRNA 4930581F22Rik regulates skeletalmuscle differentiation through the ERK/MAPK signaling pathway
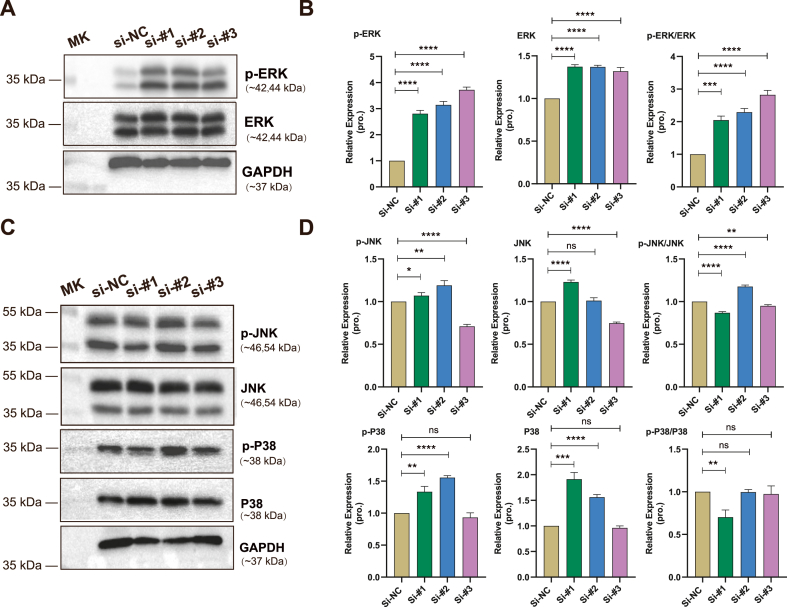
Subsequently, we attempted to elucidate the underlying mechanism by which lncRNA 4930581F22Rik deficiency impairs myogenesis. The MAPK signaling pathway is vital in living organisms and plays important roles in cell proliferation, differentiation, and apoptosis. Our previous research has uncovered the crucial importance of the MAPK signaling pathway in maintaining the cell homeostasis during skeletal muscle development. Therefore, we initially examined three branches of the MAPK pathway, ERK1/2, JNK, and P38 signaling. Western blotting results showed that only the ERK1/2 pathway showed an obviously trend of change. Moreover, analysis demonstrated a significant increase only in the phosphorylated-ERK (p-ERK) protein, with slightly alterations in total ERK protein levels (Fig. 4A and B, and Supplementary Fig. 4A), while minimal changes were observed in the phosphorylated-JNK (p-JNK) and phosphorylated-P38 (p-P38) proteins (Fig. 4C and D, and Supplementary Fig. 4B). These findings illustrate that ERK signaling may be a dominant downstream signaling pathway regulated by lncRNA 4930581F22Rik.
Fig. 4.
lncRNA 4930581F22Rik regulates skeletal muscle differentiation through the ERK/MAPK signaling pathway. (A) Western blot analysis reveals the protein expression levels of p-ERK and ERK in myoblasts transfected with siRNAs. (B) Quantification of protein expression for p-ERK and ERK in myoblasts transfected with siRNAs. (C) Western blot analysis reveals the protein expression levels of p-JNK, JNK, p-38 and P38 in myoblasts transfected with siRNAs. (D) Quantification of protein expression for p-JNK, JNK, p-38 and P38 in myoblasts transfected with siRNAs. GAPDH was used as a loading control. The molecular weight of the targeting protein is indicated as the marker (MK) showed. ns: no significant difference. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
3.5. Inhibiting ERK/MAPK signaling rescue the effects of lncRNA 4930581F22Rik on skeletal muscle differentiation
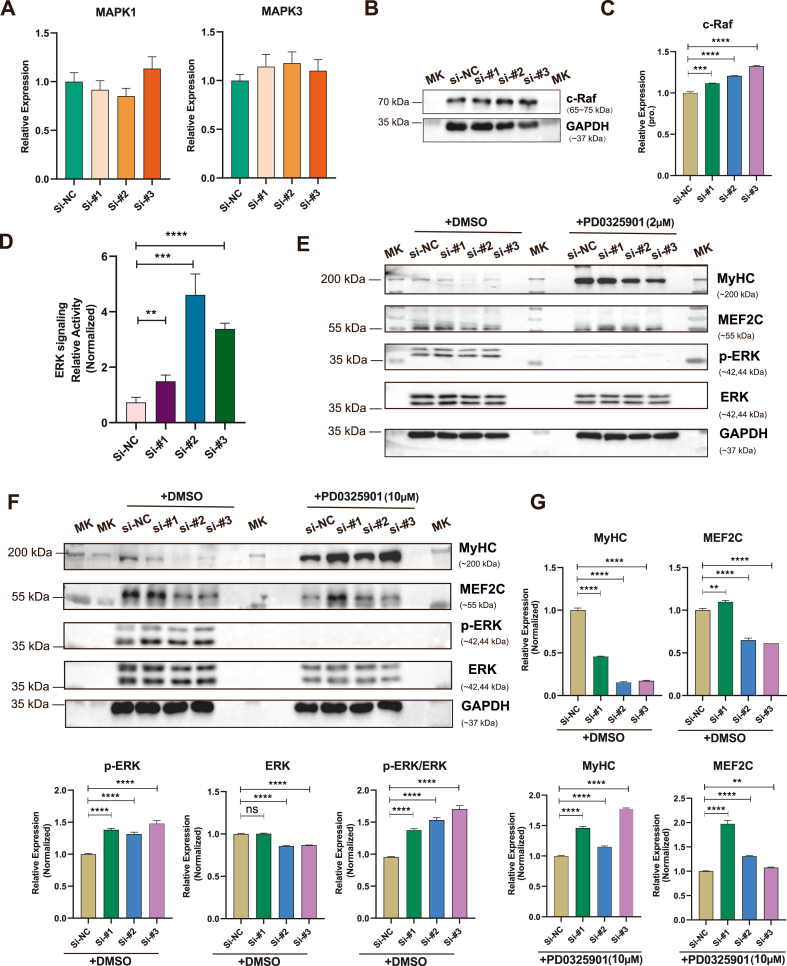
Furthermore, we examined the relationship between the lncRNA 4930581F22Rik and the ERK/MAPK pathway. We detected the RNA expression levels, and found that the RNA expression levels of MAPK1 and MAPK3 corresponding to ERK1 and ERK2 did not change (Fig. 5A). The c-Raf expression, a key upstream signaling molecule in the ERK/MAPK pathway, was notably increased following the depletion of lncRNA 4930581F22Rik (Fig. 5B and C, and Supplementary Fig. 5A). In addition, we investigated the activation of the ERK1/2 signaling pathway upon lncRNA 4930581F22Rik deprivation, and the reporter results significantly improved (Fig. 5D). To investigate the impact of inhibiting the ERK signaling pathway on cell differentiation following the suppression of lncRNA 4930581F22Rik, we exposed the siRNA-transfected cells to the ERK-specific inhibitor PD0325901. ERK phosphorylation indicated that the activity of ERK signaling was significantly inhibited. Comparing with si-NC cells, the lncRNA 4930581F22Rik-deprivating cells with no significant changes in the expression of myogenic markers MyHC and MEF2C under treated with 2 μM PD0325901. These findings suggested that the ERK signaling pathway inhibitor PD0325901 inhibited the siRNA-mediated upregulation of ERK signaling at 2 μM concentration, thus counteracting the downregulation trend of MyHC and MEF2C (Fig. 5E, Supplementary Fig. 5B, and Supplementary Fig. 6). When the siRNA-transfected cells were treated with 10 μM PD0325901, the protein expression levels of the MyHC and MEF2C were significantly increased (Fig. 5F and G, and Supplementary Fig. 5C). The PD0325901 concentration of 10 μM of ERK signaling pathway inhibitor not only inhibited the siRNA-mediated up-regulation of ERK signaling, but also reversed the downregulation of MyHC and MEF2C, showing an upregulation trend. Consistent with the fact that lncRNA 4930581F22Rik promotes skeletal muscle differentiation, the ERK1/2 signaling pathway inhibited skeletal muscle differentiation. Therefore, these findings indicate that lncRNA 4930581F22Rik play roles in the differentiation of skeletal muscle by influencing the ERK/MAPK signaling pathway.
Fig. 5.
A negative regulation between lncRNA 4930581F22Rik and ERK/MAPK pathway. (A) Quantitative real-time PCR analysis was performed to assess the expression levels of MAPK1 and MAPK3 in myoblasts transfected with siRNAs. (B) Western blot analysis shows the expression levels of c-Raf protein in myoblasts transfected with siRNAs. (C) Western blot analysis shows the expression levels of c-Raf protein in myoblasts transfected with siRNAs. (D) Luciferase reporter assay showing the effects of lncRNA 4930581F22Rik knockdown on the activity of ERK1/2 signaling pathway. (E) Western blot analysis of ERK, p-ERK, MyHC, and MEF2C in myoblasts transfected with siRNAs that were treated with 2 μM PD0325901 or DMSO. (F) Western blot analysis of ERK, p-ERK, MyHC and MEF2C in myoblasts transfected with siRNAs that were treated with 10 μM PD0325901 or DMSO. (G) Relative expression in F were calculated. The molecular weight of the targeting protein is indicated as the marker (MK) showed. ns: no significant difference. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
3.6. LncRNA 4930581F22Rik is involved in skeletal muscle injury
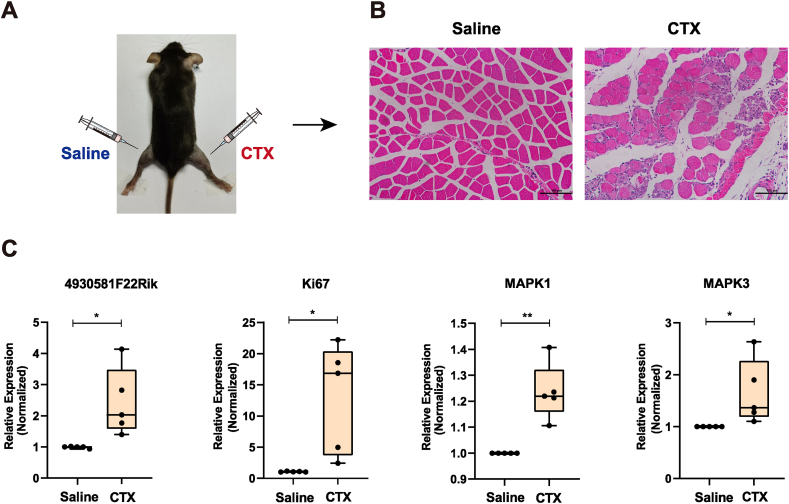
We examined the expression profile of lncRNA 4930581F22Rik during acute skeletal muscle regeneration after CTX injection to further investigate the function of lncRNA 4930581F22Rik in vivo. The right TA muscle was injected with CTX to induce muscle injury, whereas the left TA muscle was injected with an equal volum of saline to act as a negative control (Fig. 6A). Regeneration of adult muscle is supported by the activation of infiltrating macrophages and subsequent MuSCs. During the repair process, immune cells, especially macrophages, enter the damaged skeletal muscle, releasing cytokines, chemokines, and growth factors in the affected area. By activating MuSCs, altering the microenvironment, clearing cellular debris, and promoting regeneration. To assess the early stage of regenerated skeletal muscle tissues, mice were injected with CTX, and TA muscle were collected on day three. Histological analysis of the muscle through H&E staining revealed significant necrosis following CTX treatment, confirming the successful establishment of the injury and regeneration models (Fig. 6B). Expression of lncRNA 4930581F22Rik and Ki67 was measured by qPCR. The expression of lncRNA 4930581F22Rik in the CTX injection group was significantly upregulated, indicating its involvement in regeneration. Ki67 serves as a proliferation marker and is highly expressed in the CTX injection group, with approximately 18-fold upregulation, indicating that MuSCs proliferated rapidly at this stage. Muscle regeneration post-injury involves complex processes. RNA expression levels of MAPK1 and MAPK3 demonstrate an upregulation trend, suggesting their involvement in muscle differentiation and regeneration. (Fig. 6C). Discrepancies between in vivo and in vitro results are often observed. This can be attributed to the complex early-stage muscle regeneration architecture, which involves various processes such as MuSC activation, myoblast proliferation, and myotube formation. The data suggests a possible link between the lncRNA 4930581F22Rik and the early stages of muscle regeneration.
Fig. 6.
LncRNA 4930581F22Rik is implicated in the early stages of muscle injury repair. (A) Construct a muscle injury repair model (n = 5 mice). A muscle injury repair model is established by injecting saline into the left tibialis anterior muscle of mice, while CTX is injected into the right tibialis anterior muscle. (B) Immunohistochemistry results of injured muscle on day 3. (C) The RNA expression levels of lncRNA 4930581F22Rik, Ki67, MAPK1, and MAPK3 were measured after muscle injury. Values were presented as means ± SEM (n = 3). The statistical significance of differences between the two groups was calculated using unpaired t-test.
4. Discussion
Skeletal muscle is the most abundant tissue and protein reservoir. It not only governs movement but also plays a pivotal role in regulating respiration, ingestion, energy expenditure, glucose metabolism, amino acid utilization, lipid homeostasis, and maintenance of an optimal quality of life. Therefore, skeletal muscle regeneration is of paramount concern in the field of regenerative medicine. During myogenesis, proliferation and differentiation are stringently regulated, and any disruption in these processes can lead to muscle dysplasia [8]. In the present study, we demonstrated the expression pattern and potential function of lncRNA 4930581F22Rik in skeletal muscle differentiation and regeneration. LncRNA 4930581F22Rik gradually increases during myogenesis, and its knockdown, significantly impedes differentiation and impairs the expression of myogenic markers. Our findings revealed that lncRNA 4930581F22Rik modulates this process by regulating the ERK/MAPK signaling pathway. Additionally, our data indicates that lncRNA 4930581F22Rik isemerging as a potential player in the initial phase of muscle regeneration.
LncRNAs are a class of regulatory RNAs with diverse roles in cellular processes that participate in chromatin remodeling, gene imprinting, post-transcriptional regulation, and post-translational processing. The functional and regulatory mechanisms of lncRNAs remain unclear due to their poor conservation. Here, we elucidated the expression pattern and function of the little-studied lncRNA 4930581F22Rik in skeletal muscle, which is highly expressed in mouse liver and lung tissues. Its functions in skeletal muscle highlights the development and regeneration of bioprocesses. Mechanism, we have demonstrated a potential relationship between lncRNA 4930581F22Rik and the ERK/MAPK signaling cascade in this study. ERK/MAPK signaling has been well studied in extracellular stimulation response and cell proliferation. Complex interactions and cascading cytokines and signals, coordinate the balance between proliferation and differentiation [34,35]. Skeletal muscles exhibit a remarkable adaptability to environmental stimuli. However, prolonged immobility and aging can result in muscle atrophy, which subsequently leads to muscular weakness and a decline in quality of life. Consequently, the likelihood of illness and mortality significantly increases. Therefore, maintaining muscle homeostasis is of great significance. Our findings expand our knowledge of noncoding RNA functions and noncoding RNA-mediated signaling pathways.
An increasing number of studies have uncovered the roles of lncRNAs in skeletal muscle development and myopathies, including Duchenne muscular dystrophy, facial shoulder brachial muscular dystrophy, skeletal muscle hypertrophy, and amyotrophic lateral sclerosis [36]. Skeletal muscleathophy and muscle regeneration defects are more and more severe health problems in aging societies worldwide. However, effective prevention and treatment strategies are lacking. Moreover, the mechanisms regulating lncRNAs and their functions in skeletal muscle regeneration remain largely unknown. We have elucidated how the lncRNA 4930581F22Rik expression changes during differentiation and regeneration in this research. Moreover, investigating the influence of lncRNA 4930581F22Rik regulation network in muscle clinical samples could offer new perspectives for clinical diagnosis and treatment.
5. Conclusions
Thus, the present study demonstrates that upregulation of lncRNA 4930581F22Rik in myotubes promotes myogenesis via the ERK/MAPK signaling pathway. Additionally, increased expression of lncRNA 4930581F22Rik in mice with cardiotoxin-induced muscle injury, revealed that lncRNA 4930581F22Rik may play a roles in the early stages of muscle regeneration.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by grants from the Natural Science Foundation of Guangdong Province (2022A1515011413, 2021A1515010477 and 2022A1515012159), the National Natural Science Foundation of China (31701116, 82170674), Guangzhou Science and Technology Plan Projects (2023A03J0204, 202206010072, 202102010310, 2023A03J0570 and 202102020104). Guangdong Provincial Basic and Applied Basic Research Fund (2023A1515220196). And the Cultivation Project for National Natural Science Foundation of China from the Third Affiliated Hospital of Sun Yat-sen University (2024GZRPYMS01).
CRediT authorship contribution statement
Wei-Cai Chen: Writing – original draft, Investigation, Conceptualization. Wan-Xin Chen: Writing – original draft, Methodology, Investigation, Data curation. Ye-Ya Tan: Writing – review & editing, Formal analysis, Data curation. Ying-Jun Xu: Writing – original draft. Yi Luo: Methodology. Shi-Yu Qian: Supervision. Wan-Yi Xu: Investigation. Meng-Chun Huang: Methodology. Yan-Hua Guo: Supervision. Zhi-Gang Zhou: Methodology, Data curation. Qi Zhang: Supervision, Funding acquisition. Jian-Xi Lu: Writing – review & editing, Supervision, Conceptualization. Shu-Juan Xie: Writing – review & editing, Supervision, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank professor Zhen-Dong Xiao and Li-Ting Diao for their useful comments and suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30640.
Contributor Information
Wei-Cai Chen, Email: chenwc23@mail.sysu.edu.cn.
Wan-Xin Chen, Email: chenwx68@mail.sysu.edu.cn.
Ye-Ya Tan, Email: yeyatam@hotmail.com.
Ying-Jun Xu, Email: xuyj39@mail.sysu.edu.cn.
Yi Luo, Email: luoy556@mail2.sysu.edu.cn.
Shi-Yu Qian, Email: tqiansy@jnu.edu.cn.
Wan-Yi Xu, Email: 1025856820@qq.com.
Meng-Chun Huang, Email: huangmch7@mail.sysu.edu.cn.
Yan-Hua Guo, Email: gyh9927@163.com.
Zhi-Gang Zhou, Email: zgzhou@jnu.edu.cn.
Qi Zhang, Email: zhangq27@mail.sysu.edu.cn.
Jian-Xi Lu, Email: lujianxi@mail.sysu.edu.cn.
Shu-Juan Xie, Email: xieshj5@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sartori R., Romanello V., Sandri M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat. Commun. 2021;12(1):330. doi: 10.1038/s41467-020-20123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskin K.K., Winders B.R., Olson E.N. Muscle as a "mediator" of systemic metabolism. Cell Metabol. 2015;21(2):237–248. doi: 10.1016/j.cmet.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giordani L., et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell. 2019;74(3):609–621.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Bentzinger C.F., Wang Y.X., Rudnicki M.A. Building muscle: molecular regulation of myogenesis. Cold Spring Harbor Perspect. Biol. 2012;4(2) doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 6.Sartorelli V., Juan A.H. Sculpting chromatin beyond the double helix: epigenetic control of skeletal myogenesis. Curr. Top. Dev. Biol. 2011;96:57–83. doi: 10.1016/B978-0-12-385940-2.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bismuth K., Relaix F. Genetic regulation of skeletal muscle development. Exp. Cell Res. 2010;316(18):3081–3086. doi: 10.1016/j.yexcr.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28(3):225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Tidball J.G. Inflammatory processes in muscle injury and repair. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288(2):R345–R353. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 10.Pillon N.J., et al. Cross-talk between skeletal muscle and immune cells: muscle-derived mediators and metabolic implications. Am. J. Physiol. Endocrinol. Metab. 2013;304(5):E453–E465. doi: 10.1152/ajpendo.00553.2012. [DOI] [PubMed] [Google Scholar]
- 11.Lanz R.B., et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 12.Weston C.R., Davis R.J. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Wagner E.F., Nebreda A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 2009;9(8):537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 14.Edmunds J.W., Mahadevan L.C. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J. Cell Sci. 2004;117(Pt 17):3715–3723. doi: 10.1242/jcs.01346. [DOI] [PubMed] [Google Scholar]
- 15.Lu L., et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013;32(19):2575–2588. doi: 10.1038/emboj.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie S.-J., et al. Inhibition of the JNK/MAPK signaling pathway by myogenesis-associated miRNAs is required for skeletal muscle development. Cell Death Differ. 2018;25(9):1581–1597. doi: 10.1038/s41418-018-0063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie S.J., et al. Dynamic m(6)A mRNA methylation reveals the role of METTL3/14-m(6)a-MNK2-ERK signaling Axis in skeletal muscle differentiation and regeneration. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.744171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S., et al. The functional analysis of transiently upregulated miR-101 suggests a "braking" regulatory mechanism during myogenesis. Sci. China Life Sci. 2021;64:1–12. doi: 10.1007/s11427-020-1856-5. [DOI] [PubMed] [Google Scholar]
- 19.Cesana M., et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aliperti V., Skonieczna J., Cerase A. Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Noncoding RNA. 2021;7(2) doi: 10.3390/ncrna7020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L., Froberg J.E., Lee J.T. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 2014;39(1):35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mousavi K., et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinger M.E., et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4(11) doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frith M.C., et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3(1):40–48. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- 25.Hetzler K.L., et al. The homoeobox gene SIX1 alters myosin heavy chain isoform expression in mouse skeletal muscle. Acta Physiol. 2014;210(2):415–428. doi: 10.1111/apha.12168. [DOI] [PubMed] [Google Scholar]
- 26.Kallen A.N., et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan L., et al. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene. 2000;19(50):5810–5816. doi: 10.1038/sj.onc.1203965. [DOI] [PubMed] [Google Scholar]
- 28.Borensztein M., et al. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development. 2013;140(6):1231–1239. doi: 10.1242/dev.084665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinet C., et al. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development. 2016;143(6):962–971. doi: 10.1242/dev.131771. [DOI] [PubMed] [Google Scholar]
- 30.Xie S.J., et al. Characterization of long non-coding RNAs modified by m(6)A RNA methylation in skeletal myogenesis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.762669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han S., et al. Long non-coding RNA and mRNA expression analysis in liver of mice with clonorchis sinensis infection. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.754224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue F., et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan Y.-Y., et al. PERK signaling controls myoblast differentiation by regulating MicroRNA networks. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.670435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi X., Garry D.J. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20(13):1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 35.Brand-Saberi B. Genetic and epigenetic control of skeletal muscle development. Ann. Anat. 2005;187(3):199–207. doi: 10.1016/j.aanat.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Neppl R.L., Wu C.L., Walsh K. lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 2017;216(11):3497–3507. doi: 10.1083/jcb.201612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.