Abstract
Purpose
The objective of this study was to provide theoretically feasible strategies by understanding the relationship between the immune microenvironment and the diagnosis and prognosis of AML patients. To this end, we built a ceRNA network with lncRNAs as the core and analyzed the related lncRNAs in the immune microenvironment by bioinformatics analysis.
Methods
AML transcriptome expression data and immune-related gene sets were obtained from TCGA and ImmPort. Utilizing Pearson correlation analysis, differentially expressed immune-related lncRNAs were identified. Then, the LASSO-Cox regression analysis was performed to generate a risk signature consisting immune-related lncRNAs. Accuracy of signature in predicting patient survival was evaluated using univariate and multivariate analysis. Next, GO and KEGG gene enrichment and ssGSEA were carried out for pathway enrichment analysis of 183 differentially expressed genes, followed by drug sensitivity and immune infiltration analysis with pRRophetic and CIBERSORT, respectively. Cytoscape was used to construct the ceRNA network for these lncRNAs.
Results
816 common lncRNAs were selected to acquire the components related to prognosis. The final risk signature established by multivariate Cox and stepwise regression analysis contained 12 lncRNAs engaged in tumor apoptotic and metastatic processes: LINC02595, HCP5, AC020934.2, AC008770.3, LINC01770, AC092718.4, AL589863.1, AC131097.4, AC012368.1, C1RL-AS1, STARD4-AS1, and AC243960.1. Based on this predictive model, high-risk patients exhibited lower overall survival rates than low-risk patients. Signature lncRNAs showed significant correlation with tumor-infiltrating immune cells. In addition, significant differences in PD-1/PD-L1 expression and bleomycin/paclitaxel sensitivity were observed between risk groups.
Conclusion
LncRNAs related to immune microenvironment were prospective prognostic and therapeutic options for AML.
Keywords: Prognosis, Acute myeloid leukemia, Long noncoding RNA, Immune microenvironment, Predictive model
1. Introduction
Acute myeloid leukemia (AML) is one of the most aggressive hematologic malignancies, characterized by aberrant molecular heterogeneity and accumulation of immature myeloid progenitor cells in the bone marrow and peripheral blood [1]. The 5-year overall survival (OS) for adults with AML under 60 years of age receiving conventional treatment was approximately 40%, while the median survival time for elderly AML patients (>60 years) was roughly 5–10 months [2]. Chemotherapy is the “gold standard” for treating AML, but most patients experience relapse or death after initial remission. Despite extensive attempts, the overall prognosis for individuals with AML remains dismal with a 5-year OS <30% [[3], [4], [5]]. Hence, there is an urgent need to identify reliable prognostic and targeted therapeutic indicators for AML.
Immune microenvironment plays a key role in tumor development and therapeutic response in many cancers, and interactions with AML cells are important for the remission of conditions such as chemoresistance and disease recurrence, and the ability of immunological microenvironment to regulate immune damage may increase AML patients' chances of survival [6]. Immune checkpoint molecules, including programmed cell death-1 (PD-1) and programmed cell death ligand-1 (PD-L1), play important roles in oncogenesis by maintaining an immunosuppressive tumor microenvironment [7]. With the advancement of detection technology for specific immune microenvironment components [8], the therapeutic approaches to AML have been broadened, facilitating research on patient immune microenvironments and enabling researchers to overcome various immunosuppressive processes [9]. Despite the correlation between many cytogenetic abnormalities with poor prognosis and an increased risk of relapse following treatment, there are still individuals who relapse without unfavourable risk factors [1]. Thus, additional research is required to completely elucidate the molecular mechanisms of AML immune microenvironment. The widespread genomic heterogeneity revealed by current sequencing work in AML has been shown to contribute to prognosis [10], encouraging us to more accurately evaluate the prognosis of AML patients by optimizing biomarkers.
The competitive endogenous RNA (ceRNA) network has been shown to be ubiquitous in the post-transcriptional regulation of gene expression and has attracted widespread attention for its role in suggesting the pathogenesis and prognosis of AML [11]. Long non-coding RNA (lncRNA) is a 200bp-100Kbp transcript product. It binds to unusual and uncharacterized DNA conformation, also impacts AML cell proliferation, apoptosis, and cell cycle. LncRNA affects bone marrow hematopoietic cell development and may be a biomarker for AML's progression and survival [10]. In AML, it is postulated that ceRNAs compete for distinct types of RNA binding sites, thereby influencing the progression and pathogenesis of AML via mutual regulation of expression levels. LncRNAs, being a critical constituent of the ceRNA network, exert a pivotal regulatory influence on AML. Numerous investigations have demonstrated that specific lncRNAs can influence the levels of leukemia-related gene expression through ceRNA antagonism with microRNAs (miRNAs), thus regulating indirectly the inhibitory effects of miRNAs on other target RNAs. Among other biological processes, these lncRNAs may regulate cell proliferation, differentiation, apoptosis, and other critical pathogenesis-related processes in AML [12].
Accumulating evidence suggests that lncRNA is a major mediator and effector of epigenetic alterations in AML, leading to extensive exploration of the role of lncRNA in AML [13]. LncRNA is a recently discovered class of noncoding RNAs that possess mRNA-like transcripts lacking protein-coding potential [14]. This means that incorporating lncRNA into preclinical models is essential for the discovery of prognostic biomarkers. Indeed, growing evidence has revealed that lncRNAs are intimately implicated in tumorigenesis, progression, prognosis, and drug resistance/sensitivity [15], and they are emerging as key regulators in the immune system, especially in inflammatory immune responses and immune-related pathways [16]. Although research on lncRNAs in AML is still in its infancy, evidence of their involvement as essential mediators and effectors of epigenetic changes has spurred epigenetics research [13]. LncRNAs in the immune microenvironment are one of the most extensively studied kinds, regulating gene expression at the epigenetic, transcriptional, and post-transcriptional stages [15]. In AML, lncRNAs regulate the inflammatory immune response, B-cell activation, and vaccination response-related immunological pathways [16]. However, there is insufficient evidence for immune-related lncRNAs to predict AML prognosis and therapy, and their roles in the AML immune microenvironment has not received sufficient attention and warrant additional study.
For this purpose, we obtained the transcriptome expression data and clinical information for patients with AML from The Cancer Genome Atlas (TCGA) and accessed the Immunology Database and Analysis Portal (ImmPort) for immune-related gene sets. Then, a 12-immune-related lncRNA signature was constructed after the least absolute shrinkage and selection operator (LASSO) Cox regression analysis, and forest plot for each factor was used to build the multivariate Cox model using multivariate Cox analysis and stepwise regression analysis. Our following findings demonstrated the potential prognostic significance of a signature of 12 lncRNAs in AML. Fig. 1 depicts the workflow in detail.
Fig. 1.
Workflow chart.
2. Results
2.1. Construction of independent prognostic model containing 12 lncRNAs
We downloaded transcriptome data [normalized log(FPKM+1, 2)] and clinical information of 283 AML patients from TCGA repository as the main body of bioinformatic analysis. After excluding 151 cases with insufficient clinical data, 132 samples were included in the final analysis, randomly devided into training set and validation set.
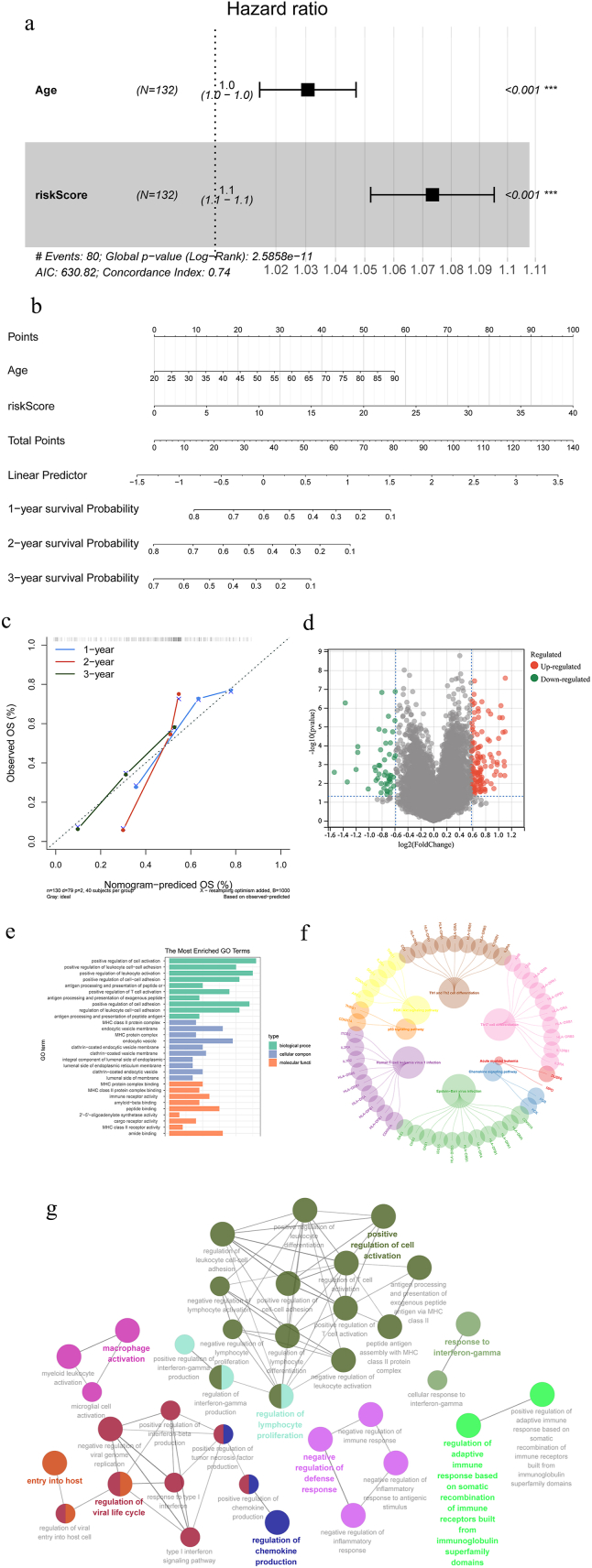
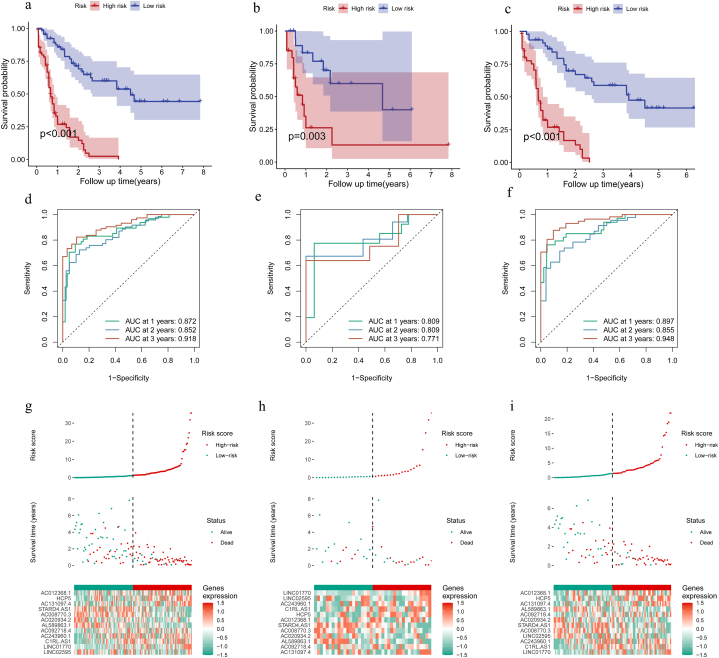
Initially, Pearson correlation analysis was conducted with the aim of finding immune-related lncRNAs with differential expression. Using |r|>0.6 and P < 0.001, 812 lncRNAs present in both databases were identified for subsequent screening (Fig. 2a and b). Long noncoding RNA enrichment analysis (LncSEA) revealed these lncRNAs implicated in tumor apoptotic and metastatic processes. After one-way Cox regression and LASSO analysis, we screened out 18 immune-related lncRNAs associated with prognostic for prediction signature construction (Fig. 2c and d), among which 12 were further identified as prognostic biomarkers for model construction via multivariate Cox analysis and stepwise regression analysis: LINC02595, HCP5, AC020934.2, AC008770.3, LINC01770, AC092718.4, AL589863.1, AC131097.4, AC012368.1, C1RL-AS1, STARD4-AS1, and AC243960.1. Forest plots for each selected lncRNA were displayed in Fig. 2e. Next, we used Kaplan-Meier curves to depict the association between risk score and survival by classifying the TCGA samples into high- and low-risk groups. Fig. 3a–c displays the statistically significant differences between the two groups (P < 0.05). Evidently, the number of death cases increased distinctly as the risk score rose. Area under receiver operating characteristic (ROC) curve (AUC) at 1-, 2-, and 3-year OS in the training set was 0.897, 0.855, and 0.948, respectively (Fig. 3d). This was confirmed in the validation set and in all samples (Fig. 3e and f), indicating the model's efficacy. Fig. 3g–i illustrated the model effect in the form of a risk curve. The above results demonstrated that the risk score is a reliable indicator of predicting OS in AML.
Fig. 2.
Determination of prognostic immune-related lncRNAs in AML. (a) Volcano map of 812 lncRNAs associated with immune-related genes. The abscissa difference represents Pearson correlation coefficient, and the ordinate represents -log10(P). Dots represent immune-related lncRNAs. The blue and red dots represent lncRNAs that meet the threshold, indicating a negative and positive correlation trend with immune-related genes, respectively; (b) bubble diagram of lncRNA functional enrichment. The color represents the enrichment results of different subclasses; (c, d) LASSO screening for characteristic genes: (c) the abscissa represents the percentage of residuals explained by the model, demonstrating the relationship between the number of characteristic genes and the percentage of residuals explained (dev), and the ordinate represents the gene coefficient; (d) the abscissa represents log (Lambda), while the ordinate represents the error of cross validation. The cross validation error at the left dashed line is the smallest. The optimal log (Lambda) value is determined based on this position (lambda. min). Then the corresponding genes and their coefficients, as well as the proportion of residual explained by the model are discovered according to the number of characteristic genes displayed; and (e) forest map of multivariate Cox analysis. The position of the black square represents the HR value of each gene, with the dashed line on the right indicating HR > 1 and on the left indicating HR < 1. The line segments on both sides of the square represent the 95% confidence interval of the HR value. HR, hazard ratio.
Fig. 3.
Construction and validation of risk models. Survival analysis of high- and low-risk groups in the training set (n = 92), (b) validation set (n = 40), and (c) all patient samples (n = 132); ROC analysis results at 1, 2, and 3 years for (d) the training set, (e) validation set, and (f) all patient samples; and risk curves at 1, 2, and 3 years for (g) training set, (h) validation set, and (i) all patient samples. ROC, receiver operating characteristic.
2.2. Verification of the accuracy of the immune-related lncRNA signature
In this section, we applied a series of strategies to guarantee the signature's accuracy as a predictive tool. First, Cox independent prognostic analysis identified age and risk score as independent prognostic indicators for AML patients over other clinical characteristics (Fig. 4a), including French American British, gender, and race (Table 1). Furthermore, the scores based on the two variables age and risk score were added to arrive at the total score (TotalPoint) for forecasting the 1-, 3-, and 5-year OS of AML. Fig. 4b-c depicted the predicted results as column line graphs, with higher scores implying a shorter OS. Using the aforementioned predictive model, a calibration curve was generated, and the concordance index was calculated to be 0.7467 (95% confidence interval [CI], 0.695-0.798). These findings supported the superior predictive ability of the risk score, which can be applied to all patients with AML.
Fig. 4.
Prognostic analysis. (a) Independent prognostic factors using Cox analysis of forest maps; (b) nomogram of the prognosis influencing factors prediction results; (c) correction curves for prediction results of prognostic factors; (d) differential volcano map; (e) GO and (f) KEGG enrichment of differential expression genes; and (g) GO_BP pathway interaction network.
Table 1.
The enrichment results for various subclasses.
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Variable | HR | 95%CI | P | HR | 95%CI | P |
| Age | 1.0348 | 1.0188-1.0511 | 0.0000 | 1.0307 | 1.0146-1.0470 | 0.0002 |
| French American British | 1.0625 | 0.9206-1.2261 | 0.4073 | – | – | – |
| Gender | 0.9624 | 0.6196-1.4949 | 0.8644 | – | – | – |
| Race | 0.8852 | 0.5469-1.4330 | 0.6198 | – | – | – |
| Risk score | 1.0792 | 1.0581-1.1008 | 0.0000 | 1.0734 | 1.0520-1.0953 | 0.0000 |
HR, hazard ratio; CI, confidence interval.
2.3. Exploration on the mechanism of immune-related lncRNA in AML
To investigate the specific mechanism of immune-related genes affecting AML, the R software “limma” package was used to evaluate differentially expressed genes (DEGs) between high- and low-risk groups using Fold Change >1.5 and P < 0.05 as the screening thresholds, and a total of 183 DEGs were discovered, including 49 down-regulated genes and 134 up-regulated genes (Fig. 4d). For exploring the enrichment of DEGs pathways, results showed that the top 10 Gene Ontology (GO) terms with the most enriched signaling pathways were: positive regulation of cell activation, positive regulation of leukocyte-cell adhesion, processing and presentation of peptide or polysaccharide antigens via major histocompatibility complex (MHC) class II antigens, positive regulation of T cell activation, processing and presentation of exogenous peptide antigens via MHC class II antigens, and immune receptor activity (Fig. 4e).
Similarly, Kyoto Encyclopedia of Gene Genomes (KEGG) analysis demonstrated these DEGs regulate immune response-related pathways such as the PI3K-Akt signaling pathway, p53 signaling pathway, Th17 cell differentiation, Th1 and Th2 cell differentiation, AML, and chemokine signaling pathways. In addition, these DEGs were engaged in Epstein-Barr virus infection and human T-cell leukemia virus 1 infection (Fig. 4f). To further explore the functions and relationships of the aforesaid DEGs in biological process, the Cytoscape software module ClueGO + CluePedia enriched 36 GO biological process terms for 183 DEGs, comprising 43 pairs of reciprocal GO term connections, as visualized in Fig. 4g.
2.4. Immune-related lncRNAs can influence immune microenvironment of AML
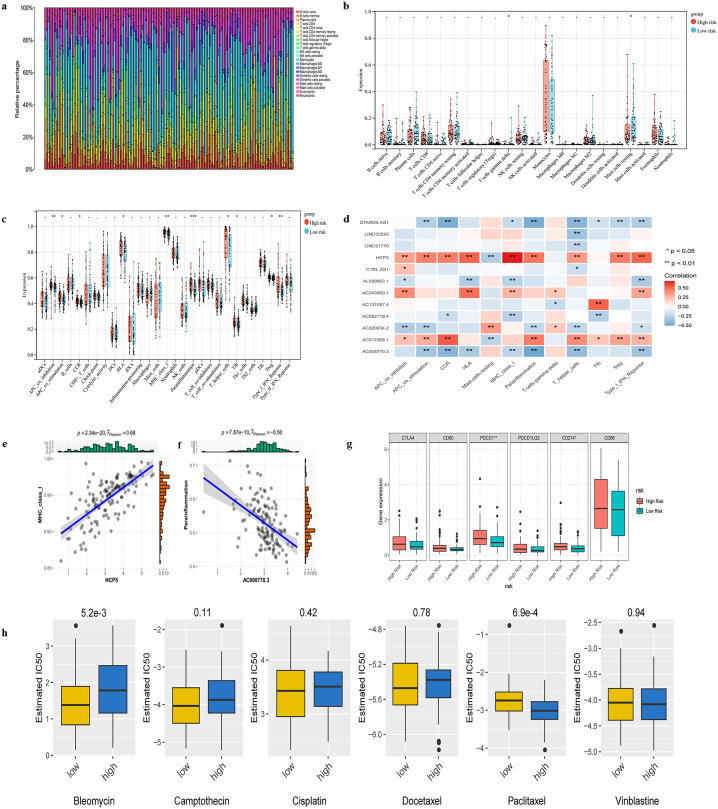
Moreover, we used single-sample gene set enrichment analysis (ssGSEA) and CIBERSORT to determine if immune-related lncRNAs have any relation to tumor-infiltrating immune cells in AML. For each TCGA-AML sample, the proportions of 22 tumor-infiltrating immune cells were determined; T cells gamma delta and mast cells resting showed substantial difference (P < 0.05) between low- and high-risk groups (Fig. 5a-b). To further investigate the infiltration of immune cells in the AML immune microenvironment, we used the CIBERSORT tool to calculate the relative abundance of diverse immune cells based on the transcriptome data of all samples. Multiple immune cell markers, such as APC coinhibition, APC costimulation, CCR, human leukocyte antigen (HLA), MHC class I, parainflammation, T helper cells, Tfh, Treg, and Type I interferon response, were found to have significantly distinct correlations with immune-related lncRNAs in low- and high-risk groups (Fig. 5c). Specifically, HCP5 showed the strongest positive association with MHC class I (r = 0.68, P = 2.34e-20), whereas AC008770.3 demonstrated the strongest negative association with parainflammation (r = −0.50, P = 7.87e-10) (Fig. 5d–f).
Fig. 5.
Immune cell screening. (a) Bar graph of immune cell types based on CIBERSORT algorithm: the abscissa represents the sample, and the ordinate represents the proportion of immune cell composition; (b) differential immune cell box graph based on CIBERSORT algorithm; (c) differential immune cell box diagram based on ssGSEA method; (d) correlation between risk model lncRNA and differential immune cells; (e-f) correlation scatter chart; and (g) differences in immune checkpoint genes among different risk groups. *P < 0.05, **P < 0.01, ***P < 0.001; and (h) results of IC50.
2.5. Analysis and exploration of subtypes based on the immune-related lncRNAs
We investigated a new subtype classifying method of AML using the risk score, and all the samples were divided into two groups in this way. Analysis of the expression levels of six common immune checkpoints between two groups revealed significant differences in PD-1 and PD-L1 expression (Fig. 5g). In the chemotherapeutic drug sensitivity analysis, there were also distinct differences in the half-maximal drug inhibitory concentration (IC50) of bleomycin and paclitaxel between risk groups (Fig. 5h).
2.6. Key ceRNA network
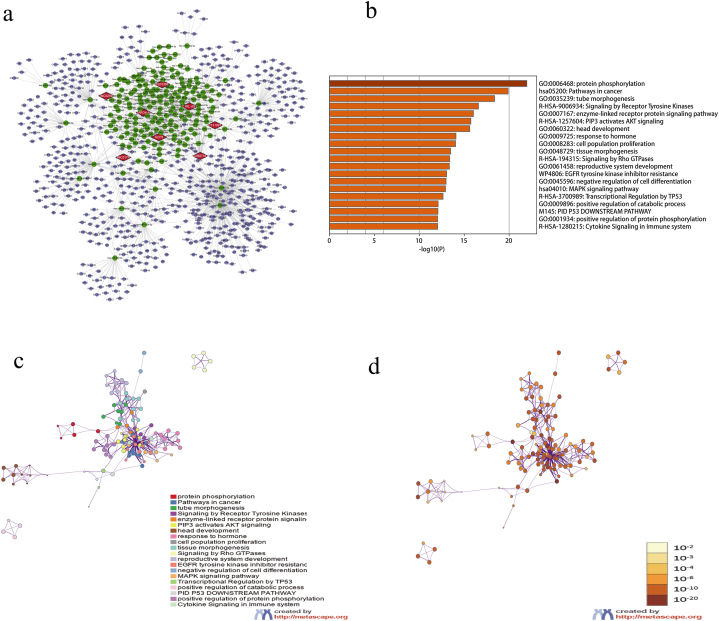
In order to find out the upstream and downstream regulatory factors of immune-related lncRNAs, we constructed the ceRNA network of 12 lncRNAs included in the signature. We eventually got 186 miRNAs and 622 mRNAs, and cytoscape software was used to visualize the interactions of them (Fig. 6a). The signaling pathway enrichment analysis of these mRNAs showed that immune-related lncRNAs may be involved in the regulation of protein phosphorylation, cancer-related pathways, MAPK signaling pathway, hormone response, and also related to miRNA in the tumor (Fig. 6b). Fig. 6c and d depicted the links between the enriched terms.
Fig. 6.
The ceRNA screening. (a) ceRNA network diagram; (b) functional enrichment column diagram of target genes; and (c-d) target gene enrichment network diagram in the ceRNA network. The color on the left represents the enrichment pathway, and the color on the right represents the P-value size.
3. Discussion
AML comprises a diverse collection of hematological malignancies, all of which have been definitively linked to mutations occurring in various genes implicated in cellular processes such as proliferation, differentiation, and apoptosis. AML cells have evolved various strategies to elude the detection and elimination by the host immune system. For instance, cancer stem cells may utilize immune evasion mechanisms to regulate the tumor immune microenvironment (TIME) and may also interact with TIME to expedite the progression of the disease [17]. Despite the fact that numerous malignancies have demonstrated the critical role of the immune microenvironment in tumorigenesis, progression, and therapeutic response, its precise function in AML remains obscure. The abnormal expression of diverse biological activities of lncRNA, an essential prognostic biomarker in the biological and pathological processes of AML, is associated with the development and clinical outcome of AML and has the potential to aid in diagnosis and prognosis [4,18]. The involvement of lncRNA in tumorigenesis and immune responses is especially noteworthy. Cell type-specific gene expression and other functional evidence support the view that lncRNA has unique biological activity in the AML immune microenvironment [19]. This highlights the importance of investigating immune-related lncRNAs and developing risk prediction models for AML prognosis. We obtained immune-related lncRNAs with differential expression in AML from relevant databases and used them to build a predictive model for AML risk prediction. Additionally, we identified upstream and downstream regulatory factors of immune-related lncRNAs and investigated how immune-related genes affect AML.
The ceRNA network mechanism significantly contributes to the classification and prognosis of tumor risk and promotes tumor occurrence and development. Despite the fact that miRNA, circRNA, and lncRNA are all implicated in the pathogenesis of AML, comprehensive analyses of the ceRNA network of circRNA-lncRNA-miRNA-mRNA in AML prognosis continue to be scantly published [19,20]. LncRNA expression is an important predictive indicator in the biological and pathological process of AML [4,18]. Some studies have used the tumor immune microenvironment classification to determine the extent of cellular infiltration in AML immune microenvironment and developed prognostic models based on this information, but these efforts have been limited by only involving cell components without considering genes [21]. This study developed and validated a differential immune-related lncRNAs-based prognostic model. The results of the verification indicate a positive correlation between the manifestation of biomarkers and the prognosis of the patient. Its significance lies in the fact that it may reveal whether the patient's present treatment regimen is associated with an improved prognosis. A study [21] devised the TIME classification to assess the extent of cell infiltration and developed and validated prognostic models in response to the identification of numerous cellular components in the immune microenvironment of AML. This was, nevertheless, restricted in scope by its focus solely on cellular components and its failure to account for the influence of heredity.developed and validated prognostic models utilizing the TIME classification to assess the extent of cell infiltration. This was, nevertheless, restricted in scope by its focus solely on cellular components and its failure to account for the influence of heredity. Zhong et al. [22] created a m6A-related lncRNA risk model for independent prediction of OS in AML patients, correlated with clinicopathological features and immune infiltration, suggesting that lncRNAs evaluated with this methodology could predict outcomes and develop immunotherapies for AML patients. The tiny number of lncRNAs they initially retrieved led to unrepresentative conclusions. Instead, we selected immune-related lncRNAs for modeling and identified their associations with tumor immune infiltration to improve AML patients' prognosis and immunotherapy response, demonstrating the importance of lncRNAs in assessing tumor immune infiltration. As shown by the validation set and all samples, high-risk patients had substantially shorter OS than low-risk individuals. The risk and calibration curves confirmed the ability to effectively predict survival outcomes. The combination of risk scores and clinicopathological variables provide individualized, effective patient outcome monitoring. Our model's predictive performance better reflects AML patients' OS, which facilitates identifying immune regulation prospects and therapeutic targets.
Through an exhaustive search of TCGA and GEO databases, we identified five of the twelve lncRNAs screened in this investigation that had previously been documented in the context of blood diseases: LINC02595, HCP5, AC092718.4, C1RL-AS1, and STARD4-AS1. HCP5, which is positively correlated with a bleak prognosis and is significantly expressed in AML, has been implicated in the pathogenesis of the disease. It is anticipated to pave the way for a novel approach to targeted therapy of AML.
The lncRNA STARD4-AS1, which is implicated in the JAK/STAT cascade, has been identified in patients with myelodysplastic syndrome [23]. While MHC_class_I, AC008770.3, and parainflammation are associated with the prognosis of malignant tumors, hematological diseases have not yet been the subject of research in this area [24,25]. Confirming the use of the remaining seven lncRNAs in hematological diseases is necessary for the development of novel approaches to AML therapy. Inferring the characteristics from the most significant positive and negative correlations between the HCP5 gene and MHC_class_I cells and the AC008770.3 gene and parainflammation cells, respectively, this study analyzed the correlation between the aforementioned immune cells and characteristic genes. HCP5 and AC008770.3 genes potentially regulate the progression of AML mediated by immune cells. Gradually elucidating the functions and mechanisms of the an increasing number of biomarkers identified in AML will facilitate a more comprehensive analysis of the immune microenvironment characteristics of AML patients and offer novel avenues for the development and implementation of therapeutic strategies.
In addition, AC008770.3 showed the strongest negative association with parainflammation in this study. Although MHC class I, parainflammation, and AC008770.3 are related to the prognosis of malignancies, there lacks relevant reports in the field of hematology [24,25]. Several other lncRNAs have previously been described in malignant diseases. Patients with myelodysplastic syndrome exhibit elevated levels of STARD4-AS1, which is implicated in the JAK/STAT cascade response [23]. LINC02595 is identified as an oncogene in colorectal cancer, which may exerts its effect on the NF‐κB signaling pathway [26]. Chen et al. [27] discovered that knockdown of AC092718.4 by siRNA substantially inhibited tumor cell proliferation and accelerated cell apoptosis, indicating its potential as a lung adenocarcinoma biomarker. C1RL-AS1 was reported associated with tumor necrosis factor, tightly associated with the regulation of tumor immune therapy, and could serve as an independent prognostic biomarker in glioblastoma multiforme [28]. Further research is required to confirm the roles of these lncRNAs in hematological disorders for gaining new insights into the development of innovative AML treatments. This will assist describe the AML immune microenvironment and provide new opportunities for medication selection and implementation.
Due to the scarcity of AML treatment options, researchers have combined ex vivo drug sensitivity with genomics, transcriptomics, and clinical annotation for large cohorts of AML patients to uncover functional genomic correlations, making transcriptomic profiling-based genetic marker identification a promising approach for tracking cancer prognosis [29]. The majority of individuals with AML have resistance to conventional chemotherapy; combination therapy with targeted molecular inhibitors seems promising. In this study, we utilized LASSO Cox regression analysis to inform the development of an immune microenvironment-related risk model comprising 12 immune-related lncRNAs. In addition, risk curves and calibration curves validated its predictive capability with regard to patient outcomes. The integration of risk scores and clinicopathological factors presents the potential for individualized prognostic monitoring applications. In contrast to previously documented models, our model exhibits superior predictive performance in discerning the OS of AML patients, elucidating potential immune regulation prospects, and facilitating the identification of druggable targets in AML patients.
High- and low-risk patients showed substantial variations in PD-1 and PD-L1 expression. Thus, individuals with poor response to inhibitors targeting PD-1 and PD-L1 may benefit from drugs that block PD-L2, CTLA4, CD80, and CD86. From this perspective, pertinent characteristics of immune checkpoints may indicate AML patients' immune status and highlight potential immunotherapeutic implications, although the underlying mechanisms require further investigation.
In addition, the drug sensitivity analysis showed risk group variations in bleomycin and paclitaxel IC50s. Current research focuses on the relationship and mechanism between lncRNAs and tumor immune microenvironment, aiming to explore the potential application and prospect of drug metabolism pathway in it [30]. Multiple mechanisms allow bleomycin and paclitaxel to achieve therapeutic objectives by inducing cell necrosis or apoptosis in AML, raising the possibility that they could be candidates for AML [31,32]; nevertheless, numerous concerns remain about the efficacy, safety, and relevance to patient prognosis.
LncRNA exerts the role of ceRNA by inhibiting its miRNA target genes [26]. The ceRNA network's function in clarifying the pathophysiology and prognosis of AML has attracted considerable attention, thanks to its widespread existence in the post-transcriptional modulation of gene expression [11]. The ceRNA network mechanism promotes tumorigenesis and progression, and contributes to tumor risk classification and prognosis. AML pathogenesis involves miRNAs, circRNAs, and lncRNAs, but comprehensive analysis of circRNA-lncRNA-miRNA-mRNA ceRNA network in AML prognosis is rarely reported [11,33]. Herein, our ceRNA network with lncRNAs as the centre indicated that immune-related lncRNAs may be involved in the regulation of protein phosphorylation, cancer-related pathways, MAPK signaling pathway, hormone response, as well as miRNA regulation in tumors.
Nevertheless, this study has certain limitations. Firstly, the range of treatment medicines is rather limited, making it impossible to compare treatment results and choose more appropriate chemotherapy drugs. Furthermore, it is imperative to identify the composition of immune cells (MHC_class_I and parainflammation) and assess the levels of specific genes (HCP5 and AC008770.3) chosen by this model when samples from AML patients become accessible. Subsequently, correlation analysis and validation should be performed.
4. Conclusion
In this study, we built a lncRNA-miRNA-mRNA ceRNA network by searching various databases, and found that lncRNAs are closely linked to the immune microenvironment of AML patients, and that specific lncRNAs have the potential to predict poor prognosis in patients with varying risks. The identified genes HCP5 and AC008770.3 could play essential roles in controlling immune cell-mediated AML development. We hope that this study's findings will help narrow down on potential prognostic biomarkers that influence risk prediction in the immune microenvironment of AML tumors, thoroughly examine the features of AML patients' immune environments, and develop a prognostic risk model based on the pertinent markers. This will assist the clinic in identifying potential targets and fresh avenues for investigation for the treatment and early detection of AML.
5. Materials and methods
5.1. Data acquisition and collection
AML RNA-seq expression level data and clinical information in TCGA were downloaded from the UCSC Xena (https://xena.ucsc.edu/) database. Sample with missing clinical information was removed during analysis. ImmPort (https://www.immport.org/home) was used to retrieve the immune-related gene set.
6. Screening of immune-related lncRNAs
In the first step, we selected common lncRNAs and conducted Pearson correlation analysis in TCGA and ImmPort dataset, and differentially expressed immune-related lncRNAs were screened by the standard correlation coefficient |r|>0.6 and P < 0.001, with 816 required lncRNAs acquired. A functional analysis of differentially expressed lncRNAs was performed using the LncSEA database.
6.1. Survival analysis and clinicopathological characteristics analysis
Data from patient samples was randomly split into training and validation sets on a 7:3 ratio to examine the association between differentially expressed immune-related lncRNAs and patient survival. Following univariate Cox proportional hazard regression (to eliminate genes with P < 0.05) and LASSO analysis, a prognostic model comprised of 12 lncRNAs was derived. Kaplan-Meier estimator was applied to draw survival curves for each lncRNA or between high- and low-risk groups, using SURVIVAL from the R package. Next, univariate and multivariate independent prognostic analysis, as well as the ROC curve analysis, were used to assess the predictive accuracy and reliability of the risk model. The log-rank P-value and hazard ratio (HR) with 95% CI were analyzed.
Additionally, column line graphs from clinical data were created by the R package “rms” to investigate the prognosis of clinicopathological characteristics and risk models. The contribution of each variable to patient survival was then determined by incorporating clinicopathological variables into a Cox independent prognostic analysis.
6.2. Functional enrichment analysis
A list of DEGs between low- and high-expression groups was created using the “limma” package of R (version 3.52.2), employing Fold Change >1.5 and P < 0.05 as criteria. In addition, GO and KEGG pathway enrichment analysis of DEGs were carried out using the R package “clusterProfiler” (version 4.0.5). The enrichment of DEGs pathways was investigated using the online tool Metascape (http://metascape.org/), along with the R software.
7. Associations of lncRNAs with immune cell tumor infiltration
Through ssGSEA in the “GSVA” R package, which reliably retrieves immune-related gene sets for individual sample [22], a violin diagram depicting the differences between high- and low-risk groups of tumor-infiltrating immune cells was generated. Pearson correlation analysis was utilized to investigate the associations between the lncRNAs employed in the model and various subsets of the immune cells.
7.1. Analysis of immune checkpoints and drug sensitivity in subgroups
In distinct risk groups of TCGA-PRAD patients, differences in the expression of immune checkpoint genes were analyzed. Six commonly reported immune checkpoints were chosen, including PD-1, PD-L1, PD-L2, CTLA4, CD80, and CD86 [7]. For drug sensitivity analysis, IC50 of six common chemotherapeutic agents—bleomycin, camptothecin, cisplatin, docetaxel, paclitaxel, and vinblastine—was calculated by pRRophetic package and compared between high- and low-risk groups.
7.2. Key ceRNA networks mining
Since lncRNAs can regulate the expression of downstream mRNAs via miRNAs, the MiRcode (http://mircode.org/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) databases were applied to predict the miRNAs interacting with immune-related lncRNAs and their target genes. Using Cytoscape version 3.9.2 software, we then merged the results from TargetScan, miRDB, and miRTarBase provided by miRWalk and maintained the target regulatory pairs that appeared in all three databases simultaneously.
7.3. Statistics analysis
Statistical analyses were conducted through R software (version 4.1.2). Wilcox test was employed to evaluate tumor-infiltrating immune cells in high- and low-risk groups. Differences in survival between high- and low-risk groups of AML patients were represented in Kaplan-Meier curves. The Pearson correlation analysis was conducted to calculate correlation coefficients. The threshold for statistical significance was set at P < 0.05.
Funding
This research did not receive any specific funding.
Ethical approval
Not Applicable.
Informed consent
Not Applicable.
CRediT authorship contribution statement
Meng Zhang: Writing – original draft, Conceptualization. Li-Li Zhang: Writing – original draft, Conceptualization. Ling-Bo Yi: Investigation, Conceptualization. Xiao-Nian Tu: Investigation, Conceptualization. Ying Zhou: Writing – original draft. Dai-Yang Li: Writing – review & editing. Han-Chun Xue: Writing – review & editing, Data curation. Yu-Xia Li: Writing – review & editing, Data curation. Zhong-Zheng Zheng: Writing – review & editing, Supervision.
Declaration of competing interest
None.
Acknowledgements
We would like to express our gratitude to authors for assistance with contribution.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30616.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Marando L., Huntly B. Molecular landscape of acute myeloid leukemia: prognostic and therapeutic implications. Curr. Oncol. Rep. 2020;22:61. doi: 10.1007/s11912-020-00918-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan T., Wang S., Feng H., Xu J., Zhang M., Yao Y., Xu K., Niu M. Preclinical evaluation of the ROCK1 inhibitor, GSK269962A, in acute myeloid leukemia. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1064470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu D., Zhang B., Wu S., Zhang Y., Xie J., Ning W., Jiang H. Prognosis and characterization of immune microenvironment in acute myeloid leukemia through identification of an autophagy-related signature. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.695865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo H., Zhang Y., Hu N., He Y., He C. Systematic construction and validation of an RNA-binding protein-associated prognostic model for acute myeloid leukemia. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.715840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Hao J.P., Uddin M.N., Wu Y., Chen R., Li D.F., Xiong D.Q., Ding N., Yang J.H., Ding X.S. Identification and validation of inferior prognostic genes associated with immune signatures and chemotherapy outcome in acute myeloid leukemia. Aging (Albany NY) 2021;13:16445–16470. doi: 10.18632/aging.203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F., Jiang Y., Wang L., Chen Y., Zhang Y., Ma M. One-chip isolation of drug-resistant acute myeloid leukemia cells with CXCR4-targeted magnetic fluorescent nanoprobes. Nanomaterials. 2022;12 doi: 10.3390/nano12101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X., Ma L., Zhang X., Huang L., Wei J. Targeting PD-1/PD-L1 pathway in myelodysplastic syndromes and acute myeloid leukemia. Exp. Hematol. Oncol. 2022;11:11. doi: 10.1186/s40164-022-00263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sendker S., Reinhardt D., Niktoreh N. Redirecting the immune microenvironment in acute myeloid leukemia. Cancers. 2021;13:1423. doi: 10.3390/cancers13061423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tettamanti S., Pievani A., Biondi A., Dotti G., Serafini M. Catch me if you can: how AML and its niche escape immunotherapy. Leukemia. 2022;36:13–22. doi: 10.1038/s41375-021-01350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong F., Yao F., Cheng Y., Liu J., Zhang N., Li S., Li M., Huang B., Wang X. m6A-related lncRNAs predict prognosis and indicate immune microenvironment in acute myeloid leukemia. Sci. Rep. 2022;12:1759. doi: 10.1038/s41598-022-05797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y., Su Y., Wang S., Liu Y., Jin L., Wan Q., Liu Y., Li C., Sang X., Yanget al L. Identification of circRNA-lncRNA-miRNA-mRNA competitive endogenous RNA network as novel prognostic markers for acute myeloid leukemia. Genes. 2020;11:868. doi: 10.3390/genes11080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin L., Li B., Wang S., Tang Y., Fahira A., Kou Y., Li T., Hu Z., Huang Z. Construction of an immune-related prognostic signature and lncRNA‒miRNA-mRNA ceRNA network in acute myeloid leukaemia. J. Leukoc. Biol. 2024 doi: 10.1093/jleuko/qiae041. [DOI] [PubMed] [Google Scholar]
- 13.Wurm A.A., Pina C. Long non-coding RNAs as functional and structural chromatin modulators in acute myeloid leukemia. Front. Oncol. 2019;9:899. doi: 10.3389/fonc.2019.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z., Liu L., Weng S., Guo C., Dang Q., Xu H., Wang L., Lu T., Zhang Y., Sunet al Z. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat. Commun. 2022;13:816. doi: 10.1038/s41467-022-28421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-Miranda G.M., Hidalgo-Miranda A., Barcenas-Lopez D.A., Nunez-Enriquez J.C., Ramirez-Bello J., Mejia-Arangure J.M., Jimenez-Morales S. Long non-coding RNA and acute leukemia. Int. J. Mol. Sci. 2019;20:735. doi: 10.3390/ijms20030735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R., Wu S., Wu X., Zhao P., Li J., Xue K., Li J. Immune-relatedlncRNAs can predict the prognosis of acute myeloid leukemia. Cancer Med. 2022;11:888–899. doi: 10.1002/cam4.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang X., Gong Y. One stone, two birds: N6-methyladenosine RNA modification in leukemia stem cells and the tumor immune microenvironment in acute myeloid leukemia. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.912526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang P., Xie M., Wei Y., Xie X., Chen D., Jiang Z. A 10-long non-coding RNA-based expression signature as a potential biomarker for prognosis of acute myeloid leukemia. Med Sci Monit. 2019;25:4999–5004. doi: 10.12659/MSM.917182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park E.G., Pyo S.J., Cui Y., Yoon S.H., Nam J.W. Tumor immune microenvironment lncRNAs. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbab504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Jing X.B., Wang Z.C., Han Q.K. HCP5, as the sponge of miR-1291, facilitates AML cell proliferation and restrains apoptosis via increasing PIK3R5 expression. Hum. Genom. 2021;15:38. doi: 10.1186/s40246-021-00340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng T., Cui L., Huang W., Liu Y., Si C., Qian T., Deng C., Fu L. The establishment of a prognostic scoring model based on the new tumor immune microenvironment classification in acute myeloid leukemia. BMC Med. 2021;19:176. doi: 10.1186/s12916-021-02047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Q., Xu H., Deng R., Li N., Mu R., Qi Z., Shen Y., Wang Z., Wen J., Zhaoet al J. Landscape of prognostic m6A RNA methylation regulators in hepatocellular carcinoma to aid immunotherapy. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.669145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szikszai K., Krejcik Z., Klema J., Loudova N., Hrustincova A., Belickova M., Hruba M., Vesela J., Stranecky V., Kundratet al D. LncRNA profiling reveals that the deregulation of H19, WT1-AS, TCL6, and LEF1-AS1 is associated with higher-risk myelodysplastic syndrome. Cancers. 2020;12:2726. doi: 10.3390/cancers12102726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S., Li X., Tang C., Kuang W. Inflammation-related long non-coding RNA signature predicts the prognosis of gastric carcinoma. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.736766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang L., Xia W., Yao L., Wu Q., Hua L., Cheng G., Wang Z., Zhao R. Long non-coding RNA profile study identifies an immune-related lncRNA prognostic signature for prostate adenocarcinoma. Int. Immunopharm. 2021;101 doi: 10.1016/j.intimp.2021.108267. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z., An Y., Wang N., Dong X., Kang H. LINC02595 promotes tumor progression in colorectal cancer by inhibiting miR-203b-3p activity and facilitating BCL2L1 expression. J. Cell. Physiol. 2020;235:7449–7464. doi: 10.1002/jcp.29650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Yu Y., Yuan Y., Chen X., Zhou F., Li Y., Wang P., Jiang X., Tian S., Ren W. A novel long noncoding RNA AC092718.4 as a prognostic biomarker and promotes lung adenocarcinoma progression. Aging (Albany NY) 2022;14:9924–9941. doi: 10.18632/aging.204426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long S., Wu B., Yang L., Wang L., Wang B., Yan Y., Jiang J., Yang B., Zhou Q., Shiet al M. Novel tumor necrosis factor-related long non-coding RNAs signature for risk stratification and prognosis in glioblastoma. Front. Neurol. 2023;14 doi: 10.3389/fneur.2023.1054686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottomly D., Long N., Schultz A.R., Kurtz S.E., Tognon C.E., Johnson K., Abel M., Agarwal A., Avaylon S., Bentonet al E. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell. 2022;40:850–864. doi: 10.1016/j.ccell.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z., Chen Y., Ma L., Chen Y., Liu J., Guo Y., Yu T., Zhang L., Zhu L., Shu Y. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol. Ther. 2022;30:3133–3154. doi: 10.1016/j.ymthe.2022.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeung M., Hurren R., Nemr C., Wang X., Hershenfeld S., Gronda M., Liyanage S., Wu Y., Augustine J., Leeet al E.A. Mitochondrial DNA damage by bleomycin induces AML cell death. Apoptosis. 2015;20:811–820. doi: 10.1007/s10495-015-1119-z. [DOI] [PubMed] [Google Scholar]
- 32.Wan Y.F., Guo X.Q., Wang Z.H., Ying K., Yao M.H. Effects of paclitaxel on proliferation and apoptosis in human acute myeloid leukemia HL-60 cells. Acta Pharmacol. Sin. 2004;25:378–384. [PubMed] [Google Scholar]
- 33.Liu Y., Cheng Z., Pang Y., Cui L., Qian T., Quan L., Zhao H., Shi J., Ke X., Fu L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019;12:51. doi: 10.1186/s13045-019-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.