ABSTRACT
Fungal phytopathogens cause significant reductions in agricultural yields annually, and overusing chemical fungicides for their control leads to environmental pollution and the emergence of resistant pathogens. Exploring natural isolates with strong antagonistic effects against pathogens can improve our understanding of their ecology and develop new treatments for the future. We isolated and characterized a novel bacterial strain associated with the species Burkholderia cenocepacia, termed APO9, which strongly inhibits Zymoseptoria tritici, a commercially important pathogenic fungus causing Septoria tritici blotch in wheat. Additionally, this strain exhibits inhibitory activity against four other phytopathogens. We found that physical contact plays a crucial role for APO9’s antagonistic capacity. Genome sequencing of APO9 and biosynthetic gene cluster (BGC) analysis identified nine classes of BGCs and three types of secretion systems (types II, III, and IV), which may be involved in the inhibition of Z. tritici and other pathogens. To identify genes driving APO9’s inhibitory activity, we screened a library containing 1,602 transposon mutants and identified five genes whose inactivation reduced inhibition efficiency. One such gene encodes for a diaminopimelate decarboxylase located in a terpenoid biosynthesis gene cluster. Phylogenetic analysis revealed that while some of these genes are also found across the Burkholderia genus, as well as in other Betaproteobacteria, the combination of these genes is unique to the Burkholderia cepacia complex. These findings suggest that the inhibitory capacity of APO9 is complex and not limited to a single mechanism, and may play a role in the interaction between various Burkholderia species and various phytopathogens within diverse plant ecosystems.
IMPORTANCE
The detrimental effects of fungal pathogens on crop yields are substantial. The overuse of chemical fungicides contributes not only to environmental pollution but also to the emergence of resistant pathogens. Investigating natural isolates with strong antagonistic effects against pathogens can improve our understanding of their ecology and develop new treatments for the future. We discovered and examined a unique bacterial strain that demonstrates significant inhibitory activity against several phytopathogens. Our research demonstrates that this strain has a wide spectrum of inhibitory actions against plant pathogens, functioning through a complex mechanism. This plays a vital role in the interactions between plant microbiota and phytopathogens.
KEYWORDS: plant fungal pathogen, Septoria, Pythium, oomycete, antifungal
INTRODUCTION
Crop losses due to fungal pathogens constitute a major challenge to food safety and the bioeconomy (1). Wheat is the most extensively cultivated crop worldwide and a vital source of calories for humans (2). However, high-density cultivation and variable climatic conditions render wheat vulnerable to multiple pathogens (3, 4). One of the most devastating of which is Zymoseptoria tritici (5–7), which causes Septoria tritici blotch (STB), a foliar disease of wheat. STB leads to a ~5%–10% decline in wheat yields every year (8), and is typically managed through the use of fungicides, crop rotation, and resistant wheat cultivars (9).
While alternative methods for controlling fungal pathogens have been developed, modern agricultural systems still rely heavily on chemical fungicides (10). However, this practice leads to environmental pollution and the emergence of fungicide-resistant strains, which ultimately diminishes the effectiveness of these chemical agents (11, 12). The discovery and development of novel natural products and inhibitors with diverse antifungal mechanisms can effectively control fungal diseases and decelerate the emergence of pathogen resistance (13, 14).
Microorganisms are a promising source for novel treatments of pathogens, as various microbes have been shown to prevent and reduce plant disease via both direct and indirect mechanisms (15–18). Direct mechanisms refer to microbe-pathogen interactions, such as the production of antifungal compounds or competition for nutrients and space, leading to reduced fungal growth. Indirect mechanisms involve microbe-host interactions, which enhance the immunity of the host plant against pathogen infections (19–21).
Burkholderia is a versatile bacterial genus in soil- and root-associated ecosystems, containing strains that produce natural products (9, 22–24), promote plant growth (25–29), and degrade pollutants (30), attributes that have facilitated its widespread presence across diverse ecological niches (31). Some species within this genus, including species within the Burkholderia cepacia complex (Bcc) (32, 33) are recognized as opportunistic human pathogens (34, 35). However, in recent years, several novel Burkholderia species with plant-beneficial traits that are rarely associated with human infection have been identified (29, 36). Of these, the soil-dwelling Burkholderia cenocepacia is particularly interesting as it encompasses strains with the capability to suppress various phytopathogens and promote plant growth (29, 37–39). Although some strains of B. cenocepacia have been characterized, the mechanism of inhibition is currently poorly understood, and many new subspecies remain unexplored. Therefore, B. cenocepacia may play an important ecological role in the plant root microbiome and shows promise as a potential source of novel antifungal compounds for controlling plant fungal diseases.
Here, we phenotypically and genotypically characterize a novel B. cenocepacia strain, designated APO9, which was isolated from the root of a healthy cucumber plant in a Pythium aphanidermatum (PA)-infested greenhouse. We found that APO9 exhibits strong antifungal activity against the destructive fungal pathogen Z. tritici and four other phytopathogens, which is reduced in the absence of physical contact. Genomic examination of APO9 showed that the genome contains nine classes of biosynthetic gene clusters (BGCs) and three types of secretion systems (types II, III, and IV). By screening a mutant library, we identified five genes with significantly reduced inhibitory effects, which are also widely present in the Burkholderia genus. These findings indicate that APO9 possesses extensive inhibitory capabilities against plant pathogens and through a complex mechanism, playing a significant role in the interactions between plant microbiota and phytopathogens.
MATERIALS AND METHODS
Strain collection
The GFP (Green fluorescent protein) -tagged Z. tritici strain K7099 used in this study was obtained from Syngenta (Basel, Switzerland). The pathogen was cultivated in potato dextrose broth medium (PDB, BD, USA) and maintained in 20% glycerol solution and skim milk at −80°C. B. cenocepacia strain APO9 was isolated from the root of a healthy cucumber plant cv. Romi (Hazera, Israel) in a Pythium aphanidermatum-infested greenhouse in Hefer Valley in central Israel. Other bacteria were previously isolated from soil at various locations in the Faculty of Agriculture in Rehovot (Hebrew University, Israel) (40).
Antifungal activity screens by kChip and off-Chip validation
The Z. tritici was streaked onto potato dextrose agar (PDA, BD, USA) plates and grown at 18°C for 4 days. Afterward, the spores were washed down with PDB, and the spore concentration was diluted to 2 × 106 cells mL−1. Bacterial strains were inoculated from −80°C stock directly into 1 mL of PDB medium in 96 deep-well plates and grown overnight at 30°C, 700 rpm (Heidolph, Germany). The starters were normalized to OD600 0.02. The normalized cultures will be utilized to generate droplets for the kChip screen system, which functions as an ultrahigh-throughput coculture platform (41, 42).
Before droplet generation, a “color code” was assigned to each bacterial and fungal culture and control medium. This code was a unique ratio of three fluorescent dyes (Thermo Fisher, AlexaFluor: 555, 594, 647, final dye concentration of 2.5 M) specific to each input. The dyes did not interfere with GFP and bacterial growth, as described by Kehe (42). Droplets were produced on a Bio-Rad QX200 Droplet Generator; 2% (wt/wt) fluorosurfactant (3M Novec 7500) was added to stabilize the droplets. Approximately 20,000 1 nL droplets were generated in fluorocarbon oil from 20 µL input of cultures for each kChip loading. For each input (~49 screen species, Z. tritici monoculture, one hygromycin and one blank control), about 5,000 droplets were generated, totaling ~260,000 droplets. Bacteria and Z. tritici cultures were combined at a ratio of 1:1 to ensure droplets contained a final concentration of OD600 = 0.01 bacterial species and 1 × 106 cells mL−1 of Z. tritici. The droplets were then pooled on the chip and incubated at 18°C for 96 h, and cultures were scanned every 24 h. The number of replicates varied for each droplet combination, ranging from 9 to 50 with an average of 20. GFP fluorescence and averaging all replicates were used to evaluate growth changes.
Off-chip validation was carried out in micrometer plates using the same concentrations as used on the kChip. APO9 coculture with Z. tritici, Z. tritici monoculture, and Gh24 coculture with Z. tritici (as a control, since it does not exhibit inhibitory activity toward Z. tritici growth) were included with three biological replicates and three technical replicates. The cultures were incubated at 18°C for 96 h, and growth changes were assessed by measuring fluorescence using the Microplate Reader (Synergy H1, BioTek, USA) every 24 h.
Microscopic examination
Samples for imaging were taken from cultures of different bacteria cocultured with Z. tritici for 96 h. Culture samples were transferred onto slides, covered with coverslips, and imaged at 60× magnification with a GFP filter using a Nikon Eclipse Ti2 microscope (Japan).
Non-contact coculture
The non-contact coculture assay was carried out in the Teflon Micro-equilibrium dialysis plate (HTD 96B; HTDialysis LLC, USA), which is separated by a polycarbonate membrane (0.1 µm, Sterlitech, USA) in the middle, allowing metabolites to move freely through the dialysis membrane while cells cannot. In brief, the dialysis membranes were soaked in sterilized deionized water for 1 h, then in 70% ethanol for 15 min, and finally washed three times in sterilized deionized water. The equilibrium dialysis plate was assembled by sandwiching conditioned membranes between rows of wells (150 µL culture per chamber) and clamped tightly to the assembled apparatus (HTD Universal Base, USA) (Fig. 2A). The split plate was sealed with sealing film (AeraSeal BS-25, Excel Scientific, USA), and conducted in a shaker (600 rpm, Heidolph, Germany) maintained at 18°C for 96 h. The chamber concentration was double the expected concentration, so that at equilibrium, both chambers contained the expected concentration (bacterial OD600 = 0.01 and 1 × 106 cells mL−1 of Z. tritici). For each experimental combination, measurements were performed in 40 replicates, and Gh11 controls were performed in six replicates. Following 96 h of incubation, aliquots were obtained from both sides, and all samples were read for OD and GFP using the plate reader (Synergy H1, BioTek, USA).
Testing of spent media, volatile, and pH
To determine whether APO9 secreted secondary metabolites to inhibit the growth of Z. tritici, a supernatant experiment was employed. Based on the time of inhibition of Z. tritici growth by APO9, we selected cultures after 24 h of incubation. The suspensions of each condition contained in the original cultures (i.e., spent cell-free culture filtrate) were prepared using the following procedure. Three starters were prepared: APO9 monoculture (OD600 = 0.01), Z. tritici monoculture (1 × 106 cells mL−1), and APO9 and Z. tritici coculture (OD600 = 0.01 and 1 × 106 cells mL−1 of Z. tritici) in PDB medium at 18°C for 24 h, resulting in a viable spent culture medium. The cells in the spent medium were centrifuged (15,000 × g for 10 min), and the spent medium obtained by following the above procedure was vacuum filtered through a 0.22 µm Millipore filter to furnish a cell-free culture medium. Z. tritici was then cultured separately in the different spent media obtained above (the ratio was two portions of spent medium and one portion of concentrated fresh PDB). We evaluated the growth of Z. tritici in both monoculture and coculture with APO9, using fresh PDB medium as the control condition. GFP fluorescence was measured after 96 h of incubation at 18°C.
To evaluate the effect of APO9 volatile organic compounds (VOCs), segmented Petri dishes (9 cm diameter) were used in the experiment. Three experimental designs were employed: the first with Z. tritici streak monoculture on both sides as a control; the second with a Z. tritici streak on one side and an APO9 streak on the other; the third with a Z. tritici streak on one side and APO9 and Z. tritici streaks on the other. The plates were immediately sealed with Parafilm and incubated at 18°C for 96 h. The effects of the VOCs were monitored visually daily.
The experiment aimed to assess the pH and GFP fluorescence of different cultures after 48 h of growth. Three cultures were grown in PDB and ALkyl Ester (AE) Broth medium, comprising APO9 coculture with Z. tritici, APO9 monoculture, and Z. tritici monoculture. Utilizing 1M HCl and 1M NaOH, we adjusted the pH of PDB and AE media to a pH value of 3.5, 4, 5, 6, 7, or 8. Z. tritici was then cultured in these differentially adjusted media for a duration of 96 h, with GFP fluorescence being monitored at 24-h intervals. The pH of each culture was measured using a pH meter (PHOENIX Instrument EC-30, Germany), while the GFP fluorescence was measured using a plate reader (Synergy H1, BioTek, USA).
Whole genome sequencing, assembly, annotation, and visualization
Genomic DNA was extracted from a single APO9 colony using the Exgene Cell SV genomic DNA extraction kit (GeneAll, Seoul, Korea) according to the manufacturer’s instructions. genomic DNA (gDNA) yield and quality were examined using a Qubit flurometer (Thermo Fisher Scientific Inc., Waltham, MA) and a Nanodrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and its integrity was verified by gel electrophoresis (1% agarose wt/vol). The gDNA was sequenced using short (Illumina MiSeq) and long (Oxford Nanopore Minion) read sequencing technologies at Genotypic Technologies (Bengaluru, India) as described below. A volume of 1 µg of DNA from each isolate was used for Nextera XT DNA library preparation using the manufacturer’s protocol (Cat# FC-131–1024), and libraries were sequenced on an Illumina HiSeq X Ten sequencer (Illumina, San Diego, USA). In tandem, sequencing was performed on an Oxford Nanopore GridION X5 sequencing (Oxford, UK) using a SpotON flow cell R9.4 (FLO-MIN106) in a 48-h sequencing protocol. The quality of the genomes was analyzed using BUSCO (43). Trimmomatic software version 0.39 (44) was used for removal of adaptors and low-quality sequences using the following parameters: ILLUMINACLIP:adapters.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. Subsequently, Unicycler version 0.4.8 (45) was used for hybrid assembly of the trimmed Illumina paired-end and Nanopore reads using defaults parameters.
Phylogenetic tree construction and metabolite BGC analysis
The genomes used in this study were downloaded from the National Center for Biotechnology Information (NCBI) databases and are listed in Tables S3 and S4. The similarity of APO9 was defined by calculating the average nucleotide identity (ANI) shared between the genomes of all available Burkholderia spp. and B. cenocepacia species using FastANI (v.1.33) (46). BactaxR (47) was used to create ANI dendrograms, and FastANI visualization of the reciprocal mappings computed among species and genera was performed to estimate the ANI value using genoPlotR (48). The 95% average nucleotide identity species threshold was generally used. The bioinformatics tool antiSMASH v.6.1.1 (49) was used to analyze the BGCs of secondary metabolites.
Construction of mutant library and evaluation of antagonistic activity of Tn5 mutants
The transposon mutant library of APO9 was generated using the EZ-Tn5 Transposase (MA126E, Lucigen, USA) following the manufacturer’s instructions. In the APO9 strain, DNA transformation was challenging due to the presence of multiple restriction and modification systems that cleave the unmodified DNA. To enhance the transformation efficiency, we added restriction inhibitors (MA180E, Lucigen, USA). After transformation, approximately 1,602 independent transposon insertion mutants were obtained, and the library was saved and stored at −80°C.
To evaluate the antagonistic properties of APO9 transposon mutants, individual mutants were grown in 200 µL of Nutrient Broth (NB) medium in 96-well plates for 24 h. Next, 5 µL of each culture was gently combined with 200 µL of Z. tritici::GFP (1 × 106 cells mL−1) and incubated at 18°C for 96 h. The growth ofZ. tritici in monoculture, in coculture with APO9, and in coculture with Gh24 served as controls. The GFP fluorescence was measured after incubation, and high green fluorescence indicated Tn5 mutants that were defective in killing, calculated as:
We cocultured Z. tritici with APO9 and 14 mutants for 4 days, followed by plating the cultures and incubating them for an additional 5 days before scanning. We then quantified the area of black fungi phenotype using ImageJ as follows: convert the images to 8-bit grayscale, invert the colors, apply thresholding using the “setThreshold (116, 255, ‘raw’)” method to segment the image, and finally, analyze and measure the area characteristics of the detected particles using “Analyze Particles,” with parameters “size = 10-Infinity circularity = 0.50–1.00 display exclude clear summarize.” Subsequently, we calculated the correlation between fluorescence values and plating results.
Mutant identification and functional prediction
We validated the insertion sites on the genome of 14 mutants that were demonstrated to have reduced inhibition efficiency. Identification of the Tn5 insertion mutants was done by inverse PCR as previously described (50). Genomic DNA was extracted (K0721, Thermo, USA) from 14 Tn5 mutants, digestion by restriction endonuclease SalI (NEB, USA) at 37°C for 6 h, and ligated overnight at 16°C using T4 DNA ligase (NEB, USA). Inverse PCR was performed with the Tri_2_F and Tri_2_R primers (Table S1) and the resulting PCR products were sequenced by Macrogen Europe (Amsterdam, The Netherlands). The Tn5 insertion points were identified by comparing the flanking sequences with the genome of APO9 and Tn5 transposon sequence, and were presented on the genomic map using Proksee. Functional prediction was performed using NCBI Blast and UniProt.
In addition, we amplified and sequenced the 16S rRNA gene to ensure that all 14 mutants were APO9 strains. Meanwhile, the Tn5 trimethoprim cassette and each specific gene in Tn5 wild type and mutant was amplified by designing site-specific primer pairs, which were designed based on the nucleotide sequences of each gene (Table S1). The size of the mutant was 629 bp larger than the wild type due to the insertion of Tn5 cassette. The PCR products were analyzed on 1.2% agarose gels.
Phylogenetic analyses and the identification of genes related to inhibitory activity
Protein identity searches for five mutant genes with significantly reduced inhibitory effects were performed with BLAST v2.14.1. All search results for sequence similarity were filtered using an E-value (expect value) threshold of ≤1e − 5, retaining only the sequence with the highest identity for each bacterial species. If the sequence identity of the highest identical sequence exceeds 80%, the gene is considered to be present in a genome. Conversely, if it falls below 80%, the gene under investigation is deemed to be absent in the genome. The genomic sequences used for these searches were obtained from the NCBI databases and are listed in Table S6. The 16S ribosomal RNA sequences were utilized to construct a phylogenetic tree using MEGA11 software by the neighbor-joining method, for the analysis of bacteria belonging to various classes.
Plate confrontation assay
In order to test the effect of APO9 on different pathogens phyla, plate confrontation plate was conducted. The pathogens Pythium aphanidermatum, Rhizoctonia solani (RS) and Fusarium oxysporum f. sp. radicis-cucumerinum (FORC) were laboratory stock, and Fusarium oxysporum f. sp. lycopersici race 2 4287 (FOL) was kindly provided by Shay Covo laboratory at the Hebrew University of Jerusalem, Israel.
For the plate confrontation assays, a 1 cm bacteria strip was evenly spread at the center of PDA plates and incubated at 30°C for 24 h, with sterile Distilled water (DW) as a control. The mycelial plug of each fungus (6 mm in diameter, taken from the margins of a 3-day-old colony of four different pathogens) was then placed on both poles of the PDA plates with the bacterial strip or control (three replicates for each treatment). The plates were further incubated at 25°C for 5 days, and the areas of mycelial growth of phytopathogens for both disks were marked. The width of the inhibition zone was recorded, including the short and the long distance (Fig. S11A). Fungal growth inhibition was evaluated by the distance of inhibition zone against the phytopathogens, calculated as:
where A1 means average area covered by phytopathogens in the presence of DDW (control), and A2 means average area covered by phytopathogens in the presence of the test bacterium.
Statistical analysis
The data were subjected to analysis of variance (ANOVA) test using SPSS version 26.0 software (IBM SPSS, USA). Differences were considered statistically significant at P < 0.05 according to Duncan’s multiple range test or Dunnett’s multiple comparison test.
RESULTS
APO9 strongly inhibits Z. tritici
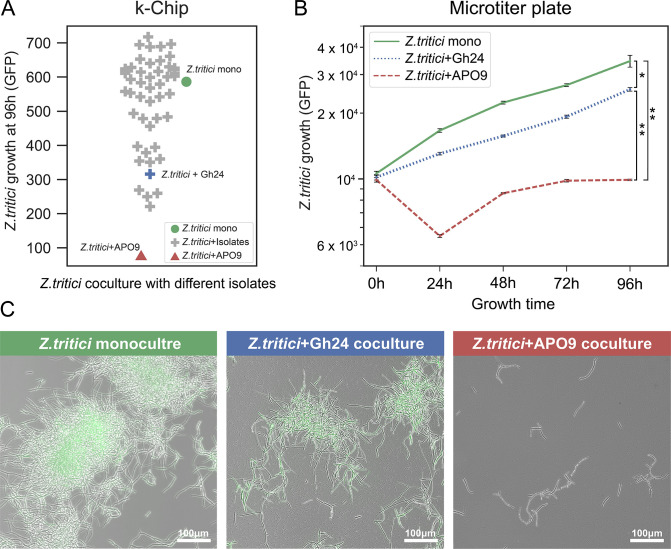
We screened 49 bacterial soil isolates (Materials and Methods) for inhibition of Z. tritici using the kChip system, and determined that APO9 strongly inhibited the growth of Z. tritici on the chip (Fig. 1A; Fig. S1). Off-chip validation in microtiter plates further confirmed this result, APO9 substantially inhibited Z. tritici with significantly lower fluorescence than Z. tritici monoculture and coculture with another bacterial strain (Gh23) that showed limited inhibition on the kChip (P < 0.01; Fig. 1B). Moreover, microscopic observation revealed that coculture with APO9 caused growth inhibition of Z. tritici, and loss of fluorescence signal was compared to the control culture (Z. tritici monoculture and coculture with Gh24; Fig. 1C).
Fig 1.
B. cenocepacia APO9 strongly inhibits Z. tritici. (A) Screening for growth inhibition of Z. tritici with 49 isolates using kChip. Z. tritici monoculture was used as the control. Growth was quantified by the change in GFP fluorescence (arbitrary units) observed after 96 h of incubation, normalized to the initial fluorescence measurements. (B) Validation of bacterial inhibition in microtiter plates. Statistical comparisons were conducted using Dunnett’s multiple comparison test to compare the fluorescence results at 96 h across different cocultures, where * denotes P < 0.05 and ** denotes P < 0.01. (C) Microscopic examination is carried out after 96 h of culturing from the microtiter plate.
Inhibition efficiency is reduced in the absence of physical contact
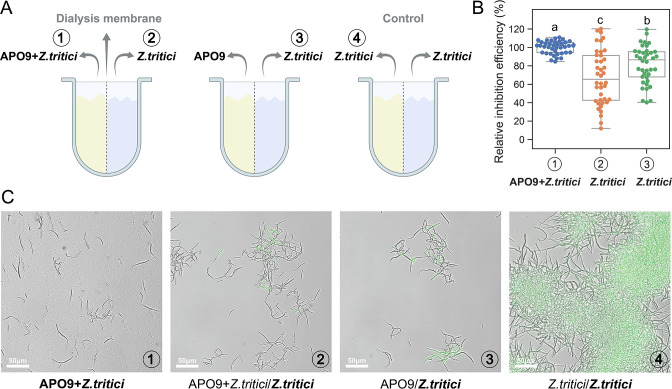
To investigate whether the inhibitory effect of APO9 is contact-dependent, we employed a split-plate assay where each well is divided into two compartments separated by a dialysis membrane that is permeable to nutrients and other small molecules, but not to bacterial and fungal cells (Fig. 2A). Using this plate, we compared the inhibitory efficiencies of cocultures with and without physical contact between APO9 and Z. tritici. We observed that the inhibitory efficiency was significantly reduced in the absence of physical contact (Fig. 2B and C; Fig. S2). The relative inhibition of the coculture without physical contact was reduced by 18.58% compared to that of the coculture with physical contact (P < 0.05) (Fig. 2B). Furthermore, when the other side of the Z. tritici monoculture was a coculture of APO9 and Z. tritici, a 32.43% reduction in inhibitory effect was observed (P < 0.05). However, these inhibitory effects were still significantly higher than those of Z. tritici monoculture and a control coculture (Gh11) (P < 0.05) (Fig. S2). Microscopic inspection revealed that there was no physical contact between APO9 and Z. tritici in coculture, displaying a faint fluorescence compared to when they were directly cocultured (Fig. 2C, 1 vs 2 and 3). These results indicated that the inhibition is likely partially contact-dependent.
Fig 2.
Inhibition efficiency is reduced in the absence of physical contact. (A) Schematic diagram of the split-plate device. (B) The relative inhibition efficiency is the inhibition efficiency after 96 h of incubation under the corresponding conditions divided by the GFP fluorescence of Z. tritici in contact coculture with APO9. Bold fonts and numbers indicate the measurement of the corresponding condition in panel A. Each dot indicates 1 of 40 replicates for each condition. Values with different letters (A–C) indicate statistically significant differences among treatments (P < 0.05), Duncan’s tests. (C) Microscopic examination after 96 h of incubation from split plates.
We next investigated whether APO9 secreted secondary metabolites or produced volatile compounds to inhibit the growth of Z. tritici. Spent media from both APO9 monoculture and APO9-Z. tritici cocultures did not exhibit antifungal activity (Fig. S3), suggesting that APO9 does not produce soluble extracellular compounds that antagonize Z. tritici. In the volatile compound assay, coculturing APO9 and Z. tritici in segmented Petri dishes showed no inhibitory activity, indicating that APO9 did not produce volatile compounds that inhibit Z. tritici (Fig. S4A and B). Nevertheless, when APO9 and Z. tritici grew on the same side, the growth of Z. tritici was inhibited, showing no spore production (Fig. S4C).
To test whether the inhibition may be related to pH modification, we measured the pH of both mono- and cocultures. In PDB medium, APO9-Z. tritici cocultures acidified the medium (pH~3.84), whereas in AE medium the medium was alkalized (pH~8.32; Fig. S5A). Nonetheless, APO9 antagonized Z. tritici in both media (Fig. S5B). We then adjusted the initial pH of the PDB and AE media to a wide range of values (pH = 3.5, 4, 5, 6, 7, 8) and cultured monocultures of Z. tritici in each of these adjusted media. Z. tritici growth was impeded in the acidic medium, but to a much lower extent than when cocultured with APO9 (Fig. S5C through F). Notably, there were viable Z. tritici cells even after 96 h of incubation in media with a pH of 3.5 (Fig. S5E and F). Collectively, these results suggest that APO9-mediated Z. tritici mortality is not solely attributed to pH fluctuations.
Bioinformatics analysis of APO9 reveals antagonistic potential
To further understand the inhibitory potential and mechanism of APO9, we performed whole genome sequencing (WGS) and analysis of its genome. The complete genome of APO9 consisted of two circular chromosomes of 6,891,196 bp and 1,052,173 bp and a plasmid of 110,035 bp which was predicted by using RFPlasmid (51) (Fig. S6). These three replicons encode for 7,225 predicted coding sequences (CDS).
To assess the phylogenetic relationship of APO9 within the genus Burkholderia, we analyzed the genome sequences of 11 B. cenocepacia and 19 B. cepacia strains (Table S3) and 38 Burkholderia spp. (Table S4) using ANI assays. WGS-based approach has become one of the most reliable methods for measuring genomic phylogenetic relationships, capable of distinguishing between distantly and closely related bacteria (52, 53), and ANI values are widely used as a gold standard for defining prokaryotic species (32, 46). We found that APO9 and B. cepacia DDS 7H-2 are positioned within the same branch, and also have a close phylogenetic relationship with B. cepacia MINF and B. cenocepacia species, as corroborated by ANI values greater than 99% (Fig. S7A and S8A). These ANI values underscore the pronounced genomic similarity between these strains (Tables S3 and S4). Moreover, the genomes of these strains all share a distinct chromosomal rearrangement that underscore the distinct genomic architecture among these strains (Fig. S7B and S8B). Hence, the phylogenetic tree constructed from ANI comparison suggests APO9 should be considered a B. cenocepacia-like strain.
In order to evaluate the capacity of APO9 to produce fungal antagonizing secondary metabolites, we annotated its genome and searched for the presence of BGCs. We identified BGCs encoding for metabolites with predicted antimicrobial activity, including a type I polyketide synthase (T1PKS), two non-ribosomal peptide synthetase (NRPS), eight terpene, one phosphonate, one NRPS-like peptide, two RIPP-like peptides (ribosomally synthesized and post-translationally modified peptides), one redox cofactor, one hserlactone, and one aryl polyene. Additionally, gene annotation revealed genes encoding for three secretion systems (types II, III, and IV) (Table S2), which may also facilitate antifungal capacity in APO9.
Screening a transposon mutant library identified five genes related to inhibition efficiency
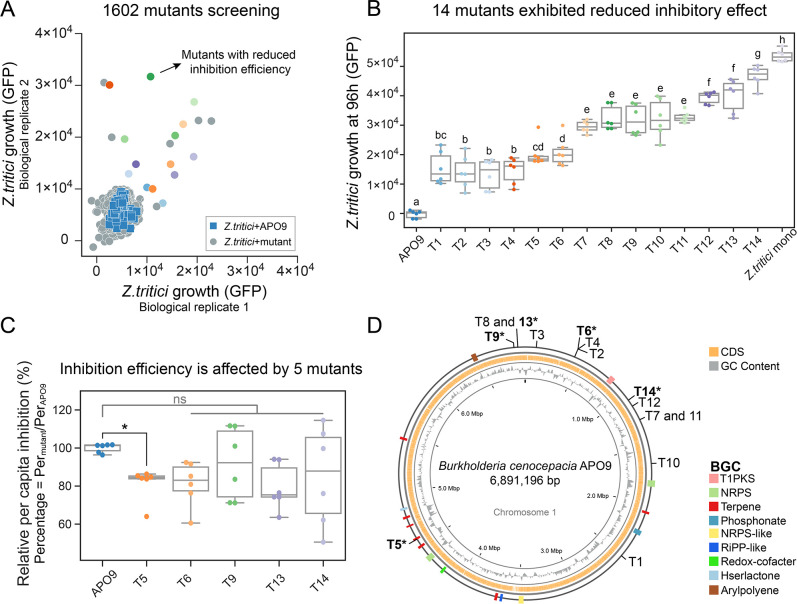
In order to experimentally identify genes related to APO9’s inhibitory activity, we constructed a total of 1,602 APO9 Tn5 transposon insertion mutants and evaluated their antagonist activities by coculturing each mutant with Z. tritici. We identified several mutants that displayed significantly reduced inhibitory effects compared to the wild-type strain (P < 0.05) (Fig. 3A). The reduced effect of these mutants was similar to that of Gh24—a species with a much lower inhibitory efficiency than the wild type (wt) APO9 (Fig. S9).
Fig 3.
Five genes related to inhibition efficiency were identified by screening a transposon mutant library. (A) Quantification of the antagonistic activity of 1,602 Tn5 mutants after coculture 96 h with Z. tritici. The x-axis and y-axis represent the two biological replicates of Z. tritici, respectively. Blue squares indicate Z. tritici cocultures with APO9 wild type. Gray circles indicate Z. tritici cocultures with mutants. The other color circles are mutants with reduced inhibition efficiency, and consistent with the final screening of the 14 mutants. (B) Fourteen mutants exhibited reduced inhibitory effect. Two biological replicates of Z. tritici and three biological replicates of mutants. Values with different letters (a–h) indicate statistically significant differences among treatments (P < 0.05), ANOVA, Duncan’s tests. (C) Five mutants exhibited lower per capita inhibition efficiency than APO9 wild type. Dunnett’s test, *P < 0.05; ns, not significant. (D) Schematic circular diagram of the APO9 genome and 14 mutant insertion sites as well as BGC, CDS, and GC contents are shown.
The antagonistic activity of these mutants was validated in six replicates, identifying 14 mutants (designated T1–T14) with significantly reduced inhibition (Fig. 3B). Microscopic examination further corroborated these results (Fig. S10). Additionally, we conducted a plating assay of the cocultures, scanned them after 5 days of incubation, and assessed the variations in inhibitory effects by examining fungal growth (Fig. S11A). The area of the black fungi phenotype was quantified using ImageJ (Materials and Methods), revealing a strong correlation between fluorescence intensity and plating results, with a Pearson correlation coefficient of 0.84 and a P-value of 8.64e − 05 (Fig. S11B). 16S rRNA gene sequencing verified that all 14 mutants were indeed APO9, ruling out potential contamination by other less inhibitory species. Successful insertion of the Tn5 transposon into the genome conferred trimethoprim resistance to the mutants. PCR amplification revealed that all 14 Tn5 mutants had a single band with a size of 629 bp, indicating successful insertion of the transposon (Fig. S12). Additionally, random transposon insertion into the genome caused the corresponding gene of the mutant to lose its original function, which resulted in varied growth of mutants (Fig. S13).
Next, we tested whether the reduced inhibition of the mutants was due to lower population size, or lower per capita activity. Five mutants had lower per capita inhibition than that of the wild type, of which T5 was significantly reduced by 18.62% (P < 0.05), and the others reduced by 18.84%, 8.32%, 20.59%, and 14.92%, respectively (ns, P > 0.05) (Fig. 3C; Fig. S14).
To identify the Tn5 transposon insertion site, we used specific primers and inverse PCR. The Tn5 insertion locations were identified by aligning to the APO9 genome and were presented on the genomic map (Fig. 3D). Among them, two pairs of mutants have the same transposon insertion points on the genome, T8 and T13 that were inserted in the same gene and T7 and T11 that were inserted in the same site. Notably, the gene disrupted by T5 was part of a terpene biosynthetic gene cluster. The accuracy of gene localization was further verified by locus-specific primers (Table S1). The amplified fragments of all mutants were larger than the corresponding wild types by around 629 bp (corresponding to the Tn5 transposon size; Fig. S15). Genome annotation and UniProt website predictions were combined to infer the functions of the related genes (Table 1; Table S5), which were involved in pathways related to amino acid synthesis, ion binding, and kinase activities, etc.
TABLE 1.
Functional annotation of five mutant genes correlated with decreased inhibitory efficiency
| Mutants | Gene annotation of insertion sites | Pathway of function involved |
|---|---|---|
| T5 | Diaminopimelate decarboxylase (DAPDC) | L-lysine biosynthesis via DAP pathway |
| T6 | ATP-phosphoribosyltransferase (ATP-PRT) | L-histidine biosynthesis |
| T9 | Rod shape-determining protein (MreD) | Involved in formation of the rod shape of the cell. And regulation of formation of penicillin-binding proteins |
| T13 | DNA topoisomerase III (Topo III) | Metal ion binding/DNA binding/chromosome separation |
| T14 | Ribosome maturation factor (RimM) | Ribosomal small subunit biogenesis |
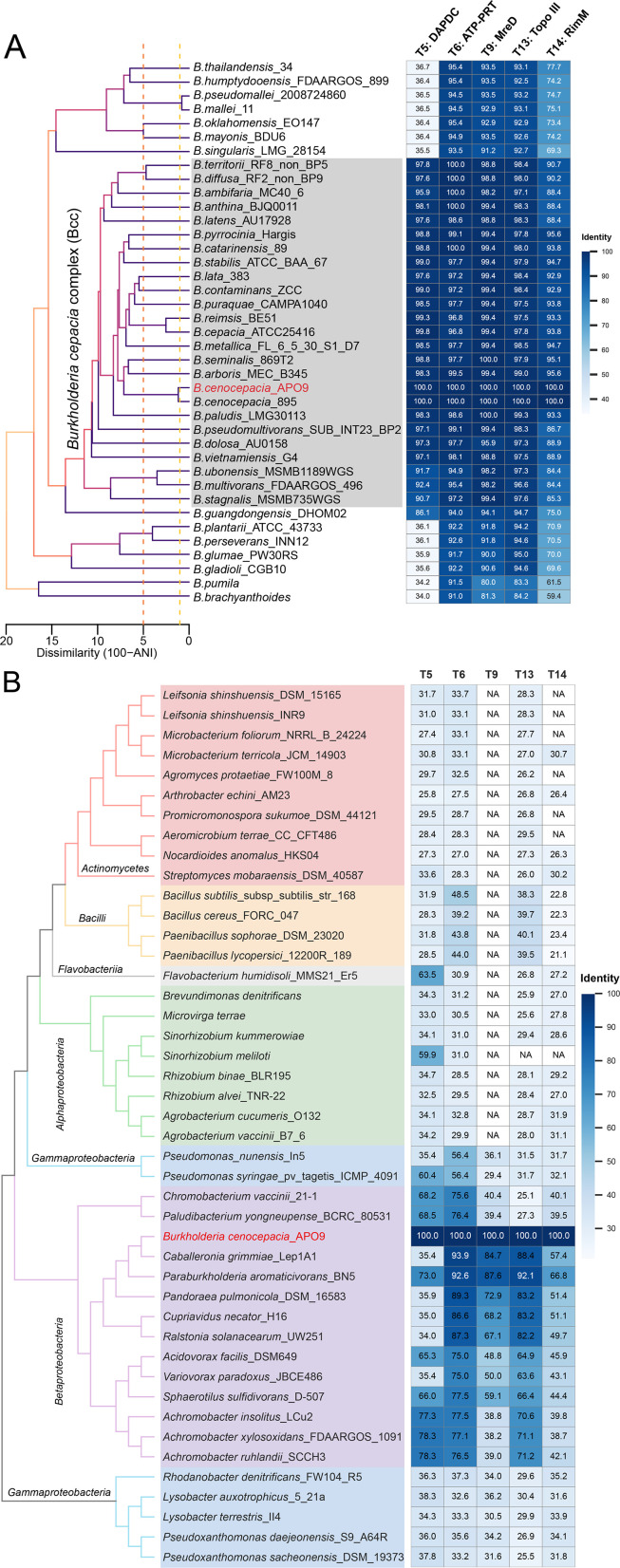
Five genes related to APO9’s antifungal activity are widely distributed in the Burkholderia genus and the class of Betaproteobacteria
In order to assess how prevalent the five genes are with significantly reduced inhibitory effects, we looked for similar genes in other species across the Burkholderia genus (Fig. 4A; Table S6). Three of these genes [ATP-phosphoribosyltransferase (ATP-PRT) (T6), MreD (T9), and Topo III (T13)] were ubiquitous across the Burkholderia, while the diaminopimelate decarboxylase (DAPDC) (T5) gene is present in 25 out of the 38 investigated strains, 24 of which are part of the Bcc (Fig. 4A, Materials and Methods). Intriguingly, the DAPDC gene was absent from strains within the Burkholderia pseudomallei complex, including B. pseudomallei, Burkholderia mallei, and Burkholderia singularis, which are the principal Burkholderia pathogens affecting both animals and humans (54). Moreover, Burkholderia strains from another clade which have been reported to induce phytopathological diseases also lack this gene, including Burkholderia glumae (55, 56), Burkholderia gladioli (57, 58), and Burkholderia plantarii (59) primarily. The ribosome maturation factor (RimM) (T14) gene exhibits similar results, having higher identity with strains in the Bcc group and reduced identity in other Burkholderia.
Fig 4.
Five genes related to suppression efficiency are widely distributed in the Burkholderia genus and the class of Betaproteobacteria. (A) The phylogenetic tree of Burkholderia genus and the identification of five genes. Horizontal distance represents ANI dissimilarity (%), with vertical dashed lines indicating ANI values of 95 (orange) and 99 (yellow). Gene identification was conducted using genome BLASTP (E-value ≤1e − 5), based on amino acid similarity. The gray area indicate the Bcc group. (B) A cladogram depicting the phylogenetic relationships of strains were determined based on 16S rRNA, and the identification of five genes. The pink bar represent the class of Actinomycetes, the yellow bar is the class of Bacilli, the gray bar is the class of Flavobacteriia, the green bar is the class of Alphaproteobacteria, the blue bar is the class of Gammaproteobacteria, and the purple bar is the class of Betaproteobacteria.
We further assessed the prevalence of these genes in more distantly related taxa. We expanded our search for homologous sequences to include 43 species across six classes and four phyla (Fig. 4B; Table S7). We discovered that no sequence similarities exceeding 80% were identified for the DAPDC (T5) and RimM (T14) genes. The identity of the MreD (T9) in Caloplaca grimmiae and Paraburkholderia aromaticivorans exceeds 80%, which are species with close evolutionary relationships to APO9. The homologous genes corresponding to T6 (ATP-PRT) and T13 (Topo III) are prevalent in most Betaproteobacteria, exhibiting high sequence identity. Conversely, beyond the Betaproteobacteria class, the amino acid identity associated with these five genes is markedly lower (Fig. 4B). The combination of these genes is prevalent within our clade, but some of them are also common in the Burkholderia genus and the class of Betaproteobacteria.
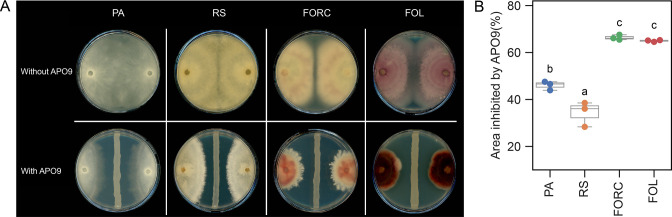
APO9 suppresses additional plant pathogens
We tested the inhibitory activity of APO9 against several additional plant pathogens, including PA, RS, FORC, and FOL. APO9 strongly inhibited all of them (Fig. 5A), with inhibitory effects of 46.01%, 34.30%, 66.38%, and 64.98%, respectively (Fig. 5B). The 14 APO9 mutants that showed reduced inhibition of Z. tritici typically also displayed lower antagonistic activity against these additional fungal pathogens, though some mutant-pathogen combinations displayed an increased inhibitory activity (Fig. S16A through F).
Fig 5.
APO9’s ability to suppress additional plant pathogens. (A) Plate confrontation assay with PA, RS, FORC, and FOL. (B) APO9 exhibited variable suppression efficiency against different phytopathogens.
DISCUSSION
Fungal diseases are recognized as a major impediment to crop growth and yield (28, 60), necessitating the development of environmentally sustainable approaches for pathogen suppression (14). In this context, plant-associated microbes (microbial inoculants) have been shown to have potential to improve plant quality and yield (28, 61, 62) by stimulating plant growth (63, 64), enhancing stress resistance (65), and protecting against pathogen infestation (66, 67). In the present study, we isolated and characterized a new B. cenocepacia-like strain that exhibited significant antifungal activity against a variety of plant fungal pathogens.
Burkholderia spp. are remarkably versatile with broad secondary metabolite production capabilities (23, 68). We analyzed APO9’s inhibition mechanism by testing whether it secreted compounds that inhibit the growth of Z. tritici. Despite the fact that APO9 had significant inhibitory activity even in the absence of physical contact with Z. tritici (Fig. 2B and C), no inhibition was observed in the presence of spent media from APO9 monoculture or APO9 cocultured with Z. tritici (Fig. S3). Moreover, a volatile compound assay revealed that APO9 exhibited some inhibitory effect against Z. tritici when incubated on the same side of Petri dish, despite having no contact between the bacteria and fungi (Fig. S4C). These results suggest that APO9 may secrete secondary metabolites that are able to cross the semipermeable membrane, thereby inhibiting the growth of Z. tritici. The lack of inhibition by APO9’s supernatant suggests that these compounds may be unstable or degrade quickly after being secreted, similar to findings regarding bacterial polyynes (69, 70). Nonetheless, the fact that inhibition efficiency is reduced in the absence of physical contact suggests that, perhaps, APO9 may also act through contact-dependent mechanisms (Table S2). These mechanisms encompass the type III secretion system, which injects the secretory molecules into the host cell (71, 72), and type IV secretion system, which requires attachment to the host cell either through direct cell-to-cell contact or via a bridge-like apparatus (73, 74). Both systems have been reported to be capable of antagonizing other microorganisms, causing tissue damage, and invading host cells (75, 76). Additionally, the contact-dependent inhibition by APO9 may involve the translocation of toxins via the secretion system, higher local concentrations of secondary metabolites, or the induction of secondary metabolite production in APO9 upon contact with Z. tritici. The inhibitory effect of APO9 on Z. tritici is both contact- and non-contact-mediated.
Microbes produce an array of natural products, which are encoded by different biosynthetic gene clusters (77). We found that the APO9 genome contains nine classes of BGCs (Fig. 3D; Fig. S6), including clusters encoding for T1PKS, NRPS, terpenes, and phosphonates. It is known that these BGCs are frequently involved in the synthesis of antibiotics, antifungal agents, siderophores, and immunosuppressants (78–81), suggesting that APO9 produces bioactive natural products that can antagonize phytopathogens. However, additional work is required to identify specific compounds produced by these clusters, and their role in inhibiting fungal pathogens.
Despite the promising potential of Burkholderia spp. for plant protection, they are not “generally recognized as safe” since some Burkholderia species are known to be opportunistic human and plant pathogens (55, 82). Therefore, direct application of APO9 as biocontrol agent is currently uncertain and is unlikely to become a viable option. Nonetheless, application of specific metabolites, as opposed to using the whole organism, offers a viable alternative, and APO9 may present a valuable reservoir for discovering novel compounds for controlling fungal plant pathogens.
Using random mutagenesis and screening, we identified five mutants impaired in different genes that displayed reduced antagonism of Z. tritici. The most significantly reduced suppressive activity was observed in a DAPDC gene mutant (T5). This gene is associated with a terpene biosynthetic gene cluster and it functions as an additional biosynthetic gene. Disruption of this gene may lead to a reduction in the biosynthetic function of the corresponding secondary metabolites. Interestingly, homologous expression of DAPDC has been reported to increase antibiotic production in Streptomyces clavuligerus (83), suggesting that it may also play a role in regulating secondary metabolite production in APO9.
The functional annotations of the genes disrupted in the other mutants with reduced inhibitory activity do not clearly reveal their role in fungal inhibition. The mutant T6, encoding an ATP-PRT, is the first enzyme in the histidine biosynthesis pathway. Since previous research has elucidated a linkage between amino acid synthesis and antibiotic production (84–86), the connection between histidine biosynthesis and APO9 antagonistic activity warrants further investigation in future research. The T13 mutation is located in a DNA topoisomerase III gene. DNA topoisomerases are essential enzymes that modulate DNA topology in cells, playing a crucial role in processes such as DNA replication, transcription, repair, and recombination (87, 88). In Escherichia coli, both topoisomerases I and III belong to the type IA topoisomerase family (89, 90). The recently described DNA topoisomerase I (topA) mutation significantly reduced type III secretion system expression in Pseudomonas aeruginosa (91), suggesting the T13 mutation (topoIII) may affect APO9 secretion systems, thus reducing the antagonism of Z. tritici. It is important to note that our mutant library is not exhaustive, and therefore, it is likely that additional APO9 genes are also involved in fungal antagonism. Constructing a comprehensive mutant library covering the entire APO9 gene set may reveal additional genes related to its suppressive activity in future studies.
Phylogenetic analysis of the five genes whose inactivation resulted in significantly reduced inhibitory activity revealed their ubiquitous presence within the Burkholderia genus. Intriguingly, these genes are seldom identified in classes beyond Betaproteobacteria. In species such as B. pseudomallei, B. mallei, B. plantarii, and B. gladioli, the homologous DAPDC (T5) gene is absent. Notably, these species are recognized as pathogens of animals or plants (54, 58, 59). The DAPDC (T5) gene is associated with the terpene biosynthesis gene cluster. The reasons for its absence in pathogens remain to be elucidated, though we conjecture this to be related to its antimicrobial activity.
Our research demonstrated that APO9 not only antagonizes Z. tritici but also suppresses multiple economically relevant soil-borne phytopathogens (Fig. 5). APO9, which was originally isolated from the root of a healthy cucumber plant in a P. aphanidermatum-infested greenhouse, might play a role in antagonizing soil-borne pathogens in agroenvironments due to its antifungal properties. Genes associated with antagonistic capacity, which are widely found in Bcc, suggest that Burkholderia species play a significant role in the interactions between plant microbiota and pathogens. The broad range of phytopathogens it suppresses suggests that instead of being tailored to a specific phytopathogen, APO9 could potentially be utilized as a source of natural product to antagonize various phytopathogens. The fact that APO9 inhibits several different pathogens through both contact-dependent and contact-independent activity suggests that it employs several different antagonistic mechanisms. This multifaceted activity may hinder the capacity of pathogens to develop resistance to APO9, akin to that achieved via combination therapy (92, 93).
It remains a challenge to uncover all the antifungal mechanisms of APO9 and their ecological implication. This research has provided a foundation for further exploration of its antifungal mechanism, and demonstrates that APO9 strongly inhibits a wide range of fungal pathogens via a complex and diverse mechanism. APO9 and other Bcc group strains which harbor the same genes related to fungal inhibition may thus play a significant role in the interactions between bacteria and fungi. Further in-depth investigations of the antifungal mechanisms of APO9 and the interactions between strains of the Bcc clade and plant-associated fungi are required to reveal the potential of these strains for developing new treatment options to combat plant pathogens, and to elucidate the role these bacterial strains play in shaping plant microbiomes and plant health.
ACKNOWLEDGMENTS
We gratefully acknowledge Shay Covo for providing the FOL strain, Jiawei Li for assistance with graphics, and members of Friedman lab for valuable discussions.
This research was supported by the ISF-NSFC joint research program (grant no. 3395/20), the Israeli Ministry of Science and Technology (grant no. 0001998), the Israeli Ministry of Agriculture and Rural Development (grant no. 20–13-0027), the Lady Davis Fellowship Trust, and the China Scholarship Council (CSC).
J.F. and T.S. designed and conducted the study. T.S. analyzed the data and drafted the manuscript. S.G. isolated APO9 strains, Y.S. provided technical assistance. O.F. and E.C. contributed to constructive discussions. All authors contributed to acquisition of data, interpretation of results and critical discussion, and approved the final version of the manuscript.
Contributor Information
Jonathan Friedman, Email: yonatan.friedman@mail.huji.ac.il.
John R. Spear, Colorado School of Mines, Golden, Colorado, USA
DATA AVAILABILITY
All raw data employed in the analyses of this study are accessible in Table S8. Comprehensive details on genome sequencing, assembly, and gene annotation are available at the following link: https://figshare.com/s/7f565216e17dc8607556.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02222-23.
Figures S1 to S16.
List of primers used in this study.
Secretion system.
ANI value of APO9 compared with Burkholderia cepacia and Burkholderia cenocepacia species.
ANI values of Burkholderia genus.
The specific genes of the 9 mutants and the pathways involved.
Evolutionary analysis of five genes within 38 species of the Burkholderia genus.
Evolutionary analysis of five genes within 43 species across 6 classes.
Original data for plotting figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Fones HN, Bebber DP, Chaloner TM, Kay WT, Steinberg G, Gurr SJ. 2020. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat Food 1:332–342. doi: 10.1038/s43016-020-0075-0 [DOI] [PubMed] [Google Scholar]
- 2. Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A. 2019. The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3:430–439. doi: 10.1038/s41559-018-0793-y [DOI] [PubMed] [Google Scholar]
- 3. Váry Z, Mullins E, McElwain JC, Doohan FM. 2015. The severity of wheat diseases increases when plants and pathogens are acclimatized to elevated carbon dioxide. Glob Chang Biol 21:2661–2669. doi: 10.1111/gcb.12899 [DOI] [PubMed] [Google Scholar]
- 4. Velásquez AC, Castroverde CDM, He SY. 2018. Plant and pathogen warfare under changing climate conditions. Curr Biol 28:R619–R634. doi: 10.1016/j.cub.2018.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kilaru S, Fantozzi E, Cannon S, Schuster M, Chaloner TM, Guiu-Aragones C, Gurr SJ, Steinberg G. 2022. Zymoseptoria tritici white-collar complex integrates light, temperature and plant cues to initiate dimorphism and pathogenesis. Nat Commun 13:5625. doi: 10.1038/s41467-022-33183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brennan CJ, Benbow HR, Mullins E, Doohan FM. 2019. A review of the known unknowns in the early stages of septoria tritici blotch disease of wheat. Plant Pathology 68:1427–1438. doi: 10.1111/ppa.13077 [DOI] [Google Scholar]
- 7. Steinberg G. 2015. Cell biology of Zymoseptoria tritici: pathogen cell organization and wheat infection. Fungal Genet Biol 79:17–23. doi: 10.1016/j.fgb.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fones H, Gurr S. 2015. The impact of septoria tritici blotch disease on wheat: an EU perspective. Fungal Genet Biol 79:3–7. doi: 10.1016/j.fgb.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mullins AJ, Murray JAH, Bull MJ, Jenner M, Jones C, Webster G, Green AE, Neill DR, Connor TR, Parkhill J, Challis GL, Mahenthiralingam E. 2019. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium burkholderia ambifaria. Nat Microbiol 4:996–1005. doi: 10.1038/s41564-019-0383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P. 2018. Navigating complexity to breed disease-resistant crops. Nat Rev Genet 19:21–33. doi: 10.1038/nrg.2017.82 [DOI] [PubMed] [Google Scholar]
- 11. Saintenac C, Cambon F, Aouini L, Verstappen E, Ghaffary SMT, Poucet T, Marande W, Berges H, Xu S, Jaouannet M, Favery B, Alassimone J, Sánchez-Vallet A, Faris J, Kema G, Robert O, Langin T. 2021. A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against septoria tritici blotch. Nat Commun 12:433. doi: 10.1038/s41467-020-20685-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Torriani SFF, Melichar JPE, Mills C, Pain N, Sierotzki H, Courbot M. 2015. Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet Biol 79:8–12. doi: 10.1016/j.fgb.2015.04.010 [DOI] [PubMed] [Google Scholar]
- 13. Droby S, Gonzalez-Estrada RR, Avila-Quezada G, Durán P, Manzo-Sánchez G, Hernandez-Montiel LG. 2022. Microbial antagonists from different environments used in the biocontrol of plant pathogens, p 227–244. In Kumar A (ed), Microbial biocontrol: food security and post harvest management. Vol. 2. Springer International Publishing, Cham. [Google Scholar]
- 14. Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360:739–742. doi: 10.1126/science.aap7999 [DOI] [PubMed] [Google Scholar]
- 15. Woo SL, Hermosa R, Lorito M, Monte E. 2023. Trichoderma: a multipurpose, plant-beneficial microorganism for ECO-sustainable agriculture. Nat Rev Microbiol 21:312–326. doi: 10.1038/s41579-022-00819-5 [DOI] [PubMed] [Google Scholar]
- 16. Bonaterra A, Badosa E, Daranas N, Francés J, Roselló G, Montesinos E. 2022. Bacteria as biological control agents of plant diseases. Microorganisms 10:1759. doi: 10.3390/microorganisms10091759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Legein M, Smets W, Vandenheuvel D, Eilers T, Muyshondt B, Prinsen E, Samson R, Lebeer S. 2020. Modes of action of microbial biocontrol in the phyllosphere. Front Microbiol 11:1619. doi: 10.3389/fmicb.2020.01619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel R, Mehta K, Prajapati J, Shukla A, Parmar P, Goswami D, Saraf M. 2022. An anecdote of mechanics for fusarium biocontrol by plant growth promoting microbes. Biol Control 174:105012. doi: 10.1016/j.biocontrol.2022.105012 [DOI] [Google Scholar]
- 19. Kurniawan O, Wilson K, Mohamed R, Avis TJ. 2018. Bacillus and Pseudomonas spp. provide antifungal activity against gray mold and alternaria rot on blueberry fruit. Biol Control 126:136–141. doi: 10.1016/j.biocontrol.2018.08.001 [DOI] [Google Scholar]
- 20. Eljounaidi K, Lee SK, Bae H. 2016. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases – review and future prospects. Biol Control 103:62–68. doi: 10.1016/j.biocontrol.2016.07.013 [DOI] [Google Scholar]
- 21. Li Z, Ye X, Liu M, Xia C, Zhang L, Luo X, Wang T, Chen Y, Zhao Y, Qiao Y, Huang Y, Cao H, Gu X, Fan J, Cui Z, Zhang Z. 2019. A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J 13:2223–2235. doi: 10.1038/s41396-019-0424-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. doi: 10.1007/s00253-016-7520-x [DOI] [PubMed] [Google Scholar]
- 23. Kunakom S, Eustáquio AS. 2019. Burkholderia as a source of natural products. J Nat Prod 82:2018–2037. doi: 10.1021/acs.jnatprod.8b01068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foxfire A, Buhrow AR, Orugunty RS, Smith L. 2021. Drug discovery through the isolation of natural products from Burkholderia. Expert Opin Drug Discov 16:807–822. doi: 10.1080/17460441.2021.1877655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An C, Ma S, Liu C, Ding H, Xue W. 2022. Burkholderia ambifaria XN08: a plant growth-promoting endophytic bacterium with biocontrol potential against sharp eyespot in wheat. Front Microbiol 13:906724. doi: 10.3389/fmicb.2022.906724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarli DA, Sánchez LA, Delgado OD. 2021. Burkholderia gladioli MB39 an antarctic strain as a biocontrol agent. Curr Microbiol 78:2332–2344. doi: 10.1007/s00284-021-02492-y [DOI] [PubMed] [Google Scholar]
- 27. Xue L, Yang C, Jihong W, Lin L, Yuqiang Z, Zhitong J, Yanxin W, Zhoukun L, Lei F, Zhongli C. 2022. Biocontrol potential of Burkholderia sp. BV6 against the rice blast fungus Magnaporthe oryzae. J Appl Microbiol 133:883–897. doi: 10.1111/jam.15605 [DOI] [PubMed] [Google Scholar]
- 28. Pal G, Saxena S, Kumar K, Verma A, Sahu PK, Pandey A, White JF, Verma SK. 2022. Endophytic Burkholderia: multifunctional roles in plant growth promotion and stress tolerance. Microbiol Res 265:127201. doi: 10.1016/j.micres.2022.127201 [DOI] [PubMed] [Google Scholar]
- 29. Ho Y-N, Huang C-C. 2015. Draft genome sequence of Burkholderia cenocepacia strain 869T2, a plant-beneficial endophytic bacterium. Genome Announc 3:e01327-15. doi: 10.1128/genomeA.01327-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu P, Zheng L-Q, Sun J-J, Liu H-M, Li S-P, Hong Q, Li W-J. 2012. Burkholderia zhejiangensis sp. nov., a methyl-parathion-degrading bacterium isolated from a wastewater-treatment system. Int J Syst Evol Microbiol 62:1337–1341. doi: 10.1099/ijs.0.035428-0 [DOI] [PubMed] [Google Scholar]
- 31. Coenye T, Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- 32. Jin Y, Zhou J, Zhou J, Hu M, Zhang Q, Kong N, Ren H, Liang L, Yue J. 2020. Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol Direct 15:6. doi: 10.1186/s13062-020-0258-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Devanga Ragupathi NK, Veeraraghavan B. 2019. Accurate identification and epidemiological characterization of Burkholderia cepacia complex: an update. Ann Clin Microbiol Antimicrob 18:7. doi: 10.1186/s12941-019-0306-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- 35. Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x [DOI] [PubMed] [Google Scholar]
- 36. Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:F1000 Faculty Rev-1007. doi: 10.12688/f1000research.8221.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho Y-N, Chiang H-M, Chao C-P, Su C-C, Hsu H-F, Guo C, Hsieh J-L, Huang C-C. 2015. In planta biocontrol of soilborne fusarium wilt of banana through a plant endophytic bacterium, Burkholderia cenocepacia 869T2. Plant Soil 387:295–306. doi: 10.1007/s11104-014-2297-0 [DOI] [Google Scholar]
- 38. Rojas-Rojas FU, Salazar-Gómez A, Vargas-Díaz ME, Vásquez-Murrieta MS, Hirsch AM, De Mot R, Ghequire MGK, Ibarra JA, Estrada-de Los Santos P. 2018. Broad-spectrum antimicrobial activity by Burkholderia cenocepacia TAtl-371, a strain isolated from the tomato rhizosphere. Microbiology (Reading) 164:1072–1086. doi: 10.1099/mic.0.000675 [DOI] [PubMed] [Google Scholar]
- 39. You M, Fang S, MacDonald J, Xu J, Yuan Z-C. 2020. Isolation and characterization of Burkholderia cenocepacia CR318, a phosphate solubilizing bacterium promoting corn growth. Microbiol Res 233:126395. doi: 10.1016/j.micres.2019.126395 [DOI] [PubMed] [Google Scholar]
- 40. Baichman-Kass A, Song T, Friedman J. 2023. Competitive interactions between culturable bacteria are highly non-additive. Elife 12:e83398. doi: 10.7554/eLife.83398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kulesa A, Kehe J, Hurtado JE, Tawde P, Blainey PC. 2018. Combinatorial drug discovery in nanoliter droplets. Proc Natl Acad Sci U S A 115:6685–6690. doi: 10.1073/pnas.1802233115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kehe J, Ortiz A, Kulesa A, Gore J, Blainey PC, Friedman J. 2021. Positive interactions are common among culturable bacteria. Sci Adv 7:eabi7159. doi: 10.1126/sciadv.abi7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manni M, Berkeley MR, Seppey M, Zdobnov EM. 2021. BUSCO: assessing genomic data quality and beyond. Curr Protoc 1:e323. doi: 10.1002/cpz1.323 [DOI] [PubMed] [Google Scholar]
- 44. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carroll LM, Wiedmann M, Kovac J. 2020. Proposal of a taxonomic nomenclature for the Bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. mBio 11:e00034-20. doi: 10.1128/mBio.00034-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guy L, Kultima JR, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. doi: 10.1093/bioinformatics/btq413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. doi: 10.1093/nar/gkab335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang M, Wang X, Ahmed T, Liu M, Wu Z, Luo J, Tian Y, Jiang H, Wang Y, Sun G, Li B. 2020. Identification of genes involved in antifungal activity of Burkholderia seminalis against Rhizoctonia solani using Tn5 transposon mutation method. Pathogens 9:797. doi: 10.3390/pathogens9100797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van der Graaf-van Bloois L, Wagenaar JA, Zomer AL. 2021. Rfplasmid: predicting plasmid sequences from short-read assembly data using machine learning. Microbial Genomics 7:000683. doi: 10.1099/mgen.0.000683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hall CM, Baker AL, Sahl JW, Mayo M, Scholz HC, Kaestli M, Schupp J, Martz M, Settles EW, Busch JD, Sidak-Loftis L, Thomas A, Kreutzer L, Georgi E, Schweizer HP, Warner JM, Keim P, Currie BJ, Wagner DM. 2022. Expanding the Burkholderia pseudomallei complex with the addition of two novel species: Burkholderia mayonis sp. nov. and Burkholderia savannae sp. nov. Appl Environ Microbiol 88:e0158321. doi: 10.1128/AEM.01583-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ham JH, Melanson RA, Rush MC. 2011. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mondal KK, Mani C, Verma G. 2015. Emergence of bacterial panicle blight caused by Burkholderia glumae in North India. Plant Dis 99:1268–1268. doi: 10.1094/PDIS-01-15-0094-PDN [DOI] [Google Scholar]
- 57. Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. 2009. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the Southern United States. Plant Dis 93:896–905. doi: 10.1094/PDIS-93-9-0896 [DOI] [PubMed] [Google Scholar]
- 58. Md Zali AZ, Ja’afar Y, Paramisparan K, Ismail SI, Saad N, Mohd Hata E, Md Hatta MA, Ismail MR, Yusof MT, Zulperi D. 2022. First report of Burkholderia gladioli causing bacterial panicle blight of rice in Malaysia. Plant Dis 107:551. doi: 10.1094/PDIS-03-22-0650-PDN [DOI] [PubMed] [Google Scholar]
- 59. Wang M, Wei P, Cao M, Zhu L, Lu Y. 2016. First report of rice seedling blight caused by Burkholderia plantarii in North and Southeast China. Plant Disease 100:645. doi: 10.1094/PDIS-07-15-0765-PDN [DOI] [Google Scholar]
- 60. Calicioglu O, Flammini A, Bracco S, Bellù L, Sims R. 2019. The future challenges of food and agriculture: an integrated analysis of trends and solutions. Sustainability 11:222. doi: 10.3390/su11010222 [DOI] [Google Scholar]
- 61. Afzal I, Shinwari ZK, Sikandar S, Shahzad S. 2019. Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol Res 221:36–49. doi: 10.1016/j.micres.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 62. Sturz AV, Nowak J. 2000. Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. App Soil Ecol 15:183–190. doi: 10.1016/S0929-1393(00)00094-9 [DOI] [Google Scholar]
- 63. Verma SK, Sahu PK, Kumar K, Pal G, Gond SK, Kharwar RN, White JF. 2021. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J Appl Microbiol 131:2161–2177. doi: 10.1111/jam.15111 [DOI] [PubMed] [Google Scholar]
- 64. Singh RK, Malik N, Singh S. 2013. Improved nutrient use efficiency increases plant growth of rice with the use of IAA-overproducing strains of endophytic Burkholderia cepacia strain RRE25. Microb Ecol 66:375–384. doi: 10.1007/s00248-013-0231-2 [DOI] [PubMed] [Google Scholar]
- 65. Lata R, Chowdhury S, Gond SK, White JF. 2018. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett Appl Microbiol 66:268–276. doi: 10.1111/lam.12855 [DOI] [PubMed] [Google Scholar]
- 66. Kim H, Mohanta TK, Park Y-H, Park S-C, Shanmugam G, Park J-S, Jeon J, Bae H. 2020. Complete genome sequence of the mountain-cultivated ginseng endophyte Burkholderia stabilis and its antimicrobial compounds against ginseng root rot disease. Biol Control 140:104126. doi: 10.1016/j.biocontrol.2019.104126 [DOI] [Google Scholar]
- 67. Ahmad T, Bashir A, Farooq S, Riyaz-Ul-Hassan S. 2022. Burkholderia gladioli E39CS3, an endophyte of Crocus sativus LINN., induces host resistance against corm-rot caused by Fusarium oxysporum. J Appl Microbiol 132:495–508. doi: 10.1111/jam.15190 [DOI] [PubMed] [Google Scholar]
- 68. Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li X-C, Jacob MR, Khan SI, Ashfaq MK, Babu KS, Agarwal AK, Elsohly HN, Manly SP, Clark AM. 2008. Potent in vitro antifungal activities of naturally occurring acetylenic acids. Antimicrob Agents Chemother 52:2442–2448. doi: 10.1128/AAC.01297-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kai K, Sogame M, Sakurai F, Nasu N, Fujita M. 2018. Collimonins A–D, unstable polyynes with antifungal or pigmentation activities from the fungus-feeding bacterium Collimonas fungivorans TER331. Org Lett 20:3536–3540. doi: 10.1021/acs.orglett.8b01311 [DOI] [PubMed] [Google Scholar]
- 71. Cornelis GR. 2006. The type III secretion injectisome. Nat Rev Microbiol 4:811–825. doi: 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- 72. Galán JE, Waksman G. 2018. Protein-injection machines in bacteria. Cell 172:1306–1318. doi: 10.1016/j.cell.2018.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LRS, Salinas RK, Guzzo CR, Farah CS. 2015. Bacterial killing via a type IV secretion system. Nat Commun 6:6453. doi: 10.1038/ncomms7453 [DOI] [PubMed] [Google Scholar]
- 74. Sgro GG, Oka GU, Souza DP, Cenens W, Bayer-Santos E, Matsuyama BY, Bueno NF, Dos Santos TR, Alvarez-Martinez CE, Salinas RK, Farah CS. 2019. Bacteria-killing type IV secretion systems. Front Microbiol 10:1078. doi: 10.3389/fmicb.2019.01078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Klein TA, Ahmad S, Whitney JC. 2020. Contact-dependent interbacterial antagonism mediated by protein secretion machines. Trends Microbiol 28:387–400. doi: 10.1016/j.tim.2020.01.003 [DOI] [PubMed] [Google Scholar]
- 76. Pena RT, Blasco L, Ambroa A, González-Pedrajo B, Fernández-García L, López M, Bleriot I, Bou G, García-Contreras R, Wood TK, Tomás M. 2019. Relationship between quorum sensing and secretion systems. Front Microbiol 10:1100. doi: 10.3389/fmicb.2019.01100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hug JJ, Krug D, Müller R. 2020. Bacteria as genetically programmable producers of bioactive natural products. Nat Rev Chem 4:172–193. doi: 10.1038/s41570-020-0176-1 [DOI] [PubMed] [Google Scholar]
- 78. Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. 2014. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111:9259–9264. doi: 10.1073/pnas.1401734111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. 2018. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558:440–444. doi: 10.1038/s41586-018-0207-y [DOI] [PubMed] [Google Scholar]
- 80. Carroll CS, Moore MM. 2018. Ironing out siderophore biosynthesis: a review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit Rev Biochem Mol Biol 53:356–381. doi: 10.1080/10409238.2018.1476449 [DOI] [PubMed] [Google Scholar]
- 81. Shi J, Xu X, Liu PY, Hu YL, Zhang B, Jiao RH, Bashiri G, Tan RX, Ge HM. 2021. Discovery and biosynthesis of guanipiperazine from a NRPS-like pathway. Chem Sci 12:2925–2930. doi: 10.1039/d0sc06135b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Syed I, Wooten RM. 2021. Interactions between pathogenic Burkholderia and the complement system: a review of potential immune evasion mechanisms. Front Cell Infect Microbiol 11:701362. doi: 10.3389/fcimb.2021.701362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Otur Ç, Kurt-Kızıldoğan A. 2020. Homologous expression of lysA encoding diaminopimelic acid (DAP) decarboxylase reveals increased antibiotic production in Streptomyces clavuligerus. Braz J Microbiol 51:547–556. doi: 10.1007/s42770-019-00202-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nasution U, van Gulik WM, Ras C, Proell A, Heijnen JJ. 2008. A metabolome study of the steady-state relation between central metabolism, amino acid biosynthesis and penicillin production in Penicillium chrysogenum. Metab Eng 10:10–23. doi: 10.1016/j.ymben.2007.07.001 [DOI] [PubMed] [Google Scholar]
- 85. Nayak S, Baliyarsingh B, Dash B, Mishra BB. 2017. Soil Bacillus-A natural source of antifungal compounds against Candida infection, p 166–176 [Google Scholar]
- 86. De Felice M, Levinthal M, Iaccarino M, Guardiola J. 1979. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol Rev 43:42–58. doi: 10.1128/mr.43.1.42-58.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang JC. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440. doi: 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- 88. Tse-Dinh Y-C. 2009. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res 37:731–737. doi: 10.1093/nar/gkn936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sharma A, Mondragón A. 1995. DNA topoisomerases. Curr Opin Struct Biol 5:39–47. doi: 10.1016/0959-440x(95)80007-n [DOI] [PubMed] [Google Scholar]
- 90. Bush NG, Evans-Roberts K, Maxwell A. 2015. DNA topoisomerases. EcoSal Plus 6. doi: 10.1128/ecosalplus.ESP-0010-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yan R, Hu S, Ma N, Song P, Liang Q, Zhang H, Li Y, Shen L, Duan K, Chen L. 2019. Regulatory effect of DNA topoisomerase I on T3SS activity, antibiotic susceptibility and quorum- sensing-independent pyocyanin synthesis in Pseudomonas aeruginosa. Int J Mol Sci 20:1116. doi: 10.3390/ijms20051116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ons L, Bylemans D, Thevissen K, Cammue BPA. 2020. Combining biocontrol agents with chemical fungicides for integrated plant fungal disease control. Microorganisms 8:1930. doi: 10.3390/microorganisms8121930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Maron B, Rolff J, Friedman J, Hayouka Z. 2022. Antimicrobial peptide combination can hinder resistance evolution. Microbiol Spectr 10:e0097322. doi: 10.1128/spectrum.00973-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S16.
List of primers used in this study.
Secretion system.
ANI value of APO9 compared with Burkholderia cepacia and Burkholderia cenocepacia species.
ANI values of Burkholderia genus.
The specific genes of the 9 mutants and the pathways involved.
Evolutionary analysis of five genes within 38 species of the Burkholderia genus.
Evolutionary analysis of five genes within 43 species across 6 classes.
Original data for plotting figures.
Data Availability Statement
All raw data employed in the analyses of this study are accessible in Table S8. Comprehensive details on genome sequencing, assembly, and gene annotation are available at the following link: https://figshare.com/s/7f565216e17dc8607556.