ABSTRACT
Cyclic di-GMP (c-di-GMP) is a crucial signaling molecule found extensively in bacteria, involved in the regulation of various physiological and biochemical processes such as biofilm formation, motility, and pathogenicity through binding to downstream receptors. However, the structural dissimilarity of c-di-GMP receptor proteins has hindered the discovery of many such proteins. In this study, we identified LspE, a homologous protein of the type II secretion system (T2SS) ATPase GspE in Lysobacter enzymogenes, as a receptor protein for c-di-GMP. We identified the more conservative c-di-GMP binding amino acid residues as K358 and T359, which differ from the previous reports, indicating that GspE proteins may represent a class of c-di-GMP receptor proteins. Additionally, we found that LspE in L. enzymogenes also possesses a novel role in regulating the production of the antifungal antibiotic HSAF. Further investigations revealed the critical involvement of both ATPase activity and c-di-GMP binding in LspE-mediated regulation of HSAF (Heat-Stable Antifungal Factor) production, with c-di-GMP binding having no impact on LspE’s ATPase activity. This suggests that the control of HSAF production by LspE encompasses two distinct processes: c-di-GMP binding and the inherent ATPase activity of LspE. Overall, our study unraveled a new function for the conventional protein GspE of the T2SS as a c-di-GMP receptor protein and shed light on its role in regulating antibiotic production.
IMPORTANCE
The c-di-GMP signaling pathway in bacteria is highly intricate. The identification and functional characterization of novel receptor proteins have posed a significant challenge in c-di-GMP research. The type II secretion system (T2SS) is a well-studied secretion system in bacteria. In this study, our findings revealed the ATPase GspE protein of the T2SS as a class of c-di-GMP receptor protein. Notably, we discovered its novel function in regulating the production of antifungal antibiotic HSAF in Lysobacter enzymogenes. Given that GspE may be a conserved c-di-GMP receptor protein, it is worthwhile for researchers to reevaluate its functional roles and mechanisms across diverse bacterial species.
KEYWORDS: c-di-GMP, receptor proteins, T2SS GspE, antifungal antibiotic
INTRODUCTION
In 1987, cyclic dimeric GMP (c-di-GMP) was first discovered as an allosteric activator of cellulose synthase (1). Subsequently, it was found to be widely present in bacteria, serving as an important second messenger signaling molecule (2–4). The intracellular concentration of c-di-GMP in bacteria is regulated by two types of enzyme: diguanylate cyclases (DGCs), which contain GGDEF/GGEEF domains to synthesize c-di-GMP, and c-di-GMP-specific phosphodiesterases (PDEs), which contain EAL or HD-GYP domains and degrade c-di-GMP (5–9). c-di-GMP is referred to as a second messenger because it does not directly exert its function like hormone-like small molecules do. Instead, it regulates various physiological and biochemical processes downstream by binding to specific c-di-GMP receptors and modulating their activities, which, in turn, leads to phenotypic changes in cell differentiation, biofilm formation, resistance to environmental stresses, and toxicity (10–12). The interplay among enzymes, receptors, and the signaling transduction between them collectively form a complex regulatory network of c-di-GMP.
Currently, the number and diversity of identified c-di-GMP receptors are relatively limited. They encompass various proteins such as PilZ, GIL, MshEN domain proteins, functionally degenerate DGCs and PDEs, transcription factors like Clp and YajQ, and riboswitches, among others (13–17). However, the number of c-di-GMP synthesis and degradation enzymes varies significantly among different bacteria, ranging from a few to even hundreds, surpassing the number of c-di-GMP receptors by far (18). With increasing evidence highlighting the specificity of c-di-GMP signaling, where certain DGCs and PDEs are specifically involved in regulating particular physiological and biochemical functions (19, 20), it implies that there should be a proportionate number of receptor proteins to facilitate signal reception and execution of regulatory functions within the c-di-GMP regulatory network. This indicates that there are likely many undiscovered c-di-GMP receptor proteins awaiting exploration. However, apart from the I-site (AXXA) in DGCs and motifs like RXXXR and [DN]XSXXG in the PilZ domain, and EXLXR motif in EAL domain, there seems to be no structural similarity among the known c-di-GMP receptor proteins, making it challenging to predict new receptor proteins solely through bioinformatics approaches (16). Currently, experimental approaches are the key methods for discovering novel c-di-GMP receptor proteins.
Lysobacter enzymogenes is a soil-derived environmentally friendly bacterium that produces the broad-spectrum antifungal and anti-oomycete antibiotic HSAF (Heat-Stable Antifungal Factor), which is active against important plant pathogens such as Valsa pyri, Fusarium graminearum, Pythium aphanidermatum, and so on (21–26). HSAF is a cyclic tetrapeptide with a molecular weight of 513.2959. Its biosynthetic gene cluster includes four redox enzymes and a hybrid PKS/NRPS (Polyketide Synthase/Non-ribosomal Peptide Synthetase) (22). HSAF inhibits the growth of target fungi by interfering with fungal sphingolipid biosynthesis (27), and this unique mechanism of action differs from currently available antifungal agents on the market. Given its potential as a bio-fungicide in agriculture, our focus was on studying the regulatory network of HSAF production to gain insights into enhancing its yields. In our previous work, we uncovered the interaction between the DGC LchD and the PDE LchP, which specifically transmits the c-di-GMP signal to the downstream receptor protein, transcription factor Clp, which modulates the binding strength of Clp to the promoter of the HSAF biosynthetic gene cluster, thus positively regulating HSAF production (28, 29). However, our data (data not shown) indicate the involvement of other DGCs and PDEs in L. enzymogenes in HSAF production, suggesting the existence of undiscovered c-di-GMP receptor proteins in the HSAF regulatory network.
In this study, we focused on L. enzymogenes strain OH11 and utilized surface plasmon resonance (SPR) experiments to identify a total of 68 c-di-GMP binding proteins, GspE, the ATPase of the bacteria type II secretion system (T2SS), included. Although GspE in Pseudomonas aeruginosa has been reported as a receptor for c-di-GMP (30), there is currently no known functional characterization of its binding to c-di-GMP. Here, we identified that LspE (Lysobacter GspE) in L. enzymogenes acts as a c-di-GMP receptor protein and is involved in antifungal antibiotic HSAF production regulation. Subsequent investigations identified specific amino acid residues lysine at position 358 (K358) and threonine at position 359 (T359) in LspE responsible for c-di-GMP binding, and mutations in these residues were found to affect the regulation of HSAF production. Meanwhile, the ATPase activity of LspE was also found to be closely associated with HSAF production. However, it was found that c-di-GMP binding does not affect the ATPase activity. In summary, our study reveals the new function of the conserved ATPase component of the T2SS, namely its role in binding c-di-GMP to participate in the regulation of antifungal antibiotic production.
RESULTS
Screening of c-di-GMP receptors in regulating HSAF production
To identify the c-di-GMP receptor proteins involved in the regulation of HSAF production in L. enzymogenes, we employed c-di-GMP as a gripper to capture proteins from the OH11 whole cell lysate using SPR. The captured proteins were then subjected to protein sequencing and BLAST analysis. Through this approach, we successfully identified 68 potential c-di-GMP receptor proteins (Listed in Table 1). Among these proteins, 14 are known c-di-GMP receptors, including 3747(FimX), 4562, 0155, 4875, 3688, 0067, 4029, 3677, 1158, 4727 (RpfG), 2762 (LchP), and 1632, which are involved in c-di-GMP synthesis or degradation (16), as well as 3639 (PilZ) (13) and 0848 (Clp) (28), which are previously reported c-di-GMP receptors. The presence of these known receptors among the identified proteins adds credibility and reliability to our data.
TABLE 1.
Table1 Potential c-di-GMP receptor proteins by SPR assaya
| Protein ID | Predicted function | Score |
|---|---|---|
| 0328 | ATP-binding cassette, subfamily B, bacterial | 1,860.74 |
| 4172 | Histidine kinase | 1,845.72 |
| 4790 | PAS domain S-box protein | 1,839.60 |
| 1890 | Para-aminobenzoate synthetase | 1,831.15 |
| 4943 | Hypothetical protein | 1,829.90 |
| 3747 (FimX) | GGDEF domain-containing protein | 1,818.89 |
| 1886 | Serine hydrolase | 1,791.40 |
| 3639 (PilZ) | PilZ domain-containing protein (13) | 1,740.59 |
| 0705 | pilR two-component system, NtrC family, response regulator PilR | 1,674.38 |
| 4562 | Two-component system response regulator WspR, GGDEF domain containing protein | 1,643.62 |
| 0155 | GGDEF domain-containing protein | 1,617.86 |
| 1020 | 3-Oxoacyl-[acyl-carrier protein] reductase | 1,594.19 |
| 3049 | Peptide chain release factor 3 | 1,583.60 |
| 0691 | ATP-dependent Clp protease ATP-binding subunit ClpB | 1,562.76 |
| 4875 | GGDEF domain-containing protein | 1,561.41 |
| 3688 | GGDEF domain-containing protein | 1,560.77 |
| 0067 | GGDEF domain-containing protein | 1,557.90 |
| 0770 | Type II secretion system ATPase GspE | 1,547.64 |
| 4029 | GGDEF domain-containing protein | 1,474.64 |
| 4656 | MerR family transcriptional regulator, copper efflux regulator | 1,458.28 |
| 3677 | GGDEF domain-containing protein | 1,390.65 |
| 2512 | PAS domain S-box protein | 1,385.58 |
| 5095 | Two-component system, OmpR family, sensor histidine kinase KdpD | 1,384.01 |
| 1158 | GGDEF domain-containing protein | 1,372.03 |
| 4727 (RpfG) | Two-component system, response regulator RpfG, HD-GYP domain containing protein | 1,369.92 |
| 3810 | Biofilm PGA synthesis N-glycosyltransferase PgaC | 1,364.51 |
| 0820 | Hypothetical protein | 1,357.96 |
| 2762 (LchP) | GGDEF and EAL domain-containing protein | 1,338.04 |
| 1363 | Hypothetical protein | 1,325.54 |
| 0229 | Hypothetical protein | 1,310.95 |
| 0910 | Hypothetical protein | 1,284.18 |
| 4558 | Chemotaxis protein methyltransferase WspC | 1,283.28 |
| 4781 | Molecular chaperone GrpE | 1,255.45 |
| 1583 | Type III secretion protein SctV | 1,242.25 |
| 1157 | LysM peptidoglycan-binding domain-containing protein | 1,224.18 |
| 3498 | Two-component system, NarL family, response regulator YdfI | 1,199.31 |
| 4096 | Hydrophobic/amphiphilic exporter-1 (mainly G-bacteria), HAE1 family | 1,179.93 |
| 0440 | Peptidyl-tRNA hydrolase, PTH1 family | 1,166.91 |
| 2012 | Hypothetical protein | 1,041.79 |
| 1012 | Shape-determining protein MreB and related proteins | 1,041.21 |
| 1632 | GGDEF domain-containing protein | 964.89 |
| 4431 | Hypothetical protein | 902.47 |
| 3194 | DMT family transporter | 854.90 |
| 2535 | Hypothetical protein | 809.27 |
| 2784 | Parallel beta-helix repeat protein | 770.13 |
| 0285 | Type IV pilus assembly protein PilY1 | 747.79 |
| 3928 | N-acetyltransferase | 734.80 |
| 1452 | Fatty acyl CoA synthetase | 732.42 |
| 1252 | Hypothetical protein | 658.70 |
| 4774 | Carbamoyl-phosphate synthase small subunit | 647.30 |
| 2394 | Hydrophobic/amphiphilic exporter-1 (mainly G-bacteria), HAE1 family | 631.31 |
| 0606 | Hypothetical protein | 623.39 |
| 1277 | Citrate-Mg2+:H + or citrate-Ca2+:H + symporter, CitMHS family | 614.70 |
| 3555 | Membrane glycosyltransferase | 600.43 |
| 4468 | Gluconate 2-dehydrogenase subunit 3 family protein | 566.59 |
| 2781 | Hypothetical protein | 530.31 |
| 0381 | GTP-binding protein | 507.00 |
| 0031 | DUF1800 domain-containing protein | 491.73 |
| 4429 | 2,3-Dihydroxybenzoate-AMP ligase | 456.72 |
| 4088 | DUF4189 domain-containing protein | 421.99 |
| 3061 | Xanthine dehydrogenase family protein molybdopterin-binding subunit | 400.97 |
| 1499 | TonB-dependent receptor | 343.01 |
| 4605 | Malate dehydrogenase (quinone) | 331.51 |
| 4739 | TonB-dependent receptor | 299.15 |
| 0463 | Membrane protein | 296.08 |
| 0848 (Clp) | CRP/FNR family transcriptional regulator, cyclic AMP receptor protein (27) | 294.43 |
| 0385 | DUF2127 domain-containing protein | 208.03 |
| 3440 | Hypothetical protein | 176.70 |
The putative c-di-GMP binding proteins were captured from the whole-cell lysate of OH11 through SPR assay. The score is the probability that the observed match is a random event. We report scores as −10*LOG10(P), where P is the absolute probability. Therefore, the higher the score, the greater the likelihood of this protein being a c-di-GMP receptor. The gray shading represents known c-di-GMP receptor proteins.
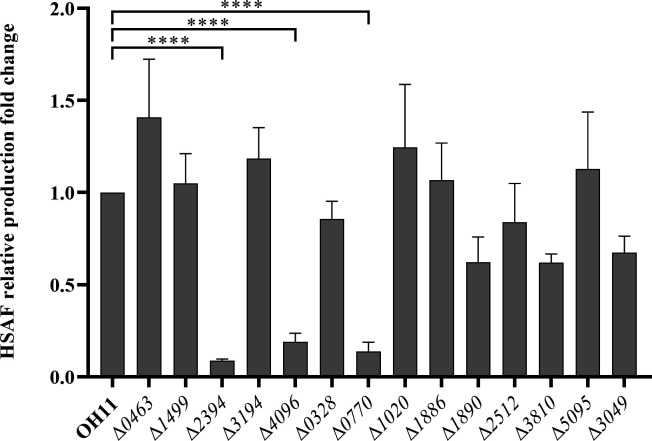
Subsequently, we generated deletion mutants of some of the c-di-GMP receptor protein-encoding genes (Fig. S1) and evaluated their HSAF yields using HPLC. The results, as depicted in Fig. 1, revealed impaired HSAF production in some of the mutants compared to the wild-type strain, including mutant Δ2394, Δ4096, and Δ0770. In this study, we focus on the protein 0770.
Fig 1.
Comparison of HSAF production in a series of potential c-di-GMP receptor protein deletion mutants. The HSAF production in all strains was detected by HPLC. The peak area corresponding to HSAF in the wild-type strain OH11 was assumed to be 1, and all other strains were compared to the wild type. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. ****P < 0.0001, relative to the wild-type OH11. Biological experiments for each treatment were performed three times and assayed in triplicate.
T2SS ATPase LspE is a c-di-GMP receptor
Protein 0770 was found to be a homologous protein of GspE, the ATPase of the Bacteria T2SS, as determined through protein blasting on the Pfam database (31) (http://pfam.xfam.org/, Fig. S2A). ATPase is an enzyme that hydrolyzes ATP into ADP and inorganic phosphate, releasing energy for cellular activities. T2SS ATPase as a c-di-GMP receptor has been identified in P. aeruginosa (30), but the functions regulated by c-di-GMP as a receptor have not been further studied. Since there were no reports on the correlation between the type II secretion system and the production of antibiotics, we renamed the 0770 protein as LspE (Lysobacter GspE) and decided to conduct further research on it.
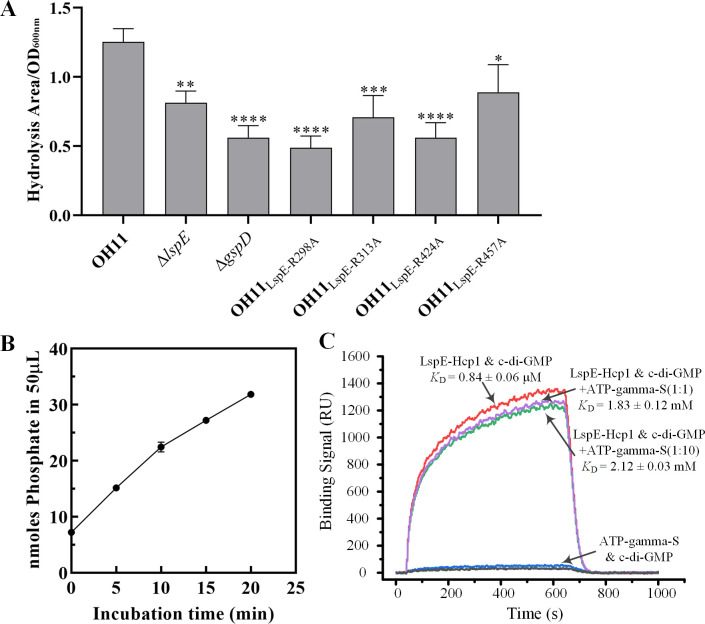
To confirm that LspE functions as an ATPase of the T2SS, we first performed a protease activity assay to compare the hydrolysis zones of LspE mutant and the wild-type strains. Here, we used the mutant of T2SS structural protein GspD as a control. Our results showed that the hydrolysis zone of the LspE and GspD mutant strain was smaller than that of the wild-type strain (Fig. 2A), indicating that LspE has the conserved function of secreting hydrolytic enzymes via the T2SS. Furthermore, we conducted an in vitro ATPase activity assay using purified LspE-His protein expressed heterologously in Escherichia coli. Unfortunately, we detected almost no ATPase activity of this protein (Fig. S2B). Previous studies by Sandkvist and colleagues reported that the GspE protein EspE in Vibrio cholerae exists as a monomer in vitro, with only a small fraction forming hexameric structures, resulting in low ATP hydrolysis activity (32, 33). It was found that Hcp1, a hexameric protein from P. aeruginosa, assists in the formation of hexameric structures of EspE and exhibits approximately 20-fold higher ATPase activity compared to the monomeric form (34). Thus, we fused the Hcp1 protein to the C-terminus of LspE with five amino acids KLASG as a linker in an attempt to enhance its ATP hydrolysis capability. Then, we detected the in vitro ATPase activity of LspE-Hcp1. Fig. 2B shows phosphate continuously released with the incubation time increasing, demonstrating the robust ATPase activity of LspE-Hcp1. These results support that LspE functions as the ATPase of the T2SS in L. enzymogenes.
Fig 2.
LspE is a T2SS ATPase and a c-di-GMP receptor. (A) The protease activity of the wild-type and LspE mutants. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, relative to the wild type. Biological experiments for each treatment were performed three times and assayed in triplicate. (B) In vitro ATP hydrolysis activity assay of LspE-Hcp1 protein. (C) SPR experiments were performed to evaluate the binding ability of c-di-GMP with LspE-Hcp1 with or without ATP-gamma-S.
Subsequently, we conducted binding experiments between the purified LspE-Hcp1 protein and c-di-GMP. SPR experiments revealed a strong binding interaction between LspE-Hcp1 and c-di-GMP, with a dissociation constant (KD) of 0.84 ± 0.06 µM (Fig. 2C). Given the structural similarity between c-di-GMP and ATP and both can bind to LspE, we also investigated whether they competitively bind to LspE. In this experiment, c-di-GMP was immobilized on photo cross-linker SPRi sensor chip, LspE-Hcp1, and a non-hydrolyzable ATP analog ATP-gamma-S flowed over the surface of chip. As shown in Fig. 2C, when the ratio of LspE-Hcp1 to ATP-gamma-S in the interaction system is 1:1, the KD value is 1.83 ± 0.12 µM, and when their ratio is 1:10, the KD value is 2.12 ± 0.03 µM. Although a slight decrease in affinity of LspE-Hcp1 for c-di-GMP was observed in the presence of the non-hydrolyzable ATP analog, such a minor difference is unlikely to be the result of competitive binding between c-di-GMP and ATP-gamma-S. It may be due to a slight conformational change in LspE-Hcp1 upon ATP-gamma-S binding. These results indicate that there is no competitive binding between c-di-GMP and ATP to LspE. Overall, our findings indicate that LspE, as an ATPase of the type II secretion system, is a c-di-GMP receptor protein.
K358 and T359 of LspE are required for c-di-GMP binding and are related to HSAF production
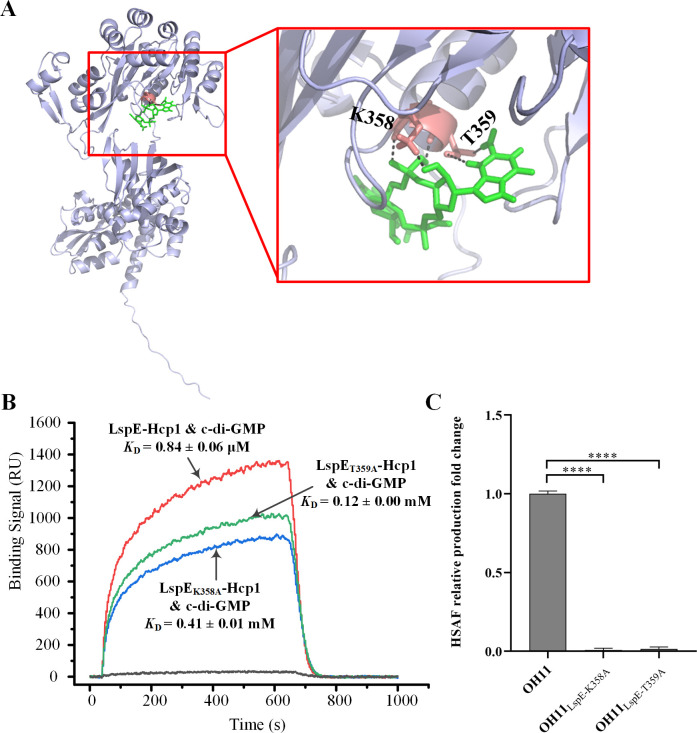
To elucidate the c-di-GMP binding site of LspE, molecular docking simulations were performed between LspE and c-di-GMP. The results indicated that the amino acids K358 and T359 might serve as the binding site for c-di-GMP (Fig. 3A). To validate this hypothesis, site-directed mutagenesis was performed on these two residues, resulting in the generation of the purified LspEK358A-Hcp1 and LspET359A-Hcp1 proteins through heterologous expression. Circular dichroism measurement was used to confirm that these two mutant proteins still correctly folded (Table S3). Subsequent SPR analysis revealed a significant increase in the dissociation constants of LspEK358A-Hcp1 and LspET359A-Hcp1 with c-di-GMP, measured as 0.41 ± 0.01 mM and 0.12 ± 0.00 mM (Fig. 3B), respectively, indicating a weakened binding affinity with c-di-GMP. These results indicated that the amino acid residues K358 and T359 are required for the binding of LspE to c-di-GMP.
Fig 3.
The amino acids lysine at position 358 (K358) and threonine at position 359 (T359) are key binding sites of LspE to c-di-GMP. (A) Molecular docking predicts that K358 and T359 are crucial binding sites for c-di-GMP. The structure of LspE generated by AlphaFold is represented in purple, and the c-di-GMP molecule is shown in green. (B) SPR experiments are conducted to measure the binding affinity of c-di-GMP to LspE-Hcp1, LspEK358A-Hcp1, and LspET359A-Hcp1, respectively. (C) HSAF production comparison between the two site-mutated strain OH11LspE-K358A and OH11LspE-T359A to the wild-type strain. The peak area corresponding to HSAF in the wild-type strain OH11 was assumed to be 1, and all other strains were compared to the wild type. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. ****P < 0.0001, relative to the wild-type OH11. Biological experiments for each treatment were performed three times and assayed in triplicate.
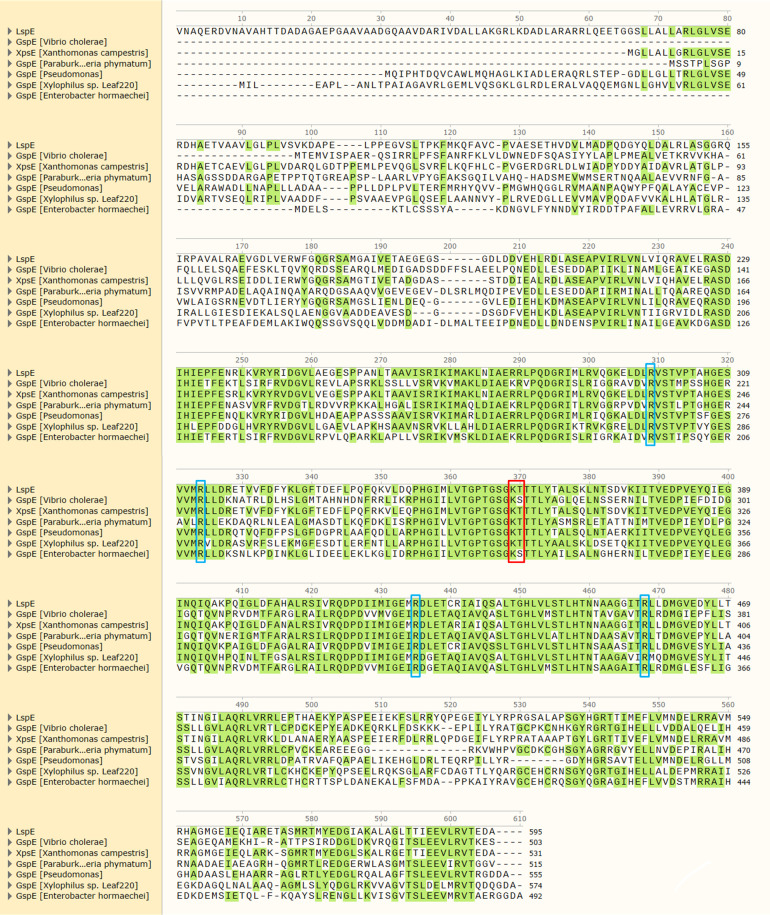
We then compared the amino acid sequences of LspE with homologous proteins from different bacteria and found that the amino acid lysine is completely conserved in all seven GspE proteins, and the amino acid threonine is also conserved, except for GspE from V. cholerae and Enterobacter hormaechei (Fig. 4, in red box). This result suggests that ATPase proteins in the T2SS of bacteria are likely to be conserved c-di-GMP receptor proteins although their specific functions in their respective host bacteria are still unknown.
Fig 4.
Sequence alignment of amino acid sequences of GspE proteins from seven different bacteria. The amino acids represented in the red boxes are identified as key binding sites of c-di-GMP with LspE in this study. The amino acids represented in the blue boxes are ATPase activity key sites reported in EspE from V. cholerae (31).
Furthermore, we introduced the mutations K358A and T359A of LspE into the genome of L. enzymogenes strain OH11, resulting in the generation of the OH11LspE-K358A and OH11LspE-T359A mutants. Extraction and detection of HSAF demonstrated a significant decrease in the production of HSAF in both OH11LspE-K358A and OH11LspE-T359A strains (Fig. 3C), indicating a positive correlation between the c-di-GMP-binding capacity of LspE and its impact on HSAF production.
The ATPase activity of LspE plays a key role in HSAF production regulation
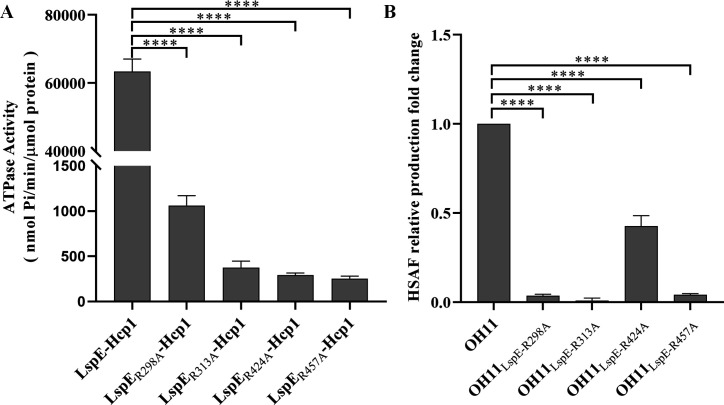
As an ATPase, is there a correlation between the ATP hydrolysis activity of LspE and HSAF production? By comparing with the amino acid residues reported to affect ATPase activity in EspE of V. cholerae (35), we identified four potential ATPase activity sites in LspE, including R298, R313, R424, and R457 (Fig. 4, in blue box). We generated their site mutation and expressed their fusion proteins with Hcp1, followed by the measurement of ATP hydrolysis activity in vitro. The results showed a significant reduction in ATPase activity in these four mutant proteins, LspER298A-Hcp1, LspER313A-Hcp1, LspER424A-Hcp1, and LspER457A-Hcp1, compared to the wild-type LspE-Hcp1 (Fig. 5A; Fig. S3), indicating that these identified sites are, indeed, ATPase activity key amino acid residues. Furthermore, we introduced point mutations in these four amino acid sites in the OH11 genome, resulting in the generation of four strains, including OH11LspE-R298A, OH11LspE-R313A, OH11LspE-R424A, and OH11LspE-R457A. As expected, these mutant strains showed a weakened protease activity compared to the wild type due to the impaired ATPase function (Fig. 2A). HSAF production was then measured in these strains. As shown in Fig. 5B, the HSAF production of OH11LspE-R424A is approximately half of that of the wild-type strain, while the other three mutant strains almost do not produce any HSAF (Fig. 5B). These findings revealed that the decreases in ATPase activity of LspE also resulted in decreases in HSAF production.
Fig 5.
The ATPase activity of LspE is closely correlated with HSAF production. (A) Mutation R298, R313, R424, and R457 of LspE, respectively, significantly reduces the ATP hydrolysis ability compared to the wild type. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. ****P < 0.0001, relative to the LspE-Hcp1. Biological experiments for each treatment were performed three times and assayed in triplicate. (B) Point mutations of the four R residues in L. enzymogenes OH11 genome severely impair the ability of the strain to produce HSAF. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. ****P < 0.0001, relative to the wild-type OH11. Biological experiments for each treatment were performed three times and assayed in triplicate.
ATP hydrolysis activity of LspE is independent of its binding to c-di-GMP
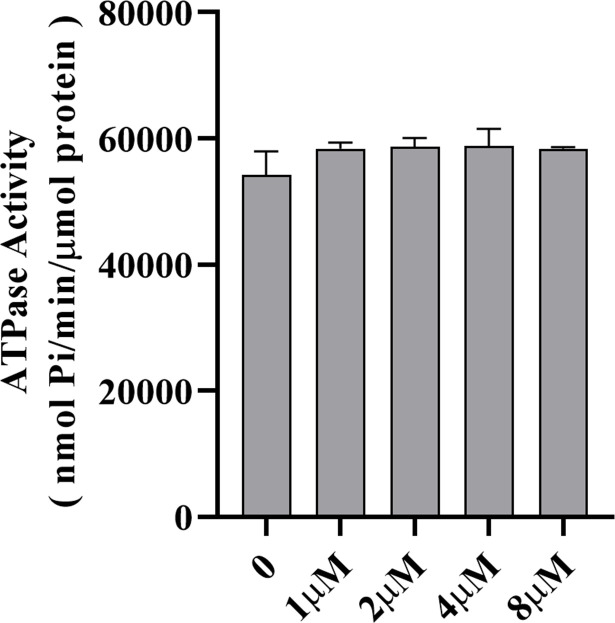
Is the ATP hydrolysis activity of LspE affected by its binding to c-di-GMP, considering that its impact on HSAF production is correlated with both its ATPase activity and c-di-GMP binding? We conducted experiments by adding a gradient concentration of c-di-GMP to the system used to measure LspE’s ATP hydrolysis ability. We incubated 1, 2, 4, and 8 µM c-di-GMP with 0.5 µM LspE-Hcp1 for 0.5 h to allow sufficient time for binding to occur. After that, we detected the ATPase activity, respectively. The results showed only a slight increase in activity compared to the control without c-di-GMP, and there was no significant difference (Fig. 6). Although the ATPase activity of LspE and its binding to c-di-GMP demonstrate consistency in their impact on HSAF production, based on the current data, these two processes do not appear to be related.
Fig 6.
c-di-GMP binding does not affect the ATPase activity of LspE. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA (Dunnett’s multiple comparisons test). The error bars indicate standard errors. Biological experiments for both treatment were performed three times and assayed in triplicate.
DISCUSSION
As one of the important secretion systems, since its discovery in the 1980s (36), the T2SS has been extensively studied in the composition, structure, and function (37–40). Comprising 12–15 proteins spanning the inner to outer membranes, the T2SS transports toxins and enzymes such as proteases (41–43), lipid-modifying enzymes (44, 45), carbohydrate-active enzymes (46, 47), phosphatases (48, 49), nucleic acid-targeting enzymes (50, 51), and more, to the extracellular space. The ATPase GspE serves as the energy supplier for the T2SS (33), and it is the main focus of our present study.
The c-di-GMP signaling molecule is widely present in bacteria, participating in various physiological and biochemical functions such as biofilm formation, motility, and virulence factor expression (4). However, there are currently only two published articles concerning the c-di-GMP signaling molecule and its relationship with the T2SS. One discovered that c-di-GMP can induce the expression of the gene cluster-encoding T2SS proteins in V. cholerae in a transcriptional factor VpsR-dependent manner (52) and the other reported that T2SS ATPase PA14_29490 in P. aeruginosa is a c-di-GMP receptor but with limited function study (30).
c-di-GMP receptor proteins are typically identified using experimental methods. One common approach is to capture the complex of c-di-GMP and receptor protein through methods such as biotin labeling or UV crosslinking and then identify the captured receptor protein by LC/MS/MS (Liquid Chromatography-Tandem Mass Spectrometry) (53, 54). Another method involves constructing a protein expression library, immobilizing proteins on nitrocellulose membrane, co-incubating with [α32P]c-di-GMP and identifying receptor proteins using the DRaCALA (Differential radial capillary action of ligand assay) method (30). The method used in the present study is more similar to the first approach but without any molecular labeling. It involves batch capturing c-di-GMP-binding proteins from the whole-cell lysate of L. enzymogenes OH11 using SPR experiments, ultimately leading to the identification of the T2SS protein GspE. Through our investigation, we determined that K358 and T359 of LspE contribute to c-di-GMP binding (Fig. 3). It is noteworthy that K358 is conserved among the GspE proteins of the seven different bacteria we compared, and T359 is conserved in five out of the seven proteins (Fig. 4). This suggests that the GspE protein of the T2SS is likely to serve as a receptor protein for c-di-GMP in different species of bacteria, making the GspE protein a conserved c-di-GMP-binding protein.
Our study has also revealed an intriguing link between the GspE protein and the production of the antifungal antibiotic HSAF in L. enzymogenes. This discovery marks the first time that the T2SS in bacteria has been associated with the production of antibiotics. HSAF has been extensively studied for over a decade, with research focusing on its gene cluster, biosynthetic pathway, and regulatory mechanisms (22, 23, 28, 29, 55–57). However, there is limited understanding of how HSAF is transported to the extracellular space. Currently, only Yue and colleagues have reported that HSAF can be released through outer membrane vesicles (OMVs) produced by L. enzymogenes strain OH11 (58). Nevertheless, with genetic modifications of strain OH11 and optimization of fermentation conditions, HSAF production has been increased to 440.26 ± 16.14 mg/L (59, 60), making it unlikely that such a large amount of HSAF can solely rely on OMVs for transport.
In our study, we observed that the deletion of lspE resulted in a significant reduction in HSAF production (Fig. 1), and mutations in the ATPase active site of LspE also led to decreased HSAF production (Fig. 5B). These findings raise the possibility that HSAF secretion to the extracellular space occurs through the T2SS. To test this hypothesis, we deleted the essential structural protein encoding gene gspD of the T2SS in L. enzymogenes strain OH11 and observed a significant decrease in HSAF production (Fig. S4), providing further evidence for the involvement of the T2SS in HSAF secretion. However, considering that the T2SS is typically responsible for transporting various enzymes to the extracellular space, and there are no direct reports on the transport of antibiotics, it is conceivable that HSAF may bind to a specific protein before being transported through the T2SS. Further investigations will be conducted in future studies to explore and validate this hypothesis.
Furthermore, we found that LspE, functioning as a c-di-GMP receptor protein, also influences HSAF production when its c-di-GMP-binding site is mutated (Fig. 3C). However, based on the current results, c-di-GMP binding does not affect the ATPase activity of LspE (Fig. 6), suggesting that LspE not only affects HSAF production through the T2SS but also independently influences HSAF production as a c-di-GMP receptor. Currently, except the report on MshE, the type IV pilus ATPase, also a c-di-GMP receptor, that its ATPase activity could be enhanced by c-di-GMP binding (30), there is still a lack of research on how this kind of c-di-GMP receptor transmits c-di-GMP signals downstream. By comparing transcriptome data (data not shown) of differentially expressed genes between LspE mutant and the wild-type OH11, we identified that LspE seems to influence the expression of multiple gene clusters, including the HSAF biosynthesis gene cluster (qRT-PCR data in Fig. S5), in a pattern similar to that of a transcriptional factor. In recent years, there has been extensive research on enzymes acting as transcriptional regulators, such as inosine monophosphate dehydrogenase in Drosophila acting as a DNA-binding transcriptional repressor to regulate cell proliferation (61), fructose-1,6-bisphosphate aldolase acting as a transcriptional regulator in Francisella (62), and AAA+ protease family protein Lon in P. syringae and E. coli identified as a DNA-binding protein (63, 64). Therefore, it is not ruled out that LspE may function as a transcriptional factor to regulate the biosynthesis of HSAF. Further investigation will be conducted to explore this possibility and uncover the role that c-di-GMP plays in this regulation. We also encourage researchers to re-examine the function and mechanism of action of the T2SS ATPase GspE in other bacteria species.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
The bacterial strains and plasmids utilized in this study were presented in Table S1. Lysobacter strains were cultured in LB medium at a temperature of 28°C unless specifically mentioned. E. coli strains were cultured in LB medium at 37°C. Antibiotics, including kanamycin (25 µg/mL), gentamicin (25 µg/mL), and ampicillin (100 µg/mL), were added when necessary.
Gene deletion and site mutation
In-frame gene deletion in L. enzymogenes OH11 was performed as described in our previous study before (65). Primers used in this study were listed in Table S2. The site mutant gene fragment of LspE was synthesized by TsingKe Biological Technology Company, Beijing. After sequencing verification, depending on the specific purpose, it was cloned into an expression vector or integrated into the genome using the gene knockout method described above.
SPR analysis
SPR analysis was performed by Betterways Inc., China. Briefly, for c-di-GMP-binding proteins capturing, c-di-GMP was first immobilized on a photo cross-linker SPRi sensor chip, followed by flowing the cell lysate of Lysobacter over the sensor chip surface. The proteins bound to c-di-GMP were captured and detected by bScreen LB 991 label-free microarray system (Berthold Technologies, Germany). Then, LC/MS/MS experiment was used to identify the types and relative abundance of proteins captured on the surface of the chip. For binding affinity of c-di-GMP to LspE and its derivatives, c-di-GMP was first immobilized on photo cross-linker SPRi sensor chip, LspE and its derivatives were diluted separately with running buffer at the concentrations of 200, 400, 800, 1,600, and 3,200 nM and injected over 600 s at a flow rate of 0.5 µL/s in each associating stage, followed by running buffer for 360 s at a flow rate of 0.5 µL/s in each dissociating stage. At the end of each associating-dissociating circle, the surface was regenerated to remove any remaining bound material with a pulse of 10 mM glycine-HCl (pH 2.5) at 20 µL/min for 30 s.
Protein expression and purification
LspE gene and its derivatives were amplified and cloned into the pET-30a (+) plasmid (Table S1). The resulting constructs were transformed into E. coli strain BL21 (DE3) (Table S1) for protein expression and purification. An overnight culture (2 mL) was transferred to 200 mL of fresh LB medium and grown at 37°C with shaking at 200 rpm until reaching an OD600nm of 0.6. Isopropyl β-d-1-thiogalactopyranoside (IPTG, Sigma) was then added into the culture to a final concentration of 0.5 mM, followed by incubation at 16°C for 16 h. The cell pellet was collected by centrifugation (6,000 rpm) at 4°C; resuspended in purification buffer (33) consisting of 20 mM Tris-HCl (pH 8), 500 mM NaCl, 5% (vol/vol) glycerol, 1 mM TCEP/HCl, 0.1 mM ATP, and 5 mM MgCl2; and sonicated using a Sonifier 250 (Branson Digital Sonifier, Branson). After centrifugation, the soluble proteins were mixed with pre-equilibrated Ni2+ resin (GE Healthcare, Shanghai, China) and washed with 75 mM imidazole. The target protein was eluted with 250 mM imidazole. The purified protein was diluted 10 times with imidazole-free purification buffer, centrifuged with Milibo ultrafiltration tube to achieve buffer replacement, and repeated twice. Finally, the protein was diluted 10 times with purified buffer supplemented with 10% (vol/vol) glycerol and centrifuged to the appropriate concentration and stored. The purity of the protein was evaluated by SDS-PAGE, and the protein concentration was determined using a BCA assay kit (TransGen Biotech, China).
HSAF extraction and detection
The extraction and quantification of HSAF were performed following previously described methods (65). Given that HSAF is hardly accumulated in the cell interior, we extracted HSAF from the whole fermentation broth. Briefly, HSAF was extracted from L. enzymogenes whole cell cultures grown in 1/10 TSB by adding an equal volume of ethyl acetate. The HSAF content was detected and quantified using high-performance liquid chromatography (HPLC). Each treatment was performed in biological triplicates and repeated three times for experimental consistency.
qRT-PCR
Strains were cultured in LB medium at 28°C overnight. One milliliter of the culture was transferred into 50 mL of 1/10 TSB, and cells were collected at OD600nm = 1.0. RNA extraction was performed using the Bacterial RNA Kit (Omega, China), and RNA concentrations were measured with a Nanodrop ND-1000 UV Spectrophotometer (Thermo Fisher, USA). cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). qRT-PCR was conducted using an Applied Biosystems 7500 system, with the 16S rRNA gene used as an internal control as previously described (29). The primers utilized in this study are listed in Table S2.
Molecular docking
The 3D structure of LspE, predicted by AlphaFold (66) (https://alphafold.com/), was obtained from UniProt and downloaded for further analysis. The obtained structure was processed using PyMOL (67), involving the removal of water molecules and unwanted heteroatoms, resulting in the modified receptor structure. For the ligand structure, the c-di-GMP molecule was checked for charge and protonation state using PyMOL. The molecular docking analysis was performed using AutoDockTools (68). Finally, the molecular docking figure was generated using PyMOL.
In vitro ATPase activity assay
ATPase activities were measured using the Malachite Green Phosphate Assay Kit (Sigma-Aldrich). The reaction mixture consisted of 0.5 µM protein, 5 mM ATP, and 5 mM MgCl2, in a buffer containing 100 mM HEPES (pH 8.5), 65 mM NaCl, and 5% (vol/vol) glycerol (34). The reactions were incubated at 37°C for different time intervals (0, 5, 10, 15, and 20 min for LspE-Hcp1; 0, 120, 180, 240, and 300 min for LspER298A-Hcp1, LspER313A-Hcp1, LspER424A-Hcp1, and LspER457A-Hcp1) and then assayed for the release of inorganic phosphate. The amount of phosphate was determined by measuring the absorbance at 650 nm and comparing it to a phosphate standard curve. The reported data were obtained from three independent samples of the same purified proteins, and each sample was assayed in duplicate.
For the reaction system supplemented with different concentrations of c-di-GMP, protein and c-di-GMP were mixed in the reaction buffer and pre-incubated at 37°C for 0.5 h. Then, the remaining reaction components were added, and the mixture was incubated for 10 min at 37°C. A blank control was prepared by replacing c-di-GMP with a reaction buffer.
Protease activity assay
LB agar plates containing 10% (wt/vol) skim milk were used to detect protease activity. Fresh bacterial cultures of strain OH11 and its derivatives were spotted onto the plate. After 60 h of incubation at 28°C, the protein hydrolysis zones on the plates were observed. One milliliter of ddH2O was used to suspend the colonies and measure the OD600nm value. Ultimately, the ratio of the hydrolysis zone area to the OD600nm of the bacteria was used to indicate the strength of protease activity. Each treatment was performed in biological triplicates and repeated three times for experimental consistency.
Statistical analysis
In this study, certain experiments were conducted in triplicates and repeated three times. The means of three replicates are presented, with error bars representing the standard errors. Statistical comparisons were performed using one-way ANOVA (Dunnett’s multiple comparisons test) with GraphPad software (GraphPad, La Jolla, CA). A significance level of P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (No. 32372622) and Jiangsu Provincial Key Technology Support Program (BE2023353).
X.G. and L.F. conceived the idea and designed the research. L.H. performed the experiments. G.B. contributed Molecular docking. X.G. and L.H. wrote the manuscript. All authors approved the final version of the manuscript.
Contributor Information
Gaoge Xu, Email: gg880712@163.com.
Fengquan Liu, Email: fqliu20011@sina.com.
Gladys Alexandre, University of Tennessee at Knoxville, Knoxville, Tennessee, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00418-24.
Tables S1 to S3; Figures S1 to S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0 [DOI] [PubMed] [Google Scholar]
- 2. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423 [DOI] [PubMed] [Google Scholar]
- 3. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 4. Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200 [DOI] [PubMed] [Google Scholar]
- 7. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bellini D, Caly DL, McCarthy Y, Bumann M, An SQ, Dow JM, Ryan RP, Walsh MA. 2014. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol 91:26–38. doi: 10.1111/mmi.12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- 10. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61:131–148. doi: 10.1146/annurev.micro.61.080706.093426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park S, Sauer K. 2022. Controlling biofilm development through cyclic di-GMP signaling. Adv Exp Med Biol 1386:69–94. doi: 10.1007/978-3-031-08491-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z, Song L, Liu X, Shen X, Li X. 2023. Bacterial second messenger c-di-GMP: emerging functions in stress resistance. Microbiol Res 268:127302. doi: 10.1016/j.micres.2023.127302 [DOI] [PubMed] [Google Scholar]
- 13. Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200 [DOI] [PubMed] [Google Scholar]
- 14. Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang YC, Chin KH, Tu ZL, He J, Jones CJ, Sanchez DZ, Yildiz FH, Galperin MY, Chou SH. 2016. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun 7:12481. doi: 10.1038/ncomms12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chou SH, Galperin MY. 2016. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hengge R. 2021. High-specificity local and global c-di-GMP signaling. Trends Microbiol 29:993–1003. doi: 10.1016/j.tim.2021.02.003 [DOI] [PubMed] [Google Scholar]
- 18. Liang ZX. 2015. The expanding roles of c-di-GMP in the biosynthesis of exopolysaccharides and secondary metabolites. Nat Prod Rep 32:663–683. doi: 10.1039/c4np00086b [DOI] [PubMed] [Google Scholar]
- 19. Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol 193:4685–4698. doi: 10.1128/JB.05483-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ha DG, Richman ME, O’Toole GA. 2014. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 80:3384–3393. doi: 10.1128/AEM.00299-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. 2014. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew Chem Int Ed 53:7524–7530. doi: 10.1002/anie.201403500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. 2011. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645. doi: 10.1021/ja105732c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother 51:64–72. doi: 10.1128/AAC.00931-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou R, Li K, Guo B, Zhao Y, Li C, Tang B, Sun W, Wang B, Chen W, Sheng C, Kan J, Zhao Y, Liu F. 2023. Antifungal compound from the predatory bacterium Lysobacter enzymogenes inhibits a plant pathogenic fungus by targeting the AAA ATPase VpVeb1. J Agric Food Chem 71:15003–15016. doi: 10.1021/acs.jafc.3c06262 [DOI] [PubMed] [Google Scholar]
- 25. Zhao Y, Qian G, Chen Y, Du L, Liu F. 2017. Transcriptional and antagonistic responses of biocontrol strain Lysobacter enzymogenes OH11 to the plant pathogenic oomycete Pythium aphanidermatum. Front Microbiol 8:1025. doi: 10.3389/fmicb.2017.01025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W, Tang B, Hou R, Sun W, Han C, Guo B, Zhao Y, Li C, Sheng C, Zhao Y, Liu F. 2024. The natural polycyclic tetramate macrolactam HSAF inhibit Fusarium graminearum through altering cell membrane integrity by targeting FgORP1. Int J Biol Macromol 261:129744. doi: 10.1016/j.ijbiomac.2024.129744 [DOI] [PubMed] [Google Scholar]
- 27. Li S, Du L, Yuen G, Harris SD. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 17:1218–1227. doi: 10.1091/mbc.e05-06-0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu G, Han S, Huo C, Chin KH, Chou SH, Gomelsky M, Qian G, Liu F. 2018. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res 46:9276–9288. doi: 10.1093/nar/gky803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu G, Zhou L, Qian G, Liu F. 2022. Diguanylate cyclase and phosphodiesterase interact to maintain the specificity of cyclic di-GMP signaling in the regulation of antibiotic synthesis in Lysobacter enzymogenes. Appl Environ Microbiol 88:e0189521. doi: 10.1128/AEM.01895-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, Galperin MY, Yildiz FH, Lee VT. 2015. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog 11:e1005232. doi: 10.1371/journal.ppat.1005232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. The protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J 14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Camberg JL, Sandkvist M. 2005. Molecular analysis of the Vibrio Cholerae type II secretion ATPase EpsE. J Bacteriol 187:249–256. doi: 10.1128/JB.187.1.249-256.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu C, Turley S, Marionni ST, Park Y-J, Lee KK, Patrick M, Shah R, Sandkvist M, Bush MF, Hol WGJ. 2013. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure 21:1707–1717. doi: 10.1016/j.str.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patrick M, Korotkov KV, Hol WGJ, Sandkvist M. 2011. Oligomerization of epse coordinates residues from multiple subunits to facilitate ATPase activity. J Biol Chem 286:10378–10386. doi: 10.1074/jbc.M110.167031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. d’Enfert C, Ryter A, Pugsley AP. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J 6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korotkov KV, Gonen T, Hol WGJ. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci 36:433–443. doi: 10.1016/j.tibs.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rondelet A, Condemine G. 2013. Type II secretion: the substrates that won't go away. Res Microbiol 164:556–561. doi: 10.1016/j.resmic.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 39. Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020 [DOI] [PubMed] [Google Scholar]
- 40. Korotkov KV, Sandkvist M. 2019. Function, and substrates of the type II secretion system. EcoSal Plus 8:ESP–0034 doi: 10.1128/ecosalplus.ESP-0034-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harding CM, Kinsella RL, Palmer LD, Skaar EP, Feldman MF. 2016. Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog 12:e1005391. doi: 10.1371/journal.ppat.1005391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park BR, Zielke RA, Wierzbicki IH, Mitchell KC, Withey JH, Sikora AE. 2015. A metalloprotease secreted by the type II secretion system links Vibrio cholerae with collagen. J Bacteriol 197:1051–1064. doi: 10.1128/JB.02329-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White RC, Gunderson FF, Tyson JY, Richardson KH, Portlock TJ, Garnett JA, Cianciotto NP. 2018. Type II secretion-dependent aminopeptidase LapA and acyltransferase PlaC are redundant for nutrient acquisition during Legionella pneumophila intracellular infection of amoebas. mBio 9:e00528-18. doi: 10.1128/mBio.00528-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson TL, Waack U, Smith S, Mobley H, Sandkvist M. 2015. Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol 198:711–719. doi: 10.1128/JB.00622-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jha G, Rajeshwari R, Sonti RV. 2005. Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. MPMI 18:891–898. doi: 10.1094/MPMI-18-0891 [DOI] [PubMed] [Google Scholar]
- 46. Chapon V, Czjzek M, El Hassouni M, Py B, Juy M, Barras F. 2001. Type II protein secretion in Gram-negative pathogenic bacteria: the study of the structure/secretion relationships of the cellulase Cel5 (formerly EGZ) from Erwinia chrysanthemi. J Mol Biol 310:1055–1066. doi: 10.1006/jmbi.2001.4787 [DOI] [PubMed] [Google Scholar]
- 47. He SY, Schoedel C, Chatterjee AK, Collmer A. 1991. Extracellular secretion of pectate lyase by the Erwinia chrysanthemi out pathway is dependent upon Sec-mediated export across the inner membrane. J Bacteriol 173:4310–4317. doi: 10.1128/jb.173.14.4310-4317.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol Microbiol 43:475–485. doi: 10.1046/j.1365-2958.2002.02759.x [DOI] [PubMed] [Google Scholar]
- 49. Putker F, Tommassen-van Boxtel R, Stork M, Rodríguez-Herva JJ, Koster M, Tommassen J. 2013. The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environ Microbiol 15:2658–2671. doi: 10.1111/1462-2920.12115 [DOI] [PubMed] [Google Scholar]
- 50. Mulcahy H, Charron-Mazenod L, Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol 12:1621–1629. doi: 10.1111/j.1462-2920.2010.02208.x [DOI] [PubMed] [Google Scholar]
- 51. Wilton M, Halverson TWR, Charron-Mazenod L, Parkins MD, Lewenza S. 2018. Secreted phosphatase and deoxyribonuclease are required by Pseudomonas aeruginosa to defend against neutrophil extracellular traps. Infect Immun 86:e00403-18. doi: 10.1128/IAI.00403-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sloup RE, Konal AE, Severin GB, Korir ML, Bagdasarian MM, Bagdasarian M, Waters CM. 2017. Cyclic di-GMP and Vpsr induce the expression of type II secretion in Vibrio cholerae. J Bacteriol 199:e00106-17. doi: 10.1128/JB.00106-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. An S, Caly DL, McCarthy Y, Murdoch SL, Ward J, Febrer M, Dow JM, Ryan RP. 2014. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog 10:e1004429. doi: 10.1371/journal.ppat.1004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. 2012. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics 75:4874–4878. doi: 10.1016/j.jprot.2012.05.033 [DOI] [PubMed] [Google Scholar]
- 55. Qian G, Hu B, Jiang Y, Liu F. 2009. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agri Sci China 8:68–75. doi: 10.1016/S1671-2927(09)60010-9 [DOI] [Google Scholar]
- 56. Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, Qian G, Liu F, Shen Y, Du L. 2015. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol 99:801–811. doi: 10.1007/s00253-014-6120-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen Y, Xia J, Su Z, Xu G, Gomelsky M, Qian G, Liu F. 2017. Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl Environ Microbiol 83:e03397-16. doi: 10.1128/AEM.03397-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yue H, Jiang J, Taylor AJ, Leite ADL, Dodds ED, Du L. 2021. Outer membrane vesicle-mediated codelivery of the antifungal HSAF metabolites and lytic polysaccharide monooxygenase in the predatory Lysobacter enzymogenes. ACS Chem Biol 16:1079–1089. doi: 10.1021/acschembio.1c00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tang B, Zhao YC, Shi XM, Xu HY, Zhao YY, Dai CC, Liu FQ. 2018. Enhanced heat stable antifungal factor production by Lysobacter enzymogenes OH11 with cheap feedstocks: medium optimization and quantitative determination. Lett Appl Microbiol 66:439–446. doi: 10.1111/lam.12870 [DOI] [PubMed] [Google Scholar]
- 60. Tang B, Sun C, Zhao Y, Xu H, Xu G, Liu F. 2018. Efficient production of heat-stable antifungal factor through integrating statistical optimization with a two-stage temperature control strategy in Lysobacter enzymogenes OH11. BMC Biotechnol 18:69. doi: 10.1186/s12896-018-0478-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kozhevnikova EN, van der Knaap JA, Pindyurin AV, Ozgur Z, van Ijcken WFJ, Moshkin YM, Verrijzer CP. 2012. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol Cell 47:133–139. doi: 10.1016/j.molcel.2012.04.030 [DOI] [PubMed] [Google Scholar]
- 62. Ziveri J, Tros F, Guerrera IC, Chhuon C, Audry M, Dupuis M, Barel M, Korniotis S, Fillatreau S, Gales L, Cahoreau E, Charbit A. 2017. The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat Commun 8:853. doi: 10.1038/s41467-017-00889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hua C, Wang T, Shao X, Xie Y, Huang H, Liu J, Zhang W, Zhang Y, Ding Y, Jiang L, Wang X, Deng X. 2020. Pseudomonas syringae dual-function protein Lon switches between virulence and metabolism by acting as both DNA-binding transcriptional regulator and protease in different environments. Environ Microbiol 22:2968–2988. doi: 10.1111/1462-2920.15067 [DOI] [PubMed] [Google Scholar]
- 64. Zehnbauer BA, Foley EC, Henderson GW, Markovitz A. 1981. Identification and purification of the Lon+ (capR+) gene product, a DNA-binding protein. Proc Natl Acad Sci U S A 78:2043–2047. doi: 10.1073/pnas.78.4.2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qian G, Xu F, Venturi V, Du L, Liu F. 2014. Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104:224–231. doi: 10.1094/PHYTO-07-13-0188-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. The Pymol molecular graphics system, version 3.0 schrödinger, LLC. 2000. https://pymol.org/support.html.
- 68. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3; Figures S1 to S5.